Abstract
Bacteria possess a repertoire of distinct regulatory systems promoting survival in disparate environments. Under in vitro conditions it was demonstrated for the human pathogen Staphylococcus aureus that the expression of most virulence factors is coordinated by the global regulator agr. To monitor bacterial gene regulation in the host, we developed a method for direct transcript analysis from clinical specimens. Quantification of specific transcripts was performed by competitive reverse transcription-PCR, and results were normalized against the constitutively expressed gene for gyrase (gyr). Using sputum from cystic fibrosis (CF) patients infected with S. aureus we examined the transcription of the effector molecule RNAIII of agr, of spa (protein A), generally repressed by agr, and of hla (alpha-toxin), generally activated by agr. In the CF lung RNAIII was expressed poorly, indicating an inactive agr in vivo. Despite the low level of RNAIII expression, spa was detectable only in minute amounts and an irregular transcription of hla was observed in all sputum samples. After subculturing of patient strains agr-deficient isolates and isolates with unusual expression profiles, i.e., not consistent with those obtained from prototypic strains, were observed. In conclusion, the agr activity seems to be nonessential in CF, and from the described expression pattern of spa and hla, other regulatory circuits aside from agr are postulated in vivo.
Over time, bacteria have evolved sophisticated regulatory circuits to modulate their gene expression in response to disparate environments (25). Our understanding of such adaptation processes during infection is limited by our ability to recreate host conditions in an experimental setting. Therefore, the sequential gene expression essential for host colonization, evasion of the immune system, tissue invasion, and maintenance in different organs remains to be determined for most bacterial infections. In recent years new approaches have been developed to identify bacterial genes which are induced during infection (15, 23). While these methods are very useful for screening new candidate genes involved in pathogenesis, until now no method has been available to discern the transcription pattern of characterized virulence genes directly during infection.
Staphylococcus aureus causes a variety of local and systemic infections in humans and is one of the most important community-acquired and nosocomially acquired pathogens. S. aureus infections are probably established via the coordinated synthesis of extracellular and cell-bound virulence factors (32). The expression of most virulence factors is controlled by the global regulator agr, which is thought to be a prime pathogenesis factor in S. aureus. The agr locus is composed of two divergent transcriptional units (RNAII and RNAIII). The RNA molecule RNAIII is the effector of the operon, which exhibits negative and positive regulatory functions (17, 29), activating extracellular proteins such as the hemolysins but repressing others, for example, protein A and coagulase. The transcription of RNAIII is highly dependent on the activation of the agr genes (agrA, agrB, agrC, and agrD) encoding RNAII. It has been shown that agrB and agrD are responsible for the synthesis of an extracellular octapeptide which operates as a quorum-sensing system (3, 18). This explains the growth-phase-dependent expression of agr-regulated genes. agrA shows sequence homology to the response regulator, while agrC corresponds to the histidine protein kinase signal transducer (28) of the classical two-component regulatory system (35). AgrC is thought to bind the octapeptide (18) and subsequently to phosphorylate AgrA. The respective promoters for RNAII and RNAIII (P2 and P3) are both thought to be autocatalytically activated by phosphorylated AgrA (28). Both promoters are also activated by a second regulatory locus, sar (8, 26). However, sar influences the expression of virulence factors not only via agr but also by independent mechanisms (7). Whereas the regulation of many S. aureus virulence factors has been studied extensively in vitro, the significance of coordinated gene expression during an actual infection is largely a matter of speculation.
Especially at risk for developing S. aureus infections are immunocompromised patients and patients with underlying diseases such as the genetic disorder cystic fibrosis (CF). Progressive pulmonary disease due to bacterial infection (i.e., by S. aureus and Pseudomonas aeruginosa) is the principal cause of morbidity and mortality in CF patients (11). In the preantibiotic era chronic S. aureus lung infections were the leading cause of death, and the bacteria are still difficult to clear efficiently from the bronchial system, despite the use of antimicrobial therapy. The role of S. aureus virulence factors in the establishment and progression of disease in patients with CF is largely unknown.
The aim of our study was to develop a method for the direct analysis of gene expression during bacterial infections. For the first time, the activity of a global regulator, agr of S. aureus, which controls the expression of major virulence factors, was monitored during infection. Here, we report on the investigation of S. aureus gene expression and regulation during chronic lung infection in CF patients. Specifically, we examined the transcription of RNAIII, the effector molecule of the agr operon. Additionally, transcription of spa (encoding protein A), a gene repressed by agr, and of hla (encoding alpha-toxin), a gene activated by agr, was monitored.
MATERIALS AND METHODS
Patients, collection, and bacteriological analysis of sputa.
A total of 12 CF patients of the Centro Fibrosi Cistica in Florence, Italy, were selected for this study. Eight patients were chronically colonized with S. aureus, and four patients were colonized with S. aureus and P. aeruginosa. Sputum samples from individual patients were repeatedly found to be positive for S. aureus over a long time (4 to 15 years), indicating chronic lung infection. Seventeen sputum samples were collected from the CF patients at their routine visits to the clinic. The sputum samples were frozen immediately in liquid nitrogen. One aliquot of each sample was stored at −70°C for RNA isolation. A second aliquot was treated with 1 M N-acetylcysteine (1:1 [vol/vol]) at 37°C for 30 min for the bacteriological analysis of the sputa. Serial dilutions of samples in 0.9% NaCl were cultured on sheep blood agar plates for quantitative bacteriology. S. aureus was identified with tube coagulase (bioMerieux, Nürtingen, Germany) and Staphaurex plus (Murex, Burgwedel, Germany). S. aureus colonies were phenotypically characterized by determining colony appearance (pigmentation) on sheep blood agar plates and assessing hemolysis on sheep blood agar (for the detection of alpha-hemolysin) and rabbit blood agar (for the detection of beta-hemolysin) plates after incubation at 37°C and again after subsequent incubation at 4°C (hot-cold hemolysis of beta-hemolysin). All isolates were typed with pulsed-field gel electrophoresis after SmaI digestion of chromosomal DNA as described previously (33). Genome types were defined using a similarity index calculated by the Dice coefficient.
Bacterial strains and growth conditions.
Strains are listed in Table 1. S. aureus was grown in CYPG (27) or on tryptic soy agar with the appropriate antibiotics. For phenotypic characterization the cells were inoculated from an overnight culture to an initial optical density at 600 nm (OD600) of 0.05 in CYPG and grown to the mid-exponential (OD600 = 0.6 at 2.5 h after inoculation), late exponential (OD600 = 2.5 at 4 h after inoculation), or postexponential (OD600 = 8 at 8 h after inoculation) phase.
TABLE 1.
S. aureus strains used in this study
RNA isolation from sputum.
Frozen sputum samples were thawed rapidly, and 200-μl aliquots were used for RNA isolation. S. aureus cells were lysed directly in 1 ml of Trizol LS reagent (Gibco BRL, Karlsruhe, Germany) with 0.5 ml of zirconia-silica beads (diameter, 0.1 mm) in a high-speed homogenizer (Savant Instruments, Farmingdale, N.Y.). RNA was isolated as directed in the instructions of the manufacturer of Trizol.
RNA isolation from culture.
Bacteria were grown until the desired growth phase. A total of 1010 cells were pelleted and lysed in 1 ml of Trizol reagent (Gibco BRL). Cell lysis and RNA isolation were performed as described above.
DNA digestion.
Contaminating DNA was degraded by digesting RNA samples with DNase. The reaction was performed with 5 mM MgCl2, 40 U of RNasin (Promega, Madison, Wis.), and 20 U of DNase I (Boehringer Mannheim, Mannheim, Germany) at room temperature for 30 min.
Slot blot hybridization.
Serial dilutions of sample RNA in 10 mM NaOH–1 mM EDTA were transferred onto a positively charged nylon membrane (Boehringer Mannheim) with a Slot-Blotter (Bio-Rad, Hercules, Calif.). Hybridization was performed using standard procedures, and the signals were detected by chemiluminescence. Slot blot hybridization was applied to detect the rare transcript gyrase. Specific primers (GenBank accession no. D10489, nucleotides [nt] 219 to 536) TTATGGTGCTGGGCAAATACA and CACCATGTAAACCACCAGATA were used to generate a digoxigenin-labeled probe by PCR labeling (Boehringer Mannheim).
In order to quantify total RNA we developed an rRNA slot blot technique using a digoxigenin-labeled oligonucleotide (GenBank accession no. X68417, nt 212 to 251) GCAGCGCGGATCCATTAAGTGACAGCAAGACGCTC specific for S. aureus 16S rRNA. Serial dilutions of known amounts of a PCR-generated rDNA fragment (GenBank no. X68417, nt 118 to 341) AACACGTGGATAACCTACCTA and ACCGTGTCTCAGTTCCAGTGT were employed as a standard on each blot to quantify the sample RNA. The signal intensity was determined with a densitometer (Cybertech, Berlin, Germany).
Quantification of specific transcripts with competitive RT-PCR.
Reverse transcription-PCR (RT-PCR) was carried out using the TITAN One Tube RT-PCR System (Boehringer Mannheim). Master mixes were prepared following the manufacturer's instructions, using primers as follows: for gyr (GenBank no. D10489, nt 219 to 536), TTATGGTGCTGGGCAAATACA and CACCATGTAAACCACCAGATA; for spa (GenBank no. J01786, nt 254 to 561), TACTTATATCTGGTGGCGTAA and GGTCGTCTTTAAGACTTTGA; for hla (GenBank no. X01645, nt 489 to 897), AGAAAATGGCATGCACAAAAA and TATCAGTTGGGCTCTCTAAAA; and for RNAIII (GenBank no. X52543, nt 1483 to 1242), GAAGGAGTGTTTCAATGG and TAAGAAAATACATAGCACTGAG. A 25-μl reaction volume was supplemented with various amounts of competitor RNA (100, 20, 4, 0.8, and 0.16 amol for the quantification of gyr, spa, and hla; 1,000, 200, 40, 8, and 1.6 amol for the quantification of RNAIII), and constant amounts of sample RNA were added (1 ng of total RNA for gyr, spa, and hla; 0.1 ng of total RNA for RNAIII). After RT for 30 min at 50°C the following temperature profile was utilized for amplification: initial denaturation at 94°C for 2 min; 35 cycles at 94°C for 30 s, 50°C (55°C for spa) for 30 s, and 68°C for 45 s (an extension of the elongation step by 5 s per cycle was programmed after 10 cycles); and a final extension at 68°C for 7 min. To exclude the possibility of DNA contamination, control samples were subjected to amplification without prior reverse transcription. Aliquots of the amplified products were analyzed on a 3% agarose gel. The concentration of specific mRNA was calculated in comparison with the competitor.
Construction of specific RNA competitor.
Sequence-modified RNA templates for competitive RT-PCR specific for gyr, spa, hla, and RNAIII were engineered by a deletion mutagenesis PCR technique using an oligonucleotide composed of two distinctly spaced target sites, thus generating deletions in the final sequence to allow discrimination between competitor and target amplicons. The following primers were used: for gyr (D10489), TTATGGTGCTGGGCAAATACATTAGTGTGGGAAATTGTCGATAAT (5′ position, nt 219) and GTACGATTTAATACCGCCCTCATA (3′ position, nt 898); for spa (J01786), TACTTATATCTGGTGGCGTAAATGCCTAACTTAAATGCTGAT (5′ position, nt 254) and TTTTTAGCTTCTGACAATAGG (3′ position, nt 791); for hla (X01645), AGAAATGGCATGCACAAAAACGAAGAAGGTGCTAACAAAA (5′ position, nt 498) and TGCAATTGGTAATCATCACGAACTC (3′ position, nt 1185); and for RNAIII (X52543), GAAGGAGTGATTTCAATGGGGATTATCGACACAGTGAA (5′ position, nt 1501) and TAAGAAAATACATAGCACTGAG (3′ position, nt 1242). The resulting PCR constructs were cloned into pCRII-TOPO (Invitrogen, Carlsbad, Calif.), transformed to XL1-Blue, and sequenced to determine the clones containing the correct modification. Using either of these recombinant pCRII-TOPO plasmid DNAs as a template, a second PCR was performed with a gene-specific primer for gyr, spa, hla, or RNAIII, with a 5′ extension encompassing the T7 phage promoter sequence (instead of using the promoter of the vector), thus generating highly transcription-competent amplicons. T7-driven in vitro transcription of single-stranded competitor RNA was performed using a standard transcription assay (Riboprobe; Promega). The reaction mixture was subjected to DNase I treatment (Boehringer Mannheim), and the RNA was recovered with phenol-chloroform extraction and isopropyl alcohol precipitation. Quantification of the transcripts was done spectrophotometrically and verified by ethidium bromide staining on agarose gels.
Northern analysis.
For Northern blot analysis, 2 μg of total RNA isolated from bacterial cultures was electrophoresed through a 1% agarose–0.66 M formaldehyde gel and blotted by alkaline transfer (Turbo Blotter; Schleicher and Schuell, Dassel, Germany) onto a positively charged nylon membrane (Boehringer Mannheim). The intensities of the 23S and 16S rRNA bands stained by ethidium bromide were verified to be equivalent in all the samples before transfer. High-stringency hybridization was performed according to the instructions given by the manufacturer of the digoxigenin labeling and detection kit (Boehringer Mannheim); signals were detected by chemiluminescence. Specific primers were used to generate digoxigenin-labeled probes by PCR labeling (Boehringer Mannheim). The following primer pairs were used: for RNAIII (nt 999 to 1510, SAAGRAB), GAAGGAGTGTTTCAATGG and TAAGAAAATACATAGCACTGAG; for spa (nt 219 to 771, SASPA), AGGTGTAGGTATTGCATCTGT and TTTTTAGCTTCTGACAATAGG, and for hla (nt 498 to 1098, SATOXA), AGAAAATGGCATGCACAAAAA and TGTAGCGAAGTCTGGTGAAAA.
PCR for detection of the agr operon.
The following primer pairs were used for the detection of agr in S. aureus isolates: for agrA (X52543, nt 3829 to 4342), CGAAGACGATCCAAAA and TTATCTAAATGGGCAATGAGT; for agrBDC (X52543, nt 1754 to 2973), CAGTTGAGGAGAGTGGTGTAAA and AAAAAGTAAGCAGTAAGATAG; and for RNAIII (X52543, nt 999 to 1510), TATATTTTAACGGCGGGTCTTCA and TTAATTAAGGAAGGAGTGATTT.
Whole-cell enzyme-linked immunosorbent assay (ELISA) for detection of protein A.
Bacteria were grown until the desired growth phase was reached. A total of 109 (OD600 = 1) bacteria were pelleted and washed twice with 0.9% NaCl. Serial dilutions of cells (1:50 to 1:156,250) in 1× phosphate-buffered saline (PBS)–0.05% Tween 20 were transferred onto a membrane filter (Millipore, Karlsruhe, Germany) with a pore diameter of 0.22 μm (at this size, antibodies can pass through, but bacteria are retained), which was fitted into a dot blotter (Bio-Rad). No vacuum was applied at this point, so the bacteria remained in solution. For the detection of cell-bound protein A, the bacteria were incubated with alkaline phosphatase (AP)-conjugated rabbit anti-mouse immunoglobulin G (1:5,000 in 1× PBS) for 2 h at 37°C. Excess antibody was removed by washing three times with PBS-Tween 20 under application of vacuum. Detection of AP was carried out in 10 ml of 0.1 M Tris-HCl–0.1 M NaCl (pH 9.5) plus 200 μl of nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (BCIP) solution (Boehringer Mannheim).
Statistical analysis.
The paired Student's t test was used for the statistical analysis of data.
RESULTS
Direct transcript analysis from clinical specimens.
In order to gain insight into the regulatory response of S. aureus during infection, specific transcripts of the virulence genes RNAIII, spa, and hla were quantified by competitive RT-PCR. Direct transcript analysis was performed with ex vivo material without subculturing the bacteria. Briefly, S. aureus cells were lysed nonenzymatically in the sputa and the RNA was isolated. Contaminating DNA was degraded, total RNA was quantified, and equal amounts were subjected to competitive RT-PCR (Fig. 1). To compare the expression profile during infection with the transcription pattern in vitro, the RNA derived from the exponential growth phase and that derived from the postexponential growth phase of sputum strains grown in culture were analyzed at the same time.
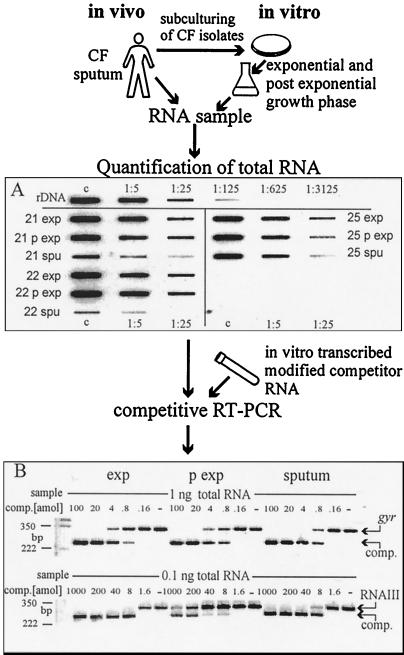
FIG. 1.
S. aureus cells were lysed nonenzymatically in the sputa, and the RNA was isolated. The RNA of sputum strains grown in culture was obtained in the same manner. An S. aureus-specific rRNA slot blot was developed to determine total RNA. Constant amounts of sample RNA were then spiked with serial dilutions of gene-specific competitor RNA for gyr, spa, hla, and RNAIII. After competitive RT-PCR the samples were separated by agarose gel electrophoresis and visualized after ethidium bromide staining. Quantification of transcripts was possible by comparison with the competitor. (A) Serial dilutions (1:1, 1:5, and 1:25) of samples were hybridized with a digoxigenin-labeled oligonucleotide specific for S. aureus 16S rRNA. Aliquots of RNA isolated in the exponential (exp) and the postexponential (p exp) growth phases of S. aureus isolates 21, 22, and 25 and aliquots of RNA isolated from sputum samples 21, 22, and 25 are shown. rDNA, rDNA PCR fragment starting with 10 fmol for the control dilution; c, control dilution (1:1). (B) Example of a gel after competitive RT-PCR. The top shows the detection of gyrase (gyr), and the bottom shows the detection of RNAIII. RNA was derived from the exponential (exp) and postexponential (p exp) growth phases of an S. aureus sputum strain and directly from the sputum. Constant amounts (1 and 0.1 ng) of sample RNA were spiked with serial dilutions of the competitors (100 to 0.16 amol and 1,000 to 1.6 amol, respectively; −, no competitor). In all three samples the same amount of gyrase was detectable. comp., competitor.
Because of low bacterial numbers in the specimens, it was necessary to establish a method for the quantification of isolated bacterial RNA. Therefore, a sensitive slot blot technique specific for S. aureus 16S rRNA was developed which discriminates between S. aureus-specific RNA and RNA derived from other organisms within the specimens (Fig. 1A). Serial dilutions of known amounts of a PCR-generated rDNA fragment of S. aureus 16S rRNA were used as a standard on each blot to allow quantification. Hybridization was performed with an oligonucleotide specific for a nonconserved region of the 16S rRNA gene. A sensitivity of 100 pg of S. aureus RNA was reached and no cross-reaction with RNA derived from P. aeruginosa was detected (data not shown).
We looked for a constitutively expressed S. aureus gene that would be suitable for use as an external reference for specific transcript quantification. In pilot experiments it could be shown by slot blot hybridization of S. aureus RNA that the transcription of the gene for gyrase (gyr) was not affected by cell density and/or agr (Fig. 2). Similarly, gyrase-specific RT-PCR revealed no significant differences in gyr expression between the different growth phases (Fig. 1B). Thus, gyr proved to be suitable for use as an external reference, and in subsequent experiments the molar ratio of specific transcripts was determined in relation to gyr.
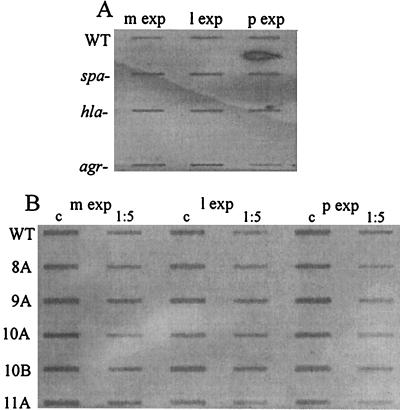
FIG. 2.
Slot blot for the detection of the transcript for gyrase. RNA was derived from the midexponential (m exp), late exponential (l exp), and postexponential (p exp) growth phases of ISP479C (agr+) (the wild type [WT]), DU5723 (spa mutant), DU1090 (hla mutant), and ISP546 (agr mutant) (A) and ISP479C (the WT strain) and the CF isolates 8A, 9A, 10A, 10B and 11A (1:1 and 1:5 dilutions) (B). Samples were hybridized with a digoxigenin-labeled gene probe specific for S. aureus gyr. c, control dilution (1:1).
To determine the reproducibility of the procedure, RNA from S. aureus strain ISP479C grown to the exponential and postexponential phase and from RNA ISP 546 (agr mutant) grown to the postexponential phase were isolated in quadruplicate from each source. Quantification of gyr with competitive RT-PCR resulted in a mean ± standard deviation of 0.475 ± 0.18 amol/ng of total RNA; again, no significant difference in gyr transcription was found between the exponential and postexponential phases (P ≤ 0.6) and between agr-positive and -negative strains (P ≤ 1). The reliability of the quantitative RT-PCR was further confirmed by comparison with Northern analysis.
In order to determine the sensitivity limit of direct transcript analysis, sputum obtained from CF patients not infected with S. aureus was inoculated with serial dilutions of S. aureus cells. RNAIII was still detectable in aliquots containing 10 bacteria; spa and hla were still detectable in aliquots containing 100 bacteria, and gyr was still detectable in aliquots containing 1,000 bacteria. The sensitivity limits reflect the different concentrations of the specific transcripts, since in some strains RNAIII transcription exceeds that of gyr by a factor of more than 500. The abundance of RNAIII has also been shown by other investigators (2), and it seems to be necessary for the optimal functioning of the agr regulon.
Bacteriological analysis of sputum samples.
A total of 17 sputum samples from 12 CF patients (Centro Fibrosi Cistica, Florence, Italy) colonized with S. aureus were assayed. Bacterial numbers were estimated to be in the range of 104 to 107 CFU/ml of sputum in all samples. The S. aureus phenotype (colony appearance, hemolytic pattern) and genome type were also assessed. Within 12 sputum samples only a single phenotype could be detected, whereas in 5 sputum samples S. aureus isolates with dissimilar phenotypes were observed upon primary subculturing on blood agar plates. Genome typing revealed that one patient was infected with two distinct genome types and another patient was infected with three distinct genome types simultaneously (Table 2). In the remaining three sputum samples the dissimilar phenotypes could be ascribed to a single genome type each.
TABLE 2.
Direct transcript analysis of five CF sputum samples containing dissimilar S. aureus phenotypesa
| Sputum sample no. | Iso | No. of CFU/ml of sputum | GT | Amt (amol/amol of gyr) ofb:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RNAIII
|
spa
|
hla
|
||||||||||
| exp | p exp | s | exp | p exp | s | exp | p exp | s | ||||
| 7 | 7B | 4 × 106 | 1 | 0 | 12.5 | 0 | 5 | 25 | 6 | 0 | 0.13 | 1 |
| 7C | 1 × 106 | 1 | 0 | 4 | 10 | 20 | 0 | 1 | ||||
| 8 | 8A | 1 × 107 | 19 | 2 | 150 | 8 | 2 | 5 | 2 | 0.02 | 0.5 | 0.03 |
| 8B | 3 × 106 | 2 | 0.2 | 2.5 | 5 | 5 | 0.08 | 0.4 | ||||
| 8C | 1 × 107 | 60 | 3 | 200 | 2 | 5 | 0.01 | 0.1 | ||||
| 9 | 9A | 8 × 105 | 20 | 0.08 | 2 | 0.2 | 0.8 | 4 | 0.4 | 0.1 | 0.6 | 0.1 |
| 9B | 4 × 105 | 20 | 1.25 | 150 | 0.5 | 0 | 0.15 | 0.15 | ||||
| 10 | 10A | 4 × 106 | 36 | 3 | 300 | 15 | 0.5 | 0.2 | 0.25 | 0.05 | 2 | 1.25 |
| 10B | 1 × 106 | 36 | 0.8 | 200 | 5 | 2.5 | 0.065 | 2 | ||||
| 13 | 13A | 1 × 104 | 48 | 40 | 250 | 100 | 0 | 0 | 2 | 0.16 | 1.6 | 1 |
| 13B | 5 × 105 | 63 | 4.5 | 100 | 0.6 | 5 | 0.12 | 0.4 | ||||
Iso, isolate; GT, genome type.
exp, exponential growth phase; p exp, postexponential growth phase; s, sputum (values are for sputum sample, regardless of isolate).
RNAIII expression during chronic lung infection in CF.
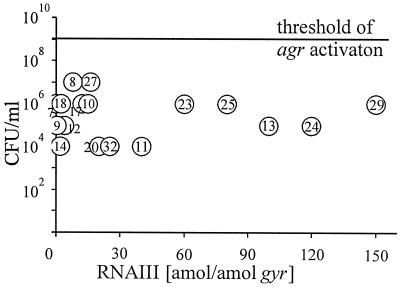
Our aim was to study the activity of the global regulator agr of S. aureus in vivo, which is thought to control the expression of major virulence factors. Interestingly, the bacteria expressed RNAIII, the effector molecule of the agr operon, poorly in all the sputum samples (Fig. 3A and Table 2). Since agr is activated by a quorum-sensing system, we plotted S. aureus cell numbers in the sputa against the RNAIII transcription in vivo (Fig. 4). No correlation between density and RNAIII expression was found. In all the sputum samples the bacterial density was below the critical threshold for agr activation in vitro (109 CFU/ml [3]). Transcription of RNAIII in the sputa was always lower than the expression of the respective S. aureus strain grown to the postexponential phase. For instance in sputum sample 10 15 amol of RNAIII/amol of gyr was detected, whereas the sputum isolates 10A and 10B produced 300 and 200 amol of RNAIII/amol of gyr, respectively, in the postexponential phase (Table 2). In 15 of the CF isolates, agr could be activated after subculturing and was characterized by a strong expression of RNAIII in the postexponential growth phase (>100 amol of RNAIII/amol of gyr [Fig. 3A]). In eight of the strains, an inactive agr regulon was found in vitro, meaning that the expression of RNAIII was either low (<50 amol of RNAIII/amol of gyr [Fig. 3A]) or not detectable.
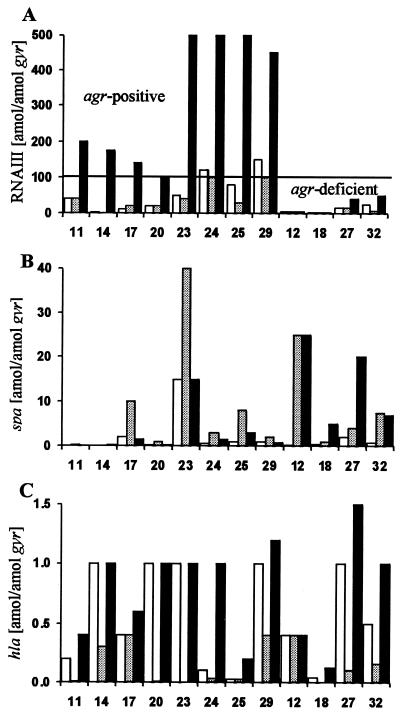
FIG. 3.
Expression of the S. aureus virulence genes RNAIII (A), spa (B), and hla (C) during chronic lung infection in CF patients (sputum [white columns]) and in 12 sputum isolates after growth in culture (exponential phase [light grey columns] and postexponential phase [black columns]). The sputum sample identification numbers are indicated below the columns. Transcripts were quantified in reference to the transcription of gyrase (in attomoles per attomole of gyr). As shown in panel A, agr-positive strains (derived from sputum sample no. 11, 14, 17, 20, 23, 24, 25, and 29) had strong expression of RNAIII in the postexponential growth phase (>100 amol of RNAIII/amol of gyr) and agr-deficient strains (derived from sputum sample no. 12, 18, 27, and 32) had expression of RNAIII that was either low (<50 amol of RNAIII/amol of gyr) or not detectable.
FIG. 4.
S. aureus cell numbers (in CFU per milliliter) in 17 CF sputum samples (sputum sample identification numbers are given within the open circles) and RNAIII transcription in vivo. The line indicates the threshold for agr activation in vitro.
Protein A expression during chronic lung infection in CF.
The cell-bound protein A is a prototypic down-regulated target molecule of agr in vitro. In the sputa, the expression of spa was greatly diminished compared to its transcription in culture (Fig. 3B and Table 2). For instance in sputum sample 25, 1 amol of spa/amol of gyr was detected, whereas the sputum strain 25 produced 8 amol of spa/amol of gyr in the exponential growth phase and 3 amol of spa/amol of gyr in the postexponential growth phase. Even in agr-deficient strains (Fig. 3B, strains 12, 18, 27, and 32) spa was only poorly expressed in vivo. However, after subculturing, these strains showed the expected lack of spa inhibition in the postexponential growth phase.
Alpha-toxin expression during chronic lung infection in CF.
The secreted protein alpha-toxin is a prototypic activated target of agr in vitro. In the sputa up to 1.25 amol of hla/amol of gyr was detected (Fig. 3C and Table 2). Also, after the sputum strains were grown to the postexponential phase, transcription of hla never exceeded 2 amol of hla/amol of gyr. The typically alpha-hemolytic laboratory strains ISP479C, Reynolds, and Becker showed the same range of hla transcription (4, 1, and 0.5 amol of hla/amol of gyr, respectively). Therefore, the abundance of the hla transcript is low compared to RNAIII and spa transcripts. Interestingly, in some agr-deficient strains, hla was transcribed both in vitro and in vivo at levels comparable to those in the agr-positive strains (up to 1.5 amol of hla/amol of gyr [Fig. 3C, strains 27 and 32]).
In vitro characterization of S. aureus sputum strains.
A total of 23 S. aureus isolates were obtained from the 17 CF sputum samples. The strains were discriminated phenotypically by colony appearance and/or hemolysis on blood agar plates (Table 3). No small colony variants were detected. To rule out the possibility that the expression pattern observed in the different sputum samples is due to a unique, CF-specific S. aureus strain, sputum isolates were typed with pulsed-field gel electrophoresis. The 23 CF isolates could be assigned to 11 genome types. Seven of these genome types were also detected in the nares of healthy controls (C. Goerke et al., unpublished results). Hence, the strain population causing infections in CF patients does not differ from the strain populations colonizing healthy individuals.
TABLE 3.
In vitro characterization of 23 S. aureus isolates from CF sputa
| Groupd and isolate | Result of hemolysisb
|
GTe | Cc | Northern analysisa
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RNAIII
|
spa
|
hla
|
|||||||||
| R | S | 4°C | exp | p exp | exp | p exp | exp | p exp | |||
| Group I | |||||||||||
| 9B | + | + | + | 20 | y | − | ++ | + | − | +/− | +/− |
| 10A | + | + | + | 36 | y | +/− | +++ | +/− | − | − | + |
| 10B | ++ | ++ | +++ | 36 | w | − | +++ | ++ | + | − | + |
| 11 | ++ | ++ | +++ | 62 | w | + | +++ | +/− | − | − | + |
| 13A | ++ | ++ | +++ | 48 | w | +/− | ++ | +/− | +/− | − | + |
| 17 | + | + | ++ | 17 | w | − | ++ | + | − | − | +/− |
| 20 | + | + | + | 36 | w | +/− | ++ | +/− | − | − | + |
| 23 | ++ | ++ | +++ | 2 | w | ++ | +++ | ++ | + | − | + |
| 24 | + | ++ | +++ | 61 | w | +/− | +++ | + | +/− | +/− | + |
| 25 | − | + | + | 2 | w | − | +++ | + | +/− | − | +/− |
| 29 | + | + | ++ | 17 | y | +/− | +++ | +/− | − | − | + |
| Group II | |||||||||||
| 7B | − | − | + | 1 | y | − | − | + | ++ | − | +/− |
| 7C | − | − | + | 1 | w | − | − | + | ++ | − | + |
| 8B | − | + | ++ | 2 | w | − | − | ++ | ++ | − | +/− |
| 9A | − | − | + | 20 | y | − | − | + | ++ | +/− | +/− |
| 12 | − | − | − | 19 | w | − | − | ++ | ++ | +/− | +/− |
| 18 | − | − | ++ | 19 | w | − | − | + | + | − | +/− |
| 27 | − | − | ++ | 1 | w | − | − | + | ++ | − | + |
| 32 | − | − | +/− | 20 | y | − | +/− | ++ | ++ | − | + |
| Group III | |||||||||||
| 8A | + | + | ++ | 19 | y | − | ++ | + | ++ | − | +/− |
| 8C | ++ | ++ | +++ | 60 | w | − | ++ | + | ++ | − | +/− |
| 13B | ++ | ++ | ++ | 63 | y | − | + | + | ++ | − | +/− |
| 14 | ++ | ++ | +++ | 36 | w | − | ++ | − | +/− | +/− | + |
| Group IV | |||||||||||
| ISP479C | ++ | ++ | ++ | w | + | +++ | +/− | − | +/− | ++ | |
| DU5723 | ++ | ++ | ++ | w | + | +++ | − | − | +/− | ++ | |
| ISP546 | − | − | − | w | − | − | + | ++ | − | − | |
For Northern analysis RNA was isolated in exponential (exp) and postexponential (p exp) growth phases and specific transcripts were probed with digoxigenin-labeled gene probes for RNAIII, spa, and hla. Signals were detected by chemiluminescence and characterized as follows: −, none; +/−, weak; +, median; ++, strong; +++, very strong.
Hemolysis was determined on rabbit (R) and sheep (S) blood agar plates and after incubating plates at 4°C. Responses were characterized as follows: −, none; +/−, weak; +, median; ++, strong; +++, very strong.
C, colony appearance, characterized as yellow (y) or white (w) colonies on sheep blood agar plates.
Group I, agr-positive strains; group II, agr-deficient strains; group III, agr-positive strains with inverse spa expression; and group IV, prototypic strains.
GT, genome type.
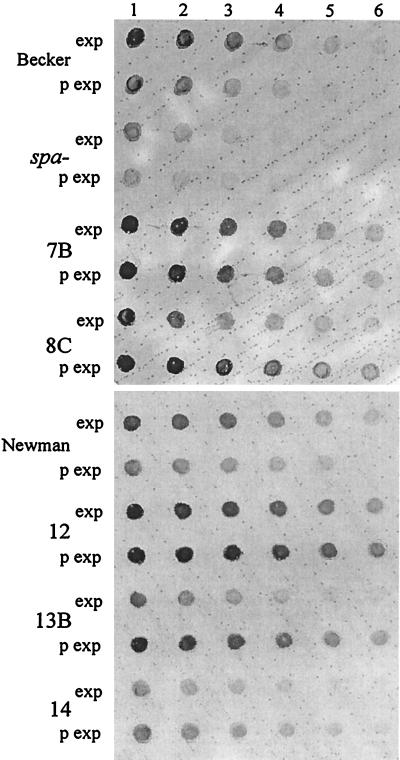
Competitive RT-PCR revealed unusual expression patterns in some sputum isolates after subculturing. To further analyze the transcription of RNAIII, spa and hla Northern blot analysis was performed on strains grown to the exponential and postexponential phase. Although this method does not allow absolute quantification of transcripts the comparison with the RT-PCR yielded the same relative results. Most of the CF isolates showed a cell density-dependent expression of spa and hla in vitro comparable to that of ISP479C and consistent with a functional agr locus (group I of Table 3). A second group encompassing agr-deficient strains (group II of Table 3) showed the typical expression pattern of an agr mutant like ISP546 (expression of spa during postexponential phase). Interestingly, such a lack of spa inhibition during the growth cycle was also found in four agr-positive strains (group III of Table 3). The four isolates do not represent a single clone, as confirmed by genome typing (Table 3). To further analyze whether the enhanced transcription of spa in the postexponential phase results in elevated cell-bound protein in those strains, we established a whole-cell protein A ELISA technique. In concordance with the transcriptional data an increase in protein A was observed in strains of group III during growth (Fig. 5). As expected, the agr-deficient strains (7B and 12) showed constitutive protein A production.
FIG. 5.
Whole-cell ELISA specific for S. aureus protein A. Serial dilutions of bacterial cells derived from the exponential (exp) and postexponential (p exp) growth phases were transferred onto a membrane filter, and protein A was detected with an AP-conjugated antibody. S. aureus strains Newman and Becker showed the typical agr-dependent protein A inhibition in the postexponential phase. The agr-deficient CF isolates 7B and 12 showed constitutive protein A production. In the agr-positive strains 8C, 13B, and 14, elevated levels of protein A could be observed in the postexponential phase. Dilutions were as follows: 1:50 (lane 1), 1:250 (lane 2), 1:1,250 (lane 3), 1:6,250 (lane 4), 1:31,250 (lane 5), 1:156,250 (lane 6).
DISCUSSION
In this study we were able to analyze the regulatory response of S. aureus during an infection in humans. In principle this approach can be used to study other infections with low bacterial numbers and/or the presence of low-abundance transcripts. Here, we show evidence that the S. aureus global regulator agr is not activated during chronic lung infection in CF patients. This was demonstrated by quantifying the regulatory molecule RNAIII in sputum samples and after growth of the sputum strains in vitro. The transcription of RNAIII in vivo was comparable to the level of expression in exponential phase and was always far below the transcription level in postexponential growth in which the RNAIII level necessary for target gene activation is reached. The significance of the agr locus as a virulence factor has been validated in various animal models. In these models agr mutants were less virulent than the corresponding wild-type strains (1, 6). Possibly the agr operon is activated only during particular types of infections and/or at certain stages of a given infection. Generally, surface proteins (protein A, fibronectin binding protein, etc.) are thought to play a role in the establishment of infection and subsequent evasion of host defense, whereas secreted proteins are necessary for progression into new ecological niches (tissue penetration) (28). In the chronic CF lung infection, S. aureus is localized in and restricted to the highly viscous mucus (36) and infection is not accompanied by systemic manifestations of S. aureus. Therefore, the lack of agr activation may be unique for S. aureus infections in patients with CF. No correlation between bacterial cell density and RNAIII expression was found in vivo. One may speculate that agr might be locally activated due to the higher concentration of the autoinducer within bacterial clusters which may occur in the viscous sputa. This may account for the residual RNAIII expression in vivo. However, single-cell assays which would allow the analysis of gene expression within such a heterogeneous bacterial population are not available so far. Recently, another particular S. aureus phenotype was described in CF patients (16). It was shown by immunofluorescence that the capsular polysaccharide type 5 of S. aureus was greatly diminished in the CF airways. This phenomenon can be explained by the inactive agr, since the synthesis of capsular polysaccharide is positively regulated by agr (10) (our unpublished observation).
Interestingly, despite the inactive agr only minute amounts of spa were detectable in vivo. In contrast, in an animal model we detected large amounts of spa by using the described method (data not shown). Therefore, the low spa transcription may be specific for S. aureus lung infection in CF patients due to specific environmental conditions and/or host signals within the lung. The role of spa as a virulence factor is highly disputed and depends on the animal model studied (5, 31). Besides the inhibitory effect of the regulatory loci agr and sar (7) on spa transcription, little is known about the influence of other signals. The simultaneous inhibition of RNAIII and spa indicates the presence of signals and additional regulators not linked to agr. Possibly the spa inhibition is caused by the increased production of sar. However, this would imply that in this case sar does not activate agr as described by Heinrichs et al. (14).
The transcript of the cytotoxic alpha-toxin was detectable in all the sputum samples. Transcription was not correlated to RNAIII expression. Again this indicates additional agr-independent regulatory mechanisms in vivo. However, we have no evidence for the actual production of the alpha-toxin protein. No damage to the epithelial tissue underlying S. aureus-containing mucus was seen in scanning and transmission electron microscopy of CF lung specimens (36), suggesting an inactivation of the cytotoxic alpha-toxin in the mucus by proteases or antibodies (13, 21). One may speculate that in chronic and localized infections alpha-toxin is not essential. This hypothesis is further accentuated by the fact that the small-colony variants of S. aureus do not produce alpha-toxin (19, 37). Small-colony variants were found in CF patients receiving trimethoprim-sulfomethoxazole (19).
After subculturing patient strains, we observed unusual expression profiles of virulence genes which were not consistent with those obtained from prototypic strains subcultured in laboratories over the decades. This illustrates the great diversity of the genus S. aureus. One set of strains (group II) showed little or no RNAIII even after growth to the postexponential phase. Like other investigators (17) we observed that spontaneous mutations of the agr locus accumulate upon repeated cultivation on agar plates. Here, we were able to show that agr-deficient strains are able to infect CF airways. The molecular basis of the agr-deficient phenotype remains to be determined. Preliminary analysis revealed that PCR fragments with the expected size and specific for RNAII and RNAIII could be generated from all strains. Since, as shown here, RNAIII is not expressed even in agr-positive strains, the agr activity seems to be nonessential in CF lung infections. The occurrence of agr-deficient strains in other types of S. aureus infections is currently under investigation. A second set of strains (group III) showed a lack of spa inhibition during the growth cycle in an agr-positive background. The inverse growth-dependent regulation of spa in these strains was not accompanied by an irregular expression of agr and sar (data not shown). The molecular basis of the in vitro expression pattern of spa (e.g., differences in the promoter region of spa or additional regulatory factors specific to these strains) remains to be determined.
In summary, we analyzed the differential expression of virulence genes during infection. The observed expression pattern, entailing low levels of RNAIII and spa transcription, may be specific for chronic lung infection in CF. One may speculate that under the condition of low bacterial densities as expected for many types of infection agr is generally inactivated and replaced with other regulatory circuits. Since this is the first report on direct transcript analysis in vivo, similar studies on other important virulence genes and regulons of S. aureus are required for further elucidation. Simplification of our method, for instance, by continuous detection of amplicons during PCR, will help to accelerate future studies. Hybridization techniques, which would help to upscale gene analysis, are limited by their low sensitivity at the moment and therefore are not yet applicable for investigations in vivo.
In recent years, new approaches identifying bacterial genes induced during infection have been developed and applied to S. aureus (9, 22, 24). While these methods are very useful for screening new candidates involved in pathogenesis, our method provides a direct approach to the evaluation of putative virulence genes involved in different infections in animals and, more importantly, in humans. A combination of these methods should advance our understanding of bacterium-host interactions.
ACKNOWLEDGMENTS
We thank D. Blaurock for critically reading the manuscript.
This work was supported by grants from Mukoviszidose e.V., Fortüne (no. 258), and the Deutsche Forschungsgemeinschaft (Wo 578/3-1).
REFERENCES
- 1.Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun. 1993;61:3879–3885. doi: 10.1128/iai.61.9.3879-3885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvidson S, Janzon L, Lofdahl S, Morfeldt E. The exoprotein regulatory region exp of S. aureus. In: Butler E O, Moseley E B, editors. Genetic transformation and expression. Intercept. Dorset, United Kingdom: Wimborne; 1989. pp. 511–518. [Google Scholar]
- 3.Balaban N, Novick R P. Autocrine regulation of toxin synthesis by Staphylococcus aureus. Proc Natl Acad Sci USA. 1995;92:1619–1623. doi: 10.1073/pnas.92.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown D R, Pattee P A. Identification of a chromosomal determinant of alpha-toxin production in Staphylococcus aureus. Infect Immun. 1980;30:36–42. doi: 10.1128/iai.30.1.36-42.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callegan M C, Engel L S, Hill J M, O'Callaghan R J. Corneal virulence of Staphylococcus aureus: roles of alpha-toxin and protein A in pathogenesis. Infect Immun. 1994;62:2478–2482. doi: 10.1128/iai.62.6.2478-2482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung A L, Eberhardt K J, Chung E, Yeaman M R, Sullam P M, Ramos M, Bayer A S. Diminished virulence of a sar-/agr- mutant of Staphylococcus aureus in the rabbit model of endocarditis. J Clin Investig. 1994;94:1815–1822. doi: 10.1172/JCI117530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung A L, Eberhardt K J, Heinrichs J H. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect Immun. 1997;65:2243–2249. doi: 10.1128/iai.65.6.2243-2249.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chien Y-T, Cheung A L. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J Biol Chem. 1998;273:2645–2652. doi: 10.1074/jbc.273.5.2645. [DOI] [PubMed] [Google Scholar]
- 9.Coulter S N, Schwan W R, Ng E Y W, Langhorne M H, Ritchie H D, Westbrock-Wadman S, Hufnagle W O, Folger K R, Bayer A S, Stover C K. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol. 1998;30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 10.Dassy B, Hogan T, Foster T J, Fournier J M. Involvement of the accessory gene regulator (agr) in expression of type 5 capsular polysaccharide. J Gen Microbiol. 1993;139:1301–1306. doi: 10.1099/00221287-139-6-1301. [DOI] [PubMed] [Google Scholar]
- 11.Davis P B, Drumm M, Konstan M W. Cystic fibrosis. Am J Respir Crit Care Med. 1996;154:1229–1256. doi: 10.1164/ajrccm.154.5.8912731. [DOI] [PubMed] [Google Scholar]
- 12.Duthie E S, Lorenz L L. Staphylococcal coagulase: mode of action and antigenicity. J Gen Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- 13.Ericsson A, Granstrom M, Möllby R, Strandvik B. Antibodies to staphylococcal teichoic acid and alpha toxin in patients with cystic fibrosis. Acta Paediatr Scand. 1986;75:139–144. doi: 10.1111/j.1651-2227.1986.tb10170.x. [DOI] [PubMed] [Google Scholar]
- 14.Heinrichs J H, Bayer M G, Cheung A L. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J Bacteriol. 1996;178:418–423. doi: 10.1128/jb.178.2.418-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 16.Herbert S, Worlitzsch D, Dassy B, Boutonnier A, Fournier J M, Bellon G, Dalhoff A, Döring G. Regulation of Staphylococcus aureus capsular polysaccharide type 5: CO2 inhibition in vitro and in vivo. J Infect Dis. 1997;176:431–438. doi: 10.1086/514061. [DOI] [PubMed] [Google Scholar]
- 17.Janzon L, Arvidson S. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9:1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji G, Beavis R C, Novick R P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kahl B, Herrmann M, Everding A S, Koch H G, Becker K, Harms E, Proctor R A, Peters G. Persistent infection with small colony variant strains of Staphylococcus aureus in patients with cystic fibrosis. J Infect Dis. 1998;177:1023–1029. doi: 10.1086/515238. [DOI] [PubMed] [Google Scholar]
- 20.Karakawa W W, Vann W. Capsular polysaccharides of Staphylococcus aureus. Semin Infect Dis. 1982;4:285–293. [Google Scholar]
- 21.Li S, Arvidson S, Mollby R. Variation in the agr-dependent expression of alpha-toxin and protein A among clinical isolates of Staphylococcus aureus from patients with septicaemia. FEMS Microbiol Lett. 1997;152:155–161. doi: 10.1111/j.1574-6968.1997.tb10422.x. [DOI] [PubMed] [Google Scholar]
- 22.Lowe A M, Beattie D T, Deresiewicz R L. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol Microbiol. 1998;27:967–976. doi: 10.1046/j.1365-2958.1998.00741.x. [DOI] [PubMed] [Google Scholar]
- 23.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 24.Mei J M, Nourbakhsh F, Ford C W, Holden D W. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 25.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morfeldt E, Tegmark K, Arvidson S. Transcriptional control of the agr dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol Microbiol. 1996;21:1227–1237. doi: 10.1046/j.1365-2958.1996.751447.x. [DOI] [PubMed] [Google Scholar]
- 27.Novick R P. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 28.Novick R P, Projan S J, Kornblum J, Ross H F, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 29.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Reilly M, de Azavedo J C, Kennedy S, Foster T J. Inactivation of the alpha-haemolysin gene of Staphylococcus aureus 8325-4 by site-directed mutagenesis and studies on the expression of its haemolysins. Microb Pathog. 1986;1:125–138. doi: 10.1016/0882-4010(86)90015-x. [DOI] [PubMed] [Google Scholar]
- 31.Patel A H, Nowlan P, Weavers E D, Foster T. Virulence of protein A-deficient and alpha-toxin-deficient mutants of Staphylococcus aureus isolated by allele replacement. Infect Immun. 1987;55:3103–3110. doi: 10.1128/iai.55.12.3103-3110.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Projan S J, Novick R P. The molecular basis of pathogenicity. In: Crossley K B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 55–82. [Google Scholar]
- 33.Schlichting C, Branger C, Fournier J M, Witte W, Boutonnier A, Wolz C, Goullet P, Döring G. Typing of Staphylococcus aureus by pulsed-field gel electrophoresis, zymotyping, capsular typing, and phage typing: resolution of clonal relationships. J Clin Microbiol. 1993;31:227–232. doi: 10.1128/jcm.31.2.227-232.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smeltzer M S, Hart M E, Iandolo J J. Phenotypic characterization of xpr, a global regulator of extracellular virulence factors in Staphylococcus aureus. Infect Immun. 1993;61:919–925. doi: 10.1128/iai.61.3.919-925.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulrich M, Herbert S, Berger J, Bellon G, Louis D, Münker G, Döring G. Localization of Staphylococcus aureus in infected airways of patients with cystic fibrosis and in a cell culture model of S. aureus adherence. Am J Respir Cell Mol Biol. 1998;19:83–91. doi: 10.1165/ajrcmb.19.1.3137. [DOI] [PubMed] [Google Scholar]
- 37.von Eiff C, Heilmann C, Proctor R A, Woltz C, Peters G, Götz F. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J Bacteriol. 1997;179:4706–4712. doi: 10.1128/jb.179.15.4706-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]