Abstract
The spread of plasmid-mediated colistin resistance has posed a serious threat to public health owing to its effects on the emergence of pandrug-resistant bacteria. In this study, we investigated the prevalence and characteristics of mcr-1-positive Escherichia coli isolated from retail meat samples in Korea. In total, 1,205 E. coli strains were isolated from 3,234 retail meat samples in Korea. All E. coli strains were subjected to antimicrobial susceptibility testing and were examined for the presence of mcr-1 gene. All mcr-1-positive E. coli (n = 10, 0.8%) from retail meat were subjected to pulse-field gel electrophoresis (PFGE) and whole-genome sequencing (WGS). The transferability of mcr-1 gene was determined by conjugation assays. The mcr-1-positive strains exhibited diverse clonal types. Our mcr-1 genes were located in plasmids belonged to the IncI2 (n = 1) and IncX4 (n = 8) types, which were reported to be prevalent in Asia and worldwide, respectively. Most mcr-1 genes from mcr-1-positive strains (9/10) were transferable to the recipient strain and the transfer frequencies ranged from 2.4 × 10-3 to 9.8 × 10-6. Our data suggest that the specific types of plasmid may play an important role in spreading plasmid-mediated colistin resistance in Korea. Furthermore, our findings suggest that the retail meat may be an important tool for disseminating plasmid-mediated colistin resistance.
Keywords: Colistin resistance, mcr-1, prevalence, IncI2 plasmid, IncX4 plasmid, retail meats
Introduction
Colistin is a cationic polypeptide antibiotic that acts against most gram-negative bacteria, including those from the family Enterobacteriaceae. Colistin is a type of polymyxin, the five known subtypes of which (A–E) lead to disruption of membrane permeability by mediating electrostatic interactions between positively charged residues of polymyxin and negatively charged lipid A components of lipopolysaccharides (LPSs) in the bacterial membrane [1]. The use of colistin, also known as polymyxin E, in clinical practice has been limited because of its nephrotoxicity and neurotoxicity [2, 3]. However, owing to recent increases in multidrug resistant gram-negative bacteria and the rapid expansion of carbapenemase-producing Enterobacteriaceae, colistin has re-emerged as the last treatment option for severe bacterial infections [4, 5].
Several studies have reported that colistin resistance is related to chromosomal mutations in two-component systems, such as PmrAB and PhoPQ, leading to modification of LPS moieties in the outer membrane [6, 7]; therefore, there was little concern regarding the spread of colistin resistance. However, since Liu and colleagues first described the plasmid-mediated transfer of colistin resistance in China in 2015 [8], the spread of the mobile colistin resistance gene has posed a serious threat to human health because of the possible emergence of bacteria resistant to all available antimicrobials [9, 10].
The mobile colistin resistance gene mcr-1 encodes a phosphoethanolamine transferase enzyme that is capable of modifying lipid A in the bacterial membrane and reducing the affinity for colistin [11]. Most mcr-1 genes are located on various types of plasmids, including IncI2-, IncX4-, IncHI2-, IncP-, IncY-, and IncFI1-type plasmids, and are easily transferred to other strains [12, 13]. These mcr-1 genes were identified globally in various species of Enterobacteriaceae, such as Escherichia coli and Salmonella enterica isolated from humans, food animals, foods, and the environment [14]. mcr-1-positive E. coli have also been found in humans and food animals in Korea [15-17]. However, few studies have reported mcr-1-bearing bacteria recovered from food samples in Korea [18].
Since 2003, the Korean government has monitored and surveyed the antimicrobial resistance of bacteria collected from foods, such as retail meats, within the framework of the National Program on Antimicrobial Resistance Management [19]. mcr-1-positive E. coli were recently identified from retail meat samples. In this study, we report the occurrence rates of mcr-1-positive E. coli from retail raw meats in Korea and their genetic characteristics.
Materials and Methods
Sample Collection and Bacterial Isolation
In total, 3,234 raw meat samples, including beef (n = 1,290), pork (n = 1,126), and chicken (n = 818), were purchased at 291 retail stores spread across all the provinces of South Korea between 2015 and 2018. Overall, an average of ~800 raw meat samples were purchased per year. The domestic meat samples were obtained from 43 reputable processing companies for beef, 32 for pork, and 18 for chicken. Among the imported meat samples, beef samples were from 5 countries, pork from 14 countries, and chicken from 4 countries (Table S1). The meat samples were kept on ice during transportation from the grocery stores to the laboratory. Twenty-five grams of each meat sample was homogenized with 225 ml EC broth (Difco, USA) using a stomacher. The EC broth was incubated under aerobic conditions at 37°C for 24 h. An aliquot of each sample was streaked onto selective medium of Eosin Methylene Blue agar (Oxoid, UK) and incubated at 37°C for 24 h. Typical E. coli colonies (green metallic sheen) were sub-cultured on nutrient agar (Difco) and confirmed using a Vitek 2 Compact microbial identification system (bioMérieux, France) or Vitek MS (bioMérieux) in accordance with the manufacturer’s instructions. One typical and well-isolated E. coli strain per meat sample was selected. If no typical growth was observed, the sample was treated as a negative sample and was discarded. All isolates were stored at -80°C in Tryptic Soy Broth (Difco), mixed with 15% glycerol.
Antimicrobial Susceptibility
All E. coli strains (n = 1,205) were subjected to antimicrobial susceptibility testing using the following antibiotics: amoxicillin/clavulanic acid (AmC), ampicillin (AMP), cefoxitin (FOX), ceftiofur (CTF), chloramphenicol (CHL), ciprofloxacin (CIP), colistin (COL), gentamicin (GEN), nalidixic acid (NAL), streptomycin (STR), tetracycline (TET), and trimethoprim/sulfamethoxazole (SXT). The minimum inhibitory concentrations (MICs) of these antimicrobials were determined by a broth-microdilution method using a commercially available Sensititre plate KRNV4F (Trek Diagnostic Systems, USA). E. coli ATCC 25922 was used as a reference strain. For screening of colistin-resistant E. coli, the breakpoint for colistin resistance was applied at greater than 2 μg/ml, by referring to the European Committee on Antimicrobial Susceptibility Testing guidelines [20]. Susceptibility results of MICs for other antibiotics were interpreted in accordance with the Clinical and Laboratory Standards Institute guidelines [21] and the National Antimicrobial Resistance Monitoring System [22].
Polymerase Chain Reaction (PCR) Amplification of mcr-1 Genes
Template DNA from E. coli isolates for PCR was prepared using an UltraClean Microbial DNA Isolation Kit (MO BIO Laboratories Inc., USA) following the manufacturer’s instructions and stored at -20°C until use. The presence of mcr-1 was detected by PCR amplification using previously described primers [8]. A DNA thermal cycler (C1000 PCR System; Bio-Rad, USA) was used for PCR amplification, with the following protocol: 94°C for 15 min; 25 cycles of 94°C for 30 sec, 58°C for 90 sec, and 72°C for 60 sec; and final extension at 72°C for 10 min. The PCR products were analyzed using 1.5% (w/v) agarose gel electrophoresis.
Conjugation Assay of mcr-1-Positive Isolates
The transmissibility of mcr-1 gene was determined by conjugation assays in accordance with previously described broth-mating methods [23]. Briefly, azide-resistant E. coli strain J53 and isolates bearing mcr genes were used as the recipient and donor, respectively. Recipient and donor strains were mixed and incubated at a ratio of 1:1 in Luria-Bertani broth (Difco) for 8 h. Aliquots of these mixtures were plated on tryptic soy agar (Difco) containing sodium azide (200 μg/ml) and colistin (4 μg/ml) and incubated at 37°C for 20 h. PCR was used to confirm that the transconjugants carried the mcr-1 gene. Conjugation frequencies were determined as the number of transconjugants per a donor cell.
Pulsed-Field Gel Electrophoresis (PFGE) of mcr-1-Positive Isolates
Genotyping of mcr-1-positive E. coli isolates was conducted by PFGE with the CHEF-Mapper system (Bio-Rad) according to the PulseNet standardized protocol (http://www.pulsenetinternational.org/protocols/). Genomic DNA was digested with XbaI (Roche Molecular Biochemicals, USA) and separated on 1.0% pulsed-field certified agarose. Running conditions were as follows: 6.0 V/cm at 14°C for 18 h, with pulse times ramped from 2.2 to 54.2 s in 0.5× Tris-borate-ethylenediaminetetraacetic acid buffer. Genomic DNA from Salmonella enterica Braenderup H9812 (ATCC BAA-664) restricted with XbaI was used as a size marker. The PFGE patterns were analyzed with BioNumerics software ver. 5. 1 (Applied Maths, Belgium) using the Dice similarity coefficient with a 1.5% position tolerance, and clustering was performed by the unweighted-pair group method with average linkages (UPGMA). The results were interpreted according to the criteria reported previously [24].
Whole-Genome Sequencing (WGS) of mcr-1-Positive Isolates
WGS and assembly were performed at ChunLab Inc. (Korea) and Senigen Inc. (Korea). High-quality genomic DNA was extracted using an UltraClean Microbial DNA Isolation Kit (MO BIO Laboratories Inc.) according to the manufacturer’s instructions. The whole genome of mcr-1-positive isolates was sequenced on an Illumina Miseq desktop sequencer (Illumina Inc., USA), with paired-end reads of 300 bp length. The sequencing library was prepared with a TruSeq DNA LT Sample Prep Kit (Illumina Inc.) for the Illumina system. A de novo assembly was performed using SPAdes genome assembler version 3.13.0 [25]. The number of assembled contigs ranged between 69 and 135, with an average sequencing coverage of 129×. Antibiotic-resistance genes, replicon typing, and multilocus sequence typing (MLST) were conducted in ResFinder 3.1, PlasmidFinder 2.0, and MLST 2.0, respectively, on the Center for Genomic Epidemiology website (http://www.genomicepidemiology.org) [26-28]. Genome sequences were compared using the BLAST Ring Image Generator (BRIG) [29].
The whole-genome sequencing data reported in this study have been deposited at GenBank with the following accession numbers: from JACABR000000000 to JACABY000000000, WVVJ00000000, and WVVM00000000 (Table S2).
Statistical Data Analysis
Statistical analysis was performed using Epitools [30]. Comparisons between groups were evaluated by Chi-square (χ2) test. Results with p values of less than 0.05 were considered significant.
Results
Prevalence of mcr-1-Postive E. coli from Retail Meat
A total of 1,205 E. coli strains were isolated from 3,234 retail meat samples in Korea between 2015 and 2018. The colistin resistance of E. coli isolated from retail meat and the mcr-1 carriage rates are shown in Table 1. Of the 1,205 E. coli isolates, 51 isolates (4.2%) were resistant to colistin. The colistin-resistant isolates were obtained from domestic meat (3.7%, 33/891) and imported meat (5.7%, 18/314). Among these isolates, the mcr-1 gene was identified in 10 E. coli isolates, including one isolate from domestic pork, one from German pork, and eight from Brazilian chicken meat. However, 41 other-colistin-resistant isolates did not have the mcr-1 gene. The prevalence rates of mcr-1-positive isolates from domestic and imported meats were 0.1% and 2.9%, respectively. Isolates from imported meat samples showed significantly higher mcr-1 carriage rates than isolates from domestic meat samples (p < 0.05).
Table 1.
Prevalence of colistin-resistant and mcr-1-positive E. coli isolates from retail meat.
| Category | Prevalence of colistin-resistant E. coli (no. of resistant isolates/no. of tested isolates) | Prevalence of mcr-1-positive E. coli, % (no. of positive isolates/no. of tested isolates) | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Domestic | Imported | Total | P valuea | Domestic | Imported | Total | P valuea | |
| Beef | 2.6 (9/343) | 2.6 (2/78) | 2.6 (11/421) | 0.9762 | 0.0 (0/343) | 0.0 (0/78) | 0.0 (0/421) | > 0.9999 |
| Pork | 6.0 (8/134) | 5.6 (6/107) | 5.8 (14/241) | 0.9048 | 0.7 (1/134) | 0.9 (1/107) | 0.8 (2/241) | 0.8728 |
| Chicken | 3.9 (16/414) | 7.8 (10/129) | 4.8 (26/543) | 0.0710 | 0.0 (0/414) | 6.2 (8/129) | 1.5 (8/543) | < 0.0001 |
| Total | 3.7 (33/891) | 5.7 (18/314) | 4.2 (51/1205) | 0.1247 | 0.1 (1/891) | 2.9 (9/314) | 0.8 (10/1205) | < 0.0001 |
aP value, difference between the proportions of strains from domestic and imported meat samples by Chi-squared test.
Antimicrobial Resistance of Colistin-Resistant E. coli
Antimicrobial resistance of 51 colistin-resistant E. coli to 12 antibiotics is shown in Table 2. Among colistin-resistant strains, 34 strains (66.7%) exhibited a multidrug resistance (MDR) phenotype, which means that the strains were resistant to three or more antibiotics belonging to different categories. The occurrence of MDR in mcr-1-positive strains was not significantly different from that in mcr-1-negative strains. Moreover, no significant difference in the occurrence of resistance to each antimicrobial agent was present between mcr-1-positive strains and mcr-1-negative strains.
Table 2.
Antimicrobial resistance of colistin-resistant E. coli.
| Antibiotics | Source | Range tested (μg/ml) | Break points (μg/ml) | MIC50a | MIC90a | Resistance % (n) | P valueb |
|---|---|---|---|---|---|---|---|
| AmC | mcr-1-positive strains (n=10) | 2/1-32/16 | ≥ 32/16 | 8/4 | 32/16 | 20.0 (2) | 0.73 |
| mcr-1-negative strains (n=41) | 8/4 | > 32/16 | 31.7 (13) | ||||
| subtoal (n=51) | 8/4 | > 32/16 | 29.4 (15) | ||||
| AMP | mcr-1-positive strains (n=10) | 2-64 | ≥ 32 | 128 | > 64 | 60.0 (6) | 0.67 |
| mcr-1-negative strains (n=41) | 8 | > 64 | 46.3 (19) | ||||
| subtoal (n=51) | 8 | > 64 | 49.0 (25) | ||||
| CIP | mcr-1-positive strains (n=10) | 0.13-16 | ≥ 4 | 0.5 | 8 | 20.0 (2) | 0.63 |
| mcr-1-negative strains (n=41) | ≤ 0.13 | 8 | 34.1 (14) | ||||
| subtoal (n=51) | ≤ 0.13 | 8 | 31.4 (16) | ||||
| CHL | mcr-1-positive strains (n=10) | 2-64 | ≥ 32 | 8 | > 64 | 30.0 (3) | 0.87 |
| mcr-1-negative strains (n=41) | 8 | > 64 | 39.0 (16) | ||||
| subtoal (n=51) | 8 | > 64 | 37.3 (19) | ||||
| COL | mcr-1-positive strains (n=10) | 2-32 | > 2 | 8 | 16 | 100.0 (10) | NDc |
| mcr-1-negative strains (n=41) | 32 | > 32 | 100.0 (41) | ||||
| subtoal (n=51) | 32 | > 32 | 100.0 (51) | ||||
| CTF | mcr-1-positive strains (n=10) | 0.5-8 | ≥ 8 | ≤ 0.5 | > 8 | 20.0 (2) | 1.00 |
| mcr-1-negative strains (n=41) | ≤ 0.5 | 8 | 17.1 (7) | ||||
| subtoal (n=51) | ≤ 0.5 | > 8 | 17.6 (9) | ||||
| FOX | mcr-1-positive strains (n=10) | 1-32 | ≥ 32 | 8 | 32 | 20.0 (2) | 1.00 |
| mcr-1-negative strains (n=41) | 8 | > 32 | 17.1 (7) | ||||
| subtoal (n=51) | 8 | > 32 | 17.6 (9) | ||||
| GEN | mcr-1-positive strains (n=10) | 1-64 | ≥ 16 | ≤ 1 | 16 | 20.0 (2) | 1.00 |
| mcr-1-negative strains (n=41) | 2 | 32 | 14.6 (6) | ||||
| subtoal (n=51) | 2 | 32 | 15.7 (8) | ||||
| NAL | mcr-1-positive strains (n=10) | 2-128 | ≥ 32 | 8 | > 128 | 30.0 (3) | 0.87 |
| mcr-1-negative strains (n=41) | 4 | > 128 | 39.0 (16) | ||||
| subtoal (n=51) | 4 | > 128 | 37.3 (19) | ||||
| STR | mcr-1-positive strains (n=10) | 16-128 | ≥ 32 | ≤ 16 | 128 | 50.0 (5) | 1.00 |
| mcr-1-negative strains (n=41) | ≤ 16 | > 128 | 48.8 (20) | ||||
| subtoal (n=51) | ≤ 16 | > 128 | 49.0 (25) | ||||
| SXT | mcr-1-positive strains (n=10) | 0.13/2.4-4/76 | ≥ 4/76 | 0.25 | > 4/76 | 30.0 (3) | 1.00 |
| mcr-1-negative strains (n=41) | ≤ 0.13/2.4 | > 4/76 | 26.8 (11) | ||||
| subtoal (n=51) | ≤ 0.13/2.4 | > 4/76 | 26.8 (14) | ||||
| TET | mcr-1-positive strains (n=10) | 2-128 | ≥ 16 | ≤ 2 | 64 | 40.0 (4) | 0.67 |
| mcr-1-negative strains (n=41) | 32 | 128 | 53.7 (22) | ||||
| subtoal (n=51) | 32 | 128 | 51.0 (26) | ||||
| MDRc | mcr-1-positive strains (n=10) | ND | ND | ND | ND | 60.0 (6) | 0.90 |
| mcr-1-negative strains (n=41) | ND | ND | ND | ND | 68.3 (28) | ||
| subtoal (n=51) | ND | ND | ND | ND | 66.7 (34) |
aMIC50 and MIC90 are the concentration at which 50% and 90% of the isolates were inhibited.
bP value, difference between the proportions of mcr-1-positive and mcr-1-negative strains among colistin-resistant E. coli by Chi-squared test.
cAbbreviations: MDR, multidrug resistance; ND, not determined.
Characteristics of mcr-1-Positive E. coli
Among mcr-1-positive strains, eight strains exhibited a resistance phenotypes to at least two and up to 10 (Table 3). In particular, two strains from domestic pork (EC2018_100) and Brazilian chicken meat (EC2017_I306) showed the extended-spectrum β-lactamase (ESBL) phenotype, which were previously reported from our group [31]. These two strains carried the blaCTX-M-55 gene and the blaCTX-M-15 gene, respectively. Meanwhile, four strains from imported meat samples harbored β-lactamase-related genes (blaTEM, blaSHV), which showed a non-ESBL phenotype and resistance to AMP. Three strains (EC2016_I15, EC2016_I115, and EC2018_100) exhibited the resistance to NAL or CIP. Although two of these strains (EC2016_I15 and EC2016_I115) did not harbor the quinolone resistance genes, the presence of point mutations in quinolone resistance-determining regions (QRDR) in chromosomal gyrA or parC genes was noted (Table S3). Resistance phenotypic results correlated with the presence of the different resistance genes for each antimicrobial family (Tables 3 and S3). All mcr-1-positive strains harbored the mdf(A) gene, but the resistance to macrolides was not determined in this study.
Table 3.
Genetic features of mcr-1-positive E. coli isolated from retail meat.
| Strain | Source | Year | Resistance phenotypea | Resistance genes | Plasmid repliconsb | mcr-1 gene transfer |
|---|---|---|---|---|---|---|
| EC2015_I58 | Chicken (Brazil) | 2015 | COL, STR | mcr-1.1, aadA1, mdf(A), qnrB19 | Col(pHAD28), IncFⅠB(K), IncFⅡ(29), IncX4 | Yes |
| EC2016_I15 | Chicken (Brazil) | 2016 | AmC, AMP, COL, FOX, NAL, STR, SXT | mcr-1.1, aph(3'')-Ib, aph(6)-Id, blaTEM-1B, mdf(A), sul2, dfrA14 | IncFⅠB(AP001918), IncFⅡ, IncX4 | Yes |
| EC2016_I103 | Pork (Germany) | 2016 | AMP, CHL, COL, TET | mcr-1.1, aadA2b, blaTEM-1B, mdf(A), qnrS1, tet(A), dfrA8 | IncR, IncX4 | Yes |
| EC2016_I115 | Chicken (Brazil) | 2016 | CIP, COL, GEN, NAL, STR, TET | mcr-1.1, aac(3)-Ⅴia, aadA1, mdf(A), sul1, tet(A) | IncFⅠB(AP001918), IncFⅡ(SE11), IncX4 | No |
| EC2016_I119 | Chicken (Brazil) | 2016 | AmC, AMP, CHL, COL, FOX, STR, SXT | mcr-1.1, aadA1, aadA2, blaTEM-1A, Inu(A), mdf(A),cmlA1, qnrB19, sul3, dfrA12 | Col(pHAD28), IncFⅠB(AP001918), IncI1-Ⅰ, IncI2, IncX1, IncX4, IncY | Yes |
| EC2016_I183 | Chicken (Brazil) | 2016 | COL | mcr-1.1, mdf(A) | IncX4 | Yes |
| EC2016_I182 | Chicken (Brazil) | 2016 | COL | mcr-1.5, aadA1, aadA2b, aph(3')-Ia, mdf(A),cmlA1, sul3 | IncFⅠB(AP001918), IncI1-Ⅰ, IncI2 | Yes |
| EC2017_I300 | Chicken (Brazil) | 2017 | AMP, COL | mcr-1.1, blaSHV-12, mdf(A) | IncFⅠB(AP001918), IncI1-I, IncFⅡ(pRSB107), IncX4, p0111 | Yes |
| EC2017_I306 | Chicken (Brazil) | 2017 | AMP, COL, CTF, TET | mcr-1.1, blaCTX-M-55, fosA3, mdf(A), tet(A) | IncFⅡ(pHN7A8), IncX4, p0111 | Yes |
| EC2018_100 | Pork (Korea) | 2018 | AMP, CHL, CIP, COL, CTF, GEN, NAL, STR, SXT, TET | mcr-1.1, aac(3)-IId, aadA1, blaCTX-M-15, blaTEM-1B, Inu(F), mdf(A),cmlA1, qnrS1, sul2, sul3, tet(A), tet(M), dfrA12 | IncFⅠB(AP001918), IncI2, IncX1 | Yes |
aUnderlining indicates resistance by transconjugants.
bBolded plasmid name means replicons found in the same contig as the mcr-1 gene.
mcr-1 genes, except EC2016_I182 strain, were found in the same contigs as replicons of the families of IncX4 (n =8) and IncI2 (n = 1), which means that mcr-1 genes were located in the IncX4 and IncI2 plasmid. Additionally, mcr-1-positive strains contained a wide variety of plasmid incompatibility group replicons, ranging from one to seven per strain.
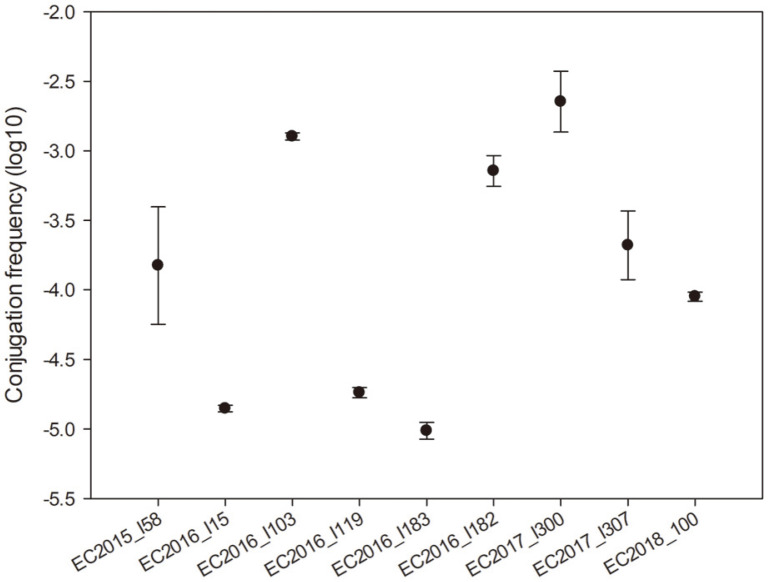
Conjugation tests showed that 9 of the 10 mcr-1-positive strains were able to transfer their colistin-resistance phenotype to E. coli J53 (Table 3). Conversely, no other resistance among these strains was cotransferred to the recipient strain except the EC2018_100 strain. The transfer frequencies ranged from 2.4 × 10-3 to 9.8 × 10-6 (Fig. 1).
Fig. 1. Conjugation frequencies of nine mcr-1-positive E. coli from retail meat.
The data represent the averages and standard deviations.
Epidemiology of mcr-1-Positive E. coli
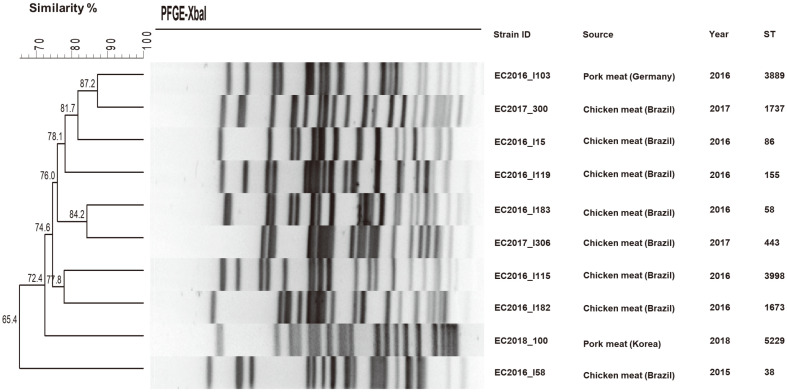
In silico MLSTs of mcr-1-positive strains were generated from draft genome sequences, and sequence types (STs) were assigned according to the E. coli Achtman scheme. MLST indicated that nine strains belonged to different STs (Fig. 2). The ST (ST58) of one strain from Brazilian chicken meat was the clonal MLST complex of ST155. Additionally, all mcr-1-positive E. coli strains showed very diverse PFGE pulsotypes.
Fig. 2. XbaI PFGE dendrogram with the corresponding MLST sequence types of mcr-1-positive E. coli strains from retail meat samples.
Based on the UPGMA algorithm, the dendrogram revealed each different PFGE pulsotype.
Discussion
In our study, we detected the plasmid-mediated colistin resistance gene mcr-1 in 10 E. coli strains from retail meat samples purchased at Korean grocery stores between 2015 and 2018. Amongst 10 mcr-1-positive E. coli strains, nine E. coli strains were recovered from the imported meat samples, and one E. coli strain was isolated from a domestic pork sample in 2018. The observed 0.1% prevalence of mcr-1-positive strains from retail domestic meat samples is comparable with the previously reported 0.1% prevalence in livestock in Korea [16]. Moreover, the prevalence of mcr-1-positive strains from the domestic meat in our study was lower than that of isolates previously reported from other countries, such as Brazil, China, Germany, Japan, the Netherlands, and Portugal [8,32-36]. Further, the prevalence of colistin-resistant E. coli strains from the domestic meat samples in our study was lower than the previously reported prevalence of isolates from meat samples in Germany and Brazil (p < 0.05) when comparing the prevalence between our study and previous studies by chi-square test [35, 36]. The national sales data for veterinary antibiotics have shown that sales of colistin are relatively low for animal breeding [37]. This may explain why the prevalence of colistin-resistant strains was also low. To the best of our knowledge, this is the first report of such a low prevalence of mcr-1-positive E. coli isolated from retail meat in Korea. Interestingly, no mcr-1-positive E. coli isolates have been found from cattle and beef in Korea [16-18,38,39]. In our study, most mcr-1-positive E. coli were isolated from Brazilian chicken meat samples. In Brazil, chicken meat was previously reported as a reservoir for mcr-1-positive E. coli [35]. This may explain the high occurrence of mcr-1-positive isolates from Brazilian chicken meat. Amongst 51 colistin-resistant isolates, 41 mcr-1-negative isolates were identified. The resistance to colistin in these isolates may be associated with chromosomal mutations in PmrAB and PhoPQ, leading to reduce the binding affinity of colistin for its target [6, 7]. However, further studies are needed to elucidate why mcr-1-negative isolates presented resistance to colistin.
The mcr-1 gene in mcr-1-positive E. coli from domestic pork was located in the IncI2-type plasmid, which was prevalent in Asia [12] and was the type of the first reported mcr-1-harboring plasmid (pHNSHP45) from porcine E. coli in China [8]. The mcr-1 genes in mcr-1-positive E. coli from other sources, such as poultry carcasses, chicken feces, chicken meat, and patients, were reported to be located in IncI2-type plasmids [15, 16, 18, 39]. Furthermore, the mcr-1-bearing IncI2 plasmid contig from domestic pork meat was similar to IncI2-type plasmids from other sources, such as livestock, humans, and chicken meat, in Korea [15, 16, 18, 39] (Fig. S1). This suggests that IncI2-type plasmid may play an important role in spreading mcr-1 gene in Korea.
The mcr-1 genes in eight mcr-1-positive E. coli from Brazilian chicken and German pork samples were located in IncX4-type plasmid, which is distributed worldwide [12]. The IncX4 plasmid type was previously found in Brazilian poultry meat samples [35, 40] and in German swine samples [41, 42]. The IncX4-type plasmid was previously detected in mcr-1-positive E. coli from a diseased swine feces sample in Korea [16, 39].
All but one of mcr-1 genes located in the IncX4-type plasmids were successfully transferred to the E. coli recipient strain. The transfer frequencies of mcr-1 genes varied, which is comparable with the previous study [39]. A previous study reported that mcr-1-bearing, IncI2-type plasmids from E. coli were transmissible to other gram-negative bacteria such as Salmonella and Klebsiella as well as E. coli with colistin resistance [18]. However, a previous study showed that the transferability of the mcr-1-bearing plasmid depended on the recipient strain or species rather than the plasmid type [43]. The reason for the nontransferability of mcr-1 gene located in the IncX4 plasmid of EC2016_I115 strain was not determined in our study.
MLST and PFGE are powerful molecular typing techniques for tracking genetic relatedness [44]. In this study, mcr-1-positive E. coli strains showed very diverse STs and PFGE pulsotypes, presumably indicating that they originated from different clones. ST5229, identified from E. coli in domestic pork meat (EC2018_100), was found in porcine E. coli in Spain and human E. coli in Hong Kong [45, 46]. ST5229 belongs to the clonal MLST complex of ST101, which was previously found in New Delhi metallo-β-lactamase-producing E. coli from humans and mcr-1-carrying E. coli from pig feces in Korea [17, 47]. STs in mcr-1-positive E. coli from imported meat were previously found from livestock, humans, and food in Asia, Europe, America, and Australia (enterobase.warwick.ac.kr) and were either rare or widespread.
A limitation of this study is the sampling design of colistin-resistant and mcr-1-positive E. coli strains. Due to the fact that just one E. coli strain per meat sample was selected, the prevalence of colistin-resistant and mcr-1-positive E. coli strains in our study could be underestimated. Despite these limitations, this study provides a comprehensive overview of mcr-1-positive E. coli diversity and common plasmid-type-bearing mcr-1 in retail meat in Korea.
In this study, we describe the prevalence and characteristics of mcr-1-positive E. coli isolated from domestic and imported meat samples in Korea. Our data showed that the prevalence of mcr-1-positive E. coli strains from retail meat was 0.8%. The mcr-1-positive strains from retail meat samples exhibited diverse STs and PFGE pulsotypes, suggesting that the strains had evolved from different E. coli clones. However, the mcr-1 genes in our study were located in the specific types of plasmid (IncI2 and IncX4) and these plasmid types bearing mcr-1 were also found in mcr-1-positive E. coli from other sources, including humans, animals, and chicken meat in Korea. These findings suggested that the specific types of plasmid may play an important role in spreading plasmid-mediated colistin resistance in Korea. The mcr-harboring plasmid may contribute to the spread of colistin resistance and the emergence of pandrug-resistant pathogens due to its high transferability to other strains. Thus, retail meat may pose a health risk to consumers and food handlers despite the low prevalence of mcr-1-positive E. coli from retail meat in Korea, if contaminated with plasmid-mediated colistin-resistant strains. Therefore, close surveillance of mcr-1-positive strains should be continued to establish a containment strategy for preventing the spread of colistin resistance throughout the food chain.
Supplemental Material
Supplementary data for this paper are available on-line only at http://jmb.or.kr.
Acknowledgments
This research was supported by grants (nos. 15161MFDS645 and 18161MFDS035) from the Ministry of Food and Drug Safety. The findings and conclusions of this article are ours and do not necessarily represent the views of the Ministry of Food and Drug Safety. We thank the members of AMR working group (J. Kim, S. Seo, and J. Park) for contribution to collecting some E. coli strains used in this study.
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
REFERENCES
- 1.Falagas ME, Kasiakou SK, Saravolatz LD. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 2005;40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 2.Javan AO, Shokouhi S, Sahraei Z. A review on colistin nephrotoxicity. Eur. J. Clin. Pharmacol. 2015;71:801–810. doi: 10.1007/s00228-015-1865-4. [DOI] [PubMed] [Google Scholar]
- 3.Yahav D, Farbman L, Leibovici L, Paul M. Colistin: new lessons on an old antibiotic. Clin. Microbiol. Infect. 2012;18:18–29. doi: 10.1111/j.1469-0691.2011.03734.x. [DOI] [PubMed] [Google Scholar]
- 4.Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 2017;30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2013;13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeannot K, Bolard A, Plesiat P. Resistance to polymyxins in gram-negative organisms. Int. J. Antimicrob. Agents. 2017;49:526–535. doi: 10.1016/j.ijantimicag.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 7.Baron S, Hadjadj L, Rolain J-M, Olaitan AO. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int. J. Antimicrob. Agents. 2016;48:583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 9.Forde BM, Zowawi HM, Harris PN, Roberts L, Ibrahim E, Shaikh N, et al. Discovery of mcr-1-mediated colistin resistance in a highly virulent Escherichia coli lineage. mSphere. 2018;3:e00486–00418. doi: 10.1128/mSphere.00486-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du H, Chen L, Tang Y-W, Kreiswirth BN. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect. Dis. 2016;16:287–288. doi: 10.1016/S1473-3099(16)00056-6. [DOI] [PubMed] [Google Scholar]
- 11.Hinchliffe P, Yang QE, Portal E, Young T, Li H, Tooke CL, et al. Insights into the mechanistic basis of plasmid-mediated colistin resistance from crystal structures of the catalytic domain of MCR-1. Sci. Rep. 2017;7:39392. doi: 10.1038/srep39392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matamoros S, Van Hattem JM, Arcilla MS, Willemse N, Melles DC, Penders J, et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 2017;7:15364. doi: 10.1038/s41598-017-15539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li R, Xie M, Lv J, Wai-Chi Chan E, Chen S. Complete genetic analysis of plasmids carrying mcr-1 and other resistance genes in an Escherichia coli isolate of animal origin. J. Antimicrob. Chemother. 2017;72:696–699. doi: 10.1093/jac/dkw509. [DOI] [PubMed] [Google Scholar]
- 14.Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill. 2016;21:30155. doi: 10.2807/1560-7917.ES.2016.21.9.30155. [DOI] [PubMed] [Google Scholar]
- 15.Kim ES, Chong YP, Park S-J, Kim M-N, Kim S-H, Lee S-O, et al. Detection and genetic features of MCR-1-producing plasmid in human Escherichia coli infection in South Korea. Diagn. Microbiol. Infect. Dis. 2017;89:158–160. doi: 10.1016/j.diagmicrobio.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 16.Lim S-K, Kang HY, Lee K, Moon D-C, Lee H-S, Jung S-C. First detection of the mcr-1 gene in Escherichia coli isolated from livestock between 2013 and 2015 in South Korea. Antimicrob. Agents Chemother. 2016;60:6991–6993. doi: 10.1128/AAC.01472-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh S-S, Song J, Kim J, Shin J. Increasing prevalence of multidrug-resistant mcr-1-positive Escherichia coli isolates from fresh vegetables and healthy food animals in South Korea. Int. J. Infect. Dis. 2020;92:53–55. doi: 10.1016/j.ijid.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 18.Kim J, Hwang BK, Choi H, Wang Y, Choi SH, Ryu S, et al. Characterization of mcr-1-harboring plasmids from pan drug-resistant Escherichia coli strains isolated from retail raw chicken in South Korea. Microorganisms. 2019;7:344. doi: 10.3390/microorganisms7090344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koo HJ, Woo GJ. Characterization of antimicrobial resistance of Escherichia coli recovered from foods of animal and fish origin in Korea. J. Food Prot. 2012;75:966–972. doi: 10.4315/0362-028X.JFP-11-003. [DOI] [PubMed] [Google Scholar]
- 20.EUCAST. Breakpoint tables for interpretation of MICs and zone diameters. Version 10.0. 2020. [Accessed Jan. 20, 2020.]. Available from http://www.eucast.org .
- 21.CLSI. CLSI supplement M100. 28th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. Performance Standards for Antimicrobial Susceptibility Testing, CLSI supplement M100. [Google Scholar]
- 22.NARMS. Antimicrobial agents used for susceptibility testing for E. coli isolates. 2020. https://www.cdc.gov/narms/antibiotics-tested.html.
- 23.Shin SW, Shin MK, Jung M, Belaynehe KM, Yoo HS. Prevalence of antimicrobial resistance and transfer of tetracycline resistance genes in Escherichia coli isolates from beef cattle. Appl. Environ. Microbiol. 2015;81:5560–5566. doi: 10.1128/AEM.01511-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 1995;33:2233–2239. doi: 10.1128/JCM.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using plasmid finder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sergeant E. Epitools epidemiological calculators. 2018. [Accessed Feb. 21, 2018]. Available from http://epitools.ausvet.com.au .
- 31.Kim S, Kim H, Kim Y, Kim M, Kwak H, Ryu S. Whole-genome sequencing-based characteristics in extended-spectrum beta-lactamase-producing Escherichia coli isolated from retail meats in Korea. Microorganisms. 2020;8:508. doi: 10.3390/microorganisms8040508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clemente L, Manageiro V, Correia I, Amaro A, Albuquerque T, Themudo P, et al. Revealing mcr-1-positive ESBL-producing Escherichia coli strains among Enterobacteriaceae from food-producing animals (bovine, swine and poultry) and meat (bovine and swine), Portugal, 2010-2015. Int. J. Food Microbiol. 2019;296:37–42. doi: 10.1016/j.ijfoodmicro.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Ohsaki Y, Hayashi W, Saito S, Osaka S, Taniguchi Y, Koide S, et al. First detection of Escherichia coli harboring mcr-1 gene from retail domestic chicken meat in Japan. Jpn. J. Infect. Dis. 2017;70:590–592. doi: 10.7883/yoken.JJID.2016.572. [DOI] [PubMed] [Google Scholar]
- 34.Schrauwen EJ, Huizinga P, van Spreuwel N, Verhulst C, Kluytmans-van den Bergh MF, Kluytmans JA. High prevalence of the mcr-1 gene in retail chicken meat in the Netherlands in 2015. Antimicrob. Resist. Infect. Control. 2017;6:83. doi: 10.1186/s13756-017-0242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monte DF, Mem A, Fernandes MR, Cerdeira L, Esposito F, Galvão JA, et al. Chicken meat as a reservoir of colistin-resistant Escherichia coli strains carrying mcr-1 genes in South America. Antimicrob. Agents Chemother. 2017;61:e02718–02716. doi: 10.1128/AAC.02718-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Irrgang A, Roschanski N, Tenhagen B-A, Grobbel M, Skladnikiewicz-Ziemer T, Thomas K, et al. Prevalence of mcr-1 in E. coli from livestock and food in Germany, 2010-2015. PLoS One. 2016;11:e0159863. doi: 10.1371/journal.pone.0159863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korea Animal Health Products Association (KAHPA), Animal and Plant Quarantine Agency (APQA), National Institute of Food and Drug Safety Evaluation (NIFDS) The sales of antimicrobials (estimation) in animals and fisheries. In National antibiotics use in food animals and monitoring of antimicrobial resistance in 2018. 2020. [Accessed March 29, 2020]. Available from http://ebook.qia.go.kr/20190918_104137 .
- 38.Yoon E-J, Hong JS, Yang JW, Lee KJ, Lee H, Jeong SH. Detection of mcr-1 plasmids in Enterobacteriaceae isolates from human specimens: Comparison with those in Escherichia coli isolates from livestock in Korea. Ann. Lab. Med. 2018;38:555–562. doi: 10.3343/alm.2018.38.6.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J-Y, Lim S-K, Choi Y, Moon D-C, Shin J, Ko KS. Whole sequences and characteristics of mcr-1-harboring plasmids of Escherichia coli strains isolated from livestock in South Korea. Microb. Drug. Resist. 2018;24:489–492. doi: 10.1089/mdr.2017.0369. [DOI] [PubMed] [Google Scholar]
- 40.Moreno LZ, Gomes VT, Moreira J, de Oliveira CH, Peres BP, Silva APS, et al. First report of mcr-1-harboring Salmonella enterica serovar Schwarzengrund isolated from poultry meat in Brazil. Diagn. Microbiol. Infect. Dis. 2019;93:376–379. doi: 10.1016/j.diagmicrobio.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 41.Roschanski N, Roesler U, Guenther S, Imirzalioglu C, Falgenhauer L, Chakraborty T, et al. Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from German swine farms. J. Antimicrob. Chemother. 2017;72:1289–1292. doi: 10.1093/jac/dkw585. [DOI] [PubMed] [Google Scholar]
- 42.Roschanski N, Falgenhauer L, Grobbel M, Guenther S, Kreienbrock L, Imirzalioglu C, et al. Retrospective survey of mcr-1 and mcr-2 in German pig-fattening farms, 2011-2012. Int. J. Antimicrob. Agents. 2017;50:266–271. doi: 10.1016/j.ijantimicag.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 43.Valérie DT, Laurent P, Patrice N. Transferability of the mcr-1 colistin resistance gene. Microb. Drug. Resist. 2017;23:813–814. doi: 10.1089/mdr.2016.0191. [DOI] [PubMed] [Google Scholar]
- 44.Noller AC, McEllistrem MC, Stine OC, Morris J, Glenn J, Boxrud DJ, Dixon B, et al. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. Clin. Microbiol. 2003;41:675–679. doi: 10.1128/JCM.41.2.675-679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El Garch F, De Jong A, Bertrand X, Hocquet D, Sauget M. mcr-1-like detection in commensal Escherichia coli and Salmonella spp. from food-producing animals at slaughter in Europe. Vet. Microbiol. 2018;213:42–46. doi: 10.1016/j.vetmic.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 46.Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microb. Infect. 2018;7:122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoo JS, Kim HM, Koo HS, Yang JW, Yoo JI, Kim HS, et al. Nosocomial transmission of NDM-1-producing Escherichia coli ST101 in a Korean hospital. J. Antimicrob. Chemother. 2013;68:2170–2172. doi: 10.1093/jac/dkt126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.