Abstract
The EUO gene of chlamydia is highly expressed early in the developmental cycle, relative to other genes, but continues to be expressed throughout the active growth phases. The precise function of EUO protein is not known, but it binds to DNA in vitro. In this study, we developed a selection and amplification scheme for identifying chlamydial genomic fragments to which EUO preferentially binds in vitro. The scheme involved mixing recombinant EUO with a Chlamydia psittaci genomic library in a pBluescript plasmid vector in vitro, trapping EUO-bound plasmid clones on filters, and amplifying the clones in Escherichia coli. After nine rounds of enrichment, the EUO binding sites of the three most highly enriched clones were identified by DNase I footprint analysis. All three clones had multiple binding sites of various sizes with no clear distinguishing feature other than they were AT-rich and were usually not located in putative promoter regions. We used limited site-specific mutagenesis to characterize the strongest binding site of the most-highly-enriched clone, which represented about 50% of the population after nine rounds. This mutagenesis identified a core binding site of 15 nucleotides (nt) whose sequence was used to find related sequences within each of the strong binding sites in the other two clones. Using the frequency of bases at specific positions within this group of sequences as a guide, we carried out trial-and-error searching with many related sequences, eliminating those which identified nonfootprinted sites. This process led us to the consensus 15-nt sequence AHGAAAWVTYTWDAY, which, when allowing two mismatches, picked out all of the strong binding sites and no nonfootprinting sites within the three enriched clones. This sequence may be useful for predicting additional possible EUO binding sites in the chlamydial genome.
Species of the genus Chlamydia are obligate intracellular parasites of humans and animals. A unique developmental cycle in which chlamydiae alternate between an infectious elementary body (EB) and the vegetative reticulate body (RB) distinguishes the genus Chlamydia from other intracellular bacteria (19, 22). The EB is metabolically inert and resistant to the hostile extracellular environment, whereas the RB divides intracellularly and cannot survive outside of a host cell. The cycle commences with entry of EBs into host cells by a phagocytic process, followed by differentiation of EBs to dividing RBs, a process that takes several hours to complete. RBs divide over a period of 1 or more days before they reorganize back into the inert EB form. Ultimately, EBs are released, usually by lysis of the host cell, and the cycle begins anew upon uptake of EBs into new host cells. Very little is known about how gene expression is regulated during the developmental cycle other than the few late-stage genes that have been identified so far appear to require the major chlamydial sigma factor for transcription initiation (9). Environmental stimuli and transcriptional activators that trigger macromolecular synthesis in EBs upon entry into host cells are completely unknown.
Wichlan and Hatch (33) cloned a Chlamydia psittaci 6BC gene, designated EUO, on the basis of a high level of expression at 1.5 h postinfection (hpi) compared to that at 24 hpi. In a subsequent study, Zhang et al. (36) found that EUO transcripts are most abundant early (1 to 2 hpi) in the developmental cycle relative to the major outer membrane protein (MOMP) gene transcripts and the 16S rRNA content of chlamydiae; however, EUO transcripts were detected until the end of the log phase of growth. Synthesis of EUO protein in chlamydiae was also found as early as 1 hpi, but EUO protein was most abundant, relative to the MOMP, during the log phase of growth. EUO protein appears to be unstable, because levels decline at late times postinfection and are completely absent in EBs (36). EUO is a minor protein and lacks significant homology to known proteins, but it does contain a putative helix-turn-helix (HTH) DNA-binding motif, as determined by the algorithm of Dodd and Egan (4). Using the late-developmental-stage-specific cysteine-rich envelope protein (crp) operon promoter region as a model, Zhang et al. (36) demonstrated that recombinant EUO (rEUO) binds to multiple AT-rich sites in DNA in vitro, with the strongest binding site located in the crp promoter between bp −60 and −9.
In the present study, we demonstrate that deletion of the putative HTH motif results in the loss of EUO binding to the crp promoter region. We also found that rEUO inhibited the in vitro transcription of the crp operon. However, rEUO also inhibited the in vitro transcription of two chlamydial genes that are expressed during the growth phases of the developmental cycle, suggesting that EUO is not a specific regulator of late-stage gene expression and that its effect on in vitro transcription may be nonspecific. We therefore attempted to identify potential target genes for EUO binding, without prejudice concerning the function of EUO. Toward this end, we developed a selection-amplification procedure that enriched for DNA sequences within a chlamydial genomic library to which rEUO preferentially binds in vitro. The strongest binding site of the most highly enriched clone was partially characterized by deletion and mutation analysis. Sequences containing the most important bases in this site were used to search the enriched clones to identify a 15-bp consensus binding sequence which matched (allowing two errors) at least one sequence in every strong binding site and none outside the binding sites.
MATERIALS AND METHODS
Preparation of rEUO.
His-tagged rEUO was purified by nickel-agarose affinity chromatography, according to the manufacturer's instructions (Novagen, Madison, Wis.), and was used in all experiments, except for gel mobility shift assays carried out with an rEUO HTH-deletion mutant (rEUO-ΔHTH). For these experiments, the rEUO gene without a His tag was amplified by PCR with the template pDGWE-47 and primers E1 and E2 (36). After EcoRI digestion and gel purification, the fragments were ligated into the EcoRI site of pBluescript KS(+) vector (Stratagene, La Jolla, Calif.). The resulting plasmid pLZ1F1 was treated with BsaAI, which digested the plasmid at a unique site within the sequence encoding the HTH motif. A linear plasmid lacking HTH sequence was then amplified by PCR from two divergent primers, DHTHEHF (5′ ACAGGGACGATGGCAGG 3′) and DHTHEHR (5′ ACAACCCTTCTCATTATC 3′), which hybridized to sequences immediately flanking the motif sequence. The amplified linear plasmid was blunt end ligated to form a circular plasmid, and the DNA insert was sequenced to confirm the in-frame deletion of the HTH motif from the EUO gene. The EcoRI fragment of the plasmid was ligated into the EcoRI site of the expression vector, pT7-5 (29), and overexpression and gel purification of rEUO-ΔHTH were performed as previously described for rEUO without a His tag (36).
In vitro transcription assay.
In vitro transcription assays were carried out by using partially purified C. psittaci 6BC RNA polymerase preparations (18) under the conditions described by Douglas and Hatch (6) with templates consisting of promoter regions of chlamydial genes cloned into the pUC19-spf′ transcription assay vector (7). The templates assayed were pALR202, which contains the promoter of the plasmid antisense transcripts gene of Chlamydia trachomatis L2 (10), pLZBMP3, which contains a 124-bp insert encoding the C. psittaci 6BC MOMP gene P3 promoter region (−85 to +39) (35) ligated into XbaI and BamHI sites of pUC19-spf′, and pLZ208C, which contains a 416-bp DNA fragment encoding the promoter region (−404 to +12) of the crp operon of C. psittaci 6BC (8) ligated into the XbaI and BamHI sites of pUC19-spf′.
Construction of a chlamydial genomic library.
The genomic DNA of C. psittaci 6BC was partially digested with Sau3AI to generate relatively small fragments with a broad distribution of end points. The endonuclease-treated DNA was fractionated by electrophoresis on a 0.8% agarose gel, and fragments from 400 to 700 bp were electroeluted from the gel (32) and ethanol precipitated. The pellet was suspended in water, and the fragments were ligated at equal molar ratio to pBluescript KS(+) vector (Stratagene), which had been previously digested with BamHI and treated with calf intestine alkaline phosphatase. Escherichia coli XL1-Blue (Stratagene) was transformed, and about 20,000 colonies were collected and combined in Luria broth (LB) medium. Plasmids from the combined genomic library were prepared by using a Midiprep kit (GmbH, Qiagen, Valencia, Calif.).
In vitro enrichment for rEUO binding sites.
A nitrocellulose filter-binding assay was used for separation of His-tag rEUO protein-DNA complexes from protein-free plasmids. Binding reactions were carried out in triplicate for 30 min at room temperature, each in a final volume of 50 μl of 1× DNA-binding buffer (36). The reaction mixtures contained 2 μg of genomic library plasmid DNA, 300 ng of rEUO protein, and 200 μg of calf thymus DNA as a nonspecific competitor. The reactions were passed through a Protran BA85 nitrocellulose filter (Schleicher and Schuell, Keene, N.H.), presoaked in 1× DNA-binding buffer for 30 min at room temperature, a procedure which retains protein-DNA complexes on the filter but not unbound double-stranded DNA (dsDNA) (23, 28). After the filter was washed three times with 0.5 ml of 1× DNA binding buffer, it was placed facedown in a 20-ml scintillation vial containing 450 μl of filter elution buffer (20 mM Tris-HCl [pH 7.8], 0.2% sodium dodecyl sulfate, 0.3 M sodium acetate) and was incubated in a water bath at 30°C for 2 h with gentle agitation. The eluate was removed to a microcentrifuge tube and extracted with phenol, and plasmid DNA was ethanol precipitated. The resulting plasmids were transformed into E. coli host XL1-Blue and amplified by overnight growth on LB agar plates containing 100 μg of carbenicillin per ml and 17 μg of tetracycline per ml. Each transformation yielded approximately 2,000 to 3,000 colonies, for a total of about 7,000 colonies. After randomly picking 20 individual colonies, the remaining colonies were collected from the plates by adding LB medium, and mini plasmid preparations (2) were performed to extract DNA, representing the population of genomic recombinant plasmids after the first round of enrichment. Mini plasmid preparations were also performed with the 20 individual colonies, and inserts were obtained by digestion with HindIII and XbaI and analyzed by 0.8% agarose gel electrophoresis. Eight additional rounds of enrichment for rEUO binding sites were performed by exactly repeating the first-round enrichment procedure, except that 20 individual colonies were picked only after rounds 5 and 9.
Two controls were carried out to test the specificity of the enrichment process. In one, rEUO was added in the binding reaction to equal molar concentrations of pBluescript KS(+) vector and pALR207. pALR207 contains a 267-bp insert in pBluescript KS(+) and encodes the crp promoter (−200 to +67), an AT-rich sequence, to which rEUO binds strongly (36). After filter binding, elution, and transformation, transformants were grown on LB plates containing IPTG (isopropyl-β-d-thiogalactopyranoside), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and antibiotics. The total number of the transformants was around 2,000 to 3,000, and the ratio of white (pALR 207) to blue colonies (empty vector) was between 5 and 8 to 1, which represented the selection force per round. In the second control, rEUO was omitted from the binding reaction described above. In this case, only about 100 colonies in approximately equal ratios of white to blue were noted, indicating that nonspecific retention of circular plasmid DNA by the filter was low (5%) compared to that of the rEUO-plasmid DNA complexes.
DNase I footprinting of enriched clones.
Overlapping linear DNA fragments, which covered the entire insert of each enriched clone, were generated by PCR amplification with [γ-32P]ATP-end-labeled oligonucleotides (10) shown in Table 1. Binding reactions were carried out in a final volume of 50 μl of 1× DNA binding buffer containing 1.8 nM 32P-dsDNA (equal to about 30 to 40 ng of dsDNA), various amounts of rEUO, and 100-fold excess of sonicated calf thymus DNA (3 to 4 μg) as competitor. DNase I treatment and analysis on DNA sequencing gels were carried out as described by Sasse-Dwight and Gralla (24).
TABLE 1.
Oligonucleotides used in sequencing and DNase I footprinting studies
| Oligonucleotide | Sequence |
|---|---|
| Common | |
| T7 | 5′ AATACGACTCACTATAG 3′ |
| T3 | 5′ ATTAACCCTCACTAAAG 3′ |
| pLZ9R8 | |
| 8T7 | 5′ TTCGTTTGTCCCGAGAATTTAAAG 3′ |
| 8T3 | 5′ CCACTAATAAGCATGCAGAG 3′ |
| 8F | 5′ GTATAGGGAAATCTACATATG 3′ |
| 8R | 5′ GAGCAATTCCTTCATTGG 3′ |
| pLZ9R17 | |
| 17T71F | 5′ GTGGTGTAATCAACTCTG 3′ |
| 17T72F | 5′ GCGTACGGTAATATCAAA 3′ |
| 17T73F | 5′ TAAGTAGATAGTATTCCG 3′ |
| 17T71R | 5′ CAGAGTTGATTACACCAC 3′ |
| 17T72R | 5′ TTTGATATTACCGTACGC 3′ |
| 17T73R | 5′ CGGAATACTATCTACTTA 3′ |
| 17T31F | 5′ CTCTGAGAAAACTCTCTC 3′ |
| pLZ9R5 | |
| 5T71F | 5′ GTTCTCTAGTCGCTACAA 3′ |
| 5T72F | 5′ GCTGACATGAGTAAGATA 3′ |
| 5T73F | 5′ ATGTAAACATCTGCTGAG 3′ |
| 5T74F | 5′ TTAGCTAAGAGATGTCAG 3′ |
| 5T75F | 5′ ATAAGGGAATATCATGAG 3′ |
| 5T71R | 5′ TTGTAGCGACTAGAGAAC 3′ |
| 5T72R | 5′ TATCTTACTCATGTCAGC 3′ |
| 5T73R | 5′ CTCAGCAGATGTTTACAT 3′ |
| 5T74R | 5′ CTGACATCTCTTAGCTTA 3′ |
| 5T31F | 5′ GAATCTGGAATATGGACA 3′ |
| 5T32F | 5′ TGTCCATACATTTCAAGC 3′ |
| 5T31R | 5′ TGTCCATATTCCAGATTC 3′ |
| 5T32R | 5′ GCTTGAAATGTATGGACA 3′ |
Gel mobility shifts of rEUO binding site mutants.
Mutant forms of the strongest binding site (site 2) of the most highly enriched clone (pLZ9R8) were generated by annealing synthetic oligonucleotides. After annealing at 65°C for 3 min and slowly cooling to room temperature, gel shifts were performed with 10 ng of 32P-end-labeled mutant dsDNA and 0, 1, 10, or 50 ng of rEUO (36).
Circular permutation (bending) assay.
The assay was carried out according to the procedure of Wu and Crothers (34). Two oligonucleotides, representing site 2 of pLZ9R8 (9R8T2/32Xb [5′ CTAGATTTTCGTATTCCAAGAAATGTCTTAACAATGT 3′] and 9R8B2/32Xb [5′ CTAGACATTGTTAAGACATTTCTTGGAATACGAAAAT 3′]), were annealed and cloned into the pBend2 vector (17), which was previously digested with XbaI. The resulting plasmid, pBend(LZ9R8/2), was sequenced on both strands with oligonucleotides HOW3 (5′ AAAATAGGCGTATCACGAGG 3′) and HOW4 (5′ CTGCGTTAGCAATTTAACTGTG 3′) to confirm the construction and then digested with MluI, NheI, SpeI, DraI, PvuII, StuI, and BamHI and treated with calf intestine alkaline phosphatase (Boehringer Mannheim, Indianapolis, Ind.). The seven digested products were extracted with phenol, precipitated with ethanol, end labeled with [γ-32P]ATP, and purified with MicroSpin G-25 columns (Pharmacia LKB Biotechnology, Piscataway, N.J.). Binding reactions were performed with 1 ng of rEUO and 1 ng of labeled DNA probes, as in the gel mobility shifts, except that samples were analyzed on 10% rather than 6% nondenaturing acrylamide gels.
DNA sequencing.
The dideoxy-chain termination DNA sequencing was performed with Sequenase kits, following the instructions of the manufacturer (U.S. Biochemical Corp., Cleveland, Ohio).
Computer analysis.
The homology searches of DNA and protein sequences of enriched clones were performed by using Blast, Fasta, Fastn, Tfasta, and BestFit of GCG software (Genetics Computer Group, Inc., Madison, Wis.) and the Chlamydial Genome Project database (26). The alignment and sequence analysis of EUO binding sites were carried out with the Motifs, BestFit, FindPatterns, PileUp, and Window programs of GCG software (3, 5); BLOCKS (16); and the Megalign program in DNASTAR (DNASTAR, Inc., Madison, Wis.).
RESULTS
The role of the HTH motif in rEUO binding to DNA.
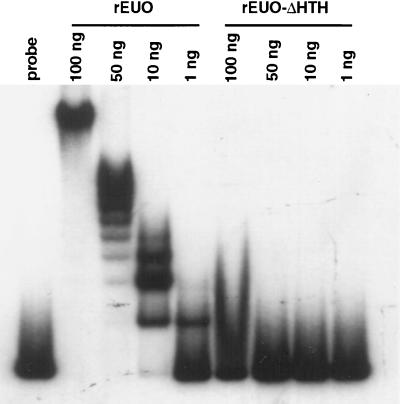
The algorithm of Dodd and Egan (4) predicts an HTH motif in EUO; however, neither the Motifs nor the BLOCKS program identifies an HTH motif in the protein. To test whether the putative HTH motif was required for DNA binding, we constructed a recombinant EUO protein (rEUO-ΔHTH) lacking only the sequence encoding the motif. As we had observed previously (36), we found multiple shifted bands at high concentrations of wild-type EUO, as determined by gel mobility shift assay (Fig. 1). This ladder pattern of shifted bands probably reflected the binding of EUO to multiple sites on the 267-bp DNA probe, the promoter region of the crp operon (8). In contrast, rEUO-ΔHTH failed to shift the probe at any concentration tested. Although the possibility that the deletion induced conformational changes that were responsible for the loss of DNA binding activity in the EUO protein cannot be eliminated, these results are consistent with the conclusion that EUO binds to DNA through an HTH motif.
FIG. 1.
Gel electrophoresis mobility shift assay with rEUO and rEUO-ΔHTH. A standard gel shift assay was carried out with the amounts of gel-purified recombinant proteins indicated above the lanes. Electrophoresis of 1 ng of free probe (−200 to +67 of the crp operon promoter region) is shown in the far-left lane.
The effect of rEUO binding on in vitro transcription.
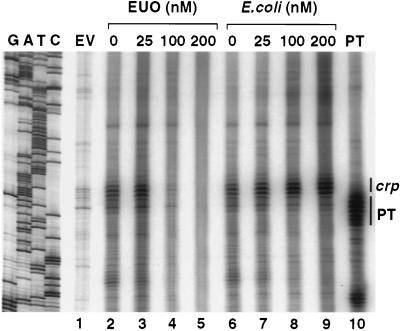
Because rEUO binds to the promoter region of the crp operon (36), the effect of rEUO on crp transcription in vitro was examined. Inhibition of transcription was noted at concentrations of rEUO of 100 nM and above (Fig. 2, lanes 2 to 5). The inhibition probably was not due to extraneous substances in the gel-purified rEUO preparation, since an irrelevant E. coli protein purified from the same gel had no effect (Fig. 2, lanes 6 to 9). However, the in vitro transcription of two other chlamydial genes, the MOMP gene of C. psittaci and the plasmid antisense transcript genes of C. trachomatis, was also inhibited by similar concentrations of rEUO (data not shown). The MOMP and plasmid antisense genes are expressed during the growth phases of the developmental cycle (21, 27); therefore, it is unlikely that rEUO is a negative regulator of these genes in vivo. The mechanism by which rEUO inhibited transcription of all three genes is not known, but may have been due to the ability of the protein to bind to multiple DNA sequences at high protein concentrations rather than to binding solely to specific regulatory elements.
FIG. 2.
The effect of rEUO on in vitro transcription from the crp operon. The templates were empty vector without insert (EV); pLZ208C, containing the C. psittaci crp operon promoter from −404 to +12 (lanes 2 to 9); and pALR202, containing the C. trachomatis L2 plasmid antisense transcripts promoter (PT) from −498 to +6. The concentrations of EUO protein (lanes 2 to 5) and an irrelevant E. coli protein (lanes 6 to 9), purified from the same gel from which rEUO was purified, are shown above the lanes; no recombinant protein was present in the EV and PT controls. The position of the in vitro-generated crp and PT transcripts are indicated on the right side. Other bands represent transcription from unknown sites within the vector or the inserts. A DNA ladder, which was used to estimate the length of the transcripts, is shown on the left side.
Enrichment for rEUO binding sites.
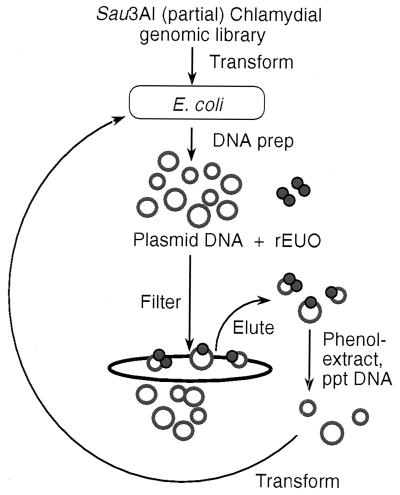
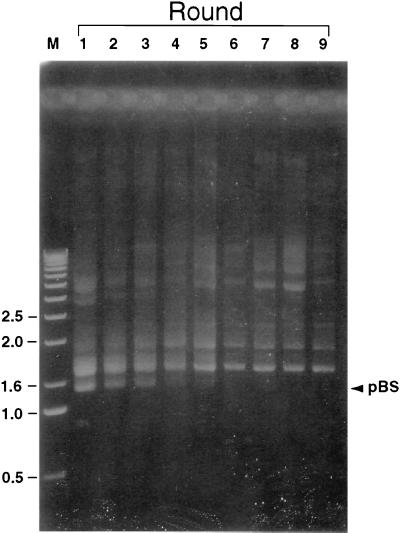
To identify preferred binding sites for EUO without prejudice concerning its function, we devised a selection-amplification system for enrichment of EUO-bound DNA fragments within the chlamydial genome. A scheme of the procedure is shown in Fig. 3. A chlamydial genomic library was constructed, consisting of 400- to 700-bp inserts from a partial Sau3AI digest ligated into pBluescript KS(+). Plasmid DNA from the library of approximately 20,000 clones was incubated with rEUO, and the protein-DNA complexes were separated from free plasmid DNA by trapping on a nitrocellulose filter. The DNA was eluted from the filters, extracted with phenol, and used to transform E. coli for the purpose of amplification. Plasmid DNA was reisolated and subjected to eight additional rounds of enrichment for rEUO-DNA complexes. The conditions in the multiple-round enrichment procedure were set up so that when pBluescript without an insert and pALR207, a pBluescript plasmid containing the crp promoter region, were incubated in equimolar amounts with rEUO, five to eight times as much pALR207 plasmid as pBluescript was recovered from the filters. Under these conditions, the retention of plasmids in the absence of rEUO was reduced by 95%. A portion of the isolated, undigested plasmid preparations after each round of enrichment was fractionated by agarose-gel electrophoresis (Fig. 4). After round 1, the library consisted of a heterogeneous population of plasmids, including a relatively large proportion of pBluescript without insert. As the enrichment processes progressed, the heterogeneity of the library diminished, with a focusing of plasmids with inserts of approximately 500 bp and some other larger species; pBluescript without insert could not be detected after round 5.
FIG. 3.
Scheme for in vitro enrichment of rEUO binding sites from a chlamydial genomic library. ppt, precipitated.
FIG. 4.
Plasmid content of the genomic library through nine rounds of enrichment. Plasmids were prepared from the entire library after each round of enrichment (rounds 1 to 9), fractionated by agarose gel electrophoresis, and stained with ethidium bromide. Linear DNA markers (M) were run in the first lane, and their sizes in kilobases are shown on the left side. The migration of monomeric pBluescript without inserts is indicated on the right side (pBS). Because the plasmids were not digested with restriction enzymes, covalently closed, nicked, and concatameric forms were present in the population.
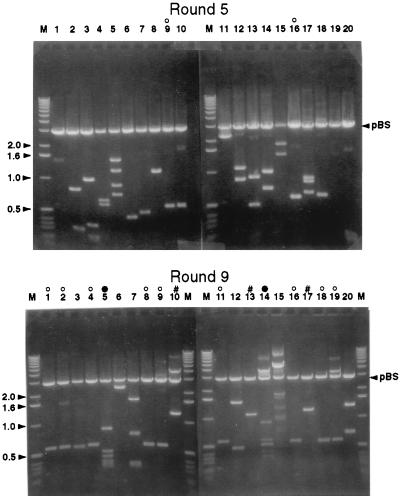
Approximately 7,000 colonies were obtained after each round of enrichment. The inserts from 20 randomly selected clones were examined by electrophoresis after rounds 5 and 9 (Fig. 5). Restriction fragment analysis and subsequent sequencing indicated that after round 9, 3 clones were present more than once among the 20 selected. The most-highly-enriched clone, pLZ9R8 (9 of 20), was found to contain a single Sau3AI fragment of 529 bp. DNA sequencing indicated that the fragment is homologous to part of the dcd gene, which encodes the C-terminal portion of deoxycytidine triphosphate deaminase (Fig. 6). This clone was also detected twice among the 20 enriched clones after round 5 (Fig. 5). Southern blot analysis was performed by dot blotting of 600 individual colonies from the C. psittaci 6BC genomic library (round 0) and 200 colonies after nine rounds to distinguish between enrichment and possible overrepresentation of pLZ9R8 in the library. Whereas 96 of the round 9 clones (48%) hybridized to a pLZ9R8 probe, only 1 of the 600 clones in the original library tested positive, confirming that pLZ9R8 was highly selected by the in vitro DNA binding and amplification process (data not shown).
FIG. 5.
Insert DNA fragments of representative clones enriched at rounds 5 and 9. The plasmids of 20 random clones picked after round 5 (top panel) or round 9 (bottom panel) were digested with HindIII and XbaI and fractionated by agarose gel electrophoresis. Lanes marked M represent linear DNA markers (in kilobases). ○, clones with an insert size of approximately 500 bp. DNA sequencing revealed that all of these clones contain the same insert, which was designated LZ9R8. #, clones with identical insert DNA fragments totaling about 1.2 kb. The insert of these clones was designated LZ9R17. ●, clones with identical inserts of 2.3 kb, which were designated LZ9R5. Migration of the linear vector (pBluescript [pBS]) is indicated on the right side.
FIG. 6.
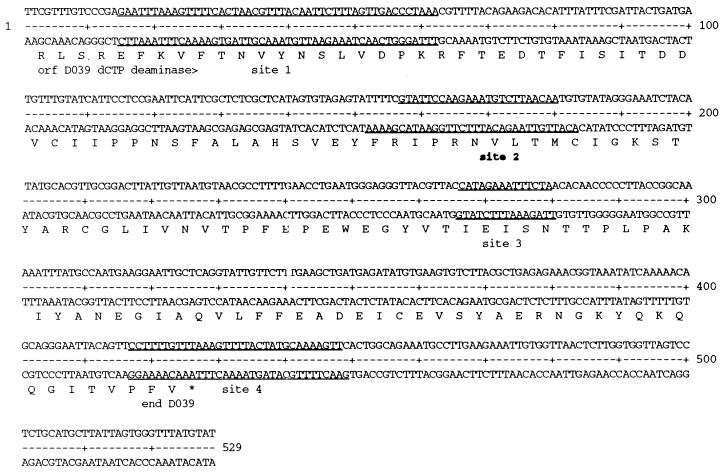
Summary of the rEUO binding sites of LZ9R8. The binding sites, as determined by rEUO protection from DNase I and numbered in order from the beginning to the end of the insert, are underlined. Site 2 (designated in boldface) was the only strong binding site. An ORF with homology to the C-terminal portion of dCTP deaminase (dcd [ORF D039 in C. trachomatis serovar D]) is shown below the DNA sequence.
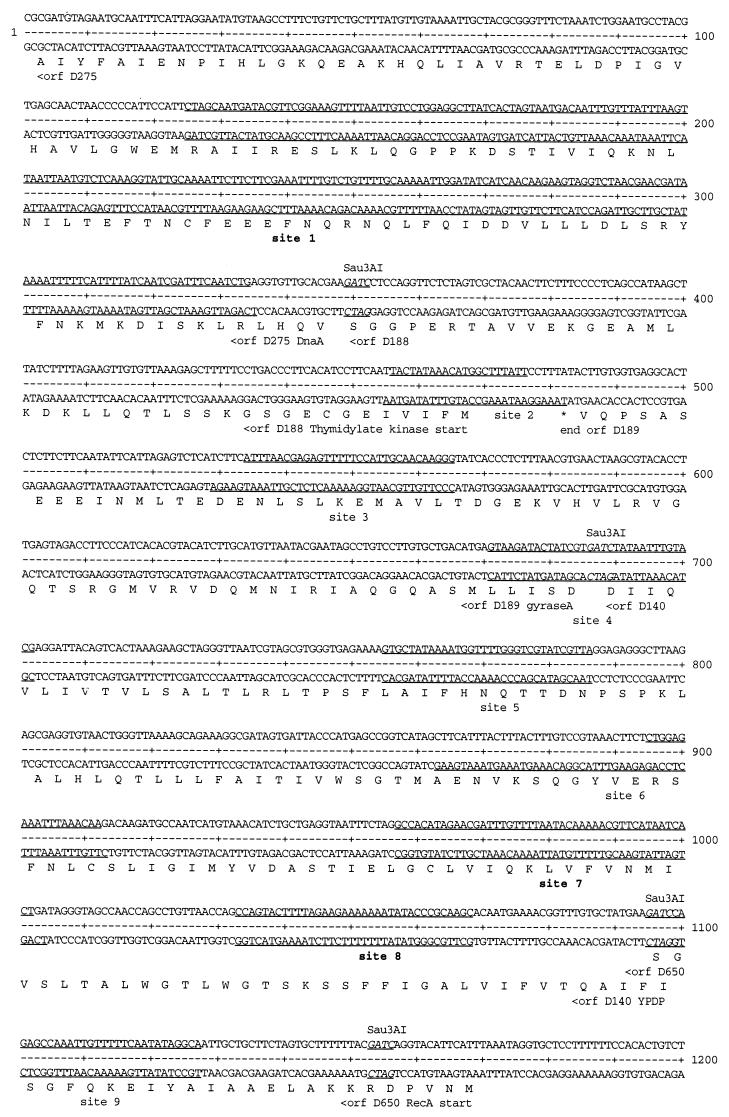
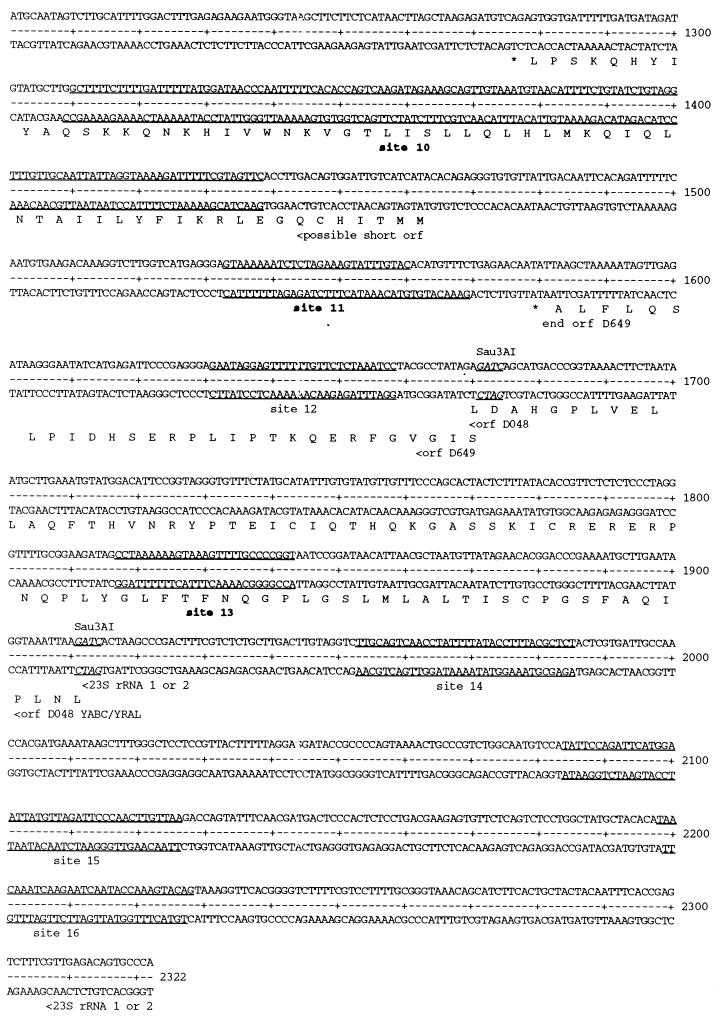
The second most highly enriched clone of round 9, pLZ9R17 (3 of 20), was generated by ligation of three Sau3AI fragments (Fig. 7) to yield a 1.2-kb insert. It encodes four partial open reading frames (ORFs) which bear homology to ORFs D263, D264 (MSBA-transport ATP-binding protein), D425, and D602 of C. trachomatis serovar D genome (26). The third enriched clone of round 9, pLZ9R5 (2 of 20; 2.3 kb), was the ligation product of six Sau3AI fragments (Fig. 8). The six fragments encode eight partial ORFs that are homologous to genes found in the C. trachomatis serovar D genome (26): dnaA (DnaA), tdk (thymidylate kinase), gyrA (gyrase A), ypdp (hypothetical protein), recA (recombination protein A), ygfA (possible formyl tetrahydrofolic acid cycloligase), yABC/yraL (possible methyltransferase), and a 23S rRNA gene.
FIG. 7.
Summary of the rEUO binding sites in LZ9R17. The binding sites are underlined, with the single strong site (site 2) designated in boldface. ORFs with homology to known proteins, including ORFs identified in the genome of C. trachomatis serovar D, are shown below the coding sequence, with arrowheads indicating the orientation of ORFs. The Sau3AI sites are underlined and in italic.
FIG. 8.
Summary of the rEUO binding sites in LZ9R5. The binding sites are underlined, with ORFs and Sau3AI sites designated as described for Fig. 7. Strong sites are designated in boldface (sites 1, 7, 8, 10, 11, and 13).
The A + T content of the C. psittaci 6BC genome is not precisely known, but the kinetics of DNA melting suggest that it is about 60% (12), similar to the genome of C. trachomatis D/UW-3/Cx (26). The A + T content of the inserts in the three most highly enriched clones ranged from 59 to 63%, similar to the expected overall content of the genome.
Southern blot analysis of plasmid DNA prepared from the combined 7,000 colonies collected after rounds 1, 5, and 9 were probed with the insert of pALR207, which encodes the crp promoter region from −200 to +67. A relatively strong signal was observed after round 1, a weak signal after round 5, and a background signal after round 9 (data not shown). These observations suggest that at least part of the crp promoter region was present in the genomic library, but was not selected by the enrichment scheme. It is possible that plasmids encoding this promoter did not replicate as well as the enriched plasmids or that in vitro binding of rEUO to the enriched inserts under the chosen selection condition was, in fact, better than binding to the crp promoter sequences.
DNase I footprinting of the enriched clones.
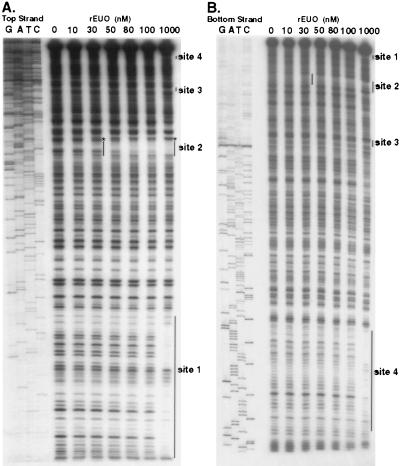
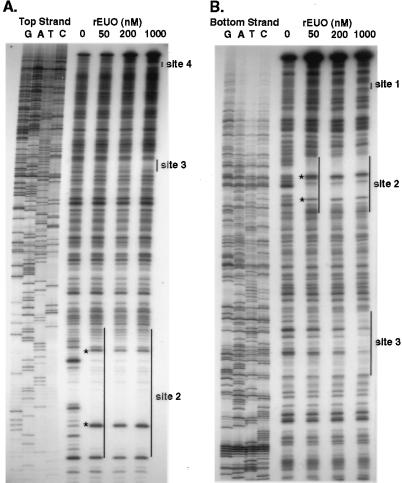
DNase I footprinting analysis was carried out to determine the sites to which rEUO binds in vitro. Inserts of pLZ9R8 (LZ9R8) and pLZ9R17 (LZ9R17) are shown as representative footprint results (Fig. 9 and 10). All three clones contained multiple protection sites, with a total of 24 being detected. Several of the sites included hypersensitive bands, including the strong binding sites in LZ9R8 (site 2, Fig. 9) and LZ9R17 (site 2, Fig. 10). The protected sites are underlined in Fig. 6 (LZ9R8), 7 (LZ9R17), and 8 (LZ9R5). The DNA strands in the sites were arbitrarily designated “top” and “bottom.” The concentration of rEUO required to protect the sites from DNase I ranged from 30 or 50 nM (high affinity, strong sites) to between 100 nM and 1 μM (low affinity, weak sites). The lengths of the footprinted regions ranged from 15 to 260 nucleotides (nt), with half falling in the 27- to 37-nt range. The shortest strong binding site found was the 24-bp site 2 of LZ9R8. Three sites were noticeably longer than the others (60, 129, and 260 nt) and probably resulted from the presence of multiple closely spaced binding sites along with cooperative extension of the footprints from an initial binding site. The sites identified in the enriched clones were located entirely within coding regions, except for site 4 of LZ9R8 and site 4 of LZ9R17, which included both coding and upstream sequences. There were three notable features among the footprinted sites. The first was their AT-rich nature; they had an average AT content of 69% (range, 59 to 75% for strong sites and 53 to 80% for weak sites). Most also contained a region within the site that was even more AT rich. Finally, all of the strong sites and 60% of the weak sites contained at least one stretch of four contiguous A's or T's. To determine whether high AT content is the sole determinant for EUO binding, we analyzed the inserts of all three clones for average AT content with the Window program in the GCG software package (3, 5) over window lengths of 15, 20, and 30 nt (not shown). Although regions with very high AT content were most often located within strong sites, other areas with similarly high AT content were also found within weak sites and nonfootprinting sites. For example, the best predictor of a strong site was a peak average AT content above 80% when calculated in 1-nt increments with a window of 20 nt. Under this search condition, only five of the eight strong sites and one weak site were identified, along with one nonfootprinting site. All strong sites reached a minimum peak average AT content of 75%; however, peaks of 75% identified as many nonfootprinted as footprinted sites. Similarly, strings of at least four consecutive A's or T's were also located in many AT-rich nonfootprinting sites. We conclude that whereas high AT content is required for rEUO binding to DNA in vitro, other factors such as specific nucleotide sequence or DNA secondary structure are responsible for discrimination between footprinting and nonfootprinting sites. We also used the computer algorithms BestFit, Pileup, and Megalign to align the sites; unfortunately, all three failed to identify a common sequence pattern within the strong or weak sites of the three enriched clones, perhaps in part because the binding sites were of such different sizes.
FIG. 9.
Representative DNase I footprints of the LZ9R8 insert by rEUO. (A) Top strand. (B) Bottom strand. The concentrations of rEUO that were used in the protection study are shown at the top of the lanes. Protected regions are indicated by vertical lines, and an asterisk indicates a hypersensitive site in the top strand. DNA sequencing ladders, produced by using the same primers that were used in the protection study, are shown on the left side of each panel.
FIG. 10.
Representative DNase I footprints of LZ9R17 insert by rEUO. (A) Top strand. (B) Bottom strand. The concentrations of rEUO are shown on top of the lanes. Protected regions are indicated by vertical lines, and hypersensitive sites are indicated by asterisks. DNA sequencing ladders are shown on the left side of each panel.
Bending of LZ9R8 site 2 DNA upon rEUO binding.
Several hypersensitive bands were noted in DNase I footprints, suggesting that EUO may induce local distortions in DNA upon binding. LZ9R8 site 2 was examined by the circular permutation assay of Wu and Crothers (34) to determine whether rEUO can induce bends in DNA. This site lacks lengthy A and T tracts and therefore is not likely to contain intrinsic bends (15). Site 2 was cloned into pBend2, and the resulting plasmid was digested with restriction enzymes to yield seven circularly permuted DNA fragments of identical size (158 bp), differing only in the position of the rEUO binding site. The seven fragments were then examined by standard gel mobility shift assay (Fig. 11). The slower migration of the fragments with the EUO binding site nearer the middle of the fragment suggests that rEUO may induce a small bend upon its binding to LZ9R8 site 2.
FIG. 11.
Circular permutation assay for rEUO-induced DNA bending. The top panel shows DNA fragments generated by restriction endonuclease digestion of pBend(LZ9R8/2) with MluI (a), NheI (b), SpeI (c), DraI (d), PvuII (e), StuI (f), or BamHI (g). The 32-bp LZ9R8 site 2 is shown as a solid box. The bottom panel shows the results of the gel shift experiment with rEUO binding and circular permutated DNA pBend(LZ9R8/2) fragments (a to g). Lane 0, rEUO not added. Fragments in which the rEUO binding sites were closer to the center were shifted more than the fragments in which the sites were closer to the ends.
Mutational analysis of LZ9R8 site 2.
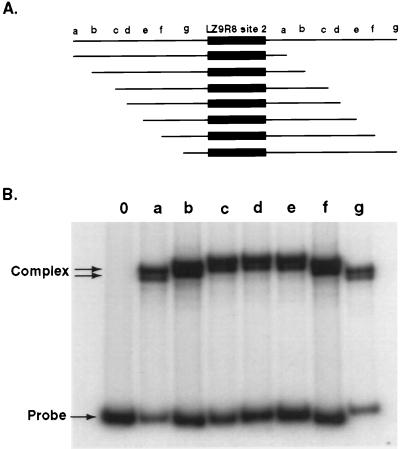
Because site 2 in LZ9R8 is the only strong site in the most highly enriched clone and is of relatively small size (24 nt, top strand; 32 nt, bottom strand), it was chosen for deletion studies in an attempt to define the minimal sequence required for rEUO binding to a given site as defined by mobility shift analysis. The results of the deletion study are summarized in Fig. 12. The minimal length for full binding affinity of rEUO required a probe (M20) of 20 bp (TCCAAGAAATGTCTTAACAA [top strand]).
FIG. 12.
Summary of rEUO binding to deletion fragments of LZ9R8 site 2. The 32-bp rEUO binding site, as determined by DNase I protection studies, is shown at the top. Gel mobility shift analysis was carried out with synthetic double-stranded oligonucleotides, deleted from either or both ends of the site, and 1, 10, or 50 ng of rEUO, as described in Materials and Methods. The lengths of fragments are represented by horizontal lines. The gel shift phenotype was scored as follows: +++, shift equal to that of the wild type at all three rEUO concentrations; ++, decreased shift at 1 ng of rEUO, wild-type shift at higher concentrations; +, decreased shift at 1 and 10 ng of rEUO, wild-type shift at 50 ng; −, no major band shift at any of three rEUO concentrations.
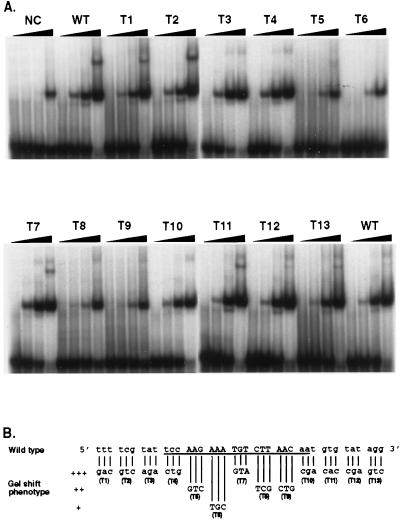
To identify the nucleotides which are critical for rEUO binding in LZ9R8 site 2, sequential, 3-bp-at-a-time mutations were made in 39-bp double-stranded oligonucleotides (site 2 and flanking sequence) and tested by gel mobility shift assay for binding affinity to rEUO. The changes were a mixture of radical (either A or T to either G or C and vice versa) and conservative (the AT content was not changed) alterations. As shown in Fig. 13, rEUO shifted the wild-type 39-bp site 2 sequence when present at 1 ng, whereas an unrelated, control 39-bp LZ9R8 sequence that failed to footprint required a 50-fold-larger amount. Only four of the mutant triplets had a decreased binding affinity for rEUO: T6 (AAA→TGC) showed the most severe decrease in binding; the other three mutant triplets, T5 (AAG→GTC), T8 (CTT→TCG), and T9 (AAC→CTG), showed a more moderate decrease. All of these down mutants were clustered in a 15-bp core region of LZ9R8 site 2 and entailed changes that made the triplets more GC rich. One mutant triplet in this region, T7 (TGT→GTA), had no effect on binding affinity to rEUO. This triplet is roughly in the center of the core and may not contain sequence required for binding; however, the overall AT content of the mutant was the same as that in the wild-type triplet, and the changes may have had a neutral effect for this reason. The 15-bp core is located inside of the 20-bp minimal binding sequence determined by deletion analysis (Fig. 12), further indicating that this region is critical for binding to rEUO.
FIG. 13.
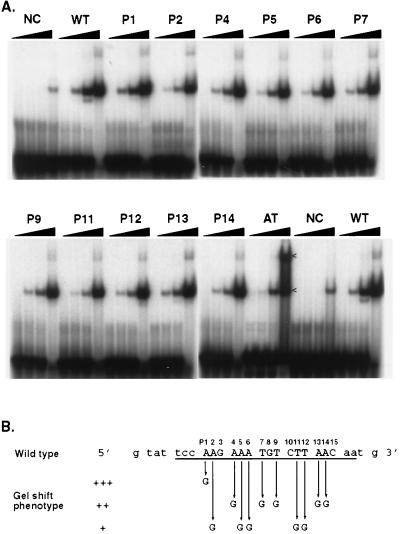
Triple-mutation analysis of pLZ9R8 site 2. (A) Gel mobility shift. The double-stranded oligonucleotides used as probes were as follows: NC, a 39-bp negative control DNA from nonfootprinting sequence of pLZ9R8 (CGATTACTGATGATGTTTGTATCATTCCTCCGAATTCAT); WT, 39 bp of wild-type LZ9R8 site 2 and surrounding sequence (see panel B); T1 to T13, 13 triple mutants of LZ9R8 site 2, as indicated in panel B. (B) Summary of the results of the triple-mutation analysis. The number of plus signs reflects the relative strength of the shift, with +++ being equivalent to wild type. The triple substitutions (T1 to T13) in the wild-type sequence are shown below the wild-type sequence.
Single-base-pair mutations were made by sequentially changing each A or T in the 15-bp core binding sequence of site 2 to a G; the total length of the fragment analyzed (26 bp) included LZ9R8 sequence on each side of the core to ensure that the mutated oligomers formed stable double strands and were of sufficient length to allow binding of rEUO. The results (Fig. 14) indicated that loss of A's at positions 2, 5, and 6 and T's at positions 11 and 12 had the greatest negative effects on binding; when G's were substituted in these positions, 10 ng of rEUO was required to produce a shift equivalent to that seen with 1 ng on the wild-type sequence. Less-negative effects were seen when G's were substituted in positions 4, 7, 9, 13, and 14, and essentially no effect was noted when a G was substituted for the A in position 1 of the wild type. A 26-bp poly(A)-poly(T) DNA (AT in Fig. 14) fragment was also tested. Interestingly, the AT fragment shifted less well than the wild-type fragment at 1 ng of rEUO. However, at a high concentration, rEUO bound better to the AT fragment, inducing a second shifted species that was much weaker for the other fragments. These results suggest that poly(A) is not the optimal binding sequence for rEUO, but it may provide multiple sites for binding.
FIG. 14.
Single-mutation analysis of pLZ9R8 site 2. (A) Gel mobility shift. The assay was carried out with 0, 1, 10, or 50 ng, as described in Materials and Methods. NC, a 26-bp negative control DNA from nonfootprinting sequence of pLZ9R8 (CATTACTGATGATGTTTGTATCATTC); WT, wild-type 26-bp LZ9R8 site 2 (see panel B); P1 to P15, single-base-pair mutations, as indicated in panel B. Arrows indicate the positions of DNA-protein complexes. (B) Summary of the results of single-mutation analysis. The number of plus signs reflects the relative strength of the shift, with +++ being equivalent to wild type.
Derivation of a consensus EUO binding site.
Since we were unable to identify a common sequence pattern within the binding sites in the enriched clones by using computer algorithms, we decided to apply the information derived from the mutagenesis studies described above to identify related sequences within each of the eight strong binding sites and then to develop a consensus from those sequences. By using the FindPatterns program (3, 5), we searched the three enriched clones with the 15-bp LZ9R8 site 2 core sequence that was identified in the mutagenesis study. It was necessary to allow six mismatches to recover at least one matching sequence within each strong site. Under this search condition, a total of 19 sequences were identified in the strong sites (Table 2). Two possible consensus sequences that can be derived from the frequency table for the 19 matches are AAGAAATGTCTTAAC and AADAAHNNTNTTDNH (by convention, D = not C; H = not G; N = G, A, T, or C; W = A or T; V = not T; and Y = C or T). The first represents the most common nucleotide at each position and is identical to the original search sequence (site 2 of LZ9R8). It is not a useful consensus, because it required four to six mismatches to identify any given strong site, and under the search condition of six mismatches, it identified many nonfootprinting sites. The second consensus was derived by choosing a single nucleotide for a given position if it were present in 14 or more of the 19 sequences (approximately 75%); otherwise, a symbol representing all of the observed bases in a given position was used. This consensus, with two mismatches, is found at least once in all eight of the strong binding sites, at least once in 13 of 16 weak binding sites, and in 26 nonfootprinting sites. We next attempted, by trial and error, to find a less degenerate sequence which would identify at least one sequence in every strong site without identifying any nonfootprinting sites, when allowing two mismatches. After searching with approximately 100 candidate sequences, we found that the sequence AHGAAAWVTYTWDAY fulfilled these criteria. Interestingly, only one sequence in each strong site, including even the long footprinting sites, was identified when this consensus sequence was used to search (allowing two mismatches) the three enriched clones (Table 3). That is, many of the original 19 sites identified by matches to the 15-nt core sequence were eliminated when the search was carried out with a sequence that specifically excluded nonfootprinting sites. In addition, one of the final consensus-matching sequences in Table 3, ATGAAAAATTTTTAT, was not present in the original 19 sites, suggesting that this approach did not unduly bias the derived consensus toward the 15-nt core sequence used for the first search. The most striking feature of the frequency table based on the identified 8 sequences is the high degree of specificity for particular nucleotides in several positions (A-GAAA--T-TT-A-), suggesting that these are the most important nucleotides required for binding of EUO to DNA.
TABLE 2.
Sequences in strong binding sites that match the LZ9R8 15-nt core with six mismatches or less
| Site or basea | Sequence or frequency | No. of mismatches |
|---|---|---|
| Sites | Sequences | |
| 8T2 | AAG AAA TGT CTT AAC | 0 |
| 8B2 | AAG ACA TTT CTT GGA | 5 |
| 17B2 | AAT AAA CGT TTT GAT | 5 |
| 5T1 | AAT TAA TGT CTC AAA | 4 |
| 5T1 | TCG GAA AGT TTT AAT | 6 |
| 5B1 | GAG ACA TTA ATT AAC | 5 |
| 5B1 | ATT AAT TAA CTT AAA | 6 |
| 5B1 | ACA AAT TGT CAT TAC | 5 |
| 5B1 | TAA AAC TTT CCG AAC | 6 |
| 5T7 | ACG ATT TGT TTT AAT | 5 |
| 5T7 | AAA AAC GTT CAT AAT | 6 |
| 5T8 | AAG AAA AAA ATA TAC | 6 |
| 5T10 | AAG ATT TTT CGT AGT | 6 |
| 5B10 | GAA AAA TCT TTT ACC | 5 |
| 5B10 | ACG AAA AAT CTT TTA | 6 |
| 5B10 | AAA AAT TGG GTT ATC | 5 |
| 5T11 | AAA AAA TCT CTA GAA | 5 |
| 5B11 | TAG AGA TTT TTT ACT | 6 |
| 5T13 | AAG TAA AGT TTT GCC | 5 |
| Bases | Frequencies | |
| G | 2 010 1 1 0 1 8 1 1 1 1 4 2 0 | |
| A | 1414 6 161412 4 3 3 2 2 2 1212 5 | |
| T | 3 1 3 2 2 5 13 615 61515 3 2 6 | |
| C | 0 4 0 0 2 2 1 2 0 10 1 0 0 3 8 |
Site designations are shown as follows. The first number refers to the clone number (e.g., 8 = LZ9R8); T and B refer to the top and bottom strands, respectively; and the final number refers to the site number within the clone.
TABLE 3.
Sequences in strong binding sites that match the consensus sequence with two mismatches or less
| Site or base | Sequence or frequency |
|---|---|
| Sites | Sequences |
| 8-2 | AAG AAA TGT CTT AAC |
| 17-2 | AAT AAA CGT TTT GAT |
| 5-1 | ATG AAA AAT TTT TAT |
| 5-7 | ACG ATT TGT TTT AAT |
| 5-8 | AAG AAA AAA ATA TAC |
| 5-10 | ACG AAA AAT CTT TTA |
| 5-11 | AAA AAA TCT CTA GAA |
| 5-13 | AAG TAA AGT TTT GCC |
| Bases | Frequencies |
| G | 0 0 6 0 0 0 0 4 0 0 0 0 3 0 0 |
| A | 8 5 1 7 7 7 4 3 1 1 0 2 2 6 2 |
| T | 0 1 1 1 1 1 3 0 7 4 8 6 3 1 3 |
| C | 0 2 0 0 0 0 1 1 0 3 0 0 0 1 3 |
DISCUSSION
A number of approaches have been taken to identify consensus binding sequences of DNA binding proteins, the most random of which are variations of the SELEX (systematic evolution of ligands by exponential enrichment) method developed by Tuerk and Gold (31). The common theme of the SELEX methodology is the generation of completely random RNA or DNA oligonucleotides as targets for in vitro protein binding, followed by multiple rounds of selection and amplification of the bound sequences. We chose to use less-random targets, namely chlamydial genome fragments present in a pBluescript library. This approach could potentially identify specific EUO binding sequences within the chlamydial genome and did not require knowing beforehand the length of DNA needed for rEUO binding. The amplification of bound plasmids was accomplished by transformation and replication of the pBluescript clones in E. coli. There were several potential limitations to this approach: large Sau3AI genomic fragments were excluded from the library, clones with large inserts presented multiple targets for binding, and the amplification process may have enriched for clones that replicate most successfully in E. coli. Nevertheless, this successfully enriched for three clones, all of which contained at least one strong and multiple weak binding sites.
A common sequence pattern among the strong binding sites of the three enriched clones was not detectable by simple inspection or computer algorithms. As an alternative, we chose to use mutagenesis to characterize the strong binding site of the most highly selected clone (site 2 of LZ9R8) and to use sequences derived from its important core region to find other related sequences in the other strong binding sites. Refinement of these sequences by elimination of those matching nonfootprinted sites led to the identification of the consensus AHGAAAWVTYTWDAY. When two mismatches are allowed, this consensus identifies one sequence in each of the eight strong sites and does not identify any nonfootprinting sites in any of the three most-highly-enriched clones. Although some additional refinement of the consensus may result from the investigation of additional enriched clones, the current sequence may already be useful for predicting additional possible EUO binding sites in the chlamydial genome. From the eight sequences identified by this consensus, the most frequent, and therefore potentially the most important, bases were A-GAAA--T-TT-A-. Interestingly, four out of the five most important nucleotides identified by the limited single-base-pair mutagenesis study are specified in this sequence.
The ability of EUO to bind to a broad spectrum of sequences and to recognize a relatively degenerate AT-rich consensus sequence is a property common to several so-called nucleoid-associated proteins. This group, which includes HU (25), H-NS (1), FIS (10), IHF (11, 13, 14), and LRP (20), are generally small, abundant, basic proteins that bind to DNA, often resulting in DNA compaction and bending. In their roles as chromosome organizers, they facilitate interaction of proteins bound to contiguous and noncontiguous DNA sites, leading to the assembly of specific nucleoprotein complexes that participate in replication, recombination, and regulation of expression of a large number of often unrelated target genes. Both H-NS and HU show less sequence specificity than FIS, IHF, and LRP and preferentially bind to curved, bent, or kinked DNA. We have not yet determined whether the strong binding sites in the enriched clones include curved or bent sequences. Like EUO, FIS and LRP have roughly 15-nt degenerate consensus sequences and, at low protein concentrations, protect regions 20 to 30 nt in length. As is the case with nucleoid-associated proteins, at high concentrations, EUO binds to multiple weak sites, extends footprint lengths (possibly by cooperative binding), and also covers long stretches of DNA. There are two striking differences between these nucleoid-associated proteins and EUO. The first is that EUO causes only a small DNA bend when bound to a single strong site, whereas FIS generates a 40° to 90° bend, the LRP monomer generates a roughly 50°C bend, the LRP dimer generates a 135° bend, and IHF generates a 140° bend. We do not know if binding of EUO to multiple adjacent sites would generate a more pronounced bend. The second difference is that EUO is a quantitatively minor protein. Even when EUO is present in peak amounts at around 12 to 18 hpi, it represents only a small fraction of 1% of the total protein (36). Therefore, it is likely that EUO can bind in vivo to only a limited number of sites at any given time. Nevertheless, its low abundance does not rule out the possibility that it may serve an organizing and regulating effect at those few sites.
The enriched clones with the largest inserts that we identified contain intergenic sequences, but DNase I-protected regions were almost exclusively within coding regions, suggesting that EUO may not interact directly with most promoter sequences. However, the enriched sequences may not represent in vivo targets. It is possible that in vivo conditions, such as the presence of other DNA binding proteins, facilitate EUO binding to a limited number of sites that have lower intrinsic affinities for EUO than do the enriched sites. In this respect, two AT-rich regions have been noted adjacent to the −35 hexamer of several putative chlamydial promoters; both of these regions have been shown by in vitro transcription studies to contribute to the strength of the chlamydial MOMP gene and rRNA operon promoters (6, 30). The upstream sequence appears to be an UP element that is most likely recognized by the carboxy-terminal domain of the α subunit of RNA polymerase (30). The second AT-rich element appears to be unique to chlamydiae. It is located in the spacer region, just downstream of −35 hexamers in many of the putative chlamydial promoters thus far identified on the basis of primer extension studies (30). It is intriguing to speculate that EUO in some way may facilitate the recognition of this element by chlamydial RNA polymerase. Investigation of the in vivo targets of EUO is needed to address this possibility.
ACKNOWLEDGMENT
This work was supported by Public Health Service grant AI 19570.
REFERENCES
- 1.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 2.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dodd I B, Egan J B. Improved detection of helix-turn-helix DNA binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolz R. GCG: database searching. Methods Mol Biol. 1994;24:101–116. doi: 10.1385/0-89603-246-9:101. [DOI] [PubMed] [Google Scholar]
- 6.Douglas A L, Hatch T P. Mutagenesis of the P2 promoter of the major outer membrane protein gene of Chlamydia trachomatis. J Bacteriol. 1996;178:5573–5578. doi: 10.1128/jb.178.19.5573-5578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erickson J W, Gross C A. Identification of the ςE subunit of Escherichia coli RNA polymerase: a second alternative ς factor involved in high temperature gene expression. Genes Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- 8.Everett K D E, Hatch T P. Sequence analysis and lipid modification of the cysteine-rich envelope proteins of Chlamydia psittaci 6BC. J Bacteriol. 1991;173:3821–3830. doi: 10.1128/jb.173.12.3821-3830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fahr M J, Douglas A L, Xia W, Hatch T P. Characterization of late gene promoters in Chlamydia trachomatis. J Bacteriol. 1995;177:4252–4260. doi: 10.1128/jb.177.15.4252-4260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fahr M J, Sriprakash K S, Hatch T P. Convergent and overlapping transcripts of the Chlamydia trachomatis 7.5-kb plasmid. Plasmid. 1992;28:247–257. doi: 10.1016/0147-619x(92)90056-g. [DOI] [PubMed] [Google Scholar]
- 11.Finkel S E, Johnson R C. The FIS protein: it's not just for DNA inversion anymore. Mol Microbiol. 1992;6:3257–3265. doi: 10.1111/j.1365-2958.1992.tb02193.x. [DOI] [PubMed] [Google Scholar]
- 12.Gerloff R K, Ritter D B, Wilson W O. Studies on thermal denaturation of DNA from various chlamydiae. J Infect Dis. 1970;121:65–69. doi: 10.1093/infdis/121.1.65. [DOI] [PubMed] [Google Scholar]
- 13.Goodrich J A, Schwartz M L, McClure W R. Searching for and predicting the activity sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF) Nucleic Acids Res. 1990;18:4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goosen N, van de Putte P. The regulation of transcription initiation by integration host factor. Mol Microbiol. 1995;16:1–7. doi: 10.1111/j.1365-2958.1995.tb02386.x. [DOI] [PubMed] [Google Scholar]
- 15.Hagerman P J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff S, Henikoff J G. Automated assembly of protein blocks for database searching. Nucleic Acids Res. 1991;19:6565–6572. doi: 10.1093/nar/19.23.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Zwieb C, Wu C, Adhya S. Bending of DNA by gene regulatory proteins: construction and use of a DNA bending vector. Gene. 1989;85:15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- 18.Mathews S A, Douglas A, Sriprakash K S, Hatch T P. In vitro transcription in Chlamydia psittaci and Chlamydia trachomatis. Mol Microbiol. 1993;7:937–946. doi: 10.1111/j.1365-2958.1993.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 19.Moulder J M. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman E B, Lin R. Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli. Annu Rev Microbiol. 1995;49:747–775. doi: 10.1146/annurev.mi.49.100195.003531. [DOI] [PubMed] [Google Scholar]
- 21.Pearce B, Fahr M J, Hatch T P, Sriprakash K S. A chlamydial plasmid is differentially transcribed during the life cycle of Chlamydia trachomatis. Plasmid. 1991;26:116–122. doi: 10.1016/0147-619x(91)90051-w. [DOI] [PubMed] [Google Scholar]
- 22.Raulston J E. Chlamydial envelope components and pathogen-host cell interactions. Mol Microbiol. 1995;15:607–616. doi: 10.1111/j.1365-2958.1995.tb02370.x. [DOI] [PubMed] [Google Scholar]
- 23.Riggs A D, Bourgeois S, Newby R F, Cohn M. DNA binding of the lac repressor. J Mol Biol. 1968;34:361–368. doi: 10.1016/0022-2836(68)90261-1. [DOI] [PubMed] [Google Scholar]
- 24.Sasse-Dwight S, Gralla J D. Footprinting of promoter-DNA complexes in vivo. Methods Enzymol. 1991;208:146–147. doi: 10.1016/0076-6879(91)08012-7. [DOI] [PubMed] [Google Scholar]
- 25.Schmid M B. More than just “Histone-like” proteins. Cell. 1990;63:451–453. doi: 10.1016/0092-8674(90)90438-k. [DOI] [PubMed] [Google Scholar]
- 26.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, Koonin E V, Davis R W. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 27.Stephens R S, Wagar E A, Edman U. Developmental regulation of tandem promoters for the major outer membrane protein gene of Chlamydia trachomatis. J Bacteriol. 1988;170:744–750. doi: 10.1128/jb.170.2.744-750.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stockley P G. Filter-binding assays. In: Kneale G G, editor. DNA-protein interactions. Totowa, N.J: Humana Press; 1994. pp. 251–262. [Google Scholar]
- 29.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan M, Gaal T, Gourse R L, Engel J N. Mutational analysis of the Chlamydia trachomatis rRNA P1 promoter defines four regions important for transcription in vitro. J Bacteriol. 1998;180:2359–2366. doi: 10.1128/jb.180.9.2359-2366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 32.Weinand U, Schwarz Z, Felix G. Electrophoretic elution of nucleic acids from gels adapted for subsequent biological tests: application for analysis of mRNAs from maize endosperm. FEBS Lett. 1978;98:319–323. doi: 10.1016/0014-5793(79)80208-2. [DOI] [PubMed] [Google Scholar]
- 33.Wichlan D G, Hatch T P. Identification of an early-stage gene of Chlamydia psittaci 6BC. J Bacteriol. 1993;175:2936–2942. doi: 10.1128/jb.175.10.2936-2942.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu H M, Crothers D M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984;308:509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- 35.Yuan Y, Zhang Y-X, Manning D S, Caldwell H D. Multiple tandem promoters of the major outer membrane protein gene (omp1) of Chlamydia psittaci. Infect Immun. 1990;58:2850–2855. doi: 10.1128/iai.58.9.2850-2855.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Douglas A L, Hatch T P. Characterization of a Chlamydia psittaci DNA binding protein (EUO) synthesized during the early and middle phases of the developmental cycle. Infect Immun. 1998;66:1167–1173. doi: 10.1128/iai.66.3.1167-1173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]