Abstract
Atlastin (ATL) GTPases undergo trans dimerization and a power strokelike crossover conformational rearrangement to drive endoplasmic reticulum membrane fusion. Fusion depends on GTP, but the role of nucleotide hydrolysis has remained controversial. For instance, nonhydrolyzable GTP analogs block fusion altogether, suggesting a requirement for GTP hydrolysis in ATL dimerization and crossover, but this leaves unanswered the question of how the ATL dimer is disassembled after fusion. We recently used the truncated cytoplasmic domain of wild-type Drosophila ATL (DATL) and a novel hydrolysis-deficient D127N variant in single turnover assays to reveal that dimerization and crossover consistently precede GTP hydrolysis, with hydrolysis coinciding more closely with dimer disassembly. Moreover, while nonhydrolyzable analogs can bind the DATL G domain, they fail to fully recapitulate the GTP-bound state. This predicted that nucleotide hydrolysis would be dispensable for fusion. Here we report that the D127N variant of full-length DATL drives both outer and inner leaflet membrane fusion with little to no detectable hydrolysis of GTP. However, the trans dimer fails to disassemble and subsequent rounds of fusion fail to occur. Our findings confirm that ATL mediated fusion is driven in the GTP-bound state, with nucleotide hydrolysis serving to reset the fusion machinery for recycling.
INTRODUCTION
Guanine nucleotide-binding (G) proteins function broadly as molecular switches that interconvert between a GTP-bound ‘on’ state and a GDP-bound ‘off’ state to regulate diverse cellular processes. The G proteins that established this paradigm include the translation initiation and elongation factors, the trimeric G proteins, and the Ras superfamily G proteins, all members of the P-loop class of NTPases. Each is turned on by heterocomplex formation with its respective guanine nucleotide-exchange factor (GEF), which accelerates GDP release and GTP binding. GTP binding induces changes in G domain switch I and switch II regions to promote interactions with one or more effectors for execution of function. The G proteins are turned off when a GTPase-activating protein (GAP) binds the G domain and complements or stabilizes the active site to accelerate GTP hydrolysis (Bourne et al., 1991).
More recently, G proteins activated by nucleotide-dependent dimerization (GADs) have been identified as a distinct functional P-loop G protein class. GADs, which include the signal recognition particle (SRP) and its receptor (SR), members of the dynamin superfamily, the septins, MnmE, and many others, appear to use a regulatory mechanism that stands apart from the more familiar G protein paradigm (Gasper et al., 2009). First, GADs have a relatively low nucleotide affinity and so they readily exchange GDP for GTP without need for a GEF; and second, GTP hydrolysis is triggered when the G domains homodimerize upon GTP binding, leading to the ordering of catalytic residues in the G domain. Thus GADs in general need neither an external GEF nor GAP (Gasper et al., 2009).
By analogy to canonical G proteins, the GADs were initially proposed to execute their biological function in the GTP-bound homodimer state with GTP hydrolysis disassembling the dimer to terminate G protein function (Gasper et al., 2009). However, to our knowledge, this has been demonstrated clearly for only one GAD, the SRP-SR complex. During secretory and membrane protein targeting to the endoplasmic reticulum (ER), formation of a GTP-bound “loose” pseudo homodimer between the G (termed NG) domains of SRP and SR delivers the ribosome nascent chain complex (RNC) bearing a signal (or membrane anchor) sequence to the ER, whereupon a series of large-scale conformational rearrangements transfer the RNC to the ER Sec61p translocase (Lee et al., 2021). Transfer coincides with reordering of switch I and II regions and conversion to a ‘tight’ dimer wherein the NG domains mutually activate one another for GTP hydrolysis, thereby terminating the SRP-SR interaction and resetting SRP and SR for reuse (Shan, 2016). In this paradigm, GTP hydrolysis is not required for ER targeting per se, but is necessary for recycling; a nonhydrolyzable GTP analog can replace GTP in an in vitro ER targeting assay, but SRP-SR can no longer be recycled because it cannot be disassembled (Connolly et al., 1991).
Whether the SRP-SR GTPase paradigm extends to other GADs is unclear. For instance, the MnmE (TrmE) G protein, which together with MnmG (GidA) plays a vital role in tRNA modification, forms a tight G domain homodimer with notable reordering of catalytic residues in the presence of the transition state analog GDP-AlF4–, Mg+2 and K+ (Scrima and Wittinghofer, 2006). However, unlike SRP-SR, a nonhydrolyzable GTP analog does not support tRNA modification in an in vitro reconstitution assay; likewise, a hydrolysis defective MnmE variant is inactive (Meyer et al., 2009). In other examples, members of the dynamin superfamily (Praefcke and McMahon, 2004; Ford and Chappie, 2019), including dynamin, guanylate binding protein, atlastin (ATL), and mitofusin, all undergo G domain dimerization in the presence of GTP, nonhydrolyzable GTP analogs and/or GDP-AlF4–. Accordingly, crystal structures with one or more of these analogs show G domain homodimers in which the switch regions and catalytic residues are optimally positioned for nucleotide hydrolysis, though some of the details differ between superfamily members (Chappie et al., 2010; Ghosh et al., 2006; Byrnes et al., 2013; Cao et al., 2017). But as with MnmE, in vitro reconstitution assays for dynamin (Sweitzer and Hinshaw, 1998; Roux et al., 2006; Bashkirov et al., 2008; Roux and Antonny, 2008; Pucadyil and Schmid, 2008; Mattila et al., 2015), mitofusin (Brandt et al., 2016; Qi et al., 2016), and ATL (Orso et al., 2009; Saini et al., 2014; Liu et al., 2015) all show a block in function when either nonhydrolyzable GTP analogs or certain hydrolysis-deficient variants are used. Thus these GADs are generally thought to require nucleotide hydrolysis to execute their function either because formation of the functional homodimer cannot be achieved without it or because execution of function requires dimer disassembly. An important caveat is that a nonhydrolyzable analog may not accurately evoke the GTP bound state. Additionally, a mutation in the active site that disrupts GTP hydrolysis may impair other steps in the reaction cycle. Nevertheless, it is entirely plausible that individual GADs differ in how GTP hydrolysis is coupled to their respective biological functions.
For the dynamin superfamily GTPases, the difference may stem from the nature of their biological function. Dynamin, mitofusin, and ATL are mechanochemical enzymes that operate under membrane load. Each performs a power strokelike conformational change concomitant with G domain dimerization that serves to remodel the underlying membrane (Ford and Chappie, 2019). Because these enzymes must carry out their reaction cycle under membrane load, the energy obtained from GTP binding may be insufficient for execution of function. Additionally, it could be argued that most GADs belong to the TRAFAC (translation factors) phylogenetic class of P-loop NTPases, whereas SRP/SR belongs to the SIMIBI (SRP, MinD, and BioD) class (Leipe et al., 2002) and could have evolved a unique regulatory mechanism. On the other hand, the microtubule subunit tubulin and its bacterial Z ring counterpart FtsZ, for whom polymerization and depolymerization is also coupled to mechanical work, form functional polymers in the GTP-bound state and disassemble following nucleotide hydrolysis (Carlier and Pantaloni, 1981; Hyman et al., 1992; Mukherjee and Lutkenhaus, 1998). Tubulin and FtsZ are Rossman-type fold GTPases evolutionarily unrelated to either the TRAFAC or the SIMIBI class of P-loop ATPases (Kull and Fletterick, 1998); Thus stabilization of the functional dimer or oligomer in the GTP-bound state and destabilization in the GDP-bound state may be a shared feature among widely diverse G proteins. Even so, the preponderance of data using nonhydrolyzable GTP analogs and hydrolysis-deficient mutant variants has been interpreted as a requirement for GTP hydrolysis for many GADs to assemble and carry out their biological function. Paradoxically, this leaves unanswered the question of how the GAD homodimers are subsequently disassembled for recycling.
To address this conundrum, we have focused our attention on the ER-localized G protein ATL, which catalyzes membrane fusion and is required for the branched morphology of the ER network (Orso et al., 2009; Hu et al., 2009; Morin-Leisk et al., 2011; Zhao et al., 2016). ATL is arguably the simplest representative GAD because it does not undergo significant higher order oligomerization and dimerizes primarily through an interaction interface that includes the G domain (Bian et al., 2011; Byrnes and Sondermann, 2011; Byrnes et al., 2013). Despite its simplicity, DATL alone is sufficient to mediate GTP-dependent membrane fusion in an in vitro reconstitution assay (Orso et al., 2009).
In recently published work, we used truncated DATL labeled with conformationally sensitive fluorescent probes in single turnover assays to find that formation of the DATL crossover dimer (Bian et al., 2011; Byrnes and Sondermann, 2011; Byrnes et al., 2013), which represents the “post power stroke” conformation and considered to reflect the postfusion state (Daumke and Praefcke, 2011; Moss, Daga and McNew, 2011; McNew et al., 2013; Hu and Rapoport, 2016; Wang and Rapoport, 2019), consistently precedes the hydrolysis of GTP (Winsor et al., 2018). This surprising finding predicted that DATL’s fusion function should also take place prior to nucleotide hydrolysis, a prediction at odds with the long-standing observation that either nonhydrolyzable GTP analogs or a hydrolysis defective variant blocks fusion (Orso et al., 2009; Saini et al., 2014; Liu et al., 2015). The discrepancy was reconciled by our observation that a GTP-dependent conformation change in DATL’s G domain, as monitored by changes in intrinsic tryptophan fluorescence within the truncated protein, cannot be fully recapitulated either by nonhydrolyzable GTP analogs or by the commonly used R48A hydrolysis defective DATL variant (Winsor et al., 2018). In addition, we identified a novel switch II catalytic mutation, D127N, which allows rapid crossover dimerization even though it slows GTP hydrolysis (Winsor et al., 2018). Thus we hypothesized that the fusion function of DATL may be executed in the GTP-bound state, with hydrolysis serving to recycle the protein for reuse.
A major limitation of our previous study was that our single turnover assays were carried out in the soluble phase using a truncated form of DATL as a proxy for full-length membrane-anchored DATL. Therefore one could argue that the observed functional dimerization without GTP hydrolysis could be due to the lack of membrane load in the system. Here we address this major caveat by directly measuring GTP hydrolysis and dimerization by full-length DATL, under membrane load, as it carries out its membrane fusion function. We use the hydrolysis-deficient D127N DATL variant to show, for the first time, that membrane-anchored DATL is capable of driving GTP-dependent membrane fusion well before hydrolyzing GTP. Furthermore, we show that delayed nucleotide hydrolysis prevents the rapid disassembly and recycling of the postfusion dimer, thereby limiting the full extent of membrane fusion. Thus the SRP-SR-like paradigm appears to extend to at least one member of the dynamin superfamily of membrane remodeling GADs.
RESULTS AND DISCUSSION
D127N DATL promotes only one round of lipid mixing
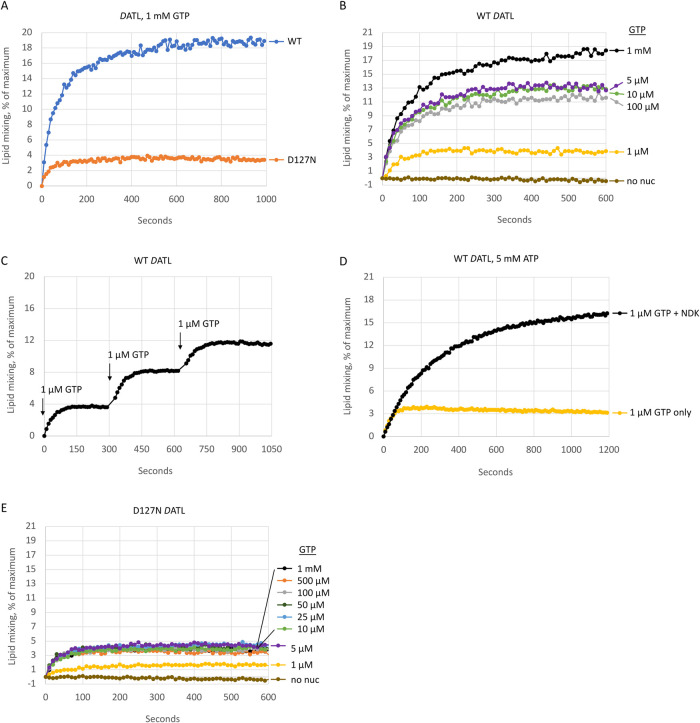
To assay membrane fusion, we utilized DATL protein purified from a HEK293-derived mammalian suspension cell line since our recent work revealed DATL from these cells to be more active than protein purified from Escherichia coli (Crosby et al., 2022). Wild type and D127N DATL were incorporated into preformed liposomes with similar efficiency (Supplemental Figure S1). As before, we used a well-established fusion assay that monitors the dequenching of fluorescently labeled lipids upon mixing and dilution with unlabeled lipids (Orso et al., 2009). When incorporated into labeled and unlabeled vesicles at a protein-to-lipid molar ratio of 1:1000 and assayed with saturating GTP, wild-type DATL catalyzed rapid and robust GTP-dependent lipid mixing with an initial rate of ∼0.3 s–1 (Figure 1A). Lipid mixing by the D127N DATL variant occurred on a similar time scale, though its initial rate was somewhat lower at ∼0.1 s–1. A more pronounced difference was that D127N DATL reached a plateau more quickly than the wild type. After the first ∼100 s, very little further increase was observed, resulting in a fivefold lower final extent of fusion (Figure 1A). This difference is similar to that seen previously when the proteins were purified from E. coli (Winsor et al., 2018).
FIGURE 1:
The hydrolysis-deficient D127N DATL variant promotes only one round of lipid mixing. (A–E) Wild type or D127N DATL reconstituted into donor and acceptor vesicles and fusion monitored over time as the dequenching of NBD-labeled lipid in donor vesicles upon mixing with unlabeled lipid in acceptor vesicles at 37°C. (A) Lipid mixing by wild type or D127N DATL after the addition of 1 mM GTP. (B) Lipid mixing by wild-type DATL after the addition of the indicated final concentrations of GTP. (C) Lipid mixing by wild-type DATL after the addition of 1 µM GTP followed by two additional spikes of 1 µM GTP at the indicated times. (D) Lipid mixing by wild-type DATL preincubated with 5 mM ATP after the addition of 1 µM GTP with or without 1 U nucleoside diphosphate kinase. (E) Lipid mixing by D127N DATL after the addition of the indicated final concentrations of GTP. All lipid mixing was performed at a 1:1000 protein/lipid ratio, a 1:2 donor/acceptor vesicle ratio, and DATL present at 0.6 µM (final concentration). All data are the average of two technical replicates and similar results obtained with at least two biological replicates.
Based on our earlier observation that the truncated soluble domain of D127N DATL undergoes dimer disassembly more slowly than the wild type (Winsor et al., 2018), a defect in disassembly and recycling that would limit fusion to only one round was a plausible explanation for the early fusion plateau observed for the D127N variant. If so, we predicted that wild-type DATL would behave similarly to the variant were GTP limited to enable only one round of GTP hydrolysis. Indeed, when GTP was reduced to 1 µM, a GTP-to-DATL molar ratio >1 but <2, wild-type DATL also plateaued early, mimicking the D127N variant at saturating GTP (Figure 1B, compare wild-type trace at 1 µM GTP with D127N trace in Figure 1A). To test whether the early plateau by the wild type under this condition was due solely to the lack of sufficient GTP to support multiround fusion, we spiked the reaction with additional GTP. Remarkably, each subsequent spike of 1 µM GTP gave rise to a stepwise increase consistent with one added round of fusion per GTP spike (Figure 1C). Also as predicted, inclusion of nucleoside diphosphate kinase and 5 mM ATP, to regenerate GTP from any GDP produced during the fusion reaction, led to sustained, multiround fusion with 1 µM GTP (Figure 1D). In the absence of a regenerating system, intermediate GTP levels produced intermediate plateaus for the wild type, as expected (Figure 1B). Perhaps surprisingly, a similar extent of fusion was observed over a wide range of 5–100 µM GTP and this was only surpassed at the substantially higher 1 mM GTP concentration (Figure 1B). However, a straightforward explanation for the lack of difference between 5 and 100 µM GTP could be gleaned from the multiround fusion data obtained with the stepwise addition of 1 µM GTP (Figure 1C). As each addition of 1 µM GTP led to a 4% of maximal increase in fusion, 5 µM GTP, sufficient for ∼ 5 rounds, would be predicted to drive 20% of maximal fusion, which corresponds to the maximal fusion signal achievable under these assay conditions. Arguably more importantly, the extent of fusion by the D127N variant was largely independent of GTP concentration; the fusion kinetic at 5 µM GTP was no different from that at 1 mM GTP (Figure 1E). Collectively, these data are consistent with the idea that the D127N variant cannot catalyze more than one round of fusion due to an inability to recycle.
Of note, the GTP concentration required to elicit the same ∼ 1 round of fusion was modestly higher for the D127N variant than for the wild type; the D127N variant required 5 µM GTP to achieve the same initial rate and final extent of fusion as the wild type at 1 µM GTP (Figure 1, B and E). This suggests a modestly reduced nucleotide and/or dimer affinity for the D127N variant. Though it remains to be rigorously established, we favor the latter explanation because previous intrinsic tryptophan fluorescence measurements revealed a similar half maximal GTP concentration required to elicit an early G domain conformation change, indicating a similar GTP affinity for the D127N variant (Winsor et al., 2018). On the other hand, the amplitude of the tryptophan fluorescence change was not identical, possibly indicative of a slightly different G domain conformation for the D127N variant that does not favor dimerization quite as strongly as the wild type (Winsor et al., 2018). Nevertheless, even at 1 µM GTP, the D127N variant promoted some fusion (Figure 1E).
D127N DATL promotes both inner and outer leaflet lipid mixing
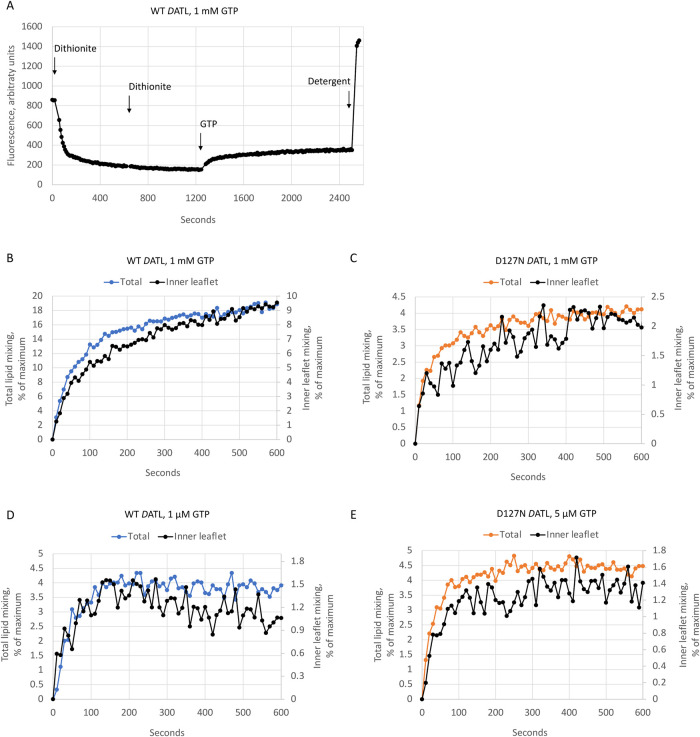
A recycling defect could fully account for the lower fusion plateau observed with D127N DATL. However, a lack of inner leaflet lipid mixing by the variant could also contribute. To address this possibility, we utilized sodium dithionite to selectively reduce the outer leaflet NBD-labeled lipids in our fusion assay to a nonfluorescent derivative (McIntyre and Sleight, 1991). Because dithionite does not readily cross intact lipid bilayers (Langner and Hui, 1993), NBD-labeled head groups in the inner leaflet should be spared, and therefore any NBD fluorescence dequenching observed after dithionite treatment should reflect inner leaflet lipid mixing (Weber et al., 1998; Xu et al., 2005; Lu et al., 2008). To ensure complete reduction of all outer leaflet NBD-lipids, dithionite was added in two rounds to achieve a flat baseline fluorescence prior to GTP addition, as shown in a sample raw fluorescence trace for the wild type (Figure 2A). Somewhat curiously, and as observed previously (Orso et al., 2009), ∼70% of the total initial fluorescence was lost from DATL proteoliposomes after dithionite quenching; more than the 50% expected were NBD-labeled lipids distributed equally between outer and inner leaflets. Protein-free liposomes lost the expected 50% of the total initial fluorescence after two rounds of dithionite quenching (Supplemental Figure S2), raising a concern that the DATL proteoliposomes might be leaky. However, two further rounds of fresh dithionite addition caused little further loss from either DATL proteoliposomes or protein-free liposomes (Supplemental Figure S2), indicating relatively stable retention of NBD fluorescence in the inner leaflet of both types of liposomes after the first two rounds of dithionite treatment. Indeed, after normalization, GTP addition triggered a rise in NBD fluorescence with the expected kinetics for inner leaflet mixing by both wild type and D127N DATL at either saturating GTP (Figure 2B for wild-type DATL and 2C for D127N DATL) or under limiting GTP conditions (Figure 2D for wild-type DATL at 1 µM GTP and 2E for D127N DATL at 5 µM GTP). Based on these results, we conclude that D127N DATL promotes inner leaflet mixing as well as the wild type. Therefore the lower final extent of fusion by the D127N variant can be attributed to a recycling defect.
FIGURE 2:
The D127N DATL variant promotes both inner and outer leaflet lipid mixing. (A) Sample raw fluorescence trace monitoring inner leaflet mixing. Sodium dithionite was added twice to wild-type DATL donor and acceptor vesicles to ensure complete outer leaflet NBD quenching prior to the start of the reaction. After obtaining a flat baseline indicating no further quenching of the outer leaflet, 1 mM GTP was added to start the reaction. After completion of fusion, 0.5% Anaope X-100 was added to obtain maximum fluorescence. (B–E) Similar inner leaflet mixing kinetics were observed for wild type (B, D) and D127N (C, E) DATL at both saturating (B, C) and limiting (D, E) GTP. For reference, the data for each are plotted relative to total (inner plus outer) lipid mixing by the same variant at the same protein and GTP concentrations. All lipid mixing was performed at a 1:1000 protein/lipid ratio and a 1:2 donor/acceptor vesicle ratio. All data are the average of two technical replicates and similar results obtained with two biological replicates.
Membrane fusion occurs without GTP hydrolysis
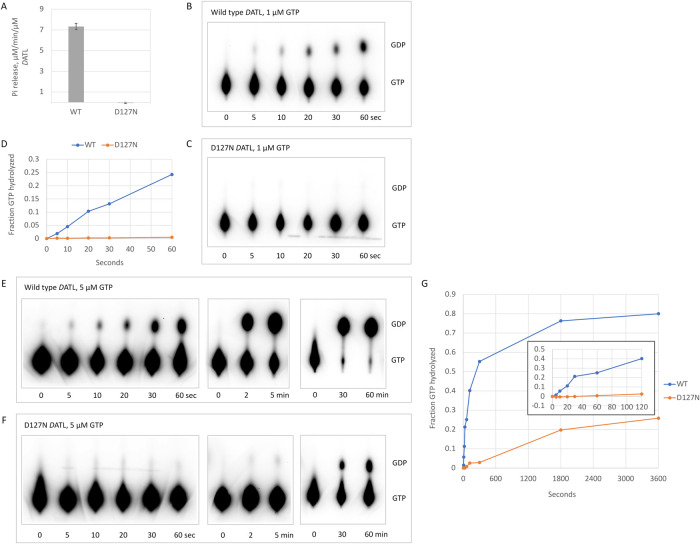
We next addressed the central question of whether the D127N variant could drive fusion in the GTP bound state prior to hydrolysis. We first used an assay that measures continuous inorganic phosphate release under saturating GTP conditions and observed a steady state rate of 7.3 µM/min/µM DATL for full-length wild-type membrane-anchored DATL. Under the same conditions, the D127N variant exhibited no detectable phosphate release (Figure 3A). This was reminiscent of the lack of steady state GTPase activity observed previously for the truncated soluble domain D127N DATL (Winsor et al., 2018).
FIGURE 3:
GTP hydrolysis by D127N DATL does not account for fusion. (A) Steady state inorganic phosphate release by full-length wild type and D127N DATL during membrane fusion. (B-G) GTP hydrolysis by full-length and D127N DATL during membrane fusion. GTP hydrolysis by wild type (B, E) or D127N (C, F) DATL at 1 µM (B, C) or 5 µM (E, F) 32P-α-GTP. (D) Quantification of B, C shows little or no hydrolysis by D127N DATL within the initial 60 s period of membrane fusion at 1 µM GTP. (G) Quantification of E, F shows little or no hydrolysis by D127N DATL within the first 120 s of membrane fusion at 5 µM GTP, though it is detected at later times (inset shows an enlarged view of the first 120 s). All assays were performed under fusion assay conditions with full-length DATL incorporated into liposomes at a 1:1000 protein/lipid ratio and DATL present at 0.6 µM (final concentration). In B, C, E, and F, reactions were acid quenched at the indicated time points and resolved by TLC. All TLC plates were exposed for 20 h to ensure maximal sensitivity for detecting GDP.
The block in the continuous phosphate release assay for D127N DATL indicated a clear defect in its GTPase cycle, but it did not distinguish an inability of D127N DATL to hydrolyze GTP from an inability to release phosphate that has been hydrolyzed. This left open the possibility that membrane-anchored D127N DATL can perform limited GTP hydrolysis, sufficient to drive at least one round of membrane fusion, though not releasing the cleaved phosphate. To examine this, we turned to an acid quenched fixed time point assay wherein GTP and any of its cleavage products that remain bound at a given time point are forcibly released by acid-induced protein denaturation. We focused on the first 60 s after GTP addition during which time the lipid mixing signal for both wild type and D127N DATL increased most rapidly (Figure 2, D and E). Additionally, we used 32P-α-GTP and thin-layer chromatography (TLC) to increase the sensitivity of the assay while maintaining the total GTP at low, limiting concentrations to minimize the number of potential reaction cycles. Finally, DATL and the lipids were maintained at precisely the same concentrations used in the fusion assay (Figure 2, D and E) so that the amount of GTP hydrolyzed could be related directly to the amount of lipid mixing.
The first set of 32P-α-GTP hydrolysis assays used 1 µM GTP and 0.6 µM DATL, the same, near stoichiometric ratio of GTP:DATL that produced ∼1 round of fusion for wild-type DATL. During the initial period, GTP hydrolysis by wild-type DATL increased linearly (Figure 3B, quantified in D), and by the end of the 60-s period, one quarter of all the GTP was hydrolyzed, which is an acceptable fraction for ∼1 round of GTP turnover. In contrast, D127N DATL exhibited little to no GTP hydrolysis during this period (Figure 3C, quantified in D), even though the variant promoted a measurable, albeit lower than wild type, level of fusion (Figure 1E). In the second set of assays, the GTP was increased to 5 µM while maintaining the same 0.6 µM DATL concentration for both wild type and D127N DATL (Figure 3, E and F). This condition was chosen because it produced a maximal fusion signal for D127N DATL (Figure 1E). Even at this higher GTP concentration, there was little to no detectable GTP hydrolyzed by D127N DATL within the first 120 s (Figure 3F, quantified in inset in G) when most of the fusion by the D127N variant was observed (Figures 1E and 2E). A significant amount of hydrolyzed GDP was observed only after 30 min (Figure 3F, quantified in G), which is well after the fusion signal for the variant had leveled off (Figures 1E and 2E). For comparison, wild-type DATL had hydrolyzed nearly all its GTP by 30 min (Figure 3E, quantified in G). The lack of any detectable GTP hydrolysis by D127N DATL until well after its fusion plateau indicates that membrane fusion does not require GTP hydrolysis.
GTP hydrolysis drives dimer disassembly
The lack of requirement for GTP hydrolysis in the fusion function of DATL strongly implied that nucleotide hydrolysis serves to disassemble the functional dimer for reuse as established for SRP-SR (Connolly et al., 1991). Consistent with this, we had observed a marked slowing of disassembly of the truncated soluble domain D127N dimer under single turnover conditions (Winsor et al., 2018). However, this remained untested for full-length DATL under the conditions of membrane fusion.
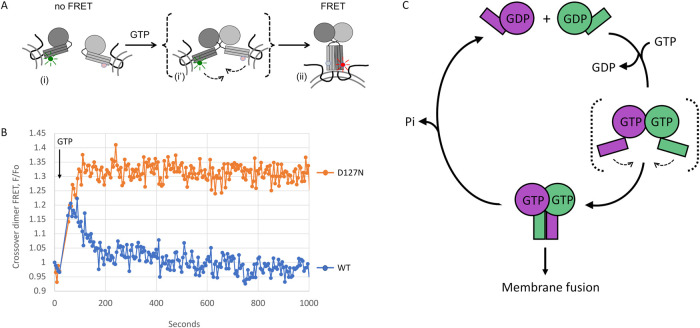
To monitor formation of the full-length functional dimer, we engineered a cysteine residue T364C on the three helical bundle (3HB) of both wild type and D127N DATL for Förster resonance energy transfer (FRET). The 3HB, which is attached to the G domain through a short flexible linker, undergoes a large power strokelike conformational rearrangement upon G domain dimerization (Bian et al., 2011; Byrnes and Sondermann, 2011; Byrnes et al., 2013). Because this brings the T364C residues on two 3HBs within ∼30 Å of the other, formation of the crossover dimer can be monitored by FRET (Figure 4A). Accordingly, we conjugated donor or acceptor FRET probes to the engineered T364C residue in full-length wild type or D127N DATL and separately incorporated donor dye- and acceptor dye-labeled protein into membrane vesicles at a 1:1000 protein/lipid ratio. Vesicles containing either donor- or acceptor-labeled DATL were then mixed as for the lipid mixing assay, though a slightly higher overall DATL concentration (2 µM) than typically used in the lipid mixing assay (0.6 µM) produced an optimal FRET signal, likely due to inactivation of some DATL protein during labeling. Upon addition of 5 µM GTP, a similar rapid increase in the FRET signal was observed for wild type and D127N DATL (Figure 4B), reflecting rapid crossover dimerization for both. However, the fate of the dimers diverged thereafter; whereas disassembly ensued quickly for wild-type DATL due to consumption of the limiting GTP in the assay (a 2:1 M ratio of GTP/DATL), the D127N DATL signal remained constant, as though locked in the assembled state for the duration of the assay. Based on the very slow rate of GTP hydrolysis by D127N DATL under these conditions (Figure 3F, quantified in inset in G), we could attribute the sustained FRET signal to a requirement for GTP hydrolysis for DATL dimer disassembly. Altogether, our data confirm a paradigm for DATL originally proposed for GADs (Gasper et al., 2009), in which the G protein executes its fusion function in the GTP-bound homodimer state with GTP hydrolysis triggering dimer disassembly to terminate the process and reset the fusion machinery for reuse.
FIGURE 4:
D127N DATL is blocked in dimer disassembly and model. (A) Schematic of FRET assay for dimerization and crossover by full-length membrane-anchored DATL. GTP-induced G domain dimerization and concomitant conformational changes bring donor dye- and acceptor dye-labeled 3HBs of opposing DATLs (i) into close parallel alignment to induce FRET (ii). A transient intermediate (i’) dimerized only through the G domain is shown in brackets. (B) D127N DATL is defective in dimer disassembly. Change in FRET acceptor fluorescence was measured by first establishing a baseline followed by 5 µM GTP addition at the timepoint indicated. FRET donor- and acceptor-labeled proteins were incorporated into vesicles at a 1:1000 protein/lipid ratio. Assays used a 1:2 donor/acceptor ratio at 2 µM final protein concentration. Data shown are the average of two technical replicate traces. (C) Model. GTP binding by ATL in opposing membranes induces trans G domain dimerization and a crossover conformational change that drives membrane fusion. GTP hydrolysis, induced only after stable dimerization has occurred, triggers subsequent dimer disassembly to enable reuse of ATL subunits for multiple rounds of fusion catalysis. A transient intermediate dimerized only through the G domain, which is speculated to initiate membrane tethering, is shown in brackets.
From extensive structural, biophysical, and biochemical studies over the past decade, a consensus model for the DATL fusion mechanism has emerged (McNew et al., 2013; Hu and Rapoport, 2016). However, the question of how the chemical energy of GTP is used to drive the fusion reaction cycle has long remained a topic of debate (McNew et al., 2013; Hu and Rapoport, 2016; Ford and Chappie, 2019). The results presented here together with our previous work (Winsor et al., 2018) resolve this debate. We propose that ATL follows the familiar precedent of G proteins, in that GTP binding is sufficient to activate its biological function, and the hydrolysis of GTP serves to disassemble the functional state to reset the G protein for the next reaction cycle (see model in Figure 4C). As in the SRP-SR paradigm, in which GTP binding-driven dimerization of SRP and SR sets off a multistep cascade of interactions and conformational changes that ultimately deliver the RNC to the Sec61p translocase (Lee et al., 2021), we propose that GTP binding-driven dimerization by DATL sets off a cascade of conformational changes and protein–lipid interactions that ultimately drive the tethering and fusion of opposing lipid bilayers. For both the SRP-SR pseudo homodimer and the DATL homodimer, a relatively late step in the conformation cascade is the reordering of switch I and II and catalytic residues which serve to activate the hydrolysis of the bound nucleotide, bringing about the demise of the active state and a return of the G protein to its original GDP-bound conformation. For SRP-SR, this late step appears to be further fine-tuned by a negative feedback regulatory mechanism such that pseudo homodimer disassembly is more likely to occur only after successful transfer (Lee et al., 2021). It is unclear whether there is an analogous fine-tuning in the ATL mechanism to increase the success rate of membrane fusion, but DATL variants that are defective in fusion due to mutations outside the G domain appear to retain a near normal GTPase rate (Saini et al., 2014), suggesting a lack of feedback regulation.
Finally, the extent to which the ATL regulatory paradigm extends to other GADs remains an open question. It is conceivable that the widely observed inhibition of biological function of GADs by nonhydrolyzable GTP analogs could be due to the widespread inability of the analogs to evoke all the conformational changes necessary for dimer function; similarly, the inhibition of function by hydrolysis defective mutant variants could be due to underappreciated additional consequences of the mutations on the GTPase cycle. However, it is also conceivable that nature has found diverse ways by which the chemical energy of GTP can be harnessed by different GADs to carry out useful work. Clear examples of such mechanistic diversity are the evolutionarily related motor proteins myosin and kinesin, which use distinct strategies for coupling cargo movement to the hydrolysis of ATP.
MATERIAL AND METHODS
Request a protocol through Bio-protocol.
Reagents and constructs
All DATL constructs used for protein expression and purification were both HA- and 6× His-tagged at the N-terminus in the pGW1-CMV vector and contained the following amino acid substitutions for the purpose of restricting reactivity with sulfydryl-reactive compounds to a single surface-exposed engineered cysteine: G343C, C350A, C429L, C452L, and C501A. We previously reported that a wild-type DATL version containing the indicated substitutions had fusion activity similar to that of the unaltered protein (Saini et al., 2014). All amino acid-substituted constructs were generated using PfuTurbo polymerase (Agilent Technologies, Santa Clara, CA)-mediated PCR mutagenesis and fully sequence confirmed (GENEWIZ, South Plainfield, NJ). All mutagenic oligos were from IDT (Coralville, IA). GTP and ATP was purchased from Sigma-Aldrich (St. Louis, MO); reconstituted to 100 mM stocks in 10 mM Tris, pH 8.0, and 1 mM EDTA; and stored at –80°C. Lipids were purchased from Avanti Polar Lipids (Birmingham, AL). 32P-α-GTP was purchased from PerkinElmer (Waltham, MA). Alexa Fluor 488/647 maleimide was from Thermo Fisher Scientific (Waltham, MA). Bovine nucleoside diphosphate kinase was from Sigma-Aldrich. Anapoe X-100, an ultrapure version of Triton X-100 packaged under argon, was from Anatrace (Maumee, OH) and used for the last steps of protein purification as well as for protein incorporation into liposomes.
Protein expression and purification
Two hundred milliters of Expi293 cells (Thermo Fisher Scientific) were cultured at 37°C and 8% CO2 per manufacturer’s protocol and transient transfections were done per manufacturer’s instructions. Two days after transfection, cells were harvested, washed once with cold PBS, and flash-frozen. All purifications steps were conducted on ice or at 4°C with chilled buffers. Cell pellets were resuspended in 20 ml lysis 1 (50 mM Tris, pH 8.0, 5 mM MgCl2, 300 mM NaCl, 10 mM imidazole, 10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 1 µg/ml pepstatin, 1 µM leupeptin, and 2 mM 2-mercaptoethanol [2-ME]) until homogenous; 5 ml of lysis 2 (50 mM Tris, pH 8.0, 5 mM MgCl2, 300 mM NaCl, 10 mM imidazole, 10% glycerol, 10% Triton X-100, 0.5 mM PMSF, 1 µg/ml pepstatin, 1 µg/ml leupeptin, and 2 mM 2-ME) was added and mixed, then diluted 1:1 with lysis 1 for a final Triton X-100 concentration of 1% and rotated for 30 min at 4°C. Samples were then centrifuged at 12,000 rpm in a F20-12 × 50 LEX rotor (Thermo Fisher Scientific) for 30 min. The supernatant was further centrifuged at 50,000 rpm in a Ti70 rotor (Beckman Coulter, Pasadena, CA) for 1 h. The final supernatant was filtered through a 0.45-µm filter and mixed with 0.25 ml Ni+2-NTA agarose (Qiagen, Hilden, Germany) overnight at 4°C. The next day the beads were poured into a column support, washed with 30 ml of wash 1 (50 mM Tris, pH 8.0, 5 mM MgCl2, 100 mM NaCl, 20 mM imidazole, 10% glycerol, 1% Triton X-100, 2 mM 2-ME) and 30 ml of wash 2 (50 mM Tris, pH 8.0, 5 mM MgCl2, 100 mM NaCl, 20 mM imidazole, 10% glycerol, 0.1% Anapoe X-100, 2 mM 2-ME). Protein was eluted in 0.25-ml fractions with elution buffer (50 mM Tris, pH 8.0, 5 mM MgCl2, 100 mM NaCl, 250 mM imidazole, 10% glycerol, 0.1% Anapoe X-100, 2 mM 2-ME). Fractions were flash-frozen and stored at –80°C. Samples of each fraction were resolved by SDS–PAGE and found to be >95% pure.
Steady GTPase assay
GTPase activity of DATL proteoliposomes were measured under steady-state conditions using the EnzChek Phosphate Assay Kit (Molecular Probes; Thermo Fisher Scientific) using a Tecan Spark plate reader (Tecan, Mannedorf, Switzerland). A standard reaction involved mixing 1 U/ml purine nucleoside phosphorylase, 0.2 mM 2-amino-6-mercapto-7-methylpurine riboside, 0.5 mM GTP and 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2 mM MgCl2, and 2 mM 2-ME in a total volume of 0.2 ml at 37°C. The reaction was started with the addition of 0.6 µM ATL (final concentration).
Radiolabeled GTPase assay
GTP hydrolysis assays used 32P-α-GTP. A standard reaction used DATL in proteoliposomes at 0.6 µM (final concentration) and the indicated final GTP concentrations (added from a stock containing 66.7 µM GTP at 2 µCi/µl 32P-α-GTP) in A100 buffer at 37°C. At indicated times, samples were quenched by addition of 1 vol of 1 M perchloric acid, neutralized with addition of 0.75 vol of 1 M KOAc, and cleared by centrifugation at 4000 rpm in a microcentrifuge for 2 min to remove denatured protein and membranes; 3 µl of each supernatant was loaded onto a PEI cellulose TLC plate (Macherey-Nagel, Düren, Germany), and nucleotides were resolved with a solution of 1 M LiCl2 and 1.6 M acetic acid. After drying, TLC plates were exposed to a phosphor screen for indicated times followed by visualization using a phosphorimager (Typhoon, Amersham, UK). The fraction of total GTP hydrolyzed to GDP in all assays were quantified in ImageJ (National Institutes of Health, Bethesda, MD).
Preparation of liposomes, lipid-mixing fusion assay
Lipids in chloroform were dried down by rotary evaporation for 1 h, hydrated by resuspension in A100 buffer (25 mM HEPES, pH 7.4, 100 mM KCl, 10% glycerol, 1 mM EDTA, and 2 mM 2-ME) at a final 10 mM lipid concentration and subjected to 11 freeze–thaw cycles in liquid N2 and 42°C water bath. Liposomes (100–300 nm diameter) were formed by extrusion through 100 nm polycarbonate filters 15× using the LipoFast LF-50 extruder (Avestin, Ottawa, Canada) and checked for size by dynamic light scattering (Zen3600, Malvern Panalytical, Malvern, UK). Purified DATL was incorporated at a 1:1000 protein/lipid ratio into labeled and unlabeled liposome populations at an effective detergent/lipid ratio of ∼0.7 by incubating protein and lipid at 4°C for 1 h followed by four 1 h detergent-removal incubations with SM-2 Bio-Beads (Bio-Rad, Hercules, CA) at 1 g beads per 70 mg Anapoe X-100. This was desalted over a 2.4-ml Sephadex G-25 column into A100 buffer, stored at 4°C, and used the same day or flash-frozen and stored at −80°C. Unlabeled liposomes consisted of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC) and 1,2-dioleoylsn-glycero-3-phospho-l-serine (PS) at an 85:15 M ratio. Labeled liposomes consisted of PC:PS:1,3-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (NBD):1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) at an 82:15:1.5:1.5 M ratio. For the fusion assay, proteoliposomes (0.6 mM total lipid final) were incubated in A100 buffer containing 5 mM MgCl2 at a 1:2 labeled/unlabeled ratio. Following a 5-min incubation at 37°C, the indicated GTP concentration or buffer was added via multichannel pipette and fluorescence dequenching of NBD monitored at 37°C in a Tecan Spark plate reader (Tecan) at 10-s intervals at 538 nm after excitation at 460 nm. After 60 min, 0.5% Anapoe X-100 was added for determination of the maximum possible dequenching signal. Data were plotted using the equation ([Fluorescence observed—Initial fluorescence]/ [Maximum fluorescence—Initial fluorescence]) *100. The slow loss of fluorescence due to photobleaching was accounted for by subtracting the minus GTP value at each time from the plus GTP value. For inner leaflet lipid mixing, the assay was set up as in the above except that the outer leaflet NBD was first reduced to the nonfluorescent derivative 7-amino-2,1,3-benzoxadiazol with the membrane-impermeable compound sodium dithionite. Dithionite was added twice, 2 and 1 mM, until a flat NBD baseline fluorescence was obtained after which the indicated GTP concentration was added and inner leaflet NBD fluorescence dequenching monitored at 10-s intervals. All lipid mixing was performed at a 1:1000 protein/lipid ratio. All lipid mixing data were the average of at least two independent traces, typically collected from a single protein prep. When repeated with an independent protein prep, the traces were similar, with only a 5–10% deviation in initial fusion rates.
FRET assay for crossover dimerization
Purified, detergent solubilized DATL was labeled with Alexa Fluor 488 (donor) and 647 (acceptor) maleimide on an engineered 3HB cysteine T364C (also C429L, C452L, C501A, C350A). For labeling, each engineered protein was desalted over 0.5-ml Zeba Spin columns 40K MWCO (Thermo Fisher) into labeling buffer (50 mM Tris, pH 7.4, 5 mM MgCl2, 100 mM NaCl, 250 mM imidazole, 10% glycerol, 50 µM TCEP, 0.1% Anaope X-100). Dye was added at a 1:1 protein/dye molar ratio and incubated for 2 h at 4°C. Labeled protein was then desalted twice as previously described to remove free dye (Winsor et al., 2018) while transferring to A100 buffer with 0.1% Anaope X-100. Proteins were then incorporated into preformed vesicles at a 1:1000 protein/lipid ratio as described for the fusion assay, and assays were performed using 5 µM GTP and 2 µM total DATL (final concentrations). This DATL concentration, higher than the 0.6 µM used for all other assays, was required to observe a robust FRET signal, likely due to some inactivation of the DATL during labeling. FRET assays used a 1:2 donor/acceptor ratio and acceptor (670 nm) fluorescence was monitored after 490-nm donor excitation at 10-s intervals in a Tecan Spark plate reader. A baseline fluorescence was established followed by GTP addition at the indicated times.
Supplementary Material
Acknowledgments
The authors thank B. Schmidt and members of the Woolford lab for generous sharing of their time and equipment. A. Vulgamott, S. Bryce and A. Linstedt provided helpful comments on the manuscript. This work was supported by a grant from the National Institute of General Medical Sciences, National Institutes of Health (R01GM107285 to T.H.L.). The authors declare no competing financial interests.
Abbreviations used:
- 2-MER
2-mercaptoethanol
- 3HB
three helical bundle
- ATL
atlastin
- DATL
Drosophila atlastin
- FRET
Förster resonance energy transfer
- GAD
G protein activated by nucleotide-dependent dimerization
- GAP
GTPase-activating protein
- GEF
guanine nucleotide-exchange factor
- G protein
GTP binding protein
- PMSF
phenylmethylsulfonyl fluoride
- RNC
ribosome nascent chain complex
- SR
signal recognition
- SRP
signal recognition particle
- TLC
thin-layer chromatography.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-05-0164) on September 21, 2022.
REFERENCES
- Bashkirov PV, Akimov SA, Evseev AI, Schmid SL, Zimmerberg J, Frolov VA (2008). GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell 135, 1276–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X, Klemm RW, Liu TY, Zhang M, Sun S, Sui X, Liu X, Rapoport TA, Hu J (2011). Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc Natl Acad Sci USA 108, 3976–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F (1991). The GTPase superfamily: conserved structure and molecular mechanism. Nature 349, 117–127. [DOI] [PubMed] [Google Scholar]
- Brandt T, Cavellini L, Kühlbrandt W, Cohen MM (2016). A mitofusin-dependent docking ring complex triggers mitochondrial fusion in vitro. eLife 5, e14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes LJ, Singh A, Szeto K, Benvin NM, O’Donnell JP, Zipfel WR, Sondermann H (2013). Structural basis for conformational switching and GTP loading of the large G protein atlastin. EMBO J 32, 369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes LJ, Sondermann H (2011). Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc Natl Acad Sci USA 108, 2216–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao YL, Meng S, Chen Y, Feng JX, Gu DD, Yu B, Li YJ, Yang JY, Liao S, Chan DC, Gao S (2017). MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion. Nature 542, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Pantaloni D (1981). Kinetic analysis of guanosine 5’-triphosphate hydrolysis associated with tubulin polymerization. Biochemistry 20, 1918–1924. [DOI] [PubMed] [Google Scholar]
- Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F (2010). G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature 465, 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T, Rapiejko PJ, Gilmore R (1991). Requirement of GTP hydrolysis for dissociation of the signal recognition particle from its receptor. Science 252, 1171–1173. [DOI] [PubMed] [Google Scholar]
- Crosby D, Mikolaj MR, Nyenhuis SB, Bryce S, Hinshaw JE, Lee TH (2022). Reconstitution of human atlastin fusion activity reveals autoinhibition by the C terminus. J Cell Biol 221, e202107070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daumke O, Praefcke GJ (2011). Structural insights into membrane fusion at the endoplasmic reticulum. Proc Natl Acad Sci USA 108, 2175–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MGJ, Chappie JS (2019). The structural biology of the dynamin-related proteins: New insights into a diverse, multitalented family. Traffic 20, 717–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A (2009). It takes two to tango: regulation of G proteins by dimerization. Nat Rev Mol Cell Biol 10, 423–429. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C (2006). How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature 440, 101–104. [DOI] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C (2009). A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138, 549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Rapoport TA (2016). Fusion of the endoplasmic reticulum by membrane-bound GTPases. Semin Cell Dev Biol 60, 105–111. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Salser S, Drechsel DN, Unwin N, Mitchison TJ (1992). Role of GTP hydrolysis in microtubule dynamics: Information from a slowly hydrolyzable analogue, GMPCPP. Mol Biol Cell 3, 1155–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull FJ, Fletterick RJ (1998). Is the tubulin/FtsZ fold related to the G-protein fold? [1]. Trends Cell Biol 8, 306–307. [DOI] [PubMed] [Google Scholar]
- Langner M, Hui SW (1993). Dithionite penetration through phospholipid bilayers as a measure of defects in lipid molecular packing. Chem Phys Lipids 65, 23–30. [DOI] [PubMed] [Google Scholar]
- Lee JH, Jomaa A, Chung SY, Fu YHH, Qian R, Sun X, Hsieh HH, Chandrasekar S, Bi X, Mattei S, et al. (2021). Receptor compaction and GTPase rearrangement drive SRP-mediated cotranslational protein translocation into the ER. Sci Adv 7, eabg0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L (2002). Classification and evolution of P-loop GTPases and related ATPases. J Mol Bio 317, 41–72. [DOI] [PubMed] [Google Scholar]
- Liu TY, Bian X, Romano FB, Shemesh T, Rapoport TA, Hu J (2015). Cis and trans interactions between atlastin molecules during membrane fusion. Proc Natl Acad Sci USA 112, E1851–E1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Zhang Y, Shin YK (2008). Supramolecular SNARE assembly precedes hemifusion in SNARE-mediated membrane fusion. Nat Struct Mol Biol 15, 700–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila JP, Shnyrova AV, Sundborger AC, Hortelano ER, Fuhrmans M, Neumann S, Müller M, Hinshaw JE, Schmid SL, Frolov VA (2015). A hemi-fission intermediate links two mechanistically distinct stages of membrane fission. Nature 524, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre JC, Sleight RG (1991). Fluorescence assay for phospholipid membrane asymmetry. Biochemistry 30, 11819–11827. [DOI] [PubMed] [Google Scholar]
- McNew JA, Sondermann H, Lee T, Stern M, Brandizzi F (2013). GTP-dependent membrane fusion. Annu Rev Cell Dev Biol 29, 529–550. [DOI] [PubMed] [Google Scholar]
- Meyer S, Wittinghofer A, Versées W (2009). G-domain dimerization orchestrates the tRNA wobble modification reaction in the MnmE/GidA complex. J Mol Biol 392. 10.1016/j.jmb.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Morin-Leisk J, Saini SG, Meng X, Makhov AM, Zhang P, Lee TH (2011). An intramolecular salt bridge drives the soluble domain of GTP-bound atlastin into the postfusion conformation. J Cell Bio 195, 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss TJ, Daga A, McNew JA (2011). Fusing a lasting relationship between ER tubules. Trends Cell Biol 21, 416–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J (1998). Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J 17, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, McNew JA, Daga A (2009). Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460, 978–983. [DOI] [PubMed] [Google Scholar]
- Praefcke GJ, McMahon HT (2004). The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol 5, 133–147. [DOI] [PubMed] [Google Scholar]
- Pucadyil TJ, Schmid SL (2008). Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell 135, 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Yan L, Yu C, Guo X, Zhou X, Hu X, Huang X, Rao Z, Lou Z, Hu J (2016). Structures of human mitofusin 1 provide insight into mitochondrial tethering. J Cell Biol 215, 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A, Uyhazi K, Frost A, Camilli PD (2006). GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature 441, 528–531. [DOI] [PubMed] [Google Scholar]
- Roux A, Antonny B (2008). The long and short of membrane fission. Cell 135, 1163–1165. [DOI] [PubMed] [Google Scholar]
- Saini SG, Liu C, Zhang P, Lee TH (2014). Membrane tethering by the atlastin GTPase depends on GTP hydrolysis but not on forming the crossover configuration. Mol Biol Cell 25, 3942–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrima A, Wittinghofer A (2006). Dimerisation-dependent GTPase reaction of MnmE: How potassium acts as GTPase-activating element. EMBO J 25, 2940–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan SO (2016). ATPase and GTPase tangos drive intracellular protein transport. Trends Biochem Sci 41, 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweitzer SM, Hinshaw JE (1998). Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell 93, 1021–1029. [DOI] [PubMed] [Google Scholar]
- Wang N, Rapoport TA (2019). Reconstituting the reticular ER network—Mechanistic implications and open questions. J Cell Sci 132, jcs227611. [DOI] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE (1998). SNAREpins: minimal machinery for membrane fusion. Cell 92, 759–772. [DOI] [PubMed] [Google Scholar]
- Winsor J, Machi U, Han Q, Hackney DD, Lee TH(2018). GTP hydrolysis promotes disassembly of the atlastin crossover dimer during ER fusion. J Cell Biol 217, 4184–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhang F, Su Z, McNew JA, Shin YK (2005). Hemifusion in SNARE-mediated membrane fusion. Nat Struct Mol Biol 12, 417–422. [DOI] [PubMed] [Google Scholar]
- Zhao G, Zhu PP, Renvoisé B, Maldonado-Báez L, Park SH, Blackstone C (2016). Mammalian knock out cells reveal prominent roles for atlastin GTPases in ER network morphology. Exp Cell Res 349, 32–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.