Abstract
Background
Nonalcoholic fatty liver disease affects about 24% of the world’s population and may progress to nonalcoholic steatohepatitis (NASH), cirrhosis, and hepatocellular carcinoma (HCC). While more common in those that are obese, NASH-HCC can develop in lean individuals. The mechanisms by which HCC develops and the role of epigenetic changes in the context of obesity and normal weight are not well understood.
Methods
In this study, we used previously generated mouse models of lean and obese HCC using a choline deficient/high trans-fat/fructose/cholesterol diet and a choline supplemented/high trans-fat/fructose/cholesterol diet, respectively, to evaluate methylation differences in HCC progression in lean versus obese mice. Differentially methylated regions were determined using reduced representation bisulfite sequencing.
Results
A larger number of differentially methylated regions (DMRs) were seen in NASH-HCC progression in the obese mice compared to the non-obese mice. No overlap existed in the DMRs with the largest methylation differences between the two models. In lean NASH-HCC, methylation differences were seen in genes involved with cancer progression and prognosis (including HCC), such as CHCHD2, FSCN1, and ZDHHC12, and lipid metabolism, including PNPLA6 and LDLRAP1. In obese NASH- HCC, methylation differences were seen in genes known to be associated with HCC, including RNF217, GJA8, PTPRE, PSAPL1, and LRRC8D. Genes involved in Wnt-signaling pathways were enriched in hypomethylated DMRs in the obese NASH-HCC.
Conclusions
These data suggest that differential methylation may play a role in hepatocarcinogenesis in lean versus obese NASH. Hypomethylation of Wnt signaling pathway-related genes in obese mice may drive progression of HCC, while progression of HCC in lean mice may be driven through other signaling pathways, including lipid metabolism.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-022-10389-7.
Keywords: Liver cancer, Non-alcoholic fatty liver disease, Methylation
Background
Nonalcoholic fatty liver disease (NAFLD) affects approximately 24% of the world’s population [1]. NAFLD encompasses a spectrum of diseases characterized by fat in the liver that may progress to nonalcoholic steatohepatitis (NASH) with inflammation and fibrosis and ultimately to cirrhosis, which results in an increased risk of developing hepatocellular carcinoma (HCC) [2]. Treatment options for advanced stage HCC are limited, so understanding the development of HCC may identify opportunities for drug interventions or opportunities for primary prevention efforts [3]. Knowledge of the role of diet in the development of HCC has great importance for understanding the mechanism by which NASH progresses to HCC.
While often associated with obesity, NAFLD may develop among lean individuals as well, especially among those that are of normal weight but metabolically obese [1]. The exact causes of lean NAFLD are not clear, but those with lean NAFLD are less likely to have obesity-related co-morbidities [4, 5]. The role of diet in the progression of NASH to HCC is not well understood. The complex pathways involved in NASH-related HCC likely involve genetic and epigenetic factors [2]. Differential methylation patterns of HCC may be useful in developing pharmacological interventions, since DNA methylation is reversible and hence susceptible to intervention [6].
It was previously found that the remodeling of DNA methylation occurs at genes in patients with NASH and fibrosis, suggesting that epigenetic signatures may be a possible biomarker for severity of disease [1]. Previous studies have identified potential causal relationships between epigenetic changes and liver carcinogenesis [6]. In HCC, hypomethylation has been found with transcriptional enhancers and hypermethylation has been found with promoter-associated CGIs and cis-regulatory elements [6]. Additionally, lower expression of phosphatidylethanolamine N-methyltransferase (PEMT) was found in individuals with lean NASH, which could be implicated in its progression [5].
To date, only a few studies have investigated the role of epigenetic changes in the progression of NASH-related HCC in lean versus obese individuals. In this study we used previously developed novel models of lean and obese NASH-HCC in mice using choline deficient (CD) and choline supplemented (CS) high trans-fat/fructose/cholesterol diets to examine differences in DNA methylation during the progression of NASH to HCC as well as differences of DNA methylation in HCC progression in lean versus obese mice.
Methods
Animals and experimental diets
This study was approved by the Institutional Animal Care and Use Committee at the University of Nebraska Medical Center (Protocol #: 17–018) and was also conducted in compliance with the ARRIVE guidelines. Male (n = 103) C57BL/6 N mice (Charles River Laboratories) were allowed to acclimate and housed as previously described beginning at 3 weeks of age [7]. 30 males were fed a choline supplemented, high trans-fat, fructose, and cholesterol diet (CS-HFFC; D18091706), 38 males were fed a choline deficient, high trans-fat, fructose, and cholesterol diet (CD-HFFC; D17071001), and 35 males were fed a low-fat control diet (CON; D16120211; Research Diets, New Brunswick, New Jersey). The estimated HCC penetrance from our previous work was used to determine this sample size [7]. The consumption of food was monitored, and the mice were regularly weighed and husbandry checks were performed as previously described [7].
Histological evaluation
All mice were monitored until the endpoint of the study (64 weeks of age). Any mice showing signs of poor health were euthanized per institutional ethical guidelines by CO2 inhalation. After harvesting tissues for analysis, exsanguination was done to confirm death. A cardiac puncture was performed to collect blood right after euthanasia was performed. Livers were excised, weighed, and observed grossly for the appearance of nodules. The tissue samples were snap frozen in liquid nitrogen and then stored at − 80 °C. The remaining tissues were fixed in 10% formalin for 2 hours and paraffin embedded at the Tissue Sciences Facilities at the University of Nebraska Medical Center. Hematoxylin and Eosin (H&E) and Masson-Trichome were used to stain tissue sections. An additional reticulin stain was added at necropsy. An experienced pathologist blindly evaluated the stained sections, scoring the sections for steatosis, ballooning, and inflammation to determine the presence of NAFLD and NASH [8]. The stained liver sections were also evaluated for the presence of regenerative nodules, dysplastic nodules, and hepatocellular carcinomas as previously described [7].
DNA extraction and RRBS
Forty samples were selected for analysis: normal liver tissue from 6 controls, NASH tissue from 4 CD-HFFC fed mice, dysplastic tissue from 7 CD-HFFC fed mice, HCC tissue from 7 CD-HFFC fed mice, NASH tissue from 2 CS-HFFC fed mice, dysplastic tissue from 8 CS-HFFC fed mice, and HCC tissue from 6 CS-HFFC fed mice. DNA was isolated from 25 mg of snap-frozen liver tissue using the Qiagen DNeasy blood and tissue kit (Qiagen, Germantown, MD). Genome wide DNA methylome analyses were carried out on DNA samples (400-500 ng) using the Diagenode Inc. (Denville, NJ) Premium Reduced Representation Bisulfite Sequencing (RRBS) kit on mouse samples. DNA concentration of the samples was analyzed using the Qubit® dsDNA Assay Kit (Thermo Fisher Scientific) and DNA quality was assessed using the Fragment Analyzer™ and the DNF-488 High Sensitivity genomic DNA Analysis Kit (Agilent).
DNA methylation data analysis
The output from RRBS was read into the methylKit (v1.16.1) package in R (v4.0.3) and analyzed by the Bioinformatics and Systems Biology Core at the University of Nebraska Medical Center [9]. Logistic regression tests were used for the differential methylation analyses and the sliding linear model (SLIM) method was used for multiple testing adjustment on 15 comparisons [10]. Generated q-values were used for representing the adjusted p-values. Significance levels were determined by the criteria q < 0.01 and percent methylation differences larger than 25%.
Annotation and functional pathway analysis
MethylKit was used to annotate the differentially methylated regions to include the distance to the corresponding gene and gene ID. UniProt and Reference sequence (RefSeq) were used to identify gene names and function for significantly differentially methylated regions [11, 12]. Gene Ontology enrichment analysis was done on the differentially methylated regions between the choline deficient and choline supplemented HCC samples and between HCC and NASH in both models to find the molecular function of regions that are over- or under-represented in the sample [13–15].
Results
Characterization of differentially methylated regions
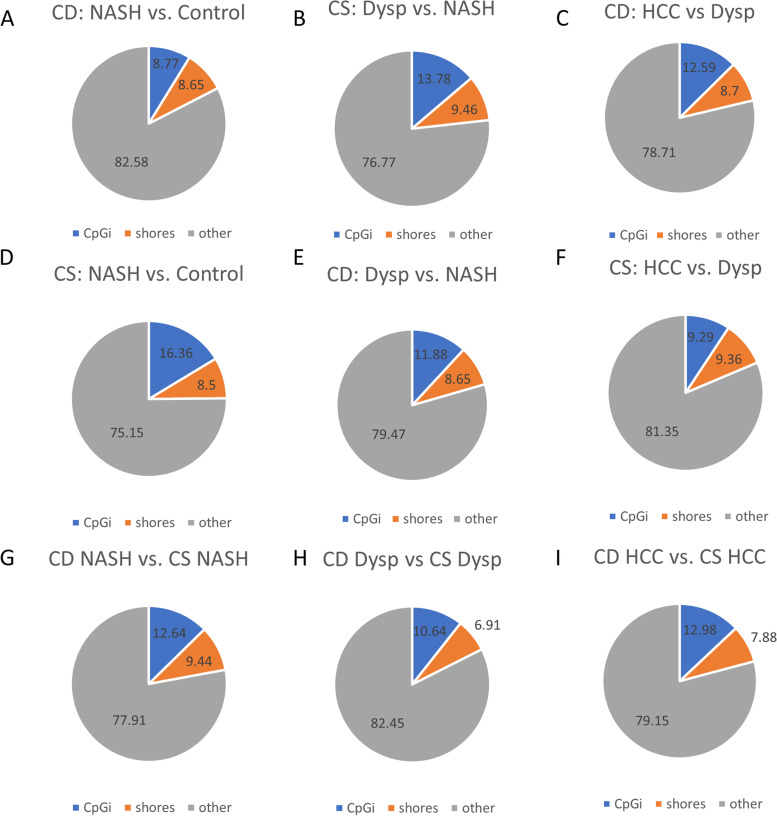
Differentially methylated regions between each comparison (diet or disease stage) were categorized as occurring either in CpG islands, shores, or other regions (regions greater than 2 kb from CpG islands). Among all comparisons shown in Fig. 1, the majority of DMRs occurred in regions other than CpG islands and shores. In the choline deficient model, there were an increasing proportion of DMRs in CpG islands through the progression of HCC and decreasing DMRs in the other regions (Fig. 1A-C). In the choline supplemented model, there was a decreasing proportion of DMRs in CpG islands and an increasing proportion of DMRs in the other regions through the progression of HCC (Fig. 1D-F). No uniform pattern of DMRs by gene region was seen between the choline deficient and supplemented models at the NASH, dysplastic, and HCC stages.
Fig. 1.
Percentages of CpG categories (islands, shores, or other) for significant differential DNA methylation loci. A. CD: NASH vs. Control. B. CD: Dysp vs. NASH C. CD: HCC vs. NASH. D. CS: NASH vs Control. E. CS: Dysp vs. NASH F. CS: HCC vs. Dysp. G. CD NASH vs. CS NASH. H. CD Dysp. vs. CS Dysp. I. CD HCC vs. CS HCC
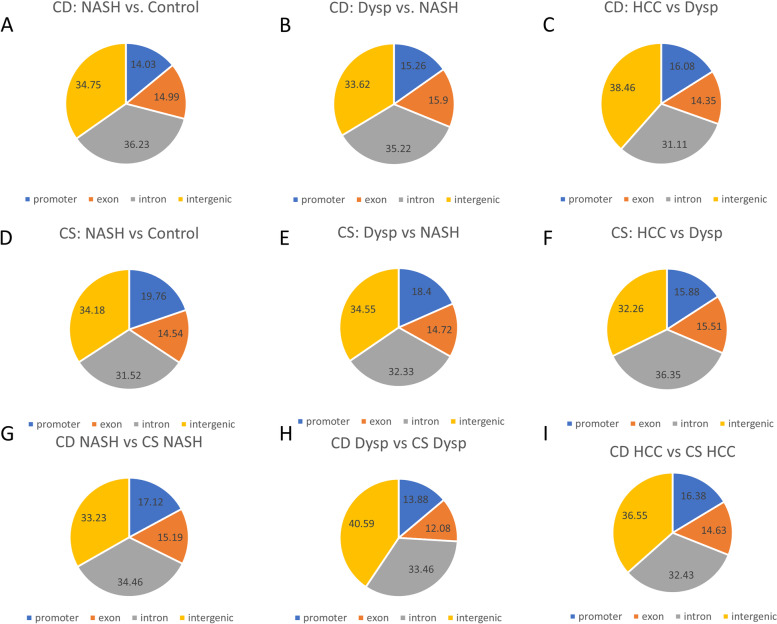
The proportion of DMRs by methylation loci (promoter, exon, intron, and intergenic regions) are shown in Fig. 2. Similar patterns were observed across the progression of HCC in the choline deficient and supplemented models. Between the comparisons of the two diet models at each stage (Fig. 2G-I), the largest difference was seen in the proportion of DMRs in intergenic regions between the NASH and dysplastic stages, with the latter showing a larger proportion of DMRs in intergenic regions.
Fig. 2.
Percentages of gene structure categories for significant differential DNA methylation loci. A. CD: NASH vs. Control. B. CD: Dysp vs. NASH C. CD: HCC vs. NASH. D. CS: NASH vs Control. E. CS: Dysp vs. NASH F. CS: HCC vs. Dysp. G. CD NASH vs. CS NASH. H. CD Dysp vs. CS Dysp. I. CD HCC vs. CS HCC
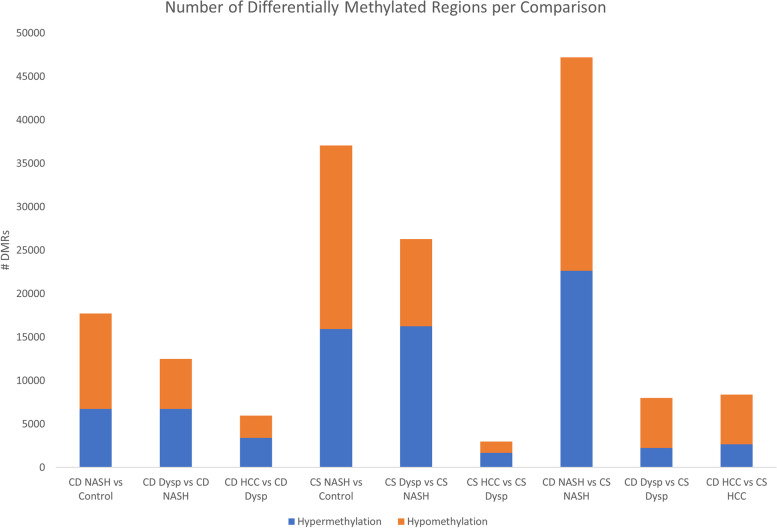
Figure 3 shows the number of differentially methylated regions per comparison, as well as the proportion of hypermethylated and hypomethylated regions per each comparison. In both diet models, the largest number of DMRs was seen between NASH and the controls; the lowest number of DMRs was seen between HCC and the dysplastic stage (Supplemental File 1). Among the disease stage progression, there was a higher number of DMRs among the choline supplemented model than the choline deficient model, except for the HCC vs Dysplastic Nodules comparison. The proportion of DMRs that were hypermethylated increased throughout the HCC progression in both models, even though the number of hypermethylated regions decreased with tumor progression. In the final three comparisons in Fig. 3 that compare the stages between the two diet models, the highest number of DMRs was seen at the NASH stage. Compared to the choline deficient model, there was a greater proportion of hypomethylated DMRs in the choline supplemented model in the NASH, dysplastic, and HCC stages.
Fig. 3.
Number of differentially methylated regions by comparison
Genomic distribution of DMRs
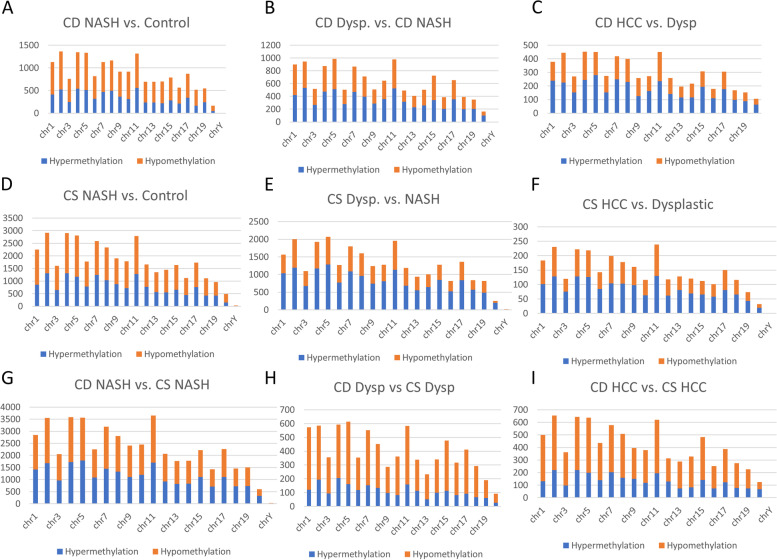
There were higher numbers of differentially methylated regions in the progression of HCC in the choline-supplemented model (Fig. 4D-F, Supplemental File 1) compared to the choline-deficient model (Fig. 4A-C) for each chromosome. While the numbers of DMRs were different, the overall pattern per chromosome was similar in both diet models. The proportion of DMRs that were hypermethylated was higher in the choline-supplemented model. Comparing each stage of progression between the two models, there was a greater proportion of hypomethylation per chromosome in the dysplastic and HCC stages compared to NASH (Fig. 4G-I, Supplemental File 1).
Fig. 4.
Number of differentially methylated regions per chromosome. A. CD: NASH vs. Control. B. CD: Dysp vs. NASH C. CD: HCC vs. NASH. D. CS: NASH vs Control. E. CS: Dysp vs. NASH F. CS: HCC vs. Dysp. G. CD NASH vs. CS NASH. H. CD Dysp vs. CS Dysp. I. CD HCC vs. CS HCC
Genes potentially modified by hyper/hypomethylation in the progression of HCC
Comparing HCC and NASH in the CD and CS models, a greater proportion of DMRs were in CpG islands and promoter regions in the CS model; while there was significantly more DMRs in the CS model compared to the CD model, the proportion of DMRs per chromosome was similar (Supplemental Fig. 1). The top ten differentially hypermethylated and hypomethylated regions with the greatest percent methylation differences between CD HCC and NASH are presented in Table 1. The regions with the largest percent methylation difference were a mix of hypermethylated and hypomethylated regions; the majority were outside of CpG islands and shores and were found mainly in exons or introns. Within the CD model, several of the largest differentially methylated regions are in genes involved in lipid metabolism (PNPLA6 and LDLRAP1), transcription (CHCHD2), and Wnt signaling pathways (JADE1), and cell migration and binding (TMEM88b, MYO5b, FSNC1, and ZDHHC12) [11, 12]. The top ten differentially hypermethylated and hypomethylated regions between CS HCC and NASH are presented in Table 2. The percent methylation difference was larger among the hypermethylated regions. There was no overlap in the regions with the highest percent methylation difference between HCC and NASH in the choline deficient and supplemented models. DMRs were located mostly outside of CpG islands and shores and were found in introns and exons. The percent methylation differences between HCC and NASH were higher in the CS model (Table 2) than in the CD model (Table 1). Within the CS model, the significantly differentially methylated regions are found in genes involved in ion channels (GJA8, KCNQ5, and LRRC8D), cell cycle regulation (PTPRE and RNF123), and cell signaling pathways (RNF217), including Wnt signaling (SOSTDC1) [11, 12]. Four of the differentially methylated regions were in genes previously found to be associated with obesity, metabolic regulation, and glycemic control (LRRC8D, SOSTCD1, MORN3, and MCF2) [16–19].
Table 1.
Top 10 hypermethylated and hypomethylated regions between HCC and NASH in the choline deficient model
| Chromosome | Start | End | Q-value | Methylation difference (%) | Gene | CpG Category | Gene Structure | |
|---|---|---|---|---|---|---|---|---|
| Hypermethylation | Chr 8 | 3517616 | 3517616 | 1.65E-48 | 76.2212 | PNPLA6 | Shore | Exon |
| Chr 17 | 32105855 | 32105855 | 8.63E-47 | 68.636 | – | Other | – | |
| Chr 11 | 99645764 | 99645764 | 5.30E-74 | 67.33949 | KRTAP4–7 | Other | – | |
| Chr 5 | 129557719 | 129557719 | 4.11E-59 | 65.9518 | CHCHD2 | Other | Exon | |
| Chr 5 | 142945496 | 142945496 | 1.39E-55 | 65.23347 | FSCN1 | Other | – | |
| Chr 3 | 41581453 | 41581453 | 2.32E-44 | 64.13043 | JADE1 | Other | Exon | |
| Chr 9 | 121792412 | 121792412 | 5.80E-53 | 62.74613 | – | Other | Intron | |
| Chr 4 | 155781218 | 155781218 | 4.83E-39 | 62.30984 | TMEM88B | Other | Intron | |
| Chr 6 | 31666012 | 31666012 | 1.21E-81 | 62.05207 | Gm13848 | Other | Exon | |
| Chr 18 | 82756567 | 82756567 | 7.26E-80 | 61.81989 | – | Other | Intergenic | |
| Hypomethylation | Chr 2 | 30100458 | 30100458 | 3.98E-61 | −68.875 | ZDHHC12 | Other | Intron |
| Chr 2 | 170791632 | 170791632 | 8.07E-49 | −67.2973 | – | Other | Intron | |
| Chr 10 | 69846110 | 69846110 | 8.28E-53 | −66.5238 | GM33416 | Other | Intron | |
| Chr 9 | 57968665 | 57968665 | 1.39E-36 | −63.029 | MYO5B | Other | Intron | |
| Chr 4 | 110051897 | 110051897 | 5.55E-47 | −62.4214 | DMRTA2 | Shore | – | |
| Chr 16 | 97823834 | 97823834 | 1.62E-34 | −61.5291 | – | Other | Intron | |
| Chr 4 | 134744349 | 134744349 | 5.94E-31 | −60.3271 | LDLRAP1 | Other | Exon | |
| Chr 7 | 36873554 | 36873554 | 7.09E-53 | −59.9929 | – | Other | – | |
| Chr 7 | 42750002 | 42750002 | 3.33E-34 | −59.3331 | DOCK3 | Other | Exon | |
| Chr 5 | 74960682 | 74960682 | 4.97E-51 | −59.0323 | Gm6116 | Other | Intron |
Abbreviation: Chr Chromosome
Table 2.
Top 10 hypermethylated and hypomethylated regions between HCC and NASH in the choline supplemented model
| Chromosome | Start | End | Q-value | Methylation difference (%) | Gene | CpG Category | Gene Structure | |
|---|---|---|---|---|---|---|---|---|
| Hypermethylation | Chr 10 | 31860051 | 31860051 | 5.08E-36 | 100 | RNF217 | Other | Intron |
| Chr 3 | 96919939 | 96919939 | 6.65E-59 | 98 | GJA8 | Other | Exon | |
| Chr 17 | 54643532 | 54643532 | 2.75E-37 | 97.71429 | Gm32055 | Other | – | |
| Chr 1 | 21988549 | 21988549 | 3.91E-54 | 96.51515 | KCNQ5 | Other | Intron | |
| Chr 5 | 142385284 | 142385284 | 2.78E-42 | 95.35104 | PTPRE | Other | – | |
| Chr 2 | 123549054 | 123549054 | 1.02E-29 | 95.26627 | MORN3 | Other | – | |
| Chr 3 | 55348282 | 55348282 | 1.08E-31 | 93.17359 | Gm40051 | Other | Intron | |
| Chr 13 | 51672327 | 51672327 | 5.01E-42 | 91.07143 | SECISBP2 | Other | Intron | |
| Chr X | 60093579 | 60093579 | 4.50E-29 | 90.47619 | MCF2 | Other | Intron | |
| Chr 16 | 29868324 | 29868324 | 8.94E-38 | 89.09953 | RNF123 | Other | – | |
| Hypomethylation | Chr 5 | 36228842 | 36228842 | 1.50E-57 | −87.4512 | PSAPL1 | Other | Intron |
| Chr 4 | 98726981 | 98726981 | 1.18E-39 | −87.4396 | L1TD1 | Other | Intron | |
| Chr 4 | 82537544 | 82537544 | 7.33E-64 | −87.2659 | – | Other | Intron | |
| Chr 7 | 108951080 | 108951080 | 1.18E-26 | −86.2069 | Gm39067 | Island | Intron | |
| Chr 11 | 82930668 | 82930668 | 8.16E-37 | −86.1035 | UNC45BOS | Shore | Exon | |
| Chr 5 | 128527390 | 128527390 | 1.68E-27 | −85.9649 | – | Other | – | |
| Chr 5 | 105759229 | 105759229 | 1.97E-27 | −84.8837 | LRRC8D | Other | Intron | |
| Chr 12 | 25240371 | 25240371 | 6.01E-20 | −84.3137 | – | Other | Intron | |
| Chr 2 | 172863024 | 172863024 | 6.24E-28 | −83.871 | – | Other | Intron | |
| Chr 12 | 36317937 | 36317937 | 8.35E-39 | −83.8572 | SOSTDC1 | Shore | Exon |
Abbreviation: Chr Chromosome
Differences in methylation patterns of hepatocellular carcinoma in lean and obese mice
Table 3 shows the top 10 differentially hypermethylated and hypomethylated regions between HCC in the CD model and HCC in the CS model. All but one of the top 10 overall regions with the highest percent difference in methylation were hypomethylation, indicating that there was a greater degree of methylation in the choline supplemented model HCC compared to the choline deficient model. Of the hypermethylated regions, half were in CpG islands and shores. Differential methylation was found mostly in exons and introns. The corresponding genes are involved in physiological process including cell signaling pathways, catabolic processes, and protein translation [11, 12]. Five hypermethylated regions were in genes previously associated with cancer (TRAP1, SLC38A3, CHRM1, EDN2, and PROX1), while six hypomethylated regions were in genes previously associated with cancer (NUMBL, ALDH1B1, FTCD, FASTKD2, FAM96A, and ARHGAP15) [20–30]. Five differentially methylated regions were in genes previously found to be associated with obesity and altered metabolic states (TRAP1, SLC38A3, PROX1, ALDH1B1, and FAM96A) [31–35].
Table 3.
Top 10 hypermethylated and hypomethylated regions between choline deficient HCC and choline supplemented HCC
| Chromosome | Start | End | Q-value | Methylation difference (%) | Gene | CpG Category | Gene Structure | |
|---|---|---|---|---|---|---|---|---|
| Hypermethylation | Chr 7 | 27658325 | 27658325 | 5.06E-41 | 68.16218 | TTC9B | Island | Exon |
| Chr 5 | 110363759 | 110363759 | 1.58E-41 | 57.24965 | LCROL1 | Island | Exon | |
| Chr 1 | 42741736 | 42741736 | 1.39E-86 | 56.99229 | LOC108167622 | Other | Intron | |
| Chr 16 | 4078449 | 4078449 | 8.78E-54 | 55.9661 | TRAP1 | Shore | – | |
| Chr 7 | 107210287 | 107210287 | 4.31E-25 | 54.50725 | RBMXL2 | Island | Exon | |
| Chr 9 | 107669835 | 107669835 | 3.47E-63 | 54.2572 | SLC38A3 | Other | Intron | |
| Chr 19 | 8679239 | 8679239 | 1.30E-35 | 54.08986 | CHRM1 | Other | Exon | |
| Chr 4 | 120124981 | 120124981 | 7.45E-29 | 54.015 | EDN2 | Other | Exon | |
| Chr 7 | 107210301 | 107210301 | 1.45E-24 | 53.93071 | RBMXL2 | Island | Exon | |
| Chr 1 | 190139252 | 190139252 | 8.04E-63 | 53.61003 | PROX1 | Other | Intron | |
| Hypomethylation | Chr 9 | 121076534 | 121076534 | 2.87E-43 | −66.7969 | OLFR843 | Other | Intron |
| Chr 7 | 27257369 | 27257369 | 6.56E-67 | −62.1354 | NUMBL | Shore | Exon | |
| Chr 10 | 76634403 | 76634403 | 3.22E-65 | −61.6133 | – | Other | Intron | |
| Chr 4 | 45785050 | 45785050 | 1.68E-54 | −60.7105 | ALDH1B1 | Other | – | |
| Chr 10 | 76584578 | 76584578 | 4.26E-46 | −59.9492 | FTCD | Other | Exon | |
| Chr 12 | 74283740 | 74283740 | 5.55E-38 | −59.9486 | FASTKD2 | Shore | Intron | |
| Chr 10 | 73300112 | 73300112 | 2.99E-43 | −59.8009 | FAM96A | Other | Intron | |
| Chr 9 | 46986808 | 46986808 | 1.95E-46 | −59.3403 | GM4791 | Other | – | |
| Chr 2 | 44162442 | 44162442 | 5.78E-45 | −59.2628 | ARHGAP15 | Other | Intron | |
| Chr 7 | 37356388 | 37356388 | 3.96E-45 | −58.4701 | – | Other | – |
Abbreviation: Chr Chromosome
Functional analyses
Gene Ontology enrichment analysis was performed on the genes indicated as having a 25% or greater methylation difference in HCC versus NASH in both the choline deficient and choline supplemented models. The DMRs in both comparisons shared some functions, including transcription activation and DNA binding in transcription (Table 4). Functions specific to the DMRs of the choline deficient model of HCC progression included hormone binding and calcium ion binding. Functions specific to the DMRs of the choline supplemented model included neuropeptide receptor activity, voltage-gated potassium channel activity, transcription repressor activity, and signaling receptor activator activity. Additionally, of the hypomethylated regions in HCC versus NASH, the molecular functions of Wnt-protein binding and frizzled binding were found to be enriched in the choline supplemented model, but not the choline deficient model (Table 5). These different functional activities of differentially methylated regions may be involved in pathways associated with the progression of HCC from NASH in the context of lean and obese mice.
Table 4.
Top statistically significant Gene Ontology molecular functions
| Model | Molecular Function | Fold Enrichment |
|---|---|---|
| CD HCC vs. NASH | Hormone binding | 3.05 |
| DNA-binding transcription activator activity, RNA polymerase II-specific | 1.88 | |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | 1.72 | |
| Calcium ion binding | 1.57 | |
| CS HCC vs. NASH | Neuropeptide receptor activity | 2.76 |
| Voltage-gated potassium channel activity | 2.03 | |
| DNA-binding transcription activator activity, RNA polymerase II-specific | 1.61 | |
| DNA binding transcription repressor activity, RNA polymerase-II specific | 1.5 | |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | 1.49 | |
| Signaling receptor activator activity | 1.39 |
Top statistically significant Gene Ontology molecular functions revealed by enrichment analysis between HCC and NASH in the choline deficient and choline supplemented mice
Table 5.
Enriched molecular functions in hypomethylated regions between HCC and NASH
| Model | Molecular Function | Fold Enrichment |
|---|---|---|
| CD HCC vs. NASH | DNA-binding transcription activator activity, RNA polymerase II-specific | 2.14 |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | 1.82 | |
| Protein binding | 1.17 | |
| CS HCC vs. NASH | Wnt-protein binding | 4.20 |
| Frizzled binding | 3.17 | |
| DNA-binding transcription activator activity, RNA polymerase II-specific | 2.21 | |
| DNA-binding transcription repressor activity, RNA polymerase II-specific | 2.07 | |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | 2.00 | |
| Receptor ligand activity | 1.52 | |
| Cation binding | 1.18 |
Top statistically significant Gene Ontology molecular functions revealed by enrichment analysis of hypomethylated regions between HCC and NASH in the choline deficient and choline supplemented mice
Discussion
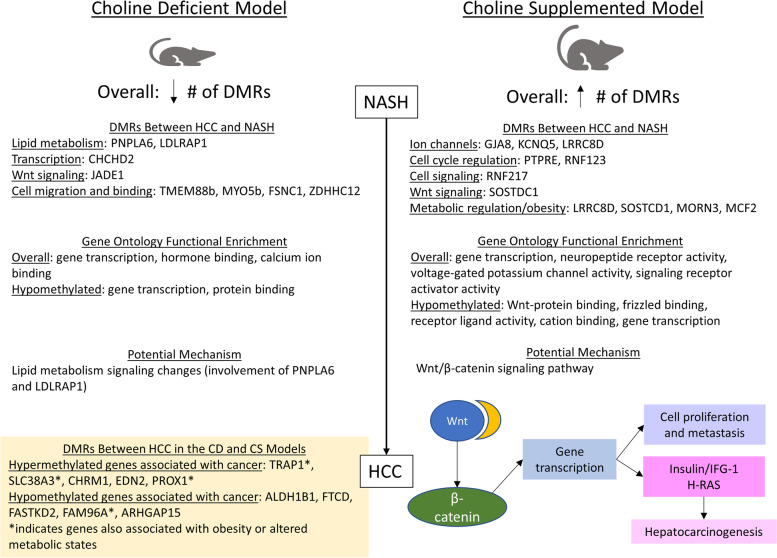
In this work, we demonstrated that significant differential methylation exists in the progression of NASH-HCC in the context of obesity (through choline supplementation) and non-obesity (through choline deficiency) in mice fed a high fat/fructose/cholesterol diet (Fig. 5). Larger numbers of DMRs were found between HCC and NASH in the obese mice (41,979) compared to the non-obese mice (11,104). In both models, the largest number of DMRs were seen at the beginning stages of disease progression (in NASH and dysplasia) and the proportion of DMRs that were hypermethylated increased with progression. DNA methylation has previously been implicated in carcinogenesis; previous studies have identified differential methylation in NASH-related HCC in both mice and humans [36, 37]. Global hypomethylation is common in carcinogenesis, with numerous methylation changes occurring early in tumor progression [38]. The increased number of DMRs and hypermethylation in the obese mice may be driven by dietary choline, which is involved in one-carbon metabolism in methylation [39]. Additionally, our previous work found that mice with HCC in the context of obesity had higher plasma fasting glucose and cholesterol levels than lean mice with HCC [40]; differences in glucose levels may contribute to the differential methylation patterns identified, which is in line with previous work that found glucose levels were associated with CpG methylation levels [41].
Fig. 5.
Summary of differential methylation in the CD and CS models. A summary of the differential methylation patterns between the choline deficient and choline supplemented models, differentially methylated genes and functions, and potential implications
In the choline deficient model, significant hypermethylation of CHCHD2 (within an exon) and FSCN1 was seen. Overexpression of both of these genes has been found to be associated with HCC and found to be an indicator of poor prognosis [42–44]. Our previous work identified faster progression of HCC and worse survival in the choline deficient model; the role of these genes in carcinogenesis may depend on the stage of progression and site of methylation. Significant differential methylation was also found in exons of two genes involved in lipid metabolism, PNPLA6 (within a CpG shore) and LDLRAP1 [11].
In the choline supplemented model, significant hypermethylation of RNF217 was found, which has also been seen with alcohol-related HCC in humans [45]. RNF217 is involved in process of ubiquitination [11]. Differential methylation was also seen in genes previously identified as tumor suppressor genes and oncogenes, including KCNQ5, PTPRE, and SOSTDC1 [46–48]. Additionally, hypermethylation was found in GJA8 (within an exon) and hypomethylation was found in PSAPL1 and LRRC8D (within introns); these three genes have all been found to be associated with hepatocarcinogenesis [49–51]. Increased methylation of MORN3 and decreased methylation of LRRC8D in the context of obesity is consistent with previous studies [16, 18]. Hypermethylation of MCF2, a proto-oncogene involved in Rho protein signal transduction, was also seen in obese patients with breast cancer [11, 19].
Comparing HCC in the choline deficient model and choline supplemented model, significant methylation differences were seen in 11 genes previously associated with cancer (hypermethylation of TRAP1, SLC38A3, CHRM1, EDN2, and PROX1; hypomethylation of ALDH1B1, FTCD, FASTKD2, FAM96A, and ARHGAP15). Of these genes, four (TRAP1, SLC38A3, PROX1, and FAM96A) have also been associated with obesity and altered metabolic states, indicating that they may be implicated in differential progression of lean versus obese NASH-HCC (Fig. 5).
No overlap exists in the regions with the highest methylation difference between HCC and NASH in the lean and obese models, indicating that differential methylation may act through different mechanisms to promote carcinogenesis in the two models. Gene ontology enrichment analysis on the DMRs between HCC and NASH in the two models revealed some shared molecular functions, including functions related to gene transcription. The choline deficient model had DMRs in genes involved with hormone binding and calcium ion binding, while the choline supplemented model had DMRs in genes involved in neuropeptide receptor activity, voltage-gated potassium channel activity, and signaling receptor activator activity. These signaling pathway molecular functions may represent potential mechanisms of HCC progression in the two models. Calcium signaling has previously been found to be enriched in NAFLD-associated HCC [52].
Looking at the functional enrichment of hypomethylated regions between HCC and NASH, which would result in overexpression of the associated genes, several important differences were seen between the two models. In the choline supplemented model, significant hypomethylation was seen in genes involved in Wnt-protein binding and frizzled binding, as well as receptor ligand activity and cation binding, which were not seen in the choline deficient model. Additionally, one gene in Table 2, SOSTDC1, was found to be hypomethylated (within a CpG shore of an exon); SOSTDC1 is involved in enhancing Wnt signaling pathways [11].
The Wnt/β-catenin signaling pathway has been implicated in several mechanisms of hepatocarcinogenesis, including growth, survival, and migration [53]. In the canonical Wnt signaling pathway, Wnt interacts with frizzled receptors and activates intracellular signaling pathways leading to the stabilization of β-catenin, which can then enter the nucleus and influence transcription of target genes [54, 55]. β-catenin may regulate the transcription of genes involved in proliferation and metastasis and may also interact with other oncogenic pathways such as insulin/IFG-1 and H-RAS to influence pathogenesis (Fig. 5) [53, 55]. Mutations in β-catenin have been found in a large proportion of liver tumors [55]. Additionally, β-catenin has been found to be involved in changing the tumor-immune microenvironment in NAFLD-associated HCC [56]. Hypomethylation of genes involved in Wnt/β-catenin signaling pathways likely contributes to the development and progression of HCC in the context of obesity. Functional enrichment of Wnt/ β-catenin was not seen in the model of lean NASH-HCC, indicating that direct involvement of the Wnt signaling pathways through methylation changes may be unique to the choline supplemented model. In the choline deficient model, differential methylation in exons of genes involved in lipid metabolism (PNPLA6 and LDLRAP1) may alter signaling pathways leading to the progression of HCC. Thus, tumor progression may involve lipid metabolism signaling changes in the lean NASH-HCC model. Differentially methylated regions may be targets for emerging epigenetic cancer therapeutics; additional work is needed to test the study results in humans and examine the effect of differential methylation on the effectiveness of available therapeutics [57]. Further research is needed to understand the underlying mechanisms through which differentially methylated genes drive NASH-HCC development, and progression and to examine differentially methylated regions in the progression of NASH-HCC in humans.
Conclusions
A large number of differentially methylated regions are seen in the progression of NASH-HCC in both lean and obese mice; differential methylation is also seen between the stages of progression in the two models. With methylation differences seen in both mice and humans, further studies could be conducted using the obese and lean mice models to elucidate the mechanisms of progression of NASH-HCC in the context of obesity and normal weight. Additionally, differentially methylated regions may be able to serve as biomarkers for cancer progression or potential therapeutics, highlighting the importance of the study of epigenetic changes in hepatocarcinogenesis.
Supplementary Information
Additional file 1: Supplemental file 1. Absolute numbers and proportions of comparisons by comparison and chromosome. Description: Absolute number of hypermethylated and hypomethylated regions, totals, and proportions of between each comparison and by chromosome number in the choline supplemented, choline deficient, and control models.
Additional file 2: Supplemental Fig. 1. Comparison of HCC vs. NASH in choline supplemented and choline deficient models. Description: Percentages of CpG categories (islands, shores, or other) and gene structure categories for significant differential DNA methylation loci, number of DMRs per comparison, and number of DMRs per chromosome. A. Percentages of CpG categories for CD: HCC vs. NASH Percentages of B. CpG categories for CS: HCC vs. NASH. C. Percentages of gene structure categories for CD: HCC vs. NASH D. Percentages of gene structure categories for CS: HCC vs. NASH. E. Number of differentially methylated regions by comparison. F. Number of DMRs per chromosome comparing CD: HCC vs. NASH. G. Number of DMRs per chromosome comparing CS: HCC vs. NASH.
Acknowledgments
This work was funded by a Fred & Pamela Buffett Cancer Center, Cancer Genes and Molecular Regulation Program (CGMRP) Pilot Grant and a project leader award from the Nebraska Prevention of Obesity Diseases through Dietary Molecules NIH grant 1P20GM104320-04. Dr. Kurt W. Fisher is also funded by National Cancer Institute (NCI), grant CA22287. We thank the Bioinformatics and Systems Biology Core at UNMC for providing the DNA methylation data analysis services, which receives support from Nebraska Research Initiative (NRI) and NIH (2P20GM103427, 5P30CA036727 and 2U54GM115458).
Abbreviations
- NAFLD
Non-alcoholic liver disease
- NASH
Non-alcoholic steatohepatitis
- HCC
Hepatocellular carcinoma
- CD
Choline deficient
- CS
Choline supplemented
- HFFC
High trans-fat, fructose, and cholesterol
- RRBS
Reduced representation bisulfite sequencing
- DMR
Differentially methylated region
Authors’ contributions
PAF designed the study and oversaw the study. EH and PAF analyzed the data, prepared the figures and tables, drafted the manuscript, and revised the manuscript. KF performed the histological evaluations of the liver tissue. All authors read and approved the final manuscript.
Funding
This work was funded by a Fred & Pamela Buffett Cancer Center, Cancer Genes and Molecular Regulation Program (CGMRP) Pilot Grant and a project leader award from the Nebraska Prevention of Obesity Diseases through Dietary Molecules NIH grant 1P20GM104320–04. The funders were not involved in the design, collection, analysis, interpretation, or writing of the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval
All methods were carried out in accordance with relevant institutional guidelines and recommendations. The study was approved by the Institutional Animal Care and Use Committee at the University of Nebraska Medical Center (Protocol #: 17–018). All methods are reported in accordance with the ARRIVE guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15(1):11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 2.Kutlu O, Kaleli HN, Ozer E. Molecular pathogenesis of nonalcoholic steatohepatitis- (NASH-) related hepatocellular carcinoma. Can J Gastroenterol Hepatol. 2018;2018:8543763. doi: 10.1155/2018/8543763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown AL, Conrad K, Allende DS, Gromovsky AD, Zhang R, Neumann CK, et al. Dietary choline supplementation attenuates high-fat-diet-induced hepatocellular carcinoma in mice. J Nutr. 2020;150(4):775–783. doi: 10.1093/jn/nxz315. [DOI] [PubMed] [Google Scholar]
- 4.Fracanzani AL, Petta S, Lombardi R, Pisano G, Russello M, Consonni D, et al. Liver and cardiovascular damage in patients with lean nonalcoholic fatty liver disease, and association with visceral obesity. Clin Gastroenterol Hepatol. 2017;15(10):1604–11.e1. doi: 10.1016/j.cgh.2017.04.045. [DOI] [PubMed] [Google Scholar]
- 5.Younes R, Bugianesi E. NASH in lean individuals. Semin Liver Dis. 2019;39(1):86–95. doi: 10.1055/s-0038-1677517. [DOI] [PubMed] [Google Scholar]
- 6.Fernández-Barrena MG, Arechederra M, Colyn L, Berasain C, Avila MA. Epigenetics in hepatocellular carcinoma development and therapy: the tip of the iceberg. JHEP Rep. 2020;2(6):100167. doi: 10.1016/j.jhepr.2020.100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vlock EM, Karanjit S, Talmon G, Farazi PA. Reduction of polyunsaturated fatty acids with tumor progression in a lean non-alcoholic steatohepatitis-associated hepatocellular carcinoma mouse model. J Cancer. 2020;11(19):5536–5546. doi: 10.7150/jca.48495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedossa P, Poitou C, Veyrie N, Bouillot JL, Basdevant A, Paradis V, et al. Histopathological algorithm and scoring system for evaluation of liver lesions in morbidly obese patients. Hepatology. 2012;56(5):1751–1759. doi: 10.1002/hep.25889. [DOI] [PubMed] [Google Scholar]
- 9.Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13(10):R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang HQ, Tuominen LK, Tsai CJ. SLIM: a sliding linear model for estimating the proportion of true null hypotheses in datasets with dependence structures. Bioinformatics. 2011;27(2):225–231. doi: 10.1093/bioinformatics/btq650. [DOI] [PubMed] [Google Scholar]
- 11.UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D4D9. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47(D1):D419–DD26. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Gene Ontology resource Enriching a GOld mine. Nucleic Acids Res. 2021;49(D1):D325–Dd34. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Xie L, Gunasekar SK, Tong D, Mishra A, Gibson WJ, et al. SWELL1 is a regulator of adipocyte size, insulin signalling and glucose homeostasis. Nat Cell Biol. 2017;19(5):504–517. doi: 10.1038/ncb3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henley KD, Gooding KA, Economides AN, Gannon M. Inactivation of the dual bmp/Wnt inhibitor Sostdc1 enhances pancreatic islet function. Am J Physiol Endocrinol Metab. 2012;303(6):E752–E761. doi: 10.1152/ajpendo.00531.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrade S, Morais T, Sandovici I, Seabra AL, Constância M, Monteiro MP. Adipose tissue epigenetic profile in obesity-related Dysglycemia - a systematic review. Front Endocrinol (Lausanne) 2021;12:681649. doi: 10.3389/fendo.2021.681649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hair BY, Troester MA, Edmiston SN, Parrish EA, Robinson WR, Wu MC, et al. Body mass index is associated with gene methylation in estrogen receptor-positive breast tumors. Cancer Epidemiol Biomark Prev. 2015;24(3):580–586. doi: 10.1158/1055-9965.EPI-14-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masgras I, Sanchez-Martin C, Colombo G, Rasola A. The chaperone TRAP1 as a modulator of the mitochondrial adaptations in Cancer cells. Front Oncol. 2017;7:58. doi: 10.3389/fonc.2017.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Xu T, Liu C, Meng G, Sun Y, Qian L, et al. Liver-enriched genes are associated with the prognosis of patients with hepatocellular carcinoma. Sci Rep. 2018;8(1):11197. doi: 10.1038/s41598-018-29237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skawran B, Steinemann D, Weigmann A, Flemming P, Becker T, Flik J, et al. Gene expression profiling in hepatocellular carcinoma: upregulation of genes in amplified chromosome regions. Mod Pathol. 2008;21(5):505–516. doi: 10.1038/modpathol.3800998. [DOI] [PubMed] [Google Scholar]
- 23.Vastrad B, Vastrad C. Bioinformatics analysis of differentially expressed genes in non alcoholic fatty liver disease using next generation sequencing data. bioRxiv. 2021:2021.12.16.472893. 10.1101/2021.12.16.472893.
- 24.Liu Y, Zhang JB, Qin Y, Wang W, Wei L, Teng Y, et al. PROX1 promotes hepatocellular carcinoma metastasis by way of up-regulating hypoxia-inducible factor 1α expression and protein stability. Hepatology. 2013;58(2):692–705. doi: 10.1002/hep.26398. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Cheng Y, Yan LL, An R, Wang XY, Wang HY. Exploring DNA methylation profiles altered in cryptogenic hepatocellular carcinomas by high-throughput targeted DNA methylation sequencing: a preliminary study for cryptogenic hepatocellular carcinoma. Onco Targets Ther. 2020;13:9901–9916. doi: 10.2147/OTT.S267812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang CK, Wang XK, Liao XW, Han CY, Yu TD, Qin W, et al. Aldehyde dehydrogenase 1 (ALDH1) isoform expression and potential clinical implications in hepatocellular carcinoma. PLoS One. 2017;12(8):e0182208. doi: 10.1371/journal.pone.0182208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Chen Z, Huang Z, Yu H, Li Y, Huang W. Formiminotransferase Cyclodeaminase suppresses hepatocellular carcinoma by modulating cell apoptosis, DNA damage, and phosphatidylinositol 3-kinases (PI3K)/Akt signaling pathway. Med Sci Monit. 2019;25:4474–4484. doi: 10.12659/MSM.916202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang Q, Tan C, Xiao F, Yin F, Liu M, Lei L, et al. Integrated profiling identifies ITGB3BP as prognostic biomarker for hepatocellular carcinoma. Bosn J Basic Med Sci. 2021;21(6):712–723. doi: 10.17305/bjbms.2021.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang WP, Gao HY. Combination therapy of hTERTR and FAM96A for hepatocellular carcinoma through enhancing apoptosis sensitivity. Exp Ther Med. 2018;15(1):641–648. doi: 10.3892/etm.2017.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drozdov I, Bornschein J, Wex T, Valeyev NV, Tsoka S, Malfertheiner P. Functional and topological properties in hepatocellular carcinoma transcriptome. PLoS One. 2012;7(4):e35510. doi: 10.1371/journal.pone.0035510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahajan N, Khare P, Kondepudi KK, Bishnoi M. TRPA1: pharmacology, natural activators and role in obesity prevention. Eur J Pharmacol. 2021;912:174553. doi: 10.1016/j.ejphar.2021.174553. [DOI] [PubMed] [Google Scholar]
- 32.Busque SM, Stange G, Wagner CA. Dysregulation of the glutamine transporter Slc38a3 (SNAT3) and ammoniagenic enzymes in obese, glucose-intolerant mice. Cell Physiol Biochem. 2014;34(2):575–589. doi: 10.1159/000363024. [DOI] [PubMed] [Google Scholar]
- 33.Escobedo N, Proulx ST, Karaman S, Dillard ME, Johnson N, Detmar M, et al. Restoration of lymphatic function rescues obesity in Prox1-haploinsufficient mice. JCI Insight. 2016;1(2):e85096. doi: 10.1172/jci.insight.85096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dogan H, Shu J, Hakguder Z, Xu Z, Cui J. Elucidation of molecular links between obesity and cancer through microRNA regulation. BMC Med Genet. 2020;13(1):161. doi: 10.1186/s12920-020-00797-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huckins LM, Hatzikotoulas K, Southam L, Thornton LM, Steinberg J, Aguilera-McKay F, et al. Investigation of common, low-frequency and rare genome-wide variation in anorexia nervosa. Mol Psychiatry. 2018;23(5):1169–1180. doi: 10.1038/mp.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dreval K, Tryndyak V, de Conti A, Beland FA, Pogribny IP. Gene expression and DNA methylation alterations during non-alcoholic steatohepatitis-associated liver carcinogenesis. Front Genet. 2019;10:486. doi: 10.3389/fgene.2019.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuramoto J, Arai E, Tian Y, Funahashi N, Hiramoto M, Nammo T, et al. Genome-wide DNA methylation analysis during non-alcoholic steatohepatitis-related multistage hepatocarcinogenesis: comparison with hepatitis virus-related carcinogenesis. Carcinogenesis. 2017;38(3):261–270. doi: 10.1093/carcin/bgx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Locke WJ, Guanzon D, Ma C, Liew YJ, Duesing KR, Fung KYC, et al. DNA methylation Cancer biomarkers: translation to the clinic. Front Genet. 2019;10:1150. doi: 10.3389/fgene.2019.01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem. 2012;23(8):853–859. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hymel E, Vlock E, Fisher KW, Farazi PA. Differential progression of unhealthy diet-induced hepatocellular carcinoma in obese and non-obese mice. PLoS One. 2022;17(8):e0272623. doi: 10.1371/journal.pone.0272623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hotta K, Kitamoto A, Kitamoto T, Ogawa Y, Honda Y, Kessoku T, et al. Identification of differentially methylated region (DMR) networks associated with progression of nonalcoholic fatty liver disease. Sci Rep. 2018;8(1):13567. doi: 10.1038/s41598-018-31886-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gundamaraju R, Lu W, Manikam R. CHCHD2: the power House's potential prognostic factor for Cancer? Front Cell Dev Biol. 2020;8:620816. doi: 10.3389/fcell.2020.620816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao Y, Su J, Zhao L, Li R, Liu K, Wang S. CHCHD2 promotes hepatocellular carcinoma and indicates poor prognosis of hepatocellular carcinoma patients. J Cancer. 2019;10(27):6822–6828. doi: 10.7150/jca.31158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, Zhang Y, Li L, Cao J, Guo Y, Wu Y, et al. Fascin actin-bundling protein 1 in human cancer: promising biomarker or therapeutic target? Molecular Therapy - Oncolytics. 2021;20:240–264. doi: 10.1016/j.omto.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Udali S, Guarini P, Ruzzenente A, Ferrarini A, Guglielmi A, Lotto V, et al. DNA methylation and gene expression profiles show novel regulatory pathways in hepatocellular carcinoma. Clin Epigenetics. 2015;7:43. doi: 10.1186/s13148-015-0077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shorthouse D, Rahrmann E, Kosmidou C, Greenwood B, Hall M, Devonshire G, Gilbertson R, Fitzgerald R, Hall B. KCNQ gene family members act as both tumor suppressors and oncogenes in gastrointestinal cancers. bioRxiv. 2020:2020.03.10.984039.
- 47.Chen B, Liao Z, Qi Y, Zhang H, Su C, Liang H, et al. miR-631 inhibits intrahepatic metastasis of hepatocellular carcinoma by targeting PTPRE. Front Oncol. 2020;10:565266. doi: 10.3389/fonc.2020.565266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu L, Wu S, Yang Y, Cai J, Zhu X, Wu J, et al. SOSTDC1 is down-regulated in non-small cell lung cancer and contributes to cancer cell proliferation. Cell Biosci. 2016;6:24. doi: 10.1186/s13578-016-0091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang M, Zeng Q, Dai S, Liang H, Dai F, Xie X, et al. Comparative analysis of hepatocellular carcinoma and cirrhosis gene expression profiles. Mol Med Rep. 2017;15(1):380–386. doi: 10.3892/mmr.2016.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bian J, Long J, Yang X, Xu Y, Lu X, Guan M, et al. Construction and validation of a prognostic signature using CNV-driven genes for hepatocellular carcinoma. Ann Transl Med. 2021;9(9):765. doi: 10.21037/atm-20-7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Giorgi V, Buonaguro L, Worschech A, Tornesello ML, Izzo F, Marincola FM, et al. Molecular signatures associated with HCV-induced hepatocellular carcinoma and liver metastasis. PLoS One. 2013;8(2):e56153. doi: 10.1371/journal.pone.0056153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen J, Tsoi H, Liang Q, Chu ES, Liu D, Yu AC, et al. Oncogenic mutations and dysregulated pathways in obesity-associated hepatocellular carcinoma. Oncogene. 2016;35(49):6271–6280. doi: 10.1038/onc.2016.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khalaf AM, Fuentes D, Morshid AI, Burke MR, Kaseb AO, Hassan M, et al. Role of Wnt/β-catenin signaling in hepatocellular carcinoma, pathogenesis, and clinical significance. J Hepatocell Carcinoma. 2018;5:61–73. doi: 10.2147/JHC.S156701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4(2):68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He S, Tang S. WNT/β-catenin signaling in the development of liver cancers. Biomed Pharmacother. 2020;132:110851. doi: 10.1016/j.biopha.2020.110851. [DOI] [PubMed] [Google Scholar]
- 56.Wong AM, Ding X, Wong AM, Xu M, Zhang L, Leung HH-W, et al. Unique molecular characteristics of NAFLD-associated liver cancer accentuate β-catenin/TNFRSF19-mediated immune evasion. J Hepatol. 2022;77(2):410–423. doi: 10.1016/j.jhep.2022.03.015. [DOI] [PubMed] [Google Scholar]
- 57.Jin N, George TL, Otterson GA, Verschraegen C, Wen H, Carbone D, et al. Advances in epigenetic therapeutics with focus on solid tumors. Clin Epigenetics. 2021;13(1):83. doi: 10.1186/s13148-021-01069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplemental file 1. Absolute numbers and proportions of comparisons by comparison and chromosome. Description: Absolute number of hypermethylated and hypomethylated regions, totals, and proportions of between each comparison and by chromosome number in the choline supplemented, choline deficient, and control models.
Additional file 2: Supplemental Fig. 1. Comparison of HCC vs. NASH in choline supplemented and choline deficient models. Description: Percentages of CpG categories (islands, shores, or other) and gene structure categories for significant differential DNA methylation loci, number of DMRs per comparison, and number of DMRs per chromosome. A. Percentages of CpG categories for CD: HCC vs. NASH Percentages of B. CpG categories for CS: HCC vs. NASH. C. Percentages of gene structure categories for CD: HCC vs. NASH D. Percentages of gene structure categories for CS: HCC vs. NASH. E. Number of differentially methylated regions by comparison. F. Number of DMRs per chromosome comparing CD: HCC vs. NASH. G. Number of DMRs per chromosome comparing CS: HCC vs. NASH.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.