Abstract
We estimated the probability of undetected emergence of the SARS-CoV-2 Omicron variant in 25 low and middle-income countries (LMICs) prior to December 5, 2021. In nine countries, the risk exceeds 50 %; in Turkey, Pakistan and the Philippines, it exceeds 99 %. Risks are generally lower in the Americas than Europe or Asia.
Keywords: Omicron, LMICs, Risk, Emergence, Covid Importation
1. Main text
The B.1.1.529 SARS-COV-2 variant was first detected and reported in South Africa on November 26th, 2021 (WHO, 2021). By December 5th, 2021, more than 40 countries reported Omicron variant cases. Most of these countries are developed and high-income countries, including the United Kingdom, the United States, and Netherlands (GISAID, 2021,). However, low and middle income countries (LMICs) may be less likely to detect a new variant (Sisa et al., 2021) and more vulnerable to catastrophic public health outcomes than high-income countries, because of lower capacity for COVID-19 testing, vaccination, and medical treatment (Helmy et al., 2016, Siow et al., 2020). Only a few LMICs have direct flights from the countries in Southern Africa where Omicron was initially detected (Flightradar24, 2021) and many enacted border policies to reduce Omicron importation risks from these countries (Mallapaty, 2021). However, LMICs were at risk for Omicron importations from large international destinations outside of Southern Africa in which Omicron emerged in late 2021.
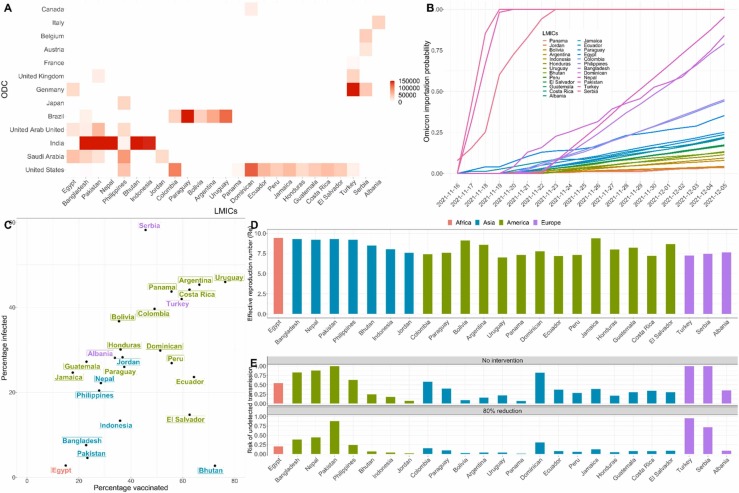
We analyzed the risks of the Omicron variant importation in 25 LMICs in which Omicron was not reported as of December 5, 2021: Bangladesh, Nepal, Philippines, Colombia, Egypt, Pakistan, Paraguay, Turkey, Serbia, Bolivia, Argentina, Uruguay, Bhutan, Indonesia, Albania, Jordan, Panama, Dominican Republic, Ecuador, Peru, Jamaica, Honduras, Guatemala, Costa Rica, and El Salvador. We first estimated the daily travel volume to each country from 13 large countries in which Omicron had already been detected, based on data from Facebook Data for Good (Fig. A) (Data for Good Tools and Data., 2021). We estimated the prevalence of Omicron in each of the 13 Omicron detected countries (ODCs) assuming that only 2.5 % of early cases were identified and reported (Davis et al., 2021, GISAID, 2021), and then estimated the probability of travel-based introductions into each LMIC by December 5, 2021 (see Supporting Information). The European LMICs (Serbia and Turkey), which are highly connected to Western European countries that reported Omicron cases by November 2021, have the highest estimated risks, followed by the Asian LMICs (Pakistan, Bangladesh, and Nepal), with high inflows of travelers from South Africa (via connecting flights), the UK, and India. LMICs in the Americas (Colombia, Dominican Republic, and Paraguay) are primarily at risk for importations from the US and Brazil. We estimate that 6 of the 25 studied LMICs had over a 50 % chance of having received at least one travel-based Omicron importation from ODCs by December 5, 2021 (Figure B).
To assess the risk of Omicron transmission following importation, we estimate the immunity-based effective reproduction number () in each LMIC as of December 5, 2021, based on reported vaccination levels (Mathieu et al., 2021), estimates of infection-acquired immunity (World Health Organization, 2021) (Figure C), and recent estimates for the transmissibility and immune-evasiveness of the Omicron variant (Miller et al., 2012) (see Supporting Information). Recent studies have estimated that Omicron has double the reproduction number of the Delta variant, suggesting a basic reproduction number () of 11.88 (95%CI: 9.16–14.61) (Figure SI.3) (Chen et al., 2021). The same study also suggests that SARS-CoV-2 vaccines are 50 % less effective (VE) against Omicron compared to Delta (Figure SI.5) (Chen et al., 2021). Given the low vaccination coverage and high infectivity of Omicron, we estimate that the immunity-based of Omicron on December 5, 2021 ranges from 7.0 to 9.4 across the 25 studied LMICs, without additional public health interventions (Figure D).
Combining our estimates of Omicron importation and transmission risks, we estimate the probability of undetected Omicron transmission in LMICs by early December (Figure E- details see Supporting Information). Among the studied LMICs, Turkey (99.99 %), Pakistan (99.95 %), and Serbia (99.81 %) have the highest estimated risk, followed by Nepal (87.98 %), Bangladesh (84.86 %), and the Dominican Republic (82.21 %). If these countries implement non-pharmaceutical interventions that reduce transmission by 80 %, the probability of undetected emergence declines by 12.02–80.77 % across the 25 LMICs (Figure E). Given the high socioeconomic costs of travel restrictions and some non-pharmaceutical interventions, many of these LMICs did not take measures to prevent introductions or slow spread (Torres-Rueda et al., 2021). Our analyses suggest that SARS-CoV-2 variants like Omicron can rapidly emerge in LMICs and spread for weeks before detection.( Fig. 1).
Fig. 1.
: Risks of Omicron emergence in 25 low- and middle-income countries by December 5, 2021. A. Cumulative number of travelers to each of the 25 LMICs reported by Facebook Data for Good from November 16 to December 5, 2021 from 13 countries in which Omicron had already been detected by December 5 (ODCs): Belgium, Italy, United Kingdom, Germany, Canada, Austria, Japan, Brazil, United State, United Arab Emirates, Saudi Arabia, France, India (Data for Good Tools and Data., 2021). B. The probability of receiving at least one Omicron importation via travelers from the 13 ODCs between November 16 and December 5, 2021. C. Estimated vaccination and prior infection rates for each LMIC, assuming a 40 % infection reporting rate for all countries (Irons and Raftery, 2021). D. Estimated effective reproduction number for the Omicron variant in each LMIC, assuming that Omicron is twice as transmissible as the Delta variant and that vaccines have 50% lower efficacy against Omicron in comparison to Delta. E. Comparisons of the estimated risks of the undetected Omicron transmission in LMICs with and without intervention that reduces transmission by 80%.
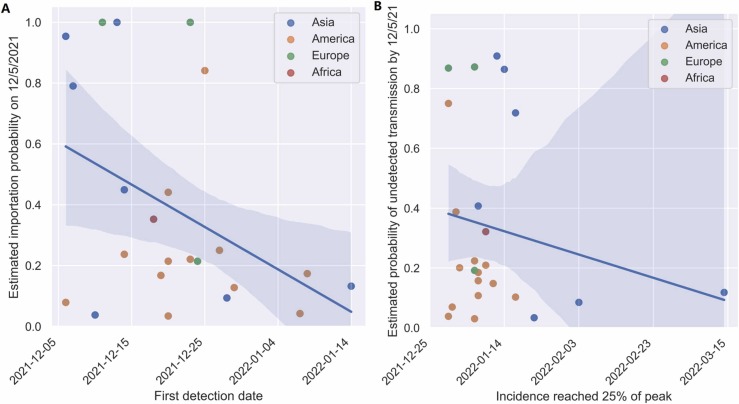
In November 2022, we retrospectively compared our early estimates of Omicron importation and transmission risks with reported data and found a significant correlation between our estimated risks of importation and the timing the first case was detected (p < 0.05) and a non-significant but positive correlation between our estimates of transmission risks and the time at which incidence reached 25 % of the winter Omicron peak ( Fig. 2 and Table SI.10). Across all 25 LMICs, the average time between the first detection and incidence reaching the 25 % mark was 19.88 days, with a standard deviation of 1.51 days. Given the short window between detection and wide transmission, LMIC countries with limited surveillance capabilities should consider initiating control measures when threatening variants are first identified in other countries.
Fig. 2.
Retrospective comparison of estimated to observed Omicron importation and transmission events. (A) The probability of receiving at least one Omicron importation by December 5, 2021 compared to the first Omicron detection dates in LMICs. (B) Estimated risks of undetected Omicron transmission compared to the date on which reported incidence reached 25% of the eventual peak incidence in LMICs (Ritchie et al., 2020). Solid lines and shading represent the fitted regression lines and 95% confidence intervals, respectively.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank financial support from the National Institutes of Health (grant no. R01 AI151176-Suppl), the Centers for Disease Control and Prevention (grant no. U01IP001136-Suppl) and the Council of State and Territorial Epidemiologists (grant no. NU38OT000297).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.epidem.2022.100660.
Appendix A. Supplementary material
Supplementary material
.
Data availability
The authors do not have permission to share data.
References
- Chen, Jiahui , Rui Wang , Nancy Benovich Gilby , and Guo-Wei Wei , 2021. Omicron (B. 1.1. 529): Infectivity, vaccine breakthrough, and antibody resistance." arXiv preprint arXiv:2112.01318. [DOI] [PMC free article] [PubMed]
- Data for Good Tools and Data.” 2021. Dataforgood.facebook.com. https://dataforgood.facebook.com/dfg/tools.
- Davis Jessica T., Matteo Chinazzi, Nicola Perra, Kunpeng Mu, Marco Ajelli, Dean Natalie E., Corrado Gioannini, et al. Cryptic transmission of SARS-CoV-2 and the first COVID-19 wave. Nature. 2021:1–9. doi: 10.1038/s41586-021-04130-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flightradar24, 2021 Live flight tracker - real-time flight tracker map Flightradar24. Accessed December 12, 2021. https://www.flightradar24.com/data/airports.
- GISAID - HCov19 Variants.” 2021. Www.gisaid.org. https://www.gisaid.org/hcov19-variants/.
- Helmy Mohamed, Awad Mohamed, Mosa Kareem A. Limited resources of genome sequencing in developing countries: challenges and solutions. Appl. Transl. Genom. 2016;9:15–19. doi: 10.1016/j.atg.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons Nicholas J., Raftery Adrian E. Estimating SARS-CoV-2 infections from deaths, confirmed cases, tests, and random surveys. arXiv Prepr. arXiv. 2021;2102:10741. doi: 10.1073/pnas.2103272118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty, Smriti, 2021. "Omicron-variant border bans ignore the evidence, say scientists." Nature. [DOI] [PubMed]
- Mathieu E., Ritchie H., Ortiz-Ospina E., et al. A global database of COVID-19 vaccinations. Nat. Hum. Behav. 2021 doi: 10.1038/s41562-021-01122-8. [DOI] [PubMed] [Google Scholar]
- Miller Joel C., Anja C.Slim, Erik M.Volz. Edge-based compartmental modelling for infectious disease spread. J. R. Soc. Interface 9. 2012;no. 70:890–906. doi: 10.1098/rsif.2011.0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah Ritchie, Edouard Mathieu, Lucas Rodés-Guirao, Cameron Appel, Charlie Giattino, Esteban Ortiz-Ospina, Joe Hasell, Bobbie Macdonald, Diana Beltekian and Max Roser , 2020 - Coronavirus Pandemic (COVID-19). Published online at OurWorldInData.org. Ret.
- Siow Wen, Ting Mei Fong, Liew Babu Raja, Shrestha Faisal, Muchtar Kay Choong. Managing COVID-19 in resource-limited settings: critical care considerations. Crit. Care. 2020;24(1):1–5. doi: 10.1186/s13054-020-02890-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisa Ivan, Fornasini Marco, Teran Enrique. COVID-19 research in LMICs. Lancet. 2021;398:1212–1213. doi: 10.1016/S0140-6736(21)01605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rueda Sergio, Sweeney Sedona, Bozzani Fiammetta, Naylor Nichola R., Baker Tim, Pearson Carl, Rosalind Eggo, et al. Stark choices: exploring health sector costs of policy responses to COVID-19 in low-income and middle-income countries. BMJ Glob. Health. 2021;6(12) doi: 10.1136/bmjgh-2021-005759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2021. “Update on Omicron.” Www.who.int. November 28, 2021. https://www.who.int/news/item/28–11-2021-update-on-omicron.
- World Health Organization. 2021. “WHO COVID-19 Dashboard.” Covid19.Who.int. World Health Organization. 2021. https://covid19.who.int/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The authors do not have permission to share data.