Abstract
Colonization of the intestine by oral microbes has been linked to multiple diseases such as inflammatory bowel disease (IBD) and colon cancer, yet mechanisms allowing expansion in this niche remain largely unknown. Veillonella parvula, an asaccharolytic, anaerobic, oral microbe that derives energy from organic acids, increases in abundance in the intestine of patients with IBD. Here, we show that nitrate, a signature metabolite of inflammation, allows V. parvula to transition from fermentation to anaerobic respiration. Nitrate respiration, through the narGHJI operon, boosted Veillonella growth on organic acids and also modulated its metabolic repertoire, allowing it to use amino acids and peptides as carbon sources. This metabolic shift was accompanied by changes in carbon metabolism and ATP production pathways. Nitrate respiration was fundamental for ectopic colonization in a mouse model of colitis, as a Veillonella narG deletion mutant colonized significantly less than a wild-type strain during inflammation. These results suggest that Veillonella harnesses conditions present during inflammation to colonize in the intestine.
INTRODUCTION
The oral cavity harbors the second largest and most diverse microbiota within the human body1,2 and, along with the gut, forms the mouth-gut axis3. Interactions between microbes of these niches and the host are implicated in the pathogenesis of gastrointestinal diseases and chronic inflammation3,4. The presence of prototypical oral microbes, including Veillonella and Fusobacterium, in the intestinal mucosa is associated with multiple pathologies including IBD5–8, colorectal cancer9–11, and cystic fibrosis12–14. Ingestion of saliva is the main introduction of oral microbes into the gut environment, although oral microbes are usually poor gut colonizers and do not cause disease in healthy individuals15. Klebsiella species isolated from saliva of IBD patients, however, were identified as pathobionts whose ectopic intestinal colonization induces type 1 T helper (Th1) cells4.
Molecular mechanisms associated with shifts in oral and gut microbes during inflammation are poorly understood. Gut inflammation is accompanied by changes in a vast number of metabolites that can drive expansion or depletion of species16–18. For instance, inflammation elevates levels of reactive oxygen and nitrogen species (ROS and RNS) that kill select microbes, particularly strict anaerobes in close proximity to the epithelium. Inflammation therefore results in an altered gut milieu that could promote colonization by oral microbes.
Much of our understanding of molecular strategies adopted by microbes to invade the inflamed gut originates from model Proteobacteria species19–22. Escherichia coli gains a colonization advantage during inflammation through its ability to use formate as a carbon and energy source when coupled with oxygen respiration20 and to use nitrate as a terminal electron acceptor19. Salmonella enterica serovar Typhimurium utilizes ethanolamine—unlike most gut microbes—and lactate as carbon sources when coupled with tetrathionate and oxygen respiration, respectively22,23. Under anaerobic conditions, Salmonella Typhi uses nitrate via a periplasmic nitrate reductase to grow in the intestinal lumen24,25. Less is known, however, about the physiology and ecology of strict anaerobes, owing to the lack of genetic tools to manipulate these organisms and the inherent complexity of the gut environment.
Veillonella are gram-negative, obligate anaerobes present in the human oral cavity that obtain energy by fermenting short-chain organic acids. They are normally found in biofilms and tend to co-aggregate with lactic acid bacteria26–29 that produce lactate as a by-product, which serves as the main carbon and energy source for Veillonella26,30,31. In this study, we show that Veillonella, a prototypical member of the oral microbiome commonly enriched in the intestines of IBD patients7,32, is capable of transitioning from fermentation to anaerobic respiration. We demonstrate that nitrate, a metabolite enriched during inflammation, is used by Veillonella strains as a terminal electron acceptor to expand their metabolic landscapes and boost growth. Using a reductionist approach, we elucidate the metabolic pathways that accompany this transition and enable Veillonella to colonize and persist in the inflamed gut. Our findings provide a framework to understand the increased abundance of Veillonella species in patients with IBD and other inflammatory conditions.
RESULTS
Gut inflammation is associated with Veillonella enrichment
Veillonella is implicated in the disease course of IBD patients7,32. To further illustrate Veillonella enrichment in the context of inflammation, we used longitudinal gut microbiome data from treatment-naive, pediatric ulcerative colitis (UC)32 and Crohn’s disease (CD)33 studies. Abundances of V. parvula and V. dispar increased with increasing levels of fecal calprotectin, an inflammation marker, in UC patients (Fig. 1a). Comparing CD cases to non-IBD subjects revealed a similar increase with disease (Fig. 1b). We therefore hypothesized that gut injury and inflammation provides favorable conditions for expansion of Veillonella species.
Figure 1. The abundance of nitrate-respiring Veillonella is increased during inflammatory bowel diseases.
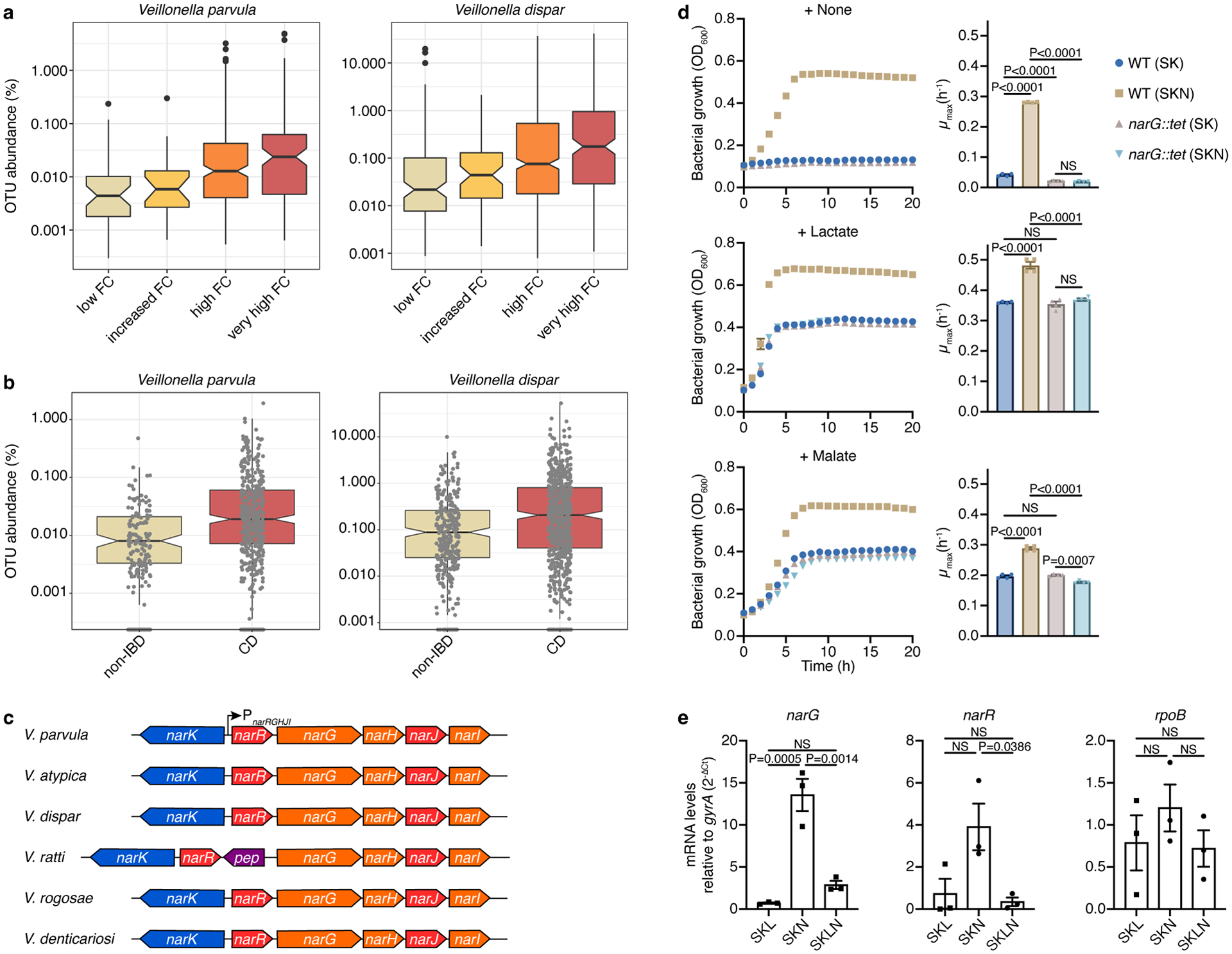
a) Abundances of V. parvula (n=487 samples) and V. dispar (n=1,006 samples) from a pediatric UC cohort increase with higher levels of inflammation, as measured by fecal calprotectin (FC) concentration: low (<100 mcg/g), increased (100–200), high (200–3,000), or very high (>3,000). b) V. parvula (n=529 samples) and V. dispar (n=846 samples) abundances are increased in new-onset CD patients. c) Organization of the narK-narR-narGHJI cluster in Veillonella species. The cluster in V. ratti additionally includes a gene encoding a putative peptidase. d) Growth (measured by optical density at 600nm, OD600) of wild-type (WT) and narG::tet V. parvula was studied for 20 h in SK medium with or without nitrate (SKN and SK, respectively) and lactate or malate as the carbon source. Maximum-specific growth rates (μmax) were calculated for each growth curve (n=4 biological replicates). e) Comparative analysis of expression profiles of the narG, narR, and rpoB genes by real-time quantitative reverse transcription PCR (RT-qPCR) during growth on SK medium supplemented with lactate (SKL), nitrate (SKN), or both (SKLN). gyrA was the internal reference. WT cells were collected at the mid-exponential phase (n=3 biological replicates). Boxplots (a,b) display median, 25th and 75th percentiles, where a shows outliers and b shows all data points; data were analyzed by two-sided Wilcoxon test. Data (d,e) represent mean±SEM (standard error of the mean), analyzed by one-way ANOVA and Tukey’s honestly significant difference (HSD) test. NS, not significant.
Nitrate respiration expands Veillonella’s metabolic landscape
Veillonella is a member of the Fimicutes phylum that uses a fermentative metabolism to extract carbon and energy from only a few organic acids34. In certain strains, nitrate reductase activity was previously observed35–38. Given that nitrate is increased during inflammation and harnessed by some facultative anaerobic Proteobacteria in the gut19,24, we hypothesized that inflammation-associated nitrate is used by Veillonella to expand in the inflamed gut. An unbiased search for the presence of genetic clusters involved in nitrate reduction within the membrane-bound respiratory (Nar), periplasmic dissimilatory (Nap), and cytoplasmic assimilatory (Nas) nitrate reductase families39 confirmed the increased representation of these clusters in Proteobacteria compared to other gut microbes (Extended Data Fig. 1a). Moreover, homologs of these genes are uncommon in oral Firmicutes and Bacteroidetes as well as gut Firmicutes, Bacteroidetes, and Actinobacteria (Extended Data Fig. 1a–c). Veillonella species, however, contained a nar family nitrate reduction cluster (Fig. 1c; Extended Data Fig. 1c). As no other nitrate reductase homologs were detected in Veillonella genomes, this suggests that Veillonella can potentially perform anaerobic respiration using nitrate as the terminal electron acceptor.
The membrane-bound respiratory nitrate reductase cluster in V. parvula comprises two divergently-located operons that contain genes encoding a potential NarGHI quinol:nitrate oxidoreductase and NarK nitrate/nitrite transporter (narRGHJI and narK operons) (Fig. 1c). This cluster exhibits similar organization in most Veillonella species, suggesting a common origin and function (Fig. 1c). We used the genetically tractable V. parvula SKV3840 as the wild-type (WT) strain to assess the role of nitrate on growth. While a complex medium (SK) alone was unable to support V. parvula growth, the addition of lactate or malate stimulated growth rates and final cell densities (Fig. 1d), as seen previously with pyruvate36. When the medium was supplemented with nitrate (SKN), we observed growth without any additional carbon source, suggesting that alternative carbon sources present in SK medium are used by Veillonella in the presence of nitrate (Fig. 1d). This phenotype was reproducible in other rich growth media (Extended Data Fig. 2a). Moreover, we observed higher cell densities and growth rates when nitrate was present with either lactate or malate as the carbon source (Fig. 1d). Nitrate utilization, therefore, enhances growth on organic acids and allows Veillonella to grow on non-canonical carbon sources.
To confirm the involvement of the narGHJI gene cluster in nitrate utilization, we generated a narG::tet deletion, which also likely inactivated narHJI genes. Genetic complementation with narGHJI genes was unsuccessful, but whole-genome sequencing analysis revealed no uncharacterized mutations in narG::tet. The narG::tet strain was unable to grow on SKN alone and boost growth with lactate and malate, demonstrating a defect in utilizing nitrate (Fig. 1d). To further test the role of NarGHI, we generated a narI::chl deletion lacking the γ-chain of the respiratory nitrate reductase. As expected, narI::chl fully phenocopied narG::tet, and its complementation with an ectopic copy of narI driven from the PnarRGHJI promoter fully restored the WT phenotype (Extended Data Fig. 2b).
Transcription of the narRGHJI operon was also induced in response to nitrate (Fig. 1e). Notably, several clinical Veillonella isolates exhibited similar growth phenotypes in response to nitrate, suggesting similar metabolic strategies (Extended Data Fig. 2c). V. parvula reduced nitrate to nitrite, and at least one fraction of nitrite was further reduced to ammonia or incorporated into biomass (Extended Data Fig. 3a), consistent with the dual assimilatory and respiratory nature of nitrate reduction previously observed in V. parvula36. Growth was not enhanced when ammonium or nitrite was used instead of nitrate, irrespective of the carbon source (Extended Data Fig. 3b,c). Altogether, we conclude that Veillonella species containing this cluster are able to use nitrate for respiration in a NarGHI-dependent manner.
Nitrate respiration allows Veillonella growth on amino acids
Nitrate respiration allowed V. parvula to grow on SKN medium. Dissection of the carbon sources showed that yeast extract was essential for growth; however, when yeast extract was present, omitting casitone significantly reduced growth (Fig. 2a). We therefore hypothesized that V. parvula is capable of using amino acids and/or peptides as carbon sources.
Figure 2. Nitrate respiration expands the metabolic repertoire of Veillonella.
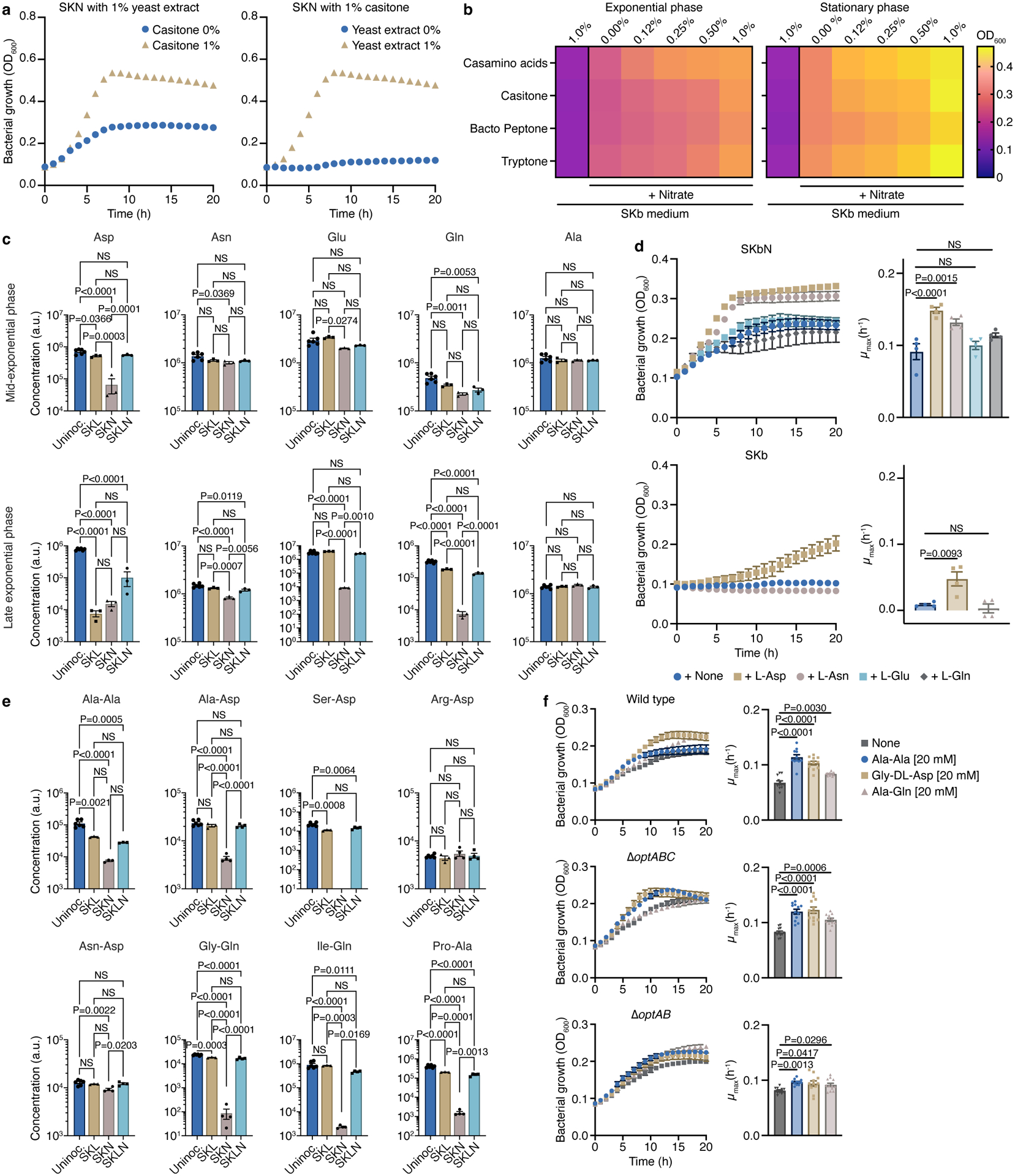
WT growth in a) SKN lacking casitone or yeast extract (n=3) and b) SK base (SKb) with or without nitrate and increasing concentrations of casamino acids, casitone, bacto peptone, or tryptone (n=4). c) Amino acid composition in uninoculated SK (n=6) and spent SKL, SKN, and SKLN (n=3) media at mid- and late exponential phases. d) WT growth in SKb plus nitrate (SKbN) and SKb with or without the indicated amino acids (25mM) (n=4). e) Dipeptide consumption in uninoculated SK (n=6) and spent SKL, SKN, and SKLN (n=3) media at the late exponential phase. f) WT (n=12), oppABC (n=12), and optAB (n=11) growth in SKbN with or without the indicated dipeptides (20 mM). Growth rates (μmax) were calculated for each growth curve (d,f). Data (c-f) represent mean±SEM, analyzed by one-way ANOVA and Tukey’s HSD test. NS, not significant. N numbers denote biological replicates.
To assess contributions of various carbon sources, we used the SK base (SKb) medium: 0.25% yeast extract plus SK salts. During nitrate respiration, a dose-dependent growth increase resulted from supplementation with various protein hydrolysates (Fig. 2b). Casamino acids, which contain primarily free amino acids, led to higher optical densities compared to more complex sources at most concentrations tested (Fig. 2b). Casamino acids and casitone also positively impacted growth of other Veillonella strains during nitrate respiration (Extended Data Fig. 4a). During fermentation (when nitrate was absent), V. parvula failed to grow in SKb supplemented with any of the protein hydrolysates (Fig. 2b), while addition of lactate or malate stimulated growth (Extended Data Fig. 4b).
We then analyzed the spent SK medium by LC-MS to investigate whether amino acids or peptides were depleted during growth, noting significant decreases in the amount of amino acids at different growth phases (Fig. 2c; Extended Data Fig. 5a,b; Supplemental Data File 1). Asp was consumed during the mid-exponential phase, while Glu, Gln and Asn were depleted during the late exponential phase only when lactate (a preferred carbon source) was absent. Solely Asp and Asn were capable of enhancing the biomass and growth rate of V. parvula during nitrate respiration, although some growth was also observed after a significant lag phase with Asp in the absence of nitrate (Fig. 2d). Asp also stimulated the growth of other Veillonella strains in the presence of nitrate (Extended Data Fig. 4a). Utilization of Glu and Gln, by contrast, likely requires other metabolic intermediates.
Many dipeptides were also consumed during the late exponential phase, mainly in the absence of lactate (Fig. 2e; Supplementary Data Files 1,2). Of the dipeptides we annotated and analyzed, those with long and charged amino acids such as Glu, Lys, or Arg at either terminus or the respective presence of Trp and Pro on the N- and C-terminus exhibited reduced consumption. When Ala-Ala, Gly-Asp, or Ala-Gln were tested as carbon sources in SKbN medium, growth increased with Gly-Asp (Fig. 2f). Deletion of proton-dependent Opt family peptide transporters (optA::chl optB::tet double mutant)41 but not ATP-dependent Opp oligopeptide transporters42 eliminated this effect (Fig. 2f at 10h), further implicating the use of dipeptides for growth during nitrate respiration. The V. ratti nitrate reductase cluster contains a putative peptidase-encoding gene (Fig. 1c), highlighting the connection between nitrate respiration and peptide consumption for growth.
Metabolic changes associated with nitrate respiration
To study metabolic strategies associated with fermentation and respiration as well as use of amino acids/peptides during nitrate respiration, we examined profiles of Veillonella’s major by-products: propionate and acetate43. Propionate is generated through decarboxylation of succinate, the by-product of fumarate reduction (Fig. 3a,b)44. Propionate concentrations were low during nitrate respiration regardless of the carbon source present (Fig. 3c,d), consistent with reduced succinate pathway activity previously observed in response to nitrate45 and the reduced requirement of fumarate as the terminal electron acceptor. Lactate fermentation, however, resulted in elevated propionate levels (Fig. 3c,d).
Figure 3. Metabolic changes associated with nitrate respiration.
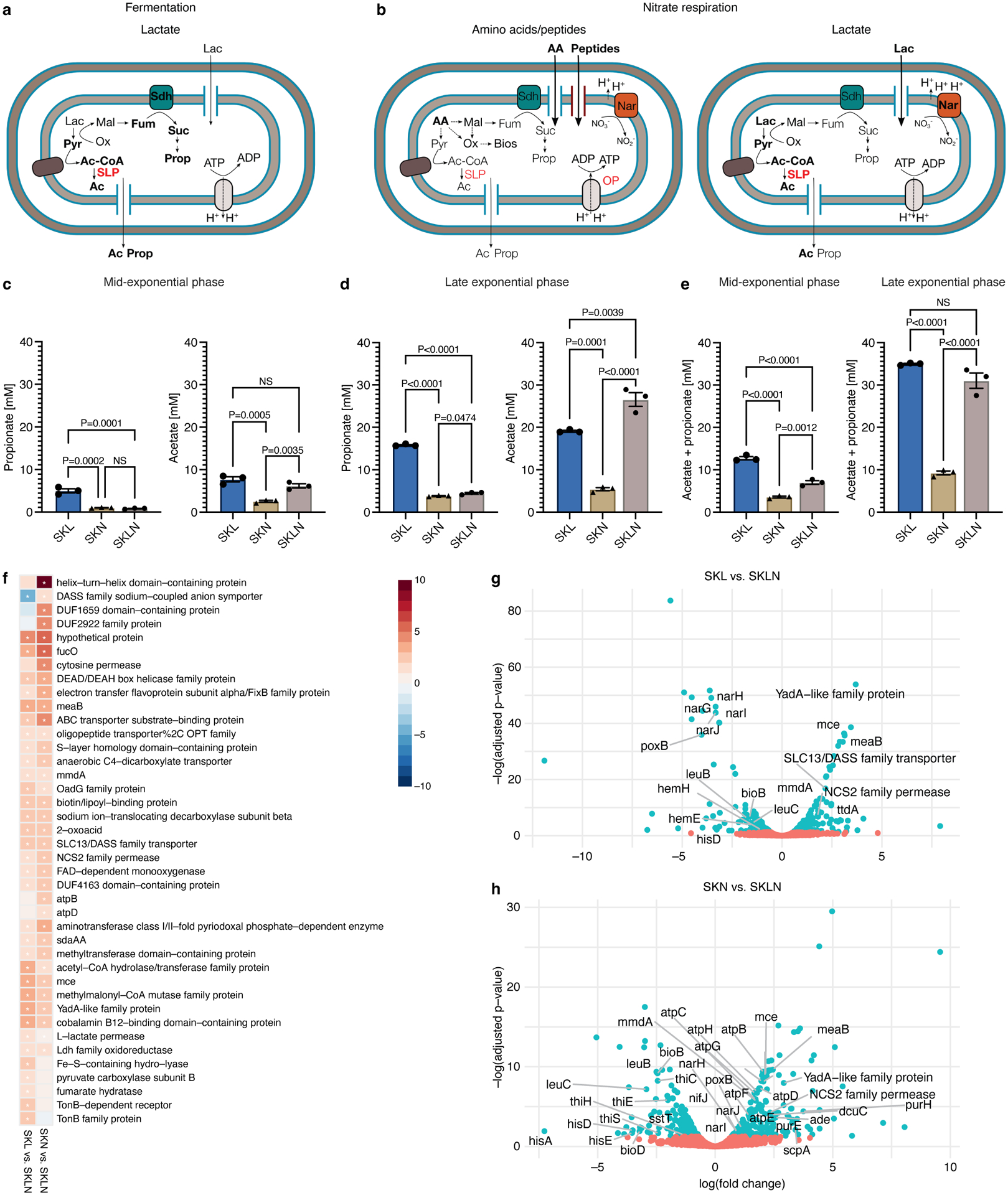
Veillonella metabolism during a) lactate fermentation and b) nitrate respiration. Propionate and acetate concentrations in SKL, SKN, and SKLN media during c) mid-exponential (n=3) and d) late exponential (n=3) phases. Propionate and acetate concentrations in uninoculated SK medium: 0.230±0.047mM and 0.885±0.042mM, respectively (n=6). e) Total amount of propionate plus acetate (n=3). f) Transcriptomics of WT cells growing in SK with nitrate and/or lactate (n=3), ordered by statistical significance. Heatmap indicates log(fold-change). Blue, downregulated in SKL or SKN. Red, upregulated in SKL or SKN. *, significance at false discovery rate (FDR) <0.05. Differential gene expression in cells growing in g) SKL vs. SKLN and h) SKN vs. SKLN. Points represent genes colored by significance: Red, FDR>0.05. Blue, FDR<0.05. Select genes illustrate metabolic pathways of interest. Data (c-e) represent mean±SEM, analyzed by one-way ANOVA and Tukey’s HSD test. NS, not significant. N numbers denote biological replicates.
Acetate is produced via oxidation and decarboxylation of pyruvate, which results in the formation of both ATP and presumably reducing power (Fig. 3a,b)43,46–49. During fermentation and nitrate respiration with lactate present, acetate concentration was elevated (Fig. 3c,d). In contrast, during nitrate respiration with amino acids/peptides (SKN), acetate concentration was dramatically decreased. Reduced acetate production during growth with amino acids/peptides compared to growth with lactate reflects not only a more gluconeogenic metabolism, but also a decrease in the generation of ATP via pyruvate (Fig. 3b). Diminished formation of by-products (propionate plus acetate) during respiratory growth with amino acids/peptides also implies that a comparatively larger fraction of carbon is likely available for biomass synthesis (Fig. 3e).
We next sought to understand which genes are differentially expressed in response to different growth conditions. First, we compared expression profiles of cells growing with lactate during fermentation (SKL) and nitrate respiration (SKLN), which represent likely environments encountered in the oral cavity and inflamed gut, respectively (Fig. 3f,g). Nitrate respiration resulted in upregulation of the nitrate reductase cluster, as well as pathways associated with synthesis of leucine, histidine, and heme; it also resulted in downregulation of fumarate hydratase and the methylmalonyl pathway (8-fold), which presumably led to decreased propionate formation from fumarate and succinate during anaerobic respiration. During nitrate respiration, poxB, which encodes a pyruvate:ubiquinone oxidoreductase that channels electrons to the respiratory chain, was upregulated 16-fold. Next, we compared the transcriptomes of nitrate-respiring cells growing with amino acids/peptides (SKN) or lactate (SKLN) to analyze responses to distinct carbon sources during nitrate respiration (Fig. 3f,h). With amino acids/peptides, genes involved in utilization or transport of purines, amino acids, and peptides (optB, opp1) were upregulated, consistent with our previous observation that amino acids/peptides support Veillonella growth when nitrate is present (Fig. 2). Notably, the atp operon was also upregulated with amino acids/peptides, indicative of a more determinant role for oxidative phosphorylation (OP) in this condition.
Nitrate respiration affects ATP synthesis in Veillonella
To understand how nitrate respiration expands the metabolic repertoire of V. parvula and boosts its growth with preferred carbon sources, we assessed the roles of the proton motive force (PMF) and ATP synthesis pathways during various growth conditions. To study the PMF, we used the DiOC2 dye that is transported into cells as a function of the electrochemical gradient, thus reflecting changes in membrane potential. Regardless of the carbon source, the presence of nitrate led to a marked increase in membrane potential in WT cells, which was dependent on a functional NarGHI complex (Fig. 4a). Treatment with the ionophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP) dissipated the gradient (Extended Data Fig. 6a). The importance of the electrochemical gradient was further assessed using a second uncoupler of OP: 2,4-dinitrophenol (2,4-DNP; Fig. 4b). During lactate fermentation, 2,4-DNP treatment reduced biomass and growth rate, reflecting the necessity of an active PMF. In the presence of nitrate, addition of 2,4-DNP reduced biomass and growth rates with lactate and amino acids/peptides. The growth rate reduction was more dramatic during amino acid/peptide than lactate utilization, suggesting that growth at the expense of these carbon sources occurs under a more elevated membrane potential (Fig. 4b, left in Fig. 3b).
Figure 4. Nitrate respiration-mediated changes in membrane potential are harnessed for substrate transport.
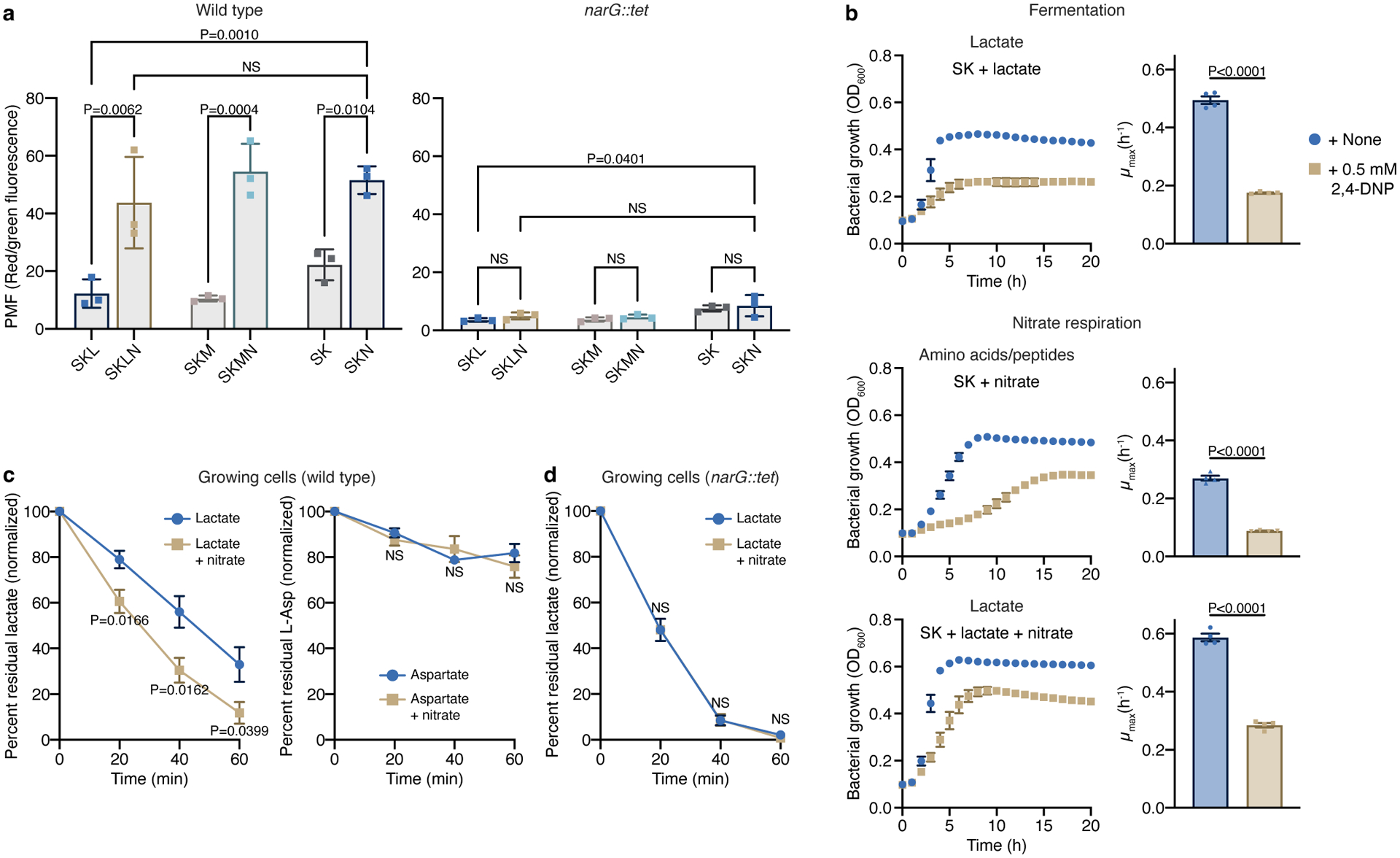
a) Measurement of the proton motive force (PMF) in WT and narG::tet cells growing on SK media supplemented or not with lactate or malate before and 40 min after addition of 40 mM nitrate (n=3 biological replicates). b) Growth of V. parvula in the presence of 2,4-DNP in SK and SKL media supplemented or not with nitrate (n=4 biological replicates). Growth rates (μmax) were calculated for each growth curve. c) Lactate (n=6 biological replicates) and aspartate (n=3 biological replicates) consumption by WT growing cells was determined in the spent medium 0, 20, 40, and 60 min after the addition of these carbon sources. d) Lactate consumption was assessed as in c using the narG::tet strain (n=4 biological replicates). Data (a-d) represent mean±SEM, analyzed by one-way ANOVA and Tukey’s HSD test (a) or two-sided t-test (b-d) . NS, not significant.
Dissipation of the proton gradient reduced growth in all conditions, implying that the PMF is critical for substrate transport via proton-dependent transporters or ATP synthesis via OP. To study the role of PMF, we first assessed lactate and Asp transport (Fig. 4c,d; Extended Data Fig. 6b,c). In resting cells without nitrate, both lactate and Asp were scarcely consumed; addition of nitrate, however, increased lactate but not Asp uptake (Extended Data Fig. 6b). Growing cells consumed lactate, which was further stimulated by nitrate (Fig. 4c). This enhancement was dependent on nitrate respiration, as the difference due to nitrate addition disappeared in narG::tet cells (Fig. 4d). It is, however, uncertain why narG::tet cells in the absence of nitrate consumed lactate more rapidly than WT cells. Asp was slightly consumed by growing cells, but nitrate addition had no effect (Fig. 4c). Using computational and functional analyses, we identified L-lactate permease (LldP), a homolog of the proton-dependent LldP in E. coli, as the lactate transporter in Veillonella (Extended Data Fig. 6d). Thus, nitrate respiration directly promotes Veillonella growth by facilitating the uptake of nutrients transported in a PMF-dependent manner.
We then assessed the importance of the PMF on ATP synthesis using a strain lacking ATP synthase (atpBEFHAGDC::tet, hereafter atp::tet), a multicomplex enzyme that couples ATP synthesis to proton transport into the cytoplasm (Fig. 5a). Cells lacking ATP synthase were unable to grow with lactate during fermentation, but grew nearly as well as WT during nitrate respiration (Fig. 5b). Nitrate allowed for atp::tet growth, yet no ATP could be generated via OP, impling that with lactate, ATP is primarily produced via substrate-level phosphorylation (SLP). The PMF generated with nitrate, therefore, must be harnessed to facilitate substrate uptake. The atp::tet strain failed to grow under nitrate respiration with amino acids/peptides, reflecting a potential requirement for ATP synthase under these conditions (Fig. 5b). Deletion strains lacking the ε (atpC::chl) or β and ε (atpDC::chl) subunits of ATP synthase displayed similar phenotypes (Extended Data Fig. 7a).
Figure 5. The conditional dispensability of ATP synthase unveils mechanistic aspects associated with fermentation and nitrate respiration.
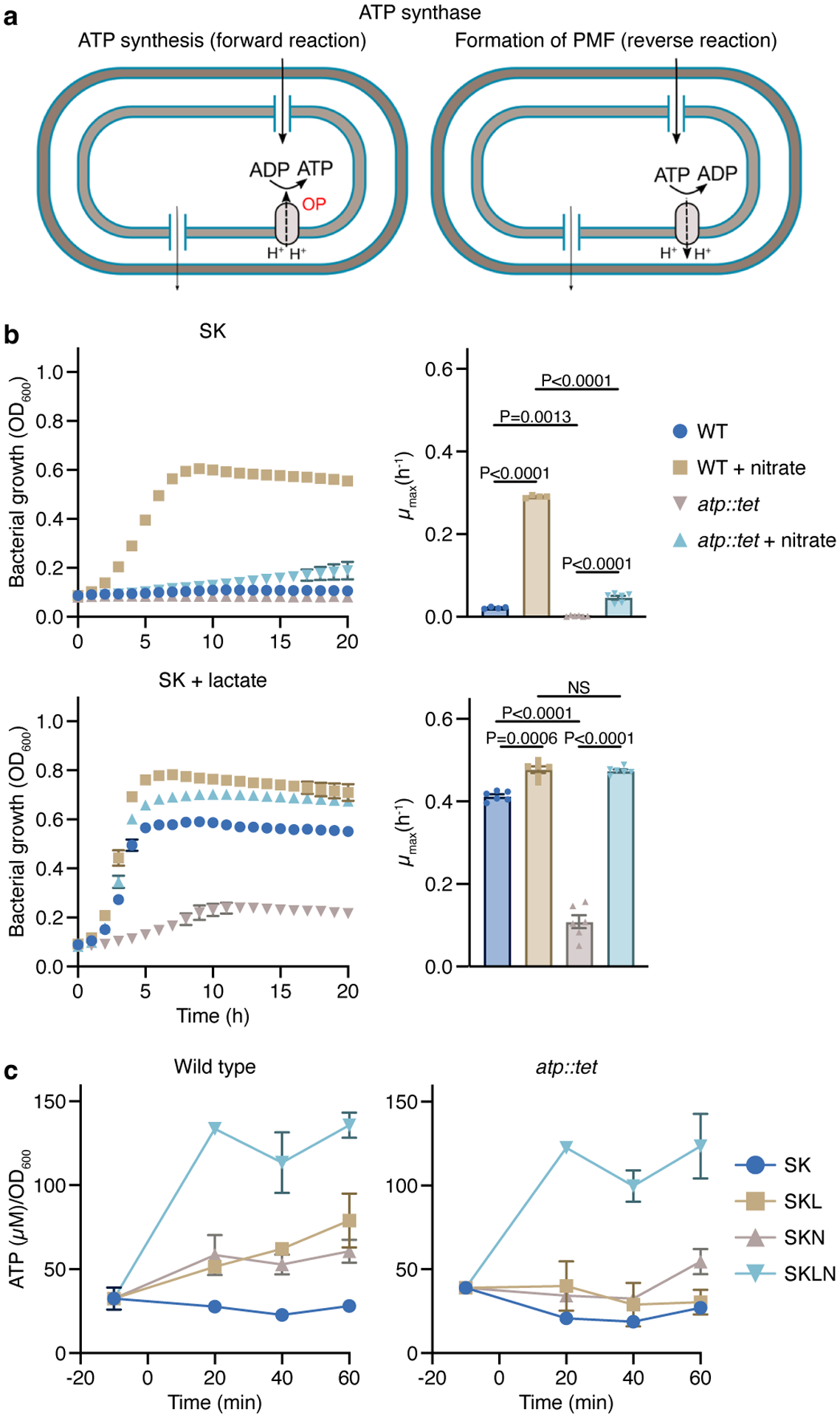
a) The ATP synthase is a multiprotein complex that couples ATP formation to the proton motive force (forward reaction) or generates an electrochemical potential using ATP hydrolysis (reverse reaction). b) Growth curves of V. parvula lacking the ATP synthase (atp::tet) in the presence or absence of nitrate and/or lactate compared to WT V. parvula (n=4 or 6 biological replicates). Growth rates (μmax) were calculated for each growth curve. c) Measurements of ATP in WT and atp::tet cells in media with and without lactate and/or nitrate (n=3 biological replicates). Data (b,c) represent mean±SEM, analyzed by one-way ANOVA and Tukey’s HSD test. NS, not significant.
To further understand the role of the PMF and the contributions of SLP and OP to ATP synthesis during fermentation and respiration, we measured ATP levels in WT and atp::tet cells. Cells were grown on SKLN medium to mid-exponential phase, then washed and resuspended in SK medium. ATP was measured before and after supplementation with lactate and/or nitrate. Lactate addition significantly increased ATP levels in WT but not atp::tet cells. Supplementation with lactate plus nitrate dramatically elevated ATP levels in both WT and atp::tet cells, further confirming that ATP is mostly produced via SLP when lactate is present (Fig. 5c). Supplementation with lactate plus nitrate produced only minor differences in ATP levels between the strains, demonstrating that the growth deficiency of atp::tet observed during lactate fermentation (Fig. 5b) was not related to its capacity to produce ATP but instead to limited lactate uptake (Extended Data Fig. 6c). This also suggests that during lactate fermentation, the reverse reaction of ATP synthase is likely required to generate the PMF and energize proton-dependent lactate uptake (Fig. 5a). With nitrate supplementation, atp::tet cells produced approximately half the ATP produced by WT cells (Fig. 5c at 20 and 40min). OP, therefore, is required for ATP synthesis when Veillonella utilizes amino acids/peptides for growth. This is consistent with greater induction of the atp operon when cells were grown with amino acids/peptides (Fig. 3f,g; Extended Data Fig. 7b).
Veillonella expansion during inflammation is nitrate-dependent
We next assessed the importance of this metabolic switch on the capability of Veillonella to colonize the inflamed intestine. First, we examined whether epithelial injury and colitis led to V. parvula expansion in a gnotobiotic mouse model. We treated mice pre-colonized with altered Schaedler flora (ASF) with either 3% dextran sulfate sodium (DSS) or control solution for 8 days and orally gavaged the mice with WT or narG::tet (2×109 colony-forming units, CFUs) on day four (Fig. 6a). Inflammation was evident in DSS-treated mice by inflammatory cytokine production, neutrophil expansion, and decreased body weight and colon length (Fig. 6b,c; Extended Data Fig. 8a–d). Inflammation also resulted in a dramatic increase in Veillonella abundance in stool, as determined by qPCR, while absolute 16S rRNA counts were unaffected (Fig. 6d, Extended Data Fig. 9a). Our previous in vitro experiments did not reveal any growth defects for narG::tet when nitrate was absent (Fig. 1b), allowing us to test the importance of nitrate respiration in Veillonella expansion during inflammation. The narG::tet strain also expanded during inflammation, but to a significantly lower abundance than WT (Fig. 6d). Similar results were obtained by counting CFUs in stool and colonic tissue from control- and DSS-treated mice (Extended Data Fig. 9b,c). narG was upregulated, providing further evidence for in vivo nitrate utilization (Fig. 6e). In mice colonized with WT Veillonella, DSS treatment resulted in a 2-fold increase in lactate concentration in colonic tissue (Extended Data Fig. 9d). However, we observed no upregulation of Veillonella genes associated with lactate metabolism or induced by lactate in our RNA-sequencing data (lldD, lldP, hisA, argB) in DSS-treated mice (Extended Data Fig. 9e).
Figure 6. Nitrate reductase activity is required for efficient colonization in a mouse model of inflammation.
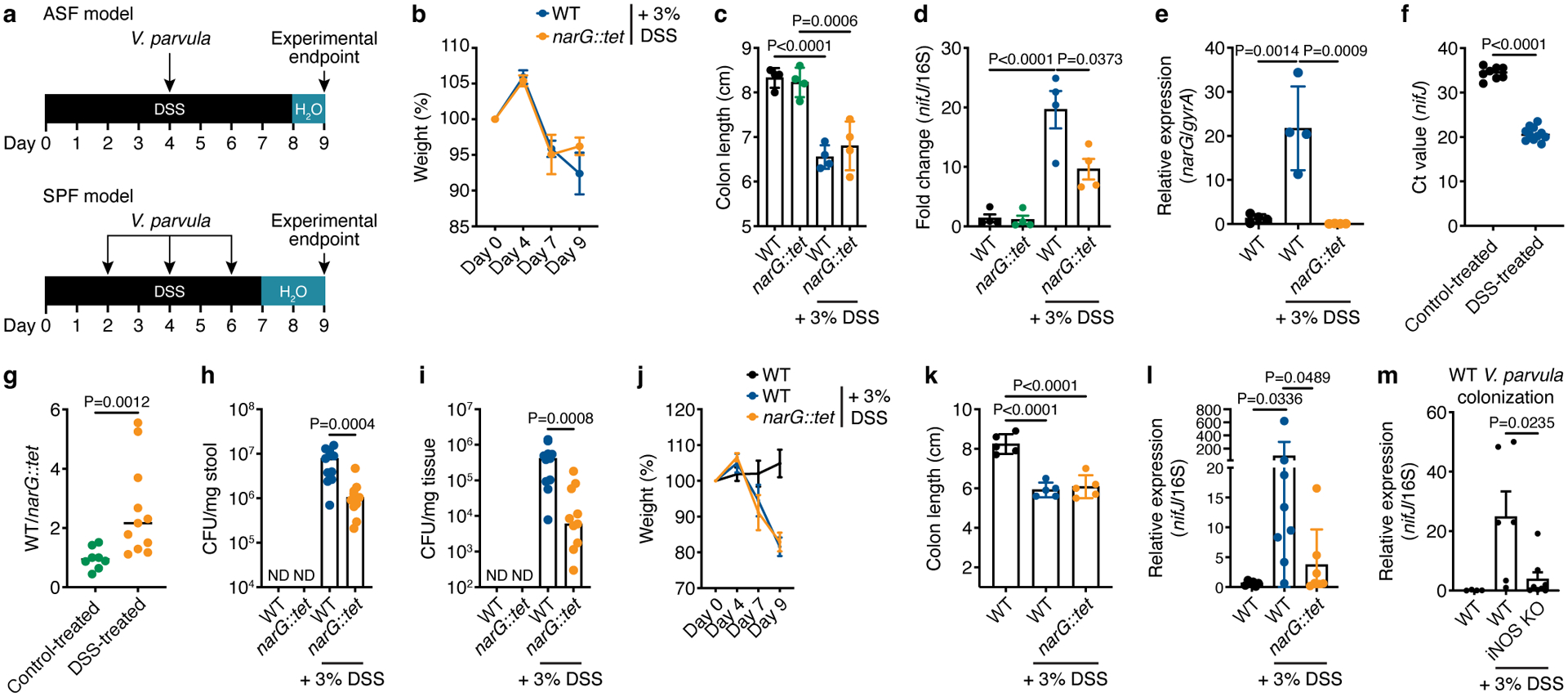
a) Experimental design of mouse models of colitis. Assessments of b) weight change over DSS treatment course and c) colon length at experimental endpoint (n=5). d) Relative abundance of Veillonella in stool of ASF mice, calculated using the ΔΔC(t) method comparing V. parvula nifJ and 16S rRNA genes (n=4). e) Relative expression of the narG gene in WT Veillonella colonizing stool of control- and DSS-treated ASF mice, with narG::tet-colonized mice serving as a negative control (n=4). gyrA was used as an internal control. f) Ct values of the Veillonella nifJ gene measured by qPCR in stool of WT-colonized ASF mice with and without DSS treatment (n=8–11). g) qPCR analysis of the relative colonization of WT and narG::tet strains in stool of ASF mice inoculated with a 1:1 mixture of each strain with and without DSS treatment (n=8–11). The y-axis represents the fold-change difference in abundance of each strain. Colony-forming units (CFUs) measured from h) stool and i) colonic tissue of ASF mice inoculated 1:1 with WT and narG::tet strains (n=8–11). ND, not detected. Weight change j) and colon length k) in SPF mice (n=5). l) Colonization of WT and narG::tet strains in stool of SPF mice measured by qPCR, comparing Ct values of nifJ and 16S rDNA (n=6–8). m) Colonization of WT Veillonella in stool of WT and iNOS KO DSS-treated SPF mice compared to control-treated WT mice measured by qPCR (n=4–8). Data represent two independent experiments and show mean (b-g, j-m) ± SEM (b-e, j-m) or median (h,i) values. Data was analyzed by one-way ANOVA and Tukey’s HSD test or Mann-Whitney U-test. NS, not significant. N numbers denote mice.
To further assess nitrate respiration in vivo, we performed a competition experiment in which ASF mice received a 1:1 ratio of WT(chl) and narG::tet strains at the same dose and time point as the single colonization experiment. WT(chl) is an otherwise identical WT strain except for a chloramphenicol resistance gene in the intergenic region of narK and hemZ to distinguish between the Veillonella strains. WT(chl) colonized to significantly higher levels than narG::tet in the inflamed gut, as assessed by qPCR (Fig. 6g). Reduced fitness of narG::tet during inflammation was further confirmed by assessing CFUs in stool and colonic tissue from control- and DSS-treated mice, where we observed a 10-fold difference in colonization between WT(chl) and narG::tet (Fig. 6h,i; Supplementary Fig. 1). WT Veillonella were at least 100-fold more abundant in DSS- than control-treated ASF mice (Fig. 6h,i), suggesting that inflammation facilitates engraftment or provides additional cues for expansion.
We then asked whether Veillonella was able to colonize mice with a more complex gut microbiome, reflecting the human gut that harbors other nitrate utilizers, and adapted our single colonization model to mice from a specific-pathogen-free (SPF) environment (Fig. 6a). Given higher colonization resistance in these mice, we orally gavaged them with WT or narG::tet three times (days 2, 4 and 6) before the experimental endpoint. DSS-treated mice again displayed decreased weight and colon length (Fig. 6j,k). The ability of WT Veillonella to expand in the intestine increased during inflammation and surpassed that of narG::tet (Fig. 6l). Veillonella colonization in the intestine of SPF mice was variable and required additional inoculations, reflecting increased resistance of a complex murine microbiome to a human-derived oral microbe.
To gain further insight into the role of inflammatory nitrate in Veillonella expansion, we compared Veillonella colonization in WT and inducible nitric oxide synthase (iNOS) knockout (KO) SPF mice. Previous studies indicated that free nitrate in the gut during inflammation is largely derived from iNOS activity in immune cells50. DSS treatment showed equal effects on the colon lengths of WT and iNOS KO mice (Extended Data Fig. 9f), but Veillonella abundance in the stool of iNOS-deficient mice was significantly reduced compared to WT mice (Fig. 6m), phenocopying colonization with narG::tet (Fig. 6d,l). Collectively, these results demonstrate that Veillonella utilizes inflammatory nitrate derived from iNOS activity in host cells to ectopically colonize the gut.
DISCUSSION
Inflammation is an overarching factor that determines the ability of extraintestinal microbes to colonize the gut. One transverse factor resulting in expansion of these microbes is emergence of new electron acceptors that enable anaerobic respiration. Yet, the molecular and physiological implications of these molecules in non-model microbes has been poorly explored. Our mechanistic study unveiled key strategies that V. parvula, and potentially other Veillonella species, use to exploit metabolic signatures of inflammation and ectopically colonize the intestine.
Ectopic colonization of Veillonella observed in IBD patients indicates that inflammation provides appropriate environmental signals for its enrichment. Veillonella is aerotolerant and thus well adapted to oxygenic regions of the inflamed gut and to the presence of ROS, potentially increasing its success in these conditions. Moreover, we found that, unlike other gut Firmicutes, Veillonella possesses a respiratory nitrate reductase that enables anaerobic respiration. Nitrate respiration not only boosted Veillonella growth on organic acids, but also allowed amino acids and peptides to be used as carbon and energy sources. A mouse model of colitis confirmed the importance of nitrate respiration for colonizing the inflamed gut, as inactivation of V. parvula narG or murine iNOS impaired colonization. Transition from the oral cavity to the inflamed gut, therefore, may be driven by this fermentation-to-respiration switch. A potential limitation of this observation is the abundance of other nitrate utilizers, such as E. coli, in the mouse may not be reflective of that in the human gut.
The switch to nitrate respiration augments the proton gradient, which enhances proton-dependent transport and increases ATP generation via OP. Additionally, it reduces use of fumarate and the succinate pathway45. Our findings support a model in which nitrate respiration allows Veillonella growth on amino acids and peptides via the generation of extra ATP through OP as well as through a reduction in carbon waste in the forms of acetate and propionate. Similarly, the increased electrochemical gradient likely energizes peptide and amino acid transport via proton-coupled transporters, such as OptB41. Increased formation of ATP via ATP synthase using the energy of the PMF compensates for reduced energy formation via both SLP and the Na+-driven gradient.
During inflammatory conditions in vivo, nitrate respiration increases Veillonella growth and hence its ability to compete and establish in the gut environment. Expansion of Veillonella’s metabolic repertoire enables utilization of more diverse carbon sources, conferring an extra ability to colonize alternative niches. Small molecules (>3kDa) released from the death of gut epithelial cells were shown to promote growth of multiple intestinal Enterobacteriaceae51. In other oral microbes, such as P. gingivalis, the use of amino acids and peptides represents a virulence factor that allows colonization of inflamed tissue52. Veillonella might also potentially consume lactate that accumulates in tissues during inflammation53 and is consistently observed to be enriched in IBD patients17. Although OP was less important in the presence of lactate, we hypothesize that it is also harnessed for ATP synthesis in settings where lactate is abundant. Additionally, increased PMF during nitrate respiration can be used to enhance the uptake efficiency of molecules mobilized via proton-dependent transporters, therefore facilitating growth in the gut environment. Discerning the relevant carbon sources in vivo and their implications for colonization remain to be explored.
V. parvula and other oral microbes are linked with multiple inflammatory diseases32. Our approach not only allowed us to confirm the positive correlation between inflammation and Veillonella abundance observed in IBD patients, but also to unravel the mechanistic strategies that underlie the transition from fermentation to nitrate respiration and allow for successful colonization of the inflamed gut. The capacity of Veillonella to alternate between different metabolic pathways highlights the versatility of the bacterial world, and additional microbes enriched during inflammatory conditions likely use parallel strategies. The fact that Veillonella, as well as some Proteobacteria, use inflammatory nitrate to boost growth implies that reducing inflammation or the abundance of these microbes may interrupt the positive feedback between inflammation and dysbiosis.
MATERIALS AND METHODS
Analysis of human gut microbiome samples
The PROTECT cohort generated gut microbial profiles from pediatric UC patients32. We extracted the abundances of all operational taxonomic units (OTUs) assigned to Veillonella species from the published OTU abundance profiles (V. dispar OTU 342427, V. parvula OTU 518743, V. parvula OTU 3889756). Fecal calprotectin (FC) values were binned into low (<100 mcg/g), increased (100–200), high (200–3,000), and very high (>3,000). A two-sided Wilcoxon test was applied to compare the abundance of V. parvula and V. dispar across the 4 bins. Only samples where the respective OTU was present were included: V. dispar OTU 342427 was present in 1,006 samples, V. parvula OTU 518743 in 874 samples, and V. parvula OTU 3889756 in 487 samples.
The RISK study profiled pediatric CD patients33. We extracted the abundances of all Veillonella OTUs at species level from the published abundance profiles (V. parvula OTU 1755, V. dispar OTU 39) and compared their abundance in CD patients to non-IBD controls using a two-sided Wilcoxon test. Only samples where the respective OTU was present were included: V. parvula OTU 1755 was present in 529 samples, V. dispar OTU 39 in 846 samples.
Strains and culture media
Veillonella parvula SKV38 was used as the wild-type strain for our experiments. We also used V.parvula DSM2008, V. dispar LD205, V. atypica RJX1392, V. atypica RJX1514, and V. ratti 84 (Supplementary Table 1). Cells were grown routinely in SK agar (composition in g/L: yeast extract 10, casitone 10, Na2HPO4 0.4, NaCl 2.0) supplemented or not with DL-lactate (25 mM) and/or potassium nitrate (20 mM) unless otherwise indicated. The media with both lactate and nitrate added we called SKLN. Cells were incubated for 24 h under anaerobic conditions at 37°C. The growth of SKV38 was also assessed in GIFU anaerobic medium (HiMedia, Cat# M1801), Difco™ Reinforced Clostridium medium (BD Cat# 218081), DifcoTM Columbia broth (BD, Cat# 294420), MRS broth (Sigma, Cat# 69966), and BHI medium (HiMedia Cat# M210). Antibiotics were added to the medium when appropriate: 5 μg ml−1 tetracycline, 5 μg ml−1 chloramphenicol, 20 μg ml−1 erythromycin for V. parvula, and 100 μg ml−1 ampicillin and 100 μg ml−1 spectinomycin for E. coli. X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) was dissolved in DMSO (40 mg/ml), and added to plates for white/blue selection at a final concentration of 0.1 mg/ml.
Molecular biology techniques
The assembly of DNA fragments was routinely performed using Golden Gate cloning. For this, we used the plasmid pDRT004, which was constructed in this study, and allowed the modular assembly of DNA fragments. This plasmid contained BsaI and BbsI restriction sites, respectively, surrounding a lacZ gene for blue/white selection and the ampicillin resistance gene. For gene knockout, two fragments of DNA flanking the gene of interest were amplified by PCR (~0.5 – 1.0 kb), and assembled with an antibiotic resistance gene (tet, ery, or chl) in pDRT004. The tet resistance gene was obtained from the pBSLJ1 plasmid, whereas the chl and ery genes under control of the gyrA promoter were synthesized as DNA fragments (IDT Technologies) (Supplementary Table 2) and cloned into pDRT004 as pDRT004 + narG(chl) and pDRT004 + narG(ery) for routine amplification of the resistance genes using primers DR096 and DR097. A typical 20 μl Golden Gate assembly reaction contained 2 μl of T4 DNA ligase buffer, 100 ng of pDRT004, equimolar amounts of the resistance gene (i.e. erythromycin, chloramphenicol or tetracycline whose transcription is driven from the PgyrA promoter) and the two homology regions, 1.5 μl of BsaI HFv2 (NEB), 0.5 μl of T4 DNA ligase (NEB), and ultrapure water up to 20 μl. The reaction was incubated at 37°C for 1 h, and 60°C for 5 min. The Golden Gate reaction was then transformed into NEB Turbo competent E. coli, and the white colonies were selected for further experiments. For transformation of V. parvula SKV38, the assembled DNA fragments encompassing the homology regions and the resistance gene were amplified by PCR, and the PCR products were used for transformation. Mutations were verified by PCR and Sanger sequencing. Multiple clones were selected to assess phenotypes. In all cases, the DNA fragments were amplified using the Phusion™ high-fidelity DNA polymerase (Thermo F530S). All primers were purchased from Genewiz (USA), and the sequences are provided in Supplementary Table 3.
Veillonella parvula natural transformation
For genetic manipulation of V. parvula, we used natural transformation in SKL medium. Briefly, cells were grown overnight on SKLN medium agar plates, and cells from multiple colonies were resuspended to an OD600 ~ 0.400 in SK medium. A volume of 45 μl of the bacterial suspension was combined with 250–500 ng of DNA in a final volume of 50 μl. The DNA for this reaction was obtained by PCR amplification of the assembled vectors, as described above. Three 15 μl drops were then placed on SKL solid medium (previously autoclaved and poured under aerobic conditions) and incubated for 24 h at 37 °C under anaerobic conditions. Next day, one drop was resuspended in 50 μl of SK medium and plated on SKLN medium supplemented with the appropriate antibiotics (i.e. tetracycline 5 μg ml−1, chloramphenicol 5 μg ml−1, erythromycin 20 μg ml−1). Colonies were visible after 24–48 h of incubation. Colonies were restreaked on SKLN medium containing antibiotics, and the mutations were verified by PCR and sequencing. DL-lactate (25 mM), nitrate (20 mM), and/or antibiotics were added after autoclaving. Plates were air dried for 48 h at 37 °C under aerobic conditions. For double or triple deletion mutants, the same protocol described above was used, but a deletion strain was used as the recipient strain for transformation. The antibiotic resistance profile, PCR, and sequencing were carried out to verify the genotypes.
Growth experiments
To study the impact of organic acids, amino acids, dipeptides, and peptones on growth of SKV38, we used the SK base broth (composition in g/L: yeast extract 2.5 (Sigma), Na2HPO4 0.4 (Sigma), NaCl 2.0 (Sigma)). KNO3 (40 mM) (Sigma) was included or not as required. The composition of this medium was chosen as it permits the growth of V. parvula up to OD600~0.2, but further growth can be achieved by supplementation with other carbon sources. For the experiments, cells were grown overnight in SK with 40 mM nitrate. The next day, cells were washed twice in SK salts (composition in g/L: Na2HPO4 0.4, NaCl 2.0), and resuspended in the same buffer. The optical density of the suspension was measured, and the cultures inoculated at an initial OD600 of 0.025. Growth was monitored for 24 h by following OD600 in a BioTek Synergy H1. Sodium nitrite (Sigma), ammonium chloride (Sigma) were also used. The following carbon sources were used over the course of the experiment: L-lactate (Sigma), L-(−)-malic acid disodium salt (Sigma), sodium succinate (Sigma), sodium fumarate (Sigma), sodium pyruvate (Sigma). For assessment of the carbon sources in the other Veillonella strains, the SK base medium was modified as follows (in g/L): yeast extract 5.0, Na2HPO4 0.4, NaCl 2.0.
Maximum-specific growth rate (μmax) values were determined from absorbance growth curves using the Exponential model (Eq. 1)54, where ABSt is absorbance at time t, k is a constant, μABS is the μmax determined from absorbance measurements using the Exponential model (h−1), and t is time (h). Absorbance values were taken during exponential growth.
| (Eq. 1) |
Evaluation of growth with amino acids and complex peptides
To determine the capability of SKV38 to grow on amino acids, we used the SK base medium. The following amino acids were studied final concentration of 50 mM from stocks of 200 mM at pH ~ 7.0: L-aspartic acid (Sigma), L-glutamic acid potassium salt (Sigma), L-asparagine (Sigma), L-glutamine (Sigma). Casaminoacids (0.5%) (Calbiochem), Bacto™-Casitone (0.5%) (BD), tryptone (Sigma), Bacto™ peptone (BD), and yeast extract (Sigma) were also included for comparison. For this experiment, cells were grown on SK medium supplemented with 20 mM KNO3. The next day, cells were collected by centrifugation at 10,000 × g, washed twice with 1X SK buffer, and resuspended in 1X SK buffer; OD600 was recorded and adjusted to 0.025. The experiment was monitored by spectrophotometry over a period of 24 h.
Gene expression analysis: RT-qPCR
Cells (1.5 ml) were harvested at the specific time points, centrifuged at 15,000 × g for 2 min under anaerobic conditions, the supernatant discarded, and finally the pellet resuspended in 0.6 ml of Trizol (Zymo). The suspensions were stored at −80 °C for further analysis. The cellular suspension was next combined with 1 g of zirconium beads and placed on a Tissue Lyser for cell disruption. RNA was then harvested using the Direct-zol RNA kit (Zymo) with in-column DNase treatment following manufacturer’s instructions. RNA was eluted in 50 μl of ultrapure water. Synthesis of cDNA was performed using the iScript™ cDNA synthesis kit (Bio-Rad) following the instructions of the manufacturer. qPCR was performed using the iTaq Universal SYBR Green Supermix (Bio-Rad). Primers were designed using Primer3 v. 0.4.0 (https://primer3.ut.ee) and listed in Supplementary Table 4. The gene gyrA was used as reference.
Phylogenetic analyses of nitrate reductase genes
To test for detection of nitrate reductase genes in the human microbiome, we searched for species containing the gene in the assembled gene catalog from Pasoli et al (2019)55, which consists of 9,428 metagenomes from different body sites, ages, countries, and lifestyles. The E. coli narG and napA, and Klebsiella oxytoca nasA amino acid sequences were queried against the gene catalog using USEARCH v8.1 “search_global” command56. Genes with at least 45% amino acid similarity and that originated from either stool or oral cavity samples were used for downstream analysis. To place the genomes containing a nitrate reductase gene on a phylogenetic tree, we used PhyloPhlAn v3.0.51 with default settings including all genomes containing at least one of the nitrate reductase genes57.
Generation and analysis of RNA-sequencing data
Illumina cDNA libraries were generated using a modified version of the RNAtag-seq protocol58. Briefly, 500–1000 ng of total RNA was fragmented, depleted of genomic DNA, dephosphorylated, and ligated to DNA adapters carrying 5’-AN8–3’ barcodes of known sequence with a 5’ phosphate and a 3’ blocking group. Barcoded RNAs were pooled and depleted of rRNA using the RiboZero rRNA depletion kit (Illumina). Pools of barcoded RNAs were converted to Illumina cDNA libraries in 2 main steps: (1) reverse transcription of the RNA using a primer designed to the constant region of the barcoded adaptor with addition of an adapter to the 3’ end of the cDNA by template switching using SMARTScribe (Takara Bio)59; and (2) PCR amplification using primers whose 5’ ends target the constant regions of the 3’ or 5’ adaptors and whose 3’ ends contain the full Illumina P5 or P7 sequences. cDNA libraries were sequenced on the Illumina NovaSeq 6000 platform to generate paired end reads and collected using NovaSeq Control Software 1.6.
Sequencing reads from each sample in a pool were demultiplexed based on their associated barcode sequence using custom scripts. Up to 1 mismatch in the barcode was allowed provided it did not make assignment of the read to a different barcode possible. Barcode sequences were removed from the first read as were terminal Gs from the second read that may have been added by SMARTScribe during template switching. Reads were aligned to NZ_LR778174.1 using the Burrows-Wheeler Alignment tool (BWA) version 0.7.15-r1142-dirty60 and read counts were assigned to genes and other genomic features using custom scripts. Differential expression analysis was conducted with edgeR v3.1561. Visualization of raw sequencing data and coverage plots in the context of genome sequences and gene annotations was conducted using GenomeView v260062.
Metabolite analysis
WT cells were grown until mid- and late exponential phase in SK medium supplemented or not with lactate and/or nitrate until they reached an OD600 ~0.3–0.4. A volume of 1.5 ml of the culture was then collected, centrifuged at 15,000 × g for 3 min, and 1.0 ml of the supernatant collected. This volume was then diluted 10 times and filter-sterilized for further analysis. For determination of organic acids, amino acids, and dipeptides, we used two liquid chromatography tandem mass spectrometry (LC-MS) methods designed to measure polar metabolites63. Briefly, amino acids and dipeptides were estimated using hydrophilic interaction liquid chromatography (HILIC) and an LC-MS system comprised of a Shimadzu Nexera X2 U-HPLC (Shimadzu Corp., Marlborough, MA) coupled to a Q Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Metabolites were extracted from media samples (10 μL) using 90 μL of acetonitrile/methanol/formic acid (74.9:24.9:0.2 v/v/v) containing stable isotope-labeled internal standards (valine-d8, Sigma-Aldrich, St. Louis, MO; phenylalanine-d8, Cambridge Isotope Laboratories, Andover, MA). The samples were centrifuged (9,000 × g, 4°C, 10 min), and the supernatants were injected directly onto a 150 × 2 mm, 3 μm Atlantis HILIC column (Waters, Milford, MA). MS analyses were carried out using electrospray ionization in the positive ion mode using full scan analysis over 70–800 m/z at 70,000 resolution. For the measurement of propionate HILIC and MS in negative ionization mode were conducted using an Shimadzu Nexera X2 U-HPLC (Shimadzu Corp., Marlborough, MA) coupled to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Metabolites were extracted from media (30 μL) using 120 μL of 80% methanol containing inosine-15N4, thymine-d4 and glycocholate-d4 internal standards (Cambridge Isotope Laboratories, Andover, MA). The samples were centrifuged (10 min, 9,000 × g, 4°C), and the supernatants were injected directly onto a 150 × 2.0 mm Luna NH2 column (Phenomenex, Torrance, CA). MS analyses were carried out using electrospray ionization in the negative ion mode using full scan analysis over m/z 70–750 at 70,000 resolution and 3 Hz data acquisition rate. Additional MS settings were: ion spray voltage, −3.0 kV; capillary temperature, 350°C; probe heater temperature, 325 °C; sheath gas, 55; auxiliary gas, 10; and S-lens RF level 50. Compound identification was carried out by matching the retention time and m/z of feature in the data with an in-house collection of authentic standards including a library of 336 dipeptides (Anaspec AS-65126–336, Fremont, CA). Absolute quantitation of propionate was conducted using calibration curves spiked into media with stable isotope-labeled 13C propionate (Cambridge Isotope Labs CLM-771 and CLM-627, Tewksbury, MA). Acetate (Sigma, MAK086), aspartate (Sigma, MAK095), glutamate (Sigma, MAK004), and nitrate / nitrite (Sigma, 482655) were determined using enzymatic assays, following the procedure recommended by the manufacturer. Raw data were processed using TraceFinder version 5.0 (Thermo Fisher Scientific, Waltham, MA) and Progenesis QI version 1.0 (Nonlinear Dynamics, Newcastle upon Tyne, UK). Extracted ion chromatograms for peaks of interest were generated and inspected using Thermo Scientific Xcalibur v. 4.3.
Measurement of the proton motive force
The membrane potential was assessed using the BacLight™ Bacterial Membrane Potential Kit (Invitrogen). Briefly, WT cells were grown on SK + 50 mM DL-lactate + 40 mM KNO3 or SK + 50 mM L-malate + 40 mM KNO3 broth until they reached mid-exponential phase (OD ~ 0.3–0.4). Next, cells were washed twice in SK medium, and resuspended at the same optical density in 1) SK + 50 mM DL-lactate, 2) SK + 50 mM DL-lactate + 40 mM nitrate, 3) SK + 50 mM L-malate, 4) SK + 50 mM L-malate + 40 mM nitrate, 5) SK, and 6) SK + 40 mM nitrate. After 45 min of incubation, cells were collected, diluted in 1 ml of filtered PBS (1/50 dilution), and treated or not with 10 μl of CCCP. Next, 10 μl of the DiOC2 dye were added, the reaction incubated for 30 min, and fluorescence measured by flow cytometry.
2,4 – DNP experiments
Cells were grown overnight in SK medium supplemented with 20 mM nitrate and 25 mM lactate. The next day, the culture was centrifuged at 10,000 × g and rinsed twice with SK medium. Growth curves were then initiated in the presence of different concentrations of 2,4-dinitrophenol (2,4-DNP). The stock of 2,4-DNP was prepared at 100 mM in DMSO.
Lactate and aspartate consumption in growing and resting cells
The dynamics of sodium L-lactate (Sigma) and L-Asp (Sigma) consumption in growing and resting cells were studied in SKV38 WT cells. For this, cells were grown overnight on SK medium supplemented with 25 mM DL-lactate and 20 mM KNO3. The next day, a new culture was started in SK + 50 mM DL-lactate + 40 mM KNO3 medium, inoculated with 1/40 of the ON culture, and incubated for ~3–4 h until cells reached an OD600 = 0.3–0.4 under anaerobic conditions. Cells were collected by centrifugation at 10,000 × g, washed twice with phosphate-buffered saline (PBS), and resuspended in RC buffer (0.1 M potassium phosphate buffer pH 6.5, 20 mM MgSO4, 20 mM beta-mercaptoethanol) for resting cells64 or SK medium for growing cells. Absorbance was measured and adjusted to a final OD600 = 2.0 in a volume of 500 μl. After 40 min of incubation to deplete the pool of ATP, L-lactate, L-aspartate, and/or nitrate were added to the reaction. Samples of 20 μl were taken at specific time points, diluted with 480 μl of MiliQ water, centrifuged at 15,000 × g for 3 min, and the supernatant used for analytical determinations. Lactate was determined using the Lactate Determination Kit (Sigma, Cat# MAK064), and aspartate was determined using the Aspartate Determination Kit (Sigma, Cat# MAK095) following the instructions of the manufacturer.
ATP measurements
To measure ATP levels, cells were grown up to the mid-exponential phase, washed twice, and resuspended in the SK medium at an OD600 = 0.300. Cells were incubated for 40 min at 37°C to deplete intracellular ATP, and then lactate and/or nitrate added at the defined concentrations. ATP and optical density (600 nm) were simultaneously monitored after −10, 0, 20, 40, and 60 min. For this, 250 μl samples were combined with 100 μl ice-cold 3 M perchloric acid and incubated on ice for 2 h. Sterile medium was used as a control. Then, the pH was neutralized by the addition of 100 μl of 3 M KOH and 120 μl of 1.0 M Tes-NaOH buffer pH 7.0. The reaction was mixed by vortexing and centrifuged at 15,000 × g at 4 °C for 3 min. A volume of 400 μl of the supernatants were collected and kept at −20 °C for ATP determination. ATP determination was performed using the ATP Determination Kit (Invitrogen, Cat #A22066) based on the luciferase assay and following the manufacturer’s recommendations. Luminescence was measured after 3 min of incubation. The concentration of ATP in the cells was determined by using a calibration curve with increasing concentrations of ATP.
Mouse experiments
All mouse experiments were performed according to protocols approved by the Institutional Animal Care and Use Committees (IACUC) of the Broad Institute and complied with all relevant ethical regulations. Mice were fed an autoclavable diet ad libitum (Lab Diet,; cat# 0001326) and housed with a 12-hour light/dark cycle at an ambient temperature between 18–24°C and 30–70% relative humidity. Mice were randomized into experimental groups; the number of mice utilized in each experiment is indicated in the figure legends.
Gnotobiotic mice colonized with altered Schaedler flora (ASF) used in experiments were C57BL/6N females or males aged 6–10 weeks and maintained in the germ-free facility at the Broad Institute. These mice were maintained with ASF for several generations. ASF mice in the DSS groups were given 3% dextran sulfate sodium salt (Mr approx. 40,000 kDa) in drinking water. After 4 days of DSS treatment, mice were orally gavaged with 100 μL of Veillonella culture at 2 × 1010 CFU/mL and continued DSS treatment for an additional 4 days post-gavage. DSS was removed and replaced with water for one day before sacrifice. For the specific-pathogen-free (SPF) model, we utilized male C57BL/6J mice from the Jackson Laboratories aged 8–10 weeks. We used both the WT strain (strain # 000664) and a strain deficient in iNOS activity (strain #002609; B6.129P2-Nos2tm1Lau/J). SPF mice were gavaged with Veillonella as above iteratively on days 2, 4 and 6. SPF mice were given DSS as above for 7 days; DSS was removed and replaced with water for two days before sacrifice. Mice not receiving DSS treatment were gavaged with the appropriate V. parvula SKV38 WT or narG::tet strain at the same time and the same amount as DSS-treated mice. Veillonella cultures were prepared by setting up fresh cultures without antibiotics that were grown until OD600 reached 0.4–0.5 (~4 hours). Cells were washed in PBS and concentrated three times in an anaerobic chamber and finally diluted to reach OD600 = 10, which is equivalent to 2 × 1010 CFU/mL in both WT and narG::tet strains.
Lactate quantification in mouse tissue
Lactate quantification in mouse tissues was performed using a colorimetric assay kit (abcam Cat# ab65331), as per the manufacturer’s instructions. For this, 10–20 mg of colonic tissue were homogenized in 1 ml of PBS, and 50 μl collected for lactate determination. Lactate values were normalized per mg of mouse tissue.
Lymphocyte isolation and flow cytometry
Epithelial cells were first stripped via mechanical disruption in RPMI (Gibco) containing 2 mM EDTA (Corning) and 1% fetal bovine serum (Gibco) for 45 min. Lamina propria lymphocytes were then obtained via subsequent mechanical disruption of remaining tissue in RPMI (Gibco) containing 0.5 mg/mL Collagenase VIII (Sigma) and 20% fetal bovine serum (Gibco) for 1 hr. The following conjugated antibodies were used to identify cellular markers: CD45 BUV805 (30-F11) (Bd Biosciences), Ly6G BV605 (1A8) (Biolegend), and Ly6C efluor450 (HK1.4) (Thermo-Fischer). All antibodies were used at a 1/400 dilution. Cells were analyzed on a Cytek Aurora cytometer (Cytek Biosciences). FlowJo 10.8.0. was used for analysis.
Cytokine assays
For ex vivo quantification of cytokine secretion, mesenteric lymph node cells were isolated in complete tissue culture media (RPMI, 10% FBS, 1% glutamine and 1% of 1:1 penicillin/streptomycin) and cultured in 96-well plates at 100,000 cells per well in 200 uL for 18 hours at 37°C, 5% carbon dioxide. The resulting supernatants were used to determine the amount of each cytokine using murine-specific cytokine bead array kit for mouse inflammatory cytokines (Biolegend), relative to a standard curve, according to the manufacturer’s recommendations.
Quantification of viable bacteria
Stool and colonic tissue collected from ASF mice colonized with Veillonella were weighed and homogenized in a buffer of 0.05% cysteine in PBS. Homogenates were serially diluted from 10−3 to 10−8 in sterile, deep-well 96-well plates. To assess colonization of Veillonella, resulting dilutions were plated on SKLN media with 5 μg/mL chloramphenicol for the WT strain and 10 μg/mL tetracycline for the narG::tet strain. Plates were incubated at 37°C under anaerobic conditions for 2 days. Colonies were counted and sent for 16S rRNA sequencing to confirm Veillonella was counted for each mouse. Colonies were considered below the limit of detection (nd) if no countable colonies were present at the 10−3 dilution factor.
Veillonella colonization assessment by qPCR
Stool from ASF and SPF mice colonized with Veillonella were collected and frozen immediately on dry ice. DNA was extracted using the PowerFecal Pro DNA extraction kit (Qiagen) following the manufacturer’s instructions. All extracted DNA was normalized to the same concentration between different samples and experimental conditions before being used. The assessment of Veillonella abundance was performed using the iTaq Universal SYBR Green Supermix (Bio-Rad) and primers listed in Supplementary Table 4. The 16S rRNA gene was used as a reference gene for abundance. PCR was performed on an Applied Biosystems 7500 machine; cycles consisted of 95 °C for 15 min and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Relative expression compared to 16S rRNA was calculated using the ΔΔC(t) method. In the competition experiment, relative expression was calculated by directly comparing Ct values of the chl and tet genes in the WT(chl) and narG::tet strains, respectively.
Statistical analysis
For each experiment, cell sampling and the number of independent replicates are indicated in figure legends, along with the corresponding statistical test used. Power analysis was not formally conducted, however sample sizes were chosen based on previous experience and published literature. Parametric distributions of the data were tested and statistical tests used appropriately depending on the presence of a normal distribution. Data were analyzed with either a Student’s t-test to compare two conditions with normal distributions, or a Mann-Whitney U-test for two conditions without normal distributions. For assessing statistical significance among three or more groups, a one-way ANOVA and Tukey’s honestly significant difference (HSD) test or Dunnett’s test was utilized as indicated. The n number reported in each figure legend refers to biological replicates. Data collection and analysis were not blinded due to the nature of the experimental designs in this study, but unbiased quantifications were applied to obtain the results. No data points were excluded from the analysis. Statistical analysis was performed with assistance from R versions 3.5 and 4.1 or GraphPad Prism version 9.0. Statistical significance is given as P values in the figures, where NS (not significant) indicates P ≥ 0.05. Results are expressed as the mean value with standard error of the mean (SEM), unless otherwise indicated
Extended Data
Extended Data Figure 1. Phylogenetic analysis of nitrate reductases.
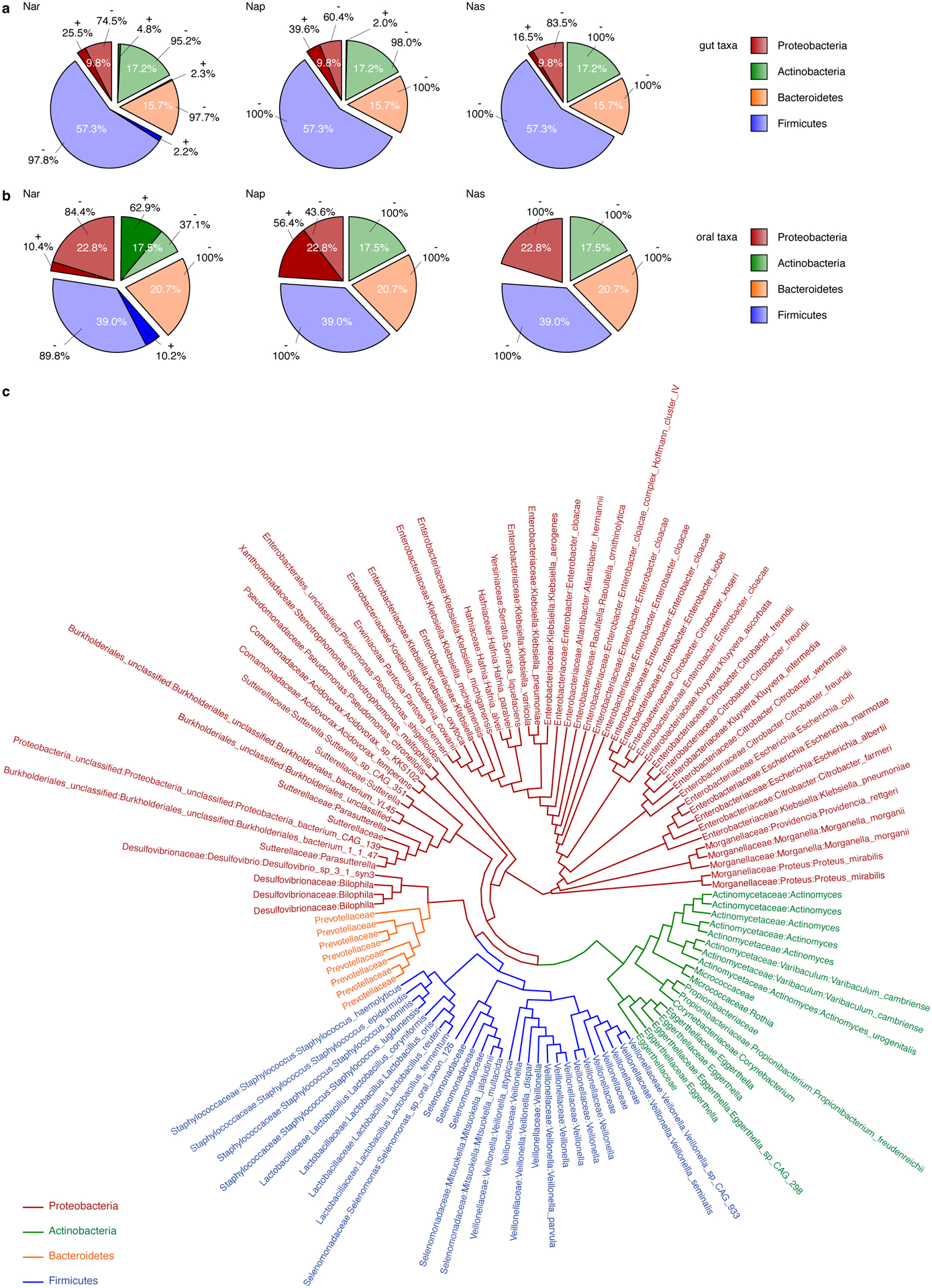
Analysis of the Nar, Nap, and Nas nitrate reductase families in human a) gut and b) oral microbes, using the NarG and NapA protein sequences of E. coli, and NasA of K. pneumoniae as templates. The sequences were searched in the assembled gene catalog from Pasolli et al. (2019)55. c) Phylogenetic tree of NarG in gut microbes reveals that Veillonella is one of the few genera within the Firmicutes with a putative nitrate reductase. Genes with at least 45% amino acid similarity were used for analysis.
Extended Data Figure 2. Nitrate-stimulated growth of various Veillonella strains, and narI::chl complementation experiments.
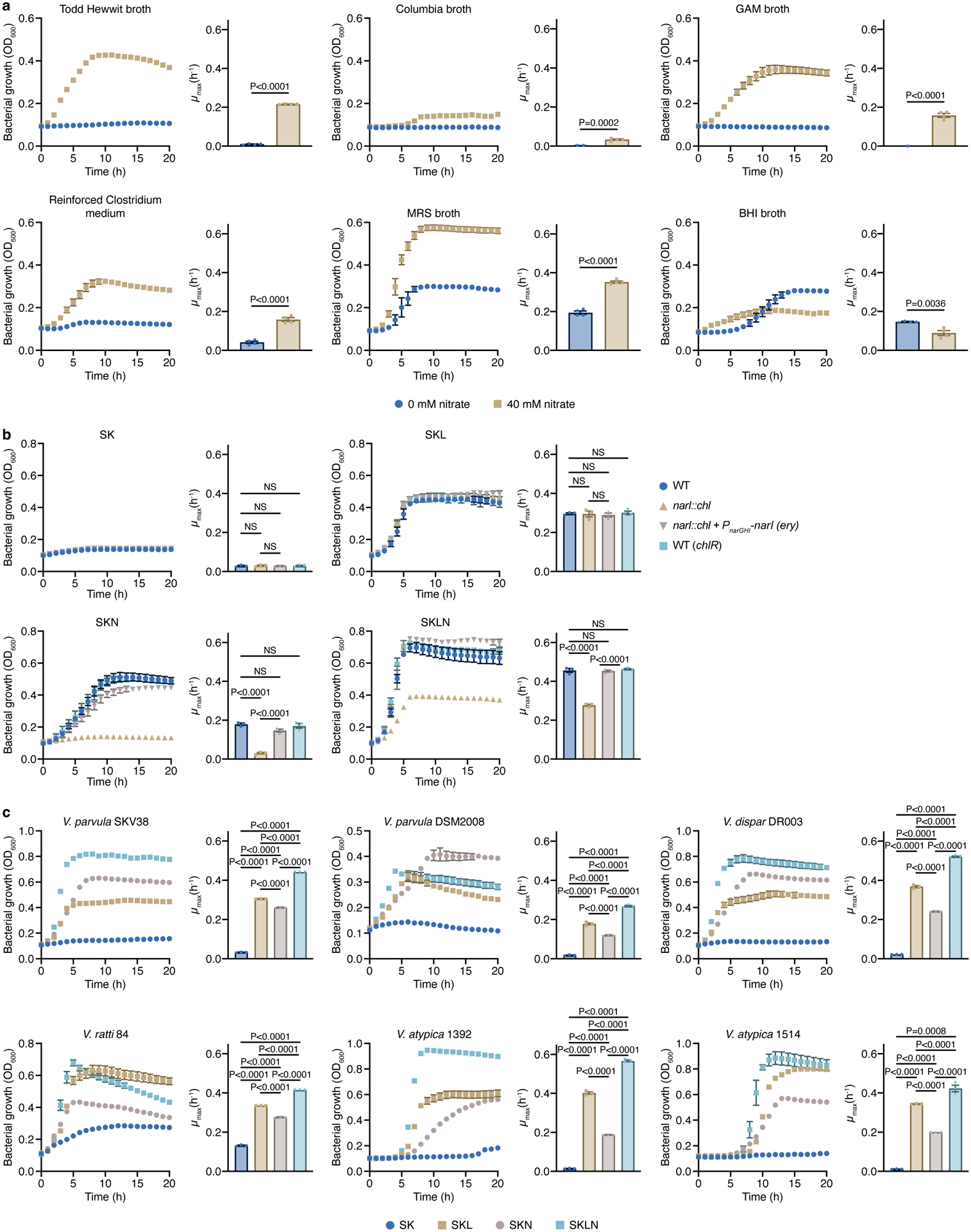
a) The growth of V. parvula SKV38 was assessed in different complex media supplemented or not with 40 mM nitrate (n=4 biological replicates). b) Growth curves of WT and narI::chl strains in SK, SKL, SKN, and SKLN media. Cells lacking the NarI protein are unable to grow in SKN, and nitrate does not stimulate its growth when lactate is present (n=3 biological replicates). c) The growth of various Veillonella strains was assessed in SK, SKL, SKN, and SKLN media (n=3 biological replicates). Growth rates (μmax) were calculated for each growth curve. Data (a-c) represent mean ± SEM, analyzed by one-way ANOVA and Tukey’s HSD test. NS, not significant.
Extended Data Figure 3. Nitrate metabolism in Veillonella.
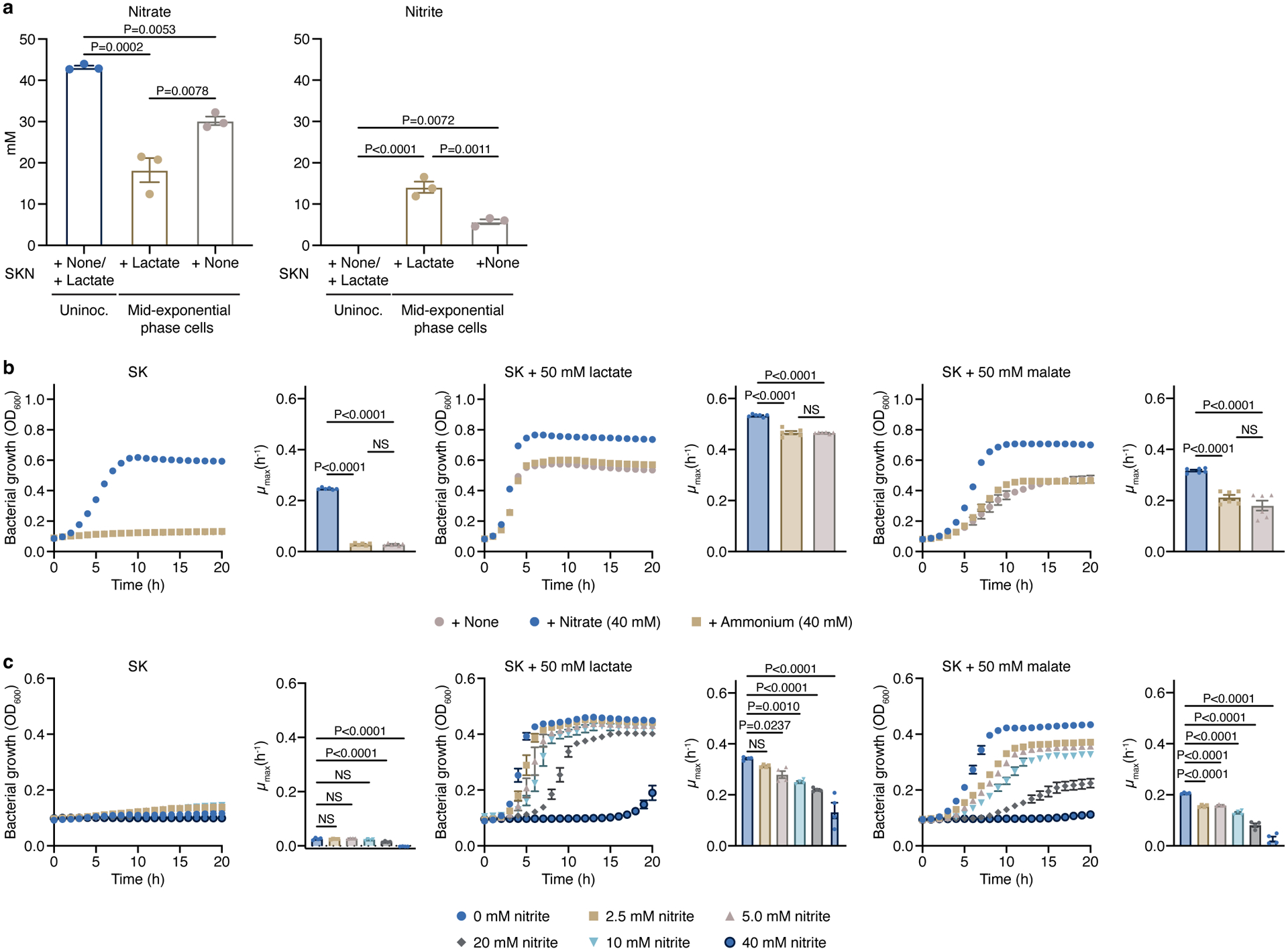
a) Levels of nitrate and nitrite were measured at the mid-exponential phase in cells growing on SKN medium supplemented or not with lactate as the carbon source (n=3 biological replicates). The measured concentrations of nitrate and nitrite in the uninoculated SK medium were 43.161 ± 0.410 mM and <0.050 mM, respectively (n=3 biological replicates). b) Ammonium (n=6 biological replicates) or c) nitrite (n=4 biological replicates) were used to determine if the positive effect of nitrate on growth of V. parvula was due to nitrate being used as a source of nitrogen or an electron acceptor, respectively. Growth rates (μmax) were calculated for each growth curve. Data represent mean ± SEM, analyzed by one-way ANOVA and Tukey’s HSD test (a,b) or Dunnett’s test (c). NS, not significant.
Extended Data Figure 4. Amino acids and peptides stimulate the growth of different Veillonella strains.
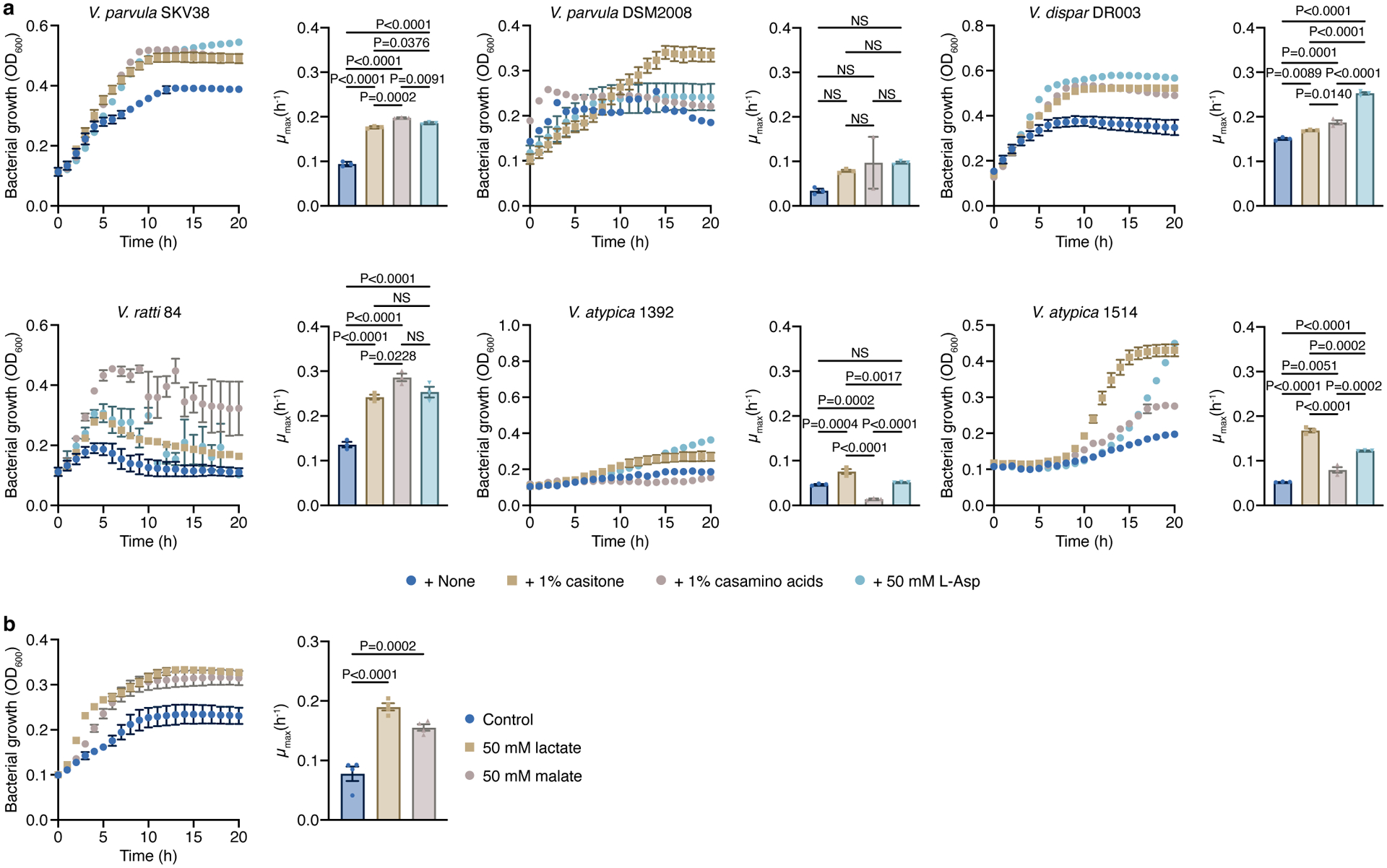
a) The growth of various Veillonella strains was studied in SKbN with 0.5% yeast extract, and with and without 1% casitone, 1% casamino acids, or 50 mM L-aspartic acid (n=3 biological replicates). b) The addition of lactate or malate stimulates growth of V. parvula in SKb medium (n=4 biological replicates). Growth rates (μmax) were calculated for each growth curve. Data represent mean ± SEM, analyzed by one-way ANOVA and Tukey’s HSD test (a) or Dunnett’s test (b). NS, not significant.
Extended Data Figure 5. LC-MS analysis of amino acids in unspent and spent media.
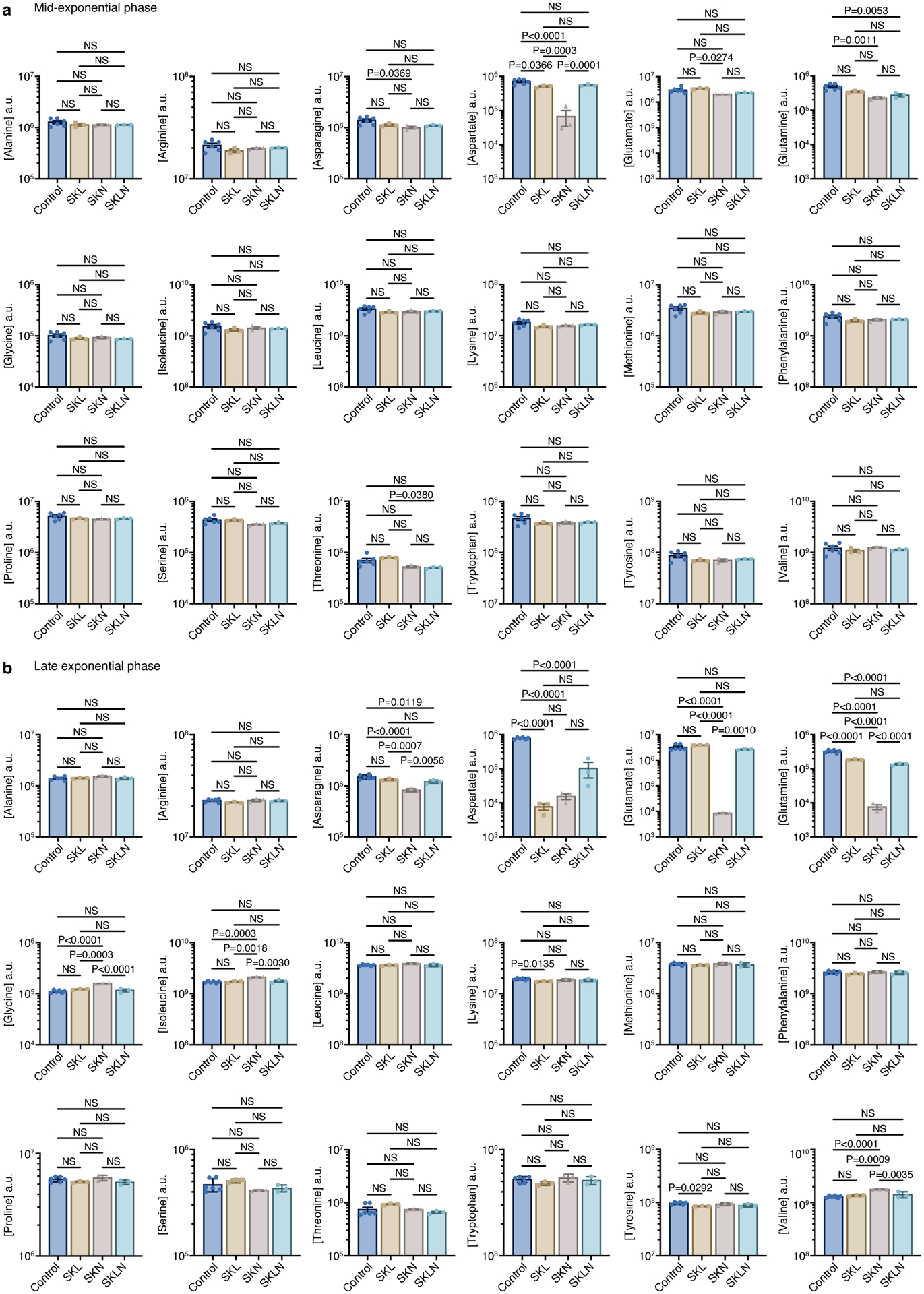
LC-MS analysis of the unspent (n=6 biological replicates) and spent SKL, SKN, and SKLN media (n=3 biological replicates) was used to study amino acid consumption. Supernatants were collected at the a) mid-exponential and b) late exponential phases. Data represent mean ± SEM, analyzed by one-way ANOVA and Tukey’s HSD test. NS, not significant.
Extended Data Figure 6. Role of the PMF in the production of ATP and transport, and phenotype of the L-lactate permease (lldP::chl) mutant.
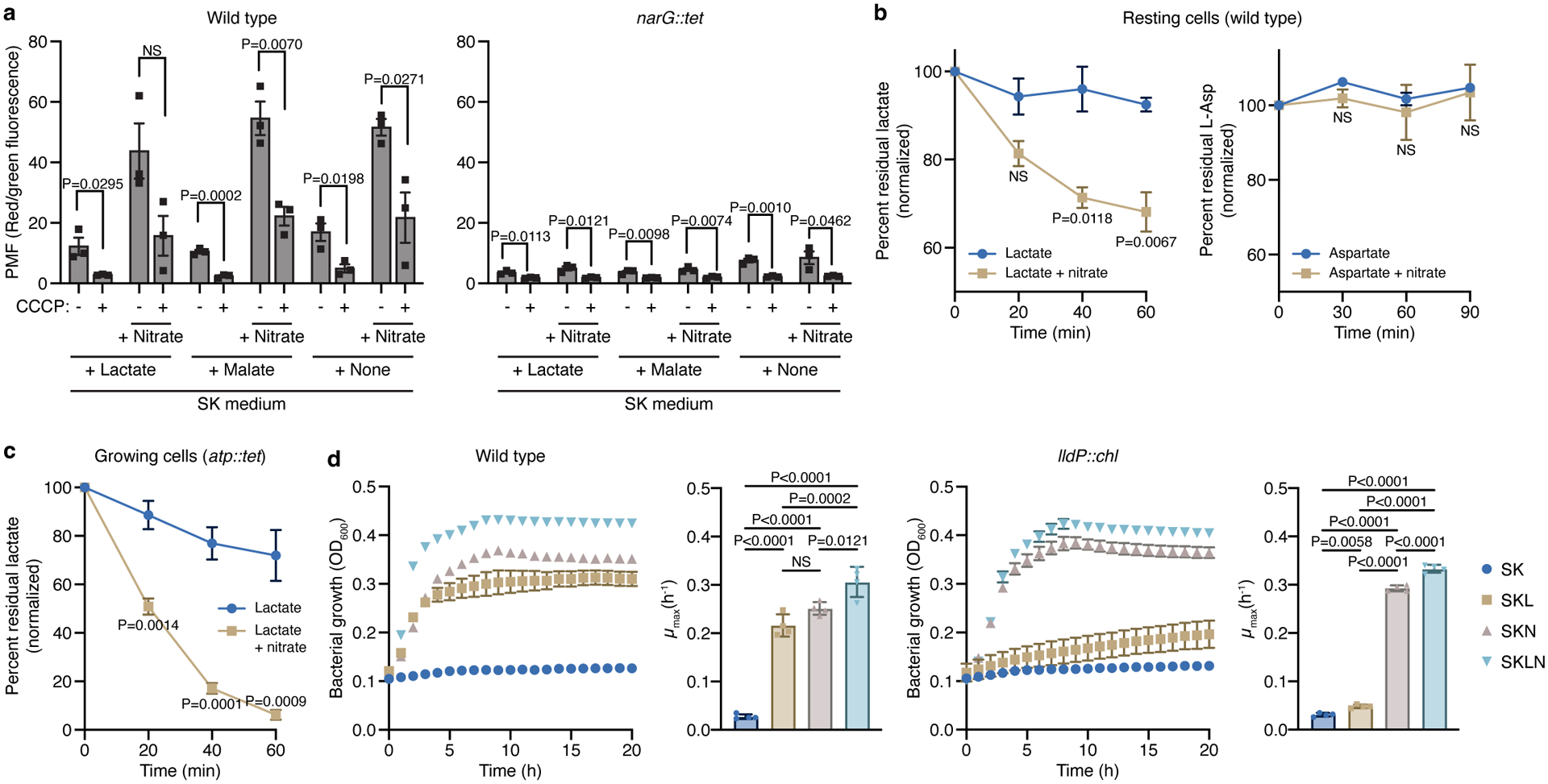
a) Measurement of the proton motive force (PMF) in WT and narG::tet cells growing on SK with and without lactate or malate, and with or without CCCP. The PMF was measured before and 40 min after addition of 40 mM nitrate. CCCP was added to confirm that the observed changes were associated with the proton gradient (n=3 biological replicates). b) Lactate (n=3 biological replicates) and aspartate (n=2 biological replicates) consumption were measured in resting WT cells resuspended in resting cell (RC) buffer with or without nitrate. c) Lactate consumption (n=4 biological replicates) was measured in growing atp::tet cells with or without nitrate. Both metabolites were measured in the supernatant using colorimetric techniques. d) Growth experiments with WT Veillonella and a strain lacking LldP (lldP::chl) in SK, SKL, SKN, and SKLN media (n=4 biological replicates). Cells lacking the LldP transporter are nearly unable to grow with lactate as the carbon source. Growth rates (μmax) were calculated for each growth curve. Data (a-d) represent mean ± SEM, analyzed by two-sided t-test (a, b, and c), or one-way ANOVA and Tukey’s HSD test (growth rates in d). NS, not significant.
Extended Data Figure 7. Phenotype of cells lacking components of the ATP synthase, and expression analysis of the atp operon.
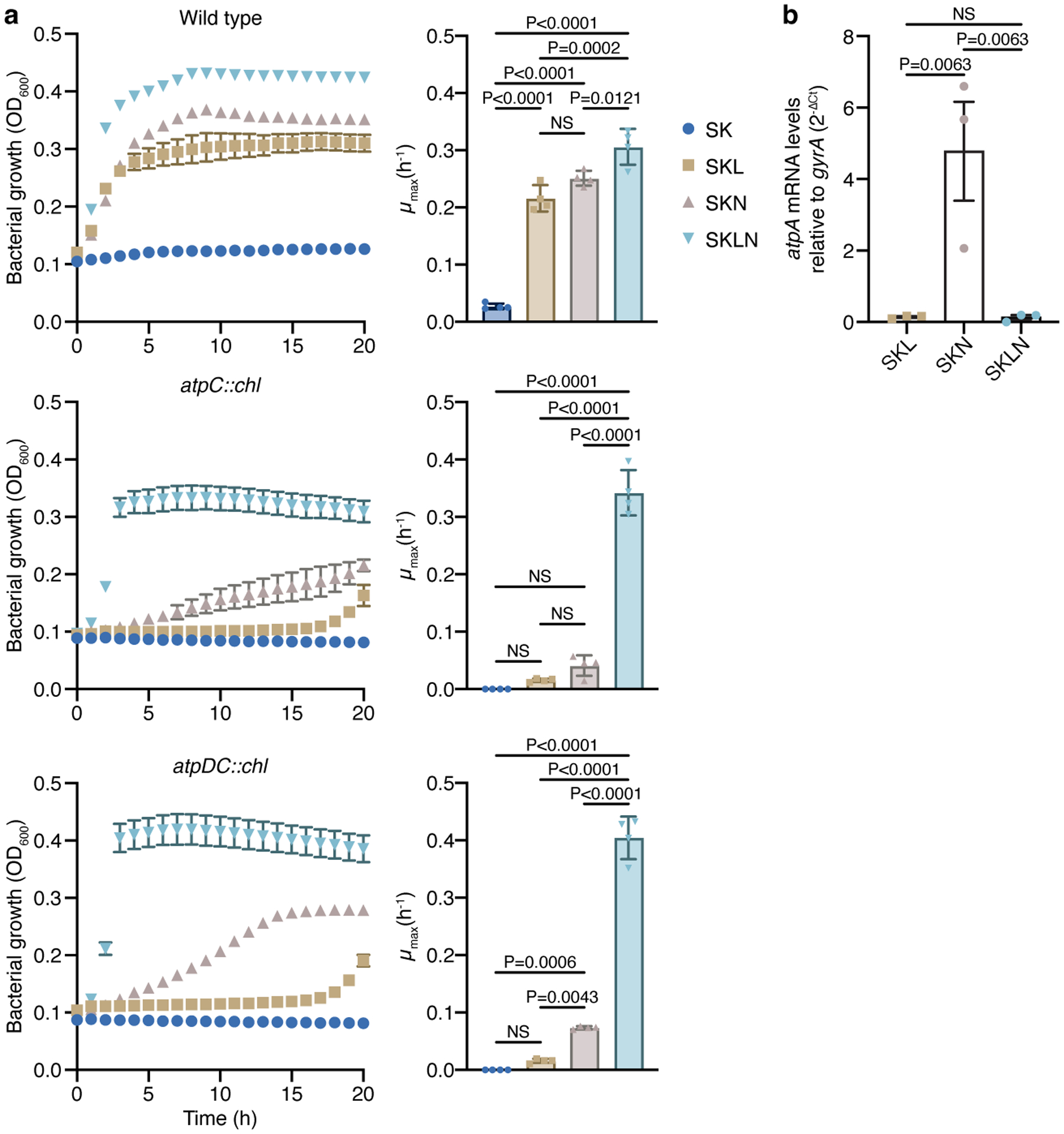
a) Growth experiments with WT Veillonella and the AtpC (atpC::chl) and AtpD-AtpC (atpDC::chl) deletion strains in SK, SKL, SKN, and SKLN media (n=4 biological replicates). These deletion strains exhibited a phenotype similar to the one seen in the atp::tet strain. Growth rates (μmax) were calculated for each growth curve. b) Transcriptional analysis of the atpA gene in cells growing in SKL, SKN, and SKLN media (n=3 biological replicates). Data represent mean ± SEM, analyzed by one-way ANOVA and Tukey’s HSD test. NS, not significant.
Extended Data Figure 8. Validation of the DSS-treated mouse model.
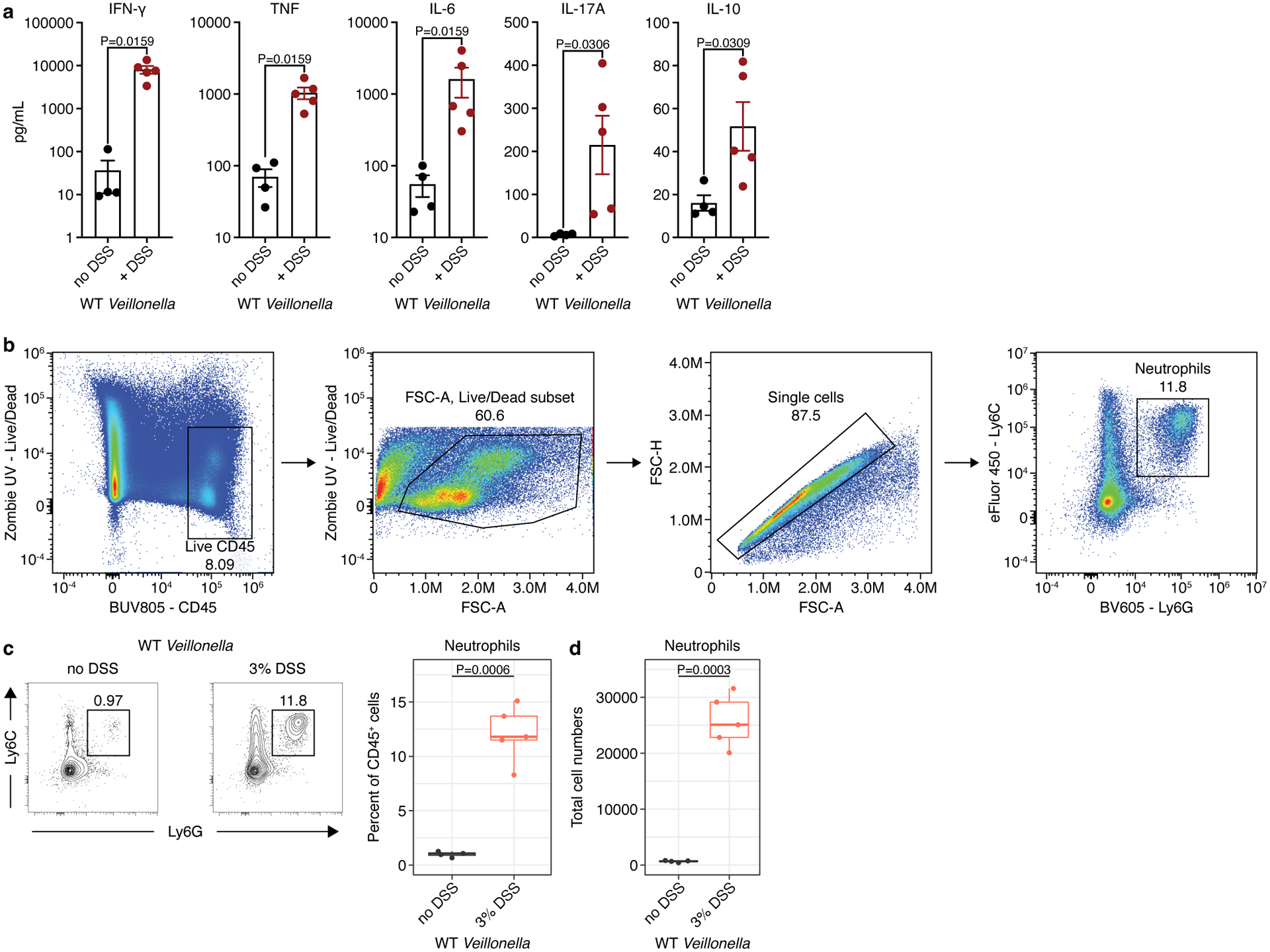
a) Measurements of IFN-γ, TNF, IL-6, IL-17A, and IL-10 in the mesenteric lymph nodes of WT-colonized ASF mice without (n=4) and with (n=5) DSS treatment. b) Lamina propria cells were isolated from colonic tissue and analyzed via flow cytometry. From left to right: Cells were first gated for positive expression of the pan immune cell marker CD45 while excluding dead cells staining for the viability dye Zombie UV. Next, gating was further cleaned by gating on FSC-A to exclude debris and FSC-A versus FSC-H to exclude doublets. Finally neutrophils were gated based on double positive expression of the two surface protein markers Ly6G and Ly6C. c) Left: representative contour plots showing the proportion of neutrophils based on positive expression of Ly6C and Ly6G on live CD45+ lymphocytes isolated from mouse colonic lamina propria. Right: Frequency of neutrophils among total CD45+ colonic lamina propria lymphocytes. d) Total number of neutrophils isolated from mouse colons based on gating in c. The data in a, c, and d are representative of two independent experiments. For c and d, boxplots display the first and third quartiles with a thick line representing the median; n=4 mice without DSS, and n=5 mice with DSS treatment. A Student’s t-test was used in a, c, and d for statistical analyses.
Extended Data Figure 9. Colonization of Veillonella in a mouse model of colitis.
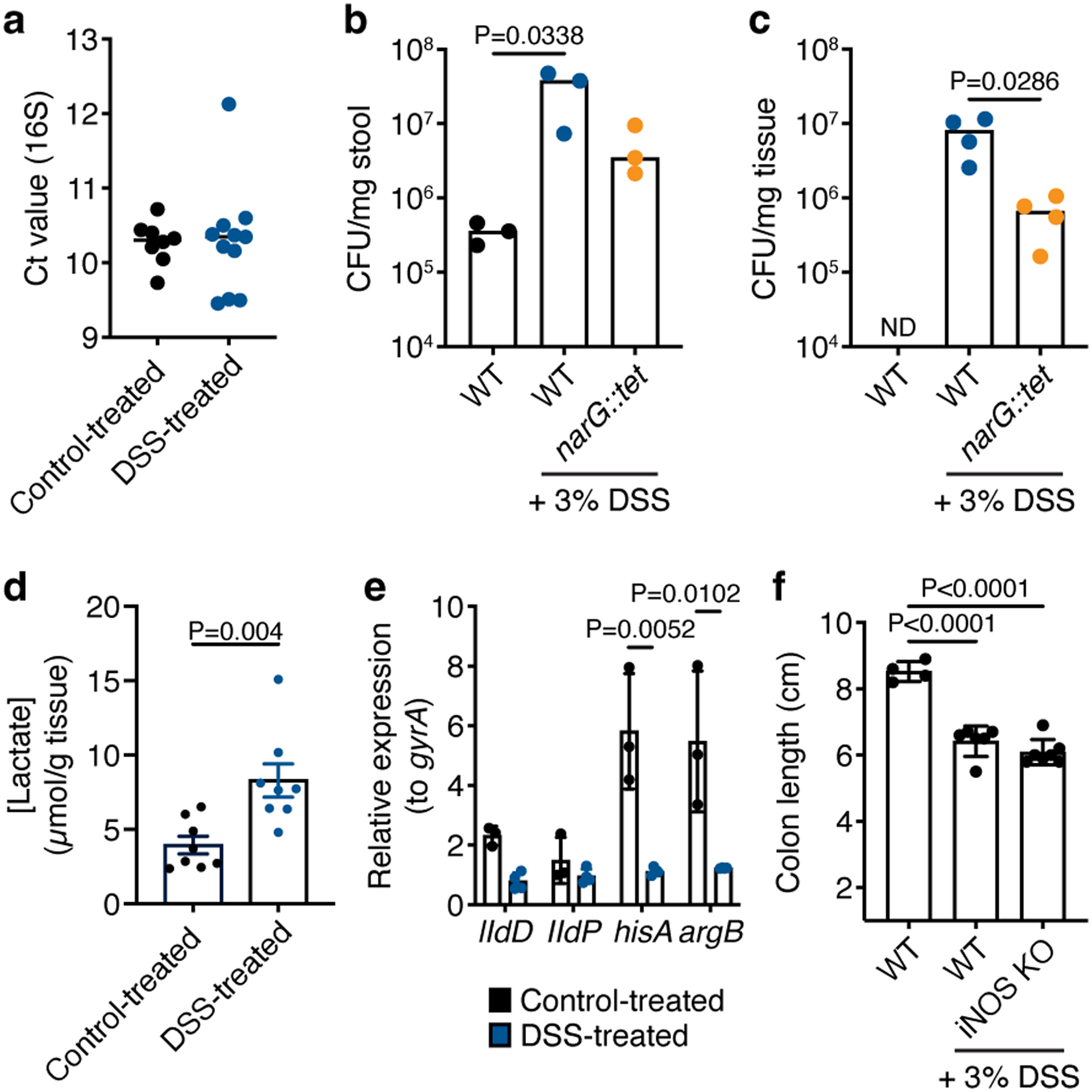
a) Raw Ct values of the Veillonella 16S rRNA gene measured by qPCR in the stool of WT-colonized ASF mice with and without DSS treatment (n=8–11 mice). b) Absolute abundance of Veillonella in the stool of ASF mice as measured by CFU per mg of stool on selective SKLN media (n=3 mice). c) Absolute abundance of Veillonella in the colonic tissue of ASF mice as measured by CFU per mg of tissue on selective SKLN media (n=4 mice). d) Concentration of lactate in the gut of control- and DSS-treated ASF mice colonized with WT Veillonella (n=8 mice). e) Relative expression of the IIdD, lldP, hisA and argB genes in WT Veillonella colonizing the stool of mice with and without DSS treatment (n=3–4). The gyrA gene was used as an internal control. f) Colon lengths of WT and iNOS KO SPF mice (n=4–7). Data represent two independent experiments and show mean (a, d-f) ± SEM (d-f) or median (b,c) values. Data were analyzed by one-way ANOVA and Tukey’s HSD test or Mann-Whitney U-test.
Supplementary Material
Supplementary Data File 2. LC-MS analysis of unspent and spent SKL, SKN, and SKLN media at the late exponential phase used to study dipeptide consumption
Supplementary Data File 1. LC-MS data
Supplementary Table 2. Resistance genes used for Veillonella parvula mutants
Supplementary Table 3. Primers for mutants
Supplementary Table 4. Primers for qPCR
ACKNOWLEDGMENTS
We deeply thank Justin L. Merritt at Oregon Health & Science University for the SKV38 strain of Veillonella parvula and the pBSLJ1 plasmid, as well as José Vargas Asencio at MIT for helpful discussions, and Theresa Reimels for editorial assistance. RNA-sequencing libraries were constructed and sequenced at the Broad Institute of MIT and Harvard by the Microbial ‘Omics Core and Genomics Platform, respectively. The Microbial ‘Omics Core also provided preliminary analysis for all RNA-sequencing data. We thank the gnotobiotic animal facility at the Broad Institute, specifically Carlos Umana and Allan Discua for helping with experiments and Tyler Caron for providing support for running the facility. This work was supported by grants from the National Institutes of Health (P30 DK043351), Center for Microbiome Informatics and Therapeutics, and Crohn’s and Colitis Foundation to R.J.X and from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, 426120468) to M.S.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
DATA AVAILABILITY
RNA-sequencing data is available on the Sequence Read Archive (SRA) under BioProject ID PRJNA838968. Source data are provided with this paper. Human microbiome data was accessed through data deposited on the SRA under BioProject ID PRJNA436359.
CODE AVAILABILITY
No custom code was generated or used in this study.
REFERENCES
- 1.Lamont RJ, Koo H & Hajishengallis G The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol 16, 745–759, doi: 10.1038/s41579-018-0089-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dewhirst FE et al. The human oral microbiome. J Bacteriol 192, 5002–5017, doi: 10.1128/JB.00542-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitamoto S et al. The Intermucosal Connection between the Mouth and Gut in Commensal Pathobiont-Driven Colitis. Cell 182, 447–462 e414, doi: 10.1016/j.cell.2020.05.048 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atarashi K et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 358, 359–365, doi: 10.1126/science.aan4526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gevers D et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392, doi: 10.1016/j.chom.2014.02.005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaw KA et al. Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med 8, 75, doi: 10.1186/s13073-016-0331-y (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ananthakrishnan AN et al. Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host Microbe 21, 603–610 e603, doi: 10.1016/j.chom.2017.04.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santoru ML et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep 7, 9523, doi: 10.1038/s41598-017-10034-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geng J et al. Co-occurrence of driver and passenger bacteria in human colorectal cancer. Gut Pathog 6, 26, doi: 10.1186/1757-4749-6-26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kostic AD et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215, doi: 10.1016/j.chom.2013.07.007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flemer B et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut 67, 1454–1463, doi: 10.1136/gutjnl-2017-314814 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tunney MM et al. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am J Respir Crit Care Med 177, 995–1001, doi: 10.1164/rccm.200708-1151OC (2008). [DOI] [PubMed] [Google Scholar]
- 13.Rogers GB et al. characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16s ribosomal DNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol 42, 5176–5183, doi: 10.1128/JCM.42.11.5176-5183.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers GB, Skelton S, Serisier DJ, van der Gast CJ & Bruce KD Determining cystic fibrosis-affected lung microbiology: comparison of spontaneous and serially induced sputum samples by use of terminal restriction fragment length polymorphism profiling. J Clin Microbiol 48, 78–86, doi: 10.1128/JCM.01324-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seedorf H et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell 159, 253–266, doi: 10.1016/j.cell.2014.09.008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hove H & Mortensen PB Influence of intestinal inflammation (IBD) and small and large bowel length on fecal short-chain fatty acids and lactate. Dig Dis Sci 40, 1372–1380, doi: 10.1007/BF02065554 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Franzosa EA et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 4, 293–305, doi: 10.1038/s41564-018-0306-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schicho R et al. Quantitative metabolomic profiling of serum, plasma, and urine by (1)H NMR spectroscopy discriminates between patients with inflammatory bowel disease and healthy individuals. J Proteome Res 11, 3344–3357, doi: 10.1021/pr300139q (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winter SE et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711, doi: 10.1126/science.1232467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes ER et al. Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host Microbe 21, 208–219, doi: 10.1016/j.chom.2017.01.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winter SE et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429, doi: 10.1038/nature09415 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillis CC et al. Dysbiosis-Associated Change in Host Metabolism Generates Lactate to Support Salmonella Growth. Cell Host Microbe 23, 54–64 e56, doi: 10.1016/j.chom.2017.11.006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thiennimitr P et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108, 17480–17485, doi: 10.1073/pnas.1107857108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez CA, Rivera-Chavez F, Byndloss MX & Baumler AJ The Periplasmic Nitrate Reductase NapABC Supports Luminal Growth of Salmonella enterica Serovar Typhimurium during Colitis. Infect Immun 83, 3470–3478, doi: 10.1128/IAI.00351-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez CA et al. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. mBio 3, doi: 10.1128/mBio.00143-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes CV, Kolenbrander PE, Andersen RN & Moore LV Coaggregation properties of human oral Veillonella spp.: relationship to colonization site and oral ecology. Appl Environ Microbiol 54, 1957–1963, doi: 10.1128/aem.54.8.1957-1963.1988 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mashima I & Nakazawa F The interaction between Streptococcus spp. and Veillonella tobetsuensis in the early stages of oral biofilm formation. J Bacteriol 197, 2104–2111, doi: 10.1128/JB.02512-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Hoeven JS, Toorop AI & Mikx RH Symbiotic relationship of Veillonella alcalescens and Streptococcus mutans in dental plaque in gnotobiotic rats. Caries Res 12, 142–147, doi: 10.1159/000260324 (1978). [DOI] [PubMed] [Google Scholar]
- 29.Mikx FH & Van der Hoeven JS Symbiosis of Streptococcus mutans and Veillonella alcalescens in mixed continuous cultures. Arch Oral Biol 20, 407–410, doi: 10.1016/0003-9969(75)90224-1 (1975). [DOI] [PubMed] [Google Scholar]
- 30.Periasamy S & Kolenbrander PE Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J Bacteriol 192, 2965–2972, doi: 10.1128/JB.01631-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delwiche EA, Pestka JJ & Tortorello ML The veillonellae: gram-negative cocci with a unique physiology. Annu Rev Microbiol 39, 175–193, doi: 10.1146/annurev.mi.39.100185.001135 (1985). [DOI] [PubMed] [Google Scholar]
- 32.Schirmer M et al. Compositional and Temporal Changes in the Gut Microbiome of Pediatric Ulcerative Colitis Patients Are Linked to Disease Course. Cell Host Microbe 24, 600–610 e604, doi: 10.1016/j.chom.2018.09.009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kugathasan S et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet 389, 1710–1718, doi: 10.1016/S0140-6736(17)30317-3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogosa M The Genus Veillonella. I. General Cultural, Ecological, and Biochemical Considerations. J Bacteriol 87, 162–170, doi: 10.1128/jb.87.1.162-170.1964 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doel JJ, Benjamin N, Hector MP, Rogers M & Allaker RP Evaluation of bacterial nitrate reduction in the human oral cavity. Eur J Oral Sci 113, 14–19, doi: 10.1111/j.1600-0722.2004.00184.x (2005). [DOI] [PubMed] [Google Scholar]
- 36.Inderlied CB & Delwiche EA Nitrate reduction and the growth of Veillonella alcalescens. J Bacteriol 114, 1206–1212, doi: 10.1128/jb.114.3.1206-1212.1973 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitsui T, Saito M & Harasawa R Salivary nitrate-nitrite conversion capacity after nitrate ingestion and incidence of Veillonella spp. in elderly individuals. J Oral Sci 60, 405–410, doi: 10.2334/josnusd.17-0337 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Wicaksono DP, Washio J, Abiko Y, Domon H & Takahashi N Nitrite Production from Nitrate and Its Link with Lactate Metabolism in Oral Veillonella spp. Appl Environ Microbiol 86, doi: 10.1128/AEM.01255-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreno-Vivian C, Cabello P, Martinez-Luque M, Blasco R & Castillo F Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J Bacteriol 181, 6573–6584, doi: 10.1128/JB.181.21.6573-6584.1999 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knapp S et al. Natural Competence Is Common among Clinical Isolates of Veillonella parvula and Is Useful for Genetic Manipulation of This Key Member of the Oral Microbiome. Front Cell Infect Microbiol 7, 139, doi: 10.3389/fcimb.2017.00139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reuss O & Morschhauser J A family of oligopeptide transporters is required for growth of Candida albicans on proteins. Mol Microbiol 60, 795–812, doi: 10.1111/j.1365-2958.2006.05136.x (2006). [DOI] [PubMed] [Google Scholar]
- 42.Berntsson RP et al. The structural basis for peptide selection by the transport receptor OppA. EMBO J 28, 1332–1340, doi: 10.1038/emboj.2009.65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Vries W, Rietveld-Struijk RM & Stouthamer AH ATP formation associated with fumarate and nitrate reduction in growing cultures of Veillonella alcalescens. Antonie Van Leeuwenhoek 43, 153–167, doi: 10.1007/BF00395670 (1977). [DOI] [PubMed] [Google Scholar]
- 44.Galivan JH & Allen SH Methylmalonyl coenzyme A decarboxylase. Its role in succinate decarboxylation by Micrococcus lactilyticus. J Biol Chem 243, 1253–1261 (1968). [PubMed] [Google Scholar]
- 45.de Vries W, van Wijck-Kapteyn WM & Oosterhuis SK The presence and function of cytochromes in Selenomonas ruminantium, Anaerovibrio lipolytica and Veillonella alcalescens. J Gen Microbiol 81, 69–78, doi: 10.1099/00221287-81-1-69 (1974). [DOI] [PubMed] [Google Scholar]
- 46.McCormick NG, Ordal EJ & Whiteley HR DEGRADATION OF PYRUVATE BY MICROCOCCUS LACTILYTICUS I. : General Properties of the Formate-Exchange Reaction. J Bacteriol 83, 887–898, doi: 10.1128/jb.83.4.887-898.1962 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Griffith MJ & Nishimura JS Acetate kinase from Veillonella alcalescens. Purification and physical properties. J Biol Chem 254, 442–446 (1979). [PubMed] [Google Scholar]
- 48.Pelroy RA & Whiteley HR Regulatory properties of acetokinase from Veillonella alcalescens. J Bacteriol 105, 259–267, doi: 10.1128/jb.105.1.259-267.1971 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bowman CM, Valdez RO & Nishimura JS Acetate kinase from Veillonella alcalescens. Regulation of enzyme activity by succinate and substrates. J Biol Chem 251, 3117–3121 (1976). [PubMed] [Google Scholar]
- 50.Kolios G, Valatas V & Ward SG Nitric oxide in inflammatory bowel disease: a universal messenger in an unsolved puzzle. Immunology 113, 427–437, doi: 10.1111/j.1365-2567.2004.01984.x (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson CJ et al. Microbes exploit death-induced nutrient release by gut epithelial cells. Nature 596, 262–267, doi: 10.1038/s41586-021-03785-9 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Madej M et al. Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis. Nat Microbiol 5, 1016–1025, doi: 10.1038/s41564-020-0716-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pucino V et al. Lactate Buildup at the Site of Chronic Inflammation Promotes Disease by Inducing CD4(+) T Cell Metabolic Rewiring. Cell Metab 30, 1055–1074 e1058, doi: 10.1016/j.cmet.2019.10.004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dalgaard P & Koutsoumanis K Comparison of maximum specific growth rates and lag times estimated from absorbance and viable count data by different mathematical models. J Microbiol Methods 43, 183–196, doi: 10.1016/s0167-7012(00)00219-0 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Pasolli E et al. Extensive Unexplored Human Microbiome Diversity Revealed by Over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell 176, 649–662 e620, doi: 10.1016/j.cell.2019.01.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar RC Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461, doi: 10.1093/bioinformatics/btq461 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Segata N, Bornigen D, Morgan XC & Huttenhower C PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun 4, 2304, doi: 10.1038/ncomms3304 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shishkin AA et al. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat Methods 12, 323–325, doi: 10.1038/nmeth.3313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu YY, Machleder EM, Chenchik A, Li R & Siebert PD Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques 30, 892–897, doi: 10.2144/01304pf02 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760, doi: 10.1093/bioinformatics/btp324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, doi: 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abeel T, Van Parys T, Saeys Y, Galagan J & Van de Peer Y GenomeView: a next-generation genome browser. Nucleic Acids Res 40, e12, doi: 10.1093/nar/gkr995 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mascanfroni ID et al. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-alpha. Nat Med 21, 638–646, doi: 10.1038/nm.3868 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ng SK & Hamilton IR Lactate metabolism by Veillonella parvula. J Bacteriol 105, 999–1005, doi: 10.1128/jb.105.3.999-1005.1971 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data File 2. LC-MS analysis of unspent and spent SKL, SKN, and SKLN media at the late exponential phase used to study dipeptide consumption
Supplementary Data File 1. LC-MS data
Supplementary Table 2. Resistance genes used for Veillonella parvula mutants
Supplementary Table 3. Primers for mutants
Supplementary Table 4. Primers for qPCR
Data Availability Statement
RNA-sequencing data is available on the Sequence Read Archive (SRA) under BioProject ID PRJNA838968. Source data are provided with this paper. Human microbiome data was accessed through data deposited on the SRA under BioProject ID PRJNA436359.


