Summary
Acinar cells are the principal secretory unit of multiple exocrine organs. A single cell layered, lumenized acinus forms from a large cohort of epithelial progenitors that must initiate and coordinate three cellular programs of acinar specification, namely, lineage progression, secretion, and polarization. Despite this well-known outcome, the mechanism(s) regulating these complex programs are unknown. Here, we demonstrate that neuronal-epithelial cross-talk drives acinar specification through neuregulin (NRG1)-ERBB3-mTORC2 signaling. Using single-cell and global RNA-sequencing of developing murine salivary glands, we identified NRG1-ERBB3 to precisely overlap with acinar specification during gland development. Genetic deletion of Erbb3 prevented cell lineage progression and the establishment of lumenized, secretory acini. Conversely, NRG1 treatment of isolated epithelia was sufficient to recapitulate the development of secretory acini. Mechanistically, we found NRG1-ERBB3 regulates each developmental program through an mTORC2 signaling pathway. Thus, we reveal a neuronal-epithelial (NRG1/ERBB3/mTORC2) mechanism to orchestrate the creation of functional acini.
Keywords: ERBB3, mTOR, neuregulin, neuronal-epithelial communication, organogenesis, specification, secretory, acinus
Graphical Abstract

eTOC Blurb
May and Mattingly et al. demonstrate that nerve-derived NRG1 activates ERBB3 enriched on acinar progenitors to drive acinar specification during salivary gland development. Specifically, they show that NRG1-ERBB3 regulates acinar lineage progression, polarization and secretion via mTORC2 to orchestrate the creation of functional acini.
Introduction
Exocrine organs such as the pancreas, lacrimal, mammary and salivary glands, consist of a diverse range of cell types that are patterned to synthesize, secrete and transport a variety of essential proteins. These cells act in a multitude of functions, from protecting the host against foreign microbes to enabling food digestion and tissue maintenance1. Acinar cells are one of the major cell types within most exocrine tissues and are a paradigmatic example of a polarized, secretory cell. Alignment of acinar cell architecture creates a highly organized single layered epithelium, enabling release of secretory products into a central lumen (Figure 1A). For acini to obtain this unique function and architecture, gene signatures that drive acinar cell lineage progression, secretory function and polarization programs must be established to guide a progenitor through the developmental stages of acinar differentiation and, ultimately, functional maturation.
Figure 1. The spatiotemporal dynamics of ERBB3 expression correlates with acinar cell programming.
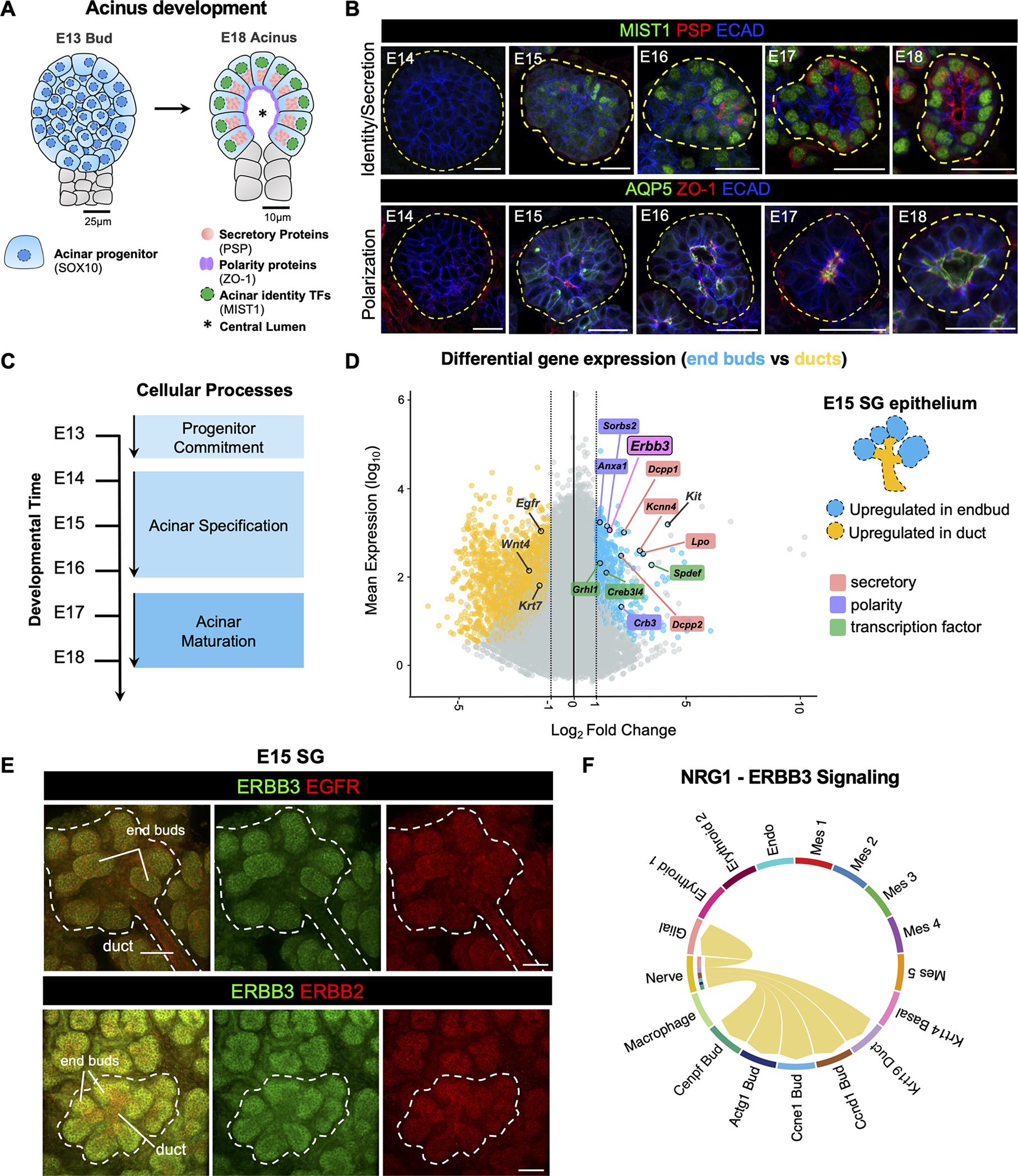
(A) Schematic of SG acinus during embryonic development. (B) Immunostaining of SG E14-E18 end buds for MIST1, PSP, ECAD, AQP5, and ZO1. End buds (E14) and acini (E15-E18) are outlined with dashed lines. Scale bar = 25μm. (C) Embryonic timeline of acinar cell processes as defined by expression of gene signatures and protein. (D) MA plot displaying relative gene expression of selected TFs (green), secretory (red), polarity (purple) genes and Kit (grey) enriched in KIT+ acinar end bud progenitors (blue points) vs. enrichment of duct genes (grey) in KIT− duct cells (gold points) in E15 SGs. (E) E15 SGs immunostained for ERBB3 (green) and EGFR (red, top) and ERBB2 (red, bottom). (F) Chord diagram showing Nrg1-Erbb3 ligand receptor interactions from nerves to epithelial cells and glial cells, determined by E14 SG scRNAseq.
In many secretory organs2–4, acinar cells are formed during development from a distal pool of multipotent epithelial progenitors (termed end bud, tip or cap cells) expressing the transcription factor (TF) SOX10 (Figure 1A). Using the murine salivary gland (SG) as the model system, we previously showed that commitment of SOX10+ progenitors, often termed proacinar progenitors, to the acinar lineage starts several days after organ initiation (i.e. embryonic day (E)11.5)3, and proceeds with the expression of well-known acinar cell markers, such as Aqp55 and Bhlha15/MIST16 during later stages of embryonic development. By the end of the embryonic period, a compact acinus emerges with a well-defined lumen that is outlined by apically restricted proteins, such as tight junctions (e.g., zonula occludens 1 (ZO-1)) and mature secretory proteins (e.g., parotid secretory protein (PSP)) (Figure 1A)7. Although multiple genetic models and ex vivo assays utilizing exocrine organs have revealed relevant growth factor signaling pathways and TFs that regulate epithelial progenitor cell maintenance8–11 and promote acinar cell lineage commitment3,12,13, the signaling pathways that initiate and coordinate the genetic and cellular programs encompassing acinar specification have not been determined.
We addressed this question by first mapping the unique initiation time and progression of acinar secretory, polarity and lineage programs during development (defined as acinar specification). On the molecular level, we linked a neuronal-epithelial signaling pathway, neuregulin (NRG1)-ERBB3, to acinar progenitors that initiates the 3 developmental programs during their specification. Our analyses of conditional Erbb3 knock-out mice and ex vivo culture assays confirmed that the programmatic building of a fully developed acinus is dependent on NRG1 to activate ERBB3 via the downstream mTORC2 pathway and a core gene network regulating all 3 programs, thus, showcasing an important role for neuronal-epithelial crosstalk in acinus formation.
Results
Acinar specification gene signatures follow a spatiotemporal profile during early SG morphogenesis
The precise timing of program initiation and progression during acinus formation has not been resolved. We first mapped the unique spatiotemporal profile of the 3 specification programs during murine SG development through detection of major regulators of acinar cell lineage (TF and master regulator of secretory processes, MIST1/Bhlha1514), cell polarity (tight junction protein ZO1), and secretion (water channel AQP5) and function (secreted protein PSP15), (Figure 1A–B). As shown in Figure 1B, E14 end buds were devoid of all 4 protein types, however, by E15, cells expressing AQP5, ZO1 and MIST1 were visible, suggesting the E14-E15 transition is heavily associated with the initiation of all 3 programs. The next phase, E15 to E16, was tightly associated with cell lineage progression and acinus polarization, as shown by the increase in the number of end bud cells expressing MIST1, and both AQP5 and ZO1 transitioning from global membrane expression at E15, to apical expression at E16, followed by total restriction to the luminal surface by E17/E18 (Figure 1B). Although the secretory program was active by E15 (as noted by expression of MIST1 and AQP5), acinar progenitors did not begin producing the secreted protein PSP until E16, suggesting the E15 to E16 transition is associated with the initiation of secretory protein biosynthesis, which precedes final secretory cell maturation.
To further define gene signature cohorts encompassing the various programs of acinus formation, and the precise timing at which they initiate, we performed bulk RNA-sequencing of embryonic SGs in 24-hour intervals between E13 and E18 (Figure 1B,C, Data S1). Major transcriptional differences were identified between E14 and E18, as seen by principal component analysis (PCA) (Figure S1A), with the most extensively altered gene cohorts occurring between E14-E15 and E15-E16. Indeed, hierarchical gene clustering confirmed two main clusters of genes being upregulated at E14-E15 (Figure S1B, Cluster A, Data S1) and E16 (Figure S1B, Cluster B, Data S1). Gene Ontology (GO) analysis indicated genes upregulated at E14-E15 (Cluster A) to be involved with structural development and early acinar cell lineage, while genes increasing expression at E16 (Cluster B) were associated with transport processes (Figure S1C). Consistent with this notion, manual exploration of genes known for their involvement in cell polarity (Sorbs2, Cldn4/8/10, and Crb3) and acinar cell lineage (TFs: E2f1, Grhl1/2, Klf5, Ehf, and Hes1) were upregulated by E14, and very few secretory-related genes (Dccp1 and Kcnn4) were expressed at this time point (Figure S1D). Most acinar secretory protein genes (Aqp5, Ano1, Agr2, Bpifa2, Dcpp2, Tesc, Lpo, Smgc, Muc19, Muc5b), alongside secretory-related TFs (Bhlha15, Spdef, Creb3l1/4, Ascl3), and several polarity genes (Ocln and Cav1) were upregulated at E15. More developed secretory proteins (Pip and Prol1) and mature acinar lineage TFs (Xbp1, Caz1, Etv1) were upregulated later at E16 (Figure S1D). Thus, acinar specification initiates at E14, as indicated by upregulation of gene signatures of acinar cell lineage progression and polarity programs and continues throughout E15-E16 in conjunction with the secretory program (Figure 1C).
ERBB3 is enriched in end bud progenitors during acinar progenitor cell specification
To identify potential signaling regulators that drive acinar specification at our recently defined time-point, we re-evaluated our previously published RNAseq analysis of FACS sorted E15 end bud (KIT+) and duct (KIT−) cells (GSE89896)16. Differential gene expression analysis revealed 535 genes as significantly upregulated in end bud cells (Figure 1D, blue datapoints), whereas 1,807 genes were upregulated in ductal cells (Figure 1D, yellow datapoints, log2foldchange≥1; p adj<0.05, Data S1). Previously defined transcripts for genes of the secretory and polarity programs, as well as acinar progenitor-related TFs were confirmed as upregulated in the end bud population at this time point, in contrast to previously correlated ductal markers, Krt717 and Wnt418 (Figure 1D). Strikingly, the only additional growth factor receptor identified within end bud cells was Erbb3 (Figure 1D). Immunostaining indicated exclusive expression of ERBB3 protein in E15 end buds (Figure 1E), illustrating a unique temporal and spatial overlap with end bud progenitors during acinar specification. Due to the requirement for ERBB3 to heterodimerize with other ERBB3 family members for signal transduction, putative co-receptors were also evaluated. Similar to previous reports19, ERBB3 co-receptors EGFR and ERBB2 were expressed throughout the epithelium, confirming both receptors to be co-expressed with ERBB3 in the E15 end buds (Figure 1E)20. Thus, these data illustrate end buds contain two types of ERBB3 co-receptors, ERBB2 and EGFR, which allow activation of ERBB3 targets and downstream signaling pathways.
Next, to define putative ERBB3 ligand-receptor interactions with progenitors during acinar specification, we applied an in silico bioinformatics approach. Using a recently published E14 SG single-cell RNA-sequencing (scRNAseq) dataset7, we compared gene expression profiles between the 4 end bud sub-populations (Actg1+, Ccne1+, Cenpf+, and Ccnd1+), the Krt14+ basal and Krt19+ differentiated duct population, various mesenchymal and erythroid cell groups, endothelial cells, macrophages, glial cells, and a Tubb3+ nerve population (Figure S1E). Surprisingly, rendering the R program CellChat21 to analyze cell-cell communication patterns, only one ligand for ERBB signaling, Nrg1, was identified (Nrg2 transcripts were not significantly expressed by any cell type) and was solely enriched in the SG nerve population (Figure S1F), a finding consistent with the known neuronal production of NRG122,23. Putative correlations between NRG1 and ERBB3 involved a well-recognized neuronal-glial relationship (ERBB3 is required for Schwann cell survival24), as well as a neuronal-epithelial interaction (Figure 1F). Although Erbb3 transcripts were detected in Krt19 duct cells, little to no protein was observed in ducts (Figure 1E, Figure S1F), indicating that the neuronal NRG1-ERBB3 pathway is uniquely designated towards end bud progenitors.
Based on these data and our previous studies authenticating SG neurons as essential regulators of early epithelial morphogenesis, duct formation and stem cell expansion12,25,26, we hypothesized that a unique NRG1-ERBB3-mediated neuronal-epithelial crosstalk specifically influences acinar specification in end bud progenitors.
ERBB3 is required for initiating and driving the specification of end bud progenitors to establish polarized, secretory acini
Considering the enrichment of ERBB3 in developing acini at this critical time point, we next investigated whether it acts as a regulator of acinar cell lineage progression, polarity and secretory programs through genetic deletion using the Cre/LoxP system. Erbb3 was specifically deleted in Keratin-14 (Krt14)-expressing epithelial cells (these give rise to end bud and duct cells12,27) of Krt14CreERT2/+;Erbb3fl/fl mice at the initiation of SG development (E10.5 and E11.5), an outcome confirmed by the loss of acinar progenitor-related ERBB3 protein at E16 (Figure S2A). Henceforth, Krt14CreERT2+/+;Erbb3fl/fl embryos are referred to as wild-type (WT) and Krt14CreERT2/+;Erbb3fl/fl embryos as Erbb3 knockout (Erbb3 KO).
Analysis of WT and Erbb3 KO glands at the late stage of acinar specification (E16) clearly showed ERBB3 is essential for acinus formation. Although a branched network of end buds and ducts was observed in SG epithelia devoid of epithelial Erbb3, the morphology of the E16 Erbb3 KO glands mimicked that of immature ~E14 controls, as shown by the significant reduction in end bud number (>50%) and an increase in end bud size (>50%) (Figure 2A). Transcriptional profiling of E16 WT and Erbb3 KO tissue was performed to determine alterations in gene signatures of the 3 acinar specification programs. PCA analysis of RNA-Seq data revealed Erbb3 KO SGs to cluster separately from WT tissue (Figure S2B), indicative of significant changes in gene expression upon Erbb3 deletion. Of the 1,452 protein coding genes that were differentially expressed between both groups (log2foldchange≥0.5; p adj<0.05), 737 were decreased, while 715 were increased in Erbb3 KO compared to WT (Figure S2C, Data S2). Out of our defined cohort of genes regulating the acinar secretory program, the majority were dependent on Erbb3 signaling, with transcripts of Bpifa2/PSP, Pip, Smgc, Muc5b, Lpo, Agr2, Muc19, Tesc, and Dcpp2 being significantly reduced and all other genes trending towards lower expression (Figure 2B and Figure S2C–D). Similarly, expression of genes associated with cell polarity were reduced in Erbb3 KO glands, specifically Sorbs2, Cldn8, Cldn10, and Cav1 (Figure 2B and Figure S2C–D). In addition, a substantial number of TFs associated with acinar cell lineage progression were significantly downregulated upon Erbb3 deletion, including TFs modulating early-stages of specification (E14), such as Ehf and Grhl1 (Figure 2B and Figure S2C–D), and those regulating intermediate to late stages from E15 to E16, including Bhlha15/MIST1, Etv1, Creb3l4 and Casz1 (Figure 2B and Figure S2C–D). However, not all TFs were reduced in gene expression. The early-stage TFs E2f17 and Sox103, that are actively expressed before E14, were both significantly increased in the absence of Erbb3 (Figure 2B and Figure S2C–D). Given Sox10 also regulates acinar cell fate3, such an outcome implies that the role of ERBB3 during acinus development lies majorly in the regulation of acinar cell specification through the polarity and secretory program as opposed to the acquisition of the initial cell fate. Finally, we confirmed that there were no significant reductions in E16 ductal-specific genes7 (Figure S2E), indicating the ductal system develops independently of ERBB3.
Figure 2: ERBB3 is required for initiating and driving the specification of end bud progenitors to establish polarized, secretory acini.

(A) Brightfield images of wild-type (WT) and Erbb3 KO SGs at E16. Quantification of end bud diameter (left) and number of end buds per gland (right) (n = 3–5 mice per genotype; Mean ± SD; Student’s t test. *P<0.05, **P<0.01). (B) Heatmaps displaying differential expression of secretory and polarity genes and TFs. Levels of gene expression shown as relative z-score (red = upregulated, blue = downregulated). n=4 SMG+SLG/genotype n=4 mice/genotype. Asterisks indicate significantly differentially expressed genes (log2FoldChange ≥0.5, p adj<0.05). (C) WT and Erbb3 KO SGs immunostained for ECAD, MIST1, and PSP. Acini outlined with dashed lines. Scale bar = 50μm. Quantification of average MIST1+ and PSP+ cells as a percentage of acinar cells in E16 WT and Erbb3 KO SGs. n= 2 SMG+SLG/embryo, n=3–4 mouse embryos. Mean ± SD; Student’s t test. *P<0.05, ***P<0.001. (D) WT and Erbb3 KO SGs immunostained for ECAD, AQP5, and ZO-1. Acini outlined with dashed lines. Representative trace of measured intensity values across an acinus shown by yellow dashed line. Scale bar = 50μm. Graphs depict fluorescence intensity profiles of AQP5 and ZO-1 over trace for E16 WT and Erbb3 KO SGs. (E) WT and Erbb3 KO SGs immunostained for SOX10 and ECAD. Acini outlined with dashed lines. Scale bar = 50μm. Quantification of average SOX10+ cells as a percentage of all end bud cells in E16 WT and Erbb3 KO SGs. n=3–4 SGs per genotype. Mean ± SD; Student’s t test. *P<0.05.
We next confirmed alterations in gene expression at the protein level through immunofluorescent analysis. Consistent with the gene expression data, a dramatic reduction in the number of end bud cells expressing secretory program-related proteins was observed in Erbb3-deficient SGs. This included the intermediate stage TF and master regulator of acinar cell lineage and function Bhlha15/MIST1 (3±3.1% of Erbb3 KO cells compared to 39±6.4% of WT cells (p=0.0003)), and secreted protein Bpifa2/PSP (1±1% of Erbb3 KO end bud cells compared to 18±3.1% of WT end bud cells (p=0.0008)) (Figure 2C), thus indicating a failure in the acquisition of the secretory program. End bud polarization was also extensively disrupted through the ablation of Erbb3. In contrast to the strict deposition of water channel AQP5 and tight junction protein ZO-1 in the apical membrane of E16 WT end bud cells (Figure 2D, arrows in graph), both proteins were diffusely expressed throughout the Erbb3 KO end bud cell membranes (Figure 2D, * in graph), mirroring their cellular location at an earlier developmental time point (E15) (compare to Figure 1B). The immature state of the Erbb3 KO end buds was further supported by the striking increase in Sox10 transcripts (Figure 2B) and the expansion of SOX10+ cells in Erbb3 KO end buds (82±6.8%) compared to WT (65±1.2%, p=0.04, Figure 2E), indicating that end bud cells deficient in Erbb3 retain their initial acinar cell fate but do not enter the multiple programs of specification.
Overall, these results demonstrate that ERBB3 is essential for initiating and driving acinar specification of end bud progenitors to establish polarized, secretory acini.
NRG1 activation of ERBB3 ex vivo is sufficient to recapitulate the development of secretory, polarized acini
To demonstrate the direct involvement of NRG1-ERRB3 signaling in acinar specification, E13 SG epithelia were isolated and cultured in the sole presence of recombinant NRG1 over 4 days (Figure 3A), a timeframe chosen to mirror early to late stages of acinus formation (E13 to E17). Strikingly, NRG1-mediated activation of ERBB3 was sufficient to not only induce epithelial cell survival (as previously shown in the mammary gland28 and intestine29) but to also result in the formation of highly branched epithelia that are architecturally indistinguishable from freshly isolated E17 glands (Figure 3B, Figure S3A). Moreover, the kinetics of NRG1-stimulated specification matched in vivo gland development (Figure 3B). As with differentiating in vivo tissue, end bud expansion (E13+1 day, ~E14) was followed by the expression of secretory markers MIST1 and PSP at E13+2 days mimicking in vivo E15 SGs (Figure 3B). NRG1-mediated culture also induced progression through late acinar specification stages, as shown by enhanced MIST1 and PSP protein expression being similar to that found in vivo (E16–17, Figure 3B). Transcriptomic profiling on 48–96 hours cultures was performed to further investigate NRG1-induced molecular changes. PCA analysis showed each epithelial culture to cluster as separate entities, indicative of distinct gene expression profiles (Figure S3B). Secretory genes Dcpp1, Dcpp2 and Agr2 were the first to show heightened expression with NRG1 stimulation at 48 hours of culture (~E15), while most secretory markers (Kcnn4, Aqp5, Bpifa2, Tesc, Lpo, Smgc, Pip, Prol1) showed extensive upregulation by 72 hours (~E16) and maintained expression up to 96 hours (~E17, Figure 3C). In line with the progression of the acinar cell lineage, NRG1 increased expression of the early (E14) developmental TFs involved in cell specification, namely, Grhl2, Klf5, Ehf, Spdef, Creb3l1 and Crebl4 by 48 hours (Figure 3C). However, most TFs showed reduced expression by 96 hours, indicating early acinar cell lineage-related TFs are upregulated by ERBB3 but not necessarily maintained long-term (Figure 3C). Intermediate (E15) to late (E16) acinar lineage-related TFs Bhlha15, Ascl3, Xbp1, Casz1 and Etv1 were detected by 48 hours in culture, and progressively increased expression up to 96 hours (Figure 3C), confirming NRG1 activates both the acinar secretory and TF programs.
Figure 3. NRG1 activation of ERBB3 is sufficient to recapitulate the development of specified, secretory, polarized acini.

(A) Schematic of E13 SG epithelia dissection and in vitro culture with NRG1. (B) (Left) Schematic of a differentiated acinus showing secretory program proteins MIST1 and PSP. (Right) E13 SG epithelia cultured with NRG1 over 96 hours immunostained for ECAD, MIST1, and PSP. Images of each day in culture are shown with corresponding images of E14-E17 SGs. Scale bar = 50μm. (C) Heatmaps displaying differential expression of secretory genes and TFs in NRG1 epithelia cultures. Levels of gene expression shown as relative z-score (red = upregulated, blue = downregulated). (D) (Left) Schematic of an acinus showing polarity program proteins AQP5 and ZO-1. (Right) Immunostaining of E13 SG epithelia cultured with NRG1 for 96 hours. Images of each day in culture are shown with corresponding images of E14-E17 SGs. Scale bar = 50μm. (E) Immunostaining of E13 SG epithelia cultured with NRG1 for 120 hours and freshly isolated E18 SGs for ECAD, AQP5, ZO-1 and aPKC. Scale bar = 5μm. (F) Cluster map showing master TFs E2f1, Irf6 and Xbp1 and putative downstream genes in the 3 acinar programs. Acini outlined with dashed lines in B, D and E.
As with differentiating in vivo tissue, initiation of microlumen formation and expression of AQP5 and ZO-1 proteins occurred at 48 hours (~E15 in vivo; Figure 3D). This was followed by microlumen fusion at 72 hours (~E16) with the remodeling of epithelial cells to create a single layered acinus by 96 hours (~E17, Figure 3D). Polarity was further confirmed in a longer 120 hour/5 day culture by atypical protein kinase C (aPKC), a serine/threonine kinase essential to establish and maintain apical identity30. aPKC was tightly localized to the apical membranes of NRG1-stimulated epithelial end buds at day 5, phenocopying the polarized architecture of an E18 acinus (Figure 3E). On the transcriptional level, NRG1 also progressively increased expression of all polarity signature genes over time, except for Cldn4 that increased as early as 48 hours (Figure 3C). Thus, NRG1-ERBB3 signaling is able to induce and progress the polarity program during the entire acinar specification stage. Together, these data confirm NRG1 activates the acinar polarity and secretory programs, as well as the TF cluster related to acinar lineage progression, and does so at a rate equivalent to in vivo development.
As ERBB3 is catalytically inactive and requires dimerization to EGFR to enable downstream signaling, we next tested if EGFR signaling alone could drive acinar specification. To do so, we cultured E13 SG epithelia in the presence of heparin binding EGF (HB-EGF), a ligand enriched in developing SG that specifically binds and activates EGFR but not ERBB331. We found HB-EGF to induce epithelial growth and morphogenesis over a 96-hour period (Figure S3C). However, in contrast to NRG1, HB-EGF-treated epithelia underwent ductal elongation but not acinar cell specification, as revealed by unpolarized acini and absence of AQP5 and ZO1 at apical membranes (Figure S3C). Thus, activation of EGFR signaling alone is not sufficient to drive acinar specification.
To further predict what TFs regulate the NRG1-induced genes within the secretory, polarity and cell lineage progression programs, a bioinformatic (gProfiler) approach was taken wherein all upregulated genes (log2foldchange≥0.2) over the E13+4 day culture period were analyzed for TF regulatory binding motifs (Data S3). Three NRG1-mediated TFs, Irf6, E2f1 and Xbp1, were identified to regulate multiple genes in GO clusters related to cell migration, anatomical structure morphogenesis and organ development (Figure S3D). Analysis of genes within our defined acinar specification programs (Figure S1D) predicted the involvement of these 3 TFs in the regulation of acinar cell secretion (e.g. Pip, Dcpp1), polarity (e.g. Cldn10, Ocln) and lineage and function (e.g. Bhlha15, Etv1), as outlined by the mapped network (Figure 3F). Together, our findings suggest that NRG1-ERBB3 signaling is sufficient to drive acinar specification from end bud progenitors by establishing cell lineage progression, secretion, and polarity programs through a master regulatory network of IRF6, E2F1 and XBP1.
NRG1 rapidly upregulates a network of TFs involved in cell differentiation and genes related to mTOR signaling
To identify transcriptional regulators immediately downstream of NRG1-ERBB3 signaling at early stages of acinar cell development, we analyzed E13 and E14 end buds cultured with or without (control) NRG1 for 4 hours via RNAseq (Figure 4A). PCA showed extensive global transcriptome differences between control and NRG1-treated end buds at both time points, indicative of a large alteration in gene expression with NRG1 treatment (Figure S4A). Furthermore, hierarchical gene clustering across samples indicated greater similarity between E13 and E14 NRG1-treated end buds, suggestive of similar gene targets at these time points (Figure 4B, Data S4). However, compared to their individual stage-related control, E13 NRG1-treated glands upregulated 936 more genes than E14 NRG1-treated glands, suggesting ERBB3 signaling has a higher transcriptomic impact on E13 progenitor cells (Figure 4B, p adj<0.05). For further analysis, we extracted the 2,248 genes that were upregulated in both E13 and E14 NRG1-treated glands, as compared to their individual control (Figure 4C, Data S4), and subjected these to PANTHER32 and ChEA333. Through this analysis we identified all TFs within the NRG1-mediated upregulated gene cohort (Data S4) to be associated with Biological Processes of tissue development, cell differentiation and regulation of cell differentiation (Figure 4D). Specifically, TFs related to cell differentiation and regulation of cell differentiation were investigated for their predictive binding motifs (gProfiler). Twelve regulators (E2f1, E2f4, Irx1, Etv4, Etv5, Myc, Egr1, Egr2, Tfap2c, Xbp1, Hey1, and Yy1) generated a network of TFs that were upregulated at 4 hours by NRG1 in both E13 and E14 end buds (Figure 4E). All, except for Irx1 and Yy1, were significantly upregulated above the control (≥log2foldchange 0.5, p adj<0.05, green contour). Several of those master TFs have been linked to the induction of various epithelial cell lineages (E2F434, IRX135, EGR236, TFAP2C37, HEY138, YY139), including acinar cells (E2F140, EGR236, XBP141,42) as well as epithelial cell fate (ETV4, ETV5)43. Analysis of E14 SG single cell populations7 revealed widespread expression of these TFs across all clusters, however enrichment of most TFs was evident in the epithelial end bud populations (Figure S4B). Other highly upregulated and relevant downstream TF targets of the 12 NRG1-mediated regulators included Irf6, a factor we identified through our in silico analysis of long term cultures, as well as Etv1 and Id4, the latter of which is uniquely expressed in E14 end buds (Figure S4C) and has been shown to repress specific lineages to allow initiation of others44,45. In addition to regulating a whole cluster of TFs, four of the 12 NRG1-mediated regulators (E2F1, E2F4, EGR1 and EGR2) also presented binding motifs in genes of the secretory (Aqp5, Kcnn4, Dcpp1/2) and polarity program (Cldn10 and Sorbs2) (Figure 4E), illustrating aspects of both acinar programs are rapidly initiated after NRG1 stimulation.
Figure 4. NRG1 rapidly upregulates a network of transcription factors involved in cell differentiation and genes related to mTOR signaling.

(A) Schematic of E13 and E14 SG epithelia dissection and in vitro culture with NRG1. (B) Hierarchical clustering of differentially expressed genes from E13 and E14 epithelial cultured for 4 hours with NRG1 or without (control = C). Plot highlights upregulated genes (green) and down-regulated genes (red) with NRG1 stimulation. (C) Venn diagram representing upregulated genes in E13 and E14 NRG1-stimulated end buds. (D) GO Biological Processes for all up-regulated TFs in E13 and E14 NRG1 stimulated epithelia. (E) (right) Cluster map of 12 master TFs (highlighted by the red box) with binding motifs in downstream TFs in the 4 hour NRG1 E13/14 cultures. (left) Subset of the master regulator TFs putatively regulating genes in the acinar secretory and polarity programs. (F) GO KEGG results of genes up regulated with NRG1 stimulation in both E13 and E14 epithelia. (G) Cluster map showing E2f1 and Irf6 with downstream targets involved in mTOR signaling. Green icons indicate log2FoldChange≥0.5, padj<0.05 in NRG1 E13/14 samples vs. WT (D-F).
Next, we analyzed the 2,248 genes that were upregulated in response to 4 hours of NRG1 treatment for potential downstream signaling pathways using gProfiler. The NRG1-mediated upregulated genes were primarily related to KEGG pathways of rna transport, cell cycle and metabolism (Figure 4F). Notably, many upregulated targets downstream of NRG1 were clustered to the mTOR pathway. Therefore, we further investigated which NRG1-mediated TFs regulated this network. Strikingly, enrichment analysis predicted both E2F1 and IRF6 to have binding motifs in genes related to the mTOR pathway, such as Deptor, Kras, Pik3r3 and Chuk (Figure 4G). Furthermore, fifty percent (17/34) of E2f1 and Irf6 predicted downstream gene targets were highly upregulated by NRG1 at 4 hours (≥log2foldchange 0.5, green contour, Figure 4B). Thus, based on these data, we conclude that NRG1 rapidly increases genes of a transcriptional master network, including Irf6, E2f1, and Xbp1, related to cell differentiation and that these may regulate specification programs via mTOR signaling.
ERBB3 activates the mTOR pathway during acinar specification
To determine whether ERBB3-mediated mTOR signaling was involved in regulating the secretory, polarity and acinar cell lineage programs, we utilized gene set enrichment analysis (GSEA46) to investigate KEGG-enriched biological pathways related to Erbb3 loss. In E16 Erbb3 KO SGs, multiple pathways were downregulated compared to WT (Figure 5A), including those related to exocrine secretion (Mucin Type O-Glycan Biosynthesis, Salivary Secretion, Calcium Signaling) and polarization (Tight Junction, Regulation of Actin Cytoskeleton). We found a reduction in PI3K-AKT signaling, indicative of Erbb3’s known affinity to bind and recruit PI3K and activate downstream AKT phosphorylation47. Furthermore, the reduction in expression of genes associated with the mTOR signaling pathway suggested that it is via this pathway that PI3K/AKT is signaling in Erbb3-enriched end buds48. In addition, a GSEA enrichment score indicated multiple genes related to mTOR signaling to be downregulated in Erbb3 KO SGs (Figure 5A), including Pik3r3, Mtor and Rictor (Data S2).
Figure 5. ERBB3 activates the mTOR pathway during acinar specification.

(A) KEGG pathways down-regulated in E16 Erbb3 KO SG compared to WT. Data was generated from GSEA analysis from RNASeq data described (Figure 3B) and organized by normalized enrichment score. (B) E15 SG immunostained for pS6K or pAKTSer473 with ECAD. Scale bar = 25μm. (C) E16 WT and Erbb3 KO SG immunostained for ECAD, pAKTSer473, and SOX10. Scale bar = 50μm. Quantification of pAKT+ cells is shown as a percentage of total ECAD+ cells per field of view for each phenotype. n=3–4 SG per genotype. Mean ± SD; Student’s t test (unpaired, two-tailed), ***P<0.001. (D) Schematic representation of NRG1-stimulated E16 SGs. (E) Western blot analysis of ERBB3, 4E-BP1, and AKT, with each protein’s phosphorylated forms in NRG1-stimulated E16 SGs (0–60 minutes). Representative blots are shown. n=4–5 SG per time-point. (F) NRG1-stimulated E16 SGs (15 minutes) immunostained for SOX10 and pAKTSer473. Acini are outlined with dashed lines in B, C and F. Scale bar = 50μm.
As mTOR mediates its functions through the assembly of two structurally distinct multiprotein complexes, mTORC1 and mTORC249, we analyzed E15 end buds for spatial expression of downstream mTORC1 target pS6K50, and mTORC2 activated pAKT (pAKTSer473)51. As shown in Figure 5B, pAKTSer473 was exclusively associated with end buds, whereas pS6K was distributed throughout the epithelium, suggesting that mTORC2 is uniquely activated in developing end buds. Considering this exclusive expression of pAKTSer473, we asked if mTORC2 signaling is downstream of ERBB3 by analyzing pAKTSer473 expression in the E16 Erbb3 KO. As predicted, the number of pAKTSer473-expressing cells in Erbb3 KO end buds decreased by ~80% compared to WT (Figure 5C), confirming the presence of an Erbb3-mediated mTOR pathway in acinar progenitors.
To evaluate ERBB3’s active role in stimulating mTOR, we cultured E16 glands with NRG1 for 0 to 60 minutes (Figure 5D), and quantified changes in the level of mTORC1 target p4E-BP152 and mTORC2 target pAKTSer473. Successful stimulation of ERBB3, as noted by Tyr1289 phosphorylation, occurred within 5 minutes of NRG1 addition (Figure 5E and Supplementary Figure 5A). In addition, levels of both p4E-BP1 and pAKTSer473 significantly increased at 15 minutes and remained activated up to 60 minutes, indicating ERBB3 enables rapid activation of mTORC1 and mTORC2 pathways (Figure 5E–F and Figure S5A). EGFR was also significantly phosphorylated within 15 minutes of ERBB3 stimulation, which attests to an active downstream signaling after ERBB3-EGFR dimerization (Figure S5B).
Overall, these data reveal that NRG1-ERBB3 rapidly activates an mTOR pathway during the specification of acinar progenitors within the end buds.
ERBB3-mediated mTOR signaling is essential for acinar progenitor cell specification
Considering the mTOR pathways are directly activated by ERBB3 signaling, we investigated whether activation of mTOR is necessary for initiating acinar cell specification through chemical inhibition. E13 epithelia were cultured with NRG1 in the presence or absence of mTORC1/2 inhibitor Torin-153 for 2, 3, and 5 days (Figure 6A), and analyzed for changes in gene and protein expression. At 2 days of culture, we found a profound decrease in the number of MIST1+ cells in Torin-1-treated epithelia (>90% reduction) compared to DMSO controls (Figure 6B–C). End buds treated with Torin-1 also contained 50% more SOX10+ cells, suggesting inhibiting mTOR signaling downstream of NRG1 prevents end bud progenitors from initiating the acinar secretory program but they retain their acinar progenitor fate (Figure 6B–C).
Figure 6. ERBB3-mediated mTOR signaling is essential for acinar specification.

(A) Schematic of E13 SG epithelia isolation and culture mTOR inhibitor. (B) 48-hour control and Torin-1 E13 epithelia immunostained for MIST1, pAKTSer473, and SOX10. Acini are outlined with dashed lines. Scale bar = 50μm. (C) Quantification of MIST1+ and SOX10+ cells as a percentage of total epithelial cells per field of view for 48 hours control and Torin-1 epithelia. n=6 rudiments per treatment, averaged from three images. Data are shown as mean ± SD; Student’s t test. **P<0.01, ***P<0.001. (D) Brightfield images of control and Torin-1 E13 epithelia cultured with NRG1 for 120 hours (left, scale bar = 200μm). Cultures were immunostained for ECAD, AQP5, ZO-1, MIST1, and PSP. Scale bar = 50μm. (E) Heatmaps displaying differential expression of acinar secretory program, polarity program and cell lineage TF genes in control and Torin-1 cultured epithelia (72 hours). Levels of gene expression shown as relative z-score (red = upregulated, blue = downregulated). Asterisks indicate significantly differentially expressed genes (log2FoldChange≥0.5, padj<0.05).
Over the next 3 days, end bud formation, maturation of end bud architecture and the synthesis of polarity proteins were drastically reduced in tissue cultured with Torin-1 (Figure 6D). Furthermore, unlike control epithelia which displayed a single layer of acinar cells wrapped around a polarized lumen enriched in apical AQP5 and ZO-1, Torin-1-treated end buds were composed of multiple cell layers showing basolateral expression of AQP5, loss of ZO-1 and the absence of a single contiguous lumen, indicative of impaired morphogenesis (Figure 6D). Although a few MIST1+ cells were visible in Torin-1-treated end buds by 5 days, possibly due to a residual low level of mTOR, there was a severe disruption in secretory cell maturation as shown by the complete absence of PSP (Figure 6D).
RNAseq analysis of epithelia cultured for 3 days further revealed that inhibition of mTOR1/2 signaling by Torin-1 had a profound effect on global gene expression (Figure S6A). A total of 1,578 genes were differentially expressed between the two culture conditions (≥log2foldchange 0.5; p adj< 0.05), with 888 genes showing decreased expression and 690 increased with Torin-1 treatment compared to untreated controls (Figure S6B, Data S5). Moreover, signature genes in all 3 acinar programs were impacted. There was a significant reduction in genes related to secretory function, including Aqp5, Bpifa2, Tesc, Lpo, Smgc, Pip and Prol1 in Torin-1 treated tissue (Figure 6E). The major polarity program associated genes Cldn10 and Crb3 were significantly reduced, and other signature genes showed a trending reduction with Torin-1 compared to control (Figure 6E). Finally, Torin-1 significantly reduced the expression of TFs highly relevant to E15 to E16 acinar lineage progression (Bhlha15, Spdef, Creb3l4, Xbp1, Etv1) compared to controls (Figure 6E). Crucially, these outcomes recapitulate the deficiencies seen in Erbb3 KO SGs, further suggesting that the mTORC1/2 pathways are necessary for acinar cell specification in vivo.
Finally, given the enriched localization of pAKTSer473 in the end buds, and the reduction in pAKTSer473-expressing acinar progenitors in the Erbb3 KO, we next tested whether mTORC2, as opposed to mTORC1, was the primary driver of acinar specification in response to ERBB3 signaling. NRG1-treated epithelia were cultured with the specific mTORC1 inhibitor rapamycin and changes in cell lineage, polarization, and secretory programs were assessed after 5 days. While the overall growth of SG epithelia was reduced with mTORC1 inhibition, acinar specification of rapamycin-treated epithelial end buds resembled those of DMSO controls, with a single layer of MIST1+/PSP+ cells surrounding a polarized lumen enriched in AQP5 and ZO1 (Figure S6C). Thus, these outcomes further support mTORC2 as the downstream regulator of NRG1-ERBB3-mediated acinar specification.
Altogether, these data demonstrate that NRG1-ERBB3 signaling is essential for the development of secretory acini through the regulation of the 3 specification programs and mediates this, in part, through an mTORC2 signaling pathway.
Discussion
Many fundamental questions regarding the mechanisms driving specification of various epithelial cell types and organized tissue architecture are still unanswered, especially that of the secretory acinar cell. Neuronal-epithelial interactions regulate multiple processes including tissue function54,55, stem cell maintenance56,57 and organ regeneration58–60. In the SG, we have previously defined a neuronal-epithelial signaling complex that is required to maintain and expand epithelial progenitors via acetylcholine/CHRM1/EGFR signaling25 and to establish SG tubulogenesis via the VIP/VIPR/cAMP-PKA-dependent pathway26. Here, we show a neuronal-driven mechanism that regulates tissue architecture during later stages of development, this time focused on the initiation and progression of acinar specification in end bud progenitors. Considering widespread NRG1 expression in neuronal populations23 and ERBB3 expression in a variety of epithelial cell types61–63, this nerve-derived signaling may also regulate secretory cell specification in other organs.
The ability of NRG1-ERBB3 signaling to promote the expansion and differentiation of progenitor cells is supported by previous studies showing NRG1 to drive stem cell fate and progenitor proliferation in developing and regenerating epithelial organs, such as the lung64, eye65, intestine61,66 and mammary gland28,67,68. Our study highlights multifunctional roles for NRG1-ERBB3 signaling alone is sufficient to (1) expand and differentiate progenitor cells and (2) promote the extensive reorganization and specification required for glandular function. Cell specific functions of the NRG1-ERBB3 cascade have been elucidated in adult epithelial organs, including the mammary glands where NRG1 activates luminal cell maturation during pregnancy69 and the intestine where NRG1 maintains adult cell stemness in the intestine66. Thus, NRG1-ERBB3 signaling in adult organs contributes to limited outcomes as compared to developing tissues. Furthermore, in these organs, NRG1 has been shown to be derived from non-neuronal cell types i.e., basal cells in the mammary gland69 and mesenchymal cells in the intestine66. This suggests non-neuronal NRG1 is capable of supporting a number of processes in adult organ systems, however highly innervated tissues may also rely on neuronal NRG1 to regulate development, homeostasis and/or regeneration. Further work is needed to understand the specific role of neuronal-derived NRG1 in tissue outcomes. Moreover, whether ERBB3 is limited to specific cell types during development or switches to alternative cell types in the adult needs to be determined.
Several studies report TF regulation downstream of NRG1 in adult organs in endocardium70, ischemic stroke71, brain ischemia72, Schwann cells73, and cancer cells74. However, no studies to date have proposed a role for NRG1 in transcriptionally regulating acinar specification. We report that NRG1 significantly upregulates a cluster of 12 master regulators within 4 hours, all of which are known to be involved in cell differentiation and/or linked to the acinar lineage (e.g., EGR2, XBP1). While many of these may directly or indirectly be involved in initiating the acinar specification programs, 3 TFs (IRF6, E2F1, and XBP1) also aligned as master regulators during 96 hours of culture with NRG1, strengthening their relevance within NRG1-mediated acinar formation. Supporting evidence already indicates that E2f1−/− mice produce less salivary volume, and SGs with fewer acini40. XBP1 plays a crucial role in the development of mature secretory cells of many organs41,75–78, and regulates Bhlha15/MIST179. Lastly, deletion of Irf6 in mice also leads to a significant reduction in SG and pancreas Bhlha15/MIST1 expression, implicating the need for IRF6 in acinar cell lineage commitment80. It is noteworthy that none of those TFs are exclusively regulated by NRG1-ERBB3, but it is plausible that NRG1 enhances their gene expression to activate downstream gene clusters to guide progenitors towards acinar cell specification. It will be of future interest to evaluate whether each TF is essential to induce the acinar programs individually or as a cluster.
Lastly, our study establishes a link between ERBB3 and mTOR in a developmental setting. In disease settings, such as head and neck cancer81, lung adenocarcinomas82, gastric cancer83 and meningiomas84, ERBB3 has been linked to mTOR to induce proliferation. However, here we demonstrate a role for ERBB3-mTOR in driving cell specification and tissue architecture. Specifically, we show that mTOR signaling acts downstream of ERBB3 to orchestrate the specification of the epithelial end bud into a secretory acinus. Furthermore, our findings indicate that specification occurs via mTORC2, rather than mTORC1. mTORC1/2 regulate developmental processes in multiple epithelial organs including the skin85,86, mammary gland87 and prostate88, however, similar to the outcomes shown here, mTORC1 and 2 can mediate distinct functions within the same organ87,89. Whether NRG1-ERBB3 activates mTORC2 exclusively to drive acinar specification or if it pairs with other signaling cascades, remains to be determined.
Thus, altogether we propose an in-depth regulatory network for acinar specification directed by NRG1-ERBB3-mTORC2 that allows for the lineage progression of the epithelial end bud into a secretory acinus.
Limitations of Study
Various transcriptional regulators of the NRG1-ERBB3-mTOR pathway discovered in silico were based on bioinformatic analysis of sequencing data and require further investigation at the functional level. In addition, it is still undetermined whether NRG1-ERBB3 exclusively activates mTORC2 to drive acinar specification or whether other signaling pathways are involved. The in vitro epithelial culture approach used here depletes other potential regulators of acinus development that may act in parallel with NRG1. Future work will also be required to validate findings on the role of neuronal-derived NRG1 in secretory cell specification in other tissues.
STAR Methods section
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sarah M. Knox (sarah.knox@ucsf.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This paper does not report any original code. Additional information on code used to analyze data reported is available from the lead contact upon request. RNA-sequencing data has been deposited in the Gene Expression Omnibus (GEO) database (GSE214976).
Experimental Model and Subject Details
Animals and animal handling
All procedures were approved by the UCSF Institutional Animal Care and Use Committee (IACUC) and adhered to the NIH Guide for the Care and Use of Laboratory Animals. All animals were housed 5 per cage (where possible) in a barrier facility in standard ventilated cages with free access to water and food with a 12-hr light cycle (6 am-6pm). Animals were handled according to an approved institutional animal care and use committee (IACUC) protocol number [AN186940-01]. Breeding was carried out in duos or trios. The Erbb3-floxed (MGI: 3690340)90 mouse line was a gift from Rebecca Cook, Vanderbilt University School of Medicine. The Krt14CreERT2 (MGI: 2177426)91 mouse line was purchased from The Jackson Laboratory. Mouse lines were used as described previously90,91. All transgenic mice were maintained on a C57BL/6 background. E13 to E18 embryos were obtained from outbred wild-type timed pregnant CD1 female mice purchased from Envigo. The embryonic stages used are discussed and detailed in each result description.
For generation of embryonic SG deficient in Erbb3, a Krt14CreERT2fl/+; Erbb3fl/fl male mice were crossed to Erbb3fl/fl female mice and noon of the day when vaginal plugs were first recorded was determined as 0.5 days post coitum (E0.5). Timed pregnant females were administered 11.25 mg/30 g of tamoxifen (Sigma-Aldrich) by oral gavage at E10.5 and E11.5 and were euthanized by CO2 inhalation at E16.
Method Details
Ex vivo SG epithelial rudiment culture
In all ex vivo epithelial rudiment culture experiments, SG were derived from CD1 timed pregnant mice at E13. E13 SG epithelia were freshly isolated as previously described11,12. Briefly, the SG epithelium was separated from the mesenchyme using dispase II (Gibco) treatment followed by mechanical dissection and cultured in 15μl of laminin-1 (R&D Systems) on a nucleopore filter floating above 200 μL of serum-free media (DMEM/F12; Thermo Fisher Scientific) containing penicillin-streptomycin (Gibco), 50μg/ml holo-transferrin (HT), 150μg/ml ascorbic acid (AsA) and 200ng/ml human recombinant NRG1 isoform SMDF (R&D Systems), or 20ng/ml HB-EGF (R&D Systems) for up to 120 h. For inhibitor experiments, epithelia were cultured at 37°C in a humidified 5% CO2/95% air atmosphere in complete media in the presence of 0.2% DMSO (Sigma-Alrdrich), 5μM of mTORC1 inhibitor rapamycin (Biotang Inc.), or 1μM mTORC1/2 inhibitor Torin-1 (Cell Signaling Technologies). In all experiments, media was refreshed daily.
Ex vivo SG organ culture
SG were dissected from E16 wild type (Krt14CreERT2fl/+; Erbb3fl/+) and Erbb3 deficient (Krt14CreERT2fl/+; Erbb3f/fl) embryos and placed directly on a nucleopore filter over serum-free media (DMEM/F12, HT, AsA), as described above. SG explants were left to equilibrate at 37°C in a humidified 5% CO2/95% air atmosphere for 60 minutes before 400 ng/ml NRG1 was added to media. Explants were then cultured for 0–60 minutes at 37°C prior to fixation for immunofluorescent analysis.
Section immunostaining
Intact SG were fixed in 4% paraformaldehyde for 20 minutes, washed 3 times with PBS and then dehydrated by incubating with increasing concentrations of sucrose (12.5–25%) overnight at 4°C. Tissue was embedded in OCT (Tissue-Tek) and sectioned using a Leica cryostat. Cryosections were kept at −20°C until immunofluorescent analysis and OCT tissue blocks were kept at −80°C. For staining, tissue sections were left to equilibrate at room temperature for 10 min and underwent immunostaining as described below.
Whole mount immunostaining
Whole, freshly dissected E13-E18 SGs and cultured epithelia were fixed in 4% paraformaldehyde for 20–30 min followed by permeabilizing with 0.1% Triton X in PBS (Alfa Aesar) for 10 minutes. Tissue was blocked for a minimum of 2 hours or overnight at 4°C with 10% Donkey Serum (Jackson Laboratories), 1% BSA (Sigma-Aldrich), and MOM IgG-blocking reagent (Vector Laboratories) in 0.01% PBS–Tween 20 (Thermo Fisher Scientific). Tissue was incubated with primary antibodies overnight at 4°C (Table S1). The following day, samples were washed 3 times with PBS and subsequently incubated with species specific secondary antibodies (Cy2-, Cy3-, or Cy5-conjugated secondary Fab fragment, Jackson Laboratories) at 1:300 in PBS overnight at 4°C. The following day tissue was washed 3 times with PBS and nuclei stained using Hoechst 33342 (1:1,000, Sigma-Aldrich). Tissues were then mounted on slides using Vectashield (Vector Laboratories) for imaging.
Imaging
Images were captured on an inverted Zeiss LSM 700 or Zeiss LSM 900 confocal microscope. Confocal imaging of whole mount samples or tissue sections was performed using a 10, 40 or 63X oil lens, depending on application. For high magnification acquisitions, images were acquired with pinhole set to approximately 1μm and a 1μm step size was used for Z stack collection. Maximum intensity projections of substacks specifically containing each layer were made using ImageJ.
RNA Sequencing Analysis
For bulk RNA-sequencing experiments, embryonic SG were dissected from CD1 embryos at E13-E18 and from wild type and Krt14CreERT2;Erbb3fl/fl embryos at E16. SG epithelial rudiments were collected on 46, 72, and 96 hour timepoints of ex vivo culture. Each biological replicate consisted of RNA pooled from 10 (E13 sample) and 6 SGs (E14-E18 samples) for developmental time course studies and one SG for E16 Krt14CreERT2;Erbb3fl/fl analysis. Each biological replicate consisted of RNA pooled from 6 epithelial rudiments of ex vivo SG analysis. A minimum of 3 biological replicates for whole SG studies and 2 biological replicates of epithelial studies were included. RNA was purified using the Purelink RNA Mini Kit (Invitrogen) and quantification was performed on an Agilent 2100 Bioanalyzer. RNA was further processed for transcriptome sequencing by Novogene (UC Davis, USA). In short, a total amount of 1μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. PCR products were purified (AMPure XP system) and library quality was assessed on an Agilent 2100 Bioanalyzer. Clustering of samples was performed on a cBot Cluster Generation System using PE Cluster Kit cBot-HS (Illumina) and library preparations were sequenced on an Illumina platform and paired-end reads were generated. Sequencing data were processed, and downstream analysis performed as described below.
Quantification and Statistical Analysis
Statistical analyses were performed using Prism 9 (GraphPad), and statistical details related to tests performed and sample size are specified in the figure legends.
Morphometric analysis and cell counts
For acinar diameter measurements, whole E14 or E16 SGs were immunostained for E-Cadherin (ECAD) and imaged for confocal microscopy using 320μm2, ≥50μm projections assembled from 5μm stacks. Using ImageJ software, a line was drawn across an acinus (determined by morphology) at its largest width perpendicular to its adjacent duct and expressed in micrometers. Two glands from the same embryo were averaged for each data point, with a minimum of 4 acini measured per gland (8 total). For total endbud quantification, whole glands were imaged on a Zeiss Axiovert 200 brightfield microscope and end buds quantified using ImageJ software (NIH).
For fluorescence intensity measurements, tissue sections of E16 SGs were labeled for AQP5, ZO-1, and ECAD and single 160μm2 planes were imaged. Using ImageJ software, a line was drawn across the length of an individual acinus and fluorescence intensity plotted using the “Plot Profile” analysis tool. Intensity measurements of each marker were normalized against the highest intensity data point. Intensity plots were generated in Prism 9 (GraphPad).
For cell counts of SGs, MIST+, PSP+, SOX10+, and pAKT+ cells were quantified using ImageJ software from 320μm2 (for Figure 2) or 101 μm2 (for Figure 5 and 6) images taken from SG tissue sections or whole mounted epithelial rudiments, respectively. Cells were quantified from 6μm projections from combined 2μm stacks for Figure 2, and single slices were used for other quantifications. For Figure 2, ECAD+ cells that were positive for each marker were manually counted and divided by total ECAD+ cells in each field of view using the multi-point tool. For Figure 5 and 6, cells positive for each marker were divided by total number of nuclei in each field of view. At least three independent images were quantified and averaged for each data point.
Analysis of bulk RNA-Seq data
Raw data (raw reads) of FASTQ format were firstly processed through fastp92. In this step, clean data (clean reads) were obtained by removing reads containing adapter and poly-N sequences and reads with low quality from raw data. At the same time, Q20, Q30 and GC content of the clean data were calculated. All the downstream analyses were based on the clean data with high quality. Paired-end clean reads were aligned to the mm9 reference genome using Spliced Transcripts Alignment to a Reference (STAR) software93. FeatureCounts94 was used to count the read numbers mapped of each gene. For downstream analysis, scripts were adapted from Berry et al.95 and Love et al.96. Read counts were filtered and normalized using the R package edgeR97. Hierarchical gene clustering and differential gene expression was analyzed using DESeq296 and limma98. Genes with a defined log2FoldChange and an adjusted p-value<0.05 were determined to be differentially expressed. Panther32 was used to analyze genes within each cluster to identify transcription factors, secretory and polarity genes. Gene Ontology (GO) analysis for Biological Processes were determined by gProfiler99. Heatmaps of differentially expressed gene clusters were visualized using ggplot2100 and pheatmap101 and volcano plots created using EnhancedVolcano102. Gene set enrichment analysis (GSEA) was performed using v4.1.0 of the GSEA desktop application46. Normalized Enrichment Scores (NES) were plotted using Prism 9 (GraphPad).
Transcription factor binding motif analysis
NRG1 upregulated gene lists (Figure 3I) or gProfiler-defined KEGG/GO clusters of genes (Figure 4E–F) were imported into the gProfiler program to identify predicted binding motifs of transcription factors (TFs). Predicted TFs were aligned with the NRG1 upregulated gene list to identify overlapping TFs. The predictive target genes of those NRG1-regulated TFs were then manually evaluated for log2FoldChange expression and/or overlap with genes in the defined acinar programs. In addition, transcription factors were enriched from gene lists (Figure 4E) through analysis of bioinformatical programs Panther, CheA3 and gProfiler. Connections between predictive TFs and their gene target were manually imported into Cytoscape103 to create gene networks.
Analysis of single cell RNA-seq data
Analysis and R scripts used for E14 SG singe cell RNAseq data were adapted from Hauser et al.7. Briefly, filtered gene-barcode matrices were analyzed using the Seurat v4.0104. Seurat objects were generated with CreateSeuratObject (min.cells = 3, min.features = 200) for E14 SG cells. Cells were filtered based on the distribution of number of genes (nFeature, >200,<2500) and percent mitochondrial genes (percent.mito, :>0.5,<5) per cell. Data were normalized for sequencing depth, log-transformed, and multiplied by a scale factor of 10000 using the default parameters of NormalizeData. The top 2000 variable genes within each dataset were selected based on a variance stabilizing transformation (FindVariableGenes, selection.method = ‘vst’) and used in downstream principal component analysis (PCA). Cell clusters were identified by construction of a shared nearest neighbor graph (FindNeighbors) and a modularity optimization-based clustering algorithm (FindClusters) using the PCs determined by PCA (dims = 1:10). Clustering was performed at multiple resolutions between 0.2 and 2, and optimal resolution was determined empirically based on the expression of known population markers and the FindMarkers function (resolution = 0.5). To investigate the SG cells, epithelial cell clusters were determined by expression of EpCAM and EpCAM-negative cell clusters were eliminated. Cells and clustering were visualized using Uniform Manifold Approximation and Projection (UMAP) dimensional reduction (RunUMAP). Markers for each cluster were identified with FindAllMarkers using default parameters, and cluster identity was determined based on the presence of known markers and populations previously described7. For cell-cell ligand-receptor interaction analysis, Seurat object for E14 SG single cell data was analyzed using CellChat21 using standard parameters.
Western Blotting and Analysis
E16 SGs dissected from CD1 timed pregnant female mice were lysed in radioimmunoprecipitation assay buffer (RIPA buffer) containing a cocktail of protease inhibitors. Protein concentration was measured by bicinchoninic acid (BCA) assay (Thermo Fisher Scientific). A quantity of 10μg of protein per samples was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on 4–12% Bis Tris gels (Invitrogen) and transferred to PVDF membranes. Membranes were blocked in 5% milk and incubated in primary antibodies overnight at 4 °C. After washing three times in TBST, the blots were incubated with appropriate horseradish-peroxidase-conjugated secondary antibodies and visualized with the Clarity Max Western ECL chemiluminescence detection system (Biorad). Densitometric quantification of bands was performed using ImageJ software and phospho-protein intensity was normalized by dividing by total non-phosphorylated protein intensity for each blot. Antibodies used can be found in Table S1.
Supplementary Material
Data S1. Enrichment in gene expression in developing SG from E13 to E18 and in epithelial end buds compared to ducts at E15, related to Figure 1
Data S2. Differential Gene Expression Analysis and Gene Set Enrichment Analysis of E16 Erbb3 KO and WT SGs, related to Figures 2 and 5
Data S3. gProfiler analysis of TF regulatory binding motifs of upregulated genes in E13 epithelia treated with NRG1 for 4 days, related to Figure 3
Data S4. Differential Gene Expression Analysis of NRG1 stimulated end buds vs control, related to Figure 4
Data S5. Differential Gene Expression Analysis of NRG1+Torin-1 vs NRG1+DMSO (control) E13 epithelia cultured for 72 hours, related to Figure 6
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-ECADH | Thermo Fisher Scientific | Cat# 13-1900; RRID: AB_2533005 |
| Rabbit anti-AQP5 | Millipore | Cat# AB3559; RRID: AB_2141915 |
| Mouse anti-ZO-1 | Thermo Fisher Scientific | Cat# 339194; RRID: AB_2532188 |
| Rabbit anti-MIST1 | Sigma-Aldrich | Cat# HPA047834; RRID: AB_2680172 |
| Mouse anti-MIST1 | Thermo Fisher Scientific | Cat# MA1-517; RRID: AB_2259158 |
| Guinea Pig anti-PSP | gift from Stefan Ruhl | N/A |
| Goat anti-MUC10 | Abcore | Cat# AC21-2394 |
| Sheep anti-ERBB3 | R&D Systems | Cat# AF4518; RRID: AB_2099728 |
| Rabbit anti-ERBB2 | Cell Signaling Technology | Cat# 2165; RRID: AB_10692490 |
| Rabbit anti-EGFR | Abcam | Cat# ab52894; RRID: AB_869579 |
| Goat anti-KIT | Santa Cruz Biotechnology | Cat# SC-1494; RRID: AB_631032 |
| Goat anti-SOX10 | Santa Cruz Biotechnology | Cat# SC-17342; RRID: AB_2195374 |
| Rabbit anti-Cleaved Caspase-3 | Cell Signaling Technology | Cat# 9664; RRID: AB_2070042 |
| Mouse anti-Ki67 | BD Biosciences | Cat# 550609; RRID: AB_393778 |
| Rabbit anti-PKCζ | Santa Cruz Biotechnology | Cat# SC-216; RRID: AB_632241 |
| Mouse anti-AKT | Cell Signaling Technology | Cat# 2920; RRID: AB_1147620 |
| Rabbit anti-Phospho-AKT (Ser473) | Cell Signaling Technology | Cat# 9271; RRID: AB_329825 |
| Rabbit anti-Phospho-AKT (Thr308) | Cell Signaling Technology | Cat# 13038; RRID: AB_2629447 |
| Rabbit anti-ERBB3 | Cell Signaling Technology | Cat# 12708; RRID: AB_2721919 |
| Rabbit anti-Phospho-ERBB3 (Tyr1289) | Cell Signaling Technology | Cat# 4791; RRID: AB_2099709 |
| Rabbit anti-EGFR | Cell Signaling Technology | Cat# 4267; RRID: AB_2246311 |
| Rabbit anti-Phospho-EGFR (Tyr1068) | Cell Signaling Technology | Cat# 2234; RRID: AB_331701 |
| Rabbit anti-4E-BP1 | Cell Signaling Technology | Cat# 9644; RRID: AB_2097841 |
| Rabbit anti-Phospho-4E-BP1 (Thr37/46) | Cell Signaling Technology | Cat# 2855; RRID: AB_560835 |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| Hoechst 33342 | AnaSpec | Cat# AS83218 |
| Paraformaldehyde | Thermo Fisher Scientific | Cat# AA47340-9M |
| OCT | Andwin Scientific Tissue-Tek | Cat# 14-373-65 |
| Dispase II | Gibco | Cat# 17105041 |
| Cultrex 3D Culture Matrix Laminin I | R&D Systems | Cat# 3446-005-01 |
| DMSO | Sigma-Aldrich | Cat # D2650 |
| DMEM/F12 | Thermo Fisher Scientific | Cat# 11320082 |
| Penicillin-Streptomycin | Gibco | Cat# 15140122 |
| Holo-transferrin | Sigma-Aldrich | Cat# T1283 |
| Ascorbic Acid | Sigma-Aldrich | Cat# A4544 |
| Recombinant Human NRG1 Isoform SMDF | R&D Systems | Cat# 378-SM-025 |
| Recombinant Human HB-EGF | R&D Systems | Cat# 259-HE-250 |
| Rapamycin | Biotang Inc | Cat# 50-148-809 |
| Torin-1 | Cell Signaling Technologies | Cat# 14379 |
| Critical commercial assays | ||
| Purelink RNA Mini Kit | Invitrogen | Cat# 12-183-018A |
| NEBNext® UltraTM RNA Library Prep Kit for Illumina ® | New England Lab inc. | Cat# E7530L |
| Deposited data | ||
| Raw RNA-seq data | This paper | GSE: 214976 |
| Experimental models: Organisms/strains | ||
| Tg(KRT14-cre/ERT2)1Ipc | Li et al.91 | MGI:2177426 |
| Erbb3tm1.1Squ | Qu et al.90 | MGI: 3690340 |
| Software and algorithms | ||
| fastp | Chen et al.92 | https://github.com/OpenGene/fastp |
| STAR | Dobin93 | https://github.com/alexdobin/STAR |
| FeatureCounts | Liao et al.94 | |
| The R Project for Statistical Computing | CRAN | https://www.R-project.org/ |
| R Studio | R Studio | https://www.rstudio.com/ |
| DIY Transcriptomics | Berry et al.95 | https://diytranscriptomics.com/ |
| edgeR | Robinson et al.97 | https://bioconductor.org/packages/release/bioc/html/edgeR |
| DeSeq2 | Love et al.96 | https://bioconductor.org/packages/release/bioc/html/DESeq2 |
| limma | Ritchie et al.98 | https://bioconductor.org/packages/release/bioc/html/limma |
| Panther | Mi et al.32 | http://pantherdb.org/ |
| gProfiler | Reiman et al.99 | https://biit.cs.ut.ee/gprofiler/gost |
| ggplot2 | Wickham100 | https://ggplot2.tidyverse.org/ |
| pheatmap | Kolde101 | https://github.com/raivokolde/pheatmap |
| EnhancedVolcano | Blighe et al.102 | https://github.com/kevinblighe/EnhancedVolcano |
| GSEA | Subramanian et al.46 | https://www.gsea-msigdb.org/gsea/index.jsp |
| Seurat | Hao et al.104 | https://satijalab.org/seurat/ |
| CellChat | Jin et al.21 | http://www.cellchat.org/ |
| Cytoscape | Shannon et al.103 | https://cytoscape.org/ |
| Fiji ImageJ | NIH | https://imagej.net/software/fiji/ |
| Graphpad Prism 9 | Graphpad Software, Inc | https://www.graphpad.com |
Highlights.
Nerve-derived NRG1 signals via ERBB3 enriched in acinar progenitors
Epithelial deletion of Erbb3 inhibits acinar specification
NRG1 treatment of epithelia is sufficient to recapitulate in vivo acinus formation
NRG1-ERBB3 signaling regulates acinar specification through mTORC2
Acknowledgements
The authors thank Drs. Mathew Hoffman, Licia Selleri, Julie Sneddon, Susan Fisher, and Natalia Jura for providing guidance and critical reading on the manuscript. Funding was provided by the National Institutes of Health (F31DE027607 (AJMattingly), 5R01DE024188 (SMK) and R35DE028255 (SMK)), and the National Science Foundation (2014187660 (AJMattingly)).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Inclusion and Diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a gender minority in their field of research. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community. One or more of the authors of this paper self-identifies as living with a disability. One or more of the authors of this paper received support from a program designed to increase minority representation in their field of research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brückner A, and Parker J (2020). Molecular evolution of gland cell types and chemical interactions in animals. J. Exp. Biol. 223. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Huang J, Liu Y, Dattilo LK, Huh S-H, Ornitz D, and Beebe DC (2014). FGF signaling activates a Sox9-Sox10 pathway for the formation and branching morphogenesis of mouse ocular glands. Development 141, 2691–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Athwal HK, Murphy G, Tibbs E, Cornett A, Hill E, Yeoh K, Berenstein E, Hoffman MP, and Lombaert IMA (2019). Sox10 Regulates Plasticity of Epithelial Progenitors toward Secretory Units of Exocrine Glands. Stem Cell Reports 12, 366–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dravis C, Spike BT, Harrell JC, Johns C, Trejo CL, Southard-Smith EM, Perou CM, and Wahl GM (2015). Sox10 Regulates Stem/Progenitor and Mesenchymal Cell States in Mammary Epithelial Cells. Cell Rep. 12, 2035–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larsen HS, Aure MH, Peters SB, Larsen M, Messelt EB, and Kanli Galtung H (2011). Localization of AQP5 during development of the mouse submandibular salivary gland. J. Mol. Histol. 42, 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemercier C, To RQ, Swanson BJ, Lyons GE, and Konieczny SF (1997). Mist1: A novel basic helix-loop-helix transcription factor exhibits a developmentally regulated expression pattern. Dev. Biol. 182, 101–113. [DOI] [PubMed] [Google Scholar]
- 7.Hauser BR, Aure MH, Kelly MC, Hoffman MP, and Chibly AM (2020). Generation of a Single-Cell RNAseq Atlas of Murine Salivary Gland Development. iScience 23, 101838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hart A, Papadopoulou S, and Edlund H (2003). Fgf10 maintains notch activation, stimulates proliferation, and blocks differentiation of pancreatic epithelial cells. Dev. Dyn. 228, 185–193. [DOI] [PubMed] [Google Scholar]
- 9.Afelik S, Pool B, Schmerr M, Penton C, and Jensen J (2015). Wnt7b is required for epithelial progenitor growth and operates during epithelial-to-mesenchymal signaling in pancreatic development. Dev. Biol. 399, 204–217. [DOI] [PubMed] [Google Scholar]
- 10.Chatzeli L, Gaete M, and Tucker AS (2017). Fgf10 and Sox9 are essential for the establishment of distal progenitor cells during mouse salivary gland development. Dev. 144, 2294–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinberg Z, Myers C, Heim VM, Lathrop CA, Rebustini IT, Stewart JS, Larsen M, and Hoffman MP (2005). FGFR2b signaling regulates ex vivo submandibular gland epithelial cell proliferation and branching morphogenesis. Development 132, 1223–1234. [DOI] [PubMed] [Google Scholar]
- 12.Emmerson E, May AJ, Nathan S, Cruz-Pacheco N, Lizama CO, Maliskova L, Zovein AC, Shen Y, Muench MO, and Knox SM (2017). SOX2 regulates acinar cell development in the salivary gland. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Direnzo D, Hess DA, Damsz B, Hallett JE, Marshall B, Goswami C, Liu Y, Deering T, MacDonald RJ, and Konieczny SF (2012). Induced Mist1 Expression Promotes Remodeling of Mouse Pancreatic Acinar Cells. Gastroenterology 143, 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo HYG, Jin RU, Sibbel G, Liu D, Karki A, Joens MS, Madison BB, Zhang B, Blanc V, Fitzpatrick JAJ, et al. (2017). A single transcription factor is sufficient to induce and maintain secretory cell architecture. Genes Dev. 31, 154–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mikkelsen TR, Brandt J, Larsen HJ, Larsen BB, Poulsen K, Ingerslev J, Din N, and Hjorth JP (1992). Tissue-specific expression in the salivary glands of transgenic mice. Nucleic Acids Res. 20, 2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi T, Lombaert IMA, Hauser BR, Patel VN, and Hoffman MP (2017). Exosomal MicroRNA Transport from Salivary Mesenchyme Regulates Epithelial Progenitor Expansion during Organogenesis. Dev. Cell 40, 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musselmann K, Green JA, Sone K, Hsu JC, Bothwell IR, Johnson SA, Harunaga JS, Wei Z, and Yamada KM (2011). Salivary gland gene expression atlas identifies a new regulator of branching morphogenesis. J. Dent. Res. 90, 1078–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knosp WM, Knox SM, Lombaert IMA, Haddox CL, Patel VN, and Hoffman MP (2015). Submandibular parasympathetic gangliogenesis requires sprouty-dependent Wnt signals from epithelial progenitors. Dev. Cell 32, 667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyazaki Y, Nakanishi Y, and Hieda Y (2004). Tissue interaction mediated by neuregulin-1 and ErbB receptors regulates epithelial morphogenesis of mouse embryonic submandibular gland. Dev. Dyn. 230, 591–596. [DOI] [PubMed] [Google Scholar]
- 20.Gresik EW, Kashimata M, Kadoya Y, Mathews R, Minami N, and Yamashina S (1997). Expression of epidermal growth factor receptor in fetal mouse submandibular gland detected by a biotinyltyramide-based catalyzed signal amplification method. J. Histochem. Cytochem. 45, 1651–1657. [DOI] [PubMed] [Google Scholar]
- 21.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, and Nie Q (2021). Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021 121 12, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Bates R, Yin DM, Shen C, Wang F, Su N, Kirov SA, Luo Y, Wang JZ, Xiong WC, et al. (2011). Specific Regulation of NRG1 Isoform Expression by Neuronal Activity. J. Neurosci. 31, 8491–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrenschee M, Lange C, Cossais F, Egberts JH, Becker T, Wedel T, and Bottner M (2015). Expression and function of Neuregulin 1 and its signaling system ERBB2/3 in the enteric nervous system. Front. Cell. Neurosci. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Syroid DE, Maycox PR, Burrola PG, Liu N, Wen D, Lee KF, Lemke G, and Kilpatrick TJ (1996). Cell death in the Schwann cell lineage and its regulation by neuregulin. Proc. Natl. Acad. Sci. U. S. A. 93, 9229–9234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knox S, Lombaert I, Reed X, Vitale-Cross L, Gutkind JS, and Hoffman MP (2010). Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science (80-.). 329, 1645–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nedvetsky PI, Emmerson E, Finley JK, Ettinger A, Cruz-Pacheco N, Prochazka J, Haddox CL, Northrup E, Hodges C, Mostov KE, et al. (2014). Parasympathetic Innervation Regulates Tubulogenesis in the Developing Salivary Gland. Dev. Cell 30, 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.May AJ, Cruz-Pacheco N, Emmerson E, Gaylord EA, Seidel K, Nathan S, Muench MO, Klein OD, and Knox SM (2018). Diverse progenitor cells preserve salivary gland ductal architecture after radiation-induced damage. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams MM, Vaught DB, Joly MM, Hicks DJ, Sanchez V, Owens P, Rahman B, Elion DL, Balko JM, and Cook RS (2017). ErbB3 drives mammary epithelial survival and differentiation during pregnancy and lactation. Breast Cancer Res. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Dubé PE, Washington MK, Yan F, and Polk Brent D (2012). ErbB2 and ErbB3 regulate recovery from dextran sulfate sodium-induced colitis by promoting mouse colon epithelial cell survival. Lab. Invest. 92, 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong Y (2018). aPKC: The Kinase that Phosphorylates Cell Polarity [version 1; referees: 2 approved]. F1000Research 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umeda Y, Miyazaki Y, Shiinoki H, Higashiyama S, Nakanishi Y, and Hieda Y (2001). Involvement of heparin-binding EGF-like growth factor and its processing by metalloproteinases in early epithelial morphogenesis of the submandibular gland. Dev. Biol. 237, 202–211. [DOI] [PubMed] [Google Scholar]
- 32.Mi H, Muruganujan A, Casagrande JT, and Thomas PD (2013). Large-scale gene function analysis with PANTHER Classification System. Nat. Protoc. 8, 1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keenan AB, Torre D, Lachmann A, Leong AK, Wojciechowicz ML, Utti V, Jagodnik KM, Kropiwnicki E, Wang Z, and Ma’ayan A (2019). ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 47, W212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humbert PO, Rogers C, Ganiatsas S, Landsberg RL, Trimarchi JM, Dandapani S, Brugnara C, Erdman S, Schrenzel M, Bronson RT, et al. (2000). E2F4 Is Essential for Normal Erythrocyte Maturation and Neonatal Viability. Mol. Cell 6, 281–291. [DOI] [PubMed] [Google Scholar]
- 35.Yu W, Li X, Eliason S, Romero-Bustillos M, Ries RJ, Cao H, and Amendt BA (2017). Irx1 regulates dental outer enamel epithelial and lung alveolar type II epithelial differentiation. Dev. Biol. 429, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tchegnon E, Liao C-P, Ghotbi E, Shipman T, Wang Y, McKay RM, and Le LQ (2021). Epithelial stem cell homeostasis in Meibomian gland development, dysfunction, and dry eye disease. JCI Insight 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kojima Y, Yamashiro C, Murase Y, Yabuta Y, Okamoto I, Iwatani C, Tsuchiya H, Nakaya M, Tsukiyama T, Nakamura T, et al. (2021). GATA transcription factors, SOX17 and TFAP2C, drive the human germ-cell specification program. Life Sci. Alliance 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin X, Zeng Z, Xing J, Zhang A, Jiang W, Wang W, Sun H, and Ni L (2018). Hey1 functions as a positive regulator of odontogenic differentiation in odontoblast-lineage cells. Int. J. Mol. Med. 41, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang SS, Kim YU, Lee S, Jang SW, Kim MK, Koh BH, Lee W, Kim J, Souabni A, Busslinger M, et al. (2013). Transcription factor YY1 is essential for regulation of the Th2 cytokine locus and for Th2 cell differentiation. Proc. Natl. Acad. Sci. 110, 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satoh K, Narita T, Matsuki-Fukushima M, Okabayashi K, Ito T, Senpuku H, and Sugiya H (2013). E2f1-deficient NOD/SCID mice have dry mouth due to a change of acinar/duct structure and the down-regulation of AQP5 in the salivary gland. Pflugers Arch. 465, 271–281. [DOI] [PubMed] [Google Scholar]
- 41.Lee AH, Chu GC, Iwakoshi NN, and Glimcher LH (2005). XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 24, 4368–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hess DA, Humphrey SE, Ishibashi J, Damsz B, Lee A-H, Glimcher LH, and Konieczny SF (2011). Extensive Pancreas Regeneration Following Acinar-Specific Disruption of Xbp1 in Mice. Gastroenterology 141, 1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garg A, Hannan A, Wang Q, Collins T, Teng S, Bansal M, Zhong J, Xu K, and Zhang X (2018). FGF-induced Pea3 transcription factors program the genetic landscape for cell fate determination. PLOS Genet. 14, e1007660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker LA, Holliday H, and Swarbrick A (2016). ID4 controls luminal lineage commitment in normal mammary epithelium and inhibits BRCA1 function in basal-like breast cancer. Endocr. Relat. Cancer 23, R381–R392. [DOI] [PubMed] [Google Scholar]
- 45.Holliday H, Roden D, Junankar S, Wu SZ, Baker LA, Krisp C, Chan CL, McFarland A, Skhinas JN, Cox TR, et al. (2021). Inhibitor of Differentiation 4 (ID4) represses mammary myoepithelial differentiation via inhibition of HEB. iScience 24, 102072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulze WX, Deng L, and Mann M (2005). Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol. Syst. Biol. 1, 2005.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ersahin T, Tuncbag N, and Cetin-Atalay R (2015). The PI3K/AKT/mTOR interactive pathway. Mol. Biosyst. 11, 1946–1954. [DOI] [PubMed] [Google Scholar]
- 49.Yu JSL, and Cui W (2016). Proliferation, survival and metabolism: The role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Dev. 143, 3050–3060. [DOI] [PubMed] [Google Scholar]
- 50.Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, Hara K, Tanaka N, Avruch J, and Yonezawa K (2003). The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278, 15461–15464. [DOI] [PubMed] [Google Scholar]
- 51.Hresko RC, and Mueckler M (2005). mTOR·RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J. Biol. Chem. 280, 40406–40416. [DOI] [PubMed] [Google Scholar]
- 52.Burnett PE, Barrow RK, Cohen NA, Snyder SH, and Sabatini DM (1998). RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. U. S. A. 95, 1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Q, Kang SA, Thoreen CC, Hur W, Wang J, Chang JW, Markhard A, Zhang J, Sim T, Sabatini DM, et al. (2012). Development of ATP-competitive mTOR inhibitors. Methods Mol. Biol. 821, 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garrett JR (1987). The proper role of nerves in salivary secretion: a review. J. Dent. Res. 66, 387–397. [DOI] [PubMed] [Google Scholar]
- 55.Browning KN, and Travagli RA (2014). Central Nervous System Control of Gastrointestinal Motility and Secretion and Modulation of Gastrointestinal Functions. Compr. Physiol. 4, 1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang S, Kuri P, Aubert Y, Brewster M, Li N, Farrelly O, Rice G, Bae H, Prouty S, Dentchev T, et al. (2021). Lgr6 marks epidermal stem cells with a nerve-dependent role in wound re-epithelialization. Cell Stem Cell 28, 1582–1596.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao Y, Thoresen DT, Williams JS, Wang C, Perna J, Petrova R, and Brownell I (2015). Neural Hedgehog signaling maintains stem cell renewal in the sensory touch dome epithelium. Proc. Natl. Acad. Sci. U. S. A. 112, 7195–7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stocum DL (2011). The role of peripheral nerves in urodele limb regeneration. Eur. J. Neurosci. 34, 908–916. [DOI] [PubMed] [Google Scholar]
- 59.Brownell I, Guevara E, Bai CB, Loomis CA, and Joyner AL (2011). Nerve-derived sonic hedgehog defines a niche for hair follicle stem cells capable of becoming epidermal stem cells. Cell Stem Cell 8, 552–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knox SM, Lombaert IMA, Haddox CL, Abrams SR, Cotrim A, Wilson AJ, and Hoffman MP (2013). Parasympathetic stimulation improves epithelial organ regeneration. Nat. Commun. 4, 1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le TL, Galmiche L, Levy J, Suwannarat P, Hellebrekers DMEI, Morarach K, Boismoreau F, Theunissen TEJ, Lefebvre M, Pelet A, et al. (2021). Dysregulation of the NRG1/ERBB pathway causes a developmental disorder with gastrointestinal dysmotility in humans. J. Clin. Invest. 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Polosa R, Prosperini G, Tomaselli V, Howarth PH, Holgate ST, and Davies DE (2000). Expression of c-erbB receptors and ligands in human nasal epithelium. J. Allergy Clin. Immunol. 106, 1124–1131. [DOI] [PubMed] [Google Scholar]
- 63.Liu Z, Carvajal M, Carothers Carraway CA, Carraway K, and Pflugfelder SC (2001). Expression of the receptor tyrosine kinases, epidermal growth factor receptor, ErbB2, and ErbB3, in human ocular surface epithelia. Cornea 20, 81–85. [DOI] [PubMed] [Google Scholar]
- 64.Dammann CEL, Nielsen HC, and Carraway KL (2012). Role of Neuregulin-1β in the Developing Lung. Am. J. Respir. Crit. Care Med. 167, 1711–1716. [DOI] [PubMed] [Google Scholar]
- 65.Jeong W-Y, Yoo H-Y, and Kim C-W (2018). Neuregulin-1 accelerates corneal epithelial wound healing. 10.1080/08977194.2018.1436055 35, 225–233. [DOI] [PubMed] [Google Scholar]
- 66.Jardé T, Chan WH, Rossello FJ, Kahlon TK, Theocharous M, Arackal TK, Flores T, Giraud M, Richards E, Chan E, et al. (2020). Mesenchymal Niche-Derived Neuregulin-1 Drives Intestinal Stem Cell Proliferation and Regeneration of Damaged Epithelium. Cell Stem Cell 27, 646–662.e7. [DOI] [PubMed] [Google Scholar]
- 67.Jones FE, Joseph Jerry D, Guarino BC, Andrews GC, and Stern DF (1996). Heregulin induces in vivo proliferation and differentiation of mammary epithelium into secretory lobuloalveoli. Cell Growth Differ. 7. [PubMed] [Google Scholar]
- 68.Stern DF (2008). ERBB3/HER3 and ERBB2/HER2 duet in mammary development and breast cancer. J. Mammary Gland Biol. Neoplasia 13, 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Forster N, Saladi SV, Bragt M, van, Sfondouris, Jones FE, Li Z, and Ellisen LW (2014). Basal cell signaling by p63 controls luminal progenitor function and lactation via NRG1. Dev. Cell 28, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lai D, Liu X, Forrai A, Wolstein O, Michalicek J, Ahmed I, Garratt AN, Birchmeier C, Zhou M, Hartley L, et al. (2010). Neuregulin 1 Sustains the Gene Regulatory Network in Both Trabecular and Nontrabecular Myocardium. Circ. Res. 107, 715–727. [DOI] [PubMed] [Google Scholar]
- 71.Simmons LJ, Surles-Zeigler MC, Li Y, Ford GD, Newman GD, and Ford BD (2016). Regulation of inflammatory responses by neuregulin-1 in brain ischemia and microglial cells in vitro involves the NF-kappa B pathway. J. Neuroinflammation 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Surles-Zeigler MC, Li Y, Distel TJ, Omotayo H, Ge S, and Ford BD (2018). Transcriptomic analysis of neuregulin-1 regulated genes following ischemic stroke by computational identification of promoter binding sites: A role for the ETS-1 transcription factor. PLoS One 13, e0197092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.El Soury M, Fornasari BE, Morano M, Grazio E, Ronchi G, Incarnato D, Giacobini M, Geuna S, Provero P, and Gambarotta G (2018). Soluble Neuregulin1 Down-Regulates Myelination Genes in Schwann Cells. Front. Mol. Neurosci. 0, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haskins JW, Nguyen DX, and Stern DF (2014). Neuregulin 1-activated ERBB4 interacts with YAP to induce Hippo pathway targetgenes and promote cell migration. Sci. Signal. 7, ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hasegawa D, Calvo V, Avivar-Valderas A, Lade A, Chou H-I, Lee YA, Farias EF, Aguirre-Ghiso JA, and Friedman SL (2015). Epithelial Xbp1 is required for cellular proliferation and differentiation during mammary gland development. Mol. Cell. Biol. 35, 1543–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EES, Higgins DE, Schreiber S, Glimcher LH, et al. (2008). XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134, 743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Davis KR, Giesy SL, Long Q, Krumm CS, Harvatine KJ, and Boisclair YR (2016). XBP1 Regulates the Biosynthetic Capacity of the Mammary Gland During Lactation by Controlling Epithelial Expansion and Endoplasmic Reticulum Formation. Endocrinology 157, 417–428. [DOI] [PubMed] [Google Scholar]
- 78.Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, et al. (2004). XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21, 81–93. [DOI] [PubMed] [Google Scholar]
- 79.Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, and Dynlacht BD (2007). XBP1 Controls Diverse Cell Type- and Condition-Specific Transcriptional Regulatory Networks. Mol. Cell 27, 53–66. [DOI] [PubMed] [Google Scholar]
- 80.Metwalli KA, Do MA, Nguyen K, Mallick S, Kin K, Farokhnia N, Jun G, and Fakhouri WD (2018). Interferon Regulatory Factor 6 Is Necessary for Salivary Glands and Pancreas Development. J. Dent. Res. 97, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Z, Goto Y, Allevato MM, Wu VH, Saddawi-Konefka R, Gilardi M, Alvarado D, Yung BS, O’Farrell A, Molinolo AA, et al. (2021). Disruption of the HER3-PI3K-mTOR oncogenic signaling axis and PD-1 blockade as a multimodal precision immunotherapy in head and neck cancer. Nat. Commun. 2021 121 12, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Odintsov I, Mattar MS, Lui AJW, Offin M, Kurzatkowski C, Delasos L, Khodos I, Asher M, Daly RM, Rekhtman N, et al. (2021). Novel Preclinical Patient-Derived Lung Cancer Models Reveal Inhibition of HER3 and MTOR Signaling as Therapeutic Strategies for NRG1 Fusion-Positive Cancers. J. Thorac. Oncol. 16, 1149–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cao G, Chen K, Chen B, and Xiong M (2017). Positive prognostic value of HER2-HER3 co-expression and p-mTOR in gastric cancer patients. BMC Cancer 17, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Beauchamp RL, Erdin S, Witt L, Jordan JT, Plotkin SR, Gusella JF, and Ramesh V (2021). mTOR kinase inhibition disrupts neuregulin 1-ERBB3 autocrine signaling and sensitizes NF2-deficient meningioma cellular models to IGF1R inhibition. J. Biol. Chem. 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Keppler-Noreuil KM, Parker VER, Darling TN, and Martinez-Agosto JA (2016). Somatic overgrowth disorders of the PI3K/AKT/mTOR pathway & therapeutic strategies. Am. J. Med. Genet. Part C Semin. Med. Genet. 172, 402–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Castilho R, Squarize C, and Gutkind J (2013). Exploiting PI3K/mTOR signaling to accelerate epithelial wound healing. Oral Dis. 19, 551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morrison MM, Young CD, Wang S, Sobolik T, Sanchez VM, Hicks DJ, Cook RS, and Brantley-Sieders DM (2015). mTOR Directs Breast Morphogenesis through the PKC-alpha-Rac1 Signaling Axis. PLoS Genet. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ghosh S, Lau H, Simons BW, Powell JD, Meyers DJ, De Marzo AM, Berman DM, and Lotan TL (2011). PI3K/mTOR signaling regulates prostatic branching morphogenesis. Dev. Biol. 360, 329–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ding X, Bloch W, Iden S, Rüegg MA, Hall MN, Leptin M, Partridge L, and Eming SA (2016). mTORC1 and mTORC2 regulate skin morphogenesis and epidermal barrier formation. Nat. Commun. 2016 71 7, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qu S, Rinehart C, Wu HH, Wang SE, Carter B, Xin H, Kotlikoff M, and Arteaga CL (2006). Gene targeting of ErbB3 using a Cre-mediated unidirectional DNA inversion strategy. genesis 44, 477–486. [DOI] [PubMed] [Google Scholar]
- 91.Li M, Indra AK, Warot X, Brocard J, Messaddeq N, Kato S, Metzger D, and Chambon P (2000). Skin abnormalities generated by temporally controlled RXRalpha mutations in mouse epidermis. Nature 407, 633–636. [DOI] [PubMed] [Google Scholar]
- 92.Chen S, Zhou Y, Chen Y, and Gu J (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- 95.Berry ASF, Amorim CF, Berry CL, Syrett CM, English ED, and Beiting DP (2021). An open-source toolkit to expand bioinformatics training in infectious diseases. MBio 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reimand J, Kull M, Peterson H, Hansen J, and Vilo J (2007). g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35, W193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag; New York: ). [Google Scholar]
- 101.Kolde R (2012). Pheatmap: pretty heatmaps. 726. [Google Scholar]
- 102.Blighe K, Rana S, and Lewis M (2019). EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling.
- 103.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Enrichment in gene expression in developing SG from E13 to E18 and in epithelial end buds compared to ducts at E15, related to Figure 1
Data S2. Differential Gene Expression Analysis and Gene Set Enrichment Analysis of E16 Erbb3 KO and WT SGs, related to Figures 2 and 5
Data S3. gProfiler analysis of TF regulatory binding motifs of upregulated genes in E13 epithelia treated with NRG1 for 4 days, related to Figure 3
Data S4. Differential Gene Expression Analysis of NRG1 stimulated end buds vs control, related to Figure 4
Data S5. Differential Gene Expression Analysis of NRG1+Torin-1 vs NRG1+DMSO (control) E13 epithelia cultured for 72 hours, related to Figure 6
Data Availability Statement
This paper does not report any original code. Additional information on code used to analyze data reported is available from the lead contact upon request. RNA-sequencing data has been deposited in the Gene Expression Omnibus (GEO) database (GSE214976).


