Abstract
Acne vulgaris is a common inflammatory disorder affecting more than 80% of young adolescents. Cutibacterium acnes plays a role in the pathogenesis of acne lesions, although the mechanisms are poorly understood. The study aimed to explore the microbiome at different skin sites in adolescent acne and the role of biofilm production in promoting the growth and persistence of C. acnes isolates. Microbiota analysis showed a significantly lower alpha diversity in inflammatory lesions (LA) than in non-inflammatory (NI) lesions of acne patients and healthy subjects (HS). Differences at the species level were driven by the overabundance of C. acnes on LA than NI and HS. The phylotype IA1 was more represented in the skin of acne patients than in HS. Genes involved in lipids transport and metabolism, as well as potential virulence factors associated with host-tissue colonization, were detected in all IA1 strains independently from the site of isolation. Additionally, the IA1 isolates were more efficient in early adhesion and biomass production than other phylotypes showing a significant increase in antibiotic tolerance. Overall, our data indicate that the site-specific dysbiosis in LA and colonization by virulent and highly tolerant C. acnes phylotypes may contribute to acne development in a part of the population, despite the universal carriage of the microorganism. Moreover, new antimicrobial agents, specifically targeting biofilm-forming C. acnes, may represent potential treatments to modulate the skin microbiota in acne.
Subject terms: Microbiology, Antimicrobials, Biofilms, Clinical microbiology, Microbial communities
Introduction
Acne vulgaris is a chronic inflammatory skin disorder of the pilosebaceous unit, affecting 67–95% of adolescents worldwide1–3. In adults, its prevalence and incidence have grown, especially among women4–6. The psychological impact of acne is substantial, causing a detrimental impact on life quality7,8. Acne is characterized by increased sebum production, leading to non-inflammatory (NI) comedones and inflammatory lesions (LA) such as papules, pustules, or nodules, mainly on the face and on the back and chest9. The etiology is associated with changes in sebum production under androgen control, an altered keratinization process, and an increased release of inflammatory mediators10–13. Dysbiosis and follicular colonization by Cutibacterium acnes have been linked to the pathophysiology of inflammatory acne14,15. However, the role of C. acne remains unclear due to its ubiquitous distribution both in the sebaceous areas of healthy skin and inflammatory acne lesions and its highly variable density in the skin of different individuals15. Although generally regarded as a low-virulence bacterium and a human skin commensal, C. acnes can be considered an opportunistic pathogen associated with invasive skin and soft tissue infections and implant-associated infections16–18. Indeed, C. acnes, by producing multiple virulence factors such as lipases, proteases, and the Christine Atkins Munch-Petersen (CAMP) factors, can trigger inflammation and host tissue damage19–24. Besides, C. acnes proliferation in the pilosebaceous unit can induce the upregulation of different proinflammatory cytokines by keratinocytes, sebocytes, or peripheral blood mononuclear cells25–28. C. acnes strains are categorized into the six phylotypes IA1, IA2, IB, IC, II, and III, correlated with a variable body distribution, clinical conditions, antimicrobial susceptibility profile, and inflammatory properties29–32. C. acnes IA1 is the predominant phylotype isolated from moderate to severe acne. In contrast, IB, II, and III phylotypes are more commonly isolated from soft tissues and medical implant infections33–35. Notably, IA1 isolates exhibit a more virulent profile, including a greater production of β-defensin 2 from cultured keratinocytes and higher lipase activity levels than other phylotypes associated with healthy skin36,37. These observations suggest that the apparent loss of phylotype diversity in acne patients and the dominance of IA1 virulent strains may reflect a dysbiotic shift within a follicle linked to microenvironmental changes38. In particular, biofilm production has been linked to specific C. acnes phylotypes, thus suggesting a possible association with the chronic colonization of the pilosebaceous unit observed in acne patients39–42. Previous works have demonstrated that C. acnes form biofilms in the follicles of acne patients43, suggesting that this condition may lead to a homeostatic imbalance of the microbiota15. Indeed, the chronic bacterial persistence and relapse following antibiotic therapy are strongly indicative of biofilm-related colonization44. Although biofilm is considered a primary factor that ensures C. acnes persistence during acne antibiotic treatment 45, factors that promote early bacteria adhesion and biofilm formation have not been identified yet46. This study analyzed the process of C. acnes colonization and its relation to the skin microbiota in patients with severe acne from NI and LA and healthy subjects (HS). Besides, using a metagenomic approach, we characterized the genetic background and the phylotype-dependent biofilm production of C. acnes isolates with regard to the dynamics of early adherence, the overall amount of biofilm biomass, and morphology. Finally, we also investigated the contribution of biofilm to antibiotic tolerance of C. acnes isolates.
Results
Demographic and baseline data
A total of ten patients affected by severe acne vulgaris, and ten healthy control subjects (HS), with no history of acne were included in the study. The two groups were matched for age and sex. The demographic and clinical characteristics are summarized in Table 1.
Table 1.
Demographic and clinical characteristics of acne patients at inclusion.
| Baseline demographic and clinical characteristics of acne patients | N = 10 |
| Sex, Female/Male | 6/4 |
| Age (years), Mean ± SD; Median [Min–Max] | 14.1 ± 2; 14 [11–18] |
| Dermatological examination | No signs apart from acne |
|
Facial acne severity with the GEA scale Grade 4–5 (Severe) |
10 |
Skin surface microbiota of healthy subjects and acne patients
Analysis of 2,275,010 reads (min. number of reads: 43,111; max. number of reads: 187,496) grouped into 2037 taxa. Specifically, 2,139,830 reads grouped into 916 taxa were collected after filtering out singletons and taxa with a prevalence lower than 5%. After rarefaction at a sample depth of 40,000 reads, 880 taxa were recovered, and alpha and beta diversity were evaluated. Two samples were excluded from the 10 healthy controls due to low quality and library size.
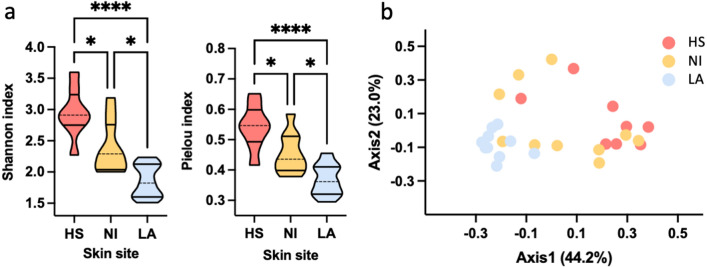
Alpha diversity of the skin of ten healthy subjects (HS) and non-inflammatory (NI) and inflammatory lesions (LA) of ten acne patients was evaluated using the Shannon diversity index and Pielou evenness index. Interestingly, a significant (P = 0.0002) decrease in Shannon index was observed in both NI (Mean ± SD = 2.39 ± 0.41) and LA (Mean ± SD = 1.86 ± 0.26) compared to HS (Mean ± SD = 2.95 ± 0.39), (Fig. 1a). Similarly, a significant (P = 0.0004) decrease in Pielou index was reported in NI (Mean ± SD = 0.45 ± 0.07) and LA (Mean ± SD, 0.37 ± 0.05) samples than in HS (Mean ± SD, 0.54 ± 0.07), (Fig. 1a). Bray Curtis beta diversity, represented as principal coordinate analysis (PCoA), corroborated previous findings showing a distinctive spatial clusterization of HS samples and subjects with LA (PERMANOVA test: P = 0.001, R2 = 0.3522) (Fig. 1b).
Figure 1.
Loss of diversity characterizes inflammatory acne. Skin microbiota was evaluated on samples collected from the skin of ten healthy subjects (HS) and non-inflammatory (NI), and inflammatory lesions (LA) of ten acne patients. (a) Alpha diversity was calculated using the Shannon diversity index (HS vs. NI, P = 0.0209; HS vs. LA, P < 0.0001; LA vs. NI, P = 0.0118) and Pielou Evenness index (HS vs. NI, P = 0.0241; HS vs. LA, P < 0.0001; LA vs. NI, P = 0.0136). Statistical differences were determined using the Kruskal–Wallis test. (b) Bray Curtis beta diversity was calculated at the genus level and represented as principal coordinate analysis (PCoA). PERMANOVA test was used to assess significance. *, P < 0.05; **, P < 0.01; ***, P < 0.001, ****, P < 0.0001.
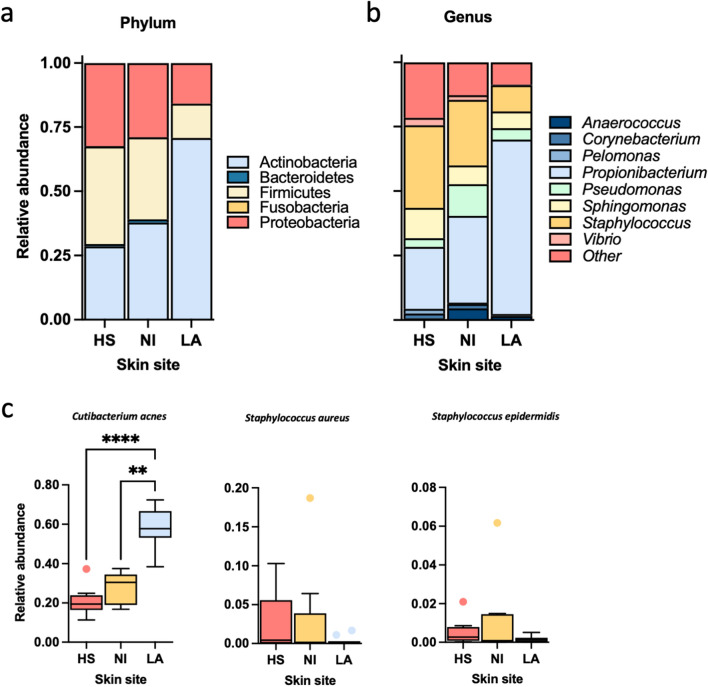
Important site-dependent differences were identified in bacterial abundance at phylum and genus level (Fig. 2). The relative abundance of Actinobacteria increased in LA specimens compared to what was observed for Firmicutes and Proteobacteria (Fig. 2a). Accordingly, the relative abundance of Actinobacteria accounted for 30% of the microbial community in HS, 38% in NI, and increased to 71%in LA (P < 0.001). Differently, Firmicutes and Proteobacteria appeared more abundant in HS (38% and 32%) and NI (32% and 29%) compared to LA (13%, 5%).
Figure 2.
Microbiota variation between healthy subjects and acne patients. Skin microbiota was evaluated on samples collected from the skin of ten healthy subjects (HS) and non-inflammatory (NI) and inflammatory lesions (AL) of 10 acne patients. Relative abundances at the phylum level (a) and top eight genera (b) were represented in a stacked bar plot. (c). Relative abundance was evaluated at the species level for Cutibacterium acnes, Staphylococcus aureus, and Staphylococcus epidermidis relative abundances. Significance was assessed by using the Kruskal Wallis static test. *, P < 0.05; **, P < 0.01; ***, P < 0.001, ****, P < 0.0001.
A point worth noting is that NI appeared more variable than HS and LA specimens, especially regarding Firmicutes and Proteobacteria abundances. The inverse relationship observed for Firmicutes and Actinobacteria phyla was confirmed by analyzing the top ten genera (Fig. 2b). The most dramatic difference was the dominance of Propionibacterium and the reduction in the relative abundance of Staphylococcus genus in LA compared to HS and NI. At the species level (Fig. 2c), Cutibacterium acnes showed a significant difference between different sites (P < 0.0001). Conversely, the relative abundance of Staphylococcus aureus and Staphylococcus epidermidis did not change significantly in HS, NI, and LA.
Typing and whole-genome sequencing of C. acnes isolates
To determine the characteristics of the culturable C. acnes, bacterial colonies were isolated from site-matched skin in healthy controls (N10) and patients with acne (N10). The WGS results of C. acnes isolates are reported in Table 2. The 20 strains were distributed among five clonal complexes (CCs), with the most frequent being CC1 (n = 9; 45.0%), followed by CC3 (n = 5; 25%) and CC4, CC5 and CC6 (n = 2; 10% each). The overall distribution of CCs showed a significant difference between HS and LA (P = 0.03). The most represented C. acnes phylotype was IA1 (n = 16; 80%) followed by type IB (n = 2; 10%) and II (n = 2; 10%) strains. The phylotype analysis revealed a significant difference in the distribution between HS and LA (P = 0.03), with the IA1 significantly more abundant in LA than in HS (P = 0.03).
Table 2.
Multilocus sequence typing (MLST) and phylotype profiles of the C. acnes strains isolated from the skin of healthy subjects (HS) and the lesional area of acne patients (LA).
| Source | Sequence typing | Clonal complex | Phylotype |
|---|---|---|---|
| HS | 6 | CC6 | II |
| HS | 4 | CC4 | IA1 |
| HS | 5 | CC5 | IB |
| HS | 1 | CC1 | IA1 |
| HS | 3 | CC3 | IA1 |
| HS | 1 | CC1 | IA1 |
| HS | 1 | CC1 | IA1 |
| HS | 5 | CC5 | IB |
| HS | 7 | CC6 | II |
| HS | 4 | CC4 | IA1 |
| LA | 1 | CC1 | IA1 |
| LA | 3 | CC3 | IA1 |
| LA | 3 | CC3 | IA1 |
| LA | 115 | CC3 | IA1 |
| LA | 1 | CC1 | IA1 |
| LA | 3 | CC3 | IA1 |
| LA | 1 | CC1 | IA1 |
| LA | 1 | CC1 | IA1 |
| LA | 1 | CC1 | IA1 |
| LA | 1 | CC1 | IA1 |
| ATCC 11827 | 1 | CC1 | IA1 |
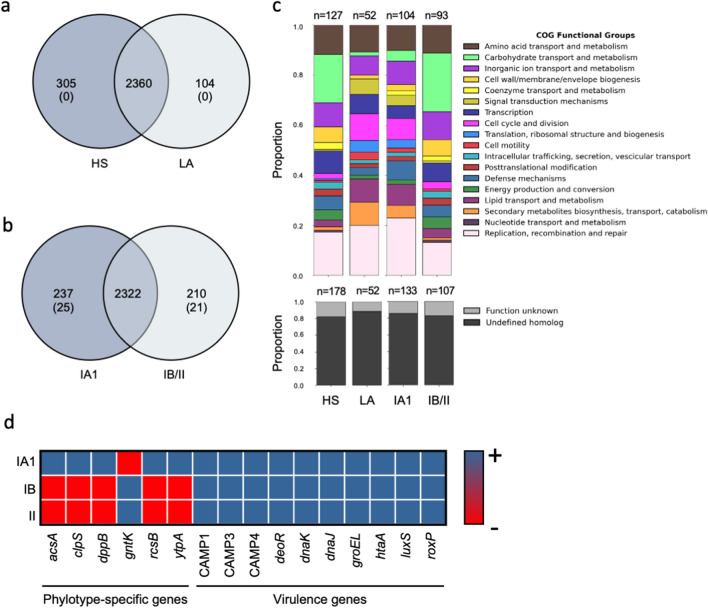
The number of putative protein-coding sequences in different genomes varied between 2332 and 2427, with an average of 2380. To assess genomic conservation across C. acnes isolates, the coding sequences were used to determine the pan-genome. This analysis revealed a total of 2769 genes representing the pan-genome of 20 C. acnes strains. To investigate the genetic determinants differentiating between different C. acnes isolates, unique genes were identified by querying the set for group-specific genes. Specifically, 305 genes occurred uniquely in C. acnes strains isolated from HS, whereas 104 unique genes were identified in C. acnes isolated from LA (Fig. 3a).
Figure 3.
Shared and unique genes in 20 C. acnes strains. Venn diagrams display the distribution of shared and unique genes between (a) C. acnes strains from healthy subjects (HS) and the lesional area of acne patients (LA) and (b) phylotypes IA1 and IB/II. Overlapping regions show the genes conserved within strains. The number between brackets represents the unique genes present in all the strains of a particular group. (c) The stacked bar chart of Cluster of Orthologous Genes (COGs) functional category proportions is based on the unique genes in all groups. The n above each group indicates the absolute count of COGs identified in each group. (d) Similarity matrix categories represent the presence (blue; +) or the absence (red, -) of C. acnes virulence genes and phylotype-specific genes.
Nevertheless, no unique genes were present in all the strains isolated from different skin sites.
The unique and core genes within phylotypes were identified to investigate further the possible clustering in C. acnes isolates (Fig. 3b). The genetic analysis revealed that the IA1 phylotype contained a higher number of unique genes (2559) than the IB/II (2532). The unique gene analyses showed that 237 genes occurred uniquely in the IA1 phylotype while 210 characterized the IB/II group (Fig. 3b). Of the 237 unique genes, 25 were present in all the type IA1 strains and 21 in the IB/II strains, representing the key genetic content distinguishing type IA1 strains from the IB/II clades (Fig. 3b). Functional categories were analyzed and reported in Fig. 3c. Notably, of the 237 unique genes present in IA1, 43.9% were assigned to specific functions, while 56.1% remained with unknown functions. Similarly, in IB/II 44.3% of the 210 unique genes were assigned to particular functions, and 55.7% had unknown functions (Fig. 3c). Interestingly, for the phylotype IA1, five unique genes with known functions were identified (acsA, clpS, dppB, rcsB, ytpA). In particular, AcsA and RcsB are positively involved in biofilm formation in different bacterial species. ClpS is an essential gene product involved in a highly conserved mechanism that targets specific proteins for destruction in prokaryotes and eukaryotes; DppB is a dipeptide ABC transporter required for bacterial growth, and the lysophospholipase YtpA is a virulence factor in C. acnes contributing to host-tissue degradation and inflammation. The type IB/II clades only displayed one unique gene with known function shared across all strains, the Shikimate kinase gntK. Putative virulence factor genes, coding for the iron acquisition protein (HtaA), the heat shock proteins (DnaK, DnaJ, and GroEL), the luxS gene, which is involved in the synthesis of the autoinducer 2, the radical oxygenase RoxP and the Christie–Atkins–Munch–Petersen (CAMP) factors 1, CAMP 2, and CAMP 4 were ubiquitous in every C. acnes strain, and not associated with specific genotypes (Fig. 3d). The co-hemolytic activity of the CAMP factor was further confirmed on blood agar plates for all C. acnes isolates. To further decipher the relationship among the phylotype IA1 strains, we generated a phylogenomic tree based on the core genome variants alignment and a phylogenetic tree based on the core pangenome allelic variations (supplemental information). The trees showed the samples clustered by clonal complex and sequence typing but not by the skin site (HS vs. LA).
Cutibacterium acnes adhesion and biofilm formation
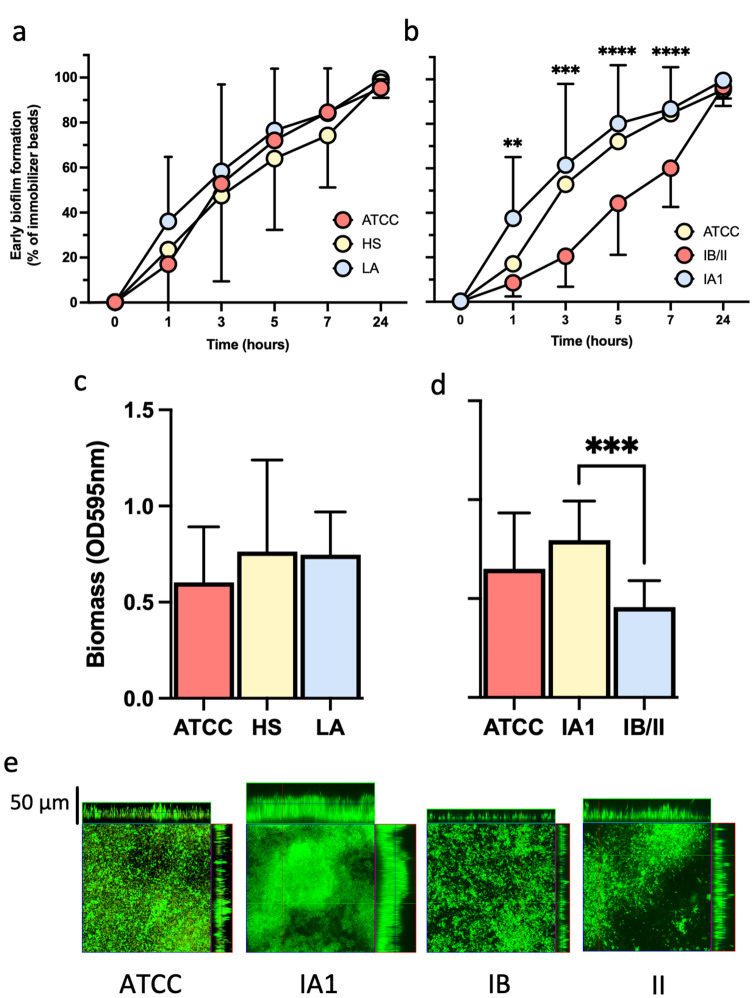
Cutibacterium acnes biofilms are frequently observed in patients with acne47. Moreover, the hierarchical clustering analysis indicated that the biofilm-related genes AcsA and RcsB were present in IA1 isolates but not in IB/II, suggesting possible differences in biofilm production. Therefore, the biofilm-forming ability of different C. acnes isolates was investigated. First, the early bacterial adhesion kinetics was measured at 0, 1, 3, 5, 7, and 24 h. No significant differences in the early biofilm formation of the C. acnes strains from HS and LA were observed during all the periods analyzed (Fig. 4a). Moreover, since IA1 colonized both HS and LA, the kinetics of early biofilm formation were analyzed on the phylotypes groups. In particular, the C. acnes strains belonging to the phylotypes IA1 were significantly faster in the early biofilm formation, than the phylotypes IB/II, after 1 (P = 0.0016), 3 (P = 0.0004), 5 (P < 0.0001) and 7 (P < 0.0001) hours. Nevertheless, after 24 h, all the strains analyzed achieved a comparable inhibition of the magnetic microparticles independently from the phylotype.
Figure 4.
Biofilm formation of C. acnes strains. a Kinetic of bacterial adhesion was measured using the BioFilm Ring Test for C. acnes strains isolated from healthy subjects (HS) and the lesional skin of acne patients (LA) and (b) according to the phylotypes IA1, IB, II. C. acnes ATCC 1182 was used as a reference control strain. The mean and corresponding standard errors for three independent experiments of duplicate samples for each time point are shown. (c,d) The amount of biofilm biomass after 72 h was measured by the crystal violet assays (optical density (OD) at 595 nm). The mean and corresponding standard errors for three independent experiments of duplicate samples for each time point are shown. (e) Confocal microscopy image of biofilm formation of different C. acnes isolates and the C. acnes ATCC 1182 after 72 h of incubation in BHI at 37 °C. Significance was assessed by using the Kruskal Wallis static test. *, P < 0.05; **, P < 0.01; ***, P < 0.001, ****, P < 0.0001.
Biofilm formation was further quantified by determining the biomass with crystal violet (CV) 72 h after incubation (Fig. 4b). The strains isolated from LA produced a comparable biomass level to those from HS and C. acnes 11827 ATCC (Fig. 4c,d). Notably, IA1 isolates formed significantly (P = 0.0004) more biofilm than the IB/II strains, suggesting that the phylotype impacted biomass production rather than the isolation site (Fig. 4d).
The morphology of biofilms was also investigated using confocal microscopy (Fig. 4e) for the reference strains C. acnes 11827 ATCC and the phylotypes IA1, IB, and II. Most of the C. acnes isolates produced a thick biofilm after 72 h, as indicated by the presence of cell aggregates and pillars of more than 20 µm (z projection). Consistent with the results obtained by microtiter plate tests, the IA1 isolates exhibited increased biomass and thickness compared to the IB and II isolates.
The biofilm matrix of C. acnes is composed of carbohydrates, proteins, and eDNA48. The impact of DNase I and proteinase K on biofilms has been used to investigate the contribution of eDNA and proteins in early adhesion and biofilm formation49. The treatment with DNase I and proteinase K caused a substantial reduction in the initial attachment to abiotic surfaces of C. acnes strains of all phylotypes (Fig. 5). However, DNase I treatment showed a significant (P < 0.001) reduction compared with proteinase K in the initial attachment of C. acnes strains suggesting that eDNA is critical for the initial adhesion. In particular, the reduction in the initial attachment to abiotic surfaces of C. acnes is significantly more pronounced in the phylotypes IB/II compared to IA1 (Fig. 5).
Figure 5.
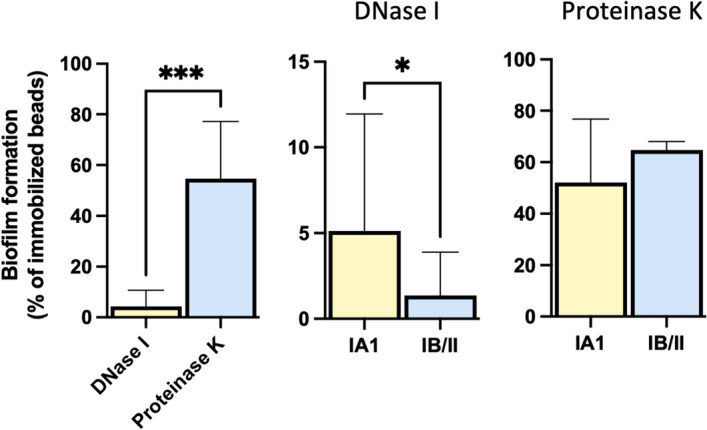
DNase I and Proteinase K reduce C. acnes adhesion. Comparison of extracellular DNA and proteins on early surface adhesion for the IA1 and IB/II phylotypes. Results are expressed as relative differences (Eq. 1) in the amounts of biofilm as measured by the BioFilm Ring Test after 6 h of incubation in the presence of DNase and Proteinase K compared with untreated control strains. Data represent means and the corresponding standard errors of two independent experiments analyzed in duplicate; *, P < 0.05; **, P < 0.01; ***, P < 0.001, ****, P < 0.0001 using the Mann–Whitney test.
Assessment of the antimicrobial susceptibility profiles
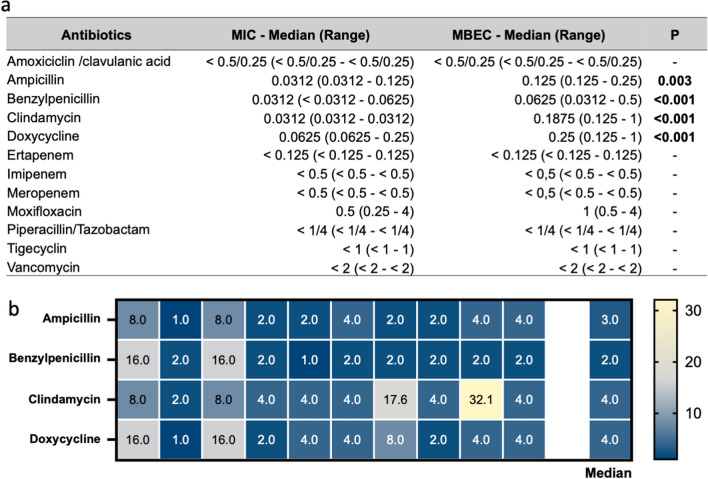
The activity of different antibiotics by the minimum inhibitory concentration (MIC) and minimal biofilm eradication concentration (MBEC) was measured in planktonic and biofilm growth on the ten C. acnes phylotype IA1 strains isolated from LA. The MIC values for each antibiotic are summarized in Fig. 6. The susceptibility profiles apparently contradicted the lack of response to several anti-C. acnes agents during acne treatment44. Since all strains were substantial biofilm producers, we next evaluated the potential correlation with the increased antibiotic tolerance. To this end, the antimicrobial susceptibility profiles were assessed in microbial isolates growing in biofilms.
Figure 6.
Antimicrobial tolerance in C. acnes isolates. (a) Antimicrobial susceptibility testing against ten C. acnes strains (phylotype IA1) isolated from the lesional skin of acne patients in planktonic and biofilm growth measured as minimum inhibitory concentration (MIC) and minimal biofilm eradication concentration (MBEC) for the indicated antibiotics. (b) Heat map showing the biofilm tolerance (BT), calculated as the ratio MBEC/MIC for ampicillin, benzylpenicillin, clindamycin, and doxycycline. Yellow indicates high BT values, and blue represents low BT for the indicated antibiotics. Statistical differences were determined using the Kruskal–Wallis test, followed by Dunn’s post hoc test for multiple comparisons.
Carbapenems, amoxicillin/clavulanic acid, piperacillin/tazobactam, and vancomycin were the most effective antibiotics against C. acnes biofilms with an MBEC comparable to the MIC values. However, biofilm-growing C. acnes exhibited a significant increase in the tolerance for ampicillin (P = 0.003), benzylpenicillin (P < 0.001), clindamycin (P < 0.001), and doxycycline (P < 0.001). Thus, the MBEC/MIC ratio, which indicates the fold increase in the antimicrobial dose needed to inhibit or kill C. acnes cells in biofilms compared to planktonic growth, was used to quantify the biofilm tolerance (BT) score (Fig. 6b). The maximum BT values were 32.1 and 17.6 reported for clindamycin, and the median BT values for the tested antimicrobials were between 2 (benzylpenicillin) and 4 (clindamycin and doxycycline).
Discussion
Cutibacterium is one of the most abundant genera in the skin, particularly in sebum reach areas50, and its proliferation has been largely associated with acne51,52. Although, the specific role in the pathophysiology of acne vulgaris remains uncertain. To account for the complexity of the skin microbial environment, we analyzed the microbiota in different skin districts of healthy and acne patients. Our study demonstrated statistically significant differences in alpha and beta diversity between the skin of the healthy subjects and inflammatory lesions of acne patients. Specifically, mean alpha diversity was significantly reduced in LA compared with HS. This data suggests that the anaerobic and lipid-rich conditions within the pilosebaceous unit of inflammatory acne lesions may provide an optimal microenvironment for C. acnes growth, thus limiting potential competitors53,54. Previous metagenomic studies revealed that the relative abundances of C. acnes do not differ significantly between acne and healthy subjects55–57. However, the difference in C. acnes distribution could be ascribed to several variables, including the sequencing procedures and sampling methods56. The sequencing strategy and primer selection for 16S rRNA gene analysis are critical factors in characterizing the compositions of bacterial skin communities58. For example, the V1–V3 regions of the 16S RNA gene had greater accuracy than the V4 region in defining genus and species level classification of major skin bacteria59. In particular, V4 sequencing resulted in an inaccurate assessment of prominent skin bacteria showing lower relative abundances of S. epidermidis and C. acnes and higher relative abundance of S. aureus compared with V1–V3 sequencing59. Based on these data, we selected the V1-V3 region, which allowed us to effectively recover Cutibacterium and Staphylococcus species defining unbiased skin microbial community profiles and identifying potential microbial biomarkers associated with acne. The site and sampling technique can also affect the outcome of the sequencing-based analysis of the skin microbiome60. Previous studies reported that a swab is a reliable technique to analyze the skin microbiome comparable to the tape-stripping method61,62. In our study, microbiome samples were collected by the swab technique directly on LA and NI sites for each patient and compared with the same area of HS, providing a narrowed characterization of the local microbial population. The possibility of collecting microbiome samples from the lesional skin may have yielded more targeted results compared to other studies applying different sampling techniques. Indeed, others collected samples from the nose microcomedones of acne patients using adhesive strips. This sampling technique is ideal for identifying a homogeneous microbial population; however, it may not specifically reflect the microbial distribution present in the different microenvironments of acne55. In our study, the Cutabacterium genus was more abundant in LA and NI compared to HS. Accordingly, a significant increase in C. acnes was observed in LA compared to NI and HS. These results suggest that, although highly prevalent in the skin of HS and patients with acne, the relative abundance of C. acnes significantly increases in inflamed LA compared to NI. Therefore, the overgrowth of C. acnes in LA can potentially explain the prevalence of the disease in a part of the population, despite the universal carriage of the microorganism. These observations support previous findings suggesting that pilosebaceous unit obstruction and increasing C. acnes proliferation are likely central factors in the onset of inflammatory acne lesions63,64. This is consistent with the increase of C. acnes counts found in acne, which, in turn, correlates with the acne flares in adolescents, with high levels of hormones and sebum production65. Indeed, teenagers with acne have significantly higher C. acnes counts than age-matched controls, demonstrating a marked increase in this microorganism, promoting inflammation through various mechanisms64,66,67. Specific host factors, such as sebum production, hormone level, the inflammatory milieu, and physical changes in the pilosebaceous unit, may contribute to acne pathogenesis, indicating that certain strains can become pathogenic in different conditions or under specific environmental stimuli16. It has been suggested that inflammatory acne results from an imbalance in the skin microbiota associated with specific C. acnes phylotypes35,68. Our study found that the clonal complexes CC1 and CC3 were more abundant in LA than in HS; instead, CC4, CC5, and CC6 were present only in HS. In addition, IA1 was significantly more represented in LA than in HS, and the phylotypes IB and II were found only in the skin of HS. Data from this study are consistent with previous reports revealing that severe inflammatory acne lesions of both the face and back are associated with loss of diversity in C. acnes phylotypes, with a high predominance of phylotype IA1 compared to healthy controls38,69,70. Fitz-Gibbon et al., analyzed the distribution of C. acnes ribotypes (RT1, RT2, RT3) among both acne and normal follicles55, reporting that CC3 and CC4 of the phylotype IA1 were significantly enriched in patients with acne but rarely found in individuals with healthy skin. In contrast, RT6, which represents a subpopulation of phylotype II, was 99% associated with healthy skin. Interestingly, the non-acne-associated IB, types II and III, are also commonly recovered from deep tissue infections and retrieved from medical devices16. The loss of diversity between the six phylotypes, characterized by a dominance of IA1 rather than C. acnes proliferation, may play a key role in triggering acne69–73. As C. acnes is the major skin colonizer, the strain-level analysis is important to help understand the role of this bacterium in acne pathogenesis and skin health. Full genome sequencing of different strains from variable environmental sources, such as acne vulgaris and implant-associated infection, has revealed the pan-genome and the genetic repertoire of C. acnes31,74–76. Moreover, metagenomic analyses showed that several distinct virulence-associated gene elements that encode antimicrobial peptides, cytotoxins, and proteases were enriched in C. acnes strains associated with acne12. The accessory genome of C. acnes is relatively small, and non-core genes frequently code for a variety of phylotype-specific functions31,74,75. The variability at the phylotype level may correlate with the commensal or pathogenic phenotype of C. acnes and its contribution to acne69. In our study, the WGS analyses showed the CAMP factors were detected across all strains. Moreover, in accordance with previous observations, we reported that major virulence determinants are widespread among C. acnes and not specifically associated with any site of isolation or the phylotype77. Likewise, we observed that luxS and roxP, essential for adherence to and colonization of the skin, were ubiquitous in every C. acnes strain and not associated with specific genotypes. The high distribution of virulence genes across C. acnes isolates was previously described suggesting that transcription regulation may be critical in the differential virulence expression among phylotypes78–80. Nevertheless, the functional group analysis showed that the phylotype IA1 was characterized by a reduced number of carbohydrate metabolism and sugar transporters genes compared with phylotypes IB/II. This data suggests that carbohydrate transport and metabolism may confer to C. acnes an ecological advantage in healthy skin but are not strictly required for the selective colonization of inflammatory acne lesions. Indeed, previous studies hypothesized a connection between high sebum levels and C. acnes colonization in acne patients81. Accordingly, we observed that lipid transport and metabolism genes are increased in phylotype IA1 strains, compared with phylotypes IB/II, independently from the isolation site. Indeed, it is unlikely that phylotype IA1 strains isolated from LA have, per se, different properties than the same phylotype strains from healthy skin. In agreement with previous publications, we found no differences in the genomes and biofilm production of phylotype IA1 strains isolated from acne and healthy skin82. Future studies are required to define how virulence genes are expressed in different phylotypes. Previous imaging analysis showed that microcomedones' structure in acne patients resembles a pouch containing lipids with clusters of bacteria, whose outer shell comprises corneocyte layers83. The extensive bacteria colonization is visible using TEM83. The metabolism of C. acnes modifies the skin's lipid composition, and bacterial lipases release free fatty acids from triglycerides. Free fatty acids being more viscous than triglycerides, can obstruct the pilosebaceous unit, which in turn becomes anaerobic, promoting the selective proliferation of phylotype IA1 that better exploits the lipid-rich microenvironments of the pilosebaceous unit during acne compared to other phylotypes83–85. The analyses of the unique genes present in different phylotypes allowed us to identify that of 2769 genes, five genes with known functions were univocally present on all IA1 isolates (acsA, clpS, dppB, rcsB, and ytpA). Notably, the ytpA gene, which encodes for a lysophospholipase that contributes to host-tissue degradation and inflammation, was present in all IA1 isolates.
To overcome these difficulties, a better understanding of the pathogenic potential of the individual sub-populations is needed. Additionally, others speculated that the persistent nature of acne vulgaris is linked to C. acnes’s colonization of the pilosebaceous unit in a biofilm, thereby eluding antibiotic eradication40,86. This theory was supported by observing that the genome of IA1 isolates contains genes such as rcsB, acsA that have a positive role in biofilm formation in other bacterial species41,87–89. Specifically, transcriptomic analysis of Helicobacter pylori revealed that several acetone metabolism genes, including acsA, are upregulated in biofilm cells89. The RcsCDB phosphorelay pathway coordinates the expressions of a large number of genes essential for maintaining cell wall integrity, cell division, stationary-phase sigma factor activity, biofilm development, motility, and virulence in Enterobacteriaceae90,91. RcsB upregulates genes promoting biofilm formation while downregulating different metabolic functions in Escherichia coli87,88. The comparative analysis of in vitro biofilm formation showed that the phylotype IA1 was significantly faster in early biofilm formation than IB and II strains in inhibiting the magnetic microparticles.
Moreover, biofilm quantification showed that IA1 strains produced significantly more mature biofilm than the other phylotypes confirming that both DNA and proteins are required for the early adhesion and biofilm formation in C. acnes39. In addition, our findings suggest that independently from the site of isolation, all the strains produce biofilm; however, phylotype IA1, which is the most prevalent in AP, resulted in the most effective. This notion is indirectly reinforced by confocal microscopy results, which clearly show the development of a thick and structurally complex biofilm matrix in the phylotype IA1. Furthermore, DNase I- and Proteinase K-sensitivity assays revealed that eDNA and proteins are central for early surface adhesion to abiotic surfaces, with eDNA playing a major role. Besides, an increased DNase I-sensitivity of phylotypes IB/II could be observed. Thus, in acne patients, a hyperseborrheic microenvironment, characterized by high availability of sebum, may result in an increased proportion of metabolically active biofilm-producing strains, providing a selective advantage to a subset of acne-associated phylotypes or sub-types, thus contributing to the proinflammatory milieu of acne lesions54,65.
Although antibiotics can reduce or eliminate C. acnes, recolonization frequently occurs within a few weeks, with limited clinical efficacy44. The high tolerance of C. acnes to antimicrobial treatments does not depend on the expression of multidrug-resistant genes since C. acnes are generally susceptible to most antimicrobials44,92. Conversely, the C. acnes capability of chronic persistence and relapse following antibiotic therapy is strongly suggestive of biofilm-related colonization9,93. These findings align with our results showing that all C. acnes isolates were highly susceptible to most antibiotics, with MIC values largely below the clinical breakpoints. However, the susceptibility profile of biofilm-growing C. acnes strains indicated a significant increase in antimicrobial tolerance against ampicillin, benzylpenicillin clindamycin, and doxycycline. For systemic treatment, oral tetracyclines (doxycycline, minocycline, oxytetracycline, or tetracycline) are the most recommended antibiotics for treating severe acne, followed by clindamycin due to relevant adverse effects9; while, topical antibiotic treatments mainly rely on clindamycin, erythromycin, and tetracycline9. These observations may explain the high tolerance of C. acnes to antimicrobial treatment and the conflicting evidence about the clinical benefit of using anti-C. acnes agents, even in cases where therapy was based on MIC results44. Besides, we found that vancomycin, piperacillin/tazobactam, and amoxicillin/clavulanic acid could effectively eradicate mature C. acnes biofilms, with BMIC similar to MIC. The most common and effective antibiotic against C. acnes in patients with prosthetic valve endocarditis was vancomycin94,95. In addition, we found that C. acnes isolates were highly susceptible to carbapenems in both planktonic and biofilm states. Previous research showed that carbapenems were highly active in vitro and in vivo against C. acnes strains isolated from postsurgical endophthalmitis96.
Our study suffers from some limitations. Indeed, the analysis herein was performed on a small group of patients. In addition, the in vitro condition for biofilm testing may not be fully representative of the real in vivo conditions of the pilosebaceous unit of acne patients. Moreover, only a single C. acnes isolate was selected from each patient. This procedure may not have captured the entire C. acnes phylotypes diversity in the facial region. However, a recent publication demonstrates that C. acnes colonies from the skin of adult subjects without acne vulgaris are often very closely related, suggesting the presence of person-specific populations97. Thus, further research will be required to elucidate whether these observations can be transferred in vivo, and any clinical findings should be interpreted with caution.
Overall, the results presented in this study indicate that the inflammatory lesions of acne patients are characterized by dysbiosis and decreased bacterial diversity. At the species level, C. acnes is significantly more abundant in LA than in NI and HS. Significant genotypic and phenotypic differences among C. acnes isolates were primarily dictated by the phylotype rather than the anatomical site of isolation. In particular, IA1 isolates were more efficient in early adhesion, biomass production, and antibiotic tolerance than other phylotypes. Thus, we hypothesize that in adolescent acne, the biochemical and physical changes of the pilosebaceous unit may promote dysbiosis, thus providing a selective advantage to a subset of metabolically active biofilm-producing phylotypes, such as IA1, that have the potential and the virulence, to outcompete commensals exacerbating the inflammatory response98. Accordingly, adolescent acne pathogenesis may not be caused by the mere presence of a disease-associated phylotype but rather by the response of the overall microbial community to microenvironmental changes of the pilosebaceous unit. Factors contributing to C. acnes growth compared to competitors during the progression from the non-inflamed to the inflamed acne lesions remain to be elucidated. Nevertheless, new antimicrobial agents targeting C. acnes biofilm matrix components may represent potential treatments to modulate the skin microbiota in adolescent acne.
Methods
Study design and patients’ enrolment
Patients with severe acne lesions were recruited among subjects at their first access to the Acne Unit at the San Gallicano Dermatological Institute. The control group was enrolled among HS undergoing mole checks in the same Institution. Informed consent was obtained from all the participants and their legal representative. Two dermatologists assessed the acne severity scores and the clinical grading. The intensity of clinical manifestations was evaluated on the face areas according to the Global European Acne severity (GEAS) criteria99. Briefly, subjects were classified as non-affected (clear) when no or very few lesions were clinically observable (0 ≤ GEAS < 1). GEA scores between 1 and 2 were referred to as the mild group, GEA scores 3 as the moderate group, and patients with GEAS 4–5 were classified as severe groups. Participants were asked to maintain their skincare routines except for prohibiting the use of cosmetics and moisturizing skincare products on the day of the enrolment. Criteria of inclusion were the absence of systemic diseases and skin disorders other than acne. The exclusion criteria were oral or topic pharmacological treatments at the first visit or up to 2 weeks before the examination, smoking, and use of sunbeds. The study was conducted according to the Helsinki declaration, and all enrolled patients signed informed consent. The Central Ethics Committee I.R.C.C.S. Lazio, the section of the Istituti Fisioterapici Ospitalieri in Rome, approved this study (Protocol 7679—21.06.2016, trials registry number 821/16).
Sample collection
The samples were collected by dermatologists with commercially available sterile swabs (COPAN swabs, Brescia, Italy) from the skin of 10 HS and the unaffected skin and pustule of 10 acne patients in duplicate to assess the presence of C. acnes and for microbiome analysis. Swabs were brought to the Microbiology and Virology Laboratory of San Gallicano Institute for culture analysis and processed immediately in an anaerobic atmosphere. Schaedler Agar plates with 5% Sheep Blood (bioMérieux) were used to isolate and cultivate C. acnes. Plates were monitored for bacterial growth after 5–7 days of anaerobic incubation at 37 °C. Up to seven representative colonies of C. acnes were preliminarily identified by colony morphology and Gram staining. Further identification of C. acnes isolates was performed by MALDI-TOF mass spectrometry. Positive isolates were subcultured on Schaedler agar plates to obtain a pure culture of C. acnes. From each culture, bacterial DNA was obtained, and sequence analysis (ABI PRISM 3130xl Genetic Analyzer) of the 16SrRNA gene was used to confirm bacterial identification100. At the same time, the phylotype was preliminarily identified by Touch-down PCR101. Each sample's most common phylotype (detected in 85–100% of the colonies) was considered the dominant type. A single representative isolate of each dominant phylotype was selected for further analysis. C. acnes isolates were frozen and stored at − 80 °C. The co-hemolytic reaction of the CAMP factor on blood agar plates (bioMérieux) was performed using S. aureus strain ATCC 25923, according to the previous method23.
Sequencing and analysis
Extracted DNA was amplified by PCR with dual-index primers targeting the V1-V3 regions of the bacterial 16S rRNA gene, using the ARROW for NGS Microbiota solution A kit (ARROW Diagnostics) according to the manufacturer’s instruction. A sterile sample tube that had undergone the same DNA extraction and PCR amplification procedures was used for quality control102. Before sequencing, amplicons were purified using the Agencourt® AMPure XP PCR purification system (Beckman Coulter, Milan, Italy), and equal amounts (10 nM) of the sample’s DNA were pooled and diluted to reach a 4-nM concentration. Finally, a 5 pM of the denatured library was used to generate sequences using the 2 × 250 cycles MiSeq Reagent kit (Illumina) on an Illumina MiSeq instrument. Sequencing data were analyzed using the MicrobAT system102,103. During MicrobAT processing, demultiplexed sequences showing reads of length less than 200 nucleotides, an average Phred quality score below 25, and at least one ambiguous base were discarded104. The resulting sequences were aligned at a 97% sequence similarity and assigned to taxonomic (e.g., species) levels at an 80% classification threshold using the Ribosomal Database Project (RDP) classifier (release 11.5)105. Species that did not meet these criteria were assigned to the corresponding group, “unclassified [genus]”. The Biological Observation Matrix (BIOM) was obtained, and the following analysis was carried out in R studio (https://www.rstudio.com/; version 4.0.2) using the phyloseq package106. Microbial community differences were measured in terms of alpha and beta diversity after reading depth rarefaction. Shannon index and Pielou index were used to evaluate alpha diversity, and significance was assessed by the Kruskal Wallis test. Bray Curtis beta diversity was calculated, and the distance matrix was represented as Principal coordinate analysis (PCoA)107. Significance was assessed by Permutational multivariate analysis of variance (PERMANOVA)108. Bacterial relative abundances at phylum and genus level between selected groups were examined.
Cutibacterium acnes typing
FASTQ files, including Quality Control, trimming, assembly, and gene annotation, were elaborated using the Bactopia suite109. The pan-genome was obtained using Roary110 on the annotated assemblies. Functional annotation was performed using EggNOG v5.0111 on the protein sequences. The Phylotypes and Clonal Complexes were determined by querying the API of PubMLST (www.pubmlst.org) using custom Python scripts. Whole-genome analysis of variants (SNPs/indels) on the IA1 strains and the phylogenomic tree was obtained as described previously112. The independent analysis of the core genome was performed using PIRATE113 on the IA1 strains to build a phylogenomic tree.
Biofilm Ring Test® (BRT)
Biofilm production was evaluated using the clinical BioFilm Ring Test (cBRT) with some modifications114. Briefly, overnight bacterial cultures grown on Schaedler agar plates + 5% sheep blood (bioMérieux, France) was used to inoculate 2 mL of 0.45% saline solution (AirLife, Carefusion, CA, USA) to the equivalent of 2.5 ± 0.3 McFarland turbidity standard. The bacterial suspension was used to inoculate a 96-well polystyrene plate with 200 μL/well. The test was performed using toner solution (TON) (Biofilm Control, Saint Beauzire, France) containing magnetic beads 1% (v/v) mixed in Brain Heart Infusion medium (BHI, Difco, Detroit, MI, USA). Sample dilutions (tenfold serial dilutions, from 1 × 10–1 to 1 × 10–3) were performed in a volume of 200 μL BHI/TON mix. The laboratory strain C. acnes ATCC 11827 was included in each test as standard reference and quality control. A well containing the BHI/TON mix without microbial cells was used in each experiment as a negative control. Plates were incubated at 37 °C without shaking (static culture) under anaerobic conditions. Biofilm formation was assessed at different time points. After incubation, wells were covered with few drops of contrast liquid (inert opaque oil used), placed for 1 min on the block carrying 96 mini-magnets (Block test), and scanned with a specifically designed plate reader (Pack BIOFILM, Biofilm Control, Saint Beauzire, France)115. Each strain was analyzed in duplicate, and experiments were repeated three times.
Assessment of C. acnes biofilm composition
The Biofilm Ring Test method was used to evaluate the early attachment in the presence of DNase I (100 μg/mL) and proteinase K (50 μg/mL)49,116. Standardized bacterial suspensions containing 1 vol % magnetic beads were supplemented with DNase (100 μg/mL) and proteinase K (50 μg/mL) and incubated at 37 °C in a 96-well microplate (200 μL/well). Negative controls contained 200 μL of sterile BHI with magnetic beads and enzymes. The plate was read after 6 h of incubation, as described above. The early adhesion in the presence of DNase and proteinase K was expressed using the relative difference (RD):
| 1 |
The analysis was performed three times in duplicate for each sample.
Evaluation of the biofilm formation with the crystal violet staining
Sterile 96-well polystyrene plates were inoculated with 200 μL of an initial bacterial suspension (105 CFU/mL) in BHI broth incubated at 37 °C for 72 h in anaerobic conditions without shaking. As described previously, biofilm formation was assayed using crystal violet staining in 96-well microtitre plates114.
Susceptibility testing
Minimum biofilm inhibitory concentration (MIC)
MICs were determined for each strain using the broth microdilution method described previously116 and adapted to the specific growth conditions of C. acnes. Specifically, C. acnes strains grown on Schaedler agar plates were inoculated into 2 mL of 0.45% saline solution (Air Life, Carefusion, CA, USA) to obtain turbidity of 0.5 ± 0.1 McFarland turbidity standard (approximately 108 CFU/mL). Samples were diluted at 1:100 in BHI, and 100 μL of bacterial suspension, were seeded into a sterile 96-multiwell polystyrene plate containing different antibiotics at variable concentrations (Corning Inc., Corning, NY, USA). Serial two-fold dilutions of the amoxicillin/clavulanic acid, ampicillin, benzylpenicillin, clindamycin, doxycycline, ertapenem, imipenem, meropenem, moxifloxacin, piperacillin/tazobactam, tigecycline, vancomycin were prepared. After the antibiotic treatment, viable cells were determined by plate counting for the CFU/mL determination. The MIC was defined as the lowest concentration of an antibiotic preventing bacterial growth. Experiments were conducted in triplicate.
Minimal biofilm eradication concentration (MBEC) assays
For each experiment, an overnight culture of C. acnes grown on a blood agar plate was used to inoculate 2 mL of 0.45% saline solution to 0.5 ± 0.1 McFarland turbidity standard. For biofilm cultures, diluted cell suspensions (approximately 106 CFU/mL) were used to inoculate a 96-well polystyrene flat-bottom plate with 100 mL BHI. After 24 h at 37 °C, in anaerobic conditions, the wells were rinsed with 0.45% saline solution to remove nonadherent bacteria. The plate was washed three times with distilled water, and the adherent cells were resuspended in 100 mL of BHI supplemented with two-fold serial dilutions of amoxicillin/clavulanic acid, ampicillin, benzylpenicillin, clindamycin, doxycycline, ertapenem, imipenem, meropenem, moxifloxacin, piperacillin/tazobactam, tigecycline, vancomycin. After overnight treatment, antibiotics were removed, and the plate was washed twice with 200 μL of sterile distilled water. Biofilms were scraped thoroughly, and the total number of viable cells was determined by serial dilution and plating on Schaedler agar plates to estimate the CFU number. The MBEC was defined as the lowest concentration of an antibiotic agent preventing bacterial growth.
MBEC/MIC-ratios were calculated to assess the biofilm tolerance (BT) score, which indicates the fold increase in the antimicrobial dose needed to inhibit or kill C. acnes cells in biofilm compared to planktonic growth116.
Biofilm Imaging
Cutibacterium acnes colonies, grown overnight on Schaedler agar plates, were used to inoculate 3 mL of 0.45% saline solution (Air Life, Carefusion, CA, USA) to obtain turbidity of 2.5 ± 0.3 McFarland turbidity standard corresponding approximately to 1 × 108 CFU/mL. Samples were diluted 1:1000 and resuspended in 1 mL of BHI in a μ-Slide, 8 well glass bottom chamber slides (Ibidi, Germany). The bacterial suspension was incubated at 37 °C for 72 h to allow biofilm formation. Subsequently, the medium was removed, and samples were washed in a 0.45% saline solution. According to supplier specifications, the biofilm cells were stained using the LIVE/DEAD BacLight kit (Life Technologies, New York, NY, USA)116 and examined with a Zeiss LSM5 Pascal Laser Scan Microscope (Zeiss, Oberkochen, Germany) Software Release 2.8 (Zeiss).
Statistics
Data were expressed as the mean ± standard error of the mean. Statistical analysis was performed using either ANOVA with Tukey post-hoc or Kruskal–Wallis with Dunn’s post-hoc tests, followed by appropriate p-value correction. For Beta diversity, PERMANOVA was used on Bray–Curtis distance matrices. P-values of 0.05 or less were considered statistically significant. IBM SPSS v.21 statistics software (IBM, Chicago, IL, USA) was used for all statistical analyses.
Supplementary Information
Author contributions
E.D. designed the study and wrote the paper. M.T., F.D., M.S., F.P., T.B., F.A., A.M., and F.P., discussed the results and implications and wrote the manuscript. F.S., I.C., G.C., F.L., M.P., B.C., and E.D. performed the experiments. M.T. and F.D. carried out the statistical analysis. B.C., A.M., A.C., and F.P. collected, supervised, and interpreted the clinical data. All authors analyzed the data, revised the manuscript critically, and approved the submitted version.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper or from the corresponding author upon reasonable request. The data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB55087 (https://www.ebi.ac.uk/ena/browser/view/ PRJEB55087).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-25436-3.
References
- 1.Tuchayi MS, et al. Acne vulgaris. Nat. Rev. Dis. Primer. 2015;1:15029. doi: 10.1038/nrdp.2015.29. [DOI] [PubMed] [Google Scholar]
- 2.Yentzer BA, Hick J, Reese EL, Uhas A, Feldman SR, Balkrishnan R. Acne vulgaris in the United States: Descriptive epidemiology. Cutis. 2010;86:94–99. [PubMed] [Google Scholar]
- 3.McConnell RC, Fleischer AB, Williford PM, Feldman SR. Most topical tretinoin treatment is for acne vulgaris through the age of 44 years: An analysis of the National Ambulatory Medical Care Survey, 1990–1994. J. Am. Acad. Dermatol. 1998;38:221–226. doi: 10.1016/S0190-9622(98)70598-5. [DOI] [PubMed] [Google Scholar]
- 4.Dreno B, Bagatin E, Blume-Peytavi U, Rocha M, Gollnick H. Female type of adult acne: Physiological and psychological considerations and management. J. Dtsch. Dermatol. Ges. J. Ger. Soc. Dermatol. 2018;16:1185–1194. doi: 10.1111/ddg.13664. [DOI] [PubMed] [Google Scholar]
- 5.Bagatin E, et al. Adult female acne: A guide to clinical practice. An. Bras. Dermatol. 2019;94:62–75. doi: 10.1590/abd1806-4841.20198203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perkins AC, Cheng CE, Hillebrand GG, Miyamoto K, Kimball AB. Comparison of the epidemiology of acne vulgaris among Caucasian, Asian, Continental Indian and African American women. J. Eur. Acad. Dermatol. Venereol. 2011;2:1054–1060. doi: 10.1111/j.1468-3083.2010.03919.x. [DOI] [PubMed] [Google Scholar]
- 7.Rocha MA, Bagatin E. Skin barrier and microbiome in acne. Arch. Dermatol. Res. 2018;310:181–185. doi: 10.1007/s00403-017-1795-3. [DOI] [PubMed] [Google Scholar]
- 8.Kubota Y, et al. Community-based epidemiological study of psychosocial effects of acne in Japanese adolescents. J. Dermatol. 2010;37:617–622. doi: 10.1111/j.1346-8138.2010.00855.x. [DOI] [PubMed] [Google Scholar]
- 9.Williams HC, Dellavalle RP, Garner S. Acne vulgaris. Lancet Lond. Engl. 2012;379:361–372. doi: 10.1016/S0140-6736(11)60321-8. [DOI] [PubMed] [Google Scholar]
- 10.Bernales Salinas A. Acne vulgaris: Role of the immune system. Int. J. Dermatol. 2021;60:1076–1081. doi: 10.1111/ijd.15415. [DOI] [PubMed] [Google Scholar]
- 11.Cong TX, et al. From pathogenesis of acne vulgaris to anti-acne agents. Arch. Dermatol. Res. 2019;311:337–349. doi: 10.1007/s00403-019-01908-x. [DOI] [PubMed] [Google Scholar]
- 12.Barnard E, Shi B, Kang D, Craft N, Li H. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci. Rep. 2016;6:39491. doi: 10.1038/srep39491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suh DH, Kwon HH. What’s new in the physiopathology of acne? Br. J. Dermatol. 2015;172:13–19. doi: 10.1111/bjd.13634. [DOI] [PubMed] [Google Scholar]
- 14.Dréno B, Dagnelie MA, Khammari A, Corvec S. The skin microbiome: A new actor in inflammatory acne. Am. J. Clin. Dermatol. 2020;21:18–24. doi: 10.1007/s40257-020-00531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramasamy S, Barnard E, Dawson TL, Li H. The role of the skin microbiota in acne pathophysiology. Br. J. Dermatol. 2019;181:691–699. doi: 10.1111/bjd.18230. [DOI] [PubMed] [Google Scholar]
- 16.Achermann Y, Goldstein EJC, Coenye T, Shirtliff ME. Propionibacterium acnes: From commensal to opportunistic biofilm-associated implant pathogen. Clin. Microbiol. Rev. 2014;27:419–440. doi: 10.1128/CMR.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zappe B, Graf S, Ochsner PE, Zimmerli W, Sendi P. Propionibacterium spp. in prosthetic joint infections: A diagnostic challenge. Arch. Orthop. Trauma Surg. 2008;128:1039–1046. doi: 10.1007/s00402-007-0454-0. [DOI] [PubMed] [Google Scholar]
- 18.Zeller V, et al. Propionibacterium acnes: An agent of prosthetic joint infection and colonization. J. Infect. 2007;55:119–124. doi: 10.1016/j.jinf.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Li ZJ, et al. Propionibacterium acnes activates the NLRP3 inflammasome in human sebocytes. J. Investig. Dermatol. 2014;134:2747–2756. doi: 10.1038/jid.2014.221. [DOI] [PubMed] [Google Scholar]
- 20.Brüggemann H, et al. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science. 2004;305:671–673. doi: 10.1126/science.1100330. [DOI] [PubMed] [Google Scholar]
- 21.Valanne S, et al. CAMP factor homologues in Propionibacterium acnes: A new protein family differentially expressed by types I and II. Microbiol. Read Engl. 2005;151:1369–1379. doi: 10.1099/mic.0.27788-0. [DOI] [PubMed] [Google Scholar]
- 22.Lwin SM, Kimber I, McFadden JP. Acne, quorum sensing and danger. Clin. Exp. Dermatol. 2014;39:162–167. doi: 10.1111/ced.12252. [DOI] [PubMed] [Google Scholar]
- 23.Nakatsuji T, et al. Propionibacterium acnes CAMP factor and host acid sphingomyelinase contribute to bacterial virulence: Potential targets for inflammatory acne treatment. PLoS ONE. 2011;6:e14797. doi: 10.1371/journal.pone.0014797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bek-Thomsen M, Lomholt HB, Scavenius C, Enghild JJ, Brüggemann H. Proteome analysis of human sebaceous follicle infundibula extracted from healthy and acne-affected skin. PLoS ONE. 2014;9:e107908. doi: 10.1371/journal.pone.0107908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thiboutot DM, Layton AM, Anne EE. IL-17: A key player in the P. acnes inflammatory cascade? J. Investig. Dermatol. 2014;134:307–310. doi: 10.1038/jid.2013.400. [DOI] [PubMed] [Google Scholar]
- 26.Dessinioti C, Katsambas AD. The role of Propionibacterium acnes in acne pathogenesis: Facts and controversies. Clin. Dermatol. 2010;28:2–7. doi: 10.1016/j.clindermatol.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Kim J. Review of the innate immune response in acne vulgaris: Activation of Toll-like receptor 2 in acne triggers inflammatory cytokine responses. Dermatol. Basel Switz. 2005;211:193–198. doi: 10.1159/000087011. [DOI] [PubMed] [Google Scholar]
- 28.Kistowska M, et al. IL-1β drives inflammatory responses to Propionibacterium acnes in vitro and in vivo. J. Investig. Dermatol. 2014;134:677–685. doi: 10.1038/jid.2013.438. [DOI] [PubMed] [Google Scholar]
- 29.Dagnelie MA, Khammari A, Dréno B, Corvec S. Cutibacterium acnes molecular typing: Time to standardize the method. Clin. Microbiol. Infect. 2018;24:1149–1155. doi: 10.1016/j.cmi.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 30.McDowell A, et al. An expanded multilocus sequence typing scheme for Propionibacterium acnes: Investigation of «pathogenic», «commensal» and antibiotic resistant strains. PLoS ONE. 2012;7:e41480. doi: 10.1371/journal.pone.0041480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scholz CFP, Jensen A, Lomholt HB, Brüggemann H, Kilian M. A novel high-resolution single locus sequence typing scheme for mixed populations of Propionibacterium acnes in vivo. PLoS ONE. 2014;9:e104199. doi: 10.1371/journal.pone.0104199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayslich C, Grange PA, Dupin N. Cutibacterium acnes as an opportunistic pathogen: An update of its virulence-associated factors. Microorganisms. 2021;9:303. doi: 10.3390/microorganisms9020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salar-Vidal L, et al. Genomic analysis of Cutibacterium acnes strains isolated from prosthetic joint infections. Microorganisms. 2021;9:1500. doi: 10.3390/microorganisms9071500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dréno B, et al. Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: A brief look at the latest updates. J. Eur. Acad. Dermatol. Venereol. 2018;2:5–14. doi: 10.1111/jdv.15043. [DOI] [PubMed] [Google Scholar]
- 35.Paugam C, et al. Propionibacterium acnes phylotypes and acne severity: An observational prospective study. J. Eur. Acad. Dermatol. Venereol. 2017;31:e398–e399. doi: 10.1111/jdv.14206. [DOI] [PubMed] [Google Scholar]
- 36.Borrel V, et al. Adaptation of acneic and non acneic strains of Cutibacterium acnes to sebum-like environment. MicrobiologyOpen. 2019;8:e00841. doi: 10.1002/mbo3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Neill AM, Gallo RL. Host–microbiome interactions and recent progress into understanding the biology of acne vulgaris. Microbiome. 2018;6:177. doi: 10.1186/s40168-018-0558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDowell A, Nagy I, Magyari M, Barnard E, Patrick S. The opportunistic pathogen Propionibacterium acnes: Insights into typing, human disease, clonal diversification and CAMP factor evolution. PLoS ONE. 2013;8:e70897. doi: 10.1371/journal.pone.0070897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuehnast T, et al. Comparative analyses of biofilm formation among different Cutibacterium acnes isolates. Int. J. Med. Microbiol. 2018;308:1027–1035. doi: 10.1016/j.ijmm.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Brandwein M, Steinberg D, Meshner S. Microbial biofilms and the human skin microbiome. NPJ Biofilms Microbiomes. 2016;2:3. doi: 10.1038/s41522-016-0004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jahns AC, Eilers H, Ganceviciene R, Alexeyev OA. Propionibacterium species and follicular keratinocyte activation in acneic and normal skin. Br. J. Dermatol. 2015;172:981–987. doi: 10.1111/bjd.13436. [DOI] [PubMed] [Google Scholar]
- 42.Jahns AC, Alexeyev OA. Three dimensional distribution of Propionibacterium acnes biofilms in human skin. Exp. Dermatol. 2014;23:687–689. doi: 10.1111/exd.12482. [DOI] [PubMed] [Google Scholar]
- 43.Alexeyev OA, et al. Pattern of tissue invasion by Propionibacterium acnes in acne vulgaris. J. Dermatol. Sci. 2012;67:63–66. doi: 10.1016/j.jdermsci.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Zaenglein AL, et al. Guidelines of care for the management of acne vulgaris. J. Am. Acad. Dermatol. 2016;74:945–973.e33. doi: 10.1016/j.jaad.2015.12.037. [DOI] [PubMed] [Google Scholar]
- 45.Dessinioti C, Katsambas A. Propionibacterium acnes and antimicrobial resistance in acne. Clin. Dermatol. 2017;35:163–167. doi: 10.1016/j.clindermatol.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 46.Mongaret C, et al. Cutibacterium acnes biofilm study during bone cells interaction. Microorganisms. 2020;8:E1409. doi: 10.3390/microorganisms8091409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brandwein M, Steinberg D, Meshner S. Microbial biofilms and the human skin microbiome. NPJ Biofilms Microbiomes. 2016;2:3. doi: 10.1038/s41522-016-0004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jahns AC, Eilers H, Alexeyev OA. Transcriptomic analysis of Propionibacterium acnes biofilms in vitro. Anaerobe. 2016;42:111–118. doi: 10.1016/j.anaerobe.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 49.Tasse J, et al. Association between biofilm formation phenotype and clonal lineage in Staphylococcus aureus strains from bone and joint infections. PLoS ONE. 2018;13:e0200064. doi: 10.1371/journal.pone.0200064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukherjee S, et al. Sebum and hydration levels in specific regions of human face significantly predict the nature and diversity of facial skin microbiome. Sci. Rep. 2016;6:36062. doi: 10.1038/srep36062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakatsuji T, et al. Antibodies elicited by inactivated Propionibacterium acnes-based vaccines exert protective immunity and attenuate the IL-8 production in human sebocytes: Relevance to therapy for acne vulgaris. J. Investig. Dermatol. 2008;128:2451–2457. doi: 10.1038/jid.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J. Acne vaccines: Therapeutic option for the treatment of acne vulgaris? J. Investig. Dermatol. 2008;128:2353–2354. doi: 10.1038/jid.2008.221. [DOI] [PubMed] [Google Scholar]
- 53.Brown SK, Shalita AR. Acne vulgaris. Lancet Lond. Engl. 1998;351:1871–1876. doi: 10.1016/S0140-6736(98)01046-0. [DOI] [PubMed] [Google Scholar]
- 54.Liu PF, et al. Propionibacterium acnes in the pathogenesis and immunotherapy of acne vulgaris. Curr. Drug Metab. 2015;16:245–254. doi: 10.2174/1389200216666150812124801. [DOI] [PubMed] [Google Scholar]
- 55.Fitz-Gibbon S, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Investig. Dermatol. 2013;133:2152–2160. doi: 10.1038/jid.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Omer H, McDowell A, Alexeyev OA. Understanding the role of Propionibacterium acnes in acne vulgaris: The critical importance of skin sampling methodologies. Clin. Dermatol. 2017;35:118–129. doi: 10.1016/j.clindermatol.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 57.Li CX, You ZX, Lin YX, Liu HY, Su J. Skin microbiome differences relate to the grade of acne vulgaris. J. Dermatol. 2019;46:787–790. doi: 10.1111/1346-8138.14952. [DOI] [PubMed] [Google Scholar]
- 58.Kong HH. Details matter: Designing skin microbiome studies. J. Investig. Dermatol. 2016;136:900–902. doi: 10.1016/j.jid.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meisel JS, et al. Skin microbiome surveys are strongly influenced by experimental design. J. Investig. Dermatol. 2016;136:947–956. doi: 10.1016/j.jid.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boxberger M, Cenizo V, Cassir N, La Scola B. Challenges in exploring and manipulating the human skin microbiome. Microbiome. 2021;9:125. doi: 10.1186/s40168-021-01062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bjerre RD, et al. Effects of sampling strategy and DNA extraction on human skin microbiome investigations. Sci. Rep. 2019;9:17287. doi: 10.1038/s41598-019-53599-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ogai K, et al. A comparison of techniques for collecting skin microbiome samples: Swabbing versus tape-stripping. Front. Microbiol. 2018;9:2362. doi: 10.3389/fmicb.2018.02362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lavker RM, Leyden JJ, McGinley KJ. The relationship between bacteria and the abnormal follicular keratinization in acne vulgaris. J. Investig. Dermatol. 1981;77:325–330. doi: 10.1111/1523-1747.ep12482524. [DOI] [PubMed] [Google Scholar]
- 64.Leyden JJ, McGinley KJ, Vowels B. Propionibacterium acnes colonization in acne and nonacne. Dermatology (Basel, Switzerland) 1998;196:55–58. doi: 10.1159/000017868. [DOI] [PubMed] [Google Scholar]
- 65.Sardana K, Gupta T, Garg VK, Ghunawat S. Antibiotic resistance to Propionibacterium acnes: Worldwide scenario, diagnosis and management. Expert Rev. Anti Infect. Ther. 2015;13:883–896. doi: 10.1586/14787210.2015.1040765. [DOI] [PubMed] [Google Scholar]
- 66.Leyden JJ, McGinley KJ, Mills OH, Kligman AM. Propionibacterium levels in patients with and without acne vulgaris. J. Investig. Dermatol. 1975;65:382–384. doi: 10.1111/1523-1747.ep12607634. [DOI] [PubMed] [Google Scholar]
- 67.Miura Y, et al. Quantitative PCR of Propionibacterium acnes DNA in samples aspirated from sebaceous follicles on the normal skin of subjects with or without acne. J. Med. Dent. Sci. 2010;57:65–74. [PubMed] [Google Scholar]
- 68.Dreno B, et al. Skin microbiome and acne vulgaris: Staphylococcus, a new actor in acne. Exp. Dermatol. 2017;26:798–803. doi: 10.1111/exd.13296. [DOI] [PubMed] [Google Scholar]
- 69.McLaughlin J, et al. Propionibacterium acnes and acne vulgaris: New insights from the integration of population genetic, multi-omic, biochemical and host-microbe studies. Microorganisms. 2019;7:E128. doi: 10.3390/microorganisms7050128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dagnelie MA, et al. Decrease in diversity of Propionibacterium acnes Phylotypes in patients with severe acne on the back. Acta Derm. Venereol. 2018;98:262–267. doi: 10.2340/00015555-2847. [DOI] [PubMed] [Google Scholar]
- 71.Pécastaings S, et al. Characterisation of Cutibacterium acnes phylotypes in acne and in vivo exploratory evaluation of Myrtacine®. J. Eur. Acad. Dermatol. Venereol. JEADV. 2018;32:15–23. doi: 10.1111/jdv.15042. [DOI] [PubMed] [Google Scholar]
- 72.Corvec S, Dagnelie MA, Khammari A, Dréno B. Taxonomy and phylogeny of Cutibacterium (formerly Propionibacterium) acnes in inflammatory skin diseases. Ann. Dermatol. Venereol. 2019;146:26–30. doi: 10.1016/j.annder.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 73.Dagnelie MA, et al. Cutibacterium acnes phylotypes diversity loss: A trigger for skin inflammatory process. J. Eur. Acad. Dermatol. Venereol. JEADV. 2019;33:2340–2348. doi: 10.1111/jdv.15795. [DOI] [PubMed] [Google Scholar]
- 74.Tomida S, et al. Pan-genome and comparative genome analyses of Propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. MBio. 2013;4:e00003-13. doi: 10.1128/mBio.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scholz CF, et al. Genome stability of Propionibacterium acnes: A comprehensive study of indels and homopolymeric tracts. Sci Rep. 2016;6:20662. doi: 10.1038/srep20662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brüggemann H, Salar-Vidal L, Gollnick H, Lood R. A janus-faced bacterium: Host-beneficial and -detrimental roles of Cutibacterium acnes. Front. Microbiol. 2021;12:673845. doi: 10.3389/fmicb.2021.673845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cobian N, Garlet A, Hidalgo-Cantabrana C, Barrangou R. Comparative genomic analyses and CRISPR-Cas characterization of Cutibacterium acnes provide insights into genetic diversity and typing applications. Front. Microbiol. 2021;12:758749. doi: 10.3389/fmicb.2021.758749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valanne S, et al. CAMP factor homologues in Propionibacterium acnes: A new protein family differentially expressed by types I and II. Microbiology. 2005;151:1369–1379. doi: 10.1099/mic.0.27788-0. [DOI] [PubMed] [Google Scholar]
- 79.Nakatsuji T, Tang DC, Zhang L, Gallo RL, Huang CM. Propionibacterium acnes CAMP factor and host acid sphingomyelinase contribute to bacterial virulence: Potential targets for inflammatory acne treatment. PLoS ONE. 2011;6:e14797. doi: 10.1371/journal.pone.0014797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Andersson T, et al. Common skin bacteria protect their host from oxidative stress through secreted antioxidant RoxP. Sci. Rep. 2019;9:3596. doi: 10.1038/s41598-019-40471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leyden JJ, McGinley KJ, Mills OH, Kligman AM. Age-related changes in the resident bacterial flora of the human face. J. Investig. Dermatol. 1975;65:379–381. doi: 10.1111/1523-1747.ep12607630. [DOI] [PubMed] [Google Scholar]
- 82.Lomholt HB, Scholz CFP, Brüggemann H, Tettelin H, Kilian M. A comparative study of Cutibacterium (Propionibacterium) acnes clones from acne patients and healthy controls. Anaerobe. 2017;47:57–63. doi: 10.1016/j.anaerobe.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 83.Josse G, et al. High bacterial colonization and lipase activity in microcomedones. Exp. Dermatol. 2020;29:168–176. doi: 10.1111/exd.14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zouboulis CC. Propionibacterium acnes and sebaceous lipogenesis: A love–hate relationship? J. Investig. Dermatol. 2009;129:2093–2096. doi: 10.1038/jid.2009.190. [DOI] [PubMed] [Google Scholar]
- 85.Huang YC, Yang CH, Li TT, Zouboulis CC, Hsu HC. Cell-free extracts of Propionibacterium acnes stimulate cytokine production through activation of p38 MAPK and Toll-like receptor in SZ95 sebocytes. Life Sci. 2015;139:123–131. doi: 10.1016/j.lfs.2015.07.028. [DOI] [PubMed] [Google Scholar]
- 86.Burkhart CN, Burkhart CG. Microbiology’s principle of biofilms as a major factor in the pathogenesis of acne vulgaris. Int. J. Dermatol. 2003;42:925–927. doi: 10.1111/j.1365-4632.2003.01588.x. [DOI] [PubMed] [Google Scholar]
- 87.Lehti TA, Heikkinen J, Korhonen TK, Westerlund-Wikström B. The response regulator RcsB activates expression of Mat fimbriae in meningitic Escherichia coli. J. Bacteriol. 2012;194:3475–3485. doi: 10.1128/JB.06596-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sharma VK, et al. Disruption of rcsB by a duplicated sequence in a curli-producing Escherichia coli O157:H7 results in differential gene expression in relation to biofilm formation, stress responses and metabolism. BMC Microbiol. 2017;17:56. doi: 10.1186/s12866-017-0966-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hathroubi S, Hu S, Ottemann KM. Genetic requirements and transcriptomics of Helicobacter pylori biofilm formation on abiotic and biotic surfaces. NPJ Biofilms Microbiomes. 2020;6:56. doi: 10.1038/s41522-020-00167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Latasa C, et al. Salmonella biofilm development depends on the phosphorylation status of RcsB. J. Bacteriol. 2012;194:3708–3722. doi: 10.1128/JB.00361-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Howery KE, Clemmer KM, Rather PN. The Rcs regulon in Proteus mirabilis: Implications for motility, biofilm formation, and virulence. Curr. Genet. 2016;62:775–789. doi: 10.1007/s00294-016-0579-1. [DOI] [PubMed] [Google Scholar]
- 92.Oprica C, Nord CE, ESCMID Study Group on Antimicrobial Resistance in Anaerobic Bacteria European surveillance study on the antibiotic susceptibility of Propionibacterium acnes. Clin. Microbiol. Infect. 2005;11:204–213. doi: 10.1111/j.1469-0691.2004.01055.x. [DOI] [PubMed] [Google Scholar]
- 93.Coenye T, Spittaels KJ, Achermann Y. The role of biofilm formation in the pathogenesis and antimicrobial susceptibility of Cutibacterium acnes. Biofilm. 2022;4:100063. doi: 10.1016/j.bioflm.2021.100063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Banzon JM, et al. Propionibacterium acnes endocarditis: A case series. Clin. Microbiol. Infect. 2017;23:396–399. doi: 10.1016/j.cmi.2016.12.026. [DOI] [PubMed] [Google Scholar]
- 95.Verkaik NJ, Schurink CAM, Melles DC. Antibiotic treatment of Propionibacterium acnes endocarditis. Clin. Microbiol. Infect. Dis. 2018;24:209. doi: 10.1016/j.cmi.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 96.Shames R, Satti F, Vellozzi EM, Smith MA. Susceptibilities of Propionibacterium acnes ophthalmic isolates to ertapenem, meropenem, and cefepime. J. Clin. Microbiol. 2006;44:4227–4228. doi: 10.1128/JCM.00722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Conwill A, et al. Anatomy promotes neutral coexistence of strains in the human skin microbiome. Cell Host Microbe. 2022;30:171–182.e7. doi: 10.1016/j.chom.2021.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smith RN, Braue A, Varigos GA, Mann NJ. The effect of a low glycemic load diet on acne vulgaris and the fatty acid composition of skin surface triglycerides. J. Dermatol. Sci. 2008;50:41–52. doi: 10.1016/j.jdermsci.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 99.Dréno B, et al. Development and evaluation of a Global Acne Severity Scale (GEA Scale) suitable for France and Europe. J. Eur. Acad. Dermatol. Venereol. 2011;25:43–48. doi: 10.1111/j.1468-3083.2010.03685.x. [DOI] [PubMed] [Google Scholar]
- 100.Di Domenico EG, et al. Misidentification of Streptococcus uberis as a human pathogen: A case report and literature review. Int. J. Infect. Dis. IJID. 2015;33:79–81. doi: 10.1016/j.ijid.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 101.Barnard E, et al. Multiplex touchdown PCR for rapid typing of the opportunistic pathogen Propionibacterium acnes. J. Clin. Microbiol. 2015;53:1149–1155. doi: 10.1128/JCM.02460-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Posteraro P, et al. First bloodstream infection caused by Prevotella copri in a heart failure elderly patient with Prevotella-dominated gut microbiota: A case report. Gut Pathog. 2019;11:44. doi: 10.1186/s13099-019-0325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cutuli SL, et al. COVID-19 influences lung microbiota dynamics and favors the emergence of rare infectious diseases: A case report of Hafnia Alvei pneumonia. J. Crit. Care. 2021;64:173–175. doi: 10.1016/j.jcrc.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces usingPhred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 105.Cole JR, et al. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:633–642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Anderson, M. J. Permutational Multivariate Analysis of Variance (PERMANOVA). Wiley StatsRef: Statistics Reference Online 10.1002/9781118445112.stat07841 (2017).
- 109.Petit RA, Read TD. Bactopia: A flexible pipeline for complete analysis of bacterial genomes. mSystems. 2020;5:e00190-20. doi: 10.1128/mSystems.00190-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Page AJ, et al. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinform. Oxf. Engl. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huerta-Cepas J, et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:309–314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Seemann, T. snippy: Fast bacterial variant calling from NGS reads (Access 29 September 2022); https://github.com/tseemann/snippy (2015).
- 113.Bayliss SC, et al. PIRATE: A fast and scalable pangenomics toolbox for clustering diverged orthologues in bacteria. Gigascience. 2019;8:giz119. doi: 10.1093/gigascience/giz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Di Domenico EG, et al. Development of an in vitro assay, based on the BioFilm Ring Test®, for rapid profiling of biofilm-growing bacteria. Front. Microbiol. 2016;7:1429. doi: 10.3389/fmicb.2016.01429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Di Domenico EG, et al. The impact of bacterial biofilms on end-organ disease and mortality in patients with hematologic malignancies developing a bloodstream infection. Microbiol. Spectr. 2021;9:e0055021. doi: 10.1128/Spectrum.00550-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sivori F, et al. Role of extracellular DNA in dalbavancin activity against methicillin-resistant Staphylococcus aureus (MRSA) biofilms in patients with skin and soft tissue infections. Microbiol. Spectr. 2022;10:e0035122. doi: 10.1128/spectrum.00351-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper or from the corresponding author upon reasonable request. The data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB55087 (https://www.ebi.ac.uk/ena/browser/view/ PRJEB55087).