Abstract
Increasing evidence suggests a potential role of microbial colonization in the inception of chronic airway diseases. However, it is not clear whether the lung and gut microbiome dysbiosis is coincidental or a result of mutual interaction. In this study, we investigated the airway microbiome in interleukin 13 (IL-13)-rich lung environment and related alterations of the gut microbiome. IL-13- overexpressing transgenic (TG) mice presented enhanced eosinophilic inflammatory responses and mucus production, together with airway hyperresponsiveness and subepithelial fibrosis. While bronchoalveolar lavage fluid and cecum samples obtained from 10-week-old IL-13 TG mice and their C57BL/6 wild-type (WT) littermates showed no significant differences in alpha diversity of lung and gut microbiome, they presented altered beta diversity in both lung and gut microbiota in the IL-13 TG mice compared to the WT mice. Lung-specific IL-13 overexpression also altered the composition of the gut as well as the lung microbiome. In particular, IL-13 TG mice showed an increased proportion of Proteobacteria and Cyanobacteria and a decreased amount of Bacteroidetes in the lungs, and depletion of Firmicutes and Proteobacteria in the gut. The patterns of polymicrobial interaction within the lung microbiota were different between WT and IL-13 TG mice. For instance, in IL-13 TG mice, lung Mesorhizobium significantly affected the alpha diversity of both lung and gut microbiomes. In summary, chronic asthma-like pathologic changes can alter the lung microbiota and affect the gut microbiome. These findings suggest that the lung-gut microbial axis might actually work in asthma.
Keywords: Interleukin-13, asthma, microbiota, gastrointestinal microbiome, microbial interactions
Introduction
Asthma is a complex, heterogeneous disease that can be characterized by different cellular, inflammatory, and molecular phenotypes [1]. Decades of research have illustrated a complex and pleiotropic role of interleukin (IL)-13, a key effector cytokine of Th2 inflammation, in allergic disease [2, 3]. For instance, transgenic (TG) mice with lung-specific overexpression of IL-13 presented with hypertrophy of airway epithelial cells, mucus hypersecretion, eosinophilic lung inflammation, and subepithelial fibrosis [4].
Recent advances in the exploration of microbiota by culture-independent sequencing have led to an extended knowledge of the role of commensal microorganisms in asthma. Indeed, microbes play an important role in shaping normal and pathologic immune responses in both lung and gut. For example, increasing evidence suggests a potential role of microbial colonization in the inception of asthma [5]. The composition of the microbiota of asthmatic patients may be modulated by diet [6], antibiotics [7], and infections [8], especially early in life. In addition, recent studies in animal models have focused on the gut microbiota as an important driver of airway allergic inflammation [9-11] and/or oral supplementation of beneficial species (e.g., Bifidobacterium and Lactobacillus) to reduce Th2 cytokine production [12-15]. However, it is difficult to ascertain whether asthma-dependent changes in epithelial structures subsequently induce lung or gut dysbiosis.
In recent years, special focus has been placed upon the microbiota of the airway and gut in the pathogenesis of asthma [16, 17]. This has led to the coining of terms such as the “lung-gut axis.” However, the occurrence of communication between the lung and gut is still under investigation. Despite the emerging study on this lung-gut microbial axis in asthma, it is not clear whether this association is a result of a simple correlation or a causal relationship linked to the pathomechanisms of the disease. If the latter is true, airway microbial dysbiosis might induce alterations in the gut microbiota over time, which in turn would affect other organ immune responses. Several studies have demonstrated that chronic inflammatory airway diseases altered the airway microbiome and had a profound effect on the gut microbiota [12, 14, 18, 19]. However, the causality between emergence of lung diseases and alterations of the gut microbiota is still largely unexplored.
Here, we attempted to characterize the airway microbiome of well-characterized, IL-13-overexpressing TG mice and investigated related secondary alterations in the gut microbiota. Next, we performed an integrative analysis of the lung and gut microbiome of IL-13 TG mice to explore the microbial interactions between lung and gut.
Materials and Methods
Lung and Gut Microbiome Specimens from IL-13-Overexpressing TG Mice
The transgenic mice were kindly provided by Professor Jack A. Elias of Brown University [20, 21]. To evaluate the respiratory effector functions of IL-13, we used constitutional IL-13 TG mice with Clara cell 10-kDa protein (CC10) promoter to target the expression of IL-13 in the airway since constitutive IL-13 TG mice can express the transgene without doxycycline, which can affect commensals. Bronchoalveolar lavage (BAL) fluid, lung tissue, and stool were collected for microbiome analysis from 10-week-old, IL-13-overexpressing TG mice (n = 30) and their C57BL/6 wild-type (WT) littermates (n = 30). Mice were euthanized and two volumes of 1 ml of PBS were infused, gently aspirated, and pooled. Samples were centrifuged at 3,000 ×g for 10 min to recover cells, while the supernatants were collected and stored at -70°C for further analysis. Cell pellets were resuspended in PBS and total cell counts were determined using a hemocytometer. The study protocols were approved by the IACUC of Seoul National University Hospital (Approval No. SNUH-IBC-1606-018-004).
DNA Extraction and 16S rRNA Gene Sequencing
Genomic DNA was extracted from 1 mL of BAL fluid, two right lobes of lung, and 500 mg of cecum samples using a commercial microbial DNA isolation kit (FastDNA SPIN Kit, MP Biomedicals, USA) according to the manufacturer’s instruction. PCR amplification was performed using the primers 341F (5'- TCGTCGGCA GCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3') and 805R (5'-GTCTCGTGGGCT CGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3') and 2X KAPA HiFi HotStart ReadyMix (KAPA Biosystems, USA). PCR reactions were carried out as follows: 3 min at 95°C, 25 cycles of 30 sec at 95°C, 30 sec at 55°C, 30 sec at 72°C, 5 min at 72°C, and hold at 4°C. A second PCR for the indexing process was performed using the Nextera XT Index Kit (Illumina, USA). The amplicon was purified using a TopQXSEP MagBead matrix (Celemix, Korea) and then sequenced using the MiSeq v3 platform (Illumina).
Data Analysis
To test differences between the IL-13 TG group and the WT group, the Mann‐Whitney test was used for nonparametric data while an unpaired t-test was used for parametric data. Sequencing data were analyzed using the QIIME pipeline referring to the SILVA 16S rRNA gene database. To evaluate the alpha diversity of bacterial communities, observed operational taxonomic units (OTUs) the Shannon index, the Chao1 index, and the Inverse Simpson’s diversity index were calculated. The beta-diversity of the microbiome was calculated using Principal coordinate analysis (PCoA) and permutational multivariate ANOVA (PERMANOVA) by R package vegan. The differences in taxa abundance were calculated using the Wilcoxon test (p < 0.05). Linear discriminant analysis (LDA) effect size (LEfSe) was applied to the OTU table (Kruskal-Wallis test, p < 0.05; Wilcoxon test, p < 0.05; LDA > 2.0) [22]. Co-occurrence between bacterial OTUs was calculated in terms of Pearson’s correlation coefficients and the results were visualized as a network using Cytoscape. The Benjamini-Hochberg correction for multiple comparisons was applied, and a false discovery rate (FDR) < 0.05 was selected.
Results
Lung Inflammation in IL-13-Overexpressing Mice and Preparation of Samples
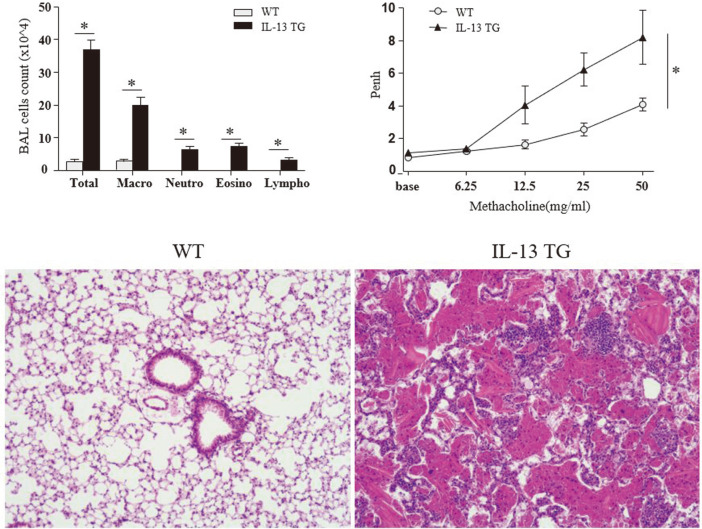
To examine the airway inflammatory changes occurring in IL-13 TG mice, we analyzed the extent of airway hyperresponsiveness (AHR) in their lung tissue samples and compared it with that of WT mice. Histopathological analyses revealed that IL-13 TG mice exhibited lung tissue inflammation, in particular eosinophilic infiltration and AHR, which are both typical features of asthma (Fig. 1). Moreover, counting of BAL-derived cells showed a significantly increased total BAL cell count in IL-13 TG mice, associated with an increased number of macrophages, eosinophils, neutrophils, and lymphocytes (p < 0.05).
Fig. 1. Comparision of total and differential cell count (upper left), airway hyperresponsiveness (upper right), and, histological features of wild-type mice and IL-13-overexpressing transgenic mice.
*P-value < 0.05.
Assessment of Bacterial Diversity in Lungs and Gut
We compared bacterial diversity of BAL fluids and lung lysates to identify the best method of tissue sampling most accurately reflecting the lower airway microbiota (Suppl. A). However, the amount of bacterial biomass from lung lysates was too low to conduct 16S rRNA gene amplification. Thus, we chose BAL fluids for the current experiment.
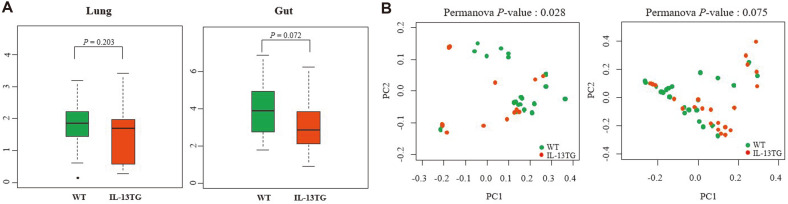
We first compared alpha diversity indicators among groups, which included the number of observed OTUs, the Shannon index, the Chao1 index, and the Inverse Simpson’s diversity index. Microbial communities of BAL samples from the WT mice displayed a higher Shannon index (1.73 ± 0.64) than those from the IL-13 TG mice (1.65 ± 0.90); however, the difference was not significant (P = 0.203) (Fig. 2A). Moreover, analysis of alpha diversity of the gut microbiome demonstrated a slight decrease in Shannon index in the IL-13 TG mice with respect to the WT mice, but this difference did not reach statistical significance (WT: 3.98 ± 1.29, TG: 3.26 ± 1.50, P = 0.072). However, when plotting matrices of weighted UniFrac distances, the beta diversity of the lung microbiota of IL-13 TG mice was significantly lower than that of WT mice (P = 0.028; PERMANOVA) (Fig. 2B). In contrast, the beta diversity of the gut microbiota between the two groups showed a slight, but not significant, difference (P = 0.075; PERMANOVA).
Fig. 2. Differences in microbiome diversity between IL-13 TG and WT mice.
(A) Alpha diversity (Shannon index). (B) Beta diversity.
Effect of Lung-Specific IL-13 Overexpression on Lung and Gut Commensals
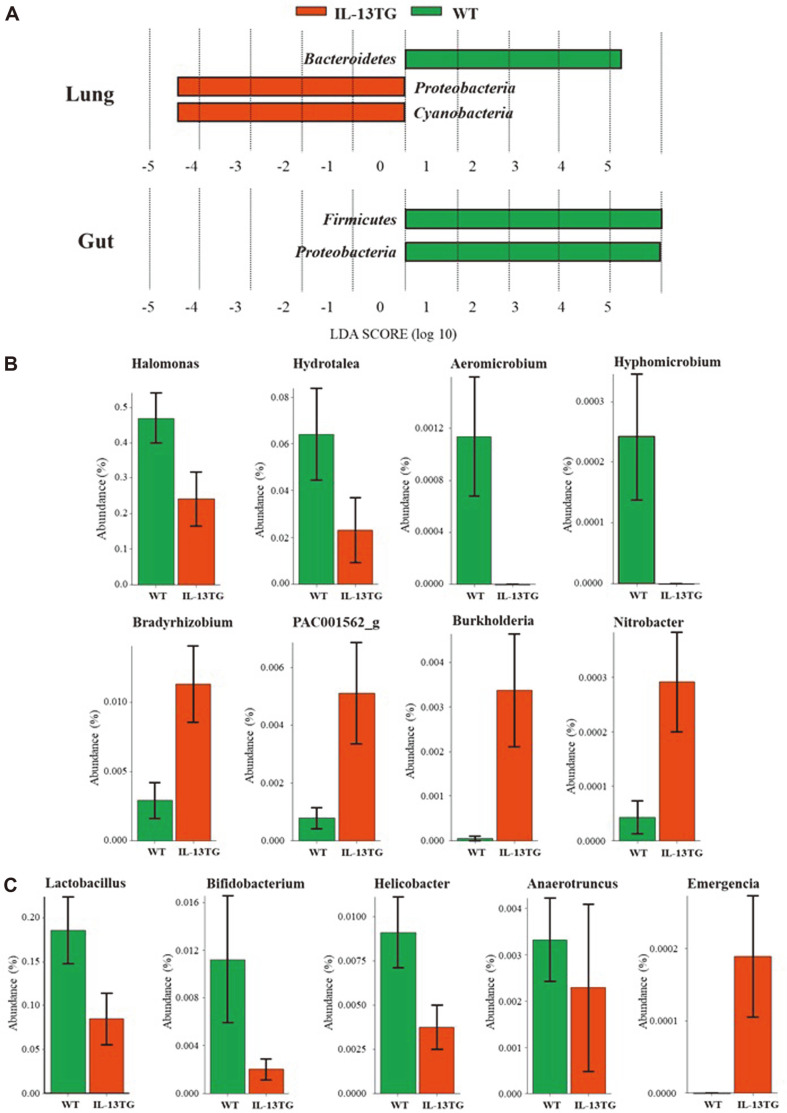
We next examined differences in the composition of microbial communities in terms of phyla and genera between IL-13 TG and WT mice. Interestingly, we found significant, highlighted differences in the abundance of certain microbial taxa between the two groups in both lungs and gut (Fig. 3A).
Fig. 3.
(A) Analysis of differences in the composition of the microbiome between IL-13 TG and WT mice by linear discriminant analysis (LDA) effect size (LEfSe). (B) Relative abundance (%) of bacterial genera displaying significantly different abundance in the airways of the two groups. (C) Relative abundance (%) of bacterial genera displaying significantly different abundance in the gut of the two groups.
At first, microbiomes were compared at the phylum level. BAL samples of IL-13 TG mice showed reduced abundance of Bacteroidetes, and an increased proportion of Proteobacteria and Cyanobacteria compared to those of WT mice. On the other hand, in cecum samples, the abundance of Firmicutes and Proteobacteria observed in WT mice decreased in mice with lung-specific IL-13 overexpression. Interestingly, the relative abundance of Proteobacteria was differentially associated with the lung and gut microbiomes. At the species level, Halomonas, Burkholderia, Acinetobacter, Klebsiella, and Bradyrhizobium were increased in the lung in the IL-13 TG mice group compared to controls. On the other hand, the proportions of Helicobacter and Desulfovibrio were decreased in the gut in the IL-13 TG mice, compared to the WT mice.
At the genus level, a total of eight OTUs showed different abundance between BAL samples of WT and IL-13-overexpressing mice (Fig. 3B). In particular, we observed that the abundance of Halomonas, Hydrotalea, Aeromicrobium, and Hyphomicrobium decreased in the IL-13 TG group with respect to the WT group. Conversely, the proportion of Bradyrhizobium, Burkholderia, PAC-001562, and Nitrobacter increased in the IL-13 TG group.
In cecum samples, the abundance of four genera, i.e., Lactobacillus, Bifidobacterium, Helicobacter, and Anaerotruncus, decreased, whereas only the genus Emergencia showed increased abundance in the IL-13 TG group with respect to the WT group (Fig. 3C).
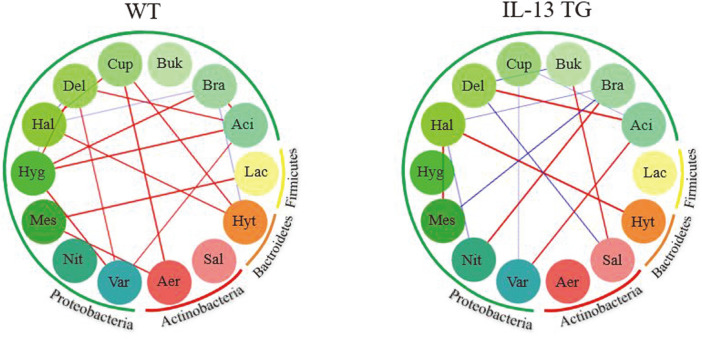
Polymicrobial Interaction Patterns in Lung and Gut Communities
To gain insights into the interaction within lung and gut microbiomes, we performed a bacterial OTU network analysis using CoNet (co-occurrence network interference; http://psbweb05.psb.ugent.be/conet/index.php). Specifically, we calculated Pearson’s correlation coefficients to explore the effect of the IL-13 cytokine on the microbial composition within the BAL and cecum microbiome after filtering by using the top 25 percentile genera (Fig. 4., See Supplement B for all relevant genera data). In the resulting network, each node corresponds to one microbial genus while the color of the edges indicates a positive (red) or negative (blue) relationship. Interestingly, genus-genus interactions within the lung community were considerably different between WT mice and IL-13 TG mice. Indeed, the IL-13 TG mice displayed more competitive interactions presenting negative correlation than WT mice. There was not much interaction within the gut microbiome in either group. Within gut microbial communities, IL-13 TG mice showed a positive correlation between the abundances of Bacteroides and Lactobacillus, while in the WT group, the abundances of the genera Clostridium and Adlercreutzia presented a positive correlation (Suppl. C).
Fig. 4. Polymicrobial interaction network between airway microbial communities at the genus level. Red and blue edges represent positive and negative correlations, respectively.
(Aer: Aeromicrobium, Sal: Salinispora, Hyt: Hydrotalea, Lac: Lactobacillus, Aci: Acidoborax, Bra: Bradyrhizobium, Bur: Burkholderia, Cup: Cupriavidus, Del: Delftia, Hal: Halomonas, Hyg: Hydrogenophaga, Mes: Mesorhizobium, Nit: Nitrobacter, Var: Variovorax)
Microbial Interactions Between the Lung and Gut Communities in IL-13 TG Mice
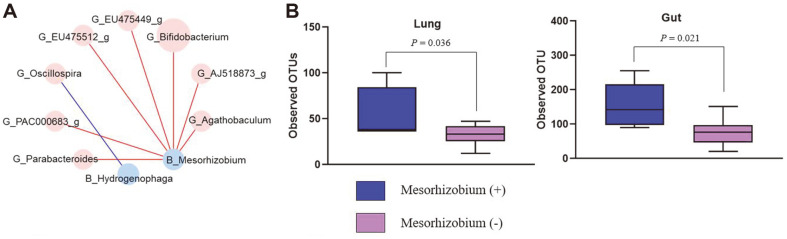
Next, we examined the FDR after correction of p-values for multiple comparisons using the Benjamini-Hochberg method (Fig. 5A and Supplement D). While there were no obvious relationships between the bacterial composition of BAL and cecum samples in the WT group, two genera in BAL samples and eight genera in stool formed significantly correlated microbial pairs in IL-13 TG mice. Primarily, the genus Mesorhizobium, displaying reduced abundance in BAL samples of the IL-13 TG group, was positively associated with Bifidobacterium, Agathobaculum, and Parabacteroides in the gut. We additionally compared microbial diversity based on the presence of Mesorhizobium in the IL-13 TG group. Notably, OTU richness in the lung and gut communities was significantly reduced in the absence of Mesorhizobium in IL-13 TG (P = 0.036 vs. P = 0.021, respectively) (Fig. 5B).
Fig. 5.
(A) Results of multiple comparisons (corrected by the Benjamin-Hochberg method) in lung and gut microbiota of IL-13 TG mice. (B) Bacterial diversity based on the presence of Mesorhizobium in both lung and gut microbiota of IL-13 TG mice.
Discussion
In this study, we investigated the effects of IL-13-induced type 2 inflammation on the lung and gut microbiota. Lung-specific IL-13 TG mice showed altered beta diversity and composition of lung microbiome. In addition to lung microbiome, gut microbiome also presented relative deficiency in the phyla Firmicutes and Proteobacteria in IL-13 TG mice. Thus, we herein provide direct evidence that chronic lung inflammation induced by IL-13 can cause dysbiosis not only of the lung microbiota but also of the gut microbiota. Taken together, these findings suggest the potential lung-gut microbiota interaction in a murine asthma model.
The cytokine IL-13 is considered a central effector of allergic asthma which is both necessary and sufficient to induce key features of asthma such as eosinophilic inflammation, mucus production, and airway hyperresponsiveness [23]. In addition, IL-13 leads to subepithelial fibrosis, which contributes to airway remodeling in severe asthma [24]. Comprehensive studies of targeted and inducible pulmonary overexpression of IL-13 in TG mice have defined the role of IL-13 in the cellular and molecular responses to allergic asthma [4, 20, 21]. However, there is scarce evidence on whether an IL-13-rich microenvironment can induce dysbiosis of the lung and/or gut microbiome.
In human, chronic airway inflammation and environmental factors such as diet and antibiotics can shift the lung microbiota towards decreased abundance of beneficial microbiota accompanied by outgrowth of pathogenic microbiota [7,16]. This perturbation in microbial diversity and composition is referred to as lung dysbiosis. Comparing healthy controls with asthmatics, phylum-level abundance has demonstrated dysbiosis characterized by increased Proteobacteria and decreased Bacteroidetes in asthmatic airways, which were similar to the findings in this study [25]. We also found eight OTUs showing different abundance in BAL between WT mice and IL-13-TG mice, suggesting future research to evaluate the role of those microbes in the development of lung pathology. Interestingly, a previous study reported that Halomonas decreased in the BAL of patients with eosinophilic asthma [26]. Similarly, Halomonas reduced in the BAL of IL-13 TG mice. Burkholderia is a genus of Proteobacteria and significantly increased in the BAL of IL-13 TG mice compared to WT mice. The proportion of Burkholderia in the upper airway microbiome was increased in asthmatics with chronic rhinosinusitis and asthmatics with at least one emergency room visit [27].
A recent study suggested the microbiome interactions are as important as the individual species in shaping fitness traits by mapping gut bacteria against the fruit fly model [28]. Comparison of pairwise correlation between groups showed more negatively correlated microbial pairs in the lungs of IL-13 TG which indirectly implies the competitive condition in the chronic inflammatory microenvironment.
Murine and human studies linked antibiotics use in critical developmental window time to gut dysbiosis and increased asthma risk [29-31]. These data suggest that gut dysbiosis also plays a critical role against chronic airway inflammation by regulating innate and adaptive immune response [32]. We found that the abundance of bacteria producing short-chain fatty acids (SCFAs) upon fiber fermentation, such as Lactobacillus and Bifidobacterium species, was typically reduced in the gut of IL-13 TG mice, as compared to WT mice. SCFAs, such as acetate, propionate and butyrate, are the most abundant products of anaerobic fermentation of dietary fiber and important metabolites for the maintenance of intestinal homeostasis [33]. Notably, a recent study revealed that Lactobacillus supplementation could reduce allergic inflammation and modulate the activity of gut microbes by inducing butyrate biosynthesis in house dust mite (HDM)-sensitized mice [34]. Therefore, the role of SCFAs in the lung-gut microbial interactions remains to be further investigated.
It is well established that disturbances in the gut microbiota during early life can cause allergic asthma, as confirmed by experimental and epidemiological studies [11, 35, 37]. Indeed, an increasing number of studies have revealed that the gut microbiota can influence lung immunity [11, 29]. Thus far, previous studies indicated the primary direction of crosstalk from the gut to the lungs [31,37,38]. On the other hand, whether the lung microbiota affects its gut counterpart is less clear at present. Only one study investigated the influence of acute lung injury on the gut microbiota by lipopolysaccharide instillation [39]. Although the underlying mechanism of microbial communication along the gut-lung axis is not fully understood, several factors are proposed, such as epigenetic modification by bacterial SCFAs [6, 40, 41], enhanced Th 17 inflammation by segmented filamentous bacteria [42], and induction of the NF-κB-dependent pathway via activation of Toll-like receptors by microbes [32]. Our study showed the impact of chronic Th2 lung inflammation on the lung and gut microbiome and should provide new insights into disease pathogenesis and potential therapeutic strategies.
Considering that the lung and gut are structurally similar with identical embryonic origin, these two distant mucosal sites might interact via immune response. Accordingly, previous epidemiologic studies found that individuals with asthma were more likely to develop chronic gastrointestinal comorbidities such as inflammatory bowel disease [43-45]. These findings support the hypothesis that structural and functional alterations in lung epithelium could contribute to gut dysbiosis and vice versa. The lung-gut axis is defined as a shared mucosal immune system along which extensive communication between the lung and the gut takes place, including microbial and immunological changes [17]. Using IL-13 TG mice, we found that chronic lung inflammation induces alterations in the gut microbiota, suggesting a relationship between the lung-gut axis and asthma. However, further investigation is warranted to determine whether gut dysbiosis depends on lung-gut microbial crosstalk or if chronic lung inflammation influences the gut mucosa or the composition of the gut microbiome by inducing inflammation in gut mucosa.
In this study, we found that the lung-gut microbiome interaction showed a different pattern in IL-13 TG mice compared to those of WT mice. To understand the complex and diverse nature of the microbiome, we performed network-based analysis and found that certain genera in airway were associated with abundance of gut microbiota. Particularly, the Mesorhizobium, a genus of gram-negative soil bacteria [46], was significantly associated with alpha diversity not only of lung but also of gut in IL-13 TG mice. Although the exact role of Mesorhizobium in asthma was not validated in this study, it is interesting to find that there are certain key lung taxa that interact with microbes in the gut and are associated with microbiome abundance in both lung and gut.
Although our data have demonstrated that asthma could alter the microbiome and the polymicrobial interaction between lung and gut, there are still some unsolved issues. First, it is unclear whether IL-13 contributes to gut dysbiosis in a direct or indirect way. IL-13 TG mice simulate chronic asthma which may accompany long standing hypoxia. Further research exploring the effect of IL-13 blockade on lung and/or gut dysbiosis would be helpful to demonstrate the direct effect of IL-13 on the microbiome. Second, this study is limited in ascertaining the directionality of the lung-gut microbial interactions. Further research is needed to determine the direction of lung-gut interactions by transplanting fecal or lung microbiome from IL-13 TG into healthy mice. Third, we did not directly evaluate the SCFAs from cecal samples in IL-13 TG mice for verification of reduction of SCFA-producing bacteria. Finally, we cannot conclude that there is a direct causal-effect relationship between increase in the Mesorhizobium and dysbiosis in both lung and gut, although we showed that these genera altered the diversity of both lung and gut in IL-13 TG mice.
In conclusion, our data demonstrated that IL-13-driven type 2 lung inflammation induces not only lung but also gut dysbiosis. Further investigation of microbial communication along the lung-gut axis might provide novel insight into this mechanism in asthma.
Supplemental Material
Supplementary data for this paper are available on-line only at http://jmb.or.kr.
Acknowledgments
We thank Dr. Jack A. Elias and Dr. Chun Geun Lee (Brown University, RI, USA) for providing IL-13-overexpressing transgenic mice. This work was supported by a National Research Foundation of Korea (NRF) grant (No. 2015R1C1A2A01054797) funded by the Korea government (MSIT).
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
REFERENCES
- 1.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat. Med. 2012;18:716–25. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 2.Wynn TA. IL-13 effector functions. Annu. Rev. Immunol. 2003;21:425–456. doi: 10.1146/annurev.immunol.21.120601.141142. [DOI] [PubMed] [Google Scholar]
- 3.Ingram JL, Kraft M. IL-13 in asthma and allergic disease: asthma phenotypes and targeted therapies. J. Allergy Clin. Immunol. 2012;130:829–842. doi: 10.1016/j.jaci.2012.06.034. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, Wang J, et al. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J. Clin. Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu AH. Revisiting the hygiene hypothesis for allergy and asthma. J. Allergy Clin. Immunol. 2015;136:860–865. doi: 10.1016/j.jaci.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 7.Wypych TP, Marsland BJ. Antibiotics as instigators of microbial dysbiosis: implications for asthma and allergy. Trends Immunol. 2018;39:697–711. doi: 10.1016/j.it.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herbst T, Sichelstiel A, Schär C, Yadava K, Bürki K, Cahenzli J, et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am. J. Respir. Crit. Care Med. 2011;184:198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 11.Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, et al. Early life antibiotic‐driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karimi K, Inman MD, Bienenstock J, Forsythe P. Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am. J. Respir. Crit. Care Med. 2009;179:186–193. doi: 10.1164/rccm.200806-951OC. [DOI] [PubMed] [Google Scholar]
- 13.MacSharry J, O'Mahony C, Shalaby KH, Sheil B, Karmouty-Quintana H, Shanahan F, et al. Immunomodulatory effects of feeding with Bifidobacterium longum on allergen-induced lung inflammation in the mouse. Pulm. Pharmacol. Ther. 2012;25:325–334. doi: 10.1016/j.pupt.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 14.Harb H, Van Tol E, Heine H, Braaksma M, Gross G, Overkamp K, et al. Neonatal supplementation of processed supernatant from Lactobacillus rhamnosus GG improves allergic airway inflammation in mice later in life. Clin. Exp. Allergy. 2013;43:353–364. doi: 10.1111/cea.12047. [DOI] [PubMed] [Google Scholar]
- 15.Zhang B, An J, Shimada T, Liu S, Maeyama K. Oral administration of Enterococcus faecalis FK-23 suppresses Th17 cell development and attenuates allergic airway responses in mice. Int. J. Mol. Med. 2012;30:248–254. doi: 10.3892/ijmm.2012.1010. [DOI] [PubMed] [Google Scholar]
- 16.Marsland BJ, Trompette A, Gollwitzer ES. The gut-lung axis in respiratory disease. Ann. Am. Thorac. Soc. 2015;12:S150–S156. doi: 10.1513/AnnalsATS.201503-133AW. [DOI] [PubMed] [Google Scholar]
- 17.Budden KF, Gellatly SL, Wood DL, Cooper MA, Morrison M, Hugenholtz P, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 2017;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- 18.MacSharry J, O'Mahony C, Shalaby KH, Sheil B, Karmouty-Quintana H, Shanahan F, et al. Immunomodulatory effects of feeding with Bifidobacterium longum on allergen-induced lung inflammation in the mouse. Pulm. Pharmacol. Ther. 2012;25:325–334. doi: 10.1016/j.pupt.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Ai C, Zhang Q, Ren C, Wang G, Liu X, Tian F, et al. Genetically engineered Lactococcus lactis protect against house dust mite allergy in a BALB/c mouse model. PLoS One. 2014;9:e109461. doi: 10.1371/journal.pone.0109461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng T, Zhu Z, Wang Z, Homer RJ, Ma B, Riese RJ, Jr, et al. Inducible targeting of IL-13 to the adult lung causes matrix metalloproteinase- and cathepsin-dependent emphysema. J. Clin. Invest. 2000;106:1081–1093. doi: 10.1172/JCI10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Z, Ma B, Zheng T, Homer RJ, Lee CG, Charo IF, et al. IL-13-induced chemokine responses in the lung: role of CCR2 in the pathogenesis of IL-13-induced inflammation and remodeling. J. Immunol. 2002;168:2953–2962. doi: 10.4049/jimmunol.168.6.2953. [DOI] [PubMed] [Google Scholar]
- 22.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:1–18. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wills-Karp M. Interleukin-13 in asthma pathogenesis. Immunol. Rev. 2004;202:175–190. doi: 10.1111/j.0105-2896.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 24.Takayama G, Arima K, Kanaji T, Toda S, Tanaka H, Shoji S, et al. Periostin: a novel component of subepithelial fibrosis of bronchial asthma downstream of IL-4 and IL-13 signals. J. Allergy Clin. Immunol. 2006;118:98–104. doi: 10.1016/j.jaci.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 25.Huang YJ. Asthma microbiome studies and the potential for new therapeutic strategies. Curr. Allergy Asthma. Rep. 2013;13:453–461. doi: 10.1007/s11882-013-0355-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sverrild A, Kiilerich P, Brejnrod A, Pedersen R, Porsbjerg C, Bergqvist A, et al. Eosinophilic airway inflammation in asthmatic patients is associated with an altered airway microbiome. J. Allergy Clin. Immunol. 2017;140:407–417. doi: 10.1016/j.jaci.2016.10.046. [DOI] [PubMed] [Google Scholar]
- 27.Yang HJ, LoSavio PS, Engen PA, Naqib A, Mehta A, Kota R, et al. Association of nasal microbiome and asthma control in patients with chronic rhinosinusitis. Clin. Exp. Allergy. 2018;48:1744–1747. doi: 10.1111/cea.13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gould AL, Zhang V, Lamberti L, Jones EW, Obadia B, Korasidis N, et al. Microbiome interactions shape host fitness. Proc. Natl. Acad. Sci. USA. 2018;115:E11951–E11960. doi: 10.1073/pnas.1809349115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell SL, Gold MJ, Willing BP, Thorson L, McNagny KM, Finlay BB. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. 2013;4:158–164. doi: 10.4161/gmic.23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J. Allergy Clin. Immunol. 2001;107:129–134. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- 31.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016;22:1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L, Fang Z, Liu X, Hu W, Lu W, Lee Y-k, et al. Lactobacillus reuteri attenuated allergic inflammation induced by HDM in the mouse and modulated gut microbes. PLoS One. 2020;15:e0231865. doi: 10.1371/journal.pone.0231865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bønnelykke K, et al. Childhood asthma after bacterial colonization of the airway in neonates. N. Engl. J. Med. 2007;357:1487–1495. doi: 10.1056/NEJMoa052632. [DOI] [PubMed] [Google Scholar]
- 36.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barfod KK, Roggenbuck M, Hansen LH, Schjørring S, Larsen ST, Sørensen SJ, et al. The murine lung microbiome in relation to the intestinal and vaginal bacterial communities. BMC Microbiol. 2013;13:303. doi: 10.1186/1471-2180-13-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooke KR, Hill GR, Gerbitz A, Kobzik L, Martin TR, Crawford JM, et al. Hyporesponsiveness of donor cells to lipopolysaccharide stimulation reduces the severity of experimental idiopathic pneumonia syndrome: potential role for a gut-lung axis of inflammation. J. Immunol. 2000;165:6612–6619. doi: 10.4049/jimmunol.165.11.6612. [DOI] [PubMed] [Google Scholar]
- 39.Sze MA, Tsuruta M, Yang S-WJ, Oh Y, Man SP, Hogg JC, et al. Changes in the bacterial microbiota in gut, blood, and lungs following acute LPS instillation into mice lungs. PLoS One. 2014;9:e111228. doi: 10.1371/journal.pone.0111228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McAleer JP, Kolls JK. Contributions of the intestinal microbiome in lung immunity. Eur. J. Immunol. 2018;48:39–49. doi: 10.1002/eji.201646721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 2015;6:7320. doi: 10.1038/ncomms8320. [DOI] [PubMed] [Google Scholar]
- 42.Bradley CP, Teng F, Felix KM, Sano T, Naskar D, Block KE, et al. Segmented filamentous bacteria provoke lung autoimmunity by inducing gut-lung axis Th17 cells expressing dual TCRs. Cell Host Microbe. 2017;22:697–704.:e694. doi: 10.1016/j.chom.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yazar A, Atis S, Konca K, Pata C, Akbay E, Calikoglu M, et al. Respiratory symptoms and pulmonary functional changes in patients with irritable bowel syndrome. Am. J. Gastroenterol. 2001;96:1511–1516. doi: 10.1111/j.1572-0241.2001.03748.x. [DOI] [PubMed] [Google Scholar]
- 44.Kuenzig ME, Bishay K, Leigh R, Kaplan GG, Benchimol EI, Crowdscreen SR review Team. Co-occurrence of asthma and the inflammatory bowel diseases: a systematic review and meta-analysis. Clin. Transl. Gastroenterol. 2018;9:188. doi: 10.1038/s41424-018-0054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vieira WA, Pretorius E. The impact of asthma on the gastrointestinal tract (GIT) J. Asthma Allergy. 2010;3:123–130. doi: 10.2147/JAA.S10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marta L, Ana A, Solange O. Legume growth-promoting rhizobia: an overview on the Mesorhizobium genus? Microbiol. Res. 2014;20:2–17. doi: 10.1016/j.micres.2013.09.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.