Abstract
The malaria parasite affects millions of people each year, lives and multiplies in two different hosts, and synthesizes a large number of proteases and heat shock proteins (HSPs) for its survival. We describe here the characterization of a metalloprotease activity which resides in the small HSP (PVHSP28) of the common but noncultivable human malaria parasite Plasmodium vivax. The protein is expressed by erythrocytic stages of the parasite. It is expressed as a ∼55-kDa polypeptide which is then processed to the 28-kDa mature protein. The latter was found to be an active protease in gelatin zymography. This protease showed its optimal activity at 37°C (pH 7.6). It also retained its proteolytic activity at higher temperatures of up to 55°C. The enzyme belongs to the metalloprotease class, as its proteolytic activity was most effectively blocked by 1,10-phenanthroline and was restored to a maximal level by the addition of zinc metal ions. Inhibitors for the cysteine, serine, and aspartate classes of proteases were ineffective against this enzyme. A homology search indicates that PVHSP28 probably belongs to a new class of HSPs which possess the metalloprotease signature sequence.
Malaria is one of the common infectious diseases in tropical areas. It affects a large number of people, causing two to three million deaths each year (40). The disease remains uncontrolled to date, as the parasite is rapidly developing resistance towards the existing antimalarial drugs and showing a very high rate of antigenic variation. This makes the available antimalarial drugs ineffective and creates problems for development of universal and effective malaria vaccines. Furthermore, the complex life cycle of and the number of antigens expressed by the parasite have further complicated this task. Therefore, identification of new target molecules is required to develop any new therapeutic agent.
The life cycle of the malaria parasite includes its transfer from a poikilothermic to a homeothermic host and a number of specialized invasive stages. During its transfer from one host to another as well as during malaria fever, the parasite faces a temperature shift. The response to this is the synthesis of a large number of heat shock proteins (HSPs) by the parasite. These HSPs may provide protection to the parasite during its exposure to different temperatures, similar to the case for the HSPs of other organisms, but their biological function by and large remains unknown (7, 20, 26, 38). Several HSPs belonging to high-molecular-weight families have been identified in the cultivable human malaria parasite Plasmodium falciparum without assignment of a definitive biological role (4, 23, 25, 41, 42).
The malaria parasite also produces a large number of proteases (6). These proteases are essential for the parasite's survival, since they play various important roles in areas such as host cell invasion, nutrition and growth, processing of precursor proteins, etc. (1, 3, 5, 18, 33). The specific protease inhibitors interfere with the protease functions and also affect the normal parasite growth in vitro (2, 10, 11, 15, 17, 29). The proteases are therefore considered potential drug targets (30).
Earlier, we have described the cloning and sequencing of the gene for a small HSP (PVHSP28) from the common but noncultivable human malaria parasite Plasmodium vivax (13). Here, we describe the characterization of the encoded protein and show the presence of metalloprotease activity in it. This unique HSP of the parasite, containing a metalloprotease activity, seems to belong to a special class of HSP.
MATERIALS AND METHODS
Polyclonal antibodies.
The transfer of the 188-bp EcoRI insert of lambda gt11 clone Pv9 into the plasmid pIH902, its expression, and purification of the recombinant maltose-binding fusion protein were carried out as described previously (32, 36). This fusion protein was used to raise polyclonal antibodies in rabbits. These antibodies were used for indirect immunofluorescence assay, Western blot analysis, and immunoprecipitation. The position of the polypeptide encoded by the 188-bp part of the PVHSP28 gene corresponds to amino acids (aa) 288 to 344 in a 482-aa protein. This is downstream from the HEXXH signature sequence (aa 157 to 161) in PVHSP28 (13).
Western blot analysis.
The total parasite antigen was prepared from P. vivax-infected erythrocytes as described previously (32). Briefly, heparinized blood was subjected to Ficoll-Hypaque centrifugation to remove peripheral blood mononuclear cells. The remaining cells were passed through a Percoll gradient to purify the trophozoite- and schizont-containing erythrocytes. These purified infected erythrocytes were treated with saponin to free the parasites and then centrifuged. The pellet containing free parasites was solubilized by being boiled for 2 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 0.1% bromophenol blue, 10% glycerol, and 5% β-mercaptoethanol). After SDS-PAGE (12% acrylamide), proteins were transferred onto nitrocellulose paper for 1.5 h at room temperature using a semidry electrotransfer method (39). The nitrocellulose filter was reacted with anti-Pv9 antibody (1:100 dilution) and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G secondary antibodies (Amersham Corp., Arlington Heights, Ill.) at a 1:15,000 dilution. Detection was performed by using the enhanced chemiluminescence system from Amersham and following their instructions.
In vitro expression of the PVHSP28 gene.
The entire open reading frame of the PVHSP28 gene from the previously described pBST clone (13) was amplified by PCR using the primers 5′-GCGTGAATTCATGACCGCAAG-3′ and 5′-AGAGCACTGCAGATATCGTGTG-3′. The PCR product was cloned into pGEMT (Promega Corp., Madison, Wis.). The clones were sequenced to confirm the orientation. The recombinant and nonrecombinant (pGEM5Z) plasmids were transcribed with T7 RNA polymerase and translated in a reaction mixture containing 40 μl of rabbit recticulocyte lysate from Promega and 20 μCi of [35S]methionine (specific activity, 1,000 Ci per mmol) according to the manufacturer's instructions. The reaction mixtures were incubated at 30°C for the desired times. Five microliters of each reaction mix was removed and resolved by SDS–12% PAGE. The gel was dried under vacuum at 80°C and exposed to X-ray film.
Immunoprecipitation.
P. vivax-infected erythrocytes were purified from patients' blood by Percoll gradient centrifugation and lysed with saponin, as described above (32). The pellet was solubilized in 12 volumes of SDS buffer (0.15 M NaCl, 0.05 M Tris-HCl [pH 7.4], 0.5% Nonidet P-40, 0.05% SDS) and centrifuged at 12,000 × g for 3 min. The supernatant (100 μg of protein) was immunoprecipitated with protein A of Staphylococcus aureus and the above-mentioned anti-Pv9 antibodies according to the method of Gottesman and Cabral (19). For the in vitro-translated product, 50 μl of the mixture was used for immunoprecipitation. The immunoprecipitated pellets were dissolved in SDS loading buffer (5% SDS, 0.01% bromophenol blue, 10% glycerol, and 0.12 M Tris-HCl, pH 6.8) and electrophoresed by gelatin-PAGE as described below.
Gelatin-PAGE zymography.
Samples were solubilized in SDS loading buffer lacking β-mercaptoethanol and electrophoresed through a 12% polyacrylamide gel copolymerized with 0.1% gelatin (22). After electrophoresis at 4°C, the SDS was removed from the gel by incubating it in 2.5% Triton X-100 for 1 h at room temperature. The gel was then incubated in 40 mM Tris buffer (pH 7.4) for 72 h at 37°C, followed by staining with Coomassie blue. The protease activity was detectable as a clear zone against the blue background.
Casein hydrolysis assay.
The protease activity of in vitro-produced PVHSP28 was also measured by casein hydrolysis assay with a Quanti Cleave protease assay kit according to the instructions of the manufacturer (Pierce Co., Rockford, Ill.). All of the assays were performed in microtiter plates. Briefly, a solution of 100 μg of succinylated casein in a 100-μl volume (prepared in 40 mM Tris-HCl buffer [pH 7.4] at a concentration of 1 mg/ml) was added to the wells of the left half of the plate, whereas an equal volume (100 μl) of the buffer was added to the wells of the right half. A different amount of PVHSP28 immunoprecipitate (dissolved in 40 mM Tris-HCl buffer, pH 7.4) was added to each well. Equal amounts of immunoprecipitated PVHSP28 were added to blank buffer wells to blank out the background contributed by the proteins present in the immune complex. Immunoprecipitated samples from an in vitro translation mixture of vector pGEM5Z alone (without the PVHSP28 gene) served as controls. The samples were incubated at 37°C for 30 min, and then 50 μl of diluted (1:149) trinitrobenzenesulfonic acid was added to each well and incubated for 20 min at room temperature. The color development was measured at a wavelength of 405 nm using a Multiscan reader (Molecular Devices Corp., Sunnyvale, Calif.).
Effect of protease inhibitors on enzymatic activity.
A variety of inhibitors were incubated at 37°C for 30 min with equal amounts of immunoprecipitated PVHSP28 from 15 μl of in vitro reaction mixture prior to the addition of the substrate (8). In gelatin zymography, inhibitors were also added to the 10 ml of Tris-HCl buffer in which gels were incubated after electrophoresis. The final concentrations of inhibitors used were as follows: leupeptin, 10 μM in water; pepstatin A, 10 μM in ethanol; N-α-p-tosyl-l-phenylalanine chloromethyl ketone (TPCK), 0.1 μg/μl in ethanol; phenylmethylsulfonyl fluoride (PMSF), 1 mM in isopropanol; thioglycolic acid, 1 mM in water; 1,10-phenanthroline, 1 mM in Tris-HCl buffer (pH 7.2); β-mercaptoethylamine, 1 mM in water; and EDTA, 1 mM in water. For each inhibitor solvent control was used as described (8). The gelatin zymography and casein hydrolysis assay were performed as described above except that in zymography gels were incubated for 1 week.
Reactivation of protease activity by metal ions.
Immunoprecipitated PVHSP28 was preincubated with the inhibitor 1,10-phenanthroline (1 mM) at 37°C for 30 min in 40 mM Tris-HCl (pH 7.4), and then the different metal ions (ZnCl2, CaCl2, MnCl2, and MgCl2) were added to the reaction mixture at a concentration of 100 μM each and incubated further for 30 min at 37°C (31). Protease activity was measured by the casein hydrolysis method as described above. The proteolytic activity of PVHSP28 without addition of any inhibitor or metal ions was taken as 100%.
RESULTS
Determination of the size of PVHSP28 in P. vivax.
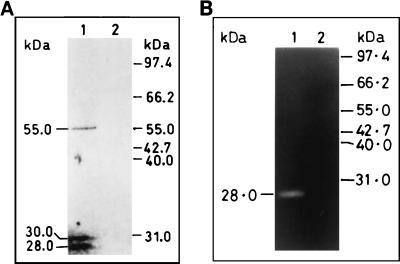
The polyclonal anti-Pv9 antibodies, raised against the recombinant protein encoded by the 188-bp part of the gene (36), and total blood stage parasite antigens were used to evaluate the size of PVHSP28 on Western blots. The antibody reacted with three bands (∼55, 30, and 28 kDa) in the parasite lysate, while preimmune serum did not show any reaction (Fig. 1A). The antibody is specific to the parasite, as it does not react with any of the host cell antigens in Western blotting, zymography, or immunofluorescence assay (data not shown). The observed size of the ∼55-kDa polypeptide is close to the calculated size of 51.2 kDa for PVHSP28 (13). The slight difference could be due to posttranslational modifications, since PVHSP28 contains several myristylation sites and one N-glycosylation site. The origin of the low-molecular-mass bands of 30 and 28 kDa is discussed below.
FIG. 1.
Identification of PVHSP28 protein and protease in extracts of P. vivax. (A) Total parasite proteins obtained from P. vivax-infected erythrocytes were electrophoresed, transblotted, and probed with anti-Pv9 rabbit antibodies (lane 1) and preimmune serum (lane 2). (B) Immunoprecipitates from P. vivax lysate, obtained by using anti-Pv9 antibodies (lane 1) and preimmune serum (lane 2), were used to detect the protease band in gelatin-PAGE.
Detection of protease activity in PVHSP28.
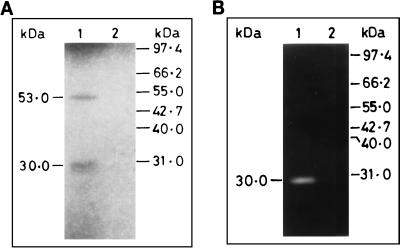
Interestingly, the amino acid sequence of PVHSP28 contains a signature motif (HEXXH) for metalloprotease (13). Therefore, attempts were made to confirm if the protease activity was indeed present in PVHSP28, because there are certain proteins which contain the HEXXH sequence but do not possess the protease activity (31). To identify protease activity in PVHSP28, immunoprecipitation was undertaken using the above-mentioned anti-Pv9 antibodies and total parasite lysate. This antibody immunoprecipitated all three bands as seen in Western blotting, but only the lowermost band of 28 kDa showed gelatin degradation activity on zymography (Fig. 1B). This was also confirmed from the in vitro expression studies. In these studies, the in vitro-expressed product of the PVHSP28 gene also showed a high-molecular-mass band of ∼53 kDa and a low-molecular-mass band of 30 kDa (Fig. 2A). Of them, only the low-molecular-mass band showed a protease activity (Fig. 2B). The protease activity was specific to the insert-derived polypeptide, as the vector alone (without the insert) did not show any such band. Nevertheless, there was a size difference between the protease bands of the parasite- and in vitro-expressed products. The reasons for the differences in the number and size of bands between in vitro and in vivo products are discussed in Discussion.
FIG. 2.
In vitro expression and proteolytic activity in PVHSP28. The open reading frame of the PVHSP28 gene was used for in vitro expression with a rabbit reticulocyte lysate system. The translated products were immunoprecipitated with anti-Pv9 antibodies and analyzed by SDS-PAGE followed by autoradiography (A) as well as by gelatin-PAGE for zymography (B). The immunoprecipitates from the in vitro-translated products from a PVHSP28-encoding recombinant plasmid (lane 1) and vector alone (lane 2) were analyzed.
In vitro processing of PVHSP28.
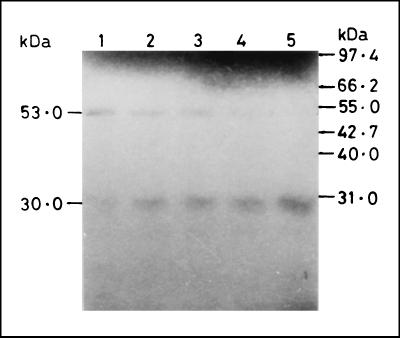
To prove that the lower band of mature protease is derived from the high-molecular-weight band, pulse-chase experiments or N-terminal sequencing studies are required. These experiments would require in vitro culture or a large number of parasites, respectively. Since the parasite is noncultivable and the source of material from patients is scanty, in vitro processing studies were designed as an alternate strategy. For this purpose, the entire coding region of the PVHSP28 gene was transcribed and translated in vitro by using the rabbit reticulocyte lysate system, which showed two bands of ∼53 and 30 kDa as described above. These bands were recognized by the anti-PV9 antibodies. That the low-molecular-mass mature protease band was derived from the upper band was evident from an experiment with incubation for different time periods (Fig. 3). The results showed that in the beginning (30-min incubation) the 53-kDa band was more prominent. The amount of this band decreased with longer incubation periods, whereas the intensity of the 30-kDa band increased correspondingly. At 90 min of incubation, almost all of the 53-kDa band had disappeared while the 30-kDa band showed its maximum intensity, indicating that the lower band of mature protease was the processed product of the higher-molecular-mass band.
FIG. 3.
In vitro processing of PVHSP28. Transcription and translation with a reticulocyte lysate containing [35S]methionine were performed with a PVHSP28 gene-bearing plasmid construct. Aliquots were removed every 15 min thereafter up to 90 min of incubation. Products were resolved by SDS–12% PAGE under reducing conditions and processed for autoradiography. The precursor and processed PVHSP28 products are indicated. Lane 1, 30 min; lane 2, 45 min; lane 3, 60 min; lane 4, 75 min; lane 5, 90 min. No incorporation was seen at 15 min of incubation.
Effect of protease inhibitors on enzymatic activity.
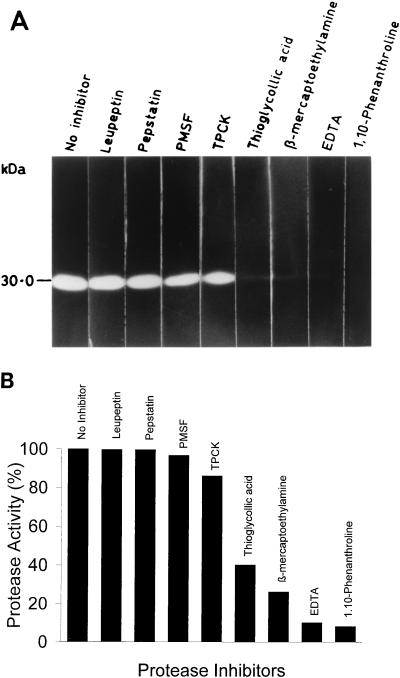
The sensitivity of the in vitro-expressed enzyme to the classic protease inhibitors was assayed quantitatively and qualitatively by using casein and gelatin substrates, respectively (8). The results are shown in Fig. 4. The most effective inhibition of the enzyme activity was observed with 1 mM 1,10-phenanthroline (92% in the casein assay) as compared with other metal chelators. However, PMSF, pepstatin, leupeptin, and TPCK had a minimal or no inhibitory effect on this protease. Inhibition by metal chelators suggests a requirement for metal ions for optimal enzyme activity. This was found to be true when the maximum enzymatic activity was restored by the addition of zinc ions along with the inhibitor 1,10-phenanthroline (Table 1).
FIG. 4.
Effect of protease inhibitors on protease activity. (A) Immunoprecipitated PVHSP28 from an in vitro translation reaction was incubated with protease inhibitors for 30 min at 37°C and then electrophoresed on a gelatin gel. Individual lanes of the gel were kept with the indicated inhibitors in Tris-Cl buffer, pH 7.4. Gels were developed as described in Materials and Methods. (B) Hydrolysis of succinylated casein by PVHSP28 was studied in the presence of inhibitors. Each assay was carried out in triplicate, and the results shown are the mean percent inhibition, taking 0% inhibition as that for the control without any inhibitor. The concentrations of protease inhibitors were as follows: 1,10-phenanthroline, 1 mM; EDTA, 1 mM; β-mercaptoethylamine, 1 mM; thioglycolic acid, 1 mM; TPCK, 0.1 μg/μl; PMSF, 1 mM; pepstatin, 10 μM; and leupeptin, 10 μM.
TABLE 1.
Effect of metal ions on the P. vivax metalloprotease activitya
| Ion | Proteinase activity (%) in presence of 1 mM 1,10-phenanthrolineb
|
|
|---|---|---|
| Without ion | With ion (100 μM) | |
| ZnCl2 | 3.3 | 95.0 |
| CaCl2 | 1.5 | 53.0 |
| MgCl2 | 4.28 | 9.0 |
| MnCl2 | 5.4 | 78.0 |
Casein hydrolysis assay was used to determine the protease activity in the immunoprecipitate of the in vitro-translated product of the PVHSP gene.
The proteinase activity without 1,10-phenanthroline was considered 100% activity.
DISCUSSION
We have detected a protease activity in the small HSP of P. vivax (PVHSP28), whose gene we cloned earlier (13). It is synthesized in the proenzyme form and then processed to the mature 28-kDa protease. The enzyme is optimally active at neutral pH and 37°C. It is also stable at higher temperatures of up to 55°C. It was found to be a metalloprotease, since 1,10-phenanthroline inhibited its activity to a maximum degree (92%) as compared to those inhibitors which were specific for three other classes of proteases. Furthermore, the protease activity, inhibited by 1,10-phenanthroline, was restored by the addition of metal ions. This metalloprotease required zinc ions for its optimal activity as compared to Mg2+, Mn2+, and Ca2+. This is in concurrence with the observation that zinc binds to the metalloprotease motif HEXXH, which is also present in PVHSP28 (13, 24).
Our in vitro expression experiments suggest that this malarial metalloprotease is synthesized as the larger inactive proenzyme, which is then processed to a smaller mature protease. A similar processing phenomenon has been described earlier for the cysteine and subtilisin-like proteases of P. falciparum (3, 34). The production of mature enzyme therefore requires posttranslational modifications which vary from organism to organism (35). This could explain the discrepancies in the number and size of bands between parasite- and in vitro-expressed proteins. PVHSP28 is known to contain putative glycosylation, myristylation, and phosphorylation sites (13). Therefore, any variation in these posttranslational modifications by the rabbit reticulocyte lysate system from that of the parasite would lead to the synthesis of a protein with a different size. This might explain the differences observed between the proenzyme forms of the parasite (∼55 kDa) and the in vitro-derived products (∼53 kDa). Also, the comparison of the sizes of the processed (proteolytically active) forms of PVHSP28 in the parasite (28 kDa) and the in vitro translation reaction (30 kDa) indicates that rabbit reticulocyte lysate processes the PVHSP28 differently. This is quite possible if the protein folding and proteolytic cleavage occur differently in the in vitro system. A similar analogy has been drawn in several cases where the same protein was processed differently when different expression systems were used, e.g., expression of the P. falciparum cysteine protease in baculovirus and the Trypanosoma cruzi cysteine protease in Escherichia coli (12, 35). Nevertheless, the identity of the extra band in the parasite lysate is yet to be established, i.e., whether it is an intermediate form of this metalloprotease or a cross-reacting antigen.
To the best of our knowledge, this is the first malarial HSP found to possess the protease activity and thus to have some biological function. So far, only HSC70 of Plasmodium knowlesi was found to have some regulatory function to inhibit actin polymerization after forming a complex with 32/34-kDa actin-binding protein (38). It is quite possible that several or all of the malarial HSPs indeed have some biological function. Different parasite HSPs may have different biological functions, since all HSPs do not possess the protease activity as found in PVHSP28 and the actin polymerization activity associated with HSC70.
It is quite interesting that the malarial parasite produces an HSP molecule which could have a dual function. In case of PVHSP28, the parasite may use both properties of the molecule, similar to the case for a recently described HSP of E. coli (37). It is tempting to speculate that PVHSP28 could bind the misfolded denatured proteins, like other HSPs, to allow them to refold to their native state. At the same time it degrades those proteins which could not be refolded to their native state by using its own protease activity. The latter possibility is supported by the fact that the protease activity of PVHSP28 is still retained at higher temperatures. Alternatively, it is quite possible that PVHSP28 cleaves the inactive preproteins to the functional mature proteins, similar to the case for its yeast homolog STE24 (16).
The malaria parasite synthesizes a large number of HSPs and proteases, but to date none of the described proteases have been found to belong to the HSP class and vice versa. Furthermore, this protease-containing HSP belongs to the family of low-molecular-weight HSPs and not to the already characterized high-molecular-weight HSP family. A recent database search indicates that there could be a separate class of HSPs which possess metalloprotease activity (Fig. 5). The majority of these HSPs are from prokaryotes, except for yeast and human HSPs and this malarial HSP. This parasite molecule (PVHSP28) therefore could belong to a special class of HSPs and is the first metalloprotease of P. vivax. In fact, the parasite showed enhanced expression of PVHSP28 at 39°C compared to 37°C (unpublished data).
FIG. 5.
Amino acid sequence alignment of different domains of the P. vivax HSP (PVHSP28) with other proteins obtained by using the FASTA program and the EMBL database. HTPX, small heat shock protein containing the metalloprotease motif; HTPXEco, protein from E. coli (accession no. P23894); HTPXHin, protein from Haemophilus influenzae (accession no. P44840); HTPXAae, protein from Aquifex aeolicus (accession no. 2984218); HTPXPho, protein from Pyrococcus horikoshii (accession no. BAA 30357); HTPXMja, protein from Methanococcus jannaschii (accession no. H64509); HTPXSgo, protein from Streptococcus gordonii (accession no. 2407215); HTPXMtu, protein from Mycobacterium tuberculosis (accession no. CAB08997); HTPXBsu, protein from Bacillus subtilis (accession no. CAB 13222); HTPXHpy, protein from Helicobacter pylori (accession no. AA007972.1); HTPXMth, protein from Methanobacterium thermoautotrophicum (accession no. 2621646); HTPXAfl, protein from Archaeoglobus fulgidus (accession no. 2650402); HTPXSsp, protein from a Synechocystis sp. (accession no. BAA18200); STE24, yeast homologue of HTPX (accession no. NP005848.1); HSTE24, human homologue of HTPX (accession no. 300769 and AF064867). The amino acid residues identical to those in the PVHSP of P. vivax are shown in boldface. The amino acids involved in metal binding are marked with an asterisk, and those involved in the catalytic activity are marked with a plus sign. Dashes indicate the absence of an amino acid residue at that position. The shaded amino acids represent the consensus (HEXXH and EXXXD) metalloprotease motif. Numbers on left of each sequence indicate the amino acid residue number in that protein.
Metalloproteases are expressed by several pathogens, such as Vibrio, Listeria, Legionella, Leishmania, Enterococcus, and Pseudomonas spp., etc., and play a vital role in the pathogenesis of the disease by degrading a wide range of host molecules (8, 9, 21, 27). The malaria parasite also degrades host proteins for its entry into the host cell, degrades nutrients for its survival and growth, and ruptures the host cell for its exit (3, 5, 18, 33). It also cleaves its own precursor proteins to produce functionally active molecules. Therefore, it requires a large number of proteases to perform these functions. Many (at least 25) malarial proteases belonging to all four classes have indeed been detected in the parasite, but only a few of them from P. falciparum have been characterized (3, 5, 6, 14, 18, 33). The PVHSP28 reported here may have more than one role to play in the parasite, because it is not only an HSP but also a protease. This protease is also stable at higher temperatures, similar to the case for GP63 of Leishmania (8, 28). PVHSP28 therefore could play a significant role in the survival of the parasite during its transfer from mosquito to human and during malaria fever. Further studies are being carried out in the laboratory on the homologous molecule from the cultivable human malaria parasite P. falciparum. This will facilitate the characterization of this heat shock metalloprotease in greater detail, elucidating the substrate specificity and contributing to the development of newer antimalarial drugs.
ACKNOWLEDGMENTS
This work was supported by grants from the Department of Biotechnology (Government of India) and the Council of Scientific and Industrial Research (to Y.D.S.). A Senior Research Fellowship (to J.M.F.) was from CSIR.
REFERENCES
- 1.Barale J C, Blisnick T, Fujioka H, Alzari P M, Aikawa M, Braun-Breton C, Langsley G. Plasmodium falciparum subtilisin-like protease 2, a merozoite candidate for the merozoite surface protein 1-42 maturase. Proc Natl Acad Sci USA. 1999;96:6445–6450. doi: 10.1073/pnas.96.11.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackman M J, Chappel J A, Shai S, Holder A A. A conserved parasite serine protease processes the Plasmodium falciparum Merozoite Surface Protein-1. Mol Biochem Parasitol. 1993;62:103–114. doi: 10.1016/0166-6851(93)90182-w. [DOI] [PubMed] [Google Scholar]
- 3.Blackman M J, Fujioka H, Stafford W H L, Sajid M, Clough B, Fleck S L, Aikawa M, Grainger M, Hackett F. A subtilisin-like protein in secretory organelles of Plasmodium falciparum merozoites. J Biol Chem. 1998;273:23398–23409. doi: 10.1074/jbc.273.36.23398. [DOI] [PubMed] [Google Scholar]
- 4.Bonnefoy S, Attal G, Langsley G, Tekaia F, Mercereau-Puijalon O. Molecular characterization of the heat shock protein 90 gene of the human malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 1994;67:157–170. doi: 10.1016/0166-6851(94)90105-8. [DOI] [PubMed] [Google Scholar]
- 5.Braun-Breton C, Blisnick T, Jouin H, Barale J C, Rabilloud T, Langsley G, Pereira Da Silva L H. Activation of a Plasmodium falciparum protease correlated with merozoite maturation and erythrocyte invasion. Proc Natl Acad Sci USA. 1988;89:9647–9651. [Google Scholar]
- 6.Braun-Breton C, Pereira da Silva L H. Malarial proteases and red blood cell invasion. Parasitol Today. 1993;9:92–96. doi: 10.1016/0169-4758(93)90212-x. [DOI] [PubMed] [Google Scholar]
- 7.Bukau B, Horwich A. The HSP70 and HSP60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 8.Chaudhuri G, Chaudhuri M, Pan A, Chang K P. Surface acid proteinase (gp63) of Leishmania maxicana. A metalloenzyme capable of protecting liposome encapsulated proteins from phagolysosomal degradation by macrophages. J Biol Chem. 1989;264:7483–7489. [PubMed] [Google Scholar]
- 9.Dreyfus L A, Iglewski B H. Purification and characterization of an extra cellular protease of Legionella pneumophila. Infect Immun. 1986;51:736–743. doi: 10.1128/iai.51.3.736-743.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dulzewski A R, Rangachari K, Wilson R J M, Gratzer W B C. Plasmodium falciparum: protease inhibitors and inhibition of erythrocyte invasion. Exp Parasitol. 1986;62:416–422. doi: 10.1016/0014-4894(86)90050-0. [DOI] [PubMed] [Google Scholar]
- 11.Dutta G P, Banyal H S. In vitro susceptibility of erythrocytes of Presbytisentellus (Indian Langur) to Plasmodium knowlesi and blocking of merozoite invasion process by certain protease inhibitors. Indian J Exp Biol. 1981;19:9–11. [PubMed] [Google Scholar]
- 12.Eakin A E, Mills A A, Harth G, McKerrow J H, Craik C S. The sequence, organization, and expression of the major cysteine protease (cruzain) from Trypanosoma cruzi. J Biol Chem. 1992;267:7411–7420. [PubMed] [Google Scholar]
- 13.Fakruddin J M, Biswas S, Sharma Y D. Identification of Plasmodium vivax heat shock protein which contains a metalloprotease sequence motif. Mol Biochem Parasitol. 1997;90:387–390. doi: 10.1016/s0166-6851(97)00173-4. [DOI] [PubMed] [Google Scholar]
- 14.Florent I, Derhy Z, Allary M, Monsigny M, Mayer R, Schrevel J. A Plasmodium falciparum aminopeptidase gene belonging to the M1 family of zinc-metallopeptidases is expressed in erythrocytic stages. Mol Biochem Parasitol. 1998;97:149–160. doi: 10.1016/s0166-6851(98)00143-1. [DOI] [PubMed] [Google Scholar]
- 15.Francis S E, Gluzman I Y, Oksman A, Knickerbocker A, Muller R, Bryant M I, Sherman D R, Russel D G, Goldberg D E. Molecular characterization and inhibition of a Plasmodium falciparum aspartic hemoglobinase. EMBO J. 1994;13:306–317. doi: 10.1002/j.1460-2075.1994.tb06263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimura-Kamada K, Nouvet F J, Michaelis S. A novel membrane-associated metalloprotease, Ste 24P, is required for the first step of NH2-terminal processing of the yeast alpha factor precursor. J Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gantt S M, Myung J M, Briones M R, Li W D, Corey E J, Omura S, Nussenzweig V, Sinnis P. Proteosome inhibitors block development of Plasmodium spp. Antimicrob Agents Chemother. 1998;42:2731–2738. doi: 10.1128/aac.42.10.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gluzman I Y, Francis S E, Oksman A, Smith C E, Duffin K L, Goldberg D E. Order and specificity of the Plasmodium falciparum hemoglobin degradation pathway. J Clin Investig. 1994;93:1602–1608. doi: 10.1172/JCI117140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottesman M M, Cabral F. Purification and characterization of a transformation-dependent protein secreted by cultured murine fibroblasts. Biochemistry. 1981;20:1659–1665. doi: 10.1021/bi00509a039. [DOI] [PubMed] [Google Scholar]
- 20.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 21.Hase C C, Finkelstein R A. Cloning and nucleotide sequence of the Vibrio cholerae hemagglutinin/protease (HA/protease) gene and construction of an HA/protease-negative strain. J Bacteriol. 1991;173:3311–3317. doi: 10.1128/jb.173.11.3311-3317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heussen C, Dowdle E B. Electrophoretic analysis of plasminogen activators in polyacrylamide gels containing sodium dodecyl sulfate and copolymerized substrates. Anal Biochem. 1980;102:196–202. doi: 10.1016/0003-2697(80)90338-3. [DOI] [PubMed] [Google Scholar]
- 23.Holloway S P, Min W, Inselburg J S. Isolation and characterization of a chaperonin-60 gene of the human malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 1994;64:25–32. doi: 10.1016/0166-6851(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 24.Jongeneel C V, Bouvier J, Bairoch A. A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett. 1989;242:211–214. doi: 10.1016/0014-5793(89)80471-5. [DOI] [PubMed] [Google Scholar]
- 25.Kumar N, Syin C A, Carter R, Quakyi I, Miller L H. Plasmodium falciparum gene encoding a protein similar to the 78-kDa rat glucose-regulated stress protein. Proc Natl Acad Sci USA. 1988;85:6277–6281. doi: 10.1073/pnas.85.17.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindquist S, Craig E. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 27.Makinen P L, Clewell D B, An F, Makinen K K. Purification and substrate specificity of a strongly hydrophobic extracellular metalloendopeptidase (“gelatinase”) from Streptococcus faecalis (strain OGI-10) J Biol Chem. 1989;264:3325–3334. [PubMed] [Google Scholar]
- 28.Morihara K. Pseudolysin and other pathogen endopeptidases of thermolysin family. Methods Enzymol. 1995;248:242–253. doi: 10.1016/0076-6879(95)48017-x. [DOI] [PubMed] [Google Scholar]
- 29.Nankya-Kitaka M F, Curley G P, Gavigan C S, Bell A, Dalton J P. Plasmodium chabaudi chabaudi and P. falciparum: inhibition of aminopeptidase and parasite growth by bestatin and nitrobestatin. Parasitol Res. 1998;84:552–558. doi: 10.1007/s004360050447. [DOI] [PubMed] [Google Scholar]
- 30.Olliaro P L, Gottlieb M, Wirth D F. Plasmodium falciparum proteinases: targeted drug development. Parasitol Today. 1996;12:413. [Google Scholar]
- 31.Rawlings N D, Barrett A J. Evolutionary families of metallopeptidases. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 32.Ray P, Ansari M A, Sharma Y D. Plasmodium vivax immune responses in a cross-section of the population in the Delhi area of India. Am J Trop Med Hyg. 1994;51:436–443. [PubMed] [Google Scholar]
- 33.Rosenthal P J, McKarrow J H, Aikawa M, Nagasawa H, Leech J H. A malarial cysteine proteinase is necessary for hemoglobin degradation by Plasmodium falciparum. J Clin Investig. 1988;82:1560–1566. doi: 10.1172/JCI113766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenthal P J, Nelson R G. Isolation and characterization of a cysteine protease gene of Plasmodium falciparum. Mol Biochem Parasitol. 1992;51:143–152. doi: 10.1016/0166-6851(92)90209-3. [DOI] [PubMed] [Google Scholar]
- 35.Salas F, Fichmann J, Lee G K, Scott M D, Rosenthal P J. Functional expression of falcipain, a Plasmodium falciparum cysteine proteinase, supports its role as a malarial hemoglobinase. Infect Immun. 1995;63:2120–2125. doi: 10.1128/iai.63.6.2120-2125.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma Y D, Sharma V P, Ray P, Laal S, Sawant S D, Verma S. Isolation and serological characterization of a Plasmodium vivax recombinant antigen. Infect Immun. 1991;59:1922–1926. doi: 10.1128/iai.59.6.1922-1926.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spiess C, Beil A, Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat-shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- 38.Tardieux I, Baines I, Mossakowska M, Ward G E. Actin-binding proteins of invasive malaria parasites and the regulation of actin polymerization by a complex of 32/34-kDa proteins associated with heat-shock protein 70kDa. Mol Biochem Parasitol. 1998;93:295–308. doi: 10.1016/s0166-6851(98)00044-9. [DOI] [PubMed] [Google Scholar]
- 39.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh J A. Disease problem in the Third World. Ann NY Acad Sci. 1989;569:1–16. doi: 10.1111/j.1749-6632.1989.tb27354.x. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe J. Cloning and characterization of heat-shock protein Dnaj homologues from Plasmodium falciparum and comparison with ring infected erythrocyte surface antigen. Mol Biochem Parasitol. 1997;88:253–258. doi: 10.1016/s0166-6851(97)00073-x. [DOI] [PubMed] [Google Scholar]
- 42.Yang Y F, Tan-ariya P, Sharma Y D, Kilejian A. The primary structure of a Plasmodium falciparum polypeptide related to heat shock proteins. Mol Biochem Parasitol. 1987;26:61–67. doi: 10.1016/0166-6851(87)90130-7. [DOI] [PubMed] [Google Scholar]