Summary
Human elongating multi-lineage organized (EMLOC) gastruloid technology captures key aspects of trunk neurodevelopment including neural integration with cardiogenesis. We generate multi-chambered, contractile EMLOC gastruloids with integrated central and peripheral neurons using defined culture conditions and signaling factors. hiPSC colonies are primed by activating FGF and Wnt signaling pathways for co-induced lineages. EMLOC gastruloids are then initialized with primed cells in suspension culture using timed exposure to FGF2, HGF, IGF1, and Y-27632. Cardiogenesis is stimulated by FGF2, VEGF, and ascorbic acid.
For complete details on the use and execution of this protocol, please refer to Olmsted and Paluh (2022).1
Subject areas: Bioinformatics, Developmental biology, Neuroscience, Stem Cells, Organoids
Graphical abstract
Highlights
-
•
2D hiPSCs primed for trunk mesoderm, endoderm, and neural differentiation
-
•
2D to 3D aggregate formation with committed neural and cardiac polarized domains
-
•
Embryo-like cardiogenesis, heart tube-to-chamber morphogenesis, and specialization
-
•
Model of trunk central and peripheral neurogenesis integrated with cardiogenesis
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Human elongating multi-lineage organized (EMLOC) gastruloid technology captures key aspects of trunk neurodevelopment including neural integration with cardiogenesis. We generate multi-chambered, contractile EMLOC gastruloids with integrated central and peripheral neurons using defined culture conditions and signaling factors. hiPSC colonies are primed by activating FGF and Wnt signaling pathways for co-induced lineages. EMLOC gastruloids are then initialized with primed cells in suspension culture using timed exposure to FGF2, HGF, IGF1, and Y-27632. Cardiogenesis is stimulated by FGF2, VEGF, and ascorbic acid.
Before you begin
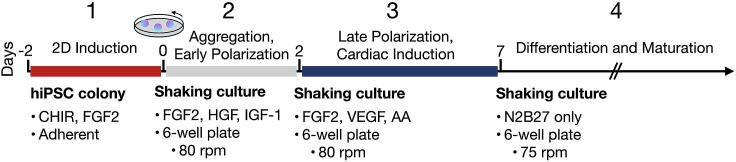
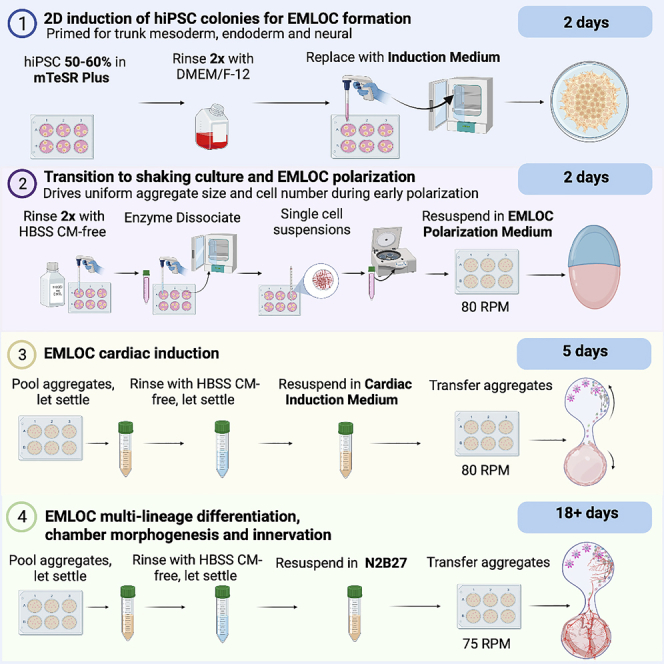
This protocol (Figure 1) describes the generation of EMLOCs from the hiPSC line H3.1.1 derived from fibroblasts of a self-designated Hispanic-Latino donor. This low passage hiPSC line was originally published with other ethnically diverse lines.2,3 The protocol is defined by four general stages: (1) 2D induction of hiPSC colonies, (2) transition to shaking culture and EMLOC early polarization, (3) EMLOC cardiac induction, and (4) EMLOC multi-lineage differentiation, chamber morphogenesis and innervation. In order to begin the EMLOC formation protocol, it is necessary to establish robust hiPSC cultures on Matrigel-coated cultureware. The following detailed steps provide instructions on cultureware preparation and hiPSC culture initiation and maintenance. The recipe for N2B27 basal medium used during all steps of EMLOC formation and differentiation is also provided.
Figure 1.
Overview of EMLOC formation and multi-lineage differentiation protocol
Timeline of EMLOC formation and differentiation in four general stages: (1) 2D induction of hiPSC colonies, (2) transition to shaking culture and EMLOC early polarization, (3) EMLOC cardiac induction, and (4) EMLOC multi-lineage differentiation, chamber morphogenesis and innervation.
Coat cultureware with hESC-qualified Matrigel
Timing: 24 h
Note: Description is for a 35 mm culture dish.
-
1.Handle and store hESC-qualified Corning Matrigel according to manufacturer’s instructions with attention to preparing and storing aliquots:
-
a.Thaw 5 mL parafilm-wrapped Matrigel on ice placed within a 4°C fridge.
-
b.The next day, prepare 100 μL aliquots of thawed, undiluted Matrigel on ice.
-
c.Store remaining aliquots at −20°C for later use.
-
a.
-
2.Prepare freshly coated cultureware:
-
a.Obtain a fresh 35 mm culture dish.
-
b.UV sterilize the culture dish with the top off for 30 min in a laminar flow tissue culture hood.
-
c.Pre-chill dish at −20°C, ∼20 min prior to use after 30 min of UV sterilization.
-
d.Thaw a single aliquot (100 μL) of Matrigel on ice ∼45 min prior to use.
-
e.Dilute Matrigel stock 1:100 in ice cold DMEM/F-12 (e.g., 20 μL/2 mL).
-
f.Add 1 mL of diluted Matrigel per dish.
-
g.Incubate dish at 37°C in a humidified incubator with 5% CO2 for at least 1 h prior to use to polymerize matrix coating.
-
a.
CRITICAL: Steps should be carried out on ice to prevent premature gelling and/or non-uniform coating of matrix.
Human induced pluripotent stem cell (hiPSC) culture
Timing: ∼3–7 days
-
3.Thaw hiPSCs cryopreserved in mFreSR cryopreservation medium:
-
a.Prewarm mTeSR Plus pluripotency medium to 25°C without a water bath for 15 min.
-
b.Transfer 1 vial containing 1 mL of cell suspension from liquid nitrogen storage to 37°C water bath.
-
c.While cell suspension is thawing, remove DMEM/F-12 from the freshly coated Matrigel plate, rinse 1× with 1 mL DMEM/F-12, and replace with 2.5 mL mTeSR Plus hiPSC pluripotency medium containing 1× penicillin-streptomycin.
-
i.mTeSR Plus is supplemented 1× with penicillin-streptomycin unless otherwise specified in this protocol.
-
i.
-
d.Carefully administer 1 mL of cells in thawed mFreSR dropwise to fresh mTeSR Plus (3.5 mL total) under a laminar flow tissue culture biosafety hood, attempting to distribute cells evenly over the surface area of the dish.
-
e.Incubate seeded cells at 37°C in a humidified incubator with 5% CO2 for 24 h.
-
a.
-
4.
The next morning, visually inspect for stem cell colony adherence (Figure 2A).
-
5.Perform medium change:
-
a.Rinse the cells 2× with 1 mL of fresh DMEM/F-12 prewarmed in 37°C water bath.
-
b.Replace medium with 2 mL mTeSR Plus prewarmed to 25°C and return to incubator for cell expansion.
-
a.
-
6.Follow cells by visual inspection to ensure proper colony density and maintenance of undifferentiated cells.
-
a.Replace fresh mTeSR Plus every other day or as needed until the cultures reach proper confluency for routine passaging (∼70% confluency) or to begin induction steps (50%–60% confluency).
-
b.Well-defined, undifferentiated colony edges and minimal colony-colony contact is ideal.
-
a.
-
7.
For advancing to EMLOC formation, passage hiPSCs 1:6 into Matrigel-coated 6-well plates when hiPSC cultures are stable (∼60% confluency).
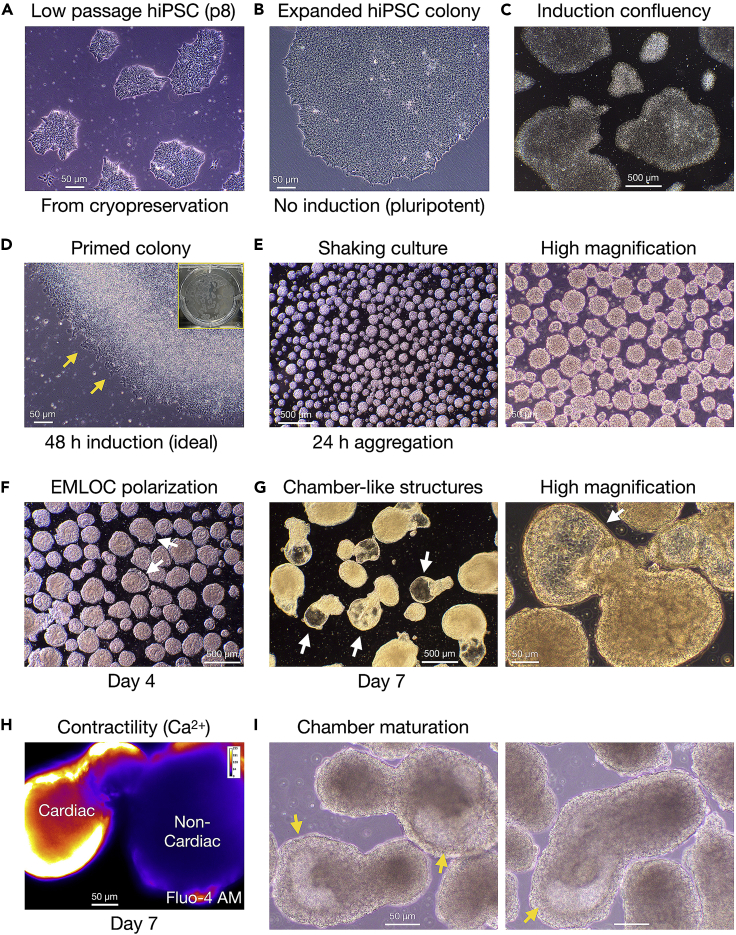
Figure 2.
Workflow of EMLOC induction, polarization, and maturation stages
(A) H3.1.1 low passage hiPSCs broken out on Matrigel after cryopreservation.
(B) Expanded hiPSC colony in mTeSR Plus pluripotency medium.
(C) Low magnification image of hiPSC cultures in mTeSR Plus at adequate confluency to begin 2D induction (∼60%).
(D) Stage 1: Induction of 2D hiPSC colonies in Induction Medium (N2B27 basal medium with 3 μM CHIR 99021 and 40 ng/mL FGF2) at 48 h. By visual inspection, optimal colony induction is characterized by slight raised-edge character just as cells begin to migrate away from the colony border (yellow arrows). Inset is white light image of primed colonies just before generation of single cell suspensions.
(E) Stage 2: Transition to shaking culture in EMLOC Polarization Medium. Single cell suspensions are generated and transferred to low-adhesion 6-well plates (2 × 106 cells/well). Spontaneous aggregation (size range ∼50–100 μm) at 24 h. Low 5× magnification is shown (left) with high magnification image (right).
(F) Stage 3: Early EMLOC polarization in Cardiac Induction Medium with cardiac crescent formation (white arrows) at day 4 post-aggregation.
(G) Polarized EMLOCs with contractile chamber-like structures (white arrows) at low magnification (left) and high magnification (right).
(H) Contracting cardiogenic region by live-cell calcium imaging using Fluo-4 AM dye in day 7 EMLOC.
(I) Stage 4: maturation of EMLOC chamber-like structures (yellow arrows). Two fields are shown. Individual scale bars are provided for all images.
Passaging human induced pluripotent stem cells
Timing: ∼24 h
Note: Description is for passaging from a 35 mm culture dish to 6-well plate.
-
8.Remove media from the well to be passaged and immediately add 1 mL of Gentle Cell Dissociation Reagent (GCDR) to the empty well.
-
a.Incubate at 25°C for ∼3 min.
-
a.
CRITICAL: The GCDR incubation time requires cell line-specific optimization according to manufacturer’s instructions.
Note: The incubation time here allows cells to be released in small ‘colony patches’ and not as single cells.
-
9.
Gently aspirate the GCDR without dislodging the cells.
-
10.
Immediately add 3 mL mTeSR Plus to the well being passaged.
-
11.To dislodge colonies, use a 5 mL serological pipette oriented orthogonally to the plane of the plate:
-
a.Perform a side-to-side scraping motion over the entire area of the well.
-
b.Rotate the plate 90 degrees and repeat the side-to-side scraping motion to ensure that the bulk of cells are dislodged from the substrate.
-
c.Pipet the suspension up and down 2× in order to further reduce clump size using a P-1000 blue tip.
-
a.
-
12.Gently transfer 0.5 mL of dislodged cells to each well of a Matrigel-treated 6-well plate already containing 1 mL of mTeSR Plus:
-
a.Cells should be added dropwise quickly and serially to obtain an even distribution in each well (1.5 mL total).
-
b.Detailed protocol steps for Matrigel coating are described above for a 35 mm culture dish. Each well of a 6-well plate can be treated equivalently to a single 35 mm dish.
-
a.
-
13.
Incubate the plate for 24 h, undisturbed at 37°C to allow colonies to settle and adhere.
-
14.The next day, visually inspect cultures for adherence of small colonies. If adherent, perform medium change:
-
a.Remove the 1.5 mL mTeSR Plus.
-
b.Rinse 2× in 1 mL DMEM/F-12 to remove non-adherent cell debris.
-
c.Add 2 mL fresh mTeSR Plus to each well.
-
d.Return the plate to the incubator.
-
a.
-
15.Add fresh media changes every 2 days until the colonies are expanded and cultures are ∼50%–60% confluent (Figures 2B and 2C).
-
a.This confluency percentage is optimal for cell induction to generate EMLOCs.
-
a.
Preparation of N2B27 basal medium
Timing: ∼30 min
-
16.
Prepare 1:1 DMEM/F-12:Neurobasal Plus medium appropriately supplemented to the following final concentrations: 2% (v/v) B-27 Plus, 1% (v/v) N-2, 1× GlutaMAX, 1× MEM Non-Essential Amino Acids, 1× penicillin-streptomycin. Recipe for 500 mL is shown in table below.
-
17.
Use a 0.2 μm pore filter to sterilize this solution and store at 4°C. Prewarm working volumes to 25°C as needed.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-SOX2 (dilution: 1:500) | R&D Systems | Cat. No.: AF2018; RRID: AB_355110 |
| Goat anti-GATA4 (dilution: 1:500) | R&D Systems | Cat. No.: AF2606; RRID: AB_2232177 |
| Goat anti-GATA6 (dilution: 1:500) | R&D Systems | Cat. No.: AF1700; RRID: AB_2108901 |
| Goat anti-FOXA2 (dilution: 1:250) | R&D Systems | Cat. No.: AF2400; RRID: AB_2294104 |
| Rabbit anti-β-tubulin III/TUJ1 (TUBB3) (dilution 1:2,000) | BioLegend | Cat. No.: 802001; RRID: AB_2564645 |
| Mouse anti-cardiac Troponin T (dilution: 1:500) | R&D Systems | Cat. No.: MAB1874 |
| Goat anti-CDH1/E-cadherin (dilution: 1:250) | R&D Systems | Cat. No.: AF648; RRID: AB_355504 |
| Rabbit anti-CDH2/N-cadherin (dilution: 1:1,000) | Cell Signaling Technologies | Cat. No.: 13116; RRID: AB_2687616 |
| Chemicals, peptides, and recombinant proteins | ||
| hESC-qualified Matrigel | Corning | Cat. No.: 08-774-552 |
| mTeSR Plus | STEMCELL Technologies | Cat. No.: 05825 |
| Gentle Cell Dissociation Reagent | STEMCELL Technologies | Cat. No.: 07174 |
| Neurobasal Plus Medium | Thermo Fisher Scientific | Cat. No.: A3582901 |
| DMEM/F-12 | Thermo Fisher Scientific | Cat. No.: 11320033 |
| N-2 supplement (100×) | Thermo Fisher Scientific | Cat. No.: 17502048 |
| B-27 supplement (50×) | Thermo Fisher Scientific | Cat. No.: 17504044 |
| GlutaMAX | Thermo Fisher Scientific | Cat. No.: 35050061 |
| MEM Non-Essential Amino Acids | Thermo Fisher Scientific | Cat. No.: 11140050 |
| Penicillin-Streptomycin | Thermo Fisher Scientific | Cat. No.: 15140122 |
| CHIR 99021 | Tocris Bioscience | Cat. No.: 4423 |
| bFGF/FGF2 | R&D Systems | Cat. No.: 233-FB |
| HGF | R&D Systems | Cat. No.: 294-HG |
| IGF-1 | R&D Systems | Cat. No.: 291-G1 |
| Y-27632 | Tocris Bioscience | Cat. No.: 1254 |
| VEGF 165 | R&D Systems | Cat. No.: 293-VE |
| L-ascorbic acid | Tocris Bioscience | Cat. No.: 4055 |
| Accutase | STEMCELL Technologies | Cat. No.: 07920 |
| Anti-Adherence Rinsing Solution | STEMCELL Technologies | Cat. No.: 07010 |
| HBSS CM-free | Thermo Fisher Scientific | Cat. No.: 14175079 |
| Experimental models: Cell lines | ||
| H3.1.1 (original human fibroblasts: foreskin of 1-day-old Hispanic-Latino male, karyotype XY) | Paluh lab | Chang et al.2; Tomov et al.3 |
| Other | ||
| 35 mm tissue culture-treated culture dish | CELLTREAT | Cat. No.: 229635 |
| 6-well plate | CELLTREAT | Cat. No.: 229105 |
| Disposable Vacuum Filter/Storage Systems | Thermo Fisher Scientific | Cat. No.: 430773 |
| 100 mm petri dish | Fisher | Cat. No.: S33580A |
| CHEMcell VERSA-ORB2 orbital shaker | Chemglass Life Sciences | Cat. No.: CLS-4021-100 |
Materials and equipment
N2B27 Basal Medium (500 mL)
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM/F-12 | N/A | 235 mL |
| Neurobasal Plus | N/A | 235 mL |
| B-27 Plus | 2% | 10 mL |
| N-2 | 1% | 5 mL |
| GlutaMAX (100×) | 1× | 5 mL |
| MEM Non-Essential Amino Acids (100×) | 1× | 5 mL |
| Penicillin-Streptomycin (100×) | 1× | 5 mL |
| Total | N/A | 500 mL |
∗Filter sterilize; medium can be stored at 4°C for up to 2 weeks.
Induction Medium
| Reagent | Final concentration | Amount |
|---|---|---|
| N2B27 | N/A | 10 mL |
| FGF2 | 40 ng/mL (100 μg/mL stock) | 4 μL |
| CHIR 99021 | 3 μM (10 mM stock) | 3 μL |
| Total | N/A | 10 mL |
∗Prepare fresh with supplements.
∗∗48 h total, replace full medium at 24 h.
EMLOC Polarization Medium
| Reagent | Final concentration | Amount |
|---|---|---|
| N2B27 | N/A | 10 mL |
| FGF2 | 10 ng/mL (100 μg/mL stock) | 1 μL |
| IGF1 | 2 ng/mL (100 μg/mL stock) | 200 nL |
| HGF | 2 ng/mL (100 μg/mL stock) | 200 nL |
| Y-27632 | 50 μM (10 mM stock) | 50 μL |
| Total | N/A | 10 mL |
∗Prepare fresh with supplements.
∗∗Replace one-half volume of aggregation medium at 24 h; one-half volume replacement medium contains 2× the concentration of FGF2, IGF1, and HGF to maintain steady-state levels, and excludes Y-27632.
Cardiac Induction Medium
| Reagent | Final concentration | Amount |
|---|---|---|
| N2B27 | N/A | 9.9 mL |
| FGF2 | 30 ng/mL (100 μg/mL stock) | 3 μL |
| Ascorbic acid | 0.5 mM (50 mM stock) | 100 μL |
| VEGF | 5 ng/mL (100 μg/mL stock) | 0.5 μL |
| Total | N/A | 10 mL |
∗Prepare fresh with supplements.
∗∗Full medium change to Cardiac Induction Medium at 48 h post-aggregation; EMLOCs are maintained in Cardiac Induction Medium for 5 days total, with 1× medium change after 72 h or longer from first addition (5 d post-aggregation).
Step-by-step method details
2D induction of hiPSC colonies for EMLOC formation (stage 1)
Timing: 2 days (for steps 1 to 6)
The short induction time at this step yields mesendodermal-like cellular starting material that importantly is also primed for neural differentiation. Protocols for trunk-biased, uniformly committed neuromesodermal progenitors (NMPs) typically rely on more sustained exposure to FGF and CHIR signaling (e.g., 4–5 d induction period).4,5 The induction factors for 2D adherent colonies and for subsequent aggregate formation and polarization were originally identified by the Gouti laboratory to generate neuromuscular trunk organoids using human stem cells.6 These factors were then optimized for elongating multi-lineage organized (EMLO) gastruloids by our laboratory.7 However, the original EMLO gastruloid protocol in that study was not optimized for cardiogenesis.7
Note: hiPSC colonies at ∼50%–60% confluency in single wells of a 6-well plate are ready for 2D induction (Figures 2B and 2C).
-
1.
Supplement working volume (2 mL per well) of N2B27 with 3 μM CHIR 99021, 40 ng/mL FGF2 (referred to herein as Induction Medium) and prewarm to 25°C without a water bath.
Note: FGF proteins influence caudalization to trunk phenotype in part through initiation of HOX gene expression.8,9 CHIR 99021 activates Wnt/β-catenin signaling via GSK3 inhibition.10 Together these factors promote generation of axial progenitors.4
Note: Medium supplementation during stages in EMLOC formation are summarized below:
CRITICAL: Cell-line dependent induction with CHIR 99021 and FGF2 must be optimized both for concentration and time of exposure. In our hands, the ideal CHIR 99021 concentration window for line H3.1.1 is between 3–3.5 μM for 2–3 d of induction. By visual inspection, optimal colony induction is characterized by slight raised-edge character just as cells begin to migrate away from the colony border (Figure 2D). Also important is consistency of cell numbers being induced so that factor concentrations are appropriate.
Note: See troubleshooting 1.
-
2.
Remove the mTeSR Plus medium and rinse cells 2× with DMEM/F-12.
-
3.
Add 2 mL of Induction Medium to the hiPSC wells that will be used to generate EMLOCs.
-
4.
Return plate to incubator.
-
5.
After 24 h, exchange the medium with 2 mL fresh Induction Medium.
-
6.
Return to incubator for an additional 24 h.
Transition to shaking culture and EMLOC polarization (stage 2)
Timing: 2 days (for steps 7 to 14)
Approximately 48 h after induction as adherent 2D colonies, cultures are primed to generate single cell suspensions to form 3D aggregates by orbital shaking. This transition helps to drive formation of uniformly and appropriately sized aggregates and introduces additional mechanical cues during the early polarization stage. Initial starting aggregates with small cell number (∼50–100 μm diameter aggregates; 50–100 cells each) are critical to establish the necessary axis length for local signaling and polarization.11 This size is achieved uniformly and spontaneously in our hands using line H3.1.1 and the culture conditions described.
Note: 48 h after the initial induction as 2D colonies in Induction Medium, cells can be used to generate 3D aggregates.
-
7.Prepare a fresh 6-well plate by treatment with 1 mL Anti-Adherence Rinsing Solution per well for 10 min.
-
a.Remove the Anti-Adherence Rinsing Solution from the wells and rinse 2× with 1 mL HBSS (CM-free). On the second rinse, do not remove the HBSS to prevent drying.
-
b.Remove the Induction Medium from the wells containing primed colonies and rinse 2× with 1 mL HBSS (CM-free).
-
a.
-
8.For enzymatic dissociation to single cells, dilute Accutase 1:1 with HBSS (CM-free) and add 1 mL per well. Incubate at 37°C for 5–10 min (cell line-dependent incubation time).
-
a.After incubation, gently remove dissociation solution and add 1 mL of N2B27 (no supplements) to the empty well.
-
b.Dislodge the cells using a side-to-side scraping motion over the entire surface area of the well with a serological pipette oriented orthogonally to the surface.
-
c.Triturate the suspension manually with a P-1000 pipette ∼6× to generate a single cell suspension.
-
a.
Note: See troubleshooting 2.
-
9.Combine 2–3 Accutase-treated hiPSC wells.
-
a.Add the entire volume to a 15 mL conical tube and centrifuge at 350 × g for 5 min to pellet the single cells.
-
a.
-
10.Aspirate the supernatant from the cell pellet and resuspend in EMLOC Polarization Medium (N2B27 basal medium supplemented with 10 ng/mL FGF2, 2 ng/mL IGF1, 2 ng/mL HGF, 50 μM ROCK inhibitor Y-27632) at the appropriate cell density.
-
a.Remove remaining HBSS from treated wells and transfer cell suspension to the pretreated low adherence well plate.
-
a.
Note: ROCK inhibitor Y-27632 promotes single cell survival and potentiates primed cells towards neural crest.12 IGF1 and HGF, together with FGF2, have been shown to enhance mesodermal progenitor expansion during early differentiation.13
CRITICAL: The combined suspensions used to generate aggregates for EMLOCs should have ∼2 × 106 cells total in 2 mL medium (1 × 106 cells/mL). Too many cells will interfere with correct ratio of factors. Too few cells will prevent any aggregate formation. Total cell number per well may range from 2 × 106 to 4 × 106 cells depending on cell line in 2 mL total volume.
Note: See troubleshooting 3.
-
11.
Place the plate on an orbital shaker at 80 rpm clockwise in a humidified incubator with 5% CO2.
-
12.
Visually inspect the orbital shaking cultures at 24 h post-aggregation.
Note: At this stage, round aggregates of similar size distribution (∼50–100 μm) should be visible (Figure 2E). To change the culture medium, pool the aggregates in a 15 mL conical tube and allow them to settle by gravity for ∼10 min.
Note: Do not let cultures over-settle to avoid aggregate fusion.
CRITICAL: If there are aggregates present that are much larger than the rest, manually remove the larger aggregates with a P-1000 blue tip.
-
13.
Aspirate one-half volume of the media and replace with fresh, modified EMLOC Polarization Medium (N2B27 supplemented with 20 ng/mL FGF2, 4 ng/mL IGF1, 4 ng/mL HGF without ROCK inhibitor Y-27632).
Note: See troubleshooting 4.
Note: The concentration of the growth factors is doubled to maintain the same overall concentration in the wells (assuming the original recombinant proteins are still present).
-
14.
Return the culture to the orbital shaker at 80 rpm clockwise in a humidified incubator with 5% CO2.
Note: The rotational speed is key to achieving the initial size of the starting aggregates and may require cell line-specific optimization.
EMLOC cardiac induction (stage 3)
Timing: 5 days (for steps 15 to 19)
EMLOC cardiogenesis is stimulated with defined angiocrine and cardiogenic factors in combination (FGF2, VEGF 165, ascorbic acid) to recapitulate morphological hallmarks such as thin walled, dilated chamber-like structures with spontaneous contractility. These factors were previously shown to stimulate cardiogenesis in mouse gastruloids and are applied here, adapting our original EMLO protocol to induce human cardiogenesis with retained neurogenesis within the multi-lineage gastruloid framework.7,14
-
15.48 h post-aggregation, initiate cardiac induction by pooling aggregates in a 15 mL conical tube and allow them to settle by gravity for 10 min.
-
a.Completely aspirate the EMLOC Polarization Medium and rinse with 10 mL HBSS (CM-free).
-
b.Let the aggregates re-settle and aspirate the HBSS (CM-free).
-
a.
-
16.
Resuspend in Cardiac Induction Medium (N2B27 supplemented with 5 ng/mL VEGF, 30 ng/mL FGF2, 0.5 mM ascorbic acid).
Note: VEGF regulates the development of the vascular endothelium and endocardium through the activation of Akt signaling in endothelial cells,15 and ascorbic acid promotes cardiac differentiation by enhancing the proliferation of cardiac progenitor cells via MEK-ERK1/2 signaling.16
-
17.
Return cells to the orbital shaker at 80 rpm clockwise in a humidified incubator with 5% CO2.
Note: See troubleshooting 5, 6, and 7.
-
18.
Replace Cardiac Induction Medium with fresh medium at 4–5 d post-aggregation.
Note: It may be useful to replace procardiogenic media only once to allow intra-aggregate cell-cell and paracrine signaling.
-
19.
Visually monitor cultures for early polarization and cardiac crescent formation (Figure 2F) and formation of contractile chamber-like structures (Figure 2G).
Note: Calcium-mediated cardiomyocyte contractility can be verified by live-cell calcium imaging with Fluo-4 AM dye (Figure 2H).1
EMLOC multi-lineage differentiation, chamber morphogenesis and innervation (stage 4)
Timing: 18+ days (for steps 20 to 23)
After 7 d post-aggregation, Cardiac Induction Medium is replaced with non-supplemented N2B27. This is intended to permit neurogenesis and cardiogenesis without further lineage restriction, favoring aggregate-derived signaling factors and self-organization.
Note: The maximal duration for continued maintenance and development of EMLOC analysis has not yet been determined beyond 25 d from induction.
-
20.
At 7 d post-aggregation, collect aggregates in a 15 mL tube and let settle at 37°C for 10 min.
-
21.Remove and replace medium:
-
a.Remove medium and rinse with HBSS (CM-free).
-
b.Let re-settle and remove the HBSS (CM-free).
-
c.Exchange the medium to non-supplemented N2B27 and re-distribute EMLOCs evenly to new cultureware freshly treated with Anti-Adherence Rinsing Solution.
-
a.
-
22.
Place on orbital shaker at 75 rpm clockwise in a humidified incubator with 5% CO2.
-
23.
Replenish N2B27 basal media every 3–5 d as needed to maintain the maturing EMLOCs for the remainder of the protocol.
Expected outcomes
The EMLOC formation and multi-lineage differentiation protocol occurs in four general stages (Figure 1). Stage 1 of hiPSC induction to EMLOC-primed cells is performed using intact 2D colonies beginning at ∼50%–60% confluency (Figures 2B and 2C). The induction period using N2B27 supplemented with CHIR 99021 and FGF2 occurs over 48 h, but may require longer exposure to between 48–72 h depending on cell line. By 48 h, optimal colony induction by visual inspection is characterized by slight raised-edge character just as cells begin to migrate away from the colony border (Figure 2D). Induced cells should co-express neuroectodermal biomarker SOX2 and mesendodermal biomarker FOXA2 by immunofluorescence (Figure 3A). At this point, 2D cultures are primed and ready to proceed to Stage 2, transition to shaking culture and EMLOC early polarization.
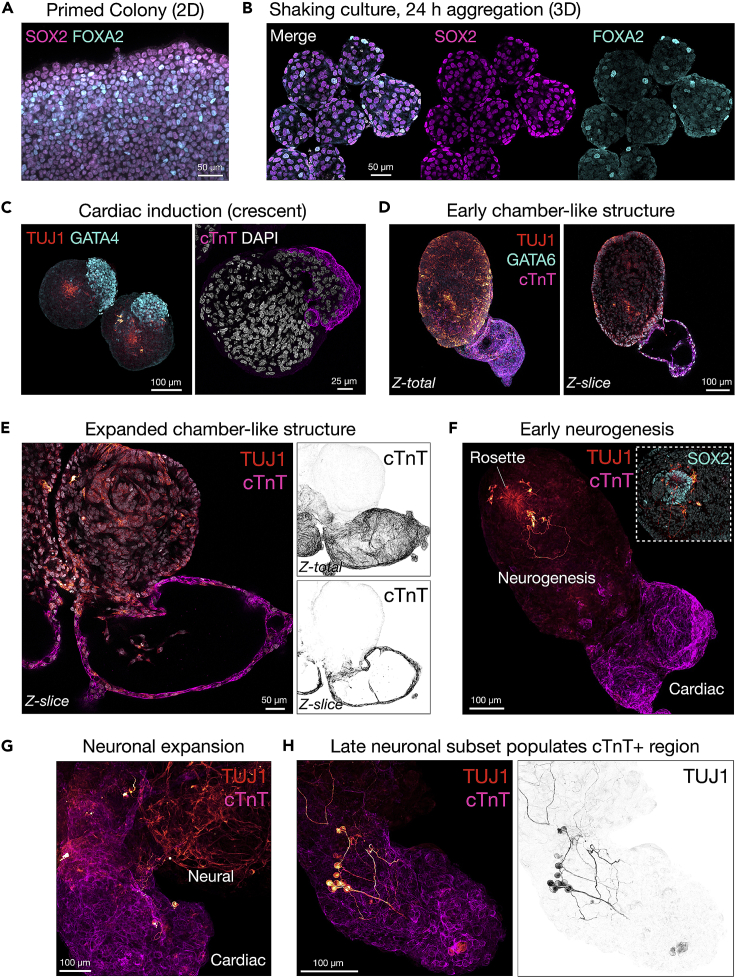
Figure 3.
Polarization of biomarkers at key steps in EMLOC formation by immunofluorescence
(A) Stage 1 (Induction Medium): immunofluorescence image of primed 2D hiPSC colony induced by CHIR 99021 and FGF2 in N2B27 basal medium. Co-expression of neuroectodermal biomarker SOX2 and mesendodermal biomarker FOXA2 is shown.
(B) Stage 2 (EMLOC Polarization Medium): Individual spherical aggregates at 24 h post-aggregation in shaking culture. Expression pattern of SOX2 and FOXA2 is shown as merge (+DAPI) and separate channels. Seven aggregates are visible.
(C) Stage 3 (Cardiac Induction Medium): day 4 EMLOCs with early polarized cardiac crescent (GATA4 left; cTnT right). Very early neurogenesis (TUJ1) is visible from single rosettes (left).
(D) Early chamber-like structures (cTnT) and distinct neurogenesis compartment (TUJ1). EMLOC is co-stained with GATA6. Maximally projected Z-stack (Z-total) is shown with single Z-slice to highlight morphology.
(E) Expanded chamber-like structure (cTnT) and distinct neurogenesis compartment (TUJ1). Z-total (top right) and single Z-slice (bottom right) of inverted cTnT channel is shown.
(F) Stage 4 (N2B27, no supplements): early neurogenesis from single rosette (TUJ1) with maturing chamber-like structure (cTnT). Inset is high magnification image showing SOX2+ rosette. Early neurons are diverted away from the cardiac region.
(G) Neuronal expansion (TUJ1) in neurogenesis compartment and early interaction with cTnT+ cardiac region.
(H) A late neuronal subset (TUJ1) begins to populate the cTnT+ cardiac region. Inverted TUJ1 channel is shown (right). Individual scale bars are provided for all images.
Stage 2 is characterized by exposure to FGF2, IGF1 and HGF in N2B27 basal medium for 48 h during aggregation of 2D primed colonies to 3D spherical aggregates. This necessitates dissociation of primed colonies to a single cell suspension using diluted Accutase. Prior to aggregation on orbital shaking culture in low-adhesion 6-well plates, a satisfactory single cell suspension should be verified by visual inspection using a tissue culture microscope. Single cells are applied at 2–4 × 106 cells/well depending on cell line in 2 mL of EMLOC Polarization Medium. After 24 h on the orbital shaker (80 rpm), spherical aggregates with size distribution of ∼50–100 μm should be present (Figure 2E). A critical step is to ensure appropriate size, and therefore cell number per initial aggregate (50–100 cells). Aggregates at 24 h should retain co-expression of SOX2 and FOXA2 as shown (Figure 3B), but do not yet express GATA6. A one-half volume media change to EMLOC Polarization Medium (no ROCK inhibitor) is made at 24 h with attempts to maintain steady-state levels of the recombinant proteins FGF2, IGF1 and HGF. After an additional 24 h (48 h post-aggregation), aggregates should have increased by ∼50% in size before proceeding to Stage 3.
At Stage 3, early EMLOC aggregates undergo cardiac induction by exposure to the angiocrine and cardiogenic factors FGF2, VEGF and ascorbic acid in N2B27 basal medium. By 48 h after exposure to the Cardiac Induction Medium (day 4 post-aggregation), polarized EMLOCs are oblong with cardiac crescent structures evident on visual inspection (Figure 2F) and identified by immunofluorescence of the biomarkers GATA4 and cTnT (Figure 3C). The cardiac crescent elongates to become the contractile chamber-like structure by day 7 (Figures 2G and 3D), which retains GATA4 and cTnT expression in addition to GATA6. The border of the cardiac crescent with non-cardiac EMLOC components can also be visualized by the N-cadherin, CDH2. Contractility can be quantified in culture (beat frequency, percentage of beating aggregates) and by live-cell calcium imaging with Fluo-4 AM dye (Figure 2H). In our original study, we quantified EMLOC formation efficiency as percentage of EMLOCs with contractile regions at day 7, which ranged from ∼60%–80% across experiments.1 Once contractile chamber-like structures emerge and are frequently identified in culture, cardiac induction factors can be withdrawn. Importantly, early neurogenesis from SOX2+ neural rosettes distal to the cardiac compartment should be occurring at day 7, and can be identified as early as day 4 (Figures 3C and 3F). At this stage, newly differentiated neurons are diverted away from the cardiac region, potentially due to influence by extracellular matrix-derived factors (Figure 3F). Self-organized anterior foregut tissue (FOXA2, CDH1) posterior to chamber-like structures may also be identified during Stage 3. Beyond day 7, cultures are maintained in N2B27 basal medium without supplements to permit further EMLOC multi-lineage differentiation, cellular diversification, and maturation (Stage 4).
The contractile, polarized EMLOCs undergo robust neurogenesis during Stage 4 and were maintained in the original publication to day 25. By this time point, the cTnT+ cardiac region should be populated by a subset of the neurons generated (Figure 3H). Throughout this stage of maturation, further events with developmental relevance can be observed such as expansion and consolidation of the multi-layered chamber wall with ongoing morphogenesis (Figure 2I). The range of lineages present in EMLOCs along with cellular diversity and maturation state can be interrogated by immunofluorescence in addition to single cell RNA-Seq. We originally performed the RNA-Seq at two time points (days 7 and 16). In our dataset, the predominant lineages to be expected are cardiac (cardiomyocytes, epicardium, cardiac fibroblasts, etc.), vascular endothelial, trunk nervous tissue (spinal cord neural progenitors, peripheral neurons, Schwann cell glia), anterior foregut, and genitourinary epithelium with intermediate mesoderm-derived metanephric mesenchyme. In contrast to EMLOs that primarily generate motor neurons, EMLOCs generate predominantly autonomic neurons (∼70%; PHOX2B/ASCL1) with a subset of peripheral sensory (25%; POU4F1) and motor neurons (5%; MNX1). Extensive gene biomarker lists based on cell stage and lineage are also provided in our original manuscript related to this protocol.1 The protocol for dissociation of EMLOCs to single cells for sequencing is also described therein.
Limitations
This protocol describes the generation of EMLOCs with the Hispanic-Latino H3.1.1 hiPSC line.1 In Olmsted and Paluh,7 we demonstrated the use of this line and others to reproducibly generate enteric gut formation with neural integration in an elongating multi-lineage organized (EMLO) gastruloid model. In Tomov et al.3 we comprehensively compared multiple ethnically-diverse hiPSC lines generated in collaboration with our laboratory,2 including cardiac differentiation and narrow effective windows in CHIR 99021 concentration gradient that are cell-line specific.3 These publications may be helpful when replicating the protocol with a new hiPSC line, ideally at low passage number.
Troubleshooting
Problem 1
It is unclear whether proper induction of cellular starting material has been achieved. Refer to Stage 1, #1.
Potential solution
Optimize CHIR 99021 and FGF2 induction time and concentration. Perform immunofluorescence on adherent colonies to evaluate co-expression of neuroectodermal biomarker SOX2 and mesendoderm biomarker FOXA2/HNF-3β (Figure 3A).
Problem 2
If after the 10 min incubation with 1:1 Accutase:HBSS (CM-free) dissociation solution, the cells have detached and are floating. Refer to Stage 2, #8.
Potential solution
Optimize time of exposure for cell line. For the current experiment, salvage the cells: add 1 mL N2B27 (no supplements), detach remaining cells by orienting a serological pipette orthogonally to the plane of the plate and perform a side-to-side scraping motion over the entire surface area of the well. Triturate the cell suspension manually and centrifuge at 350 × g for 5 min to remove dissociation solution. Resuspend the cell pellet in supplemented media.
Problem 3
Single cells do not aggregate in shaking culture. Refer to Stage 2, #10.
Potential solution
Cell number per well is likely too low prior to aggregation. Combine 2 or 3 wells to achieve at least 2 × 106 cells in 2 mL medium. Typically, the combination of primed cultures from 2 wells is sufficient but this too is cell line dependent.
Problem 4
Aggregates do not polarize. Refer to Stage 2, #13.
Potential solution
Use immunofluorescence to evaluate co-expression of the pluripotency marker SOX2 (posterior; region will mature to have neural identity), FOXA2/HNF-3β and GATA4 (anterior; region will mature through cardiac crescent to have primarily cTnT+ identity) (Figures 3B and 3C). The detailed protocol for immunofluorescence of 3D aggregates is described in a separate manuscript (Olmsted and Paluh7). Cell number in the initial aggregate is also key to establish appropriate length scale (50–100 cells/aggregate). Cell number can be interrogated by direct and/or indirect methods: (1) determine cell number directly by fixation of a subset of aggregates 24 h after transitioning to orbital shaker, staining with DAPI and counting cells from imaging data using confocal microscope-generated Z-stacks. Determine the average cell number; (2) indirect bulk method to determine the concentration of aggregates from a subset of the population by hemocytometer, dilute in buffer to achieve a known absolute number of aggregates, dissociate to single cells, and count again with hemocytometer or an automated cell counting system. Divide the total number of cells by the number of aggregates used to determine an average value at this stage. A final measure to take is to reduce the aggregate density by 2× in order to increase accessibility to polarization factors. This is done by doubling the total culture volume by splitting aggregates 1:2 to new wells in 2 mL per well.
Problem 5
EMLOCs do not contract. Refer to Stage 3, #19.
Potential solution
Verify early polarization biomarkers and cardiogenic biomarker expression cTnT at the cardiac crescent formation stage (Figures 3B and 3C). Performing less media changes in Cardiac Induction Medium may be beneficial. Purchase fresh stocks of induction factors and/or optimize ascorbic acid concentration. Attempt L-ascorbic acid phosphate Mg salt from different supplier (Wako, cat # 013-12061).14 Prepare fresh reagents. See problem 4 above.
Problem 6
Orbital shaking ruptures the contractile region or causes separation of cell compartments. Refer to Stage 3, #19.
Potential solution
Decrease the shaking speed. 75 rpm is recommended since reduced rpm below this level can cause aggregate fusion. 6-well plate cultures can also be transitioned to 100 mm petri dishes pretreated with Anti-Adherence Rinsing Solution (STEMCELL Technologies) in 7–8 mL of culture media.
Problem 7
Neurogenesis in contractile EMLOCs has not been validated. Refer to Stage 3, #19.
Potential solution
Evaluate expression of neuronal biomarker TUJ1 and cardiac marker cTnT via immunofluorescence at day 7 and a later time point (Figures 3D–3H).
Resource availability
Lead contact
Further information and reasonable requests for resources should be directed to and will be fulfilled by the Lead Contact, Dr. Janet L. Paluh (paluhj@sunypoly.edu).
Materials availability
This study applies a unique reagent that is a Hispanic-Latino low passage hiPSC line previously generated and initially characterized with other ethnically-diverse hiPSC lines.2,3 The ethnically diverse hiPSC lines are being made available via WiCell (Madison, Wisconsin).
Acknowledgments
This research was funded by SUNY Polytechnic SEED 917035-21 and CATN2 MIP awards. It used published lines developed and initially characterized through previous grants awarded to the Paluh laboratory for New York State Stem Cell research (NYSTEM) and Spinal Cord Injury research board (NYSCIRB) research. Analysis by scRNAseq was originally performed at the SUNY Buffalo Genomics and Bioinformatics Core . Figures and the graphical abstract were generated by combined use of Keynote and BioRender.
Author contributions
All authors contributed equally to the writing of the STAR protocol. Z.T.O. and M.B.P.-E. composed figures from data and generated graphical images.
Declaration of interests
Application for patent for which J.L.P. and Z.T.O. are co-inventors has been filed by the State University of New York Research Foundation (SUNYRF) with the US Patent Office on the EMLOC technology and detailed methods. U.S. patent filings: 63/311,498 and 63/419,507.
Data and code availability
Single cell RNA-Seq datasets corresponding to the original publication (Olmsted and Paluh1) are available through GEO (GSE194356). The technology for generation of EMLOCs is covered under U.S. provisional patent filing: 63/311,498.
References
- 1.Olmsted Z.T., Paluh J.L. A combined human gastruloid model of cardiogenesis and neurogenesis. iScience. 2022;25:104486. doi: 10.1016/j.isci.2022.104486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang E.A., Tomov M.L., Suhr S.T., Luo J., Olmsted Z.T., Paluh J.L., Cibelli J. Derivation of ethnically diverse human induced pluripotent stem cell lines. Sci. Rep. 2015;5:15234. doi: 10.1038/srep15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomov M.L., Olmsted Z.T., Dogan H., Gongorurler E., Tsompana M., Otu H.H., Buck M., Chang E.A., Cibelli J., Paluh J.L. Distinct and Shared Determinants of cardiomyocyte contractility in multi-lineage competent ethnically diverse human iPSCs. Sci. Rep. 2016;6:37637. doi: 10.1038/srep37637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gouti M., Tsakiridis A., Wymeersch F.J., Huang Y., Kleinjung J., Wilson V., Briscoe J. In vitro generation of neuromesodermal progenitors reveals distinct roles for wnt signaling in the specification of spinal cord and paraxial mesoderm identity. PLoS Biol. 2014;12:e1001937. doi: 10.1371/journal.pbio.1001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olmsted Z.T., Stigliano C., Badri A., Zhang F., Williams A., Koffas M.A.G., Xie Y., Linhardt R.J., Cibelli J., Horner P.J., Paluh J.L. Fabrication of homotypic neural ribbons as a multiplex platform optimized for spinal cord delivery. Sci. Rep. 2020;10:12939. doi: 10.1038/s41598-020-69274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faustino Martins J.M., Fischer C., Urzi A., Vidal R., Kunz S., Ruffault P.L., Kabuss L., Hube I., Gazzerro E., Birchmeier C., et al. Self-organizing 3D human trunk neuromuscular organoids. Cell Stem Cell. 2020;27:172–186. doi: 10.1016/j.stem.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Olmsted Z.T., Paluh J.L. Co-development of central and peripheral neurons with trunk mesendoderm in human elongating multi-lineage organized gastruloids. Nat. Commun. 2021;12:3020. doi: 10.1038/s41467-021-23294-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bel-Vialar S., Itasaki N., Krumlauf R. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development. 2002;129:5103–5115. doi: 10.1242/dev.129.22.5103. [DOI] [PubMed] [Google Scholar]
- 9.Goto H., Kimmey S.C., Row R.H., Matus D.Q., Martin B.L. FGF and canonical Wnt signaling cooperate to induce paraxial mesoderm from tailbud neuromesodermal progenitors through regulation of a two-step epithelial to mesenchymal transition. Development. 2017;144:1412–1424. doi: 10.1242/dev.143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naujok O., Lentes J., Diekmann U., Davenport C., Lenzen S. Cytotoxicity and activation of the Wnt/beta-catenin pathway in mouse embryonic stem cells treated with four GSK3 inhibitors. BMC Res. Notes. 2014;7:273. doi: 10.1186/1756-0500-7-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den Brink S.C., Baillie-Johnson P., Balayo T., Hadjantonakis A.K., Nowotschin S., Turner D.A., Martinez Arias A. Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells. Development. 2014;141:4231–4242. doi: 10.1242/dev.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim K., Ossipova O., Sokol S.Y. Neural crest specification by inhibition of the ROCK/Myosin II pathway. Stem Cell. 2015;33:674–685. doi: 10.1002/stem.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chal J., Pourquié O. Making muscle: skeletal myogenesis in vivo and in vitro. Development. 2017;144:2104–2122. doi: 10.1242/dev.151035. [DOI] [PubMed] [Google Scholar]
- 14.Rossi G., Broguiere N., Miyamoto M., Boni A., Guiet R., Girgin M., Kelly R.G., Kwon C., Lutolf M.P. Capturing cardiogenesis in gastruloids. Cell Stem Cell. 2021;28:230–240.e6. doi: 10.1016/j.stem.2020.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madonna R., de Caterina R. VEGF receptor switching in heart development and disease. Cardiovasc. Res. 2009;84:4–6. doi: 10.1093/cvr/cvp270. [DOI] [PubMed] [Google Scholar]
- 16.Cao N., Liu Z., Chen Z., Wang J., Chen T., Zhao X., Ma Y., Qin L., Kang J., Wei B., et al. Ascorbic acid enhances the cardiac differentiation of induced pluripotent stem cells through promoting the proliferation of cardiac progenitor cells. Cell Res. 2012;22:219–236. doi: 10.1038/cr.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Single cell RNA-Seq datasets corresponding to the original publication (Olmsted and Paluh1) are available through GEO (GSE194356). The technology for generation of EMLOCs is covered under U.S. provisional patent filing: 63/311,498.