Abstract
Porphyromonas gingivalis is an asaccharolytic and anaerobic bacterium that possesses a complex proteolytic system which is essential for its growth and evasion of host defense mechanisms. In this report, we show the purification and characterization of prolyl dipeptidyl peptidase IV (DPPIV) produced by this organism. The enzyme was purified to homogeneity, and its enzymatic activity and biochemical properties were investigated. P. gingivalis DPPIV, like its human counterpart, is able to cleave the N terminus of synthetic oligopeptides with sequences analogous to those of interleukins 1β and 2. Additionally, this protease hydrolyzes biologically active peptides including substance P, fibrin inhibitory peptide, and β-casomorphin. Southern blot analysis of genomic DNA isolated from several P. gingivalis strains reveal that a single copy of the DPPIV gene was present in all strains tested.
Porphyromonas gingivalis is an anaerobic, asaccharolytic periodontopathogen that is unable to take up free amino acids and therefore utilizes only short oligopeptides as carbon and energy sources (13). In this context, it is likely that to meet this fastidious nutritional requirement, P. gingivalis has evolved a complex and diverse cell surface-associated proteolytic system composed of several unique peptidases (34). Some of these enzymes have been shown to not only play a role in the evasion of host defense mechanisms but also indirectly participate in the pathological destruction of periodontal tissues during the progression of periodontitis (40). The best-characterized enzymes of this system are gingipains R and K, arginine- and lysine-specific cysteine proteinases, respectively (34). These enzymes contribute significantly to the development and maintenance of pathological processes within the infected periodontal pocket through their ability to (i) activate the kallikrein-kinin system (22), (ii) release neutrophil chemotactic activity from the native and oxidized C5 component of complement pathway (14), (iii) activate factor X, protein C, and prothrombin (21, 23), (iv) process or degrade cytokines, including interleukin 6 (IL-6) (4, 15), IL-8 (32, 42), gamma interferon (41), and tumor necrosis factor alpha (10), (v) degrade fibrinogen and some plasma proteins (37), (vi) activate neutrophils through cleavage of proteinase-activated receptor 2 (28), and (vii) cleave and inactivate the C5a receptor on phagocytes (25). The other group of P. gingivalis cysteine proteinases comprise the prtT gene product (30) and periodontain, a recently purified enzyme with the ability to cleave and inactivate α1-proteinase inhibitor (33). Another gene, tpr, coding for a papain-like proteinase, has also been cloned, sequenced and expressed in Escherichia coli (8). Although the Tpr protease has never been purified from P. gingivalis, characterization of a recombinant enzyme and study of an isogenic mutant lacking a functional tpr gene indicates that Tpr is present on the cell surface, has broad endopeptidase activity, and is expressed in a negatively controlled manner by the increased availability of peptides but not free amino acids (29). Other members of the P. gingivalis proteolytic system are cysteine proteases with gelatinolytic activity (27) and a serine endopeptidase (19); however, in comparison to other proteinases, these enzymes are superficially characterized.
Collagen type I is a major constituent of collagen fibers which account for approximately 60% of the gingival connective tissue volume and can be degraded to large fragments by both human and bacterial collagenases. The collagenolytic activity of P. gingivalis and other periodontopathogens has been previously described (7, 39), but its contribution to collagen degradation at the periodontal lesion is doubtful. Instead, the bulk of evidence indicates that matrix metalloproteases, especially neutrophil collagenase (MMP-8), are responsible for collagen fiber cleavage (24), which makes the fragments susceptible to further degradation by endopeptidases released by plaque bacteria. This concerted action would likely generate a pool of collagen-derived oligopeptides rich in proline and hydroxyproline residues which are resistant to further degradation by most proteases. However, hydrolysis of such peptides may be particularly important in providing nutrients for plaque bacteria in general, and especially for asaccharolytic organism such as P. gingivalis. For this reason, we focused on a specialized group of P. gingivalis peptidases capable of hydrolyzing peptide bonds containing proline residues. In our previous report (3), we presented the purification, characterization, and cloning of prolyl tripeptidyl peptidase A (PtpA), an enzyme which liberates tripeptides from the N-terminal region of substrates containing proline residues in the third position. More recently, a P. gingivalis homologue of human angiotensin-converting enzyme which is able to cleave oligopeptides after internal proline residue has also been described (2). Clearly, these two enzymes, together with a glycyl-prolyl surface-associated protease (16), are part of the proteolytic machinery of P. gingivalis, which is involved in the degradation of proline-containing peptides. In this system, glycyl-prolyl peptidase, which recently has been found to be a homologue of human prolyl dipeptidyl peptidase IV (DPPIV) (CD26) (26), may have an important function because of the ability of P. gingivalis to thrive on dipeptides as the sole source of carbon (38). This serine protease was previously partially purified and characterized, but conflicting data on its molecular mass and biochemical properties were reported (1, 6, 16). In addition, the detection of three DPPIV-related genes expressed in P. gingivalis (3) suggests that a rigorous purification of this enzyme(s) is necessary to ensure the separation from other related peptidases. In this report, we describe a procedure for obtaining homogenous preparations of DPPIV from P. gingivalis. In addition, we characterize the enzyme with regard to specificity, stability, and inhibition by both protease class-specific and peptide inhibitors.
MATERIALS AND METHODS
Source and cultivation of bacteria.
The bacterial strains of P. gingivalis, HG66, W83, W50, and ATCC 33277, used in this study were grown as described previously (11). P. gingivalis DPPIV was purified from strain HG66, the kind gift of Roland Arnold (University of North Carolina, Chapel Hill).
Localization of DPPIV activity.
Cultures of P. gingivalis HG66, W83, W50, and ATCC 33277, at different phases of growth, were subjected to the following fractionation procedure. Cells were removed by centrifugation (10,000 × g, 30 min), washed twice with 10 mM Tris–150 mM NaCl (pH 7.4), resuspended in 50 mM Tris (pH 7.6), and disintegrated by ultrasonication in an ice bath at 1500 Hz for five cycles (5 min of sonication/5-min brake). Unbroken cells and large debris were removed by centrifugation (10,000 × g, 30 min), and the supernatant was subjected to ultracentrifugation (150,000 × g, 120 min), yielding a pellet containing bacterial membranes and a supernatant which was considered a membrane-free cell extract. All fractions were assayed for amidolytic activity against H-Gly-Pro-p-nitroanilide (pNA).
Enzyme purification.
All purification steps were performed at 4°C except for fast protein liquid chromatography (FPLC) separations, which were carried out at room temperature. Cells were harvested by centrifugation for 30 min at 6,000 × g, washed with 50 mM phosphate buffer (pH 7.4), and finally resuspended in this buffer (150 ml per 50 g [wet weight]). Triton X-100 (10% [vol/vol] solution) was added to a final concentration of 0.05%; after 120 min of gentle stirring, the suspension was centrifuged (28,000 × g, 60 min). Proteins in the supernatant were precipitated with cold acetone (−20°C), which was added to a final concentration of 60%, and collected by centrifugation; the protein pellet was redissolved in 50 mM potassium phosphate buffer (pH 7.0) and extensively dialyzed against 20 mM potassium phosphate–0.02% sodium azide (pH 7.0). The dialyzed fraction was clarified by centrifugation (28,000 × g, 30 min) and applied onto a hydroxyapatite column equilibrated with 20 mM potassium phosphate (pH 7.0) at a flow rate of 20 ml/h. The column was washed until the A280 baseline fell to zero. A gradient of 20 to 300 mM potassium phosphate was then applied, and fractions (5 ml) were collected and assayed for activity against Gly-Pro-pNA and Ala-Phe-Pro-pNA. The hydroxyapatite chromatography step resulted in a separation of these two activities. Fractions active against Gly-Pro-pNA were pooled, saturated with 1 M ammonium sulfate, and clarified by centrifugation. The solution was then loaded onto a phenyl-Sepharose HP column equilibrated with 50 mM potassium phosphate–1 M ammonium sulfate (pH 7.0) (buffer A) at a flow rate of 30 ml/h. The column was first washed with buffer A and then subjected to a descending gradient from 1 to 0.5 M ammonium sulfate, which resulted in the elution of several proteins. The rest of the bound proteins were eluted with buffer A containing no ammonium sulfate. The active fractions were pooled, extensively dialyzed against 20 mM Tris-HCl (pH 8.0) (buffer B), and applied to a MonoQ FPLC column equilibrated with buffer B. The column was washed with buffer B at 1.0 ml/min, and the remaining bound proteins were eluted with a gradient of 0 to 300 mM NaCl.
Enzyme activity assays.
The activity of DPPIV was determined using 1 mM H-Gly-Pro-pNA in 200 μl of 0.2 M HEPES (pH 7.5) at 37°C. The reaction was followed for specific time intervals in a thermostated microplate reader (SpectraMax; Applied Biosystems), and the release of p-nitroaniline was monitored at 405 nm. Other p-nitroanilide substrates were used in the same manner.
Protein determination.
Protein concentration was determined using a bicinchoninic acid BCA reagent kit (Sigma, St. Louis, Mo.) according to the manufacturer's protocol.
SDS-PAGE.
The sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) system designed by Schagger and von Jagow (36) was used throughout this study. For amino-terminal sequence analysis, proteins resolved by SDS-PAGE were electrotransferred to a polyvinylidene difluoride membrane (31). Amino acid sequence analysis was performed with an Applied Biosystems 4760A sequencer, using the program designed by the manufacturer.
Southern blot analysis.
The DPPIV gene was amplified from P. gingivalis HG66 DNA by PCR. The primers 5′-AATGGATCCGGAAAGATTGICGAAACAAAAAC-3′ and 5′-CGCGGATCCCCGGATGGAGAAACACTATAC-3′ were designed based on the data published by Kiyama et al. (26). The PCR product containing the DPPIV open reading frame was subcloned into plasmid vector pQE16 (Qiagen, Chatsworth, Calif.) and subjected to sequencing. DNA from P. gingivalis HG66, W83, W50, and ATCC 33277 was isolated using a Puregene kit (Gentra Systems, Minneapolis, Minn.). Purified DNA was subsequently digested with restriction enzymes, and restriction fragments were separated on 0.7% agarose gel. After electrophoresis, gels were incubated first in denaturation solution (0.5 M NaOH, 1.5 M NaCl) for 30 min and then in renaturation buffer (1 M Tris-HCl [pH 7.5], 1.5 M NaCl) for 1 h. DNA was blotted on a Hybond-XL membrane (Amersham Pharmacia Biotech, Little Chalfont, England) by capillary transfer in 10× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Blots were hybridized with a DNA fragment corresponding to the DPPIV open reading frame radiolabeled by random priming using a High Prime kit (Boehringer Mannheim, Mannheim, Germany) in a modified Church and Gilbert hybridization buffer (12). The positions of restriction fragments corresponding to the DPPIV gene were visualized by autoradiography.
Enzyme specificity.
The determination of substrate specificity was based on the separation of the products of peptide hydrolysis by reverse-phase chromatography. The peptide substrates were first incubated at an enzyme substrate ratio of 1:50 for either 1 or 24 h in 200 mM HEPES (pH 7.5) at 37°C, and the reaction was stopped by acidification with trifluoroacetic acid. The mixtures were separated on a Waters (Milford, Mass.) reverse-phase μBondapak C18 column (3.9 by 300 mm) column using an acetonitrile gradient (0 to 60% in 0.075% trifluoroacetic acid in 50 min). Each peak detected at 220 nm was collected, lyophilized, redissolved in 50% (vol/vol) methanol–0.1% acetic acid, and subjected to identification by mass spectrometry.
RESULTS
Enzyme localization, purification, and initial characterization.
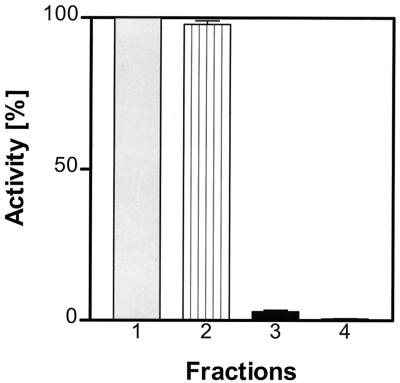
Analysis of amidolytic activity against H-Gly-Pro-pNA in several fractions of P. gingivalis HG66, W50, W83, and ATCC 33277 clearly indicates that the majority of this enzymatic activity is present on the cell surface of all strains tested (Fig. 1). Less than 5% of the activity was found in the culture medium before ultracentrifugation, but after this step it was apparent that all activity was pelleted and therefore is likely to be vesicle associated. Cell-bound enzyme was detached from bacterial surfaces by treatment with a low concentration (0.05%) of Triton X-100. This procedure repeatedly released 80 to 85% of the activity in a soluble form. Subsequent acetone precipitation of proteins in the Triton X-100 fraction successfully separated the activity from pigments which remained in solution. This step results in a significant loss of the amidolytic activity against Gly-Pro-pNA, which, however, may reflect the separation of DPPIV from other proteinases that are able to cleave this substrate. After dialysis, the redissolved protein fraction was applied to the hydroxyapatite chromatography, and at this step substantial separation of the DPPIV activity from both the PtpA and bulk protein was achieved (Fig. 2A). Further purification by subsequent chromatography steps on phenyl-Sepharose (Fig. 2B) and MonoQ (Fig. 2C) columns resulted in the final isolation of a homogenous enzyme. The yield of both protein and activity recovered by this purification procedure is summarized in Table 1.
FIG. 1.
Distribution of Gly-Pro-pNA-hydrolyzing activity in different fractions of P. gingivalis cells. Columns: 1, whole culture; 2, washed cell suspension; 3, culture supernatant; 4, vesicle-free supernatant.
FIG. 2.
Purification of DPPIV from the acetone precipitate of the P. gingivalis cell extract. Absorbance at 280 nm (▵), amidolytic activity against Ala-Phe-Pro-pNA (⧫), and Gly-Pro-pNA (●) are shown. (A) Separation of DPPIV on hydroxyapatite (100 ml) equilibrated with 20 mM potassium phosphate buffer (pH 7.0). The amidolytic activity against Gly-Pro-pNA was separated at this step from that found for the hydrolysis of Ala-Phe-Pro-pNA. (B) Separation of DPPIV obtained from the previous step on phenyl-Sepharose HP (25 ml) equilibrated with 50 mM potassium phosphate–1 M ammonium sulfate (pH 7.0) at a flow rate of 30 ml/h. (C) Separation of DPPIV on a MonoQ FPLC column. OD 280, optical density at 280 nm.
TABLE 1.
Purification of P. gingivalis DPPIV
| Step | Vol (ml) | Protein (mg) | Total activity (Ua) | Sp act (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Triton X-100 extract after centrifugation | 200 | 1200 | 80,000 | 66 | 1 | 100 |
| Acetone precipitate | 50 | 600 | 52,000 | 86 | 1.3 | 65 |
| Hydroxyapatite chromatography | 45 | 18 | 30,000 | 1,666 | 25 | 37 |
| Phenyl-Sepharose | 30 | 8 | 15,000 | 1,875 | 28 | 18 |
| MonoQ | 12 | 2 | 9,000 | 4,500 | 68 | 1.1 |
One unit is defined as mOD per minute per 10 μl.
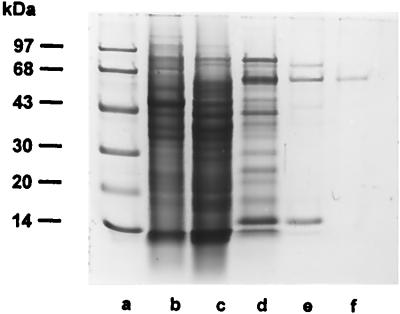
SDS-PAGE analysis of the purified enzyme revealed the presence of a single protein band with an apparent molecular mass of 69 kDa (Fig. 3, lane f) which, after electroblotting onto the nitrocellulose membrane, was subjected to amino-terminal sequence analysis. The sequence obtained, NH2-H-S-Y-R-A-A-V-Y-D-R-D-V-R-G(R)-N-L-V-K-P-L(Q)-S-E-H-V-G-G, was identified within the translated sequence of the DPPIV gene and indicated that the purified 69-kDa enzyme was derived from the original gene product through proteolytic cleavage of a 13-kDa amino-terminal peptide.
FIG. 3.
SDS-PAGE of fractions obtained during the purification of P. gingivalis DPPIV. Lane a, molecular mass markers (phosphorylase b, 97 kDa; bovine serum albumin, 68 kDa; ovalbumin, 43 kDa; carbonic anhydrase, 30 kDa; soybean trypsin inhibitor, 20 kDa; α-lactalbumin, 14 kDa); lane b, Triton X-100 extract of P. gingivalis; lane c, acetone precipitate from Triton X-100 extract of P. gingivalis (15 μg of protein loaded); lane d, hydroxyapatite column eluate (15 μg of protein loaded); lane e, phenyl-Sepharose column eluate (5 μg of protein loaded); lane f, MonoQ column eluate (1 μg of protein loaded).
pH optimum and stability.
Using the amidolytic activity assay with H-Gly-Pro-pNA as a substrate, it was found that the purified enzyme had a broad pH optimum ranging from pH 6.5 to 8.0. DPPIV had no activity in buffers with pH below 6.5 or over 8.5 (Fig. 4) and was stable in 0.2 M HEPES (pH 7.6) for 12 h at 4°C. However, it lost 50% of its activity after 1 week at this temperature. The proteinase showed no appreciable loss of activity when kept frozen at −80°C for 1 month, but it retained only 40% of its activity when kept at −20°C over the same period of time. After 3 h of incubation at 37 and 45°C, the activity was reduced to 50 and 25%, respectively. The optimum temperature for the hydrolysis of Gly-Pro-pNA was determined to be 37°C.
FIG. 4.
pH optimum of the DPPIV activity against Gly-Pro-pNA.
Inhibition profile.
DPPIV activity was not affected by class-specific synthetic inhibitors of cysteine proteinases or metalloproteinases (Table 2). In contrast, preincubation of the enzyme with diisopropylfluorophosphate (DFP) or Pefabloc resulted in the total loss of activity, supporting its classification as a serine proteinase. Surprisingly, however, 3,4-dichloroisocumarin did not affect enzyme activity, and both phenylmethanesulfonyl fluoride (PMSF) and prolinal were poor inhibitors of this enzyme. In contrast, enzyme activity was sensitive to inactivation by detergents (1% SDS and 1% Triton X-100) and heavy metal ions including Zn2+ and Hg2+. However, there was no substantial difference in activity when the assay was performed in the presence or absence of reducing agents. Human plasma inhibitors, such as α1-proteinase inhibitor, α1-antichymotrypsin, and α2-macroglobulin, did not affect enzyme activity, nor were any cleaved by DPPIV (data not shown).
TABLE 2.
Inhibition profile of P. gingivalis DPPIV
| Inhibitor | Concn | % of residual activity |
|---|---|---|
| DFP | 1 mM | 0 |
| PMSF | 10 mM | 20 |
| Pefabloc | 1 mg/ml | 3 |
| 3,4-Dichloroisocoumarin | 1 mM | 100 |
| Iodoacetamide | 0.1 mM | 110 |
| N-ethylmaleimide | 5 mM | 100 |
| 1,10-Orthophenanthroline | 1 mM | 97 |
| EDTA | 10 mM | 100 |
| Leupeptin | 0.1 mM | 100 |
| Antipain | 0.1 mM | 100 |
| Prolinal | 0.1 mM | 20 |
| Cysteine | 10 mM | 105 |
| Gly-Pro | 10 mM | 200 |
| Ala-Pro | 10 mM | 30 |
| Val-Pro | 10 mM | 0 |
| Ala-Pro-Gly | 10 mM | 0 |
| Zn2+ | 1 mM | 1 |
| Hg2+ | 1 mM | 3 |
| SDS | 1% | 0 |
| Triton X-100 | 1% | 0 |
The Gly-Pro dipeptide did not inhibit DPPIV activity; rather, this dipeptide acted as a potent stimulator of its amidolytic activity. On the other hand, Ala-Pro only slightly inhibited P. gingivalis DPPIV activity, in contrast to Val-Pro, which at 10 mM totally inactivated the proteinase activity.
Substrate specificity.
Among several chromogenic substrates tested, including H-Gly-Pro-pNA, H-Arg-Pro-pNA, H-Ala-Pro-pNA, H-Ala-Ala-pNA, H-Ala-Phe-Pro-pNA, H-Ala-Phe-pNA, Z-Gly-Pro-pNA, Z-Ala-Pro-pNA, and H-Pro-pNA, only the first three were rapidly hydrolyzed by DPPIV; weak activity was also found toward H-Ala-Ala-pNA, indicating that the purified proteinase has a typical dipeptidyl peptidase IV activity. To further confirm specificity, several synthetic peptides composed of 6 to 34 amino acid residues and containing at least one proline residue were tested as substrates for DPPIV. Out of 20 peptides tested, only those with a proline or an alanine residue in the second position from the amino-terminal end were cleaved (Table 3), the significant exception being peptides with adjacent proline or hydroxyproline residues (peptides 5, 10, and 11). Apart from these two limitations, the peptide bond -Pro↓Yaa- or -Ala↓Yaa- was cleaved in all peptides with the general formula NH2-Xaa-Pro/Ala-Yaa-(Xaa)n, where Xaa represents any amino acid residue and Yaa represents any residue except proline or hydroxyproline, regardless of the chemical nature of the amino acids and the length of the peptide. In all cases, the reaction was completed in less than 1 h and prolonged incubation for 24 h did not affect the pattern of cleavage, confirming the absolute requirement for a proline or alanine residue at the second position from the N terminus. In addition, these data indicate that the preparation of P. gingivalis DPPIV was free of any contamination with either aminopeptidase, other dipeptidyl peptidases, PtpA, or endopeptidase activities. The lack of the latter activity in the DPPIV preparation was further demonstrated by the inability of this enzyme to cleave collagen, gelatin, or azocasein regardless of either enzyme concentration or time of incubation.
TABLE 3.
Cleavage specificity of P. gingivalis DPPIV on synthetic peptides
| Peptide | Cleavage sitea |
|---|---|
| 1 (substance P) | H-Arg-Pro↓Lys-Pro↓Gln-Gln-Phe-Phe-Gly-Leu-Met-OH |
| 2 | H-Arg-Pro↓Lys-Pro↓Gln-Gln-Phe-OH |
| 3 | H-Ser-Pro↓Tyr-Ser-Ser-Glu-Thr-Thr-OH |
| 4 (IL-1β N terminus) | H-Ala-Pro↓Val-Arg-Ser-Leu-Asn-Cys-Thr-Leu-Arg-Asp-Ser-Asn-Asn-Lys-OH |
| 5 | H-Gly-Pro↓Arg-Pro-Pro-Glu-Arg-His-Gln-Ser-OH |
| 6 (IL-1β N terminus) | H-Ala-Pro↓Val-Arg-Ser-Leu-OH |
| 7 | H-Leu-Pro↓Asp-Leu-Asp-Ser-Ser-Leu-Ala-Ser-Ile-Gln-Glu-Leu-Leu-Ser-Pro-Gln-Glu-Pro-Pro-Arg-Pro-Pro-Glu-OH |
| 8 (β-casomorphin) | H-Tyr-Pro↓Phe-Pro↓Gly-Pro↓Ile-OH |
| 9 (IL-2 N terminus) | H-Ala-Pro↓Thr-Ser-Ser-Ser-Thr-Lys-Lys-Thr-Arg-OH |
| 10 | H-Val-Pro-Pro-Gly-Glu-Asp-Ser-Lys-OH |
| 11 | H-Arg-Pro-Hyp-Gly-Phe-Ser-Pro-Phe-Arg-OH |
| 12 | H-Lys-Arg-Pro-Pro-Gly-Phe-Ser-Pro-Phe-Arg-OH |
| 13 | H-Pro-Asn-Pro-Asn-Gln-Gly-Asn-Phe-Ile-OH |
| 14 | H-Gln-Lys-Gln-Met-Ser-Asp-Arg-Arg-Glu-Asn-Asp-Met-Ser-Pro-Ser-Asn-Asn-Val-Val-Pro-Ile-His-Val-Pro-Pro-Thr-Thr-Glu-Asn-Lys-Pro-Lys-Val-Gln-OH |
| 15 | H-Phe-Leu-Arg-Glu-Pro-Val-Ile-Phe-Leu-OH |
| 16 | H-Gly-Ile-Arg-Pro-Tyr-Glu-Ile-Leu-Ala-OH |
| 17 | H-Cys-Leu-Ser-Ser-Gly-Thr-Leu-Pro-Gly-Pro-Gly-Asn-Asp-Ala-Ser-Arg-Glu-Leu-Glu-Ser-OH |
| 18 | H-Trp-Ala↓Gly-Gly-Asp-Ala-Ser-Gly-Glu-OH |
| 19 | H-Ile-Ala↓Arg-Arg-His-Pro-Tyr-Phe-Leu-OH |
| 20 | H-Lys-Ile-Ala-Gly-Tyr-His-Leu-Glu-Leu-OH |
↓ indicates cleavage site.
We have also investigated the capability of P. gingivalis DPPIV to cleave biologically active peptides including substance P, fibrin inhibitory peptide, and β-casomorphin as well as the oligopeptides representing the N-terminal parts of IL-1β and IL-2 (Table 3). All of these peptides were suitable substrates for DPPIV.
DPPIV expression in various P. gingivalis strains.
Southern blot analysis of genomic DNA isolated from several P. gingivalis strains showed that a single copy of the DPPIV gene was present in all of the strains tested (Fig. 5). In each case, the gene was functional as indicated by enzyme activity assays, which showed the same localization and similar levels of the DPPIV activity in every P. gingivalis strain examined.
FIG. 5.
Southern blot analysis of DPPIV genes in various P. gingivalis strains. DNA isolated from P. gingivalis W83, HG66, W12, 33277, and 405B1 was digested with restriction enzymes as indicated and hybridized with a full-length DPPIV open reading frame from P. gingivalis HG66.
DISCUSSION
In this study we present a simple and efficient method for the purification of DPPIV from Triton X-100 outer membrane extracts of P. gingivalis HG66. The use of low concentrations of the detergent as a solubilization agent has been found to be a very effective method to obtain not only DPPIV but also PtpA (3), aminopeptidase P (unpublished data), and other exopeptidases produced by this bacterium. Such treatment results in a high recovery of these enzymes without any significant loss of activity. In the case of DPPIV, the purification steps yielded 2 mg of active proteinase from 100 g (wet weight) of bacterial pellet. The purified enzyme migrated as a single band on SDS-PAGE, and its N-terminal sequence was located within the primary structure of the translated product of the DPPIV gene. Apparently, the purified enzyme is truncated at the amino terminus due to the action of an arginine-specific proteinase, most likely gingipain R. Taking into account that the N termini of DPPIV homologues from both eukaryotic and prokaryotic organisms contain membrane anchorage domains, it is likely that the N-terminal truncation noted here occurred during the isolation procedure.
The inhibition by typical serine protease inhibitors like DFP, Pefabloc, and PMSF, as well as resistance to sulfhydryl group-blocking reagents and chelating agents, classifies this enzyme as a serine protease. Based on both the amino acid sequence around the putative active-site residue (26) and the specificity profile, the P. gingivalis form of DPPIV can be considered a member of the S9 family of serine proteinases, which comprises various enzymes of bacterial and eukaryotic origin (5). However, the P. gingivalis enzyme displays an interesting inhibition profile compared with well-studied mammalian homologues. In contrast to human DPPIV, for which Gly-Pro dipeptide is a competitive inhibitor (9), the amidolytic activity of the P. gingivalis enzyme is significantly stimulated by this dipeptide. This difference may represent an important bacterial adaptation to the natural environment which is potentially rich in Gly-Pro dipeptides released from degraded collagen fibers. Additionally, these results may suggest differences in architecture of the binding site of human and bacterial enzymes which is further reflected by their different specificities and kinetic properties.
Substrate specificity studies revealed that DPPIV sequentially removes dipeptides from the N terminus of a polypeptide chain when either proline or alanine is present in the penultimate position (P1), the significant exception being peptides that contain proline or hydroxyproline residue in the P1′ position. Many of the natural cytokines, including IL-1β, IL-2, IL-3, RANTES, SDF-1, and granulocyte-macrophage colony-stimulating factor, possess proline in the second position within the polypeptide chain, and their activity strongly depends on the retention of an intact N-terminal segment of the molecule. In this report, we have shown that P. gingivalis DPPIV, like its human counterpart (20), is able to modify the N termini of synthetic oligopeptides with sequences analogous to those of IL-1β and IL-2. To evaluate the physiological relevance of these results, however, additional studies with native cytokines should be performed. This is especially important since it has already been shown that human DPPIV can proteolytically inactivate RANTES and SDF-1 by cleavage of an N-terminal dipeptide (35).
Previously, three independent groups have reported the purification of proteinases with glycyl-prolyl activity from P. gingivalis. Unfortunately, the relationship of these enzymes to the enzyme described in this paper can be evaluated only by comparing general properties because no sequence data are available. From such an analysis, it is apparent that at least two of the previously described glycyl-prolyl peptidases are different from DPPIV. One enzyme, with a molecular mass of 29 kDa, gelatinolytic activity, a pH optimum around 6.5, and an activity which is insensitive to treatment with 20 mM SDS, differs in all parameters from DPPIV (17) and therefore cannot be the enzyme described in this report. A second peptidase with a molecular mass of approximately 160 kDa was partially purified and not fully characterized (1). Only the surface-located 80-kDa protease described by Barua et al. (6) has characteristics in common with DPPIV. The two enzymes have similar inhibition profiles, molecular masses, and pH optimum but differ in the ability to cleave Gly-Phe-pNA. We have already purified and characterized the enzyme responsible for this activity and identified it as a separate dipeptidyl peptidase which is the product of a distinct gene (unpublished data).
The cell surface localization of P. gingivalis DPPIV renders it a good candidate for the involvement in the direct degradation of host proteins. Working in concert with other proline-specific peptidases, DPPIV may not only serve nutritional purposes but also be involved in the metabolism of vasoactive peptides, cleavage of hormones and neuropeptides, and proteolysis of cytokines and salivary proline-rich peptides. In this way, DPPIV could contribute significantly to the deregulation of inflammatory processes during the course of periodontal infection.
ACKNOWLEDGMENTS
This work was supported by grant 6 PO4A 047 17 from the Committee of Scientific Research (Poland) (to J.P. and A.B.) and National Institutes of Health grant DE 09761 (to J.T.).
We thank Dorota Panek for superb technical assistance and Adam Dubin, Pawel Mak, and Tomasz Dec for their help.
REFERENCES
- 1.Abiko Y, Hayakawa M, Murai S, Takiguchi H. Glycylprolyl dipeptidylaminopeptidase from Bacteroides gingivalis. J Dent Res. 1985;64:106–111. doi: 10.1177/00220345850640020201. [DOI] [PubMed] [Google Scholar]
- 2.Awano, S., H. Mochizuki, T. Ansai, and T. Takehara. Identification and characterisation of endothelin converting enzyme-like endopeptidase in Porphyromonas gingivalis 381. J. Dent. Res., in press.
- 3.Banbula A, Mak P, Bugno M, Silberring J, Dubin A, Nelson D, Travis J, Potempa J. Prolyl tripeptidyl peptidase from Porphyromonas gingivalis. A novel enzyme with possible pathological implications for the development of periodontitis. J Biol Chem. 1999;274:9246–9252. doi: 10.1074/jbc.274.14.9246. [DOI] [PubMed] [Google Scholar]
- 4.Banbula A, Bugno M, Kuster A, Heinrich P C, Travis J, Potempa J. Rapid and efficient inactivation of IL-6 by lysine and arginine specific proteinases (gingipains) from Porphyromonas gingivalis. Biochem Biophys Res Commun. 1999;261:598–602. doi: 10.1006/bbrc.1999.1075. [DOI] [PubMed] [Google Scholar]
- 5.Barrett A J, Rawlings N D, Woessner J F. Handbook of proteolytic enzymes. London, England: Academic Press; 1998. [Google Scholar]
- 6.Barua P K, Neiders M E, Topolnycky A, Zambon J J, Birkedal-Hansen H. Purification of an 80,000-Mr glycylprolyl peptidase from Bacteroides gingivalis. Infect Immun. 1989;57:2522–2528. doi: 10.1128/iai.57.8.2522-2528.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birkedal-Hansen H, Taylor R E, Zambon J J, Barwa P K, Neiders M E. Characterization of collagenolytic activity from strains of Bacteroides gingivalis. J Periodontal Res. 1988;23:258–264. doi: 10.1111/j.1600-0765.1988.tb01369.x. [DOI] [PubMed] [Google Scholar]
- 8.Bourgeau G, Lapointe H, Peloquin P, Mayrand D. Cloning, expression, and sequencing of a protease gene (tpr) from Porphyromonas gingivalis W83 in Escherichia coli. Infect Immun. 1992;60:3186–3192. doi: 10.1128/iai.60.8.3186-3192.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandt W, Lehmann T, Thondorf I, Born I, Schutkowski M, Rahfeld J U, Neubert K, Barth A. A model of the active site of dipeptidyl peptidase IV predicted by comparative molecular field analysis and molecular modelling simulations. Int J Pept Protein Res. 1995;46:494–507. doi: 10.1111/j.1399-3011.1995.tb01605.x. [DOI] [PubMed] [Google Scholar]
- 10.Calkins C C, Platt K, Potempa J, Travis J. Inactivation of tumor necrosis factor-alpha by proteinases (gingipains) from the periodontal pathogen, Porphyromonas gingivalis. Implications of immune evasion. J Biol Chem. 1998;273:6611–6614. doi: 10.1074/jbc.273.12.6611. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z, Potempa J, Polanowski A, Wikstrom M, Travis J. Purification and characterization of a 50-kDa cysteine proteinase (gingipain) from Porphyromonas gingivalis. J Biol Chem. 1992;267:18896–18901. [PubMed] [Google Scholar]
- 12.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:991–995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dashper S G, Kandasamy S, O'Brien-Simpson N, Reynolds E C. Amino acid and peptide uptake by Porphyromonas gingivalis. J Dent Res. 1998;77:1133. [Google Scholar]
- 14.Discipio R G, Daffern P J, Kawahara M, Pike R, Travis J, Hugli T E, Potempa J. Cleavage of human complement component C5 by cysteine proteinases from Porphyromonas (Bacteroides) gingivalis. Prior oxidation of C5 augments proteinase digestion of C5. Immunology. 1996;87:660–667. doi: 10.1046/j.1365-2567.1996.478594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fletcher J, Nair S, Poole S, Henderson B, Wilson M. Cytokine degradation by biofilms of Porphyromonas gingivalis. Curr Microbiol. 1998;36:216–219. doi: 10.1007/s002849900297. [DOI] [PubMed] [Google Scholar]
- 16.Grenier D, McBride B C. Surface location of a Bacteroides gingivalis glycylprolyl protease. Infect Immun. 1989;57:3265–3269. doi: 10.1128/iai.57.11.3265-3269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grenier D, McBride B C. Isolation of a membrane-associated Bacteroides gingivalis glycylprolyl protease. Infect Immun. 1987;55:3131–3136. doi: 10.1128/iai.55.12.3131-3136.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grenier D, Chao G, McBride B C. Characterization of sodium dodecyl sulfate-stable Bacteroides gingivalis proteases by polyacrylamide gel electrophoresis. Infect Immun. 1989;57:95–99. doi: 10.1128/iai.57.1.95-99.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinode D, Hayashi H, Nakamura R. Purification and characterization of three types of proteases from culture supernatants of Porphyromonas gingivalis. Infect Immun. 1991;59:3060–3068. doi: 10.1128/iai.59.9.3060-3068.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann T, Faust J, Neubert K, Ansorge S. Dipeptidyl peptidase IV (CD 26) and aminopeptidase N (CD 13) catalyzed hydrolysis of cytokines and peptides with N-terminal cytokine sequences. FEBS Lett. 1993;336:61–64. doi: 10.1016/0014-5793(93)81609-4. [DOI] [PubMed] [Google Scholar]
- 21.Hosotaki K, Imamura T, Potempa J, Kitamura N, Travis J. Activation of protein C by arginine-specific cysteine proteinases (gingipains-R) from Porphyromonas gingivalis. Biol Chem. 1999;380:75–80. doi: 10.1515/BC.1999.009. [DOI] [PubMed] [Google Scholar]
- 22.Imamura T, Potempa J, Pike R N, Travis J. Effect of free and vesicle-bound cysteine proteinases of Porphyromonas gingivalis on plasma clot formation: implications for bleeding tendency at periodontitis sites. Infect Immun. 1995;63:1999–2003. doi: 10.1128/iai.63.12.4877-4882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imamura T, Potempa J, Tanase S, Travis J. Activation of blood coagulation factor X by arginine-specific cysteine proteinases (gingipain-Rs) from Porphyromonas gingivalis. J Biol Chem. 1997;272:16062–16067. doi: 10.1074/jbc.272.25.16062. [DOI] [PubMed] [Google Scholar]
- 24.Ingman T, Tervahartiala T, Ding Y, Tschesche H, Haerian A, Kinane D F, Konttinen Y T, Sorsa T. Matrix metalloproteinases and their inhibitors in gingival crevicular fluid and saliva of periodontitis patients. J Clin Periodontol. 1996;23:1127–1132. doi: 10.1111/j.1600-051x.1996.tb01814.x. [DOI] [PubMed] [Google Scholar]
- 25.Jagels M A, Travis J, Potempa J, Pike R, Hugli T E. Proteolytic inactivation of the leukocyte C5a receptor by proteinases derived from Porphyromonas gingivalis. Infect Immun. 1996;64:1984–1991. doi: 10.1128/iai.64.6.1984-1991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiyama M, Hayakawa M, Shiroza T, Nakamura S, Takeuchi A, Masamoto Y, Abiko Y. Sequence analysis of the Porphyromonas gingivalis dipeptidyl peptidase IV gene. Biochim Biophys Acta. 1998;1396:39–46. doi: 10.1016/s0167-4781(97)00225-x. [DOI] [PubMed] [Google Scholar]
- 27.Lawson D A, Meyer T F. Biochemical characterization of Porphyromonas (Bacteroides) gingivalis collagenase. Infect Immun. 1992;60:1524–1529. doi: 10.1128/iai.60.4.1524-1529.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lourbakos A, Chinni C, Thompson P, Potempa J, Travis J, Mackie E J, Pike R N. Cleavage and activation of proteinase-activated receptor-2 on human neutrophils by gingipain-R from Porphyromonas gingivalis. FEBS Lett. 1998;435:45–48. doi: 10.1016/s0014-5793(98)01036-9. [DOI] [PubMed] [Google Scholar]
- 29.Lu B, McBride B C. Expression of the tpr protease gene of Porphyromonas gingivalis is regulated by peptide nutrients. Infect Immun. 1998;66:5147–5156. doi: 10.1128/iai.66.11.5147-5156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madden T E, Clark V L, Kuramitsu H K. Revised sequence of the Porphyromonas gingivalis prtT cysteine protease/hemagglutinin gene: homology with streptococcal pyrogenic exotoxin B/streptococcal proteinase. Infect Immun. 1995;63:238–247. doi: 10.1128/iai.63.1.238-247.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 32.Mikolajczyk-Pawlinska J, Travis J, Potempa J. Modulation of interleukin-8 activity by gingipains from Porphyromonas gingivalis: implications for pathogenicity of periodontal disease. FEBS Lett. 1998;4:282–286. doi: 10.1016/s0014-5793(98)01461-6. [DOI] [PubMed] [Google Scholar]
- 33.Nelson D, Potempa J, Kordula T, Travis J. Purification and characterization of a novel cysteine proteinase (periodontain) from Porphyromonas gingivalis. Evidence for a role in the inactivation of human alpha1-proteinase inhibitor. J Biol Chem. 1999;274:12245–12251. doi: 10.1074/jbc.274.18.12245. [DOI] [PubMed] [Google Scholar]
- 34.Potempa J, Travis J. Porphyromonas gingivalis proteinases in periodontitis, a review. Acta Biochim Pol. 1996;43:455–465. [PubMed] [Google Scholar]
- 35.Proost P, De Meester I, Schols D, Struyf S, Lambeir A M, Wuyts A, Opdenakker G, De Clercq E, Scharpe S, Van Damme J. Amino-terminal truncation of chemokines by CD26/dipeptidyl-peptidase IV. Conversion of RANTES into a potent inhibitor of monocyte chemotaxis and HIV-1-infection. J Biol Chem. 1998;273:7222–7227. doi: 10.1074/jbc.273.13.7222. [DOI] [PubMed] [Google Scholar]
- 36.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 37.Scott C F, Whitaker E J, Hammond B F, Colman R W. Purification and characterization of a potent 70-kDa thiol lysyl-proteinase (Lys-gingivain) from Porphyromonas gingivalis that cleaves kininogens and fibrinogen. J Biol Chem. 1993;268:7935–7942. [PubMed] [Google Scholar]
- 38.Tang-Larsen J, Claesson R, Edlund M B, Carlsson J. Competition for peptides and amino acids among periodontal bacteria. J Periodontal Res. 1995;30:390–395. doi: 10.1111/j.1600-0765.1995.tb01292.x. [DOI] [PubMed] [Google Scholar]
- 39.Toda K, Otsuka M, Ishikawa Y, Sato M, Yamamoto Y, Nakamura R. Thio-dependent collagenolytic activity in culture media of Bacteroides gingivalis. J Periodontal Res. 1984;19:372–381. doi: 10.1111/j.1600-0765.1984.tb01010.x. [DOI] [PubMed] [Google Scholar]
- 40.Travis J, Pike R, Imamura T, Potempa J. Porphyromonas gingivalis proteinases as virulence factors in the development of periodontitis. J Periodontal Res. 1997;32:120–125. doi: 10.1111/j.1600-0765.1997.tb01392.x. [DOI] [PubMed] [Google Scholar]
- 41.Yun P L, DeCarlo A A, Hunter N. Modulation of major histocompatibility complex protein expression by human gamma interferon mediated by cysteine proteinase-adhesin polyproteins of Porphyromonas gingivalis. Infect Immun. 1999;67:2986–2995. doi: 10.1128/iai.67.6.2986-2995.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Dong H, Kashket S, Duncan M J. IL-8 degradation by Porphyromonas gingivalis proteases. Microb Pathog. 1999;26:275–280. doi: 10.1006/mpat.1998.0277. [DOI] [PubMed] [Google Scholar]