Abstract
Eukaryotic cells possess considerable internal complexity, differentiating them from prokaryotes. Eukaryogenesis, an evolutionary transitional period culminating in the last eukaryotic common ancestor (LECA), marked the origin of the eukaryotic endomembrane system. LECA is reconstructed as possessing intracellular complexity akin to modern eukaryotes. Construction of endomembrane compartments involved three key gene families: coatomer, BAR-domain proteins, and ESCRT. Each has a distinct evolutionary origin, but of these coatomer and BAR proteins are eukaryote specific, while ESCRT has more ancient origins. We discuss the structural motifs defining these three membrane-coating complexes and suggest that compared with BAR and ESCRT, the coatomer architecture had a unique ability to be readily and considerably modified, unlocking functional diversity and enabling the development of the eukaryotic cell.
THE EUKARYOTIC BAUPLAN AND THE ROLE OF COATING COMPLEXES
A hallmark of eukaryotic cells is elaborate internal membrane-bound compartments, collectively termed organelles, that facilitate segregation of specific biochemical functions (Rout and Field, 2017). While many prokaryotes do have membranous organelles, these are considerably less diverse, limited to one or a handful in each species. Although the genetic origins of the eukaryotic endomembrane system descended from prokaryotes, eukaryotes share few structures and mechanisms with prokaryotic organelles (Figure 1A) (Grant et al., 2018). Moreover, it is likely that the last common ancestor of all eukaryotes (LECA) possessed an intracellular membranous organelle cohort essentially as elaborate or even exceeding those possessed by their modern descendants (Rout and Field, 2017)
FIGURE 1:
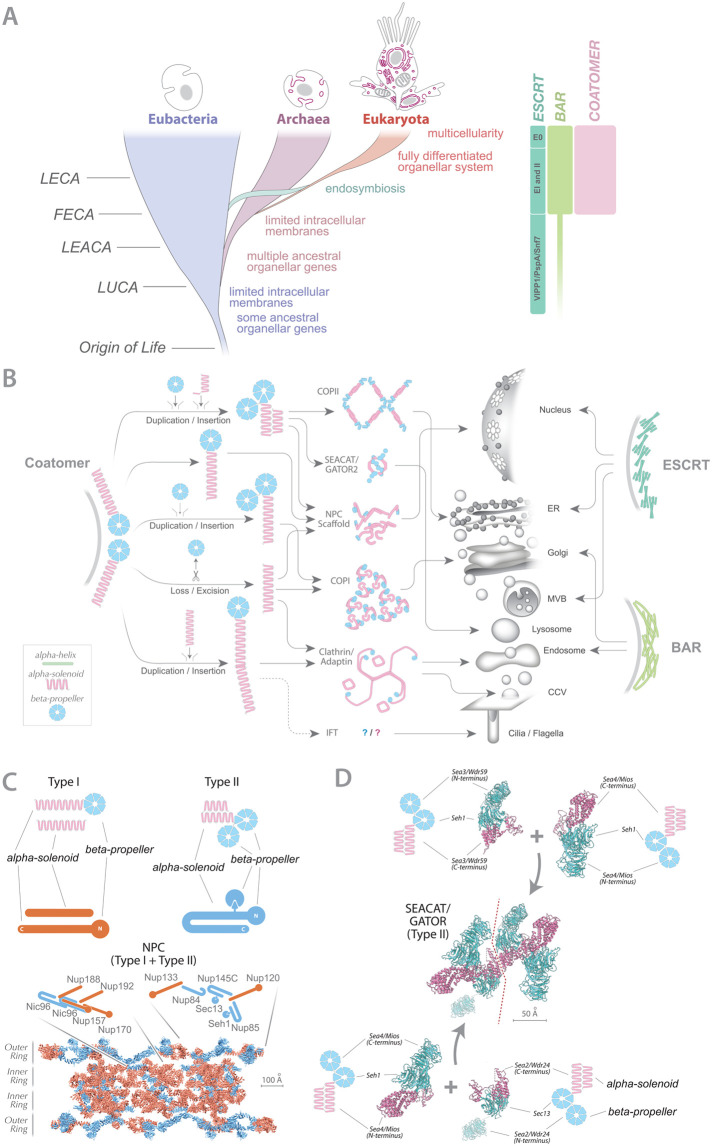
Coats, compartments, and eukaryotic origins. (A) Left: Eukaryotes arose as a branch of the Archaea. The three major lineages of life are shown together with conceptualized diagrams of cell structure; for Eukaryota, we show a choanocyte and amoebocyte from a sponge, among the earliest forms of multicellularity. Major evolutionary events are also shown together with probable timings for origins of membrane coats. LECA, last eukaryotic common ancestor; FECA, first eukaryotic common ancestor; LEACA, last eukaryotic archaeal common ancestor; LUCA, last universal common ancestor. (A) Right: approximate relative timings of origins of the coating complexes discussed here. See text for other details. (B) Structural characteristics, evolutionary paths, and intracellular locations for major functions for coatomers, BARs, and ESCRTs. Schematics for cellular compartments are at center right in gray. Arrows indicate approximate locations where individual complexes operate in the cell. Data are a composite from multiple cells/lineages, and not all locations for these complexes are shown for simplicity. (C) Neofunctionalization of coatomer architectures to form the NPC. Type I (red) and type II (blue) coatomer architectures are shown, with idealized structures above (see key to B). A section of the Saccharomyces cerevisiae NPC structure viewed from the side is shown at the bottom, illustrating only the type I- and type II-related subunits, similarly colored red (Type I) and blue (Type II). Note the intermixing of types within the overall structure. (D) Stepwise conceptual assembly of SEACAT/GATOR2 from paralogous and identical duplications of a basic type II coatomer architecture. Schematic structures of the progenitor coatomer units are shown (see key to B) next to ribbon representations of subunits, with beta-propellers in cyan and alpha-solenoids in magenta. The beta propeller of Wrd59 is shown as translucent as its structure has not yet been solved.
The basic functions of protein families involved in organellogenesis are an ability to bend and mold membranes, and at least three perform this function in eukaryotes, all of which were present in the LECA. The first, BAR-domain proteins are alpha-helical bundles and polymerize to form scaffolds of varying curvatures. The second, the ESCRT complex, operates by polymerization of alpha-helical subunits into ring or helical structures. Finally, the coatomer system predominantly comprises arrangements of extended alpha-helical solenoids flanked or associated with beta-propellers. Despite the ability of BAR, ESCRT, and coatomer to fulfill similar mechanistic roles, their evolutionary origins, subsequent evolutionary trajectories, and the modern functions of each are very distinct (Figure 1B) (Field et al., 2011; Rout and Field, 2017).
Given this, there is potential redundancy among BAR, ESCRT, and coatomer complexes in terms of their ability to deform membranes. Hence we ask, was there competition among these membrane-coating complexes to obtain the newly emerging functionalities required in the cell as it elaborated and diversified its internal membrane organization? Alternatively, was there cooperativity among them in generating membrane-bounded organelles, or even mutual indifference, as their unique architectures propelled their evolution along very separate routes? We also consider the structural plasticity within BAR, ESCRT and coatomer protein families and specifically the architecture of proteins within each family and how these subunits assemble to form a membrane coat. From this, we ask if certain inherent properties of the underlying coatomer architecture, in contrast with those of the BAR and ESCRT complexes, were the driving factors behind expansion of coatomer into a wide variety of different coating architectures.
THE EVOLUTIONARY ORIGINS OF COATING COMPLEXES
The ESCRT complex consists of several subcomplexes known as E0, I, II, and III in metazoa, with an EIII-associated complex that incorporates the Vps4 ATPase, a regulator for ESCRT assembly. The majority of EIII and EIII-associated subunits likely arose by paralogous expansion and, except for E0, the entire system was present in the LECA (Leung et al., 2008). In metazoan ESCRT supports endosomal trafficking, protein turnover, cytokinesis, repair of the nuclear envelope, and plasma membrane fissures among many other roles (Figure 1B). ESCRT-mediated membrane bending involves a core set of Snf7-domain (CHMP in metazoan) proteins, but recruitment to individual subcellular sites in metazoa requires specific receptors, for example, LEM2 for nuclear events, HRS/STAM for late endosomes, and CEP55 during cytokinesis (Vietri et al., 2020). Importantly, Snf7-domain/CHMP proteins are structurally conserved, albeit that distinct CHMP paralogues can bend membrane with specific topology; for example, CHMP4 forms flat spirals, but CHMP1B forms positively curved spirals (McCullough et al., 2015). Thus new ESCRT roles have arisen largely by adding differential targeting modules to a conserved core and bringing essentially the same coating complex to different membranes.
Snf7-domain proteins have a clear prokaryotic origin and were likely present in the earliest cells (Figure 1A). Vipp1/PspA, found in chloroplasts and bacteria, respectively, form rings and rods in vitro (Junglas et al., 2021; Liu et al., 2021) and possess an alpha-helical structure remarkably homologous to the eukaryotic Snf7 domain. Further, Archaeal Snf7 homologues function in cytokinesis (Samson et al., 2011) and, while representation of ESCRT homologues across the Archaea is patchy (Caspi and Dekker, 2018), a role in cell division persists into eukaryotes including in metazoa (McCullough et al., 2018). Hence the core membrane-deforming components of ESCRT have a very ancient origin and remained essentially structurally invariant for billions of years.
BAR-domain proteins can be categorized into one of three subfamilies: classical (or N-) BAR or the F- and I-BAR domain subfamilies. Significantly, N- and F-BAR polymers subtend a positive curvature, while I-BAR polymers are flat or negatively curved (Carman and Dominguez, 2018; Simunovic et al., 2019). BAR-domain proteins participate in a wide variety of membrane events, including collaboration in clathrin-mediated transport, organelle shaping, and as components of the retromer complex (Figure 1B) (Carman and Dominguez, 2018; Simunovic et al., 2019). While, like ESCRT, the core-coating structure remains similar across the family (Figure 1B), most BAR proteins also possess additional domains, conferring specificity and additional functionality. At least some BAR-domain proteins were considerable LECA components and originated at that time, although with evidence for lineage-specific expansion later in eukaryotic evolution (Figure 1A) (Koumandou et al., 2011). The presence of BAR-domain proteins in bacteria is sparse and is possibly a lateral gene transfer from a eukaryotic donor (Phillips et al., 2021). Significantly, neither the ESCRT nor the BAR protein families appear malleable in terms of architecture, with comparatively subtle variation among paralogues facilitating altered membrane curvature but with polymerized assemblies retaining highly similar structures and are frequently homopolymers (Carman and Dominguez, 2018; McCullough et al., 2018).
The final system we consider is the coatomer family. The major features of this family are proteins with beta-propeller domains at the N-terminus and alpha-solenoid domains at the C-terminus as well as components comprised of either alpha-solenoids or beta-propellers alone; both domains are formed from highly repetitive motifs and can form long and flexible subunits. In contrast with ESCRT or BAR proteins, the coatomer subunit architecture displays notable structural plasticity, allowing for highly diverse architectures (Figure 1, B–D) (Bethune and Wieland, 2018; Dacks and Robinson, 2017; Rout and Field, 2017). The origins of the many different coatomer complexes from a progenitor “protocoatomer” complex have been described by us elsewhere (Devos et al., 2004; Field and Rout, 2019; Rout and Field, 2017).
Coatomer architectures have been evolutionarily molded from this original protocoatomer into two major classes; type I, typified by clathrin-coated vesicles and COPI transport vesicles, and type II, typified by COPII transport vesicles. Coatomers also form assemblies with other roles, including intraflagellar transport complexes, nuclear pore complexes (NPCs) and the SEACAT (Seh1-associated) complex involved in nutrient sensing (Figure 1B) (Field et al., 2011; Rout and Field, 2017; van Dam et al., 2013). Indeed, coatomers were clearly instrumental in the formation and maintenance of the majority of defining eukaryotic organelles, including the endoplasmic reticulum, Golgi complex, lysosomes, endosomes, cilia, and the nuclear envelope. The coatomer paralogues that populate most of these organelles were established prior to the LECA (Figure 1A) (Rout and Field, 2017). There is currently no evidence for coatomer being present in the Archaea, suggesting an origin initiated post the first eukaryotic common ancestor (FECA; Figure 1A) (Field and Rout, 2019; Rout and Field, 2017).
ADAPTABILITY AND CONSTRAINTS IN COATING COMPLEX STRUCTURE
Functional expansions can be mediated by gene duplications and subsequent sequence divergence among the resulting paralogues (Kuzmin et al., 2021). If the original paralogue is retained, we have suggested that this represents a mechanism to evolve new membrane coats and hence compartments (Dacks and Field, 2007). However, there is significant constraint, as any heteropolymer containing both old and new paralogues must still assemble a functional coat, and structurally incompatible paralogues are likely to be a selective disadvantage. For BAR and ESCRT membrane-deforming subunits, which are both based on reflexed and heavily intertwined alpha-helices (Figure 1B) (Carman and Dominguez, 2018; Simunovic et al., 2019), this most likely provides considerable restraint to novelty and consistent with the high conservation of Snf7/CHMP and BAR-domain protein and polymer structures (Figure 1B).
The coatomer complexes, stand in sharp contrast with such assemblies in terms of their constituent folds. Coatomer beta-propellers generally limit themselves to seven repeated blades in a circle and associate with other subunits either by constituting the N-terminus to an alpha-solenoid C-terminus, or indirectly via donation of one of their blades to an alpha-solenoid protein but again near the latter’s N-terminus (e.g., Valenstein et al., 2022) (Figure 1B). The alpha-solenoids are a simple linear repetition of an alpha-helical zigzag and, by contrast, demonstrate great variability in architecture, but generally though indels, kinks, or hairpin turns along the length of the alpha-solenoid rod (Figure 1B). These relatively repetitive and linear coatomer structures form assembly contacts largely limited to the ends of each subunit, which means that deletions or insertions within their repetitive sections—and particularly the alpha-solenoids—are likely better tolerated in coassembly with the original version. As a result, a hybrid coatomer coat would be more likely to retain function and not act as a dominant negative compared with BAR and ESCRT (Dacks and Robinson, 2017; Field and Rout, 2019). This gives the coatomer family a clear path to neofunctionalization and is supported by the enormous morphological diversity within members of the family (Figure 1B) (Traub, 2009; Sochacki et al., 2017; Bethune and Wieland, 2018).
Membrane-bending ESCRT proteins arose in bacteria and therefore suffered for a consderable time or no competition from either BAR or coatomer complexes, neither of which evolved for over a billion years prior to eukaryogenesis. We suggest this is compelling evidence that, despite participation in an increasing number of functions, ESCRT was unable to fulfill the roles required to diversify intracellular architecture to the level of modern eukaryotes. Additionally, the limited roles of BAR-domain proteins since their origin prior to the LECA suggest a similar constraint, albeit with considerable expansions in some modern lineages where a role in facilitating post-LECA innovations is recognized (Suetsugu et al., 2010).
EXAMPLES OF COATOMER NEOFUNCTIONALIZATION
How adaptable have coatomer complexes proven? We consider two examples, the NPC and the SEACAT/GATOR2 complex. Detailed structural information has recently been obtained for both structures, allowing us to examine whether and how their evolution utilized the considerable structural flexibility of the basic coatomer architecture (Kim et al., 2018; Akey et al., 2022; Bley et al., 2022; Fontana et al., 2022; Mosalaganti et al., 2022; Petrovic et al., 2022; Valenstein et al., 2022; Zhu et al., 2022).
The NPC is a hub for protein and RNA transport across the nuclear envelope, an organizer for RNA processing and the nuclear interior (Devos et al., 2004; Hayama et al., 2017; D’Angelo, 2018; Holzer and Antonin, 2018; Lin and Hoelz, 2019; De Magistris, 2021; Fernandez-Martinez and Rout, 2021; Paci et al., 2021). To meet these many demands, the NPC has some of the more extreme neofunctionalizations of coatomer architecture necessitating multiple duplications, alterations, and amalgamations in their constituent coatomer-related components. It is far from the original role of a membrane-deforming complex (Figure 1C).
Well over half of the mass of the NPC is composed of coatomer proteins (Devos et al., 2006; Hayama et al., 2017; Rout and Field, 2017; Beck et al., 2018; Kim et al., 2018; Field and Rout, 2019; Fernandez-Martinez and Rout, 2021). The NPC formed through an amalgamation of both type I and type II coating complexes; this complex formed early in the evolution of the NPC (Field and Rout, 2019), and subunits subsequently duplicated and diverged several times to form the inner and outer ring complexes of the NPC scaffold, resulting in both type I- and type II-derived coatomer proteins intertwined throughout the scaffold, a major elaboration not seen in other coatomer complexes to date (Figure 1C) (Field and Rout, 2019).
This flexible approach to construction extends to a high degree of heterogeneity in architecture, both within and between species, much of this generated by the addition or subtraction of outer rings on the nuclear and cytoplasmic NPC faces (Kim et al., 2018; Akey et al., 2022; Bley et al., 2022; Fontana et al., 2022; Makarov et al., 2021; Mosalaganti et al., 2018 #1180; Mosalaganti et al., 2022; Petrovic et al., 2022; von Appen et al., 2015; Zhu et al., 2022; Zimmerli et al., 2021). Thus while their protocoatomer ancestors were largely restricted to forming vesicle coat complexes, NPCs have been extensively modified and adapted for differential roles between organisms.
The SEACAT complex is a subcomplex of the larger SEA complex and an excellent exemplar of the protocoatomer hypothesis in general (Devos et al., 2004) and more specifically proposals here concerning extreme adaptability. The SEA complex was first identified in yeast and then in mammalian cells and as likely present in the LECA and is a major regulator of the stress response regulator mTORC1, conveying the status of arginine and leucine levels (Dokudovskaya et al., 2011; Bar-Peled et al., 2013; Loissell-Baltazar and Dokudovskaya, 2021; Valenstein et al., 2022). We proposed that SEACAT was evolutionarily related to coatomers despite being a regulator and not a membrane-coating complex, as SEACAT subunits have predicted structural homology to coatomer, and the complex incorporates bona fide coatomer subunits Sec13 and Seh1, taken from COPII/NPC and the NPC, respectively (Dokudovskaya et al., 2011; Field and Rout, 2019). Indeed a recently solved mammalian SEACAT complex structure (Valenstein et al., 2022) elegantly confirms this proposal, revealing that almost the entire scaffold comprises iterations of type II coatomer subunits. Furthermore, as with the NPC, it is possible to reconstruct how the complex may have arisen from a simple type II coatomer that duplicated, diverged, and multimerized into a full octameric complex as seen today (Figure 1D). These examples of coatomer adaptability compared with BAR and ESCRT family proteins underscore a potential particular to coatomer complexes for wide plasticity in architecture and function.
IMPLICATIONS
To conclude, we suggest that ESCRT, despite a presence in the earliest cells, was unable to unlock the functions required for eukaryogenesis, and that this block was likely due to structural inflexibility. BAR-domain proteins arose at around the same time as coatomers but are less structurally diverse than coatomer. Hence neither ESCRT nor BAR-domains competed with coatomer for the functions the latter took on, as they were simply unable to fulfill these roles. Significantly, as expanded intracellular complexity arose, driven by the expansion and diversification of the coatomer family, new functional opportunities were created that could be populated by adaptations of the BAR and ESCRT families. For example, the BAR-derived retromer complex, associated with the Golgi complex, could only have arisen after the COPI and COPII coatomer complexes formed the Golgi apparatus, while roles in nuclear envelope repair and NPC surveillance undertaken by ESCRT required prior evolution of the nuclear envelope and NPC. Thus a process of cooperation to further elaborate endomembrane functions operated to generate the full complexity of the LECA and modern eukaryotes, but it appears that coatomer alone unlocked the original and full eukaryotic bauplan.
Acknowledgments
Work in our laboratories is supported by the Wellcome Trust (204697/Z/16/Z to MCF) and the National Institutes of Health (P41 GM109824, R01 GM112108, and R01 CA228351 to M.P.R.).
Abbreviations used:
- BAR
Bin, Amphiphysin and Rvs
- CEP55
centrosomal protein of 55 kDa
- CHMP
charged multivesicular body protein
- ESCRT
endosomal sorting complex required for transport
- F-BAR
FCH-BAR, or EFC for extended FCH homology preceding the BAR
- FECA
first eukaryotic comon ancestor
- GATOR2
Seh1-associated GAP activity to TOR 2
- HRS/STAM
hepatocyte growth factor-regulated tyrosine kinase substrate and signal transducing adaptor molecule
- I-BAR
inverse BAR
- LEACA
last eukaryotic archaea common ancestor
- LECA
last eukaryotic common ancestor
- LEM2
LAP2, emerin, MAN1 domain protein 2
- LUCA
last universal common ancestor
- mTORC1
mammalian target of rapamycin complex 1
- N-BAR
N-terminal amphipathic helix preceding the BAR
- NPC
nuclear pore complex
- PspA
phage shock protein A
- SEACAT
SEA subcomplex activating TORC1
- Snf7
sucrose nonfermenting protein 7
- Vipp1
vesicle-inducing protein in plastids1.
Footnotes
REFERENCES
- Akey CW, Singh D, Ouch C, Echeverria I, Nudelman I, Varberg JM, Yu Z, Fang F, Shi Y, Wang J, et al. (2022). Comprehensive structure and functional adaptations of the yeast nuclear pore complex. Cell 185, 361–378.e325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM (2013). A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Mosalaganti S, Kosinski J (2018). From the resolution revolution to evolution: structural insights into the evolutionary relationships between vesicle coats and the nuclear pore. Curr Opin Struct Biol 52, 32–40. [DOI] [PubMed] [Google Scholar]
- Bethune J, Wieland FT (2018). Assembly of COPI and COPII Vesicular Coat Proteins on Membranes. Annu Rev Biophys 47, 63–83. [DOI] [PubMed] [Google Scholar]
- Bley CJ, Nie S, Mobbs GW, Petrovic S, Gres AT, Liu X, Mukherjee S, Harvey S, Huber FM, Lin DH, et al. (2022). Architecture of the cytoplasmic face of the nuclear pore. Science 376, eabm9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman PJ, Dominguez R (2018). BAR domain proteins-a linkage between cellular membranes, signaling pathways, and the actin cytoskeleton. Biophys Rev 10, 1587–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi Y, Dekker C (2018). Dividing the archaeal way: the ancient Cdv cell-division machinery. Front Microbiol 9, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA (2018). Nuclear pore complexes as hubs for gene regulation. Nucleus (Austin, Tex.) 9, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacks JB, Field MC (2007). Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J Cell Sci 120, 2977–2985. [DOI] [PubMed] [Google Scholar]
- Dacks JB, Robinson MS (2017). Outerwear through the ages: evolutionary cell biology of vesicle coats. Curr Opin Cell Biol 47, 108–116. [DOI] [PubMed] [Google Scholar]
- De Magistris P (2021). The great escape: mRNA export through the nuclear pore complex. Int J Mol Sci 22, 11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP (2004). Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol 2, e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A (2006). Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci USA 103, 2172–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokudovskaya S, Waharte F, Schlessinger A, Pieper U, Devos DP, Cristea IM, Williams R, Salamero J, Chait BT, Sali A, et al. (2011). A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol Cell Proteomics 10, M110 006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Martinez J, Rout MP (2021). One ring to rule them all? structural and functional diversity in the nuclear pore complex. Trends Biochem Sci 46, 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Rout MP (2019). Pore timing: the evolutionary origins of the nucleus and nuclear pore complex. F1000Res 8, 16402.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Sali A, Rout MP (2011). Evolution: On a bender–BARs, ESCRTs, COPs, and finally getting your coat. J Cell Biol 193, 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana P, Dong Y, Pi X, Tong AB, Hecksel CW, Wang L, Fu TM, Bustamante C, Wu H (2022). Structure of cytoplasmic ring of nuclear pore complex by integrative cryo-EM and AlphaFold. Science 376, eabm9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CR, Wan J, Komeili A (2018). Organelle formation in bacteria and archaea. Annu Rev Cell Dev Biol 34, 217–238. [DOI] [PubMed] [Google Scholar]
- Hayama R, Rout MP, Fernandez-Martinez J (2017). The nuclear pore complex core scaffold and permeability barrier: variations of a common theme. Curr Opin Cell Biol 46, 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer G, Antonin W (2018). Nuclear pore complexes: global conservation and local variation. Curr Biol 28, R674–R677. [DOI] [PubMed] [Google Scholar]
- Junglas B, Huber ST, Heidler T, Schlosser L, Mann D, Hennig R, Clarke M, Hellmann N, Schneider D, Sachse C (2021). PspA adopts an ESCRT-III-like fold and remodels bacterial membranes. Cell 184, 3674–3688.e3618. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, Raveh B, Herricks T, Slaughter BD, Hogan JA, Upla P, et al. (2018). Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumandou VL, Klute MJ, Herman EK, Nunez-Miguel R, Dacks JB, Field MC (2011). Evolutionary reconstruction of the retromer complex and its function in Trypanosoma brucei. J Cell Sci 124, 1496–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin E, Taylor JS, Boone C (2021). Retention of duplicated genes in evolution. Trends Genet 38, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KF, Dacks JB, Field MC (2008). Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic 9, 1698–1716. [DOI] [PubMed] [Google Scholar]
- Lin DH, Hoelz A (2019). The structure of the nuclear pore complex (an update). Annu Rev Biochem 88, 725–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Tassinari M, Souza DP, Naskar S, Noel JK, Bohuszewicz O, Buck M, Williams TA, Baum B, Low HH (2021). Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. Cell 184, 3660–3673.e3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loissell-Baltazar YA, Dokudovskaya S (2021). SEA and GATOR 10 years later. Cells 10, 2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov AA, Padilla-Mejia NE, Field MC (2021). Evolution and diversification of the nuclear pore complex. Biochem Soc Trans 49, 1601–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Clippinger AK, Talledge N, Skowyra ML, Saunders MG, Naismith TV, Colf LA, Afonine P, Arthur C, Sundquist WI, et al. (2015). Structure and membrane remodeling activity of ESCRT-III helical polymers. Science 350, 1548–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Frost A, Sundquist WI (2018). Structures, functions, and dynamics of ESCRT-III/Vps4 membrane remodeling and fission complexes. Annu Rev Cell Dev Biol 34, 85–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turonova B, Zimmerli CE, Buczak K, Schmidt FH, Margiotta E, Mackmull MT, et al. (2022). AI-based structure prediction empowers integrative structural analysis of human nuclear pores. Science 376, eabm9506. [DOI] [PubMed] [Google Scholar]
- Mosalaganti S, Kosinski J, Albert S, Schaffer M, Strenkert D, Salomé PA, Merchant SS, Plitzko JM, Baumeister W, Engel BD, Beck M (2018). In situ architecture of the algal nuclear pore complex. Nat Commun 9, 2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paci G, Caria J, Lemke EA (2021). Cargo transport through the nuclear pore complex at a glance. J Cell Sci 134, jcs247874. [DOI] [PubMed] [Google Scholar]
- Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, Nie S, Mobbs GW, Stevens TA, Liu X, et al. (2022). Architecture of the linker-scaffold in the nuclear pore. Science 376, eabm9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DA, Zacharoff LA, Hampton CM, Chong GW, Malanoski AP, Metskas LA, Xu S, Bird LJ, Eddie BJ, Miklos AE, et al. (2021). A bacterial membrane sculpting protein with BAR domain-like activity. Elife 10, e60049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Field MC (2017). The evolution of organellar coat complexes and organization of the eukaryotic cell. Annu Rev Biochem 86, 637–657. [DOI] [PubMed] [Google Scholar]
- Samson RY, Obita T, Hodgson B, Shaw MK, Chong PL, Williams RL, Bell SD (2011). Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol Cell 41, 186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic M, Evergren E, Callan-Jones A, Bassereau P (2019). Curving cells inside and out: roles of BAR domain proteins in membrane shaping and its cellular implications. Annu Rev Cell Dev Biol 35, 111–129. [DOI] [PubMed] [Google Scholar]
- Sochacki KA, Dickey AM, Strub MP, Taraska JW (2017). Endocytic proteins are partitioned at the edge of the clathrin lattice in mammalian cells. Nat Cell Biol 19, 352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Toyooka K, Senju Y (2010). Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin Cell Dev Biol 21, 340–349. [DOI] [PubMed] [Google Scholar]
- Traub LM (2009). Clathrin couture: fashioning distinctive membrane coats at the cell surface. PLoS Biol 7, e1000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenstein ML, Rogala KB, Lalgudi PV, Brignole EJ, Gu X, Saxton RA, Chantranupong L, Kolibius J, Quast JP, Sabatini DM (2022). Structure of the nutrient-sensing hub GATOR2. Nature 607, 610–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam TJ, Townsend MJ, Turk M, Schlessinger A, Sali A, Field MC, Huynen MA (2013). Evolution of modular intraflagellar transport from a coatomer-like progenitor. Proc Natl Acad Sci USA 110, 6943–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri M, Radulovic M, Stenmark H (2020). The many functions of ESCRTs. Nat Rev Mol Cell Biol 21, 25–42. [DOI] [PubMed] [Google Scholar]
- von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, et al. (2015). In situ structural analysis of the human nuclear pore complex. Nature 526, 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli CE, Allegretti M, Rantos V, Goetz SK, Obarska-Kosinska A, Zagoriy I, Halavatyi A, Hummer G, Mahamid J, Kosinski J, Beck M (2021). Nuclear pores dilate and constrict in cellulo. Science 374, eabd9776. [DOI] [PubMed] [Google Scholar]
- Zhu X, Huang G, Zeng C, Zhan X, Liang K, Xu Q, Zhao Y, Wang P, Wang Q, Zhou Q, et al. (2022). Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex. Science 376, eabl8280. [DOI] [PubMed] [Google Scholar]