Abstract
Phenylalanine ammonia-lyase (PAL) catalyzes the reversible deamination of phenylalanine to cinnamic acid and ammonia. Algae have been considered as biofactories for PAL production, however, biochemical characterization of PAL and its potency for myristicin biotransformation into MMDA (3-methoxy-4, 5-methylenedioxyamphetamine) has not been studied yet. Thus, PAL from Anabaena flos-aquae and Spirulina platensis has been purified, comparatively characterized and its affinity to transform myristicin was assessed. The specific activity of purified PAL from S. platensis (73.9 μmol/mg/min) and A. flos-aquae (30.5 μmol/mg/min) was increased by about 2.9 and 2.4 folds by gel-filtration comparing to their corresponding crude enzymes. Under denaturing-PAGE, a single proteineous band with a molecular mass of 64 kDa appeared for A. flos-aquae and S. platensis PAL. The biochemical properties of the purified PAL from both algal isolates were determined comparatively. The optimum temperature of S. platensis and A. flos-aquae PAL for forward or reverse activity was reported at 30°C, while the optimum pH for PAL enzyme isolated from A. flos-aquae was 8.9 for forward and reverse activities, and S. platensis PAL had maximum activities at pH 8.9 and 8 for forward and reverse reactions, respectively. Luckily, the purified PALs have the affinity to hydroaminate the myristicin to MMDA successfully in one step. Furthermore, a successful method for synthesis of MMDA from myristicin in two steps was also established. Gas chromatography-mass spectrometry (GC-MS) analysis was conducted to track the product formation.
Keywords: Phenylalanine ammonia-lyase, purification, properties, myristicin, MMDA
Introduction
Phenylalanine ammonia-lyase (PAL, E.C. 4.3.1.5) belongs to the ammonia lyases family [1], and catalyzes the deamination (forward) and hydroamination (reverse) reactions of phenylalanine as substrate [2, 3]. The forward reaction involves deamination of L-phenylalanine (L-Phe) to trans-cinnamic acid (t-Ca) and ammonia, while, the reversal reaction involves biotransformation of trans-cinnamic acid to L-phenylalanine [4, 5], as illustrated in Fig. 1A. PAL is widely distributed in plants, acting as a key enzyme in phenylpropanoid pathway [6, 7], controlling the production of secondary metabolites such as flavonoids, coumarins, lignins, stilbenes and phytoalexins [7]. PAL has also been reported found in fungi, yeasts and bacteria [4]. Fungal PAL can utilize L-phenylalanine as a carbon and nitrogen source [5, 8]. Bacterial PAL was reported to be implemented in biosynthesis of secondary metabolites as cinnamamide in Streptomyces verticillatus [9], 3,5-dihydroxy-4-isopropylstilbene in Photorhabdus luminescens and enterocin in Streptomyces maritimus [1, 10]. Few studies were documented to the PAL from algae, except Anabaena variabilis, Nostoc punctiforme [11], Dunaliella marina [4] and Anacystis nidulans [12]. Animal [4] and human [13, 14] tissues are free of PAL. Recently, there is a growing interest in exploring the biochemical properties of PAL because of its clinical and commercial applications [15]. Chemically modified PAL has been used as a therapy for phenylketonuria (PKU) [9, 16], which is an inborn disorder of phenylalanine metabolism caused by deficiency of phenylalanine hydroxylase (PAH), causing cognitive development loss and mental retardation as a result of hyperphenylalaninemia [17, 18]. Also, PAL has been used for anticancer activity due to its selectivity for phenylalanine by inhibiting the growth of neoplasms in vitro and in lymphoblastic leukemia [4, 15, 19]. Moreover, reverse pathway of PAL meets the great requirements for L-phenylalanine in the food and pharmaceutical industries [15]. As an essential amino acid, L-phenylalanine [20] serves as a supplement for human nutrition and as a precursor for the synthesis of artificial sweetener aspartame [21-23].
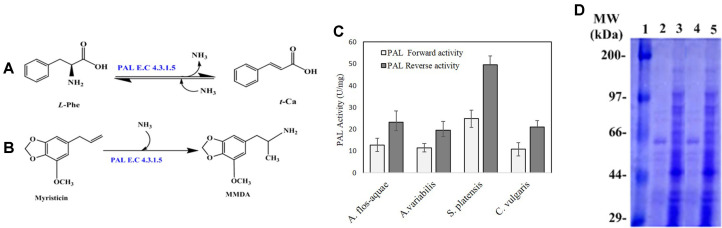
Fig. 1. Forward and reverse pathways catalyzed by PAL (A), Proposed biotransformation of myristicin to MMDA by PAL (B), Forward and reverse activities of PAL from different algal isolates (C), SDS-PAGE of the purified PAL from S. platensis and A. flos-aquae (D).
Lane 1, broad range marker (Cat# sc-2361, 6-200 kDa, Santa Cruz Biotechnology, Inc. USA), lane 2 and lane 4 are purified PAL from S. platensis and A. flos-aquae, respectively, lane 3 and lane 5 are crude extracts of S. platensis and A. flos-aquae, respectively.
Myristicin (1-allyl-5-methoxy-3,4-methylenedioxybenzene), is a naturally occurring phenylpropanoid derivative [24, 25] found in nutmeg [26], cinnamon, parsley, basil, carrot [27], dill [28] and various plants belonging to the Umbelliferae [29]. Myristicin has various applications in the food and cosmetic industries, in treatment of anxiety, stomach cramps, diarrhea, and cholera in addition to having antibacterial, anti-inflammatory, hepatoprotective, anti-cholinergic [27, 30] and insecticidal [31, 32] activities. Metabolic conversion of myristicin into 3-methoxy-4, 5-methylenedioxy-amphetamine (MMDA) had been studied in rats [33] and humans [34, 35]. MMDA is a potent psychoactive drug [36-38] that has about twice higher activities than those of mescaline [33], with similar activity to 3,4,5-trimethoxyamphetamine (TMA) [39]. MMDA attenuates anxiety and loneliness feelings, suppresses appetite [40] and elevates the mood [35, 41]. MMDA is used for treatment of autism, major depressive disorder, anxiety [42, 43], alcohol dependence, Alzheimer’s, attention-deficit hyperactivity disorder (ADHD), and parkinsonism-dystonia infantile and schizophrenia [43-45]. MMDA has been used as a psychotherapy adjunct [46], as a useful drug for neurosis treatment [35]. Moreover, another study reported at the American Psychological Association’s annual meeting in 2018 that MMDA is used for treatment of social anxiety, depression, and post-traumatic stress disorder [47]. However, production of MMDA is the main technical challenge for its various pharmaceutical applications. MMDA has been synthesized commercially from myristicin by Shulgin [48] and Clark et al., [49], however, the price of MMDA is about 17.8 folds higher than myristicin [50]. Thus, searching for alternative novel methods for production of MMDA with higher yield and low price is an objective for many researchers. Production of MMDA via biotransformation of myristicin with PAL seems to be a novel hypothetical approach for higher yield of MMDA, as proposed in Fig. 1B. From literature, PAL of algal sources was reported to have a higher turnover number, catalytic activity and broad substrate specificity, than plant and fungal and bacterial sources of PAL (https://www.brenda-enzymes.org/enzyme.php?ecno=4.3.1.5). Thus, the objective of this work was to purify PAL from different algal sources, in addition to exploring the potentiality of purified PAL to transform myristicin as substrate into MMDA, using one-step enzymatic synthesis. To the best of our knowledge, nothing has been reported on the production of PAL by green algae like Chlorella vulgaris and blue-green algae such as Anabaena flos-aquae and Spirulina platensis except Anabaena variabilis var. kashiensis (Bharadwaja) [5, 10]. In view of this, the second aim of this research is to discover new microbial sources of PAL production. Thus, our report investigates the extraction, purification, and characterization of PAL from A. flos-aquae and S. platensis. Moreover, investigation of myristicin biotransformation by purified PAL is the latest aim.
Materials and Methods
Algal Strains and Culture Conditions
Fifty microbial isolates (twenty fungal isolates, twenty bacterial isolates, and ten algal isolates) were collected from the microbiology laboratory of the Faculty of Science, Zagazig, Zagazig University, Egypt. The fungal isolates were grown on potato dextrose broth media [51], while the bacterial isolates were grown on nutrient broth media [52]. Anabaena flos-aquae and A. variabilis were grown on blue-green medium (BG-11) [53], Spirulina platensis was grown on Zarrouk medium [54], and Chlorella vulgaris was grown on diatom culture medium (DM) [55]. The fungal cultures were incubated for 10 days at 30°C, while bacterial cultures were incubated at 5 days at 37°C. Algal cultures were maintained at 27°C ± 1°C, except S. platensis, which was incubated at 31 ± 0.5°C under white fluorescent illumination of 30 to 40 μEm-2s-1 provided by fluorescent tubes (Philips Trulite, Col 82). The cultures were exposed to light: dark photoperiod (12:12) and aerated with air current through an electric pump (20 l/h). The pH of media was adjusted to 7.1 (BG-11), 9.5 (Zarrouk medium) and 6.9 (DM) by 0.1N HCl and /or NaOH using a Jenway 3510 pH meter.
Algal Harvesting and Extraction of Phenylalanine Ammonia Lyase (PAL)
The algal cells were harvested by centrifugation at 10,000 ×g (4°C) for 15 min at mid-logarithmic phase, washed three times with sterile distilled water and the algal masses were stored at -20°C. Twenty grams of frozen algal biomass of each strain were separately pulverized in liquid nitrogen and suspended into 50 ml of 100 mM cold Tris-HCl buffer (pH 8.9), with 0.2% Na2-EDTA and 125 μl β-mercaptoethanol. The extracts were centrifuged at 10,000 ×g for 10 min at 4°C and the supernatants were used as crude extracts for enzyme assay, protein estimation and purification.
Phenylalanine Ammonia Lyase (PAL) Assay
The deaminating and hydroaminating activities of the crude enzyme were determined [1, 56, 57] with slight modifications. Briefly, for deaminating activity, assay was performed using 50 μM L-phenylalanine, 250 μl of enzyme extract, in 2 ml of 100 mM Tris-HCl buffer (pH 8.9), and incubated at 37°C for 30 min. The reaction was stopped by addition of 500 μl of 1N HCl and the absorbance of cinnamic acid was measured at 270 nm. The concentration of cinnamic acid was calculated with regard to standard curve. For hydroamination assay, the reaction (2 ml) contained 100 mM Tris-HCl buffer (pH 8.9), 200 μl of 1 mM trans-cinnamic acid in 5 M NH4OH and 200 μl of enzyme preparation and was incubated at 37°C for 30 min. Formation of L-phenyl-alanine as a byproduct was quantitatively estimated at 257 nm (Rigol ultra-3660 UV-VIS spectrophotometer). One unit (U) of PAL was expressed by the amount of enzyme that catalyzes the formation of 1 μM of trans-cinnamic acid/L-phenylalanine per min under optimal assay conditions. The specific activity (U/mg) was expressed by the enzyme activity in unit per milligram of protein.
The concentration of proteins was estimated by Folin’s reagent according to Lowry et al. [58], using bovine serum albumin as standard.
Purification of PAL by Gel-Filtration and Ion-Exchange Chromatography
The crude enzyme from the selected algal isolates was purified using gel-filtration and ion-exchange chromatographic approaches [59-64]. The crude PAL was precipitated with two-fold chilled acetone, and incubated at -20°C for 30 min. The mixture was centrifuged at 6,000 ×g for 15 min at 4°C, and the supernatant was decanted. The protein pellets were dissolved in 5 ml of 100 mM cold Tris-HCl buffer (pH 8.9) as described above, followed by centrifugation at 10,000 ×g for 5 min at 4°C. Crude extract (3.5 and 8 mg protein/ml) of PAL from S. platensis and A. flos-aquae, respectively, was applied to a previously equilibrated Sephadex G-200 column (30 cm × 2 cm), with 100 mM cold Tris-HCl buffer (pH 8.9). The column was equilibrated with 100 mM Tris-HCl (pH 8.9) at flow rate 0.5 ml/min, and the fractions (1 ml) were eluted. The activity of PAL and protein content were measured for each fraction. The molecular homogeneity of the most active fractions of PAL were checked by SDS-PAGE, the most active and molecularly homogenous fractions were pooled, collected and concentrated by dialysis (Dialysis Membrane, Size 20, Cat# 546-00051, Wako Chemicals, USA) with 100 mM Tris-HCl buffer (pH 8.9) at 4°C, till reduction of the total volume to 2 ml. The pooled fractions of PAL were further purified by ion-exchange chromatography with DEAE-cellulose. The partially purified PAL was loaded to the top of DEAE-cellulose column previously equilibrated with 100 mM Tris-HCl buffer. The enzyme was eluted on the same buffer with gradient concentrations of NaCl (100-500 mM). The activity of PAL and protein concentration for each fraction were analyzed as described above. The molecular homogeneity of the active fractions was checked by SDS-PAGE analysis, and the molecularly active homogenous fractions were pooled, gathered and concentrated by dialysis, prior to further biochemical analyses.
Subunit Structure and Molecular Mass of Purified PAL
The subunit structure and molecular mass of the purified PAL from the algal species were determined by the SDS-PAGE and native-PAGE [65, 66], according to Laemmli [66] with slight modifications [51, 64]. The protein samples (50 μl) were boiled in dissociation loading buffer for 5 min, and then loaded into the wells of stacking gel. The gel running was conducted at 100 mA for 40 min (Bio-Rad, Model 2000/200). After running, the gel was immersed in Coomassie brilliant blue stain with gentle shaking at 50 rpm, then, the gel was washed by de-staining. The molecular weight of the appeared protein bands was calculated from the inference of protein ladder (Cat# sc-2361, 6-200 kDa, Santa Cruz Biotechnology Inc., USA).
Biochemical Properties of Purified PAL
The biochemical properties of the purified PAL such as reaction temperature, thermal stability, reaction pH, pH stability, substrate specificity and effect of various compounds and metals were studied as described previously [51, 62-64]. Michalis-Menten constant (Km), maximum velocity (Vmax), turnover number (kcat) and catalytic efficiency (kcat/Km) are the common kinetic parameters that were determined towards the substrates [60].
Enzymatic Synthesis of MMDA with PAL Using Myristicin as Substrate
Myristicin was isolated and purified from wild Daucus pumilus (Gouan) and its chemical structure was validated from the spectrometric analyses [67, 68]. The biotransformation of myristicin into 3-Methoxy-4, 5-methylenedioxyamphetamine (MMDA) was illustrated in Fig. 1B. The proposed mechanism of the hydroamination reactions of myristicin to MMDA using PAL has been postulated by Lovelock [1]. The enzymatic reaction contained 30 mM myristicin (dissolved in acetone) in Tris-HCl buffer (pH 8.0), and 500 μl of PAL preparation in 5 ml total volume, and the reaction was incubated at 37°C for 1 h with shaking at 120 rpm. Blanks of enzyme free substrate and substrate free enzyme were used as baseline. The reaction pH was adjusted to pH 12 with the addition of 10N NaOH and the product was extracted with diethyl ether three times (20 ml each). Upper phase was combined and concentrated. The concentrated residue was dissolved in 500 μl of methanol and analyzed using gas chromatography-mass spectrometry (GC-MS).
Chemical Synthesis of MMDA
Chemical synthesis of MMDA was performed by hydrohalogenation followed by amination reactions according to Clark, et al., [49] with slight modification. Briefly, hydrohalogenation of myristicin was carried out by addition of 48% of hydrogen bromide (HBr) with stirring at room temperature, then the reaction was terminated by cooling in ice, and the hydrohalogenation product was extracted with diethyl ether. The ether layers were collected, washed with distilled water and concentrated by rotary evaporator at 50°C and 100 rpm. The resultant oil was dissolved in methanol, stirred with 35% NH4OH at room temperature to catalyze the amination reaction. After evaporation of the mixture, the resulting oil was dissolved in 10% HC1 and washed with ether. The pH was adjusted to 12 with the addition of NaOH pellets. The aqueous basic solution was extracted with ether, and the pooled ether extracts were evaporated till dryness under reduced pressure. The hydrohalogenation and/or amination reactions were monitored by precoated thin-layer chromatography (TLC) plates (silica gel 60, GF254 (60-250 mesh), Merck, Germany) using a solvent system (hexane: methylene chloride (1:1)) and p-anisaldehyde sulfuric acid spray according to Gerlacha et al. [69], as visualizing agent. The TLC plates were heated for about 5 min at 100°C, and resultant oil from hydrohalogenation and/or amination reactions was analyzed directly by GC-MS.
GC-MS analysis of synthesized MMDA. The chemical and enzymatic synthesized MMDA were analyzed by Agilent 6890 gas chromatograph equipped with an Agilent mass spectrometric detector, with a direct capillary interface fused with silica capillary column PAS-5 ms (30 mm × 0.32 mm × 0.25 μm film thickness). A 1 μl sample in methanol was injected to the GC-MS [49], with helium as carrier gas at a flow rate of 1 ml/min. The solvent delay was 3 min and the mass spectrometric detector was operated in electron impact ionization mode with an ionizing energy of 70 e.v. scanning from m/z 50 to 500. The ion source temperature was 230°C and the electron multiplier voltage (EM voltage) was maintained at 1,250 v. The GC was manually tuned using perfluorotributyl amine, the temperature program was started at 70°C then elevated to 150°C at a rate of 15°/min and from 150°C to 250°C at a rate of 25°/min with 6 min hold time. The detector and injector temperature were set at 280°C and 250°C, respectively. The putative names of the target compounds from the spectroscopic data were identified from Wiley and NIST spectral libraries.
Results and Discussion
Screening for Phenylalanine Ammonia-Lyase from Different Microbial Sources
Among 50 microbial isolates (20 fungal isolates, 20 bacterial isolates and 10 algal isolates) (Supplementary Data), four algal isolates, namely; A. flos-aquae, A. variabilis, S. platensis and C. vulgaris were selected for their promising yield of PAL. The crude PAL from the four algal isolates displayed visual forward and reverse PAL catalytic reactions on phenylalanine and cinnamic acid as substrates, respectively. The forward activities of PAL from A. flos-aquae, A. variabilis, S. platensis and C. vulgaris were 12.8, 11.5, 24.8, and 10.8 μmol/mg/min, while the reverse activities were 23.3, 19.6, 49.6, and 21.0 μmol/mg/min, respectively (Fig. 1C). These results revealed that S. platensis and A. flos-aquae displayed the highest PAL activities, thus, the enzyme from both sources has been further purified and characterized comparatively.
Purification, Molecular Subunit Structure of PAL from Selected Algal Isolates
The PAL was purified from the cultures of S. platensis and A. flos-aquae by gel-filtration and ion-exchange chromatographic approaches. The purification profile of PAL from S. platensis and A. flos-aquae was summarized in Table 1. The specific activities of PAL from S. platensis and A. flos-aquae were increased by about 1.7 and 1.3 folds than their corresponding crude enzymes with an overall yield of 73.3 and 87.1 %, respectively, upon acetone precipitation. By Sephadex G200 column, the specific activities of PAL from S. platensis and A. flos-aquae were increased by 2.9 and 2.4 folds with overall yield of 13.2 and 15.9%, respectively. With the ion-exchange chromatography, the specific activities of PAL from S. platensis and A. flos-aquae were increased by 4.7 and 3.5 folds comparing to their crude enzymes, respectively. Consequently, the activity of PAL from S. platensis was higher than A. flos-aquae by 1.3 folds with the last purification step. The active fractions from gel-filtration and ion-exchange chromatography were assessed based on their colorimetric activity and molecular homogeneity by denaturing PAGE. Prior to biochemical characterization, the most active and molecularly homogenous fractions were gathered and concentrated by dialysis with polyethylene glycol.
Table 1.
Overall purification profile of PAL from S. platensis and A. flos-aquae
| S. platensis | A. flos-aquae | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Purification step | Total activity (U) | Total protein (mg) | Specific activity (U/mg) | Purification fold | Recovery (%) | Total activity (U) | Total protein (mg) | Specific activity (U/mg) | Purification fold | Recovery (%) |
| Crude | 10222 | 409.9 | 24.9 | 1 | 100 | 7598.6 | 588.9 | 12.9 | 1 | 100 |
| Acetone precipitate | 7485 | 175.6 | 42.6 | 1.7 | 73.3 | 6619.1 | 400.1 | 16.5 | 1.2 | 87.1 |
| Gel filtration | 1353 | 18.3 | 73.9 | 2.9 | 13.4 | 1210.7 | 39.64 | 30.5 | 2.3 | 15.9 |
| Ion-exchange chromatography | 985 | 9.8 | 98.8 | 3.9 | 9.6 | 850 | 22.8 | 54.7 | 4.3 | 11.2 |
The subunit structure of PAL from S. platensis and A. flos-aquae was assessed by gel electrophoresis. SDS-PAGE analysis (Fig. 1D) showed a single protein band of molecular weight 64 kDa, revealing the homogeneity of the purified S. platensis and A. flos-aquae PAL. The molecular mass of recovered PAL is in coincidence with those reported for cyanobacterial PAL from A. variabilis and Nostoc punctiforme [10]. However, PAL from Trichosporon cutaneum [70] and Rhodotorula glutinis [11] showed 79 and 75 kDa, respectively.
Biochemical Characterization of PAL from S. platensis and A. flos-aquae
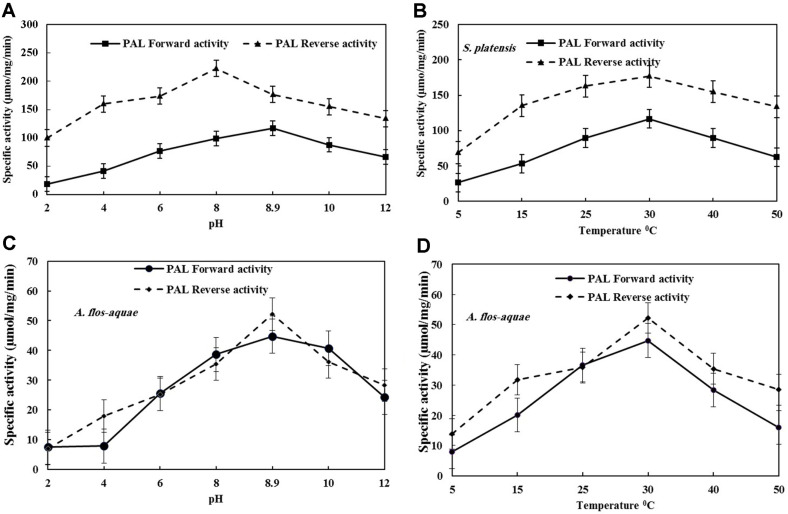
Optimum reaction temperature and pH. The effect of reaction temperature and pH on the activity of PAL from both algal isolates was assessed. From the results (Fig. 2), the purified PAL from S. platensis had a maximum forward (116.7 μmol/mg/min) and reverse activity (176.7 μmol/mg/min) at 30°C incubation temperature. Also, at 30°C, the maximum activity of A. flos-aquae PAL was 44.7 and 52.2 μmol/mg/min for forward and reverse reactions. Being partially consistent, Moffitt et al., [10] reported that the optimum temperature of A. variabilis PAL was recorded at 40°C, while the optimal activity of PAL of tobacco and sunflower was reported at 35 and 55°C, respectively, and the optimum temperature of PAL of Rhizoctonia ranged from 44-46°C [4].
Fig. 2. Effect of different pH and temperature on specific activity of purified PAL from S. platensis (A and B) and A. flos-aquae (C and D), respectively.
Each data point shows the average of at least three replicates. The standard errors are represented by vertical bars.
The highest activity of PAL of S. platensis was recorded at pH of 8.9 and 8 of forward and reverse reactions, respectively. Moreover, PAL derived from A. flos-aquae exhibited maximum activity of forward and reverse reactions at pH of 8.9. The optimal activity of PAL at this pH from both algal isolates were consistent with those reported for PAL production by A. variabilis [10] and within the optimum pH range for PAL [4, 8] from various microbial sources. Furthermore, PAL from the yeasts Trichosporon cutaneum [70] and Rhodotorula glutinis [11] had optimum pH at 8-9, respectively.
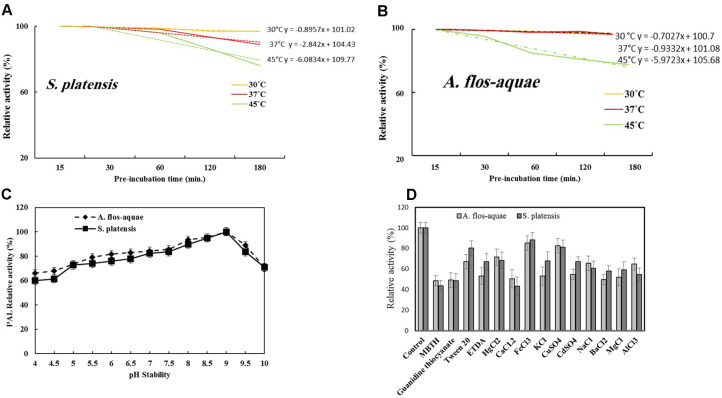
Thermal stability, pH stability, inhibitors and activators of PAL. The thermal stability of PAL from both algal isolates was determined by pre-incubation of enzyme without substrate at various temperatures (30, 37, and 45ºC). The residual enzyme activity was measured by the forward assay after 15, 30, 60, 120, and 180 min for each temperature degree. The profile of thermal stability of PALs was shown in Fig. 3. The half-life times of PAL from S. platensis at 30, 37, and 45°C were 9.5, 3.2, and 1.6 h, respectively. Meanwhile, PAL from A. flos-aquae had half-life times of 12, 9.0, and 1.5 h at 30, 37, and 45°C, respectively. Therefore, PAL from A. flos-aquae has a relatively higher stability than from S. platensis.
Fig. 3. Thermal stability (A&B), pH stability (C), compounds and metals (D) of PAL from S. platensis and A. flos-aquae, respectively.
The pH stability of PAL was assessed by pre-incubation of enzyme without substrate at different pH ranges (pH range 4-10) using potassium phosphate buffer at 4°C for 2 h, then the residual activity was determined as described above. PALs from both algal isolates displayed a higher stability at pH range 8-9, with dramatic reduction to its activity at acidic pH and higher alkaline pH (Fig. 3C). Zhu et al. stated that activity of Rhodotorula glutinis PAL was stable at pH range of 6-10 [11].
The effect of various compounds and metals on activity of PAL was estimated by incubating the enzyme with each compound at 1mM final concentration for 2 h at 4°C, then measuring the residual activity by the standard assay. Tested compounds such as 3-Methyl-2-benzo-thiazolinone hydrazone (MBTH), guanidine thiocyanate, Tween 20 and ethylenediaminetetraacetic acid (EDTA) and different metals such as Na+, K+, Ca2+, Cu2+, Cd2+, Ba2+, Mg2+, Hg2+, Fe3+, and Al3+ were used. By addition of compounds, the residual activity of PAL from S. platensis was maximally reported in presence of FeCl3 (88.2%), followed by CuSO4 (81.1%) and Tween 20 (80.5%). On the other hand, 85.4% of PAL activity from A. flos-aquae remained after incubation with FeCl3, followed by 82.7% with CuSO4 and HgCl2 (71.4%) (Fig. 3D). It was reported that PAL activity from Cistanche deserticola was inhibited by Hg2+, Zn2+, and pb2+, while Co2+, Fe3+, and Cu2+ had no significant inhibitory effect [6]. Moreover, Mg2+ and Ba2+ were reported to be a slight activator for PAL activity [4].
Substrate specificity and kinetics of the purified PAL. The specificity of purified PALs from the two algal isolates towards different amino acids had been evaluated based on the standard forward assay. PALs from S. platensis and A. flos-aquae have affinity only toward tyrosine by 18.32% and 51.15%, respectively, comparing to phenylalanine (Table 2). PALs from the two algal isolates have no activity on other amino acids such as methionine, glycine, asparagines, ornithine, lysine, arginine, alanine, cysteine and valine. These results are also consistent with those reported by MacDonald and D’Cunha [8].
Table 2.
Substrate Specificity of purified PAL from A. flos-aquae and S. platensis
| Substrate | Anabaena flos-aquae | Spirulina platensis | ||
|---|---|---|---|---|
|
| ||||
| Specific activity (U/mg) | Relative activity (%) | Specific activity (U/mg) | Relative activity (%) | |
| Phenylalanine | 2.5 | 100 | 10.6 | 100 |
| Methionine | 0 | 0 | 0 | 0 |
| glycine | 0 | 0 | 0 | 0 |
| asparagine | 0 | 0 | 0 | 0 |
| ornithine | 0 | 0 | 0 | 0 |
| lysine | 0 | 0 | 0 | 0 |
| Tyrosine | 1.3 | 51.2 | 1.9 | 18.4 |
| Arginine | 0 | 0 | 0 | 0 |
| Alanine | 0 | 0 | 0 | 0 |
| Cysteine | 0 | 0 | 0 | 0 |
| Valine | 0 | 0 | 0 | 0 |
The catalytic and kinetic parameters of the enzyme towards phenylalanine and cinnamic acid were summarized in Table 3. Different concentrations of L-phenylalanine and trans-cinnamic acid were tested and the activity of enzymes was measured by forward and reverse assay, respectively. PAL from both algal isolates displayed a higher affinity and velocity towards trans-cinnamic acid than L-phenylalanine. S. platensis PAL has a higher turnover number (kcat) and catalytic efficiency (kcat/Km) (1.25 s-1 and 2.03 ms-1s-1, respectively) than A. flos-aquae PAL (32× 10-2 s-1 and 1.28 ms-1s-1, respectively) toward trans-cinnamic acid as a substrate. Furthermore, turnover number and catalytic efficiency for S. platensis PAL (24.4 × 10-2 s-1 and 14.9 × 10-2 ms-1s-1, respectively) was higher toward L-phenylalanine than A. flos-aquae PAL (6.6× 10-2 s-1 and 4.4× 10-2 ms-1s-1, respectively). Whereas, the wild-type cyanobacterial PAL such as A. variabilis and Nostoc punctiforme had a higher turnover number (kcat 4.3 s-1 and 1.96 s-1, respectively) and catalytic efficiency (kcat/Km 72.2 ms-1s-1 and 43.8 ms-1s-1, respectively) using L-phenylalanine as substrate [10] comparing to our algal isolates. Moreover, PAL from Streptomyces maritimus had a smaller turnover number (kcat 0.0048 s-1) and catalytic efficiency (kcat/Km 2.1 × 10-3μM-1s-1) toward L-phenylalanine [71].
Table 3.
Kinetic parameters of PAL from S. platensis and A. flos-aquae for L-phenylalanine (L-PA) and Trans-cinnamic acid (Trans-CA)
| Substrate | S. platensis | A. flos-aquae | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Km (mM) |
Vmax (mM/min) |
Kcat (s−1) |
Kcat/Km (mMs−1) |
Km (mM) |
Vmax (mM/min) |
Kcat (s−1) |
Kcat/Km (mMs−1) |
|
| L-PA | 1.64 | 0.01 | 0.24 | 0.15 | 1.5 | 0.01 | 0.06 | 0.04 |
| Trans-CA | 0.61 | 0.04 | 1.25 | 2.1 | 0.3 | 0.04 | 0.32 | 1.28 |
Synthesis of MMDA
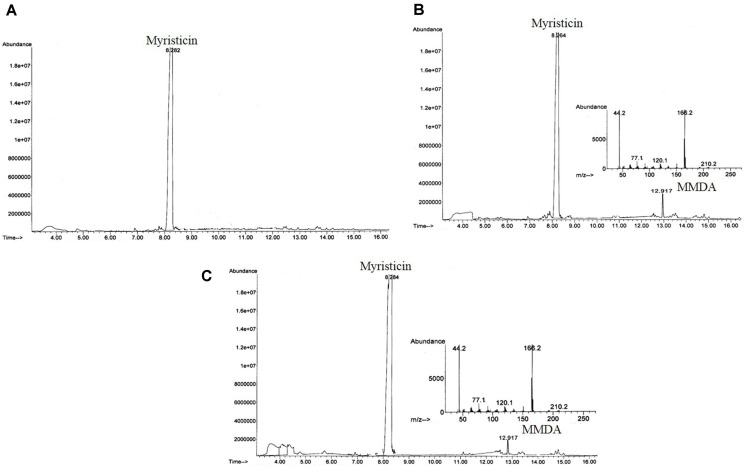
Enzymatic synthesis of MMDA by PAL from S. platensis and A. flos-aquae. The transformation of myristicin with crude PAL of S. platensis and A. flos-aquae into MMDA was assessed, as described in Materials and Methods. After incubation of the reaction mixture, the enzyme activity was stopped, and the initial concentration of myristicin and developed MMDA was quantified. The enzymatic byproducts were checked by GC-MS analysis using chemically synthesized MMDA as authentic. As shown (Fig. 4), a new peak at 12.917 min for MMDA was observed other than the myristicin peak at 8.26-8.28 min as an enzymatic byproduct of S. platensis and A. flos-aquae PAL, respectively. The enzymatic yield of MMDA was 15.3 and 10.6% for PAL of S. platensis and A. flos-aquae using myristicin substrate, respectively. Similarly, PAL from A. variabilis [72] and Rhodotorula glutinis [20] transformed trans-cinnamic acid into L-phenylalanine with a yield of about 73% and 70%. In another work, Rhodotorula glutinis PAL produced L-phenylalanine methyl ester from trans-cinnamyl methyl ester in a biphasic system [73]. Furthermore, several reports investigated the hydroamination activity of PAL of microbial [74, 75] or plant origin [76-78].
Fig. 4. Result of biotransformation of myristicin by PAL. GC chromatogram of blank (A) and reaction of myristicin with PAL from S. platensis (B) and A. flos-aquae (C) with mass fragmentation pattern of MMDA.
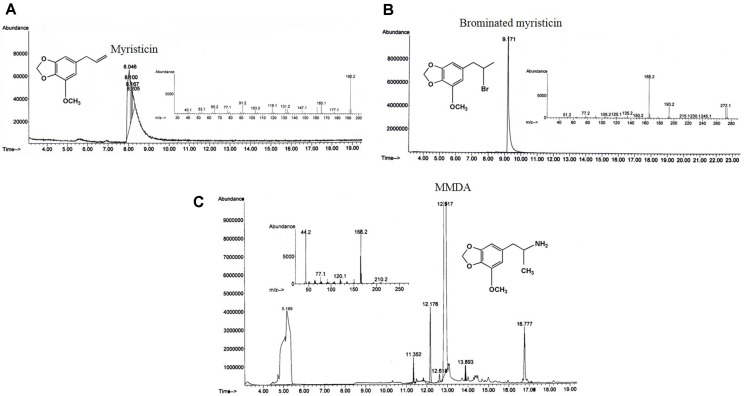
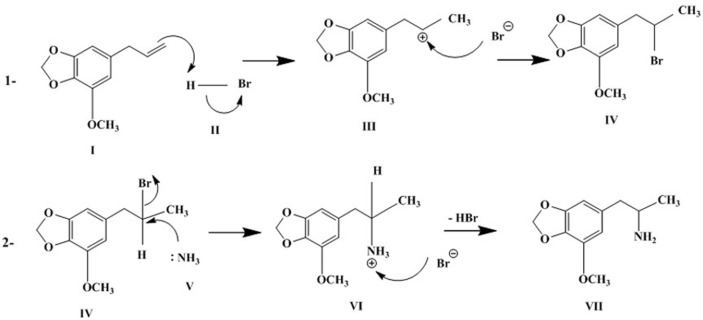
Chemical synthesis of MMDA. From the GC-MS analysis (Fig. 5), a successful synthesis of MMDA from myristicin was accomplished. These results were similar to that reported about the synthesis of MMDA derivatives from nutmeg oil [49]. This is the first report dealing with the chemical manufacturing of MMDA from the natural product myristicin via hydrohalogenation and amination reactions. As can be seen from Fig. 6, mechanisms of hydrohalogenation and amination reactions were discussed in detail. Briefly, the electrophilic addition of hydrogen bromide to the side chain, propene group of myristicin gave 2-bromo-1-(3-Methoxy-4,5-methylenedioxyphenyl)-2-propane (brominated myristicin) through the formation of the most stable secondary carbocation. In addition, amination reaction involves bimolecular nucleophilic substitution of ammonia with the intermediate, the brominated myristicin at 9.171 min to produce a desired product, MMDA at 12.917 min. Moreover, the yield of the brominated myristicin and MMDA was 99.22% and 63.63%, respectively.
Fig. 5. Gas chromatography-mass spectrometry analysis of starting material, intermediate and product during chemical synthesis of MMDA; A, B and C: chromatograms and mass spectrum peaks of myristicin, brominated myristicin and MMDA, respectively.
Fig. 6. Schematic representation of chemical synthesis of MMDA from myristicin.
1: hydrohalogenation reaction; 2: amination reaction I: myristicin; II: hydrogen bromide; III: myristicin carbocation; IV: brominated myristicin; V: ammonia; VI: ammonium cation of MMDA; VII: MMDA
Rationality and yield of MMDA from enzymatic and chemical methods. Biotransformation is the modification of a definite substance to its structurally related product by animal tissues, plants [79] and microorganisms [80, 81], as designated by white biotechnology [82]. Enzymes of microbial origin are preferred over those from animal or plant due to their economic production in a short period of time, high stability under extreme environments and feasibility of their purification [79, 83]. Free enzymes, whole cells and immobilized cells/enzymes are generally used as biological catalysts [80, 82]. High reaction enantio-, stereo- and regioselectivity, minimal byproduct yield, mild reaction conditions and eco-friendliness are the advantages of microbial transformation [82, 84]. Utilization of biological catalysts instead of chemicals in chemical reactions offers great contribution to green chemistry [81, 85, 86] including industrial sectors like pharmaceuticals [80, 81, 87], cosmetics, and food [82]. From the results (Figs. 4 and 5), the actual yield of chemically synthesized MMDA was higher than those of S. platensis and A. flos-aquae PAL byproducts by about 5.9 and 4.15 folds, using the same concentration of myristicin as substrate. However, the multiple required steps and difficulty of purification of MMDA are the main hurdles that limit the chemical approach. Although enzymatic MMDA has a lower yield, the recovery process makes it more practical and commercially feasible than chemical approaches. Therefore, further optimization of the PAL reaction process for conversion of myristicin into MMDA are ongoing to achieve the maximum yield of MMDA.
In conclusion, A. flos-aquae, A. variabilis, S. platensis and C. vulgaris were used as a new source of PAL. Purification and characterization of PAL from S. platensis and A. flos-aquae were performed. Furthermore, chemical synthesis of MMDA from myristicin with a yield of 63.63% was carried out. Additionally, MMDA biosynthesis from myristicin by PAL was studied as an eco-friendly route for the first time. Hydroamination of myristicin with the purified PAL from S. platensis and A. flos-aquae to MMDA was successfully established in a one-step process with a yield of 15.3 and 10.6%, respectively. Consequently, our future work will focus on the over expression and directed mutagenesis of S. platensis and A. flos-aquae PAL to get deeper insights into this enzyme active site structure and to increase the product yield.
Supplementary material
Supplementary data for this paper are available on-line only at http://jmb.or.kr.
Acknowledgments
We thank Prof. Dr. Eatedal Hassan Abdel-Aal, Faculty of Pharmacy, Zagazig University for her continuous guidance and advice during the chemical synthesis of MMDA.
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
References
- 1.Lovelock S. Ph.D.Thesis. The University of Manchester; United Kingdom: 2014. The development of novel biocatalysts for the asymmetric hydroamination of alkenes. [Google Scholar]
- 2.Koukol J, Conn E. The metabolism of aromatic compounds in higher plants. J. Biol. Chem. 1961;236:2692–2698. [PubMed] [Google Scholar]
- 3.Havir EA, Hanson KR. L-Phenylalanine ammonia-lyase. II. Mechanism and kinetic properties of the enzyme from potato tubers. Biochemistry. 1968;7:1904–1914. doi: 10.1021/bi00845a039. [DOI] [PubMed] [Google Scholar]
- 4.Hyun MW, Yun YH, Kim JY, Kim SH. Fungal and plant phenylalanine ammonia-lyase. Mycobiology. 2011;39:257–265. doi: 10.5941/MYCO.2011.39.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lovelock SL, Turner NJ. Bacterial Anabaena variabilis phenylalanine ammonia lyase: a biocatalyst with broad substrate specificity. Bioorg. Med. Chem. 2014;22:5555–5557. doi: 10.1016/j.bmc.2014.06.035. [DOI] [PubMed] [Google Scholar]
- 6.Hu GS, Jia JM, Hur YJ, Chung YS, Lee JH, Yun DJ, et al. Molecular characterization of phenylalanine ammonia lyase gene from Cistanche deserticola. Mol. Biol. Rep. 2011;38:3741–3750. doi: 10.1007/s11033-010-0489-0. [DOI] [PubMed] [Google Scholar]
- 7.Tuan PA, Park NI, Li X, Xu H, Kim HH, Park SU. Molecular cloning and characterization of phenylalanine ammonia-lyase and cinnamate 4-hydroxylase in the phenylpropanoid biosynthesis pathway in garlic (Allium sativum) J. Agric. Food Chem. 2010;58:10911–10917. doi: 10.1021/jf1021384. [DOI] [PubMed] [Google Scholar]
- 8.MacDonald MJ, D'Cunha GB. A modern view of phenylalanine ammonia lyase. Biochem. Cell. Biol. 2007;85:273–282. doi: 10.1139/O07-018. [DOI] [PubMed] [Google Scholar]
- 9.Hemmati S. Phenylalanine ammonia-lyase through evolution: A bioinformatic approach. Trends Pharm. Sci. 2015;1:10–14. [Google Scholar]
- 10.Moffitt MC, Louie GV, Bowman ME, Pence J, Noel JP, Moore BS. Discovery of two cyanobacterial phenylalanine ammonia lyases: kinetic and structural characterization. Biochemistry. 2007;46:1004–1012. doi: 10.1021/bi061774g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu L, Cui W, Fang Y, Liu Y, Gao X, Zhou Z. Cloning, expression and characterization of phenylalanine ammonia-lyase from Rhodotorula glutinis. Biotechnol. Lett. 2013;35:751–756. doi: 10.1007/s10529-013-1140-7. [DOI] [PubMed] [Google Scholar]
- 12.Löffelhardt W, Kindl H. Formation of benzoic acid and p-hydroxybenzoic acid in the blue green alga Anacystis nidulans: A thylakoid-bound enzyme complex analogous to the chloroplast system. Z Naturforsch C. 1976;31:693–699. doi: 10.1515/znc-1976-11-1212. [DOI] [PubMed] [Google Scholar]
- 13.Fritz RR, Hodgins D, Abell C. Phenylalanine ammonia-lyase. Induction and purification from yeast and clearance in mammals. J. Biol. Sci. 1976;251:4646–4650. [PubMed] [Google Scholar]
- 14.MacDonald MC, Arivalagan P, Barre DE, MacInnis JA, D'Cunha GB. Rhodotorula glutinis Phenylalanine/tyrosine ammonia lyase enzyme catalyzed synthesis of the methyl ester of para-hydroxycinnamic acid and its potential antibacterial activity. Front. Microbiol. 2016;7:281. doi: 10.3389/fmicb.2016.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui JD, Qiu JQ, Fan XW, Jia SR, Tan ZL. Biotechnological production and applications of microbial phenylalanine ammonia lyase: a recent review. Crit. Rev. Biotechnol. 2014;34:258–268. doi: 10.3109/07388551.2013.791660. [DOI] [PubMed] [Google Scholar]
- 16.Sarkissian CN, Kang TS, Gámez A, Scriver CR, Stevens RC. Evaluation of orally administered PEGylated phenylalanine ammonia lyase in mice for the treatment of Phenylketonuria. Mol. Genet. Metab. 2011;104:249–254. doi: 10.1016/j.ymgme.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim W, Erlandsen H, Surendran S, Stevens RC, Tyring SK, Matalon R, et al. Trends in enzyme therapy for phenylketonuria. Molecular Therapy. 2004;10:220–224. doi: 10.1016/j.ymthe.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Levy HL. Phenylketonuria: old disease, new approach to treatment. PNAS. 1999;96:1811–1813. doi: 10.1073/pnas.96.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovacs K, Banoczi G, Varga A, Szabo I, Holczinger A, Hornyanszky G, et al. Expression and properties of the highly alkalophilic phenylalanine ammonia-lyase of thermophilic Rubrobacter xylanophilus. PLoS One. 2014;9:e85943. doi: 10.1371/journal.pone.0085943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada S, Nabe K, Izuo N, Nakamichi K, Chibata I. Production of L-phenylalanine from trans-cinnamic acid with Rhodotorula glutinis containing L-phenylalanine ammonia-lyase activity. Appl. Environ. Microbiol. 1981;42:773–778. doi: 10.1128/AEM.42.5.773-778.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans CT, Hanna K, Payne C, Conrad D, Misawa M. Biotransformation of trans-cinnamic acid to L-phenylalanine: optimization of reaction conditions using whole yeast cells. Enzyme. Microb. Technol. 1987;9:417–421. doi: 10.1016/0141-0229(87)90137-2. [DOI] [Google Scholar]
- 22.Evans CT, Conrad D, Hanna K, Peterson W, Choma C, Misawa M. Novel stabilization of phenylalanine ammonia-lyase catalyst during bioconversion of trans-cinnamic acid to L-phenylalanine. Appl. Microbiol. Biotechnol. 1987;25:399–405. doi: 10.1007/BF00253308. [DOI] [Google Scholar]
- 23.Kot AM, Błażejak S, Kurcz A, Gientka I, Kieliszek M. Rhodotorula glutinis-potential source of lipids, carotenoids, and enzymes for use in industries. Appl. Microbiol. Biotechnol. 2016;100:6103–6117. doi: 10.1007/s00253-016-7611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sohilait HJ, Kainama H. Synthesis of Myristicin Ketone (3, 4-Methylenedioxy-5-Methoxyphenyl)-2-Propanone from Myristicin. Science. 2015;3:62–66. doi: 10.11648/j.sjc.20150303.15. [DOI] [Google Scholar]
- 25.Sudradjat SE, Timotius KH, Mun'im A, Anwar E. The isolation of Myristicin from nutmeg oil by sequences distillation. J. Young Pharm. 2018;10:20–23. doi: 10.5530/jyp.2018.10.6. [DOI] [Google Scholar]
- 26.Carolina A, Maman M. Larvicidal activity of essential oils from the leaves and fruits of nutmeg (Myristica fragrans Houtt) against Aedes aegyptis (Diptera: Culicidae) Turkish JAF Sci. Technol. 2016;4:552–556. doi: 10.24925/turjaf.v4i7.552-556.705. [DOI] [Google Scholar]
- 27.de Cássia da Silveira e Sá R, Andrade LN, dos Reis Barreto de Oliveira R, de Sousa DP. A review on anti-inflammatory activity of phenylpropanoids found in essential oils. Molecules. 2014;19:1459–1480. doi: 10.3390/molecules19021459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jana S, Shekhawat G. Anethum graveolens: An Indian traditional medicinal herb and spice. Pharmacogn. Rev. 2010;4:179–184. doi: 10.4103/0973-7847.70915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Jumaily EF, Al-Amiry MH. Extraction and Purification of Terpenes from Nutmeg (Myristica fragrans) J. Al-Nahrain Univ. Sci. 2012;15:151–160. doi: 10.22401/JNUS.15.3.21. [DOI] [Google Scholar]
- 30.Lee JY, Park W. Anti-inflammatory effect of myristicin on RAW 264.7 macrophages stimulated with polyinosinic-polycytidylic acid. Molecules. 2011;16:7132–7142. doi: 10.3390/molecules16087132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee HS, Jeong TC, Kim JH. In vitro and in vivo metabolism of myristicin in the rat. J. Chromatogr. B Biomed. Sci. Appl. 1998;705:367–372. doi: 10.1016/S0378-4347(97)00531-8. [DOI] [PubMed] [Google Scholar]
- 32.Mao W, Zangerl AR, Berenbaum MR, Schuler MA. Metabolism of myristicin by Depressaria pastinacella CYP6AB3v2 and inhibition by its metabolite. Insect Biochem. Mol. Biol. 2008;38:645–651. doi: 10.1016/j.ibmb.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 33.Braun U, Kalbhen D. Evidence for the biogenic formation of amphetamine derivatives from components of nutmeg. Pharmacology. 1973;9:312–316. doi: 10.1159/000136402. [DOI] [PubMed] [Google Scholar]
- 34.Rahman N, Fazilah A, Effarizah M. Toxicity of Nutmeg (Myristicin): A Review. Int. J. Adv. Sci. Eng. Inf. Technol. 2015;5:212–215. [Google Scholar]
- 35.Shulgin A, Sargent T, Naranjo C. Animal pharmacology and human psychopharmacology of 3-methoxy-4, 5-methylenedioxyphenylisopropylamine (MMDA) Pharmacology. 1973;10:12–18. doi: 10.1159/000136416. [DOI] [PubMed] [Google Scholar]
- 36.Snyder SH, Weingartner H, Faillace LA. DOET (2, 5-dimethoxy-4-ethylamphetamine) and DOM (STP)(2, 5-dimethoxy-4-methylamphetamine), new psychotropic agents: their effects in man. Arch. Gen. Psychiatry. 1970;24:50–55. doi: 10.1001/archpsyc.1971.01750070052006. [DOI] [PubMed] [Google Scholar]
- 37.Benzenhöfer U, Passie T. Rediscovering MDMA (ecstasy): the role of the American chemist Alexander T. Shulgin. Addiction. 2010;105:1355–1361. doi: 10.1111/j.1360-0443.2010.02948.x. [DOI] [PubMed] [Google Scholar]
- 38.Shulgin A. Psychopharmacological Agents. 1976. Psychomimétic agents ch. 4 in Maxwell Gordon; pp. 59–146. [DOI] [Google Scholar]
- 39.Idle J. Christmas gingerbread (Lebkuchen) and Christmas cheer-review of the potential role of mood elevating amphetamine-like compounds formed in vivo and in furno. Prague Med. Rep. 2005;106:27–38. [PubMed] [Google Scholar]
- 40.Nozaki M, Vaupel D, Bright L, Martin W. A pharmacological comparison of 3-methoxy-4, 5-methylenedioxyamphetamine and LSD in the dog. Drug Alcohol Depend. 1978;3:153–163. doi: 10.1016/0376-8716(78)90037-6. [DOI] [PubMed] [Google Scholar]
- 41.Snow O. Amphetamine Syntheses: Overview & Reference Guide for Professionals. Thoth Press; 1998. pp. 1–278. [Google Scholar]
- 42.Passie T, Benzenhöfer U. MDA, MDMA, and other "mescaline‐like" substances in the US military's search for a truth drug (1940s to 1960s) Drug Test Anal. 2018;10:72–80. doi: 10.1002/dta.2292. [DOI] [PubMed] [Google Scholar]
- 43.El-Sayed ASA, Khalaf SA, Ahmed HA. Characterization of homocysteine g-lyase from submerged and solid fermented cultures of Aspergillus fumigatus JX006238. J. Microbiol. Biotechnol. 2013;23:499–510. doi: 10.4014/jmb.1208.08070. [DOI] [PubMed] [Google Scholar]
- 44.El-Sayed ASA, Yassin M, Ibrahim H. Co-Immobilization of L-methioninase and glutamate dehydrogenase on polyacrylamide and chitosan for continuous production of L-homoalanine. Biotechnol. Appl. Biochem. 2015;62:514–522. doi: 10.1002/bab.1299. [DOI] [PubMed] [Google Scholar]
- 45.El-Sayed ASA, Fujimoto S, Yamada C, Suzuki H. Enzymatic synthesis of γ-glutamylglutamine, a stable glutamine analogue by γ-glutamyl transpeptidase from Escherichia coli K-12. Biotechnol. Lett. 2010;32:1877–1881. doi: 10.1007/s10529-010-0364-z. [DOI] [PubMed] [Google Scholar]
- 46.Naranjo C. The healing journey. Ballantine Books; 1974. pp. 1–235. [Google Scholar]
- 47.El-Sayed ASA, Shouman SA, Nassrat H. Pharmacokinetics, immunogenicity and anticancer efficiency of Aspergillus flavipes L-methioninase. Enzyme Microb. Technol. 2012;51:200–210. doi: 10.1016/j.enzmictec.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 48.Shulgin AT. 3-Methoxy-4, 5-methylenedioxy amphetamine, a new psychotomimetic agent. Nature. 1964;201:1120–1121. doi: 10.1038/2011120a0. [DOI] [PubMed] [Google Scholar]
- 49.Clark CR, DeRuiter J, Noggle FT. Analysis of 1-(3-methoxy-4, 5-methylenedioxyphenyl)-2-propanamine (MMDA) derivatives synthesized from nutmeg oil and 3-methoxy-4, 5-methylenedioxybenzaldehyde. J .Chromatogr. Sci. 1996;34:34–42. doi: 10.1093/chromsci/34.1.34. [DOI] [Google Scholar]
- 50.El-Sayed ASA, Abdel-Azim S, Ibrahim H, Yassin MA, Abdel-Ghany S, Esener S, Ali GS. Biochemical stability and molecular dynamic characterization of Aspergillus fumigatus cystathionine g-Lyase in response to various reaction effectors. Enzyme Microb. Technol. 2015;81:31–46. doi: 10.1016/j.enzmictec.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 51.El-Sayed AS, Shindia AA, AbouZaid AA, Yassin AM, Ali GS, Sitohy MZ. Biochemical characterization of peptidylarginine deiminase-like orthologs from thermotolerant Emericella dentata and Aspergillus nidulans. Enzyme Microb. Technol. 2019;124:41–53. doi: 10.1016/j.enzmictec.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 52.Giri AV, Anandkumar N, Muthukumaran G, Pennathur G. A novel medium for the enhanced cell growth and production of prodigiosin from Serratia marcescens isolated from soil. BMC Microbiol. 2004;4:11. doi: 10.1186/1471-2180-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stanier R, Kunisawa R, Mandel M, Cohen-Bazire G. Purification and properties of unicellular blue-green algae (order Chroococcales) Bacteriol. Rev. 1971;35:171. doi: 10.1128/MMBR.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zarrouk C. Ph. D. Thesis. University of Paris; France: 1966. Contribution a I'etude d'une cyanobacterie: Influence de divers facteurs physiques et chimiques et la photosynthese de Spirulina maxima (Setchell et Gardener) Geitler. [Google Scholar]
- 55.Beakes GW, Canter HM, Jaworski GH. Zoospore ultrastructure of Zygorhizidium affluens and Z. planktonicum, two chytrids parasitizing the diatom Asterionella formosa. Can J. Botechnol. 1988;66:1054–1067. doi: 10.1139/b88-151. [DOI] [Google Scholar]
- 56.Raju S, Sowmya S, Alexander Jebakumar P, Guruprasad R. Potent activator N-arylenamine-3-chloro-4-fluoroaniline for Phenylalanine Ammonia Lyase extracted from Plectranthus amboinicus. Int. J. Adv. Sci. Tech. Res. 2014;4:56–63. [Google Scholar]
- 57.Varga A, Bata Z, Csuka P, Bordea DM, Vertessy BG, Marcovici A, et al. Anovel phenylalanine ammonia-lyase from Kangiella Koreensis. Studia Universitatis Babes-Bolyai. Chemia. 2017;62:293–308. [Google Scholar]
- 58.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 59.Scopes RK. Protein purification: principles and practice. Springer Science & Business Media, Springer New York; 2013. pp. 1–397. [Google Scholar]
- 60.El‐Sayed A, Shindia A. Characterization and immobilization of purified Aspergillus flavipesl‐methioninase: continuous production of methanethiol. J. Appl. Microbiol. 2011;111:54–69. doi: 10.1111/j.1365-2672.2011.05027.x. [DOI] [PubMed] [Google Scholar]
- 61.El-Sayed AS, Ibrahim H, Sitohy MZ. Co-immobilization of PEGylated Aspergillus flavipes L-methioninase with glutamate dehydrogenase: a novel catalytically stable anticancer consortium. Enzyme Microb. Technol. 2014;54:59–69. doi: 10.1016/j.enzmictec.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 62.El‐Sayed AS, Hassan MN, Nada HM. Purification, immobilization, and biochemical characterization of l‐arginine deiminase from thermophilic Aspergillus fumigatus KJ 434941: Anticancer activity in vitro. Biotechnol. Prog. 2015;31:396–405. doi: 10.1002/btpr.2045. [DOI] [PubMed] [Google Scholar]
- 63.El-Sayed AS, Shindia AA, Diab AA, Rady AM. Purification and immobilization of l-arginase from thermotolerant Penicillium chrysogenum KJ185377. 1; with unique kinetic properties as thermostable anticancer enzyme. Arch. Pharm. Res. 2014 doi: 10.1007/s12272-014-0498-y. [Online ahead of print] [DOI] [PubMed] [Google Scholar]
- 64.El-Sayed AS, Ruff LE, Ghany SEA, Ali GS, Esener S. Molecular and spectroscopic characterization of Aspergillus flavipes and Pseudomonas putida L-methionine γ-lyase in vitro. Appl. Biochem. Biotechnol. 2017;181:1513–1532. doi: 10.1007/s12010-016-2299-x. [DOI] [PubMed] [Google Scholar]
- 65.Lim H-W, Park S, Lim C. Purification and properties of phenylalanine ammonia-lyase from leaf mustard. Mol. Cells. 1997;7:715–720. [PubMed] [Google Scholar]
- 66.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature . 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 67.You CX, Jiang HY, Zhang WJ, Guo SS, Yang K, Lei N, et al. Contact toxicity and repellency of the main components from the essential oil of Clausena anisum-olens against two stored product insects. J. Insect Sci. 2015;15:87. doi: 10.1093/jisesa/iev071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Passreiter CM, Akhtar Y, Isman MB. Insecticidal activity of the essential oil of Ligusticum mutellina roots. Z. Naturforsch C. 2005;60:411–414. doi: 10.1515/znc-2005-5-608. [DOI] [PubMed] [Google Scholar]
- 69.Gerlacha AdCL, Gadeac A, da Silveirab RMB, Clerca P, Lohézic-le Dévéhatc F. The Use of anisaldehyde sulfuric acid as an alternative spray reagent in TLC analysis reveals three classes of compounds in the genus Usnea adans.(Parmeliaceae, lichenized Ascomycota) Preprints. 2018;2018:2018020151 (doi: 10.20944/preprints201802.0151.v1) doi: 10.20944/preprints201802.0151.v1. [DOI] [Google Scholar]
- 70.Goldson-Barnaby A, Scaman CH. Purification and characterization of Phenylalanine ammonia lyase from Trichosporon cutaneum. Enzyme Res. 2013;2013:670702. doi: 10.1155/2013/670702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Louie GV, Bowman ME, Moffitt MC, Baiga TJ, Moore BS, Noel JP. Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases. Chem. Biol. 2006;13:1327–1338. doi: 10.1016/j.chembiol.2006.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weise NJ, Parmeggiani F, Ahmed ST, Turner NJ. Discovery and investigation of mutase-like activity in a phenylalanine ammonia lyase from Anabaena variabilis. Top Catal. 2018;61:288–295. doi: 10.1007/s11244-018-0898-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.D'cunha GB, Satyanarayan V, Nair PM. Novel direct synthesis of L-phenylalanine methyl ester by using Rhodotorula glutinis phenylalanine ammonia lyase in an organic-aqueous biphasic system. Enzyme Microb. Technol. 1994;16:318–322. doi: 10.1016/0141-0229(94)90173-2. [DOI] [Google Scholar]
- 74.Weise NJ, Ahmed ST, Parmeggiani F, Galman JL, Dunstan MS, Charnock SJ, et al. Zymophore identification enables the discovery of novel phenylalanine ammonia lyase enzymes. Sci. Rep. 2017;7:13691. doi: 10.1038/s41598-017-13990-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rowles I, Groenendaal B, Binay B, Malone KJ, Willies SC, Turner NJ. Engineering of phenylalanine ammonia lyase from Rhodotorula graminis for the enhanced synthesis of unnatural l-amino acids. Tetrahedron. 2016;72:7343–7347. doi: 10.1016/j.tet.2016.06.026. [DOI] [Google Scholar]
- 76.Dreßen A, Hilberath T, Mackfeld U, Billmeier A, Rudat J, Pohl M. Phenylalanine ammonia lyase from Arabidopsis thaliana (AtPAL2): a potent MIO-enzyme for the synthesis of non-canonical aromatic alpha-amino acids: Part I: comparative characterization to the enzymes from Petroselinum crispum (PcPAL1) and Rhodosporidium toruloides (RtPAL) J. Biotechnol. 2017;258:148–157. doi: 10.1016/j.jbiotec.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 77.Bartsch S, Wybenga GG, Jansen M, Heberling MM, Wu B, Dijkstra BW, et al. Redesign of a phenylalanine aminomutase into a phenylalanine ammonia lyase. ChemCatChem. 2013;5:1797–1802. doi: 10.1002/cctc.201200871. [DOI] [Google Scholar]
- 78.Bartsch S, Bornscheuer UT. Mutational analysis of phenylalanine ammonia lyase to improve reactions rates for various substrates. Protein Eng. 2010;23:929–933. doi: 10.1093/protein/gzq089. [DOI] [PubMed] [Google Scholar]
- 79.Sabu A, Nampoothiri KM, Pandey A. Microbial Enzymes and Biotransformations. 2005. L-Glutaminase as a therapeutic enzyme of microbial origin; pp. 75–90. [DOI] [Google Scholar]
- 80.Smitha M, Singh S, Singh R. Microbial biotransformation: a process for chemical alterations. J. Bacteriol. Mycol. Open Access. 2017;4:00085. doi: 10.15406/jbmoa.2017.04.00085. [DOI] [Google Scholar]
- 81.Hegazy M-EF, Mohamed TA, ElShamy AI, Abou-El-Hamd HM, Mahalel UA, Reda EH, et al. Microbial biotransformation as a tool for drug development based on natural products from mevalonic acid pathway: a review. J. Adv. Res. 2015;6:17–33. doi: 10.1016/j.jare.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Patra S. Ph.D.Thesis. University of Mysore; 2007. Biotransformation of caffeine to value added products. [Google Scholar]
- 83.Gopinath SC, Anbu P, Arshad M, Lakshmipriya T, Voon CH, Hashim U, et al. Biotechnological processes in microbial amylase production. Biomed. Res. Int. 2017;2017:1272193. doi: 10.1155/2017/1272193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schauer F, Borriss R. Biocatalysis and Biotransformation. In: Tkacz JS, Lange L, editors. Advances in Fungal Biotechnology for Industry, Agriculture, and Medicine. Springer US; Boston, MA: 2004. pp. 237–306. [DOI] [Google Scholar]
- 85.Milner SE, Maguire AR. Recent trends in whole cell and isolated enzymes in enantioselective synthesis. Review Accounts. 2012:321–382. doi: 10.3998/ark.5550190.0013.109. [DOI] [Google Scholar]
- 86.Li Z, Held M, Panke S, Schmid A, Mathys R, Witholt B. Methods and reagents for green chemistry: an introduction. John Wiley & Sons, Inc; 2007. Biocatalysis for Industrial Green Chemistry; pp. 281–298. [DOI] [Google Scholar]
- 87.Boaventura MAD, Lopes RF, Takahashi JA. Microorganisms as tools in modern chemistry: the biotransformation of 3-indolylacetonitrile and tryptamine by fungi. Braz. J. Microbiol. 2004;35:345–347. doi: 10.1590/S1517-83822004000300014. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.