Abstract
Single-stranded DNA gaps are frequent structures that accumulate on newly synthesized DNA under conditions of replication stress. The identification of these single-stranded DNA gaps has been instrumental to uncover the mechanisms that allow the DNA replication machinery to skip intrinsic replication obstacles or DNA lesions. DNA fiber assays provide an essential tool for detecting perturbations in DNA replication fork dynamics genome-wide at single molecule resolution along with identifying the presence of single-stranded gaps when used in combination with S1 nuclease. However, electron microscopy is the only technique allowing the actual visualization and localization of single-stranded DNA gaps on replication forks. This chapter provides a detailed method for visualizing single-stranded DNA gaps at the replication fork by electron microscopy including psoralen cross-linking of cultured mammalian cells, extraction of genomic DNA, and finally enrichment of replication intermediates followed by spreading and platinum rotary shadowing of the DNA onto grids. Discussion on identification and analysis of these gaps as well as on the advantages and disadvantages of electron microscopy relative to the DNA fiber technique is also included.
Keywords: Electron microscopy, DNA replication, Replication structures, ssDNA gaps, DNA replication stress
1. Introduction
Single-stranded DNA (ssDNA) discontinuities (or gaps) are genome-destabilizing structures that need to be quickly repaired (or filled) to prevent DNA breakage and genome instability. ssDNA gaps accumulate both on leading and lagging strands of DNA replication forks after treatment with a wide range of DNA-damaging agents [1-7]. Lagging strand gaps can form because of the discontinuous nature of Okazaki fragment synthesis. On the other hand, leading strand gaps form when DNA synthesis resumes downstream of a replication-blocking lesion through a process called fork repriming. Repriming involves re-initiation of DNA synthesis beyond a DNA lesion, leaving unreplicated ssDNA gaps to be filled post-replicatively, and is mediated by human Primase and DNA-directed Polymerase (PRIMPOL) in mammalian cells [8-11].
The DNA fiber approach exploits the ability of many organisms to incorporate halogenated pyrimidine nucleoside analogs into replicating DNA and provides a powerful tool to monitor genome-wide replication perturbations at single-molecule resolution [12-17]. Ongoing replication events are typically labeled with two thymidine analogs—e.g., iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU)—and individual two-color labeled DNA tracts are visualized on stretched DNA fibers by immunofluorescence. This approach can be adapted to detect ssDNA gaps on the labeled strand by taking advantage of the unique enzymatic cleavage properties of the S1 nuclease. The S1 nuclease from Aspergillus oryzae [18] is used to nick the ssDNA and convert the ssDNA gap into a double-stranded break [19]. In the presence of ssDNA gaps, treatment with the S1 nuclease leads to shorter thymidine labeled tracts, which can be detected by DNA fiber analysis [19]. This approach can detect ssDNA gaps as short as 1–3 nucleotides. However, it also has some important limitations including the inability to determine the actual size of the ssDNA gaps as well as their exact location on the newly synthesized DNA.
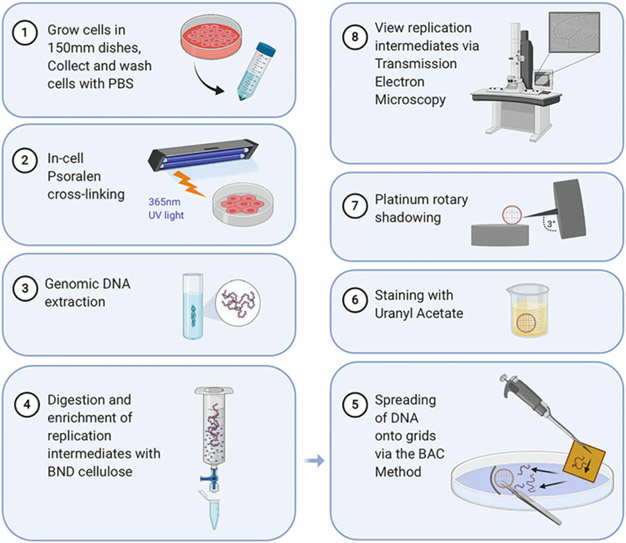
Electron microscopy (EM) is the only technique that allows direct visualization and quantification of replication intermediates [20-22]. EM has been applied to study ssDNA gaps on replication forks as it can provide unique structural insight including the size and location of the gaps on the replication forks. This chapter focuses on the experimental procedures related to the in-cell psoralen cross-linking of mammalian cell cultures, DNA extraction, enrichment, spreading, platinum shadowing, and finally viewing of the actual DNA replication intermediates and ssDNA gaps via EM (Fig. 1). Details on the carbon coating of grids for use in DNA spreading are also noted. These methods have been adapted from previously published protocols [20, 23] with emphasis on identification and analysis of ssDNA gaps within the newly synthesized DNA daughter strands. In addition, we discuss how the S1 nuclease DNA fiber assay can be used in conjunction with EM, as well as the advantages and disadvantages of using EM versus DNA fiber.
Fig. 1.
Illustrative depiction of the electron microscopy protocol summarizing the main steps
2. Materials
2.1. In-Cell Psoralen Cross-Linking and Lysis
150 mm tissue culture dish.
Cold 1× PBS (Phosphate Buffer Saline).
60 mm tissue culture dish.
200 μg/ml 4,5′,8-Trimethylpsoralen (TMP) in 100% ethanol. Care should be taken when handling TMP-containing solutions, including appropriate personal protection equipment along with use in conjunction with a certified fume hood and proper disposal, due to its potential DNA-damaging properties.
UV Cross-linker, 365 nm.
Flat frozen ice pack(s).
Flat metal plate cooled to −20 °C, no larger than the width of the interior of the cross-linker. The cold metal plate will be placed on top of the frozen ice packs and used to cool the samples, in the 60 mm petri dishes, in order to prevent heating during the incubation and irradiation times during cross-linking.
Cold Lysis Buffer stock solution: 1.28 M Sucrose, 40 mM Tris–HCL pH 7.5, 20 mM MgCl2, 4% Triton X-100.
2.2. Genomic DNA Extraction
Cold 1× PBS.
Digestion Buffer: 800 mM guanidine-HCl, 30 mM Tris–HCl pH 8.0, 30 mM EDTA pH 8.0, 5% Tween-20, 0.5% Triton X-100. Store at RT.
20 mg/ml proteinase K: made up fresh in digestion buffer.
Chloroform/Isoamyl Alcohol 24:1. Chloroform is highly toxic. Any handling of chloroform-containing solutions should be used within a certified fume hood.
100% Isopropanol.
70% Ethanol.
Nalgene Oak Ridge High-Speed Centrifuge Tubes, 50 ml, specifically designed for chloroform extractions (Thermo Fisher).
1× TE Buffer.
2.3. DNA Digestion and Enrichment of Replication Intermediates
PvuII-HF Restriction Enzyme.
10 mg/ml RNase A.
Low Salt Buffer: 10 mM Tris–HCl pH 8.0, 300 mM NaCl. Store at RT.
High Salt Buffer: 10 mM Tris–HCl pH 8.0, 1 M NaCl. Store at RT.
1.8% Caffeine Solution: made up in High Salt Buffer, incubated at 50 °C, and requires thorough mixing and vortexing in order to completely dissolve into solution. Store at RT for up to 4 months. Make new if precipitate has formed.
100 mg/ml BND Cellulose: made up in Low Salt Buffer; mix thoroughly by vortexing before use. Store at 4 °C for up to 2 months.
Poly-Prep Chromatography Columns, 2 ml.
Amicon Ultra 0.5 ml Centrifugal Filter Tubes.
1× TE Buffer.
2.4. Carbon Coating of the Grids
25 × 25 mm mica sheet (Electron Microscopy Sciences) pre-coated with carbon 12 nm in thickness. Pre-coated sheets may be obtained through any departmental core equipped with a coating machine with a carbon gun. Carbon-coated mica sheets may be stored for up to 5 months.
Scotch tape solution: 30 cm of clear Scotch brand tape in 100 ml of chloroform. After thoroughly mixing, the adhesive on the tape will dissolve into the chloroform, leaving just the backing. Store at RT for up to 1 year.
Copper 400 mesh square grids (Electron Microscopy Sciences).
Filter paper circles: diameter 45 mm.
Filter paper circles: diameter 90 mm.
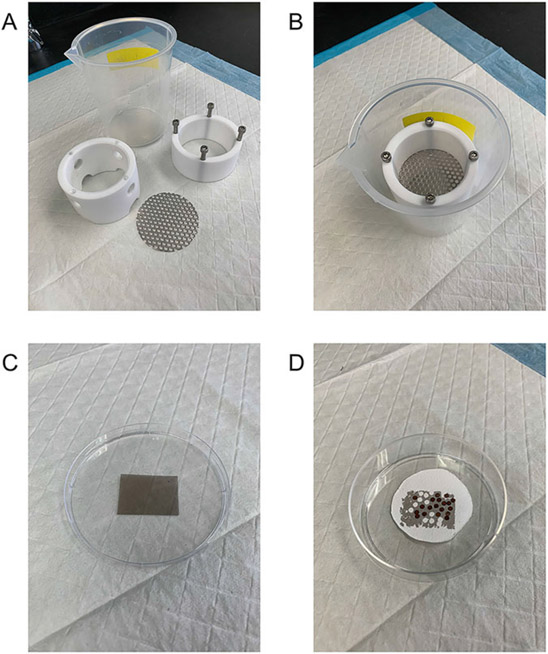
Apparatus capable of holding approximately 100 ml of double distilled water in an area 90 mm wide and 180 mm deep. These may be custom produced in any machine shop and can be made up of two Teflon or plastic rings that can be separated with a removable metal wire mesh middle to allow for water flow and placement of grids (Fig. 2a,b).
Fig. 2.
(a, b) Apparatus used to apply a 12 nm carbon layer on top of the grids. (c) Piece of carbon-coated mica sheet from which the carbon layer is taken. (d) Completed carbon-coated grids
2.5. Spreading of the DNA
Double distilled water.
33.3 μg/ml ethidium bromide: made up fresh in sterile double distilled water. Sterile water should be used in conjunction with highly purified ethidium bromide in order to eliminate any particulates in solution that could become adhered to the grids and interfere with the visualization of DNA molecules. Ethidium bromide is highly toxic and should be used within a certified fume hood due to its DNA-damaging properties.
100% formamide.
0.2% w/v benzyl-dimethyl-alkylammonium chloride (BAC): made up in formamide. Store at RT for up to 1 year.
1:10 BAC diluted in 1× tris EDTA (TE) buffer: made up fresh before use.
100% ethanol.
5 mM uranyl acetate (UrAc): made up in 5 mM HCl. Store at 4 °C for up to 1 year. UrAc has some sensitivity to light and should be stored in the dark or thoroughly covered. UrAc has a mild radioactivity level of 0.37–0.51 μCi/g, and appropriate personal protective equipment should be worn along with careful handling of the powdered form in a certified fume hood.
1:10 UrAc diluted in 100% ethanol.
150 mm tissue culture dish.
Graphite powder.
Mica sheet: cut to approximately 12.5 mm × 10 mm, freshly cleaved before use.
Super fine tip tweezers.
2.6. Platinum Shadowing of Grids
High vacuum coater: capable of low angle (3 degrees) rotary shadowing, at least one platinum gun, quartz crystal monitor, rotary stage and holder for the copper grids.
Platinum rod inserts for gun.
2.7. Visualization of DNA Via Transmission Electron Microscope
Transmission electron microscope (JEOL 1400 or equivalent) with preferably bottom mounted camera system (AMT XR401 High Sensitivity sCMOS Camera for TEM or equivalent).
Camera software to allow for saving images as a .tiff file format.
2.8. Analysis of Single-Stranded DNA Gaps at the Replication Fork
ImageJ or similar software to measure the length of ssDNA gaps.
Excel software program to measure the presence of ssDNA gaps in a given sample.
3. Methods
3.1. In-Cell Psoralen Cross-Linking and Lysis
Asynchronous mammalian cells are grown in three 150 mm tissue culture plates, in 10–20 ml growth media, reaching a confluency of approximately 60–80% for each sample. Amount of growth media may vary due to treatment conditions; care should be taken to collect cells into one 50 ml tube to minimize loss of cells.
Scrape each plate, and collect cells into 50 ml tube in their growth media.
Add 5 ml 1× PBS, scraping each plate again then collecting cells into their same respective 50 ml tubes.
Spin cells down for 5 min, at 900 × g at 4 °C.
Remove media via vacuum, and add 10 ml cold 1× PBS to wash (see Note 1).
Remove PBS via vacuum, and add 10.5 ml cold 1×PBS, resuspend cells, and transfer to a 60 mm tissue culture dish. At this point, 1 ml of cells+PBS may be removed to a 1.5 ml tube and spun down, and the pellet can be frozen at −80 °C for future processing (e.g., protein or RNA extraction). If further processing is not necessary, 9.5 ml cold 1× PBS may be used to resuspend the cell pellet.
Place 60 mm dish on the pre-cooled metal block, and place on top of the frozen ice pack(s).
In hood, add 500 μl TMP stock solution to each dish in a circular pattern to evenly spread throughout sample. Cover with the dish top.
Place entire setup in dark for 5 min.
Take ice packs and metal block with samples on top, and place in the cross-linker. Program cross-linker to highest setting, 9999, and press start.
Repeat step 8 an additional two times for a total of 3 TMP additions and cross-linking cycles (see Note 2).
Transfer cross-linked cells to fresh 15 ml tube. Add 1 ml 1× PBS to each 60 mm dish to wash and collect any cells left behind to their respective tubes. Repeat two more times for a total of three 1 ml PBS washes of the 60 mm dish.
Spin cells down for 5 min, at 900 × g at 4 °C.
Remove PBS + TMP, and collect for appropriate hazardous waste disposal. Resuspend cell pellet in 10 ml cold 1× PBS to wash.
Remove PBS via vacuum, and add another 10 ml cold 1× PBS to wash. Repeat once more for a total of three PBS washes.
Remove PBS via vacuum, and thoroughly resuspend pellet in 2 ml cold 1× PBS.
Dilute lysis buffer stock solution 1:4 with cold double distilled water.
Add 8 ml cold lysis buffer to 2 ml cell pellet+PBS, and invert several times to mix (do not vortex), and incubate on ice for 10 min.
Spin down lysed cells for 15 min at 300 × g at 4 °C.
Remove lysis+PBS via vacuum, and resuspend pellet in 4 ml cold lysis buffer to wash.
Spin down cells for an additional 15 min, at 300 × g at 4 °C.
Remove lysis buffer completely via vacuum, and freeze pellet at −80 °C overnight.
3.2. Genomic DNA Extraction
On ice, completely resuspend frozen nuclei pellet in 200 μl cold 1× PBS, using a cutoff pipet tip.
Add 5 ml digestion buffer, and mix via pipet (do not vortex).
Add 200 μl 20 mg/ml proteinase K solution, and incubate at 50 °C for 1.5 h in a water bath.
Remove samples and let cool to RT.
In hood, add 5 ml chloroform/isoamyl alcohol, vortex immediately four times to thoroughly mix the two layers, and pour into a 50 ml Oak Ridge centrifuge tube.
In hood, balance tubes with digestion buffer by using a scale to achieve exact measurements.
Spin for 30 min at 9000 × g at 4 °C.
Promptly remove tubes from centrifuge. There should now be a clear separation of both layers with a milky interface in the middle. In the hood, carefully transfer the upper layer to a new oak ridge centrifuge tube, using a cutoff pipet tip. Care should be taken to not disturb nor take up the white, cloudy interface. Additional chloroform extractions may be performed until interface is clear (see Note 3). Volume should be approximately 4 ml. Collect and appropriately discard the bottom layer as chloroform hazardous waste.
Add same volume, approximately 4 ml, of 100% isopropanol. Cap tube, and vigorously swirl to thoroughly mix in order to precipitate the DNA. Long white strands of DNA should be visible at this point and be concentrated into a single clump.
Balance tubes with isopropanol by using a scale to achieve exact measurements, and spin for 10 min at 9000 × g at 4 °C.
Carefully pipet off the isopropanol, without disturbing the DNA pellet. Pellet may be semi-transparent and could be difficult to see.
Wash pellet with 5 ml 70% ethanol.
Spin for 5 min at 9000 × g at 4 °C.
Pipet off most of the ethanol, without disturbing the pellet, and place in 50 °C water bath for 15 min or until dry. Keep tube uncapped and water bath top removed to facilitate drying. Pellet will be clear when dry and difficult to see.
Add 200 μl 1× TE buffer to pellet, cap tube, place in 50 ml tube holder, and gently rock overnight at RT to completely dissolve pellet.
Transfer dissolved DNA into a 1.7 ml tube using a cutoff pipet tip. Measure concentration with 1 μl on a NanoDrop. Measurement should be taken at least three times and average used as the final concentration.
3.3. DNA Digestion and Enrichment of Replication Intermediates
Digest 20 μg of genomic DNA with the PvuII HF restriction enzyme, along with appropriate buffer (see Note 4). Place at 37 °C for 4 h. Add 1 μl 10 mg/ml RNase to the reaction mix when 1 h remaining in the reaction. Volume of reaction mix should total 250 μl (see Note 5).
Add 2.0 ml 100 mg/ml BND cellulose resin (0.1 ml BND suspension/1 μg digested DNA) to a chromatography column, with a cutoff pipet tip, and allow liquid to flow through. Washing of the resin takes approximately 1 h and can begin 1 h before completion of the digestion.
Wash the resin 6× with 1 ml high salt buffer. Allow all liquid to run through column before adding next wash. Gently resuspend resin each time after the addition of buffer. Take care not to allow resin to dry as it may interfere with its interaction with the DNA when applied.
Wash column 6× with 1 ml low salt buffer. Gently resuspend resin each time after the addition of buffer (see Note 6).
After digestion is complete, adjust reaction mix to 300 mM NaCl final concentration by adding 5 M NaCl. Adjust to final volume of 600 μl by adding low salt buffer. Salt adjustment for PvuII HF restriction enzyme: 250 μl sample, 14.7 μl 5 M NaCl, and 335.3 μl low salt buffer (see Note 7).
Close bottom of column, and add the 600 μl NaCl adjusted sample to resin; incubate for 30 min at RT to allow for full binding of DNA to the BND cellulose. Gently resuspend the resin bed with a cutoff pipet tip every 10 min.
Remove cap and collect sample in a 1.5 ml tube. Save as “flowthrough DNA” in case of defective DNA binding to resin.
Wash DNA bound resin 2× with 1 ml high salt buffer. Resuspend resin after each addition. Collect flowthrough in a 2.5 ml tube, and save as “linear dsDNA.”
Close bottom of column, and add 600 μl of pre-warmed 50 °C 1.8% caffeine/high salt buffer in order to elute the remaining DNA molecules from the BND cellulose. Incubate for 10 min at RT. Gently resuspend resin bed after 5 min.
Remove cap and collect flowthrough into a 1.5 ml tube as the “enriched replication intermediate DNA.”
The DNA sample may now be purified and concentrated using Amicon Ultra size exclusion columns. Place the collection 600 μl enriched DNA sample into the column, which is placed into the collection tube, and spin for 8 min at 7600 × g.
Discard flowthrough and wash column with 200 μl 1× TE. Spin for 5 min at 7600 × g.
Discard flowthrough, and wash column one final time with 200 μl 1× TE. Spin for 3.5 min at 7600 × g.
Discard flowthrough, turn column upside down into a new collection tube, and use the short spin cycle to force the bound DNA from the column and into the collection tube.
Remove column, and the remaining 20–30 μl of 1× TE now contains the purified DNA enriched for replication intermediates.
Optional: 1 μl purified, enriched DNA may be loaded onto a 0.8% agarose gel to determine DNA quantity. 1 μl of the flowthrough and linear dsDNA can be loaded as well, as a control to confirm majority of DNA is obtained from the final elution (see Note 8).
Concentrate DNA samples using a standard speed vac for approximately 10 min on high. Final volume should be around 15–20 μl. Seal tubes containing DNA with parafilm to avoid evaporation of sample.
3.4. Carbon Coating of the Grids
Arrange 30 of the 400 mesh copper grids, shiny side down, on a piece of filter paper, and place in a glass petri dish.
In a certified fume hood, cover grids with the chloroform/tape mixture, using a dropper, and let dry. Repeat for a total of three chloroform/tape evaporation treatments (see Note 9). Remove filter paper containing sticky grids to a new covered petri dish.
Place a pre-coated 25 × 25 mm carbon-coated mica sheet (Fig. 2c) in a petri dish lined with a moist piece of filter paper at 37 °C for 30 min.
Set up grid coating apparatus (Fig. 2a,b) by placing the 90 mm × 180 mm chamber in a 500 ml beaker. Place a new 45 mm piece of filter paper inside the bottom of the chamber, and weigh down. Fill entire beaker with double distilled water to the top of the chamber. Remove weight on filter paper.
Place grids onto the filter paper, located inside the water-filled chamber, sticky side up, taking care to arrange the grids in a tight-packed area, the size of the mica sheet, without over-lapping. Use super fine tip tweezers to handle the grids.
Take the carbon-coated mica sheet, carbon side up, parallel to the water, and carefully lower, starting with one side, onto the top of the water, inside the center of the chamber, where the grids are located below. Use a hooked super fine tip tweezers in order to angle the carbon-coated mica sheet into the water.
Proceed to slowly turn the mica sheet perpendicular to the surface of the water, angling down, in order to separate the carbon from the mica sheet. As the mica sheet is moved in a downward fashion, under the water, the carbon will remain at the surface. When the carbon is completely free from the mica sheet, slowly raise the mica sheet out of the water, being careful to not break or bring up the carbon at the surface.
With the carbon now floating at the surface of the water, directly above the grids, use a vacuum to slowly remove the water from the beaker, guiding the carbon over the grids as the water lowers, taking care not to break the fragile carbon sheet.
As the water lowers and empties, guide the carbon on top of the grids, making sure all are covered with carbon (Fig. 2d).
After the water is gone, remove the filter paper, containing the carbon-coated grids, onto a dry piece of filter paper, located in a petri dish.
Slightly cover the still wet carbon-coated grids, with the top of the dish, and allow to dry at least overnight before using. Carbon-coated grids may be used for up to 5 months.
3.5. Spreading of DNA
In a certified fume hood, place 20 μl of the working ethidium bromide solution onto a piece of parafilm, creating a drop onto which the carbon-coated grid, carbon side down, is placed on top (Fig. 3a). Incubate for 20 min at RT. Cover with a large petri dish to avoid evaporation.
Take grids from drop, gently remove the excess ethidium bromide by dabbing the grid on a piece of filter paper, and place carbon side down on top of a new piece of filter paper to dry.
Utilizing an enclosed space free of air currents (see Note 10), set up all components required for spreading.
Pour 20 ml double distilled water into a 150 mm tissue culture dish.
Mix an amount of formamide (2–3.5 μl) equal to the amount of DNA being spread and 0.5 μl BAC working solution in the bottom of a 1.5 ml tube. Add an equal amount of DNA to the side of the 1.5 ml tube, keeping the two mixes separate until immediately before spreading (see Note 11). Volume of DNA used depends on the concentration of sample. Typical amount used is 3 μl of enriched replication intermediate DNA, but can be adjusted if concentrations are too high/low.
Position a freshly cleaved 12.5 mm × 10 mm piece of mica, with tweezers, into the 150 mm dish containing 20 ml water at a 45 degree angle against the side of the dish, partially submerged into the water.
Sprinkle a small amount of graphite powder close to the mica sheet, forming a wall around it (Fig. 3b).
Shortly spin sample containing the formamide, BAC, and DNA in a minifuge to mix all components together (spin for 5–10 s).
Pipet up the mix, and place droplet directly above the mica sheet, right above the water line, to allow DNA mix to slide down the mica sheet and spread out over the surface of the water (Fig. 3c), concentrating along the edge of the graphite powder wall (see Note 12). As the DNA mix hits the water, it will force the graphite powder wall to expand outwards as it travels across the surface of the water. Thus, the DNA will be most prominently located near the graphite’s edge (Fig. 3d).
Using a fine tip tweezers, position the grid as parallel as possible to the water, and lightly touch the surface, nearest to the graphite wall, taking care not to take up the graphite itself.
Immediately remove grid from the water, and, using the tweezers, hold the grid into the diluted uranyl acetate/ethanol solution for 15 s, and then dip the grid into 100% ethanol, to wash away any excess uranyl acetate. Place grid carbon side up on a piece of filter paper to dry.
Wipe down tweezers to remove excess ethanol (see Note 13). A second grid can now be obtained starting from step 10, taking care to collect DNA from a different area, near the graphite wall, in order to collect a maximum amount of DNA (see Note 14).
Grids are now ready for platinum shadowing.
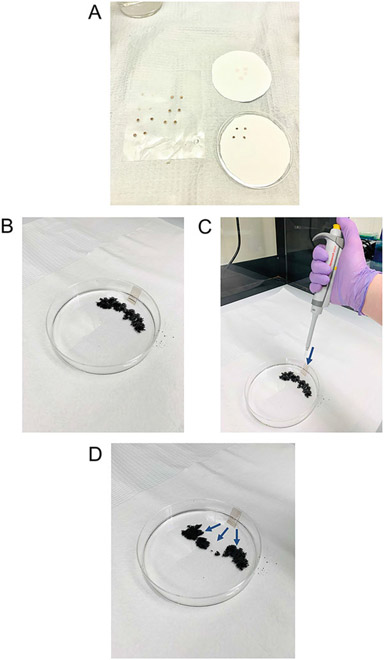
Fig. 3.
Series of images representing important steps of the spreading process. (a) Carbon-coated grids are incubated with ethidium bromide, excess is removed by dabbing onto a piece of filter paper, and the grid is dried carbon side down onto a fresh piece of filter paper. (b) A 150 mm cell-culture dish is prepared for spreading by placing a mica sheet, at an angle, against the side of the dish, containing 20 ml of distilled water and then sprinkling granite powder into the water. (c) A drop of DNA is pipetted onto the mica sheet, very near to the surface of the water, and allowed to slide down into the water. (d) The DNA is then spread onto the surface of the water (blue arrows) accumulating at the edge of the granite powder wall
3.6. Platinum Shadowing of Grids
Secure grids to a rotary stage of a high vacuum coater (Fig. 4a).
Obtain a vacuum pressure of approximately 4.5 × 10-6 mbar, and set shadowing at an angle of 3 degrees, with a platinum deposition of 3.5 nm. Grids should be rotated during shadowing to obtain even distribution of platinum.
Shadowed grids are now ready to be viewed by TEM and may be stored in an appropriate EM specimen holder at RT indefinitely.
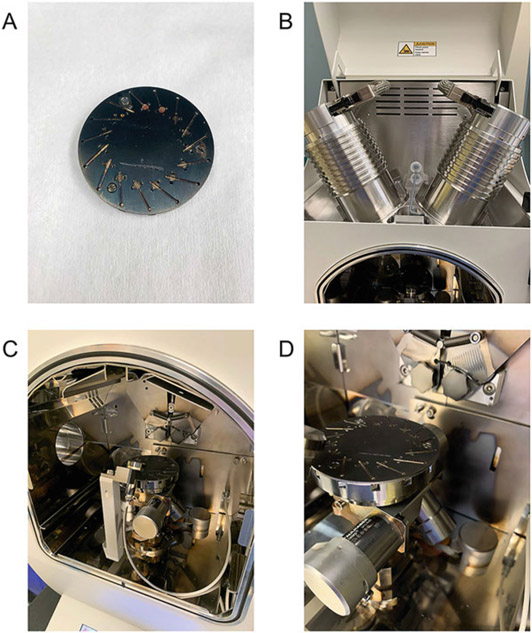
Fig. 4.
Crucial components of the Leica EM ACE600 coater used in the rotary platinum shadowing. (a) Rotary stage capable of securing up to 15 grids. (b) Carbon gun (left) and essential platinum gun (right). (c, d) Positioning of the rotary stage inside the coater right alongside the quartz monitor, located directly in front of stage, responsible for calculating the specific amount of platinum output
3.7. Visualization of DNA Via Transmission Electron Microscope
Ideal TEM parameters for the visualization of DNA include a lanthanum boride (LaB6) filament necessary for maximum brightness and sharpness of the image, along with a preferably bottom mount high-resolution camera for high-quality images. Imaging of the DNA begins at approximately 10,000× with higher magnification images being visualized at 100,000×.
Due to the BAC spreading method, in conjunction with uranyl acetate staining and platinum shadowing, DNA will appear as a long darkened, cylindrical fiber in high contrast to the lighter carbon granular background. Double-stranded DNA molecules average a thickness of around 7 nm, whereas lighter, thinner single-stranded DNA average a thickness in the order of 2–3 nm [24]. Due to various circumstances, DNA staining or amount may be less than ideal. In these cases, spreading and shadowing can be repeated in order to obtain better quality grids. If success is not achieved, re-enrichment of the genomic DNA may be performed followed by new spreadings and shadowing.
Quantity of DNA on the grid is of great importance and depends on the concentration of the sample, size of the spreading surface, and quality of the carbon grid. Spreading of the DNA may need to be repeated in order to achieve an ideal amount of DNA on a grid. Certain parts of the same grid may be better than others. Optimal concentration of DNA on the grid should allow a good amount of empty space between each DNA molecule in order to clearly distinguish replication intermediates from random crossings of DNA fibers. Having an ideal concentration of DNA reduces the likelihood of crossover events, ensuring that any meeting of three or four-stranded structures represent a true junction.
Most of the DNA present in the sample is linear, and only a small percentage can be classified as a replication intermediate. Correct identification of these intermediates requires several established criteria. Replication forks are initially identified by visualizing a three-way junction or the meeting of three DNA fibers at one contact point (Fig. 5). These fibers represent a moment in time when the DNA is actively being replicated, one arm being the parental strand and the other two arms the daughter strands. If the identified three-way junction structure corresponds to a replication fork, the daughter strands should be of equal or similar length because the PvuII restriction enzyme will cut the two daughter strands in the same location (see Subheading 3.8 for more details). To confirm that the junction is a seamless joining of three strands, the junction needs also to be imaged at high magnifications of 100,000× or greater. ssDNA gaps may also be visualized at this point as lighter, thinner sections located within the DNA fibers. These gaps are predominately found on the daughter strands as well as at the junction itself.
A smaller percentage of these replication intermediates can be classified as a reversed replication fork, characterized by a four-way junction. As with three-way junctions, reversed forks contain one parental strand and two daughter strands of similar length, along with an additional, typically shorter, fourth strand, indicative of the reversed arm. Particular attention is paid to the junction in order to confirm the presence of an open junction and rule out a possible crossover of DNA fragments. Often times the junction may be collapsed, and other indicators such as daughter strand symmetry, presence of single-stranded DNA at the junction, or the entire structure itself must be taken into account. For more details on the EM analysis of reversed fork structures, please see [20].
Approximately 80–120 images should be taken per sample. This allows for a small percentage of DNA molecules that may be discarded because of the lack of appropriate characteristics of a true replication intermediate upon later analysis. These images should be saved as .tiff files for the computational analysis performed with analytical software such as ImageJ. Standard statistical analysis using Microsoft Excel can be applied along with graphical representations of chosen parameters to visualize differences between samples.
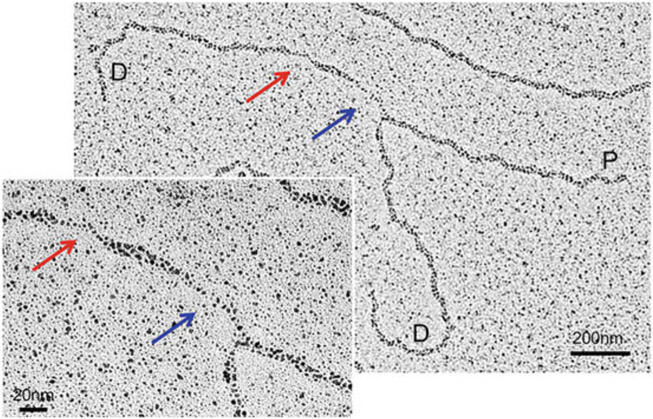
Fig. 5.
Representative image of a replication fork containing two daughter strands (D) of equal length and a single parental strand (P). The red arrow indicates the ssDNA gap within the daughter strand, and the blue arrow is indicating a ssDNA gap at the junction
3.8. Analysis of ssDNA Gaps at the Replication Fork
Once images are acquired, detailed analysis can commence using ImageJ and Microsoft Excel. Classification of a three-way junction, representing a replication fork, is made by looking at the symmetry of the daughter strands and quality of the junction. Symmetry is determined by measuring the lengths of each strand. At least two strands should be of equal length due to DNA digestion with the PvuII restriction enzyme. This symmetry is due to the same cut site being equidistant on both daughter strands. Certain circumstances may affect the symmetrical length of the daughter strands including incomplete digestion, DNA breakage due to handling, and responses to DNA damage. These instances should only affect a small percentage of three-way junctions. The meeting point at which the three strands join should be carefully analyzed using high magnification images. Visualization of the high magnification image allows to confirm the cohesive connection of all three strands. Some replication forks may contain areas of single-stranded DNA at the junction. ssDNA appears much lighter and thinner than the duplex strand. The presence of a single-stranded region at the junction is a further indicator of a replication fork.
Once a replication fork is established, careful inspection of the thickness of the daughter strand filaments allow for the identification of regions of single-stranded DNA. These ssDNA gaps can be pinpointed by monitoring changes in the filament thickness (double-stranded DNA molecules have an average thickness of approximately 7 nm, whereas single-stranded DNA has an average thickness of approximately 2–3 nm). Therefore, ssDNA gaps can be easily located as they appear thinner and lighter when compared to the surrounding double-stranded DNA. The presence or absence of these gaps, along with their quantity, can be statistically analyzed in Microsoft Excel. For example, the frequency of ssDNA gap accumulation can be calculated as a function of the specific treatment conditions or of any genetic modification made to the cell. The lengths of the gap may also be measured using ImageJ.
4. Notes
All growth media should be thoroughly removed via washing, so it does not absorb any of the monochromatic light during cross-linking.
For the cross-linking step, a 365 nm bulb should be used. The power should be set at 6.2 mW/cm2 and confirmed with a UV meter. Several factors, including the sample distance from bulbs or bulb life, may interfere with this total irradiation power, so irradiation cycle times may be adjusted to obtain ideal cross-links.
Care should be taken not to take up any of the cloudy white interphase where extracted proteins are located. Subsequent chloroform extractions might be performed in order to eliminate this unwanted middle layer. However, loss of DNA, located in the upper layer, may occur during transports to new tubes. Additional extractions should only be considered if it is too difficult to remove the top layer without taking up too much of the milky middle layer. Additional spin time may also be used to more clearly separate the two layers.
DNA is digested before enrichment in order to isolate the replication forks for easy viewing and identification in the field of vision of electron microscopy. The PvuII restriction enzyme is used because of its ideal cutting frequency of genomic DNA, resulting in DNA fragments in the order of 30–50 base pairs, allowing for high-resolution imaging of a single fork. In addition, PvuII cleavage leaves blunt ends, in contrast to ssDNA overhangs, and these blunt ends will not compete for binding to the BND cellulose.
It is important to remove any RNA contained in the sample before loading onto the BND cellulose, so that it does not interfere with the enrichment of the replication intermediate DNA.
Extensive washings of the BND cellulose must be performed in order to remove any fine particulate material from the BND suspension that may coat the grids and interfere with detection of the DNA molecules.
Salt adjustments are different for each restriction enzyme. The listed amounts are specific to PvuII. Refer to the restriction enzyme insert for respective adjustments.
If a specific quantity of DNA is desired, it is necessary to use an agarose gel to run a small amount of the acquired purified DNA alongside an appropriate marker. Quantification of DNA via NanoDrop is not recommended due to caffeine’s interference with standard spectrometry readings.
In the chloroform/tape mixture, the chloroform will dissolve the sticky residue from the backing of the tape. When this mixture is applied to the grids, the sticky residue will adhere to the surface of the grids, while the chloroform will evaporate away. This provides a sticky base to which the carbon may adhere. If the correct tape is not used or too much tape is used in the chloroform solution, it can cause the grids to acquire an unwanted cloudy coating on top. If there is not enough sticky residue, the carbon will not properly adhere to the grids and slough away when spreading the DNA.
Spreading of the DNA onto the surface of the water is highly sensitive to air currents and vibrations in the environment. Therefore, it is recommended to perform this procedure in an isolated location or in an enclosed area, in order to avoid any disruptions to the DNA layer on top of the water.
Formamide is a partially denaturing reagent and helps to unfold the DNA molecules during spreading. This is essential to reveal a clear linear image of the dsDNA and subsequent replication intermediates. However, the DNA and formamide should remain separate until right before spreading, because too much exposure to the formamide could cause uncoupling of the two DNA strands.
It is crucial for the mica sheet to be freshly cleaved in order to provide a charged surface for the DNA to repel against, providing the necessary momentum into the water and across the surface. Because of this momentum, DNA will be concentrated at the edge of the graphite wall where the grid should be placed to pick up the highest amount.
Care should be taken to remove excess ethanol on the tweezers as it can upset the surface in which the DNA is located and decrease uptake on the grid.
It is recommended that no more than two grids be used per one spreading. The DNA spread on the surface will slowly sink to the bottom, and very little will remain to pick up on a third grid.
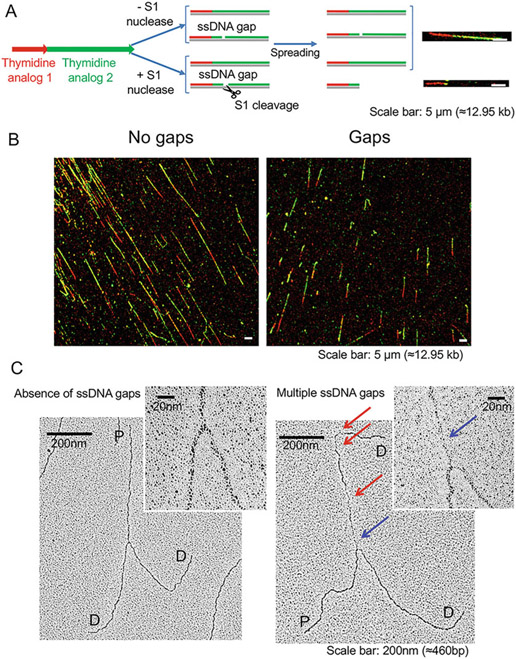
EM allows direct visualization of ssDNA gaps behind replication forks and can be combined with the modified DNA fiber assay utilizing the S1 nuclease to further characterize ssDNA gap accumulation throughout the genome (Figs. 5 and 6). For a general comparison of the EM and DNA fiber techniques, see [22]. Briefly, the DNA fiber assay starts with the incorporation of thymidine analogs into the replicating DNA, followed by spreading of the DNA onto charged microscope slides and staining with fluorescent antibodies in order to visualize the thymidine-labeled DNA tracts on a microscope [12-17]. The resolution of a DNA fiber experiment is typically limited to a few kilobases of the stretched DNA. The modified DNA fiber approach used for ssDNA gap detection takes advantage of the unique ssDNA cutting properties of the S1 nuclease. The S1 nuclease is added to the reaction after pulse labeling with the thymidine analogs to cleave the thymidine-labeled DNA containing ssDNA gaps. If gaps are present in the DNA, S1 cleavage will lead to shorter tract lengths relative to S1-untreated controls [19]. The S1 DNA fiber approach can detect ssDNA gaps as short as a few nucleotides or even nicks due to the ability of S1 nuclease to cleave substrates that contain single nucleotide nicks [18]. The EM technique has a higher resolution compared to the DNA fiber approach. However, unlike the S1 DNA fiber approach, it cannot detect ssDNA gaps shorter than 40–60 nucleotides [25]. A major advantage of the EM technique is that it shows the actual location of the ssDNA gaps on the replication forks and it allows for the measurement of the size of the ssDNA gaps [26]. This information is extremely useful to determine whether ssDNA gaps are equally distributed on the two parental strands of a replication fork and whether the size of the gaps might change under different experimental conditions. Moreover, the EM technique allows quantification of the number of gaps on an individual fork and to distinguish between ssDNA gaps that are present at replication fork junctions versus ssDNA gaps located behind the junctions. These distinctions cannot be made using the DNA fiber approach. On the other hand, an important drawback of the EM technique is that the ssDNA gaps must be located within approximately 500 nucleotides from the fork junction in order to be detected due to the relatively short size of the DNA fragments analyzed by EM (1–2 kilobases). Conversely, the S1 DNA fiber approach can potentially detect ssDNA gaps that are several kilobases away from the fork junction given that a typical thymidine-labeling scheme allows the incorporation of the thymidine analog for 20–30 kilobases. Table 1 summarizes the advantages and disadvantages of the EM and DNA fiber techniques. Taken together, both these techniques complement each other and can each contribute to a more thorough detection and ultimately confirmation of the presence of ssDNA gaps in the genome.
Fig. 6.
(a) Schematic representation of the DNA fiber assay utilizing the S1 nuclease. (b) Fluorescent microscopy images of red and green labeled ongoing forks, using the S1 nuclease, resulting in either a longer tract length due to no gaps (left) or short tract lengths due to gaps (right). (c) Electron microscopy images detailing a replication fork that has no gaps (left) or multiple gaps, indicated by the red arrows, and a single gap at the junction indicated by the blue arrow (right)
Table 1.
Summary of the advantages and disadvantages of the electron microscopy and S1 nuclease DNA fiber assays
| Assay | Advantages | Disadvantages |
|---|---|---|
| Electron Microscopy |
|
|
| S1 Nuclease DNA Fiber Assay |
|
|
Acknowledgments
We thank Alice Meroni and Annabel Quinet for their help with the preparation of the figures and for their comments on the manuscript. This work was supported by the NCI under grant numbers R01CA237263 and R01CA248526 and by the DOD BCRP Expansion Award BC191374 to A.V. This research was supported by the Alvin J. Siteman Cancer Center Siteman Investment Program (supported by The Foundation for Barnes-Jewish Hospital, Cancer Frontier Fund) and the Barnard Foundation to A.V. Figures were created with BioRender.com.
References
- 1.Diamant N, Hendel A, Vered I, Carell T, Reissner T, de Wind N, Geacinov N, Livneh Z (2012) DNA damage bypass operates in the S and G2 phases of the cell cycle and exhibits differential mutagenicity. Nucleic Acids Res 40(1):170–180. 10.1093/nar/gkr596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elvers I, Johansson F, Groth P, Erixon K, Helleday T (2011) UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic Acids Res 39(16):7049–7057. 10.1093/nar/gkr420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jansen JG, Tsaalbi-Shtylik A, Hendriks G, Verspuy J, Gali H, Haracska L, de Wind N (2009) Mammalian polymerase zeta is essential for post-replication repair of UV-induced DNA lesions. DNA Repair (Amst) 8(12):1444–1451. 10.1016/j.dnarep.2009.09.006 [DOI] [PubMed] [Google Scholar]
- 4.Lehmann AR (1972) Post-replication repair of DNA in ultraviolet-irradiated mammalian cells. No gaps in DNA synthesized late after ultraviolet irradiation. Eur J Biochem 31(3):438–445. 10.1111/j.1432-1033.1972.tb02550.x [DOI] [PubMed] [Google Scholar]
- 5.Lopes M, Foiani M, Sogo JM (2006) Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21(1):15–27. 10.1016/j.molcel.2005.11.015 [DOI] [PubMed] [Google Scholar]
- 6.Meneghini R (1976) Gaps in DNA synthesized by ultraviolet light-irradiated WI38 human cells. Biochim Biophys Acta 425(4):419–427. 10.1016/0005-2787(76)90006-x [DOI] [PubMed] [Google Scholar]
- 7.Quinet A, Vessoni AT, Rocha CR, Gottifredi V, Biard D, Sarasin A, Menck CF, Stary A (2014) Gap-filling and bypass at the replication fork are both active mechanisms for tolerance of low-dose ultraviolet-induced DNA damage in the human genome. DNA Repair (Amst) 14:27–38. 10.1016/j.dnarep.2013.12.005 [DOI] [PubMed] [Google Scholar]
- 8.Bianchi J, Rudd SG, Jozwiakowski SK, Bailey LJ, Soura V, Taylor E, Stevanovic I, Green AJ, Stracker TH, Lindsay HD, Doherty AJ (2013) PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol Cell 52(4):566–573. 10.1016/j.molcel.2013.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.García-Gómez S, Reyes A, Martínez-Jiménez MI, Chocrón ES, Mourón S, Terrados G, Powell C, Salido E, Méndez J, Holt IJ, Blanco L (2013) PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell 52(4):541–553. 10.1016/j.molcel.2013.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mourón S, Rodriguez-Acebes S, Martínez-Jiménez MI, García-Gómez S, Chocrón S, Blanco L, Méndez J (2013) Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat Struct Mol Biol 20(12):1383–1389. 10.1038/nsmb.2719 [DOI] [PubMed] [Google Scholar]
- 11.Wan L, Lou J, Xia Y, Su B, Liu T, Cui J, Sun Y, Lou H, Huang J (2013) hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO Rep 14(12):1104–1112. 10.1038/embor.2013.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bensimon A, Simon A, Chiffaudel A, Croquette V, Heslot F, Bensimon D (1994) Alignment and sensitive detection of DNA by a moving interface. Science 265(5181):2096–2098. 10.1126/science.7522347 [DOI] [PubMed] [Google Scholar]
- 13.Jackson DA, Pombo A (1998) Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140(6):1285–1295. 10.1083/jcb.140.6.1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merrick CJ, Jackson D, Diffley JF (2004) Visualization of altered replication dynamics after DNA damage in human cells. J Biol Chem 279(19):20067–20075. 10.1074/jbc.M400022200 [DOI] [PubMed] [Google Scholar]
- 15.Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, Bensimon A (1997) Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science 277(5331):1518–1523. 10.1126/science.277.5331.1518 [DOI] [PubMed] [Google Scholar]
- 16.Parra I, Windle B (1993) High resolution visual mapping of stretched DNA by fluorescent hybridization. Nat Genet 5(1):17–21. 10.1038/ng0993-17 [DOI] [PubMed] [Google Scholar]
- 17.Techer H, Koundrioukoff S, Azar D, Wilhelm T, Carignon S, Brison O, Debatisse M, Le Tallec B (2013) Replication dynamics: biases and robustness of DNA fiber analysis. J Mol Biol 425(23):4845–4855. 10.1016/j.jmb.2013.03.040 [DOI] [PubMed] [Google Scholar]
- 18.Vogt VM (1973) Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem 33(1):192–200. 10.1111/j.1432-1033.1973.tb02669.x [DOI] [PubMed] [Google Scholar]
- 19.Quinet A, Carvajal-Maldonado D, Lemacon D, Vindigni A (2017) DNA fiber analysis: mind the gap! Methods Enzymol 591:55–82. 10.1016/bs.mie.2017.03.019 [DOI] [PubMed] [Google Scholar]
- 20.Neelsen KJ, Chaudhuri AR, Follonier C, Herrador R, Lopes M (2014) Visualization and interpretation of eukaryotic DNA replication intermediates in vivo by electron microscopy. Methods Mol Biol 1094:177–208. 10.1007/978-1-62703-706-8_15 [DOI] [PubMed] [Google Scholar]
- 21.Neelsen KJ, Lopes M (2015) Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat Rev Mol Cell Biol 16(4):207–220. 10.1038/nrm3935 [DOI] [PubMed] [Google Scholar]
- 22.Vindigni A, Lopes M (2017) Combining electron microscopy with single molecule DNA fiber approaches to study DNA replication dynamics. Biophys Chem 225:3–9. 10.1016/j.bpc.2016.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopes M (2009) Electron microscopy methods for studying in vivo DNA replication intermediates. Methods Mol Biol 521:605–631. 10.1007/978-1-60327-815-7_34 [DOI] [PubMed] [Google Scholar]
- 24.Vollenweider HJ, Sogo JM, Koller T (1975) A routine method for protein-free spreading of double- and single-stranded nucleic acid molecules. Proc Natl Acad Sci U S A 72(1):83–87. 10.1073/pnas.72.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mijic S, Zellweger R, Chappidi N, Berti M, Jacobs K, Mutreja K, Ursich S, Ray Chaudhuri A, Nussenzweig A, Janscak P, Lopes M (2017) Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat Commun 8(1):859. 10.1038/s41467-017-01164-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V (2010) Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17(11):1305–1311. 10.1038/nsmb.1927 [DOI] [PMC free article] [PubMed] [Google Scholar]