Objective:
To explore the potential mechanism of triptolide in diabetic nephropathy (DN) treatment using network pharmacology.
Methods:
The main targets of triptolide were screened using the TCMSP, DrugBank, and NCBI databases, and gene targets of DN were searched using the DrugBank, DisGeNET, TTD, and OMIM databases. All of the above targets were normalized using the UniProt database to obtain the co-acting genes. The co-acting genes were uploaded to the STRING platform to build a protein-protein interaction network and screen the core acting targets. Gene ontology and Kyoto encyclopedia of genes and genomes analyses of the core targets were performed using Metascape. Molecular docking validation of triptolide with the co-acting genes was performed using the Swiss Dock platform.
Results:
We identified 76 potential target points for triptolide, 693 target points for DN-related diseases, and 24 co-acting genes. The main pathways and biological processes involved are lipids and atherosclerosis, IL-18 signaling pathway, TWEAK signaling pathway, response to oxidative stress, hematopoietic function, and negative regulation of cell differentiation. Both triptolide and the active site of the core target genes can form more than 2 hydrogen bonds, and the bond energy is less than -5kJ/mol. Bioinformatics analysis showed that triptolide had a regulatory effect on most of the core target genes that are aberrantly expressed in DKD.
Conclusion:
Triptolide may regulate the body’s response to cytokines, hormones, oxidative stress, and apoptosis signaling pathways in DN treatment by down-regulating Casp3, Casp8, PTEN, GSA3B and up-regulating ESR1, and so forth.
Keywords: diabetic nephropathy, mechanism of action, network pharmacology, triptolide
1. Introduction
Diabetic nephropathy (DN) is a major microvascular complication of diabetes mellitus (DM) and the leading cause of end-stage renal disease worldwide.[1] DN is characterized by varying degrees of proteinuria and a rapid decline in renal function in the later stages, and is currently treated mainly with symptomatic treatments such as glycemic control, lipid regulation, and edema reduction.
Tripterygium wilfordii hook f., a traditional Chinese herbal medicine, can dispel wind and dampness, promote blood circulation, relieve pain, and activate blood circulation to reduce swelling. Triptolide (TP) is an epoxidized diterpene lactone compound isolated from Tripterygium wilfordii, also known as triptolide and triptolide alcohol, and is one of the main active ingredients of Tripterygium wilfordii and its toxic component. Recent studies have found that TP has anti-inflammatory, anti-tumor, and immunosuppressive effects and is widely used in tumor, rheumatological, cardiovascular, and renal diseases.[2] Clinical experience has shown that TP could reduce proteinuria in DN, stabilize renal function, and have better efficacy than renin angiotensin blockers. However, the pathogenesis of DN is complex, and there are many targets of TP action; therefore, the specific mechanism and drug mechanism of TP in treating DN are not explicit.
Network pharmacology is a new discipline that selects a specific signal and designs multitarget drug molecules. Barabási first proposed the concept of network biology in 2004,[3] and Hopkins formally proposed the definition of network pharmacology in 2007.[4] By 2020, the total number of relevant articles published on the China Knowledge Network and PubMed reached 1944 in 1 year, which has become a popular tool for research on the basis and mechanism of traditional Chinese medicine pharmacology.[5] This study aimed to analyze the targets and mechanisms of the action of TP in treating DN by network pharmacology and conduct molecular docking validation to provide a theoretical basis and experimental direction for subsequent cell and molecular experiments and promote its clinical application.
2. Materials and methods
2.1. Major databases and softwares
TCMSP, NCBI, DrugBank, DisGeNET, TTD, OMIM, Uniprot, Venny 2.1, Cytoscape 3.9, String platform, Metascape, Auto Dock Vina, GEO database, and so forth.
2.2. Drug and disease targets acquisition
The targets of TP were searched in the TCMSP, DrugBank, and NCBI databases with “Triptolide” as the search term, and the species was limited to “Homo sapiens.” The targets of DN were searched in the DrugBank, DisGeNET, TTD, and OMIM databases with “diabetic nephropathy” as the search term. All the above targets were gene normalized using the UniProt database, and the drug targets and disease targets were integrated and de-weighted to obtain the TP and DN target sets.
2.3. Construction of protein–protein interaction (PPI) networks for the targets of action of triptolide
A PPI network was built using the string platform. The co-acting targets of TP and DN were collected using Venny 2.1 software. These were imported into the String platform to work a PPI network, set the protein species to “Homo sapiens,” hide the isolated proteins in the network, and the other parameters were kept at the default settings. The PPI network graph data were exported in TSV format. The data were imported into Cytoscape, and a network analyzer was used to analyze and calculate the network topology characteristic value of the intersection PPI and filter the data to obtain the core acting targets. Simultaneously, the protein functional sub-modules were determined by further analysis of the PPI network using the MCODE plug-in.
2.4. Pathway enrichment analysis
The Metascape platform integrates multiple authoritative databases and supports annotation, enrichment analysis, and construction of PPI networks for batch genes or proteins, with more powerful functions, timely updates, and reliable data. The core acting targets were analyzed using the Metascape database for gene ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG). The results were saved and passed through R software for visualization.
2.5. Molecular docking of TP with core acting targets
Molecular docking is an effective method to verify the relationship between molecules and targets. Molecular docking was performed using TP as the ligand and the core targets of TP for DN treatment as the receptor. The 3-dimensional structure of TP was downloaded from PubChem CID, and the core acting target structures were downloaded from the PDB database and routinely processed using Chem-Bio Drew. Then, Pymol software was used to remove redundant protein structures, remove other irrelevant ligands such as water molecules, and import Auto Dock Vina software to select the semi-flexible docking mode with docking parameters as default. A binding energy of less than 0 kJ/mol indicated that the ligand molecule could spontaneously bind to the receptor protein. And the binding energy less than –5.0 kJ/mol indicates good binding, and the smaller the binding energy, the better the docking.
2.6. Expression of co-acting targets in disease or triptolide intervention
Most of the therapeutic effects of drugs on diseases are achieved by directly or indirectly regulating the genes that are aberrantly expressed in the disease. Diabetic Kidney Disease and Triptolide were used as the key words to search in the GEO database, and the species was limited to “Homo sapiens.” Appropriate datasets were selected for bioinformatics analysis to clarify the abnormal expression of each co-acting target in DKD, as well as to clarify the effect of TP intervention on gene expression of co-acting targets. If the expression changes of the same gene are opposite in these 2 cases, there is a high degree of certainty that TP will work through that gene; the opposite is less certain.
3. Results
3.1. The case for drugs and disease targets
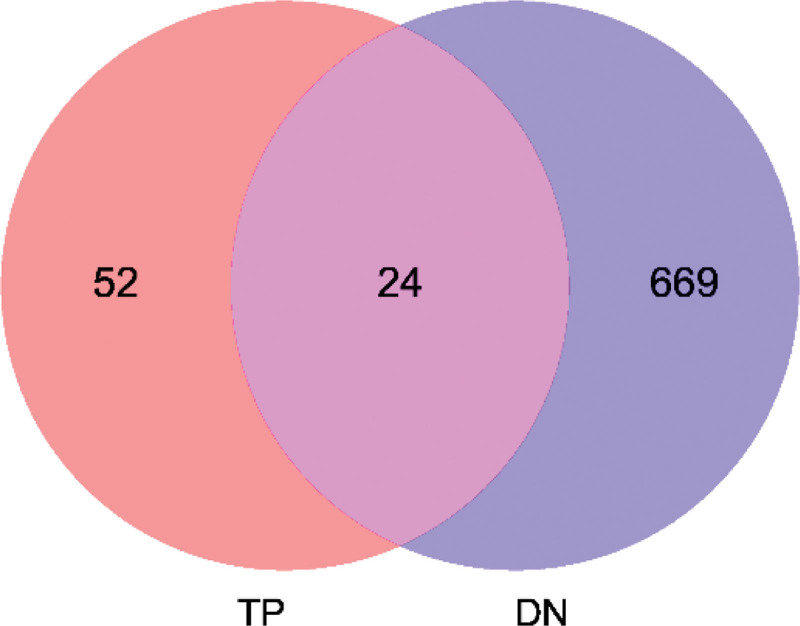
A total of 76 drug target genes, 693 disease target genes, and 24 target gene intersections were identified (Fig. 1 and Table 1).
Figure 1.
The intersection of drug and disease targets. Note: The red part indicates the drug targets and the blue part indicates the disease targets.
Table 1.
Compound-disease common targets.
| No | Uniprot ID | Protien names | Gene names |
|---|---|---|---|
| 1 | P01024 | Complement C3 | C3 |
| 2 | P42574 | Caspase-3 | CASP3 |
| 3 | Q14790 | Caspase-8 | CASP8 |
| 4 | P24864 | G1/S-specific cyclin-E1 | CCNE1 |
| 5 | P38936 | Cyclin-dependent kinase inhibitor 1 | CDKN1A |
| 6 | P61073 | C-X-C chemokine receptor type 4 | CXCR4 |
| 7 | P03372 | Estrogen receptor | ESR1 |
| 8 | P49841 | Glycogen synthase kinase-3 beta | GSK3B |
| 9 | Q16665 | Hypoxia-inducible factor 1-alpha | HIF1A |
| 10 | P11021 | Endoplasmic reticulum chaperone BiP | HSPA5 |
| 11 | P05231 | Interleukin-6 | IL6 |
| 12 | O60674 | Tyrosine-protein kinase JAK2 | JAK2 |
| 13 | P05412 | Transcription factor AP-1 | JUN |
| 14 | Q16236 | Nuclear factor erythroid 2-related factor 2 | NFE2L2 |
| 15 | P00749 | Urokinase-type plasminogen activator | PLAU |
| 16 | P60484 | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | PTEN |
| 17 | P35354 | Prostaglandin G/H synthase 2 | PTGS2 |
| 18 | Q04206 | Transcription factor p65 | RELA |
| 19 | P42224 | Signal transducer and activator of transcription 1-alpha/beta | STAT1 |
| 20 | P40763 | Signal transducer and activator of transcription 3 | STAT3 |
| 21 | P01137 | Transforming growth factor beta-1 proprotein | TGFB1 |
| 22 | P01375 | Tumor necrosis factor | TNF |
| 23 | P04637 | Cellular tumor antigen p53 | TP53 |
| 24 | P15692 | Vascular endothelial growth factor A | VEGFA |
ESR = estrogen receptor, IL6 = interleukin 6, JAK = Janus kinase, STAT3 = signal transducer and activator of transcription 3, TNF = tumor necrosis factor, TP = triptolide, VEGFA = vascular endothelial growth factor A.
3.2. PPI network and core target genes
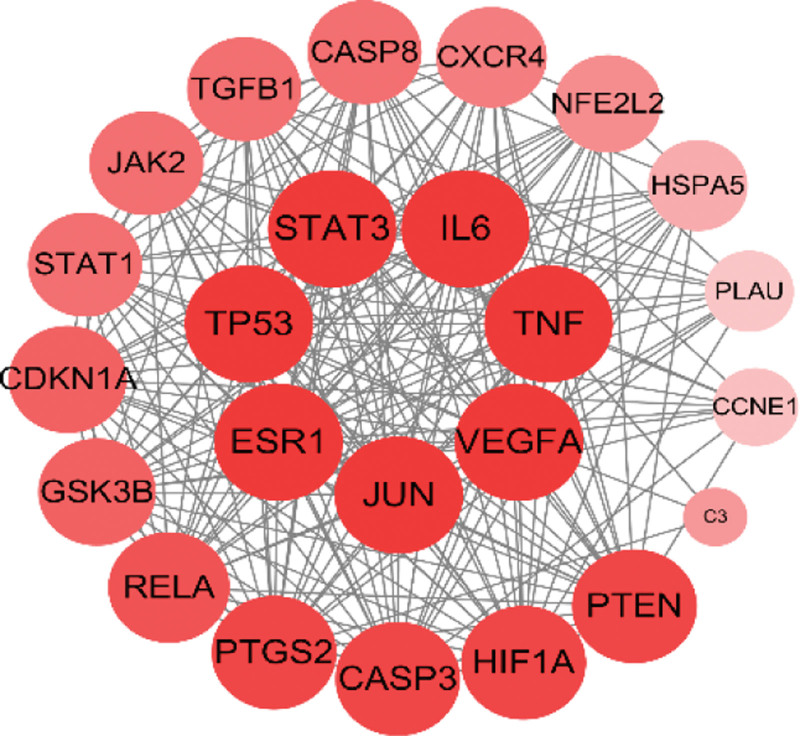
The co-acting targets were uploaded to the STRING platform to obtain the PPI network map. The results showed that there were 24 nodes in the network, with 220 interactions, and the average degree value was 18.3. Seven genes, including vascular endothelial growth factor A (VEGFA), tumor necrosis factor (TNF), Interleukin 6 (IL6), signal transducer and activator of transcription 3 (STAT3), cellular tumor antigen p53 (TP53), estrogen receptor (ESR1), and Transcription factor AP-1 (JUN), were associated with the highest degree value of 22; the C3 gene had the lowest degree value of 4. By combining the average degree value and median degree value, genes with degree values greater than 15 were selected as the core acting targets of TP for DN treatment. The intersection network diagram is shown in Figure 2, where the color from dark to light and the area from large to small represent the degree values from large to small. The PPI network was further analyzed using MCODE of Cytoscape with default screening parameters. Only one tightly connected protein submodule was obtained, containing 21 nodes and 195 interactions, except for C3, PLAU, and CCNE1, with a score of 19.5.
Figure 2.
PPI network diagram of the co-acting targets. PPI = protein–protein interaction.
3.3. Visualization of enrichment analysis of pathway of TP for DN
Gene enrichment analysis was performed using the Metascape platform, including KEGG pathway analysis, biological process, cellular component, and molecular function GO analysis.
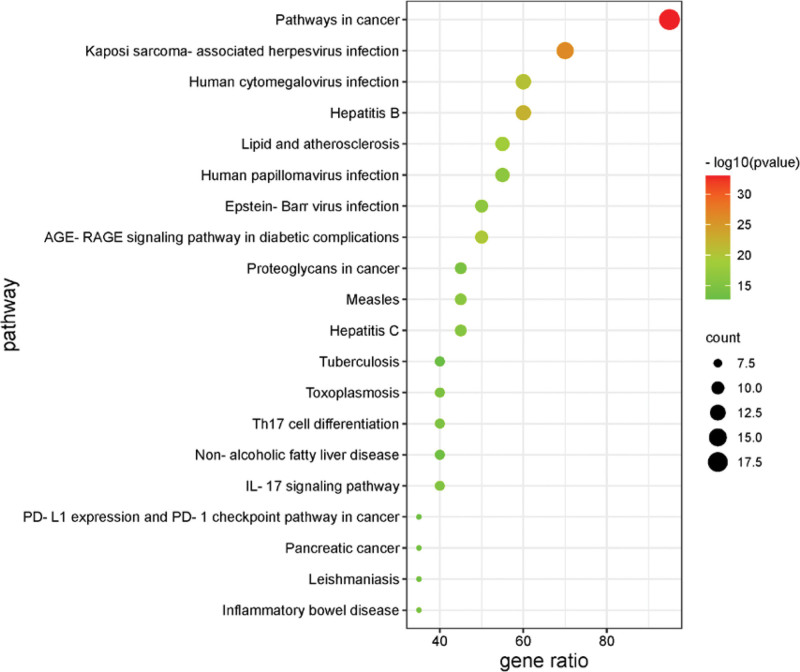
After KEGG pathway analysis, the top 20 genes of Homo sapiens were sorted according to the number of enriched genes. The bubbles were plotted in R. The bubbles from red to green represent the -Log10 (P) values from large to small; the bubble area represents the number of genes in the pathway, and the horizontal axis represents the ratio of genes in the pathway to all genes. The results are shown in Figure 3.
Figure 3.
KEGG pathway analysis of the core acting targets. KEGG = Kyoto encyclopedia of genes and genomes.
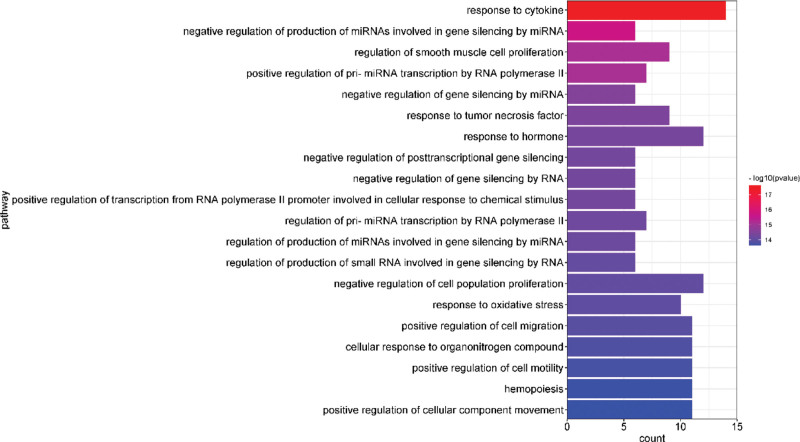
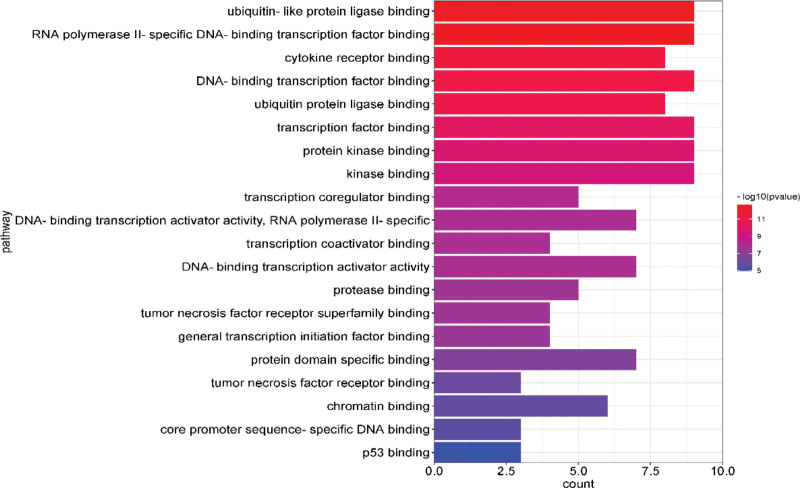
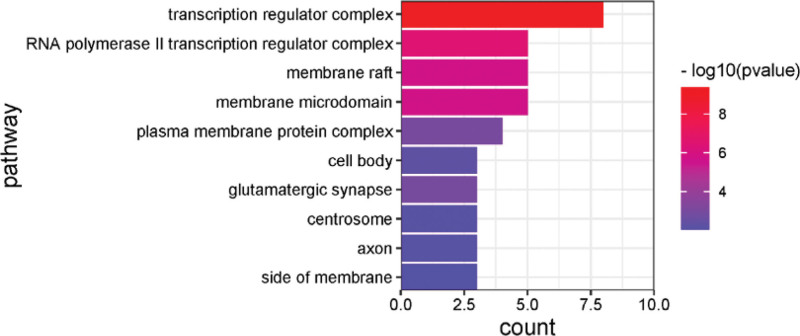
After GO analysis, the top 20 results with the -Log10(P) value were selected for visualization, and biological process, cellular component, and molecular function were plotted in bar graphs with the bar color from dark to light representing the -Log10(P) values from large to small and the bar length representing the gene count of that pathway. The results are shown in Figure 4–6.
Figure 4.
Enrichment gene ontology–biological process (GO–BP) terms for analysis of the core acting targets. BP = biological process, GO = gene ontology.
Figure 6.
Enrichment gene ontology–molecular function (GO–MF) terms for analysis of the core acting targets. GO = gene ontology, MF = molecular function.
Figure 5.
Enrichment gene ontology–cellular components (GO–CC) terms for analysis of the core acting targets. CC = cellular component, GO = gene ontology.
3.4. Molecular docking of TP with core acting targets
The active sites of the core acting targets with a higher degree were able to form more than 2 hydrogen bonds, and the binding energies were all less than -5 kJ/mol, indicating good binding activity. Among them, the binding energy of tretinoin with signal transducer and activator of transcription1 was the lowest, at -7.83 kJ/mol, with the most active binding ability. Details are presented in Table 2.
Table 2.
Molecular docking results of TP with core acting targets.
| Target | Binding energy (kJ/mol) | Target | Binding energy (kJ/mol) |
|---|---|---|---|
| STAT1 | –7.83 | TP53 | –7.42 |
| CASP8 | –7.78 | PTGS2 | –7.38 |
| CXCR4 | –7.77 | RELA | –7.20 |
| PTEN | –7.60 | CASP3 | –7.20 |
| ESR1 | –7.58 | IL6 | –7.16 |
| GSK3B | –7.58 | CDKN1A | –7.13 |
| TNF | –7.50 | HIF1A | –6.86 |
| JAK2 | –7.49 | STAT3 | –6.82 |
| TGFB1 | –7.48 | VEGFA | –6.54 |
| NFE2L2 | –7.45 | JUN | –6.54 |
ESR = estrogen receptor, JAK = Janus kinase, STAT3 = signal transducer and activator of transcription 3, TNF = tumor necrosis factor, TP = triptolide, VEGFA = vascular endothelial growth factor A.
3.5. Expression of co-acting targets in disease or triptolide intervention
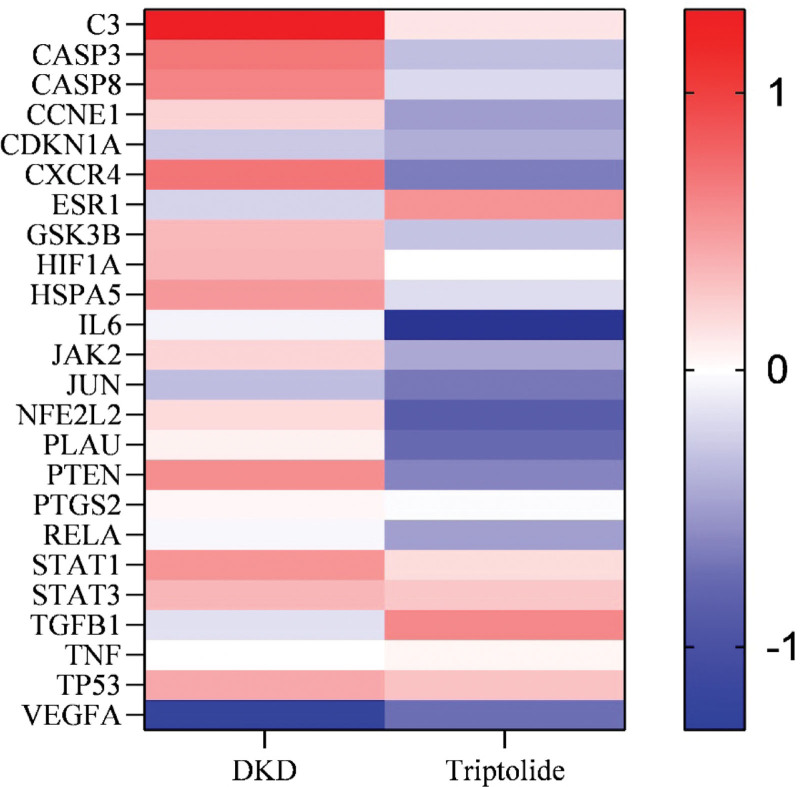
The GSE30122 dataset was selected for bioinformatics analysis to clarify the aberrant expression of co-acting targets in DKD, and the GSE93206 dataset was selected for bioinformatics analysis to determine the effect of TP intervention on the expression of co-acting targets by condition qualification. Red indicates gene up-regulation, blue indicates gene down-regulation, and the color from dark to light represents the absolute value of Log2FC from large to small, that is the expression difference is from large to small. The results are shown in Figure 7.
Figure 7.
Red indicates gene up-regulation, blue indicates gene down-regulation, and the color from dark to light represents the absolute value of Log2FC from large to small, i.e., the expression difference is from large to small.
4. Conclusion
By 2019, 463 million adults aged 20 to 79 years worldwide had DM, approximately 15% to 25% of patients with type I diabetes and 30% to 40% of patients with type II diabetes developed renal complications and may progress to end-stage renal disease.[6] However, the pathogenesis of DN is not yet clear, and the treatment is not comprehensive enough.
4.1. Analysis of core acting target results
In the PPI network analysis in this study, VEGFA, TNF, IL6, STAT3, TP53, ESR1, and JUN were tied for first place with a degree value of 22. VEGFA regulates vascular permeability and it is a key regulator of angiogenesis during the growth of solid tumors.[7,8] It has been found that VEGF expression levels are significantly higher in renal epithelial cells and distal tubular epithelial cells in DN patients compared to the general population,[9] which may be associated with increased renal vascular permeability leading to proteinuria. TNF is produced by monocytes and macrophages during acute immune responses and can promote cell proliferation, differentiation, cytokine production, apoptosis, and necrosis[10]; a close relationship between TNF and the development of DM has been demonstrated.[11] IL6 is one of the most important inflammatory cytokines, while IL6 can bind to specific receptors to stimulate the Janus kinase/signal transducer and activator of transcription signaling pathway,[12] plays a vital role in peripheral metabolic organs such as the adipose, pancreas, and immune system, and is a new potential therapeutic target for diabetes.[13] STAT3 can affect cellular autophagy through different subcellular localization patterns. For example, cytoplasmic STAT3 inhibits cellular autophagy by sequestering eukaryotic initiation factor 2-α kinase 2 and other signaling molecules associated with autophagy.[14–16] In addition, mitochondrial translocation of STAT3 inhibits oxidative stress-induced autophagy and may effectively protect the mitochondria from mitotic degradation.[17–19] TP53 encodes proteins that respond to various cellular stresses and regulate the expression of target genes to induce cell cycle arrest, apoptosis, senescence, DNA repair, or metabolic changes[20–22]; for example, by downregulating the anti-apoptotic gene product Bcl-2, upregulates the pro-apoptotic gene product Bax, and other pathways to participate in the regulation of the apoptosis pathway.[23] The ESR1 inhibits nuclear factor kappa B (NF-κB) -mediated transcription of the IL6 promoter by reducing NF-κB DNA-binding activity and displaces RELA/p65 and associated co-regulators from the promoter,[24] thereby regulating inflammation, immunity, and stress responses. AP-1 (AP1), a transcription factor involved in JUN, is a recognized integrator of extracellular signals[25,26] and plays a vital role in the treatment of various inflammatory lesions, transplant rejection, fibrosis, and organ damage.[27] The remaining core target genes were mostly associated with inflammatory responses, cellular autophagy, immunity, oxidative stress, apoptosis, cellular senescence, and tumors.
4.2. Analysis of pathway and biological process
A total of 109 KEGG pathways were obtained in this study, and the enrichment results showed that TP treatment of DN mainly involved the following types of signaling pathways: DM-related, immune-related, and cell survival-related pathways.
DM-related pathways include the AGE-RAGE signaling pathway in diabetic complications and insulin resistance. The AGE-RAGE signaling pathway activates NF-κB and causes the expression and release of large amounts of IL6 and TNF, which eventually causes chronic cell activation and tissue damage.[28,29] In podocytes, the AGE-RAGE signaling pathway can stimulate VEGF, which increases vascular permeability and causes proteinuria.[30] It also stimulates the production of TGF-β1, which leads to glomerular extracellular matrix production and tubular epithelial mesenchymalization[31–33]; induces enhanced MCP-1 expression and promotes glomerulosclerosis and tubulointerstitial fibrosis[34]; stimulates angiotensin II production or overexpression at renal component AT1 receptors to accelerate the progression of DM[35]; AGE-RAGE signaling pathway activates nicotinamide adenine dinucleotide phosphate oxidase, which in turn activates mitogen-activated protein kinase, extracellular regulated protein kinases, extracellular signal-regulated kinase [ERK1/2] ERK1/2 and P38 and other mediated signaling pathways via reactive oxygen species, resulting in the activation of NF-κB by phosphorylation.[36–38] TP may reduce renal damage and decrease urinary protein levels in DN patients through the AGE-RAGE signaling pathway and has a positive effect on blood glucose in DN patients through insulin resistance
Immune-related pathways include the Interleukin 17 (IL-17) signaling pathway, the Toll-like receptor signaling pathway, Th1 and Th2 cell differentiation, and the NF-κB signaling pathway. The IL-17 signaling pathway can activate NF-κB and induce NF-κB-dependent cytokines to upregulate inflammatory gene expression[39]; IL-17 has pathogenic functions in immune-mediated glomerular diseases.[40] IL-17 has also been found to limit the growth of fungi in the kidney, prevent kidney tissue damage, and protect kidney function during the transmission of candidiasis through the kallikrein (Klk) Kallikrein- Kinin system (KKS).[41] The Toll-like receptor signaling pathway (TILs signaling pathway) is one of the earliest determinants of immune activation, which can activate TGF-β1, TNF, AT1, NF-κB, etc, through cellular exogenous, intrinsic, and specific responses[42,43] to induce a series of immune responses and thus have an impact on DN. Th1 and Th2 cell differentiation is influenced by various factors, such as antigen type and concentration, co-stimulatory molecules, cytokine concentration, immunoreactive hormones, transcription factors, and type of antigen-presenting cells. Disruption of the Th1/ Th2 balance can induce a range of autoimmune diseases and adversely affect pancreatic β-cells,[44] thereby aggravating the condition of DN patients. The NF-κB signaling pathway includes 3 main pathways: the canonical, alternative, and atypical pathways. After activation, it can combine with other pathways, such as the TILs signaling pathway, to cause inflammation and fibrosis in the kidneys.[45,46] TP may treat DN by inhibiting local inflammation and excessive immunity in the kidney through the IL-17 signaling pathway, Toll-like receptor signaling pathway, Th1 and Th2 cell differentiation, NF-κB signaling pathway, etc.
Cell survival-related pathways include the TNF, apoptosis, NF-κB, PI3K-Akt, mTOR, and IL-17 signaling pathways. The TNF signaling pathway induces podocyte apoptosis by inducing overexpression of retinoic acid receptor responder 1 in podocytes.[47] While mediating renal inflammation and fibrosis, the NF-κB signaling pathway also aggravates renal injury and proteinuria by inducing apoptosis through oxidative stress.[48] The PI3K/AKT signaling pathway is related to proliferation, differentiation, and apoptosis. It connects downstream mTOR signaling to the PI3K-AKT-mTOR pathway, which can induce autophagy and increase the renal oxidative stress response.[49,50] The mTOR pathway can also exacerbate glomerulosclerosis by promoting DN mouse thylakoid proliferation in combination with other factors and pathways,[51] which has been confirmed by in vitro experiments on human renal thylakoid cells.[52] In addition, the IL-17 signaling pathway activates the mitogen-activated protein kinase pathway, which includes extracellular signal-regulated kinase, p38, and JUN N-terminal kinase, to induce apoptosis.[39] TP may reduce renal injury and decrease proteinuria by reducing apoptosis and autophagy in renal cells by inhibiting the TNF, NF-κB, PI3K-Akt, mTOR, and other signaling pathways.
4.3. Analysis of co-acting targets analysis
Analysis of the retrieved datasets found that TP inhibited the overexpressed CASP3, CASP8, CCNE1, CXCR4, GSK3B, HSPA5, Janus kinase 2, NFE2L2, PLAU, PTEN and PTGS2 in DKD, while upregulating ESR1 and TGFB1, which were repressed in DKD, promoting each gene restores its proper expression level, thereby treating DKD. The expression trends of some other genes under TP intervention were consistent with the trends in DKD, suggesting that TP is less likely to treat DKD through these genes. However, human is a complex whole, and the pathways and organismal responses associated with each gene are complex and variable. Therefore, a consistent trend does not mean that regulating the expression of these genes is ineffective, and further experimental verification is still needed. In addition, different disease stages, disease states, and individual responses to gene expression may also vary widely, and the effects of drugs on them will also be different. Perhaps this can be used to explain the poor efficacy of some patients in clinical practice.
4.4. Summary and prospect
At present, the construction of a drug-disease molecular network is rarely reported in DN. The application of molecular network science can help to further reveal the mechanism of TP for DN treatment and provide direction for further research on pharmacological, toxicological, and pharmacodynamic substance bases. In this study, the mechanism of TP in the treatment of DN was predicted based on network pharmacology and molecular docking, and it was found that the mechanism was mainly related to the reduction in the inflammatory response, immune suppression, inhibition of apoptosis, and inhibition of glomerulosclerosis. In addition, TP may have ameliorating effects on insulin resistance. However, this study only predicted the mechanism of TP for DN from the available internet data, lacking experimental and clinical validation, and failed to clarify the treatment protocol for the prescribed dose and duration of TP for DN. Therefore, further experimental validation and validation in a large sample, multicenter, randomized, controlled clinical trials are still needed.
Author contributions
YG designed the study and wrote the paper. YL and ZG performed the research and analyzed the data.
Conceptualization: Ying Gao.
Data curation: Yingying liu.
Formal analysis: Ying Gao, Zhaoan Guo.
Writing – original draft: Ying Gao, Zhaoan Guo.
Abbreviations:
- DM =
- diabetic mellitus
- DN =
- diabetic nephropathy
- ERK =
- extracellular signal-regulated kinase
- ESR =
- estrogen receptor
- GO =
- gene ontology
- IL6 =
- interleukin 6
- KEGG =
- Kyoto encyclopedia of genes and genomes
- NF-κB =
- nuclear factor kappa B
- PPI =
- protein–protein interaction
- STAT3 =
- signal transducer and activator of transcription 3
- TNF =
- tumor necrosis factor
- TP =
- triptolide
- VEGFA =
- vascular endothelial growth factor A
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
The study was supported in part by the following grant: National Natural Science Foundation of China (No. 81874440).
The authors have no conflicts of interest to disclose.
How to cite this article: Gao Y, Guo Z, Liu Y. Analysis of the potential molecular biology of triptolide in the treatment of diabetic nephropathy: A narrative review. Medicine 2022;101:48(e31941).
Contributor Information
Ying Gao, Email: 2021100075@sdutcm.edu.cn.
Yingying Liu, Email: liuyiingjiayouya@163.com.
References
- [1].Lv W, Booz GW, Wang Y, et al. Inflammation and renal fibrosis: recent developments on key signaling molecules as potential therapeutic targets. Eur J Pharmacol. 2018;820:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wang H, Sijie T, Wang X, et al. Analysis of research trends and hotspots of tripterygium wilfordii polyglycosides. China Pharmaceutical Herald. 2021;18:18–24. [Google Scholar]
- [3].Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–13. [DOI] [PubMed] [Google Scholar]
- [4].Hopkins AL. Network pharmacology. Nat Biotechnol. 2007;25:1110–1. [DOI] [PubMed] [Google Scholar]
- [5].Chen J, Chen QL. Current status and considerations of network pharmacology in traditional Chinese medicine research. J Shanghai Univ Trad Chin Med. 2021;35:1–6 + 13. [Google Scholar]
- [6].Kim SJ, Natesan V. Diabetic. Nephropathy: a review of risk factors, progression, mechanism, and dietary management. Biomol Ther (Seoul). 2021;29:365–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Peach CJ, Mignone VW, Arruda MA, et al. Molecular pharmacology of VEGF-A isoforms: binding and signalling at VEGFR2. Int J Mol Sci. 2018;19:1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J Intern Med. 2013;273:2114–27. [DOI] [PubMed] [Google Scholar]
- [9].Eremina V, Jefferson JA, Kowalewska J, et al. VEGF inhibition and renal thrombotic microangiopathy. N Engl J Med. 2008;358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Haitham TI, Naismith JH. TNF alpha and the TNF receptor superfamily: structure-function relationship(s). Microsc Res Tech. 2000;50:184–95. [DOI] [PubMed] [Google Scholar]
- [11].Parameswaran N, Patial S. Tumor necrosis factor-α signaling in macrophages. Crit Rev Eukaryot Gene Expr. 2010;20:87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Murray PJ. The JAK-STAT signaling pathway: input and output integration. J Immunol. 2007;178:2623–9. [DOI] [PubMed] [Google Scholar]
- [13].Dodington DW, Desai HR, Woo M. JAK/STAT - emerging players in metabolism. Trends Endocrinol Metab. 2018;29:55–65. [DOI] [PubMed] [Google Scholar]
- [14].Shen S, Niso-Santano M, Adjemian S, et al. Cytoplasmic STAT3 represses autophagy by inhibiting PKR activity. Mol Cell. 2012;48:667–80. [DOI] [PubMed] [Google Scholar]
- [15].Shen S, Niso-Santano M, Adjemian S, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–71. [DOI] [PubMed] [Google Scholar]
- [16].Oh H-M, Yu C-R, Dambuza I, et al. STAT3 protein interacts with Class O forkhead transcription factors in the cytoplasm and regulates nuclear/cytoplasmic localization of FoxO1 and FoxO3a proteins in CD4(+) T cells. J Biol Chem. 2012;287:30436–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].You L, Wang Z, Li H, et al. The role of STAT3 in autophagy. Autophagy. 2015;11:729–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pietrocola F, Izzo V, Niso-Santano M, et al. Regulation of autophagy by stress-responsive transcription factors. Semin Cancer Biol. 2013;23:310–22. [DOI] [PubMed] [Google Scholar]
- [20].Marcel V, Perrier S, Aoubala M, et al. Δ160p53 is a novel N-terminal p53 isoform encoded by Δ133p53 transcript. FEBS Lett. 2010;584:4463–8. [DOI] [PubMed] [Google Scholar]
- [21].Hu W, Chen S, Thorne RF, et al. TP53, TP53 target genes (DRAM, TIGAR), and autophagy. Adv Exp Med Biol. 2019;1206:127–49. [DOI] [PubMed] [Google Scholar]
- [22].Navalkar A, Paul A, Sakunthala A, et al. Oncogenic gain of function due to p53 amyloids occurs through aberrant alteration of cell cycle and proliferation. J Cell Sci. 2022;135:jcs259500. [DOI] [PubMed] [Google Scholar]
- [23].Lohmüller M, Roeck BF, Szabo TG, et al. The SKP2-p27 axis defines susceptibility to cell death upon CHK1 inhibition. Mol Oncol. 2022;16:2771–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Molli PR, Singh RR, Lee SW, et al. MTA1-mediated transcriptional repression of BRCA1 tumor suppressor gene. Oncogene. 2008;27:1971–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–6. [DOI] [PubMed] [Google Scholar]
- [26].Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. Cell Sci. 2004;117:5965–73. [DOI] [PubMed] [Google Scholar]
- [27].Bejjani F, Evanno E, Zibara K, et al. The AP-1 transcriptional complex: local switch or remote command?. Biochim Biophys Acta Rev Cancer. 2019;1872:11–23. [DOI] [PubMed] [Google Scholar]
- [28].Liu F, Fu Y, Wei C, et al. The expression of GPR109A, NF-kB and IL-1β in peripheral blood leukocytes from patients with type 2 diabetes. Ann Clin Lab Sci. 2014;44:443–8. [PubMed] [Google Scholar]
- [29].Matsumori A. Novel biomarkers of inflammation for the management of diabetes: immunoglobulin-free light chains. Biomedicines. 2022;10:666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Stavas J, Filler G, Jain D, et al. Renal autologous cell therapy to stabilize function in diabetes-related chronic kidney disease: corroboration of mechanistic action with cell marker analysis. Kidney Int Rep. 2022;7:1619–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pankewycz OG, Guan J-X, Bolton WK, et al. Renal TGF-beta regulation in spontaneously diabetic NOD mice with correlations in mesangial cells. Kidney Int. 1994;46:748–58. [DOI] [PubMed] [Google Scholar]
- [32].Chen Y-T, Jhao P-Y, Hung C-T, et al. Endoplasmic reticulum protein TXNDC5 promotes renal fibrosis by enforcing TGF-β signaling in kidney fibroblasts. J Clin Invest. 2021;131:e143645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Maity S, Das F, Kasinath BS, et al. TGFβ acts through PDGFRβ to activate mTORC1 via the Akt/PRAS40 axis and causes glomerular mesangial cell hypertrophy and matrix protein expression. J Biol Chem. 2020;295:14262–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Xu L, Sharkey D, Cantley LG. Tubular GM-CSF promotes late MCP-1/CCR2-mediated fibrosis and inflammation after ischemia/reperfusion injury. J Am Soc Nephrol. 2019;30:1825–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fukami K, Yamagishi S-I, Kaifu K, et al. Telmisartan inhibits AGE-induced podocyte damage and detachment. Microvasc Res. 2013;88:79–83. [DOI] [PubMed] [Google Scholar]
- [36].Wang X, Meng L, Zhao L, et al. Resveratrol ameliorates hyperglycemia-induced renal tubular oxidative stress damage via modulating the SIRT1/FOXO3a pathway. Diabetes Res Clin Pract. 2017;126:172–81. [DOI] [PubMed] [Google Scholar]
- [37].Temviriyanukul P, Lertmongkolaksorn T, Supasawat P, et al. Phikud Navakot extract attenuates lipopolysaccharide-induced inflammatory responses through inhibition of ERK1/2 phosphorylation in a coculture system of microglia and neuronal cells. J Ethnopharmacol. 2022;296:115440. [DOI] [PubMed] [Google Scholar]
- [38].Do HTT, Bui BP, Sim S, et al. Anti-inflammatory and anti-migratory activities of isoquinoline-1-carboxamide derivatives in LPS-treated BV2 microglial cells via inhibition of MAPKs/NF-κB pathway. Int J Mol Sci. 2020;21:2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Awane M, Andres PG, Li DJ, et al. NF-kappa B-inducing kinase is a common mediator of IL-17-, TNF-alpha-, and IL-1 beta-induced chemokine promoter activation in intestinal epithelial cells. J Immunol. 1999;162:5337–44. [PubMed] [Google Scholar]
- [40].Krohn S, Nies JF, Kapffer S, et al. IL-17C/IL-17 receptor E signaling in CD4+ T cells promotes TH17 cell-driven glomerular inflammation. J Am Soc Nephrol. 2018;29:1210–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ramani K, Garg AV, Jawale CV, et al. The Kallikrein-Kinin system: a novel mediator of IL-17-driven anti-Candida immunity in the kidney. PLoS Pathog. 2016;12:e1005952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fitzgerald KA, Kagan JC. Toll-like receptors and the control of immunity. Cell. 2020;180:1044–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Xia P, Wu Y, Lian S, et al. Research progress on Toll-like receptor signal transduction and its roles in antimicrobial immune responses. Appl Microbiol Biotechnol. 2021;105:5341–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ding Z, Cheng R, Yang Y, et al. The succinoglycan riclin restores beta cell function through the regulation of macrophages on Th1 and Th2 differentiation in type 1 diabetic mice. Food Funct. 2021;12:11611–24. [DOI] [PubMed] [Google Scholar]
- [45].Liao Y, Tan R-Z, Li J-C, et al. Isoliquiritigenin attenuates UUO-induced renal inflammation and fibrosis by inhibiting Mincle/Syk/NF-kappa B signaling pathway. Drug Des Devel Ther. 2020;14:1455–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Peng X, Wang Y, Li H, et al. ATG5-mediated autophagy suppresses NF-κB signaling to limit epithelial inflammatory response to kidney injury. Cell Death Dis. 2019;10:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chen A, Feng Y, Lai H, et al. Soluble RARRES1 induces podocyte apoptosis to promote glomerular disease progression. J Clin Invest. 2020;130:5523–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhang H-F, Wang J-H, Wang Y-L, et al. Salvianolic acid a protects the kidney against oxidative stress by activating the Akt/GSK-3β/Nrf2 signaling pathway and inhibiting the NF-κB signaling pathway in 5/6 nephrectomized rats. Oxid Med Cell Longev. 2019;2019:2853534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tu QD, Jin J, Hu X, et al. Curcumin improves the renal autophagy in rat experimental membranous nephropathy via regulating the PI3K/AKT/mTOR and Nrf2/HO-1 signaling pathways. Biomed Res Int. 2020;2020:7069052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chiou TT, Chau YY, Chen JB, et al. Rapamycin attenuates PLA2R activation-mediated podocyte apoptosis via the PI3K/AKT/mTOR pathway. Biomed Pharmacother. 2021;144:112349. [DOI] [PubMed] [Google Scholar]
- [51].Lu X, Li L, Suo L, et al. Single-cell RNA sequencing profiles identify important pathophysiologic factors in the progression of diabetic nephropathy. Front Cell Dev Biol. 2022;10:798316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Han F, Xue M, Chang Y, et al. Triptolide suppresses glomerular mesangial cell proliferation in diabetic nephropathy is associated with inhibition of PDK1/Akt/mTOR pathway. Int J Biol Sci. 2017;13:1266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]