Abstract
Staphylococcus aureus is one of the most common microorganisms and causes foodborne diseases. In particular, biofilm-forming S. aureus is more resistant to antimicrobial agents and sanitizing treatments than planktonic cells. Therefore, this study aimed to investigate the anti-biofilm effects of cell-free supernatant (CFS) of Saccharomyces cerevisiae isolated from cucumber jangajji compared to grapefruit seed extract (GSE). CFS and GSE inhibited and degraded S. aureus biofilms. The adhesion ability, auto-aggregation, and exopolysaccharide production of CFS-treated S. aureus, compared to those of the control, were significantly decreased. Moreover, biofilm-related gene expression was altered upon CFS treatment. Scanning electron microscopy images confirmed that CFS exerted anti-biofilm effects against S. aureus. Therefore, these results suggest that S. cerevisiae CFS has anti-biofilm potential against S. aureus strains.
Keywords: Anti-biofilm, Saccharomyces cerevisiae, cell-free supernatant, grapefruit seed extract, Staphylococcus aureus
Introduction
Foodborne diseases caused by pathogens pose a great threat to human health. According to the Center for Disease Control and Prevention (CDC), foodborne diseases are caused by Salmonella spp., Clostridium perfringens, Campylobacter spp., and Staphylococcus aureus, the most common microorganisms found in food eaten in the United States [1].
Most foodborne pathogens can form biofilms on various surfaces, such as stainless steel, glass, polyurethane, and rubber [2,3]. Biofilms formed by foodborne pathogens have become a major problem in meat, dairy, and fish processing, as well as in the ready-to-eat (RTE) food industry. Biofilm formation is an important factor in the cross-contamination and persistence of foodborne pathogens [4]. It is difficult to remove foodborne pathogens once they form biofilms because biofilm cells are more resistant to antimicrobial agents and sanitizing treatments than planktonic cells [5]. Biofilms are surrounded by extracellular polymeric substances, which are composed of exopolysaccharides (EPS), proteins, lipids, and extracellular DNA [6]. Extracellular polymeric substances protect bacterial cells from the effects of antimicrobial agents and assist in retaining the structural integrity of the biofilm.
S. aureus is a major foodborne pathogen that causes 241,148 illnesses per year [7] and forms biofilms on food and food contact surfaces [8]. Moreover, biofilm formation by S. aureus plays a crucial role in staphylococcal infections by protecting the colony from the host immune system and antimicrobial treatment [9]. Therefore, the prevention of S. aureus biofilm formation has become important for countering foodborne staphylococcal outbreaks.
Since antibiotics are limited in their ability to inhibit S. aureus biofilms, many studies on natural antimicrobial agents and compounds produced using probiotics have been performed [10-13]. For example, some natural preservatives, such as grapefruit seed extract (GSE), have been used together with antimicrobial and anti-biofilm agents against Gram-positive and Gram-negative bacteria [14]. Moreover, some probiotics consisting of lactic acid bacteria have been reported to inhibit biofilm formation by Listeria monocytogenes [4], Pseudomonas aeruginosa [15], Bacillus licheniformis [16], Vibrio spp. [17], and methicillin-resistant S. aureus (MRSA) [18].
Saccharomyces cerevisiae has been reported that the probiotic yeast isolated from fermented food such as kefir, wine, and cucumber jangajji (Korean fermented food) [19,20]. The positive effects of S. cerevisiae, including antioxidant, anti-inflammatory, toxin eradication, and antagonistic activities, have been investigated [21-23]. In particular, the supernatant and lysate of S. cerevisiae cultures exerted an anti-biofilm effect against S. aureus, as demonstrated by the expression of the α-hemolysin and enterotoxin A genes (hla and sea). However, the effects of S. cerevisiae on the cell surface characteristics, EPS production, and expression of biofilm-related genes in S. aureus have not yet been investigated.
Therefore, the present study aimed to investigate the anti-biofilm effects of cell-free supernatant (CFS) of S. cerevisiae isolated from cucumber jangajji against S. aureus. The anti-biofilm mechanisms were evaluated by investigating the differences in cell surface characteristics (adhesion ability, auto-aggregation, and hydrophobicity), EPS production, and biofilm-related gene expression compared to treatment with GSE. Moreover, S. aureus biofilms formed on glass coupons were observed by scanning electron microscopy (SEM).
Materials and Methods
Strains and Growth Conditions
Five strains of S. cerevisiae (KU200270, KU200278, KU200280, KU200281, and KU200284) isolated from cucumber jangajji were used in this study [19,20]. Nine strains of S. aureus (P130, P131, P86, P87, P88, P89, ATCC 6538, ATCC 12692, and ATCC 25923) were screened to investigate their biofilm-forming capacity. S. cerevisiae strains were cultured in yeast mold (YM) broth (Difco Laboratories, USA) at 25°C for 48 h and S. aureus was cultured in tryptic soy broth (TSB; Difco Laboratories) at 37°C for 24 h. All strains were stored at -80°C in a 20%glycerol solution.
CFS Preparation
S. cerevisiae strains were inoculated in YM broth and cultures were agitated at 150 rpm at 25°C for 48 h. After incubation, S. cerevisiae culture medium was centrifuged at 15,420 ×g at 4°C for 15 min and the pH was adjusted to 6.5 ± 0.3 using 1 M NaOH. Then, the supernatant was filtered through a syringe filter (0.45-μm pore size)(Advantec, Japan) and stored at -80°C.
Evaluation of Anti-Biofilm Effect by Minimum Inhibitory Concentration (MIC)
To investigate the MIC of GSE (ES Food, Korea), the double broth dilution method was used [14]. CFS and GSE were dissolved in YM broth and YM broth with 0.1% Tween 80, respectively, and serially diluted two-fold. S. aureus were cultured in TSB at 37°C for 24 h and then diluted in TSB to obtain a final concentration of 105 colony-forming units (CFU)/ml. Bacterial cultures (50 μl) in TSB supplemented with 50 μl of CFS or GSE in YM broth were transferred to 96-well polystyrene plates (SPL, Korea), which were incubated at 37°C for 24 h. After incubation, the lowest concentrations of CFS and GSE that could inhibit visible S. aureus growth were defined as MICs.
Biofilm Inhibition Assay and Biofilm Degradation Assay
Biofilm inhibition and degradation by CFS were investigated by crystal violet (CV) assay [24]. To this purpose, the strains were treated with CFS and GSE at 1/2 × MIC based on the MIC related to each strains.
To examine the biofilm inhibition effects of CFS and GSE, bacterial suspension (50 μl) treated with CFS or GSE (50 μl) were transferred to a 96-well polystyrene plate and incubated at 37°C for 24 h; the control was treated with YM broth. Following incubation, cell suspensions were removed, and the wells were washed twice with 150 μl of distilled water (DW). The biofilm cells were dried at 37°C for 20 min and then stained using 1% CV solution (150 μl) for 30 min. The CV solution was removed, and the plate was washed twice with cold water. The biofilm cells were treated with dissolving solution (150 μl; 30% methanol and 10% acetic acid) to measure the optical density (OD) of the CV solution at 570 nm using a microplate reader (Molecular Devices, USA).
To examine the biofilm degradation effects of CFS and GSE, bacterial suspensions (100 μl) were transferred to a 96-well polystyrene plate and incubated at 37°C for 24 h. Following incubation, the bacterial suspensions were removed and treated with CFS or GSE (100 μl). The control was treated with YM broth. After incubation at 37°C for 24 h, biofilm quantification was performed as described above.
The biofilm inhibition and degradation rate (%) was calculated using the following equation:
Biofilm inhibition and degradation rate (%) = (1-ODtreatment/ODcontrol) × 100
Adhesion Ability to the Glass Surface
The adhesion ability to S. aureus strains to the glass surface was determined according to Islam et al. [25] with some modifications. Briefly, bacterial cultures (5 ml) diluted in TSB to obtain a final concentration of 106 CFU/ml were transferred to a glass tube and mixed with CFS or GSE (5 ml) at 1/2 × MIC; the control was treated with YM broth. Then, the glass tubes were placed at an angle of 30° and incubated at 37°C for 24 h. Following incubation, the planktonic cells was removed, and the tube was washed with phosphate buffered solution (PBS, pH 7.4; Hyclone, USA). The adhered cells were washed with PBS and re-suspended in fresh PBS (10 ml). The absorbance of the adhered cells was measured at 600 nm (ODadhered cell). Then, the adhered cells were mixed with the planktonic cells and washed with PBS, and the absorbance of the cell mixture was measured at 600 nm (ODcell mixture). The adhesion ability (%) was calculated using the following equation:
Adhesion ability (%) = (ODadhered cell/ODcell mixture) × 100
Auto-Aggregation Ability Assay
The auto-aggregation ability of S. aureus was assayed according to the methods described by Song et al. [14] with some modifications. Briefly, bacterial cells treated with CFS or GSE at 1/2 × MIC, or YM broth were incubated at 37°C for 24 h. Following incubation, the cell suspensions were centrifuged at 17,709 ×g at 4°C for 5 min and washed twice with PBS. The washed cells were resuspended in fresh PBS, and the OD600 was adjusted to 0.5 ± 0.05 (ODinitial). Absorbance-adjusted cell suspensions (4 ml) were incubated at 37°C for 8 h. After incubation, the absorbance of the upper layer was measured at 600 nm (ODtreated). Auto-aggregation (%) was calculated using the following equation:
Auto-aggregation (%) = (1- ODtreated /ODinitial) × 100
Hydrophobicity Assay
Hydrophobicity was investigated according to a previously described protocol [24]. Absorbance-adjusted cells were prepared as described above (ODinitial). Then, cell suspensions (2 ml) were mixed with chloroform (0.5 ml), and the tubes were vortexed for 2 min. Following incubation for 15 min, the absorbance of the aqueous layer was measured at 600 nm (ODtreated). Hydrophobicity (%) was calculated using the following equation:
Hydrophobicity (%) = (1- ODtreated/ODinitial) × 100
EPS Production
To examine EPS production, the phenol-sulfuric acid method was performed as described by Chiba et al. [26]. Bacterial cultures diluted in TSB to obtain a final concentration of 107 CFU/ml were mixed with CFS or GSE (5 ml) at 1/2 × MIC and incubated at 37°C for 24 h. The control was treated with YM broth. After incubation, cell suspensions were centrifuged at 8,000 ×g at 25°C for 10 min and then 1.5 M NaCl (1 ml) was added. Then, the cell suspension was re-centrifugated at 5,000 ×g at 25°C for 10 min, and the supernatants (60 μl) were mixed with 5%phenol (60 μl) and sulfuric acid (4 ml). After incubation at 30°C for 10 min, OD490 was measured. EPS production (%) was calculated using the following equation:
EPS quantification (%) = (ODtreatment/ODcontrol) × 100
Biofilm-Related Gene Expression
S. aureus ATCC 12692 was cultured in TSB at 37°C for 24 h, and the bacterial cultures were diluted in TSB to obtain a final concentration of 106 CFU/ml. The diluted cell suspensions were treated with CFS of S. cerevisiae KU200278 or GSE at 1/2 × MIC; the control was treated with YM broth. After treatment, total RNA was extracted using TRIzol (Invitrogen, Thermo Fisher Scientific, USA) and complementary DNA (cDNA) was synthesized using the cDNA synthesis kit. Then, reverse transcription-polymerase chain reaction real-time PCR (RT-PCR) was performed to evaluate the expression of biofilm-related genes using specific primers (Table 2). The results were calculated using the ΔΔCt method.
Table 2.
Sequences of primers used for real-time PCR analysis.
| Gene | Primers sequence (5’to 3’) | Reference |
|---|---|---|
| icaA | F: CTGGCGCAGTCAATACTATTTCGGGTGTCT | [31] |
| R: GACCTCCCAATGTTTCTGGAACCAACATCC | ||
| icaD | F: ACCCAACGCTAAAATCATCG | [30] |
| R: GCGAAAATGCCCATAGTTTC | ||
| icaR | F: CAATAATCTAATACGCCTGAG | [28] |
| R: AGTAGCGAATACACTTCATCT | ||
| agrA | F: TGATAATCCTTATGAGGT GCTT | [31] |
| R: CACTGTGACTCGTAACGAAAA | ||
| sarA | F: CAAACAACCACAAGTTGTTAAAGC | [31] |
| R: TGTTTGCTTCAGTGATTCGTTT | ||
| sigB | F: AGTGTTAGAAGCAATGGAAATG | [32] |
| R: CGATACGCTCACCTGTCTCT | ||
| 16S rRNA | F: ACTCCTACGGGAGGCAGCAG | [31] |
| R: ATTACCGCGGCTGCTGG |
SEM Analysis
To investigate morphological changes in treated bacterial cells, SEM analysis of S. aureus ATCC 12692 treated with CFS of S. cerevisiae KU200278 or GSE at 1/2 × MIC on a glass coupon was performed. Prior to microscopic analysis, S. aureus ATCC 12692 was cultured in TSB at 37°C for 24 h, and then the grown bacterial culture was diluted in TSB to obtain a final concentration of 106 CFU/ml. To estimate the biofilm inhibition effects of CFS and GSE, bacterial cultures (2 ml) were treated with CFS or GSE on glass coupons. The biofilm degradation effects of CFS and GSE were determined on glass coupons using pre-formed biofilms treated with CFS or GSE. The control was treated with YM broth. Following treatment, planktonic cells were washed twice with PBS, and biofilm cells were fixed with 2.5% glutaraldehyde in PBS at 4°C for 1 h. Then, the fixed samples were washed twice with PBS and dehydrated using ethanol at graded concentrations (50%, 70%, 80%, 90%, and 100%) for 15 min (for each concentration). After dehydration, ethanol was replaced with isoamyl acetate, and the samples were freeze-dried for 48 h. To observe the biofilm structure, the samples were coated with gold (15 mV for 1.5 min) and observed using a field emission scanning electron microscope (FESEM; SU8010; Hitachi High-Technologies Co., Japan).
Statistical Analysis
All experiments were performed in triplicate. Statistical analysis was performed by SPSS version 18.0 (SPSS Inc., USA). The results were presented as the mean ± standard error. Significant differences among the mean values were evaluated by one-way analysis of variance (ANOVA).
Results and Discussion
MIC of CFS for S. aureus
Among the nine S. aureus strains, three strains (ATCC 6538, ATCC 12692, and ATCC 25923) showed high biofilm-forming capacity (data not shown) and were hence used for further study. The antimicrobial effects of CFS and GSE against S. aureus were confirmed; however, CFS did not inhibit the growth of S. aureus. On the other hand, the MIC of GSE that exerts antimicrobial effects against S. aureus ATCC 6538, ATCC 12692, and ATCC 25923 was 100, 50, and 200 μg/ml, respectively (Table 1). According to Heggers et al. [27], GSE damages the cell membrane, degrades cytoplasmic components, and finally induces the cell death. S. cerevisiae was reported on anti-biofilm effects by diverse enzyme, peptides, and antibiotics [12]. Conversely, the anti-biofilm effects of S. cerevisiae have been reported to depend on the activity of diverse enzymes, peptides, and antibiotics [12]. Therefore, S. cerevisiae CFS is supposed to show better anti-biofilm effects compared to GSE.
Table 1.
Minimum inhibitory concentration (MIC) of grapefruit seed extract against S. aureus.
| Strain | MIC (μg/ml) |
|---|---|
| S. aureus ATCC 6538 | 100 |
| S. aureus ATCC 12692 | 50 |
| S. aureus ATCC 25923 | 200 |
All experiments were performed in triplicates.
Effect of CFS on Biofilm Inhibition and Degradation
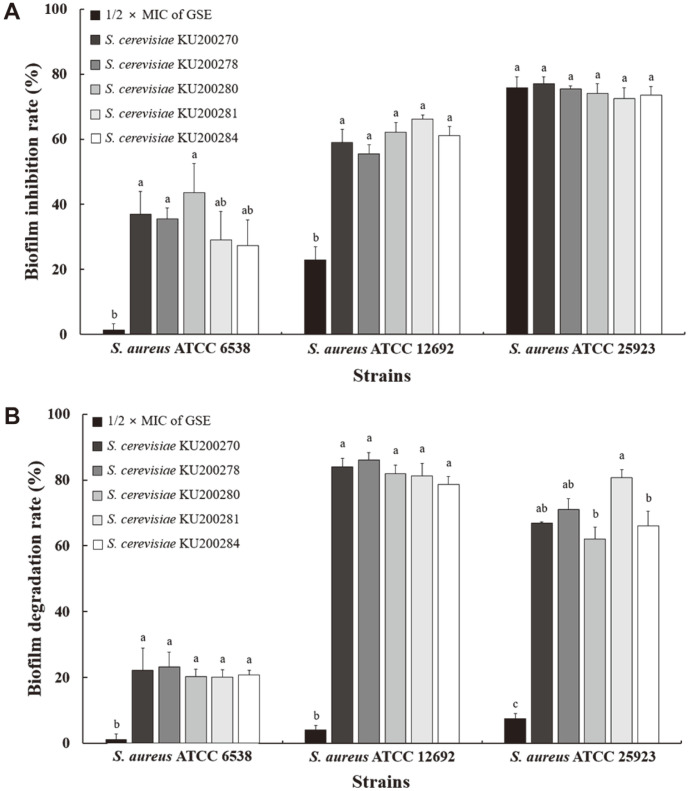
To compare the effect of CFS and GSE on bacteria, cells were treated with CFS or GSE at 1/2 × MIC [24,28]. The inhibitory effects of CFS and GSE on S. aureus biofilm are shown in Fig. 1A. CFS and GSE significantly inhibited biofilm formation by the three S. aureus strains (p < 0.05). Interestingly, the biofilm inhibition rates of CFS-treated S. aureus ATCC 6538 and ATCC 12692 ranged from 27.27% to 43.66% and from 55.55% to 66.29%, respectively. On the other hand, GSE treatment inhibited S. aureus ATCC 6538 and ATCC 12692 biofilm formation by 1.39%and 22.96%, respectively. No significant difference was observed between CFS- and GSE-dependent biofilm inhibition of S. aureus ATCC 25923 (p > 0.05). The effects of CFS and GSE on S. aureus biofilm degradation are shown in Fig. 1B. The mature S. aureus biofilms were significantly degraded by both CFS and GSE (p < 0.05). In particular, CFS degraded S. aureus biofilms at a rate ranging from 20.01% to 86.04%, whereas GSE degraded the biofilms at a rate ranging from 1.16% to 7.53%.
Fig. 1. Effects of cell-free supernatant (CFS) of S. cerevisiae strains on S. aureus biofilms.
(A) Biofilm inhibition effect of grapefruit seed extract (GSE) at 1/2 × minimum inhibitory concentration (MIC) and CFS; (B) biofilm degradation effect of GSE at 1/2 × MIC and CFS. The MIC of GSE against S. aureus ATCC 6538, S. aureus ATCC 12692, and S. aureus ATCC 25923 was 50, 25, and 100 μg/ml, respectively. All experiments were performed in triplicates, and reported values represent the mean ± standard error. a-cDifferent letters in the same bar indicate significant differences (p < 0.05).
Similar to our results, previous reports showed that the supernatant and lysate of S. cerevisiae cultures inhibited S. aureus and P. aeruginosa biofilm formation [12, 29]. Moreover, the supernatant and lysate of S. cerevisiae exerted anti-biofilm effects against S. aureus, inhibiting biofilm formation of methicillin-sensitive S. aureus (MSSA) and MRSA strains by 48% and 69%, respectively. Mannoproteins, components of the S. cerevisiae cell wall, also inhibited biofilm formation and dispersion. Moreover, the anti-biofilm effects of GSE against S. aureus and Escherichia coli were also investigated [14]. Notably, 1/2 × MIC GSE-treated S. aureus and E. coli displayed inhibited biofilm formation and maturation. In particular, GSE at 1/2 × MIC showed inhibition rates of S. aureus and E. coli biofilm of 44.2% and 29.8%, respectively, and biofilm degradation rates of 35.2% and 36.9%, respectively.
Cell Surface Characteristics and EPS Production of CFS-treated S. aureus
To investigate the anti-biofilm mechanisms of CFS, the adhesion ability, auto-aggregation ability, hydrophobicity, and EPS production of S. aureus were examined. Table 3 shows the characteristics of S. aureus strains treated with CFS of S. cerevisiae KU200278, which showed the most effective anti-biofilm potential among CFSs from the yeast strains or 1/2 × MIC GSE. Adhesion, auto-aggregation, hydrophobicity, and EPS production are related to bacterial attachment and biofilm formation rate [30, 31].
Table 3.
Effects of cell-free supernatant (CFS) of S. cerevisiae KU200278 on cell surface characteristics (adhesion ability, auto-aggregation ability, and hydrophobicity) and exopolysaccharide (EPS) production of S. aureus.
| Treatment | Control | GSE at 1/2 × MIC | CFS |
|---|---|---|---|
| Adhesion ability (%) | |||
| S. aureus ATCC 6538 | 88.79±0.30b | 89.86±0.80c | 58.29±0.66a |
| S. aureus ATCC 12692 | 94.32±0.97b | 98.38±0.48c | 64.02±0.16a |
| S. aureus ATCC 25923 | 85.28±0.00c | 48.42±0.56a | 72.95±0.53b |
| Auto-aggregation (%) | |||
| S. aureus ATCC 6538 | 51.03±0.13c | 47.52±0.48b | 32.10±0.76a |
| S. aureus ATCC 12692 | 73.54±0.36c | 69.15±0.31b | 53.52±0.56a |
| S. aureus ATCC 25923 | 65.05±0.57c | 60.57±0.55b | 31.56±0.49a |
| Hydrophobicity (%) | |||
| S. aureus ATCC 6538 | 92.36±0.00a | 93.14±0.24b | 92.97±0.06ab |
| S. aureus ATCC 12692 | 94.03±0.00a | 94.11±0.17a | 94.08±0.13a |
| S. aureus ATCC 25923 | 87.04±0.16a | 89.73±0.16b | 87.46±0.14a |
| EPS quantification (%) | |||
| S. aureus ATCC 6538 | 100±0.45c | 41.96±0.45b | 35.27±1.18a |
| S. aureus ATCC 12692 | 100±1.00c | 58.00±0.50b | 26.50±0.50a |
| S. aureus ATCC 25923 | 100±0.55c | 63.44±1.16b | 41.22±0.82a |
1/2 × minimum inhibitory concentration (MIC) of grapefruit seed extract (GSE) (S. aureus ATCC 6538, 50 μg/ml; S. aureus ATCC 12692, 25 μg/ml; S. aureus ATCC 25923, 100 μg/ml).
All experiments were performed in triplicates, and reported values represent the mean ± standard error.
a-cValues with different letters in the same column are significantly different (p < 0.05).
CFS treatment of all tested strains and GSE treatment of S. aureus ATCC 25923, compared with the control treatment (85.28%-94.32%), significantly decreased the adhesion ability to the glass surface (48.42%-72.95%)(p < 0.05). However, GSE treatment of S. aureus ATCC 6538 and ATCC 12692 significantly increased the adhesion ability (p < 0.05). On the other hand, auto-aggregation and EPS production of S. aureus treated with CFS or GSE at 1/2 × MIC, compared to those of the control, significantly decreased (p < 0.05). Specifically, S. aureus strains treated with CFS showed significantly reduced auto-aggregation and EPS production with respect to strains treated with GSE at 1/2 × MIC (p < 0.05). However, the effects of treatments on S. aureus hydrophobicity differed depending on the strain. Nevertheless, CFS did not significantly affect the hydrophobicity of any of the tested strains (p > 0.05).
Biofilm formation is followed by adhesion to the surface, auto-aggregation, attachment via hydrophobic interactions, and EPS production [3, 32, 33]. Therefore, CFS and GSE affected all steps of biofilm formation: 1) the initial step of adhesion, attachment, and biofilm formation of S. aureus; 2) the step of attachment by auto-aggregation and establishment of hydrophobic interactions; and 3) the step of maturation by EPS production.
Effect of CFS on Expression of Biofilm-Related Gene
The effects of CFS on the expression of biofilm-related genes of S. aureus were determined by qRT-PCR. Polysaccharide intercellular adhesin (PIA), a component of the polysaccharide slime of S. aureus, is synthesized by the intercellular adhesion (ica) operon and plays an important role in biofilm formation and adhesion [34]. The ica operon consists of the icaABCD cluster and icaR; among operon components, icaA is the essential regulator of icaD expression, which mediates PIA biosynthesis [32].
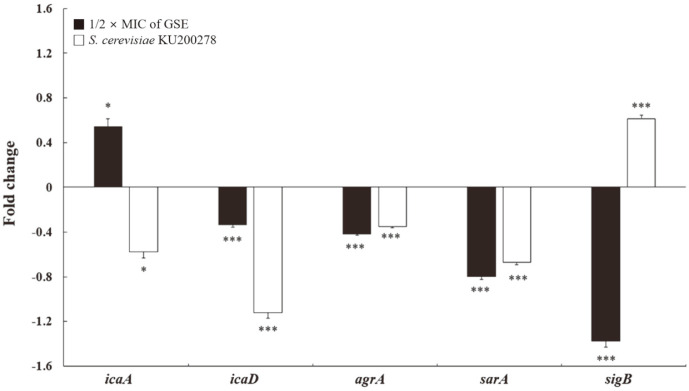
As shown in Fig. 2, CFS and GSE at 1/2 × MIC altered the expression of ica operon genes in S. aureus. Indeed, the expression of icaA and icaD in CFS-treated S. aureus was significantly downregulated (p < 0.05), whereas the expression of icaA in GSE-treated S. aureus was significantly upregulated (p < 0.05). However, the expression of icaR in S. aureus treated with GSE or CFS did not significantly change compared with the control (data not shown). Therefore, CFS altered icaA and icaD expression by decreasing biofilm formation and adhesion ability.
Fig. 2. Effects of cell-free supernatant (CFS) of S. cerevisiae KU200278 on gene expression of S. aureus ATCC 12692.
All experiments were performed in triplicates, and reported values represent the mean ± standard error (*p < 0.05, **p < 0.01, ***p < 0.001). Grapefruit seed extract (GSE) at 1/2 × minimum inhibitory concentration (MIC); icaA and icaD, intercellular adhesion; agrA, accessory gene regulator; sarA, Staphylococcal accessory regulator; sigB, modulator of the expression of the virulence gene agrA and sarA.
The virulence genes accessory gene regulator (agrA) and staphylococcal accessory regulator (sarA) also affect biofilm formation [8]. In fact, agrA and sarA regulate the quorum sensing system and the secretion of adhesion factors, thereby inhibiting initial adhesion and cell-cell interactions, respectively [31,32]. Notably, sigB modulates the expression of agrA and sarA during stress response [32].
Fig. 2 shows that CFS or GSE treatment triggered the downregulation of agrA and sarA expression in S. aureus. In particular, agrA expression was significantly downregulated by 0.35-fold and 0.42-fold upon CFS and GSE treatment, respectively. Moreover, sarA expression was significantly downregulated by 0.67-fold and 0.79-fold upon CFS and GSE treatment, respectively. However, sigB expression was upregulated by 0.61-fold upon CFS treatment, while being downregulated by 1.37-fold upon GSE treatment. Therefore, agrA and sarA downregulation and sigB upregulation could affect the inhibition and degradation of S. aureus biofilm.
SEM Analysis of S. aureus ATCC 12692 on Glass Surface
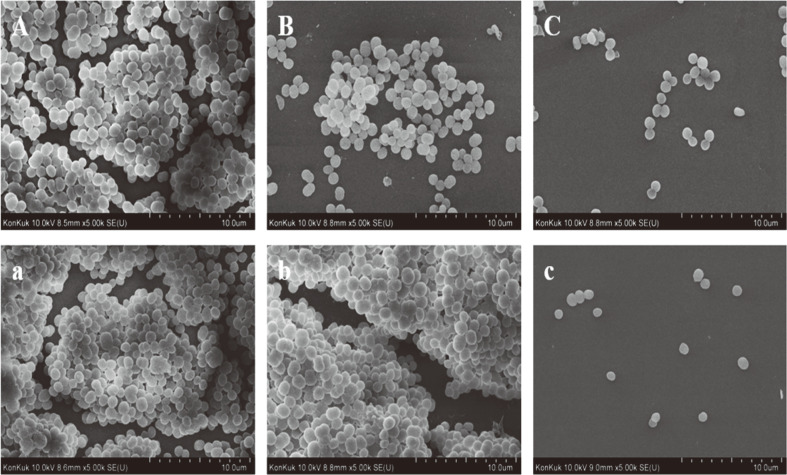
To confirm the morphological changes of S. aureus ATCC 12692 treated with CFS of S. cerevisiae KU200278, SEM analysis was performed (Fig. 3). The biofilm inhibition and degradation effects of CFS and GSE were also investigated. In the control S. aureus groups, complex and clumped biofilm structures with thick and multiple layers were formed (Figs. 3A and 3a). On the other hand, CSF-treated biofilms were single layered and showed reduced attachment of S. aureus (Figs. 3B and 3b). Moreover, the images of S. aureus treated with GSE at 1/2 × MIC were well correlated with the measured anti-biofilm effects. Indeed, GSE inhibited biofilm formation by decreasing cell attachment and aggregation (Fig. 3C). Nevertheless, GSE treatment, unlike CFS treatment, did not trigger S. aureus biofilm degradation (Fig. 3c). In fact, GSE-treated mature biofilms showed clumped, complex, and multi-layered structures. These results indicated that CFS from S. cerevisiae KU200278 showed considerable anti-biofilm potential against S. aureus.
Fig. 3. Scanning electron microscopy analysis of S. aureus ATCC 12692 treated with cell-free supernatant (CFS) of S. cerevisiae KU200278 on glass coupons (× 10,000 magnification).
(A-C) Biofilm inhibition; (A) control; (B) treatment with grapefruit seed extract (GSE) at 1/2 × minimum inhibitory concentration (MIC); (C) CFS treatment; (a-c), biofilm degradation; (a) control; (b) treatment with GSE at 1/2 × MIC; (c) CFS treatment.
In conclusion, this study investigated the anti-biofilm effect of CFS of a S. cerevisiae isolate against S. aureus. CFS significantly inhibited biofilm formation and degraded mature biofilms of S. aureus strains. Notably, the adhesion ability, auto-aggregation ability, and EPS production of CFS-treated S. aureus, compared to those of the control, were significantly decreased. Furthermore, RT-PCR showed that CFS altered biofilm-related gene expression. Finally, SEM images confirmed that CFS inhibited and degraded S. aureus biofilms. Altogether, these results suggested that CFS of S. cerevisiae isolated from cucumber jangajji showed anti-biofilm potential against MSSA and MRSA strains.
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
REFERENCES
- 1.Center for Disease Control and Prevention (CDC) Foodborne germs and illnesses. 2020. [Accessed Mar. 18, 2020.]. Available from https://www.cdc.gov/foodsafety/foodborne-germs.html .
- 2.Meeslip N, Mesil N. Effect of microbial sanitizers for reducing biofilm formation of Staphylococcus aureus and Pseudomonas aeruginosa on stainless steel by cultivation with UHT milk. Food Sci. Biotechnol. 2019;28:289–296. doi: 10.1007/s10068-018-0448-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song H, Lee SY. Resistance of pathogenic biofilms on glass fiber filters formed under different conditions. Food Sci. Biotechnol. 2020;29:1241–1250. doi: 10.1007/s10068-020-00773-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hossain MI, Mizan MFR, Ashrafudoulla M, Nahar S, Joo HJ, Jahid IK, et al. Inhibitory effects of probiotic potential lactic acid bacteria isolated from kimchi against Listeria monocytogenes biofilm on lettuce, stainless-steel surfaces, and MBEC™ biofilm device. LWT-Food Sci. Technol. 2020;118:108864. doi: 10.1016/j.lwt.2019.108864. [DOI] [Google Scholar]
- 5.Nguyen HDN, Yang YS, Yuk HG. Biofilm formation of Salmonella Typhimurium on stainless steel and acrylic surfaces as affected by temperature and pH level. LWT-Food Sci. Technol. 2014;55:383–388. doi: 10.1016/j.lwt.2013.09.022. [DOI] [Google Scholar]
- 6.Cao Y, Naseri M, He Y, Xu C, Walsh LJ, Ziora ZM. Non-antibiotic antimicrobial agents to combat biofilm-forming bacteria. J. Glob. Antimicrob. Resist. 2020;21:445–451. doi: 10.1016/j.jgar.2019.11.012. [DOI] [PubMed] [Google Scholar]
- 7.Center for Disease Control and Prevention (CDC) Burden of Foodborne Illness: Findings. 2011. [Accessed Nov. 05, 2018]. Available from https://www.cdc.gov/foodborneburden/2011-foodborne-estimates.html .
- 8.Farha AK, Yang QQ, Kim G, Zhang D, Mavumengwana V, Habimana O, et al. Inhibition of multidrug-resistant foodborne Staphylococcus aureus biofilms by a natural terpenoid (+)-nootkatone and related molecular mechanism. Food Control. 2020;112:107154. doi: 10.1016/j.foodcont.2020.107154. [DOI] [Google Scholar]
- 9.Olia AHG, Ghahremani M, Ahmadi A, Sharifi Y. Comparison of biofilm production and virulence gene distribution among community-and hospital-acquired Staphylococcus aureus isolates from northwestern Iran. Infect. Genet. Evol. 2020;81:104262. doi: 10.1016/j.meegid.2020.104262. [DOI] [PubMed] [Google Scholar]
- 10.Pontes EKU, Melo HM, Nogueira JWA, Firmino NCS, de Carvalho MG, Catunda FEA, Cavalcant TTA. Antibiofilm activity of the essential oil of citronella (Cymbopogon nardus) and its major component, geraniol, on the bacterial biofilms of Staphylococcus aureus. Food Sci. Biotechnol. 2019;28:633–639. doi: 10.1007/s10068-018-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olszewska MA, Gędas A, Simões M. Antimicrobial polyphenol-rich extracts: applications and limitations in the food industry. Food. Res. Int. 2020;134:109214. doi: 10.1016/j.foodres.2020.109214. [DOI] [PubMed] [Google Scholar]
- 12.Saidi N, Owlia P, Marashi SMA, Saderi H. Inhibitory effect of probiotic yeast Saccharomyces cerevisiae on biofilm formation and expression of α-hemolysin and enterotoxin A genes of Staphylococcus aureus. Iran J. Microbiol. 2019;11:246–254. doi: 10.18502/ijm.v11i3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan X, Gu S, Cui X, Shi Y, Wen S, Chen H, Ge J. Antimicrobial, anti-adhesive and anti-biofilm potential of biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum against Staphylococcus aureus CMCC26003. Microb. Pathog. 2019;127:12–20. doi: 10.1016/j.micpath.2018.11.039. [DOI] [PubMed] [Google Scholar]
- 14.Song YJ, Yu HH, Kim YJ, Lee NK, Paik HD. Anti-biofilm activity of grapefruit seed extract against Staphylococcus aureus and Escherichia coli. J. Microbiol. Biotechnol. 2019;29:1177–1183. doi: 10.4014/jmb.1905.05022. [DOI] [PubMed] [Google Scholar]
- 15.Cui T, Bai F, Sun M, Lv X, Li X, Zhang D, Du H. Lactobacillus crustorum ZHG 2-1 as novel quorum-quenching bacteria reducing virulence factors and biofilms formation of Pseudomonas aeruginosa. LWT-Food Sci. Technol. 2020;117:108696. doi: 10.1016/j.lwt.2019.108696. [DOI] [Google Scholar]
- 16.Wang N, Yuan L, Sadiq FA, He G. Inhibitory effect of Lactobacillus plantarum metabolites against biofilm formation by Bacillus licheniformis isolated from milk powder products. Food Control. 2019;106:106721. doi: 10.1016/j.foodcont.2019.106721. [DOI] [Google Scholar]
- 17.Kaur S, Sharma P, Kalia N, Singh J, Kaur S. Anti-biofilm properties of the fecal probiotic Lactobacilli against Vibrio spp. Front. Cell. Infect. Microbiol. 2018;8:120. doi: 10.3389/fcimb.2018.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braïek, OB, Merghni A, Smaoui S, Mastouri M. Enterococcus lactis Q1 and 4CP3 strains from raw shrimps: Potential of antioxidant capacity and anti-biofilm activity against methicillin-resistant Staphylococcus aureus strains. LWT-Food Sci. Technol. 2019;102:15–21. doi: 10.1016/j.lwt.2018.11.095. [DOI] [Google Scholar]
- 19.Hong JY, Lee NK, Yi SH, Hong SP, Paik HD. Physicochemical features and microbial community of milk kefir using a potential probiotic Saccharomyces cerevisiae KU200284. J. Dairy Sci. 2019;102:10845–10849. doi: 10.3168/jds.2019-16384. [DOI] [PubMed] [Google Scholar]
- 20.Lee NK, Hong JY, Yi SH, Hong SP, Lee JE, Paik HD. Bioactive compounds of probiotic Saccharomyces cerevisiae strains isolated from cucumber jangajji. J. Funct. Foods. 2019;58:324–329. doi: 10.1016/j.jff.2019.04.059. [DOI] [Google Scholar]
- 21.de Lima MDSF, de Souza KMS, Albuquerque WWC, Teixeira JAC, Cavalcanti MTH, Porto ALF. Saccharomyces cerevisiae from Brazilian kefir-fermented milk: An in vitro evaluation of probiotic properties. Microb. Pathog. 2017;110:670–677. doi: 10.1016/j.micpath.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Fakruddin MD, Hossain MN, Ahmed MM. Antimicrobial and antioxidant activities of Saccharomyces cerevisiae IFST062013, a potential probiotic. BMC Complement. Altern. Med. 2017;17:64. doi: 10.1186/s12906-017-1591-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moslehi-Jenabian S, Lindegaard L, Jespersen L. Beneficial effects of probiotic and food borne yeasts on human health. Nutrients. 2010;2:449–473. doi: 10.3390/nu2040449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu HH, Song YJ, Yu HS, Lee NK, Paik HD. Investigating the antimicrobial and antibiofilm effects of cinnamaldehyde against Campylobacter spp. using cell surface characteristics. J. Food Sci. 2020;85:157–164. doi: 10.1111/1750-3841.14989. [DOI] [PubMed] [Google Scholar]
- 25.Islam B, Khan SN, Haque I, Alam M, Mushfiq M, Khan AU. Novel anti-adherence activity of mulberry leaves: inhibition of Streptococcus mutans biofilm by 1-deoxynojirimycin isolated from Morus alba. J. Antimicrob. Chemother. 2008;62:751–757. doi: 10.1093/jac/dkn253. [DOI] [PubMed] [Google Scholar]
- 26.Chiba A, Sugimoto S, Sato F, Hori S, Mizunoe Y. A refined technique for extraction of extracellular matrices from bacterial biofilms and its applicability. Microb. Biotechnol. 2015;8:392–403. doi: 10.1111/1751-7915.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heggers JP, Cottingham J, Gusman J, Reagor L, McCoy L, Carino E, et al. The effectiveness of processed grapefruit-seed extract as an antibacterial agent: II. Mechanism of action and in vitro toxicity. J. Altern. Complement. Med. 2002;8:333–340. doi: 10.1089/10755530260128023. [DOI] [PubMed] [Google Scholar]
- 28.Kang J, Jin W, Wang J, Sun Y, Wu X, Liu L. Antibacterial and anti-biofilm activities of peppermint essential oil against Staphylococcus aureus. LWT-Food Sci. Technol. 2019;101:639–645. doi: 10.1016/j.lwt.2018.11.093. [DOI] [Google Scholar]
- 29.Ghorbani Z, Owlia P, Marashi MA, Saderi H. Effect of supernatant and cell lysate extracts of Saccharomyces cerevisiae on biofilm and alginate production by Pseudomonas aeruginosa. Iran J. Med. Microbiol. 2018;12:189–198. doi: 10.30699/ijmm.12.3.189. [DOI] [Google Scholar]
- 30.Kim BR, Bae YM, Hwang JH, Lee SY. Biofilm formation and cell surface properties of Staphylococcus aureus isolates from various sources. Food Sci. Biotechnol. 2016;25:643–648. doi: 10.1007/s10068-016-0090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kouidhi B, Zmantar T, Hentati H, Bakhrouf A. Cell surface hydrophobicity, biofilm formation, adhesives properties and molecular detection of adhesins genes in Staphylococcus aureus associated to dental caries. Microb. Pathog. 2010;49:14–22. doi: 10.1016/j.micpath.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Vijayakumar K, Bharathidasan V, Manigandan V, Jeyapragash D. Quebrachitol inhibits biofilm formation and virulence production against methicillin-resistant Staphylococcus aureus. Microb. Pathog. 2020;149:104286. doi: 10.1016/j.micpath.2020.104286. [DOI] [PubMed] [Google Scholar]
- 33.Ates O. Systems biology of microbial exopolysaccharides production. Front. Bioeng. Biotech. 2015;3:200. doi: 10.3389/fbioe.2015.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu M, Wu X, Li J, Liu L, Zhang R, Shao D, Du X. The specific anti-biofilm effect of gallic acid on Staphylococcus aureus by regulating the expression of the ica operon. Food Control. 2017;73:613–618. doi: 10.1016/j.foodcont.2016.09.015. [DOI] [Google Scholar]
- 35.Bai JR, Zhong K, Wu YP, Elena G, Gao H. Antibiofilm activity of shikimic acid against Staphylococcus aureus. Food Control. 2019;95:327–333. doi: 10.1016/j.foodcont.2018.08.020. [DOI] [Google Scholar]
- 36.Cui H, Zhang C, Li C, Lin L. Inhibition mechanism of cardamom essential oil on methicillin-resistant Staphylococcus aureus biofilm. LWT-Food Sci. Technol. 2020;122:109057. doi: 10.1016/j.lwt.2020.109057. [DOI] [Google Scholar]