Abstract
Trichoderma reesei is the major filamentous fungus used to produce cellulase and there is huge interest in promoting its ability to produce higher titers of cellulase. Among the many factors affecting cellulase production in T. reesei, the mycelial phenotype is important but seldom studied. Herein, a close homolog of the Neurospora crassa COT1 kinase was discovered in T. reesei and designated TrCOT1, which is of 83.3% amino acid sequence identity. Functional disruption of Trcot1 in T. reesei by RNAi-mediated gene silencing resulted in retarded sporulation on potato dextrose agar and dwarfed colonies on minimal medium agar plates containing glucose, xylan, lactose, xylose, or glycerol as the sole carbon source. The representative mutant strain, SUS2/Trcot1i, also displayed reduced mycelia accumulation but hyperbranching in the MM glucose liquid medium, with hyphal growth unit length values decreased to 73.0 μm/tip compared to 239.8 μm/tip for the parent strain SUS2. The hyperbranching phenotype led to slightly but significantly increased cellulase secretion from 24 to 72 h in a batch culture. However, the cellulase production per unit of mycelial biomass was much more profoundly improved from 24 to 96 h.
Keywords: Trichoderma reesei, Trcot1, hyperbranching, RNAi, cellulase, biofuel
Introduction
Recalcitrant cellulose is a major component of plant cell wall polysaccharides, which are widely regarded as the most abundant renewable bioresource for production of bioethanol and bio-based chemicals [1]. Cellulase is a biocatalyst used to decompose cellulose into fermentable glucose and simple cello-oligosaccharides (such as cellobiose)[2]. Cellulase had long been thought to be only composed of cellobiohydrolase, endo-glucanase, and β-glucosidase [3]. In recent years, however, lytic polysaccharide mono-oxygenase (formerly GH61 endo-glucanase) was discovered to be able to boost cellulose degradation by canonical glycoside hydrolase cellulase [4]. Trichoderma reesei (anamorph Hypocrea jecorina) is a filamentous fungus serving as the main industrial microbial workhorse to produce cellulase [5]. However, its ability to produce cellulase does not parallel with the rigorous demand for low-cost cellulase in production of cellulosic ethanol and biochemicals. Therefore, there is an urgent need to improve the ability of T. reesei to produce higher titers of cellulase through which the cost of cellulase can be reduced.
Tremendous efforts have been carried out to improve cellulase production in T. reesei, through both rational genetic engineering [6] and random mutagenesis [7, 8]. Since it is well known that the regulation of cellulase expression in T. reesei takes place primarily at the stage of transcription [9], many trials have dealt with engineering the promoters or transcription factors [10, 11], i.e. the cis-and trans-elements essential for transcription, respectively. This strategy also expands to manipulating key components in signaling pathways affecting the transcription of cellulase [12, 13]. However, there are also many other aspects beyond transcription regulation that can impact cellulase secretory expression in T. reesei. For filamentous fungi, the secretory production of proteins is impressively related to the cell morphology due to the cell polarity [14]. Particularly, enzymes are reported to be primarily secreted at the site of actively growing hyphal tips [15] and septa [16] in filamentous fungi. Although the loci of secretion in T. reesei have not been reported, a highly branched hyphal morphology appears to be directly related to enhanced cellulase production in a mutant strain [17]. It can thus be assumed that the hyphal tip of T. reesei is likely also an important locus of protein secretion. Based on this hypothesis, engineering T. reesei into a hyperbranching phenotype with more hyphal tips would, therefore, be beneficial for cellulase production.
Formation of hyphal tips (i.e. development of cell polarity) in filamentous fungi is a complex physiological process requiring orchestrated regulation by multiple pathways and associated genes [18-21]. Although in T. reesei there was no literature reporting a gene regulating the mycelial development, in other fungi, many genes have already been documented for their role in formation of hyphal tips. Knocking out COT1 encoding a serine/threonine protein kinase that belongs to the NDR family is accompanied with higher branching events in Neurospora crassa [22]. BarA, an acyl-CoA-dependent ceramide synthase, regulates the mycelium polarity of Aspergillus nidulans [23]. Deletion of the pcl2 gene encoding a secretion pathway-specific (KEX2-like) endo-protease increases the hyphal branching frequency in Aspergillus oryzae [24]. Notably, these factors belong to much differing biochemical pathways and some of them, if not all, have homologous genes in T. reesei. This raises the possibility that by manipulating expression of the homologs of these regulatory genes, specifically RNAi-mediated gene silencing of the Trcot1 (a homologous gene of the N. crassa cot-1) in this study, a phenotypic change may be observed in T. reesei and the consequent effects on secretory cellulase expression can be evaluated.
Materials and Methods
Strains, Plasmids, and Culture Conditions
The Escherichia coli Trans1 strain (Transgen, China) was used for plasmid construction and propagation throughout this study. The T. reesei SUS2 strain is an auxotroph of a mutant of QM9414 with an enhanced cellulase-producing ability [25] and is maintained in our lab. The plasmid pAPA, with the direct repeats of ampicillin resistance genes, was constructed for looping out the pyr4 selection marker gene in the T. reesei transformants when needed and has been described earlier [25]. E. coli was cultured in Luria-Bertani (LB) medium with appropriate concentrations of ampicillin at 37°C when needed. The T. reesei strains were grown in the minimal medium (MM, containing (NH4)2SO4, 5.0 g/l; KH2PO4, 15 g/l; MgSO4, 0.6 g/l; CaCl2, 0.6 g/l; FeSO4·7H2O, 0.005 g/l; MnSO4·H2O, 0.0016 g/l; ZnSO4·7H2O, 0.0014 g/l; CoCl2, 0.002 g/l) supplemented with a certain kind of carbon sources (2% glucose for mycelial growth; or 2% Avicel cellulose for cellulase induction). For sporulation, T. reesei were grown on a potato dextrose agar (PDA) plate at 28°C.
Plasmid Construction
The homologous gene of the N. crassa COT1 in T. reesei was identified by a BLAST search of the T. reesei genome. A total of ten homologous genes were found for N. crassa COT1 with amino acid sequence identities ranging from 39.3% to 83.3%. The gene with the highest homology was designated Trcot1 (Trire2:78909). The T. reesei SUS1 genomic DNA was extracted using a fungal DNA genome extraction kit (TianGen). The pdc1 and eno1 promoters and a cot1- gene fragment (49-570 bp of the first exon) were all amplified from the genomic DNA of SUS1 using the primer pairs of Pdc1-F/R, Eno1-F/R, and cot1-F/R (Table 1). To silence the Trcot1 gene, the pdc1 and eno1 promoters were first ligated head-to-head in the BamHI and EcoRI restriction sites of the plasmid pAPA to obtain pAPA-pdc1p-eno1p using the Gibson assembly method [26]. This intermediate plasmid was linearized using EcoRI and ligated with the Trcot1 gene fragment using the same method. This resulted in pCot1i for use in silencing the corresponding Trcot1 gene.
Table 1.
Primers used in this study.
| Primer | Sequences (5′→3′)a |
|---|---|
| cot1-F | CGCAGCTACAGCACAATCGAATTCGTCCTTGAAGAAGTCT |
| cot1-R | CTGAAATAGCTTCAAAGCCAACGATCGAGCCT |
| Pdc1-F | ATCACTAGTTCTAGAGCGGCCGCCGATGAAAGCCTTGCA |
| Pdc1-R | CTGAAATAGCTTCAAAGAATTCGATTGTGCTGTAGCT |
| Eno1-F | AGCTACAGCACAATCGAATTCTTTGAAGCTATTTCA |
| Eno1-R | TCATTACCAATTGGCGCGCCTTCTCAAATACCGCA |
| YZ-cot1-F | CTTCTTCATCAACCACC |
| YZ-cot1-R | GCCAGGCCGTCACCAGC |
| qActin-F | TGAGAGCGGTGGTATCCACG |
| qActin- R | GGTACCACCAGACATGACAATGTTG |
| qcot1-F | CTTGCCGGGTGGAGATTTGA |
| qcot1- R | TCGCTGTTTGCAAGGTCAGT |
| RTQcbh1F | GCTGCCGGTGCGGCTTGAAC |
| RTQcbh1R | CTGGCCATTGATGAACTTCAGATCGC |
| RTQcbh2F | CGTCAAATTGTCGTGGAA |
| RTQcbh2R | ACTGAGCATTGGCACACTT |
| RTQegl2F | CTACCTCAACAAGCTCATCAA |
| RTQegl2R | TCTTCAACGGAGGATAAACC |
| RTQbgl1F | AGTGACAGCTTCAGCGAG |
| RTQbgl1R | GGAGAGGCGTGAGTAGTTG |
aThe underlined regions are for homologous recombination.
Transformation of T. reesei
The plasmid pCot1i was introduced into the T. reesei SUS2 strain by poly-ethylene glycol (PEG)-mediated chemical transformation [27]. Briefly, SUS2 was cultured in MM supplemented with 2%glucose and 10 mM uridine at 28°C for 18 h. The young mycelia were collected by filtration, and mixed with 10 mg/ml of lysing enzyme from Trichoderma harzianum (Sigma-Aldrich, USA). Then the mixture was incubated at 28°C with gentle shaking until large amounts of protoplasts were released. Next, 10 μg of the pCot1i plasmid was transformed into the SUS2 protoplasts and spotted on MM-glucose plates without addition of uridine at 28°C for 5-7 days. The transformants were checked by diagnostic PCR for integration of the expressing cassettes into the chromosome using the primer pairs YZ-cot1-F/R (Table 1).
Morphological Analysis
For comparison of the growth behavior on agar plates, the spores from the parent strain SUS2 and representative transformant (2 × 107/ml) were individually spotted on PDA and solid MM medium supplemented with 2% (w/v) glucose, xylan, lactose, cellobiose, xylose, or glycerol as the sole carbon source and the plates were incubated at 28°C for 72 h. The colony diameters were measured. The morphology of the Trcot1-silenced transformant was also compared to its parent strain in a liquid MM-glucose (2%) medium. Spores (1 × 107/ml) were incubated in 250-ml Erlenmeyer flasks and the culture (50 ml) was shaken on a rotary shaker (180 rpm) for 48 h in the MM medium supplemented with 2% (w/v) glucose. The Lhgu (hyphal growth unit length) value was measured to quantitate the incidence of mycelial branching for the parental strain and the representative transformant. The Lhgu value is defined as the ratio of the total mycelium length divided by the number of tips, which is therefore an indicator of the average length of each hyphal branch (mm/tip) [24]. The hyphal length, i.e. distance from the branch point to septa, and the number of branches per apical or subapical compartment were measured under 40× magnification. The total length of hyphae and the number of hyphae were counted using ImageJ software (https://imagej.nih.gov/ij/). For all morphological measurements of submerged cultivations, more than 100 hyphae were measured per sample.
Induction of Cellulase Expression in T. reesei
For induction of cellulase in shake flask fermentation, fresh spores (1 × 107) were inoculated into 50 ml of liquid MM-glucose (2%) and shaken at 28°C for 48 h. The cot1-silenced transformants grew slowly and had to be cultured for 3 d for biomass accumulation. At the end of this pre-culture, the mycelia were collected and washed twice with MM with no carbon source. Then, 2 g of the mycelia were transferred into 100 ml of MM supplemented with 2% Avicel (MM-Avicel) for cellulase induction. The culture was continued at 28°C for 6 d to induce production of cellulases. From 24 to 144 h post induction, 2 ml of the culture supernatants was periodically collected for assay of the cellulase activity, extracellular protein concentration, and mycelial biomass. The inducing culture contains insoluble cellulose Avicel, which prohibited us from directly measuring the mycelia weight. Therefore, we determined the mycelia protein according to the method described by Jayaraman [28] as a representative of the fungal biomass.
Assay of Enzymatic Activities and Protein Concentration
For the overall cellulase activity, the reaction used one strip of Whatman No.1 filter paper (6 × 1 cm) as the substrate and 100 μl of the culture supernatant as the crude enzyme in 50 mM acetate buffer (pH 4.8). The mixture (1 ml in total) was incubated at 50°C for 1 h. The released reducing sugars were determined using the 3,5-dinitro-salicylic acid (DNS) method [29] and the OD540 of the reactions was measured. One unit of the overall cellulase activity was defined as the amount of enzyme that released 1 μmol of reducing sugar per hour under the assay conditions. To determine the endo-glucanase activity, 900 μl of 1% (w/v) sodium carboxymethyl cellulose (CMC-Na) was instead used as the substrate. The reaction was incubated at 50°C for 10 min. One unit of endo-glucanase activity was defined as the amount of enzyme that released 1 μmol of reducing sugar per minute under the assay conditions. The β-glucosidase activity was determined using p-nitrophenol-β-D-glucopyranoside (pNPG) as the substrate. The reaction consisted of 100 μl of appropriately diluted enzymes and 400 μl of 1.25 mM pNPG dissolved in McIlvaine buffer (200 mM Na2HPO4, 100 mM citric acid, pH 4.8). The mixture was incubated at 50 °C for 10 min. One unit of β-glucosidase activity was defined as the amount of enzyme that released 1 μmol of p-nitrophenol in one minute. The protein concentration of the fermentation supernatants was determined using a BCA-200 Protein Assay Kit (Pierce, USA) following the instruction of the manufacturer.
Quantitative Reverse Transcription PCR Analysis
For quantitative reverse transcription PCR (qRT-PCR), the mycelia of SUS2 strain and its Trcot1-silenced transformant cultured in MM-glucose (2%) for 24 h were collected. The mycelia were quickly frozen in liquid nitrogen and pulverized using a pestle and mortar. Using the TRIzol reagent (Thermo Fisher Scientific, USA), the total RNA was isolated from the pulverized mycelia. One microgram of the total RNA was reverse-transcribed to cDNA using a First Strand cDNA Maxima Synthesis Kit (TOYOBO, China). Then, using the actin gene as an endogenous reference gene, qRT-PCR was performed in an Applied Biosystems QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems, USA) using a TransScript Green One-Step SuperMix (TransGen). The primers used in qRT-PCR were listed in Table 1. The qRT-PCR was performed with the following steps: initial denaturation at 95°C for 10 min, then 40 cycles of 94°C for 30 sec, 60°C for 20 sec, and 72°C for 20 sec.
Results
Construction of the Plasmid for RNAi-Mediated Silencing of Trcot1
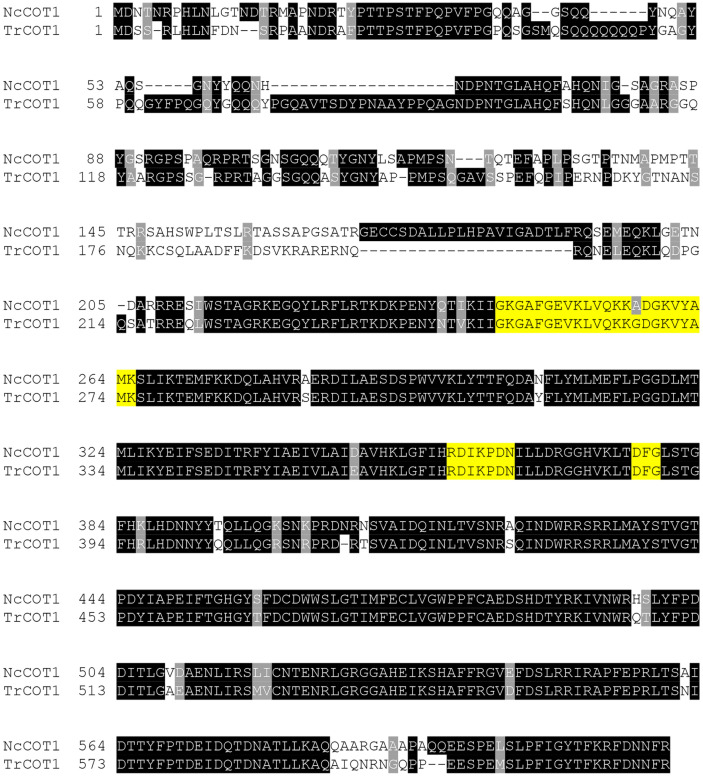
Through a BLAST search, a close homolog of NcCOT1 was identified in the T. reesei genome. An amino acid sequence alignment of TrCOT1 with NcCOT1 was given in Fig. 1. TrCOT1 is highly similar to NcCOT1, sharing an amino acid sequence identity of 83.3%. The functional sites critical for catalysis are conserved in TrCOT1. These include the glycine-rich loop containing the MgATP binding site (Gly253 -Lys275), the catalytic loop (Arg368-Asn374), and the carboxyl groups participating in substrate recognition (Asp387-Gly389) [30, 31]. These suggested that TrCOT1 is also a serine/threonine kinase and could play a regulatory role in T. reesei.
Fig. 1.
The amino acids shaded in black are conserved residues, while those in grey indicate similar ones. The amino acis in yellow indicate the glycine-rich loop containing the MgATP binding site (Gly253 -Lys275), the catalytic loop (Arg368-Asn374), and the carboxyl groups participating in substrate recognition (Asp387-Gly389).
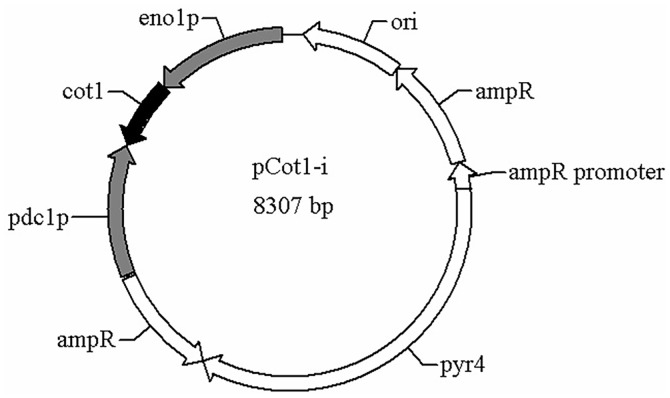
For analysis of the role of Trcot1 in regulating the morphological development of mycelia, a strategy via RNAi-mediated repression of gene expression was employed. The strong and constitutive pdc1 and eno1 promoters amplified from the T. reesei genomic DNA were assembled to obtain an intermediate plasmid, in which a gene fragment from the first exon of the Trcot1 gene (49-570 bp) was inserted between the two promoters (Fig. 2). The head-to-head dual promoters allow the transcription of the Trcot1 gene from two opposite directions, leading to formation of a double-stranded RNA in vivo. This double-stranded RNA is supposed to be recognized by RNA-induced silencing complex (RISC) and cleaved by Dicer proteins into small RNA fragments with lengths of 21-25 nt, which direct the cleavage of the target mRNA(s) [32]. The integrity of the recombinant plasmid was verified by both restriction digestion and DNA sequencing (data not shown).
Fig. 2.
Schematic diagram of the plasmid pCot1i used for RNAi-mediated gene silencing of Trcot1 in T. reesei. The key elements are as following: pdc1p, the pdc1 promoter; eno1p, the eno1 promoter; cot1, the cot1 gene fragment; pyr4, the expressing cassette for the pyr4 gene; ampr, the ampicillin resistance gene; ori, the plasmid replication origin motif.
The RNAi-Silenced Transformant Was Largely Different in Colony Morphology
The plasmid for RNAi-mediated gene silencing of Trcot1 (pCot1-i) was transformed into T. reesei and the integration of the RNAi cassette in the chromosome of transformants was verified by PCR (data not shown). From 48 h of these PCR-selected transformants, after 20 h they exhibited an appreciable change of the colony morphology on PDA plates compared with their parent strain, suggesting that the silencing of Trcot1 can affect the growth of these strains. One representative transformant, designated SUS2/Trcot1i, was selected for further analyses. Total RNA was extracted from the mycelia collected after 24 h of culture in the MM-glucose medium and the transcript levels of Trcot1 of the parent strain and the transformant were quantified by RT-qPCR. The transcript abundance of Trcot1 in SUS2/TrCot1i was 39.8 % lower than that of SUS2 (Table 2). This clearly indicated that the expression of the Trcot1 was significantly repressed by RNAi-mediated gene silencing.
Table 2.
Relative transcript abundance of selected genes in SUS2 and SUS2/Trcot1i.
| Gene | Relative transcript abundance | |
|---|---|---|
|
| ||
| SUS2 | SUS2/Trcot1i | |
| cot1 | 1.0 | 0.6±0.1 |
| cbh1 | 1.0 | 1.1±0.1 |
| cbh2 | 1.0 | 1.1±0.2 |
| egl2 | 1.0 | 1.1±0.1 |
| bgl1 | 1.0 | 1.1±0.1 |
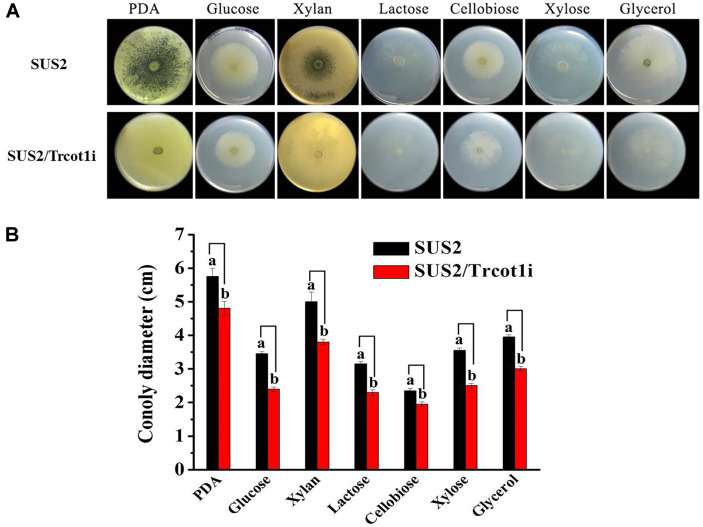
As shown in Fig. 3A, the sporulation of the transformant SUS2/Trcot1i on the PDA plate was much slower than that of the host strain. The colony diameters of SUS2/Trcot1i on the MM-agar plates containing glucose, xylan, lactose, xylose, or glycerol as the sole carbon source were smaller than that of the wild type. Although the colony diameter did not change on the MM-cellobiose medium, the mycelial density was apparently lower than that of the wild type. The quantitation of the colony diameters of the two strains on different culture medium was given in Fig. 3B, which indicated 17.0~30.4% reduction in the colony size in MM with different carbon source. These results demonstrated that the growth of the transformant was significantly retarded.
Fig. 3.
Effects of Trcot1 silencing on SUS2/Trcot1i growth on solid agar plates. (A) The colony morphology; (B) quantitation of the colony diameters. PDA: potato dextrose agar. Glucose, xylan, lactose, cellobiose, xylose, and glycerol are minimal media containing one of these carbohydrates as the sole carbon source. Different letters (a and b) for each same-culture medium mean that there are significant differences between the colony diameters (p < 0.05).
The RNAi-Silenced Transformant Displayed a Hyperbranching Phenotype
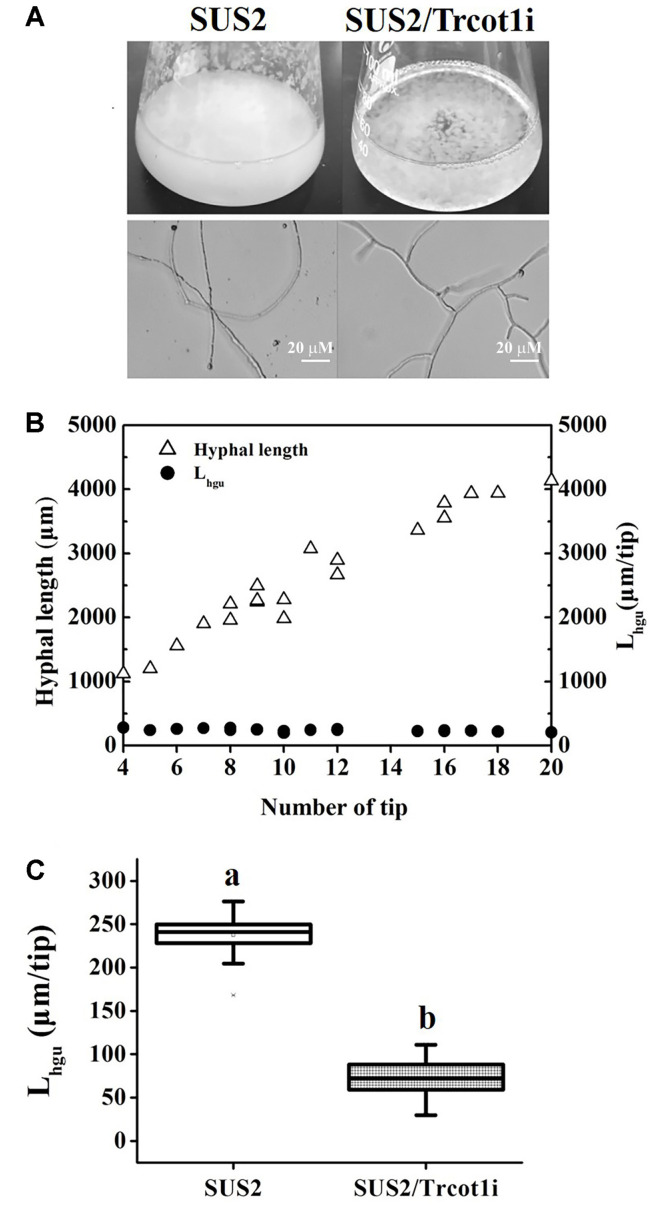
In submerged shake flask fermentation, SUS2 accumulated large amounts of mycelia after 48 h of culture. However, many fewer mycelia were observed for SUS2/Trcot1i (Fig.4A). The mycelia of SUS2/Trcot1i formed compact globules suspended in the culture broth. Observing the mycelia under microscope suggested that SUS2/Trcot1i had a hyperbranching phenotype (Fig. 4A). In order to ascertain whether the interference of Trcot1 expression would indeed result in a hyperbranching phenotype, the Lhgu value was measured in exponentially growing batch cultivations of SUS2/Trcot1i and its parent strain. Lhgu is an indicator of the degree of mycelium branching, with a lower Lhgu value indicating a more branched phenotype [24]. Using the SUS2 as an example, it was demonstrated that, although the total lengths of different mycelial units in the same strain were not the same, the corresponding Lhgu values remained as a constant (Fig. 4B), indicating that the Lhgu value could be used to measure and compare the branching morphological characteristics of different T. reesei strains. It was thus discovered that the RNAi-cassettes-bearing transformant SUS2/Trcot1i had a much smaller Lhgu value of 73.0 μm/tip than that of the SUS2 (239.8 μm/tip, Fig. 4C). The results undoubtedly demonstrated that the Trcot1-silenced strain was comparably more highly branched.
Fig. 4.
Effects of Trcot1 silencing on hyphal branching. (A) T. reesei grown in shake flasks containing the MM-glucose liquid medium as observed by naked eyes and under a microscope. Each scale bar represents 20 μm. (B) Demonstration of Lhgu as a constant value using SUS2 as a model strain. C: a comparison of Lhgu values of the SUS2 and SUS2/Trcot1i. Different letters (a and b) mean that there are significant differences between the Lhgu values (p < 0.05).
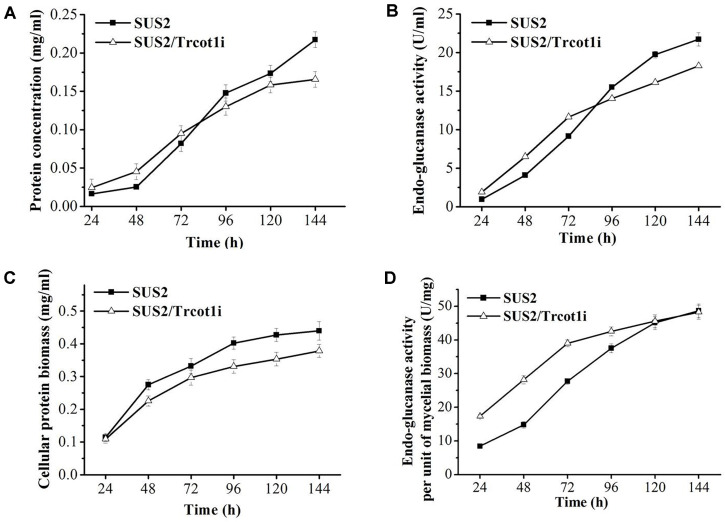
Cellulase Production in the RNAi-Silenced Transformant Since the hyperbranching phenotype of filamentous fungi has been reported to be positively related with higher amounts of secreted enzymes [8, 33], it was therefore asked if the secreted cellulase and extracellular protein concentration would increase in the RNAi-silenced hyperbranching transformant SUS2/Trcot1i. In the early stage of the culture (from 24 to 72 h), the extracellular protein concentration and the endo-glucanase activity of SUS2/Trcot1i were slightly higher than those of the parent strain SUS2. After 72 h, the protein concentration and endo-glucanase activity of SUS2/Trcot1i were outperformed by the SUS2 parent strain (Figs. 5A and 5B). SUS2/Trcot1i grew slower than its parent strain in solid and liquid media (Figs. 3 and 4A). Therefore, it was asked if the cellulase production per unit of mycelial biomass had changed. SUS2/Trcot1i clearly showed a considerable decrease in mycelial biomass accumulation (Fig. 5C). From 24 h to 144 h of cultivation, the mycelia biomass of SUS2/Trcot1i (as reflected by concentration of NaOH-extracted soluble cellular protein) amounted to 0.11~0.38 mg/ml, while the values were 0.11~0.44 mg/ml for the parent strain SUS2. As a result, the endo-glucanase activity per unit of mycelial biomass of SUS2/Trcot1i was 17.3~42.6 U/mg from 24 h to 96 h post cellulose induction, significantly higher than those of SUS2 (8.4~37.5 U/mg) (Fig. 5D). It was determined by RT-qPCR that the transcript abundance of the major cellulase genes cbh1, cbh2, egl2 and bgl1 was not affected in SUS2/Trcot1i (Table 2).
Fig. 5.
Cellulase expression in SUS2 and SUS2/Trcot1i. (A) Extracellular protein concentration. (B) Endo-glucanase activity. (C) The mycelial biomass as reflected by NaOH-extracted cellular protein concentration. (D) Endo-glucanase activity per mg of mycelial protein biomass.
Discussion
Secretory expression of cellulase in T. reesei is a very complex process involving coordinated regulation of transcription, translation, secretion, proteolysis, and post-translation modifications. Adding to this complexity is the multicellular nature of T. reesei as a filamentous fungus. Unlike the unicellular microbes such as Saccharomyces cerevisiae and Pichia pastoris, the cellular morphology of T. reesei has a large impact on secretory protein expression. The establishment of cell polarity, by itself, is also a complicated process entailing genes distributed in diverse pathways [34]. This wealth of associated genes provides a good opportunity to find a gene which, upon functional disruption, may lead to a hyperbranching phenotype.
In N. crassa, a genome-wide gene knockout mutant library is commercially available, which allows rapid screening of a mutant with desirable phenotype due to specified regulatory function [35]. This resulted in identification of gul-1 as a regulatory gene of cell polarity in N. crassa [36]. In contrast, there is no such mutant library in T. reesei. Therefore, we instead chose one regulatory gene from N. crassa with the defined function in determining cell polarity and investigated if disrupting the expression of its homologous gene in T. reesei would affect the morphology of the cell. The RNAi-mediated gene silencing strategy was used instead of gene knockout because the strain used in this study is resistant to homologous gene replacement and CRISPR/Cas9-mediated gene disruption (data not shown). One additional advantage is that RNAi can be conveniently used for a gene when its knockout leads to cell death.
The N. crassa COT1 belongs to the NDR kinase, which is important in regulating cell cycle and morphogenesis [37]. NcCOT1 genetically interacts with the mitogen-activated protein kinases MAK1 and MAK2 [22] and may modulate actin dynamics [38]. Both protein kinases have close homologs in T. reesei (Trire2:82351 for MAK1 and Trire2:121539 for MAK2, sharing amino acid sequence identities of 90.3% and 94.5%, respectively). In general, disruption of Trcot1 reduced the size of the mutant colony on solid plates on a rich medium (PDA) and the minimal media containing one of the sugar or glycerol carbon sources (Fig. 3). The appreciable change of mutant colonies strongly suggested that the cell morphology has been modified and this was ascertained by microscopic observation of the fungal mycelia cultured in liquid medium. The SUS2/Trcot1i mutant was of a hyperbranching phenotype and its biomass accumulation was lower than that of the wild type. Since the residues important for catalysis are well conserved in TrCOT1 (Fig. 1), it could be concluded that TrCOT1 indeed plays a role in regulating the cell polarity in T. reesei. Interestingly, there are nine more homologs of NcCOT1 in T. reesei with amino acid sequence identities of 39.3%-46.9%. All of them are annotated as putative protein kinases. However, whether they play a regulatory role in T. reesei and, specifically in cell morphogenesis, remains to be unveiled.
In T. reesei, the relationship of a gene function to the hyperbranching phenotype is seldom investigated. Our study thus provides an example of how to obtain a hyperbranching phenotype through genetic engineering in T. reesei. Although functional disruption of cot-1 leads to hyperbranching in N. crassa, it is not known if this phenotype is associated with more cellulase production [37]. In A. oryzae, the pcl2-deleted mutant has a hyperbranching phenotype, corresponding to an enhanced capability to secrete more enzymes [24]. However, disruption of a chitin synthase B increased the hypha branching but not a-amylase production in A. oryzae [39]. In our study, the hyperbranching phenotype slightly but significantly increased endo-glucanase secretion from 24 to 72 h. More importantly, the endo-glucanase activity per unit of mycelial biomass was much improved in the Trcot1-silenced strain from 24 to 96 h. The retarded fungal growth negatively impacted on apparent cellulase production in SUS2/Trcot1i.
The discrepancy in effects of hyperbranching on secretory protein production from our study and those of other researchers suggested that the choice of different regulatory genes may have profound effects on the ultimate protein secretion, even when the desired hyperbranching phenotype is successfully obtained. This further indicated that the genetic background of a strain could interact with the mutation. The beneficial effects incurred by increased hyphal tip numbers may be compromised by significantly decreased number of mycelia cells. To overcome this undesirable effect, on one hand, the genes involved in regulating cell polarity distributed in diverse pathways can be carefully screened until a gene whose manipulation (functional disruption or overexpression) leads to hyper-branching while not negatively affecting mycelial biomass accumulation. On the other hand, genetic engineering of other cellular genes to improve biomass accumulation, while maintaining the hyperbranching phenotype induced by Trcot1 silencing, is another route towards successfully obtaining cellulase hyperproducers.
Acknowledgments
This study was supported by the Central Public-interest Scientific Institution Basal Research Fund of China (No. Y2019XK03), a grant from the National Natural Science Foundation of China (31672458), and the National Chicken Industry Technology System of China (CARS-41).
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
REFERENCES
- 1.Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 2008;54:559568. doi: 10.1111/j.1365-313X.2008.03463.x. [DOI] [PubMed] [Google Scholar]
- 2.Galazka JM, Tian CG, Beeson WT, Martinez B, Glass NL, Cate J HD. C ellodex rtin t ransport i n yeast f or improved biofuel production. Science. 2010;330:84–86. doi: 10.1126/science.1192838. [DOI] [PubMed] [Google Scholar]
- 3.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris PV, Welner D, McFarland KC, Re E, Navarro Poulsen JC, Brown K, et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry. 2010;49:3305–3316. doi: 10.1021/bi100009p. [DOI] [PubMed] [Google Scholar]
- 5.Cherry JR, Fidantsef AL. Directed evolution of industrial enzymes: an update. Curr. Opin. Biotechnol. 2003;14:438–443. doi: 10.1016/S0958-1669(03)00099-5. [DOI] [PubMed] [Google Scholar]
- 6.Zhang JJ, Zhang GX, Wang W, Wei DZ. Enhanced cellulase production in Trichoderma reesei RUT C30 via constitution of minimal transcriptional activators. Microb. Cell Fact. 2018;17(1):75. doi: 10.1186/s12934-018-0926-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong YH, Yu HN, Wang XL, Lu Y, Wang TH. Towards a novel efficient T-DNA-based mutagenesis and screening system using green fluorescent protein as a vital reporter in the industrially important fungus Trichoderma reesei. Mol. Biol. Rep. 2011;38:4145–4151. doi: 10.1007/s11033-010-0534-z. [DOI] [PubMed] [Google Scholar]
- 8.Durand H, Clanet M, Tiraby G. Genetic improvement of Trichoderma reesei for large scale cellulase production. Enzyme Microb. Technol. 1988;10:341–346. doi: 10.1016/0141-0229(88)90012-9. [DOI] [Google Scholar]
- 9.Foreman PK, Brown D, Dankmeyer L, Dean R, Diener S, Dunn-Coleman NS, et al. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J. Biol. Chem. 2003;278:31988–31997. doi: 10.1074/jbc.M304750200. [DOI] [PubMed] [Google Scholar]
- 10.Rassinger A, Gacek-Matthews A, Strauss J, Mach RL, MachAigner AR. Truncation of the transcriptional repressor protein Cre1 in Trichoderma reesei Rut-C30 turns it into an activator. Fungal Biol. Biotechnol. 2018;5:15. doi: 10.1186/s40694-018-0059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, Zhao X, Bai F. Improvement of cellulase production in Trichoderma reesei Rut-C30 by overexpression of a novel regulatory gene Trvib-1. Bioresour. Technol. 2018;247:676–683. doi: 10.1016/j.biortech.2017.09.126. [DOI] [PubMed] [Google Scholar]
- 12.de Paula RG, Antonieto ACC, Carraro CB, Lopes DCB, Persinoti GF, Peres NTA, et al. The duality of the MAPK signaling pathway in the control of metabolic processes and cellulase production in Trichoderma reesei. Sci. Rep. 2018;8(1):14931. doi: 10.1038/s41598-018-33383-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen YM, Shen YL, Wang W, Wei DZ. Mn2+ modulates the expression of cellulase genes in Trichoderma reesei Rut-C30 via calcium signaling. Biotechnol. Biofuels. 2018;11:54. doi: 10.1186/s13068-018-1055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riquelme M, Sanchez-Leon E. The Spitzenkorper: a choreographer of fungal growth and morphogenesis. Curr. Opin. Microbiol. 2014;20:27–33. doi: 10.1016/j.mib.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Riquelme M, Bredeweg EL, Callejas-Negrete O, Roberson RW, Ludwig S, Beltran-Aguilar A, et al. The Neurospora crassa exocyst complex tethers Spitzenkorper vesicles to the apical plasma membrane during polarized growth. Mol. Biol. Cell. 2014;25:1312–1326. doi: 10.1091/mbc.e13-06-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayakawa Y, Ishikawa E, Shoji J, Nakano H, Kitamoto K. Septum-directed secretion in the filamentous fungus Aspergillus oryzae. Mol. Microbiol. 2011;81:40–55. doi: 10.1111/j.1365-2958.2011.07700.x. [DOI] [PubMed] [Google Scholar]
- 17.He RL, Li C, Ma LJ, Zhang DY, Chen SL. Effect of highly branched hyphal morphology on the enhanced production of cellulase in Trichoderma reesei DES-15. 3 Biotech. 2016;6(2):214. doi: 10.1007/s13205-016-0516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson-Hayes L, Hill TW, Loprete DM, Fay LM, Gordon BS, Nkashama SA, et al. Two GDP-mannose transporters contribute to hyphal form and cell wall integrity in Aspergillus nidulans. Microbiology. 2008;154:2037–2047. doi: 10.1099/mic.0.2008/017483-0. [DOI] [PubMed] [Google Scholar]
- 19.Dreyer J, Eichhorn H, Friedlin E, Kurnsteiner H, Kuck U. A homologue of the Aspergillus velvet gene regulates both cephalosporin C biosynthesis and hyphal fragmentation in Acremonium chrysogenum. Appl. Environ. Microbiol. 2007;73:3412–3422. doi: 10.1128/AEM.00129-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gatherar IM, Pollerman S, Dunn-Coleman N, Turner G. Identification of a novel gene hbrB required for polarised growth in Aspergillus nidulans. Fungal Genet. Biol. 2004;41:463–471. doi: 10.1016/j.fgb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Lindsey R, Cowden S, Hernandez-Rodriguez Y, Momany M. Septins AspA and AspC are important for normal development and limit the emergence of new growth foci in the multicellular fungus Aspergillus nidulans. Eukaryot. Cell. 2010;9:155–163. doi: 10.1128/EC.00269-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maerz S, Ziv C, Vogt N, Helmstaedt K, Cohen N, Gorovits R, et al. The nuclear Dbf2-related kinase COT1 and the mitogen-activated protein kinases MAK1 and MAK2 genetically interact to regulate filamentous growth, hyphal fusion and sexual development in Neurospora crassa. Genetics. 2008;179:1313–1325. doi: 10.1534/genetics.108.089425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Du L, Yuen G, Harris SD. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell. 2006;17:1218–1227. doi: 10.1091/mbc.e05-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.te Biesebeke R, Record E, van Biezen N, Heerikhuisen M, Franken A, Punt PJ, et al. Branching mutants of Aspergillus oryzae with improved amylase and protease production on solid substrates. Appl. Microbiol. Biotechnol. 2005;69:44–50. doi: 10.1007/s00253-005-1968-4. [DOI] [PubMed] [Google Scholar]
- 25.Gao F, Hao Z, Sun X, Qin L, Zhao T, Liu W, et al. A versatile system for fast screening and isolation of Trichoderma reesei cellulase hyperproducers based on DsRed and fluorescence-assisted cell sorting. Biotechnol. Biofuels. 2018;11:261. doi: 10.1186/s13068-018-1264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 27.Penttila M, Nevalainen H, Ratto M, Salminen E, Knowles J. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene. 1987;61:155–164. doi: 10.1016/0378-1119(87)90110-7. [DOI] [PubMed] [Google Scholar]
- 28.Jayaraman J, Cotman C, Mahler HR, Sharp CW. Biochemical correlates of respiratory deficiency. VII. Glucose repression. Arch. Biochem. Biophys. 1966;116:224–251. doi: 10.1016/0003-9861(66)90029-4. [DOI] [PubMed] [Google Scholar]
- 29.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 30.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 31.Taylor SS. cAMP-dependent protein kinase. Model for an enzyme family. J. Biol. Chem. 1989;264:8443–8446. [PubMed] [Google Scholar]
- 32.Li L, Chang SS, Liu Y. RNA interference pathways in filamentous fungi. Cell. Mol. Life Sci. 2010;67:3849–3863. doi: 10.1007/s00018-010-0471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiedler MRM, Cairns TC, Koch O, Kubisch C, Meyer V. Conditional expression of the small GTPase ArfA impacts secretion, morphology, growth, and actin ring position in Aspergillus niger. Front. Microbiol. 2018;9:878. doi: 10.3389/fmicb.2018.00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris SD. Cell polarity in filamentous fungi: shaping the mold. Int. Rev. Cytol. 2006;251:41–77. doi: 10.1016/S0074-7696(06)51002-2. [DOI] [PubMed] [Google Scholar]
- 35.Coradetti ST, Craig JP, Xiong Y, Shock T, Tian C, Glass NL. Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc. Natl. Acad. Sci. USA. 2012;109:7397–7402. doi: 10.1073/pnas.1200785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin L, Sun Z, Li J, Chen Y, Liu Q, Sun W, et al. Disruption of gul-1 decreased the culture viscosity and improved protein secretion in the filamentous fungus Neurospora crassa. Microb. Cell Fact. 2018;17(1):96. doi: 10.1186/s12934-018-0944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yarden O, Plamann M, Ebbole DJ, Yanofsky C. cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 1992;11:2159–2166. doi: 10.1002/j.1460-2075.1992.tb05275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seiler S, Vogt N, Ziv C, Gorovits R, Yarden O. The STE20/germinal center kinase POD6 interacts with the NDR kinase COT1 and is involved in polar tip extension in Neurospora crassa. Mol. Biol. Cell. 2006;17:4080–4092. doi: 10.1091/mbc.e06-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller C, McIntyre M, Hansen K, Nielsen J. Metabolic engineering of the morphology of Aspergillus oryzae by altering chitin synthesis. Appl. Environ. Microbiol. 2002;68:1827–1836. doi: 10.1128/AEM.68.4.1827-1836.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]