Abstract
The biogenesis of lipid droplets (LDs), key organelles for cellular lipid storage and homeostasis, remains poorly understood. Seipin is essential to normal LD biogenesis but exactly how it regulates LD initiation remains to be elucidated. Our previous results suggested that seipin may bind anionic phospholipids such as PI(3)P. Here, we investigate whether PI(3)P is functionally linked to seipin and whether PI(3)P can also impact LD biogenesis. In seipin-deficient cells, there were enlarged PI(3)P puncta where its effector, DFCP1, also appeared to congregate. Reducing cellular PI(3)P partially rescued the defective LD initiation caused by seipin deficiency. Increasing PI(3)P impeded the lipidation of nascent LDs. We further demonstrated that DFCP1 localized to LDs and facilitated the efficient lipidation of nascent LDs. However, the normal function and localization of DFCP1 were disrupted when cellular PI(3)P homeostasis was perturbed. Our results thus identify PI(3)P as a novel regulator of LD initiation and suggest that PI(3)P may impact the biogenesis of LDs through DFCP1.
INTRODUCTION
Lipid droplets (LDs) are evolutionarily conserved organelles present in nearly all organisms. LDs store neutral lipids, prevent cellular lipotoxicity, and regulate a number of cellular processes such as membrane and lipid trafficking, protein storage and degradation, and bacterial and viral infection (Walther et al., 2017; Gao et al., 2019b; Olzmann and Carvalho, 2019; Bosch et al., 2020). Moreover, the accumulation of enlarged LDs in adipocytes, hepatocytes, and macrophages is associated with common metabolic disorders including obesity, hepatic steatosis, and cardiovascular diseases. In most cells, LDs make up a hydrophobic core of triacylglycerols (TAGs) and/or sterol esters (SEs), which are enclosed by a monolayer of amphipathic lipids. LDs originate from highly curved regions of the endoplasmic reticulum (ER), but the molecular mechanisms governing their biogenesis remain poorly understood (Walther et al., 2017; Gao et al., 2019b; Santinho et al., 2020).
Seipin, an evolutionarily conserved protein residing in the ER, plays a critical role in the initiation and expansion of LDs, as well as in adipogenesis (Szymanski et al., 2007; Fei et al., 2008, 2011a; Cui et al., 2011; Gao et al., 2019b). We and others first discovered the role of seipin in LD formation through genetic screens in yeast: yeast cells lackingseipin/Fld1p/Sei1p are characterized by the presence of many small, clustered LDs and a few “supersized” LDs (Szymanski et al., 2007; Fei et al., 2008). Seipin deficiency in mammalian cells impeded the lipidation of initial LDs, formed supersized LDs after prolonged oleate treatment, and disrupted normal ER-LD contacts (Pagac et al., 2016; Salo et al., 2016; Wang et al., 2016; Chung et al., 2019). Importantly, loss-of-function mutations of seipin in humans cause the most severe form of congenital generalized lipodystrophy, Berardinelli-Seip congenital lipodystrophy type 2 (BSCL2) (Magre et al., 2001; Cui et al., 2011; Fei et al., 2011a).
The molecular function of seipin has been under intensive investigation in recent years. A few studies have found a role for seipin in glycerophospholipid metabolism. In particular, changes in the level and localization of phosphatidic acid (PA) have been observed in seipin-deficient cells (Fei et al., 2011b; Sim et al., 2012; Jiang et al., 2014; Han et al., 2015; Wolinski et al., 2015; Soltysik et al., 2021; Du and Yang, 2021), possibly due to enhanced activities of the ER-localized glycerol-3-phosphate acyltransferases (GPATs) (Pagac et al., 2016; Gao et al., 2020). Other recent studies suggest that seipin functions to maintain ER-LD contacts, to promote TAG nucleation in the ER in collaboration with LDAF1, and/or to prevent LD ripening through ER-LD contact (Szymanski et al., 2007; Grippa et al., 2015; Salo et al., 2016, 2019; Wang et al., 2016; Chung et al., 2019; Santinho et al., 2020; Klug et al., 2021; Arlt et al., 2022). An integral membrane protein of the ER with both N- and C-termini facing the cytosol, seipin has two transmembrane domains and a large, evolutionarily conserved luminal loop (Lundin et al., 2006). We and others reported the cryogenic electron microscopy (cryo-EM) structures of the luminal domains of human, fly, and yeast seipin (Sui et al., 2018; Yan et al., 2018). Human seipin exists as an undecamer, and the conserved luminal domain of seipin forms an eight-stranded β-sandwich fold (Yan et al., 2018). Surprisingly, the purified full-length human seipin can bind phosphatidylinositol 3-phosphate (PI(3)P) (Yan et al., 2018). PI(3)P in the ER is a well-established facilitator of autophagosome biogenesis (Axe et al., 2008; Mizushima et al., 2011; Hurley and Young, 2017; Mercer et al., 2018; Melia et al., 2020), but there has been no reported link between PI(3)P and LD formation. Here, we demonstrate that reducing PI(3)P or overexpressing DFCP1 restored normal lipidation of initial LDs in seipin-deficient cells. Importantly, disrupting PI(3)P balance in the ER of normal cells also led to aberrant LD formation. Our results thus identify PI(3)P and DFCP1 as novel regulators of the biogenesis of LDs.
RESULTS
Increased PI(3)P aggregation in seipin-deficient cells
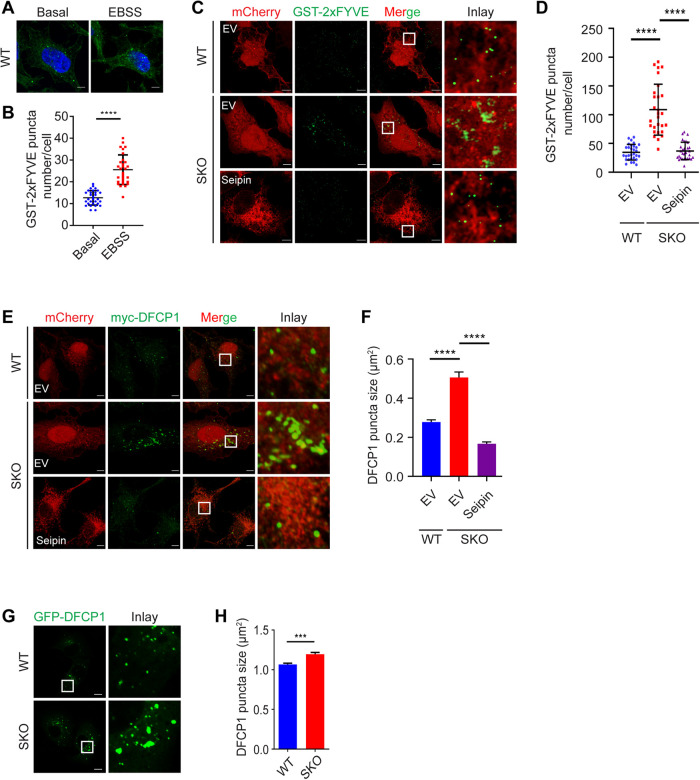
Our previous results suggested that seipin may bind PI(3)P in vitro (Yan et al., 2018). Here, we decided to examine the functional relationship between seipin and PI(3)P in vivo. First, we used a well-established probe for PI(3)P, GST-2xFYVE (for conserved in Fab1, YOTB, Vac1 and EEA1) (Gillooly et al., 2000), to detect PI(3)P in wild-type (WT) and seipin knockout (SKO) HeLa cells (Yan et al., 2018). As a control to confirm the specificity of GST-2xFYVE, we treated cells with amino acid–free media (Earle’s balanced salt solution [EBSS]) for 15 min, a condition known to increase ER PI(3)P. Indeed, the number of GST-2xFYVE–positive puncta increased dramatically in WT cells upon starvation (Figure 1, A and B). Interestingly, there were significantly more GST-2xFYVE puncta in SKO than in WT cells under normal growth conditions, and this increase was rescued by expressing WT seipin (Figure 1, C and D). We then used another well-established PI(3)P-binding probe, DFCP1 (Axe et al., 2008), to further examine PI(3)P in WT and SKO HeLa cells. DFCP1 is an ER-associated protein with two PI(3)P-binding FYVE domains. In WT cells, myc-DFCP1 formed very small punctate structures (Figure 1E) (Axe et al., 2008; Li et al., 2019). In SKO cells, the myc-DFCP1 structures appeared highly irregular and significantly enlarged (Figure 1, E and F). The total level of DFCP1 did not increase under seipin deficiency (Supplemental Figure S1A), suggesting that these enlarged structures are likely due to aggregation of DFCP1. Importantly, expressing WT seipin reduced the size of these DFCP1-positive structures in SKO cells (Figure 1, E and F). To validate the DFCP1 phenotype, we also generated SKO U2OS cell lines. Consistent with HeLa SKO cells, enlarged DFCP1 puncta were observed in U2OS SKO cells (Figure 1, G and H).
FIGURE 1:
PI(3)P is increased in seipin-deficient (SKO) cells. (A) HeLa cells grown in normal media or EBSS for 1 h were permeabilized with 0.05% saponin and treated with purified GST-2xFYVE and immunostained with anti-GST antisera. (B) Number of small GST-2xFYVE puncta per cell. Mean ± SD, n = 30 cells over three biological experiments, ****, p < 0.0001, calculated by unpaired Welch’s t test. (C) WT and SKO HeLa cells expressing mCherry-EV or mCherry-seipin were permeabilized with 0.05% saponin, fixed, and then treated with 100 µg/ml purified GST-2xFYVE for 1 h. (D) The GST-2xFYVE puncta per cell for A were counted. Mean ± SD, n = 28–30 cells over three biological experiments, ****, p < 0.0001, by one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test. (E) Irregular and enlarged myc-DFCP1 puncta in SKO cells. WT and SKO HeLa cells expressing myc-DFCP1 and mCherry-EV or mCherry-seipin were fixed and stained with anti-myc antisera. (F) Quantification of the size of myc-DFCP1 puncta as shown in E. Mean ± SEM, n > 400 puncta over three biological experiments, ****, p < 0.0001; ns, no significance, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (G) GFP-DFCP1 puncta in SKO U2OS cells grown in normal media. (H) Quantification of GFP-DFCP1 puncta size as shown in G. Mean ± SEM, n > 1224 puncta, ***, p < 0.001, calculated by unpaired Welch’s t test. Scale bars represent 5 μm.
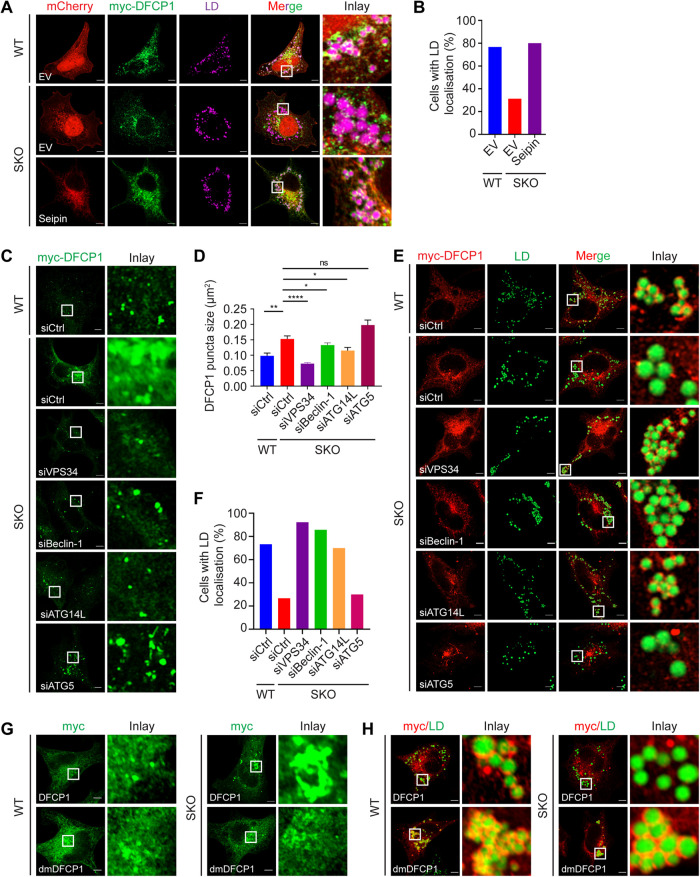
Reduced DFCP1-LD association under seipin deficiency
We and others previously reported that DFCP1 localizes to LDs (Gao et al., 2019a; Li et al., 2019) in WT cells. Strikingly, its LD association was severely impaired in SKO cells (Figure 2, A and B). WT seipin restored the LD association of DFCP1 (Figure 2, A and B). To confirm that the change in DFCP1 distribution in SKO cells is indeed related to PI(3)P, we focused on the class III PI3-kinase complex I (PIK3C3-C1), which produces PI(3)P in the ER, especially during the induction of autophagy (Hurley and Young, 2017). PIK3C3-C1 comprises VPS34 (the catalytic subunit), VPS15/P150, Beclin-1, and ATG14L. ATG14L, which is unique to PIK3C3-C1, interacts with Beclin-1 directly and recruits PIK3C3-C1 to the ER (Matsunaga et al., 2010; Hurley and Young, 2017). Knocking down ATG14L, Beclin-1, VPS34 but not ATG5 (which is essential for autophagy but not PI(3)P production) reduced the size of DFCP1-positive puncta in SKO cells (Figure 2, C and D; Supplemental Figure S1, A–D). Likewise, knocking down ATG14L, Beclin-1 or VPS34 but not ATG5 restored LD association by DFCP1 in SKO cells (Figure 2, E and F). To further confirm the involvement of PI(3)P, we also examined the distribution of a PI(3)P-binding mutant of DFCP1, dmDFCP1 (a double mutation inactivating both FYVE domains of DFCP1) (Axe et al., 2008). Distinct from WT DFCP1, dmDFCP1 did not form large irregular puncta in SKO cells (Figure 2G). Moreover, the LD association of dmDFCP1 was not impacted by seipin deficiency (Figure 2H). In fact, the dmDFCP1 mutant appeared to have stronger interaction with LDs than DFCP1 in WT HeLa cells (Figure 2H), suggesting that PI(3)P binding may disrupt LD association by DFCP1. Together, these data indicate that there is increased PI(3)P in seipin-deficient cells, causing abnormal DFCP1 distribution.
FIGURE 2:
The association of DFCP1 with LDs is regulated by seipin and PI(3)P. (A) Loss of LD association by myc-DFCP1 in SKO cells. WT and SKO HeLa cells expressing myc-DFCP1 and mCherry-EV or mCherry-seipin were treated with 400 µM oleate for 6 h, fixed, and stained with anti-myc antisera. LDs were then stained with HCS LipidTOX Deep Red Neutral Lipid Stain (purple). (B) Percentage of cells with LD association (continuous encircling structures around LDs) by myc-DFCP1 as shown in D, n = 30 cells, data from a representative experiment. (C) Knocking down ATG14L, Beclin-1, or VPS34 but not ATG5 reduced the size and number of DFCP1 puncta in SKO cells. WT and SKO HeLa cells were transfected with the indicated siRNAs, fixed, and stained with anti-myc antisera. (D) Quantification of the area of DFCP1 puncta as shown in C. Mean ± SEM, n > 154 puncta over three biological experiments, *, p < 0.05; **, p < 0.01; ****, p < 0.0001, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (E) Knocking down ATG14L, Beclin-1, or VPS34 but not ATG5 restored LD association by DFCP1 in SKO cells. WT and SKO cells were transfected with the indicated siRNAs. The cells were then treated with oleate for 6 h, fixed, and stained with anti-myc antisera. LDs were stained with BODIPY493/503 (green). (F) Percentage of cells with LD association by DFCP1 as shown in E. n = 30 cells, data from a representative experiment. (G) Distribution of myc-DFCP1 and myc-dmDFCP1 in WT and SKO HeLa cells. (H) Distribution of myc-DFCP1 and myc-dmDFCP1 (both in red) in WT and SKO HeLa cells treated with 400 µM oleate for 16 h. LDs were stained with BODIPY493/503 (green). Scale bars represent 5 μm.
Reducing PI(3)P in seipin-deficient cells restores normal LD biogenesis
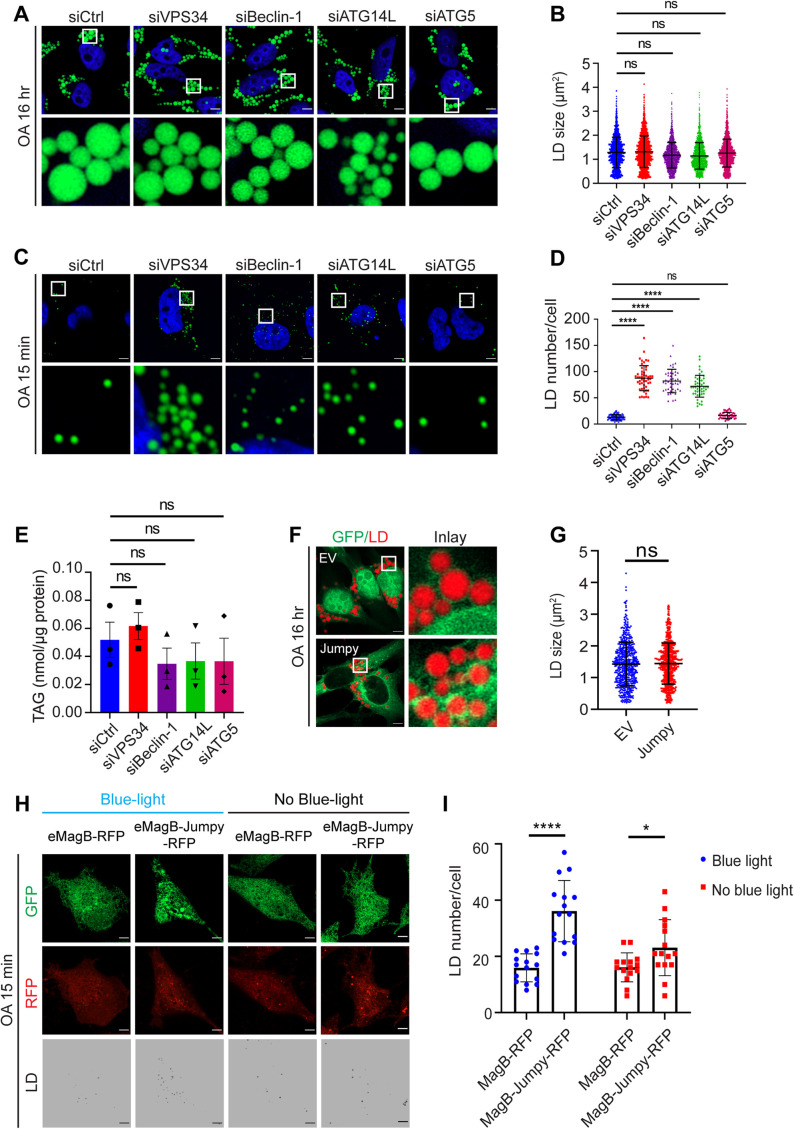
Because seipin binds PI(3)P and PI(3)P is increased in SKO cells, we wondered whether seipin’s role in LD formation is related to PI(3)P. Seipin deficiency causes two well-known phenotypes in mammalian cells: 1) Accumulation of a few highly enlarged/’supersized’ LDs after prolonged oleate loading; 2) Delayed maturation of initiating LDs during the early minutes of oleate treatment (Szymanski et al., 2007; Fei et al., 2008; Pagac et al., 2016; Salo et al., 2016; Wang et al., 2016; Yan et al., 2018; Chung et al., 2019). Either or both phenotypes could be related to the accumulation of PI(3)P in SKO cells. To examine these, we focused on components of PIK3C3-C1. Knocking down VPS34, Beclin-1, ATG14L, or ATG5 barely reduced LD size in SKO cells treated with oleate for 16 h (Figure 3, A and B) and had negligible impact on TAG mass (Supplemental Figure S1E). By contrast, knocking down components of PIK3C3-C1, but not ATG5, significantly increased the formation of BODIPY-positive LDs after 15 min oleate treatment in SKO cells, without impacting TAG (Figure 3, C–E) (Chung et al., 2019). The broad PI(3)K inhibitor wortmannin, but not the autophagy/H+-ATPase inhibitor bafilomycin A1 (BafA1), moderately decreased LD size in SKO cells treated with oleate for 16 h, without impacting TAG mass (Supplemental Figure S1, F–H). Notably, wortmannin, but not BafA1, significantly increased early LD lipidation in SKO cells after 15 min oleate treatment (Supplemental Figure S1, I and J). Finally, treating cells with a VPS34 inhibitor (VPS34IN1) also increased the lipidation of nascent LDs (Supplemental Figure S1, K and L).
FIGURE 3:
Reducing PI(3)P synthesis restored normal lipidation of nascent LDs in seipin-deficient cells. (A) SKO cells were treated with the indicated siRNAs and then incubated with 400 µM oleate for 16 h. LDs were stained with BODIPY493/503 (green). (B) Quantification of the size of LDs as shown in A. The diameters of LDs were measured. Mean ± SD, n = 1700–1765 LDs over three biological experiments, ns, no significance, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (C) SKO cells were treated with the indicated siRNAs and then starved in DMEM containing 1% LPDS for 16 h. Cells were then treated with 400 µM oleate for 15 min and LDs stained with BODIPY493/503 (green). (D) Quantification of the number of LDs per cell as shown in C. Mean ± SD, n > 30 cells over three biological experiments, ****, p < 0.0001; ns, no significance, calculated by one-way ANOVA. (E) Total TAG after 15 min oleate treatment as shown in C. Results reflect three independent experiments. Mean ± SEM, ns, no significance, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (F) Overexpression of a PI-3 phosphatase, Jumpy, and LD size. SKO HeLa cells transfected with EV-GFP or Jumpy-GFP were incubated with 400 µM oleate for 16 h, and LDs were stained with HCS LipidTOX Deep Red Neutral Lipid Stain (red). (G) Quantification of the size of LDs as shown in F. The diameters of LDs were measured: Mean ± SD, n = 550–580 LDs, ns, no significance, calculated using an unpaired Welch’s t test. (H) Transient activation of Jumpy at the ER increased the lipidation of nascent LDs in SKO HeLa cells. SKO cells were transfected with ER-EGFP-eMagA and eMagB-RFP or eMagB-Jumpy RFP. Cells were staved in 1% LPDS in DMEM for 16 h and then treated with 400 µM oleate for 15 min. Blue light was shone for 200 s. (I) Quantification of the number of LDs per cell as shown in C. Mean ± SD, n = 15 cells over three biological experiments, *, p < 0.05; ****, p < 0.0001, calculated by two-way ANOVA with Sidak’s multiple comparison test. Scale bars represent 5 μm.
To further ascertain the role of PI(3)P in seipin function, we also examined the effects of reducing PI(3)P through increasing PI(3)P hydrolysis in SKO cells. For this purpose, we employed a PI-3 phosphatase, the myotubularin-related protein (MTMR) family member 14, also known as Jumpy, to reduce PI(3)P. Jumpy has been shown to inhibit autophagy initiation presumably through reducing the cytoplasmic level of PI(3)P (Vergne et al., 2009). As with blocking PI(3)P synthesis (e.g., knocking down Beclin-1, VPS34 and ATG14), overexpressing Jumpy reduced the size of DFCP1-positive structures (Supplemental Figure S1M) but did not reduce LD size in SKO cells treated with oleate for 16 h (Figure 3, F and G). We then employed an optogenetic approach to control the activity of Jumpy temporally and spatially to examine its effect on early LD lipidation (Benedetti et al., 2020). Jumpy was fused with MagB, and the fusion protein was largely cytosolic but rapidly recruited to the ER upon blue light activation by binding to ER-localized MagA (Supplemental Figure S1, N and O). The transient recruitment of Jumpy to the ER increased the number of BODIPY-positive LDs in SKO cells treated with oleate for 15 min (Figure 3, H and I). Together, these results suggest that an increase of ER PI(3)P may underpin at least some of the abnormalities of LD formation under seipin deficiency, especially the lipidation of nascent LDs.
DFCP1 is required for the lipidation of nascent LDs
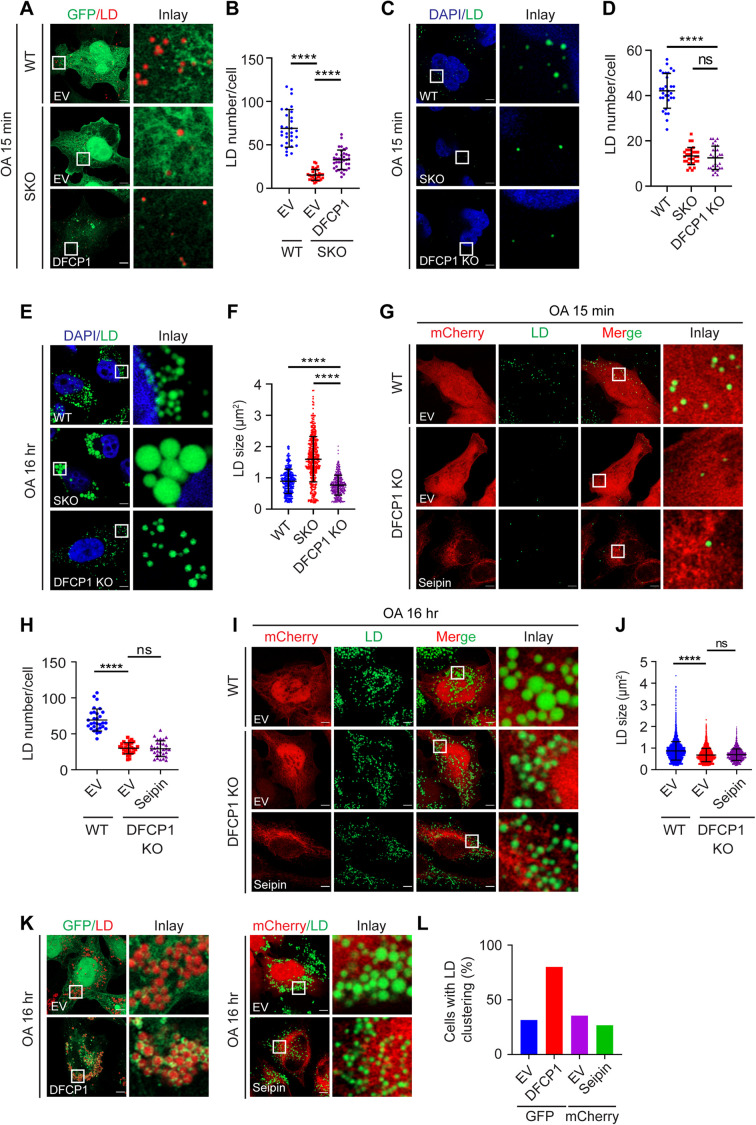
How does PI(3)P regulate the lipidation of initial LDs? We focused on the putative PI(3)P effector, DFCP1, because we and others have previously shown that DFCP1 associates with LDs and regulates LD formation (Gao et al., 2019a; Li et al., 2019). The appearance of irregular DFCP1 puncta and the dissociation of DFCP1 from LDs in SKO cells (Figures 1 and 2) suggest that DFCP1 may become trapped by excessive PI(3)P and thereby dysfunctional in SKO cells. If this is the case, then overexpressing DFCP1 should alleviate the delay in LD lipidation of SKO cells. Indeed, overexpressing DFCP1 moderately but significantly increased the formation of BODIPY-positive LDs in SKO cells 15 min post–oleate treatment (Figure 4, A and B). However, overexpressing DFCP1 had a limited effect on LD morphology in SKO cells treated with oleate for 16 h (Supplemental Figure S2, A and B), suggesting that DFCP1 may primarily function at the initial lipidation step during LD biogenesis. To test this hypothesis, we generated a DFCP1 knockout (DFCP1-KO) HeLa cell line by CRISPR. Highly reminiscent of SKO cells, there is a striking delay in the lipidation of nascent LDs in DFCP1-KO cells (Figure 4, C and D). Distinct from SKO cells, however, the sizes of LDs were moderately but significantly decreased in DFCP1-KO cells after prolonged oleate treatment (Figure 4, E and F), consistent with results from knockdown studies (Gao et al., 2019a; Li et al., 2019). Thus, both seipin and DFCP1 play critical roles in the initial stages of LD biogenesis but have rather opposite effects on LD morphology after prolonged oleate treatment.
FIGURE 4:
DFCP1 is required for normal lipidation of nascent LDs. (A) Overexpression of DFCP1 increased BODIPY-positive LDs in SKO cells. SKO HeLa cells expressing GFP-EV or GFP-DFCP1 were starved in DMEM containing 1% LPDS for 16 h and then treated with 400 µM oleate for 15 min. and LDs were stained with HCS LipidTOX Deep Red Neutral Lipid Stain (red). (B) Quantification of the number of LDs per cell as shown in A. Mean ± SD, n = 30 cells over three biological experiments, ****, p < 0.0001, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (C) Defective lipidation of nascent LDs during seipin deficiency (SKO) or DFCP1 deficiency (DFCP1 KO). WT, SKO, and DFCP1 KO HeLa cells were treated as described in A. Cells were then stained with BODIPY493/503 (green). (D) Quantification of the number of LDs per cell as shown in C. Mean ± SD, n = 30 cells over three biological experiments, ****, p < 0.0001; ns, no significance, calculated by one-way ANOVA. (E) The size of LDs increased in SKO but decreased in DFCP1 KO cells after prolonged oleate treatment. WT, SKO, and DFCP1 KO HeLa cells were treated with 400 µM oleate for 16 h and then stained with BODIPY493/503 (green). (F) Quantification of LD size as shown in. The diameters of LDs were measured: Mean ± SD, n = 450–500 LDs over three biological experiments, ****, p < 0.0001, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (G) Overexpressing seipin cannot restore lipidation of nascent LDs in DFCP1 KO cells. WT and DFCP1 KO HeLa cells expressing mCherry or mCherry-seipin were treated as described in A. Cells were then stained with BODIPY493/503 (green). (H) Quantification of the number of LDs per cell as shown in G. Mean ± SD, n = 30 cells over three biological experiments, **** p < 0.0001; ns, no significance, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (I) Overexpressing seipin cannot restore the size of LDs in DFCP1 KO cells after prolonged oleate treatment. WT and DFCP1 KO HeLa cells expressing mCherry or mCherry-seipin were treated with 400 µM oleate for 16 h. Cells were then stained with BODIPY493/503 (green). (J) Quantification of LD size as shown in I. The diameters of LDs were measured: n = 500–550 LDs over three biological experiments, ****, p < 0.0001; ns, not significance, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (K) Overexpressing DFCP1, not seipin, promotes the clustering of LDs. HeLa cells expressing EV-GFP, DFCP1-GFP, or mCherry-EV, mCherry-seipin were treated with 400 µM oleate for 16 h, and LDs were stained with HCS LipidTOX Deep Red Neutral Lipid Stain or BODIPY493/503, respectively. (L) Percentage of cells with LD clustering (structures insufficiently separated by Watershed function in ImageJ analysis) as shown in K. n = 30 cells over three biological experiments. Data from a representative experiment. Scale bars represent 5 μm.
These findings prompted us to further explore the functional and physical relationship between seipin and DFCP1. No synergistic defects were observed in initial LD lipidation in cells deficient in both DFCP1 and seipin (Supplemental Figure S2, C–E). Moreover, overexpressing seipin in DFCP1-deficient cells failed to rescue LD lipidation (Figure 4, G and H). The size of LDs decreased in DFCP1 KO cells after prolonged oleate treatment, which was not rescued by overexpressing seipin (Figure 4, I and J). While seipin and DFCP1 are functionally linked, they do not seem to interact strongly: they coimmunoprecipitated when both were overexpressed (Supplemental Figure S2F), but negligible interaction was detected between endogenous proteins (Supplemental Figure S2G). Overexpressing DFCP1, but not seipin, drastically increased the clustering of LDs (Figure 4, K and L). Finally, a loss of seipin, but not DFCP1, increased the formation of PLIN3 puncta (Supplemental Figure S2, H and I), which represent the earliest LDs (Chung et al., 2019). Together, these data suggest that both seipin and DFCP1 are required for the efficient lipidation of nascent LDs, although seipin and DFCP1 may have rather distinct molecular functions.
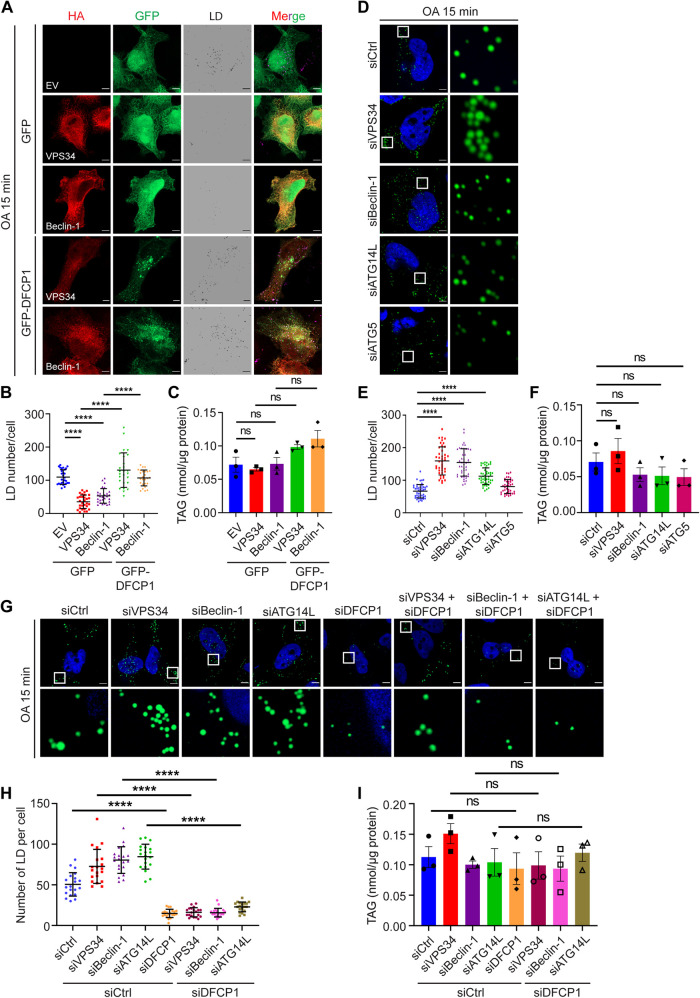
PI(3)P impacts LD formation through DFCP1
Together our data suggest that seipin deficiency disrupts the cellular distribution of PI(3)P, which affects the normal distribution and function of DFCP1 and hence causes a delay in the lipidation of nascent LDs. If this is the case, then increasing ER PI(3)P in normal cells should also impede the lipidation of nascent LDs by trapping DFCP1. Adopting a similar strategy to studying SKO cells (Figure 3), we focused on components of the PIK3C3-C1 in WT cells. Overexpressing VPS34 or Beclin-1 increased cytoplasmic DFCP1 puncta/PI(3)P (Supplemental Figure S3A), but this did not extend to dmDFCP1. Overexpressing VPS34 or Beclin-1 also impeded the lipidation of initial LDs (Figure 5, A and B). This was almost completely rescued by simultaneously overexpressing DFCP1 (Figure 5, A and B), with the level of TAG remaining unchanged during these experiments (Figure 5C). By contrast, knocking down VPS34, Beclin-1 or ATG14, but not ATG5, promoted the lipidation of initial LDs without impacting the level of TAG or DFCP1 (Figure 5, D–F, and Supplemental Figure S1, A–D). Importantly, the positive effect of PIK3C3-C1 deficiency on LD initiation depends on a functional DFCP1 (Figure 5, G–I). Consistent with knocking down components of PIK3C3-C1, wortmannin but not BafA1 increased initial LD formation (Supplemental Figure S3, B and C). The effect of changing PI(3)P on LD formation after prolonged oleate treatment is rather subtle. Overexpressing or knocking down components of PI3KC3-C1 had only a moderate impact on the size of mature LDs and the level of TAG in WT cells after prolonged oleate treatment (Supplemental Figure S3, D–I). Both wortmannin and BafA1 moderately reduced LD size after prolonged oleate treatment (Supplemental Figure S3, J and K) without significantly altering TAG levels in WT cells (Supplemental Figure S3L). Together, these data suggest that PI(3)P has a general function in regulating the lipidation of initial LDs, most likely through controlling the distribution and function of DFCP1.
FIGURE 5:
PI(3)P impedes normal lipidation of nascent LDs through trapping DFCP1. (A) Overexpressing VPS34 or Beclin-1 impaired lipidation of nascent LDs, which were rescued by coexpressing DFCP1. HeLa cells expressing various indicated plasmids were starved in 1% LPDS in DMEM and then treated with 400 µM oleate for 15 min before LD staining with HCS LipidTOX Deep Red Neutral Lipid Stain (gray). (B) Quantification of the number of LDs per cell as shown in A. Mean ± SD, n = 25–30 cells over three biological experiments, **** p < 0.0001, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (C) Total TAG after 15 min oleate treatment as shown in A. Results reflect three independent experiments. Mean ± SEM, ns, no significance, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (D) Knocking down components of PI3KC3-C1, but not ATG5, promoted lipidation of nascent LDs. HeLa cells treated with various siRNAs as indicated were treated as described in A. Cells were then stained with BODIPY493/503 (green). (E) Quantification of the number of LDs per cell as shown in D. Mean ± SD, n = 40–47 cells over three biological experiments, **** p < 0.0001, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (F) Total TAG after 15 min oleate treatment as shown in D. Results reflect three independent experiments. Mean ± SEM, ns, no significance, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (G) DFCP1 is required for the enhanced lipidation of nascent LDs under PI3KC3-C1 deficiency. HeLa cells treated with various indicated siRNAs were treated as described in A. LDs were then stained with BODIPY493/503 (green). (H) Quantification of the number of LDs per cell as shown in G. Mean ± SD, n = 20 cells over three biological experiments, **** p < 0.0001, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (I) Total TAG after 15 min oleate treatment as shown in G. Results reflect three independent experiments. Mean ± SEM, ns, no significance, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. Scale bars represent 5 μm.
PI(3)P and seipin may coordinate the biogenesis of LDs and autophagosomes
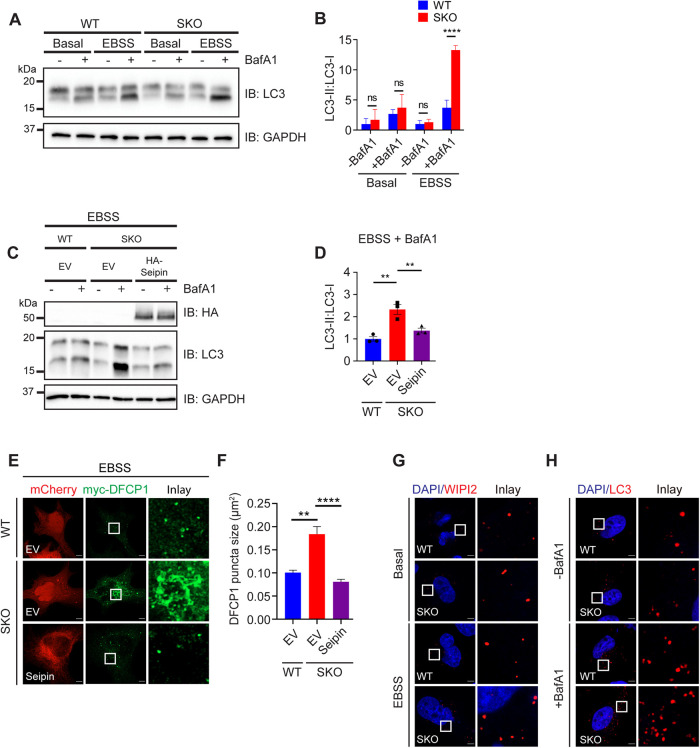
PI(3)P in the ER is a key early signal for autophagy initiation (Mizushima et al., 2011). Because seipin deficiency increased the clustering of cytoplasmic PI(3)P, we wondered whether seipin may also regulate autophagy. The ratio of LC3-II over LC3-I under seipin deficiency increased during starvation-induced autophagy, especially after BafA1 treatment (Figure 6, A and B). Moreover, the increased LC3-II in SKO cells was rescued by reexpressing WT seipin (Figure 6, C and D). We also used an RFP-GFP tandem–tagged LC3 (RFP-GFP-LC3) probe to detect autophagic flux. This probe emits yellow signals (GFP plus RFP) in the cytosol and on autophagosomes but only red signals in autolysosomes because GFP is more easily quenched and/or degraded in the lysosome than RFP (Kimura et al., 2007). There was less green signal in SKO cells after EBSS treatment (Supplemental Figure S4, A and B), suggesting increased autophagic flux. There were also enlarged DFCP1 puncta upon EBSS treatment in SKO cells, which was rescued by reexpressing WT seipin (Figure 6, E and F), consistent with Figure 1. Moreover, there were more WIPI2 (a well-established mediator of autophagosome initiation) puncta in SKO cells (Figure 6G and Supplemental Figure S4C) (Mercer et al., 2018). There were also increased LC3 puncta in SKO HeLa cells, and the LC3 puncta increased further upon BafA1 treatment (Figure 6H and Supplemental Figure S4D). Finally, increased LC3 puncta were also observed in SKO cells generated from U2OS cells (Supplemental Figure S4, E and F). Together, these data suggest that the formation of autophagosomes is increased in SKO cells.
FIGURE 6:
Seipin and PI(3)P may coordinate the initiation of autophagosomes and LDs. (A) WT and SKO cells were incubated in normal media (basal) or EBSS for 1 h with or without 100 nM BafA1 treatment. Protein extracts were isolated and blotted with antisera against LC3 and GAPDH. (B) LC3-II/LC3-I ratio as shown in A. Results reflect three independent experiments. ****, p < 0.0001, calculated by two-way ANOVA with Sidak’s multiple comparisons test. (C) WT and SKO HeLa cells expressing the indicated plasmids were grown in EBSS for 1 h and treated with or without 100 nM BafA1. Protein extracts were isolated and blotted with antisera against HA, LC3, and GAPDH. EV, vector control. (D) LC3-II/LC3-I ratio as shown in C. Results reflect three independent experiments. Mean ± SEM, **, p < 0.01, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (E) Fluorescence imaging showing myc-DFCP1 puncta in WT and SKO cells under EBSS. WT and SKO cells expressing the indicated plasmids were grown in EBSS for 1 h. Seipin and EV were tagged with mCherry. (F) Size of myc-DFCP1 puncta as shown in E. Mean ± SEM, n > 350 puncta, ** p < 0.01; ****, p < 0.0001; ns, no significance, calculated by one-way ANOVA with Dunnett’s multiple comparisons test. (G) WT and SKO HeLa cells were grown in basal media or EBSS (1 h), fixed, and stained with antisera against endogenous WIPI2. (H) WT and SKO HeLa cells were grown in EBSS for 1 h and treated with or without 100 nM BafA1. Cells were then fixed and stained with antisera against endogenous LC3-II. Scale bars represent 5 μm.
DISCUSSION
The biogenesis of LDs is an important yet challenging subject in cell biology. Here, we identify the well-known signaling lipid PI(3)P as a novel regulator of LD biogenesis and establish a role for DFCP1 in the lipidation of nascent LDs. Together, our data shed light on the molecular events underlying the biogenesis of LDs and implicate PI(3)P and DFCP1 as key molecules in coordinating the biogenesis of both autophagosomes and LDs.
Seipin controls the distribution of PI(3)P and DFCP1
Loss of seipin function has two well-defined LD phenotypes: a delay in the lipidation of nascent LDs and the appearance of large or “supersized” LDs after prolonged oleate treatment (Szymanski et al., 2007; Fei et al., 2008; Salo et al., 2016, 2019; Wang et al., 2016; Chung et al., 2019). The prevailing model posits that seipin, together with LDAF1, may physically trap TAG molecules to facilitate TAG nucleation and define sites of LD initiation (Chung et al., 2019; Klug et al., 2021; Arlt et al., 2022). Moreover, seipin may also help establish ER-LD contact and ensure optimal TAG transfer from the ER to growing LDs (Salo et al., 2016, 2019; Wang et al., 2016). However, past work from our lab and others suggests that seipin functions to maintain the proper distribution of phospholipids, such as phosphatidic acid (Fei et al., 2011b; Han et al., 2015; Wolinski et al., 2015; Pagac et al., 2016; Yan et al., 2018; Gao et al., 2019b; Soltysik et al., 2021). Surprisingly, our present data suggest that seipin regulates the distribution of PI(3)P in the ER. Because reducing PI(3)P rescued the defective lipidation of initial LDs in SKO cells, our results suggest that abnormal PI(3)P distribution in the ER may affect the activity of its putative effectors, especially DFCP1 (see below), causing defective LD initiation under seipin deficiency.
DFCP1 is required for the lipidation of initiating LDs
DFCP1 is a well-established PI(3)P-binding protein known for marking omegasomes, structural features present during the initial stages of autophagosome formation (Axe et al., 2008). However, DFCP1 is not essential for autophagy and its primary cellular function remains to be determined (Axe et al., 2008). We and others previously showed that DFCP1 associated with LDs but its effect on LD growth after prolonged oleate treatment was fairly moderate (Gao et al., 2019a; Li et al., 2019). Here, using a recently developed assay (Chung et al., 2019), we observed a striking defect in the lipidation of initial LDs in DFCP1-deficient cells. Thus, one of the primary functions of DFCP1 is likely to facilitate lipid delivery to nascent LDs. Notably, the LD lipidation defects are nearly identical in seipin- and DFCP1-deficient cells. Moreover, overexpressing DFCP1 rescued the LD lipidation defect associated with seipin loss, but not vice versa. It remains to be determined exactly how DFCP1 may facilitate the lipidation of initial LDs. One possibility is through maintaining the contact between the ER and LDs (Li et al., 2019). In this regard, it is noteworthy that the ER-LD contact is also impaired under seipin deficiency (Salo et al., 2016). Therefore, in seipin-deficient cells, DFCP1 may be trapped in PI(3)P-rich structures and separated from LDs, thereby failing to maintain proper ER-LD contact and causing a delay in the lipidation of initial LDs. Thus, our data suggest that the defective lipidation of initial LDs in seipin-deficient cells may, at least in part, be attributed to the altered distribution of PI(3)P and a dysfunctional DFCP1.
Whereas DFCP1 and seipin are connected via their control of the lipidation of initial LDs, they appear to have rather distinct biochemical functions. For instance, supersized LDs are commonly seen in SKO cells after prolonged oleate treatment. By contrast, the size of LDs decreased in DFCP1 KO cells after prolonged oleate treatment. Thus, seipin may control multiple steps in LD formation through different mechanisms. For instance, the formation of supersized LDs in SKO cells could be related to its role in controlling PA levels and distribution (Fei et al., 2011b; Pagac et al., 2016; Soltysik et al., 2021). Moreover, seipin is known to regulate calcium homeostasis, sphingolipid metabolism, and adipogenesis (Gao et al., 2019b), and seipin can also regulate the distribution of PI(3)P as demonstrated in this study. Thus, DFCP1 appears to have a rather restricted role in early LD growth, possibly by helping establish and/or maintain ER-LD contact. Seipin, on the other hand, has a much broader role in cell/lipid biology that extends beyond LD dynamics.
PI(3)P is a novel regulator of LD biogenesis
Our data also unveil a negative regulatory role for PI(3)P in LD biogenesis. Besides seipin deficiency, increasing or reducing PI(3)P through manipulating PIK3C3-C1 activity also compromised or promoted the lipidation of initial LDs, respectively. Importantly, these effects were reversed by overexpressing or knocking down DFCP1. Therefore, PI(3)P and DFCP1 have a general role in regulating LD biogenesis, independent of seipin. PI(3)P is a well-established regulator of autophagy, and our present results suggest that PI(3)P also regulates the biogenesis of LDs. Although LDs and autophagosomes have distinct functions in cells, the de novo biogenesis of LDs and the biogenesis of autophagosomes share some similarities (Joshi et al., 2017; Gao et al., 2019b). Both organelles originate from subdomains of the ER, and both lack coat proteins. Importantly, the formation of both organelles is enhanced under stressful conditions, including starvation and ER stress (Taylor and Parks, 1979; Yang et al., 1996; Zhang et al., 2003; Bernales et al., 2006; Ogata et al., 2006; Fei et al., 2009; Mizushima et al., 2011; Madeira et al., 2015; Rambold et al., 2015; Nguyen et al., 2017). Autophagy can promote LD biogenesis after prolonged starvation, but it remains unclear if and how the initiation of these two organelles at the early stages of starvation are linked. The molecular mechanism underlying the complex relationship between LDs and autophagosomes is unknown and would be an important topic for future studies.
In summary, our data identify PI(3)P and DFCP1 as novel regulators of the lipidation of initial LDs and suggest that PI(3)P may coordinate the biogenesis of both LDs and autophagosomes. Future work is required to dissect exactly how seipin regulates PI(3)P distribution and how DFCP1 regulates LD growth, as well as the complex relationship between seipin, DFCP1, and PI(3)P during LD biogenesis.
MATERIALS AND METHODS
Cell lines
HeLa and U2OS cells were obtained from the American Type Culture Collection. Both cell lines were grown in DMEM (#21969035; Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (PS). Cells were maintained at 37°C with 5% CO2 and tested for mycoplasma contamination every 6 mo. Where noted, cells were treated with wortmannin (#W1628; Sigma-Aldrich) and bafilomycin A1 (BafA1) (#B1793; Sigma-Aldrich) or VPS34-IN1 (#532628; Sigma-Aldrich) dissolved in dimethyl sulfoxide and used at a final concentration as indicated in the figure legends. To induce autophagy, cells were washed three times in phosphate-buffered saline (PBS) before incubation in EBSS media (#24010043; Thermo Fisher Scientific) for 1 h with or without BafA1.
Antibodies, chemicals, and plasmids
Antibodies, chemicals, and plasmids with source and catalogue numbers are described in Supplemental Table S1. Primers used for cloning are listed in Supplemental Table S2.
For plasmid construction, all PCRs were performed using Q5 High Fidelity Polymerase (#M0491; New England Biolabs) and restriction enzymes from New England Biolabs. For site-directed mutagenesis, PCRs were performed using KOD Hot Start Polymerase (#71086; Merck Millipore).
Transfection and RNA interference
Plasmid transfections were carried out using the Lipofectamine LTX and PLUS reagent (#15338100; Thermo Fisher Scientific) following the manufacturer’s protocol. Small interfering RNA (siRNA) transfection were carried out using the Lipofectamine RNAiMAX reagent (#13778150; Thermo Fisher Scientific) following the manufacturer’s protocol.
Generation of knock-in HeLa cells with CRISPR/Cas9a-mediated genome editing
C-terminally mCherry-tagged Plin3 or C-terminally super-folder GFP (sfGFP)-tagged seipin knock-in (KI) cells were generated via CRISPR/Cas9a gene editing. HeLa cells were simultaneously transfected using the Lipofectamine LTX and PLUS reagent with Megamer Single-Stranded DNA Fragments (Integrated DNA Technologies) containing homology arms of ∼100 base pairs upstream and downstream of the target site (Plin3) or a donor plasmid containing ∼600 base pairs upstream and downstream of the target site (Seipin) along with a single guide RNA (sgRNA) targeting upstream of the stop codon. The Single-Stranded DNA Fragments, donor template, and sgRNA information are described in Supplemental Tables S2 and S3. Cells were then selected with 1 μg/μl puromycin (#A1113803; Thermo Fisher Scientific) for 48 h, and single-cell fluorescence-activated cell sorting (FACS) sorting was performed using the BD FACSMelody cell sorter. A laser (561 or 488 nm) was used to sort mCherry- and sfGFP-positive cells, respectively.
Generation of KO cells with CRISPR/Cas9a-mediated genome editing
SKO Hela cells were generated as described (Yan et al., 2018). DFCP1 KO HeLa cells were generated by CRISPR/Cas9a gene editing. sgRNAs were designed using http://crispor.tefor.net/ and subcloned into the PX459 vector (#62988; Addgene). Cells were transfected and 48 h posttransfection were treated with 1 µg/ml puromycin for 72 h. The drug-resistant clones were then sorted using the BD Influx cell sorter to form single colonies. To screen DFCP1 KO clones, cell lysates were collected and Western blot analysis was performed to verify depletion of DFCP1 protein using DFCP1 antibody (#85156; Cell Signaling Technology).
For generating SKO U2OS cells, corresponding DNA oligos were synthesized, annealed, and subcloned into the PX330 vector through BbsI sites. The plasmids were transiently transfected in U2OS cells, and cells expressing plasmids were selected with puromycin (1 µg/ml) for 3 d. Single clones were then isolated and collected for genome DNA extraction and PCR amplification of a region containing sgRNA targeting sites. The amplified products were cloned into T-vectors and transformed into Escherichia coli. Twenty to thirty E. coli single clones were sequenced for each SKO clone. Finally, SKO single clones were further verified by the LD phenotype.
Fluorescence microscopy
Confocal microscopy was performed using a Zeiss LSM 900 confocal microscope equipped with GaAsP detectors and a ZeissAiryscan 2 detector. A 63×/1.4 or 40×/1.3 oil immersion objective was applied for imaging at room temperature (RT). For LD studies, cells were treated with 400 μM oleate-coupled bovine serum albumin (BSA). Cells were then washed in PBS, followed by fixation with 4% paraformaldehyde in PBS for 15 min at RT. After fixation, cells were washed three times in PBS. Where noted, cells were stained with 1 μg/ml BODIPY493/503 (#D3922; Thermo Fisher Scientific) or HCS LIPDTOX Deep Red Neutral Stain (#H34477; Thermo Fisher Scientific) for 15 min or 1 h, respectively. Coverslips were mounted onto glass slides using ProLong Gold Antifade Mountant with 4′,6-diamidino-2-phenylindole (DAPI) (#P36941; Thermo Fisher Scientific).
For immunofluorescence studies, cells were permeabilized with 0.2% Triton X-100 in PBS for 15 min postfixation with 4% paraformaldehyde. Cells were then blocked with 3% (wt/vol) BSA in PBS for 1 h and probed with primary antibodies as noted in the figure legends before incubation with Alexa Fluor secondary antibodies (Thermo Fisher Scientific) in 3% (wt/vol) BSA for 1 h each. Coverslips were then mounted onto glass slides using ProLong Gold Antifade Mountant with DAPI.
To detect early Plin3KI-mCherry puncta, HiLo microscopy was performed using Zeiss ELYRA. A Plan-Apochromat 63×/1.4 oil lens was used with 0.2% of 561-nm laser excitations. Images were taken with exposure time of 500 ms on an Andor Ultra 897 EMCCD camera (gain at 200) in HiLo mode (angle = 58°). No collected samples were excluded from analysis.
For optogenetic experiments, cells were seeded onto 35 mm glass bottom gridded dishes (MatTek) and transfected with eMag plasmids as noted in the figure legends. Twenty-four hours posttransfection, cells were lipid starved in 1% lipoprotein depleted fetal bovine serum (LPDS) in DMEM for 16 h. Media were changed to Fluorobrite DMEM (#A1896701; Thermo Fisher Scientific) supplemented with 1% LPDS, and blue light illumination was carried out at 37°C on Zeiss900 LSM with a 20×/0.8 objective. Tile scans of 469 μm × 469 μm were illuminated with 488 nm laser pulses for 200 s (260 ms pulses per tile) before equal volumes of media containing 800 μM oleate-coupled BSA were added, making a final concentration of 400 μM oleate-coupled BSA. Fifteen minutes post–oleate treatment, cells were fixed with 4% paraformaldehyde in PBS for 15 min. Cells were washed with PBS three times before LDs were stained with HCS LIPIDTOX Deep Red for 1 h and imaged with a 63×/1.4 oil objective. No collected samples were excluded from analysis.
PI(3)P detection by GST-2xFYVE
GST-2xFYVE peptides were purified by a GST-fusion protein purification protocol. Briefly, BL21 bacteria were cultured to OD600 ∼ 0.6 before induction with 0.1 mM isopropyl β-d-1-thiogalactopyranoside for 4 h at 37°C. Cells were centrifuged at 4000 × g for 10 min at 4°C, and the bacterial pellet was frozen at –80°C. Bacterial pellets were then resuspended in GST lysis buffer (50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1 mM EDTA, pH 8.0, 1 mM dithiothreitol, and complete protease Inhibitor Cocktail tablet, EDTA-Free) and sonicated on ice. Lysate was centrifuged at 12,000 × g for 30 min at 4°C before incubation with glutathione Sepharose 4B beads (#GE17-0756-01; Sigma-Aldrich) binding for 1 h at 4°C. Beads were washed with GST lysis buffer and eluted with GST lysis buffer supplemented with 50 mM l-glutathione reduced. The supernatant was collected and further dialyzed for final purification in 75 mM KCl, 30 mM HEPES, pH 7.4, 5 mM MgCl2.
To detect PI(3)P, cells were grown on coverslips and fixed with 4% paraformaldehyde for 15 min before incubation with permeabilization/blocking buffer for 1 h (5% goat serum and 0.05% saponin or 5% goat serum and 0.2% Triton X-100). Cells were then incubated with 100 μg/ml GST-2xFYVE for 1 h in the respective permeabilization/blocking buffer as stated in the figure legends. Cells were washed with PBS three times and probed with GST antibody followed by incubation with Alexa Fluor secondary antibody for 1 h each. Coverslips were then mounted onto glass slides using ProLong Gold Antifade Mountant with DAPI.
Coimmunoprecipitation
For immunoprecipitation, cells were lysed in coimmunoprecipitation lysis buffer (25 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% dodecyl-β-d-maltoside, and cOmplete, EDTA-free Protease Inhibitor Cocktail) and incubated on ice for 10 min. Cell lysates were centrifuged at 18,000 × g for 10 min at 4°C, and supernatant was collected. For Supplemental Figure S4, E and F, 1 or 3 mg of protein lysates, respectively, was incubated with 25 μl of GFP-trap magnetic agarose beads (#gtma-10; ChromoTek) for 16 h at 4°C. The protein–beads mixture was washed three times with 500 μl of washing buffer (25 mM HEPES, pH 7.4, 150 mM NaCl, 0.1% NP-40) and eluted with 2× Laemmli sample buffer at RT before SDS–PAGE and Western blot.
SDS–PAGE and Western blot
For Western blot analysis, cells were washed twice with ice-cold PBS and lysed using 1% Triton X-100 lysis buffer (1% Triton X-100, 0.1% SDS, 10 mM Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA, 10% glycerol, and complete protease Inhibitor Cocktail tablet, EDTA-Free). Cell lysates were then incubated on ice for 10 min, followed by centrifugation at 18,000 × g for 10 min at 4°C. Protein concentrations were measured using the BCA Protein assay kit (Sigma-Aldrich), and cell lysates were mixed with 2× Laemmli sample buffer before SDS–PAGE. Electrophoresis was carried out at 150 V for 1 h, and gels were transferred to nitrocellulose membrane at 100 V for 1 h. Membranes were incubated with 5% (wt/vol) skim milk in tris-buffered saline with Tween®20 (TBST) for 1 h at RT, followed by incubation with appropriate primary antibodies overnight at 4°C. Membranes were washed three times with TBS-T and subsequently incubated with appropriate horseradish peroxidase–conjugated secondary antibodies (Jackson Immuno Research) in TBS-T for 1 h. Membranes were washed three times with TBS-T before imaging and analysis by enhanced chemiluminescence (#WBKLS0500; Merck Millipore) and BioRadChemiDoc XRS+ imager.
TAG measurement
Cells were plated onto 100 mm dishes and transfected as noted in the figure legends. Forty-eight hours posttransfection, cells were treated with 400 μM oleate-coupled BSA as indicated in the figure legends. Triglyceride levels were determined using the Triglyceride Assay Kit (Abcam) following the manufacturer’s protocol.
Supplementary Material
Acknowledgments
This work was supported by grants 1078117, 1141938, 1141939, and 1144726 from the National Health and Medical Research Council (NHMRC) of Australia. H.Y. was also supported by an NHMRC Senior Research Fellowship (1058237) and NHMRC Investigator Grant (2009852).
Abbreviations used:
- ATG14L
autophagy related 14L
- ATG5
autophagy related 5
- BafA1
bafilomycin A1
- BSCL2
Berardinelli-Seip congenital lipodystrophy type 2
- DFCP1
double FYVE-containing protein 1
- DFCP1-KO
knockout
- EBSS
Earle’s balanced salt solution
- ER
endoplasmic reticulum
- FACS
fluorescence-activated cell sorting
- FYVE
Fab1, YOTB, Vac1 and EEA1 green
- GFP
fluorescent protein
- GPATs
glycerol-3-phosphate acyltransferases
- LC3
Microtubule-associated protein 1A/1B-light chain 3
- LDAF1
lipid droplet assembly factor 1
- LDs
lipid droplets
- LPDS
lipoprotein depleted fetal bovine serum
- MTMR
myotubularin-related protein
- OA
oleic acid
- PA
phosphatidic acid
- PBS
phosphate-buffered saline
- PI(3)P
phosphatidylinositol 3-phosphate
- PIK3C3-C1
class III PI3-kinase complex I
- RFP
red fluorescent protein
- RT
room temperature
- SEs
sterol esters
- SKO
seipin knockout
- TAGs
triacylglycerols
- TBST
tris-buffered saline with Tween®20
- VPS34
catalytic subunit of PIK3C3-C1
- VPS34IN1
VPS34 inhibitor
- WT
wild-type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-07-0279) on September 21, 2022.
REFERENCES
- Arlt H, Sui X, Folger B, Adams C, Chen X, Remme R, Hamprecht FA, DiMaio F, Liao M, Goodman JM, et al. (2022). Seipin forms a flexible cage at lipid droplet formation sites. Nat Struct Mol Biol 29, 194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT (2008). Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182, 685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti L, Marvin JS, Falahati H, Guillen-Samander A, Looger LL, De Camilli P (2020). Optimized vivid-derived magnets photodimerizers for subcellular optogenetics in mammalian cells. eLife 9, e63230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P (2006). Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol 4, e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Sanchez-Alvarez M, Fajardo A, Kapetanovic R, Steiner B, Dutra F, Moreira L, Lopez JA, Campo R, Mari M, et al. (2020). Mammalian lipid droplets are innate immune hubs integrating cell metabolism and host defense. Science 370, eaay8085. [DOI] [PubMed] [Google Scholar]
- Chung J, Wu X, Lambert TJ, Lai ZW, Walther TC, Farese RV Jr (2019). LDAF1 and seipin form a lipid droplet assembly complex. Dev Cell 51, 551–563.e557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Wang Y, Tang Y, Liu Y, Zhao L, Deng J, Xu G, Peng X, Ju S, Liu G, Yang H (2011). Seipin ablation in mice results in severe generalized lipodystrophy. Hum Mol Genet 20, 3022–3030. [DOI] [PubMed] [Google Scholar]
- Du X, Yang H (2021). Seipin regulates the formation of nuclear lipid droplets from a distance. J Cell Biol 220, e202011166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Du X, Yang H (2011a). Seipin, adipogenesis and lipid droplets. Trends Endocrinol Metab 22, 204–210. [DOI] [PubMed] [Google Scholar]
- Fei W, Shui G, Gaeta B, Du X, Kuerschner L, Li P, Brown AJ, Wenk MR, Parton RG, Yang H (2008). Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol 180, 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Shui G, Zhang Y, Krahmer N, Ferguson C, Kapterian TS, Lin RC, Dawes IW, Brown AJ, Li P, et al. (2011b). A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet 7, e1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Wang H, Fu X, Bielby C, Yang H (2009). Conditions of endoplasmic reticulum stress stimulate lipid droplet formation in Saccharomyces cerevisiae. Biochem J 424, 61–67. [DOI] [PubMed] [Google Scholar]
- Gao G, Sheng Y, Yang H, Chua BT, Xu L (2019a). DFCP1 associates with lipid droplets. Cell Biol Int 43, 1492–1504. [DOI] [PubMed] [Google Scholar]
- Gao M, Huang X, Song BL, Yang H (2019b). The biogenesis of lipid droplets: lipids take center stage. Prog Lipid Res 75, 100989. [DOI] [PubMed] [Google Scholar]
- Gao M, Liu L, Wang X, Mak HY, Liu G, Yang H (2020). GPAT3 deficiency alleviates insulin resistance and hepatic steatosis in a mouse model of severe congenital generalized lipodystrophy. Hum Mol Genet 29, 432–443. [DOI] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. (2000) Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J 19, 4577–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grippa A, Buxo L, Mora G, Funaya C, Idrissi FZ, Mancuso F, Gomez R, Muntanya J, Sabido E, Carvalho P (2015). The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J Cell Biol 211, 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Binns DD, Chang YF, Goodman JM (2015). Dissecting seipin function: the localized accumulation of phosphatidic acid at ER/LD junctions in the absence of seipin is suppressed by Sei1p(DeltaNterm) only in combination with Ldb16p. BMC Cell Biol 16, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Young LN (2017). Mechanisms of autophagy initiation. Annu Rev Biochem 86, 225–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Gao M, Wu C, He H, Guo X, Zhou Z, Yang H, Xiao X, Liu G, Sha J (2014). Lack of testicular seipin causes teratozoospermia syndrome in men. Proc Natl Acad Sci USA 111, 7054–7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AS, Zhang H, Prinz WA (2017). Organelle biogenesis in the endoplasmic reticulum. Nat Cell Biol 19, 876–882. [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T (2007). Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460. [DOI] [PubMed] [Google Scholar]
- Klug YA, Deme JC, Corey RA, Renne MF, Stansfeld PJ, Lea SM, Carvalho P (2021). Mechanism of lipid droplet formation by the yeast Sei1/Ldb16 Seipin complex. Nat Commun 12, 5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao YG, Li D, Zhao H, Huang J, Miao G, Feng D, Liu P, Li D, Zhang H (2019). The ER-localized protein DFCP1 modulates ER-lipid droplet contact formation. Cell Rep 27, 343–358.e345. [DOI] [PubMed] [Google Scholar]
- Lundin C, Nordstrom R, Wagner K, Windpassinger C, Andersson H, von Heijne G, Nilsson I (2006). Membrane topology of the human seipin protein. FEBS Lett 580, 2281–2284. [DOI] [PubMed] [Google Scholar]
- Madeira JB, Masuda CA, Maya-Monteiro CM, Matos GS, Montero-Lomeli M, Bozaquel-Morais BL (2015). TORC1 inhibition induces lipid droplet replenishment in yeast. Mol Cell Biol 35, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magre J, Delepine M, Khallouf E, Gedde-Dahl T Jr, Van Maldergem L, Sobel E, Papp J, Meier M, Megarbane A, Bachy A, et al. (2001). Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat Genet 28, 365–370. [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T (2010). Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol 190, 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia TJ, Lystad AH, Simonsen A (2020). Autophagosome biogenesis: from membrane growth to closure. J Cell Biol 219, e202002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TJ, Gubas A, Tooze SA (2018). A molecular perspective of mammalian autophagosome biogenesis. J Biol Chem 293, 5386–5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y (2011). The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27, 107–132. [DOI] [PubMed] [Google Scholar]
- Nguyen TB, Louie SM, Daniele JR, Tran Q, Dillin A, Zoncu R, Nomura DK, Olzmann JA (2017). DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev Cell 42, 9–21.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. (2006). Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 26, 9220–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Carvalho P (2019). Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol 20, 137–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagac M, Cooper DE, Qi Y, Lukmantara IE, Mak HY, Wu Z, Tian Y, Liu Z, Lei M, Du X, et al. (2016). SEIPIN regulates lipid droplet expansion and adipocyte development by modulating the activity of glycerol-3-phosphate acyltransferase. Cell Rep 17, 1546–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Cohen S, Lippincott-Schwartz J (2015). Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, mitochondrial fusion dynamics. Dev Cell 32, 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo VT, Belevich I, Li S, Karhinen L, Vihinen H, Vigouroux C, Magre J, Thiele C, Holtta-Vuori M, Jokitalo E, Ikonen E (2016). Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J 35, 2699–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo VT, Li S, Vihinen H, Holtta-Vuori M, Szkalisity A, Horvath P, Belevich I, Peranen J, Thiele C, Somerharju P, et al. (2019). Seipin facilitates triglyceride flow to lipid droplet and counteracts droplet ripening via endoplasmic reticulum contact. Dev Cell 50, 478–493.e479. [DOI] [PubMed] [Google Scholar]
- Santinho A, Salo VT, Chorlay A, Li S, Zhou X, Omrane M, Ikonen E, Thiam AR (2020). Membrane curvature catalyzes lipid droplet assembly. Curr Biol 30, 2481–2494. [DOI] [PubMed] [Google Scholar]
- Sim MF, Dennis RJ, Aubry EM, Ramanathan N, Sembongi H, Saudek V, Ito D, O’Rahilly S, Siniossoglou S, Rochford JJ (2012). The human lipodystrophy protein seipin is an ER membrane adaptor for the adipogenic PA phosphatase lipin 1. Mol Metabol 2, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltysik K, Ohsaki Y, Tatematsu T, Cheng J, Maeda A, Morita SY, Fujimoto T (2021). Nuclear lipid droplets form in the inner nuclear membrane in a seipin-independent manner. J Cell Biol 220, e202005026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui X, Arlt H, Brock KP, Lai ZW, DiMaio F, Marks DS, Liao M, Farese RV Jr, Walther TC (2018). Cryo-electron microscopy structure of the lipid droplet-formation protein seipin. J Cell Biol 217, 4080–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski KM, Binns D, Bartz R, Grishin NV, Li WP, Agarwal AK, Garg A, Anderson RG, Goodman JM (2007). The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci USA 104, 20890–20895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor FR, Parks LW (1979). Triaglycerol metabolism in Saccharomyces cerevisiae. Relation to phospholipid synthesis. Biochim Biophys Acta 575, 204–214. [DOI] [PubMed] [Google Scholar]
- Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V (2009). Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J 28, 2244–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Chung J, Farese RV Jr (2017). Lipid droplet biogenesis. Annu Rev Cell Dev Biol 33, 491–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Becuwe M, Housden BE, Chitraju C, Porras AJ, Graham MM, Liu XN, Thiam AR, Savage DB, Agarwal AK, et al. (2016). Seipin is required for converting nascent to mature lipid droplets. eLife 5, e16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolinski H, Hofbauer HF, Hellauer K, Cristobal-Sarramian A, Kolb D, Radulovic M, Knittelfelder OL, Rechberger GN, Kohlwein SD (2015). Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim Biophys Acta 1851, 1450–1464. [DOI] [PubMed] [Google Scholar]
- Yan R, Qian H, Lukmantara I, Gao M, Du X, Yan N, Yang H (2018). Human SEIPIN binds anionic phospholipids. Dev Cell 47, 248–256.e244. [DOI] [PubMed] [Google Scholar]
- Yang H, Bard M, Bruner DA, Gleeson A, Deckelbaum RJ, Aljinovic G, Pohl TM, Rothstein R, Sturley SL (1996). Sterol esterification in yeast: a two-gene process. Science 272, 1353–1356. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Chieu HK, Low CP, Zhang S, Heng CK, Yang H (2003). Schizosaccharomyces pombe cells deficient in triacylglycerols synthesis undergo apoptosis upon entry into the stationary phase. J Biol Chem 278, 47145–47155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.