Abstract
Background:
Colorectal carcinoma (CRC) represents life-threatening problems worldwide. IQ motif containing GTPase activating protein 1 (IQGAP1) is acting as oncogenesis regulators. RNAi is proposed as promising cancer therapeutics.
Objective:
The objective of this work to explore the consequences of the IQGAP1 silence as a goal for treating CRC using the HCT166 cells as a model for human colon cancer. Methods: RNAi technology was used to design a short specific sequence of RNA (shRNA) to silence the IQGAP1 oncogene. The impact of IQGAP1 silencing on IQGAPs, Ras, IL-8, and TRAIL was investigated. Furthermore, the effect of IQGAP1 silencing on cell viability, proliferation, apoptosis, and invasive capacity was investigated.
Results:
The present results revealed that IQGAP1 shRNA-treated HCT166 cells showed no invasive capacity compared to the control cells. The silencing of IQGAP1 induced remarkable downregulation of IQGAP1, RAS (H&K), IL-8, CXCR1, CXCR2, NF-kB, BCL-2, and apoptosis of HCT166 cells. On the contrary, IQGAP2, IQGAP3, DR4, DR5, CASP-3, and BAX genes were significantly up-regulated.
Conclusion:
The IQGAP1 regulates the expression of IQGAPs, Ras, IL-8 receptors, and the apoptotic network. Therefore, the silence of IQGAP1 is a promising strategy for colon cancer therapy.
Key Words: Colon cancer, shRNA, IQGAP1, Cell Migration, Apoptosis, Metastasis
Introduction
Colorectal cancer (CRC) is one of global health-threatening problem. More than half-million cases are recognized annually. The incorrect lifestyle and xenobiotics exposure are predisposing factors for CRC. However, healthy lifestyle, early detection, screening of individuals at cancer high-risk for CRC, and discovery of novel CRC therapy are important factors to decrease the mortality rate of CRC (Chen et al., 2012 ). The new surgical techniques in combination with radiotherapy-chemotherapy are utilized in the treatment of CRC. Meanwhile, these strategies did not succeed in improving the overall survival rate of patients with CRC (Manceau et al., 2012 and Lugli et al., 2012). Therefore, further studies for discovering novel approaches to curing CRC with improved survival rate are necessary to address this issue.
The oncogenesis, mediated by the malfunction of the signal transducers, proto-oncogenes, tumor suppressor genes, and apoptotic mechanisms (Hanahan and Weinberg (2011). Earlier documents suggested that overexpression of isoleucine glutamine motif containing GTPase-activating proteins (IQGAPs) can stimulate mitogen-activated protein kinase 1 and β-catenin signaling cascades and this facilitates malignant transformation (Bashour et al., 1997). The mechanisms by which the IQGAP1 elaborate CRC progression is not fully assumed. Extensive effort is required to clarify this subject. Studies reported that the connections of IQGAP1 with multiple cellular proteins were shown to regulate cellular functions (Bashour et al., 1997). The upregulation of IQGAP1 was predicted in varieties of humanoid cancer, including female tumor (Dong et al., 2006), pulmonary cancer (Nakamura et al., 2005 and Zhao et al., 2014), pancreatic cancer (Wang et al., 2014 ), hepatic tumors (Chen et al., 2010), CRC (Nabeshima et al., 2002 and Hayashi et al., 2010), gastric cancer (Takemoto et al., 2001 and Walch et al., 2008), esophageal carcinomas (Wang et al., 2014) and thyroid cancer (Liu et al., 2010). However, there is not enough data about the roles of IOGAP1 in CRC. Additional studies are necessary to explore the role of IOGAP1in colon cancer. In this context, Jianyu et al. (2018) demonstrated that IQGAP1 was aberrantly overexpressed in CRC. Furthermore, IQGAP1 integrates many signal transduction pathways which contribute to oncogenesis. In this regard, IQGAP1 actin-binding modulates actin dynamics and the cell cycle program (Jianyu et al. 2018). In addition, interactions of IQGAP1 with extracellular signals affect the intracellular processes that involved cell differentiation, proliferation, cancer transformation (Nabeshima et al., 2002 and Johnson et al., 2009).
Collectively, IQGAP1 plays a vigorous role in cell adhesion, migration, proliferation, angiogenesis, and metastasis. The presence of IQGAP1 in the cell membrane may decrease adherers’ junction function, favoring dissociation and metastasis of the tumor cells (Takemoto et al., 2001). Jianyu et al., (2018) and his coworkers recommended that the knockdown of IQGPA1 attenuate cellular growth and colony formation ability of CRC cells. These effects are mediated through concurrent suppression of extracellular signal-regulated kinase (ERK) 1/2 and β-catenin expression phosphorylation Jianyu et al., (2018). The results of Jianyu and these colleagues are hopeful for novel therapy discovery for treating CRC. Therefore, this work was conducted to launch the role of IQGAP in colon cancer progression and its relation with Ras, and TRAIl families. The expression IQGAP1, IQGAP2, IQGAP3, RAS (H&K), IL-8, CXCR1, CXCR2, NF-kB, BCL-2, and apoptotic genes were investigated. Likewise, the effect of silence of IQGAP1 using the short hairpin RNA (shRNA) as an innovative therapeutic target for curing CRC was investigated. The HCT166 utilized as a surrogate model for colon cancer.
Materials and Methods
IQGAP1 knockdown
IQGAP1 knockdown followed the protocol reported in our previous study with little modifications by Zoheir et al., (2016). The IQGAP1 sequence (NC_000015.10) was obtained from the NCBI gene bank. The oligonucleotides of the short hairpin RNA (shRNA) were annealed and inserted into the BamHI and EcoRI sites of the RNAi-Ready PSIREN-Retroozs Green Vector (BD Biosciences, Clontech, CA). This plasmid encoding variant 1 of the human IQGAP1 was obtained from Santa Cruz Biotechnology Company, CA, USA (Cat. No.sc-35700). Colorectal Carcinoma Cells were then transfected with the vector, and the transfected cells were grown in the continuous presence of 200 µg /ml neomycin (G418).
Cell Cultivation
The Human Colorectal Carcinoma Cells (HCT166) was purchased from American Type Culture Collection (ATCC) and cultivated in RPMI 1640 medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Gibco), 100 µg /mL streptomycin, and 100 U/mL penicillin at 37°C in a humidified 5% CO2 atmosphere.
Cell Line Authentication
The authenticity of Colorectal Carcinoma Cells (HCT166) used in this study, as cell lines which prone to change by cross-contamination or infection. The routine authentication process had 3 steps.
1) cell morphology was assessed (cell size, shape, and performance) of the HCT166 cell line, to confirm the health of these cells before the experiments. The early signs of bacterial and/or fungal contamination were also detected in this way.
2) HCT166 cell passaging was performed in their log growth phase, to ensure the use of healthy cells, by evaluating their proliferation. Growth Curve Analysis was done to determine population doubling times.
3) Mycoplasma contamination was detected using a PCR-based approach for detecting and identifying multiple Mycoplasma strains.
IQGAP1 silencing
HCT166 cells at 70 % confluence were transfected with IQGAP1-siRNA Transfection Reagent (sc-29528; Santa Cruz Biotechnology, CA, USA) according to the manufacturer’s instructions. Following, incubate for 24, 48, or 72 h with siRNA vector. The structure of human IQGAP1 siRNA is shown in (Table 1), the cells were lysed, and total RNA was extracted and analyzed by real-time PCR (q-PCR), however, proteins were determined by western blot techniques, Furthermore, the cell viability and migratory potential of these cells were determined by MTT and cell migration assays.
Table 1.
The Sequence of Human IQGAP1 siRNA and Primers Used in the Study. Note: all sequences are in 5′ → 3′ orientation
| Gene | Sense | Antisense |
|---|---|---|
| IQGAP1 | sc-35701A: CCACAGUGAUCUUGCUGAAtt | UUCAGCAAGAUCACUGUGGtt |
| siRNA (m) | sc-35701B: CCACAGUGAUCUUGCUGAAtt | UUCAGCAAGAUCACUGUGGtt |
| sc-35701 SH | sc-35701B:CCACAAAGAUGAAGUUGUAtt | UACAACUUCAUCUUUGUGGtt |
| IQGAP1 | ATGGATGGGATGAAGCACAGAG | CAGGACAGAGCCATAGTGCG |
| (IQGAP2 | AGATCTTCGGGCGGCTAGG | TAGAGCCATAGCGGGGTCTC |
| IQGAP3 | CAGCCTATGAACGCCTCACA | TATCTGAGGGGCCAATCCCA |
| MRAS | CTGACCCGGTTCTGGACACAG | TTCTGCTTACAGGTGGCTTCC |
| KRAS | ATTGGTGAGGGAGATCCGAC | CTGCATGCACCAAAAACCCC |
| NF-KB | GGGCAGGAAGAGGAGGTTTC | TATGGGCCATCTGTTGGCAG |
| DR4 | CCGCGGCCACACCCAGAAAGT | GTACATGGGAGGCAAGCAAACAAA |
| DR5 | GCGCCCACAAAATACACCGACGAT | GCAGCGCAAGCAGAAAAGGAG |
| CASP-3 | TCTGGTTTTCGGTGGGTGTG | GTCGGCCTCCACTGGTATTT |
| BAX | CTGGATCCAAGACCAGGGTG | CCTTTCCCCTTCCCCCATTC |
| CXCR1 | CCACGCGTCCGCATAAATC | CCAGCAGATGCTCTGACTCC |
| CXCR2 | GGGTACAGTGCTATTCTGCC | GTACGCAGGGTGAATCCGTA |
| p53 | ATTGGCCAGACTGCCTTCC | TGGTGTTGTTGGACAGTGCT |
| BCL-2 | CCTTTGTGGAACTGTACGGC | CCGGCCAACAACATGGAAAG |
| IL-8 | AGTTTTTGAAGAGGGCTGAGA | ACCAAGGCACAGTGGAACAA |
| B actin | TTGCCGACAGGATGCAGAA | GCCGATCCACACGGAGTACT |
Cell proliferation (MTT) assay
HCT166 cells were seeded in 96-well plates and treated with 0, 2.5, or 5µM of IQGAP1-siRNA vector, and incubated for 24, 48, 72, and 144 h. Control cells were treated with PBS. After the incubation, the cells were incubated with 1 mg/mL of MTT reagent, 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, at 37°C for 4 h, after which it was discarded. The formed Formosan crystals were dissolved using 100 mL of DMSO, followed by incubation and shaking. Finally, colorimetric analysis using multiple readers was performed, and the absorbance was measured at 540 nm (Victor3; PerkinElmer, Wellesley, MA, USA).
Flow Cytometer Analysis
This analysis was performed using Annexin V FITC and Propidium Iodide (PI), to measure the rates of apoptosis. HCT166 cells were treated with 0, 2.5, or 5µM of IQGAP1- siRNA for 24 or 48 h. Annexin V-stained flipped phospholipids in HCT166 cell membranes and Propidium Iodide (PI)-stained that damaged DNA was used in the determination of early and late apoptosis, respectively, after treatment compared to control. Annexin V and PI were purchased from (Thermo Fisher Scientific). The apoptotic analysis was dedicated to differentiating between early and late apoptotic cells, as well as necrotic cells. The apoptosis of the treated and untreated cells was analyzed by flow cytometer instrument (Attune® Acoustic Focusing Cytometer, Life Technologies) and analyzed by its software.
In vitro Scratch Assay
The effect of different doses of IQGAP1-siRNA vector on the migration of HCT166 cells was assessed using the in vitro scratch assay described in Nature protocols with slight modifications of Liang group protocol. Briefly, HCT166 cells were treated with the vector and incubated overnight. A “scratch” was then created in a cell monolayer with the help of a sterile 200 µl pipette tip. Debris from the cells was removed by changing the medium and the cells were incubated at 37°C. The images were captured at 0, 24, 48, and 72h to monitor the migration of the cells to close the scratch area.
Photographs were taken using a charge-coupled device camera attached to an inverted phase-contrast microscope (Olympus, Tokyo, Japan) at a power of 10X. For statistical analyses, the images were taken from three separate “scratch areas” under the same magnification and the images were both compared with controls. Furthermore, a gap distance between the “scratch area” was measured and graphs were plotted for cell migration. All assays were performed in triplicate on at least two separate occasions.
Quantitative qPCR
The procedure for the reverse transcription-polymerase chain reaction (qPCR) was essential as usual. The forward and reverse primers corresponding to human genes of our study were shown in (Table 1). β-Actin mRNA was amplified as a housekeeping gene. All RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Total RNA was reverse transcribed in 20 Ul reaction systems using Superscript First-Strand Synthesis Kit for RT-PCR (Invitrogen) under conditions described by the supplier.
Western Blot Analysis
All primary antibodies and peroxidase-conjugated secondary antibodies used for western blotting were purchased from Santa Cruz Biotechnology, CA, USA. While cleaved caspase-3 (cat # 9661) was purchased from (Cell Signaling Technology, Inc., UK ). In this experiment, all primary antibodies were diluted in the ratio of (1:500) and the secondary antibodies were diluted in the ratio of (1:5000).
Protein concentrations of whole-cell lysates were determined using the Protein Assay Kit. Supernatant proteins, 50 µg from each sample, were separated by SDS-10% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Polyvinylidene Difluoride (PVDF) membrane (Bio-Rad, Hercules, CA, USA ) by standard procedures. The membranes were hybridized with primary antibodies followed by incubation with appropriate secondary antibodies. The antibody-bound proteins were visualized by treatment with the chemiluminescence detection reagent (Amersham Biosciences, UK ) according to the manufacturer’s instructions, followed by exposure to X-ray film (Kodak X-Omat). The same membranes were re-probed with the anti-β actin antibody. And it was used as an internal control for protein loading.
Statistical Analysis
All assays were repeated in three independent experiments, and only representative data are presented. The comparisons between treated and untreated cells were made using a two-tailed Student’s t-test, and values of P < 0.05 and P < 0.01 were considered statistically significant and highly significant respectively. Statistical calculations were done by using a statistical computer program: SPSS (Statistical Package for the Social Science; SPSS Inc., Chicago, IL, USA).
Results
Suppression of IQGAP1 Expression and HCT166 Growth
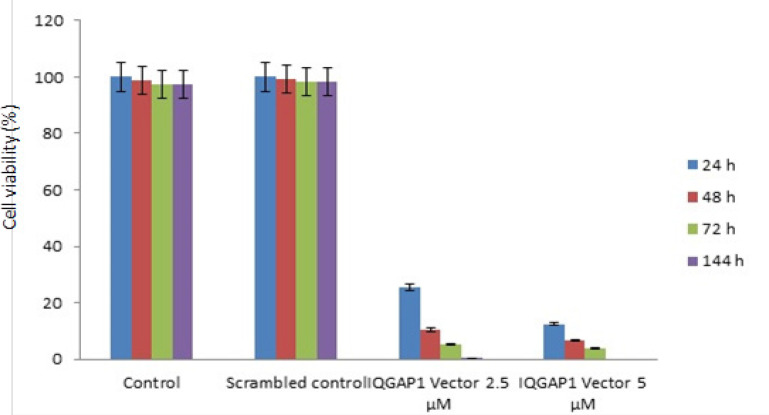
To test the effect of IQGAP1 knockdown on HCT166 cell growth, MTT and flow cytometer assays were performed and cell proliferation curves were generated. As shown in (Figure 1), cell proliferation was inhibited in IQGAP1 knockdown HCT166 cells. The data presented in (Figure 1) show the percentage of HCT166 cell viability at 24 h and 48 h. These results show that the treatment of HCT166 cells with 2.5 and 5 µM of the siRNA significantly (P < 0.01) decreased cell viability to around 80% and 90% respectively, compared with that in the controls. The treatment of cells with 2.5 µM and 5 µM of siRNA for 72 and 144 h led to a significant (P < 0.01) decrease in cell viability, to approximately 95% and 100%, respectively. A dose-dependent decrease in cell viability of HCT166 cells was observed after siRNA treatment.
Figure 1.
Showing Cell Proliferation by MTT Assay. HCT166 cells were incubated with 5μM of IQGAP1 shRNA vector for 24 48, 72 and 72 h
IQGAP1 Knockdown and HCT166 Apoptosis
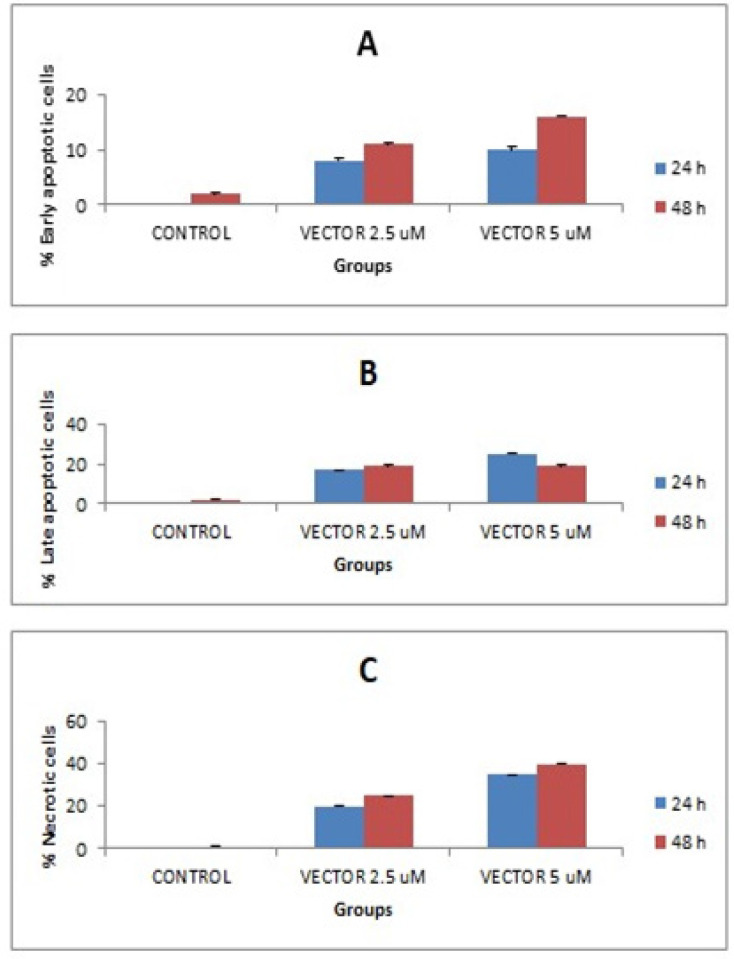
The underlying mechanisms of IQGAP1-siRNA knockdown on HCT166 cells were determined using flow cytometry. In the present study, early apoptotic, late apoptotic, and necrotic cells were nominated to be stained with Annexin V, Annexin V/PI, and PI, respectively. The present data indicated that there is an increased number of early apoptotic cells after 24 h of incubation with 2.5 or 5 μM, while late apoptosis was observed after 48 h of incubation with 2.5 and 5 μM of IQGAP1-siRNA vector (P < 0.01, compared with the control). Late Apoptosis at 48 h and 72 h of incubation was associated with an increase in necrotic cell numbers relative to that in the control see Figure 2 A-C. The number of these cells was shown to be higher than that of early apoptotic HCT166 cells after 24 h of incubation with 2.5 and 5 μM (P < 0.05).
Figure 2.
Healthy Population of HCT166 Cells was Counted Using Flow Cytometry up on Treatment with 5μM of IQGAP1 shRNA Vector for 24 and 48 h. The experiment was repeated 3 independent times (n=3).
Suppression of IQGAP1 and HCT166 the invasive capacity
The current results indicated that the scratch area of IQGAP1-siRNA treated HCT166 cells remained wider, in comparison with the control, and the control cells were able to invade the formed gap earlier than IQGAP1- siRNA-treated HCT166 cells. At 24 h, 48 h, and 72 h, the remaining gap area was 95.33%, 92.23%, and 90.07%, respectively, of the original gap width, when HCT166 cells were treated with 2.5 μM of IQGAP1- siRNA. The control gap areas were 36.92% and 16.92% of the original gap width, at 24 h and 48 h respectively while there was no gap observed after 72 h of incubation, Figure 3 depicts these results. When HCT166 cells were treated with 5 μM of siRNA, the gaps were 98.18%, 96.1%, and 95.22% of the original gap width, at 24 h, 48 h, and 72 h, respectively. The present data suggested that the suppression of IQGAP1 leads to a decrease in cellular migratory potential. That indicates an important role of IQGAP1 in the metastasis of CRC.
Figure 3.
Wound Healing Assay of IQGAP1siRNA on HCT166 Cells. (After 48 h)
IQGAP1 Knockdown drives IQGAPs/ RAS/ TRAIL/ Apoptotic members in Genetic and Proteomic levels
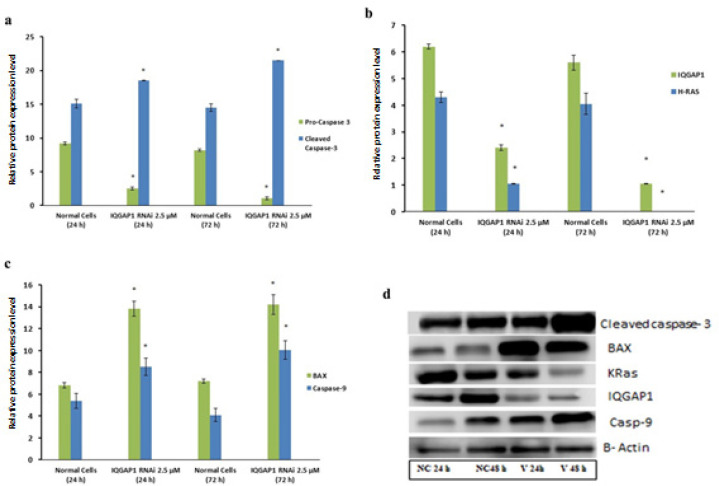
The current study used QRT-PCR and western blot assays to measure mRNA and protein expression levels, respectively, of IQGAP1, IQGAP2, IQGAP3, HRAS, KRAS, DR4, DR5, CXCR1, CXCR2 and, IL-8, CASP-3 (pro- and cleaved), CASP- 9, NF-KB, P53, BAX and BCL-2 in HCT166 cells. The examined knockdown efficiency of the IQGAP1 siRNA vector and the treated HCT166 cells showed significantly lower IQGAP1 mRNA and protein levels than the untreated cells, figures 4 displays these data. Moreover, the IQGAP1 silencing significantly decreased IQGAP1 and IQGAP3 levels and increased IQGAP2, DR4, DR5 levels, and pro- caspase-3. The induction of IQGAP2 expression was associated with the downregulation of HRAS, KRAS, and B-cell leukemia/lymphoma 2 (BCL-2), while IQGAP1 silencing led to the upregulation of cleaved caspase-3 (CASP-3), NF-KB, P53, CASP-9, and Bcl2-associated X (BAX) mRNA.
Figure 4.
Expressions of mRNA for Definite Genes after HCT166 Cell Treatment with IQGAP1 shRNA Vector. (a-f) * indicated significantly differences between means at P <0.05 and error bars represents standard error of mean (SEM). Means comparisons were performed by using Duncan’s multiple range test
Gene expression data were confirmed through western bot analysis where the protein expression of both IQGAP1 KRas and pro-caspase 3 was decreased while cleaved Casp-3, Casp-9, and BAX were elevated in treated cells when compared to control cells, Figure 5 indicates the western bot analysis.
Figure 5.
Protein Expression of IQGAP1, KRAS, BAX, casp-9 and cleaved casp-3 Using Western Blot Analysis
Discussion
Nowadays, RNAs are used as medications to cure hereditary diseases, autoimmune diseases, cancer, infectious diseases, and other diseases, particularly; mRNA is exploited instead of protein medication (Van et al., 2019). Therefore, this study was conducted to assume the possible role of IQGAP1 silence in colon cancer treatment. Particularly, IQGAP1 might regulate oncogenic properties, invasion, and metastasis of cancer cells. RNAi a promising cancer therapy. Importantly, downregulation of IQGAP1 expression was observed in MIR-124-restoration cells with simultaneous reduction of phosphorylated-ERK1/2 and β-catenin (Jianyu et al., 2018).
The present results revealed that the silencing of IQGAP1 induced remarkable downregulation of IQGAP1, RAS (H&K), IL-8, CXCR1, CXCR2, NF-kB, BCL-2, and apoptosis of HCT166 cells. On the contrary, IQGAP2, IQGAP3, DR4, DR5, CASP-3, and BAX genes were significantly up-regulated. These results are in agreement with, Jianyu et al., (2018) who described a potential mechanism underlying the MIR-124/IQGAP1 link in CRC progression. Moreover, the same authors demonstrated that the silencing of MIR-124 may depress IQGAP1 expression, leading to increased activity of ERK1/2 and β-catenin signaling (Jianyu et al., 2018). In addition, numerous immune histochemical studies have demonstrated that IQGAP1 was overexpressed in several forms of cancer and an aberrant membrane accumulation was observed. It was observed that IQGAP1 silencing inhibited β-catenin-mediated proliferation in SW1116 cells (Fukata et al., 1999). Similarly, another study demonstrated that there is a negative regulatory feedback loop between β-catenin and IQGAP1 on the proliferation of CRC (Watanabe et al., 2004). The current findings supported the role of IQGAP1 in colon carcinoma invasion. Likewise, it was stated that IQGAP1 was upregulated in tumor cells compared to healthy cells (Fukata et al., 1999). In this concern, Johnson and Henderson approved that, IQGAP1 is a crucial aspect of oncogenesis. The overexpression of IQGAP1 in tumors upregulates the signaling pathways involved in cell transformation, proliferation, and metastasis (Johnson et al., 2009).
Carcinogenesis is a multifactorial procedure and accompanied by a mutation in many tumor-associated genes. Furthermore, overexpression of the cellular proto-oncogenes is connected to malignant transformation (Johnson , and Henderson 2012). Herein, CRC progression might have associated with abnormalities in oncogenes and tumor suppressor genes. Particularly, the IQGAP1 overexpression elicits biological roles in human CRC. The finding of the present study supports this hypothesis. As well, the expression of IQGAP1 was up-regulated in CRC tissues compared with normal tissues (Jianyu et al., 2018). In this context, the suppression of IQGAP1 expression RNAi represents a gorgeous opportunity for CRC cure. The present results showed that the IQGAP1 protein and mRNA expression levels were reduced significantly after stable transfection with IQGAP1- shRNA. Moreover, MTT assays demonstrated that IQGAP1 silencing led to a significant decrease in cell proliferation. These findings are in alignment with the previous investigations that stated that IQGAP1 knockdown could attenuate cell growth (Li et al., 2005).
For additional clarification of IQGAP1 role in CRC, the cell migration and invasion assays were achieved to assess the belongings of IQGAP1 silencing on the migratory and invasive potential of HCT166 cells. The existing data displayed that reduced IQGAP1 expression repressed cell migration and invasion ability. Additionally, there is an inverse correlation of IQGAP1 expression levels with oncogenic properties of colon cancer cell lines. IQGAP1 expression affected apoptosis. The current data are consistent with numerous former investigations revealed that IQGAP1 shows central roles in the migration, invasion, and metastasis of different types of tumor cells (Li et al., 2005).
The conversion of epithelial-into-mesenchymal is a critical event in oncogenesis. This change is characterized by loss of epithelial cell adhesion and the cells are acquiring more migratory and invasive features. Subsequently, the cancer cells leaving their original site to invade the surrounding tissue, and metastasized (Kölbl et al., 2016). Herein, the understanding of epithelial-into-mesenchymal transformation may lead to the development of novel interventions to combat cancer metastasis. This transformation process is associated with the altered expression of many proteins (Kölbl et al., 2016). The current investigations demonstrated that IQGAP1 knockdown reduced migration (invasive) capacity for CRC cells. Therefore, IQGAP1 knockdown could reduce the migration and invasion of tumor cells, herein, prevent cancer metastasis. These observations are in harmony with several studies that reported the role of IQGAP1 in the modulation of cell adhesion (Kölbl et al., 2016). Moreover, the blocking of IQGAP1 expression by RNAi reduces the production of E-cadherin that responsible for cell-cell adhesion to different types of cancers (Liu et al., 2010). Collectively, present information specified that IQGAP1 silencing resulted in inhibition of invasion and metastasis.
The standard equilibrium between oncogenes like Ras and suppressor genes like, CXCR1, CXCR2 DR4, DR5, CASP-3, and BAX is essential for maintaining normal cellular proliferation (Liu et al., 2010 and Zoheir 2016). In this work, the crosstalk between IQGAP1 and these genes, this supporting our observation of the invasive capacity of colon cancer cells. These findings are aligning with the prior findings of IQGAP1 in other cancer types (Liu et al., 2010). Additionally, IQGAP1controls diverse signal transduction involved in malignant changes and metastasis. It is cooperating with Rac1, E-cadherin, β-catenin, RAF, MEK1/2, and ERK1/2 MAPK cascade kinases, all of these cascades own tumorigenic roles (Wang et al., 2013). As well, previous studies highlighted the reciprocal expression pattern of IQGAP1/IQGAP2 in cancers (Liu et al., 2010 and Colin et al., 2010).
Overall, the present findings suggest that IQGAP1 is positively associated with the proliferative and metastatic abilities of colon cancer cells. Therefore, the suppression of IQGAP1-mediated ERK activation is a possible route via which IQGAP2 restricts the oncogenic properties of colon cancer cells. The present study highlights the candidature of IQGAP1 as a novel strategy to inhibit oncogenic signaling CRC. The findings of this study prove the benefit application of IQGAP1 as a biomarker of colon cancer and as a target for CRC therapy. However, RNA-based therapeutics still have restrictions such as degradation by ribonucleases, immunogenicity. These limitations could be overcome by chemical modification of RNA and fabrication of smart delivery cargoes. Furthermore, studies are to address this issue using the animal model for CRC.
In conclusion, this study confirmed the IQGAP1 expression is upregulated in colorectal tumors. Moreover, the ratio of IQGAP1/IQGAP2, Ras, TRAIL families, and caspases could elicit a vital role in colon cancer. Therefore, IQGAP1 might be used as a novel tumor biomarker for clinical diagnostics of colon cancer. The silencing IQGAP1 shRNA reduces the cancerous properties of the HCT166 cell line. Herein, IQGAP1 might utilize a candidate for the development of colon anti-cancer therapeutics. Furthermore, studies are to address this issue using the animal model for CRC.
Author Contribution Statement
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript; KZ designed this study. KM, AA, MA, KM and AD participated in the conduct of the study. KM, AD, MA, KM., and K.Z conducted the experiments, KZ supplied critical reagents, KZ and AA wrote the manuscript.
Acknowledgments
This study was funded by National Research Centre Supporting Project, National Research Centre, Dokki, Egypt.
Declarations/ Consent for Publication
Not Applicable” in this section.
Conflicts of interest/Competing interests
The authors declared no potential conflicts of interest
Compliance with Ethical Standards
The experimental protocol used in the study was approved (NRCE-CBD1032019) by the Animal Care and Use Committee of National Research Centre, Egypt.
Availability of data and material
Data is available for all
References
- Bashour AM, Fullerton AT, Hart MJ, Bloom GS. IQGAP1, a Rac- and Cdc42-binding protein, directly binds and cross-links microfilaments. J Cell Biol. 1997;137:1555–1566. doi: 10.1083/jcb.137.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zhu HH, Zhou LF, Wu SS, Wang J, Chen Z. IQGAP1 is overexpressed in hepatocellular carcinoma and promotes cell proliferation by Akt activation. Exp Mol Med. 2010;42:477–483. doi: 10.3858/emm.2010.42.7.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WQ, Zheng RS, Zhang SW, Li N, Zhao P, Li GL, Wu LY, He J. Report of incidence and mortality in china cancer registries, 2008. Chin J Cancer Res. 2012;24:171–80. doi: 10.1007/s11670-012-0171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin D, Hema K, Dmitri V, Zhigang L, Robert D, David B. Valentina A IQGAP1 and IQGAP2 are reciprocally altered in hepatocellular carcinoma. BMC Gastroenterol. 2010;10:125. doi: 10.1186/1471-230X-10-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P, Nabeshima K, Nishimura N. Overexpression and diffuse expression pattern of IQGAP1 at invasion fronts are independent prognostic parameters in ovarian carcinomas. Cancer Lett. 2006;243:120–12. doi: 10.1016/j.canlet.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Fukata M, Kuroda S, Nakagawa M. Cdc42 and Rac1 regulate the interaction of IQGAP1 with beta-catenin. J Biol Chem. 1999;274:26044–50. doi: 10.1074/jbc.274.37.26044. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Nabeshima K, Aoki M. Overexpression of IQGAP1 in advanced colorectal cancer correlates with a poor prognosis-critical role in tumor invasion. Int J Cancer. 2010;126:2563–74. doi: 10.1002/ijc.24987. [DOI] [PubMed] [Google Scholar]
- Jianyu F, Wenjing Z, Yanting W, Ping W, Qiang G, Yu Z. miR 124 inhibits cell growth through targeting IQGAP1 in colorectal cancer. Mole Med Rep. 2018:5270–5278. doi: 10.3892/mmr.2018.9518. [DOI] [PubMed] [Google Scholar]
- Johnson M, Sharma M, Henderson BR. IQGAP1 regulation and roles in cancer. Cell Signal. 2009;21:1471–8. doi: 10.1016/j.cellsig.2009.02.023. [DOI] [PubMed] [Google Scholar]
- Johnson MA, Henderson BR. The scaffolding protein IQGAP1 co-localizes with actin at the cytoplasmic face of the nuclear envelope: Implications for cytoskeletal regulation. Bio architecture. 2012:138–142. doi: 10.4161/bioa.21182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölbl AC, Jeschke U, Andergassen U. The significance of epithelial-to-mesenchymal transition for circulating tumor cells. Int J Mol Sci. 2016;17:1308. doi: 10.3390/ijms17081308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li F, Li-Ling J, Wang X, Xu Z, Sun K. STK15 gene overexpression, centrosomal amplification, and chromosomal instability in the absence of STK15 mutations in laryngeal carcinoma. Cancer Invest. 2005;8:660–664. doi: 10.1080/07357900500359836. [DOI] [PubMed] [Google Scholar]
- Liu Z, Liu D, Bojdani E, El-Naggar A, Vasko V, Xing M. 2010) IQGAP1 plays an important role in the invasiveness of thyroid cancer. Clin Cancer Res. 16:6009–18. doi: 10.1158/1078-0432.CCR-10-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli A, Karamitopoulou E, Zlobec I. Tumour budding: a promising parameter in colorectal cancer. Br J Cancer. 2012;106:1713–7. doi: 10.1038/bjc.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau G, Karoui M, Werner A, Mortensen NJ, Hannoun L. Comparative outcomes of rectal cancer surgery between elderly and nonelderly patients: A systematic review. Lancet Oncol. 2012;13:525–53. doi: 10.1016/S1470-2045(12)70378-9. [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Shimao Y, Inoue T, Koono M. Immunohistochemical analysis of IQGAP1 expression in human colorectal carcinomas: its overexpression in carcinomas and association with invasion fronts. Cancer Lett. 2002;1:101–9. doi: 10.1016/s0304-3835(01)00742-x. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Fujita K, Nakagawa H. Expression pattern of the scaffold protein IQGAP1 in lung cancer. Oncol Rep. 2005;3:427–31. [PubMed] [Google Scholar]
- Takemoto H, Doki Y, Shiozaki H. Localization of IQGAP1 is inversely correlated with intercellular adhesion mediated by E-cadherin in gastric cancers. Int J Cancer. 2001;6:783–788. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1121>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Van Hoecke L, Roose K. How mRNA therapeutics are entering the monoclonal antibody field. J Transl Med. 2019;1:54. doi: 10.1186/s12967-019-1804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch A, Seidl S, Hermannstädter C, Rauser S, Deplazes J, et al. Combined analysis of Rac1, IQGAP1, Tiam1 and E-cadherin expression in gastric cancer. Mod Pathol. 2008;5:544–52. doi: 10.1038/modpathol.2008.3. [DOI] [PubMed] [Google Scholar]
- Wang XX, Li XZ, Zhai LQ, Liu ZR, Chen XJ, Pei Y. Overexpression of IQGAP1 in human pancreatic cancer. Hepatobiliary Pancreas. Dis Int. 2013;5:540–54. doi: 10.1016/s1499-3872(13)60085-5. [DOI] [PubMed] [Google Scholar]
- Wang XX, Wang K, Li XZ. Targeted knockdown of IQGAP1 inhibits the progression of esophageal squamous cell carcinoma in vitro and in vivo. PLoS One. 2014;5:96501. doi: 10.1371/journal.pone.0096501. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Watanabe T, Wang S, Noritake J, Sato K, Fukata M, Takefuji M. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev Cell. 2004;7:871–83. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Zhao H, Xie C, Lin X. C. 2014) expression of IQ-domain GTPase-activating protein 1 (IQGAP1) and Disheveled (Dvl) is correlated with poor prognosis in non-small cell lung cancer. PloS One. 12:e113713. doi: 10.1371/journal.pone.0113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoheir KM, Abd-Rabou AA, Harisa GI, Kumar A, Ahmad SF, et al. IQGAP1 Gene Silencing Induces Apoptosis and Decreases the Invasive Capacity of Human Hepatocellular Carcinoma Cells. Tumour Biol. 2016: 27488117. doi: 10.1007/s13277-016-5283-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data is available for all