Abstract
Type I interferons (IFNs) are among the most powerful tools that host cells deploy against intracellular pathogens. Their effectiveness is due to both the rapid, directly antiviral effects of IFN-stimulated gene products and to the effects of type I IFN on responding immune cells. Type I IFN signaling through its receptor, IFNAR, is tightly regulated at multiple steps in the signaling cascade, including at the level of IFNAR downstream effectors, which include the kinase JAK1 and the transcriptional regulator STAT1. Here, we found that tumor necrosis factor receptor (TNFR)–associated factor 3 (TRAF3) enhanced the activation of JAK1 and STAT1 specifically in CD4+ T cells by preventing recruitment of the negative regulatory phosphatase PTPN22 to the IFNAR complex. The balance between signals through IFNAR and other cytokine receptors influenced CD4+ T cell differentiation and function during infections. Our work reveals TRAF3 and PTPN22 as key regulators of CD4+ T cell activation by type I IFNs.
INTRODUCTION
Type I interferons (IFNs) are potent antiviral cytokines that are primarily produced in response to the activation of pattern recognition receptors (1, 2). Signaling through the ubiquitously expressed type I IFN receptor (IFNAR) leads to the robust and rapid increased expression of interferon-stimulated genes (ISGs) whose products affect cell growth, survival, differentiation, and the promotion of a core program of antiviral effector molecules (3). The production of type I IFNs downstream of pattern recognition receptor activation and IFNAR signaling has largely been studied in myeloid cells, as part of the early innate immune response. However, type I IFN also modulates the phenotype and function of adaptive immune cells through the cell type–dependent induction of ISGs (4). The effects of type I IFNs on T cells are highly context-dependent; the phenotypic result is determined by additional signals and the differentiation status of the T cell receiving the signal (5). These divergent outcomes suggest complex regulation of IFNAR signaling in CD4+ T cells, but the specific mechanisms are unknown. Our study addresses this important gap in knowledge by describing a distinct role for tumor necrosis factor receptor (TNFR)–associated factor 3 (TRAF3) in promoting IFNAR signaling in CD4+ T cells by preventing the recruitment of negative regulators.
Type I IFN initiates signaling by binding to the IFNAR2 subunit of the IFNAR complex, which in turn recruits the IFNAR1 subunit. Proximity of these IFNAR subunits then leads to transactivation of the Janus kinase (JAK) family members JAK1 and TYK2 (6, 7). The JAK family members phosphorylate tyrosine residues in the cytoplasmic domains of IFNAR1 and IFNAR2, which enables the recruitment and activation of transcriptional activators called signal transducers and activators of transcription (STATs) (8). In canonical IFNAR signaling, activated STAT1 and STAT2 heterodimerize and bind to interferon regulatory factor 9 (IRF9), forming a complex that translocates to the nucleus to activate the transcription of ISGs (9). IFNAR signaling has potent downstream phenotypic effects; thus, it is tightly controlled by inducible and constitutively expressed proteins (10). Among the constitutively expressed regulators are the protein tyrosine phosphatases PTP1B and PTPN2, which dephosphorylate TYK2 and JAK1, respectively (11-13). Inducible suppressor of cytokine signaling (SOCS) proteins decrease IFNAR signaling at later times; SOCS1 targets JAK1, TYK2, and STAT1 to inhibit antiviral responses induced by type I IFN (14-16).
TRAF3 is a widely expressed adaptor protein whose function varies by cell and receptor type. Lineage-specific deletion of TRAF3 revealed distinct roles for TRAF3 in Toll-like receptor, cytokine receptor, and antigen receptor signaling in both B and T lymphocytes (17-25). In conventional T cells, TRAF3 is necessary for the normal magnitude of signaling by the T cell antigen receptor (TCR) and TCR costimulatory receptors of the TNFR superfamily. Mice lacking TRAF3 in all mature T cells (T-Traf3−/−) have normal T, B, and myeloid cell numbers but mount poor CD4+ and CD8+ T cell responses to infection by the intracellular bacterium Listeria monocytogenes and cannot produce a T cell–dependent humoral response to immunization (22). Many of the functional defects exhibited by TRAF3-deficient T cells originate with defective TCR signaling, which we described in T-Traf3−/− mouse T cells and which was corroborated in human patients with one mutated TRAF3 allele (22, 26). A major role of TRAF3 in promoting TCR function is to interfere with negative regulators of early TCR signals. TRAF3 limits the localization of protein tyrosine phosphatase nonreceptor type 22 (PTPN22) and the negative regulatory kinase Csk to the TCR complex, thereby enhancing TCR signaling (18). Furthermore, TRAF3 associates with the TCR signaling protein linker of activated T cells (LAT) and restrains Dok1, a negative regulator of LAT, through a mechanism involving PTP1B (25).
Modulation of PTP localization and association with key signaling proteins are recurrent themes in the TRAF3-mediated regulation of cytokine and antigen receptors. For example, TRAF3 facilitates the association of the negative regulator PTPN22 with JAK1 at the interleukin-6 (IL-6) receptor in B cells to decrease signaling (17). In thymic regulatory T (Treg) cells, in contrast to its role in promoting TCR signaling, TRAF3 curbs IL-2 receptor signaling by enabling the association of the negative regulator PTPN2 with JAK1 and JAK3 (23). However, TRAF3 promotes IL-15 receptor signaling in invariant natural killer (iNK) T cells (27). Thus, TRAF3 can both enhance and inhibit cytokine receptor signaling in a highly cell- and context-dependent manner.
The sequence of naïve T cell activation typically involves “signal 1” (from the TCR), “signal 2” (costimulation by CD28), and “signal 3” (signaling through cytokine receptors). In T cells, type I IFN can act as a signal 3 cytokine, promoting proliferation and the acquisition of effector functions (28-30). In the absence of the previous signals 1 and 2, type I IFN can exert pro-apoptotic and antiproliferative effects in CD8+ T cells (14, 31-33). TRAF3 participates in signals 1 and 2 of T cell activation and cytokine signaling in other contexts, but its participation in signal 3, cytokine-mediated activation of T cells, is not well defined (22, 24, 25, 34). The outcome of acute viral infections, including that by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), depends on the appropriate induction of and response to type I IFNs, particularly by adaptive immune cells (35, 36). Here, we investigated the role of TRAF3 in the T cell–specific regulation of IFNAR signaling. We report that TRAF3-deficient CD4+ T cells had reduced early IFNAR signaling events and that these early defects had long-lasting effects on the downstream consequences of IFNAR signaling. We provide evidence that the decrease in signaling in the absence of TRAF3 was due to aberrant recruitment of PTPN22, which can target JAK1, to the IFNAR signaling complex. Given the conflicting reports about the regulation of IFNAR signaling by PTPN22 in different cell types (37, 38), our findings suggest that TRAF3 may control PTP access to cytokine receptors in a cell type–specific manner to prevent dysregulation.
RESULTS
T cell–specific TRAF3 deficiency inhibits early IFNAR signaling events
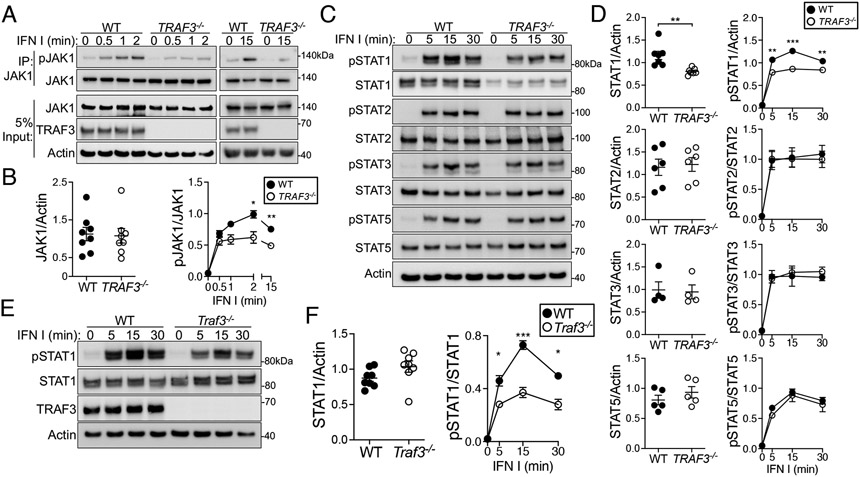
To determine how TRAF3 deficiency affected T cell IFNAR signaling, we assessed the early activation of JAK1, which becomes phosphorylated on the activating tyrosine residues Tyr1034 and Tyr1035 (Tyr1034/1035) upon type I IFN engagement of IFNAR (7). Activating phosphorylation of JAK1 was significantly reduced after type I IFN stimulation in a subclone of the CD4+ HuT28 human T cell lymphoma cell line lacking TRAF3, compared to the parental, wild-type (WT) cells (Fig. 1, A and B). Activated JAKs phosphorylate STAT family members, all of which may be activated by IFNAR signaling (39). We examined STAT1, STAT2, STAT3, and STAT5, and found that phosphorylation of STAT1 at its activating tyrosine residue (Tyr701) was significantly decreased in TRAF3-deficient HuT28 cells and TRAF3-deficient primary mouse CD4+ T cells compared to that seen in WT cells (Fig. 1, C to F and fig. S1A). Activating phosphorylation of STAT2, STAT3, and STAT5 did not differ according to TRAF3 status (Fig. 1, C and D). We confirmed the TRAF3-dependence of the STAT1 activation defect by adding TRAF3 back to TRAF3-deficient HuT28 cells, which restored STAT1 activation proportional to the amount of TRAF3 expressed in the cells (fig. S1, B and C). We found that total STAT1 protein was decreased in abundance in TRAF3-deficient HuT28 cells, but this was not observed in TRAF3-deficient primary mouse CD4+ T cells (Fig. 1, C to F), so this observation may be specific to the transformed status of HuT28 cells. We therefore normalized pSTAT1 Tyr701 in HuT28 cells and their derivatives to a loading control (actin or GAPDH) throughout to clearly convey the total amount of activated STAT1 in the cells. Our laboratory has described several signaling pathways that are altered in TRAF3-deficient B cells, but we had not investigated whether TRAF3 deficiency affected B cell IFNAR signaling. In contrast to T cells, mouse B cells with and without TRAF3 displayed similar activating STAT1 phosphorylation upon type I IFN stimulation (fig. S1D). Additionally, differences in IFN-γ-mediated activation of STAT1 in TRAF3-deficient HuT28 cells were not statistically significant (fig. S2A). These data are consistent with the overall findings of our group and others that TRAF3 performs distinct functions in different cell types, and this may be an underlying mechanism of differential IFNAR regulation among different immune cell types.
Fig. 1. Effect of TRAF3 deficiency upon early IFNAR-mediated signaling in T cells.
(A to C) WT and TRAF3−/− HuT28 human T cells were treated with type I IFN (1000 U/ml) for the indicated times. (A) JAK1 was immunoprecipitated from cell lysates and its activation (as assessed by detection of phosphorylation at Tyr1034/1035) was analyzed by Western blotting. (B) Quantification of the relative amounts of total and phosphorylated JAK1 from four biological replicates. (C) Representative Western blotting analysis of the phosphorylation and activation of STAT1 (pTyr701), STAT2 (pTyr690), STAT3 (pTyr705), and STAT5 (pTyr694). (D) Quantification of the relative amounts of the indicated total and phosphorylated STAT proteins from six biological replicates. (E and F) Mouse CD4+ T cells were treated with type I IFN (1000 U/ml) for the indicated times. (E) Representative Western blotting analysis of STAT1 activation (pTyr701). (F) Quantification of the relative amounts of total and phosphorylated STAT1 from seven biological replicates; n = 7 mice per group. Blots are representative of four (A), six (B), and seven (E) independent experiments with similar results. *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired t test (for basal total protein) or two-way ANOVA with Sidak’s multiple comparisons test (for time courses). Error bars represent the SEM.
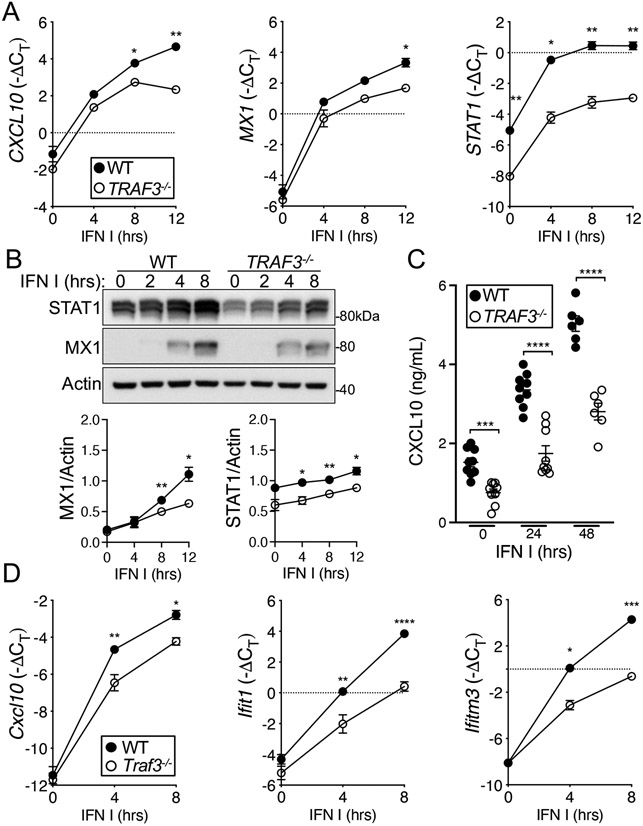
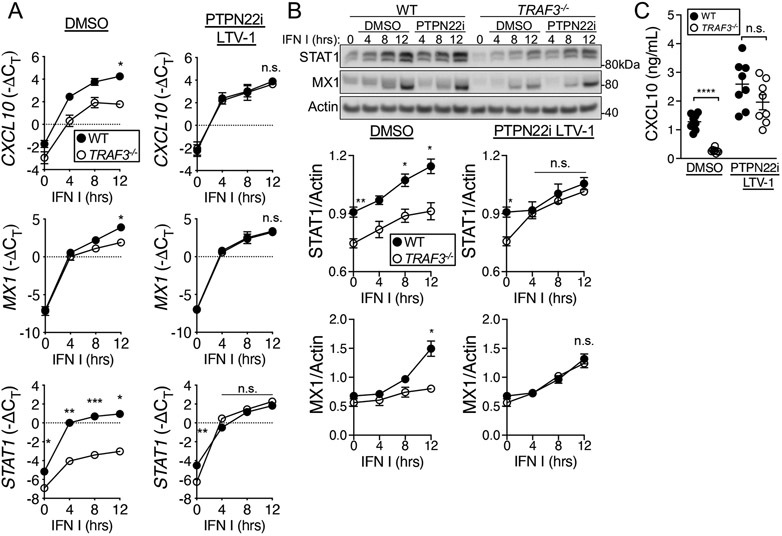
We next interrogated the downstream consequences of the defective IFNAR signaling in TRAF3-deficient T cells. Unphosphorylated STAT1 (USTAT1) controls the transcription of a number of genes in resting cells, including some that are also increased in expression upon type I IFN stimulation (40). Consistent with this, TRAF3-deficient HuT28 cells, which have reduced STAT1 protein abundance compared to that in WT cells (Fig. 1, C and D), showed decreased transcription of the STAT1 target genes CXCL10, MX1, and STAT1 both under basal conditions and upon type I IFN stimulation (Fig. 2A). Additionally, we saw decreases in the amounts of the corresponding proteins both under basal conditions and upon type I IFN stimulation in TRAF3−/− HuT28 cells (Fig. 2, B and C). In TRAF3-deficient primary mouse CD4+ T cells, STAT1 target genes were less robustly induced compared to WT cells upon type I IFN stimulation (Fig. 2D). The decreased transcription of these STAT1 target genes under basal conditions observed in HuT28 lymphoma-derived human T cells was not observed in mouse primary cells, which may reflect the similar amounts of total STAT1 in Traf3+/+ and Traf3−/− primary T cells (Fig. 1, E and F). Our target genes were carefully chosen to include genes with IFN-γ–activated sequence (GAS) regulatory sites, which are bound by STAT1 homodimers, and IFN-stimulated response elements (ISREs), which are bound by ISGF3, in the complex formed by STAT1, STAT2, and IRF9. The transcriptional defect in TRAF3-deficient T cells was apparent in genes with ISRE, GAS, or both in their regulatory regions, but we found no evidence to suggest that this was due to differences in the abundances of IRF9 or other IRFs (fig. S2B), which control the transcriptional response to IFNs. Together, these data from HuT28 human T cells and mouse CD4+ T cells indicate that TRAF3 deficiency interferes with normal signaling through IFNAR and the activation of downstream transcriptional programs.
Fig. 2. Effect of TRAF3 deficiency on ISG transcription in T cells.
(A to C) WT and TRAF3−/− HuT28 human T cells were stimulated with type I IFN (1000 U/ml) for the indicated times. (A) Induction of the expression of CXCL10, MX1, and STAT1 during type I IFN treatment, relative to that of a housekeeping gene [−ΔCT = -(CTgene of interest- CT housekeeping)], was determined by RT-qPCR analysis. (B) Top: Representative Western blotting analysis of STAT1 and MX1 proteins in the indicated cells. Bottom: Quantification of the normalized amounts of STAT1 and MX1 from seven biological replicates. (C) The amounts of CXCL10 protein secreted by the indicated cells were determined by ELISA. Each point represents an individual biological replicate. (D) Relative expression of Cxcl10, Ifitm3, and Ifit1 in type I IFN–stimulated control (WT) and Traf3−/− mouse CD4+ T cells. Graphs include biological replicates from three (A and D) and seven (B and C) independent experiments with similar results. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by two-way ANOVA with Sidak’s multiple comparisons test. Error bars represent the SEM.
Role of phosphatase recruitment in IFNAR signaling in TRAF3-deficient T cells
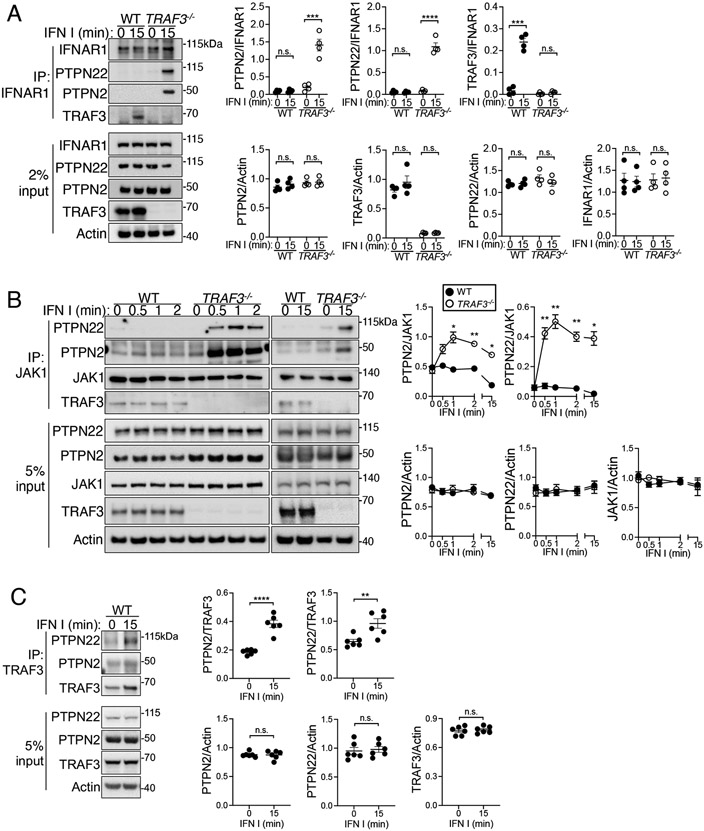
We next investigated the regulation of early signaling events at the IFNAR complex. IFNAR signaling is negatively regulated at various levels by IFN-induced and constitutively present phosphatases (10). Among these are PTPN2 and PTPN22, both of which associate with TRAF3 in T cells to regulate signaling through the IL-2R and TCR, respectively (23, 24, 34). Given these precedents for regulation of receptor-associated phosphatase localization by TRAF3, we focused on the association of PTPN2 and PTPN22 as potential targets of the TRAF3-mediated optimization of IFNAR signaling. We observed the association of PTPN2 and PTPN22 with IFNAR1 in type I IFN–stimulated TRAF3−/− HuT28 cells, but not in WT cells (Fig. 3A). We also observed the association of TRAF3 with IFNAR1 in response to type I IFN (Fig. 3A). The relative amounts of PTPN22 and PTPN2 in whole-cell lysates were not affected by the loss of TRAF3, suggesting that the presence of PTPN2 and PTPN22 at the IFNAR signaling complex in TRAF3-deficient HuT28 cells was due to preferential recruitment or a failure to dissociate, rather than to a change in total protein abundance. PTPN2 and PTPN22 target members of the JAK family of kinases for dephosphorylation, which reduces JAK activation (11, 17, 38), so we next interrogated whether TRAF3 deficiency affected the association of these phosphatases with JAK1. At short (30 s to 2 min) and longer (15 min) time points after type I IFN stimulation, more PTPN2 and PTPN22 were associated with JAK1 in TRAF3−/− HuT28 cells than in WT HuT28 cells (Fig. 3B). In WT cells, both PTPN2 and PTPN22 associated with TRAF3 after type I IFN stimulation (Fig. 3C). Together, these data suggest that in TRAF3-sufficient cells, TRAF3 associates with PTPN2 and PTPN22, altering or preventing their recruitment to components of the IFNAR signaling complex.
Fig. 3. TRAF3 deficiency alters phosphatase recruitment to the IFNAR complex.
(A to C) Left: WT and TRAF3-deficient HuT28 human T cells were stimulated with type I IFN I (1000 U/ml) for the indicated times. The indicated proteins were immunoprecipitated (IP) from cell lysates, and associated proteins or whole-cell lysates (input) were analyzed by Western blotting. Right: Quantification of the relative amounts of the indicated proteins from multiple replicates. Western blots are representative of three (A), eight (B), and six (C) biological replicates from the same number of independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; n.s., not significant by unpaired t test (A and C) or two-way ANOVA with Sidak’s multiple comparisons test (B). Error bars represent the SEM.
PTPN22 inhibition restores IFNAR signaling in TRAF3-deficient T cells
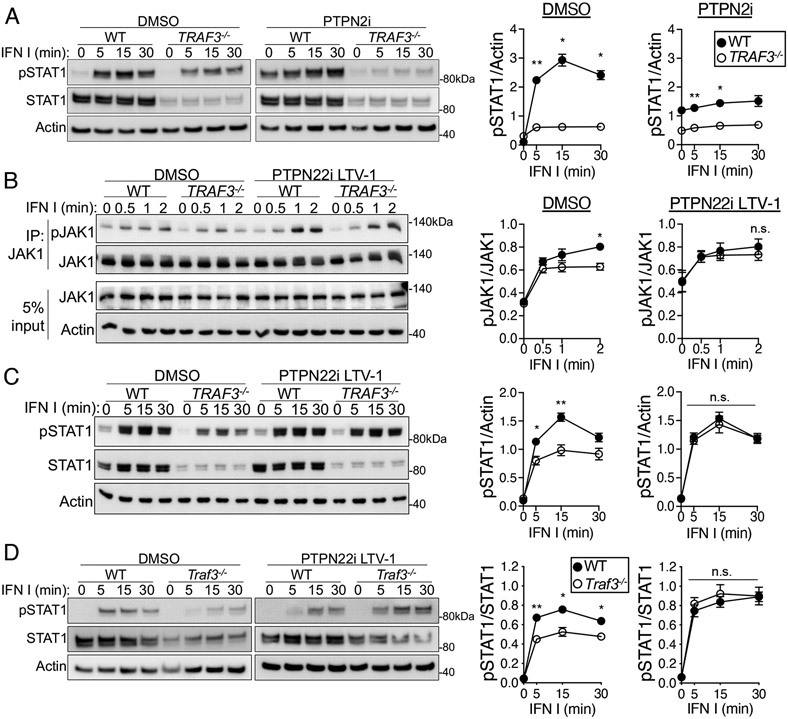
We hypothesized that the decreased IFNAR signaling in TRAF3-deficient cells was due to the enhanced recruitment of the phosphatases PTPN2, PTPN22, or both to the IFNAR signaling complex. To test this hypothesis, we treated cells with competitive inhibitors to block phosphatase activity before and during treatment with type I IFN I and then assessed IFNAR signaling events and downstream consequences. PTPN2 inhibition increased the basal amount of pSTAT1 Tyr701 in both WT and TRAF3-deficient HuT28 cells but did not increase type I IFN–stimulated pSTAT1 Tyr701 abundance to levels observed in WT cells (Fig. 4A and fig. S3A). Inhibition of PTPN22 with two small-molecule inhibitors rescued both the JAK1 activation defect in TRAF3−/− HuT28 cells and the abundance of pSTAT1 Tyr701 in TRAF3-deficient HuT28 cells (Fig. 4, B and C, and fig. S3, B to D). In mouse CD4+ T cells, type I IFN–induced pSTAT1 Tyr701 abundance was also restored to that of WT cells with PTPN22 inhibition (Fig. 4D). We predicted that the restoration of JAK-STAT signaling in TRAF3-deficient cells to the extent observed in WT cells would also restore downstream deficits in IFNAR-mediated function, and indeed, treatment of TRAF3−/− HuT28 cells with a PTPN22 inhibitor rescued the type I IFN–induced transcription of the ISGs CXCL10, MX1, and STAT1 (Fig. 5A). Consistent with the transcriptional data, STAT1 and MX1 protein abundance and CXCL10 production in WT and TRAF3−/− HuT cells were not significantly different when PTPN22 activity was inhibited before the cells were stimulated with type I IFN (Fig. 5, B and C).
Fig. 4. Role of PTPN22 in the regulation of IFNAR signaling by TRAF3 in T cells.
(A to C) WT and TRAF3-deficient HuT28 human T cells were pretreated with a PTPN22 inhibitor (PTPN22i, 10 μM LTV-1), a PTPN2 inhibitor (PTPN2i, 1 nM SF-1670), or an equivalent concentration of DMSO and then were stimulated with type I IFN (1000 U/ml) for the indicated times in the presence of inhibitor or DMSO. (A) Representative Western blotting analysis of pSTAT1 Tyr701 (left) and quantification of the relative amounts of pSTAT1 from four biological replicates (right). (B) Representative Western blotting analysis of pJAK1 Tyr1034/1035 (left) and quantification of the relative amounts of pJAK1 from five biological replicates (right). (C) Representative Western blotting analysis of pSTAT1 Tyr701 (left) and quantification of the relative amounts of pSTAT1 from four biological replicates (right). (D) Representative Western blotting analysis of pSTAT1 Tyr701 in mouse CD4+ T cells (left) that were treated as described in (A) with PTPN22i and type I IFN (left) and quantification of the relative amounts of pSTAT1 from four biological replicates (right), with n = 4 mice per group. Graphs include replicates from four (C and D) and five (B) independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by two-way ANOVA with Sidak’s multiple comparisons test. Error bars represent the SEM.
Fig. 5. Role of PTPN22 in the regulation of ISG expression by TRAF3 in T cells.
(A to C) WT and TRAF3-deficient HuT28 human T cells were pre-treated with 10uM LTV-1 (PTPN22i) or an equivalent concentration of DMSO and then were stimulated with type I IFN (1000 U/ml) for the indicated times in the presence of inhibitor or DMSO. (A) Induction of CXCL10, MX1, and STAT1 transcription, relative to that of a housekeeping gene [−ΔCT = -(CTgene of interest- CT housekeeping)] was determined by RT-qPCR analysis of three biological replicates. (B) Top: Representative Western blotting analysis of the accumulation of STAT1 and MX1 protein over time under the indicated conditions. Bottom: Quantification of the relative amounts of STAT1 and MX1 proteins from six biological replicates. (C) Quantification of the amounts of CXCL10 secreted by the indicated cells 24 hours after treatment with type I IFN was determined by ELISA analysis of samples from eight biological replicates. Graphs include replicates from three (A), six (B), and eight (C) independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by two-way ANOVA with Sidak’s multiple comparisons test. Error bars represent the SEM.
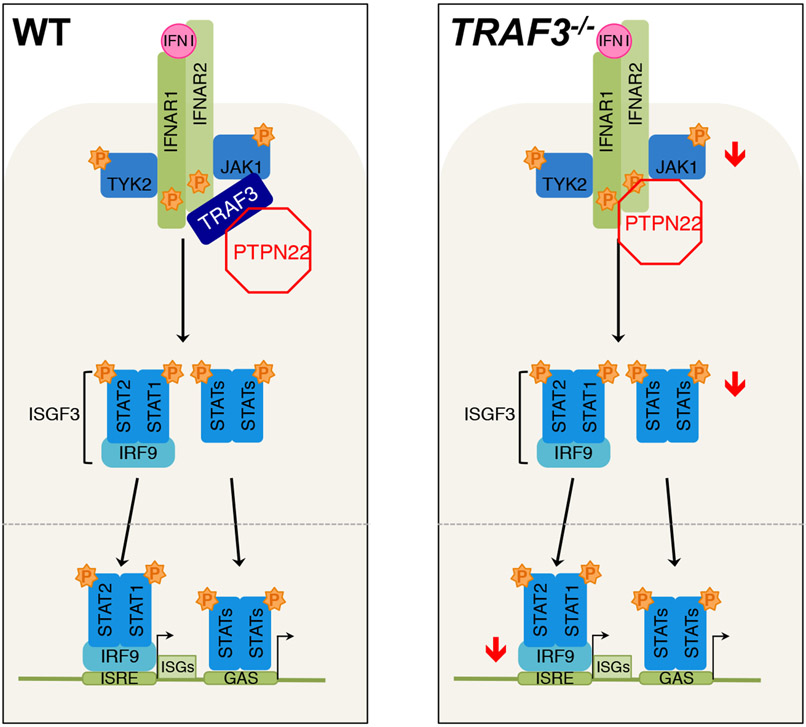
Together, these data support a model in which TRAF3 prevents the PTPN22-mediated inhibition of IFNAR signaling in T cells (Fig. 6). We showed that TRAF3-deficient T cells had dampened early signaling events in response to type I IFN compared to that in WT control cells, and this deficiency was also reflected in downstream transcriptional activation, particularly of STAT1 target genes. We propose that the decreased signaling and transcriptional response is due to the unchecked recruitment of the negative regulatory phosphatase PTPN22 to the IFNAR signaling complex in the absence of TRAF3. Inhibition of PTPN22 activity partially restored IFNAR signaling in TRAF3-deficient cells, which suggests that the inappropriate activity, localization, or both of this phosphatase in the absence of TRAF3 is the cause of defective IFNAR signaling in T cells lacking TRAF3. These findings reveal a role for TRAF3 in maintaining optimal signaling of the IFNAR receptor in CD4+ T cells and highlight PTPN22 as a key negative regulator of the IFNAR signaling complex in T cells.
Fig. 6. Model of the TRAF3-mediated regulation of IFNAR signaling in T cells.
In TRAF3-sufficient CD4+ T cells (left), TRAF3 prevents the recruitment of PTPN22 to the receptor complex, and downstream signaling and ISG transcription occurs normally. In CD4+ T cells without TRAF3 (right), PTPN22 is present at the IFNAR complex and impedes full activation of JAK1 and STAT1 and consequent ISG transcription.
DISCUSSION
Appropriate T cell responses are important for protection and recovery from infection, but inappropriate responses can be detrimental during autoimmune processes or in certain infectious contexts. Robust T cell activation depends on the receipt of three types of stimuli: signals through the TCR (signal 1), costimulatory receptors (signal 2), and cytokine receptors (signal 3). We previously reported thatTRAF3 participates in the regulation of signals 1 and 2 (22, 24, 25); here, we focused on the response to the important signal 3 cytokine, type I IFN. We revealed a role for TRAF3 in blocking the negative regulation of IFNAR signaling in T cells by the tyrosine phosphatase PTPN22. PTPN22, also known as LYP in humans and PEP in mice, is a tyrosine phosphatase that regulates effector functions and signaling in many immune cell types (41). PTPN22 variants with altered function can cause dysregulation of these effector functions, contributing to the pathogenesis of autoimmune diseases (41, 42). The most common among these variants is R620W, which increases phosphatase activity but reduces the PTPN22-TRAF3 association (37, 43). Our data from experiments with phosphatase-inactivating inhibitors show that PTPN22 phosphatase activity is required to restore IFNAR signaling to near WT levels in TRAF3-deficient cells.
TRAF3 promotes T cell–mediated immune responses through the inhibition of negative regulators, such as PTPN22, at the TCR as well. After TCR stimulation, TRAF3 sequesters PTPN22 in the cytoplasm, preventing it from dephosphorylating the early TCR signaling effector kinase Lck (24). TRAF3 also sequesters the TCR-inhibitory kinase CSK in the cytoplasm during TCR signaling, preventing it from inactivating Lck. TRAF3 is recruited to the TCR complex through the adaptor protein LAT, and it also inhibits the negative regulation of LAT (25). The cumulative effect of this segregation of negative regulators by TRAF3 is prolonged TCR signaling. The data shown here support a role for TRAF3 in preventing the inappropriate association of PTPN2 and PTPN22 with the IFNAR complex. TRAF3 additionally impedes signaling by the T cell costimulatory TNFR superfamily member GITR, through an incompletely understood mechanism that does not implicate phosphatase recruitment (44). As discussed earlier, TRAF3 also limits Treg cell differentiation by regulating the strength of IL-2R signaling on pre-Treg cells (23). Together, these findings indicate that TRAF3 overall enhances T cell activation, both by restraining or impeding the negative regulation of activating receptors in T cells, as well as restraining the functions of negative regulatory receptors, such as GITR, and the development of Treg cells. These effects of TRAF3 in T cells stand in contrast to its negative regulation of activating receptors in B cells. For example, at the IL-6R in B cells, TRAF3 enables PTPN22-mediated limitation of IL-6–induced JAK1 and STAT3 activation, and thereby restrains plasma cell differentiation (17). It will be interesting to determine whether TRAF3 impedes the access of PTPN22 or other phosphatases to additional receptors in T cells, and whether or to what extent its regulation of IFNAR involves the regulation of PTPN22 recruitment.
The main difference we observed between the primary mouse and transformed human TRAF3-deficient T cell phenotypes is the effect of TRAF3 on total STAT1 abundance. We showed here that a TRAF3-deficient human T cell lymphoma–derived cell line had reduced STAT1 abundance compared to that of WT cells, but this was not observed in mouse primary CD4+ T cells. One possible explanation for this discrepancy is low-level STAT1 activation leading to a differential increase in STAT1 abundance between WT and TRAF3-deficient HuT28 cells. IFN-γ is made in small quantities by resting HuT28 cells, and IFN-γ activates STAT1, which amplifies the transcription of STAT1. Such increases in STAT1 activity can last for days after IFN exposure (40). Our data suggest that STAT1 activation after IFN-γ receptor signaling was decreased in TRAF3-deficient HuT28 cells (fig. S1); therefore, this basal activation may be sufficient to result in a difference in total STAT1 abundance. We would expect normal STAT1 activation to result in an increase in STAT1 abundance in TRAF3-sufficient HuT28 cells, but not in TRAF3-deficient HuT28 cells. This hypothesis is particularly attractive because (i) we would not expect substantial or repeated exposure of T cells in a specific-pathogen-free mouse to IFN-γ, thus, we would not expect there to be substantial differences in total STAT1 abundance between TRAF3-sufficient and TRAF3-deficient T cells; and (ii) our previous work established that compared to TRAF3-sufficient cells, TRAF3-deficient mouse CD4+ T cells have enhanced activation of STAT1 in response to IFN-γ (23). We would therefore expect any incidental exposures to IFN-γ to stimulate an increase in STAT1 abundance in the TRAF3-deficient mouse CD4+ T cells compared to that in WT mouse CD4+ T cells. Indeed, we saw a trend toward slightly increased STAT1 abundance in the TRAF3-deficient mouse CD4+ T cells (Fig. 1F).
An intriguing question is what roles PTPN2 and PTPN22 play in IFNAR signaling in WT CD4+ T cells. Published data are incomplete and conflicting as to how and where these phosphatases regulate IFNAR signaling, and it is unclear whether there are cell type–specific differences that may explain some of the discrepancies. Thymocytes from PTPN2-deficient mice exhibit enhanced STAT1 activation after stimulation with type I IFN, but the effect on JAK1 activation was only reported in response to IFN-γ stimulation (11). PTPN22-deficient, bone marrow–derived macrophages have both enhanced and unchanged type I IFN–mediated STAT1 activation according to different reports (37, 38). We showed here that neither PTPN2 nor PTPN22 was recruited to the IFNAR signaling complex within 15 min of the initiation of signaling, except in T cells lacking TRAF3. This suggests that neither phosphatase is a key early negative regulator of T cell IFNAR signaling in the presence of TRAF3. Inhibition of PTPN2 and PTPN22 also failed to enhance the extent of type I IFN–induced JAK1-STAT1 phosphorylation compared to that in unmanipulated WT T cells, further supporting the presence of TRAF3 as playing a key role in preventing early phosphatase activity. JAK1 is a well-established target of PTPN2 and is present at multiple receptor complexes that use JAK-STAT signaling. It seems likely that a key part of the function of TRAF3 is to prevent the inappropriate localization of PTPN2 to JAK1 in response to type I IFN signaling. The data from our PTPN2 inhibitor experiments did not provide evidence that PTPN2 inhibition detectably enhanced type I IFN–mediated STAT1 activation in T cells, but this apparent inconsistency with the limited published data may be due to additional pathway changes in PTPN2-deficient cells or to the different cell types used.
Finally, it is important to consider the implications of our findings that TRAF3 modulates T cell IFNAR signaling in the context of an in vivo immune response. We are unable to obtain clearly interpretable data about this using currently available technologies because TRAF3 is an important regulator of TCR signaling, resulting in markedly defective in vivo T cell responses to infection and immunization in mice with a T cell–specific TRAF3 deficiency (22). Additionally, as discussed earlier, the inhibition of IL-2R signaling by TRAF3 results in an increased proportion of thymic-derived Treg cells in these mice, further dampening their immune responses (23). Discerning the effect of IFNAR signaling changes on the T cell response to an infection in vivo would be confounded by this suboptimal TCR signaling and downstream T cell activation, as well as by the increased numbers of Treg cells. Based upon data showing the importance of type I IFN signals for T cells (30, 45), we predict that a CD4+ T cell selectively lacking TRAF3 involvement in IFNAR signaling would exhibit reduced proliferation and effector functions that are dependent upon ISGs (for example, chemokine production). To conclude, this work highlights the importance of TRAF3 for signaling through IFNAR in CD4+ T lymphocytes and provides insights into how PTPN2 and PTPN22 are prevented from premature interference with IFNAR signaling in T cells.
MATERIALS AND METHODS
Mice and cell lines
HuT78 cells transfected to stably express CD28 (HuT28.11, “Hut28”) were a gift from Dr. Arthur Weiss, University of California, San Francisco (46). PTPN22- and TRAF3-deficient HuT28 cells were generated with CRISPR-Cas9 technology as previously described (24). TRAF3-deficient HuT28 cells were stably transfected with a plasmid encoding human TRAF3 under the control of the RSV promoter and containing a puromycin-resistance cassette. Single cells were plated after electroporation with plasmid, and clones that grew under puromycin selection (1.5 μg/ml, Gibco) were expanded for further analysis. HuT28 cells, knockout, and TRAF3-add back lines were cultured in complete medium (RPMI 1640 medium supplemented with 100 U/ml penicillin, 100 U/ml streptomycin, 2 mM L-glutamine, 10 μM β-mercaptoethanol, and 10% fetal calf serum). For experiments with cell lines, a biological replicate refers to an independent experiment performed on a different day from those of other replicates. Generation of B cell– and T cell–specific TRAF3 knockout mice has been previously described (22, 47). Briefly, CD4-Cre mice were crossed to mice containing a floxed Traf3 allele to delete Traf3 in all mature T cells from the CD4+CD8+ developmental stage onward. In figures, Traf3−/− denotes cells from CD4-Cre+Traf3fl/fl animals, and littermate control (referred to as WT for simplicity) denotes cells from CD4-Cre−Traf3fl/fl animals. The same breeding strategy with CD19-Cre mice instead of CD4-Cre mice was used to generate B cell–specific TRAF3 knockout mice. Mouse splenic and lymph node B or CD4+ T cells were isolated by negative selection (StemCell Technologies, cat# 19854 and 19852, respectively) according to the manufacturer’s instructions. Male and female mice were used, and mice were age- and sex-matched within each experiment. The Institutional Animal Care and Use Committee approved all procedures in this study under animal protocols 1062397 and 1091535.
Type I IFN stimulation, inhibitor treatment, and cell lysis
Primary mouse CD4+ T cells and HuT28 cells were treated with 1000 U/mL human IFN-α hybrid (PBL assay science, 11200-2) in complete medium for the times indicated in the figures. Cells were lysed by vortexing in 1X Radio Immunoprecipitation Assay (RIPA) buffer (Cell Signaling Technology, #9806) containing 1 mM PMSF for whole-cell lysate analysis or Immunoprecipitation (IP) lysis buffer [40 mM Tris (pH 7.5), 0.5% Triton-X-100, 100 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 2 mM Na3VO4, and EDTA-free cOmplete mini protease inhibitor cocktail (Roche)] for co-immunoprecipitation. Where indicated in the figures, 10 μM PTPN22 inhibitor LTV-1 (Calbiochem, cat# 540218), 20 μM PTPN22 inhibitor NC1 (Aobius, cat# AOB13736), or 1 nM PTPN2 inhibitor SF-1670 (Tocris, cat# 5020), or an equal volume of DMSO was included in the culture medium for 45min (for LTV-1 and NC1) or 2 hours (for SF-1670) before and during stimulation of the cells with type I IFN (37, 48).
Co-immunoprecipitations
Whole-cell lysates were prepared as described earlier and then were pre-cleared for 10 min with protein G Dynabeads (Invitrogen). Antibody recognizing one of the following targets was then added: JAK1 (Millipore Sigma #05-1154, 1:1000 dilution), TRAF3 (Santa Cruz Biotechnology #sc-1828, 1:1000, or CST #4729, 1:100), or IFNAR1 (abcam #ab45172, 1:500). Lysates were incubated with the immunoprecipitating antibody for 2 hours at 4°C, and the antibody-bound targets were then captured with protein G Dynabeads for 20 min at 4°C. Dynabeads were washed three times with IP washing buffer [20 mM Tris (pH 7.5), 1% Triton-X-100, 40 mM NaCl] and then resuspended in IP lysis buffer for Western blotting analysis.
Western blotting
Proteins were separated on Bis-Tris polyacrylamide gels and transferred to a polyvinylidene difluoride membrane with the XCell II blotting system (Invitrogen) according to the manufacturer’s recommendations. Membranes were blocked with 5% nonfat milk and incubated with primary antibody overnight at 4°C. Primary antibodies against the following proteins were used for Western blotting analysis: Cell Signaling Technologies: JAK1 (#3344), pJAK1 Tyr1034/1035 (#3331), STAT1 (#9172), pSTAT1 Tyr701 (#9167), STAT2 (#4594), pSTAT2 Tyr690 (#88410), STAT3 (#4904), pSTAT3 Tyr705 (#9145) STAT5 (#25656), pSTAT5 Tyr694 (#4322), MX1 (#37849), PTPN22 (#14693), PTPN2 (#59835), TRAF3 (#4729), IRF1 (#8478), IRF9 (#28492), SOCS1 (#68631); Santa Cruz Biotechnology: β-Actin (#sc-47778), GAPDH (#sc-47724), IRF7 (#sc-74472). After incubation with primary antibody, the membranes were washed and incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Jackson Immunoresearch Labs, #115-035-003) or HRP-conjugated goat anti-rabbit IgG (Jackson Immunoresearch Labs, #111-035-144) for 2 hours at room temperature. Blots were developed with SuperSignal West Pico or Femto substrate (Thermo Fisher Scientific) and imaged with a low-light imaging system (LAS400, Fuji Medical Imaging). Densitometry of Western blots was performed with ImageStudio Lite software (LI-COR Biosciences). The density of a band for a protein of interest was divided by the density of the band for a loading control (actin or GAPDH) from the same sample to normalize protein abundance.
qPCR assays
RNA was extracted with the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s instructions. cDNA was synthesized with Superscript III First Strand Synthesis SuperMix (Invitrogen). Quantitative PCR was performed with cDNA, primer pairs for target genes, and PerfeCta SYBR Green Fastmix (Quanta Biosciences). Reactions were run in a QuantStudio 6 Flex (Applied Biosystems) with a standard cycling protocol (90°C for 2 min; 40 cycles of 95°C for 15 s, 60°C for 1 min). Quantitative PCR data were analyzed with the 2−ΔΔCt method where the ΔΔ cycle threshold (Ct) = [(CT gene of interest-CT housekeeping gene) stimulated - (CT gene of interest-CT housekeeping gene) unstimulated]. Where indicated on the Y axis, the ratio of the CT of the gene of interest to the CT of the housekeeping gene is graphed. Actb was chosen as a housekeeping gene for mouse samples and HPRT (49) was used for human samples. The following primers for qPCR were synthesized by Iowa DNA Technologies, all sequences are listed 5’→3’: mouse Actb, Fwd: CATTGCTGACAGGATGCAGAAGG; Rev: TGCTGGAAGGTGGACAGTGAGG. Mouse Cxcl10, Fwd: ATCATCCCTGCGAGCCTATCCT; Rev: GACCTTTTTTGGCTAAACGCTTTC. Mouse Ifit1, Fwd: TACAGGCTGGAGTGTGCTGAGA; Rev: CTCCACTTTCAGAGCCTTCGCA. Mouse Ifitm3, Fwd: TTCTGCTGCCTGGGCTTCATAG; Rev: ACCAAGGTGCTGATGTTCAGGC. Human CXCL10, Fwd: CGATTCTGATTTGCTGCCTTAT; Rev: GGCTTGCAGGAATAATTTCAAGT. Human MX1, Fwd: GGCTGTTTACCAGACTCCGACA; Rev: CACAAAGCCTGGCAGCTCTCTA. Human HPRT (49), Fwd: GGACTAATTATGGACAGGACTG; Rev: GCTCTTCAGTCTGATAAAATCTAC.
The human STAT1 (PPH00811C) primer mix was purchased from Qiagen.
ELISAs
Human CXCL10 was detected in the culture medium of type I IFN–stimulated HuT28.11 cells with the Human CXCL10/IP-10 DuoSet ELISA (R&D systems, catalog #DY266-05). Mouse Cxcl10 was detected in the culture medium of type I IFN–stimulated CD4+ T cells with the Mouse Cxcl10/IP-10 DuoSet ELISA (R&D systems, catalog #DY466-05).
Statistical analysis
Data were graphed and statistical tests performed with GraphPad Prism software. Time courses were analyzed with mixed-effects analysis or two-way ANOVA with Sidak’s multiple comparisons post-hoc test. Comparisons not involving multiple time points were analyzed with unpaired t tests.
Supplementary Material
Acknowledgments:
We would like to thank A. Weiss (UCSF) for providing the HuT28.11 cell line. We are also grateful to members of the Bishop laboratory for thoughtful input and technical assistance throughout work on this project.
Funding:
This work was funded by National Institutes of Health grant R01 AI123107 (to G.A.B.), National Institutes of Health grant T32 AI007260 (to E.L.H.), National Institutes of Health grant T32 HL007344 (to E.L.H.), Holden Comprehensive Cancer Center through its National Institutes of Health grant P30CA086862 (to G.A.B.), and Senior Research Career Scientist award IK6 BX005392 from the Department of Veterans Affairs, Office of Research and Development (to G.A.B.).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Isaacs A, Lindenmann J, Andrewes CH, Virus interference. I. The interferon. Proceedings of the Royal Society of London. Series B - Biological Sciences 147, 258–267 (1957)doi: 10.1098/rspb.1957.0048). [DOI] [PubMed] [Google Scholar]
- 2.Isaacs A, Cox RA, Rotem Z, Foreign nucleic acids as the stimulus to make interferon. Lancet 2, 113–116 (1963); published online EpubJul 20 ( [DOI] [PubMed] [Google Scholar]
- 3.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM, A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011); published online EpubApr 28 ( 10.1038/nature09907). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider WM, Chevillotte MD, Rice CM, Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol 32, 513–545 (2014) 10.1146/annurev-immunol-032713-120231). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Boxel-Dezaire AHH, Rani MRS, Stark GR, Complex Modulation of Cell Type-Specific Signaling in Response to Type I Interferons. Immunity 25, 361–372 (2006); published online Epub2006/09/01/ ( 10.1016/j.immuni.2006.08.014). [DOI] [PubMed] [Google Scholar]
- 6.Colamonici O, Yan H, Domanski P, Handa R, Smalley D, Mullersman J, Witte M, Krishnan K, Krolewski J, Direct binding to and tyrosine phosphorylation of the alpha subunit of the type I interferon receptor by p135tyk2 tyrosine kinase. Mol Cell Biol 14, 8133–8142 (1994); published online EpubDec ( 10.1128/mcb.14.12.8133-8142.1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novick D, Cohen B, Rubinstein M, The human interferon alpha/beta receptor: characterization and molecular cloning. Cell 77, 391–400 (1994); published online EpubMay 6 ( 10.1016/0092-8674(94)90154-6). [DOI] [PubMed] [Google Scholar]
- 8.Yan H, Krishnan K, Lim JT, Contillo LG, Krolewski JJ, Molecular characterization of an alpha interferon receptor 1 subunit (IFNaR1) domain required for TYK2 binding and signal transduction. Mol Cell Biol 16, 2074–2082 (1996); published online EpubMay ( 10.1128/mcb.16.5.2074). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Improta T, Schindler C, Horvath CM, Kerr IM, Stark GR, Darnell JE Jr., Transcription factor ISGF-3 formation requires phosphorylated Stat91 protein, but Stat113 protein is phosphorylated independently of Stat91 protein. Proc. Natl. Acad. Sci. U. S. A 91, 4776–4780 (1994); published online EpubMay 24 ( 10.1073/pnas.91.11.4776). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arimoto KI, Miyauchi S, Stoner SA, Fan JB, Zhang DE, Negative regulation of type I IFN signaling. J. Leukoc. Biol, (2018); published online EpubJan 22 ( 10.1002/jlb.2mir0817-342r). [DOI] [PubMed] [Google Scholar]
- 11.Simoncic PD, Lee-Loy A, Barber DL, Tremblay ML, McGlade CJ, The T cell protein tyrosine phosphatase is a negative regulator of janus family kinases 1 and 3. Curr. Biol 12, 446–453 (2002); published online EpubMar 19 ( 10.1016/s0960-9822(02)00697-8). [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Han X, Tang Y, Wu Y, Qu B, Shen N, miR-744 enhances type I interferon signaling pathway by targeting PTP1B in primary human renal mesangial cells. Sci. Rep 5, 12987 (2015); published online EpubAug 11 ( 10.1038/srep12987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myers MP, Andersen JN, Cheng A, Tremblay ML, Horvath CM, Parisien JP, Salmeen A, Barford D, Tonks NK, TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J. Biol. Chem 276, 47771–47774 (2001); published online EpubDec 21 ( 10.1074/jbc.C100583200). [DOI] [PubMed] [Google Scholar]
- 14.Van De Wiele CJ, Marino JH, Whetsell ME, Vo SS, Masengale RM, Teague TK, Loss of interferon-induced Stat1 phosphorylation in activated T cells. J Interferon Cytokine Res 24, 169–178 (2004); published online EpubMar ( 10.1089/jir.2004.24.169). [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, Miyakawa Y, Fox N, Kaushansky K, Interferon-alpha directly represses megakaryopoiesis by inhibiting thrombopoietin-induced signaling through induction of SOCS-1. Blood 96, 2093–2099 (2000); published online EpubSep 15 ( [PubMed] [Google Scholar]
- 16.Piganis RA, De Weerd NA, Gould JA, Schindler CW, Mansell A, Nicholson SE, Hertzog PJ, Suppressor of cytokine signaling (SOCS) 1 inhibits type I interferon (IFN) signaling via the interferon alpha receptor (IFNAR1)-associated tyrosine kinase Tyk2. J. Biol. Chem 286, 33811–33818 (2011); published online EpubSep 30 ( 10.1074/jbc.M111.270207). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin WW, Yi Z, Stunz LL, Maine CJ, Sherman LA, Bishop GA, The adaptor protein TRAF3 inhibits interleukin-6 receptor signaling in B cells to limit plasma cell development. Science signaling 8, ra88 (2015); published online EpubSep 1 ( 10.1126/scisignal.aaa5157). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mambetsariev N, Lin WW, Stunz LL, Hanson BM, Hildebrand JM, Bishop GA, Nuclear TRAF3 is a negative regulator of CREB in B cells. Proc. Natl. Acad. Sci. U. S. A 113, 1032–1037 (2016); published online EpubJan 26 ( 10.1073/pnas.1514586113). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mambetsariev N, Lin WW, Wallis AM, Stunz LL, Bishop GA, TRAF3 deficiency promotes metabolic reprogramming in B cells. Sci. Rep 6, 35349 (2016); published online EpubOct 18 ( 10.1038/srep35349). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi JH, Sun SC, Tumor Necrosis Factor Receptor-Associated Factor Regulation of Nuclear Factor κB and Mitogen-Activated Protein Kinase Pathways. Front. Immunol 9, 1849 (2018) 10.3389/fimmu.2018.01849). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie P, Brown L, Stunz LL, Bishop GA, TRAF3 inhibits signaling by Toll-like receptors in B lymphocytes. The FASEB Journal 22, 1066.1065–1066.1065 (2008) 10.1096/fasebj.22.1_supplement.1066.5). [DOI] [Google Scholar]
- 22.Xie P, Kraus ZJ, Stunz LL, Liu Y, Bishop GA, TNF receptor-associated factor 3 is required for T cell-mediated immunity and TCR/CD28 signaling. J. Immunol 186, 143–155 (2011); published online EpubJan 1 ( 10.4049/jimmunol.1000290). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi Z, Lin WW, Stunz LL, Bishop GA, The adaptor TRAF3 restrains the lineage determination of thymic regulatory T cells by modulating signaling via the receptor for IL-2. Nat. Immunol 15, 866–874 (2014); published online EpubSep ( 10.1038/ni.2944). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallis AM, Wallace EC, Hostager BS, Yi Z, Houtman JCD, Bishop GA, TRAF3 enhances TCR signaling by regulating the inhibitors Csk and PTPN22. Sci. Rep 7, 2081 (2017); published online EpubMay 18 ( 10.1038/s41598-017-02280-4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arkee T, Hostager BS, Houtman JCD, Bishop GA, TRAF3 in T Cells Restrains Negative Regulators of LAT to Promote TCR/CD28 Signaling. J. Immunol, (2021); published online EpubJun 18 ( 10.4049/jimmunol.2001220). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rae W, Sowerby JM, Verhoeven D, Youssef M, Kotagiri P, Savinykh N, Coomber EL, Boneparth A, Chan A, Gong C, Jansen MH, du Long R, Santilli G, Simeoni I, Stephens J, Wu K, Zinicola M, Allen HL, Baxendale H, Kumararatne D, Gkrania-Klotsas E, Scheffler Mendoza SC, Yamazaki-Nakashimada MA, Ruiz LB, Rojas-Maruri CM, Lugo Reyes SO, Lyons PA, Williams AP, Hodson DJ, Bishop GA, Thrasher AJ, Thomas DC, Murphy MP, Vyse TJ, Milner JD, Kuijpers TW, Smith KGC, Immunodeficiency, autoimmunity, and increased risk of B cell malignancy in humans with TRAF3 mutations. Science immunology 7, eabn3800 (2022)doi: 10.1126/sciimmunol.abn3800). [DOI] [PubMed] [Google Scholar]
- 27.Yi Z, Stunz LL, Bishop GA, TNF receptor associated factor 3 plays a key role in development and function of invariant natural killer T cells. J. Exp. Med 210, 1079–1086 (2013); published online EpubJun 3 ( 10.1084/jem.20122135). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brinkmann V, Geiger T, Alkan S, Heusser CH, Interferon alpha increases the frequency of interferon gamma-producing human CD4+ T cells. The Journal of Experimental Medicine 178, 1655 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marrack P, Kappler J, Mitchell T, Type I Interferons Keep Activated T Cells Alive. The Journal of Experimental Medicine 189, 521 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Starbeck-Miller GR, Xue H-H, Harty JT, IL-12 and type I interferon prolong the division of activated CD8 T cells by maintaining high-affinity IL-2 signaling in vivo. The Journal of Experimental Medicine 211, 105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gil MP, Salomon R, Louten J, Biron CA, Modulation of STAT1 protein levels: a mechanism shaping CD8 T-cell responses in vivo. Blood 107, 987–993 (2006); published online EpubFeb 1 ( 10.1182/blood-2005-07-2834). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanabe Y, Nishibori T, Su L, Arduini RM, Baker DP, David M, Cutting edge: role of STAT1, STAT3, and STAT5 in IFN-alpha beta responses in T lymphocytes. J. Immunol 174, 609–613 (2005); published online EpubJan 15 ( 10.4049/jimmunol.174.2.609). [DOI] [PubMed] [Google Scholar]
- 33.Gimeno R, Lee CK, Schindler C, Levy DE, Stat1 and Stat2 but not Stat3 arbitrate contradictory growth signals elicited by alpha/beta interferon in T lymphocytes. Mol Cell Biol 25, 5456–5465 (2005); published online EpubJul ( 10.1128/mcb.25.13.5456-5465.2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bishop GA, Warren WD, Berton MT, Signaling via major histocompatibility complex class II molecules and antigen receptors enhances the B cell response to gp39/CD40 ligand. Eur. J. Immunol 25, 1230–1238 (1995); published online EpubMay ( 10.1002/eji.1830250515). [DOI] [PubMed] [Google Scholar]
- 35.Acharya D, Liu G, Gack MU, Dysregulation of type I interferon responses in COVID-19. Nature Reviews Immunology 20, 397–398 (2020); published online Epub2020/07/01 ( 10.1038/s41577-020-0346-x). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crouse J, Kalinke U, Oxenius A, Regulation of antiviral T cell responses by type I interferons. Nat. Rev. Immunol 15, 231–242 (2015); published online EpubApr ( 10.1038/nri3806). [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Shaked I, Stanford SM, Zhou W, Curtsinger JM, Mikulski Z, Shaheen ZR, Cheng G, Sawatzke K, Campbell AM, Auger JL, Bilgic H, Shoyama FM, Schmeling DO, Balfour HH Jr., Hasegawa K, Chan AC, Corbett JA, Binstadt BA, Mescher MF, Ley K, Bottini N, Peterson EJ, The autoimmunity-associated gene PTPN22 potentiates toll-like receptor-driven, type 1 interferon-dependent immunity. Immunity 39, 111–122 (2013); published online EpubJul 25 ( 10.1016/j.immuni.2013.06.013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmes DA, Suto E, Lee WP, Ou Q, Gong Q, Smith HR, Caplazi P, Chan AC, Autoimmunity-associated protein tyrosine phosphatase PEP negatively regulates IFN-α receptor signaling. J. Exp. Med 212, 1081–1093 (2015); published online EpubJun 29 ( 10.1084/jem.20142130). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matikainen S, Sareneva T, Ronni T, Lehtonen A, Koskinen PJ, Julkunen I, Interferon-alpha activates multiple STAT proteins and upregulates proliferation-associated IL-2Ralpha, c-myc, and pim-1 genes in human T cells. Blood 93, 1980–1991 (1999); published online EpubMar 15 ( [PubMed] [Google Scholar]
- 40.Cheon H, Stark GR, Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc. Natl. Acad. Sci. U. S. A 106, 9373–9378 (2009); published online EpubJun 9 ( 10.1073/pnas.0903487106). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bottini N, Peterson EJ, Tyrosine phosphatase PTPN22: multifunctional regulator of immune signaling, development, and disease. Annu. Rev. Immunol 32, 83–119 (2014) 10.1146/annurev-immunol-032713-120249). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, Mustelin T, A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat. Genet 36, 337–338 (2004); published online EpubApr ( 10.1038/ng1323). [DOI] [PubMed] [Google Scholar]
- 43.Vang T, Congia M, Macis MD, Musumeci L, Orrú V, Zavattari P, Nika K, Tautz L, Taskén K, Cucca F, Mustelin T, Bottini N, Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat. Genet 37, 1317–1319 (2005); published online Epub2005/12/01 ( 10.1038/ng1673). [DOI] [PubMed] [Google Scholar]
- 44.Li H, Hostager BS, Arkee T, Bishop GA, Multiple mechanisms for TRAF3-mediated regulation of the T cell costimulatory receptor GITR. J. Biol. Chem 297, 101097 (2021); published online EpubSep ( 10.1016/j.jbc.2021.101097). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osokine I, Snell LM, Cunningham CR, Yamada DH, Wilson EB, Elsaesser HJ, de la Torre JC, Brooks D, Type I interferon suppresses de novo virus-specific CD4 Th1 immunity during an established persistent viral infection. Proceedings of the National Academy of Sciences 111, 7409–7414 (2014) 10.1073/pnas.1401662111). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stein PH, Fraser JD, Weiss A, The cytoplasmic domain of CD28 is both necessary and sufficient for costimulation of interleukin-2 secretion and association with phosphatidylinositol 3'-kinase. Mol Cell Biol 14, 3392–3402 (1994); published online EpubMay ( 10.1128/mcb.14.5.3392-3402.1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie P, Stunz LL, Larison KD, Yang B, Bishop GA, Tumor necrosis factor receptor-associated factor 3 is a critical regulator of B cell homeostasis in secondary lymphoid organs. Immunity 27, 253–267 (2007); published online EpubAug ( 10.1016/j.immuni.2007.07.012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le HTT, Cho YC, Cho S, Inhibition of protein tyrosine phosphatase non-receptor type 2 by PTP inhibitor XIX: Its role as a multiphosphatase inhibitor. BMB Rep 50, 329–334 (2017); published online EpubJun ( 10.5483/bmbrep.2017.50.6.003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valadan R, Hedayatizadeh-Omran A, Alhosseini-Abyazani MN, Amjadi O, Rafiei A, Tehrani M, Alizadeh-Navaei R, Data supporting the design and evaluation of a universal primer pair for pseudogene-free amplification of HPRT1 in real-time PCR. Data in Brief 4, 384–389 (2015); published online Epub2015/09/01/ ( 10.1016/j.dib.2015.06.009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.