Abstract
Pkd2 is the fission yeast homologue of polycystins. This putative ion channel localizes to the plasma membrane. It is required for the expansion of cell volume during interphase growth and cytokinesis, the last step of cell division. However, the channel activity of Pkd2 remains untested. Here, we examined the calcium permeability and mechanosensitivity of Pkd2 through in vitro reconstitution and calcium imaging of pkd2 mutant cells. Pkd2 was translated and inserted into the lipid bilayers of giant unilamellar vesicles using a cell-free expression system. The reconstituted Pkd2 permeated calcium when the membrane was stretched via hypoosmotic shock. In vivo, inactivation of Pkd2 through a temperature-sensitive mutation pkd2-B42 reduced the average intracellular calcium level by 34%. Compared with the wild type, the hypomorphic mutation pkd2-81KD reduced the amplitude of hypoosmotic shock–triggered calcium spikes by 59%. During cytokinesis, mutations of pkd2 reduced the calcium spikes, accompanying cell separation and the ensuing membrane stretching, by 60%. We concluded that fission yeast polycystin Pkd2 allows calcium influx when activated by membrane stretching, representing a likely mechanosensitive channel that contributes to the cytokinetic calcium spikes.
INTRODUCTION
Polycystins are evolutionarily conserved calcium-permissive cation channels. Loss-of-function mutations of human polycystins lead to one of the most common genetic disorders, autosomal dominant polycystic kidney disorder (ADPKD), which is diagnosed in 1 in 1000 live births (Hughes et al., 1995; Mochizuki et al., 1996). Homologues of polycystins have been found in most metazoans, including fruit flies and worms (Barr and Sternberg, 1999; Gao et al., 2003), as well as unicellular organisms such as social amoebae (Lima et al., 2014), green algae (Huang et al., 2007), and fission yeast (Palmer et al., 2005).
The fission yeast polycystin homologue Pkd2 is an essential protein required for both cell division and growth. Pkd2 localizes to the plasma membrane throughout the cell cycle (Morris et al., 2019). During interphase, Pkd2 is enriched at the cell tips, where the putative channel promotes the extension of cylindrical fission yeast cells (Sinha et al., 2022). During cytokinesis, Pkd2 moves to the equatorial plane, regulating contractile ring constriction and cell separation (Morris et al., 2019). Without Pkd2, the daughter cells fail to separate. Additionally, Pkd2 antagonizes the activity of the yeast Hippo signaling pathway SIN (septation initiation network) by modulating its activity and localization during cytokinesis (Sinha et al., 2022). Although fission yeast cytokinesis is accompanied by a temporary increase in intracellular calcium concentration through cytokinetic calcium spikes (Poddar et al., 2021), it remains unclear if Pkd2 permeates calcium in this process and how it is activated.
Reconstitution experiments using bottom-up in vitro expression of transmembrane proteins, including ion channels, have become a powerful approach to investigating their functions, compared with using purified proteins expressed from cells. For expression of certain proteins in cells, growth retardation or lysis of the host cells and low endogenous expression levels contribute to poor production of heterologous recombinant proteins (Laohakunakorn et al., 2020). Despite advances in protein purification, several limitations exist with in vitro reconstitution of membrane proteins with complicated structures, such as improper protein function, compromised membrane integrity due to residual detergents, and poor control over the orientation of protein insertion (Knol et al., 1998; Rigaud and Levy, 2003; Wingfield, 2015; Jia and Jeon, 2016; Shen et al., 2016). Cell-free expression (CFE) systems coupling transcription and translation reactions outside the cellular environment have shown the potential to overcome the barriers mentioned above and can be a robust strategy for protein synthesis and investigation (Rigaud and Levy, 2003; Chong, 2014; Lu, 2017; Gregorio et al., 2019; Khambhati et al., 2019). For example, the bacterial mechanosensitive channel MscL is expressed by encapsulating CFE reactions in giant unilamellar vesicles (GUVs) and has been shown to sense physical stimuli (Majumder et al., 2017). Since bacterial lysate lacks membranous components, eukaryotic CFE systems are gaining increasing attention for in vitro production of membrane proteins (Dondapati et al., 2014). Their innate endogenous microsomal structures enable newly synthesized membrane proteins to insert directly into the natural endoplasmic reticulum (ER)–based lipid bilayers without detergents. This eukaryotic CFE-based approach significantly reduces the potential for membrane protein denaturation and favors their proper folding in vitro.
In this study, we first expressed the putative channel Pkd2 in a HeLa-based CFE system and reconstituted it in a lipid bilayer. To determine the orientation of Pkd2 in the membrane, we used a pronase digestion assay with Streptomyces griseus–derived pronase (Xu et al., 1988). We applied different osmotic pressures to GUVs coexpressing Pkd2 and G-GECO, a fluorescent calcium-sensitive reporter. To determine whether Pkd2 regulates calcium influx in vivo, we employed a GCaMP-based calcium indicator and single-cell imaging to quantify the intracellular calcium level of two pkd2 mutants. We then induced fast expansion of the plasma membrane using microfluidics-applied osmotic shock and measured the calcium spikes in the pkd2 mutant cells compared with the wild type. Finally, we quantified the constriction and separation calcium spikes, which accompany compression and expansion of the plasma membrane respectively during cytokinesis, in the pkd2 mutant cells in comparison to the wild type. Overall, our study would demonstrate Pkd2 as a likely mechanosensitive channel that promotes calcium influx during cytokinesis.
RESULTS
Expressing Pkd2 by using mammalian cell-free expression
We used a CFE system to synthesize this putative channel Pkd2 in vitro. We expressed full-length Pkd2 tagged with superfold GFP (sfGFP) at the C-terminus in a HeLa cell extract–based system. To monitor expression yield, we quantified the fluorescence of Pkd2-sfGFP over 3 h (Supplemental Figure S1). The fluorescence increased gradually and reached a plateau after 2 h. We concluded that Pkd2 is efficiently expressed in our cell-free system.
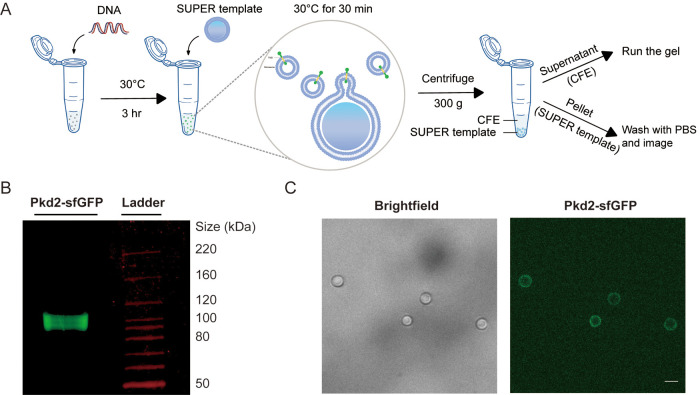
We then determined if the in vitro synthesized Pkd2 can be reconstituted as a transmembrane protein in supported lipid bilayers with extra reservoir (SUPER) templates (Figure 1A). The excess lipid bilayer membranes were generated on silica beads by the rupture and fusion of small unilamellar vesicles (SUVs) carrying lipids with negative charges at high ionic strength (Pucadyil and Schmid, 2008, 2010; Majumder et al., 2018). We incubated SUPER templates with the in vitro translated Pkd2-sfGFP and isolated them by low-speed centrifugation. The supernatant fraction contained most of the CFE reaction, while the pellet fraction included the SUPER templates for different assays (Figure 1A). The expressed protein appeared as a single band of ∼105 kDa on an SDS–PAGE gel (Figure 1B), which was consistent with the predicted molecular weight of the fusion protein (MWPkd2 = 80 kDa and MWGFP = 27 kDa). The yield was roughly 15 µg from a 10 µl reaction. The lipid-coated beads incubated with the pellet fraction became fluorescent after being washed with PBS (Figure 1C), thus confirming that Pkd2-sfGFP was incorporated into the SUPER templates.
FIGURE 1:
Localization of cell-free expressed Pkd2 in SUPER template. (A) Schematic illustrating the use of CFE for in vitro protein production and testing the incorporation of membrane proteins using SUPER templates. SUPER templated beads are added to the CFE reaction expressing Pkd2 protein fused to sfGFP at the C-terminus and incubated together. CFE reaction and SUPER templates are then isolated by low-speed centrifugation for running a gel or imaging, respectively. (B) Fluorescence gel image of cell-free expressed Pkd2-sfGFP added to SUPER templates. (C) Brightfield and fluorescence micrograph of SUPER templates incubated with cell-free expressed Pkd2-sfGFP. Beads were washed with PBS before imaging. Scale bar: 5 µm.
Since several Pkd2 homologues, including the human polycystins, localize to the ER and the plasma membrane of the cilia (Hughes et al., 1995; Gonzalez-Perrett et al., 2001; Protchenko et al., 2006), we investigated whether Pkd2 similarly translocates into endogenous microsomal structures. We applied a high-speed airfuge assay to CFE-expressed Pkd2, labeled with fluorescently tagged lysine, Green lysine, to isolate supernatant and pellet fractions; the latter presumably contained microsomes (Supplemental Figure S2A). Green lysine is a lysine-charged tRNA labeled with the fluorophore BODIPY-FL at the epsilon amino group and can be incorporated into synthesized proteins during in vitro translation reactions. We found that the majority of Pkd2 was in the pellet, while the amount of Pkd2 in the supernatant decreased as the washing cycles was increased (Supplemental Figure S2B). This indicated that ER fragments might recognize translocons and membrane protein chaperones to promote Pkd2 insertion into microsomes. When the pellet or supernatant fractions were added to the SUPER templates, only the beads incubated with the pellet fraction were fluorescent (Supplemental Figure S2C). We concluded that in vitro–expressed Pkd2 translocate into microsomal fragments, which subsequently fuse with the SUPER templates.
Reconstituted Pkd2 responds to osmotic pressure to permeate calcium
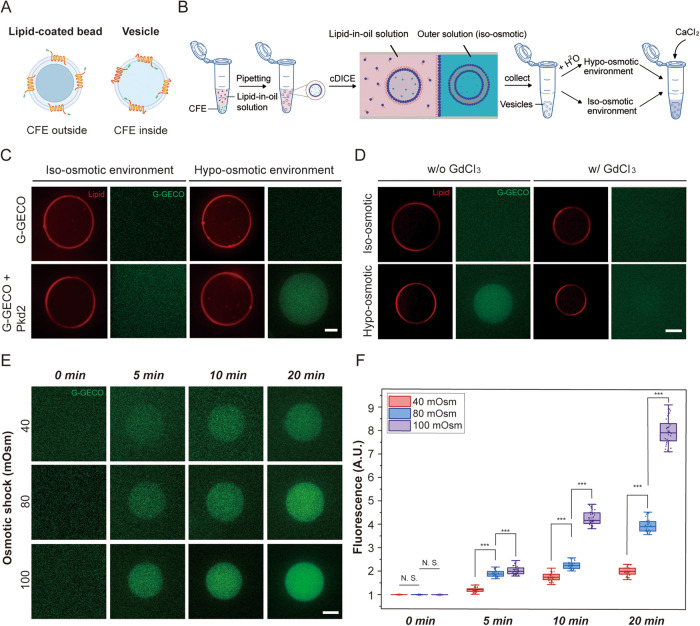
We next determined the orientation of Pkd2 in the lipid bilayer membranes on SUPER templates using an image-based pronase protection assay. Unlike the human polycystins, the structure of fission yeast Pkd2 has not been experimentally determined. AlphaFold (Jumper et al., 2021) and other transmembrane helices projection software predicted that Pkd2 possesses an N-terminal extracellular domain and a putative C-terminal cytoplasmic tail (Supplemental Figure S3A). Depending on the orientation of Pkd2-sfGFP in the lipid bilayer, sfGFP will either be exposed to pronase or be protected from degradation (Supplemental Figure S3B). Based on our previous work on reconstituting the nuclear envelope proteins SUN1 and SUN2 in SUPER templates (Majumder et al., 2018), we predicted that Pkd2 would insert into the excess lipid bilayer membranes with its C-terminus orienting outward. We observed that the fluorescence of in vitro–translated Pkd2-sfGFP disappeared following pronase treatment (Supplemental Figure S3C). We concluded that Pkd2 is inserted into microsomal fragments in an orientation that positions its C-terminus on the cytosolic side. For the GUVs used in our experiment, the C-terminus of Pkd2-sfGFP would be in the lumen of the vesicle, an orientation consistent with their predicted topology in cells (Figure 2A), since CFE reactions were encapsulated in vesicles.
FIGURE 2:
Pkd2 reconstituted in GUVs responds to osmotic pressure. (A) Schematic illustrating the directional insertion of cell-free expressed Pkd2 protein into the lipid bilayer membrane of SUPER templated beads and GUVs. Pkd2 channels are oriented with their C-termini protruding away from the lipid-coated beads and with their C-termini pointing inside the vesicles. (B) Schematic illustrates the formation of vesicles encapsulating cell-free expressed proteins using cDICE, followed by applying osmotic shock to Pkd2-containing GUVs. Vesicles were formed under isoosmotic conditions and then milli-Q water was added to the outer solution to create a hypoosmotic environment. A 100-mM CaCl2 stock solution was added to the hypoosmotic external solution to a final concentration of 10 mM. (C) Representative fluorescence micrographs of vesicles encapsulating 1-mM EGTA and cell-free expressed Pkd2 and G-GECO at t = 10 min after osmotic shock. Plasmid concentrations of Pkd2 and G-GECO were fixed at 1 nM. The final concentration of Ca2+ in the hypoosmotic external solution was 10 mM. The aqueous external solution was made by diluting the external solution stock (HEPES: MgCl2: KCl: glucose [in mM] = 15:3:150:50) with milli-Q water. The osmolarity difference between isoosmotic and hypoosmotic solutions was kept at 100 mOsm. (D) Representative fluorescence micrographs of vesicles expressing Pkd2 and G-GECO with addition of GdCl3 for blocking the force-activated function of Pkd2 channels under osmotic shock. GdCl3 was added to the outer solution with the final concentration fixed at 1 mM. The same method and solutions/conditions described in C was used to apply osmotic presure to the vesicles. The images were taken 15 min after the application of osmotic shock. Vesicles expressing Pkd2 and G-GECO without the addition of GdCl3 served as a control. (E) Representative fluorescence micrographs of vesicles encapsulating cell-free expressed Pkd2 and G-GECO under different hypoosmotic environments at specified time points. The concentrations of EGTA, Pkd2, G-GECO, and Ca2+ were the same as indicated in C. (F) Box plot depicting the fluorescence intensities of vesicles under different osmotic conditions and at different times. At least 30 vesicles were analyzed for each condition. All experiments were repeated three times under identical conditions. Scale bars: 10 µm. *** p < 0.001.
We next determined whether reconstituted Pkd2 alone is calcium-permissive. We encapsulated CFE-expressed Pkd2 in GUVs generated using continuous droplet interface crossing encapsulation (cDICE) (Bashirzadeh et al., 2021; Van de Cauter et al., 2021) and monitored calcium entry into GUVs (Figure 2B). A genetically encoded calcium fluorescent reporter, G-GECO, was used to detect calcium entry into vesicles (Majumder et al., 2017). There was no G-GECO fluorescence in GUVs under isoosmotic conditions with or without Pkd2 expression (Figure 2C), indicating that CFE-expressed Pkd2 is mostly nonpermeable to calcium without a stimulus.
To determine whether Pkd2 can become calcium-permeable upon mechanical stimulus, we stretched the membrane of Pkd2-expressing GUVs with hypoosmotic shock. We added water to the external solution of the GUVs. The G-GECO fluorescence in Pkd2-embedded GUVs gradually increased under a hypoosmotic condition of 100 mOsm (Figure 2C). In comparison, the fluorescence changed very little under the similar condition in those GUVs without Pkd2 (Figure 2C). The fluorescence increase was proportional to calcium concentrations in the external solution (Supplemental Figure S4). When gadolinium chloride (GdCl3), a nonspecific stretch-activated ion channel blocker, was added to the external solution, it blocked the fluorescence increase of G-GECO inside those Pkd2-expressing GUVs under the same conditions (Figure 2D). As expected, the peak fluorescence intensities of GUVs increased proportionally to the strength (40–100 mOsm) and duration (0–20 min) of hypoosmotic shock (Figure 2, E and F). We concluded that Pkd2 is calcium-permeable under the mechanical stimulus of membrane stretching.
Intracellular calcium level was lower in pkd2 mutants
Since the human polycystin PC-2 is found in the ER (Cai et al., 1999), we first determined whether Pkd2 also localizes to the fission yeast ER. We examined the cells expressing both Pkd2-GFP and the ER marker mCherry-ADEL with fluorescence microscopy (Supplemental Figure S5A). Although both fluorescence proteins localized to the cell periphery, this is most likely due to Pkd2 localizing to the plasma membrane (Morris et al., 2019), which is separated by less than 100 nm from the yeast cortical ER (Zhang et al., 2010). However, Pkd2-GFP localized to the equatorial plane during cytokinesis, while the ER marker did not. Moreover, Pkd2-GFP was absent from the perinuclear ER and the ER tethers, both of which were marked by mCherry-ADEL (Supplemental Figure S5A). We concluded that the calcium-permeable Pkd2 primarily localizes to the plasma membrane.
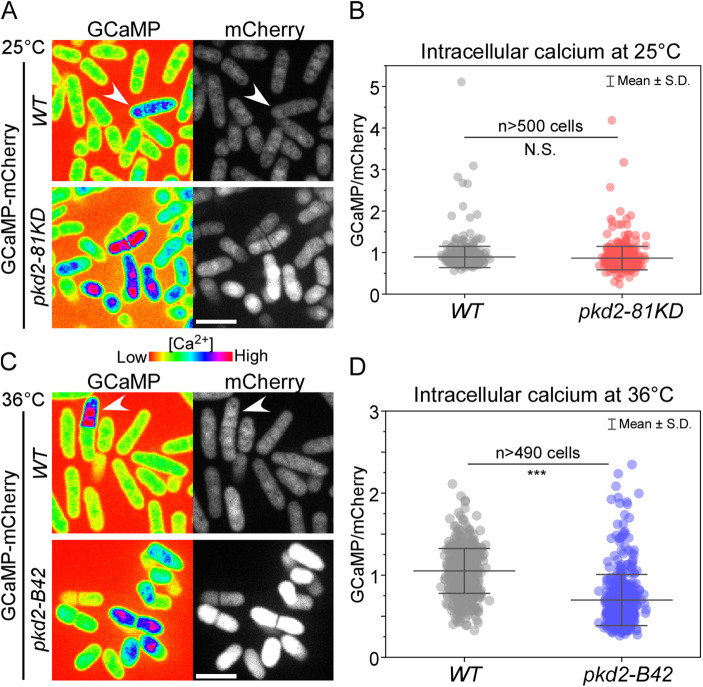
To determine if the plasma membrane–bound Pkd2 regulates calcium homeostasis in fission yeast cells, we measured the calcium level of pkd2-81KD, a hypomorphic mutant with growth and cytokinesis defects even at the permissive temperature (Morris et al., 2019). We employed the ratiometric indicator GCaMP-mCherry to measure the intracellular calcium level of single cells (Poddar et al., 2021) by quantifying the ratio of the fluorescence intensity of GCaMP to that of mCherry (Figure 3A). At 25°C, the intracellular calcium level of mutant cells (n > 500 cells) did not change significantly from that of wild type cells (Figure 3B). The pkd2 mutant cells often lost their volume temporarily in a phenotype called deflation (Morris et al., 2019; Sinha et al., 2022). However, the calcium levels of deflated and nondeflated pkd2-81KD cells were not statistically different. Next, we measured the calcium level of a novel temperature-sensitive pkd2 mutant at the restrictive temperature (Sinha et al., 2022). At 36°C, the average calcium level of pkd2-B42 cells was 34% lower than that of wild type cells (Figure 3, C and D). To rule out the possibility that reduced calcium concentration was an indirect result of either cytokinesis or growth defects of the pkd2 mutant, we examined two other temperature-sensitive mutants, sid2-250 and orb6-25. The former fails in cytokinesis, and the latter is defective in cell growth (Balasubramanian et al., 1998; Verde et al., 1998). In comparison to pkd2-B42, the intracellular calcium concentration of the sid2-250 mutant cells was only slightly lower (by 17%) than that of wild type cells at 36°C (Supplemental Figure S5, B and C). This was despite their much stronger cytokinesis defect than that of pkd2-B42, evident in the substantially increased cell length (Supplemental Figure S5B). In comparison to the wild type, orb6-25 almost doubled its intracellular calcium level, with a far higher number of cells exhibiting elevated calcium concentrations (Supplemental Figure S5, B and C). Last, we determined whether over-expression of Pkd2 interfered with intracellular calcium regulation. We overexpressed Pkd2 by replacing its endogenous promoter with a strong inducible promoter, 3nmt1 (Maundrell, 1990). Under the inducing condition, intracellular calcium levels of mutant cells were similar to those of wild type cells (Supplemental Figure S5, D and E). Therefore, overexpression of Pkd2 alone is not sufficient to alter intracellular calcium levels. Overall, we concluded that Pkd2 plays an important role in maintaining intracellular calcium levels.
FIGURE 3:
Intracellular calcium level was reduced in pkd2 mutant cells. The intracellular calcium level of wild type (WT) and pkd2 mutant cells at either the permissive temperature of 25°C, A and B, or the restrictive temperature of 36°C, C and D. (A, C) Representative fluorescence micrographs of wild type (top) and pkd2 mutant (bottom) cells expressing GCaMP-mCherry. Arrowhead: a cell with elevated calcium level. (B, D) Scatter interval plots of intracellular calcium level of the wild type and the pkd2 mutant cells. The average calcium level of pkd2-81KD cells was not significantly different from that of wild type cells (p > 0.05). In contrast, the average calcium level of pkd2-B42 cells was 34% lower than that of the wild type at 36°C. All data are pooled from three biological repeats. N.S.: no significant difference (p > 0.05), *** p < 0.001. Two-tailed Student’s t tests with unequal variance were used. Scale bars: 10 µm.
Osmotic shock–induced calcium spikes were reduced in pkd2 mutants
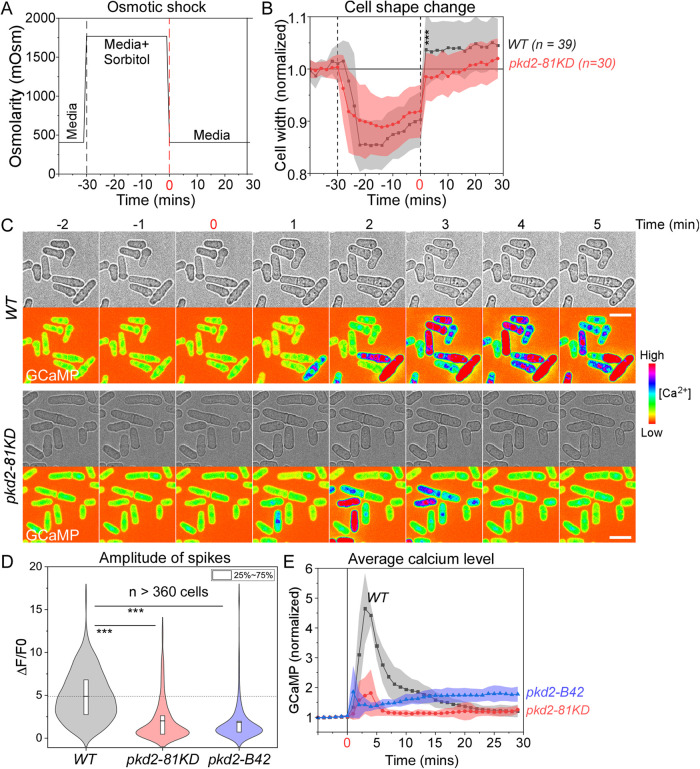
We next examined whether Pkd2 promotes calcium spikes triggered by plasma membrane stretching in vivo. In yeast, an abrupt drop in extracellular osmolarity triggers a sharp increase in intracellular calcium levels (Batiza et al., 1996). Such calcium spikes, accompanied by cell volume expansion, raise the intracellular calcium level by up to 5-folds (Poddar et al., 2021). These spikes can be captured at the single-cell level in a microfluidic device.
We first determined whether such calcium spikes depend on influx from the medium, a process that plasma membrane–localized Pkd2 likely regulates. We trapped the wild type cells in a microfluidic imaging chamber infused with the isosmotic EMM media (Figure 4A; Supplemental Figure S6A). After switching to EMM plus 1.2 M sorbitol for 30 min, we dropped the extracellular osmolarity more than 1300 mOsm by switching back to EMM (Figure 4A; Supplemental Figure S6A). This shock caused the average width of wild type cells to increase significantly (Figure 4B). As expected, this was accompanied by calcium spikes (Figure 4C). In comparison, removing calcium from EMM media during hypoosmotic shock reduced average amplitude of calcium spikes by 40% (Supplemental Figure S6, C, E, and F). Similarly, 2 mM EGTA in EMM media resulted in a 52% decrease in the amplitude of calcium spikes (Supplemental Figure S6, D–F) as compared to the wild type (Supplemental Figure S6, B, E and F). We concluded that extracellular calcium contributes significantly to calcium spikes induced by hypoosmotic shock.
FIGURE 4:
Pkd2 mutations reduced calcium spikes triggered by hypoosmotic shock. (A) Time course of the osmolarity of the extracellular environment in the microfluidics chamber. Time zero: application of hypoosmotic shock through replacing EMM plus 1.2-M sorbitol with EMM media. (B) Time course of the cell width changes during the hypoosmotic shock. Cloud represents standard deviations. Both wild type (WT) and pkd2-81KD cells expanded their cell width significantly after shock, but the wild type expanded more than the pkd2-81KD mutant. (C) Time-lapse micrographs of wild type and pkd2-81KD cells expressing GCaMP. The hypoosmotic shock triggered calcium spikes. (D) Violin plot comparing calcium spikes amplitude of wild type, pkd2-81KD, and pkd2-B42 cells. (E) Time course of normalized GCaMP fluorescence during hypoosmotic shock. Cloud represents standard deviations. All data are pooled from at least three biological repeats. *** p < 0.001. Two-tailed Student’s t tests with unequal variance were used. Scale bars: 10 µm.
We then determined whether Pkd2 contributes to osmotic shock–induced calcium spikes. We measured the spikes in pkd2-81KD mutant cells stimulated with hypoosmotic shock. Like wild type cells, pkd2-81KD mutant cells expanded their width after shock, but comparably less (Figure 4B). Peak amplitude of the calcium spikes in pkd2-81KD cells was 59% lower than in wild type cells (Figure 4, C and D). The peak amplitude of the calcium spikes in pkd2-B42 cells was similarly reduced by 62% (Figure 4D). Additionally, following the shock, the calcium level in pkd2-81KD returned to baseline sooner than the wild type (Figure 4E). We concluded that Pkd2 contributes significantly to calcium influx triggered by hypoosmotic shock in vivo.
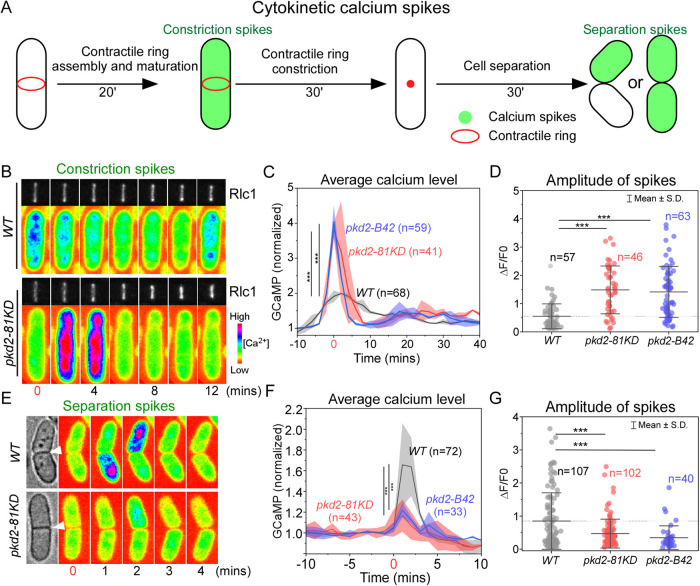
Pkd2 mutations reduced the separation calcium spikes during cytokinesis
Besides the stress-induced calcium transients, two separate calcium spikes accompany fission yeast cytokinesis (Poddar et al., 2021). The constriction spike is concurrent with the cleavage furrow ingression, during which mechanical force by the actomyosin contractile ring compresses the plasma membrane (Figure 5A). The other, termed the separation spike, is accompanied by cell separation, a process requiring stretching of the plasma membrane (Figure 5A).
FIGURE 5:
Pkd2 mutations reduced the separation calcium spikes. (A) Diagram of the time course of cytokinetic calcium spikes in fission yeast cells. (B–D) Mutations of pkd2 enhanced the constriction calcium spikes. (B) Time-lapse micrographs of a wild type (WT) and a pkd2-81KD cell expressing both GCaMP (pseudo-colored) and Rlc1-tdtomato (gray). Number: the time after the start of the contractile ring constriction. (C) Time courses of the average GCaMP fluorescence of the wild type and two pkd2 mutants (pkd2-81KD and pkd2-B42) cells throughout the contractile ring constriction. Cloud represents SD. (D) Dot plot of the peak amplitude of the constriction spikes. Line: average. (E–G) Mutations of pkd2 reduced the separation calcium spikes. (E) Time-lapse micrographs of separating wild type (WT) and pkd2-81KD cells expressing GCaMP (pseudo-colored). Number: the time after the cell separation. Arrowhead: cell separation. (F) Time course of the average GCaMP fluorescence of the wild type and two pkd2 mutants (pkd2-81KD and pkd2-B42) cells. Cloud represents SD. (G) Dot plot of the peak amplitude of the separation calcium spikes. Line: average. All the data are pooled from at least three independent biological repeats. *** p < 0.001 (Two-tailed Student’s t test).
First, we determined whether Pkd2 contributes to the constriction spike by comparing such spikes in the pkd2 mutant cells with those in the wild type cells. We carried out time-lapse calcium imaging in cells expressing the actomyosin contractile ring marker Rlc1-Tdtomato together with GCaMP throughout the ring assembly and constriction (Figure 5B). As we have shown before (Poddar et al., 2021), the GCaMP fluorescence of the wild type cells increased by an average of twofold within 4 min after the start of ring constriction. In comparison, the fluorescence increased by more than 4-fold in either pkd2-81KD or pkd2-B42 cells following ring constriction (Figure 5C). Next, we extracted individual calcium spikes using computer-assisted image analysis and quantified their peak amplitudes (see Materials and Methods). The average peak amplitude of constriction spikes in the pkd2 mutant cells increased more than 2.5-fold, compared to those of the wild type cells (Figure 5D). We conclude that Pkd2 is not essential for the constriction spikes during cytokinesis.
Next, we determined whether Pkd2 contributes to the separation spike. We applied similar methods to compare the separation spikes of the pkd2 mutant cells with those of the wild type. We tracked the last step of cytokinesis, when two daughter cells physically separate and their new ends emerge, through bright-field microscopy coupled with calcium imaging (Figure 5E). Consistent with our previous observations (Poddar et al., 2021), the GCaMP fluorescence of the wild type cells increased ∼1.7-fold within 2 min of cell separation in either both daughter cells or just one. In comparison, the fluorescence increased only 1.2-fold in either pkd2-81KD or pkd2-B42 cells (with ∼30% reduction; Figure 5F). Next, we extracted individual calcium spikes in these separating cells. The average peak amplitude of the separation spikes of pkd2-81KD and pkd2-B42 mutant cells decreased by 45% and 60%, respectively, compared with those of the wild-type cells (Figure 5G). We conclude that Pkd2 contributes significantly to the separation calcium spikes at the end of cytokinesis.
DISCUSSION
In this study, we determined the calcium permeability and activation mechanism of the putative fission yeast channel Pkd2. In vitro reconstitution established Pkd2 as calcium-permeable under membrane tension. Calcium imaging of pkd2 mutant cells demonstrated this essential protein’s critical role in regulating calcium homeostasis and adaptation to hypoosmotic shock. Most importantly, our results identified Pkd2 as the first channel that promotes the calcium transients during fission yeast cytokinesis.
In vitro reconstituted Pkd2 in GUVs allowed the passage of calcium ions in a force-dependent manner. Mechanical activation of Pkd2 was proportional to the extent of membrane stretching, suggesting that only a small fraction of the transmembrane protein was activated at the lower applied force. Compared with calcium spikes in the yeast cells, the calcium influx mediated by Pkd2 in vitro was relatively slow. This is likely due to the small amount of reconstituted Pkd2 in the membrane, which slows the calcium influx. Nevertheless, our minimal system demonstrated that Pkd2 is calcium-permeable in response to membrane stretching.
The calcium permeability of reconstituted Pkd2 is consistent with the significantly reduced intracellular level of calcium in pkd2 mutant cells. Our in vivo data have provided the first line of evidence that Pkd2 mediates calcium homeostasis in fission yeast cells. Neither cytokinesis nor cell growth defect can account for the drastically reduced intracellular calcium concentration of the pkd2-B42 cells. The cytokinesis mutant sid2-250 exhibited only slightly reduced calcium concentration, likely due to its frequent lysis (Jin et al., 2006) that could lead to calcium leakage from the cytoplasm. In contrast, pkd2-B42 mutant cells do not lyse (Sinha et al., 2022). The growth mutant orb6-25 substantially increased the intracellular calcium concentration, most likely because it inhibits the cell volume expansion without affecting the calcium influx. It remains to be determined whether fission yeast Pkd2 is also permeable to other cations such as potassium, sodium, and organic cations similar to human polycystins (Anyatonwu and Ehrlich, 2005; Kleene and Kleene, 2017; Liu et al., 2018; Ha et al., 2020).
The calcium spikes triggered by hypoosmotic shock likely come from calcium influx and internal release. Removal of extracellular calcium did not quench calcium spikes completely, suggesting that the calcium release from the internal sources must contribute to the calcium spikes. These could come from either the ER or vacuoles, the main intracellular calcium storage of yeast cells (Pittman, 2011). Since the yeast cortical ER is adjoined to the plasma membrane, it is possible that a very small fraction of Pkd2 may localize to the ER similar to the human polycystin PC2.
Consistent with the mechanosensitivity in vitro, Pkd2 also plays a critical role in adaptation to hypoosmotic shock when the tension of the plasma membrane increases. There has been a long-standing hypothesis that stretch-activated yeast channels likely contribute to osmotic adaptation, whose identities nevertheless remained unknown (Batiza et al., 1996). Considering its localization on the plasma membrane and its force sensitivity, Pkd2 likely allows the direct influx of calcium that contributes to adaptation after hypoosmotic shock. However, it is worth noting that mutations of pkd2 reduced calcium spikes even more than the removal of external calcium following hypoosmotic shock. This strongly suggests that Pkd2 regulates not only calcium influx, but also internal release of calcium during the spiking events.
The function of Pkd2 in regulating osmotic adaptation bears some similarities to that of mechanosensitive MscS channels, but there are some critical differences. Msy1 and Msy2 are fission yeast homologues of the small bacterial conductance mechanosensitive channel MscS (Nakayama et al., 2012). Like Pkd2, Msy1 and Msy2 play a crucial role in adaptation to hypoosmotic shock. However, unlike Pkd2, they localize to the ER (Nakayama et al., 2012). More surprisingly, deletion of both fission yeast MscS channels leads to enhanced calcium spikes following hypoosmotic shock (Nakayama et al., 2012), contrary to the phenotype of pkd2-81KD mutant cells. The potential interlink between polycystin and MscS channels will require further analysis.
The force-sensitive nature of the putative channel Pkd2, combined with its localization on the plasma membrane, makes it an ideal candidate to sense membrane tension and regulate turgor pressure homeostasis during cell growth. The key phenotype of pkd2 mutants is their failure to maintain turgor pressure required for both tip extension and cell separation (Sinha et al., 2022). This putative channel could play a critical role in maintaining turgor pressure during cell growth as the cell volume expands. Pkd2 is a potential candidate for the known mechanosensitive channel regulating the turgor pressure of fission yeast (Zhou and Kung, 1992).
The reduced separation calcium spikes in the pkd2 mutant cells strongly suggest that the putative Pkd2 channel may contribute directly to the calcium influx during cell separation. The calcium transients in cytokinesis have been observed in both animal embryotic cells and unicellular fission yeast cells (Fluck et al., 1991; Chang and Meng, 1995; Poddar et al., 2021). In both, there are two waves of calcium transients or spikes, one at the time of cleavage furrow ingression and the other following cell separation. However, the source of calcium and the mechanism of such sudden calcium spikes have remained unknown. Most importantly, the potential calcium channels that may have contributed to these cytokinetic calcium transients have not been identified. Our study strongly suggests Pkd2 as a key channel required for this sudden increase of intracellular calcium during cytokinesis. Pkd2 is likely activated by the membrane stretching as a result of the daughter cells increasing their volume and forming the new ends. In contrast to the separation spikes, the constriction spikes increased in the pkd2 mutant cells, suggesting that other channels may contribute to this calcium spike. Nevertheless, the increased constriction spike is consistent with the accelerated ring constriction observed in the pkd2 mutant cells (Morris et al., 2019).
The calcium permeability of Pkd2 is similar to that of mammalian polycystins, but its mechanosensitivity is different. Like Pkd2, human polycystin channels also regulate intracellular calcium levels (Liu et al., 2018; Wang et al., 2019). Moreover, between the two mammalian homologues, polycystin-1 is sensitive to mechanical stimulus (Forman et al., 2005). However, the mammalian polycystin channels mostly localize to primary cilia, where they are activated by mechanical force from fluid flow (Nauli et al., 2003).
Our results support the hypothesis that Pkd2 is a calcium-permissible ion channel activated by membrane stretching. It mediates calcium influx during two distinct biological processes, in response to osmotic shock and in cell separation. Both require temporary increase of cell volume and concurrent plasma membrane stretching. This likely activates the putative Pkd2 channel and allows the calcium influx. The mechanosensitivity of Pkd2 and its calcium permeability may underlie its essential function during both cell growth and cytokinesis, which we discovered in our previous studies (Morris et al., 2019; Sinha et al., 2022). Nevertheless, further single-channel electrophysiology studies will be required to determine the ion channel activity of Pkd2 and its gating mechanism.
MATERIALS AND METHODS
DNA construct
Pkd2-sfGFP was constructed through High-Fi DNA Assembly (NEB). All PCR reactions were carried out with Q5 High-Fidelity DNA Polymerase (NEB #M0491). The cDNA of Pkd2 was amplified from the plasmid Pkd2-EGFP-N1 (Lab stock) using the forward primer AACCCTCAAAAGACAAGACCATGAGGCTTTGGAGAAGCCC and the reverse primer AAGAATTCGTCGACCTCGAGACGAAAAGCATTGTTAGGTA. The vector pAV0714 (Vjestica et al., 2020) was amplified using the forward primer TACCTAACAATGCTTTTCGTCTCGAGGTCGACGAATTCTT and the reverse primer GGGCTTCTCCAAAGCCTCATGGTCTTGTCTTTTGAGGGTT. The PCR products were then digested with DpnI for 1 h at 37°C and purified with Macherey-Nagel NucleoSpin Gel and PCR Clean-Up kit (NC0389463). The purified fragments were assembled through HiFi DNA Assembly (NEB, E2621S) to generate the Pkd2-sfGFP construct (QC-V199).
To generate Pkd2-His6 and Pkd2-sfGFP-His6 for the HeLa CFE reaction, the SUN1FL-His6 construct in pT7-CFE1-Chis (Majumder et al., 2018) was used as a template for Gibson assembly cloning. Initially, Pkd2 was amplified from QC-V199 using the primers Pkd2–Forward: CCACCACCCATATGGGATCCGAATTCATGAGGCTTTGGAGAAGCCC and Pkd2–Reverse: CTCGAGTGCGGCCGCGTCGACTTAACGAAAAGCATTGTTAGGTAATGG with Phusion High-Fidelity DNA Polymerase. The DNA of Pkd2–sfGFP was amplified from QC-V199 using the primers Pkd2–sfGFP–Forward: CACCCATATGGGATCCGAATTCATGAGGCTTTGGAGAAGCCCAC and Pkd2–sfGFP–Reverse: CGAGTGCGGCCGCGTCGACCTTATAAAGCTCGTCCATTCCGTGAG. The next step was to insert Pkd2 or Pkd2-sfGFP into pT7-CFE1-CHis downstream from the T7 promoter construct by replacing SUN1FL with Pkd2 in the pT7-CFE1-CHis construct (Thermo Fisher Scientific). To remove SUN1FL from the pT7-CFE1-SUN1FL-His6 construct (Majumder et al., 2018) as the backbone, we used primers pT7–CFE–Forward: GAATGGACGAGCTTTATAAGGTCGACGCGGCCGCACTC and pT7–CFE–Reverse: GCTTCTCCAAAGCCTCATGAATTCGGATCCCATATGGGTGGTG with Phusion High-Fidelity DNA Polymerase for PCR amplification. Afterward, the resulting PCR products, Pkd2, Pkd2-sfGFP, and pT7CFE-CHis, were digested with DpnI for 1 h at 37°C and subsequently purified with a QIAquick Gel Extraction Kit (Qiagen #28704). They were ligated with homemade Gibson Master Mix (Supplemental Table S1) to create pT7-CFE1-Pkd2-CHis and pT7-CFE1-Pkd2-sfGFP-CHis constructs.
CFE reaction
We used the 1-Step Human Coupled IVT Kit (Thermo Fisher Scientific #88881) to produce Pkd2 protein in vitro. The reaction was carried out based on the manufacturer’s protocol. Briefly, 1 µl plasmid DNA (∼500 ng/µl) was used for one 10-µl reaction. G-GECO plasmid was used in a previous study (Majumder et al., 2017). CFE reactions were carried out at 30°C for 3 h. Pkd2-sfGFP expression was measured on a fluorescence plate reader (Biotek Synergy H1).
SUPER template generation
SUPER templated beads were generated following a published protocol (Neumann et al., 2013). For SUPER template formation, 25 µl of small unilamellar vesicle (SUV) solution was fused with 2 µl of 5-µm silica beads (Bangs Laboratories) in the presence of 1 M NaCl. The final SUPER templated beads were washed with PBS twice by centrifuging at 200 × g for 2 min and then resuspended in 30 µl of milli-Q water at a final concentration of ∼9.6 × 106 beads/ml. The SUPER template stock can be stored at room temperature for 3 h.
For SUV generation, 75% 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC), 24.9% cholesterol, and 0.1% Rhod-PE for a final concentration of 1 mM were mixed and dried under vacuum for 1 h. One milliliter of milli-Q water was then added, and the tube was thoroughly vortexed. The mixture was then passed through a liposome extruder (T&T Scientific, Knoxville, TN) with a 100-nm porous membrane 11 times to generate SUVs.
Vesicle encapsulation system
Vesicles were generated by modifying the continuous droplet interface crossing encapsulation (cDICE) method. The device contains a rotor chamber made with clear resin using a 3D printer (Formlabs) mounted on the servo motor of a benchtop stir plate. The procedure involves an inner solution (IS), outer solution (OS), and lipid-in-oil solution (LOS). HeLa-based CFE reactions with the addition of 5% OptiPrep (to increase the density to aid sedimentation of GUVs) were prepared as the IS. OS stock (115 mM HEPES, 23 mM MgCl2, 1.15 M KCl, 770 mM glucose) was diluted with Milli-Q water to the same osmolarity matching that of the IS. The LOS consists of 40% DOPC, 30% DOPE, 29.9% cholesterol, and 0.1% Rhod-PE in mole percentage with a total lipid concentration of 0.4 mM thoroughly mixed with the desired volume of 1:4 mineral oil:silicone oil by vortexing for at least 10 s. The water-in-oil emulsion was first generated by vigorously pipetting CFE reactions in 500 µl of LOS ∼10 times. Then 700 µl of aqueous OS, 5 ml of LOS, and the water-in-oil emulsion were sequentially added into the cDICE chamber rotating at 700 rpm. After 2 min of rotation, vesicles accumulating in the OS near the chamber wall could be gently collected from the capped hole near the outer edge of the chamber.
Airfuge fractionation assay
After the CFE reaction was completed, it was collected in a 1.5-ml microcentrifuge tube and then mixed well with 30 μl of extraction buffer (20 mM HEPES-KOH, pH 7.5, 45 mM potassium acetate, 45 mM KCl, 1.8 mM magnesium acetate, 1 mM dithiothreitol [DTT]). Then 40 μl of the mixture was transferred to an ultracentrifuge tube and centrifuged at around 100,000 × g for 15 min at room temperature using an airfuge (Beckman Coulter). After the centrifugation, 20 µl of the supernatant was carefully recovered and transferred to a 1.5-ml microcentrifuge tube without disturbing the pellet, and the remaining 20 µl of pellet fraction was resuspended by pipetting up and down to thoroughly mix before transferring to another microcentrifuge tube. The centrifugation cycles mentioned above can be repeated multiple times, as shown in Supplemental Figure S2A. To investigate the protein incorporation, 2 µl of SUPER templated beads were added and incubated with the supernatant and pellet fractions respectively for 30 min at room temperature and then centrifuged at 300 × g for 3 min. After the centrifugation, SUPER templated beads were visible as a small white pellet, and the remaining supernatant was collected as the final pellet fraction. The SUPER template pellets were washed twice with PBS by centrifuging at 200 × g for 2 min and then resuspended in 30 µl of milli-Q water at a final concentration of ∼9.6 × 106 beads/ml. Following the recovery of fractions, the amount of cell-free expressed Pkd2 in each fraction can be determined by visualizing fluorescence proteins on an SDS–PAGE gel.
Pronase digestion assay
Lyophilized S. griseus pronase (Roche) was dissolved in Milli-Q water to a stock concentration of 6 mg/ml and stored at 4°C for a maximum of 3 d. After 1 h of incubation of CFE reactions with SUPER templates, the beads were pelleted by centrifugation at 300 g for 3 min. The supernatant was then gently removed and collected for fluorescence gel imaging. The remaining bead pellets were washed twice with 1 ml of PBS (Ca2+ and Mg2+-free, pH 7.5) by centrifugation at 200 × g for 2 min, followed by resuspension in 20 μl of PBS. Next, 10 μl of the SUPER templated beads in PBS was incubated with 5 μl of pronase stock solution (6 mg/ml) at room temperature for 15 min. The final concentration of pronase was 2 mg/ml, and the other 10 μl of beads were used for observing the protein incorporation as a control. Confocal fluorescence images were taken 15 min after the addition of pronase.
Confocal fluorescence microscopy and in-gel imaging of in vitro reconstituted Pkd2
All images were acquired using an oil immersion 60×/1.4 NA Plan-Apochromat objective with an Olympus IX-81 inverted fluorescence microscope (Olympus, Japan) controlled by MetaMorph software (Molecular Devices, USA) equipped with a CSU-X1 spinning disk confocal head (Yokogawa, Japan), AOTF-controlled solid-state lasers (Andor, Ireland), and an iXON3 EMCCD camera (Andor). Images of sfGFP and lipid fluorescence were acquired with 488-nm laser excitation at an exposure time of 500 ms and with 561-nm laser excitation at an exposure time of 100 ms, respectively. Each acquired image contained ∼5 lipid bilayer vesicles or ∼10 lipid-coated beads that had settled upon a 96-well glass-bottom plate or a coverslip, respectively. Three images were taken at different locations across a well or coverslip for an individual experiment. Three independent repeats were carried out for each experimental condition. Samples were always freshly prepared before each experiment.
FluoroTect Green lysine-tRNA (green lysine) was purchased from Promega. In-gel imaging of Pkd2-sfGFP or Pkd2 was carried out on a Sapphire biomolecular imager (Azure Biosystems). Samples were not heated to retain in-gel sfGFP and green lysine fluorescence.
Image analysis
To quantify the fluorescence inside the lipid bilayer vesicles, all images were analyzed using MATLAB. All data was included for analysis without blinding. Since all the vesicles were labeled with rhodamine PE, the edges/boundaries of vesicles were first detected and isolated, corresponding to the red fluorescence rings using the function “imfindcircles” embedded in MATLAB. Averaged background–intensity measurements were then performed for each image by the average fluorescence (of all pixels), excluding the area of vesicles defined by the code in MATLAB from the previous step. For quantification, the final fluorescence intensity of each vesicle was obtained by averaging the fluorescence of all the pixels inside the vesicles after subtracting the average background intensity. For the box plots marking the first and third quartile and the median in Figure 2F, each data point represents the fluorescence of one vesicle after normalization with respect to the average background subtracted fluorescence intensity of vesicles corresponding to the cell-free expressed proteins at time zero under each condition. Since there are two independent variables, time and osmotic condition/osmolarity, statistical analysis was performed using two-way ANOVA followed by Dunnett’s post-hoc test for all data among all groups throughout the whole experiment. The quantitative data was compared/analyzed between the individual groups at a certain time followed by a two-tailed t-test with a significance level of 0.05. p < 0.05 was considered statistically significant. p values are indicated as * p < 0.05; ** p < 0.01; *** p < 0.001.
Yeast genetics and cell culture
We followed the standard protocols for yeast cell culture and genetics (Moreno et al., 1991). Tetrads were dissected with a Spore+ micromanipulator (Singer, UK). All the fission yeast strains used in this study are listed in Supplemental Table S2.
Microscopy of fission yeast cells
For microscopy, fission yeast cells were first inoculated in a YE5S medium for 2 days at 25°C. Then 1 ml of the exponentially growing cell culture, at a density between 5 × 106/ml and 1.0 × 107/ml, was harvested by centrifugation at 4000 rpm for 1 min. It was washed three times with synthetic EMM medium ([Ca2+] = 107 µM) and resuspended in 1 ml of EMM before proceeding for microscopy. Then 20 µl of the resuspended cells were spotted in a 10-mm Petri dish with a glass coverslip (#1.5) at the bottom (D35-10-1.5N, Cellvis). The coverslip was precoated with 50 µl of 50 µg/ml lectin (Sigma, L2380) and allowed to dry overnight at 4°C. The cells were allowed to attach to the coverslip for 10 min at room temperature before addition of another 2 ml EMM in the Petri dish.
We employed a spinning-disk confocal microscope for fluorescence microscopy using an Olympus IX71 unit equipped with a CSU-X1 spinning-disk unit (Yokogawa, Japan). The motorized stage (ASI, USA) included a Piezo Z Top plate for acquiring Z-series. The images were captured on an EMCCD camera (IXON-897, Andor) controlled by iQ3.0 (Andor). Solid-state lasers of 488 and 561 nm were used at a power of no more than 2.5 mW. Unless specified, we used a 60× objective lens (Olympus, Plan Apochromat, NA = 1.40). A Z-series of eight slices at a spatial distance of 1 µm was captured at each time point. The microscopy was carried out in a designated room maintained at 22 ± 2°C. To minimize environmental variations, we typically imaged both control and experimental groups in randomized order on the same day.
We employed a CellASIC ONIX2 system controlled by a desktop computer through ONIX software (EMD Millipore) to apply osmotic shock. Using a yeast haploid microfluidics plate (Y04C, EMD Millipore), we pushed the cells into the imaging chamber at a pressure of 34–55 kPa for a minimum of 2 min using EMM media. The trapped cells were equilibrated in EMM for 10 min at a pressure of 10 kPa. The same perfusion pressure was applied afterward for the media exchange (EMM, EMM-calcium and EMM+2 mM EGTA).
Calcium imaging of fission yeast cells and data analysis
To measure the intracellular calcium level of single fission yeast cells, we quantified the fluorescence intensity of cells expressing GCaMP-mCherry (Poddar et al., 2021). GCaMP and mCherry fluorescence were calculated from the average intensity projection of Z-series after the background subtraction. To measure the intracellular calcium level, we quantified the average fluorescence intensity on a line drawn along the long axis of a cell. GCaMP-to-mCherry fluorescence ratio gave a more accurate measurement than GCaMP alone for quantifying the calcium levels because it took into consideration the intracellular concentration of the calcium reporter. A small fraction of the pkd2 mutant cells appeared to be deflated temporarily, as we reported previously (Morris et al., 2019; Sinha et al., 2022). They were included in our measurements as well. Whenever a temperature shift was required, cells were imaged at 36°C for 4 h after incubation.
For time-lapse measurement of calcium spikes, we quantified the fluorescence intensity of cells expressing GCaMP. The GCaMP fluorescence was quantified from the average intensity projection of the Z-series on a line along the long axis of a cell throughout osmotic shock. The fluorescence intensities were background-subtracted and normalized to the average value before application of osmotic shock. Amplitudes of a calcium spike were defined as ΔF/F0. ΔF equals Fmax, the maximum value during the first 10 min after osmotic shock, minus the baseline value F0, calculated as the average of the five data points before osmotic shock.
Computation-assisted quantification of cytokinetic calcium spikes
We applied custom-written software to extract the parameters of cytokinetic calcium spikes from time-lapse microscopy. Specifically, the analysis of the separation spikes was performed in two separate segments, before and after cell separation. The first step of this analysis involves removal of background noise to increase the signal to noise ratio (SNR) by preprocessing the image sequences extracted from the videos. The images were passed through a median filter, which smoothed them while preserving their spatiotemporal resolution. The background fluorescence was removed by subtracting the temporal average of each median-filtered pixel value from the median-filtered images at each time point. A binary mask was generated by further subtracting the average pixel value of each frame and binarizing the images using Otsu’s thresholding method. The signal from individual cells were identified by applying the binary mask on the median-filtered background-subtracted images and the average fluorescence intensity value per pixel was calculated for each frame. For postcell separation, the two daughter cells were analyzed separately. The aforementioned steps were also applied on the daughter cells separately, that is, median filtering followed by background subtraction and application of the binary mask. Additionally, we applied a size-based filter to digitally eliminate any other fragments of cells that may be present within the field of view and interfere with the analysis. This filtering was done by detecting the objects (cells) and calculating their area. Any object with size below a preset cutoff was eliminated from Analysis. The cutoff size was manually calculated based on the average of more than 20 different cells.
The time-dependent average fluorescence intensity (per pixel in each frame) was recorded for each cell both before and after the cell separation. Temporal averaging of the signal was performed in order to eliminate any minute fluctuations within the data and intensity values lower than a threshold value was nullified. The locations of the peaks are defined by the point of inversion of the derivative from positive to negative. These locations (frame number/timepoint) and the average fluorescence intensity values corresponding to these time points were recorded and used for analysis.
The temporal average of the baseline fluorescence intensity value (per pixel) was calculated by applying the binary mask on the original images without any preprocessing. The baseline for the parent cell and the two daughter cells were calculated separately. The video processing and analysis was performed by implementing our custom developed code in MATLAB (MathWorks, MA).
Supplementary Material
Acknowledgments
This work has been supported by National Institutes of Health Grants R21GM134167 and R01EB030031 to AL. It has also been supported by the National Science Foundation Grant 2144701 and the National Institutes of Health Grants R15GM134496 and R01GM144652 to QC. FZ has been supported by the University of Toledo Undergraduate Summer Research and Creative Activities Program. We thank the Chen lab members for their technical support. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare no competing interests.
Abbreviations used:
- ADPKD
autosomal dominant polycystic kidney disease
- cDICE
continuous Droplet Interface Crossing Encapsulation
- CFE
cell-free expression
- ER
endoplasmic reticulum
- GUV
giant unilamellar vesicle
- sfGFP
superfold GFP
- SIN
septation initiation network
- SUPER
SUPported lipid bilayers with Extra Reservoir
- SUV
small unilamellar vesicle.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-07-0248) on October 6, 2022.
REFERENCES
- Anyatonwu GI, Ehrlich BE (2005). Organic cation permeation through the channel formed by polycystin-2. J Biol Chem 280, 29488–29493. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, McCollum D, Chang L, Wong KC, Naqvi NI, He X, Sazer S, Gould KL (1998). Isolation and characterization of new fission yeast cytokinesis mutants. Genetics 149, 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr MM, Sternberg PW (1999). A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature 401, 386–389. [DOI] [PubMed] [Google Scholar]
- Bashirzadeh Y, Wubshet N, Litschel T, Schwille P, Liu AP (2021). Rapid encapsulation of reconstituted cytoskeleton inside giant unilamellar vesicles. J Vis Exp 177, 63332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batiza AF, Schulz T, Masson PH (1996). Yeast respond to hypotonic shock with a calcium pulse. J Biol Chem 271, 23357–23362. [DOI] [PubMed] [Google Scholar]
- Cai Y, Maeda Y, Cedzich A, Torres VE, Wu G, Hayashi T, Mochizuki T, Park JH, Witzgall R, Somlo S (1999). Identification and characterization of polycystin-2, the PKD2 gene product. J Biol Chem 274, 28557–28565. [DOI] [PubMed] [Google Scholar]
- Chang DC, Meng C (1995). A localized elevation of cytosolic free calcium is associated with cytokinesis in the zebrafish embryo. J Cell Biol 131, 1539–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S (2014). Overview of cell-free protein synthesis: historic landmarks, commercial systems, and expanding applications. Curr Protoc Mol Biol 108, 16 30 11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondapati SK, Kreir M, Quast RB, Wustenhagen DA, Bruggemann A, Fertig N, Kubick S (2014). Membrane assembly of the functional KcsA potassium channel in a vesicle-based eukaryotic cell-free translation system. Biosens Bioelectron 59, 174–183. [DOI] [PubMed] [Google Scholar]
- Fluck RA, Miller AL, Jaffe LF (1991). Slow calcium waves accompany cytokinesis in medaka fish eggs. J Cell Biol 115, 1259–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman JR, Qamar S, Paci E, Sandford RN, Clarke J (2005). The remarkable mechanical strength of polycystin-1 supports a direct role in mechanotransduction. J Mol Biol 349, 861–871. [DOI] [PubMed] [Google Scholar]
- Gao Z, Ruden DM, Lu X (2003). PKD2 cation channel is required for directional sperm movement and male fertility. Curr Biol 13, 2175–2178. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perrett S, Kim K, Ibarra C, Damiano AE, Zotta E, Batelli M, Harris PC, Reisin IL, Arnaout MA, Cantiello HF (2001). Polycystin-2, the protein mutated in autosomal dominant polycystic kidney disease (ADPKD), is a Ca2+-permeable nonselective cation channel. Proc Natl Acad Sci USA 98, 1182–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio NE, Levine MZ, Oza JP (2019). A user’s guide to cell-free protein synthesis. Methods Protoc 2, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha K, Nobuhara M, Wang Q, Walker RV, Qian F, Schartner C, Cao E, Delling M (2020). The heteromeric PC-1/PC-2 polycystin complex is activated by the PC-1 N-terminus. eLife 9, e60684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Diener DR, Mitchell A, Pazour GJ, Witman GB, Rosenbaum JL (2007). Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J Cell Biol 179, 501–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J, Ward CJ, Peral B, Aspinwall R, Clark K, San Millan JL, Gamble V, Harris PC (1995). The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat Genet 10, 151–160. [DOI] [PubMed] [Google Scholar]
- Jia B, Jeon CO (2016). High-throughput recombinant protein expression in Escherichia coli: current status and future perspectives. Open Biol 6, 160196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin QW, Zhou M, Bimbo A, Balasubramanian MK, McCollum D (2006). A role for the septation initiation network in septum assembly revealed by genetic analysis of sid2-250 suppressors. Genetics 172, 2101–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khambhati K, Bhattacharjee G, Gohil N, Braddick D, Kulkarni V, Singh V (2019). Exploring the potential of cell-free protein synthesis for extending the abilities of biological systems. Front Bioeng Biotechnol 7, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene SJ, Kleene NK (2017). The native TRPP2-dependent channel of murine renal primary cilia. American Journal of Physiology. Renal Physiology 312, F96–F108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knol J, Sjollema K, Poolman B (1998). Detergent-mediated reconstitution of membrane proteins. Biochemistry 37, 16410–16415. [DOI] [PubMed] [Google Scholar]
- Laohakunakorn N, Grasemann L, Lavickova B, Michielin G, Shahein A, Swank Z, Maerkl SJ (2020). Bottom-up construction of complex biomolecular systems with cell-free synthetic biology. Front Bioeng Biotechnol 8, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima WC, Vinet A, Pieters J, Cosson P (2014). Role of PKD2 in rheotaxis in Dictyostelium. PLoS One 9, e88682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Vien T, Duan J, Sheu SH, DeCaen PG, Clapham DE (2018). Polycystin-2 is an essential ion channel subunit in the primary cilium of the renal collecting duct epithelium. eLife 7, e33183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y (2017). Cell-free synthetic biology: engineering in an open world. Synth Syst Biotechnol 2, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S, Garamella J, Wang YL, DeNies M, Noireaux V, Liu AP (2017). Cell-sized mechanosensitive and biosensing compartment programmed with DNA. Chem Commun (Camb) 53, 7349–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S, Willey PT, DeNies MS, Liu AP, Luxton GWG (2018). A synthetic biology platform for the reconstitution and mechanistic dissection of LINC complex assembly. J Cell Sci 132, 219451. [DOI] [PubMed] [Google Scholar]
- Maundrell K (1990). nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem 265, 10857–10864. [PubMed] [Google Scholar]
- Mochizuki T, Wu G, Hayashi T, Xenophontos SL, Veldhuisen B, Saris JJ, Reynolds DM, Cai Y, Gabow PA, Pierides A, et al. (1996). PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science (New York, N.Y) 272, 1339–1342. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods in Enzymology 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Morris Z, Sinha D, Poddar A, Morris B, Chen Q (2019). Fission yeast TRP channel Pkd2p localizes to the cleavage furrow and regulates cell separation during cytokinesis. Mol Biol Cell 30, 1791–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama Y, Yoshimura K, Iida H (2012). Organellar mechanosensitive channels in fission yeast regulate the hypo-osmotic shock response. Nature Communications 3, 1020. [DOI] [PubMed] [Google Scholar]
- Nauli SM, Alenghat FJ, Luo Y, Williams E, Vassilev P, Li X, Elia AE, Lu W, Brown EM, Quinn SJ, et al. (2003). Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nature Genetics 33, 129–137. [DOI] [PubMed] [Google Scholar]
- Neumann S, Pucadyil TJ, Schmid SL (2013). Analyzing membrane remodeling and fission using supported bilayers with excess membrane reservoir. Nat Protoc 8, 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CP, Aydar E, Djamgoz MB (2005). A microbial TRP-like polycystic-kidney-disease-related ion channel gene. Biochem J 387, 211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman JK (2011). Vacuolar Ca(2+) uptake. Cell Calcium 50, 139–146. [DOI] [PubMed] [Google Scholar]
- Poddar A, Sidibe O, Ray A, Chen Q (2021). Calcium spikes accompany cleavage furrow ingression and cell separation during fission yeast cytokinesis. Mol Biol Cell 32, 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protchenko O, Rodriguez-Suarez R, Androphy R, Bussey H, Philpott CC (2006). A screen for genes of heme uptake identifies the FLC family required for import of FAD into the endoplasmic reticulum. J Biol Chem 281, 21445–21457. [DOI] [PubMed] [Google Scholar]
- Pucadyil TJ, Schmid SL (2008). Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell 135, 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucadyil TJ, Schmid SL (2010). Supported bilayers with excess membrane reservoir: a template for reconstituting membrane budding and fission. Biophys J 99, 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud JL, Levy D (2003). Reconstitution of membrane proteins into liposomes. Methods Enzymol 372, 65–86. [DOI] [PubMed] [Google Scholar]
- Shen PS, Yang X, DeCaen PG, Liu X, Bulkley D, Clapham DE, Cao E (2016). The structure of the polycystic kidney disease channel PKD2 in lipid nanodiscs. Cell 167, 763–773.e711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha D, Ivan D, Gibbs E, Chetluru M, Goss J, Chen Q (2022). Fission yeast polycystin Pkd2p promotes cell size expansion and antagonizes the Hippo-related SIN pathway. J Cell Sci 135, 259046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Cauter L, Fanalista F, van Buren L, De Franceschi N, Godino E, Bouw S, Danelon C, Dekker C, Koenderink GH, Ganzinger KA (2021). Optimized cDICE for efficient reconstitution of biological systems in giant unilamellar vesicles. ACS Synth Biol 10, 1690–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Wiley DJ, Nurse P (1998). Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc Natl Acad Sci USA 95, 7526–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vjestica A, Marek M, Nkosi PJ, Merlini L, Liu G, Berard M, Billault-Chaumartin I, Martin SG (2020). A toolbox of stable integration vectors in the fission yeast Schizosaccharomyces pombe. J Cell Sci 133, 240754. [DOI] [PubMed] [Google Scholar]
- Wang Z, Ng C, Liu X, Wang Y, Li B, Kashyap P, Chaudhry HA, Castro A, Kalontar EM, Ilyayev L, et al. (2019). The ion channel function of polycystin-1 in the polycystin-1/polycystin-2 complex. EMBO Rep 20, e48336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield PT (2015). Overview of the purification of recombinant proteins. Curr Protoc Protein Sci 80, 6 1 1-6 1 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Cramer WA, Peterson AA, Hermodson M, Montecucco C (1988). Dynamic properties of membrane proteins: reversible insertion into membrane vesicles of a colicin E1 channel-forming peptide. Proc Natl Acad Sci USA 85, 7531–7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Vjestica A, Oliferenko S (2010). The cortical ER network limits the permissive zone for actomyosin ring assembly. Curr Biol 20, 1029–1034. [DOI] [PubMed] [Google Scholar]
- Zhou XL, Kung C (1992). A mechanosensitive ion channel in Schizosaccharomyces pombe. EMBO J 11, 2869–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.