Abstract
Background
Antibodies against human neutrophil antigen (HNA)-3a are associated with severe cases of transfusion-related acute lung injury (TRALI). The HNA-3 system is located on choline transporter-like 2 (CTL-2) protein. CTL-2 is encoded by the gene SLC44A2 and a single-nucleotide polymorphism c.461G>A results in two antigens: HNA-3a and HNA-3b. Three HNA-3 genotypes/ phenotypes exist: HNA-3aa, HNA-3bb, and HNA-3ab. Two different pathways of anti-HNA-3a TRALI have been described: a two-hit neutrophil-dependent pathway and a one-hit neutrophil-independent pathway. However, it is not clear whether HNA-3ab heterozygous patients have a lower risk of anti-HNA-3a-mediated TRALI compared to HNA-3aa homozygous patients.
Materials and methods
Healthy volunteers were genotyped for HNA-3 by real-time polymerase chain reaction, and phenotyped for HNA-3a by granulocyte immunofluorescence test (GIFT) and granulocyte agglutination test (GAT) against two donor sera containing anti-HNA-3a antibodies. The two sera were also used in in vitro models of human pulmonary microvascular endothelial cell (HLMVEC) cytotoxicity to investigate pathways of TRALI development.
Results
For both anti-HNA-3a sera, GIFT results matched the genotype, with a lower GIFT ratio for HNA-3ab neutrophils compared to HNA-3aa neutrophils, whereas GAT results showed no difference in agglutination. HLMVEC cytotoxicity was not observed in a one-hit neutrophil-independent model but was observed in a two-hit neutrophil-dependent model. Differences in cytotoxicity were observed between the two anti-HNA-3a sera used. Consistent with reduced HNA-3a antigen density as measured by GIFT, HNA-3ab neutrophils mediated less HLMVEC cytotoxicity than HNA-3aa neutrophils.
Conclusion
HNA-3 genotype and HNA-3a antigen expression impacted the severity of anti-HNA-3a-mediated HLMVEC cytotoxicity in a two-hit neutrophil-dependent model of TRALI. Different HNA-3a antibodies might also impact the magnitude of HLMVEC cytotoxicity.
Keywords: transfusion-related acute lung injury (TRALI), human neutrophil antigens, neutrophils, lung vascular endothelium
INTRODUCTION
Transfusion-related acute lung injury (TRALI) is a life-threatening complication mostly associated with transfusion of blood components containing either anti-human leukocytes antigens (HLA) or anti-human neutrophil antigens (HNA) antibodies1,2. The HNA-3 system localizes on choline transporter-like protein 2 (CTL-2) which is expressed on multiple cell types including neutrophils, lymphocytes, platelets3 and pulmonary endothelium4. CTL-2 is an 80 KDa transmembrane protein, containing five extracellular loops, with the HNA-3 antigen located on the first loop. CTL-2 is encoded by the SLC44A2 gene, and a single nucleotide polymorphism (SNP) at position c.461G>A3 results in a p.Arg154Gln amino-acid substitution. This leads to two CTL-2 variants which express either the HNA-3a (Arg) and HNA-3b (Gln) antigen3. Therefore, three different HNA-3 genotypes are possible: HNA-3aa homozygous, HNA-3ab heterozygous and HNA-3bb homozygous, being found in approximately 55–64%, 30–40% and 5% of the Caucasian population, respectively3. Alloantibodies against HNA-3a5–8, but not HNA-3b9, have been linked to severe TRALI. TRALI is commonly thought to develop via a two-hit mechanism10. The patient’s clinical condition is the first hit, and this predisposes them to TRALI development via activation of their pulmonary endothelium and the recruitment of primed and adherent neutrophils to the lungs. The blood transfusion is the second hit. Antibodies or BRMs present in the blood component activate the primed adherent neutrophils resulting in the release of reactive oxygen species (ROS) and enzymes that damage the pulmonary endothelium resulting in TRALI. However, the precise cellular pathways for TRALI development remain uncertain. Six different pathways have been described, two of which apply to anti-HNA-3a-mediated TRALI11. First, a two-hit neutrophil activation pathway that develops as described above10. However, a one-hit neutrophil-independent pathway has also been described12. Transfused anti-HNA-3a (first hit) binds cognate antigen on pulmonary endothelial cells resulting in their activation, ROS release and increased endothelial monolayer permeability precipitating TRALI12. These two different pathways highlight that anti-HNA-3a-mediated TRALI is not yet fully understood. Furthermore, as approximately one third of HNA-3a positive individuals have heterozygous expression3, it is important to understand whether they are at less risk of TRALI compared to HNA-3aa homozygous individuals. Therefore, we investigated the relation between HNA-3 genotype, HNA-3a antigen density and neutrophil agglutination. We also investigated whether anti-HNA-3a antibodies induced endothelial cytotoxicity via a neutrophil-independent one-hit pathway and/ or via a neutrophil-dependent two-hit pathway. Finally, we investigated whether neutrophil HNA-3 genotype/phenotype would impact the magnitude of anti-HNA-3a-mediated endothelial cytotoxicity.
MATERIALS AND METHODS
Ethics, blood collection and sera preparation
This study was by the Australian Red Cross Lifeblood Human Ethics Committee (300115).
This study was approved by the Australian Red Cross Lifeblood Ethics Committee (approvals 2008#12 and 2012#05). As a source of leukocytes or neutrophils, or for HNA-3 genotyping, 10–15 mL of whole blood was collected from healthy volunteers into ethylenediamine tetra-acetic acid (EDTA) tubes (Becton Dickinson [BD], Franklin Lakes, NJ, USA).
Two sera were obtained from blood donors previously implicated in TRALI. Granulocyte immunofluorescence test (GIFT) and granulocyte agglutination test (GAT) confirmed the presence of anti-HNA-3a antibodies and excluded the presence of other leukocyte-reactive antibodies. These sera were designated as anti-HNA-3a serum #1 and anti-HNA-3a serum #2. Sera were of a comparable titre (anti-HNA-3a serum #1: GIFT=n/8, GAT=n/16 serum; anti-HNA-3a serum #2: 2 GIFT=n/16 GAT=n/16).
Control serum employed in GIFT and GAT was an AB serum known not to contain either HNA or HLA antibodies. Control sera employed in TRALI in vitro models was prepared from 10 mL of whole blood collected from 5 healthy volunteers into serum-separator tubes (BD). Blood was left for 30 min at room temperature (RT) then centrifuged for 15 min at 3,000 × g. Sera were pooled, aliquoted, and stored at −30°C. Absence of anti-HLA and anti-HNA antibodies was determined by GIFT and GAT.
Leukocyte harvest, neutrophil isolation, and source of endothelial cells
Leukocytes used in GIFT and GAT with anti-HNA-3a sera#1 were harvested from whole blood (n=56). Cells were fixed in 0.5 X IO Test3 Fixative solution (Beckman Coulter, Brea, CA, USA) at 37°C for 4 min. Red cells were lysed in 1X IO Test3 Lysing solution (Beckman Coulter) at 37°C for 10 min. Cells were centrifuged at 200 × g for 5 min, washed in PBS/EDTA/BSA 0.2% buffer (Sigma Aldrich, St Louis, MO, USA) and the final pellet was resuspended in PBS/EDTA/BSA 0.2% buffer (Sigma Aldrich).
Neutrophils used in the CTL-2 expression (n=10), GIFT (n=19) and GAT (n=15) experiments with anti-HNA-3a sera #2, as well as in the in vitro TRALI models (n=20) were isolated from whole blood using Easy-Sep direct human neutrophil isolation kit (Stemcell Technologies, Vancouver, Canada) according to manufacturer’s instructions.
Primary human lung microvascular endothelial cells were sourced from a commercial supplier (n=3 donors; HLMVEC-L, Merck KGaA, Darmstadt, Germany). These were stored in liquid nitrogen, and upon thawing were cultured in microvascular endothelial cell growth medium (Merck) according to manufacturer’s instructions.
HNA-3 genotyping
Genomic DNA was extracted from whole blood (n=56) and HLMVECs (n=3) using Qiacube/QiaAmp manual extraction kits (Qiagen, Hilden, Germany) according to manufacturer’s instructions. HNA genotype was determined by in-house Taqman Real-Time PCR using Applied Biosystems Viia7 (Biosystems, Waltham, MA, USA) and QuantStudio6 instruments (ThermoFisher Scientific, Waltham, MA, USA). The encoding allele HNA-3a and HNA-3b frequencies were calculated as the ratio between the number of times the allele of interest was observed and the total number of copies of all the alleles at that particular genetic locus.
CTL-2 expression
Isolated neutrophils (2.5×105) were washed with PBS/EDTA/BSA 0.2% buffer (Sigma Aldrich) and incubated with 0.2 mg/mL of rabbit anti-human CTL-2 polyclonal antibody (ab221759, Abcam, Cambridge, UK) for 1 hour at room temperature (RT). An Alexa Fluor 488 goat anti-rabbit secondary antibody, (A32731, Invitrogen, Waltham, MA, USA), a CD16 FITC conjugated (0302802 BD Bioscience, Franklin Lakes, NJ, USA) and a CD177 APC-conjugated (ab77230 Abcam) monoclonal antibodies were added to the tube and incubated for 30 min RT in the dark. Neutrophils were gated on CD16 and CD177 expression and CTL-2 Median Fluorescent Intensity (MFI) were recorded by flow cytometry (FACSCanto II, BD Bioscience). Samples incubated with secondary antibody only were used as negative control.
GIFT and GAT assay
GIFT and GAT were employed as they are the gold standard for the detection of HNA-3 antibodies, as no specific commercial antibodies are available13. For GIFT, 25×104 fixed leukocytes or neutrophils were mixed 1:1 volume (25 μL) with control or test sera in a 96-well plate (Nunc). The plate was incubated (30°C, 30 min), centrifuged to pellet cells, inverted to drain, and leukocytes were washed with PBS/EDTA/BSA 0.2%. Goat anti-human F(ab′) fragment IgG-PE and IgM-AF647 (both Jackson Immunoresearch, West Grove, PA, USA) were diluted n/100 and n/400 respectively in PBS/EDTA/BSA 0.2% buffer and added to the plate which was then incubated (RT in the dark, 30 min). Cells were then washed in PBS/EDTA/BSA 0.2% and analysed by flow cytometry (BD FACSCanto II). GIFT ratio was determined as the ratio between the serum MFI value and the average MFI value from three independent wells incubated with negative control serum.
For GAT, sera were diluted (n/2) and added (5 μL) to harvested leukocytes (5×103), gently mixed and incubated at 30°C in a dry incubator for 3–5 hours. Plates were read on an inverted phase contrast microscope, and the strength of agglutination was graded on a scale of 0–4+.
In vitro models of TRALI
Four different in vitro models adapted from Silliman et al.10 were used (Table I). HNA-3aa homozygous HLMVECs were passaged according to manufacturer’s instructions and used at the 4th to 6th passage after initial thawing. In each well of a 12-well plate (Nunc), 5×104 HLMVECs were cultured with microvascular endothelial cell growth medium (Merck) to 80–90% confluence. In neutrophil-independent models, confluent HLMVECs were either left untreated (one-hit model) or stimulated with 2 μg/mL lipopolysaccharide (LPS, E. Coli O111-B4, Sigma Aldrich; two-hit model) for 6.5 hours at 37°C in 5% CO2. In neutrophil-dependent models, confluent HLMVEC were either left untreated (one-hit model) or stimulated with 2 μg/mL LPS (two-hit model) for 6 hours at 37°C in 5% CO2, EasySep-isolated neutrophils (1×106) were added (1:10 neutrophil:HLMVEC ratio) and incubated (30 mins at 37°C in 5% CO2). In all models, HLMVEC were then treated with either media, control serum or either of the two anti-HNA-3a sera (10% final volume) for 30 min. Plates were then quickly inverted onto absorbent towel and warmed medium was added. Wells were stained one by one with trypan blue (final dilution 1:20, Cat#15250061, ThermoFisher Scientific) and cells counted or photographed within 2 min.
Table I.
Pathway of TRALI tested and employed insults
| TRALI Pathway | 1st hit | 2nd hit |
|---|---|---|
| Neutrophil-independent pathway (Bayat et al. 201312) | ||
| One-hit model | Media (control) Control serum Anti-HNA-3a serum #1 Anti-HNA-3a serum #2 |
N/A |
| Two-hit model | LPS | Media (control) Control serum Anti-HNA-3a serum #1 Anti-HNA-3a serum #2 |
| Neutrophil-dependent pathway (Silliman et al. 200710) | ||
| One-hit model | Media (control) PMA (control) Control serum Anti-HNA-3a serum #1 Anti-HNA-3a serum #2 |
N/A |
| Two-hit model | LPS | Media (control) PMA (control) Control serum Anti-HNA-3a serum #1 Anti-HNA-3a serum #2 |
LPS: lipopolysaccharide; HNA: human neutrophil antigen; PMA: phorbol 12-myristate 13-acetate; TRALI: transfusion-related acute lung injury.
In preliminary experiments, two independent scorers used a published method10,14 to quantify HLMVEC viability. As an alternative, 3–5 fields (ROI=1.454 mm2 each field) per well were acquired using an Olympus IX73 microscope (Shinjuku, Tokyo, Japan) with 10X objective and DP80 Olympus camera (Olympus), and viable HLMVECs/field were counted with ImageJ software (NIH, Bethesda, MD, USA). Results were comparable between the two methods, so the latter was used in subsequent experiments.
Statistics
Figures and statistical analyses were performed using Prism (GraphPad Software, San Diego, CA). Data distribution normality was assessed by D’Agostino & Pearson, Anderson-Darling and Shapiro-Wilk tests. Non-normally distributed data were reported as median and interquartile range (IQR) and were analyzed with either Kruskal-Wallis test followed by Dunn’s post hoc test (GIFT using anti-HNA-3a serum#1), or Mann-Whitney test (GAT with both sera). Normally distributed data (CTL-2 expression, GIFT against anti-HNA-3 serum #2 and HLMVEC cytotoxicity assay), were reported as mean ± SE, and analyzed with repeated measures one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test (GIFT against anti-HNA-3a serum#2, and HLMVEC viability). p<0.05 was considered significant.
RESULTS
Genotyping for HNA-3
HNA-3a genotype was determined for 56 healthy volunteers. Thirty-one were HNA-3aa homozygous (55%), twenty-one were HNA-3ab heterozygous (37.5%), and four were HNA-3bb homozygous (7.1%). Therefore, the calculated allele frequencies were 0.741 for HNA-3a and 0.258 for HNA-3b. Genotyping was also used to identify HNA3-aa homozygous HLMVECs to use in TRALI models.
Different HNA-3 genotypes resulted in different levels of neutrophil HNA-3a antigen expression but not CTL-2 expression
HNA-3 antigens are generated by a p.Arg154Gln amino-acid exchange in the first extracellular loop of the CTL-2 protein15. We used flow cytometry and a polyclonal anti-human CTL-2 (i.e., not specific for either HNA-3a or HNA-3b variant) to investigate whether neutrophil surface expression of CTL-2 varied with different HNA-3 genotypes. Relative CTL-2 expression (MFI) was similar regardless of HNA-3 genotype (n=10: HNA-3aa: n=4; HNA-3ab: n=3; HNA-3bb: n=3; Figure 1A). CTL-2 expression on HNA-3aa genotyped HLMVECs was demonstrated using flow cytometry (Online Supplementary Content, Figure S1A) and ELISA (Online Supplementary Content, Figure S1B). These HLMVECs also bound each of the two anti-HNA-3a sera, but not an anti-HNA-3b serum, in a modified GIFT (Online Supplementary Content, Figure S1C).
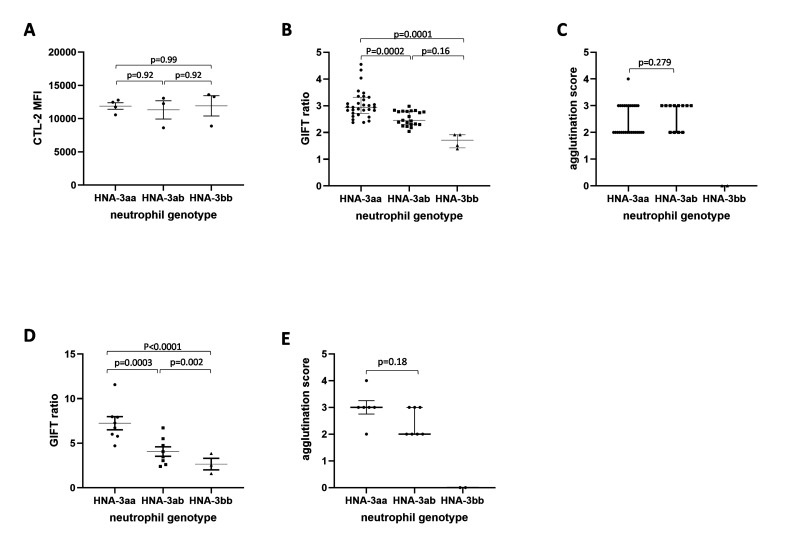
Figure 1. GIFT and GAT phenotyping.
(A) CTL-2 protein expression level was evaluated on surface of freshly isolated neutrophils. Easy-Sep isolated neutrophils were incubated with rabbit polyclonal anti-human CTL-2 antibody (0.2 mg/mL) for 60 min. After 2 washes, cells were incubated with an anti-Rabbit 488-conjugated antibody for 30 min. Following incubation, samples were analysed on FACSCanto and MFI was recorded. Total leukocytes were isolated from 56 healthy volunteers and GIFT (B) and GAT (C) were performed with one anti-HNA-3 serum (anti-HNA-3a serum #1). GIFT (D) and GAT (E) were repeated on Easy-Sep isolated neutrophils from 19 and 15 healthy volunteers, respectively, using a second anti-HNA-3a serum (anti-HNA-3a serum #2). CTL-2 and GIFT against anti-HNA serum #2 data are presented as mean ± SE. GIFT against anti-HNA-3 serum #1 and GAT data are presented as median and IQR. P <0.05 was considered statistically significant.
We used GIFT against anti-HNA-3a Serum#1 (Figure 1B) to measure neutrophil HNA-3a antigen density (n=56). Neutrophils from HNA-3aa homozygous volunteers had a higher GIFT ratio compared to HNA-3bb volunteers (2.94 IQR 0.6 (2.72, 3.32) vs 1.71 IQR 0.49 [1.421, 1.915]; p=0.0001). Neutrophils from HNA-3ab heterozygous volunteers had an intermediate phenotype, with a GIFT ratio (2.455 IQR 0.49 [2.3, 2.793]) lower than for homozygous HNA-3aa neutrophils (p=0.0002), but higher than for homozygous HNA-3bb neutrophils (p=0.163). These results suggested that HNA-3ab heterozygous neutrophils had a lower HNA-3a antigen density on their surface compared to HNA-3aa homozygous neutrophils. We then used the GAT to investigate whether the differences in HNA-3a antigen density impacted the ability of anti-HNA-3a serum #1 to induce agglutination of heterozygote HNA-3ab granulocytes. No difference in agglutination was observed between homozygous HNA-3aa and heterozygous HNA-3ab granulocytes (2 IQR 1 [2, 3] vs 3 IQR 1 [2, 3] respectively; p=0.279; Figure 1C). As expected, no agglutination was observed for homozygous HNA-3bb granulocytes (Figure 1C).
We confirmed these results using anti-HNA-3a serum #2 tested against neutrophils (GIFT: n=19, Figure 1D; and GAT: n=15; Figure 1E). Again, HNA-3aa homozygous neutrophils had a higher GIFT ratio compared to HNA-3bb homozygous neutrophils (7.24±0.78 vs 2.66±0.79, p<0.0001). HNA-3ab heterozygous neutrophils (4.07±0.56; Figure 1D) again showed an intermediate phenotype, lower than HNA-3aa homozygous neutrophils (p=0.0003), but higher than HNA-3bb homozygous neutrophils (p=0.002). Both HNA-3aa homozygous and HNA-3ab heterozygous neutrophils agglutinated in the presence of anti-HNA-3a serum#2, while HNA-3bb homozygous neutrophils didn’t (Figure 1E). Again, despite the differences observed in the GIFT, no difference in GAT agglutination score was recorded between HNA-3aa homozygous and HNA-3ab heterozygous neutrophils (p=0.18).
We also phenotyped neutrophils with different HNA genotypes against an anti-HNA-3b serum (GIFT: n=10: HNA-3aa: n=4; HNA-3ab: n=4; HNA-3bb: n=2; Online Supplementary Content, Figure S2A). HNA-3bb homozygous neutrophils had the highest GIFT ratio compared to HNA-3aa homozygous neutrophils (4.25±2.58 vs 0.75±0.05, p=0.003, Online Supplementary Content, Figure S2A). HNA-3ab heterozygous neutrophils (2.08±0.49; Online Supplementary Content, Figure S2A) had a GIFT ratio lower than HNA-3bb homozygous neutrophils (p=0.0477), but not higher than HNA-3aa homozygous neutrophils (p=0.10). In GAT, (n=8: HNA-3aa: n=3; HNA-3ab: n=3; HNA-3bb: n=2; Online Supplementary Content, Figure S2B), HNA-3bb homozygous neutrophils had increased agglutination compared to HNA-3aa homozygous neutrophils (4 IQR 0 (4, 4) vs 0 IQR 0 (0, 0) respectively; p=0.0043), but not compared to HNA-3ab heterozygous neutrophils (3 IQR 2 [1, 3]; p=0.1159).
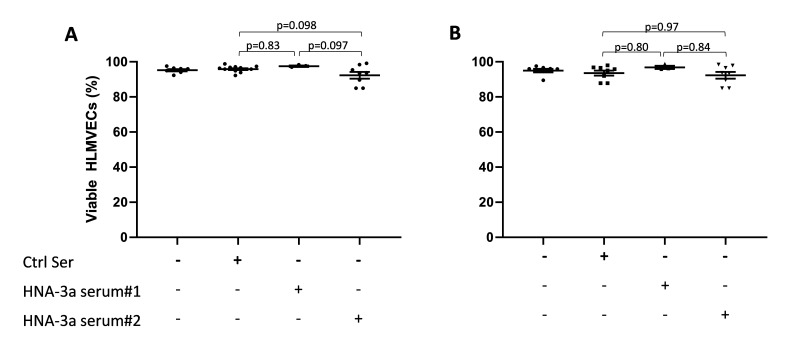
No evidence of neutrophil-independent anti-HNA-3a-mediated endothelial damage
Two different pathways have been reported for anti-HNA-3a mediated TRALI11. We first investigated whether there was any evidence that the direct endothelial cell activation pathway12 resulted in HLMVEC cytotoxicity. In the one hit model (i.e., without LPS stimulation), neither of our anti-HNA-3a sera induced HLMVEC cytotoxicity (anti-HNA-3a serum #1: 97.48±0.39% viability, and anti-HNA-3a serum#2: 92.28±1.93% viability vs control sera 95.78±0.49% viability; p=0.83 and p=0.098 respectively; Figure 2A). We therefore investigated whether a first hit (LPS) would predispose anti-HNA-3a-mediated HLMVEC cytotoxicity. However, even in this two-hit model, neither of our anti-HNA-3a sera induced HLMVEC cytotoxicity (anti-HNA-3a serum #1: 96.77±0.93% viability and anti-HNA-3a serum#2: 92.3±1.91% viability vs control sera 93.54±1.43% viability; p=0.80 and p=0.97 respectively; Figure 2B). Doubling the concentration of anti-HNA-3a sera to 20% did not result in HLMVEC cytotoxicity, suggesting that the lack of cytotoxicity was not due to an insufficient concentration of anti-HNA-3a antibodies in the serum (data not shown).
Figure 2. In vitro models of neutrophil-independent anti-HNA-3a-mediated endothelial cytotoxicity.
(A–B) Human lung microvascular endothelial cells (HLMVECs) were grown to 80–90% confluence in 12-well plates and left untreated (one hit model; A) or stimulated with 2 μg/mL lipopolysaccharide (LPS; two hit model; B) for 6 hours, followed by the addition of either buffer, control serum (Ctrl Ser) or anti-HNA-3a sera (anti-HNA-3a serum #1 or anti-HNA-3a serum #2). All sera were 10% final volume. After 30 min plates were flicked, cells were stained with Trypan blue and images were acquired. Viable HLMVECs were counted with ImageJ software. Data are presented as mean ± SE. p<0.05 was considered statistically significant.
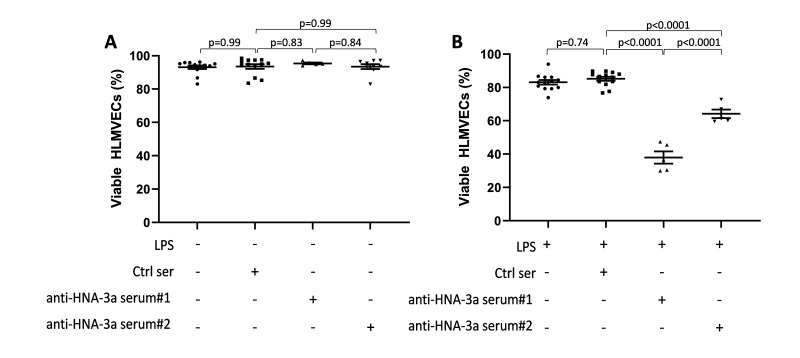
HNA-3aa homozygous neutrophil-dependent anti-HNA-3a-mediated endothelial damage required two hits
Silliman et al. reported a two-hit neutrophil-mediated pathway for anti-HNA-3a-mediated TRALI10. Therefore, we investigated whether either of our two sera could induce HNA-3aa homozygous neutrophil-dependent HLMVEC cytotoxicity. In a one-hit model (i.e., without LPS), neither sera induced HLMVEC cytotoxicity in the presence of HNA-3aa homozygous neutrophils (anti-HNA-3a serum #1: 95.41±0.53% viability and anti-HNA-3a serum#2: 93.48±1.46% viability vs control sera 93.56±1.39% viability; p=0.83 and p=0.99 respectively; Figure 3A). In contrast, in the two-hit model (i.e. with LPS), both our sera induced HLMVEC cytotoxicity which was anti-HNA-3a specific as it was not observed in response to control serum (anti-HNA-3a serum #1: 37.97±3.66% viability and anti-HNA-3a serum #2: 64.18±2.52% viability vs control sera 85.2±1.20% viability; p<0.0001 for both; Figure 3B). HLMVEC cytotoxicity induced by anti-HNA-3a serum#1 was greater than that induced by anti-HNA-3a serum#2 (p<0.0001). No further increase in HLMVEC cytotoxicity was observed when the concentration of anti-HNA-3a sera was doubled to 20% (data not shown). These results suggested that in this experimental setting, anti-HNA-3a mediated HLMVEC cytotoxicity was dependent on pre-stimulation with LPS as a first hit, the presence of HNA-3aa homozygous neutrophils, and exposure to anti-HNA-3a antibodies as a second hit.
Figure 3. In vitro models of neutrophil-dependent anti-HNA-3a-mediated endothelial cytotoxicity.
A–B: Human lung microvascular endothelial cells (HLMVECs) were grown to 80–90% confluence in 12-well plates and left untreated (one hit model; A) or stimulated with 2 μg/mL lipopolysaccharide (LPS; two hit model; B) for 6 hours, followed by the addition of 1×106 HNA-3aa neutrophils that were allowed to settle for 30 min. Then in both models, cells were treated with either buffer, control serum (Ctrl Ser) or anti-HNA-3a sera (anti-HNA-3a serum#1 or anti-HNA-3a serum #2). All sera were 10% final volume. After 30 min plates were flicked, cells were stained with Trypan blue and images were acquired. Viable HLMVECs were counted with ImageJ software. Data are presented as mean ± SE. p<0.05 was considered statistically significant.
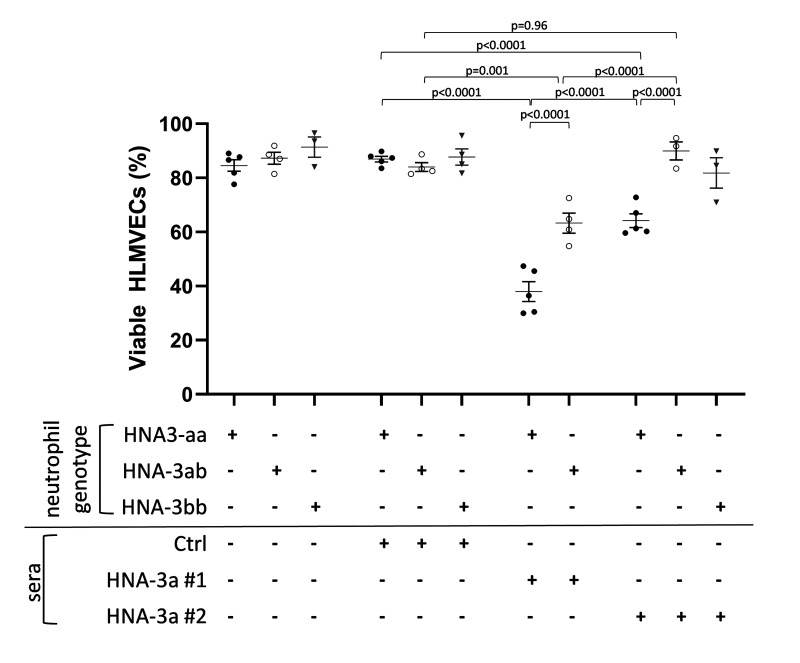
Reduced anti-HNA-3a-mediated endothelial damage was observed in the presence of HNA-3ab heterozygous neutrophils
As we had earlier demonstrated that the reduced expression of HNA-3a on HNA-3ab heterozygous neutrophils did not impact anti-HNA-3a mediated agglutination in the GAT, we investigated whether this would or would not impact anti-HNA-3a mediated HLMVEC cytotoxicity in our two-hit neutrophil-dependent model (Figure 4). When co-cultured with HNA-3ab heterozygous neutrophils, anti-HNA-3a serum#1 induced greater HLMVEC cytotoxicity than control serum (63.26±3.71% viability vs 84.00±1.61%; p=0.001). However, the anti-HNA-3a mediated HLMVEC cytotoxicity observed with HNA-3ab heterozygous neutrophils was less than that observed with HNA-3aa homozygous neutrophils (63.26±3.71% viability vs 37.97±3.66% viability; p<0.0001). In contrast, anti-HNA-3a serum #2 mediated HLMVEC cytotoxicity was evident only in the presence of HNA-3aa homozygous neutrophils (64.18±2.52% viability vs control sera: 86.95±1.045% viability; p<0.0001), and not in the presence of HNA-3ab heterozygous neutrophils (89.95±3.36% viability vs control sera 84.00±1.61%; p=0.98). These results showed that the inclusion of heterozygous HNA-3ab neutrophils in our two-hit neutrophil-dependent model was associated with less anti-HNA-3a-mediated HLMVEC cytotoxicity than when homozygous HNA-3aa neutrophils were used.
Figure 4. Two hit in vitro model of neutrophil-dependent anti-HNA-3-mediated endothelial cytotoxicity.
Human lung microvascular endothelial cells (HLMVECs) were grown to 80–90% confluence in 12-well plates and were stimulated with 2 μg/mL LPS for 6 hours, followed by the addition of buffer, freshly isolated HNA-3aa neutrophils (black dots), HNA-3ab neutrophils (white dots), or HNA-3bb neutrophils (grey triangles) that were allowed to settle for 30 min. Then cells were treated with either buffer, control serum (Ctrl serum) or anti HNA-3a sera (anti-HNA-3a serum#1 or anti-HNA-3a serum #2). All sera were 10% final volume. After 30 min plates were flicked, cells were stained with Trypan blue and 3–7 fields per well were acquired. Viable HLMVECs were counted with ImageJ software. Data are presented as mean ± SE. p<0.05 was considered statistically significant.
DISCUSSION
Anti-HNA-3a antibodies are associated with severe cases of TRALI5,6,8. Two different pathways for anti-HNA-3a-mediated TRALI are described10–12; however, regardless of the pathway, it is unclear whether HNA-3ab heterozygote patients are at decreased risk of anti-HNA-3a mediated TRALI compared to HNA-3aa homozygote patients. Our calculated allele frequencies of 0.741 for HNA-3a and 0.258 for HNA-3b were comparable to those reported in Caucasian (0.79 and 0.21 respectively)9,15–17, and Han Chinese (0.74 and 0.26 respectively)15,18 populations. While neutrophil CTL-2 expression levels were comparable across the three different HNA-3 genotypes, we found concordance between HNA-3 genotype and GIFT phenotype of neutrophils, with a lower HNA-3a antigen density on HNA-3ab heterozygous neutrophils compared to HNA-3aa homozygous neutrophils. Conversely, GAT showed no difference in agglutination between the two HNA-3a positive genotypes. Consistent with genotype and GIFT results, anti-HNA-3a-mediated HLMVEC cytotoxicity was lower in a two-hit neutrophil-dependent model in the presence of HNA-3ab heterozygous neutrophils compared to HNA-3aa homozygous neutrophils. This suggests a possible link between HNA-3 genotype and anti-HNA-3a-mediated TRALI severity.
We used two different anti-HNA-3a sera to assess the relationship between HNA-3 genotype and antigen expression. For both sera, we observed that neutrophils from individuals genotyped as HNA-3aa homozygous had a higher GIFT ratio than neutrophils from individuals genotyped as HNA-3ab heterozygous. This suggested a higher neutrophil expression of the HNA-3a antigen for HNA-3aa individuals compared to HNA-3ab individuals, and was in accordance with those reported previously9. Conversely, when tested against an anti-HNA-3b serum, HNA-3aa homozygous neutrophils had a lower GIFT ratio compared to HNA-3bb homozygous neutrophils while HNA-3ab heterozygous neutrophils had an intermediate phenotype. GAT measures the ability of IgG-sensitized granulocytes to form aggregates via a two-step process involving antibody binding and sensitisation of cells followed by chemotaxis, formation of homotypic interactions and agglutinates19,20. GAT is recognized as the most reliable serological assay for the detection of anti-HNA-3a antibodies13. Similar to our GIFT results, the GAT demonstrated that HNA-3aa homozygous neutrophils agglutinated in the presence of anti-HNA-3a serum but not anti-HNA-3b serum, while HNA-3bb homozygous neutrophils agglutinated in the presence of anti-HNA-3b serum but not anti-HNA-3a serum. However, in contrast to our GIFT results, we did not see reduced agglutination in the GAT for HNA-3ab heterozygous neutrophils compared to HNA-3aa homozygous neutrophils when exposed to anti-HNA-3a sera, or to HNA-3bb homozygous neutrophils when exposed to anti-HNA-3b serum. This was also in contrast to GAT results reported previously, where examples were shown of HNA-3ab cells reacting weakly with both anti-HNA-3a and anti-HNA-3b sera9. It is possible that while neutrophils from HNA-3ab heterozygotes express less HNA-3a antigen than HNA-3aa homozygotes, they still express sufficient HNA-3a antigen for anti-HNA-3a mediated sensitization, chemotaxis, and aggregation to occur. This might also be the case for their expression of HNA-3b antigen and their reaction to anti-HNA-3b. So why might our GAT results have differed from those reported previously9? Reil et al. tested several anti-HNA-3a and anti-HNA-3b sera9, while we only tested two, so it is possible that sera-specific properties might have contributed to the different findings. Procedural differences (antibody incubation time and temperature) may also have contributed to the discrepant results.
We adapted an in vitro model of TRALI14,21,10 to investigate the two potential pathways reported for anti-HNA-3a-mediated TRALI10–12. We first used HNA-3aa genotyped HLMVECs to investigate the one-hit neutrophil-independent pathway12 but did not observe any anti-HNA-3a-mediated HLMVEC cytotoxicity in the absence of neutrophils, even if LPS was added as a first hit to model a two-hit neutrophil-independent pathway. The differences between our results and those previously reported12 might relate to differences in experimental design. We used human sera rather than purified IgG12, and we measured HLMVEC cytotoxicity rather than endothelial barrier permeability12. As either of these outcome measures could cause extravascular fluid leakage clinically, further investigations are required to examine the role of antibody format (i.e., sera vs purified IgG) and whether either of our two sera might induce neutrophil-independent endothelial barrier permeability. We also investigated the neutrophil-dependent two-hit pathway10. Both of our anti-HNA-3a sera induced HLMVEC cytotoxicity in the presence of neutrophils from HNA-3aa individuals in our two-hit neutrophil-dependent model. As previously reported10,22, this HLMVEC cytotoxicity followed a two-hit pathway, as it was only evident when we included LPS as a first hit. Given our contrasting GIFT and GAT results, we wondered on the implications these would have for anti-HNA-3a mediated TRALI. Might HNA-3ab heterozygous patients be at reduced risk compared to HNA-3aa homozygous patients because of their lower neutrophil HNA-3a expression? Or might both HNA-3aa homozygous and HNA-3ab heterozygous patients have a similar risk because they all express HNA-3a and their neutrophils have similar agglutination responses to anti-HNA-3a antibodies? So, we also investigated the impact of neutrophil HNA-3 genotype. We observed that anti-HNA-3a sera #1 mediated HLMVEC cytotoxicity was reduced in the presence of HNA-3ab heterozygous neutrophils compared to HNA-3aa homozygous neutrophils. However, anti-HNA-3a serum #2 induced HLMVEC cytotoxicity in the presence of HNA-3aa homozygous neutrophils, but not HNA-3ab heterozygous neutrophils. These results suggested a gene-dose effect whereby the severity of HLMVEC cytotoxicity was linked to the level of HNA-3a expression. This is similar to what Reil et al. reported for GIFT and GAT9. Taken together, these results support the hypothesis that HNA-3ab heterozygous patients might be at less risk of anti-HNA-3a mediated TRALI compared to HNA-3aa homozygous patients because of the lower expression of HNA-3a on their cells. We also observed that exposure to anti-HNA-3a serum #2 in the presence of HNA-3aa homozygous neutrophils resulted in reduced HLMVEC cytotoxicity compared to exposure to anti-HNA-3a serum #1 and was comparable to that observed with anti-HNA-3a serum #1 in the presence of HNA-3ab heterozygous neutrophils. Doubling the dose of the anti-HNA-3a serum #2 did not further increase HLMVEC cytotoxicity. So even though the two anti-HNA-3a sera were of a comparable titre we observed differences in the severity of HLMVEC cytotoxicity that they mediated. Anti-HNA-3a antibodies can be classified as type I or type II based upon differences in their CTL-2 protein binding site23. One of our sera might contain type I anti-HNA-3a antibodies, while the other might contain type II anti-HNA-3a antibodies. Furthermore, CTL-2 undergoes a conformational change following priming24 that might favour binding of either type I or type II anti-HNA-3a antibodies. Future experiments could use constructs expressing different CTL-2 binding sites12 to determine whether our two sera contain different types of anti-HNA-3a antibodies. Furthermore, sera already characterised to contain either type I or type II anti-HNA-3a antibodies could be used in our in vitro TRALI model to determine the effect of anti-HNA-3a antibody type. Finally, it wasn’t possible to directly compare GIFT intensity of the two sera because when assessing anti-HNA-3a serum #1, total leukocytes were used and neutrophils were gated via surface marker expression, whereas when assessing anti-HNA-3a serum #2, isolated neutrophils were used. As HNA-3a is expressed on all leukocytes25, the reduced GIFT intensity observed with anti-HNA-3a serum #1 compared with anti-HNA-3a serum #2 (Figure 1D) likely reflects binding of anti-HNA-3a to the other leukocytes present in the cell suspension. Future experiments could look to directly compare GIFT intensity between anti-HNA-3a sera containing either type I or type II antibodies.
CONCLUSIONS
Differences in neutrophil HNA-3 genotype/phenotype impacted the magnitude of anti-HNA-3a-mediated HLMVEC cytotoxicity in our two-hit neutrophil-dependent TRALI model. This suggests that HNA-3ab heterozygous patients might be at lower risk of anti-HNA-3a-mediated TRALI compared to HNA-3aa homozygous patients. Differences in the magnitude of HLMVEC cytotoxicity observed between the two sera used in our study, suggest that antibody type might also impact anti-HNA-3a-mediated TRALI. Further analyses are required to better understand how anti-HNA-3a-mediated TRALI develops and this could lead to additional TRALI mitigation strategies that will improve transfusion safety.
Supplementary Information
ACKNOWLEDGMENTS
The Authors acknowledge Fenny Chong and Kelly Rooks for organizing the blood collections and performing the phlebotomies.
Footnotes
FUNDING AND RESOURCES
Australian Governments fund Australian Red Cross Lifeblood for the provision of blood, blood products and services to the Australian Community.
AUTHORSHIP CONTRIBUTION
SC designed the study, performed in vitro TRALI assays, analysed the data and wrote the manuscript. MB performed GIFT and GAT experiments, HLMVEC and neutrophil genotyping and CTL-2 experiments. PH and ND performed GIFT and GAT experiments. FR performed CTL-2 expression experiments and some Serum#1 in vitro TRALI assays. AJS and FTT contributed to the in vitro TRALI assay. MMD contributed to study design and reviewed the manuscript. JPT designed the study, reviewed the data and the manuscript.
All Authors declare no conflicts of interest.
REFERENCES
- 1.Popovsky MA, Abel MD, Moore SB. Transfusion-related acute lung injury associated with passive transfer of antileukocyte antibodies. Am Rev Respir Dis. 1983;128:185–189. doi: 10.1164/arrd.1983.128.1.185. [DOI] [PubMed] [Google Scholar]
- 2.Popovsky MA, Moore SB, Wick MR, Devine P, Pineda AA, Taswell HF. A blood bank consultation service: principles and practice. Mayo Clin Proc. 1985;60:312–314. doi: 10.1016/s0025-6196(12)60538-2. [DOI] [PubMed] [Google Scholar]
- 3.Muschter S, Berthold T, Greinacher A. Developments in the definition and clinical impact of human neutrophil antigens. Curr Opin Hematol. 2011;18:452–460. doi: 10.1097/MOH.0b013e32834babdd. [DOI] [PubMed] [Google Scholar]
- 4.Silliman CC, Bercovitz RS, Khan SY, Kelher MR, LaSarre M, Land KJ, et al. Antibodies to the HLA-A2 antigen prime neutrophils and serve as the second event in an in vitro model of transfusion-related acute lung injury. Vox Sang. 2014;107:76–82. doi: 10.1111/vox.12129. [DOI] [PubMed] [Google Scholar]
- 5.Nordhagen R, Conradi M, Drömtorp SM. Pulmonary reaction associated with transfusion of plasma containing anti-5b. Vox Sang. 1986;51:102–107. doi: 10.1111/j.1423-0410.1986.tb00223.x. [DOI] [PubMed] [Google Scholar]
- 6.Kopko PM, Marshall CS, MacKenzie MR, Holland PV, Popovsky MA. Transfusion-related acute lung injury: report of a clinical look-back investigation. JAMA. 2002;287:1968–1971. doi: 10.1001/jama.287.15.1968. [DOI] [PubMed] [Google Scholar]
- 7.Reil A, Keller-Stanislawski B, Gunay S, Bux J. Specificities of leucocyte alloantibodies in transfusion-related acute lung injury and results of leucocyte antibody screening of blood donors. Vox Sang. 2008;95:313–317. doi: 10.1111/j.1423-0410.2008.01092.x. [DOI] [PubMed] [Google Scholar]
- 8.Gottschall JL, Triulzi DJ, Curtis B, Kakaiya RM, Busch MP, Norris PJ, et al. The frequency and specificity of human neutrophil antigen antibodies in a blood donor population. Transfusion. 2011;51:820–827. doi: 10.1111/j.1537-2995.2010.02913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reil A, Wesche J, Greinacher A, Bux J. Geno- and phenotyping and immunogenicity of HNA-3. Transfusion. 2011;51:18–24. doi: 10.1111/j.1537-2995.2010.02751.x. [DOI] [PubMed] [Google Scholar]
- 10.Silliman CC, Curtis BR, Kopko PM, Khan SY, Kelher MR, Schuller RM, et al. Donor antibodies to HNA-3a implicated in TRALI reactions prime neutrophils and cause PMN-mediated damage to human pulmonary microvascular endothelial cells in a two-event in vitro model. Blood. 2007;109:1752–1755. doi: 10.1182/blood-2006-05-025106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tung J-P, Chiaretti S, Dean MM, Sultana AJ, Reade MC, Fung YL. Transfusion-related a cute lung injury (TRALI): Potential pathways of development, strategies for prevention and treatment, and future research directions. Blood Rev. 2022;53:100926. doi: 10.1016/j.blre.2021.100926. [DOI] [PubMed] [Google Scholar]
- 12.Bayat B, Tjahjono Y, Sydykov A, Werth S, Hippenstiel S, Weissmann N, et al. Anti-human neutrophil antigen-3a induced transfusion-related acute lung injury in mice by direct disturbance of lung endothelial cells. Arterioscler Thromb Vasc Biol. 2013;33:2538–2548. doi: 10.1161/atvbaha.113.301206. [DOI] [PubMed] [Google Scholar]
- 13.Lucas G, Rogers S, de Haas M, Porcelijn L, Bux J. Report on the Fourth International Granulocyte Immunology Workshop: progress toward quality assessment. Transfusion. 2002;42:462–468. doi: 10.1046/j.1525-1438.2002.00053.x. [DOI] [PubMed] [Google Scholar]
- 14.Wyman TH, Bjornsen AJ, Elzi DJ, Smith CW, England KM, Kelher M, et al. A two-insult in vitro model of PMN-mediated pulmonary endothelial damage: requirements for adherence and chemokine release. Am J Physiol Cell Physiol. 2002;283:C1592–1603. doi: 10.1152/ajpcell.00540.2001. [DOI] [PubMed] [Google Scholar]
- 15.Flesch BK, Reil A. Molecular genetics of the human neutrophil antigens. Transfus Med Hemother. 2018;45:300–309. doi: 10.1159/000491031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greinacher A, Wesche J, Hammer E, Furll B, Volker U, Reil A, et al. Characterization of the human neutrophil alloantigen-3a. Nat Med. 2010;16:45–48. doi: 10.1038/nm.2070. [DOI] [PubMed] [Google Scholar]
- 17.Huvard MJ, Schmid P, Stroncek DF, Flegel WA. Frequencies of SLC44A2 alleles encoding human neutrophil antigen-3 variants in the African American population. Transfusion. 2012;52:1106–1111. doi: 10.1111/j.1537-2995.2011.03396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia W, Bayat B, Sachs U, Chen Y, Shao Y, Xu X, et al. The frequencies of human neutrophil alloantigens in the Chinese Han population of Guangzhou. Transfusion. 2011;51:1271–1277. doi: 10.1111/j.1537-2995.2010.02979.x. [DOI] [PubMed] [Google Scholar]
- 19.Fung YL, Minchinton RM, Fraser JF. Neutrophil antibody diagnostics and screening: review of the classical versus the emerging. Vox Sang. 2011;101:282–290. doi: 10.1111/j.1423-0410.2011.01511.x. [DOI] [PubMed] [Google Scholar]
- 20.Simon SI, Rochon YP, Lynam EB, Smith CW, Anderson DC, Sklar LA. Beta 2-integrin and L-selectin are obligatory receptors in neutrophil aggregation. Blood. 1993;82:1097–1106. [PubMed] [Google Scholar]
- 21.Khan SY, Kelher MR, Heal JM, Blumberg N, Boshkov LK, Phipps R, et al. Soluble CD40 ligand accumulates in stored blood components, primes neutrophils through CD40, and is a potential cofactor in the development of transfusion-related acute lung injury. Blood. 2006;108:2455–2462. doi: 10.1182/blood-2006-04-017251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silliman CC. The two-event model of transfusion-related acute lung injury. Crit Care Med. 2006;34:S124–131. doi: 10.1097/01.CCM.0000214292.62276.8E. [DOI] [PubMed] [Google Scholar]
- 23.Bougie DW, Peterson JA, Kanack AJ, Curtis BR, Aster RH. Transfusion-related acute lung injury-associated HNA-3a antibodies recognize complex determinants on choline transporter-like protein 2. Transfusion. 2014;54:3208–3215. doi: 10.1111/trf.12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berthold T, Muschter S, Schubert N, Wesche J, Ameling S, Teumer A, et al. Impact of priming on the response of neutrophils to human neutrophil alloantigen-3a antibodies. Transfusion. 2015;55:1512–1521. doi: 10.1111/trf.12898. [DOI] [PubMed] [Google Scholar]
- 25.Browne T, Dearman RJ, Poles A. Human neutrophil antigens: Nature, clinical significance and detection. Int J Immunogenet. 2021;48:145–156. doi: 10.1111/iji.12514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.