Abstract
Endoderm gives rise to the gut tube in the early embryo. We differentiated human induced pluripotent stem cells (hiPSCs) to embryonic endoderm to identify a “tipping point” at which the developing system did not recover from perturbations caused by exposure to all-trans retinoic acid (ATRA). Differentiating hiPSC-derived endoderm exposed to five concentrations of ATRA between 0.001 and 10 μM at 6 h, 96 h, or 192 h was assessed for forkhead box A2 (FOXA2) protein expression and global gene transcript expression. A tipping point of 17 ± 11 nM was identified where patterns of differentially expressed genes supported a developmental trajectory shift indicating a potential teratogenic outcome. This concentration is between women’s endogenous ATRA blood plasma levels and teratogenic levels of circulating isotretinoin, an ATRA isomer used to treat acne. Taken together, these data suggest that this approach is a sensitive method to identify a point of departure for chemicals that impact early embryonic development.
Keywords: All-trans retinoic acid, Endoderm, FOXA2, Tipping point, Differentiation, iPSC, SOX17, EOMES
1. Introduction
The ToxCast™ high throughput screening (HTS) program [1–4] is a powerful approach to identify molecular targets of environmental contaminants. By applying systems biology modeling approaches to the HTS data, effects on cells, tissues, and organ systems can be deduced. Although modeling approaches allow extrapolation from adult cell lines to developmental processes, pathways involved in embryonic development are not directly represented in ToxCast™. To address this, we investigated the suitability of a differentiation assay with human induced pluripotent stem cells (hiPSCs) for modeling tissue differentiation during human embryonic development (Fig. 1). Endogenesis, the formation of endoderm, was selected due to the role of endoderm in giving rise to visceral organs derived from the gut tube including thyroid, thymus, lung, pancreas, liver, gastrointestinal system, prostate, and the visceral yolk sac. The critical role of endogenesis in proper embryonic development establishes this system as a model to screen and identify chemicals that may disrupt embryonic patterning leading to birth defects.
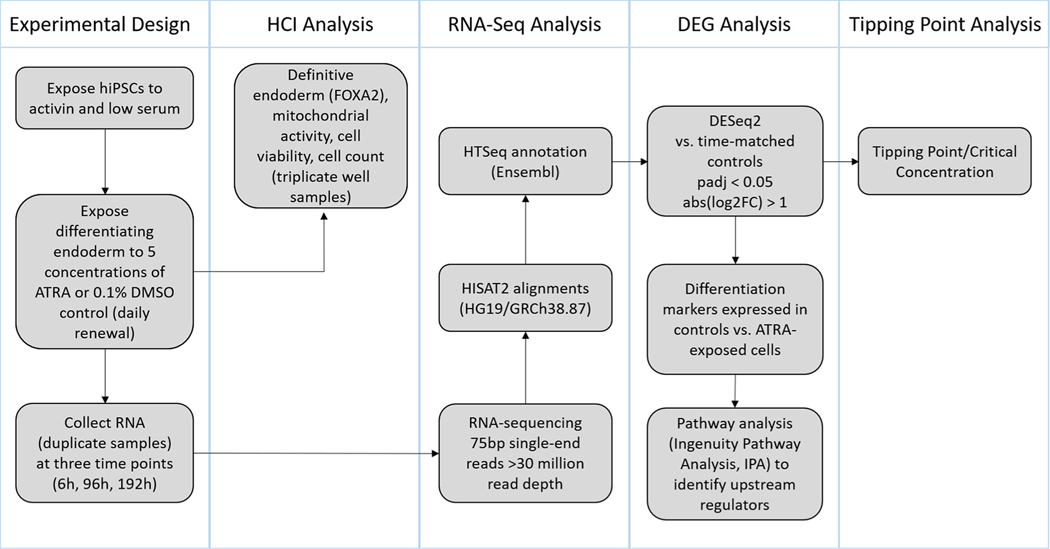
Fig. 1.
Study overview: Human induced pluripotent stem cells (hiPSCs) were exposed to a range of biologically relevant ATRA concentrations followed by high content imaging and RNA-sequencing analyses to determine a toxicological tipping point and characterize the effects of ATRA on differentiating endoderm.
Endogenesis occurs during the primitive streak stage of gastrulation when epiblast-derived ectodermal cells migrate ventrally to invade the hypoblast-derived extraembryonic (yolk sac) endoderm while transitioning to an embryonic endodermal state via the process of epithelial to mesenchymal transition (EMT) [5,6]. These events mark the establishment of the three germ layers at the earliest stage of embryogenesis in week three of human development (gestational age week 5) [7]. The anterior primitive streak gives rise to embryonic endoderm [5], in turn leading to formation of the gut tube, comprising foregut, midgut, and hindgut. The foregut is the source of anterior organs: thyroid, thymus, lungs, stomach, liver, and pancreas; the midgut becomes the small intestine; the hindgut becomes the large intestine. In addition to the gut tube-derived organs, the anterior primitive streak also gives rise to extra-embryonic endoderm, also known as yolk sac or visceral endoderm, to be incorporated into the gut epithelium [8]. The posterior primitive streak gives rise to mesoderm also through an EMT [9].
The establishment of the three germ layers from the primitive streak begins with ectoderm, which is the first layer to form and often referred to as the default cell fate from which mesoderm or endoderm (mesendoderm) is induced [10,11]. There are several well-established molecular signaling events that facilitate germ layer specification. Fibroblast growth factor (FGF) expression favors ectoderm differentiation while inhibiting endoderm formation [12]; while Wnt, bone morphogenetic protein 4 (BMP4), and Nodal/activin suppress ectodermal specification in favor of mesendoderm formation [12,13] during EMT [14]. The Nodal signaling pathway is essential for mesendoderm formation [15,16]. Nodal belongs to a subclass of transforming growth factor-beta (TGFβ) signaling molecules specific to chordates [17]. Nodals target activin (a TGFβ ligand) receptors during mesendoderm specification [17] to transduce signals from the plasma membrane to the nucleus through SMAD protein complexes, which interact with transcription factors to regulate Nodal-responsive target genes [18]. The level of Nodal/activin determines endoderm versus mesoderm fate, with higher levels driving endodermal specification. Conversely, BMP4 expression favors mesoderm formation, while suppressing endoderm formation [19]. While the signaling molecules driving germ layer formation were first elucidated using classical development models (i.e., Xenopus, zebrafish, and chick embryos) and subsequently validated using mammalian models, additional insights into the molecular mechanisms of mammalian embryonic development have been elucidated using rodent and human embryonic stem cells (ESCs) [20–22].
Kubo et al. (2004) were one of the first groups to differentiate definitive (embryonic) endoderm from mouse ESCs by adding activin to the cell culture medium [21]. D’Amour et al. (2005) were the first group to derive endoderm from human ESCs using activin and low serum and reported 80% endoderm based on expression of SRY-box 17 (SOX17) [20]. They subsequently obtained 90% definitive endoderm population by filtering cells by CXC motif chemokine receptor 4 (CXCR4) cell surface protein expression using fluorescence-activated cell sorting (FACS). To confirm definitive endoderm specification, D’Amour et al. (2005) compiled a comprehensive set of endodermal marker transcripts, including SOX17, CXCR4, and forkhead box A2 (FOXA2). They noted that FOXA2 is also a protein expressed in axial mesoderm, but co-expression of FOXA2 and SOX17 indicates definitive endoderm. They also identified primitive streak markers: brachyury (T; TBXT), WNT3, FGF4, and the EMT marker snail (SNAI1), which represses E-cadherin (CDH1). Mesoderm markers included forkhead box F1 (FOXF1), mesenchyme homeobox 1 (MEOX1), vascular endothelial growth factor receptor 2 (KDR), BMP4, and CXC motif chemokine ligand 12 (CXCL12). Ectoderm markers included SRY-box 1 (SOX1) and Zinc finger of the cerebellum 1 (ZIC1). Many of these markers were also used by Loh et al. (2014), who built upon the D’Amour et al. and other protocols to differentiate endoderm from human ESCs (hESCs), as well as hiPSCs [22]. The hESC experiments confirmed classical developmental studies that reported Wnt, FGF, Nodal, BMP4, and TGFβ as critical components for endodermal specification [22]. The extensive background work by these two groups [20,22] and other sources [23–28] was used to construct a list of developmental markers for the present study that was condensed to 97 key markers representing the developmental stages from primitive streak; through endoderm, mesoderm, and ectoderm; to the beginning of rudimentary tissue formation (Supplemental Table 1).
In addition to establishing an in vitro system representative of early embryonic development, a second objective of the current study was to identify a perturbation in the system following exposure to a known teratogen expected to disrupt endogenesis. This work builds upon a study described by Kameoka et al. (2014) demonstrating that ATRA perturbs SOX17+ embryonic endoderm differentiation from human pluripotent stem cells (hPSCs) [29]. Moreover, Kameoka et al. used hPSCs to screen chemicals for teratogenic potential. We build on this work by moving beyond half maximal inhibitory concentrations (IC50s) based on reduced SOX17 expression to calculate a transcriptome-wide “tipping point”. Toxicological tipping points represent the chemical concentration where biological systems (e.g., tissues, organs, or cultured cells) are perturbed beyond the point of recovery [30]. In the present study, they were calculated from time series transcriptomic data that include at least three time points and multiple treatment concentrations to capture ‘perturbation’ (i.e., deviation from a default differentiation trajectory) and ‘recovery’ (i.e., return to the default differentiation trajectory). Because endogenous retinoic acid (RA) is critical for embryogenesis [31,32], and exogenous RA exposure causes birth defects in organs and structures derived from all germ layers [33–35], we selected all-trans retinoic acid (ATRA) to identify the point at which differentiating endoderm is perturbed beyond the ability to recover (i.e., the tipping point). Here, perturbation represents deviation from the default endodermal differentiation trajectory. Since cultured embryonic stem cells are inducible to ectoderm by adding RA [36], which has been described as a posteriorizing agent [35,37,38], we hypothesized that ATRA would perturb endodermal differentiation in favor of alternative cell fates such as ectoderm or mesoderm. To investigate this hypothesis, a range of ATRA concentrations was used to evaluate the gene biomarker set across multiple timepoints to perform tipping point analysis of endoderm differentiation in an in vitro hiPSC model.
2. Materials and methods
2.1. Cell culture and chemical exposure
Cell culture and exposures were conducted by Vala Sciences, Inc. (San Diego, CA). An induced pluripotent stem cell line (catalog#: ABP-SC-HDFAIPS; Allele Biotechnology and Pharmaceuticals, Inc.; San Diego, CA) derived from adult human female fibroblasts using mRNA reprogramming was maintained according to the manufacturer’s protocol. Cells were cultured and passaged in a flask coated with growth factor reduced Matrigel (Corning, Inc.; Corning, NY) and mTeSR (STEMCELL Technologies, Inc., Cambridge, MA) was used for feeder-free cell maintenance. Cells were seeded with ROCK inhibitor on Matrigel (1:20 dilution) coated 384-well uClear Plates (Catalog #781091; Greiner Bio-One; Monroe, NC) at 1.5k cells/well and medium was changed (without ROCK inhibitor) at 24 h post seeding. At 48 h post seeding, differentiation was initiated by switching the cell culture medium from mTeSR to RPMI-B27 differentiation medium supplemented with 100 ng/ml activin. (Note: These are defined media that do not contain RA or Vitamin A; thus, endogenous production of RA is not expected in this cell culture system.) At 48 h post seeding, cells were exposed to ATRA (Sigma Aldrich; St. Louis, MO) in differentiation medium (50 μl exposure volume) at each of the following five nominal concentrations: 0.001, 0.01, 0.1, 1, 10 μM, or 0.1% DMSO and renewed daily up to 192 h, depending on the sampling timepoint. For high content imaging (HCI) analysis, cells were stained and fixed at each of three ATRA exposure timepoints: 6 h, 96 h, and 192 h. For RNA-sequencing analysis, duplicate cell samples collected at each timepoint were pelleted and snap frozen prior to RNA extraction.
2.2. High content imaging analysis
Mitotracker Red CMXRos (Thermo Fisher M7512) for mitochondrial activity staining and ImageIT-Dead Green (Thermo Fisher I10291) for dead cell staining were applied to cells one half hour before fixation of the cells. After fixation in 4% formaldehyde, cell cultures were washed with PBS. Hoechst dye was added during secondary antibody incubation (2 h room temperature in the dark) to visualize cell nuclei. FOXA2 primary antibody (rabbit monoclonal anti-FOXA2 [EPR4466]; Abcam ab108422) at 1:100 dilution in blocking buffer (5% Goat serum, 1% BSA in PBS, 0.2% fish gelatin) with Goat anti-rabbit Alexa Fluor 647 secondary antibody (Thermo Fisher A21245) at 1:1000 dilution in PBS without magnesium and calcium (Thermo Fisher 14190136 or equivalent) was used to measure FOXA2 expression. Plates were scanned four fields per well using a 20X objective (Nikon, Plan APO 20X NA 0.75 (0.32 μm/Pixel)) and images were acquired using a high content analysis IC200 cytometer (Vala Sciences, Inc.; San Diego, CA). Images were collected in four channels: nucleus (Hoechst 33342; Thermo Fisher H3570), ImageIT-Dead (AF488; ex/em (nm): 495/519), Mitotracker (AF548), and FOXA2 (AF647; ex/em (nm): 567/669). Image analysis was conducted in CellProfiler v2.2.0 [39] using default parameters and the following image intensity thresholds to identify cells expressing a given marker: automatic thresholding for Hoechst and FOXA2; 50% of maximum image intensity for Mitotracker; and 75% of maximum image intensity for ImageIT-Dead. Object diameter range was set at 30–80 pixel units based on manual cell measurements. Total cell counts were based on object-level identification of Hoechst-stained nuclei. This number was used to calculate the percentage of Hoechst-stained cells positive for FOXA2, ImageIT-Dead (converted to viable cells), or Mitotracker labeled subpopulations. Well-level summary data analyzed across three experimental well replicates were used for statistical analysis. The data were processed in R v3.4.0 with ggplot2 (ggpubr v0.1.7) [40,41] to produce plots with standard error (n = 3). Significant differences in the percentage of cells expressing a given marker compared to time-matched DMSO controls were calculated using one-way analysis of variance (ANOVA) with Tukey post-hoc test (padj < 0.05; honest significant difference adjustment method). Prior to conducting the ANOVA test, Bartlett’s test of homogeneity of variance and Shapiro-Wilk normality test were employed to confirm assumptions for ANOVA (p < 0.05). Data not meeting these assumptions were analyzed with the nonparametric Kruskal-Wallis test (p < 0.05) and Dunn post-hoc test (padj < 0.05; Benjamini-Hochberg adjustment method).
2.3. RNA-Sequencing
2.3.1. Ribo-depletion, NGS library preparation
RNA from two biological replicates per treatment was extracted from snap-frozen cell pellets by ARQ Genetics (Bastrop, TX) using a mirVana™ miRNA Isolation kit, with phenol (Ambion/Thermo Fisher Scientific; Waltham, MA) according to the manufacturer’s protocol. Next, total RNA was processed for library construction by Cofactor Genomics (http://cofactorgenomics.com; St. Louis, MO) according to the following procedure. Species-specific rRNA-probes were hybridized to total RNA to remove nuclear-encoded and mitochondrial contaminating ribosomal RNA from the sample. The resulting ribo-depleted RNA was then fragmented. First-strand cDNA synthesis was performed using reverse transcriptase and random primers in the presence of Actinomycin D, followed by second-strand cDNA synthesis with DNA polymerase I and RNase H. Double-stranded cDNA was end-repaired and A-tailed for subsequent adaptor ligation. Indexed adaptors were ligated to the A-tailed cDNA. Enrichment by PCR was performed to generate the final cDNA sequencing library. Libraries were sequenced at a read depth of at least 30 million single-end 75 base pair reads on an Illumina NextSeq500 (San Diego, CA) following the manufacturer’s protocols.
2.3.2. Quality control, alignment, clustering, normalization, and expression comparison
Data analysis and visualization were performed at the U.S. Environmental Protection Agency. Raw and processed data files were deposited in the Gene Expression Omnibus (GEO): accession number GSE131921. Raw sequence data in FASTQ format were assessed for quality (FastQC, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and ribosomal RNA content (sortmeRNA [42]). Reads were aligned to the human genome (GRCh38) with Ensemble (v84) gene annotations using HISAT2 (version 2.0.5) default parameters, with the exception of the “downstream transcript assembly” flag [43]. The index for the Ensemble reference genome was created using HISAT2 build and the corresponding FASTA files obtained from Ensembl. Multiple alignments per read were allowed, but only uniquely aligned reads were used in downstream analysis. HTSeq v0.9.0 [44] was used to count uniquely mapped reads for each gene ID in Ensembl (v84) with parameters: stranded = no, intersection = nonempty. These counts are the basis for expression comparison and statistics generation. For each gene with row median counts > 5, the fold change compared to time-matched DMSO control (treatment/control) was calculated for each sample using DESeq2 (v1.15.3) [45]. Counts were then filtered to select differentially expressed genes (DEGs) with padj value < 0.05 (Benjamini-Hochberg adjustment method) and absolute value of log2(fold change) > 1. Gene expression changes were visualized in Partek Genomics Suite (v6.6) with bidirectional hierarchical clustering of rows (genes) using default settings (Euclidean, average linkage clustering method).
2.4. Pathway analysis
Ingenuity® Pathway Analysis (IPA®) (Qiagen Inc., https://www.qiagenbioinformatics.com) was used to investigate the gene networks involved in the observed phenotypic changes. IPA builds causal networks based on direct or indirect gene relationships curated from the literature and stored in the Ingenuity Knowledge Base [46]. The core analysis infers upstream regulator networks based on supplied gene expression data. A total of 9997 differentially expressed genes representing the union of all treatments were uploaded into the software application, resulting in 7731 genes that mapped to the Ingenuity Knowledge Base. These genes were filtered further by absolute value of log2 expression ratio ≥1.5, resulting in 4610 genes. Next, a core analysis was run using default settings (i.e., direct and indirect relationships). These steps were repeated for control samples for which log2(fold change) had been calculated for genes at 96 h versus 6 h, 192 h versus 6 h, and 192 h versus 96 h. The union of DEGs for the control samples totaled 10816, of which 8382 were mapped to the Ingenuity Knowledge Base. Applying the −1.5 to 1.5 expression log2 ratio cutoff resulted in 4691 genes for the DMSO core analysis.
2.5. Tipping point analysis
We used tipping point analysis [30] to identify the critical concentration at which the hiPSC system did not recover from perturbations due to ATRA. Tipping points were analyzed using differential gene expression to quantify the overall hiPSC system perturbation and trajectory over time. We assume that the DMSO-treated cells follow a normal developmental trajectory and that any genes that are differentially expressed due to ATRA treatment represent a deviation from this trajectory. Thus, we quantified the transcriptomic trajectories of the developing cells produced by increasing concentrations of ATRA and measured the rate at which these trajectories recovered (called the negative velocity). We have shown previously that recovering trajectories have a negative velocity whereas non-recovering trajectories have a positive velocity (further details are provided in [30]). Our hypothesis is that the developmental tipping point occurs when the hiPSC system, in response to increasing doses of ATRA, transitions from recovering to non-recovering trajectories. Our analysis of ATRA-induced tipping points is described below.
2.5.1. Data processing
For tipping point analysis, we considered DEGs following exposure to 0.001, 0.01, 0.1, and 1 μM ATRA normalized and filtered as described in section 2.3.2. Responses for the highest concentration (10 μM) were omitted from the tipping point analysis due to substantial (> 50%) but statistically insignificant cytotoxicity at this concentration in the HCI analysis. This decision was supported by the cytotoxicity-associated burst phenomenon [47] which estimates the cytotoxicity lower bound limit for RA at 3.2 μM [Invitrodb v3.1] (https://epa.figshare.com/articles/ToxCast_Database_invitroDB_/6062623/3) [48]. Gene expression measurements that did not meet a condition for differential expression (|log2 FC| > 1 and padj-value < 0.05) were set to 0. This revealed 8340 differentially expressed genes with at least one measurement that met criteria for differential expression across three time points and four concentrations.
2.5.2. System trajectories and dynamics
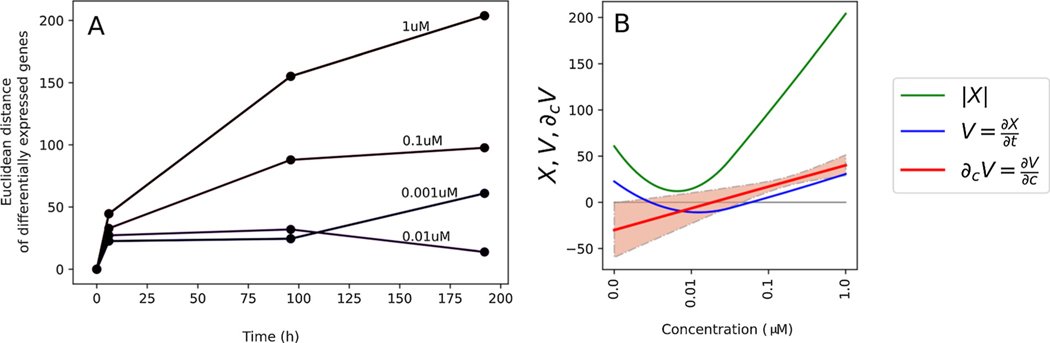
For each concentration and time point, we define the hiPSC system perturbation (X), produced by ATRA, as a vector comprising 8340 DEGs. As previously published [30], a scalar perturbation was calculated as the Euclidean norm . We assumed that the scalar perturbation is a continuous function over concentration and time X = f(c,t). Therefore, a temporal sequence of scalar perturbations was defined as a trajectory . A trajectory represents the dynamic behavior of the cell culture after a chemical treatment. We assumed that before chemical application the system was in a normal developmental state (not perturbed), i.e., f (c,0) = 0. For each concentration, we monitored a temporal trend of a trajectory as a rate of change of the scalar perturbation across the time . This value was designed as the “velocity” of the system.
2.5.3. Quantifying system recovery across concentrations
We assumed that the velocity is a continuous function of time (t) and concentration (c). To calculate “tipping points” we investigated the concentration derivative of velocity, . Concentrations that produce recovery have negative derivative , whereas concentrations that do not produce recovery have positive derivative . A specific concentration at which the system shows a “tipping point” in recovery is denoted as the Ccr, and it corresponds to the condition . Therefore, using 192 h and 96 h time points we estimated by interpolating a B-spline through points at 0.001, 0.01, 0.1, and 1 μM. After sampling 50 subsets of the concentration – velocity pairs, for which was fitted, and solved, we constructed a distribution of Ccr and estimated the 95% confidence interval of Ccr, assuming they were distributed normally.
3. Results
3.1. High content imaging analysis
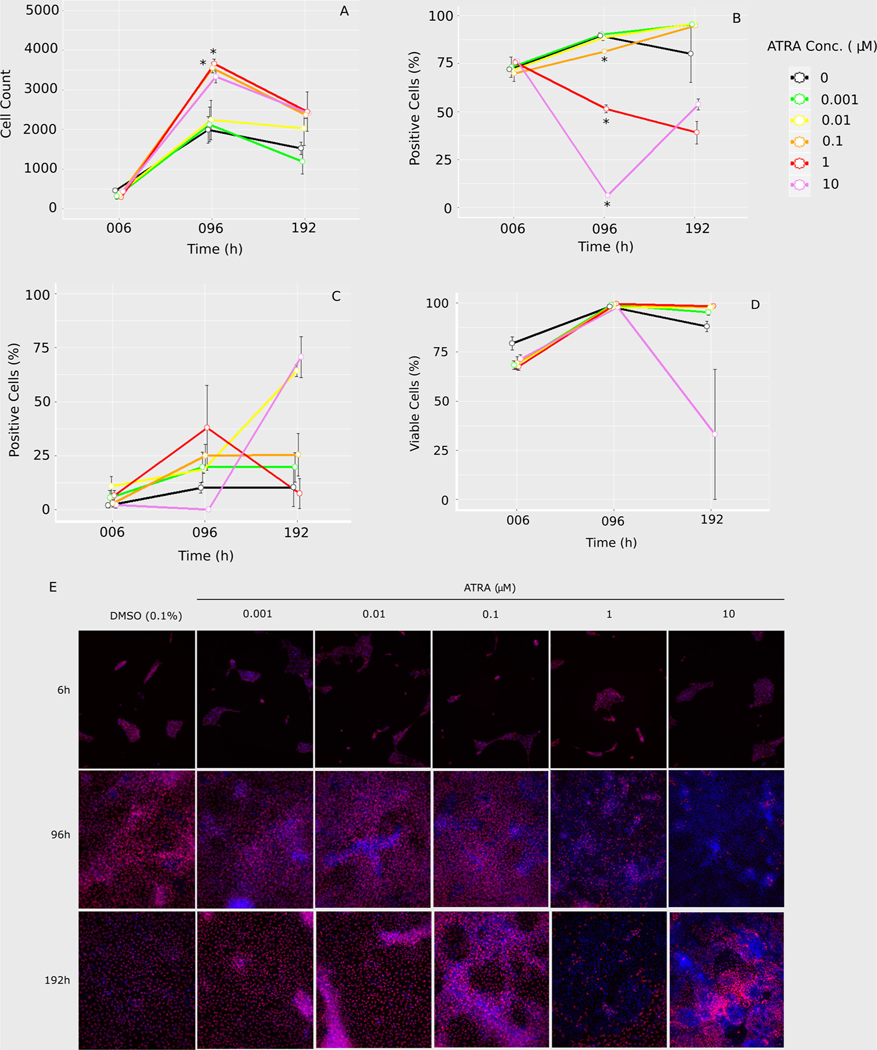
HCI analysis provided insights into cell state, activity, and quality (Fig. 2; Table 1). ATRA inhibited hiPSC endoderm differentiation at exposures above 0.01 μM based on FOXA2 suppression (and subsequent confirmation based on a decrease in SOX17 expression in the RNAseq dataset). Cell counts were low at 6 h, with no differences between ATRA treatments (Fig. 2A). By 96 h, cells had reached confluence and all treatments had increased cell counts, with significantly more cells than controls for ATRA exposures at 0.1 and 1 μM (Table 1). Cell numbers leveled off by 192 h, at which point there was no significant difference between treatments and DMSO controls. At this time, cells appeared to be overly confluent in some samples (Fig. 2E). FOXA2 protein expression at 6 h did not differ from controls for any treatment (Fig. 2B). However, there was a significant, concentration-dependent decrease in FOXA2 cells exposed to ATRA above 0.01 μM at 96 h (Table 1). By 192 h, FOXA2 expression was not significantly different from controls for any ATRA exposure, due in part to a wide degree of variance in controls at this timepoint. Images at 192 h supported the trends in FOXA2 expression for 1 μM (decreased) and 10 μM (increased) compared to the previous timepoint for the respective concentrations (Fig. 2E). Additionally, there appeared to be rudimentary structure formation in the 10 μM samples (Fig. 2E). For both mitochondrial activity (Fig. 2C) and cell viability (Fig. 2D), there was no significant difference between controls and treated cells at any timepoint, nor did controls differ across time. However, impacts on mitochondrial activity were inconclusive due to wide variation in replicates. Moreover, while not statistically significant, there was a substantial amount of cell death/cytotoxicity at 10 μM ATRA exposure. Taken together, the HCI data support that ATRA interfered with endogenesis in a time and concentration dependent manner.
Fig. 2.
Four channel high content imaging analysis: A) cell counts were based on nucleus staining (Hoechst) and used as the overlay for calculating the percentage of positive cells for B) FOXA2 expression, C) mitochondrial activity, or D) cell viablilty for each timepoint and treatment. Error bars indicate standard error for three well replicates (Cell Profiler v2.2.0). E) Representative images of Hoechst (blue) and FOXA2 (red) co-expression (purple) for each treatment. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Table 1.
High content imaging results for cell count, FOXA2 expression, mitochondrial activity, and cell viability comparing treatment concentrations to A) time-matched controls and B) DMSO controls over time. Tukey Multiple Comparison of Means (post-hoc); padj (honest significant difference adjustment method).
| A | |||
|---|---|---|---|
| Concentration (μM) | 006 hours | 096 hours | 192 hours |
|
| |||
| Cell count | |||
| 0.0001 | NS | NS | NS |
| 0.001 | NS | NS | NS |
| 0.01 | NS | NS | NS |
| 0.1 | NS | padj = 0.041 | NS |
| 1 | NS | padj = 0.028 | NS |
| 10 | NS | NS | NS |
| FOXA2 | |||
| 0.0001 | NS | NS | NS |
| 0.001 | NS | NS | NS |
| 0.01 | NS | NS | NS |
| 0.1 | NS | padj = 0.009 | NS |
| 1 | NS | padj < 0.001 | NS |
| 10 | NS | padj < 0.001 | NS |
| Mitochondrial activity | |||
| 0.0001 | NS | NS | NS |
| 0.001 | NS | NS | NS |
| 0.01 | NS | NS | NS |
| 0.1 | NS | NS | NS |
| 1 | NS | NS | NS |
| 10 | NS | NS | NS |
| Viable cells | |||
| 0.0001 | NS | NS | NS |
| 0.001 | NS | NS | NS |
| 0.01 | NS | NS | NS |
| 0.1 | NS | NS | NS |
| 1 | NS | NS | NS |
| 10 | NS | NS | NS |
|
| |||
| B | |||
| 006 vs 096 hours | 006 vs 192 hours | 096 vs 192 hours | |
|
| |||
| Cell count | padj = 0.006 | padj = 0.031 | NS |
| FOXA2 | NS | NS | NS |
| Mitochondrial activity | NS | NS | NS |
| Viable cells | NS | NS | NS |
NS =not significant.
3.2. RNA-seq analysis
3.2.1. Principal components analysis
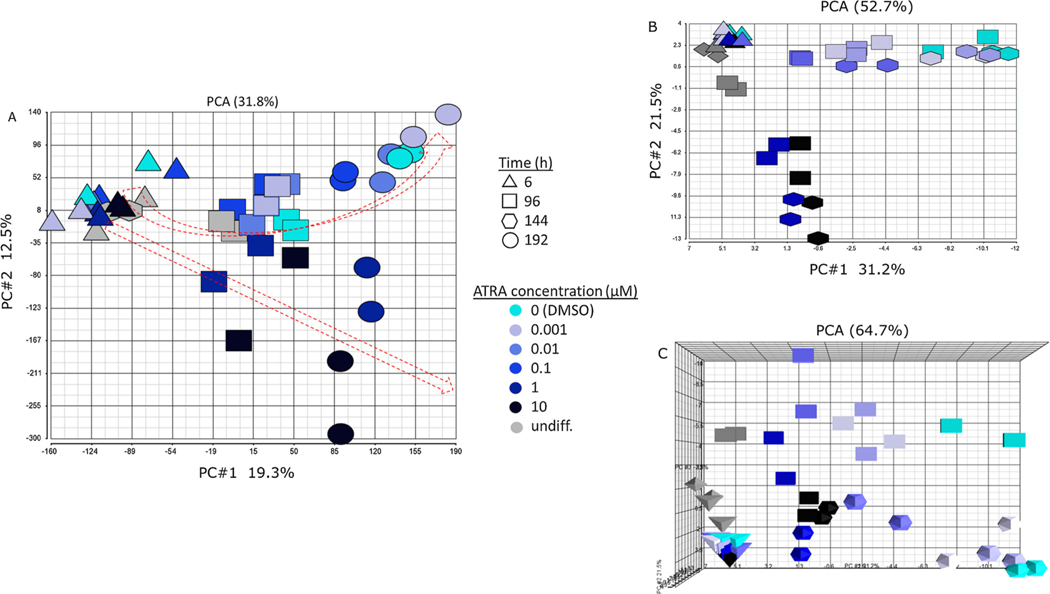
RNA-sequencing analysis resulted in read counts for 58,051 detected genes, which are represented in the principal components analysis (PCA) in Fig. 3A. Each point in this PCA indicates the raw counts for a treatment replicate (two replicates each), with increasing concentration indicated by color (light to dark) and time indicated by shape (6 h = triangle, 96 h = square, 144 h = hexagon, 192 h = circle). There are only two points for the 144 h timepoint. These were undifferentiated samples intended for the 192 h timepoint, but the undifferentiated cells began to die by day six, requiring their early collection at 144 h. The 144 h samples clustered with the 6 h samples, reflecting similar phenotypic states at an early stage of the differentiation trajectory. Exposure duration (time) increases on the x-axis (PC#1, 19.3%), while concentration decreases on the y-axis (PC#2, 12.5%). Filtering the genes to exclude those with < 5 counts (median) resulted in a similar PCA and trajectories (not shown). The red arrows indicate a diverging trajectory of the samples over time and concentration, with the divergence appearing to begin at 96 h and at concentrations above 0.1 μM. A second PCA of the 97 developmental marker genes in Supplemental Table 1 reflected a more distinct trajectory shift in cells exposed to ATRA concentrations above 0.1 μM (Fig. 3B). The first two principal components of this PCA explained 52.7% of the gene expression changes, in contrast to 31.8% in the full transcriptome-based PCA (Fig. 3A). In the 97 developmental marker-based PCA, differentiation was reflected by PC#1 (31.2%) and concentration was reflected by PC#2 (21.5%) (Fig. 3B). Addition of a third principal component (PC#3, 12%) reflected exposure duration for the 97 developmental marker genes (Fig. 3C).
Fig. 3.
Principal components analysis (PCA): A) unfiltered count data for 58,051 aligned genomic fragments (two replicates per treatment). PC#1 (19.3%) reflects increasing time from left to right; PC#2 (12.5%) reflects decreasing concentration from bottom to top. B) Principal components analysis of unfiltered count data for 97 developmental marker genes (two replicates per treatment). PC#1 (31.2%) reflects differentiation from left to right; PC#2 (21.5%) reflects decreasing concentration from bottom to top. C) Addition of a third principal component to the 97 developmental marker PCA viewed from above. PC#3 reflects differences in exposure duration (time).
3.2.2. Differential gene expression analysis
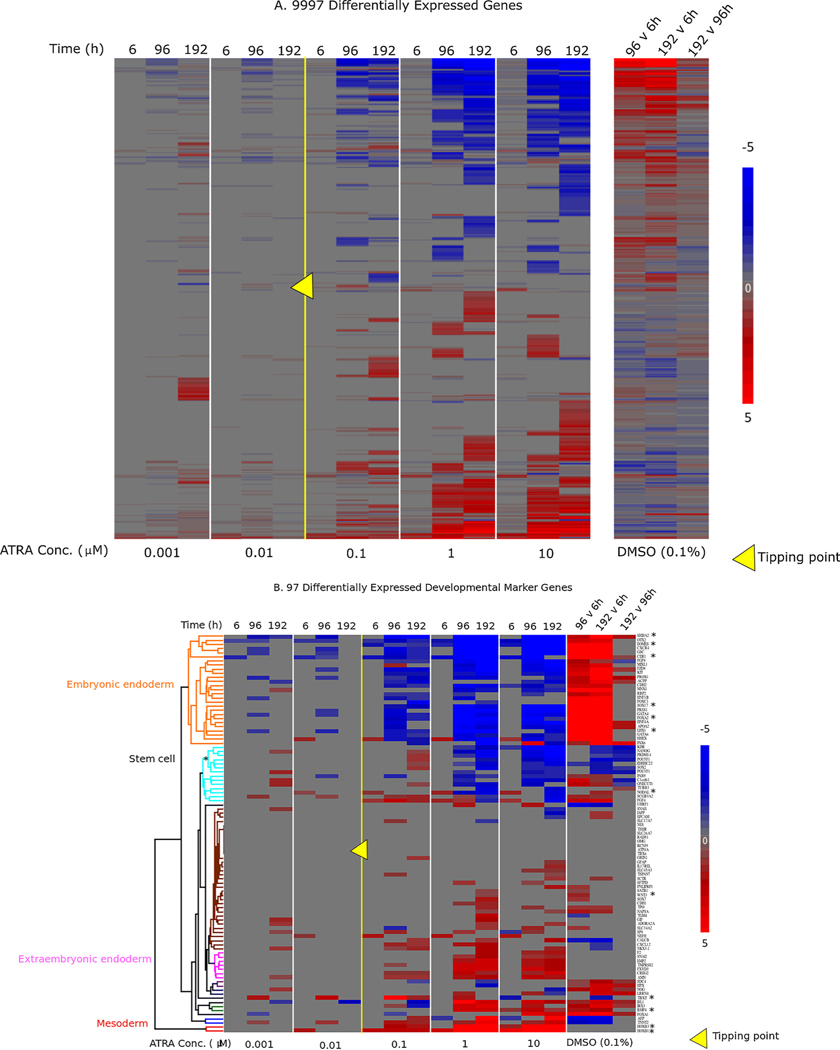
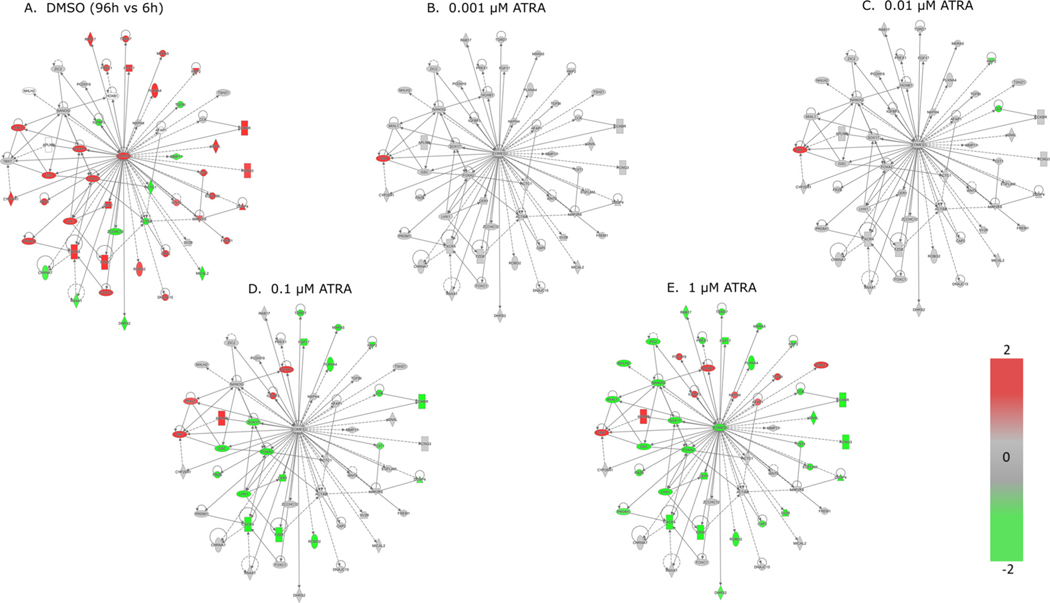
ATRA exposure induced transcriptomic changes in the full dataset (i.e., all treatments and timepoints) consistent with a divergence away from endoderm to posterior primitive streak and mesoderm. There were 24,592 genes with median counts > 5. Further filtering by absolute value of log2(fold change) versus time-matched control > 1, and padj < 0.05, resulted in 9997 DEGs (Fig. 4A). Overall, bidirectional hierarchical clustering of genes (rows) resulted in two sets of genes that either increased in expression (red) or decreased in expression (blue) at 96 h or 192 h in ATRA exposure concentrations above 0.01 μM. These global gene expression changes were also reflected in a smaller subset of 97 developmental marker genes selected from the literature to represent stages of embryonic development from the primitive streak to rudimentary organ formation and including the three germ layers (Fig. 4B; Supplemental Table 1). In control cells exposed to 0.1% DMSO, the expression of embryonic endoderm markers (e.g., FOXA2, SOX17, and GATA binding protein 4 (GATA4)) increased between 6 h and 96 h, then remained at an increased level at 192 h compared to 96 h controls (Fig. 4B). These expression changes confirmed the differentiation of most unexposed cells to an embryonic endodermal cell fate. In cells exposed to ATRA concentrations above 0.01 μM, the expression of embryonic endoderm gene markers was decreased at 96 h, concomitant with an increase in expression of primitive streak and mesodermal gene markers (e.g., homeobox B1 (HOXB1), homeobox B3 (HOXB3), and T-box transcription factor T (TBXT)) and extraembryonic endoderm genes (e.g., transmembrane protease, serine 2 (TMPRSS2) and amnionless (AMN)). These genes were not differentially expressed in controls at 96 h versus 6 h, suggesting a shift in differentiation state responding to ATRA exposure. Also, the respective fold changes in these genes suggests that the shift toward mesendoderm was more sensitive to ATRA exposure, with the greatest increase in fold change of TBXT in 0.1 μM ATRA at 96 h, whereas a greater fold change in gene expression of most mesodermal and extraembryonic endoderm genes was observed at 1 μM ATRA at 96 h.
Fig. 4.
Bidirectional hierarchical clustering of differentially expressed genes: A) 9997 differentially expressed genes with median counts >5, abs(log2FC) > 1, and padj <0.05 (Benjamini-Hochberg adjustment method). Bidirectional hierarchical clustering was based only on gene expression changes in ATRA-exposed cells. Each of the five panels on the left represents an ATRA concentration, with each of the three timepoints represented as columns within each panel. Columns in the right panel show DMSO control expression values at 96 h vs. 6 h, 192 h vs. 6 h, and 192 h vs. 96 h (DMSO gene order manually matched to ATRA-driven clustering). B) Subset of 97 developmental marker genes (Supplemental Table 1). Bidirectional hierarchical clustering was based on all treatments, including DMSO controls shown in this figure. Column labeling is the same as in A. ATRA concentrations above 0.01 μM reduced expression of embryonic endoderm markers compared to time-matched controls, while increasing expression of mesoderm and extraembryonic endoderm markers. In both A and B, red indicates an increase in expression (max : 5); blue indicates decrease (min : −5); grey indicates no change, compared to time-matched controls for ATRA or indicated comparisons for DMSO. Yellow triangle indicates tipping point between 0.01 and 0.1 μM. For figure B, asterisk denotes genes that are highlighted in Fig. 7B. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
3.3. Tipping point analysis
The initial HCI analysis suggested a tipping point between 0.01 and 0.1 μM. However, a tipping point was not confirmed with the HCI data because the significant differences between treated and control samples did not persist at 192 h. Further qualitative evidence of a tipping point was observed in the expression of the developmental marker genes in Fig. 4B. For example, several genes (e.g., CXCR4, goosecoid homeobox (GSC), GATA4) were decreased at concentrations below 0.01 μM compared to time-matched DMSO controls at 96 h, but not 192 h, indicating recovery. However, these genes were decreased at 96 h and their decreased expression persisted at 192 h at concentrations above 0.01 μM. To calculate a definitive tipping point for the RNA-seq dataset, system trajectory analysis was conducted on the 8340 DEGs for concentrations at or below 1 μM. The tipping point analysis (i.e., velocity calculation at 192 h incorporating all samples over a 192 h timecourse) resulted in a tipping point of 17 ( ± 11 SD) nM (Fig. 5), consistent with the inferred tipping point based on FOXA2 protein expression (Fig. 2B) and differential gene expression (Fig. 4). Fig. 5A depicts scalar perturbations for each concentration across time. Generally, perturbations increased in a concentration-dependent manner, except for 0.001 μM versus 0.01 μM at 192 h. The greater number of perturbations in the 0.001 μM treatment is consistent with a subset of genes that were significantly increased compared to time-matched controls at 192 h in 0.001 μM, but not 0.01 μM samples in Fig. 4A. KEGG pathway analysis (Partek Genomics Suite) of these genes indicated an association with steroid biosynthesis (data not shown).
Fig. 5.
Tipping point analysis: A) Scalar perturbation (y-axis) at each time point (x-axis) is a magnitude of DEG calculated using the Euclidean norm. Each line corresponds to a different ATRA concentration. B) Tipping point calculation at 192 h. Vertical axis: scalar perturbation X (green), velocity V (blue), and derivative of velocity with respect to concentration ∂cV (i.e., uncertainty analysis (red)). Critical concentration is defined at ∂cV = 0. Critical concentration (tipping point) was calculated to be 17 ( ±11 SD) nM, defined as the point on the x-axis at which the ∂cV intersects 0 on the y-axis. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
3.4. Pathway analysis
Tipping point analysis and observed gene and protein expression changes indicated that 96 h was the time at which the differentiating cells exposed to ATRA concentrations at and above 0.01 μM ATRA shifted their trajectory. We therefore focused on this timepoint to identify potential upstream regulators implicated in the disruption of the developmental trajectory. We initially focused on the DMSO controls to assess the transcriptional activity at 96 h compared to cells at 6 h prior to significant FOXA2 suppression by ATRA. The upstream regulator analysis predicted activation of progesterone (chemical), eomesodermin (EOMES), dexamethasone (chemical), and tretinoin (chemical); and predicted inhibition of SOX2 and topotecan (chemical) (Table 2).
Table 2.
Upstream regulator analysis (Ingenuity Pathway Analysis) for DMSO (96 h versus 6 h) and 0.1 μM at 96 h. Only the top predicted upstream regulators are shown (six for DMSO and three for 0.1 μM ATRA at 96 h). Upstream regulators that were not assigned a predicted activation state are omitted from this table.
| Concentration | Upstream Regulator | Expr Log Ratio | Predicted Activation State | Activation Z-Score | P-value of Overlap |
|---|---|---|---|---|---|
|
| |||||
| DMSO (96 h v 6 h) | progesterone | Activated | 2.148 | 1.33E-15 | |
| DMSO (96 h v 6 h) | EOMES | 6.906 | Activated | 3.685 | 7.46E-15 |
| DMSO (96 h v 6 h) | SOX2 | 0 | Inhibited | −2.731 | 7.95E-14 |
| DMSO (96 h v 6 h) | topotecan | Inhibited | −3.923 | 1.85E-12 | |
| DMSO (96 h v 6 h) | dexamethasone | Activated | 3.013 | 4.58E-12 | |
| DMSO (96 h v 6 h) | tretinoin | Activated | 2.51 | 1.01E-11 | |
| 0.1 μM (96 h) | EOMES | −1.017 | Inhibited | −2.549 | 1.22E-15 |
| 0.1 μM (96 h) | dexamethasone | Inhibited | −2.512 | 1.62E-13 | |
| 0.1 μM (96 h) | LHX1 | −2.049 | Inhibited | −2.774 | 2.81E-11 |
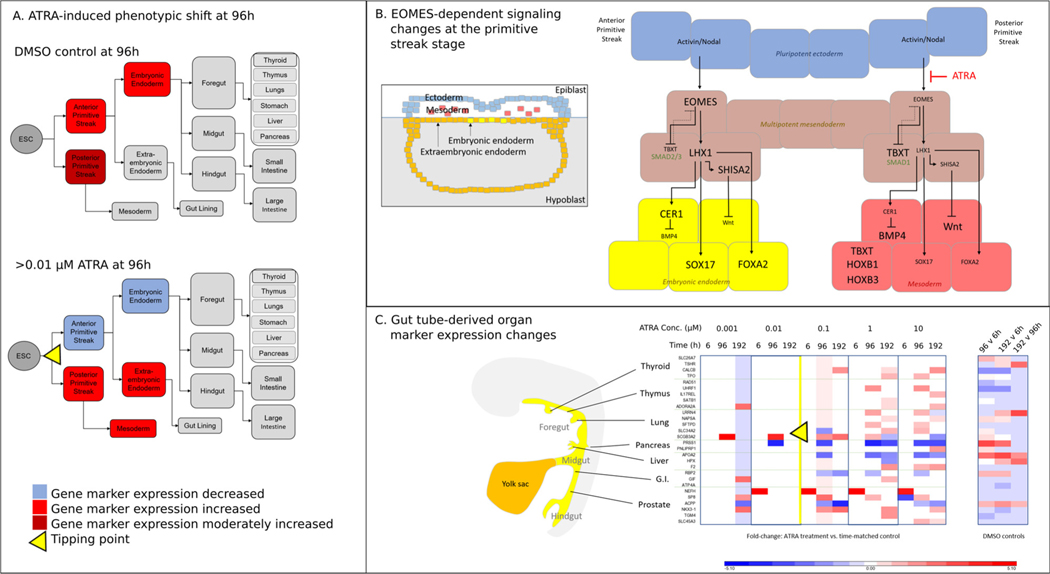
Next, upstream regulators were evaluated for ATRA exposures just above the tipping point (i.e., 0.1 μM) at 96 h to identify pathways associated with perturbed differentiation states compared to time-matched controls. The top predicted upstream regulator for the 0.1 μM treatment group relative to time-matched controls was EOMES (Table 2), which we selected for further pathway analysis. We explored the molecular targets associated with EOMES inhibition by ATRA more closely by constructing a relationship network for the 0.1 μM ATRA treatment (41 genes), as well as 42 genes (including 13 additional unique molecular targets) associated with its activation in DMSO controls at 96 h versus 6 h (54 genes total) (Fig. 6). We next overlaid the gene expression changes for DMSO control samples (96 h versus 6 h gene expression fold-change; Fig. 6A), as well as concentrations versus time-matched controls for ATRA exposure concentrations below (0.001 and 0.01 μM) and above (0.1 and 1 μM) the tipping point at 96 h (Fig. 6B–E). Consistent with endodermal differentiation, most EOMES-dependent genes were increased in controls at 96 h and remained at these levels at concentrations below the tipping point; however, these genes were significantly decreased relative to time-matched controls at the concentrations above the tipping point. Three of these genes (cholecystokinin (CCK), TBXT, and special AT-rich sequence-binding protein 2 (SATB2)) were differentially expressed near the tipping point (0.01 μM; Fig. 6C), but not in DMSO controls at 96 h versus early differentiating cells at 6 h (Fig. 6A), suggesting a potential role in the observed differentiation trajectory shift that appeared to begin at 96 h.
Fig. 6.
Ingenuity pathway analysis of upstream regulator, EOMES, and associated molecular targets at 96 h: A) DMSO fold change at 96 h v 6 h; B – E) ATRA fold change versus time-matched control at 96 h. Red = increase; green= decrease; grey = no change (i.e., gray nodes have same expression values as controls in A). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
4. Discussion
4.1. Summary
The appearance of three germ layers begins during gastrulation when the primitive streak forms at week three of human gestation. Disruption of signaling between these germ layers can lead to embryo death or malformations. We selected hiPSC-derived differentiating endoderm as an in vitro model of early embryonic development to explore its potential for use in tipping point analysis to screen ToxCast™ chemicals for developmental toxicity. By tracking a selected set of gene markers for endodermal, mesodermal, ectodermal, and primitive streak stages of development, we followed the effects of a known teratogen, ATRA, during cellular differentiation representing early embryonic development. This suite of 97 markers (Supplemental Table 1) was sufficient to capture a shift in endodermal differentiation from embryonic endoderm towards mesoderm and extraembryonic endoderm. This trajectory shift was evident in the FOXA2 HCI data beginning at 96 h at concentrations above 0.01 μM, which aligned with a nominal tipping point of 17 nM that was calculated from the DEGs that were perturbed at 96 h and did not recover (i.e., return to DMSO expression levels) by 192 h. In vivo, loss of embryonic endoderm in Sox17-null mutant mouse embryos results in abnormal gut tube development and associated embryo mortality [49].
4.2. Control samples progressed through primitive streak to differentiating endoderm
Gene markers indicated that the primitive streak was the earliest developmental stage represented in the control cell culture (Fig. 4B). Many of these primitive streak markers (i.e., FOXA1, FOXA2, FZD8) also serve as embryonic endoderm markers as development progresses [22]. Adding activin (a Nodal ligand) and low serum was expected to activate Nodal signaling and induce endogenesis in hiPSCs. Endodermal differentiation was confirmed in control samples by the protein and mRNA expression of the endodermal marker, FOXA2, in both the HCI and RNA-seq experiments, respectively. The co-expression of SOX17 and FOXA2 mRNA, in addition to other endodermal specific gene markers (Supplemental Table 1), confirmed that the majority of cells in the control samples were successfully induced along an endodermal differentiation trajectory, consistent with previous studies using activin and low serum [20]. Moreover, primitive streak/mesendoderm and mesodermal markers (e.g., TBXT, HOXB1, HOXB3) were not increased in controls at 96 h compared to 6 h, suggesting the cells did not progress beyond the posterior primitive streak stage of the mesodermal lineage.
4.3. ATRA shifted the endodermal trajectory toward mesoderm and extraembryonic endoderm
ATRA exposure induced a distinct shift from embryonic endoderm based on attenuation of the gene markers that were increased in control samples. This attenuation of embryonic endoderm markers at 96 h was concomitant with an increase in the mesendodermal marker, Brachyury (T, TBXT) [50], which had a log2(fold change) increase of 4.99 at 0.1 μM, and a concentration-dependent increase in extraembryonic endoderm and mesoderm markers compared to time-matched controls (Fig. 4B). At 1 μM ATRA, the fold change increase of primitive streak/mesodermal markers HOXB1 (log2FC = 8.04) and HOXB3 (log2FC = 6.72) was among the highest in the 97 developmental marker gene set. The effects of ATRA on the HOXB genes are consistent with the known importance of RA in embryonic anteroposterior patterning. For example, RA shifts the expression of HOX genes anteriorly [51]. In addition to mesodermal markers, several extraembryonic gene markers were also increased compared to controls at 96 h in 1 μM samples (e.g., TMPRSS2, FXYD domain-containing ion transport regulator 3 (FXYD3), cellular repressor of E1A stimulated genes 2 (CREG2), AMN) (Fig. 4B). Taken together, expression patterns of developmental marker genes suggest that increasing concentrations of ATRA above the tipping point altered the embryonic endoderm differentiation trajectory first through a mesendodermal phenotype followed by a shift toward primitive streak/mesodermal and extraembryonic endodermal trajectories (Fig. 7A).
Fig. 7.
Hypothetical differentiation trajectory shift following ATRA exposure: A) ATRA concentrations above the tipping point shift the endodermal differentiation trajectory from embryonic endoderm to primitive streak/mesoderm and extraembryonic endoderm. B) The observed shift in developmental trajectory hypothetically occurs at the primitive streak stage of development through mesenchymal to epithelial transition mediated by EOMES repression downstream of Nodal activation. The gene regulatory relationships shown here are based on known interactions from the literature. In controls, EOMES mediates differentiation of embryonic endoderm (yellow). In ATRA-exposed cells, reduced EOMES expression causes a shift in the developmental trajectory of multipotent mesendoderm (brown) away from endoderm towards mesoderm (red). For clarity, some relationships are not shown. C) Gene expression changes of gut-tube derived organ markers exposed to ATRA or DMSO (red = increase; blue = decrease). Order of appearance manually matched to anteroposterior location of the respective organ origins in the gut tube. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
4.4. EOMES-dependent signaling regulates the differentiation trajectory shift
Expression patterns of developmental marker genes indicated that the cell fate shift in response to ATRA occurs when cells have reached a primitive streak level of differentiation, with different effects on the posterior versus anterior primitive streak. The primitive streak forms at the caudal end of the embryo, with the posterior primitive streak appearing first before extending to form the anterior primitive streak. The reduction in embryonic endoderm concomitant with an increase in primitive streak/mesodermal and extraembryonic endoderm marker expression led us to hypothesize that a cell fate shift in response to ATRA exposure above the tipping point of 17 nM could be through perturbation of EMT signaling, possibly through a shift in the mesendodermal cell phenotype in favor of primitive streak, mesoderm or extraembryonic endoderm [52] via impacts on Wnt3, TGF-β, FGF, BMP, and Nodal signaling [14].
To investigate our hypothesis that perturbed EMT underlies the observed cell fate trajectory shift, we looked at Wnt3, FGF4, Nodal, BMP4, as well as functional markers of EMT: E-cadherin (CDH1 down), N-cadherin (CDH2 up), and Snail (SNAI2 up) [53] (Fig. 4B). Interestingly, only FGF4 was increased in expression just above the tipping point (0.1 μM) at 96 h, while WNT3, BMP4, and NODAL were unchanged compared to time-matched controls. When cells were exposed to 1 μM ATRA, WNT3 expression was unchanged, NODAL expression decreased, while FGF4 and BMP4 increased at 96 h. At this same concentration and timepoint, CDH2 decreased in response to ATRA; whereas, it was increased in controls at 96 h compared to 6 h. In contrast, CDH1 and SNAI2 expression levels were increased at 96 h in 1 μM samples but not differentially expressed in controls or lower ATRA concentrations. Taken together, these gene expression changes suggest perturbation of EMT by ATRA in favor of mesoderm, possibly through a reversal of EMT (i.e., mesenchymal to epithelial transition [52,53]), but do not provide a clear picture of the signaling events underlying this process.
To more thoroughly explore the signaling events occuring at concentrations above the tipping point, we used pathway analysis to identify upstream regulators impacted by ATRA exposure. EOMES [54] was one of the top upstream regulators predicted for both the DMSO and ATRA treatments. EOMES is a mesendodermal marker whose activation leads to endoderm formation, while its suppression leads to a mesodermal cell fate [55], leading us to focus on its molecular targets to investigate how ATRA impacts primitive streak derived cell specification above the tipping point (Fig. 6). Notably, another mesendodermal marker and EOMES target, TBXT [55], was one of the only genes perturbed by ATRA near the tipping point (0.01 μM) (Fig. 6C). TBXT promotes gene transcription involved in mesoderm formation and embryonic development [56]. It’s considered both a primitive streak/mesendoderm and mesoderm gene marker [22,50]. EOMES inhibits TBXT expression [55], consistent with its prediction as an upstream regulator in our dataset. Higher concentrations of ATRA (i.e., 1 μM) caused a decrease of EOMES expression (Fig. 6E) and maintained an increase in TBXT expression that was also observed at lower concentrations. Notably, EOMES is also a TBXT target; thus, a feedback loop between the two transcription factors may be involved in this system [56].
Taken together, the gene expression changes suggest that ATRA exposure initiates a process favoring mesodermal-biased EMT downstream of EOMES induction by activin/Nodal. The observation that TBXT was one of the most sensitive genes induced downstream of EOMES led us to hypothesize that the increase in TBXT expression compared to time-matched controls led in turn to decreased EOMES and other target gene expression compared to time-matched controls at ATRA concentrations above the tipping point. It is important to note that TBXT is a mesendoderm marker that peaks in expression levels at 12 h post induction in differentiating human embryonic stem cells [50]. Thus, it is possible that TBXT expression was elevated at 6 h in both DMSO controls and ATRA treated cells and remained at this elevated level at 96 h in controls. One could confirm this by comparing differentiating control samples to undifferentiated cells at the 6 h timepoint. However, the undifferentiated cells in the present study were intended as a quality control feature and were not cultured in DMSO, precluding their use for this purpose. Including these earlier timepoints and appropriate undifferentiated controls in future studies will help distinguish the effects of TBXT on signaling events downstream of EOMES. This will be important because the timing of TBXT expression changes directs germ layer formation. For example, Kameoka et al. (2014) characterized endoderm differentiation by concomitant increase in TBXT and EOMES expression at 24 h post induction [29]. TBXT could also be involved in the expression changes of several of the EOMES-regulated [55,57] endodermal markers that decreased at 0.1 μM. For example, the following genes are also regulated by TBXT: SOX17, FOXA2, GSC, and LIM homeobox 1 (LHX1) [50,56].
TBXT can induce or repress the expression of a given gene involved in germ layer specification depending on the context [56]. For example, the presence of SMAD1, a TBXT cofactor involved in BMP signaling, facilitates mesodermal induction, while its absence facilitates TBXT-mediated endoderm induction [56]. Interestingly, SMAD1 expression was increased compared to controls at 1 μM ATRA, but no other concentration tested. This was also the concentration at which the EMT markers CDH1 and SNAI2 were increased, as well as being the lowest concentration at which EOMES expression was significantly decreased. EOMES is a TBXT target gene [56]; therefore, its decreased expression concomitant with TBXT increase is suggestive of a negative feedback loop between the two transcription factors. While TBXT was the only DEG at 0.01 μM in the selected EOMES target network (Fig. 6C), several genes were differentially expressed at 0.1 μM, including increased expression of TBXT, Mix-like homeobox protein 1 (MIXL1), apelin receptor (APLNR), HOXB1, and insulin-like growth factor-binding protein 3 (IGFBP3); while numerous genes were decreased, including the endodermal marker genes FOXA2, SOX17, CXCR4, and GSC (Fig. 6D). These expression changes are likely downstream of TBXT elevation and EOMES depression.
In addition to TBXT, LHX1 could have a pivotal role in mediating the ATRA-induced trajectory shift above the tipping point. LHX1 is expressed downstream of EOMES and shares many target genes including SOX17 and FOXA2 (Fig. 6). Moreover, in mice Lhx1 activates Shisa2, a Wnt antagonist [57] that was decreased in ATRA-exposed treatments and increased in controls. Taken together, we propose that an increase in Wnt and BMP4 signaling mediated by EOMES, TBXT, and LHX1 favors mesoderm induction when differentiating endoderm is exposed to ATRA concentrations above the tipping point. A simplified network depicting these events is outlined in Fig. 7B. Experimental gene expression changes compared to time-matched controls for genes shown in Fig. 7B are depicted in Supplemental Fig. 1.
4.5. ATRA may shift differentiating endoderm towards mesoderm by disrupting TGFβ/SMAD signaling
Retinoic acid regulates embryonic development through ligand activation of retinoic acid receptor (RAR)/retinoid X receptor (RXR) heterodimers, which bind to retinoic acid response elements (RAREs) in the promoters of target genes to activate downstream gene transcription [35,58,59]. To identify potential ATRA target genes in our system, we first considered the role of RA-mediated gene activation at the primitive streak stage of embryonic development. Once the mesoderm forms, it acts as a source of embryonic RA to direct anteroposterior patterning by regulating HOX gene expression [51]. However, RA must be absent at the time of primitive streak formation for proper body axis patterning [60]. In vivo, maternal sources of RA must be degraded by embryonic cytochrome p450 26 (CYP26) for proper Nodal-mediated body axis patterning to occur [60], suggesting that the presence of ATRA may disrupt primitive streak formation by perturbing NODAL activation. The upstream regulator identified in this dataset, EOMES, has an activin response element in its promoter, supporting the role for Nodal signaling disruption by ATRA [61]. Faial et al. (2015) observed that mesoderm/posterior primitive streak genes increased in expression through BMP4 signaling and noted that antagonism of activin may have similar effects for some EOMES target genes [56]. Other evidence supporting ATRA impacts on Nodal signaling include increased TGFβ2 expression by ATRA [62] and evidence that RARs can interact directly with SMADs. For example, ATRA inhibits phosphorylation of SMAD2/3 [63]. Moreover, Pendaries et al. (2003) identified a direct interaction between RARγ and SMAD3 [64]. Overall, there is evidence that ATRA can disrupt Nodal signaling downstream of ligand activation, which is the simplest explanation of its effects in this cell culture system. Another consideration is whether saturation of ATRA metabolizing enzymes contributed to the tipping point of this system. While expression changes of representative genes (e.g., CYP26A1) did not appear to reflect metabolic saturation (data not shown), enzyme kinetic measurements beyond the scope of this study would be required to better understand the role of ATRA metabolism in this system.
4.6. Relevance
Nominal exposure concentrations were selected to roughly represent a range of potential ATRA exposures in pregnant women. Endogenous ATRA blood plasma levels in women range from 1.16 to 2.22 ng/ml (3.86–7.39 nM) [65]. Lehman and Franz (1996) measured higher levels of ATRA compared to its isomer, 13-cis-RA (isotretinoin) [65]. Isotretinoin is teratogenic in humans at therapeutic doses (0.5–1 mg/kg/day [66]), which correspond roughly to circulating levels of 262–546 ng/ml [67] (872–1817 nM). While isotretinoin is teratogenic in mice and humans, it does not transfer to the placenta as readily as ATRA [68]. Thus, the teratogenicity of 13-cis-RA in humans and mice is likely due to its 4-oxo-isotretinoin metabolite or isomerization to ATRA [69]. Due to differences in metabolism and placental transfer of 13-cis-RA and ATRA, we focused on steady state ATRA plasma levels, rather than total RA, as the best measurement of comparison to our ATRA dosing schemes in vitro. Because defined media that do not contain Vitamin A were used, endogenous production of RA is not expected in this cell culture system. Therefore, the nominal exposure concentrations are expected to approximate the amount of RA reaching the cells. The range of concentrations selected was broad enough to allow the derivation of a tipping point for perturbation of endodermal differentiation.
Women’s blood plasma levels of endogenous ATRA (3.86–7.39 nM) are slightly above the lowest concentration tested in our study (0.001 μM). This similarity is remarkable considering differences are expected between circulating ATRA levels and nominal in vitro exposure concentrations due to differences in cellular uptake and exposure volume. Interestingly, the 0.001 μM nominal exposure concentration induced a subset of genes associated with steroid biosynthesis at 192 h. RA can increase the expression of steroid receptors (e.g., estrogen and vitamin D) in cultured osteosarcoma cells [70]. We therefore hypothesize that in our system, 0.001 μM is near the endogenous level required to initiate steroid mediated developmental processes that were not otherwise detected in DMSO controls at the 192 h stage of cellular differentiation. Endogenous ATRA levels near 0.001 μM would not be expected to negatively impact a developing embryo, consistent with the identification of a tipping point an order of magnitude above endogenous levels. Individuals taking high doses of vitamin A [71] or patients taking isotretinoin for acne vulgaris reach blood plasma levels of RA within the ranges tested in this study. These circulating levels (0.87–1.82 μM) [67] are teratogenic and are within the range of nominal concentrations that caused a shift in the endodermal differentiation trajectory above the tipping point (17 nM) identified in our study. Moreover, when considering pharmacokinetic data from cynomolgus monkeys indicating 40% placental transfer [72], the expected teratogenic ATRA concentrations reaching the fetus (i.e., 0.35–0.73 μM) more closely match the concentrations above the tipping point identified in this study.
Notably, the molecular tipping point identified in this study is not only lower than circulating teratogenic levels, but it is also lower than the IC50 value, 0.07 μM, reported by Kameoka et al. (2014) based on decreased SOX17 expression in hPSCs exposed to ATRA [29], suggesting the transcriptomic tipping point approach may be a more sensitive method to identify a point of departure for differentiating stem cells. While there are differences between the present study and the Kameoka et al. study (e.g., details in the endoderm/mesendoderm differentiation protocols), we would not expect major differences in the tipping point to arise from methodological variations. For example, increasing the amount of activin that is added may result in a shift in the timing of the differentiation trajectory (i.e., identification of a tipping point at an earlier timepoint) without an impact on the tipping point concentration. Additionally, increasing the number of sampling concentrations and timepoints may have the effect of refining the tipping point concentration.
The perturbed endodermal differentiation trajectory identified in our study is hypothesized to impact gut tube-derived organ formation (Fig. 7C). Endogenous RA is required for patterning of the gut tube into organs, with lower levels specifying more anterior organs such as thyroid, and higher levels specifying more posterior organs such as lung or pancreas [73]. While the results of this study do not indicate a clear shift towards posterior organ formation, general impacts on gene markers of gut-tube derived organs are evident. For example, the lung marker secretoglobin family 3A member 2 (SCGB3A2) and the pancreas marker cationic trypsinogen (PRSS1) were among the genes with the highest fold-changes (increased and decreased, respectively) at 0.01 μM ATRA exposure at 96 h versus time-matched controls. Perturbations in gut tube development are predicted to lead to abnormal gut tube-derived organ formation and birth defects. This is evidenced in the work by Wang et al. (2006), who reported defects in foregut-derived lung, liver, and stomach formation in retinaldehyde dehydrogenase 2 (Raldh2) mutants which lacked RA during early embryonic development [74]. In the absence of externally supplemented RA, these organs failed to develop. RA supplementation delayed embryonic mortality and only partially rescued the foregut defects, confirming that controlled RA signaling and maintenance of proper RA levels are essential for proper embryonic development.
5. Conclusions
Taken together, we have characterized an in vitro model of early embryonic development that identified a deleterious dose of RA relevant to human health considerations. ATRA concentrations above the tipping point shift the endodermal differentiation trajectory from embryonic endoderm to mesoderm or extraembryonic endoderm, potentially through a shift in EMT at the primitive streak stage. These perturbations are expected to disrupt gut tube development. This developmental model can be used to screen for developmental toxicants that perturb RA signaling by measuring transcriptomic changes in a selected set of gene markers (Supplemental Table 1) and key HCI markers such as FOXA2, SOX17, TBXT, EOMES, LHX1, and BMP4. TBXT was a particularly robust gene marker that was increased in ATRA samples at 96 h, and decreased in DMSO controls at 96 h (versus 6 h) that would serve well as an HCI marker. Inclusion of SOX17 in HCI experiments will help confirm and characterize the endodermal differentiation trajectory. Future work to screen for developmental toxicants that disrupt Nodal signaling by mimicking ATRA will involve construction of an adverse outcome pathway (AOP) based on Fig. 7B with ligand activation of RARs as the molecular initiating event (MIE).
Supplementary Material
Acknowledgements
Vala Sciences, Inc. performed the chemical exposures and sample collection under U.S. Environmental Protection Agency contract #EPD13054. Cofactor Genomics performed the library construction and RNA-sequencing. All data analysis was conducted at the U.S. Environmental Protection Agency. We thank Shuyun (Lily) Feng for assistance with documenting methods details. We thank David Belair, Logan Everett, John Cowden, and the anonymous reviewers for constructive reviews of the manuscript.
Funding
This work was funded by the Chemical Safety for Sustainability (CSS) Research Program (U.S. EPA/ORD).
Footnotes
Disclaimer
The views expressed in this manuscript are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.reprotox.2019.10.001.
References
- [1].Dix DJ, et al. , The ToxCast program for prioritizing toxicity testing of environmental chemicals, Toxicol. Sci 95 (1) (2007) 5–12. [DOI] [PubMed] [Google Scholar]
- [2].Judson RS, et al. , In vitro screening of environmental chemicals for targeted testing prioritization: the ToxCast project, Environ. Health Perspect 118 (4) (2010) 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kavlock R, et al. , Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management, Chem. Res. Toxicol 25 (7) (2012) 1287–1302. [DOI] [PubMed] [Google Scholar]
- [4].Richard AM, et al. , ToxCast chemical landscape: paving the Road to 21st century toxicology, Chem. Res. Toxicol 29 (8) (2016) 1225–1251. [DOI] [PubMed] [Google Scholar]
- [5].Lawson KA, Meneses JJ, Pedersen RA, Clonal analysis of epiblast fate during germ layer formation in the mouse embryo, Development 113 (3) (1991) 891–911. [DOI] [PubMed] [Google Scholar]
- [6].Shook D, Keller R, Mechanisms, mechanics and function of epithelial-mesenchymal transitions in early development, Mech. Dev 120 (11) (2003) 1351–1383. [DOI] [PubMed] [Google Scholar]
- [7].Hill MA, Embryology Week 3, Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Week_3 (January 30) (2019).
- [8].Viotti M, Nowotschin S, Hadjantonakis AK, SOX17 links gut endoderm morphogenesis and germ layer segregation, Nat. Cell Biol 16 (12) (2014) 1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tam PP, Beddington RS, The formation of mesodermal tissues in the mouse embryo during gastrulation and early organogenesis, Development 99 (1) (1987) 109–126. [DOI] [PubMed] [Google Scholar]
- [10].Hemmati-Brivanlou A, Melton D, Vertebrate embryonic cells will become nerve cells unless told otherwise, Cell 88 (1) (1997) 13–17. [DOI] [PubMed] [Google Scholar]
- [11].Ozair MZ, Kintner C, Brivanlou AH, Neural induction and early patterning in vertebrates, Wiley Interdiscip. Rev. Dev. Biol 2 (4) (2013) 479–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Poulain M, et al. , Zebrafish endoderm formation is regulated by combinatorial Nodal, FGF and BMP signalling, Development 133 (11) (2006) 2189–2200. [DOI] [PubMed] [Google Scholar]
- [13].Piccolo S, et al. , The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals, Nature 397 (6721) (1999) 707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kalluri R, Weinberg RA, The basics of epithelial-mesenchymal transition, J. Clin. Invest 119 (6) (2009) 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Conlon FL, et al. , A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse, Development 120 (7) (1994) 1919–1928. [DOI] [PubMed] [Google Scholar]
- [16].Feldman B, et al. , Zebrafish organizer development and germ-layer formation require nodal-related signals, Nature 395 (6698) (1998) 181–185. [DOI] [PubMed] [Google Scholar]
- [17].Schier AF, Nodal signaling in vertebrate development, Annu. Rev. Cell Dev. Biol 19 (2003) 589–621. [DOI] [PubMed] [Google Scholar]
- [18].Schmierer B, Hill CS, TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility, Nat. Rev. Mol. Cell Biol 8 (12) (2007) 970–982. [DOI] [PubMed] [Google Scholar]
- [19].Winnier G, et al. , Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse, Genes Dev. 9 (17) (1995) 2105–2116. [DOI] [PubMed] [Google Scholar]
- [20].D’Amour KA, et al. , Efficient differentiation of human embryonic stem cells to definitive endoderm, Nat. Biotechnol 23 (12) (2005) 1534–1541. [DOI] [PubMed] [Google Scholar]
- [21].Kubo A, et al. , Development of definitive endoderm from embryonic stem cells in culture, Development 131 (7) (2004) 1651–1662. [DOI] [PubMed] [Google Scholar]
- [22].Loh KM, et al. , Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations, Cell Stem Cell 14 (2) (2014) 237–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Davenport C, et al. , Anterior-posterior patterning of definitive endoderm generated from human embryonic stem cells depends on the differential signaling of retinoic acid, wnt-, and BMP-Signaling, Stem Cells 34 (11) (2016) 2635–2647. [DOI] [PubMed] [Google Scholar]
- [24].Uhlen M, et al. , Proteomics. Tissue-based map of the human proteome, Science 347 (6220) (2015) 1260419. [DOI] [PubMed] [Google Scholar]
- [25].Yasunaga M, et al. , Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells, Nat. Biotechnol 23 (12) (2005) 1542–1550. [DOI] [PubMed] [Google Scholar]
- [26].Burke Z, Oliver G, Prox1 is an early specific marker for the developing liver and pancreas in the mammalian foregut endoderm, Mech. Dev 118 (1–2) (2002) 147–155. [DOI] [PubMed] [Google Scholar]
- [27].Cai J, et al. , Assessing self-renewal and differentiation in human embryonic stem cell lines, Stem Cells 24 (3) (2006) 516–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gouveia A, et al. , Patterning in time and space: HoxB cluster gene expression in the developing chick embryo, Cell Cycle 14 (1) (2015) 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kameoka S, et al. , A high-throughput screen for teratogens using human pluripotent stem cells, Toxicol. Sci 137 (1) (2014) 76–90. [DOI] [PubMed] [Google Scholar]
- [30].Shah I, et al. , Using ToxCast data to reconstruct dynamic cell state trajectories and estimate toxicological points of departure, Environ. Health Perspect 124 (7) (2016) 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lohnes D, et al. , Function of the retinoic acid receptors (RARs) during development (I). Craniofacial and skeletal abnormalities in RAR double mutants, Development 120 (10) (1994) 2723–2748. [DOI] [PubMed] [Google Scholar]
- [32].Mendelsohn C, et al. , Function of the retinoic acid receptors (RARs) during development (II). Multiple abnormalities at various stages of organogenesis in RAR double mutants, Development 120 (10) (1994) 2749–2771. [DOI] [PubMed] [Google Scholar]
- [33].Kochhar DM, Limb development in mouse embryos. I. Analysis of teratogenic effects of retinoic acid, Teratology 7 (3) (1973) 289–298. [DOI] [PubMed] [Google Scholar]
- [34].Lorente CA, Miller SA, Vitamin A induction of cleft palate, Cleft Palate J. 15 (4) (1978) 378–385. [PubMed] [Google Scholar]
- [35].Piersma AH, Hessel EV, Staal YC, Retinoic acid in developmental toxicology: teratogen, morphogen and biomarker, Reprod. Toxicol 72 (2017) 53–61. [DOI] [PubMed] [Google Scholar]
- [36].Bain G, et al. , Embryonic stem cells express neuronal properties in vitro, Dev. Biol. (Basel) 168 (2) (1995) 342–357. [DOI] [PubMed] [Google Scholar]
- [37].Molotkova N, et al. , Requirement of mesodermal retinoic acid generated by Raldh2 for posterior neural transformation, Mech. Dev 122 (2) (2005) 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Durston AJ, et al. , Retinoic acid causes an anteroposterior transformation in the developing central nervous system, Nature 340 (6229) (1989) 140–144. [DOI] [PubMed] [Google Scholar]
- [39].Kamentsky L, et al. , Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software, Bioinformatics 27 (8) (2011) 1179–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].R Core_Team, R: a Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria, 2017. https://www.R-project.org/. [Google Scholar]
- [41].Kassambara A, ggpubr:’ ggplot2′ Based Publication Ready Plots, R package version 0.1.7 (2018) https://CRAN.R-project.org/package=ggpubr.
- [42].Kopylova E, Noe L, Touzet H, SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data, Bioinformatics 28 (24) (2012) 3211–3217. [DOI] [PubMed] [Google Scholar]
- [43].Kim D, Langmead B, Salzberg SL, HISAT: a fast spliced aligner with low memory requirements, Nat. Methods 12 (4) (2015) 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Anders S, Pyl PT, Huber W, HTSeq–a Python framework to work with high-throughput sequencing data, Bioinformatics 31 (2) (2015) 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol. 15 (12) (2014) 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kramer A, et al. , Causal analysis approaches in ingenuity pathway analysis, Bioinformatics 30 (4) (2014) 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Judson R, et al. , Editor’s highlight: analysis of the effects of cell stress and cytotoxicity on in vitro assay activity across a diverse chemical and assay space, Toxicol. Sci 152 (2) (2016) 323–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].NCCT, Toxicology, EPA’s National Center for Computational (2018): ToxCast Database (invitroDB). figshare. Dataset, (2019).
- [49].Kanai-Azuma M, et al. , Depletion of definitive gut endoderm in Sox17-null mutant mice, Development 129 (10) (2002) 2367–2379. [DOI] [PubMed] [Google Scholar]
- [50].Tsankov AM, et al. , Transcription factor binding dynamics during human ES cell differentiation, Nature 518 (7539) (2015) 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Schubert M, et al. , A retinoic acid-Hox hierarchy controls both anterior/posterior patterning and neuronal specification in the developing central nervous system of the cephalochordate amphioxus, Dev. Biol. (Basel) 296 (1) (2006) 190–202. [DOI] [PubMed] [Google Scholar]
- [52].Baum B, Settleman J, Quinlan MP, Transitions between epithelial and mesenchymal states in development and disease, Semin. Cell Dev. Biol 19 (3) (2008) 294–308. [DOI] [PubMed] [Google Scholar]
- [53].Li R, et al. , A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts, Cell Stem Cell 7 (1) (2010) 51–63. [DOI] [PubMed] [Google Scholar]
- [54].Yi CH, et al. , Identification, mapping, and phylogenomic analysis of four new human members of the T-box gene family: EOMES, TBX6, TBX18, and TBX19, Genomics 55 (1) (1999) 10–20. [DOI] [PubMed] [Google Scholar]
- [55].Teo AK, et al. , Pluripotency factors regulate definitive endoderm specification through eomesodermin, Genes Dev. 25 (3) (2011) 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Faial T, et al. , Brachyury and SMAD signalling collaboratively orchestrate distinct mesoderm and endoderm gene regulatory networks in differentiating human embryonic stem cells, Development 142 (12) (2015) 2121–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Costello I, et al. , Lhx1 functions together with Otx2, Foxa2, and Ldb1 to govern anterior mesendoderm, node, and midline development, Genes Dev. 29 (20) (2015) 2108–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gudas LJ, Retinoids and vertebrate development, J. Biol. Chem 269 (22) (1994) 15399–15402. [PubMed] [Google Scholar]
- [59].Leid M, et al. , Retinoic acid signal transduction pathways, Ann. N. Y. Acad. Sci 684 (1993) 19–34. [DOI] [PubMed] [Google Scholar]
- [60].Uehara M, et al. , Removal of maternal retinoic acid by embryonic CYP26 is required for correct Nodal expression during early embryonic patterning, Genes Dev. 23 (14) (2009) 1689–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Ryan K, et al. , The Xenopus eomesodermin promoter and its concentration-dependent response to activin, Mech. Dev 94 (1–2) (2000) 133–146. [DOI] [PubMed] [Google Scholar]
- [62].Han GR, et al. , All-trans-retinoic acid increases transforming growth factor-beta2 and insulin-like growth factor binding protein-3 expression through a retinoic acid receptor-alpha-dependent signaling pathway, J. Biol. Chem 272 (21) (1997) 13711–13716. [DOI] [PubMed] [Google Scholar]
- [63].Lee SH, et al. , The effects of retinoic acid and MAPK inhibitors on phosphorylation of Smad2/3 induced by transforming growth factor beta1, Tuberc. Respir. Dis. (Seoul) 82 (1) (2019) 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Pendaries V, et al. , Retinoic acid receptors interfere with the TGF-beta/Smad signaling pathway in a ligand-specific manner, Oncogene 22 (50) (2003) 8212–8220. [DOI] [PubMed] [Google Scholar]
- [65].Lehman PA, Franz TJ, A sensitive high-pressure liquid chromatography/particle beam/mass spectrometry assay for the determination of all-trans-retinoic acid and 13-cis-retinoic acid in human plasma, J. Pharm. Sci 85 (3) (1996) 287–290. [DOI] [PubMed] [Google Scholar]
- [66].Agarwal US, Besarwal RK, Bhola K, Oral isotretinoin in different dose regimens for acne vulgaris: a randomized comparative trial, Indian J. Dermatol. Venereol. Leprol 77 (6) (2011) 688–694. [DOI] [PubMed] [Google Scholar]
- [67].Almond-Roesler B, et al. , Monitoring of isotretinoin therapy by measuring the plasma levels of isotretinoin and 4-oxo-isotretinoin. A useful tool for management of severe acne, Dermatology 196 (1) (1998) 176–181. [DOI] [PubMed] [Google Scholar]
- [68].Kraft JC, et al. , Low teratogenicity of 13-cis-retinoic acid (isotretinoin) in the mouse corresponds to low embryo concentrations during organogenesis: comparison to the all-trans isomer, Toxicol. Appl. Pharmacol 87 (3) (1987) 474–482. [DOI] [PubMed] [Google Scholar]
- [69].Nau H, Teratogenicity of isotretinoin revisited: species variation and the role of alltrans-retinoic acid, J. Am. Acad. Dermatol 45 (5) (2001) S183–7. [DOI] [PubMed] [Google Scholar]
- [70].Suzuki S, et al. , Effects of retinoic acid on steroid and vitamin D3 receptors in cultured mouse osteosarcoma cells, Bone 14 (1) (1993) 7–12. [DOI] [PubMed] [Google Scholar]
- [71].Eckhoff C, Nau H, Vitamin A supplementation increases levels of retinoic acid compounds in human plasma: possible implications for teratogenesis, Arch. Toxicol 64 (6) (1990) 502–503. [DOI] [PubMed] [Google Scholar]
- [72].Tzimas G, et al. , Retinoid metabolism and transplacental pharmacokinetics in the cynomolgus monkey following a nonteratogenic dosing regimen with all-trans-retinoic acid, Teratology 54 (5) (1996) 255–265. [DOI] [PubMed] [Google Scholar]
- [73].Bayha E, et al. , Retinoic acid signaling organizes endodermal organ specification along the entire antero-posterior axis, PLoS One 4 (6) (2009) e5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wang Z, et al. , Retinoic acid regulates morphogenesis and patterning of posterior foregut derivatives, Dev. Biol. (Basel) 297 (2) (2006) 433–445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.