Abstract
Insufficient infiltration of T cells severely compromises the antitumor efficacy of adoptive cell therapy (ACT) against solid tumors. Here, we present a facile immune cell surface engineering strategy aiming to substantially enhance the anti-tumor efficacy of Th9-mediated ACT by rapidly identifying tumor-specific binding ligands and improving the infiltration of infused cells into solid tumors. Non-genetic decoration of Th9 cells with tumor-targeting peptide screened from phage display not only allowed precise targeted ACT against highly heterogeneous solid tumors but also substantially enhanced infiltration of CD8+ T cells, which led to improved antitumor outcomes. Mechanistically, infusion of Th9 cells modified with tumor-specific binding ligands facilitated the enhanced distribution of tumor-killing cells and remodeled the immunosuppressive microenvironment of solid tumors via IL-9 mediated immunomodulation. Overall, we presented a simple, cost-effective, and cell-friendly strategy to enhance the efficacy of ACT against solid tumors with the potential to complement the current ACT.
Keywords: Adoptive cell therapy (ACT), Phage display, Cell surface engineering, Th9 cell, Solid tumor
Graphical abstract
Highlights
-
•
Rapid identifying of osteosarcoma targeting ligand by phage display technology.
-
•
Improved the anti-tumor efficacy of ACT by decorating the surface of T-cell membranes with tumor-specific binding ligands.
-
•
Th9 cells possess the potential to remodel the tumor microenvironment via the secretion of IL-9.
1. Introduction
Adoptive cell therapy (ACT) has emerged as one of the most powerful cancer immunotherapy strategies due to its specificity, and persistence ([1,2]). By transfusing autologous or allogenic immune cells to the cancer-bearing patients after being expanded in vitro, ACT is capable of inducing robust anti-tumor effects to effectively eliminate cancer cells. As such, CD19 targeting chimeric antigen receptor (CAR)-T cells has been approved by the U.S. Food and Drug Administration (FDA) in 2017 due to their extraordinary clinical responses against B cell hematologic malignancies ([3,4]). Besides, other types of immune cells including tumor-infiltrating lymphocytes (TILs) and genetically engineered T cell receptor (TCR)-T cells have also been extensively explored for their clinical application in melanoma, lung cancer, and synovial sarcoma ([[5], [6], [7]]). Although ACT has shown impressive successes in treating hematologic malignancies due to its easy access to tumor cells expressing universal tumor-specific antigens (TSA) in the bloodstream, its translation to solid tumors has been largely limited as multiple clinical trials showed unsatisfactory therapeutic efficacy ([[8], [9], [10]]).
Multiple obstacles causing poor antitumor efficacy of ACT therapy against solid tumors have been identified as solid tumors pose unusual challenges to such therapy due to their unique physiological characteristics: 1) physical barrier, mainly produced by stromal and extracellular matrix, together with mismatched chemokine receptor-chemokine axis as well as distorted blood vascular in solid tumors ([[10], [11], [12]]); 2) lack of common tumor associated antigens (TAA) owing to tumor heterogeneity ([10,13]); 3) highly immunosuppressive and hostile microenvironment for T cell survival and activation ([10,14]). Such complexity of the solid tumor inevitably led to insufficient infiltration of transferred immune cells to the targeted tumor sites and compromised therapeutic effects, as the successful arrival of transfused immune cells at tumor sites is the prerequisite for tumoricidal effects. Numerous strategies have been developed to augment ACT infiltration to improve the efficacy of ACT-based solid tumor therapy. For instance, a combination of ACT and material-based photothermal therapy has been explored to improve the infiltration of transferred cells since the physical barrier and tumor cells in solid tumors are directly destroyed after being heated ([15]). Another study by Craddock et al. aimed to repair the mismatched chemokine receptor-chemokine axis by genetic engineering of T cells or administration of oncolytic viruses armed with chemokines ([[16], [17], [18], [19]]). However, these strategies are limited by various side effects, such as uncertain safety concerns raised by viral transduction or oncolytic viruses, high-cost delivery systems as well as unideal efficacy ([20]). Thus, a simple, cost-effective, and safe strategy to improve the infiltration of adoptively transferred cells is necessary.
Recently, Th9 cell has been identified as a new subset of CD4+ T cells capable of eradicating advanced tumors, mainly depending on interleukin-9 (IL-9) and interleukin-21 (IL-21) secretion ([[21], [22], [23]]). Compared to conventional CD4+ Th subsets, including Th1, Th2, and Th17 cells, Th9 cells represent a unique subpopulation, which is less exhausted, highly cytolytic, as well as hyperproliferative, therefore endowed these cells with superior immunotherapeutic potential ([24]). Moreover, in comparison to CD8+ T cells, Th9 cells are promising potential candidates in treating so-called “cold tumors”, such as osteosarcoma (OS), pancreatic adenocarcinoma (PDAC), and prostate cancer (PCa), due to their ability to recruit/activate other immune cells, as well as their durable phenotype in vivo ([23]). However, the applications of Th9 cells in conjugation with other immunotherapeutic approaches have rarely been reported. Thus, the combination of Th9 cells with other cancer treatment modalities could be critical to enhancing their therapeutic efficacy in solid tumors.
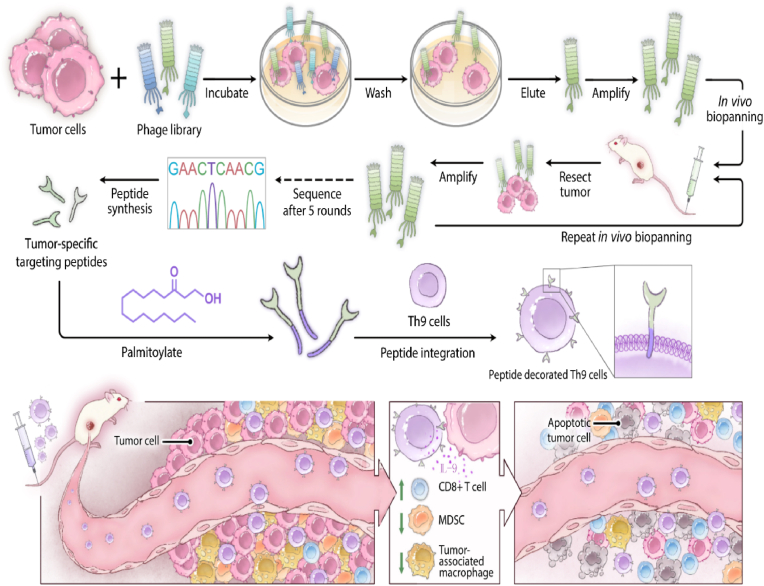
In this work, we aimed to substantially enhance the anti-tumor efficacy of Th9-mediated ACT by improving the infiltration of infused cells into solid tumors via a facile cell surface engineering strategy. Specifically, Th9 cells were non-covalently decorated with an osteosarcoma-targeting peptide screened by phage display biopanning technique. The tumor-targeting peptide was tagged onto the Th9 cell membrane with a palmitic acid (PA) tail via its hydrophobic interaction with the lipid bilayer (Fig. 1). Decoration of Th9 cells with the targeting peptide not only maintained its distribution on the cell surface without disrupting the physiological function but also led to a targeted accumulation of Th9 cells to osteosarcoma cells both in vitro and in vivo. Accordingly, infusion of the peptide decorated Th9 cells substantially improved their infiltration into osteosarcoma, which further resulted in greater antitumor efficacy against osteosarcoma. Mechanistically, peptide decoration of Th9 cells led to a dramatic increase of intratumor CD8+ T cells while a substantial decrease of myeloid-derived suppressor cell (MDSC) and tumor-associated macrophage (TAM), which together improved the antitumor efficacy of Th9 in the context of osteosarcoma management (Fig. 1). Collectively, we presented a simple, cost-effective cell surface engineering strategy to improve the infiltration of adoptively transferred cells into solid tumors in response to their intrinsic physical barrier and hostile immunosuppression microenvironment. Due to its broad applicability to a variety of cell types including TIL, TCR-T, and CAR-T, we speculate that this strategy is an important complement to current immune therapy to help further improve their efficacy against solid tumor management.
Fig. 1.
Schematic illustration of the working strategy. Phage display biopanning technology screens the tumor-specific binding peptides via 1 round of in vitro biopanning and 5 rounds of in vivo biopanning. Then, the identified tumor-specific peptide is anchored to the Th9 cells membrane by PA, a hydrophobic tail synthesized at the C-terminus. The transfusion of the tumor-specific peptide decorated Th9 cells further improves the infiltration of CD8+ T cells and decreases the infiltration of MDSC and TAM via the secretion of IL-9 to inhibit tumor development.
2. Results
2.1. Screen and verification of osteosarcoma targeted peptides by phage display
Current target therapies for osteosarcoma were unsatisfactory due to genomic alterations and few recurrent mutations of osteosarcoma ([25]). To tackle this problem, we first identified osteosarcoma-targeting peptides by a combination of 1 round of in vitro and 5 rounds of in vivo phage display biopanning strategies (Fig. 2A). The output phages from in vivo round 3 were sequenced and the sequencing data revealed 8 potential high-binding peptides from total 26 clones (Fig. 2B). Among all 8 candidates, 3 most frequent peptides: QYWNIQNGPYGG (8/26), QMGFMTSPLHSV (6/26), and ASEWQHRYMFEF (6/26) (termed Peptide 1, Peptide 2, and Peptide 3, respectively) were selected as potential targeting peptides for K7M2 cells (Fig. 2C). These targeting peptides were synthesized with the addition of four lysine amino acids (KKKK) as a linker to the C-terminal to allow further labeling and modification. The mass spectrum and chromatogram of the peptides confirmed the successful synthesis of these peptides (Fig. S1A and S1B).
Fig. 2.
Selection of tumor-targeted peptides via phage display screening. (A) Schematic description of the evolutionary selection of osteosarcoma K7M2 cells specific peptides by a combination of in vitro and in vivo phage display biopanning against K7M2 cells and tumor tissues. (B and C) Summary of peptide sequences (B) and frequencies (C) in each round of phage display against K7M2 tumors.
To screen the peptide with the highest targeting potency against osteosarcoma, the binding ability between the candidate peptides and tumor cells was tested both in vitro and in vivo. All peptides, including Peptide 1, 2, 3, and the control peptide (Peptide C), were labeled with Cy5.5 dyes to visualize their presence after being used against K7M2 cells. As revealed in Fig. 3A, only the Peptide 1 group showed a strong Cy5.5 fluorescence signal while other groups only showed dim background. The mean fluorescence intensity (MFI) of Peptide 1 (35.65) was 6.92 and 14.32 fold stronger than that of Peptide 2 (5.51) and Peptide 3 (2.49), respectively (Fig. 3B). Next, we co-cultured Peptide 1-Cy5.5 with mouse osteosarcoma cell K7M2, mouse Lewis lung carcinoma cell LLC, and mouse embryonic fibroblasts cell 3T3 respectively. Similarly, we also found that Peptide 1 was only specifically bound to K7M2 but not to the other two cell lines, indicating that Peptide 1 possessed a unique affinity to K7M2 cells (Fig. 3C and D).
Fig. 3.
In vitro confocal microscopy and in vivo fluorescence imaging to test the affinity and specificity of candidate peptides. (A) Confocal microscopy to determine the affinity of Cy5.5-peptides with Alexa Fluor 647-phalloidin-labeled K7M2 cells at the concentration of 10 μM for 30 min. Scale bar, 50 μm. (B) Fluorescence intensity of K7M2 cells stained with Cy5.5-peptides. (C) Confocal microscopy to determine the specificity of P1. Cy5.5-P1 was incubated with Alexa Fluor 647-phalloidin-labeled K7M2 cells, 3T3 cells, and LLC cells respectively at the concentration of 10 μM for 30 min. Scale bar, 50 μm. (D) Fluorescence intensity of Cy5.5-P1 incubated with K7M2 cells, 3T3 cells, and LLC cells respectively. (E)In vivo living imaging detected the accumulation of Cy5.5-peptides in K7M2 tumors 24 h after intravenous injection. (F) Fluorescence intensity of saline, Cy5.5, Cy5.5-P1, Cy5.5-P2, and Cy5.5-P3 in the K7M2 tumor site. Data were analyzed by one-way ANOVA test. n = 3 samples per group; mean ± SEM; ns indicates no significant difference,* indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001, **** indicates p < 0.0001.
Then, the in vivo targeting ability of candidate peptides was assessed using a mouse osteosarcoma model bearing K7M2 tumor. Peptides-Cy5.5 and Cy5.5 were intravenously injected into the mouse and the distribution of injected peptides in different organs and their accumulation in tumors were monitored by IVIS Spectrum Animal Imaging System. As shown in Fig. 3E, the signal of Cy5.5 was detected in ears, liver, feet, and tumors. However, the accumulation of Peptide 2 (35.90) and Peptide 3 (34.20) in tumors was less than that of Cy5.5 (46.97), while the accumulation of Peptide 1 (82.67) in tumors was 1.76-fold higher than that of Cy5.5 (Fig. 3F). Collectively, Peptide 1 demonstrated excellent targeting capability to K7M2 cells both in vitro and in vivo, thus was chosen as the K7M2 targeting peptide (KTP) to be decorated on selected immune cells to facilitate infused cell infiltration into osteosarcoma.
2.2. Th9 decoration with osteosarcoma-specific peptide via palmitic acid tail
Poor T cell trafficking into tumor tissue represents one of the major obstacles to broadening the application of ACT for more solid tumor types. One of the current approaches focuses on the genetic engineering of T cells to overcome the mismatched chemokine receptor-chemokine axis and physical barriers ([10]). However, ununiformed expression of engineered proteins on account of the heterogeneous expression of their epitopes, labor-intensive manipulations, and permanent and unpredictable physiological change to T cells restrict the application of these strategies ([26,27]). Thus, novel approaches capable of quickly modifying T cell with tumor-specific ligands without complicated cell manipulation is highly desirable to promote T-cell infiltration into tumor beds. Here, we present a facile cell surface engineering technique to effectively label T cells with desired ligands leveraging the cell membrane anchoring feature of palmitic acid (PA) ([28,29]). To realize such a purpose, PA was grafted to the linker (KKKK) of the KTP via EDC/NHS chemistry, then FITC was also introduced onto the peptide by reacting with ε-NH2 of the fourth lysine residue at the C-terminus for tracking and imaging (Fig. S2). To validate the successful decoration of immune cells with targeting peptides, Th9 cells were incubated with PA-KTP-FITC or KTP-FITC for 10 min at 37 °C to allow them to anchor onto the cell membrane (Fig. 4A). As shown in the flow cytometry spectrum, the FITC fluoresce intensity on Th9 cells gradually increased as the concentration of PA-KTP-FITC and KTP-FITC increased (Fig. 4B). Specifically, when the concentration of peptides went up to 0.5 mg/ml, nearly all Th9 cells (99.22%) were coated by KTP in the PA-KTP-FITC group, but only 50.02% of FITC positive Th9 cells in the KTP-FITC group (Fig. S3A), indicating PA facilitated uniform, dose dependent coating of KTP onto Th9 cell membranes. Besides, when the concentration of KTP-PA was kept below 0.5 mg/mL, we did not find any significant cytotoxicity caused by such cell surface modification at various culture times (Fig. 4C). Moving forward, the feeding concentration of PA-KTP was set at 0.1 mg/ml. To visualize the KTP coating of the cell membrane under the selected concentration, Th9 cells were observed by confocal microscopy after PA-KTP-FITC or KTP-FITC incubation. As evidenced by confocal images, PA-KTP-FITC colocalized with the Th9 cell membrane labeled by PKH26, while no such colocalization was observed when Th9 cells were incubated with KTP-FITC, indicating that KTP could be anchored to the membrane of Th9 cells with the modification of PA (Fig. 4D).
Fig. 4.
Generation and characterization of Th9-KTP cells. (A) Schematic illustration of flow cytometry detection of Th9 cells after incubation with PA-KTP-FITC or KTP-FITC. (B) Flow cytometry analysis of Th9 incubated with PA-KTP-FITC or KTP-FITC with different concentrations. (C) Viability of Th9 cells after incubation with different concentrations of PA-KTP-FITC or KTP-FITC for different times: 10 min, 1 h, and 10 min followed by a 24 h recovery. (D) Confocal microscopy images of Th9 cells incubated with PA-KTP-FITC or KTP-FITC for 10 min with a concentration of 0.1 mg/ml. Scale bar, 20 μm or 5 μm. (E)In vitro expansion of Th9/Th9-KTP cells after stimulation with anti-CD3e and anti-CD28 antibodies. Naïve and unstimulated T cells were used as control. (F) The percentage of CD4+ IL-9+ T cells cultured under Th9 differentiation medium for 4 days with the incubation of PA-KTP-FITC for 10 min on Day 0 or Day 4. (G) Confocal microscopy images of Th9-KTP-FITC cells after in vitro culture for 1 h, 12 h,24 h, and 48 h. Scale bar, 5 μm. Data were analyzed by one-way ANOVA test or Kruskal-Wallis test (F). n = 3 samples per group; mean ± SEM; ns indicates no significant difference,* indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001, **** indicates p < 0.0001. MFI: mean fluorescence intensity.
Next, we evaluated whether the decoration of KTP on Th9 cells would affect their physiological functions as immunotherapeutics. As shown in Fig. 4E, the proliferation of Th9 cells was slightly inhibited on day 2 and day 3 after PA-KTP-FITC decoration under TCR stimulation (with anti-CD3/CD28). However, both Th9 and Th9-KTP cells still significantly proliferated compared to Th0 cells without TCR stimulation. Moreover, IL-9 secretion of Th9-KTP was similar to unmodified Th9 cells, suggesting that Th9 cells maintained their differentiation property after KTP decoration (Fig. 4F and Fig. S3B). Together, these results suggest that PA-KTP-FITC could be efficiently anchored to the Th9 cell membrane without disturbing the physiological function, proliferation, and viability of Th9 cells.
We then assessed the stability of KTP on the Th9 cell membrane by extending culture time after PA-KTP-FITC incubation. KTP remained on the Th9 cell membrane within 12 h of culture. After that, PA-KTP-FITC illustrated by green fluorescence gradually started to fade out. At 48 h, it was nearly invisible, indicating that KTP was detached from the cell membrane (Fig. 4G). Quantification of FITC intensity showed a similar trend but KTP decoration was at least stable for 12 h, which is sufficient to allow the decorated Th9 cells to arrive at the tumor sites (Fig. S3C).
2.3. Th9-KTP cells exhibit a superior binding ability to K7M2 cells in vitro
The validation of KTP affinity to K7M2 and successful KTP decoration on Th9 cells motivated us to explore if such ligand decoration on the cell surface would promote interactions between Th9 cells and K7M2 cells. Th9-ZsGreen (green) cells were generated from B6-G/R mice and then incubated with PA-KTP to form Th9-ZsGreen-KTP. Th9-ZsGreen and Th9-ZsGreen-KTP cells were added to K7M2-mCherry (magenta) culture respectively to observe real-time T cell-K7M2 interactions by the living cell workstation (Movie S1 and Movie S2). Although we used the same Th9 cells for both groups (0 h), the number of Th9 cells in contact with tumor cells increased significantly in the Th9-KTP group (1–5 h), while there was no obvious change in the Th9 group (Fig. 5A and B). Quantitatively, the number of conjugated T cells/per tumor cell in the Th9-ZsGreen-KTP group gradually increased from 2 to 13.17 within 5 h, while the number of conjugated T cells/per tumor cell in the Th9-ZsGreen group fluctuated between 4 and 5.5 (Fig. 5C), suggesting KTP substantially enhanced the binding affinity of Th9 cells toward K7M2 cells in vitro. In the next set of experiments, an extra washing step was added to exclude the non-specific binding of Th9 to K7M2 cells. Th9-ZsGreen and Th9-ZsGreen-KTP cells were co-cultured with K7M2-mcherry for 24 h and then thoroughly washed by PBS 3 times to remove the loosely bound T cells. After washing, most Th9-ZsGreen cells were removed in both groups (Fig. 5D), but KTP modification permitted more Th9-ZsGreen cells (3.5/tumor cell) bound to tumor cells. The average number of Th9 cells with KTP decoration bound to each K7M2 cell was about 7-fold more than unmodified Th9-ZsGreen cells (0.6/tumor cell), which again confirmed the presence of KTP on Th9 cells effectively improved the interactions between Th9 cells and tumor cells (Fig. 5E). Taken together, these results suggest that decoration of immune cell surface with high-affinity ligand could significantly enhance the binding affinity of Th9 cells against corresponding tumor cells.
Fig. 5.
KTP decoration enhanced the binding affinity of Th9 cells to K7M2 cells. (A) The dynamic binding of Th9-ZsGreen-KTP and Th9-ZsGreen with K7M2-mCherry cells using confocal live-cell images. Scale bar, 50 μm. (B and C) The curve (B) and quantification (C) of binding ZsGreen-Th9-KTP and ZsGreen-Th9 cells per K7M2-OVA cell within 5 h (n = 3 samples per group). (D and E) Confocal image (D) and quantification (E) of Th9-ZsGreen-KTP and Th9-ZsGreen bonded to K7M2-mCherry cells before or after being thoroughly washed by PBS. Scale bar, 50 μm (n = 4 samples per group). Data were analyzed by one-way ANOVA test. mean ± SEM; ns indicates no significant difference,* indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001, **** indicates p < 0.0001.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.bioactmat.2022.11.022
The following is the supplementary data related to this article:
1
2
2.4. Th9-KTP cells exhibit stronger infiltration ability and superior anti-osteosarcoma capacity
Given the potent binding affinity of Th9-KTP to K7M2 cells, we investigated whether the KTP decoration could improve the infiltration ability of Th9 cells in vivo. Ovalbumin (OVA)-expressing K7M2 (K7M2-OVA) osteosarcoma were established by subcutaneous inoculation of K7M2-OVA into wild-type Blab/c mice. DO11.10 mice-derived Th9-OVA cells, which are specific for OVA antigen expressed by K7M2-OVA cells, were labeled with carboxyfluorescein diacetate succinimidyl ester (CSFE) to indicate the exogenously transferred Th9-OVA cells. CFSE+ Th9-OVA cells and CFSE+ Th9-OVA-KTP cells were intravenously transferred into K7M2-OVA bearing mice. The infiltrated CFSE+ Th9-OVA cells into the tumors were detected through flow cytometry 4 h after injection (Fig. 6A). The infiltration of CFSE+ T cells in the Th9-OVA cell group was 22.30%, while the infiltration of CFSE+ T cells in the Th9-OVA-KTP cell group was 51.75% (Fig. 6B and C). Thus decoration of Th9-OVA with KTP substantially improved Th9-OVA cells infiltration into tumor sites by 2.3 folds. To validate the enhanced homing capacity of Th9-KTP cells, we detected the accumulation of DIR-stained Th9 cells in luciferase (Luci)-expressing K7M2-OVA tumor by IVIS Spectrum Animal Imaging System 24 h after tail vein infusion (Fig. 6A). Bioluminescence imaging of luciferase-transfected tumors confirmed the existence and position of established tumors (Fig. 6D, left). As shown in Fig. 6D, the infused T cells (right) were mainly accumulated in the liver and tumor tissue, but in mice receiving Th9-OVA-KTP cells, increased localization of Th9 cells was observed in tumor sites as compared to the mice receiving undecorated Th9-OVA cells or Th9-OVA-CP cells. Statistically, decoration of Th9-OVA with CP did not increase infiltration of Th9 cells in tumors as no significant differences were detected between Th9-OVA and Th9-OVA-CP treatment. However, KTP decoration of Th9-OVA cells substantially increased their infiltration into the tumor by 2.7 times (Fig. 6E), suggesting KTP decoration approach was highly effective in promoting Th9 infiltration into solid tumors.
Fig. 6.
KTP decoration enhanced the infiltration and anti-tumor efficacy of Th9-OVA cells in K7M2-OVA tumor-bearing mice. (A) Schematic illustration of flow cytometry detection and IVIS detection of infiltrated Th9-OVA cells. (B and C) Representative density plot (B) and quantification (C) of exogenous transferred Th9 cells (CFSE+ CD4+) in TILs isolated from K7M2-OVA tumors, 4 h after the injection of CFSE-labeled Th9-OVA or Th9-OVA-KTP cells (2ⅹ10^6 cells) (n = 3 sample per group). (D)In vivo bioluminescence images of mice bearing K7M2-OVA tumors (left) and in vivo DIR fluorescence images of Th9 cells accumulation in tumor sites (right) 24 h postinjection of DIR-labeled Th9-OVA or Th9-OVA-KTP cells (2ⅹ10^6 cells) (n = 4 mice per group). (E) Quantification of the average radiant efficiency of DIR-labeled homing Th9 cells. (F) Schematic illustration of the tumor experimental timeline and treatment placement whereby mice received either the injection of Th9-OVA or Th9-OVA-KTP cells. (G and H) Tumor growth curves (G) and the tumor volume (H) of K7M2-OVA tumor-bearing Balb/c mice after intravenous injection of PBS, Th9-OVA cells, Th9-OVA-CP cells, or Th9-OVA-KTP cells (3ⅹ10^6 cells on day 7 and day14) (n = 6 mice per group). Data were analyzed by one-way ANOVA test (E and H) or unpaired t-test (C). mean ± SEM; ns denote no significant difference,* indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001,**** indicates p < 0.0001.
Cytokine release syndrome (CRS), a systemic inflammatory response aroused by cytokines derived from adoptively transferred T cells, is the most common side effect of CAR-T therapy ([30]). CRS may also lead to reversible damage to healthy organs, such as the cardiac and liver ([31]). Thus, it is essential to evaluate the safety of Th9 cells-based ACT therapy in the context of tumor immune therapy. The intravenous injection of Th9-OVA, Th9-OVA-CP, and Th9-OVA-KTP cells (Th9 cells-based ACT therapy) did not significantly change the level of CRS-related cytokines in serum, such as IL-6, TNF-α, IFN-γ, IL-1β, IL-18, as compared to the PBS treatment (Fig. S4A). The levels of aspartate aminotransferase (AST) and alanine transaminase (ALT), which are markers of liver injury, were also not significantly changed in the mice serum after receiving Th9 ACT compared to the control group (Fig. S4B). Th9 cells did not cause cardiac damage as indicated by the levels of lactate dehydrogenase (LDH) and creatine kinase (CK) (Fig. S4B). Besides, we did not observe obvious histopathological damage in the heart, liver, spleen, lung, and kidneys after the Th9 cells-based treatment (Fig. S4C). Taken together, these results suggested that Th9 cells-based ACT therapy would not cause systemic adverse effects in our experimental conditions and can be safely utilized against tumors.
Subsequently, we investigated whether improved infiltration of Th9-OVA cells caused by KTP decoration could lead to better anti-tumor effects of Th9 cells. 1.0ⅹ106 K7M2-OVA cells were subcutaneously inoculated to Blab/c mice on day 1. One week later, tumor-bearing mice were regrouped according to tumor volume to ensure that the initial tumor volume was consistent between groups. On day 7 and day 14, 3.0ⅹ106 in vitro differentiated tumor-specific Th9-OVA cells, Th9-OVA-CP cells, and Th9-OVA-KTP cells were intravenously injected into tumor-bearing mice, respectively (Fig. 6F). Th9-OVA and Th9-OVA-CP cells treatment inhibited tumor growth and reduced tumor size over 15 days as compared to PBS-treated mice, while adoptive transfer of Th9-OVA-KTP presented the slowest rate of tumor growth and smallest tumor size (Fig. 6G and H). In addition, Th9-OVA-KTP infusion resulted in severe tumor cell damage and significantly inhibited tumor cell proliferation as indicated by Ki-67 staining when compared to Th9-OVA and Th9-OVA-CP treatment (Fig. S4D). In summary, these results indicated that KTP decoration of Th9-OVA cells significantly improved their infiltration ability toward tumor sites and thereby aroused better anti-osteosarcoma efficacy.
2.5. The anti-tumor effects of Th9-KTP cells rely on CD8+ T cells and IL-9
Although the anti-tumor effect of Th9 cells has been confirmed in a variety of tumors, the anti-tumor mechanisms of Th9 cells were debated ([24,[32], [33], [34], [35]]). Therefore, we explored the underlying mechanism of KTP decorated Th9 cells against osteosarcoma. To confirm whether tumor-specific Th9 cells could directly kill K7M2 cells, a 1:1 mixture of K7M2-OVA and K7M2 cells, labeled with different concentrations of CFSE (K7M2-OVA: CFSEhigh; K7M2: CFSElow), were co-cultured with Th9-OVA or not for 24 h. Once Th9-OVA was able to directly kill the tumor cells, the ratio of CFSEhigh would be reduced in the experimental group. Thus, we detected the ratio of CFSEhigh peak and CFSElow peak in the experimental group and control group by flow cytometry. The result showed the ratio of CFSEhigh peak and CFSElow peak was not influenced by Th9-OVA cells (Fig. S5A), indicating that Th9 cells exerted their antitumor effect via an indirect mode instead of a direct killing mode. To dissect which type of immune cells may be involved in the indirect antitumor effect of Th9 cells in osteosarcoma, we extracted the tumor-infiltrating immune cells from osteosarcoma 2 days after Th9 transferring and analyzed the composition of immune cells by flow cytometry. No difference was observed in the proportion of CD4+ T cells, dendritic cells (DC), and nature killing cells (NK) in osteosarcoma after Th9-OVA cells transfusion (Fig. 7A and Fig. S5B and C). However, we noticed that the transfusion of Th9 cells improved the infiltration of CD8+ T cells (Fig. 7A and B and Fig. S5D), which was consistent with the results observed by others ([35]). Since MDSC (CD11C+ Gr1+) and TAM (CD11b+ F4/80+) are major populations responsible for the immune suppressive microenvironment in solid tumors, we next evaluated their population changes within the tumor. Interestingly, we found that the infiltration of MDSC and TAM was decreased when treated with Th9 cells (Fig. 7A and B). Besides, the ratio of CD8/TAM and CD8/MDSC was increased after Th9 cell treatment, indicating that Th9 cells alleviated the immunosuppressive microenvironment of osteosarcoma (Fig. 7B). Next, we conjectured that the improved infiltration of Th9 cells in tumor sites would further influence the infiltration of CD8+ T cells, MDSC, and TAM. Consistent with our speculation, we observed an increased proportion of intratumor CD8+ T cells in tumor-specific Th9-KTP cells treated mice compared to mice receiving tumor-specific Th9 and Th9-CP cells (Fig. 7C and D). In addition, in comparison to the Th9-OVA and Th9-OVA-CP group, the amounts of MDSC and TAM were further decreased after Th9-OVA-KTP transfusion (Fig. 7C and D). Since M2 (CD11b+ F4/80+ CD206+)-polarized macrophage has a central role in promoting osteosarcoma metastasis and invasion and closely related to poor prognosis ([36]), we evaluated the change of M1 and M2 population after Th9 infusion. First, we found that M2 macrophage was predominant in osteosarcoma as compared to M1 macrophage, which was consistent with a recent RNA sequencing analysis of osteosarcoma ([37]). More importantly, Th9 cell-based ACT treatment significantly reduced the M2 macrophage population in osteosarcoma, while M1 macrophage population almost remined the same level after Th9 ACT (Fig.S5D and E). We also noticed that Th9-OVA-KTP infusion was more efficient in decreasing M2 macrophage when compared to Th9-OVA and Th9-OVA-CP (Fig.S5D and E), suggesting the employment of tumor targeting peptide improved Th9 infiltration into tumor sites, which further led to stronger immunomodulatory effects. However, the overall ratio of M2/M1 in different groups was not significantly influenced which could be explained by the overwhelmingly dominant number of M2 macrophage in osteosarcoma. These data underscored the potential of Th9-OVA-KTP cells to convert “cold tumors” into “hot tumors”, and that CD8+ T cells may play an important role in the anti-osteosarcoma effect of Th9-OVA-KTP cells. To establish the role of CD8+ T cells, we depleted CD8+ T cells with the anti-CD8a antibody while the control group was administrated with IgG2b. As shown in Fig. 7E, tumor-bearing mice receiving Th9-KTP treatment significantly inhibited tumor growth, while CD8+ T cell depletion revived the tumor growth. However, the mice treated with PBS and CD8a depletion showed more drastic tumor growth curves and larger tumor volume as compared to the mice treated with Th9-OVA and anti-CD8a, indicating that the CD8+ T cells-independent mechanisms might be involved in the anti-tumor effect of Th9-OVA-KTP cells (Fig. 7E and F).
Fig. 7.
The anti-osteosarcoma efficacy of tumor-specific Th9-OVA-KTP cells relied on CD8+T cells and IL-9. (A and B) Flow cytometry analysis (A) and quantification (B) of the percentage of CD4+ T cells, CD8+ T cells, MDSC (Gr1+CD11c+), and TAM (F4/80+CD11b+), and the ratio of CD8/MDSC as well as the ratio of CD8/TAM in immune cells isolated from subcutaneous K7M2-OVA tumor-bearing mice 3 days after injection of PBS or Th9-OVA cells (3ⅹ10^6 cells) (n = 3 mice per group). (C and D) Flow cytometry (C) analysis and quantification (D) of the percentage of CD8+ T cells, MDSC, and TAM in TILs isolated from subcutaneous K7M2-OVA tumor-bearing mice 3 days after reinfusion of PBS, Th9-OVA cells, Th9-OVA-CP cells, and Th9-OVA-KTP cells (3ⅹ10^6 cells) (n = 4 mice per group). (E and F) Tumor growth curves (E) and the tumor volume (F) of K7M2-OVA tumor-bearing Balb/c mice after intravenous injection of PBS and Th9-OVA-KTP cells (3ⅹ10^6 cells on day 7 and day14) with the administration of anti-CD8 or IgG2b antibodies every 3 days since 1day before the inoculation of tumor cells (n = 6 mice per group). (G and H) Tumor growth curves (G) and the tumor volume (H) of K7M2-OVA tumor-bearing Balb/c mice after intravenous injection of PBS and Th9-OVA-KTP cells (3ⅹ10^6 cells on day 7 and day14) with the administration of anti-IL-9 or IgG2a antibodies every 3 days since 1day before the inoculation of tumor cells (n = 6 mice per group). (I, J, K, and L) Flow cytometry analysis (I) and quantification of the percentage of CD8+ T cells (J), MDSC (K), and TAM (L) in TILs isolated from subcutaneous K7M2-OVA tumor-bearing Balb/c mice treated as (G and H) (n = 3 or n = 4 mice per group). Data were analyzed by one-way ANOVA test (D, F, H, J, K, and I) or unpaired t-test (B). mean ± SEM; ns denote no significant difference,* indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001,**** indicates p < 0.0001.
Recent studies emphasized that Th9-induced tumor eradication relied on IL-9 and IL-9 had an important impact on the composition of immune cells in tumors, including CD8+ T cells ([32,35,38,39]). Thus, we sought to determine whether IL-9 was involved in the tumor rejection of osteosarcoma after the tumor-specific Th9-KTP transfusion by using an IL-9 neutralizing antibody (α-IL-9) while the control group was administrated with IgG2a. First, we found tumor inhibition induced by Th9-KTP cells was remarkably abrogated after IL-9 neutralization (Fig. 7G and H). In addition, we noticed that tumor growth was not influenced by the administration of α-IL-9 after PBS treatment. The tumor growth and tumor volume of the Th9-KTP + anti-IL9 group were similar to that of the PBS or PBS+anti-IL9 group, suggesting that the levels of Th9 cells and IL-9 in osteosarcoma were limited under physiological conditions, thus highlighting the potential benefits of Th9 based therapy (Fig. 7G and H). Next, we analyzed the immune composition in tumors after α-IL-9 administration. We observed a significantly decreased number of CD8+ T cells as well as substantially increased numbers of TAM and MDSC in mice treated with Th9-OVA-KTP along with the administration of α-IL-9, compared to the mice treated with Th9-OVA-KTP alone (Fig. 7I and J, K, and L). However, the numbers of these immune cells were not influenced in mice receiving PBS or PBS+anti-IL-9 treatment. In conclusion, these data suggest that the anti-osteosarcoma effect of Th9-KTP relied on the secretion of IL-9 and IL-9 was responsible for the remodeling of the immune composition in osteosarcoma.
2.6. Peptide decorated T cells strategy was also applicable to γδ T cells
To broaden the application of this strategy, we further tested this strategy in human γδ T cells. OTP (TPPRVPLLTFGS), a human osteosarcoma Hos targeting peptide, identified by phage display has been verified in our previous work ([40]). OTP was decorated to γδ T cells following the same procedures to produce Th9-KTP cells. To determine the conjugation efficiency of OTP, we incubated γδ T with different concentrations of PA-OTP-FITC or OTP-FITC (0.01–0.5 mg/ml) for 10min and then detected by flow cytometry. As shown in Fig. S6A and B, the MFI of FITC gradually improved along with the increased concentrations of the peptides, while PA-OTP exhibited a stronger conjugation ability to γδ T cells. Similar to KTP, we did not observe obvious hazardous effects of OTP decoration on γδ T cells (Fig. S4C). Thus, the work concentration of PA-OTP was also set at 0.1 mg/ml, the same as PA-KTP, in the following experiment. Similar to that of PA-KTP-FITC, PA-OTP-FITC was successfully anchored to the membrane of γδ T cells as indicated by the FITC co-localizing with PKH26 (Fig. 8A). In addition, we also tested the bonding persistence of PA-OPT in γδ T cells. The OTP-FITC signal bound to the T cell membrane gradually attenuated over time until it became faint and invisible at 48 h (Fig. S6D). Next, we explored whether PA-OTP was capable of improving the infiltration of γδ T cells to tumor sites. The result showed that when decorated by PA-OTP, more γδ T cells were accumulated in tumor sites compared to undecorated γδ T cells (Fig. 8B). Statistically, the average radiant efficiency of homing γδ T cells labeled by DIR in the γδ T-OTP group was about 1.5 times of the γδ T group (Fig. 8C). More importantly, we observed γδ T cells transfusion inhibited tumor growth as compared to the control group, while mice treated with γδ T-OTP cells exhibited the slowest rate of tumor growth and minimal tumor size and tumor weight (Fig. 8D and E, and F). Besides, γδ T-OTP cells infusion significantly inhibited tumor cell proliferation as indicated by Ki-67 staining when compared to γδ T cells treatment (Fig. S6E). Collectively, these experiments suggested that this approach can be translated to other immune cells, such as human γδ T cells, and resulted in enhanced infiltration of the immune cells in tumors to reach a better anti-tumor effect.
Fig. 8.
Human γδ T-OTP cells display enhanced intratumor accumulation and anti-tumor efficacy in HOS tumor-bearing mice. Human γδ T cells were isolated from PBMC. (A) Confocal microscopy images of γδ T cells incubated with PA-OTP-FITC or OTP-FITC for 10 min with a concentration of 0.1 mg/ml. Scale bar, 20 μm or 5 μm. (B)In vivo bioluminescence images of mice bearing HOS-Luci tumors and in vivo DIR fluorescence images of γδ T cells accumulation in tumor sites 24 h postinjection of DIR-labeled γδ T or γδ T-OTP cells (2ⅹ10^6 cells) (n = 5 mice per group). (C) Quantification of the average radiant efficiency of DIR-labeled homing γδ T cells. (D and E) Tumor growth curves (D) and the tumor volume (E) of Hos tumor-bearing Balb/c-nude mice after intravenous injection of PBS, γδ T cells, or γδ T-OTP cells (3ⅹ10^6 cells on day 7 and day14) (n = 6 mice per group). (F) The photographs and tumor weight of tumor tissues collected on day 25 (n = 6 mice per group). Data were analyzed by one-way ANOVA test. mean ± SEM; ns denote no significant difference,* indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001,**** indicates p < 0.0001.
3. Discussion
The emergence of chimeric antigen receptor (CAR)-T cells enable genetically engineered T cells to target specific antigens overexpressed on cancer cells, however poor infiltration of transferred T cells severely limits the therapeutic efficacy of ACT therapy when used against solid tumors ([10]). To tackle this problem, current strategies focus on reducing the physical barrier, repairing the mismatched chemokine receptor-chemokine axis mismatched, or alleviating the hostile tumor microenvironment in solid tumors ([11,[15], [16], [17],41]). However, these approaches pose significant challenges to be translated into clinics as they inevitably involve complicated technological and pharmaceutical interventions. Even for CAR-T therapy, the extremely high cost and serious side effects associated with cytokine storm also limited its broader clinical applications. Thus, we aimed to develop a facile, cost-effective, translatable approach to overcome these limitations in order to further expand the clinical application of ACT. In particular, we focused on highly heterogeneous cancers such as osteosarcomas, which lack of recurrent tumor-specific binding ligands for target therapy. In response, we demonstrated a novel strategy to rapidly identify tumor-targeting ligands and effectively deliver therapeutic immune cells to tumor sites via grafting tumor-specific targeting peptides, screened by phage display biopanning technology, to the surface of T cell membranes via a non-genetic facile engineering approach. Our data validated that this strategy is capable of efficiently improving the infiltration of adoptively transferred T cells since the tumor-specific targeting peptide decoration improved the binding affinity of T cells toward tumor cells, which resulted in substantially improved therapeutic efficacy of Th9-based ACT against osteosarcoma. Therefore, this approach can be readily leveraged to realize effective immunotherapy targeted to highly heterogeneous solid tumors via rapid binding ligand screened by phage display. Besides, this strategy is cost-saving, cell-friendly, easy to manufacture, and applicable to various immune cells. Hence, we speculate that our approach possesses the extraordinary potential to overcome the insufficient infiltration of ACT, which might serve as an important complement to the current immune therapy modality.
OS is the most frequent primary malignant solid tumor of bones characterized by genomic instability and heterogeneity ([42]). Generally, a high level of genomic instability highlights the potential of generating neoantigens or neo-epitopes to be applied in immunotherapies. However, it is difficult to find suitable neoantigens in osteosarcoma, due to few of the genomic alterations ultimately expressed as neoantigens ([43]). Moreover, most of the antigens expressed in osteosarcoma are also found on healthy tissues ([42]), thus their applications in clinical practice are limited by “on target-off tumor” effects. Therefore, a specific ligand targeting osteosarcoma is of great significance to solve the current dilemma of osteosarcoma immunotherapy. Phage display biopanning technology utilizes a phage library expressing billions of unique polypeptides to screen the target-binding peptides based on direct interaction between displayed peptides and unknown targets ([44,45]). The high flexibility, efficiency, and safety of phage display enable its broad application in identifying antibodies and cancer cell targeting molecules. Harnessing this technique, we successfully identified a K7M2-specific targeting peptide by phage display biopanning after 6 rounds of in vitro and in vivo screening. Our data demonstrated that this peptide exhibited superior binding affinity to K7M2 cells as indicated by 6–14 times in vitro binding efficiency and 2.2 times in vivo targeting efficiency compared to control peptides. Given the successful identification of osteosarcoma-specific ligands using phage display, we expect this rapid and cost-effective approach can be translated to many other solid tumors that lack targetable neoantigens.
The tumor-specific targeting peptide was conjugated to the T cell membrane by a nongenetic modification of PA, a hydrophobic tag, based on hydrophobic interaction between the cell membrane and PA tail. This approach showed great efficiency to anchor peptides to Th9 cell membranes. Compared to genetic modification of cell membrane, PA modification eliminates the risk of insertion tumorigenesis and the permanent changes to cells caused by the disruption of irrelevant genes ([46]). Cell viability and in vitro differentiation and expansion studies proved that this approach was safe and would not disturb the physiological function of the Th9 cells. Besides, our results proved that with the modification of PA, peptides were conjugated to the cell membrane for at least 12 h, ensuring the successful arrival of T cells to tumor sites. Furthermore, this approach was successfully translated to γδ T cells and improved the infiltration ability of γδ T cells to tumor sites, indicating the potential availability to other immune cells. In addition, this method possessed other advantages, including ease of operation, rapidity, and low cost, highlighting the potential for broader application in clinical practice.
Mechanistically, the improved antitumor effects of Th9-KTP cells after arrival in the tumor bed in our osteosarcoma model rely on IL-9-mediated adaptive immunity. We observed the infiltration of DC and NK in osteosarcoma was not influenced after the infusion of tumor-specific Th9 cells. Besides, the tumor development was not restrained in nude mice with impaired T cell function (data not shown). These data suggest that innate immunity may not be responsible for Th9-induced osteosarcoma inhibition. On the other side, we observed a significant increase of CD8+ T cells after Th9-KTP infusion, suggesting that the infiltration of CD8+ T cells was improved with the treatment of Th9-KTP cells. Correspondingly, depletion of CD8+ T cells via anti-CD8 antibodies also largely abrogated the antitumor effects of Th9-KTP cells. These observations imply that the anti-osteosarcoma effect of Th9-KTP cells may be exerted via CD8+ T cell-mediated adaptive immunity instead of innate immunity. Lu et al. reported that recruitment of CD8+ T cells may be achieved by DC cell recruitment in the context of Th9 cell-mediated antitumor treatment ([35]). Unfortunately, we did not observe an increase of DC cell infiltration after Th9 cell infusion in our study. Thus, we speculate that Th9 and Th9-KTP cells may improve the proliferation or survival of CD8+ T cells in osteosarcoma. Since such effects on CD8+ T cells are highly dependent on IL-9 regulation ([47]), we also validated the connection between IL-9 and CD8+ T cells. When IL-9 was neutralized with anti-IL-9 antibodies, the enhanced CD8+ T cell infiltration was reversed and the anti-tumor effects of Th9-KTP cells were completely abolished, suggesting the enhanced antitumor effect of CD8+ T cell may be attributed to IL-9 secreted by infused Th9 cells. Put together, the enhanced anti-osteosarcoma function of Th9-KTP cells may be attributed to the increased number of CD8+ T cells entering the tumor, which is closely related to the secretion of IL-9 by Th9 cells.
Besides, Th9 cells possess the potential to remodel the tumor microenvironment since the transfusion of Th9 and Th9-KTP cells significantly decreased the amounts of immunosuppressive cells such as MDSC and TAM. Given the fact that the overwhelming immunosuppressive microenvironment, represents a major reason for the failure of immune checkpoint inhibitors (ICI) therapy in osteosarcoma ([42]) as such a hostile microenvironment leads to insufficient immune cell infiltration and T cell exhaustion. The scavenger effect of Th9 against MDSC and TAM in solid tumors might be of great significance when combined with other cancer immunotherapy approaches. For instance, the combination of Th9 cell-based ACT and ICI therapy may have a synergistic effect to improve the anti-osteosarcoma effect, which was initialized by the Th9 cells to disrupt the balance between immunosuppressive cells and effector cells, and ultimately converted osteosarcoma into “hot tumors” by recruiting antitumor immune cells. However, we also noticed that others observed no significant change in the numbers of TAM and MDSC with the administration of α-IL-9 antibodies in melanoma ([35]). Thus, whether this phenomenon is osteosarcoma specific or broadly existed in solid tumors needs to be further explored since osteosarcoma is characterized by a high immunosuppressive phenotype as compared to other solid tumors, such as melanoma and lung cancer [37,43].
4. Material and methods
4.1. Material
Female Balb/c (5-to-8 week-old) mice were purchased from SLAC company (Shanghai, China). Immune compromised Nod/SCID mice (5-to-8 week-old) were purchased from Nanjing Animal Model Institute (Nanjing, China). DO11.10 mice were donated by Dr. Fang Zhang (Medical School of Nanjing University, Nanjing, China) and B6-G/R mice were donated by Professor Junjie Gao (Shanghai Sixth People's Hospital, Shanghai Jiaotong University, Shanghai, China). Mice were handled in a specific pathogen-free animal breeding facility and fed with sterile fodder and acidified water. All animal studies were approved by the Animal Care and Use Committee of the School of Medicine, Zhejiang University. The protocol for use of human blood was approved by the Institutional Review Board of the Second Affiliated Hospital of Zhejiang University (2020-071), and written informed consents were obtained from the patients.
4.2. Cell culture
K7M2, HOS, 3T3, and LLC cells were purchased from the Cell Bank of Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). All Cell line was identified by the Cell Collection via DNA STR genotyping. The K7M2-Luci-OVA cell, K7M2-mCherry, and HOS-Luci cells were constructed in our lab. K7M2, HOS, 3T3, and LLC cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Rockville, MD, USA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA), and 1% penicillin/streptomycin. Cells were maintained at 37 °C in 5% CO2.
4.3. In vitro/in vivo phage display
A linear 12-amino acid peptide library (Ph.D.-12 phage display peptide library, New England Biolabs, Inc.) was used for biopanning according to our previous work ([40]). First, 1 × 1012 transducing units of the phage library were co-cultured with mouse primary fibroblasts for negative selection. Then the supernatant was collected and added to K7M2 cells for positive selection. 1 h later, K7M2 cells were washed with a washing buffer five times to remove the unbound phages. Then, the cell-bound phages were eluted by the elution buffer. The selected phages were amplified by infecting E. coli host strain ER2738 for the next round of selection. After 1 round of in vitro biopanning, the remaining phages were intravenously injected into K7M2-bearing mice. 1 h later, the mice were sacrificed and the redundant phages in the blood were washed away through heart perfusion. Then the bearing tumor was incised and the tumor-targeted phages were extracted from the tumor homogenate and amplified by infecting E. coli host strain ER2738.
4.4. Synthesis and Dye labeling of peptides
Four additional lysines (KKKK) were added to the C-terminal of peptides as a linker region and manufactured by KS-V Peptide Biological Technology Co. Ltd (Hefei, China). The conjugation of Cy5.5 dye to each peptide was performed according to our previous work ([40]). Briefly, 1 × 10−3 M Cy5.5-maleimide was mixed with 1 × 10−3 M peptide saline solution at the ratio of 1:4 and stirred at room temperature for 3 h. Unconjugated peptides were removed using Slide-A-Lyzer Dialysis Cassettes (1000 MWCO, Thermo Fisher Scientific, Waltham, MA, USA). The peptides with palmitoylation were synthesized by KS-V Peptide Biological Technology Co. Ltd (Hefei, China), based on Fmoc/tBu solid-phase peptide synthesis strategy. The palmitic acid was installed on the ε-NH2 of first C terminal lysine residues.
4.5. In vitro differentiation of T cells
Naive T cells (CD4+CD62LhiCD44lo) were sorted from the spleen and lymph nodes of Balb/c mice, DO11.10 mice or B6-G/R mice according to experiment requirements by Easy™ Mouse CD4+ T cell isolation Kit (19852 A, Stem cell Technologies, Canda) and Easy™ Mouse CD4 Positive T cell isolation Kit II (18952 A, Stem cell Technologies, Canda). Sorted naive CD4+ T cells were routinely 98% pure. The sorted naive CD4+ T cells were stimulated with plate-bound anti-CD3 (145-2C11, 2 μg/ml, Bio X cell, USA) and anti-CD28 (PV-1, 2 μg/ml, Bio X cell, USA) antibodies, and polarized into Th9 cells with TGF-β1 (130-095-067, 2 ng/ml, MiltenyiBiotech, Germany), IL-4 (BE0045, 20 ng/ml, Bio X Cell, USA), and anti-IFN-γ (BE0054,10 μg/ml, Bio X cell, USA). Cells were maintained in RPMI 1640 at 37 °C in 5% CO2, supplemented with 10% FBS and 1% penicillin/streptomycin. Th9 cells were harvested on day 4 and the purity was detected by flow cytometry analysis. Human γδ T cell in vitro differentiation was conducted according to our previous work ([48]). Briefly, peripheral blood mononuclear cells were isolated by Ficoll gradient centrifugation and cultured in RPMI 1640, supplemented with 10% FBS and 1% penicillin/streptomycin and 1 μM zoledronate (Zometa; Novartis, Switzerland) and recombinant human IL-2 (400 IU/ml; PeproTech, USA) at 37 °C in 5% CO2. The complete culture medium was supplemented every 3 days. 12–15 days later, the γδ T cells were harvested and the purity was detected by flow cytometry analysis.
4.6. Peptide coating and cytotoxicity
T cells were seeded on 48-well plates (1.0 × 106 cells/well), then PA-peptide-FITC and peptide-FITC were added (0.01–0.5 mg/ml) to the T cells and incubated for 10 min. Then the unconjugated peptides were washed away. The coating rates were indicated by the FITC signal and quantized by Flow cytometry analysis. In addition, T cells were dyed with PKH 26 following manufacturer instructions and then incubated with 0.1 mg/ml PA-peptide-FITC or Peptide-FITC for 10 min. Finally, the T cells were fixed by cold methyl alcohol for 15 min at 4 °C and incubated with DAPI for 5 min, and observed under an FV3000 confocal microscope. For stability assessment, T cells were treated with PA-peptide-FITC as described above and observed by an FV3000 confocal microscope at each time point (1 h, 12 h, 24 h, 48 h).
The cytotoxicity of the peptides to Th9 cells was evaluated, including survival, proliferation, and polarization. For survival analysis, Th9 cells were cultured under the Th9 cell polarization condition medium containing TGF-β1 (2 ng/ml), IL-4 (20 ng/ml), and anti-IFN-γ (10 μg/ml). On day 4, Th9 cells were harvested and incubated with 0–0.5 mg/ml PA-KTP-FITC or KTP-FITC for 10 min (acute effects) and 1 h (extended-incubation effects) at 37 °C and then washed thoroughly in PBS 3 times. For the long-term effects, Th9 cells were harvested on day 3 and cultured for another 24 h after the T cells were incubated with 0–0.5 mg/ml PA-KTP-FITC or KTP-FITC for 10 min and washed in PBS 3 times ([28]). The survival rate was assessed by living-dead staining with PB-FVD and detected by flow cytometry.
For the proliferation assay, naïve CD4+ T (Day 0) cells were incubated with or without 0.1 mg/ml PA-KTP-FITC for 10 min at 37 °C and then washed thoroughly in PBS 3 times. Then 1.0 × 106 naïve CD4+ T cells were seeded on 48-well plates coated with anti-CD3 (2 μg/ml) and anti-CD28 (2 μg/ml) antibodies and cultured in the Th9 cell polarization condition medium. 1.0 × 106 naïve CD4+ T cells seeded on 48-well plates without anti-CD3 (2 μg/ml) and anti-CD28 (2 μg/ml) antibodies and cultured in the RPMI 1640 medium were regarded as the negative control. At day 0, 1, 2, and 3 the cells were collected and counted by flow cytometry. Fold expansion = number of cells (Th9, Th9-KTP+anti-CD3/CD28)/number of cells (No-anti-CD3/CD28).
For the polarization assay, naïve CD4+ T cells were incubated with or without 0.1 mg/ml PA-KTP-FITC for 10 min at 37 °C and then washed thoroughly in PBS 3 times, and cultured in the Th9 cell polarization condition medium. Th9-KTP or Th9 cells were harvested on day 4 and detected the level of CD4+ IL-9+ by flow cytometry.
The cytotoxicity of the peptides to γδ T cells was indicated by the survival rate. Human γδ T cells were cultured in RPMI 1640, supplemented with 10% FBS and 1% penicillin/streptomycin and 1 μM zoledronate and recombinant human IL-2 (400 IU/ml). On day 14, γδ T cells were harvested and incubated with 0–0.5 mg/ml PA-OTP-FITC or OTP-FITC for 10 min at 37 °C and then washed thoroughly in PBS 3 times. For the long-term effects, γδ T cells were harvested on day 13 and cultured for another 24 h after the γδ T cells were incubated with 0–0.5 mg/ml PA-OTP-FITC or OTP-FITC for 10 min and washed in PBS 3 times. The survival rate was assessed by living-dead staining with Fixable Viability Dye (FVD) and detected by flow cytometry.
4.7. In vivo fluorescence imaging
The Cy5.5-labeled peptides were constructed by ourselves. 100 μL of 5 × 10−5 M Cy5.5-peptides and Cy5.5 only were intravenously injected into established tumor-bearing mice. 24 h later, mice were anesthesia with a 4% isoflurane/oxygen mixture and scanned by the IVIS Spectrum Animal Imaging System (PerkinElmer, USA). For the T cell homing experiment, T cells were incubated with 0.1 mg/ml PA-peptides for 10 min and then stained by 1,1-dioctadecyl-3,3,3,3 tetramethylindotricarbocyaine iodide (DiR) (5 μM, Caliper life sciences, USA) for 20 min at 37 °C. After being thoroughly washed in PBS 3 times, T cells were delivered intravenously in 100ul PBS. Besides, the tumor cells used in the T cell homing experiment were transfected with luciferase to indicate the existence of tumors. D-luciferin (50 mg/kg, Yeasen, China) suspended in PBS was intraperitoneally injected for 10 min before imaging. IVIS Spectrum Animal Imaging System was used to evaluate the homing ability of T cells 24 h after T cells transfusion.
4.8. In vitro tumor-specific Th9 cell cytotoxicity
OVA-specific naïve CD4+ T cells were generated from OVA-specific DO11.10 mice, and in vitro polarized to Th9 cells in the Th9 cell polarization condition medium for 4 days. K7M2-OVA and K7M2-WT tumor cells were labeled with different concentrations of CFSE (Thermo Fisher Scientific, Waltham, CA, USA). K7M2-OVA cells were labeled with 5 μM CFSE (CFSEhigh) while K7M2-WT cells were labeled with 0.5 μM CFSE (CFSElow) for 10 min at 37 °C. Then the tumor cells were mixed at a 1:1 ratio, and seeded into 96-well plates (5ⅹ104 cells/well; 2.5ⅹ104 K7M2-OVA cells and 2.5ⅹ104 K7M2-WT). Th9 cells (5ⅹ105) were added and incubated for 24 h. A mixture of tumor cells cultured alone was regarded as the control. 24 h later, the cells were collected and stained by FVD and CD4 antibodies to dissect the surviving tumor cells (FVD− CD4− CFSE+) and detected by flow cytometry. Ratio = % [CFSEhigh] peak/% [CFSElow] peak. Tumor-Specific Lysis = [1-(Control ratio/Experimental ratio)] × 100.
4.9. Tumor experiments
The mice were used for tumor inoculation at the age of 6–8 weeks. To assess the antitumor effect of Th9 cells and KTP-Th9 cells, 1 × 106 K7M2-Luci-OVA cells were subcutaneously injected into Balb/c mice on day 1. On day 7 and day 14, 3 × 106 Do11.10 derived Th9 cells were intravenously injected into tumor-bearing mice respectively. To evaluate the roles of CD8+ T cells and IL-9 in the anti-tumor effects of Th9 cells, anti-CD8a (10 mg/kg, YTS 169.4, Bio X Cell, USA), and anti-IL-9 (10 mg/kg, 9C1, Bio X Cell, USA) were intraperitoneally delivered to mice every 3 days at 1 day before tumor inoculation. Rat IgG2b (10 mg/kg, LTF-2, Bio X Cell, USA) and mouse IgG2a (10 mg/kg, C1.18.4, Bio X Cell, USA) were used as isotype control. To assess the antitumor effect of γδ T cells and OTP-γδ T cells, 1 × 106 HOS-luci cells were subcutaneously injected into immune-compromised Balb/c mice on day 1. On day 7 and day 14, 3 × 106 γδ T cells and γδ T-OTP cells were intravenously injected into tumor-bearing mice respectively. The tumor volumes were monitored by caliper and calculated by the formula V = a × b2/2 (a: the maximal diameter of the tumor, b: the minimal diameter).
4.10. Tumor infiltration Lymphacyte analysis
The mice were sacrificed 48 h after T cells transfusion. The tumors were incised and enzymatically digested with 1 mg/ml collagenase IV (CLS-4, Worthington Biochemical Corp. USA), 0.5 mg/ml DNase I (DN25, Sigma-Aldrich, USA), and 25 μg/ml hyaluronidase (Sigma-Aldrich, USA) at 37 °C for 1.5 h. Then the TILs were isolated by Percoll (17089101, GE Healthcare, Sweden) gradient purification following manufacturer instructions.
4.11. Histological staining
PBS, Th9-OVA, Th9-OVA-CP, Th9-OVA-KTP, γδ T cells, and γδ-OTP T cells were intravenously injected into tumor-bearing mice on day 7 and day 14.2 days after the second transfusion, the mice were sacrificed to collect the tumor tissue and important organs, including heart, liver, spleen, lung, and kidney. Then, the tissues were fixed in 4% of formalin solution for 24 h and embedded in paraffin. For H&E staining, the organ tissues were cut into 5-μm-thick slices and manufactured according to a standard protocol. As for immunohistochemistry (IHC) analysis, the tumor tissues were cut into 5-μm-thick slices and incubated with blocking fluid for 30 min. Then the slices were incubated with rabbit-anti-Ki-67 (ab15580, Abcam, USA) or rabbit-anti-CD8a (ab217344, Abcam, USA) overnight at 4 °C, followed by a thorough wash, and then incubated with goat anti-rabbit IgG HRP.
4.12. Toxicity evaluation of Th9 cells and serum analysis
100μl PBS, 3 × 106/100μl Th9-OVA, Th9-OVA-CP, and Th9-OVA-KTP cells were intravenously injected into mice. 48 h later, the mice were sacrificed and the blood was collected. Then the serum was divided from the blood by centrifuging at 1500×g for 10 min. The levels of IL-6, IL-1β, IL-18, IFN-γ, and TNF-α were measured by multiplex secretome analysis (ABplex Mouse 6-Plex Custom panel) according to manufacturer's instructions with the assistance of ABclonal Technology. The serum activities of AST, ALT, CK, and LDH were measured by ALT, AST, CK, and LDH Reagent Kits (Biobase, Shandong, China).
4.13. Immunofluorescence
K7M2, 3T3, and LLC cells were seeded into a 24-well plate (1.0 × 106 cells per well), blocked with 10% BSA for 1 h at 37 °C, and then stained with Cy5.5-peptides and Cy5.5 only (10 × 10−6 M) for 30 min at 37 °C in the dark. After being washed 3 times by PBS, the cells were fixed in 4% paraformaldehyde for 20 min at room temperature and then incubated with Alexa Fluor 647-phalloidin for 1 h and DAPI for 5 min at 37 °C.
For live-cell imaging, naïve CD4+ T-ZsGreen (green) cells were generated from B6-G/R mice and cultured in the Th9 cell polarization condition medium for 4 days. Then Th9-ZsGreen (green) was incubated with 0.1 mg/ml PA-KTP for 10 min at 37 °C. 5.0 × 104 K7M2-mCherry (magenta) cells were plated on 35-mm glass-bottom petri dishes (P35g-1.5-14-C, MatTek, USA). Then, 5.0 × 105 Th9-ZsGreen-KTP or Th9-ZsGreen were added to the dishes.
4.14. Cell imaging and analysis
Fixed cell images were acquired on an FV3000 confocal microscope with a 40 × water objective or a 100 × oil immersion objective. Live-cell images were acquired on a Nikon A1 microscope using a 40 × water objective in a temperature-controlled chamber (37 °C, 5% CO2).
Digital images were acquired on a Nikon A1 microscope with Nikon NIS Elements software. For 3D imaging, 20- to 40-μm Z-stack images, examined in 1-μm steps, from the upper surface to bottom surface were processed. The images were recorded at the interval of every 5 min for 5 h. All images were assembled and analyzed using FV315-SW or LAS X.
4.15. CFSE labeling and transfer
KTP-Th9, Th9 cells were incubated with 5 μM CFSE in PBS at room temperature for 10 min at 37 °C in dark, and then washed extensively. 1.0ⅹ106 CFSE-labeled-Th9 cells or CFSE-labeled-Th9-KTP were intravenously transferred into mice bearing K7M2-OVA osteosarcoma. All mice were sacrificed 4 h later, and all lymphocytes were extracted through TIL experiment. CFSE+ Th9 cells were detected by flow cytometry.
4.16. Flow cytometry
Isolated tumor-infiltrating immune cells and in vitro differentiated Th9 cells were stained with antibodies against CD3 (17A2), MHC-II (M5/114.15.2) CD8 (53–6.7), CD4 (RM4-5), IL-9 (RM9A4), CD45 (30-F11), NK1.1 (S17016D), CD11c (N418), CD11b (M1/70), CD86 (GL-1), CD206 (C068C2), and Gr1 (R86-8C5) (all from eBioscience or BioLegend, USA). For intracellular staining, cells were stimulated for 4 h by PMA (P1585, 50 ng/ml, Sigma–Aldrich, USA), ionomycin (I3909, 1 μg/ml, Sigma–Aldrich, USA), and brefeldin A solution (00-4506-51, eBioscience, USA) and were then manufactured following an intracellular staining protocol. Next, cells were carried out with fixation/permeabilization buffer solution (00-5123-43 and 00-5223-56, eBioscience, USA). Then, stained cells were analyzed by Novocyte Flow cytometry, and data were analyzed using NovoExpress (Agilent Technologies, Inc, USA).
4.17. Statistics
All the data were performed the Shapiro-Wilk test to confirm their distribution. For the normally distributed data, an unpaired T-test was used for means comparison between the two groups. One-way analysis of variance (ANOVA) was used for means comparison of multiple individual datasets. For the non-normally distributed data, the Mann-Whitney test was used for means comparison between the two groups. The Kruskal-Wallis test was used for the means comparison of multiple individual datasets.
Fluorescence intensity was analyzed by Image J software. All other statistical analysis was performed by GraphPad Prism software (Version 8.0). Results were presented as means ± SEM, and a P-value <0.05 was considered statistically significant and marked by asterisks (N·S., no significance; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
Funding
This work was supported by National Natural Science Foundation of China (81872173, 82072959, 31870959, and 82102855) and Zhejiang Province Natural Science Foundation (LY20H160018, LD21H160002, and LY19H160045).
Ethics approval
The experimental protocol was established, according to the ethical guidelines of the Helsinki Declaration and was approved by the Zhejiang University School of medicine the Second Affiliated Hospital (2020–71). Written informed consent was obtained from individual or guardian participants.
CRediT authorship contribution statement
Tao Chen: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. Yucheng Xue: Methodology, Writing – review & editing. Shengdong Wang: Methodology, Funding acquisition. Jinwei Lu: Investigation. Hao Zhou: Investigation. Wenkan Zhang: Investigation. Zhiyi Zhou: Investigation. Binghao Li: Investigation. Yong Li: Validation. Zenan Wang: Software. Changwei Li: Formal analysis. Yinwang Eloy: Software. Hangxiang Sun: Validation. Yihang Shen: Investigation. Mohamed Diaty Diarra: Validation. Chang Ge: Formal analysis. Xupeng Chai: Software. Haochen Mou: Formal analysis. Peng Lin: Conceptualization. Xiaohua Yu: Conceptualization, Supervision, Writing – review & editing, Funding acquisition. Zhaoming Ye: Conceptualization, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgments
T. Chen., Y. Xue., SD. W. contributed equally to this work.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2022.11.022.
Contributor Information
Peng Lin, Email: linpengg@zju.edu.cn.
Xiaohua Yu, Email: xiaohua.yu@zju.edu.cn.
Zhaoming Ye, Email: yezhaoming@zju.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Rosenberg S.A., Restifo N.P. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finck A.V., Blanchard T., Roselle C.P., Golinelli G., June C.H. Engineered cellular immunotherapies in cancer and beyond. Nat. Med. 2022;28:678–689. doi: 10.1038/s41591-022-01765-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neelapu S.S., et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 2017;377:2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maude S.L., et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018;378:439–448. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kristensen N.P., et al. Neoantigen-reactive CD8+ T cells affect clinical outcome of adoptive cell therapy with tumor-infiltrating lymphocytes in melanoma. J. Clin. Invest. 2022;132 doi: 10.1172/JCI150535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robbins P.F., et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J. Clin. Oncol. 2011;29:917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Creelan B.C., et al. Tumor-infiltrating lymphocyte treatment for anti-PD-1-resistant metastatic lung cancer: a phase 1 trial. Nat. Med. 2021;27:1410–1418. doi: 10.1038/s41591-021-01462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirtane K., Elmariah H., Chung C.H., Abate-Daga D. Adoptive cellular therapy in solid tumor malignancies: review of the literature and challenges ahead. J Immunother Cancer. 2021;9 doi: 10.1136/jitc-2021-002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong M., Clubb J.D., Chen Y.Y. Engineering CAR-T cells for next-generation cancer therapy. Cancer Cell. 2020;38:473–488. doi: 10.1016/j.ccell.2020.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Newick K., O'Brien S., Moon E., Albelda S.M. CAR T cell therapy for solid tumors. Annu. Rev. Med. 2017;68:139–152. doi: 10.1146/annurev-med-062315-120245. [DOI] [PubMed] [Google Scholar]
- 11.Narayan V., et al. PSMA-targeting TGFβ-insensitive armored CAR T cells in metastatic castration-resistant prostate cancer: a phase 1 trial. Nat. Med. 2022;28(4):724–734. doi: 10.1038/s41591-022-01726-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sackstein R., Schatton T., Barthel S.R. T-lymphocyte homing: an underappreciated yet critical hurdle for successful cancer immunotherapy. Lab. Invest. 2017;97:669–697. doi: 10.1038/labinvest.2017.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leko V., Rosenberg S.A. Identifying and targeting human tumor antigens for T cell-based immunotherapy of solid tumors. Cancer Cell. 2020;38:454–472. doi: 10.1016/j.ccell.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu G., Rui W., Zhao X., Lin X. Enhancing CAR-T cell efficacy in solid tumors by targeting the tumor microenvironment. Cell. Mol. Immunol. 2021;18:1085–1095. doi: 10.1038/s41423-021-00655-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Q., et al. Photothermal therapy promotes tumor infiltration and antitumor activity of CAR T cells. Adv. Mater. 2019;31 doi: 10.1002/adma.201900192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craddock J.A., et al. Enhanced tumor trafficking of GD2 chimeric antigen receptor T cells by expression of the chemokine receptor CCR2b. J. Immunother. 2010;33:780–788. doi: 10.1097/CJI.0b013e3181ee6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., et al. Targeting brain lesions of non-small cell lung cancer by enhancing CCL2-mediated CAR-T cell migration. Nat. Commun. 2022;13:2154. doi: 10.1038/s41467-022-29647-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moon E.K., et al. Intra-tumoral delivery of CXCL11 via a vaccinia virus, but not by modified T cells, enhances the efficacy of adoptive T cell therapy and vaccines. OncoImmunology. 2018;7 doi: 10.1080/2162402X.2017.1395997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evgin L., et al. Oncolytic virus-mediated expansion of dual-specific CAR T cells improves efficacy against solid tumors in mice. Sci. Transl. Med. 2022;14:eabn2231. doi: 10.1126/scitranslmed.abn2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng C., et al. Engineering nano-therapeutics to boost adoptive cell therapy for cancer treatment. Small Methods. 2021;5 doi: 10.1002/smtd.202001191. [DOI] [PubMed] [Google Scholar]
- 21.Dardalhon V., et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(-) effector T cells. Nat. Immunol. 2008;9:1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veldhoen M., et al. Transforming growth factor-beta 'reprograms' the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat. Immunol. 2008;9:1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 23.Angkasekwinai P., Dong C. IL-9-producing T cells: potential players in allergy and cancer. Nat. Rev. Immunol. 2021;21:37–48. doi: 10.1038/s41577-020-0396-0. [DOI] [PubMed] [Google Scholar]
- 24.Lu Y., et al. Th9 cells represent a unique subset of CD4(+) T cells endowed with the ability to eradicate advanced tumors. Cancer Cell. 2018;33:1048–1060 e1047. doi: 10.1016/j.ccell.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gianferante D.M., Mirabello L., Savage S.A. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat. Rev. Endocrinol. 2017;13:480–491. doi: 10.1038/nrendo.2017.16. [DOI] [PubMed] [Google Scholar]
- 26.Roy S., Cha J.N., Goodwin A.P. Nongenetic bioconjugation strategies for modifying cell membranes and membrane proteins: a review. Bioconjugate Chem. 2020;31:2465–2475. doi: 10.1021/acs.bioconjchem.0c00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milone M.C., O'Doherty U. Clinical use of lentiviral vectors. Leukemia. 2018;32:1529–1541. doi: 10.1038/s41375-018-0106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kean T.J., et al. Development of a peptide-targeted, myocardial ischemia-homing, mesenchymal stem cell. J. Drug Target. 2012;20:23–32. doi: 10.3109/1061186X.2011.622398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang B., et al. Peptide modified mesenchymal stem cells as targeting delivery system transfected with miR-133b for the treatment of cerebral ischemia. Int. J. Pharm. 2017;531:90–100. doi: 10.1016/j.ijpharm.2017.08.073. [DOI] [PubMed] [Google Scholar]
- 30.Brudno J.N., Kochenderfer J.N. Toxicities of chimeric antigen receptor T cells: recognition and management. Blood. 2016;127:3321–3330. doi: 10.1182/blood-2016-04-703751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipe D.N., Shafer S. CAR-T and checkpoint inhibitors: toxicities and antidotes in the emergency department. Clin. Toxicol. 2021;59:376–385. doi: 10.1080/15563650.2021.1880008. [DOI] [PubMed] [Google Scholar]
- 32.Schanz O., et al. Tumor rejection in Cblb (-/-) mice depends on IL-9 and Th9 cells. J Immunother Cancer. 2021;9 doi: 10.1136/jitc-2021-002889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purwar R., et al. Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells. Nat. Med. 2012;18:1248–1253. doi: 10.1038/nm.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vegran F., et al. The transcription factor IRF1 dictates the IL-21-dependent anticancer functions of TH9 cells. Nat. Immunol. 2014;15:758–766. doi: 10.1038/ni.2925. [DOI] [PubMed] [Google Scholar]
- 35.Lu Y., et al. Th9 cells promote antitumor immune responses in vivo. J. Clin. Invest. 2012;122:4160–4171. doi: 10.1172/JCI65459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Q., et al. All-trans retinoic acid prevents osteosarcoma metastasis by inhibiting M2 polarization of tumor-associated macrophages. Cancer Immunol Res. 2017;5:547–559. doi: 10.1158/2326-6066.CIR-16-0259. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Y., et al. Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat. Commun. 2020;11:6322. doi: 10.1038/s41467-020-20059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pei S., et al. BFAR coordinates TGFbeta signaling to modulate Th9-mediated cancer immunotherapy. J. Exp. Med. 2021;218 doi: 10.1084/jem.20202144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy S., et al. EGFR-HIF1alpha signaling positively regulates the differentiation of IL-9 producing T helper cells. Nat. Commun. 2021;12:3182. doi: 10.1038/s41467-021-23042-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin P., et al. Tumor customized 2D supramolecular nanodiscs for ultralong tumor retention and precise photothermal therapy of highly heterogeneous cancers. Small. 2022;18(21) doi: 10.1002/smll.202200179. [DOI] [PubMed] [Google Scholar]
- 41.Hou A.J., Chen L.C., Chen Y.Y. Navigating CAR-T cells through the solid-tumour microenvironment. Nat. Rev. Drug Discov. 2021;20:531–550. doi: 10.1038/s41573-021-00189-2. [DOI] [PubMed] [Google Scholar]
- 42.Kansara M., Teng M.W., Smyth M.J., Thomas D.M. Translational biology of osteosarcoma. Nat. Rev. Cancer. 2014;14:722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 43.Wu C.C., et al. Immuno-genomic landscape of osteosarcoma. Nat. Commun. 2020;11:1008. doi: 10.1038/s41467-020-14646-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., et al. Phage display technology and its applications in cancer immunotherapy. Anti Cancer Agents Med. Chem. 2019;19:229–235. doi: 10.2174/1871520618666181029140814. [DOI] [PubMed] [Google Scholar]
- 45.Saw P.E., Song E.W. Phage display screening of therapeutic peptide for cancer targeting and therapy. Protein Cell. 2019;10:787–807. doi: 10.1007/s13238-019-0639-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vormittag P., Gunn R., Ghorashian S., Veraitch F.S. A guide to manufacturing CAR T cell therapies. Curr. Opin. Biotechnol. 2018;53:164–181. doi: 10.1016/j.copbio.2018.01.025. [DOI] [PubMed] [Google Scholar]
- 47.Wang C., et al. Th9 cells are subjected to PD-1/PD-L1-mediated inhibition and are capable of promoting CD8 T cell expansion through IL-9R in colorectal cancer. Int. Immunopharm. 2020;78 doi: 10.1016/j.intimp.2019.106019. [DOI] [PubMed] [Google Scholar]
- 48.Wang S., et al. Valproic acid combined with zoledronate enhance γδ T cell-mediated cytotoxicity against osteosarcoma cells via the accumulation of mevalonate pathway intermediates. Front. Immunol. 2018;9:377. doi: 10.3389/fimmu.2018.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2