Abstract
Experimental and computational studies pinpoint rate-limiting step(s) in metastasis governed by Rac1. Using ovarian cancer cell and animal models, Rac1 expression was manipulated, and quantitative measurements of cell–cell and cell–substrate adhesion, cell invasion, mesothelial clearance, and peritoneal tumor growth discriminated the tumor behaviors most highly influenced by Rac1. The experimental data were used to parameterize an agent-based computational model simulating peritoneal niche colonization, intravasation, and hematogenous metastasis to distant organs. Increased ovarian cancer cell survival afforded by the more rapid adhesion and intravasation upon Rac1 overexpression is predicted to increase the numbers of and the rates at which tumor cells are disseminated to distant sites. Surprisingly, crowding of cancer cells along the blood vessel was found to decrease the numbers of cells reaching a distant niche irrespective of Rac1 overexpression or knockdown, suggesting that sites for tumor cell intravasation are rate limiting and become accessible if cells intravasate rapidly or are displaced due to diminished viability. Modeling predictions were confirmed through animal studies of Rac1-dependent metastasis to the lung. Collectively, the experimental and modeling approaches identify cell adhesion, rapid intravasation, and survival in the blood as parameters in the ovarian metastatic cascade that are most critically dependent on Rac1.
INTRODUCTION
Conquering disease recurrence in ovarian cancer remains a critical challenge in treating this deadliest of gynecologic malignancies (Kuroki and Guntupalli, 2020). The overall 5-yr survival rate of ovarian cancer patients across all stages is 49% and has not changed dramatically in 20 years (National Cancer Institute, 2022).
Because approximately 80% of patients present with advanced disease that is largely localized in the peritoneal and abdominal cavity (National Cancer Institute, 2022), more limited attention has been placed on distant metastases and their role in disease recurrence. Only since 2010 has there been systematic reporting of distant metastases in the Surveillance, Epidemiology, and End Results (SEER) database, which has enabled analyses of their contributions to patient survival (Gockley et al., 2017; Deng et al., 2018; Gardner et al., 2020; Wang et al., 2021). Multiple analyses of the SEER database over different years and including thousands of stage IV ovarian cancer patients concur that metastases (expressed as % of patients) are found in the liver (38–57%), lung (28–38%), distant lymph nodes (29%), bone (4%), and brain (1%) (Gockley et al., 2017; Deng et al., 2018; Gardner et al., 2020; Wang et al., 2021). Distant metastases increase chemoresistance and disease relapse and reduce 5-yr survival to 28% of patients from an average of 48.6% (Gardner et al., 2020). The locations of distant metastases were identified as independent prognostic factors affecting overall survival (Gardner et al., 2020). Patients with the more common liver and lung metastases had median survival times of 30 and 26 mo, respectively, while survival was worse for those with rarer bone (11 mo) or brain (7 mo) metastases. Because therapy responsiveness was found to be tied to the type of distant metastases, it was recommended that treatment and management plans should be individualized. Nevertheless, ovarian cancer is still largely considered a peritoneal disease and treated with frontline surgery and chemotherapy. Therefore, there is opportunity for improving outcomes by identifying and targeting key factors in the metastatic cascade that promote ovarian cancer spread to distant metastatic sites.
Transcoelomic tumor spread, mediated by peritoneal fluid circulation, most commonly leads to metastases throughout the peritoneal cavity and parenchymal invasion of abdominal organs (Tan et al., 2006). However, metastases via hematogenous routes also occur, as demonstrated through a parabiosis model (Pradeep et al., 2014). The first site of peritoneal metastasis is frequently the omentum, a highly vascularized, adipose tissue that covers the bowel and abdominal cavity. Circulating tumor cells (CTCs) have been detected in ovarian cancer patients and likely seed the distant sites that were recently recognized to worsen survival odds through hematogenous or lymphatic circulation (Obermayr et al., 2013; Gasparri et al., 2016; Gockley et al., 2017; Deng et al., 2018; Zhang et al., 2018; Gardner et al., 2020). The most common metastases in the liver can occur either via parenchymal invasion (23%) or through hematogenous spread (16%) and can be distinguished radiologically (O’Neill et al., 2017). Hematogenous spread to the liver is found associated with high-grade tumors (grade 3) and advanced-stage disease (III or IV). Hematogenous, but not parenchymal, invasion leads to significantly shorter survival times, 63 versus 145 mo (O’Neill et al., 2017). Dissemination to the second most common distant niche occurs exclusively through tumor cells entering the blood and exiting and colonizing the lung. Therefore, elucidation of the factors and mechanisms that contribute to ovarian cancer metastases due to cell dissemination via the blood is important to ensure appropriately targeted patient therapies.
Rho family GTPases are important regulators of cancer-relevant cellular functions including actin cytoskeleton dynamics, cell adhesion and migration, and proliferation (Sahai and Marshall, 2002). In particular, the Rho family Rac1 GTPase is classified as an oncoprotein and has been shown to be mutant, overexpressed, or hyperactive in many human cancers (Schnelzer et al., 2000; Bid et al., 2013; Guo et al., 2015a; Ji et al., 2015). Our group (Guo et al., 2015a), along with Leng et al. (2015), first demonstrated Rac1 overexpression and activation in ovarian cancer patient samples. Consistent with widespread alteration of Rac1 in solid tumors (De et al., 2019), we found that Rac1 and the splice variant Rac1b are overexpressed in ovarian cancer based on The Cancer Genome Atlas (TCGA) data (Hudson et al., 2018). High RAC1, but not RAC1b, mRNA levels predict reduced ovarian cancer patient survival (Hudson et al., 2018). However, there remains a gap in knowledge as to which Rac1-regulated cell functions are most important in ovarian cancer metastasis.
The goal of this study is to identify steps in the ovarian cancer metastatic cascade that are dependent on Rac1 signaling and expression. The ovarian metastatic cascade begins with tumor cell detachment from the primary tumor as either single cells or multicellular aggregates, called spheroids (Lengyel, 2010; Al Habyan et al., 2018). Ovarian cancer cells and spheroids can then attach and invade tissues and organs in the peritoneal cavity including the favored omentum that functions in peritoneal fluid and metabolic and immune homeostasis (Meza-Perez and Randall, 2017; Liu et al. , 2021). Such parenchymal invasion involves clearance of the mesothelial cells covering the omentum and other peritoneal surfaces and organs. Once ovarian tumor cells have invaded the omentum, they can gain access to niche lymph and vascular systems (Krist et al., 1998; Etzerodt et al., 2020; Nowak and Klink, 2020) for tumor expansion and blood-mediated dissemination to distant metastatic sites such as the liver and lung, among others (Gockley et al., 2017; Deng et al., 2018; Gardner et al., 2020; Wang et al., 2021). It is well established that activated Rac1 GTPase has normal roles in cell adhesion, coordinated cell migration, and intravasation and extravasation of leukocytes (Wu et al., 2009; Wang et al., 2010; Ridley, 2015). What is less well studied is whether and how Rac1-regulated migration and intravasation of tumor cells into circulation contributes to dissemination and colonization of distant niche sites.
Because experimental tracking of metastatic dissemination involves rare events and is technically difficult, the present study couples experimental measurements with computational modeling to elucidate the role of Rac1 in ovarian cancer metastasis. Computational models have been used to investigate factors that contribute to cell motility and metastasis (Chen et al., 2011), tumor-induced angiogenesis (Bauer et al. , 2007), and drug delivery routes for ovarian cancer treatment (Winner et al., 2016). Here, experimental data are used to parameterize an agent-based model using the program NetLogo (Wilensky, 1999) to make predictions about ovarian cancer cell metastasis and dissemination. An agent-based computational modeling approach was selected because it allows us to encode characteristics of individual cells that vary with different levels of RAC expression and then observe how that variation affects dissemination into circulation and distant metastases. While all models are necessarily simplifications of a very complex reality, our models provide visual and quantitative representations of cellular adhesion, invasion, and movement, allowing us to generate quantitative hypotheses about mechanisms that contribute to ovarian cancer cell metastases. Animal studies of Rac1-dependent metastasis to the lung affirmed the modeling predictions.
RESULTS
Cell models with Rac1 overexpression and knockdown to elucidate the role of Rac1 in ovarian cancer metastasis
For the present studies, we focused on dissecting the roles of Rac1 in metastasis based on strong evidence of Rac1 overexpression and hyperactivation in ovarian cancer patient samples and the importance of Rac1 in ovarian cancer patient outcomes (Guo et al., 2015a; Leng et al., 2015). Our team previously established that Rac1 inhibition blocks ovarian tumor cell behaviors such as extracellular matrix (ECM) adhesion and downstream PAK1 proliferation and survival signaling (Guo et al., 2015b). Evidence for the importance of Rac1 expression in available ovarian cancer cell line models was obtained using the DepMAP Portal: The Cancer Dependency MAP, Broad Institute, which provides information on the essentiality of genes based on CRISPR-Cas9 ablation (Meyers et al., 2017). Computational corrections (CERES) account for gene copy number variations in different cell lines. Rac1 cancer dependency was comparatively assessed against Cdc42 and RhoA, two other Rho GTPase family members with reported roles in cancer. As shown for 65 human ovarian adenocarcinoma cell lines, RhoA is above the median of the –1 threshold for all essential genes, suggesting that it is less critical in ovarian cancer cell line survival (Supplemental Figure S1). On the other hand, half or more of ovarian cancer cells lie below the –1 threshold upon CRISPR-Cas9 ablation of Rac1 or the highly related Cdc42 GTPase, indicative of their importance in ovarian cancer cell line viability (Supplemental Figure S1). Additionally, only a weak correlation between Rac1 and Cdc42 coexpression was detected among 316 human ovarian cystadenocarcinoma samples in TCGA (cBioPortal; Pearson and Spearman coefficients 0.15, p values 6.85E-3 and 7.90E-3, respectively), consistent with the functions of these GTPases in distinct cell behaviors. All 1005 cancer cell lines characterized in the DEPMap Portal have high log2 RAC1 mRNA expression. There is also a significant correlation between high RAC1 expression and decreased overall cancer cell viability (Supplemental Figure S2A; Pearson –0.368, Spearman –0.374 p = 1.13E-33) upon RAC1 knockdown. Based on the strong experimental lines of evidence for Rac1 essentiality in ovarian cancer (Supplemental Figure S2B; significant negative gene effect), cell lines with Rac1 overexpression (OE) or knockdown (KD) were generated to enable identification of the rate-limiting step(s) in the metastatic cascade governed by Rac1 through a combination of experimental and computational studies.
Rac1-overexpressing cell lines were generated using three well-characterized ovarian cancer cell models (SKOV3ip, OVCAR3, and OVCAR8) that were selected for their p53 mutant status, due to the fact that >90% of serous ovarian cancer patient samples are p53 mutant (Domcke et al., 2013; Tan et al., 2013; Haley et al., 2016; see also Supplemental Figure S8). The selected lines also represent human serous ovarian cancer epithelial (Epi-A subtype, OVCAR8) and mesenchymal (OVCAR3, SKOV3ip) cell subtypes as defined by gene expression analyses of 1538 human tumors and comparison to the gene expression of established cell lines (Tan et al., 2013). The Epi-A subtype is associated with better prognosis, while the mesenchymal subtype is associated with poorer prognosis, advanced stage and metastasized tumors (Tan et al., 2013). We also included cell models with in vitro cell behaviors relevant to metastasis, including cell migration, invasion, and cis-platin resistance (Haley et al., 2016). Importantly, log2 RAC1 mRNA expression in 65 ovarian adenocarcinomas is similar to that seen in all cancer cell lines (Supplemental Figure S2, A and B) and 75% exhibited compromised survival upon CRISPR-Cas9–mediated Rac1 ablation (x-axis; based on CERES gene effect < 0.6). The three cell lines used in the present study all have a negative gene effect in response to CRISPR-Cas9 knockout of Rac1 (–0.65 to –1.4) (Supplemental Figure S2B).
Even while RAC1 mRNA levels based on DepMAP were highest in OVCAR8 cells, our Western blot analyses of the three parental cell lines showed that endogenous Rac1 protein levels were highest in SKOV3ip cells and lower in OVCAR8 and OVCAR3 cells when grown in two-dimensional (2D) culture (Figure 1, A and B). Rac1 expression in OVCAR8 cells is fourfold lower than in the other two cell lines (Figure 1, A and B). Support for potential differences in Rac1 expression, stability, and/or activation among ovarian cancer cell lines is based on the fact that ovarian cancer patient samples (TCGA) show differential gene amplification of Rac1 as well as key regulatory guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) (cBioPortal). Thus, the cell lines are considered to represent a spectrum of Rac1 OE and activation status that is relevant in ovarian cancer patients. The selected cell models were used to manipulate Rac1 protein expression with plasmid-based vectors for OE and KD via CRISPR-Cas9 as further detailed below.
FIGURE 1:

Ovarian cancer cell models. Ovarian cancer cell lines with Rac1 OE or KD were generated to investigate the role of Rac1 in ovarian cancer metastasis. (A, B) Quantification of endogenous Rac1 expression in SKOV3ip, OVCAR3, and OVCAR8 cell lines. SKOV3ip cells have two- and threefold increases in endogenous Rac1 expression compared with OVCAR3 and OVCAR8 cells, respectively. (C, D) Quantification of Rac1 expression in SKOV3ip ovarian cancer cells. (E, F) Quantification of Rac1 KD by CRISPR-Cas9 in SKOV3ip ovarian cancer cell clones that were selected after CRISPR electroporation via single-cell cloning. Means ± SD (n = 2 for B, D, and F).
SKOV3ip, OVCAR3, and OVCAR8 cells were stably transfected with plasmid vectors to overexpress GFP (as a control) or N-terminally green fluorescent protein (GFP)-tagged Rac1 (GFP-Rac1). Rac1 OE in each cell line was confirmed and quantified via Western blot (Figure 1, C and D; Supplemental Figure S3, A–D). SKOV3ip GFP-Rac1 High OE cells have a 2.17-fold increase in Rac1 expression compared with parental cells (Figure 1, C and D). OVCAR3 GFP-Rac1 OE cells have a 2.8-fold increase in Rac1 expression compared with GFP control cells (Supplemental Figure S3, A and B). OVCAR8 GFP-Rac1 OE cells have a 14.3-fold increase in Rac1 expression compared with OVCAR8 GFP control cells (Supplemental Figure S3, C and D).
Rac1, like all members of the Ras superfamily of small molecular GTPases, has a compact globular structure (Hirshberg et al., 1997). A central guanine nucleotide binding pocket is flanked by two switch domains that undergo conformational changes depending on whether the protein is GTP- or GDP-bound. The switch regions are pivotal for protein–protein interactions and are differentially recognized by regulatory proteins involved in Rac1 activation (GEFs) and inactivation (GAPs), as well as GTPase effector proteins involved in downstream signaling. Because of the central importance of the switch domains in Rac1 folding, structure, and activity, exons 3 and 4, which encode the indicated sequences (Supplemental Figure S4, A and B), were targeted for genome editing using CRISPR-Cas9 to knock down Rac1.
Following initial validation of individual guides as detailed in Materials and Methods, Cas9 and guide RNA complexes targeting Rac1 exon 3 or 4 were electroporated into SKOV3ip, OVCAR3, and OVCAR8 cells, simultaneously (Supplemental Figure S4, C and D). As a negative control, Cas9 enzyme was electroporated without added guide RNAs. After electroporation, cells were seeded and expanded.
Single-cell cloning was performed to isolate Cas9 only electroporated (CRISPR-Cas9 control) cell lines or cells having stable KD of Rac1 to enable both in vitro and in vivo analyses. After expansion of clones, residual Rac1 expression levels were measured via Western blot relative to negative controls (Figure 1, E and F; Supplemental Figure S5, A and B). SKOV3ip Rac1-CRISPR KD clones 23, 26, and 27 showed 4.33-, 5.24, and 2.9-fold decreases in Rac1 expression compared with SKOV3ip CRISPR-Cas9 control cells, respectively (Figure 1, E and F). OVCAR3 Rac1 CRISPR-Cas9 KD clones (clone 1 and clone 6) showed 6.25- and 3.6-fold decreases in Rac1 expression compared with OVCAR3 CRISPR-Cas9 control cells (Supplemental Figure S5, A and B). Nevertheless, there were no noteworthy changes in 2D cell growth following KD of Rac1 in these ovarian cancer cell lines.
To verify CRISPR-Cas9 targeting of exons 3 and/or 4 as the cause of the observed reduction in Rac1 expression, DNA sequencing and thymine and adenine (TA) cloning of cDNAs were performed. For DNA sequencing, primers flanking the exons were used to amplify Rac1 exon 3 or exon 4 from CRISPR-Cas9 control and Rac1 CRISPR-Cas9 KD clones and the resulting products were individually sequenced.
Based on DNA sequencing, OVCAR3 Rac1 CRISPR-Cas9 KD clone 6 has 16 nucleotides missing within exon 3 (Supplemental Figure S6A). This causes a change in protein sequence within this exon, which is critical for protein–protein interactions (see Supplemental Figure S4). TA cloning of RAC1 cDNA was used to analyze exons 3 and 4 from contiguous strands in order to probe for potential clone heterozygosity. SKOV3ip Rac1 CRISPR-Cas9 KD clone 23 has a 5-base-pair deletion in exon 3 and a 1-base-pair deletion in exon 4, resulting in a change in protein sequence (Supplemental Figure S6B). Sequence analyses of SKOV3ip Rac1 CRISPR-Cas9 KD clone 26 resulted in nonspecific priming and failed to give sequencing results. Because Rac1 CRISPR-Cas9 KD clone 26 had the lowest Rac1 protein expression (Figure 1F) and significantly diminished mRNA expression via quantitative PCR (qPCR) (Supplemental Figure S7A), it appears to have a deletion or rearrangement extending beyond exons 3 and 4. SKOV3ip Rac1 CRISPR-Cas9 KD clone 27 has an 18-base-pair deletion within exon 3 and a 2-base-pair deletion in exon 4, resulting in a change in protein sequence and a premature stop codon (Supplemental Figure S6B).
To examine the CRISPR-Cas9 impact on the mRNA expression of RAC1 relative to closely related Rho-GTPase family members CDC42 and RHOA, qPCR analyses were performed. Relative fold changes were compared with that of SKOV3ip CRISPR-Cas9 control cells and normalized to 18s rRNA. RAC1 mRNA was significantly decreased (p ≤ 0.001) in SKOV3ip Rac1 CRISPR-Cas9 KD clones (clone 23 and clone 26), with 2.5- and 10-fold decreases, respectively. CDC42 mRNA levels remained unchanged following Rac1 CRISPR-Cas9 KD, and RHOA mRNA was modestly reduced (20%) in SKOV3ip Rac1 CRISPR-Cas9 KD clone 23 (p ≤ 0.05), but not in clone 26 (Supplemental Figure S7, A–C). Because RHOA and RAC1 mRNA levels and activities are usually regulated in opposing directions in response to cell signaling, with coregulation seldomly reported (Zhang et al., 2021a; Dong et al., 2022), this change is not considered noteworthy.
Rac1 quantitatively increases ovarian cancer cell–cell adhesion and is important for cell spreading and cell–substrate adhesion
Ovarian cancer cells manipulated to alter Rac1 expression were used to measure changes in ovarian cancer cell adhesion, spreading, and barrier formation in in vitro culture using electric cell–substrate impedance sensing (ECIS) (Giaever and Keese, 1993; Applied BioPhysics, 2021). ECIS is a real-time, impedance-based method used to study activities of cells grown in culture (Giaever and Keese, 1993; Applied BioPhysics, 2021). A small alternating current (I) is applied to the electrodes at the bottom of the ECIS array wells, creating a potential (V) across the electrode (Giaever and Keese, 1993). The ECIS instrument measures this potential, and the impedance (Z) is calculated using Ohm’s law Z = V/I. When adherent cells attach to the electrodes, they act as insulators and the impedance increases (Applied BioPhysics,, 2021). Alternating current frequencies are used to assess different cellular behaviors given that frequency affects the current path of cell-covered electrodes. At lower frequencies most of the current flows through the space between adjacent cells;this allows for measurement of cell–cell barrier formation as a function of time (Szulcek et al., 2014; Applied BioPhysics, 2021). Higher frequencies (>40,000 Hz) are used to measure cell–substrate adhesion and spreading, because under these conditions current traveling capacitively through the insulating cell membranes can be measured (Applied BioPhysics, 2021). Cell attachment to the electrodes restricts the flow of current and is detected as a decrease in capacitance (Szulcek et al., 2014).
Changes in cell adhesion and spreading were compared in Rac1 OE cells relative to control and Rac1 CRISPR-Cas9 KD cells. Cells were plated at confluence on 96-well plates containing gold electrodes. Current frequencies were varied to measure cell–cell barrier formation, cell spreading or cell–substrate adhesion. To analyze changes in cell–cell barrier formation, impedance at 4000 Hz was measured over time (Figure 2A). Resistance was quantified over the first 10 h after cells were seeded onto the gold electrodes. SKOV3ip GFP-Rac1 High OE cells had a significantly higher resistance 10 h post–cell addition compared with control cell lines (Figure 2B). This increase in resistance suggests a tighter cell–cell barrier formation. SKOV3ip Rac1 CRISPR-Cas9 KD clones had significantly lower resistance at 10 h (Figure 2B), suggesting weaker cell–cell barrier formations. The slope of the resistance was measured to examine changes in resistance over the first 10 h, to compare how fast the cell barriers were forming. SKOV3ip Rac1 GFP-High OE cells had significantly higher average resistance slopes compared with controls, indicating faster cell–cell barrier formation (Figure 2, C and D). These data demonstrate that Rac1 contributes to faster and tighter cell–cell barrier formations.
FIGURE 2:
ECIS to assess cell adhesion and spreading. ECIS was used to measure cell–cell barrier formation, cell–substrate adhesion, and cell spreading. SKOV3ip cells were plated on gold-coated electrodes. (A) Resistance kinetics plot measured at 4000 Hz over 24 h to measure cell–barrier formation. (B) Resistance quantified at 10 h. SKOV3ip Rac1 High OE cells have a significantly higher resistance, indicating a tighter cell–cell barrier formation compared with control and Rac1 CRISPR-Cas9 KD cells. (C, D) Quantification of the average slope of resistance measured over the first 10 h after cells are added to the ECIS electrode arrays. SKOV3ip Rac1 High OE cells exhibited a significantly higher average slope, indicating faster cell–barrier formation. SKOV3ip Rac1 CRISPR-Cas9 KD cells have the lowest slope, suggesting a longer time to form cell–cell adhesions. (E) Capacitance kinetics plot at 64,000 Hz over 24 h to measure cell–substrate adhesion. (F) Capacitance measured at 64,000 Hz to assess cell–substrate adhesion. SKOV3ip Rac1 CRISPR-Cas9 KD cells have the highest capacitance, indicating weaker adhesion to the gold electrode. SKOV3ip Rac1 OE and control cell lines have significantly lower capacitance, suggesting a tighter adhesion to the electrode. (G) The t1/2 max, or time to cover the electrode, was quantified to assess changes in cell spreading. SKOV3ip Rac1 CRISPR-Cas9 KD cells take significantly more time to cover the electrode compared with SKOV3ip Rac1 OE or control cells. Means ± SD (n = 3). One-way ANOVA followed by Tukey’s post-hoc test for multiple comparison was used: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To measure changes in cell–substrate adhesion, capacitance at 64,000 Hz was measured over time. Changes in capacitance represent the ability of cells to adhere to the gold electrodes; therefore a decrease in capacitance suggests an increase in cell–substrate adhesion. Adhesion is the initial plateau phase in the capacitance curve (2–8 h) (Szulcek et al., 2014). Therefore, capacitance was quantified at 8 h post–cell addition to electrodes. SKOV3ip GFP-Rac1 High OE cells had capacitance measurements similar to those of SKOV3ip parental control and CRISPR-Cas9 control cell lines (Figure 2E). However, cells with Rac1 CRISPR-Cas9 KD had higher capacitance measurements, indicating less cell–substrate adhesions. This indicates that Rac1 expression is important for cell–substrate adhesion.
Cell spreading was analyzed using capacitance measurements at 64,000 Hz (Figure 2F). Cell spreading is complete after 10–30 h, depending on seeding density (Szulcek et al., 2014). The time point t1/2 (half maximal electrode coverage) is used for statistical analysis (Wegener et al., 2000; Szulcek et al., 2014). SKOV3ip Rac1 CRISPR-Cas9 KD cells had a significantly higher t1/2 max compared with control, indicating a longer time for cell spreading to occur (Figure 2G). Interestingly, SKOV3ip GFP-Rac1 High OE cells and control cells had similar t1/2 max values, suggesting that cell spreading is occurring similarly (Figure 2G).
Taken together, these results demonstrate that Rac1 contributes to ovarian cancer cell–cell barrier formation. Additionally, while the approximately twofold Rac1 overexpression in SKOV3ip cells did not result in any increased cell–substrate adhesion or cell spreading, an approximately fivefold decrease in Rac1 expression following KD significantly reduced adhesion and cell spreading. These data are evidence that a critical threshold of Rac1 expression and activity maintains cell behaviors required for cancer cell survival and migration and is congruous with the DEPMap RAC1 gene essentiality data (Supplemental Figure S2B).
Rac1 overexpression increases ovarian cancer cell invasion
To assess the role of Rac1 in ovarian cancer cell invasion, SKOV3ip ovarian cancer cell invasion was comparatively tested using Matrigel-coated Transwell invasion chambers. After 48 h, cells were fixed, the undersides of the Matrigel-coated filters were imaged, and invaded cells were counted. SKOV3ip cells with increasing levels of Rac1 OE exhibited significantly increased cellular invasion compared with GFP control cells, 26-fold for Rac1-Low OE and 60-fold for Rac1-High OE (Figure 3, A and B). Strikingly, in SKOV3ip cells with CRISPR-Cas9 KD of Rac1, there was almost no invasion compared with CRISPR-Cas9 control cells, with CRISPR clones 23 and 26 having a 60-fold decrease in cellular invasion (Figure 3, C and D).
FIGURE 3:
Rac1 is important for ovarian cancer cell invasion. Invasion of SKOV3ip ovarian cancer cells was assessed using Matrigel Transwell inserts. SKOV3ip cells with GFP or increasing levels of GFP-Rac1 overexpression (GFP-Rac1 Low and GFP-Rac1 High) or Rac1 CRISPR-Cas9 KD were compared. (A) Representative images of SKOV3ip GFP control and GFP-Rac1 OE cellular invasion. (B) Quantification of the number of invaded ovarian cancer cells after 48 h. (C) Representative images of SKOV3ip Rac1 CRISPR-Cas9 KD cellular invasion. (D) Quantification of ovarian cancer cell invasion after 48 h. Quantification is based on three or four independent experiments with three technical Transwell insert replicates per cell line. Five random fields of view were imaged, counted, and averaged for each insert. Means ± SD (n = 3 in B, n = 4 in D). One-way ANOVA: ***p < 0.0002, ****p < 0.0001.
These results indicate that Rac1 is necessary for 3D ECM invasion of ovarian cancer cells. Moreover, increasing levels of Rac1 OE in SKOV3ip cells resulted in increased cellular invasion. The levels of Rac1 expression influence the amount of cellular invasion, which varies depending on expression level. Although CDC42 mRNA levels are not altered by Rac1 CRISPR-Cas9 KD (Supplemental Figure S7, A–C), it is notable that Cdc42, which has roles in invadopodia formation, does not obviously compensate for the loss/reduction of Rac1 in the SKOV3ip CRISPR clones. This is consistent with the weak correlation between Rac1 and Cdc42 expression reported in TCGA.
Rac1 promotes ovarian cancer spheroid mesothelial cell clearance
Ovarian cancer cells that have detached from the primary tumor float in the peritoneal fluid, where they exist as multicellular clusters called spheroids that attach at secondary sites. Organs within the peritoneum are lined with a single layer of mesothelial cells, which the tumor cells need to penetrate in order to colonize a new metastatic site (Zhang et al., 1999). Mesothelial cells are absent underneath tumors (Birbeck and Wheatley, 1965; Witz et al. , 1999). Researchers have demonstrated that ovarian cancer spheroids can move or “force” the mesothelial cells out of the way in vitro, resulting in mesothelial cell clearance, as a reliable mimic of in vivo tumor invasion (Iwanicki et al., 2011). The ability of spheroids to clear the mesothelial layer is dependent on myosin-generated forces. Integrin α5 β1 (α5β1), talin 1, and myosin II have been implicated in mediating ovarian cancer cell mesothelial clearance (Iwanicki et al., 2011). Of particular interest, Rac1 has been shown to regulate myosin II through activation of protein kinase C (PKC) and increased phosphorylation of myosin light chain (MLC) (Pasapera et al., 2015; Shibata et al., 2015). Therefore, an in vitro assay (Davidowitz et al., 2012) was used to quantitatively assess how differential expression of Rac1 impacts mesothelial adhesion and clearance as a surrogate for events required for in vivo invasion of the omental niche by ovarian cancer spheroids.
SKOV3ip parental, GFP, GFP Rac1-High OE, Rac1 CRISPR-Cas9 control, and Rac1 CRISPR-Cas9 KD clones (clones 23 and 27) cells were plated in U-bottom low-attachment plates (100 cells/well), and spheroids were allowed to form over 4 d. Spheroids were then added to wells with a confluent monolayer of primary LP-9 mesothelial cells (Coriell Institute for Medical Research) that had been labeled with CellTracker Red (Davidowitz et al., 2012). The spheroids were imaged, and the initial spheroid size was measured 1 h after addition to the mesothelial cells.
Interestingly, SKOV3ip GFP-Rac1 High OE cells formed significantly smaller and more compact spheroids compared with parental and GFP control cell lines (Figure 4, A and B). These data are consistent with the ECIS cell–cell adhesion data (Figure 2) demonstrating tighter cell–cell adhesions upon Rac1 OE.
FIGURE 4:
Rac1 OE decreases area of SKOV3ip spheroids. SKOV3ip cells Parental, GFP control, GFP-Rac1 OE, Rac1 CRISPR-Cas9 control, Rac1 CRISPR-Cas9 KD clone 23, and Rac1 CRISPR-Cas9 KD clone 27 were seeded in U-bottom plates to form spheroids (100 cells/well). Spheroids were imaged in bright field after 4 ds of incubation. (A) Representative images of spheroid formation for each cell line. (B) Quantification of spheroid area comparing every cell line to each other. Means ± SD (n = 19 in Rac1 CRISPR-Cas9 control, and n = 20 in the rest of the cell lines). One-way ANOVA, followed by Tukey’s test: ****p < 0.0001. Scale bar is 200 μm.
To assess the ability of the spheroids to clear the mesothelial cell monolayer, the clearance areas were imaged 7 and 24–25 h after spheroid addition. The spheroid mesothelial clearance area was normalized to the initial spheroid area, as described in Davidowitz et al. (2012). SKOV3ip GFP-Rac1 High OE spheroids created a significantly larger normalized clearance area compared with SKOV3ip parental and GFP control spheroids (Figure 5, A–D). Conversely, the clearance area formed by Rac1 CRISPR-Cas9 KD clone 23 spheroids was significantly smaller than the parental and Rac1 CRISPR-Cas9 control clearance areas (Figure 5, E and F).
FIGURE 5:
Rac1 OE increases mesothelial clearance area of SKOV3ip spheroids. SKOV3ip cells Parental, GFP control, High-Rac1 OE, Rac1 CRISPR-Cas9 control, and Rac1 CRISPR-Cas9 KD clone 23 were seeded in U-bottom plates to form spheroids (100 cells/well). Spheroids were transferred to CellTracker Red–labeled monolayers of LP-9 mesothelial cells. Mesothelial cell clearance was imaged at 7 h and 24 or 25 h post–spheroid addition. Clearance areas were normalized at each time point by dividing the clearance area by the spheroid area at 1 h postaddition. (A, C, and E) Representative spheroid images at 1 h postaddition in bright field and mesothelial clearance areas at 7 and 25 h post–spheroid addition visualized using a TRITC filter. (B) Quantification of normalized mesothelial clearance area (MCA) of parental and High Rac1 OE cells at 25 h post–spheroid addition. Means ± SD (n = 18 in parental, and n = 23 in Rac1 High OE). (D) Quantification of normalized 25-h MCA comparing GFP and High-Rac1 OE cells. Means ± SD (n = 29 in GFP and n = 34 in High Rac1OE). (F) Quantification of normalized MCA at 25 h comparing Parental, Rac1 CRISPR-Cas9 control, and Rac1 CRISPR-Cas9 KD clone 23. Means ± SD (n = 27 in Parental, n = 23 in Rac1 CRISPR-Cas9 control, and n = 26 in Rac1 CRISPR-Cas9 KD clone 23). One-way ANOVA, followed by a Tukey’s test was performed for all three experiments. ****p < 0.0001. Scale bars are 500 μm.
Spheroids formed using the same number of cells per well produced spheroids of various sizes based on Rac1 expression, reflecting Rac1-dependent changes in cytoskeleton, cell–cell adhesion, and spheroid compaction. Furthermore, the ability of these spheroids to clear a confluent layer of mesothelial cells was also dependent on Rac1 expression. Because Rac1 OE was accomplished by expression of a GFP-Rac1 fusion protein to allow distinction from the endogenous protein, SKOV3ip cells expressing GFP alone were initially used as a control. However, spheroids expressing GFP-only had looser adhesion relative to parental SKOV3ip cells, resulting in a significantly larger overall area. High levels of cytoplasmic GFP expression are reported to have a toxic effect in some assays. Some GFP toxicity effects include the production of oxidative stress (Ansari et al., 2016), the induction of apoptosis (Liu et al., 1999), and the potential to form toxic aggregates (Link et al., 2006). To compensate for a potential GFP toxicity, we used two further controls: a SKOV3ip CRISPR-Cas9 control line (Cas9 electroporation without guide RNA) and SKOV3ip parental cell. The resulting spheroids from these lines were comparable in size and more similar to the Rac1 CRISPR-Cas9 KD cells though larger than the Rac1 OE cells (Figure 4A).
Even though Rac1 OE cells formed smaller, more compact spheroids, they generated a larger normalized mesothelial cell clearance area compared with the control spheroids, which formed larger spheroids but had a smaller normalized clearance area (Supplemental Table S1). In addition, the normalized clearance area created by Rac1 CRISPR-Cas9 KD spheroids was significantly smaller compared with that of controls despite the initial spheroid size of both clones (Figure 5, E and F, and Supplemental Figure S9).
These data demonstrate that Rac1 is important for increased ovarian cancer cell–cell adhesion, as well as in the myosin-driven forces that promote mesothelial cell clearance. Mesothelial clearance data are consistent with the above ECIS and single-cell Matrigel invasion data, demonstrating that increased cell–cell adhesion in Rac1 OE cells also results in increased invasion of matrix and cell layers. Moreover, the mesothelial clearance data for Rac1 CRISPR-Cas9 KD clones also correlate to the ECIS results that SKOV3ip Rac1 CRISPR-Cas9 KD cells have a significantly higher t1/2 max compared with control, due to a reduction in actinomyosin-dependent cell spreading (Figure 2G).
Agent-based modeling predicts that Rac1 overexpression contributes to hematogenous tumor cell metastasis
Two agent-based computational models, Ovarian Cancer Metastasis Simulation-Single Cells (OCMetSim-Single Cells) and Ovarian Cancer Metastasis Simulation-Spheroids (OCMetSim-Spheroids), were developed to take advantage of the above measured parameters for simulating ovarian cancer metastasis to distant niche sites via hematogenous spread, which is difficult to measure experimentally (Table 1). The models encompass ovarian tumor cell adhesion and invasion and entry into circulation from the omentum. The goal of the models is to make predictions about ovarian cancer cell metastasis to distant sites based on Rac1 expression. We hypothesized that Rac1 OE would result in more ovarian cancer cells in the distant metastatic sites and at a faster rate compared with Rac1 CRISPR-Cas9 KD.
TABLE 1:
OCMetSim-Single Cells and OCMetSim-Spheroids parameters.
| Experimental parameters | Initial number of cells | Cell line | Probability of adhesion calculated from ECIS data | Probability of adhesion calculated from mesothelial clearance assay | Rate of invasion calculated from Matrigel experiments (×1000) | Rate of invasion determined from Matrigel/mesothelial clearance experiments | Probability of death in the bloodstream | Probability of death from crowding |
|---|---|---|---|---|---|---|---|---|
| Single cell model | 1000 or 5000 | Rac1 OE cells | 80% | N/A | 0.060578704 | N/A | Varied between20, 50, and 80% | Varied between0 and 50% |
| Rac1 CRISPR-Cas9 KDclone 26; KD cells | 42% | N/A | 0.001757813 | N/A | Varied between20, 50, and 80% | Varied between0 and 50% | ||
| Rac1 CRISPR-Cas9 KDclone 23; KD cells | 36% | N/A | 0.000277778 | N/A | Varied between20, 50, and 80% | Varied between0 and 50% | ||
| Spheroid model | 111 | Rac1 OE cells | N/A | 100% | N/A | 0.001 | Varied between20, 50, and 80% | Varied between 0 and 50% |
| Rac1 CRISPR-Cas9 KDclone 23; KD cells | N/A | 100% | N/A | 0.0002 | Varied between20, 50, and 80% | Varied between0 and 50% |
Agent-based modeling is a powerful tool used to make predictions about complex cell and molecular behaviors (Soheilypour and Mofrad, 2018). In agent-based modeling, agents assess internal and external factors to make decisions based on a set of rules and parameters (Bonabeau, 2002). The rules and parameters determine how the agents interact with one another as well as their environment within the simulations.
The NetLogo agent-based modeling platform (Wilensky, 1999) was used to simulate ovarian cancer cell agents moving from the peritoneum into circulation and then to distant tissue sites. NetLogo is a simple agent-based modeling platform that can visually represent cells moving through space or traveling through different tissues. In the NetLogo simulation space, ovarian cancer cells (represented as yellow agents) move between three compartments, the peritoneum, the circulation, and a distant tissue site, based on parameters collected from experimental data detailed in the preceding Results sections (Figure 6A).
FIGURE 6:

NetLogo world set up for OCMetSim-Single Cells and OCMetSim-Spheroids and parameter validation. (A) Representative images of the NetLogo simulation space setup for OCMetSim-Single Cells and OCMetSim-Spheroids. Ovarian cancer cells/spheroids (yellow agents) move between the peritoneum (black patches), circulation (red patches), and to a distant metastatic niche (brown patches with white agents) based on Rac1 expression. Green patches represent the space ovarian cancer cells can adhere to the blood vessel using the parameters set from the ECIS/mesothelial clearance experiments. Blue patches represent the invaded cells into the blood vessel based on parameter set from the Matrigel invasion/mesothelial clearance experiments. The cyan patches represent adhesion to a distant metastatic niche, and the magenta patches represent invasion into a distant metastatic niche tissue. Simulations were run to compare the rate and number of invaded cells into a distant metastatic niche with Rac1 OE or KD. (B) The invasion parameters were validated running simulations with a Poisson distribution for rate of invasion using the Matrigel experimental data from Figure 3 as described in the Results and Materials and Methods sections. The simulations confirm that a Poisson distribution for rate of invasion produces results similar to those for the experimental data.
The steps of the models are as follows. Ovarian cancer cells or spheroids floating in the peritoneal ascites fluid can adhere to the omentum, the most common secondary site of metastasis (Lengyel, 2010). After adhesion to the omentum, tumor cells can invade into the tissue, enter the circulation, travel to distant niche sites, extravasate, and promote metastasis. We start with a known number of ovarian cancer cell agents within the peritoneal cavity, monitor them as they move between the three compartments based on the variables and parameters that dictate their behavior, and quantify how many reach the distant metastatic site compartment.
In OCMetSim, ovarian cancer cells with Rac1 OE or KD float in the peritoneum and can adhere, invade the omentum, move into circulation, and then extravasate into a distant niche site, promoting disease metastasis. Parameters and variables change based on Rac1 expression, that is, experimental adhesion and invasion data with Rac1 OE or KD and hypotheses or assumptions made based on how Rac1 expression influences tumor cell behavior (Table 1). Simulations were run with parameters from Rac1 OE versus Rac1 CRISPR-Cas9 KD to demonstrate the most dramatic differences due to changes in Rac1 expression (Table 2). Rac1 OE and KD adhesion parameters were normalized to their relevant controls. Invasion parameters were calculated based on a Poisson distribution (Figure 6). Parameter calculations are further detailed below in this section.
TABLE 2:
Agent-based modeling simulation results, summarizing the main assumptions and findings from the computational experiments that were performed.
| Single cell or spheroid | Cell crowding? | Figure | Main assumptions | Results | |
|---|---|---|---|---|---|
| Experiment 1;1000 cells | Single cell | No | Figure 7, A–C | Cells died if they did not adhere | 1) Rac1 OE increased the number of cells that reached the distant tissue site compared with Rac1 CRISPR-Cas9 KD2) Rac1 OE cells reached the distant tissue site faster than Rac1 CRISPR-Cas9 KD cells |
| Experiment 2;1000 cells | Single cell | Yes | Figure 8, A and B | 1) Cells died if they did not adhere2) Cells died if crowding prevented adhesion | 1) Cell death from crowding reduced the number of KD cells that reached the distant site but not the number of Rac1 OE cells2) More Rac1 OE cells reached the distant site compared with Rac1 KD3) Rac1 OE cells reached the distant site faster than Rac1 KD |
| Experiment 2;5000 cells | Single cell | Yes | Figure 8, C and D | 1) Cells died if they did not adhere2) Cells died if crowding prevented adhesion | 1) Cell death from crowding reduced the number of Rac1 OE and KD cells that reached the distant site2) More Rac1 OE cells reached the distant site compared with Rac1 KD4) Rac1 OE cells still reached the distant site faster than Rac1 KD |
| Experiment 1a | Spheroid | No | Figure 9, A–B | 1) 100% spheroid adhesion, no spheroid death2) Spheroids died if crowding prevented adhesion | 1) The numbers of Rac1 OE and KD spheroids that reached the distant site were the same2) Rac1 OE spheroids reached the distant site faster than Rac1 CRISPR-Cas9 KD spheroids |
| Experiment 2a; 100% adhesion | Spheroid | Yes | Figure 9, C and D | 1) 100% spheroid adhesion, no spheroid death2) Spheroids died if crowding prevented adhesion | 1) Cell death from crowding reduced the number of Rac1 OE and KD spheroids in the distant site2) Rac1 OE spheroids reached the distant site faster than Rac1 CRISPR-Cas9 KD spheroids |
The first model, OCMetSim-Single Cells, was designed to simulate single ovarian cancer cells floating in the peritoneal ascites fluid and then adhering and invading into the blood vessel, based on single-cell experiments used to parameterize the model. The OCMetSim-Single Cells model was modified to simulate ovarian cancer spheroid adhesion and invasion based on mesothelial clearance assay results to develop the model OCMetSim-Spheroid. Simulation parameters and results are summarized in Tables 1 and 2, respectively.
Briefly, in simulations using OCMetSim, single ovarian cancer cells or spheroids move within the peritoneal space: Ovarian cancer cell agents are created at random x and y coordinates.
Cell agents move by random diffusion and are able to move only into spaces that are not occupied by other agents.
Once the ovarian cancer cell agents encounter the blood vessel, the probability of adhering to the blood vessel is determined by adhesion parameters:
After each ovarian cancer cell comes into contract with the vessel, it has a probability of adhering based on Rac1 expression.
ECIS experimental data were used to set the parameters for cellular adhesion of single ovarian cancer cells.
If a cell does not adhere based on the probability of adhesion calculated from the ECIS experiments, we assume that it dies and therefore is removed from the simulation.
For ovarian cancer cell spheroids, we assumed that they had 100% probability of adhesion, based on 100% observed adhesion of spheroids within 1 h following transfer to mesothelial monolayers in the in vitro mesothelial clearance experiments.
If the cells adhere, then the probability of invasion is determined by the invasion parameters:
Ovarian cancer cells will invade into the tissue with an experimentally determined probability/rate of invasion, based on Rac1 expression.
Matrigel invasion data was used to set the parameters for cellular invasion of single cells.
Mesothelial cell clearance data were used to set parameters for invasion of spheroids.
Once cells have invaded into the blood vessel, they will move in the circulation:
The ovarian cancer cells in circulation will move toward a metastatic niche site reached via hematogenous spread.
Cells in circulation will die at varying probabilities and will be removed from the simulation.
If a cell contacts the metastatic niche site, then the probability of adhesion is determined by adhesion parameters:
We assumed that the probability for adhesion to the metastatic niche is the same as that for adhesion to the blood vessel, based on Rac1 expression for either single cells or spheroids.
Once cells have adhered to the distant metastatic niche site, then the probability of invasion is determined by invasion parameters:
We assumed that the rate of invasion into the distant metastatic niche site is the same as the rate of invasion into circulation, based on Rac1 expression for either single cells or spheroids.
The dimensions for the simulation space of OCMetSim-Single Cells are 181 × 121 coordinate patches. The peritoneal compartment occupies 72% of the simulation space, the blood occupies 15%, and the distant metastatic niche site occupies 10%. The other ∼3% of the simulation space are coordinate lines designated for the adhesion and invasion parameters. To keep the scale accurate for the spheroid model, the OCMetSim-Spheroids model is scaled down by 1/3 and has 61 × 41 coordinate patches. The model is scaled down by 1/3 so that spheroid agents that occupy a single patch represent a cluster of cells consisting of a 3 × 3 spheroid with nine cells. This assumes that spheroids are nine times larger than the single cells. While the Single Cells and Spheroid models have been scaled appropriately, if differences in time (ticks) are to be compared between the two models, the timescales need to be adjusted, given the difference in grid sizes. Therefore, the timescales between the models are not directly compared in the analyses below.
For the OCMetSim-Single Cells model, ECIS experimental adhesion (Figure 2) and Matrigel invasion data (Figure 3) obtained for Rac1 OE or KD cell lines were used to set parameters for cellular adhesion and invasion. Based on the ECIS data (Figure 2), Rac1 OE cells have the highest probability of adhesion relative to parental control cells and Rac1 CRISPR-Cas9 KD cells. For the adhesion parameter of Rac1 OE cells, we set the probability of adhesion to 80% (the highest adhesion probability for cells in the Single Cell model) (Table 1). This value is representative of the adhesion probability that we see in vitro when we plate single Rac1 OE cells. Using the ECIS data, the probability of adhesion for the Rac1 CRISPR-Cas9 KD cells was set as a percent reduction relative to CRISPR-Cas9 control cells. We set the KD adhesion probability of KD clones 26 and 23 to be 42 and 36%, respectively (Table 1).
As described in further detail in Materials and Methods, a random-Poisson distribution is used for the rate of invasion into circulation and distant metastatic niche site. The model assumes that the rate of invasion of one cancer cell does not influence another. The rate of invasion was calculated from Matrigel experiments and is based on the number of invaded cells after 48 h using a total of 20,000 cells. The Poisson distribution and calculations were verified by running simulations to mimic the in vitro experimental data (Figure 6B). SKOV3ip GFP-Rac1 High OE cells have 35- to 218-fold increases in the rate of invasion compared with SKOV3ip CRISPR clones 26 and 23, respectively. The measurements for capacitance at 8 h after seeding were used to calculate the probabilities of adhesion for Rac1 OE and KD parameters. The probability was calculated compared with control cells. SKOV3ip GFP-Rac1 High OE cells have a 1.9- to 2.2-fold increase in adhesion compared with SKOV3ip Rac1 CRISPR-Cas9 KD clones 26 and 23, respectively.
Next, the experimental parameters for invasion and adhesion with Rac1 OE and KD were used to run simulations to predict the rate and frequency of ovarian cancer cell colonization of a distant niche site (Table 2). Further detail on parameters and assumptions are outlined below and in Materials and Methods.
First, we assumed that cells move independently as single cells. These simulations were run with OCMetSim-Single Cells in experiment 1: Adhesion and invasion with single cells, which incorporates cell adhesion and cell death, and invasion, were run using 1000 or 5000 cells as further described below.
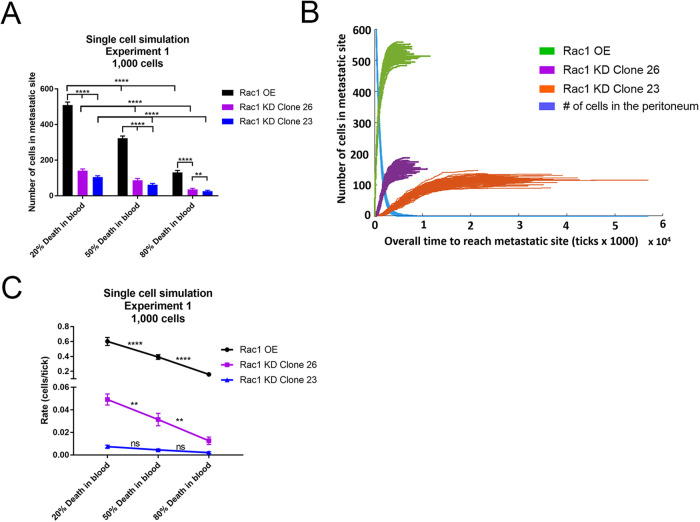
These simulations represent single ovarian cancer cells floating in the peritoneal fluid and encountering the omental tissue site. The cells can adhere and invade into this site, allowing for entry into blood vessels within the omentum, as the ometum is highly vascularized. Once the cells enter circulation, they can travel to a distant metastatic site. In these simulations we assumed that single cells would die if they did not adhere. We compared various percentages of cell death in the blood. Parameters for Rac1 OE and KD were used for cell adhesion and invasion, and the number of cells in a metastatic niche and the rate at which cells enter the niche were quantified. SKOV3ip Rac1 OE cells resulted in 3.6–4.9 times more cells in the distant metastatic niche compared with SKOV3ip Rac1 CRISPR-Cas9 KD clones (Figure 7A). As expected, the increased probability of cell death in the blood decreased the number of cells that reached the metastatic niche for all cell lines (Figure 7A).
FIGURE 7:
OCMetSim-Single Cells model predicts that Rac1 OE contributes to tumor cell metastasis to distant metastatic niche sites. (A) Single-cell simulation results comparing the number of cells that reach a distant metastatic niche with Rac1 OE or KD. Using the parameters for increased invasion and adhesion with Rac1 OE, and varying probabilities of cell death in the blood, these results demonstrate that Rac1 OE results in more ovarian cancer cells reaching a distant metastatic niche in less time, compared with two Rac1 CRISPR-Cas9 KD clones (clones 23 and 26). (B) Representative image of 100 runs with 20% death in the blood and parameters for Rac1 OE or Rac1 CRISPR-Cas9 KD, displaying the rate at which the cells enter the distant niche site. The blue line represents the decrease in the number of cells in the peritoneal compartment for Rac1 OE and KD cells over time. (C) The slope of each cell type entering the distant niche site was quantified to determine the rate at which cells entered the distant niche site. Rac1 OE cells have a higher rate at which they enter the distant niche site compared with two Rac1 CRISPR-Cas9 KD clones. Thirty simulations were run for each experiment. Error bars represent means ± SD. Two-way ANOVA followed by Tukey’s post-hoc test for multiple comparison across columns and rows was used to compare differences between cell type and probability of death in the blood: **p < 0.01, ****p < 0.0001.
To quantify the rate at which ovarian cancer cells enter the distant niche site, the rates of increase of cell number in the distant niche site over time were measured (Figure 7, B and C). Figure 7B shows 100 simulation runs for each cell type using the parameters of experiment 1 and 20% cell death in the blood. The blue decay line represents the number of cells in the peritoneal compartment over time for all three cell types. This decreases in the number of cells leaving the peritoneal space were similar for all three cell types; however, the rate at which cells entered the distant niche site are quite distinct (compare green, purple, and orange lines). Slopes of the linear portion of each of these lines were used to determine the rate at which the cells enter the metastatic site. Rac1 OE cells reached the distant metastatic niche faster than the Rac1 CRISPR-Cas9 KD cells, approximately 12 times faster than Rac1 CRISPR-Cas9 KD clone 23 and approximately 85 times faster than Rac1 CRISPR-Cas9 KD clone 26 (Figure 7C). Rac1 OE cells have a higher probability of adhesion and rate of invasion, allowing the cells to move into the tissue faster, and therefore reaching the distant metastatic niche faster than the Rac1 CRISPR-Cas9 KD cells. With increasing probability of cell death in the blood, cells entered the metastatic niche at a decreased rate. These data suggest that increasing the probability of death in the blood decreases the number of cells that reach the distant niche and the rate at which they enter the distant niche site.
Next, the effect of cell crowding was added and tested in experiment 2: Adhesion and Invasion with Crowding and Death from Crowding with single cells. We hypothesized that cells crowding on the tissue lining the blood vessel would increase cell death with less access to blood vessel sites for nutrients and loss of vessel adhesion. As a result, the number of tumor cells entering circulation would be expected to decrease.
In experiment 2, ovarian cancer single cells with Rac1 OE were compared with Rac1 CRISPR-Cas9 KD clones 26 and 23. These simulations were run with either 1000 single cells (Figure 8, A and B) or 5000 single cells (Figure 8, C and D). Based on the dynamics of cell movement in the single-cell simulations, to better understand the effects of cell crowding, the initial concentration of cells in the peritoneal compartment was tested by comparing 1000 single cells to 5000 single cells. In our simulation space, the number of sites the cancer cells can adhere to is limited. Therefore, to better implement the crowding effect, 5000 single cells were used to test saturation of these sites.
FIGURE 8:
OCMetSim-Single Cells model with added cell death from crowding predicts that cell crowding and cell death from crowding affect the number and rate at which ovarian cancer cells reach a distant niche site. (A, B) Single-cell simulations run to investigate the effects of cells dying from nonattachment caused by crowding of cells lining the blood vessel. The probability of cell death from crowding was varied and tested. Eighty percent probability of cell death in the blood was used. Rac1 CRISPR-Cas9 KD resulted in a further decrease in the number of cells that reached a distant metastatic niche. (B) The rate at which the Rac1 OE cells reached the distant niche site was decreased with crowding or crowding cell death, and the Rac1 OE reached the distant site faster than the Rac1 CRISPR-Cas9 KD cells. (C,D) Single-cell simulations with 5000 cells and cell crowding were run. (C) Crowding did not affect the number of cells that reached the distant tissue site. However, with 50% probability of dying due to cell crowding, the numbers of Rac1 OE and KD cells that reached the distant site were decreased. (D) The rate at which the Rac1 OE and Rac1 CRISPR-Cas9 KD clone 26 cells entered the distant niche site was also decreased with crowding and crowding cell death. Thirty simulations were run for each experiment. Error bars represent means ± SD. Two-way ANOVA followed by Tukey’s post-hoc test for multiple comparison across columns and rows was used to compare cell types and crowding and crowding death: *p < 0.05,***p < 0.005, ****p < 0.0001.
Fifty percent probability of cells dying due to cell crowding was compared with 0% cell death from crowding. Once cells entered the blood vessel, they had an 80% probability of cell death. Figure 8, A and B, represents the simulations with cell death due to crowding with 1000 single cells compared with no cell crowding. Adding a cell crowding parameter with 0% death from crowding into the model did not affect the number of cells that reached the distant niche site compared with no crowding for either Rac1 OE or CRISPR KD with 1000 cell simulation (Figure 8A). The added effect of crowding decreased the rate at which the Rac1 OE cells entered the distant niche site but was not seen for the Rac1 CRISPR-Cas9 KD cells (Figure 8B). When a 50% probability of cell death due to crowding was added, the number of Rac1 CRISPR-Cas9 KD cells that reached the distant site was significantly decreased for both Rac1 CRISPR-Cas9 KD clones. When 5000 single cells were tested to saturate the sites of adhesion, the added effect of cell crowding did not change the number of either Rac1 OE or CRISPR KD cells that reached the metastatic site, similar to the 1000-cell simulations (Figure 8C). However, the rate at which they entered the distant metastatic site was decreased for Rac1 OE cells and Rac1 CRISPR-Cas9 KD clone 26 (Figure 8D). Additionally, when 50% probability of cell death due to crowding was added, the number of Rac1 OE and CRISPR KD cells that reached the distant niche was statistically decreased compared with 0% death from crowding (Figure 8C). Furthermore, Rac1 OE cells reached the distant metastatic site significantly faster than the Rac1 CRISPR-Cas9 KD clones (Figure 8D).
These data suggest that the increased probability of adhesion and rate of invasion with Rac1 OE contribute to significantly more cells in the distant metastatic tissue and at a faster rate compared with Rac1 CRISPR-Cas9 KD. For simulations run with either 1000 or 5000 single cells, the crowding of cells along the blood vessel did not change the number of cells that reach the distant site via hematogenous spread. However, when 5000 cells were tested, the added effect of cell crowding and death from cell crowding changed the number and rate at which the Rac1 OE and Rac1 CRISPR-Cas9 KD clone 26 cells reached the distant niche site. These data suggest that the concentration of cells in the peritoneum is important to consider when investigating how death due to crowding of cells along the blood vessel affects how cells travel to a distant niche site. With a low concentration of cells in the peritoneal space, cells dying due to crowding did not affect cells with Rac1 OE, but the increased rate of cell death due to crowding did decrease the numbers of Rac1 CRISPR-Cas9 KD cells. This may in part be explained by Rac1 OE cells having an increased rate of invasion, allowing the OE cells to invade faster, clearing a site to allow for other cells to adhere and invade.
Additionally, with increased cell concentration and increased cell death from crowding, fewer Rac1 OE cells reach the distant niche site and at a lower rate compared with 0% cell death from crowding. These data suggest that when the number of Rac1 OE cells has saturated the available adhesion/invasion sites, the effects from cell crowding and cell death from crowding decrease the number and rate of OE cells that reach the distant site, even though they have an increased rate of invasion. Thus, higher concentrations of cells in the peritoneal space can cause cell crowding and cell death from crowding even if the cells are quickly clearly from limited intravasation sites.
Next, we modified the OCMetSim agent-based model to consider the fact that ovarian cancer cells survive preferentially as multicellular spheroids in vivo and likely attach and invade into the mesothelial cell layers that line the omentum and abdominal organs, collectively. The OCMetSim-Spheroids model simulates the ovarian cancer spheroid metastatic unit and uses parameters derived from experimental mesothelial clearance assay data as a combined measure of cell adhesion and invasion. Spheroid units likely intravasate as single-cell chains and then quickly reform clusters again in circulation, which is considered as “en bloc” intravasation in the simulations. OCMetSim-Spheroids makes predictions about how spheroids adhere and invade the omentum and invaded cells subsequently intravasate into the blood and metastasize to distant sites.
We assumed that each spheroid had a 100% chance of adhesion, based on experimental data that 100% of spheroids adhered when added to confluent mesothelial layers in vitro. This assumption is also representative of spheroids in vivo, as ovarian cancer spheroids likely have a higher probability of survival in the peritoneal space than single cancer cells. The normalized mesothelial clearance areas of Rac1 OE cells and Rac1 CRISPR-Cas9 KD cells were compared with parental controls or CRISPR-Cas9 control cells to determine the fold change in clearance area. Rac1 OE cells were five times more likely to invade the mesothelial cell layer compared with Rac1 CRISPR-Cas9 KD clones. We assumed, on the basis of minimal invasion spread of the Rac1 CRISPR-Cas9 KD clone 23 in Figure 5E that Rac1 CRISPR-Cas9 KD had no increased invasion ability in spheroid form compared with single cells. Thus, we compared spheroid invasion rate to single-cell invasion rates by equating the invasion of Rac1 CRISPR clone spheroids to that of Rac1 CRISPR clone single cells.
Using these assumptions, the experimental parameters from the mesothelial clearance assay with Rac1 OE and CRISPR KD were used to run simulations to predict the rate and frequency of ovarian cancer spheroid colonization of the distant metastatic niche in Experiment 1a: Adhesion and Invasion for spheroids, using 111 agents, each agent representing a spheroid containing nine cells, so around 1000 cells total. The probabilities for death in the blood were compared and the number of cells in the distant metastatic niche and the rate at which they enter the niche were quantified, Spheroid simulations Experiment (Figure 9, A and B). The numbers of Rac1 OE spheroids and Rac1 CRISPR-Cas9 KD clone 23 spheroids that reached the distant metastatic niche were the same, as the adhesion parameter was set to 100% for both cell lines but decreased similarly with varying cell death in the blood (Figure 9A). This was similar to the single-cell simulations with increased probability of cell death in the blood. Additionally, the rate at which the cells reached the distant niche site was significantly faster for Rac1 OE cells compared with Rac1 CRISPR-Cas9 KD clone 23 (Figure 9B).
FIGURE 9:
OCMetSim-Spheroids model predicts that Rac1 contributes to ovarian cancer spheroid intravasation into circulation and extravasation to distant niche sites. Experiment 1a: Spheroid simulation results comparing (A) the number of spheroids and (B) the rate at which they enter the distant niche site for Rac1 OE or KD clone 23. Rac1 OE or KD spheroids were given 100% probability of adhesion, based on 100% spheroid adhesion observed in the mesothelial clearance assay. (A) Varying probabilities of cells death in the blood decreased the number of ovarian cancer spheroids in the metastatic niche. (B) The rate at which the OE and KD spheroids reached the metastatic niche decreased with increasing probability of cell death in the blood; however, the Rac1 OE spheroids reached the distant site faster than the KD spheroids. Experiment 2a: Spheroid simulation results comparing cell crowding and cell death from crowding (C, D). (C) With added cell death from crowding, the number of Rac1 OE and KD cells that reached the distant site decreased. (D) With crowding and cell death due to crowding, the rate at which the Rac1 OE spheroids reached the distant site was decreased compared with no crowding. The rate at which the Rac1 CRISPR-Cas9 KD cells reached the distant site was decreased with added cell death from crowding. Thirty simulations were run for each experiment. Error bars represent means ± SD. Two-way ANOVA followed by Tukey’s post-hoc test for multiple comparison across columns and rows was used to compare cell types and crowding and crowding death: *p < 0.05, ***p < 0.005 ****p < 0.0001.
Next, the probability of spheroid crowding and its potential contribution to cells dying due to nonattachment was tested in Experiment 2a: Adhesion and Invasion with Cell Crowding and Death from Crowding for Spheroids (Figure 9, C and D). Once the spheroids entered the blood, they were given a 50% probability of cell death in the blood. It is likely that spheroids have an increased probability of survival in the blood compared with single cells. When cell death from crowding along the blood vessel was added, two times fewer Rac1 OE or Rac1 CRISPR-Cas9 KD cells reached the distant site, compared with 0% death from crowding (Figure 9C). Interestingly, the rate at which the Rac1 OE spheroids reach the distant niche site was faster compared with Rac1 CRISPR-Cas9 KD (Figure 9D). When crowding and cell death from crowding were added, the rate at which the Rac1 OE spheroids reached the distant niche site decreased compared with no crowding (Figure 9D). However, the added cell death from crowding did not affect the rate of the KD spheroids, compared with no death from crowding (Figure 9D), similarly as demonstrated in the single-cell simulations (Figure 8, B and D).
Overall, the data from both OCMetSim models show that differential Rac1 expression has an important role in ovarian cancer cell metastasis via hematogenous routes to primary niche sites such as the liver and lungs. Rac1 OE resulted in increased numbers of single cells in the distant metastatic niche and at a faster rate compared with Rac1 CRISPR-Cas9 KD. The simulations also indicate that crowding and crowding cell death of ovarian cancer cells lining the blood vessel decrease the number of single cells and spheroids that enter circulation and the rate at which they reach a distant niche site via hematogenous spread.
Rac1 overexpression increases, while knockdown decreases, ovarian cancer cell tumor burden and metastasis in vivo
Peritoneal tumor dissemination is very common in ovarian cancer and is a stepping stone to distant metastasis via lymphatic and hematogenous dissemination. To elucidate the role of Rac1 in peritoneal tumor burden, two different ovarian cancer cell lines wherein Rac1 expression was manipulated were comparatively assessed. OVCAR8 cells have low endogenous levels of Rac1 and were therefore selected to analyze the impact of Rac1 OE on tumor burden, while SKOV3ip cells natively have the highest Rac1 expression levels and were therefore selected to analyze the impact of Rac1 knockout.
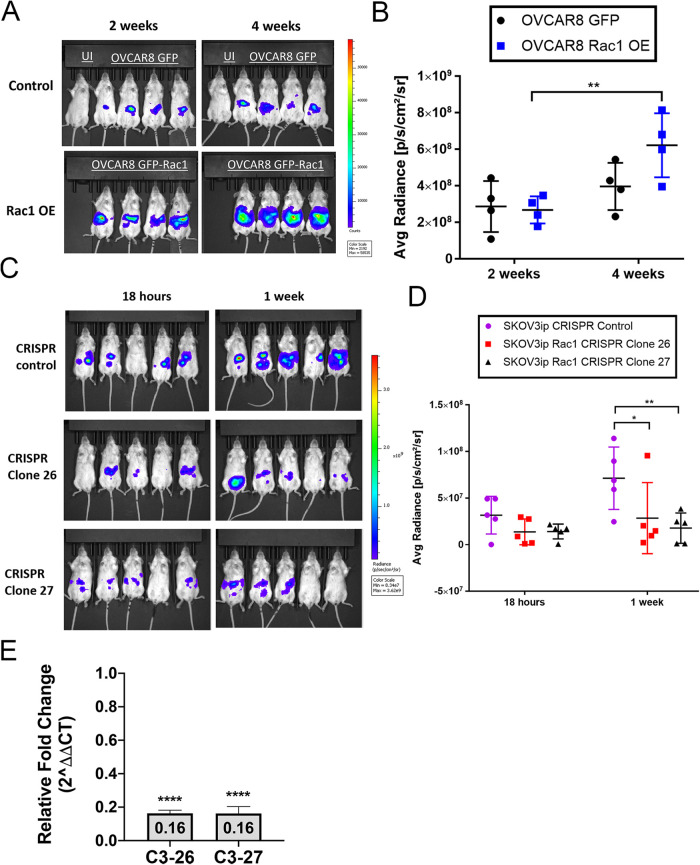
OVCAR8 and SKOV3ip cells manipulated for Rac1 expression and transduced to express luciferase were intraperitoneally (IP) injected into NSG mice. Luciferase expression was used to quantify ovarian cancer tumor burden via IVIS in vivo imaging. Mice were imaged 2- and 4-wk postinjection. The average radiance was quantified to assess tumor burden over the 4 wk. Mice injected with OVCAR8 GFP-Rac1 OE cells had a significant timedependent increase in peritoneal tumor burden compared with GFP control cells (p < 0.0043) (Figure 10, A and B). These data suggest that Rac1 contributes to increased peritoneal tumor burden.
FIGURE 10:
Rac1 overexpression increases tumor burden. OVCAR8 GFP control, OVCAR8 GFP-Rac1 OE, and SKOV3ip CRISPR-Cas9 control, SKOV3ip Rac1 CRISPR-Cas9 KD clone ovarian cancer cells transduced with luciferase were IP injected into NSG mice. Tumor burden was measured using bioluminescent IVIS imaging. (A) Representative IVIS images of NSG mice injected with OVCAR8 GFP control (top row) and OVCAR8 GFP-Rac1 OE cells (bottom row) imaged 2 and 4 wk post–IP injection. (B) Quantification of the average radiance at 2 and 4 wk postinjection. Quantification of 2D radiance using two-way ANOVA shows significant time-dependent variance; p = 0.0043 of tumor burden. N = 4 for each group. (C) Representative IVIS images of NSG mice injected with SKOV3ip CRISPR-Cas9 control cells or Rac1 CRISPR-Cas9 KD cells imaged 18 h and 1 wk post–IP injection. (D) Quantification of the average radiance at 18 h and 1 wk post-injection. Quantification of average radiance using two-way ANOVA reveals a significant decrease in tumor burden in mice injected with CRISPR KD clones (clones 26 and 27) compared with control. N = 5 for each group. *p < 0.05, **p < 0.01. (E) Omental tissue isolated from mouse xenografts of SKOV3ip ovarian cancer cells expressing luciferase. Omental tissues were isolated from mice 1 wk after being injected with 4 × 106 Rac1 CRISPR-Cas9 KD (C3-26 or C3-27) or CRISPR-Cas9 control (C4) cells, and gene expression levels of luciferase were analyzed based on targeted qPCR. Data represented are mean ± SE. These data are combined from one experiment with five biological replicates. **** indicates p value ≤ 0.0001, where values represent relative expression for Rac1 CRISPR-Cas9 KD compared with CRISPR-Cas9 control C4 (1.0) and normalized to 18s rRNA using unpaired two-tailed t test.
In a complementary experiment, SKOV3ip CRISPR-Cas9 control and Rac1 CRISPR-Cas9 KD clones (clones 26 and 27) were intraperitoneally (IP) injected into NSG mice. IVIS in vivo imaging was performed 18 h and 1 wk postinjection, and the average radiance was quantified (Figure 10, C and D). Eighteen hours postinjection no differences between Rac1 CRISPR-Cas9 KD and CRISPR-Cas9 control cells were measurable. However, the 1 wk postinjection tumor burden was significantly smaller in mice injected with Rac1 CRISPR-Cas9 KD cells from clone 26 or clone 27 as compared with the CRISPR-Cas9 control cells. Quantitative PCR for luciferase-expressing tumor cells was used as an independent measure of the effect of Rac1 knockdown on tumor burden. Two SKOV3ip Rac1 CRISPR-Cas9 KD single-cell clones (clones 26 and 27) were injected peritoneally into NSG mice, and after 1 wk omenta were collected and used to quantify luciferase gene expression as a measure of tumor cell numbers 1 wk postinjection. Compared to a CRISPR-Cas9 control there was an 80% decrease in omental implantation detected for both Rac1 CRISPR-Cas9 KD clones (Figure 10E). Collectively, these data indicate that Rac1 is an important driver of increased peritoneal tumor burden and Rac1 knockdown reduces metastatic peritoneal implantation and tumor cell growth.
Rac1 inhibition decreases ovarian tumor burden and metastasis in vivo
Rac1 dependence of ovarian cancer local and distant metastasis in vivo was further assessed by measuring tumor burden in cell models ± pharmacologic Rac1 inhibition with R-ketorolac (Grimes et al., 2021). Mice were injected with SKOV3ip cells transduced to express luciferase and treated with R-ketorolac or placebo for 4 wk. Before death, mice were injected with luciferin and bioluminescence imaging was used to monitor peritoneal and distal organ metastases. The 4-wk tumor growth period was used to mimic late-stage human ovarian cancer with hematogenous dissemination and distal metastasis. Quantification of luciferase-positive tumor cells in tissues was performed using qPCR and measurement of luciferase (LUC) gene expression levels in total RNA from distal organs. Significant decreases in LUC expression (1.85–2.3-fold) were observed in both omental (Figure 11A) and lung (Figure 11B) tissue when Rac1 was pharmacologically inhibited by R-ketorolac. The observed inhibition of lung metastases with Rac1 inhibition is congruous with the predictions of the agent-based model demonstrating decreased number of tumor cells in the metastatic site. These findings suggest further utility of the agent-based model for testing and discriminating other mechanistic factors in the ovarian metastatic cascade. Collectively, the data document a role for Rac1 in ovarian cancer metastases occurring either through parenchymal invasion via mesothelial clearance or through hematogenous dissemination and extravasation at distant organ sites.
FIGURE 11:

Pharmacologic inhibition of Rac1 inhibits local and distant metastases in vivo. Gene expression levels of targeted luciferase qPCR analyses of omental and lung tissues isolated from mouse xenografts of SKOV3ip ovarian cancer cells. Samples are from mice treated with 5 mg/kg/d R-ketorolac or placebo for 4 wk. Data represented are mean ± SE. (A) Omental tissue data are from one experiment with five biological replicates. ** indicates p value < 0.01, where values represent relative expression for R-ketorolac compared with placebo (1.0) and normalized to 18s rRNA using unpaired two-tailed t test. (B) Lung tissue data are combined from one experiment with five biological replicates. *** indicates p value < 0.001, where values represent relative expression for R-ketorolac compared with placebo (1.0) and normalized to 18s rRNA using unpaired two-tailed t test.
DISCUSSION
Rac1 GTPase acts as an oncoprotein to regulate multiple hallmarks of cancer, and elevated expression is correlated with tumor stage, blood vessel invasion, and lymph metastasis across many cancer types (Lou et al., 2018). Even while there are strong associations between Rac1 expression and patient outcomes, it is difficult to draw firm conclusions about the functional impact of Rac1 expression in specific cancers based only on such data. We present in vitro and in vivo experimental data that are used to inform computational agent-based models for the purpose of delineating the contributions of Rac1 to ovarian tumor cell behavior and metastatic spread. The model was parameterized using measured cell adhesion and invasion data and identified Rac1 as a key factor in initial peritoneal adhesion of ascitic tumor cells that have been shed from primary tumors. Rac1 is also shown to be important for mesothelial clearance and omental niche colonization. Importantly, model predictions regarding the magnitude of Rac1 importance for intravasation and tumor dissemination to distant metastatic sites by entering the blood from the peritoneum could be confirmed experimentally.
The motivation for the present study is based on our analysis of 298 stage III and IV high-grade serous ovarian cancer (HGSOC) patients with outcomes data in TCGA, which demonstrated that high total RAC1 (but not CDC42) mRNA expression is associated with worse outcomes (Hudson et al., 2018). These findings concur with those reported for a Chinese patient cohort (Leng et al., 2015). Histochemical analyses of human ovarian cancer patient samples evidenced that Rac1 and Cdc42 protein OE, but not RhoA, tracked with higher tumor grade (Guo et al., 2015a). Because GTPase expression levels were analyzed separately using pathological scoring relative to noncancerous controls, it could not be determined whether Rac1 and Cdc42 GTPases were both overexpressed in the same samples. Analyses of 316 human ovarian cystadenocarcinoma samples in TCGA (cBioPortal; Pearson and Spearman coefficients 0.15, p values 6.85E-3 and 7.90E-3, respectively) show only a weak correlation between Rac1 and Cdc42 coexpression; further supporting our focus on Rac1 in the present study. In addition to gene and protein expression parameters, we also considered Rac1 activity.
Experimental evidence demonstrates that Rac1 activity is sufficient to drive tumor-relevant activities in ovarian tumor cells. Introduction of a mutationally activated form of Rac1 (Rac1G12V) in two ovarian tumor cell lines increased invasive capacity and markers of epithelial to mesenchymal transition (EMT), functionalities viewed as central to cancer metastasis (Fang et al., 2017). Conversely, selective inhibition of Rac1 activity or KD of Rac1 expression inhibited migration and invasion of ovarian tumor cells (Guo et al., 2015b; Leng et al., 2015), and short hairpin RNA -mediated Rac1 KD reduced EMT and decreased ovarian tumor growth in a xenograft model (Leng et al., 2015). Further evidence for Rac1 GTPase activation in ovarian cancer derives from high levels of Rac1 activity measured in freshly isolated ascitic tumor cells from patients and finding a precipitous decline in Rac1 activation when cells were placed in culture for 48 h, suggesting the importance of the in vivo tumor environment for Rac1 activation (Guo et al., 2015a). The importance of Rac1 activation in tumor metastasis is forecast by increased survival of ovarian cancer patients treated with ketorolac, a drug wherein the R-enantiomer inhibits Rac1 and Cdc42 GTPases (Guo et al., 2015a,b ; Oprea et al., 2015; Grimes et al., 2021). Therefore, the current studies used OE and KD strategies to reveal the contributions of Rac1 expression and activity to ovarian cancer cell adhesion, spreading, and barrier formation. The ability of ovarian cancer spheroids to clear the mesothelial layer and adhere to underlying ECM is an essential prerequisite to implantation and invasion of peritoneal sites. A mesothelial clearance assay was used to test Rac1 importance for 3D invasion. A critical role of Rac1 in these early metastatic steps is shown by the near ablation of ovarian tumor cell invasion and mesothelial clearance after initial spheroid adhesion in the Rac1 KD models. Myosin-generated forces are necessary for ovarian cancer spheroids to clear the mesothelial layer, and myotonic dystrophy kinase-related Cdc42-binding kinases (MRCKs) are known Rac1 effector proteins (Unbekandt and Olson, 2014) further supporting the importance of activated Rac1 signaling in ovarian cancer metastatic dissemination. The in vitro findings are consistent with in vivo data showing decreased early omental implantation and tumor expansion in R-ketorolac–treated mice (Grimes et al., 2021) and greatly attenuated tumor growth in Rac1 KD xenograft models shown in the present study. The cumulative evidence based on Rac1 inhibition or manipulation of Rac1 expression levels supports the conclusion that Rac1 expression and activity are key mediators of ovarian cancer metastasis.
Because ovarian cancer has long been considered a peritoneal disease, in vivo animal studies have primarily focused on determining the mechanisms of peritoneal metastases. Hence, metastatic spread of ovarian cancer outside the peritoneal cavity is an underexplored area of research. A parabiosis study first increased awareness of the importance of hematogenous ovarian tumor dissemination (Pradeep et al., 2014). Documentation of distant metastases present at diagnosis (Gockley et al., 2017; Deng et al., 2018; Cao and Yang, 2020; Gardner et al., 2020; Wang et al., 2021) and worse patient outcomes (Gardner et al., 2020) further highlighted the importance of identifying drivers of distant metastases and provided a foundation for our study. Using agent-based modeling approaches, we provide novel insights into the role of Rac1 expression and activity on extraperitoneal dissemination. Simulations were conducted using models testing single-cell behaviors as well as multicellular spheroid behaviors. Varying probabilities of 1) cell adhesion, 2) cell death due to lack of adhesion or crowding, and 3) intravasation and extravasation based on experimental parameters derived from measurements of Rac1 OE or KD cells document Rac1 functional importance in multiple steps of the metastatic cascade. Simulations confirmed Rac1 as a key factor in 1) initial peritoneal adhesion of ascitic tumor cells shed from primary tumors, 2) mesothelial clearance, and 3) omental niche colonization. In the single-cell model simulations, Rac1 OE cells, as compared with Rac1 KO/KD, were found to enhance extraperitoneal dissemination/metastasis based on larger numbers of cells reaching the distant site and reaching the distant site at a faster rate. In the spheroid model simulations, the number of cells did not differ appreciably between Rac1 OE versus KO/KD, yet the Rac1 OE cells spread more quickly to the distant site. Notably, Rac1 OE is also predicted to increase both the rates of tumor cell dissemination and numbers of tumor cells colonizing distant metastatic sites through dissemination via circulation. Importantly, the model predicted a decrease in tumor cells upon Rac1 KD. The predictions by the model were experimentally replicated by measuring lung metastasis ± pharmacologic inhibition of Rac1 activity. This is the first experimental evidence that Rac1 inhibition may reduce extraperitoneal disease in ovarian cancer. Independent studies of distant metastasis of ovarian cancer cells (mixed 1:1 with mesenchymal stem cells) from the ovarian bursa to the lung were recently published (Atiya et al., 2021). Abdominal metastases formed 14 d postinjection, and 60–80% of animals had lung and parenchymal liver metastasis at death 30 d postinjection. The composite data highlight the importance of analyzing mechanisms of distant metastasis in ovarian cancer and demonstrate that Rac1 expression and activity are necessary for multiple steps in the metastatic cascade. Further study of the tumor microenvironment, including delineation of cells and factors that may drive Rac1 activation in support of metastasis, is considered an important next step for further elucidating the mechanisms of ovarian cancer metastasis.
Some key avenues that warrant further exploration are suggested by recent live animal imaging analyses of metastatic breast and ovarian cancer. The reported preferential metastasis of ovarian cancer cells to the highly vascularized omentum warrants further cellular and molecular analyses of this specialized lymphoid tissue (termed “milky spots” or fat-associated lymphoid clusters) (Krist et al., 1998; Nowak and Klink, 2020). A unique subset of CD163+ Tim4+ resident omental macrophages is found to be enriched in the omentum and documented to promote metastatic spread of ovarian cancer cells based on genetic and pharmacologic inhibition studies (Etzerodt et al., 2020). An obvious next step would be to test the potential role(s) of these highly specialized macrophages in promoting Rac1 activation in the tumor cells leading to increased intravasation. Furthermore, CD163+ Tim4+ macrophages cause ID8-positive mouse ovarian cancer cells to undergo epithelial-to-mesenchymal transition by reprogramming actin cytoskeletal and adhesion pathways. The described functional remodeling is similar to what we have observed with Rac1 OE/activation. Intriguingly, the presence of CD163+ Tim4+ macrophages caused spheroid compaction much like that caused by Rac1OE (see Figure 4). Such Rac1-driven clustering of cells in circulation may promote the increased cell survival under sheer stress experienced in the bloodstream during metastatic dissemination as suggested by the OCMetSim-Spheroids model and supported by in vitro studies of invasive breast cancer cells (Floerchinger et al., 2021).
Live animal imaging of breast cancer tumor cells has also evidenced tumor cell interactions with macrophages to activate specialized sites of intravasation, termed tumor microenvironment of metastasis (TMEM) doorways (Sharma et al., 2021). Model simulations of tumor cell crowding showed Rac1 OE cells to be resistant to the effects of cell crowding while higher rates of death were observed for Rac1 CRISPR-Cas9 KD cells. These findings are interesting in view of recent live animal imaging studies of the invasive MMTV-PyMT breast cancer model showing that Rac1 activity (measured using a Rac1 fluorescence resonance energy transfer sensor) is spatially regulated at distinct perivascular sites within locally invasive tumors (Floerchinger et al., 2021). Speed of extravasation and enhanced survival of disseminated breast tumor cells at distant niche sites also appear to be forecast by factors in the tumor environment (Borriello et al., 2022). Thus, the outcomes of our modeling and experimental data are well aligned with the published literature and suggest that tumor–niche cell interactions may serve to locally activate Rac1 to permit tumor cell intravasation through specialized rate-limiting sites like the described “TMEM-like doorways” in breast cancer metastasis. Facilitated exit from circulation and survival at distant sites may similarly depend on tumor–niche cell interactions. Further delineation of the molecular factors involved in tumor–niche cell interaction can also identify new nodes for therapeutic intervention to block metastasis.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell culture
SKOV3ip cells were obtained under an material transfer agreement (MTA) between the Hudson group and Anil Sood at MD Anderson, based on their mesenchymal phenotype, adaptation for growth in both cell culture and animal models, and aggressive metastatic growth. While some lines of SKOV3 cells are reported to be p53 wild type (Domcke et al., 2013), the SKOV3ip line was confirmed in our hands to be p53 null as is the case for >90% of human ovarian tumors (Supplemental Figure S8). OVCAR3 and OVCAR8 cells were obtained under an MTA from the National Cancer Institute’s (NCI’s) Biological Testing Branch of the Cancer Therapeutics Program, DCTD Tumor (and Cell) Repository. LP-9 cells (originally derived from the ascites fluid of a 26-yr-old female with ovarian cancer), G07086, were obtained from Coriell Institute for Medical Research under an authorized Assurance Agreement and are part of the NIA Aging Cell Culture Repository.
SKOV3ip, OVCAR3, and OVCAR8 ovarian cancer cells were cultured in RPMI 1640 (Corning) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. G418 was used to generate stable cell lines that overexpressed Rac1. Stable cell lines were made using Lipofectamine transfections. Plasmids were obtained from AddGene and Clontech: GFP-Rac1 WT-12980 and pEGFP-C1, C2, C3. After generation of stable cell lines, cells were sorted to collect GFP and GFP-Rac1 OE cells. To perform in vivo imaging, cells were transduced with luciferase reporter RediFect Red-FLuc-Puro (Perkin Elmer). Luciferase-transduced cells were selected via puromycin selection. Short tandem repeat (STR) profiling was performed to validate each cell line.
Rac1 CRISPR-Cas9 KD
SKOV3ip and OVCAR3 ovarian cancer parental cells were used to generate the Rac1 CRISPR-Cas9 KD cells. Guide RNAs specifically targeting exons 3 and 4 of Rac1 were used. CRISPOR (Telfor Infrastructure) was used to design and evaluate guide sequences for Rac1-CRISPR-Cas9 targeting. Guide RNAs were chosen based on CRISPOR analysis of off-target effects, predicted cleavage efficiency, and likelihood of out-of-frame deletions or frameshift mutations. The chosen guide RNAs were first tested in vitro for cleavage efficiency.
Rac1 KD was achieved using the Alt-R CRISPR-Cas9 electroporation system (IDT). crRNA:tracrRNA duplexes were formed. Ribonucleoprotein (RNP) complexes were then made with the Alt-R Cas9 enzyme (Alt-R® S.p. HiFi Cas9 Nuclease V3, IDT). The RNP complexes were simultaneously electroporated into the ovarian cancer cells using the Neon Transfection System (Invitrogen). As a negative control, cells were electroporated with the Cas9 enzyme without guide RNA. After electroporation, cells were seeded in six-well plates. Protein lysates were collected 24 h and 1 wk after electroporation to measure Rac1 protein levels via Western blot. Single-cell cloning was performed to isolate clones with the lowest Rac1 expression. Single cells were seeded in 96-well plates. Rac1 expression was measured via Western blot, and sequencing was performed to verify exon 3 and 4 targeting by CRISPR-Cas9.
Sequencing
DNA was extracted from SKOV3ip and OVCAR3 CRISPR clones, CRISPR-Cas9 control, and parental cells using a Quick-DNA MicroPrep Kit (Zymo Research Cat #D3020). DNA quality and concentration were measured on a Nanodrop. PCRs to amplified exon 3 and exon 4 products were performed using primers that flanked each exon, separately. DNA was cleaned using a Monarch PCR and DNA cleanup kit (New England BioLabs #T1030S). PCR products and primers were sent to GeneWiz Azenta Life Sciences for sequencing. Nine total samples from CRISPR-Cas9 control cells, CRISPR KD clones, and parental cell lines were analyzed. Owing to nonspecific priming in the DNA sequencing analyses, SKOV3ip clones were further analyzed for heterozygosity using the Original TA cloning kit (Invitrogen #45-0046). PCR products derived from exon 3 forward and exon 4 reverse primer sets were ligated into the pCR2.1 vector, transformed into competent Escherichia coli (DH5α from Invitrogen #18265-017), plated on Luria-Bertani medium (LB) + 100 μg/ml ampicillin (Sigma #A9518) + 40 μg/ml X-Gal (Invitrogen #15520-034) + 100 mM isopropyl β-d-1-thiogalactopyranoside (Invitrogen #15529-019) agar plates for blue/white screening selection, and single white colonies were grown in liquid LB + 100 μg/ml ampicillin (Sigma #A9518) media overnight. Plasmids were isolated from the overnight cultures using the PureLink HQ Mini Plasmid Purification kit (Invitrogen #K210001) and digested with EcoRI to release the inserted products. A ligated control insert DNA served as a negative control, and linearized pCR2.1 served as a positive control for digestion. The digestion reactions were run on agarose gels to identify insert sizes and used to select plasmids for sequence analyses. Observed fragment sizes for clone 23 were 400 base pairs, for clone 26 were uncut or had a faint 500-base-pair band, for clone 27 were 200–300 base pairs. Sequencing was performed by Genewiz Azenta Life Sciences.
The sequencing results were analyzed using data from the National Center for Biotechnology Information (NCBI) protein and nucleotide databases. Sequencing results for each cell line and exon were aligned against the Rac1 gene using the Basic Local Alignment Search Tool (BLAST; U. S. National Library of Medicine NCBI) and Geneious Prime (v2022.1.1) software to identify changes in nucleotide sequence. The nucleotides sequenced were translated into protein coding sequences to identify any frame shifts or premature stop codons.
Matrigel invasion assay
Matrigel-coated Transwell inserts with 8.0 μm polyethylene terephthalate (PET) membrane (Corning BioCoat Matrigel) were used to assess ovarian cancer cell invasion in vitro. Matrigel inserts were rehydrated in a tissue culture incubator for 2 h with warm (37°C) bicarbonate-based culture medium added to the top and bottom chambers. SKOV3ip ovarian cancer cells with Rac1 OE or KD were resuspended in serum-free RMPI media. Cells (2 × 104) were added to the upper chamber of the Matrigel-coated Transwell insert. RPMI medium (500 µl) containing 20% FBS was added to the lower chamber. After addition of cells, Transwell inserts were incubated at 37°C and ovarian cancer cell invasion was assessed. After incubation for 48 h, the chambers were removed and washed with phosphate-buffered saline (PBS). The upper chamber was cleaned using a cotton Q-tip to remove all remaining nonmigrated cells and Matrigel. The chambers were fixed with 3% paraformaldehyde for 1 h. Cells were stained with 1 mM Hoechst 33342 in PBS (Kodak, 106 1399) to count nuclei. Filters were mounted on slides and imaged using a Zeiss AxioObserver, and the transmigrated cells were counted. Quantification is based on three to four independent experiments with three technical Matrigel-coated Transwell replicates per cell line. Five random fields of view were imaged and counted on each insert.
Western blotting
Total protein was extracted from SKOV3ip cells using a Tris-Triton buffer recipe: 10 mM Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 10% glycerol, 0.1% SDS, 0.5% deoxycholate plus phosphatase inhibitors. Protein quantification was performed using the Pierce BCA Protein Assay Kit. Lysate samples (30 μg/well) were resolved by 15% SDS–PAGE, and proteins were transferred to nitrocellulose membranes. The membranes were blocked with 5% milk in PBS with Tween-20 for 1 h at room temperature. Membranes were incubated with the following antibodies overnight at 4°C: Rac1-BD 610650, α/β tubulin-Cell Signaling #2148, GAPDH-Sigma G8795. Proteins of interest were visualized with SuperSignal West Pico Chemiluminescent Substrate (Pierce) and imaged using the ChemiDoc XRS Imager (Bio-Rad). Densitometry analysis was performed using Image Lab software (Bio-Rad).
Mesothelial cell clearance assay
For spheroid formation, SKOV3ip cell lines—including parental, GFP, High Rac1 OE, Rac1 CRISPR-Cas9 control, and Rac1 CRISPR-Cas9 KD clones (clones 23 and 27)—were plated at 100 cells/well in 96-well ultra–low attachment, round bottom, Costar plates, and incubated at 37°C, with 5% CO2 for 4 d. Primary LP-9 human mesothelial cells were stained in suspension for 25 min at 37°C with 2 μM CellTracker Red CMTPX (Invitrogen C34552) in Medium 199:Ham’s F-12 Nutrient Mix (M199:F12), with no serum (supplemented with 2 mM l-glutamine, 10 ng/ml epidermal growth factor (EGF), 0.4 μg/ml hydrocortisone, and 100 μg/ml Pen Strep). Cells were then stained with 1 μM Hoechst 33342 (Kodak, 106 1399) at room temperature for 7 min. Cells were washed three times with M199:F12 supplemented with serum. Stained LP-9 cells were plated at 128,000 cells/well in a 24-well Costar plate and incubated overnight at 37°C and 5% CO2. The next day, the cells were washed three times in M199:F12 with serum. Spheroids were gently transferred to the LP-9 wells using a micropipette. Plates were incubated at 37°C with 5% CO2. Spheroids were imaged at 1 h postaddition with a Zeiss inverted microscope or Incucyte S3 live cell imaging and analysis system, using either a 4× or a 10× objective with bright field and/or a FITC filter. The mesothelial clearance area was imaged at 7 h and 24 and 25 h after spheroid addition with a TRITC filter. Images were analyzed using Fiji software. Clearance areas were normalized to compensate for differences in spheroid size to evaluate differences in cell motility and invasive forces resulting from varying Rac1 expression. To normalize, clearance areas at 7 and 25 h were divided by the spheroid size at 1 h postaddition. This analysis approach was described in Davidowitz et al. (2012) and has been used in other research articles (Iwanicki et al., 2011; Davidowitz et al., 2014; Sheets et al., 2016; Huang et al., 2020; Naffar-Abu Amara et al., 2020; Zhang et al., 2021b). To confirm statistical significance with a stricter approach, we compared normalized clearance areas to relative change-of-clearance areas. To calculate relative change, the initial spheroid area was subtracted from the clearance area at 24 or 25 h. Then this result was divided by the initial spheroid area. Both approaches yielded statistically significant differences validating the results obtained with normalized clearance areas (Figure 5 and Supplemental Figure S10). Prism 9 software (GraphPad) was used to execute statistical analyses.
ECIS measurements
Cell–cell adhesion and cell–substrate adhesion were measured using ECIS. SKOV3ip cells with Rac1 OE or KD were plated at confluence on 96-well electrode arrays, 96W10idf (Applied BioPhysics). Electrodes were stabilized with l-cysteine. Treatment with l-cysteine enhances experimental repeatability between the ECIS wells (Applied BioPhysics). The cysteine displaces unwanted molecules that have been absorbed onto the gold electrodes and creates a hydrophilic substrate that helps with cell attachment and spreading over the electrodes (Applied BioPhysics). l-cysteine (10 mM) in water (Applied BioPhysics) was added to each well and incubated for 30 min at room temperature. l-Cysteine was removed, the wells were rinsed twice with water, and then the electrodes were washed with RPMI 1640 media without FBS. Media was removed and replaced with SKOV3ip cells in prewarmed complete RPMI growth media as detailed above. Electrode arrays were placed in an ECIS Zθ instrument (Applied BioPhysics) and resistance and capacitance were measured at 4000 and 64,000 Hz, respectively, every 10 min over 72 h. Each experiment included four replicates, and three independent biological replicates were completed. All values are expressed as mean ± SD.
In vivo tumor engraftment and tumor growth measurements
All animal experiments were performed under protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the University of New Mexico Health Science Center (PI Gillette #19-200925-HSC). Female nonobese diabetic/severe combined immunodeficiency gamma (NSG) mice between the ages of 6 and 9 wk were used to measure omental engraftment and tumor burden. Luciferase-transduced ovarian cancer cells (4 × 104) were intraperitoneally injected into NSG mice. Intraperitoneal tumor burden was measured over 1–2 wk and quantified using IVIS bioluminescence imaging.
NetLogo modeling
The NetLogo platform (Northwestern University) (Wilensky, 1999) was used to build two computational models (OCMetSim-Single Cells and OCMetSim-Spheroids) to make predictions about ovarian cancer cell metastasis to distant niche sites. OCMetSim programs were developed to test the impact of Rac1 expression on adhesion and invasion of ovarian cancer cells into circulation and to metastasize to distant niche sites. Experimental data were used to set the parameters for cellular invasion and adhesion. The OCMetSim programs and user instructions can be accessed on GitHub (https://github.com/MelanieRivera44/Ovarian-Cancer-Metastasis-Simulation-OCMetSim.git).
A. Purpose of the models.
Ovarian cancer cells detach from the primary tumor and are able to adhere and invade to a distant tissue site. The omentum is a common site of initial ovarian cancer metastasis. Ovarian cancer cells in the peritoneal ascites fluid can readily adhere to the omentum and from here invade into circulation, where they can travel to distant niche sites and promote cancer metastasis. The goal of these models is to make predictions about how Rac1 expression influences ovarian cancer cell behavior and metastasis to a distant metastatic niche, via entry into circulation from the omentum. Experimental data from in vitro adhesion and invasion experiments are used to set parameters for these cellular behaviors.
B. NetLogo simulation space setup.
In the simulations, the mobile agents represent ovarian cancer single cells or spheroids. In OCMetSim-Single Cells and OCMetSim-Spheroids, simulation spaces are set up with seven differently colored patches (black, green, blue, red, cyan, magenta, and brown representing the peritoneal cavity, cell adhesion to a blood vessel, cell invasion into a blood vessel, circulating blood, cell adhesion to a distant tissue site, cell extravasation into a distant tissue site, and a distant metastatic niche site, respectively). The ovarian cancer cells move through these compartments based on a set of parameters that dictate their behavior based on Rac1 expression. Initially, cancer cells are modeled as independent cells. A second set of experiments considered how the invasion properties of spheroids would change the timing and numbers of metastasized cells that reach a distant metastatic niche.
C. Movement of ovarian cancer cells in the peritoneum.
Ovarian cancer cells start in the peritoneal compartment (black patches) at a random x-coordinate and random y-coordinate with a random heading setting. When the cells move, they do so randomly. These behaviors mimic their movement within the peritoneal fluid, floating around in random diffusion. If a cell is located at the maximum height, the cell heading will be set to 180° and the random walk will continue. The cells or spheroids check whether the patch ahead is occupied, and if this statement is false, the heading will reset to a new random position to prevent two cells or spheroids from occupying the same patch. Cells will move around the peritoneum and eventually land next to the blood vessel (red patches). When the cells are in a patch next to the blood vessel, procedure 1 or procedure 2 becomes possible.
D. Procedure 1: Cell Adhesion and Invasion of Ovarian Cancer Cells.
The models assume that the cancer cells or spheroids have a probability of adhering to the omentum and then have a rate of invasion into the tissue where they can then enter the circulation and adhere and invade to a distant tissue site. Once cells are aligned near or adjacent to the red blood patches, they have a probability of adhering to the blood vessel. The parameter for probability of adhesion is determined from the experimental ECIS data described in Figure 2. The measurements for capacitance at 8 h after seeding were used to set the parameters for Rac1 OE and KD cell adhesion probabilities. The probability was calculated compared with control cells. Rac1 OE cells have a 1.9- to 2.2-fold increase in adhesion compared with Rac1 CRISPR-Cas9 KD clones (clones 26 and 23), respectively. The model assumes that if a cell does not adhere to the blood vessel, the cell has some probability of dying because adherent cells undergo anoikis, programmed cell death, when they are not anchored. Once the cancer cells have adhered to the blood vessel, the procedure for cell invasion becomes active.
D.1. Procedure 2: Adhesion and Invasion with Crowding and Death from Crowding.
This procedure is used to investigate the effects of crowding of cells along the blood vessel. If there is a cell or spheroid that adheres to the vessel in front of another cell, the cell waiting to adhere will not adhere until the patch is empty and the current cell has invaded into the blood vessel and moved forward. This additional procedure includes the probability of cell death if the cell waiting to adhere takes too long.
E. Invasion of ovarian cancer cells into circulation.
For setting the cell invasion parameters, the Matrigel-coated Transwell experiments were used. We assumed a Poisson distribution for the rate of invasion and assumed that the stochastic invasion events were independent of one another. Using the Matrigel invasion data, we validated that this assumption made sense. The invasion data involve seeding 20,000 cells into the chambers, and the number of invaded cells is quantified after 48 h. The average number of invaded cells after 48 h for the SKOV3ip Rac1-Low OE cells is 26.11 cells. The average number of invaded cells after 48 h for the SKOV3ip Rac1-High OE cells is 56.32 cells. We calculated the average number of invaded cells in 1 h. Then we divided the number of cells invaded in 1 h by the total number of cells added (20,000 cells). This gives us the probability that a cell will invade in 1 h. This gives the Poisson distribution of probabilities that a cell will invade. We used these data to run simulations in NetLogo to validate our assumptions. One hundred simulations were run to test the probability of invasion of SKOV3ip Rac1-Low OE and Rac1-High OE cells. The data are represented in Figure 6, A and B. Our simulations resulted in an average of 25.39 invaded SKOV3ip Rac1-Low OE cells and 56.19 Rac1-High OE cells, similar to the experimental data with the Matrigel Transwell inserts. Running the simulations in NetLogo validated the parameters that we set using the experimental data and a Poisson distribution.
Additionally, we assumed that invasion into circulation was a one-step process that aggregates the invasion across the mesothelial cell layer, ECM, and endothelial cells all as one step.
F. Movement of ovarian cancer cells and spheroids in circulation.
Cells that have invaded into circulation move toward the brown patches (a distant metastatic niche compartment). We assumed that cells move in a more directed movement toward a distant metastatic niche tissue. In the blood compartment, we are assuming that a percentage of cells will die in circulation. Biologically, cancer cells in circulation will die due to anoikis, can be cleared by immune cells, or can end up in different niche sites. Probability of cell death within this compartment can be adjusted to make predictions about how cell death might influence cells traveling to distant tissue sites. If the cell survives, the cell may be able to adhere to a distant metastatic niche and the next step becomes active and dictates the commands for cell adhesion to a distant metastatic niche.
G. Adhesion of ovarian cancer cells to a distant metastatic niche site.
We assumed that the parameters for adhesion to the distant metastatic site were the same as those for adhesion to the blood vessel and change based on Rac1 expression. We also assumed that crowding of cells in this adhesion step will not affect the ability of cells to invade into the tissue because the number of cells that reach this step from the initial number in the peritoneum will be small at any given attachment site in the circulatory system. As in the above adhesion parameter, we assumed that cells would die if they did not adhere. If the cell adheres, the procedure for invasion into a distant metastatic site becomes active.
H. Invasion of ovarian cancer cells in a distant metastatic niche.
In this procedure, we are assuming a Poisson distribution for parameters of cell invasion. Cells will invade based on the parameters calculated from the experimental Matrigel invasion or mesothelial clearance experiments with SKOV3ip Rac1 OE or KD cells.
I. Parameters, assumptions, and variables
Parameters
Rate of invasion for single cells is based on Rac1 expression using Matrigel invasion data
Probability of adhesion for single cells is based on Rac1 expression using ECIS data
Adhesion and invasion probabilities for spheroids is based on Rac1 expression using mesothelial cell clearance experiments
Assumptions
Ovarian cancer cells move as single cells in experiments 1 and 2, and they are modified based on properties of spheroids in experiments 1a and 2a
The rate of invasion is a Poisson distribution
Ovarian cancer cells will die if they do not adhere to the tissue/blood vessel
The rate and probabilities for intravasation and extravasation into and out of the blood are the same
The rate of blood flow does not matter because cells will be attracted to the tissue and cells are given a probability of dying in the blood
There is no change in cell proliferation with Rac1 OE or KD
Ovarian cancer cells in the peritoneal compartment move at random
Variables
Initial number of tumor cells or spheroids
Probability of cell death in the blood
Probability of dying for crowding on the tissue or blood vessel
Statistics
Data were analyzed using GraphPad Prism 7.03 and 9 software. Experimental data are presented as the means ± SD or SE from three to four independent experiments. Two-tailed Student’s t test was used to compare differences between two groups. One-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test for multiple comparison was used for comparison of three or more groups. Two-way ANOVA followed by Sidak’s post-hoc test for multiple comparison was used to compare groups with two independent variables, that is, cell lines with differential expression of Rac1 and time. A p value < 0.05 was considered to be statistically significant. Computational modeling data are presented as the means ± SD for 30 simulation runs per group. Two-way ANOVA followed by Tukey’s post-hoc test for multiple comparison across columns and rows was used to compare differences between groups in OCMetSim simulations. A p value < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
This work was supported by an NCI fellowship to M.R. (F31CA232398), funding to M.R. and A.W.-N. through a National Institute of General Medical Sciences (NIGMS)-funded Spatiotemporal Modeling Center (P50GM085273), a NIGMS fellowship to L.T.-J. (K12 GM088021), NCATS R03 TR003324 to A.W.-N. and L.G.H., National Science Foundation (NSF) #2030037 (and a James S. McDonnell Foundation Complex Systems Scholar Award to M.E.M. This research was also partially supported by a University of New Mexico (UNM) Comprehensive Cancer Center Support Grant (NCI P30CA118100) and the following shared resources: Human Tissue Repository, Fluorescence Microscopy, Flow Cytometry, Biostatistics and Animal Models Shared Resources. Use of the Research Collaboratory for Structural Bioinformatics (RCSB PDB) is made possible through funding by the NSF (DBI-1832184) and the NIGMS (R01 GM133198). We thank Irina Lagutina for assistance with lentiviral transductions of ovarian cancer cells with luciferase and Jennifer Gillette and Mara Steinkamp for scientific advisement as members of M.R.’s thesis committee. M.R. thanks Guy Herbert in the UNM College of Pharmacy for assistance with ECIS experiments, Vanessa Surjadidjaja and Jannatul Ferdous, members of the Moses lab, for help and advice with NetLogo coding and Major Nathan Haluska for helpful modeling discussions.
Abbreviations used:
- ANOVA
analysis of variance
- Cas9
CRISPR-associated protein 9
- Cdc42
cell division protein 42
- CTCs
circulating tumor cells
- DCTD
Division of Cancer Treatment and Diagnosis, Tumor Repository
- DepMAP
The Cancer Dependency MAP
- CERES
computational corrections
- CRISPR
clustered regularly interspaced short palindromic repeats
- crRNA:tracrRNA
crispr RNA: trans-activating CRISPR RNA
- ECIS
electric cell-substrate impedance sensing
- ECM
extracellular matrix
- FBS
fetal bovine serum
- GAP
GTPase-activating protein
- GDP
guanosine 5′-diphosphate
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- GTP
guanosine 5′-triphosphate
- HRP
horseradish peroxidase
- IACUC
Institutional Animal Care and Use Committee
- IP
intraperitoneal
- IV
intravenous
- IVIS
in vivo imaging system
- KD
knockdown
- MCA
mesothelial clearance area
- NCBI
National Center for Biotechnology Information
- NCI
National Cancer Institute
- NSG
nonobese diabetic/severe combined immunodeficiency gamma
- OCMetSim
ovarian cancer metastasis simulation
- OE
overexpressing
- PAK
p21-activated kinase
- PBS
phosphate-buffered saline
- PET
polyethylene terephthalate
- P-REX1
phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 1
- q-PCR
quantitative PCR
- Rac1
Ras-related C3 botulinum
- Ras
rat sarcoma
- RhoA
Ras homologue family member A
- RNP
ribonucleoprotein
- RPMI
Roswell Park Memorial Institute
- RT-PCR
reverse transcription-PCR
- SEER
Surveillance, Epidemiology, and End Results
- TA
thymine and adenine
- TCGA
The Cancer Genome Atlas
- TRITC
tetramethyl rhodamine iso-thiocyanate.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-11-0540) on September 6, 2022.
REFERENCES
- Al Habyan S, Kalos C, Szymborski J, McCaffrey L (2018). Multicellular detachment generates metastatic spheroids during intra-abdominal dissemination in epithelial ovarian cancer. Oncogene 37, 5127–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari AM, Ahmed AK, Matsangos AE, Lay F, Born LJ, Marti G, Harmon JW, Sun Z (2016). Cellular GFP toxicity and immunogenicity: potential confounders in in vivo cell tracking experiments. Stem Cell Rev Rep 12, 553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applied BioPhysics (2021). Electric cell-substrate impedance sensing (ECIS), https://www.biophysics.com/.
- Atiya HI, Orellana TJ, Wield A, Frisbie L, Coffman LG (2021). An orthotopic mouse model of ovarian cancer using human stroma to promote metastasis. J Vis Exp 2021, doi: 10.3791/62382. [DOI] [PubMed] [Google Scholar]
- Bauer AL, Jackson TL, Jiang Y (2007). A cell-based model exhibiting branching and anastomosis during tumor-induced angiogenesis. Biophys J 92, 3105–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bid HK, Roberts RD, Manchanda PK, Houghton PJ (2013). RAC1: an emerging therapeutic option for targeting cancer angiogenesis and metastasis. Mol Cancer Ther 12, 1925–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birbeck MS, Wheatley DN (1965). An electron microscopic study of the invasion of ascites tumor cells into the abdominal wall. Cancer Res 25, 490–497. [PubMed] [Google Scholar]
- Bonabeau E (2002). Agent-based modeling: methods and techniques for simulating human systems. Proc Natl Acad Sci USA 99(Suppl 3), 7280–7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borriello L, Coste A, Traub B, Sharma VP, Karagiannis GS, Lin Y, Wang Y, Ye X, Duran CL, Chen X, et al. (2022). Primary tumor associated macrophages activate programs of invasion and dormancy in disseminating tumor cells. Nat Commun 13, 626, 10.1038/s41467-022-28076-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C, Yang X (2020). The prevalence, associated factors for lung metastases development and prognosis in ovarian serous cancer based on SEER database. Technol Cancer Res Treat 19, 1533033820983801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sprouffske K, Huang Q, Maley CC (2011). Solving the puzzle of metastasis: the evolution of cell migration in neoplasms. PLoS One 6, e17933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidowitz RA, Selfors LM, Iwanicki MP, Elias KM, Karst A, Piao H, Ince TA, Drage MG, Dering J, Konecny GE (2012). In vitro mesothelial clearance assay that models the early steps of ovarian cancer metastasis. J Vis Exp 2012, doi: 10.3791/3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidowitz RA, Selfors LM, Iwanicki MP, Elias KM, Karst A, Piao H, Ince TA, Drage MG, Dering J, Konecny GE, et al. (2014). Mesenchymal gene program-expressing ovarian cancer spheroids exhibit enhanced mesothelial clearance. J Clin Invest , 124, 2611–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De P, Aske JC, Dey N (2019). RAC1 takes the lead in solid tumors. Cells 8, doi: 10.3390/cells8050382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng K, Yang C, Tan Q, Song W, Lu M, Zhao W, Lou G, Li Z, Li K, Hou Y (2018). Sites of distant metastases and overall survival in ovarian cancer: a study of 1481 patients. Gynecol Oncol 150, 460–465. [DOI] [PubMed] [Google Scholar]
- Domcke S, Sinha R, Levine DA, Sander C, Schultz N (2013). Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun 4, 2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P, Tang X, Wang J, Zhu B, Li Z (2022). miR-653-5p suppresses the viability and migration of fibroblast-like synoviocytes by targeting FGF2 and inactivation of the Wnt/beta-catenin pathway. J Orthop Surg Res 17, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzerodt A, Moulin M, Doktor TK, Delfini M, Mossadegh-Keller N, Bajenoff M, Sieweke MH, Moestrup SK, Auphan-Anezin N, Lawrence T (2020). Tissue-resident macrophages in omentum promote metastatic spread of ovarian cancer. J Exp Med 217, e20191869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Chen H, Zhu JY, Wang W, Teng Y, Ding HF, Jing Q, Su SB, Huang S (2017). Epithelial-mesenchymal transition of ovarian cancer cells is sustained by Rac1 through simultaneous activation of MEK1/2 and Src signaling pathways. Oncogene 36, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floerchinger A, Murphy KJ, Latham SL, Warren SC, McCulloch AT, Lee YK, Stoehr J, Melenec P, Guaman CS, Metcalf XL, et al. (2021). Optimizing metastatic-cascade-dependent Rac1 targeting in breast cancer: guidance using optical window intravital FRET imaging. Cell Rep 36, 109689. [DOI] [PubMed] [Google Scholar]
- Gardner AB, Charo LM, Mann AK, Kapp DS, Eskander RN, Chan JK (2020). Ovarian, uterine, and cervical cancer patients with distant metastases at diagnosis: most common locations and outcomes. Clin Exp Metastasis 37, 107–113. [DOI] [PubMed] [Google Scholar]
- Gasparri ML, Savone D, Besharat RA, Farooqi AA, Bellati F, Ruscito I, Panici PB, Papadia A (2016). Circulating tumor cells as trigger to hematogenous spreads and potential biomarkers to predict the prognosis in ovarian cancer. Tumour Biol 37, 71–75. [DOI] [PubMed] [Google Scholar]
- Giaever I, Keese CR (1993). A morphological biosensor for mammalian cells. Nature 366, 591–592. [DOI] [PubMed] [Google Scholar]
- Gockley A, Melamed A, Bregar AJ, Clemmer JT, Birrer M, Schorge JO, Del Carmen MG, Rauh-Hain JA (2017). Outcomes of women with high-grade and low-grade advanced-stage serous epithelial ovarian cancer. Obstet Gynecol 129, 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes MM, Kenney SR, Dominguez DR, Brayer KJ, Guo Y, Wandinger-Ness A, Hudson LG (2021). The R-enantiomer of ketorolac reduces ovarian cancer tumor burden in vivo. BMC Cancer 21, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Kenney SR, Cook L, Adams SF, Rutledge T, Romero E, Oprea TI, Sklar LA, Bedrick E, Wiggins CL, et al. (2015a). A novel pharmacologic activity of ketorolac for therapeutic benefit in ovarian cancer patients. Clin Cancer Res 21, 5064–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Kenney SR, Muller CY, Adams S, Rutledge T, Romero E, Murray-Krezan C, Prekeris R, Sklar LA, Hudson LG, Wandinger-Ness A (2015b). R-ketorolac targets Cdc42 and Rac1 and alters ovarian cancer cell behaviors critical for invasion and metastasis. Mol Cancer Ther 14, 2215–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley J, Tomar S, Pulliam N, Xiong S, Perkins SM, Karpf AR, Mitra S, Nephew KP, Mitra AK (2016). Functional characterization of a panel of high-grade serous ovarian cancer cell lines as representative experimental models of the disease. Oncotarget 7, 32810–32820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshberg M, Stockley RW, Dodson G, Webb MR (1997). The crystal structure of human rac1, a member of the rho-family complexed with a GTP analogue. Nat Struct Biol 4, 147–152. [DOI] [PubMed] [Google Scholar]
- Huang YLLiang CYRitz DCoelho RSeptiadi DEstermann MCumin CRimmer NSchotzau ANunez Lopez M, et al. (2020). Collagen-rich omentum is a premetastatic niche for integrin alpha2-mediated peritoneal metastasis. eLife , 9, e59442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson LG, Gillette JM, Kang H, Rivera MR, Wandinger-Ness A (2018). Ovarian tumor microenvironment signaling: convergence on the Rac1 GTPase. Cancers (Basel) 10, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanicki MP, Davidowitz RA, Ng MR, Besser A, Muranen T, Merritt M, Danuser G, Ince TA, Brugge JS (2011). Ovarian cancer spheroids use myosin-generated force to clear the mesothelium. Cancer Discov 1, 144–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Feng X, Shi M, Cai Q, Yu Y, Zhu Z, Zhang J (2015). Rac1 is correlated with aggressiveness and a potential therapeutic target for gastric cancer. Int J Oncol 46, 1343–1353. [DOI] [PubMed] [Google Scholar]
- Krist LF, Kerremans M, Broekhuis-Fluitsma DM, Eestermans IL, Meyer S, Beelen RH (1998). Milky spots in the greater omentum are predominant sites of local tumour cell proliferation and accumulation in the peritoneal cavity. Cancer Immunol Immunother 47, 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki L, Guntupalli SR (2020). Treatment of epithelial ovarian cancer. BMJ 371, m3773. [DOI] [PubMed] [Google Scholar]
- Leng R, Liao G, Wang H, Kuang J, Tang L (2015). Rac1 expression in epithelial ovarian cancer: effect on cell EMT and clinical outcome. Med Oncol 32, 329. [DOI] [PubMed] [Google Scholar]
- Lengyel E (2010). Ovarian cancer development and metastasis. Am J Pathol 177, 1053–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link CD, Fonte V, Hiester B, Yerg J, Ferguson J, Csontos S, Silverman MA, Stein GH (2006). Conversion of green fluorescent protein into a toxic, aggregation-prone protein by C-terminal addition of a short peptide. J Biol Chem 281, 1808–1816. [DOI] [PubMed] [Google Scholar]
- Liu H, Jan M, Chou C, Chen P, Ke N (1999). Is green fluorescent protein toxic to the living cells? Biochem Biophys Res Commun 260, 712–717. [DOI] [PubMed] [Google Scholar]
- Liu M, Silva-Sanchez A, Randall TD, Meza-Perez S (2021). Specialized immune responses in the peritoneal cavity and omentum. J Leukoc Biol 109, 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou S, Wang P, Yang J, Ma J, Liu C, Zhou M (2018). Prognostic and clinicopathological value of Rac1 in cancer survival: evidence from a meta-analysis. J Cancer 9, 2571–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucato CM, Halls ML, Ooms LM, Liu HJ, Mitchell CA, Whisstock JC, Ellisdon AM (2015). The phosphatidylinositol (3,4,5)-trisphosphate-dependent Rac exchanger 1.Ras-related C3 botulinum toxin substrate 1 (P-Rex1.Rac1) complex reveals the basis of Rac1 activation in breast cancer cells. J Biol Chem 290, 20827–20840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, et al. (2017). Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet 49, 1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza-Perez S, Randall TD (2017). Immunological functions of the omentum. Trends Immunol 38, 526–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naffar-Abu Amara S, Kuiken HJ, Selfors LM, Butler T, Leung ML, Leung CT, Kuhn EP, Kolarova T, Hage C, Ganesh K, et al. (2020). Transient commensal clonal interactions can drive tumor metastasis. Nat Commun 11, 5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Cancer Institute (2022). Cancer stat facts: ovarian cancer. SEER 12. [Google Scholar]
- Nowak M, Klink M (2020). The role of tumor-associated macrophages in the progression and chemoresistance of ovarian cancer. Cells 9, 1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermayr E, Castillo-Tong DC, Pils D, Speiser P, Braicu I, Van Gorp T, Mahner S, Sehouli J, Vergote I, Zeillinger R (2013). Molecular characterization of circulating tumor cells in patients with ovarian cancer improves their prognostic significance—a study of the OVCAD consortium. Gynecol Oncol 128, 15–21. [DOI] [PubMed] [Google Scholar]
- O’Neill AC, Somarouthu B, Tirumani SH, Braschi-Amirfarzan M, Van den Abbeele AD, Ramaiya NH, Shinagare AB (2017). Patterns and prognostic importance of hepatic involvement in patients with serous ovarian cancer: a single-institution experience with 244 patients. Radiology , 282, 160–170. [DOI] [PubMed] [Google Scholar]
- Oprea TI, Sklar LA, Agola JO, Guo Y, Silberberg M, Roxby J, Vestling A, Romero E, Surviladze Z, Murray-Krezan C, et al. (2015). Novel activities of select NSAID R-enantiomers against Rac1 and Cdc42 GTPases. PLoS One 10, e0142182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasapera AM, Plotnikov SV, Fischer RS, Case LB, Egelhoff TT, Waterman CM (2015). Rac1-dependent phosphorylation and focal adhesion recruitment of myosin IIA regulates migration and mechanosensing. Curr Biol 25, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradeep SKim SWWu SYNishimura MChaluvally-Raghavan PMiyake TPecot CVKim SJChoi HJBischoff FZ, et al. (2014). Hematogenous metastasis of ovarian cancer: rethinking mode of spread. Cancer Cell 26, 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ (2015). Rho GTPase signalling in cell migration. Curr Opin Cell Biol 36, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E, Marshall CJ (2002). RHO-GTPases and cancer. Nat Rev Cancer 2, 133–142. [DOI] [PubMed] [Google Scholar]
- Schnelzer A, Prechtel D, Knaus U, Dehne K, Gerhard M, Graeff H, Harbeck N, Schmitt M, Lengyel E (2000). Rac1 in human breast cancer: overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene 19, 3013–3020. [DOI] [PubMed] [Google Scholar]
- Sharma VP, Tang B, Wang Y, Duran CL, Karagiannis GS, Xue EA, Entenberg D, Borriello L, Coste A, Eddy RJ, et al. (2021). Live tumor imaging shows macrophage induction and TMEM-mediated enrichment of cancer stem cells during metastatic dissemination. Nat Commun 12, 7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets JN, Iwanicki M, Liu JF, Howitt BE, Hirsch MS, Gubbels JA, Drapkin R, Egland KA (2016). SUSD2 expression in high-grade serous ovarian cancer correlates with increased patient survival and defective mesothelial clearance. Oncogenesis 5, e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata K, Sakai H, Huang Q, Kamata H, Chiba Y, Misawa M, Ikebe R, Ikebe M (2015). Rac1 regulates myosin II phosphorylation through regulation of myosin light chain phosphatase. J Cell Physiol 230, 1352–1364. [DOI] [PubMed] [Google Scholar]
- Soheilypour M, Mofrad MRK (2018). Agent-based modeling in molecular systems biology. Bioessays 40, e1800020. [DOI] [PubMed] [Google Scholar]
- Szulcek R, Bogaard HJ, van Nieuw Amerongen GP (2014). Electric cell-substrate impedance sensing for the quantification of endothelial proliferation, barrier function, and motility. J Vis Exp 2014, doi: 10.3791/51300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan DS, Agarwal R, Kaye SB (2006). Mechanisms of transcoelomic metastasis in ovarian cancer. Lancet Oncol 7, 925–934. [DOI] [PubMed] [Google Scholar]
- Tan TZ, Miow QH, Huang RY, Wong MK, Ye J, Lau JA, Wu MC, Bin Abdul Hadi LH, Soong R, Choolani M, et al. (2013). Functional genomics identifies five distinct molecular subtypes with clinical relevance and pathways for growth control in epithelial ovarian cancer. EMBO Mol Med 5, 1051–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unbekandt M, Olson MF (2014). The actin-myosin regulatory MRCK kinases: regulation, biological functions and associations with human cancer. J Mol Med 92, 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Wang S, Ren W (2021). Development and validation of a nomogram to predict survival outcome among epithelial ovarian cancer patients with site-distant metastases: a population-based study. BMC Cancer 21, 609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, He L, Wu YI, Hahn KM, Montell DJ (2010). Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol 12, 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener J, Keese CR, Giaever I (2000). Electric cell-substrate impedance sensing (ECIS) as a noninvasive means to monitor the kinetics of cell spreading to artificial surfaces. Exp Cell Res 259, 158–166. [DOI] [PubMed] [Google Scholar]
- Wilensky U (1999). NetLogo . Evanston, IL::Center for Connected Learning and Computer-Based Modeling, Northwestern University, https://ccl.northwestern.edu/netlogo/. [Google Scholar]
- Winner KK, Steinkamp MP, Lee RJ, Swat M, Muller CY, Moses ME, Jiang Y, Wilson BS (2016). Spatial modeling of drug delivery routes for treatment of disseminated ovarian cancer. Cancer Res 76, 1320–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witz CA, Monotoya-Rodriguez IA, Schenken RS (1999). Whole explants of peritoneum and endometrium: a novel model of the early endometriosis lesion. Fertil Steril 71, 56–60. [DOI] [PubMed] [Google Scholar]
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM (2009). A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li S, Bao J, Ge N, Hong F, Qian L (2021a). Beta-elemene inhibits non-small cell lung cancer cell migration and invasion by inactivating the FAK-Src pathway. Exp Ther Med 22, 1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yang N, Kreeger PK, Notbohm J (2021b). Topological defects in the mesothelium suppress ovarian cancer cell clearance. APL Bioeng 5, 036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Li H, Yu X, Li S, Lei Z, Li C, Zhang Q, Han Q, Li Y, Zhang K, et al. (2018). Analysis of circulating tumor cells in ovarian cancer and their clinical value as a biomarker. Cell Physiol Biochem 48, 1983–1994. [DOI] [PubMed] [Google Scholar]
- Zhang XY, Pettengell R, Nasiri N, Kalia V, Dalgleish AG, Barton DP (1999). Characteristics and growth patterns of human peritoneal mesothelial cells: comparison between advanced epithelial ovarian cancer and non-ovarian cancer sources. J Soc Gynecol Investig 6, 333–340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.