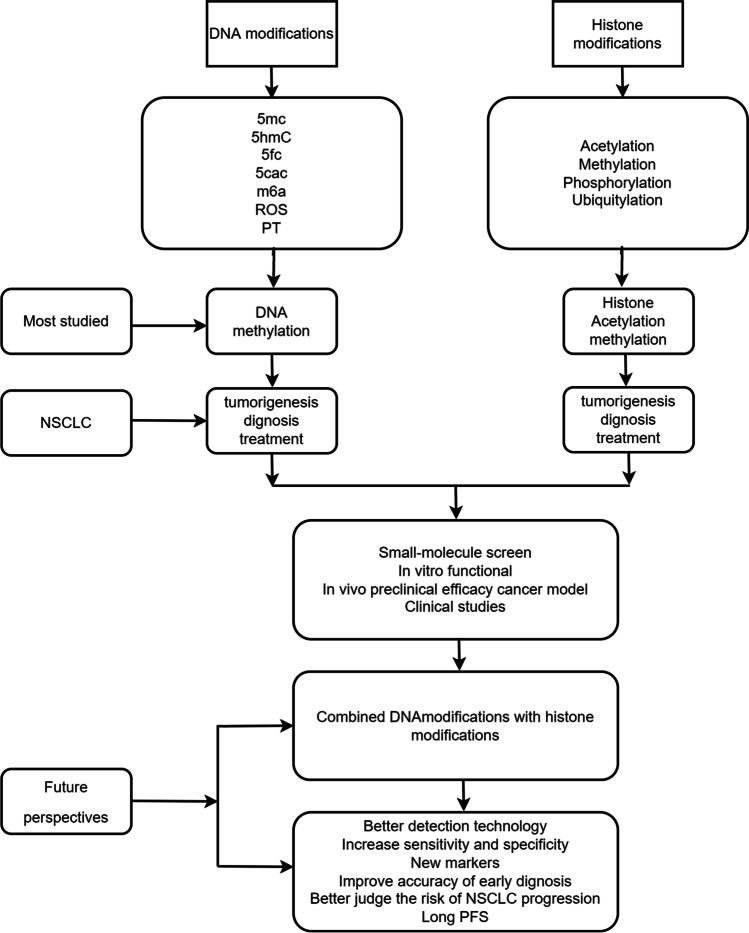
Abstract
Lung cancer has a very high mortality in females and males. Most (~ 85%) of lung cancers are non-small cell lung cancers (NSCLC). When lung cancer is diagnosed, most of them have either local or distant metastasis, with a poor prognosis. In order to achieve better outcomes, it is imperative to identify the molecular signature based on genetic and epigenetic variations for different NSCLC subgroups. We hypothesize that DNA and histone modifications play significant roles in the framework of predictive, preventive, and personalized medicine (PPPM; 3P medicine). Epigenetics has a significant impact on tumorigenicity, tumor heterogeneity, and tumor resistance to chemotherapy, targeted therapy, and immunotherapy. An increasing interest is that epigenomic regulation is recognized as a potential treatment option for NSCLC. Most attention has been paid to the epigenetic alteration patterns of DNA and histones. This article aims to review the roles DNA and histone modifications play in tumorigenesis, early detection and diagnosis, and advancements and therapies of NSCLC, and also explore the connection between DNA and histone modifications and PPPM, which may provide an important contribution to improve the prognosis of NSCLC. We found that the success of targeting DNA and histone modifications is limited in the clinic, and how to combine the therapies to improve patient outcomes is necessary in further studies, especially for predictive diagnostics, targeted prevention, and personalization of medical services in the 3P medicine approach. It is concluded that DNA and histone modifications are potent diagnostic and therapeutic targets to advance non-small cell lung cancer management from the perspective of 3P medicine.
Keywords: Epigenetics, DNA modification, Histone modification, Non-small cell lung cancer (NSCLC), Predictive diagnosis, Patient stratification, Personalized target therapy, Predictive preventive personalized medicine (PPPM / 3P medicine)
Introduction
Lung cancer-caused health problem
There were more than 2.2 million cases of newly diagnosed lung cancers worldwide and 1.9 million cases of deaths in 2020; thus, it was one of the most prevalent malignant tumors [1]. Lung cancers were grouped into two types: small cell lung cancer (SCLC; 15–20%) and non-small cell lung cancer (NSCLC; 80–85%). NSCLC includes squamous cell carcinoma (LUSC), adenocarcinoma (LUAD), and large-cell carcinoma [2]. Although many advances have been made in early diagnosis and treatment, approximately 40% of NSCLC cases have metastasis when they are first diagnosed, with poor prognosis [3].
Importance of epigenetics in lung cancer
Epigenetics, coined by Conrad Waddington, refers to heritable modifications of a cellular phenotype, which is distinct from changes in DNA sequences, including DNA methylation, gene silencing, maternal effects, and RNA modifications [4]. Epigenetic changes are responsible for regulating all DNA-based functions, including transcription, DNA repair, and replication. Typically, epigenetic dysfunction is caused by aberrant DNA and histone alterations. Several studies demonstrate that aberrant chromatin regulator expression patterns and genomic changes can induce carcinogenesis in NSCLC. The whole-genome sequencing of NSCLC produced a database of recurrent somatic mutations in several epigenetic regulators.
Roles of modifications in DNA and histone in NSCLC
Genetic information plays a significant role in the occurrence, development, and prognosis of NSCLC [4]. Modifications in DNA and histone are the most studied epigenetics, which do not change the gene sequence but change the gene expressions. DNA and histone modifications also play important roles in the processes of tumorigenesis, progression, and metastasis. Studies found that DNA and histone modifications provide potentials for early detection and diagnosis, progression, distant metastasis, therapies, prognosis, and screening recurrence of NSCLC in the framework of 3P medicine.
Importance of PPPM approach in improvement of the overall management of lung cancer-caused health problems
Despite great progressions in treatment options of lung cancer including surgery, radiotherapy, and chemotherapy, its survival improvement is still a big challenge, with a low 5-year survival rate (20%). Most (~ 85%) of lung cancers are non-small cell lung cancers (NSCLC). Upon being first diagnosed, most patients are found with local or distant metastases, with a poor prognosis [1–3], whose main reason is that the early symptoms of lung cancer are commonly ignored causing most patients to be diagnosed in middle or advanced stages [3]. Although targeted therapy and immunotherapy are very effective for NSCLC patients, only a minority of patients with driver gene mutations or PD-L1 high expression have a well-response to these treatments. Basic attributes of PPPM meet the requirements of cancer therapeutic strategy for better prevention, early diagnosis, discovery of the best possible treatment for every patient, and prediction of the patient response [5, 6]. In the context of 3P medicine, DNA and histone modifications will be helpful in early diagnosis, predicting disease development, progression, patient stratification, individualized therapy, and screening recurrence of NSCLC.
Working hypothesis in the PPPM framework of NSCLC
We hypothesize a series of molecular event and signaling pathway alterations mediated by DNA modifications and histone modifications in NSCLC. We will focus on the regulators of DNA and histone modifications, and modified substrates to discuss the significant roles of DNA and histone modifications, and the corresponding molecular pathway alterations, and discover the key molecules related to NSCLC. It will benefit the clarification of molecular mechanisms of NSCLC, discovery of new therapeutic targets/drugs, and establishment of effective biomarkers based on DNA and histone modifications, which play an important role in predictive diagnostics, targeted prevention, and personalization of medical services [7, 8]. This review aims to introduce advances in modifications in DNA and histones and their significant roles in the diagnosis, therapeutics, and prognosis of NSCLC. This review also aims to explore the connection between DNA and histone modification and the PPPM, which may provide an important contribution to improve the prognosis of NSCLC.
DNA modifications in NSCLC
Living organisms encode genetic information through four nucleobases (adenine, thymine, cytosine, and guanine) as well as modified nucleobases. DNA cytosine methylation (m5C), the most studied epigenetic alteration, controls gene expression in mammalian cells. Besides m5C, animal genomes contain a variety of DNA alterations. N6-methyladenosine (m6A) can be used by prokaryotes and eukaryotes [9]. m6A has been found in a number of mammals [10–12]. Per the earlier literature, most m6A modifications are found in mitochondrial DNA, which is then incorporated into genomic DNA via nucleotide-salvage processes [13–15]. The thymine modification 5-hydroxymethyluracil (5hmU) can be found in mammal genomes [16]. The TET enzymes in mammalian genomic DNA oxidize thymine to produce 5hmU and 5fU [16]. DNA oxidation results in a main product, 8-oxo-7,8-dihydroguanine (OG) [17], which controls gene expressions [17, 18]. Phosphothioate can modify the DNAs of bacteria and archaea, but cannot modify the DNA of eukaryotes [19].
Cytosine modification: DNA methylation
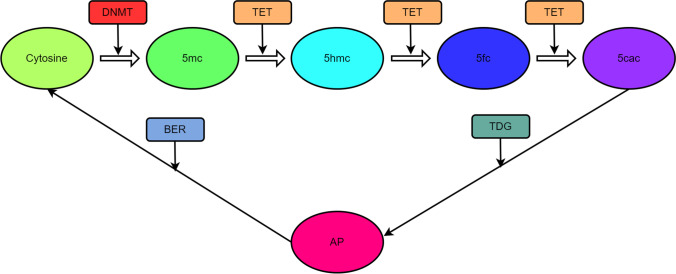
A lot of attentions were paid to cytosine methylation as a DNA alteration, which was recognized as a tuberculinic acid hydrolysis byproduct by Johnson and Coghill in 1925, although it was discovered in 1898 [20]. In the field of epigenetics, DNA methylation has received the most attention because it alters chromatin shape and transcription factor binding to control gene expression. DNA methylation is an addition of a methyl group to the cytosine 5th carbon atom by DNA methyltransferases [20]. Even if the DNA sequence is not altered, DNA methylation can mutate the genes bound to the methyl group (Fig. 1). During DNA methylation, a large number of genes can be methylated or demethylated at different times in cells [21]. In NSCLC, hypermethylation of local promoters and abnormal hypomethylation of genome-wide DNA are detected [22].
Fig. 1.
Epigenetic dysregulation is often caused by DNA and histone modifications. The purpose of this review is to highlight current progress in the identification, management, and prognosis of NSCLC based on DNA and histone alterations
Thymine modifications: 5hmu and 5fu
Numerous eukaryotes, including mammals, contain the thymine alteration 5-hydroxymethyluracil (5hmU) [16], and studies demonstrate that either passive replication-dependent loss or active TET-assisted oxidation could use 5hmC as an intermediary in DNA demethylation [16]. However, some researchers believe that DNA demethylation is not required for 5hmC to exert epigenetic activity [16].
Adenine modifications: N6-methyladenosine
N6-Methylladenosine (m6A) is commonly used to modify DNA by prokaryotes and eukaryotes [23]. It was firstly discovered to play significant roles in prokaryotic restriction-modification mechanisms and gene expression regulation [24] and was later detected in mammals [10, 11]. As a result of the nucleotide-salvage pathway, DNA polymerases incorporated m6A into genomic DNA from the RNA nucleoside of m6A, which was degraded in mitochondrial DNA [11].
DNA methylation in the mechanism of tumorigenesis
DNMT dysregulation
DNMT dysregulation plays an important role in the methylation pathway and is closely associated with cancer pathophysiology. Humans have five known DNMTs with different specificities for methylated and unmethylated DNA: DNMT-1, DNMT-2, v3A, DNMT-3B, and DNMT-3L [25]. It is possible that aberrant DNMT activities contribute to lung cancer in different ways. In view of the positive correlation between higher DNMT expression and proliferation, DNMT dysregulation can disrupt the cell cycle. Non-small cell lung cancers are found to overexpress DNMTs, such as DNMT1 upregulation, which is independently linked to poor prognosis [26]. Another study found that DNMT3a expression is a marker for prognosis, and the lack of DNMT3a is thought to facilitate tumor progression in LUAD [27]. In the mouse model of LUAD tumor progression, eliminating DNMT3a promotes tumor growth. Different histological types of lung cancer seem to specifically influence the expression levels of DNMT3a [28], and DNMT1 increased cell proliferation in EGFR-mutated NSCLC by deregulating hMLH1 and hMSH2, which disturb the cell cycle [28].
TET dysregulation
Ten-eleven translocation enzymes (TET) act on m5C to oxidize it, which reverses DNA methylation (DNAm) to erase DNA methylation; thus, this process is irreversible (Fig. 2). TET proteins have three subtypes: TET1, 2, and 3; and they have similar catalytic activities, but differ in the architecture of their domains [29]. One study demonstrated that TET1 is frequently upregulated and serves as an oncogene in NSCLC with lost p53 function and that TET1 might not be repressed by TP53-carrying transversion mutations [30].
Fig. 2.
Methylation and demethylation of DNA. DNA methyltransferases (DNMTs) methylate cytosine (C) at site C5, creating 5-methylcytosine (m5C). Through the proposed demethylation route, ten-eleven translocation enzymes catalyze the repeated oxidation of m5C to 5-(hydroxymethyl)cytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxycytosine (5caC). In order to create apyrimidinic (AP) sites, 5fC and 5caC are removed by the G/T-mismatch-specific thymine DNA glycosylase, followed by base excision repair (BER) to reinstall the cytosine
Hypermethylation with NSCLC
NSCLC as well as other lung cancers have been extensively studied in terms of hypermethylation targets and roles. The hypermethylation of many CGIs of potential tumor suppressor genes has been consistently identified in NSCLCs [31, 32]. These tumor suppressor genes play important roles in NSCLC cellular functions, including apoptosis, control of the cell cycle, adhesion, invasion, DNA repair, and modulation of signaling pathways. DNA hypermethylation markers are summarized to relate to NSCLC pathogenesis, progression, and outcome at the experimental level (Table 1). The silencing of some of these genes by DNAm continues to be confirmed when NSCLC lines are treated with methylation inhibitors [63]. Although methylation does not necessarily inactivate genes, it is proven to be useful as an epigenetic marker in NSCLC [64].
Table 1.
Summary of DNA modification markers in the pathogenesis, progression and metastasis, and prognosis of NSCLC
| DNA methylation marker | Pathway | Function | Reference |
|---|---|---|---|
| Upregulation | |||
| APC | Cell proliferation, migration, and cell adhesion | Pathogenesis and progression | [33] |
| CDH1 | Cell adhesion | Pathogenesis | [34] |
| CDH13 | Cell adhesion | Pathogenesis | [35] |
| P16 | Cell cycle regulation | Pathogenesis | [36] |
| DAPK | Apoptosis | Pathogenesis | [37] |
| DAL-1 | Progression and metastasis | [38] | |
| EPHB6 | Pathogenesis, progression, and metastasis | [39] | |
| HS3ST2 | Pathogenesis, progression, and metastasis | [40] | |
| TMEM88 | Progression and metastasis | [41] | |
| MGMT | Progression and metastasis | [42] | |
| HMLH1 | Prognosis | [43] | |
| IGFBP-3 | Prognosis | [44] | |
| RASSF1A | Cell cycle regulation, genomic-stability maintenance, apoptosis, cell migration, and invasion | Prognosis | [45] |
| TMEM196 | Prognosis | [46] | |
| GRK6 | Prognosis | [47] | |
| GSTP1 | Pathogenesis | [48] | |
| FHIT | Prognosis | [49] | |
| MLH1 | DNA repair | Pathogenesis | [50] |
| MSH2 | DNA repair | Pathogenesis | [50] |
| PTEN | Cell cycle regulation | NA | [51] |
| RUNX3 | TGF-β/Wnt signaling pathway | Pathogenesis | [52] |
| SEMA3B | Cell adhesion | Pathogenesis | [53] |
| RARβ | Cell differentiation and proliferation | Prognosis | [54] |
| RARβ2 | Cell differentiation and proliferation | Prognosis | [55] |
| SHOX2 | Cell differentiation and proliferation | Prognosis | [56] |
| TGFBR2 | Signaling | Pathogenesis | [57] |
| TSLC1 | Cell adhesion | Progression | [58] |
| Downregulation | |||
| LINE-1 | Prognosis | [59] | |
| ELMO3 | Progression and metastasis | [60] | |
| FAM83A | Prognosis | [61] | |
| MAGE | Transcriptional regulation, cancer development, and progression | Pathogenesis | [62] |
Hypomethylation with NSCLC
Lung cancers exhibit extensive global hypomethylation, particularly at repetitive sequences and segmental duplications [31]. A study found that LUAD patients with LINE-1 hypomethylation had a worse prognosis and progressed to a more advanced stage regardless of their driver gene mutations [65]. SNCG was also found to associate with advanced cancer cell invasion and migration stages [66]. Another study found that MAGE genes were overexpressed in most of NSCLCs, and such overexpression was related to the loss of methylation [67]. Lung cancer patients with MAGE overexpression had a poor prognosis and were more likely to develop metastasis and tumor growth [62]. Moreover, the increased hypomethylation of LINE-1 and Alu leads to their transcription enhancement and genomic instability increase in NSCLC [66]. Thereby, DNA hypomethylation is involved in the altered microenvironment, mutagenesis, and increased chromosomal instability.
Smoking and DNA methylation with NSCLC
A study reported that smoking was strongly associated with NSCLC; only 15–25% of NSCLC cases occurred in non-smokers [68]. Another study demonstrated that smoking was also strongly related to lung tumorigenesis via DNAm [69]. Moreover, the hypomethylation of AHRR, 6p21.33, and F2RL3 was related to smoking and the increased risk of developing NSCLC [70]. Three additional hypomethylated CpGs (cg21566642, cg05951221, and cg23387569) were also associated with lung cancer [71]. Several genes (p16, APC, and MGMT) were also found to be upregulated in smoking-associated lung cancer [72–74]. A smoking carcinogen might alter the ubiquitination level of DNMT1 and its degradation mechanisms to control DNMT1 production, which resulted in de novo hypermethylation of TSGs and ultimately caused carcinogenesis in NSCLC [75, 76]. With the increasing reactive oxygen species (ROS) and chronic inflammation, smoking also resulted in aberrant methylation and the activation of silencing complexes [77].
DNA methylation in diagnosis of NSCLC
In the past few years, clinical doctors have had increasing interests in DNA methylation detection. A study demonstrates that DNAm can be used to estimate tumor risk and has great use for tumor risk prevention [78]. DNAm markers in NSCLC early diagnosis are summarized (Table 2).
Table 2.
Summary of DNA methylation markers in lung cancer early diagnosis
| Single/combination marker | Tumor type | Sensitivity(%) | Specificity(%) | Sample type | Reference |
|---|---|---|---|---|---|
|
P16 RARB2 |
Lung cancer | 69 | 87 | Bronchial aspirates | [36] |
| RASSF1A | Lung cancer | 64 | 100 | Carcinoma tissues | [79] |
| Lung cancer | 60 | 90 | Brushing sample | ||
| RASSF1A | Lung cancer | 92 | 100 | Carcinoma tissues | [80] |
| PCDHGB6 + HOXA9 | Lung cancer | 80 | 100 | Brushing sample | |
| RASSF1A | NSCLC | 87 | 75 | Plasma(cirDNA) | [81] |
| RARB2 | Lung cancer | ||||
| SHOX2 | Squamous cell carcinoma | 60 | 90 | Plasma | [82] |
| SHOX2 | Lung cancer | 68 | 95 | Alveolar lavage fluid | [83] |
| SHOX2 | Lung cancer | 65.5 | 90 | Plasma | [56] |
| PTGER4 | Lung cancer | 56.3 | 90 | ||
| SHOX2 + PTGER4 | Lung cancer | 75.6 | 84.8 | ||
| SHOX2 | Lung cancer | 67 | 90 | Plasma | [84] |
| PTGER4 | Lung cancer | 90 | 73 | ||
| SHOX2 + RASSF1A | Lung cancer | 81 | 97.4 | Bronchoalveolar lavage fluid | [85] |
| HOXA9 | NSCLC | 55.2 | 74.3 | Plasma | [86] |
A study first reported the use of methylation detection in NSCLC early diagnosis [87]. Due to its sensitivity and stability, DNA methylation was an effective method to compensate for imaging examination defects and improve early diagnosis of lung cancer. Some examples are taken here. Studies also found that the methylated genes such as PTGER4, SHOX2, and RASSF1A were more likely to cause lung cancer [82, 88]. The analysis of alveolar lavage fluid of lung cancer patients revealed that SHOX2 gene methylation was used to distinguish benign from malignant lung tissues with specificity (95%) and sensitivity (68%) and also was used to diagnose LUAD with 82% sensitivity [83]. RASSF1A methylation was substantially correlated with NSCLC, which confirmed that RASSF1A was a tumor suppressor gene [89], and also, RASSF1A might be an indicator of invasive lung cancer because of its high specificity (93%) and sensitivity (17%), as well as its optimal performance within a screening interval of 2 years. A comprehensive analysis of tumor tissue, sputum, and blood samples from 155 lung cancer patients found that TMEM196 was methylated in lung cancer samples but not controls, and TMEM196 methylation was an independent prognostic indicator, which offered new potential clinical application of TMEM196 methylation as biomarkers for early diagnosis and prognosis in NSCLC [46]. The level of RAR2 methylation in stage III lung cancer patients was higher than that in stages I and II, and NSCLC patients had a higher level of RAR2 methylation in their plasma and cell surface-bound cirDNA; thus, cirDNA-based tests might be valuable to confirm methylated DNA markers in lung cancer diagnosis [55]. The promoter methylation index (PMI) varied significantly in lung cancers compared to healthy controls with ROC curve analysis (n = 100, AUC = 0.69, p = 0.0012), which shows that MIRA might be a viable non-invasive technique to detect early lung cancers [59]. Hypomethylation of L1RE1 was frequently detected in tumors compared to benign controls, whereas increased methylation of RARB was an independent tumor marker for high-grade lung cancer. RASSF1 hypermethylation was prevalent in NET, which might serve as an auxiliary biomarker to distinguish NSCLC from NET [90]. The combination of L1RE1 and RARB or RASSF1 methylations could serve as biomarkers to distinguish lung cancers from non-cancerous tissues [90]. HOXA9 gene methylation was proven to diagnose LUSC with sensitivity (55.2%) and specificity (74.3%) [86]. Further, the combination detection of two or more gene methylations might significantly increase the sensitivity and specificity. For instance, single-gene methylation analysis revealed that RASSF1A was sensitive in 64% of carcinoma tissues and about 60% in bronchial brushing samples [91]. However, the methylation of HOXA9, PCDHGB6, and RASSF1A was evaluated simultaneously; the sensitivity rose up to 92% in carcinoma tissues and 80% in bronchial brushing samples [91]. Thereby, dual- or multi-gene methylation detection had higher sensitivity and specificity than single-gene methylation detection in lung cancer diagnosis, which is the future direction to reduce contingencies, improve diagnostic accuracy, and lead to earlier diagnosis [64].
DNA methylation in NSCLC therapeutics
DNAm plays important roles in NSCLC treatment. With the determination of the methylation status of biomarkers, physicians are able to more effectively foresee the kind and stage of NSCLC and make a better treatment plan [86]. Several tumor suppressor genes can be demethylated to restore their normal expression and prevent the growth of NSCLC; DNMT might be a primary target of tumor epigenetic medications, with the use of DNA methyltransferase inhibitors (DNMTis) [86]. A study revealed that DNMTis in combination with histone deacetylase inhibitors were a more effective approach to treat NSCLC [92]. NSCLC can be effectively treated with immunotherapy, such as anti-programmed cell death protein 1 (PD1) and anti-programmed cell death ligand 1 (PD-L1) therapies. However, screening for suitable patients for anti-PD1/PD-L1 therapy has become a major challenge. Tumor mutation burden (TMB) is proven to be a predictor for the effectiveness of immunotherapy [93]. Previous studies demonstrate that DNAm is a good marker to predict immunotherapy efficacy, and a substantial increase occurs in DNAm aberrations and copy number variations (CNVs) in high-TMB NSCLCs, thus the association between DNAm and TMB contributes to the prediction of the immunotherapy efficacy of NSCLC [94]. A study demonstrated that hMLH1 gene methylation was a biomarker for tumor recurrence in NSCLC patients treated with cisplatin-based adjuvant cisplatin, and NSCLC patients without hMLH1 methylation benefited better from cisplatin-based adjuvant treatment [43]. Thus, hMLH1 methylation will become a biomarker that allows NSCLC patients to receive individualized treatment [43]. Also, cisplatin might be less toxic to NSCLC tumor cells when IGFBP-3 expression was decreased due to promoter-hypermethylation [44]. It is possible to identify patients who may benefit from CDDP therapy alone or in conjunction with epigenetic therapy based on IGFBP-3’s basal methylation status prior to chemotherapy [44].
Histone modifications in NSCLC
A protein octamer with four core histones (H-2A, -2B, -3, -4) surrounds the DNA to form the structure of the nucleosome in eukaryotic cells [95]. Some studies found that the histone alterations may influence the control of the transcription process [95]. Some studies found that histone modifications affect not only the transcription but also the DNA-templated processes. Histone proteins have tails, which are classic sites for post-translational modifications (PTMs). By altering DNA and histone charge density, DNA methylation and histone PTMs can affect the loosening of the nucleosome, as well as promote transcription factors and RNA polymerase to reach their targets [96–98]. The acetylation of histones was found to contribute to the development of lung cancer by stimulating gene transcription [99, 100]. Studies revealed that a number of histone deacetylase (HDAC) inhibitors were effective treatments for NSCLC in pre-clinical and clinical trials [98, 100].
Histone acetylation
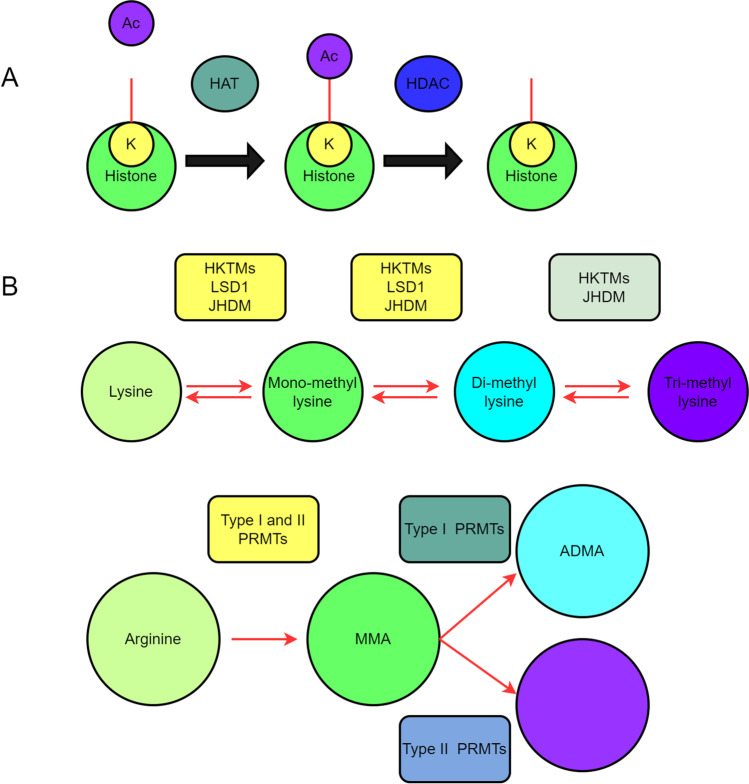
Histone acetylation is a major PTM in histones, and this modification occurs when histone deacetylase and acetyltransferase work together [101]. After the acetyl groups are removed from histones, HDACs cause compacted chromatin to reorganize, which in turn decreases gene transcription [102]. Histone lysine acetylation is generally reversible. The acetylated lysine residue can be deacetylated by histone acetyltransferases and histone deacetylases (Fig. 3A).
Fig. 3.
Regulators of histone acetylation and methylation. A Histone lysine can be acetylated in a highly reversible manner. During histone deacetylation, histone acetyltransferase enzymes (HATs/KATs) (HDACs) add acetylated lysine residues to the histone. B The histone is methylated. A histone methyltransferase adds methyl (CH3) groups to histone lysines or arginines with the help of histone demethylase. Methylation status of lysine or arginine is indicated
Histone acetylation in NSCLC
Histone modifications in NSCLC cells differ from normal cells [103]. In addition, most LUSC samples exhibit elevated levels of HDAC3 [104]. In LUAD, a poor prognosis is strongly correlated with higher levels of HDAC1 and HDAC3, and also a poor prognosis in NSCLC is associated with down-expression of HDAC2 [105, 106]. Another study found that FLIP was overexpressed in a subset of NSCLCs, FLIP inhibited caspase-8 activation to block the extrinsic apoptosis pathway. Overall survival is significantly reduced when FLIP expression is high in the cytoplasm. In NSCLC but not in healthy cells, an HDACi that targets HDAC1-3 decreases FLIP expression via the post-transcriptional mechanism and causes apoptosis [107].
Histone acetylation in NSCLC therapeutics
Single use of HDACi
Despite a substantial amount of preclinical evidence supporting their efficacy in NSCLC, HDACi has a limited clinical efficacy in clinical trials. Only three of 47 patients with advanced NSCLC who had previously received treatment responded partially to pivanex in a phase II study [108]. Vorinostat monotherapy in recurrent NSCLC patients did not show any objective tumor response, but showed obvious toxicity, including tiredness, cytopenias, and one death [109]. The completed and continuing clinical studies for NSCLC treatment based on histone acetylation are summarized (Table 3).
Table 3.
Histone modification-based completed and ongoing clinical trials for NSCLC
| Drugs | Trial phase | Patients | Outcomes | Reference |
|---|---|---|---|---|
| Completed clinical trials | ||||
| Single therapy | ||||
| Pivanex | II | 47 previously treated advanced NSCLC patients | ORR 6.4%, SD 30%, mPFS 1.5 mo, mOS 6.2 mo | [110] |
| Romidepsin | II | 19 previously treated advanced NSCLC patients | No objective responses | [108] |
| Vorinostat | II | 16 previously treated advanced NSCLC patients | No objective responses. mTTP 2.3 mo, mOS 7.1 mo | [109] |
| Entinostat | I | previously treated advanced solid tumors (n = 31; 4 NSCLC patients) | No PR or CR. One NSCLC patient had stable disease | [111] |
| Cl-994 | I | 53 previously treated solid tumors |
PR = 1 heavily pre-treated NSCLC patient SD = 3 (1 NSCLC patient) |
[112] |
| Combination with chemotherapy | ||||
| Vorinostat 400 mg/day orally or placebo + Chemotherapy (Carboplatin + Paclitaxel) | Randomized phase II | 94 previously untreated advanced NSCLC | ORR: 34% with vorinostat vs. 12.5% with placebo (p = 0.02), mPFS: 6 months vs. 4.1 months (p = 0.48), mOS: 13 months vs. 9.7 months (p = 0.17) | [113] |
| Belinostat (starting at 1000 mg/m2 dose) + Chemotherapy (Carboplatin + Paclitaxel) | I | 23 previously untreated advanced NSCLC | MTD 1400 mg/m2, ORR: 35%, mPFS: 5.7 mo | |
| Panobinostat orally 3 times a week + (Carboplatin + Etoposide) | I | 6 previously treated advanced NSCLC | No objective responses | [114] |
| Vorinostat + Carboplatin /Paclitaxel | I | 28 advanced solid tumor patients |
PR = 11 (10 NSCLC), SD = 7 Linear Pharmacokinetics |
[115] |
| Cl-994 + Gemcitabine | II | 26 NSCLC/174 patients | PR = 8, OR = 12%, MS = 194 days | [116] |
| Cl-994 + Carboplatin + Paclitaxel | I | 30 patients with advanced solid tumors | PR = 5 (3 NSCLC) | [117] |
| Decitabine + valproic acid | I | 8 patients with advanced NSCLC with prior chemotherapy | SD = 1 | [118] |
| Decitabine + vorinostat | I | 2 patients with NSCLC/44 with advanced tumors | SD = 29% | [119] |
| Azacitidine + sodium phenylbutyrate | I | 1 NSCLC/27 refractory solid tumors | SD = 1, PD = 26 | [120] |
| Hydralazine + magnesium valproate | II | 1 NSCLC/17 refractory solid tumors | PR = 4, SD = 8 | [121] |
| Combination with EGFR TKIs | ||||
| Panobinostat + Erlotinib | I | 15 Previously treated NSCLC patients | Of 12 evaluable patients, 7 had SD and 5 had PD | |
| Vorinostat + Gefitinib | I | 12 BIM deletion polymorphism/EGFR mutation double-positive NSCLC | mPFS: 5.2 mo, 6 weeks DCR: 83.3% | [122] |
| Erlotinib + Entinostat or Erlotinib + Placebo | Randomized phase II | 132 previously treated patients with stage IIIB/IV NSCLC, no prior EGFR-TKIs | ORR: 3% with EE vs. 9.2% with EP (p = 0.13), mPFS: 1.97 months with EE vs. 1.88 with EP (p = 0.98), mOS: 8.9 months with EE vs. 6.7 months with EP (p = 0.39) | [123] |
| Vorinostat + Sorafenib | I | 17 patients with advanced solid tumors | Unconfirmed PR = 2 (1 NSCLC patient) | [124] |
| Vorinostat + Erlotinib | I/II | 33 advanced NSCLC EGFR mutant patients | PFS = 8 weeks, OS = 10.3 months | [125] |
| Panobinostat + Erlotinib | I | 35 NSCLC/42 patients with advanced tumors | Disease control rate = 54%, NSCLC PR = 3, SD = 3, PFS = 4.7 months, OS = 41 months, (EGFR mutation) | [126] |
| Combination with hypomethylating agent | ||||
| Azacitidine + Entinostat | I/II | 45 advanced, refractory NSCLC | MS = 6.4 months, CR = 1, PR = 1 | [119] |
| Combination with radiation | ||||
| Vorinostat (200, 300, 400 mg/day) orally for 14 days + SRS for brain metastasis on day 3 | I | 17 NSCLC with up to 4 brain metastasis, ≤ 2 cm in size | No local failures with median follow-up of 12 months | [127] |
| Combination with ICI | ||||
| Vorinostat orally + Pembrolizumab | Phase I/Ib | ICI-naïve and ICI-pretreated advanced NSCLC patients in phase I, ICI-naïve patients only in phase Ib (n = 33) | ORR: 13%, SD: 53%, ICI-pretreated patients: ORR 12.5%, SD 42% | [128] |
| Ongoing clinical trials | ||||
| Vorinostat + Pembrolizumab | II | NCT02638090 | ||
| Entinostat + Pembrolizumab | II | NCT02638090 | ||
| Entinostat + Azacitidine + Nivolumab | II | NCT01928576 | ||
| Panobinostat + Anti PD-1 antibody PDR001 | I | NCT02890069 | ||
| Mocetinostat + Nivolumab | II | NCT02954991 | ||
| ACY-241 + Nivolumab | I | NCT02635061 | ||
| Abexinostat + Pembrolizumab | I | NCT03590054 | ||
Combination of HDACi with cytotoxic chemotherapy
There is overwhelming proof that HDACi in combination with cytotoxic chemotherapy has synergistic effects. The combination of HDACi with paclitaxel exerts synergistic anti-tumor effects by inducing hyperacetylation of p53 and tubulin, and by preventing p21 upregulation [129]. With the combination of HDACi with vinorelbine and platinum, the enhanced expressions of CHK1-2, p21, and p27 led to cell-cycle arrest and increased apoptosis [130]. A study about paclitaxel-resistant NSCLC cells found that HDAC1 was overexpressed, and combination treatment with paclitaxel and HDACi SNOH-3 options overcomes resistance to paclitaxel [131]. Based on this pre-clinical data, a randomized, double-blind, placebo-controlled study was performed to randomly assign patients to two groups and given either normal carboplatin dosages or paclitaxel plus either vorinostat or both. The response rate was greater in vorinostat group than placebo group (34% vs. 12%, p = 0.02). Progression-free survival (PFS) and overall survival (OS) were marginally better in vorinostat group although there were no statistical differences. In contrast, 3% of patients in the placebo group developed grade 4 thrombocytopenia, while 18% of patients in the vorinostat group did (p ≤ 0.05) [113]. In a phase I study, belinostat was tested through combination of paclitaxel and carboplatin, which obtained the objective response rate of 35% (partial responses), and the median PFS of 5.7 months. Although the combination of chemotherapy and HDACi has therapeutic benefits, its toxicity prevents its extensive use in clinical practice [114].
Combination of HDACi with immune checkpoint inhibition
Combined with cytotoxic chemotherapy, immune checkpoint inhibition (ICI) is becoming the norm to treat advanced NSCLC. Epigenetic control of the tumor microenvironment may be used to improve responses to ICI [132]. HDACi and ICI were initially investigated in a study that evaluated entinostat and azacitidine for dual epigenetic modulation; as a result, a total of five of the six NSCLC patients had PFS with 6 months, which was a satisfactory outcome for patients who failed to treat with ICI [133]. HDACi have been shown to prime the tumor microenvironment for response to ICI through multiple mechanisms, including upregulation of MHC expression, T cell functionality, tumor antigens, T cell chemokines, and stimulatory effects on T cells [134]. Based on gene signature mappings in NSCLC cancers and investigations of pathways induced by azacitidine in the cancer genome atlas, one found that that azacitidine promoted genetic factors related to the innate and adaptive immune systems, as well as genes implicated in immune evasion; and inhibition of both HDAC and HSP90 decreased gene transcription of PD-L1 in lung cancer cells treated with IFN-gamma, which suggests that it impacts the tumor immunosuppressive potential [135]. A combination of pembrolizumab and vorinostat, an inhibitor of the PD-1 receptor, was utilized in the advanced NSCLC patient group in a phase I/Ib study [128]. After phase I, phase Ib required patients to receive ICI-pretreatment. Every 3 weeks, 200 or 400 mg of vorinostat were given together with 200 or 400 mg of pembrolizumab as part of the therapy. No dose-limiting effects were detected. The most prevalent adverse responses were fatigue (33%) and nausea/vomiting (27%). The ICI-pretreated group contained eleven patients with stable illness, whereas three individuals showed a partial response. For future testing of this mixture in patients who have already received ICI, the findings of this exploratory trial were very positive. We are expecting the long-term results of this trial’s participants as well as the outcomes of many other ongoing trials about the combination of additional HDACi with ICI [128].
Combination of HDACi with tyrosine kinase inhibitors
The anti-EGFR antibody tyrosine kinase inhibitors (TKI) in a large percentage of advanced NSCLC tumors inhibited the activation of epidermal growth factor receptors to result in powerful responses to these medications [122]. TKI resistance ultimately develops in the overwhelming majority of patients despite their remarkable initial responses. One of these resistance mechanisms was the Bcl2-like protein 11 (commonly called BIM). In EGFR-mutated lung cancers, the proapoptotic molecule BIM may contribute to mortality [119]. In a phase I clinical study, vorinostat and gefitinib were used to treat NSCLC patients with BIM-deleting polymorphisms and EGFR mutations. Twelve EGFR-mutated NSCLC patients who previously received chemotherapy and an EGFR TKI were given escalating doses of gefitinib and vorinostat, which resulted in a combination’s well-tolerated effects, 83.3% disease control rate at 6 weeks, a modest median PFS (5.2 months), and the heartening median OS (22.1 months) [122]. Similarly, a study also demonstrated that the HDACi panobinostat in combination with the third-generation EGFR TKI osimertinib increased apoptosis induction and decreased the continued availability of cell cultures and xenograft models resistant to osimertinib, such as those with C797S alterations, through BIM upregulation [122].
Combination of HDACi with radiation therapy
Ionizing radiation can cause the single-strand breaks (SSBs), double-strand breaks (DSBs), and inter-strand crosslinks of DNA, which can produce an anti-tumor effect [136]. When DNA breaks in the double strands, DNA damage response mechanisms are activated, including recombination between homologous and non-homologous ends. Improvement of DNA double-strand breaks might contribute to the resistance to ionizing radiation. A DSB marker, HDACi, is upregulated by ionizing radiation in lung cancer cell lines [136]. RAD51, CHK1, and BRCA2 are downregulated by HDACi, which assist in repairing radiation-caused DNA damage, and the NHEJ pathway is also inhibited by Ku70/80 and XRCC4 acetylation caused by HDAC inhibition [134]. The mixture of HDACi with radiation exposure in NSCLC is now being tested in several clinical studies [137].
Histone methylation
Histone methylation is one of the most extensively studied modifications in histone, which has been shown to activate or inhibit transcription at several gene loci in NSCLC [96]. Methyltransferases and their equivalent demethylases are referred to as heterocyclic methyltransferases because they write and remove residues of histone lysine [96].
The functions of lysine methyltransferases in NSCLC
Lysine methyltransferases (KMTs) can eliminate the methyl groups at the methylated lysine residues in both histone and none histone substrates (Fig. 3B) [138, 139]. The structural organization and catalytic domain of KMTs with and without the SET domain (the SET domain is an important domain of histone methyltransferases) are used to categorize KMTs [140, 141]. EZH2 is a homolog of Drosophila En (zeste) (PRC2), an element of the Polycomb repressive complex II. Together with cofactors SUZ12 and EED, EZH2 transfers 1, 2, or 3 marks of methylation on H3K27 in a SAM-dependent manner. The EZH2 positive expression was associated with greater resistance to platinum-based treatments compared to the EZH2 negative expression in advanced NSCLC patients [142, 143]. Several malignancies are associated with dysregulation or loss of function of the MLL (KMT2) methyltransferase family [144]. Study found that MLL2 was a type of the most often altered genes in NSCLC patients in China with whole-exome sequencing, negative MLL2 mutations were present in 11.4 percent of NSCLC patients, and lack of MLL2 expression was common in NSCLC [145]. The mono- and di-methylation of H3K9 (H3K9me1, me2) was catalyzed by the G9a KMT [146]. G9a promoted the proliferation and invasion of NSCLC cells by raising H3K9me2 levels close to the CASP1 promoter and inhibiting caspase 1 [147]. A study found that the SET and MYND property 2 methylated the lysine residue in anaplastic lymphoma kinase, and activated oncogenic ALK. Crisotinib (an inhibitor of ALK) and LLY-507 (an inhibitor of SMYD2) both greatly limited the development of NSCLC cells [148]. SETD2 mutations were found in primary NSCLCs [149]. In mice, LUADs in both their early and late stages were accelerated by SETD2 deletion and subsequent H3K36me3 loss [150]. It suggests that it is conceivable for SETD2 to be used as a prognostic or diagnostic indicator for NSCLC. KMT5A, sometimes referred to as SET8, selectively methylates H4K20 and is associated with many cancer-related processes [151]. The tumor suppressor miR382 prevented carcinogenesis and metastasis in NSCLC cells by suppressing SETD8; however, restoring SETD8 might promote proliferation, migration, and invasion of NSCLC cells [152].
The functions of KDMs in NSCLC
Both lysine-specific demethylases and Jumonji (JmjC) domain-containing demethylases may be categorized as KDMs in accordance with the oxidative mechanisms of the demethylation reactions as well as the composition of the catalytic domains. LSD1, which is the first discovered and most extensively studied KDM demethylase, belongs to the LSD family that does not contain JmjC domain [153]. In NSCLC patients, overexpression of LSD1 was strongly related to a shorter overall survival time [154]. Increased tumor proliferation and EMT (epithelial-mesenchymal transition) process were brought about via LSD1 being drawn to the Kruppel-like factor 2 or Ecadherin promoters in NSCLC cells [155]. KDM2 accelerated H3K36 demethylation, which was associated with gene activation; and lung cancer was linked to KDM2A but not to its homologous gene KDM2B [156]. KDM2A was dysregulated in 54 NSCLC cell lines, and its protein level was significantly higher in primary NSCLC tissues than adjacent healthy control tissues [157]. Furthermore, KDM2A increased transcription of HDAC3 target genes while demethylating H3K36me2 at the HDAC3 promoter in order to reduce transcription of HDAC3 itself, such as NANOS1 that was related to cell invasion, and CDK6 that was related to cell cycle, in NSCLC cells with overexpressed KDM2A [158]. More than 50% of NSCLC patients were discovered to have overexpressed KDM3A, a KDM demethylase specific to the H3K9 protein [159]. By enhancing the expression of the anti-tumor miRNA let-7c and lowering the presence of the tumor-promoting EZH2, KDM3A knockdown prevented tumor growth among NSCLC cell cultures and a xenograft model [160]. KDM6A, an H3K27-specific demethylase that normally opposes EZH2, was investigated in NSCLC cells with conflicting findings about its link to lung cancer: when a spectrum of NSCLC cell lines used its inhibitor GSKJ4 to show antitumor activity, KDM6A epigenetically suppressed the TGF-induced EMT process. Based on human lung cancer tissues and transgenic NSCLC mice models, the KDM6A gene was identified as one of the important tumor suppressor genes [161].
Histone methyltransferase/demethylase inhibitors
Histone methyltransferase or demethylase inhibitors, alone or combined with other drugs, have been extensively documented to have antitumor effects for a variety of malignancies. The roles of typical histone methyltransferase/demethylase inhibitors are summarized (Table 4), along with any possible therapeutic benefits for NSCLC. In NSCLC cells that are resistant to erlotinib (NSCLC cells are resistant because erlotinib inhibits the tyrosine kinase), PRMT1 overexpression decreased E-cadherin function and facilitated EMT [162]. Considerably worse clinical outcomes were linked to significantly higher JMJD6 transcription in LUAD, which was positively associated with the extent and size of the tumor. A high JMJD6 level was also strongly correlated with pT condition, pN condition, and pathological grading, which suggests that it might be used as a prognostic and diagnostic indicator for NSCLC [163]. In NSCLC cells that are resistant to gefitinib, PAD4 expression drastically reduced (the tyrosine kinase inhibitor gefitinib is widely utilized to treat NSCLC). The overexpression of PAD4 inhibits the ETS-domain protein, which controls EMT activity and prevents gefitinib resistance [164]. Studies found that DZNep dose-dependently suppresses the development of NSCLC cell lines [165]. Lung cancers were also more responsive to other inhibitors after pharmacological EZH2 suppression, for example, DZNep or GSK126 administration may increase the anticancer impact in terms pf doxorubicin, a topoII regulator, vs BRG1 and EGFR mutated cancer of lungs [166].
Table 4.
Histone methyltransferases and demethylases with reported functions and representative methyltransferase/demethylase inhibitors in lung cancer (including NSCLC)
| Enzymes and their inhibitors | Target | Function | Reference |
|---|---|---|---|
| Histone methyltransferases | |||
| MLL2 | H3K4 | Loss of expression and deleterious mutations in NSCLC | [145] |
| SMYD2 | H3K36 | Contributed to NSCLC cell growth | [148] |
| SETD2 | H3K36 | Deleterious mutations in primary NSCLC | [149] |
| DOT1L | H3K79 | Contributed to NSCLC cell growth | [167] |
| PRMTs | Arginine on H3 and H4 | Contributed to NSCLC cell growth and overexpressed in TKI-resistant NSCLC | [164] |
| Histone demethylases | |||
| KDM2A | H3K36me2/me1 | Overexpressed in NSCLC | [157] |
| PAD4 | Arginine on H3 and H4 | Overexpression led to gefitinib resistance in NSCLC | [164] |
| KDM3 | H3K9me2/me1 | Overexpressed in NSCLC | [159] |
| EZH2 inhibitors | |||
| Clinical Trial Number | |||
| DZNep | SAH hydrolase inhibitor (Ki = 50 pM) | NA | [168] |
| GSK2816126 (GSK126) | SAM-competitive EZH2 inhibitor (IC50 = 9.9 nM | NCT02082977 | [169] |
| EPZ6438 (Tazemetostat) | SAM-competitive EZH2 inhibitor (Ki = 2.5 nM) (IC50 = 11 nM) | NCT01897571 NCT02601950 NCT02601937 | [170] |
| CPI1205 | SAM-competitive EZH2 inhibitor | NCT02395601 | [171] |
| LSD1 inhibitor | |||
| RG6016 (ORY-1001) | FAD-dependent irreversible LSD1 inhibitor (IC50 b 20 nM) | NCT02913443 | [172] |
Future perspectives
A few topics are still needed to be further investigated despite recent achievements in DNA and histone modifications. Although cell lines are a good model for cancer research, several studies have demonstrated that DNA and histone information from cell lines could not be comparable to those from actual endogenous tumor cells in cancer tissue. The bulk of DNA and histone alteration markers derived from cell lines do not have enough specificity and sensitivity to be widely used in clinical settings. In addition, progress in DNA and histone alteration studies was hampered by a lack of extremely sensitive and precise detection methods. More methods must be quickly developed for very sensitive and precise detection of DNA and histone changes. Earlier studies demonstrate that the identification of dual-gene or multi-gene alterations is more accurate and precise than the detection of single-gene mutations. Future detection of DNA and histone modifications will likely rely more heavily on double-gene or multi-gene combinations since these techniques not only reduce the contingency of findings but also enhance diagnostic precision, which is more suitable for early detection.
Moreover, there are at least ten types of modifications occurred in DNA, and many types of modifications occurred in histones [173]. The characteristics of each type of modification are different, which need different analytical approaches. Multiomics including genomics (DNA sequencing) and PTM-proteomics might be the effective methods to deeply reveal DNA and histone modifications in NSCLC, respectively [173, 174]. It would be necessary to study different types of modifications that occur in DNA, and in histones, to completely reveal the biological roles and effects of modifications in DNA and histones in NSCLC pathophysiological processes.
Conclusions and expert recommendation in the framework of 3P medicine
DNA and histone alterations are important molecular events in cancer pathophysiological processes and have become a popular focus of cancer research. This review article discussed the significance of DNA and histone alterations in NSCLC etiology, early detection, development and metastasis, therapy, and prognosis, as well as the most recent advances in the field. The in-depth investigation of DNA and histone dysregulation in NSCLC leads to identifying potential molecular targets and biomarkers for personalized treatment of NSCLC as well as improves our understanding of the mechanisms behind carcinogenesis and the development of NSCLC. Moreover, NSCLC with specific DNA and histone alteration traits may provide new, accurate classifications, which may ultimately provide more precise personalized treatment for NSCLC patients.
We recommend the emphasis on the research practice of modifications in DNA and histones in NSCLC. We propose further PPPM development and practical application of the DNA and histone modifications in NSCLC, in the following aspects:
-
(i)
Predictive medical approach: It is necessary to clarify the roles of DNA and histone modifications in the predictive approach (mechanism of tumorigenesis for NSCLC). It is possible that aberrant DNMT activity contributes to NSCLC in different ways [26, 27]. The hypermethylation and hypomethylation of many potential tumor suppressor genes have been consistently identified in NSCLCs. Several studies demonstrated that smoking is also strongly related to NSCLC tumorigenesis via DNA methylation [69]. Also, histone acetylation contributes to the development of lung cancer by stimulating gene transcription [99, 100]. Histone methylation can activate or inhibit transcription at several gene loci in NSCLC [96]. An in-depth clarification of DNA modification of these genes and histone modification might offer an effective predictive medical approach for NSCLC.
-
(ii)
Targeted prevention: The DNA and histone modifications have potential utility for targeted prevention, including reducing tumorigenesis (primary prevention) and early diagnosis (secondary prevention) of NSCLC. The study demonstrates that DNA methylation epigenetically can be used to estimate tumor risk and has great use for tumor risk prevention [78]. Previous studies have shown that DNA methylation markers can be used in NSCLC early diagnosis (Table 2). There are relatively few studies on histone modification for early diagnosis of NSCLC; however, future uses of SETD2 as a diagnostic marker for NSCLC are conceivable. It might be a potential biomarker pattern in combination with other markers for early diagnosis of NSCLC.
-
(iii)
Personalized treatments: The DNA and histone modifications have potential utility for NSCLC personalized treatments. DNA methylation also plays important roles in NSCLC treatment. By determining the methylation status of biomarkers, physicians are better able to foresee the kind and stage of NSCLC and make a better treatment plan [86]. Previous studies have shown that DNA methylation is also a good marker to predict immunotherapy efficacy [91]. A number of HDACis have shown to be an effective treatment for NSCLC in pre-clinical and clinical trials [98, 100]. Histone acetylation in NSCLC therapeutics includes a single use of HDACi, combination of HDACi with cytotoxic chemotherapy, a combination of HDACi with immune checkpoint inhibition, a combination of HDACi along with tyrosine kinase inhibitor, and a combination of HDACi with radiation. Histone methyltransferases/demethylase inhibitors, alone or in combination with other drugs, have been extensively documented to have antitumor effects for a variety of malignancies. Thereby, DNA and histone modifications are the potential effective therapeutic targets for personalized treatment of NSCLC.
Acknowledgements
The authors acknowledge the financial support from the Shandong First Medical University Talent Introduction Funds (to X.Z.), Shandong First Medical University High-level Scientific Research Achievement Cultivation Funding Program (to X.Z.), the Shandong Provincial Natural Science Foundation (ZR202103020356/ZR2021MH156 to X.Z.), Taishan Scholar Engineering Project Special Funds (to X.Z.), and the Academic Promotion Program of Shandong First Medical University (2019ZL002).
Abbreviations
- APC
Adenomatous polyposis coli
- ALK
Anaplastic lymphoma kinase
- CASP1
Silenced caspase 1
- CNVs
Copy number variations
- DSBs
Double-strand breaks
- DNMTs
DNA methyltransferases
- DNAm
DNA methylation
- ELMO3
Engulfment and cell motility 3
- EGFR
Epidermal growth factor receptor
- FHIT
Fragile histidine triad
- HR
Homologous recombination
- hMLH1
Human mutL homolog 1
- HRM
Methylation-specific high-resolution melting
- HDACis
Histone deacetylase inhibitors
- HATs
Acetyltransferases
- KLF2
Kruppel-like factor 2
- LUAD
Lung adenocarcinoma
- LOH
Loss of heterozygosity
- m6A
N6-methyladenosine
- MMP
Matrix metalloproteinase
- NSCLC
Non-small cell lung cancer
- NHEJ
Non-homologous end joining
- OG
8-Oxo-7,8-dihydroguanine
- OS
Overall survival
- PT
Phosphorothioate
- PD-1
Anti-programmed cell death protein 1
- PD-L1
Anti-programmed cell death ligand 1
- PTMs
Posttranslational modifications
- PFS
Progression-free survival
- ROS
Reactive oxygen species
- RARbeta
Retinoic acid receptor-beta
- SMYD2
SET and MYND domain-containing 2
- SCLC
Small cell lung cancer
- TMB
Tumor mutation burden
- TCGA
The Cancer Genome Atlas
- TKIs
Tyrosine kinase inhibitors
- TF
Transcription factor
- TET
Ten-eleven translocation enzymes
- TSGs
Tumor suppressor genes
- 5mC
5-Methylcytosine
- 5hmU
5-Hydroxymethyluracil
- 5fU
5-Formyluracil
- 5hmU
5-Hydroxymethyluracil
- 5fU
5-Formyluracil
- 5-Aza-dC
5-Aza-2′-deoxycytidine
- 5-aza-CdR
5-Aza-2′-deoxycytidine
Author contribution
G.Z. and Z.W. collected and analyzed literature, wrote the manuscript, they contributed equally to the manuscript. P.S. participated in collection and analysis of literature. X.Z. conceived the concept, designed the manuscript, coordinated, and critically revised the manuscript, and was responsible for the corresponding works. All authors approved the final manuscript.
Funding
This work was supported by the Shandong First Medical University Talent Introduction Funds (to X.Z.), Shandong First Medical University High-level Scientific Research Achievement Cultivation Funding Program (to X.Z.), the Shandong Provincial Natural Science Foundation (ZR202103020356/ZR2021MH156 to X.Z.), Taishan Scholar Engineering Project Special Funds (to X.Z.), and the Academic Promotion Program of Shandong First Medical University (2019ZL002).
Data availability
All data and materials are available in the current manuscript.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Code availability
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Guodong Zhang and Zhengdan Wang contributed equally to this work.
References
- 1.Sung H, Ferlay J, Siegel RL. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res. 2016;5(3):288–300. doi: 10.21037/tlcr.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamura T, Kurishima K, Nakazawa K, Kagohashi K, Ishikawa H, Satoh H, Hizawa N. Specific organ metastases and survival in metastatic non-small-cell lung cancer. Mole Clin Oncol. 2015;3(1):217–221. doi: 10.3892/mco.2014.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23(7):781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng T, Zhan X. Pattern recognition for predictive, preventive, and personalized medicine in cancer. EPMA J. 2017;8(1):51–60. doi: 10.1007/s13167-017-0083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu M, Zhan X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J. 2018;9(1):77–102. doi: 10.1007/s13167-018-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golubnitschaja O, Costigliola V. EPMA General report & recommendations in predictive, preventive and personalised medicine 2012: white paper of the European Association for Predictive Preventive Personalised Medicine. EPMA J. 2012;3(1):14. doi: 10.1186/1878-5085-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golubnitschaja O, Filep N, Yeghiazaryan K, Blom HJ, Hofmann-Apitius M, Kuhn W. Multi-omic approach decodes paradoxes of the triple-negative breast cancer: lessons for predictive, preventive and personalised medicine. Amino Acids. 2018;50(3–4):383–395. doi: 10.1007/s00726-017-2524-0. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg MVC. Bourc’his D, The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019;20(10):590–607. doi: 10.1038/s41580-019-0159-6. [DOI] [PubMed] [Google Scholar]
- 10.Koziol MJ, Bradshaw CR, Allen GE, Costa ASH, Frezza C, Gurdon JB. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat Struct Mol Biol. 2016;23(1):24–30. doi: 10.1038/nsmb.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Zhu Y, Luo GZ, Wang X, Yue Y, Wang X, Zong X, Chen K, Yin H, Fu Y, et al. Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat Commun. 2016;7:13052. doi: 10.1038/ncomms13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, Liu Y, Byrum SD, Mackintosh SG, Zhong M, et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016;532(7599):329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao Z, Wu T, Cui X, Zhu P, Tan C, Dou X, Hsu KW, Lin YT, Peng PH, Zhang LS, et al. N(6)-Deoxyadenosine methylation in mammalian mitochondrial DNA. Mol Cell. 2020;78(3):382–395.e388. doi: 10.1016/j.molcel.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musheev MU. Baumgne, The origin of genomic N(6)-methyl-deoxyadenosine in mammalian cells. Nat Chem Biol. 2020;16(6):630–634. doi: 10.1038/s41589-020-0504-2. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Lai W, Li Y, Chen S, Liu B, Zhang N, Mo J, Lyu C, Zheng J, Du YR, et al. N(6)-methyladenine is incorporated into mammalian genome by DNA polymerase. Cell Res. 2021;31(1):94–97. doi: 10.1038/s41422-020-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfaffeneder T, Spada F, Wagner M, Brandmayr C, Laube SK, Eisen D, Truss M, Steinbacher J, Hackner B, Kotljarova O, et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat Chem Biol. 2014;10(7):574–581. doi: 10.1038/nchembio.1532. [DOI] [PubMed] [Google Scholar]
- 17.Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer. 2014;14(12):786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan B, Jiang Y, Wang Y, Wang Y. Efficient formation of the tandem thymine glycol/8-oxo-7,8-dihydroguanine lesion in isolated DNA and the mutagenic and cytotoxic properties of the tandem lesions in Escherichia coli cells. Chem Res Toxicol. 2010;23(1):11–19. doi: 10.1021/tx9004264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai Y, Yuan BF, Feng YQ. Quantification and mapping of DNA modifications. RSC Chem Biol. 2021;2(4):1096–1114. doi: 10.1039/d1cb00022e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem. 1948;175(1):315–332. doi: 10.1016/S0021-9258(18)57261-6. [DOI] [PubMed] [Google Scholar]
- 21.Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392(10149):777–786. doi: 10.1016/s0140-6736(18)31268-6. [DOI] [PubMed] [Google Scholar]
- 22.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 23.Heyn H, Esteller M. An adenine code for DNA: a second life for N6-methyladenine. Cell. 2015;161(4):710–713. doi: 10.1016/j.cell.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 24.Luo GZ, He C. DNA N(6)-methyladenine in metazoans: functional epigenetic mark or bystander? Nat Struct Mol Biol. 2017;24(6):503–506. doi: 10.1038/nsmb.3412. [DOI] [PubMed] [Google Scholar]
- 25.Chen T, Li E. Establishment and maintenance of DNA methylation patterns in mammals. Curr Top Microbiol Immunol. 2006;301:179–201. doi: 10.1007/3-540-31390-7_6. [DOI] [PubMed] [Google Scholar]
- 26.Kim H, Kwon YM, Kim JS, Han J, Shim YM, Park J, Kim DH. Elevated mRNA levels of DNA methyltransferase-1 as an independent prognostic factor in primary nonsmall cell lung cancer. Cancer. 2006;107(5):1042–1049. doi: 10.1002/cncr.22087. [DOI] [PubMed] [Google Scholar]
- 27.Husni RE, Shiba-Ishii A, Iiyama S, Shiozawa T, Kim Y, Nakagawa T, Sato T, Kano J, Minami Y, Noguchi M. DNMT3a expression pattern and its prognostic value in lung adenocarcinoma. Lung Cancer. 2016;97:59–65. doi: 10.1016/j.lungcan.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 28.Gao Q, Steine EJ, Barrasa MI, Hockemeyer D, Pawlak M, Fu D, Reddy S, Bell GW, Jaenisch R. Deletion of the de novo DNA methyltransferase Dnmt3a promotes lung tumor progression. Proc Natl Acad Sci U S A. 2011;108(44):18061–18066. doi: 10.1073/pnas.1114946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filipczak PT, Leng S, Tellez CS, Do KC, Grimes MJ, Thomas CL, Walton-Filipczak SR, Picchi MA, Belinsky SA. p53-suppressed oncogene TET1 prevents cellular aging in lung cancer. Cancer Res. 2019;79(8):1758–1768. doi: 10.1158/0008-5472.CAN-18-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, Riggs AD, Pfeifer GP. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A. 2008;105(1):252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalari S, Jung M, Kernstine KH, Takahashi T, Pfeifer GP. The DNA methylation landscape of small cell lung cancer suggests a differentiation defect of neuroendocrine cells. Oncogene. 2013;32(30):3559–3568. doi: 10.1038/onc.2012.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brabender J, Usadel H, Danenberg KD, Metzger R, Schneider PM, Lord RV, Wickramasinghe K, Lum CE, Park J, Salonga D, et al. Adenomatous polyposis coli gene promoter hypermethylation in non-small cell lung cancer is associated with survival. Oncogene. 2001;20(27):3528–3532. doi: 10.1038/sj.onc.1204455. [DOI] [PubMed] [Google Scholar]
- 34.Yu Q, Guo Q, Chen L, Liu S. Clinicopathological significance and potential drug targeting of CDH1 in lung cancer: a meta-analysis and literature review. Drug Des Dev Ther. 2015;9:2171–2178. doi: 10.2147/dddt.s78537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toyooka KO, Toyooka S, Virmani AK, Sathyanarayana UG, Euhus DM, Gilcrease M, Minna JD, Gazdar AF. Loss of expression and aberrant methylation of the CDH13 (H-cadherin) gene in breast and lung carcinomas. Can Res. 2001;61(11):4556–4560. [PubMed] [Google Scholar]
- 36.Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A. 1998;95(20):11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61(1):249–255. [PubMed] [Google Scholar]
- 38.Zhang Y, Xu R, Li G, Xie X, Long J, Wang H. Loss of expression of the differentially expressed in adenocarcinoma of the lung (DAL-1) protein is associated with metastasis of non-small cell lung carcinoma cells. Tumour Biol. 2012;33(6):1915–1925. doi: 10.1007/s13277-012-0452-x. [DOI] [PubMed] [Google Scholar]
- 39.Yu J, Bulk E, Ji P, Hascher A, Tang M, Metzger R, Marra A, Serve H, Berdel WE, Wiewroth R, et al. The EPHB6 receptor tyrosine kinase is a metastasis suppressor that is frequently silenced by promoter DNA hypermethylation in non-small cell lung cancer. Clin Cancer Res. 2010;16(8):2275–2283. doi: 10.1158/1078-0432.Ccr-09-2000. [DOI] [PubMed] [Google Scholar]
- 40.Hwang JA, Kim Y, Hong SH, Lee J, Cho YG, Han JY, Kim YH, Han J, Shim YM, Lee YS, et al. Epigenetic inactivation of heparan sulfate (glucosamine) 3-O-sulfotransferase 2 in lung cancer and its role in tumorigenesis. PLoS ONE. 2013;8(11):e79634. doi: 10.1371/journal.pone.0079634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma R, Feng N, Yu X, Lin H, Zhang X, Shi O, Zhang H, Zhang S, Li L, Zheng M, et al. Promoter methylation of Wnt/β-catenin signal inhibitor TMEM88 is associated with unfavorable prognosis of non-small cell lung cancer. Cancer Biol Med. 2017;14(4):377–386. doi: 10.20892/j.issn.2095-3941.2017.0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hashimoto K, Narita Y, Matsushita Y, Miyakita Y, Ono M, Kayama T, Shibui S. Methylation status of O6-methylguanine-DNA-methyl transferase promoter region in non-small-cell lung cancer patients with brain metastasis. Clin Transl Oncol. 2012;14(1):31–35. doi: 10.1007/s12094-012-0758-6. [DOI] [PubMed] [Google Scholar]
- 43.Wu F, Lu M, Qu L, Li DQ, Hu CH. DNA methylation of hMLH1 correlates with the clinical response to cisplatin after a surgical resection in Non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8(5):5457–5463. [PMC free article] [PubMed] [Google Scholar]
- 44.Ibanez de Caceres I, Cortes-Sempere M, Moratilla C, Machado-Pinilla R, Rodriguez-Fanjul V, et al. IGFBP-3 hypermethylation-derived deficiency mediates cisplatin resistance in non-small-cell lung cancer. Oncogene. 2010;29(11):1681–1690. doi: 10.1038/onc.2009.454. [DOI] [PubMed] [Google Scholar]
- 45.Fischer JR, Ohnmacht U, Rieger N, Zemaitis M, Stoffregen C, Manegold C, Lahm H. Prognostic significance of RASSF1A promoter methylation on survival of non-small cell lung cancer patients treated with gemcitabine. Lung Cancer. 2007;56(1):115–123. doi: 10.1016/j.lungcan.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 46.Liu WB, Han F, Huang YS, Chen HQ, Chen JP, Wang DD, Jiang X, Yin L, Cao J, Liu JY. TMEM196 hypermethylation as a novel diagnostic and prognostic biomarker for lung cancer. Mol Carcinog. 2019;58(4):474–487. doi: 10.1002/mc.22942. [DOI] [PubMed] [Google Scholar]
- 47.Yao S, Wu D, Chen J, Wang P, Lv X, Huang J. Hypermethylation of the G protein-coupled receptor kinase 6 (GRK6) promoter inhibits binding of C/EBPα, and GRK6 knockdown promotes cell migration and invasion in lung adenocarcinoma cells. FEBS Open Bio. 2019;9(4):605–617. doi: 10.1002/2211-5463.12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toyooka S, Toyooka KO, Maruyama R, Virmani AK, Girard L, Miyajima K, Harada K, Ariyoshi Y, Takahashi T, Sugio K, et al. DNA methylation profiles of lung tumors. Mol Cancer Ther. 2001;1(1):61–67. [PubMed] [Google Scholar]
- 49.Z9.:61–67. S Lam, Geradts J, Ashfaq R, Virmani AK, Milchgrub S, Gazdar AF, Minna JD, 5′ CpG island methylation of the FHIT gene is correlated with loss of gene expression in lung and breast cancer. Cancer Res 2001, 61(9):3581–3585. [PubMed]
- 50.Gomes A, Reis-Silva M. Promoter hypermethylation of DNA repair genes MLH1 and MSH2 in adenocarcinomas and squamous cell carcinomas of the lung. Rev Port Pneumol. 2014;20(1):20–30. doi: 10.1016/j.rppneu.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 51.Soria JC, Lee HY, Lee JI, Wang L, Issa JP, Kemp BL, Liu DD, Kurie JM, Mao L, Khuri FR. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin Cancer Res. 2002;8(5):1178–1184. [PubMed] [Google Scholar]
- 52.Sato K, Tomizawa Y, Iijima H, Saito R, Ishizuka T, Nakajima T, Mori M. Epigenetic inactivation of the RUNX3 gene in lung cancer. Oncol Rep. 2006;15(1):129–135. [PubMed] [Google Scholar]
- 53.Kuroki T, Trapasso F, Yendamuri S, Matsuyama A, Alder H, Williams NN, Kaiser LR, Croce CM. Allelic loss on chromosome 3p21.3 and promoter hypermethylation of semaphorin 3B in non-small cell lung cancer. Cancer Res. 2003;63(12):3352–3355. [PubMed] [Google Scholar]
- 54.Virmani AK, Rathi A, Zsome 3p21.3 and promoter hypermethylation of semaphorin 3B in non-smallng KM, Thunnissen F et al. Promoter methylation and silencing of the retinoic acid receptor-beta gene in lung carcinomas. J Natl Cancer Inst 2000, 92(16):1303–1307. 10.1093/jnci/92.16.1303 [DOI] [PubMed]
- 55.Ponomaryova AA, Rykova EY, Cherdyntseva NV, Skvortsova TE, Dobrodeev AY. Zav’yalov AA, Tuzikov SA, Vlassov VV, Laktionov PP, RARβ2 gene methylation level in the circulating DNA from blood of patients with lung cancer. Eur J Cancer Prev. 2011;20(6):453–455. doi: 10.1097/CEJ.0b013e3283498eb4. [DOI] [PubMed] [Google Scholar]
- 56.Xu Z, Wang Y, Wang L, Xiong J, Wang H, Cui F, Peng H. The performance of the SHOX2/PTGER4 methylation assay is influenced by cancer stage, age, type and differentiation. Biomark Med. 2020;14(5):341–351. doi: 10.2217/bmm-2019-0325. [DOI] [PubMed] [Google Scholar]
- 57.Zhang HT, Chen XF, Wang MH, Wang JC, Qi QY, Zhang RM, Xu WQ, Fei QY, Wang F, Cheng QQ, et al. Defective expression of transforming growth factor beta receptor type II is associated with CpG methylated promoter in primary non-small cell lung cancer. Clin Cancer Res. 2004;10(7):2359–2367. doi: 10.1158/1078-0432.ccr-0959-3. [DOI] [PubMed] [Google Scholar]
- 58.Fukami T, Fukuhara H, Kuramochi M, Maruyama T, Isogai K, Sakamoto M, Takamoto S, Murakami Y. Promoter methylation of the TSLC1 gene in advanced lung tumors and various cancer cell lines. Int J Cancer. 2003;107(1):53–59. doi: 10.1002/ijc.11348. [DOI] [PubMed] [Google Scholar]
- 59.Gainetdinov IV, Kapitskaya KY, Rykova EY, Ponomaryova AA, Cherdyntseva NV, Vlassov VV, Laktionov PP, Azhikina TL. Hypomethylation of human-specific family of LINE-1 retrotransposons in circulating DNA of lung cancer patients. Lung Cancer. 2016;99:127–130. doi: 10.1016/j.lungcan.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 60.Søes S, Daugaard IL, Sørensen BS, Carus A, Mattheisen M, Alsner J, Overgaard J, Hager H, Hansen LL, Kristensen LS. Hypomethylation and increased expression of the putative oncogene ELMO3 are associated with lung cancer development and metastases formation. Oncoscience. 2014;1(5):367–74. doi: 10.18632/oncoscience.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu J, Hou M, Pei T. FAM83A is a prognosis signature and potential oncogene of lung adenocarcinoma. DNA Cell Biol. 2020;39(5):890–899. doi: 10.1089/dna.2019.4970. [DOI] [PubMed] [Google Scholar]
- 62.Jang SJ, Soria JC, Wang L, Hassan KA, Morice RC, Walsh GL, Hong WK, Mao L. Activation of melanoma antigen tumor antigens occurs early in lung carcinogenesis. Cancer Res. 2001;61(21):7959–7963. [PubMed] [Google Scholar]
- 63.Tsou JA, Hagen JA, Carpenter CL, Laird-Offringa IA. DNA methylation analysis: a powerful new tool for lung cancer diagnosis. Oncogene. 2002;21(35):5450–5461. doi: 10.1038/sj.onc.1205605. [DOI] [PubMed] [Google Scholar]
- 64.Liang R, Li X, Li W, Zhu X, Li C. DNA methylation in lung cancer patients: Opening a "window of life" under precision medicine. Biomed Pharmacother. 2021;144:112202. doi: 10.1016/j.biopha.2021.112202. [DOI] [PubMed] [Google Scholar]
- 65.Ikeda K, Shiraishi K, Eguchi A, Shibata H, Yoshimoto K, Mori T, Baba Y, Baba H, Suzuki M. Long interspersed nucleotide element 1 hypomethylation is associated with poor prognosis of lung adenocarcinoma. Ann Thorac Surg. 2013;96(5):1790–1794. doi: 10.1016/j.athoracsur.2013.06.035. [DOI] [PubMed] [Google Scholar]
- 66.Liu H, Liu W, Wu Y, Zhou Y, Xue R, Luo C, Wang L, Zhao W, Jiang JD, Liu J. Loss of epigenetic control of synuclein-gamma gene as a molecular indicator of metastasis in a wide range of human cancers. Cancer Res. 2005;65(17):7635–7643. doi: 10.1158/0008-5472.Can-05-1089. [DOI] [PubMed] [Google Scholar]
- 67.Shao T, Song P, Hua H, Zhang H, Sun X, Kong Q, Wang J, Luo T, Jiang Y. Gamma synuclein is a novel Twist1 target that promotes TGF-β-induced cancer cell migration and invasion. Cell Death Dis. 2018;9(6):625. doi: 10.1038/s41419-018-0657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smolle E, Pichler M, Non-smoking-associated lung cancer: a distinct entity in terms of tumor biology, patient characteristics and impact of hereditary cancer predisposition. Cancers (Basel) 2019, 11(2). 10.3390/cancers11020204 [DOI] [PMC free article] [PubMed]
- 69.Gao X, Zhang Y, Breitling LP, Brenner H. Tobacco smoking and methylation of genes related to lung cancer development. Oncotarget. 2016;7(37):59017–59028. doi: 10.18632/oncotarget.10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Elgizouli M. Smoking-associated DNA methylation markers predict lung cancer incidence. Clin Epigenetics. 2016;8:127. doi: 10.1186/s13148-016-0292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baglietto L, Ponzi E, Haycock P, Hodge A, Bianca Assumma M, Jung CH, Chung J, Fasanelli F, Guida F, Campanella G, et al. DNA methylation changes measured in pre-diagnostic peripheral blood samples are associated with smoking and lung cancer risk. Int J Cancer. 2017;140(1):50–61. doi: 10.1002/ijc.30431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, Fukuyama Y, Virmani AK, Zochbauer-Muller S, Tsukuda K, Sugio K, et al. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer. 2003;103(2):153–160. doi: 10.1002/ijc.10787. [DOI] [PubMed] [Google Scholar]
- 73.Kim DH, Nelson HH, Wiencke JK, Zheng S, Christiani DC, Wain JC, Mark EJ, Kelsey KT. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 2001;61(8):3419–3424. [PubMed] [Google Scholar]
- 74.Castro M, Grau L, Puerta P, Gimenez L, Venditti J, Quadrelli S. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. J Transl Med. 2010;8:86. doi: 10.1186/1479-5876-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Damiani LA, Yingling CM, Leng S, Romo PE, Nakamura J, Belinsky SA. Carcinogen-induced gene promoter hypermethylation is mediated by DNMT1 and causal for transformation of immortalized bronchial epithelial cells. Cancer Res. 2008;68(21):9005–9014. doi: 10.1158/0008-5472.Can-08-1276. [DOI] [PubMed] [Google Scholar]
- 76.Lin RK, Hsieh YS, Lin P, Hsu HS, Chen CY, Tang YA, Lee CF, Wang YC. The tobacco-specific carcinogen NNK induces DNA methyltransferase 1 accumulation and tumor suppressor gene hypermethylation in mice and lung cancer patients. J Clin Invest. 2010;120(2):521–532. doi: 10.1172/jci40706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.O’Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG islands. Cancer Cell. 2011;20(5):606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Widschwendter M, Jones A, Evans I, Reisel D, Dillner J, et al. Epigenome-based cancer risk prediction: rationale, opportunities and challenges. Nat Rev Clin Oncol. 2018;15(5):292–309. doi: 10.1038/nrclinonc.2018.30. [DOI] [PubMed] [Google Scholar]
- 79.Ma Y, Bai Y, Mao H, Hong Q, Yang D, Zhang H, Liu F, Wu Z, Jin Q, Zhou H, et al. A panel of promoter methylation markers for invasive and noninvasive early detection of NSCLC using a quantum dots-based FRET approach. Biosens Bioelectron. 2016;85:641–648. doi: 10.1016/j.bios.2016.05.067. [DOI] [PubMed] [Google Scholar]
- 80.Ren M, Wang C, Sheng D, Shi Y, Jin M, Xu S. Methylation analysis of SHOX2 and RASSF1A in bronchoalveolar lavage fluid for early lung cancer diagnosis. Ann Diagn Pathol. 2017;27:57–61. doi: 10.1016/j.anndiagpath.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 81.Ponomaryova AA, Rykova EY, Cherdyntseva NV, Skvortsova TE, Dobrodeev AY. Zav’yalov AA, Bryzgalov LO, Tuzikov SA, Vlassov VV, Laktionov PP, Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer. 2013;81(3):397–403. doi: 10.1016/j.lungcan.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 82.Kneip C, Schmidt B, Seegebarth A, Weickmann S, Fleischhacker M, Liebenberg V, Field JK, Dietrich D. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J Thorac Oncol. 2011;6(10):1632–1638. doi: 10.1097/JTO.0b013e318220ef9a. [DOI] [PubMed] [Google Scholar]
- 83.Schmidt B, Liebenberg V, Dietrich D, Schlegel T, Kneip C, Seegebarth A, Flemming N, Seemann S, Distler J, Lewin J, et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer. 2010;10:600. doi: 10.1186/1471-2407-10-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weiss G, Schlegel A, Kottwitz D. Validation of the SHOX2/PTGER4 DNA methylation marker panel for plasma-based discrimination between patients with malignant and nonmalignant lung disease. J Thorac Oncol. 2017;12(1):77–84. doi: 10.1186/1471-2407-10-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang C, Yu W, Wang L, Zhao M, Guo Q, Lv S, Hu X, Lou J. DNA methylation analysis of the SHOX2 and RASSF1A panel in bronchoalveolar lavage fluid for lung cancer diagnosis. J Cancer. 2017;8(17):3585–3591. doi: 10.7150/jca.21368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nunes SP, Diniz F, Moreira-Barbosa C, Constâncio V et al. Subtyping lung cancer using DNA methylation in liquid biopsies. J Clin Med 2019, 8 (9). 10.3390/jcm8091500 [DOI] [PMC free article] [PubMed]
- 87.Grote HJ, Schmiemann V, Geddert H, Rohr UP, Kappes R, Gabbert HE. Aberrant promoter methylation of p16(INK4a), RARB2 and SEMA3B in bronchial aspirates from patients with suspected lung cancer. Int J Cancer. 2005;116(5):720–725. doi: 10.1002/ijc.21090. [DOI] [PubMed] [Google Scholar]
- 88.Zhao QT, Guo T, Wang HE, Zhang XP, Zhang H, Wang ZK, Yuan Z, Duan GC. Diagnostic value of SHOX2 DNA methylation in lung cancer: a meta-analysis. Onco Targets Ther. 2015;8:3433–3439. doi: 10.2147/ott.S94300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu WJ, Tan XH, Guo BP, Ke Q, Sun J, Cen H. Associations between RASSF1A promoter methylation and NSCLC: a meta-analysis of published data. Asian Pac J Cancer Prev. 2013;14(6):3719–3724. doi: 10.7314/apjcp.2013.14.6.3719. [DOI] [PubMed] [Google Scholar]
- 90.Walter RFH, Rozynek P, Casjens S, Werner R, Mairinger FD, Speel EJM, Zur Hausen A, Meier S, Wohlschlaeger J, Theegarten D, et al. Methylation of L1RE1, RARB, and RASSF1 function as possible biomarkers for the differential diagnosis of lung cancer. PLoS ONE. 2018;13(5):e0195716. doi: 10.1371/journal.pone.0195716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ma Y, Li MD. Establishment of a strong link between smoking and cancer pathogenesis through DNA methylation analysis. Sci Rep. 2017;7(1):1811. doi: 10.1038/s41598-017-01856-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Topper MJ, Vaz M, Chiappinelli KB, DeStefano Shields CE, Niknafs N, Yen RC, Wenzel A, Hicks J, Ballew M, Stone M, et al. Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell. 2017;171(6):1284–1300.e1221. doi: 10.1016/j.cell.2017.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377(25):2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cai L, Bai H, Duan J, Wang Z, Gao S, Wang D, Wang S, Jiang J, Han J, Tian Y, et al. Epigenetic alterations are associated with tumor mutation burden in non-small cell lung cancer. J Immunother Cancer. 2019;7(1):198. doi: 10.1186/s40425-019-0660-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang W, Wang Q, Wan D, Sun Y, Wang L, Chen H, Liu C, Petersen RB, Li J, Xue W, et al. Histone HIST1H1C/H1.2 regulates autophagy in the development of diabetic retinopathy. Autophagy. 2017;13(5):941–954. doi: 10.1080/15548627.2017.1293768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cheng X, Structural and functional coordination of DNA and histone methylation. Cold Spring Harb Perspect Biol 2014, 6(8). 10.1101/cshperspect.a018747 [DOI] [PMC free article] [PubMed]
- 97.McGrath J, Trojer P. Targeting histone lysine methylation in cancer. Pharmacol Ther. 2015;150:1–22. doi: 10.1016/j.pharmthera.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 98.Damaskos C, Tomos I, Garmpis N, Karakatsani A, Dimitroulis D, Garmpi A, Spartalis E, Kampolis CF, Tsagkari E, Loukeri AA, et al. Histone deacetylase inhibitors as a novel targeted therapy against non-small cell lung cancer where are we now and what should we expect? Anticancer Res. 2018;38(1):37–43. doi: 10.21873/anticanres.12189. [DOI] [PubMed] [Google Scholar]
- 99.Audia JE, Campbell RM. Histone modifications and cancer. Cold Spring Harb Perspect Biol. 2016;8(4):a019521. doi: 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Petta V, Gkiozos I, Strimpakos A, Syrigos K. Histones and lung cancer: are the histone deacetylases a promising therapeutic target? Cancer Chemother Pharmacol. 2013;72(5):935–952. doi: 10.1007/s00280-013-2223-9. [DOI] [PubMed] [Google Scholar]
- 101.Li Z, Zhu WG. Targeting histone deacetylases for cancer therapy: from molecular mechanisms to clinical implications. Int J Biol Sci. 2014;10(7):757–770. doi: 10.7150/ijbs.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26(37):5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 103.Van Den Broeck A, Brambilla E, Moro-Sibilot D, Lantuejoul S, Brambilla C, Eymin B, Gazzeri S. Loss of histone H4K20 trimethylation occurs in preneoplasia and influences prognosis of non-small cell lung cancer. Clin Cancer Res. 2008;14(22):7237–7245. doi: 10.1158/1078-0432.Ccr-08-0869. [DOI] [PubMed] [Google Scholar]
- 104.Bartling B, Hofmann HS, Boettger T, Hansen G, Burdach S, Silber RE, Simm A. Comparative application of antibody and gene array for expression profiling in human squamous cell lung carcinoma. Lung Cancer. 2005;49(2):145–154. doi: 10.1016/j.lungcan.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 105.Osada H, Tatematsu Y, Saito H, Yatabe Y, Mitsudomi T, Takahashi T. Reduced expression of class II histone deacetylase genes is associated with poor prognosis in lung cancer patients. Int J Cancer. 2004;112(1):26–32. doi: 10.1002/ijc.20395. [DOI] [PubMed] [Google Scholar]
- 106.Minamiya Y, Ono T, Saito H, Takahashi N, Ito M, Motoyama S, Ogawa J. Strong expression of HDAC3 correlates with a poor prognosis in patients with adenocarcinoma of the lung. Tumour Biol. 2010;31(5):533–539. doi: 10.1007/s13277-010-0066-0. [DOI] [PubMed] [Google Scholar]
- 107.Riley JS, Hutchinson R, McArt DG, Crawford N, Holohan C, Paul I, Van Schaeybroeck S, Salto-Tellez M, Johnston PG, Fennell DA, et al. Prognostic and therapeutic relevance of FLIP and procaspase-8 overexpression in non-small cell lung cancer. Cell Death Dis. 2013;4(12):e951. doi: 10.1038/cddis.2013.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schrump DS, Fischette MR, Nguyen DM, Zhao M, Li X, Kunst TF, Hancox A, Hong JA, Chen GA, Kruchin E, et al. Clinical and molecular responses in lung cancer patients receiving Romidepsin. Clin Cancer Res. 2008;14(1):188–198. doi: 10.1158/1078-0432.Ccr-07-0135. [DOI] [PubMed] [Google Scholar]
- 109.Traynor AM, Dubey S, Eickhoff JC, Kolesar JM, Schell K, Huie MS, Groteluschen DL, Marcotte SM, Hallahan CM, Weeks HR, et al. Vorinostat (NSC# 701852) in patients with relapsed non-small cell lung cancer: a Wisconsin Oncology Network phase II study. J Thorac Oncol. 2009;4(4):522–526. doi: 10.1097/jto.0b013e3181952478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Reid T, Valone F, Lipera W, Irwin D, Paroly W, Natale R, Sreedharan S, Keer H, Lum B, Scappaticci F, et al. Phase II trial of the histone deacetylase inhibitor pivaloyloxymethyl butyrate (Pivanex, AN-9) in advanced non-small cell lung cancer. Lung Cancer. 2004;45(3):381–386. doi: 10.1016/j.lungcan.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 111.Ryan QC, Headlee D, Acharya M, Sparreboom A, Trepel JB, Ye J, Figg WD, Hwang K, Chung EJ, Murgo A, et al. Phase I and pharmacokinetic study of MS-275, a histone deacetylase inhibitor, in patients with advanced and refractory solid tumors or lymphoma. J Clin Oncol. 2005;23(17):3912–3922. doi: 10.1200/jco.2005.02.188. [DOI] [PubMed] [Google Scholar]
- 112.Prakash S, Foster BJ, Meyer M, Wozniak A, Heilbrun LK, Flaherty L, Zalupski M, Radulovic L, Valdivieso M, LoRusso PM. Chronic oral administration of CI-994: a phase 1 study. Invest New Drugs. 2001;19(1):1–11. doi: 10.1023/a:1006489328324. [DOI] [PubMed] [Google Scholar]
- 113.Ramalingam SS, Maitland ML, Frankel P, Argiris AE, Koczywas M, Gitlitz B, Thomas S, Espinoza-Delgado I, Vokes EE, Gandara DR, et al. Carboplatin and paclitaxel in combination with either vorinostat or placebo for first-line therapy of advanced non-small-cell lung cancer. J Clin Oncol. 2010;28(1):56–62. doi: 10.1200/jco.2009.24.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tarhini AA, Zahoor H, McLaughlin B, Gooding WE, Schmitz JC, Siegfried JM, Socinski MA, Argiris A. Phase I trial of carboplatin and etoposide in combination with panobinostat in patients with lung cancer. Anticancer Res. 2013;33(10):4475–4481. [PMC free article] [PubMed] [Google Scholar]
- 115.Ramalingam SS, Parise RA, Ramanathan RK, Lagattuta TF, Musguire LA, Stoller RG, Potter DM, Argiris AE, Zwiebel JA, Egorin MJ, et al. Phase I and pharmacokinetic study of vorinostat, a histone deacetylase inhibitor, in combination with carboplatin and paclitaxel for advanced solid malignancies. Clin Cancer Res. 2007;13(12):3605–3610. doi: 10.1158/1078-0432.Ccr-07-0162. [DOI] [PubMed] [Google Scholar]
- 116.Richards DA, Boehm KA, Waterhouse DM, Wagener DJ, Krishnamurthi SS, Rosemurgy A, Grove W, Macdonald K, Gulyas S, Clark M, et al. Gemcitabine plus CI-994 offers no advantage over gemcitabine alone in the treatment of patients with advanced pancreatic cancer: results of a phase II randomized, double-blind, placebo-controlled, multicenter study. Ann Oncol. 2006;17(7):1096–1102. doi: 10.1093/annonc/mdl081. [DOI] [PubMed] [Google Scholar]
- 117.Pauer LR, Olivares J, Cunningham C, Williams A, Grove W, Kraker A, Olson S, Nemunaitis J. Phase I study of oral CI-994 in combination with carboplatin and paclitaxel in the treatment of patients with advanced solid tumors. Cancer Invest. 2004;22(6):886–896. doi: 10.1081/cnv-200039852. [DOI] [PubMed] [Google Scholar]
- 118.Chu BF, Karpenko MJ, Liu Z, Aimiuwu J, Villalona-Calero MA, Chan KK, Grever MR, Otterson GA. Phase I study of 5-aza-2′-deoxycytidine in combination with valproic acid in non-small-cell lung cancer. Cancer Chemother Pharmacol. 2013;71(1):115–121. doi: 10.1007/s00280-012-1986-8. [DOI] [PubMed] [Google Scholar]
- 119.Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, Sebree R, Rodgers K, Hooker CM, Franco N, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1(7):598–607. doi: 10.1158/2159-8290.Cd-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lin J, Gilbert J, Rudek MA, Zwiebel JA, Gore S, Jiemjit A, Zhao M, Baker SD, Ambinder RF, Herman JG, et al. A phase I dose-finding study of 5-azacytidine in combination with sodium phenylbutyrate in patients with refractory solid tumors. Clin Cancer Res. 2009;15(19):6241–6249. doi: 10.1158/1078-0432.Ccr-09-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Candelaria M. Gallardo-Rincón A phase II study of epigenetic therapy with hydralazine and magnesium valproate to overcome chemotherapy resistance in refractory solid tumors. Ann Oncol. 2007;18(9):1529–1538. doi: 10.1093/annonc/mdm204. [DOI] [PubMed] [Google Scholar]
- 122.Takeuchi S, Hase T, Shimizu S, Ando M, Hata A, Murakami H, Kawakami T, Nagase K, Yoshimura K, Fujiwara T, et al. Phase I study of vorinostat with gefitinib in BIM deletion polymorphism/epidermal growth factor receptor mutation double-positive lung cancer. Cancer Sci. 2020;111(2):561–570. doi: 10.1111/cas.14260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Witta SE, Jotte RM, Konduri K, Neubauer MA, Spira AI, Ruxer RL, Varella-Garcia M, Bunn PA, Jr, Hirsch FR. Randomized phase II trial of erlotinib with and without entinostat in patients with advanced non-small-cell lung cancer who progressed on prior chemotherapy. J Clin Oncol. 2012;30(18):2248–2255. doi: 10.1200/jco.2011.38.9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dasari A, Gore L, Messersmith WA, Diab S, Jimeno A, Weekes CD, Lewis KD, Drabkin HA, Flaig TW, Camidge DR. A phase I study of sorafenib and vorinostat in patients with advanced solid tumors with expanded cohorts in renal cell carcinoma and non-small cell lung cancer. Invest New Drugs. 2013;31(1):115–125. doi: 10.1007/s10637-012-9812-z. [DOI] [PubMed] [Google Scholar]
- 125.Reguart N, Rosell R, Cardenal F, Cardona AF, Isla D, Palmero R, Moran T, Rolfo C, Pallarexpanded coho et al. Phase I/II trial of vorinostat (SAHA) and erlotinib for non-small cell lung cancer (NSCLC) patients with epidermal growth factor receptor (EGFR) mutations after erlotinib progression. Lung Cancer. 2014;84(2):161–167. doi: 10.1016/j.lungcan.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 126.Gray JE, Haura E, Chiappori A, Tanvetyanon T, Williams CC, Pinder-Schenck M, Kish JA, Kreahling J, Lush R, Neuger A, et al. A phase I, pharmacokinetic, and pharmacodynamic study of panobinostat, an HDAC inhibitor, combined with erlotinib in patients with advanced aerodigestive tract tumors. Clin Cancer Res. 2014;20(6):1644–1655. doi: 10.1158/1078-0432.Ccr-13-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Choi CYH, Wakelee HA, Neal JW, Pinder-Schenck MC, Yu HM, Chang SD, Adler JR, Modlin LA, Harsh GR, Soltys SG. Vorinostat and concurrent stereotactic radiosurgery for non-small cell lung cancer brain metastases: a phase 1 dose escalation trial. Int J Radiat Oncol Biol Phys. 2017;99(1):16–21. doi: 10.1016/j.ijrobp.2017.04.041. [DOI] [PubMed] [Google Scholar]
- 128.Gray JE, Saltos A, Tanvetyanon T, Haura EB, Creelan B, Antonia SJ, Shafique M, Zheng H, Dai W, Saller JJ, et al. Phase I/Ib study of pembrolizumab plus vorinostat in advanced/metastatic non-small cell lung cancer. Clin Cancer Res. 2019;25(22):6623–6632. doi: 10.1158/1078-0432.Ccr-19-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zuco V, De Cesare M, Cincinelli R, Nannei R, Pisano C, Zaffaroni N, Zunino F. Synergistic antitumor effects of novel HDAC inhibitors and paclitaxel in vitro and in vivo. PLoS ONE. 2011;6(12):e29085. doi: 10.1371/journal.pone.0029085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Groh T, Hrabeta J, Khalil MA, Doktorova H, Eckschlager T, Stiborova M. The synergistic effects of DNA-damaging drugs cisplatin and etoposide with a histone deacetylase inhibitor valproate in high-risk neuroblastoma cells. Int J Oncol. 2015;47(1):343–352. doi: 10.3892/ijo.2015.2996. [DOI] [PubMed] [Google Scholar]
- 131.Wang L, Li H, Ren Y, Zou S, Fang W, Jiang X, Jia L, Li M, Liu X, Yuan X, et al. Targeting HDAC with a novel inhibitor effectively reverses paclitaxel resistance in non-small cell lung cancer via multiple mechanisms. Cell Death Dis. 2016;7(1):e2063. doi: 10.1038/cddis.2015.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Beg AA, Gray JE. HDAC inhibitors with PD-1 blockade: a promising strategy for treatment of multiple cancer types? Epigenomics. 2016;8(8):1015–1017. doi: 10.2217/epi-2016-0066. [DOI] [PubMed] [Google Scholar]
- 133.Banik D, Moufarrij S, Villagra A, Immunoepigenetics combination therapies: an overview of the role of HDACs in cancer immunotherapy. Int J Mol Sci 2019, 20(9). 10.3390/ijms20092241 [DOI] [PMC free article] [PubMed]
- 134.Wrangle J, Wang W, Koch A, Easwaran H, Mohammad HP, Vendetti F, Vancriekinge W, Demeyer T, Du Z, Parsana P, et al. Alterations of immune response of non-small cell lung cancer with azacytidine. Oncotarget. 2013;4(11):2067–2079. doi: 10.18632/oncotarget.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mehndiratta S, Lin MH, Wu YW, Chen CH, Wu TY, Chuang KH, Chao MW, Chen YY, Pan SL, Chen MC, et al. N-alkyl-hydroxybenzoyl anilide hydroxamates as dual inhibitors of HDAC and HSP90, downregulating IFN-γ induced PD-L1 expression. Eur J Med Chem. 2020;185:111725. doi: 10.1016/j.ejmech.2019.111725. [DOI] [PubMed] [Google Scholar]
- 136.Samuni Y, Wink DA, Krishna MC, Mitchell JB, Goldstein S. Suberoylanilide hydroxamic acid radiosensitizes tumor hypoxic cells in vitro through the oxidation of nitroxyl to nitric oxide. Free Radic Biol Med. 2014;73:291–298. doi: 10.1016/j.freeradbiomed.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17(9):1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Moore KE, Gozani O. An unexpected journey, lysine methylation across the proteome. Biochim Biophys Acta. 2014;1839(12):1395–1403. doi: 10.1016/j.bbagrm.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Carlson SM, Moore KE, Green EM. Martation across the Proteome-wide enrichment of proteins modified by lysine methylation. Nat Protoc. 2014;9(1):37–50. doi: 10.1038/nprot.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25(13):1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood. 2011;117(25):6912–6922. doi: 10.1182/blood-2011-02-334359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xu C, Hao K, Hu H, Sheng Z, Yan J, Wang Q, Yu L. Expression of the enhancer of zeste homolog 2 in biopsy specimen predicts chemoresistance and survival in advanced non-small cell lung cancer receiving first-line platinum-based chemotherapy. Lung Cancer. 2014;86(2):268–273. doi: 10.1016/j.lungcan.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 143.Wan D, Liu C, Sun Y, Wang W, Huang K, Zheng L. MacroH2A1.1 cooperates with EZH2 to promote adipogenesis by regulating Wnt signaling. J Mol Cell Biol. 2017;9(4):325–337. doi: 10.1093/jmcb/mjx027. [DOI] [PubMed] [Google Scholar]
- 144.Denissov S, Hofemeister H, Marks H, Kranz A, Ciotta G, Singh S, Anastassiadis K, Stunnenberg HG, Stewart AF. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development. 2014;141(3):526–537. doi: 10.1242/dev.102681. [DOI] [PubMed] [Google Scholar]
- 145.Yin S, Yang J, Lin B, Deng W, Zhang Y, Yi X, Shi Y, Tao Y, Cai J, Wu CI, et al. Exome sequencing identifies frequent mutation of MLL2 in non-small cell lung carcinoma from Chinese patients. Sci Rep. 2014;4:6036. doi: 10.1038/srep06036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xue W, Huang J, Chen H, Zhang Y, Zhu X, Li J, Zhang W, Yuan Y, Wang Y, Zheng L, et al. Histone methyltransferase G9a modulates hepatic insulin signaling via regulating HMGA1. Biochim Biophys Acta Mol Basis Dis. 2018;1864(2):338–346. doi: 10.1016/j.bbadis.2017.10.037. [DOI] [PubMed] [Google Scholar]
- 147.Huang T, Zhang P, Li W, Zhao T, Zhang Z, Chen S, Yang Y, Feng Y, Li F, Shirley Liu X, et al. G9A promotes tumor cell growth and invasion by silencing CASP1 in non-small-cell lung cancer cells. Cell Death Dis. 2017;8(4):e2726. doi: 10.1038/cddis.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang R, Deng X, Yoshioka Y, Vougiouklakis T, Park JH, Suzuki T, Dohmae N, Ueda K, Hamamoto R, Nakamura Y. Effects of SMYD2-mediated EML4-ALK methylation on the signaling pathway and growth in non-small-cell lung cancer cells. Cancer Sci. 2017;108(6):1203–1209. doi: 10.1111/cas.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hao C, Wang L, Peng S, Cao M, Li H, Hu J, Huang X, Liu W, Zhang H, Wu S, et al. Gene mutations in primary tumors and corresponding patient-derived xenografts derived from non-small cell lung cancer. Cancer Lett. 2015;357(1):179–185. doi: 10.1016/j.canlet.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Walter DM, Venancio OS, Buza EL, Tobias JW, Deshpande C, Gudiel AA, Kim-Kiselak C, Cicchini M, Yates TJ, Feldser DM. Systematic in vivo inactivation of chromatin-regulating enzymes identifies Setd2 as a potent tumor suppressor in lung adenocarcinoma. Cancer Res. 2017;77(7):1719–1729. doi: 10.1158/0008-5472.Can-16-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Takawa M, Cho HS, Hayami S, Toyokawa G, Kogure M, Yamane Y, Iwai Y, Maejima K, Ueda K, Masuda A, et al. Histone lysine methyltransferase SETD8 promotes carcinogenesis by deregulating PCNA expression. Cancer Res. 2012;72(13):3217–3227. doi: 10.1158/0008-5472.Can-11-3701. [DOI] [PubMed] [Google Scholar]
- 152.Chen T, Ren H, Thakur A, Yang T, Li Y, Zhang S, Wang T, Chen M. miR-382 inhibits tumor progression by targeting SETD8 in non-small cell lung cancer. Biomed Pharmacother. 2017;86:248–253. doi: 10.1016/j.biopha.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 153.Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X, Song Y. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS ONE. 2012;7(4):e35065. doi: 10.1371/journal.pone.0035065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.He R, Zhang FH, Shen N. LncRNA FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through suppressing E-cadherin and regulating WNT pathway in non-small cell lung cancer (NSCLC) Biomed Pharmacother. 2017;95:331–338. doi: 10.1016/j.biopha.2017.08.057. [DOI] [PubMed] [Google Scholar]
- 155.Kong L, Zhang P, Li W, Yang Y, Tian Y, Wang X, Chen S, Yang Y, Huang T, Zhao T, et al. KDM1A promotes tumor cell invasion by silencing TIMP3 in non-small cell lung cancer cells. Oncotarget. 2016;7(19):27959–27974. doi: 10.18632/oncotarget.8563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 157.Wagner KW, Alam H, Dhar SS, Giri U, Li N, Wei Y, Giri D, Cascone T, Kim JH, Ye Y, et al. KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin Invest. 2013;123(12):5231–5246. doi: 10.1172/jci68642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dhar SS, Alam H, Li N, Wagner KW, Chung J, Ahn YW, Lee MG. Transcriptional repression of histone deacetylase 3 by the histone demethylase KDM2A is coupled to tumorigenicity of lung cancer cells. J Biol Chem. 2014;289(11):7483–7496. doi: 10.1074/jbc.M113.521625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cho HS, Toyokawa G, Daigo Y, Hayami S, Masuda K, Ikawa N, Yamane Y, Maejima K, Tsunoda T, Field HI, et al. The JmjC domain-containing histone demethylase KDM3A is a positive regulator of the G1/S transition in cancer cells via transcriptional regulation of the HOXA1 gene. Int J Cancer. 2012;131(3):E179–189. doi: 10.1002/ijc.26501. [DOI] [PubMed] [Google Scholar]
- 160.Zhan M, Wen F, Liu L, Chen Z, Wei H, Zhou H. JMJD1A promotes tumorigenesis and forms a feedback loop with EZH2/let-7c in NSCLC cells. Tumour Biol. 2016;37(8):11237–11247. doi: 10.1007/s13277-016-4999-9. [DOI] [PubMed] [Google Scholar]
- 161.Wu Q, Tian Y, Zhang J, Tong X, Huang H, Li S, Zhao H, Tang Y, Yuan C, Wang K, et al. In vivo CRISPR screening unveils histone demethylase UTX as an important epigenetic regulator in lung tumorigenesis. Proc Natl Acad Sci U S A. 2018;115(17):E3978–e3986. doi: 10.1073/pnas.1716589115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Iderzorig T, Kellen J, Osude C, Singh S, Woodman JA, Garcia C, Puri N. Comparison of EMT mediated tyrosine kinase inhibitor resistance in NSCLC. Biochem Biophys Res Commun. 2018;496(2):770–777. doi: 10.1016/j.bbrc.2018.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang J, Ni SS, Zhao WL, Dong XC, Wang JL. High expression of JMJD6 predicts unfavorable survival in lung adenocarcinoma. Tumour Biol. 2013;34(4):2397–2401. doi: 10.1007/s13277-013-0789-9. [DOI] [PubMed] [Google Scholar]
- 164.Duan Q, Pang C, Chang N, Zhang J, Liu W. Overexpression of PAD4 suppresses drug resistance of NSCLC cell lines to gefitinib through inhibiting Elk1-mediated epithelial-mesenchymal transition. Oncol Rep. 2016;36(1):551–558. doi: 10.3892/or.2016.4780. [DOI] [PubMed] [Google Scholar]
- 165.Kikuchi J, Takashina T, Kinoshita I, Kikuchi E, Shimizu Y, Sakakibara-Konishi J, Oizumi S, Marquez VE, Nishimura M, Dosaka-Akita H. Epigenetic therapy with 3-deazaneplanocin A, an inhibitor of the histone methyltransferase EZH2, inhibits growth of non-small cell lung cancer cells. Lung Cancer. 2012;78(2):138–143. doi: 10.1016/j.lungcan.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fillmore CM, Xu C, Desai PT, Berry JM, Rowbotham SP, Lin YJ, Zhang H, Marquez VE, Hammerman PS, Wong KK, et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015;520(7546):239–242. doi: 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim W, Kim R, Park G, Park JW, Kim JE. Deficiency of H3K79 histone methyltransferase Dot1-like protein (DOT1L) inhibits cell proliferation. J Biol Chem. 2012;287(8):5588–5599. doi: 10.1074/jbc.M111.328138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21(9):1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, Diaz E, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 170.Gulati N, Béguelin W, Giulino-Roth L. Enhancer of zeste homolog 2 (EZH2) inhibitors. Leuk Lymphoma. 2018;59(7):1574–1585. doi: 10.1080/10428194.2018.1430795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vaswani RG, Gehling VS, Dakin LA, Cook AS, Nasveschuk CG, Duplessis M, Iyer P, Balasubramanian S, Zhao F, Good AC, et al. Identification of (R)-N-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a potent and selective inhibitor of histone methyltransferase EZH2, suitable for phase I clinical trials for B-cell lymphomas. J Med Chem. 2016;59(21):9928–9941. doi: 10.1021/acs.jmedchem.6b01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Stewart CA, Byers LA. Altering the course of small cell lung cancer: targeting cancer stem cells via LSD1 inhibition. Cancer Cell. 2015;28(1):4–6. doi: 10.1016/j.ccell.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhan X, Li J, Guo Y, Golubnitschaja O. Mass spectrometry analysis of human tear fluid biomarkers specific for ocular and systemic diseases in the context of 3P medicine. EPMA J. 2021;12(4):449–475. doi: 10.1007/s13167-021-00265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li N, Desiderio DM, Zhan X. The use of mass spectrometry in a proteome-centered multiomics study of human pituitary adenomas. Mass Spectrom Rev. 2022;41(6):964–1013. doi: 10.1002/mas.21710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and materials are available in the current manuscript.