Abstract
Plastids, similar to mitochondria, are organelles of endosymbiotic origin, which retained their vestigial genomes (ptDNA). Their unique architecture, commonly referred to as the quadripartite (four-part) structure, is considered to be strictly conserved; however, the bulk of our knowledge on their variability and evolutionary transformations comes from studies of the primary plastids of green algae and land plants. To broaden our perspective, we obtained seven new ptDNA sequences from freshwater species of photosynthetic euglenids—a group that obtained secondary plastids, known to have dynamically evolving genome structure, via endosymbiosis with a green alga. Our analyses have demonstrated that the evolutionary history of euglenid plastid genome structure is exceptionally convoluted, with a patchy distribution of inverted ribosomal operon (rDNA) repeats, as well as several independent acquisitions of tandemly repeated rDNA copies. Moreover, we have shown that inverted repeats in euglenid ptDNA do not share their genome-stabilizing property documented in chlorophytes. We hypothesize that the degeneration of the quadripartite structure of euglenid plastid genomes is connected to the group II intron expansion. These findings challenge the current global paradigms of plastid genome architecture evolution and underscore the often-underestimated divergence between the functionality of shared traits in primary and complex plastid organelles.
Keywords: ancestral state reconstruction, euglenid, Euglenophyta, inverted repeat, plastid genome, secondary plastid
Introduction
Approximately 1.5 billion years ago, in the Proterozoic eon, eukaryotes acquired the ability of photosynthesis through endosymbiosis between a heterotrophic host and a photosynthetic cyanobacterial cell, which gave rise to primary plastids (Archibald 2015). This event changed life on Earth forever, as the ancient archaeplastid ancestor radiated into the diverse plastid-bearing lineages—land plants, and red and green algae, which subsequently spread the photosynthetic capabilities to other groups of eukaryotes via secondary endosymbioses (Howe et al. 2008; Keeling 2010). The integration of the cyanobacterial cells (and, later, primary plastid-bearing eukaryotic cells) with their new hosts involved massive endosymbiotic gene transfer from the symbiont's genome into the host nucleus, leading to extreme streamlining of the plastid genome (plastome, ptDNA) (Burki et al. 2014; Ponce-Toledo et al. 2019).
However reduced, plastid genomes are retained in a rather similar form across plastid-bearing lineages—they are almost invariably single, circular molecules, ranging from 50 to 200kbp in size, carrying a vestigial gene repertoire encompassing predominantly photosynthesis-related genes and a major part of host-independent gene expression apparatus (Maier and Schmitz-Linneweber 2004; de Vries and Archibald 2017, 2018). What is more, ptDNA organization is also conserved to some extent, with the quadripartite structure—comprising a small single-copy (SSC) and large single-copy (LSC) region, flanked by a pair of inverted repeats (IRs)—being the most typical (Palmer and Thompson 1982; Turmel et al. 2015, 2017; Zhu et al. 2016).
Still, as our state of knowledge on plastomes of the secondary plastid-bearing lineages expanded, it became evident that the quadripartite structure is strictly conserved mostly in land plants and green algae, while in others, the ptDNA architecture is substantially more diverse (Kamikawa et al. 2015, 2018; Oborník and Lukeš 2015; Turmel et al. 2015, 2017; Zhu et al. 2016; Han et al. 2019), including some truly spectacular outliers, such as linear, split into minicircles, or even branching forms (Smith and Keeling 2015). As a note, plastid IRs constitute a very particular case of a vast category of prokaryotic genetic elements collectively referred to as IRs, as they contain the ribosomal subunit genes (rrn16, rrn23, and rrn5), as well as transfer RNA genes and, in many taxa, protein-coding genes. In the following work, the phrase “inverted repeats” will always refer to the plastid-exclusive type of this structure (Turmel et al. 2017; Lavi et al. 2018).
The current theory states that the role of the IRs in plastomes is mainly genome stabilization—they constitute a part of the machinery for DNA repair via homologous recombination, which, in turn, is proposed to be responsible for both the lower substitution rate and less frequent genome rearrangements in IR-bearing plastid genomes in comparison with IR-deficient ones (Palmer and Thompson 1982; Maréchal and Brisson 2010; Zhu et al. 2016; Turmel et al. 2017; Jin et al. 2020). Nonetheless, studies of these phenomena have thus far been limited to the primary plastid-bearing taxa (e.g., land plants and chlorophytes), while others, even despite the abundance of genomic data on these organisms, remain rather neglected, although with a prominent exception of singular analyses of complex red plastid-bearing cryptophytes and haptophytes, pointing toward the lack of recombination between plastid-encoded rDNA copies in these groups (Hovde et al. 2014; Méndez-Leyva et al. 2019).
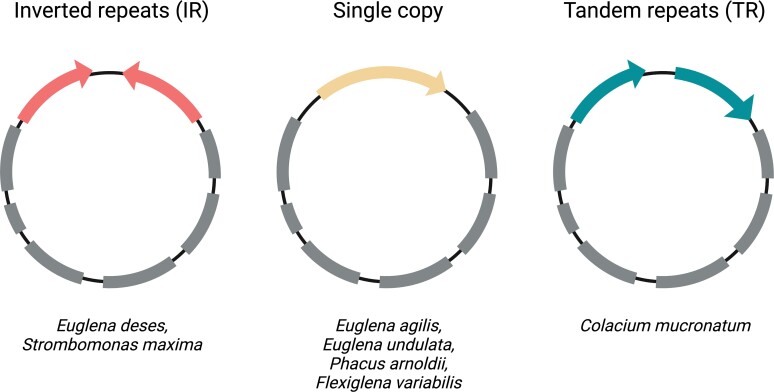
A perfect example of a secondary plastid-bearing group of algae, constituting a showcase for ptDNA architecture diversity, are the photosynthetic euglenids (Euglenophyta). This rather small (comprising below 20 genera, divided into three families—Euglenaceae, Phacaceae, and Eutreptiaceae) and relatively young (plastid acquisition is estimated to have occurred between 539 and 652 million years ago [Jackson et al. 2018]) group of algae has attracted researchers’ attention for centuries due to their immense abundance in the freshwater environments and captivating morphology (Marin et al. 2003; Novák Vanclová et al. 2020; Kostygov et al. 2021). What is more, the earliest studies of their plastid genomes revealed an array of unique hallmark traits, such as multiple tandemly repeated copies of the ribosomal operon or explosive group II intron expansion, prompting further investigation, which has so far resulted in over 30 full or partial ptDNA sequences of euglenophytes having been published up to date. These, in turn, made it possible to describe other features of divergent evolution in this group, such as the horizontal acquisition of maturase genes, but also a rather broad variability of plastid genome structure, including three main types of rDNA repeat organization: single copy, IRs, and tandem repeats (Bennett et al. 2012; Wiegert et al. 2012; Karnkowska et al. 2018; Maciszewski et al. 2022).
As transitions between organization types and their evolutionary consequences in euglenid plastid genomes remain documented, but not deeply investigated, we aimed to focus on this aspect of plastome evolution in the following study. Thus, we have selected seven species of freshwater photosynthetic euglenids (Euglenales), whose positions are close to the known nodes on their phylogenetic tree on which ptDNA structure rearrangements have most likely occurred, and sequenced their plastid genomes in order to broaden our scope of investigation for euglenids as a model group for studying plastome evolution and, having combined the new data with the aforementioned substantial body of reference, to test the long-standing hypothesis on the correlation between IR conservation and diminished mutation rate outside of the primary plastid-bearing organisms.
Results and Discussion
Plastid Genome Characteristics and Phylogeny
The basic characteristics of the seven new ptDNA sequences of freshwater euglenophytes have been shown in table 1, and their structure has been depicted on supplementary figure S1, Supplementary Material online, with schematic depiction of their rDNA organization variants shown on figure 1. As expected, based on the past studies (Bennett et al. 2012; Karnkowska et al. 2018; Maciszewski et al. 2022), the sequenced euglenid plastid genomes do not exhibit vast diversity of genetic repertoire—ranging from 88 genes in Euglena undulata to 100 in Strombomonas maxima—with the variable numbers of rDNA operon copies and group II intron maturase genes accounting for almost all of the differences in gene content between the investigated taxa.
Table 1.
Characteristics of the Seven Novel Plastid Genomes of Euglenales Presented in This Study.
| Colacium mucronatum | Euglena agilis | Euglena deses | Euglena undulata | Flexiglena variabilis | Phacus arnoldii | Strombomonas maxima | |
|---|---|---|---|---|---|---|---|
| Length (bp) | 147,974 | 107,929 | 83,748 | 92,487 | 105,412 | 84,299 | 185,621 |
| GC content | 25.6% | 26.5% | 26.5% | 27.3% | 27.3% | 25.6% | 27.5% |
| Total no. of genes | 94 | 93 | 94 | 88 | 94 | 95 | 100 |
| Protein-coding genes | 60 | 62 | 61 | 58 | 63 | 63 | 68 |
| tRNA genes | 29 | 28 | 27 | 28 | 28 | 29 | 27 |
| rRNA genes | 5 | 3 | 6 | 2 | 3 | 3 | 6 |
| Introns | 119 | 97 | 72 | 78 | 85 | 67 | 130 |
| Total intron length (bp) | 62,991 | 40,630 | 21,630 | 29,591 | 47,276 | 24,492 | 117,955 |
| Accession | OP179277 | OP179278 | OP179279 | OP179280 | OP179281 | OP179282 | OP179283 |
NOTE.—Numbers and length of introns do not include twintrons.
Fig. 1.
Schematic representation of the three rDNA operon organization types in euglenid plastomes, with species investigated in this study listed as carrying the respective structures. Arrows represent rDNA copies and orientation; undirected sections represent protein-coding genes of the large single-copy region of the plastid genome (created with BioRender.com).
In contrast, the investigated strains displayed substantial differences in total ptDNA size, ranging from approximately 83.7 kb in Euglena deses to over twice that size, 185.6 kb, in S. maxima. What is more, plastomes of S. maxima and Colacium mucronatum, both examined in our study, are currently the two largest among all euglenophytes, with the latter (147.9 kb) also exceeding the size of all previously published plastid genomes of this group. However, the number of functional genes (i.e., protein-coding genes as well as tRNA and rRNA genes) are not even a noticeable factor influencing total ptDNA size in euglenophytes—although the plastid genome of S. maxima is indeed the most gene-rich among the seven new ptDNA sequences, no such trend was visible among other examined plastomes. Instead, the total plastid genomes size differences can be almost entirely attributed to non-coding sequences—in particular, the group II introns, whose explosive expansion is among the most distinctive traits of euglenid plastids (Karnkowska et al. 2018; Maciszewski et al. 2022).
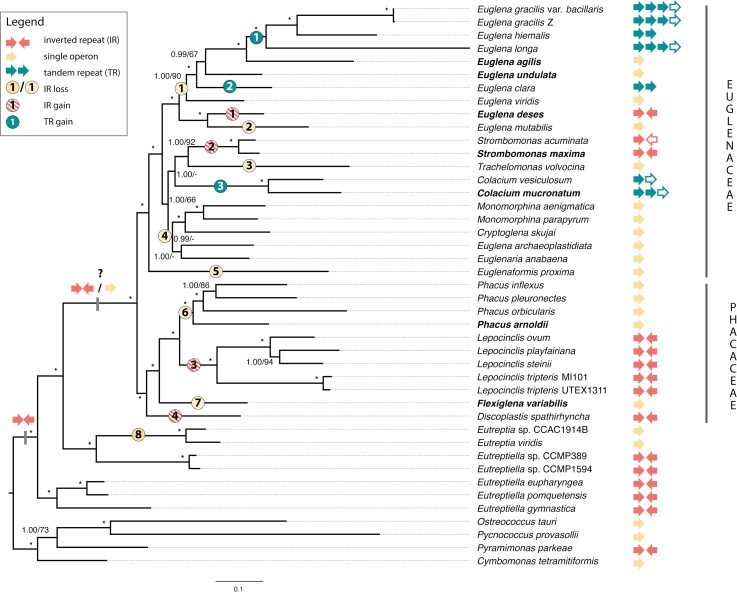
The plastid genome-based phylogeny of euglenophytes, obtained in our study (fig. 2), is almost fully congruent with the most recent nuclear and plastid rDNA-based reconstructions (Kim et al. 2015; Karnkowska et al. 2018; Maciszewski et al. 2022). Only one minor discrepancy was observed: in our reconstruction, Euglena longa is a sister taxon to a clade comprising Euglena gracilis and Euglena hiemalis, instead of being sister to only E. hiemalis. This, however, does not impact the results of our further analyses, as they relate to clades which possess identical values for traits studied in our work. Combined with the predominantly absolute or very high bootstrap support and posterior probability values for the plastid-based phylogeny presented here, as well as fully congruent topology between Bayesian and maximum likelihood reconstructions in this study and the previous works (Linton et al. 2010; Karnkowska et al. 2014, 2018; Maciszewski et al. 2022), it is reasonable to assume that the euglenophyte phylogeny shown on figure 2 is the most stable and credible one up to date. Nonetheless, certain discrepancies between phylogenies based on molecular markers of diverse origin (nuclear vs. organellar), sequence type (nucleotide vs. protein), and alignment size (single genes vs. concatenated multigene matrix) are to be expected.
Fig. 2.
Plastid-based phylogenomic tree of Euglenophyta. Species names in bold indicate organisms whose ptDNA was first sequenced in this work. rDNA operon copy number (filled arrows denote complete operon copies; empty ones denote incomplete ones) and orientation are shown on the tree tips. Ancestral IR presence in euglenophytes is marked at the corresponding node; uncertain rDNA organization is marked at the node corresponding to the last common ancestor of Euglenaceae and Phacaceae. Full dots at the tree branches denote undisputed state transitions; striped dots at the tree branches denote hypothetical, mutually exclusive scenarios of only IR gains or only IR losses within Euglenaceae and Phacaceae. Bayesian posterior probability and bootstrap support values above 50 are shown at the nodes. Asterisks (*) denote absolute probability and support (>0.99/>95).
The Conundrum of Losses Versus Gains of the rDNA Copies
As demonstrated in past studies, ptDNA organization in euglenophytes has undergone substantial diversification over time, encompassing not only the group II intron expansion, but also gains and losses of rDNA copies—the most recent reconstruction suggests three independent losses of one of the IRs: in the ancestors of genera Eutreptia and Phacus, and in the common ancestor of the family Euglenaceae (Karnkowska et al. 2018). Moreover, certain species of the genus Euglena have acquired additional copies of the ribosomal operon, situated consecutively in the same orientation in the genome, resulting in a unique genetic structure, commonly referred to as tandem repeats (see fig. 1; Hallick et al. 1993; Gockel and Hachtel 2000; Hewadikaramge and Linton 2018). An rDNA operon organization similar to euglenid TRs has only been documented in the non-photosynthetic plastid of an Apicomplexa-like parasite Piridium sociabile; the second repeat, however, constitutes only a partial 23S rRNA gene, indicating its remnant, non-functional character (Mathur et al. 2019). Strikingly, the organization of the seven new euglenid plastid genomes, when mapped onto the studied group's phylogeny (fig. 2), challenges nearly all assumptions of the previously proposed model of three IR losses and a single TR gain.
First of all, plastid genomes of Euglena agilis and E. undulata—both of which are situated within a clade of certain Euglena spp. previously proposed to possess TRs, such as E. gracilis, E. hiemalis and Euglena clara (see fig. 2)—carry a single rDNA copy, indicating either two independent gains of a TR within the genus Euglena, or alternatively (and less parsimoniously), a gain and two independent losses. Moreover, we identified yet another independent gain of tandem repeats, this time outside of the genus Euglena, specifically: in C. mucronatum, which strongly indicates that single-copy rDNAs in euglenid plastids are quite likely to undergo duplication, forming TRs. This assumption is also supported by the observation that the TR copy number is also varied among Euglena and Colacium spp.—E. clara and E. hiemalis possess two full copies, while C. mucronatum possesses “two and a half” (two full copies and an additional rrn16 gene), and E. gracilis and E. longa possess “three and a half” copies (three full copies and an additional rrn16 gene). It is also worth mentioning that our study is not the first to obtain a ptDNA sequence of a representative of the genus Colacium; however, the published sequence from Colacium vesiculosum is incomplete, with the missing part most likely including a part of a ribosomal operon repeat, which would make any conclusions on IR/TR evolution based on that sequence at least dubious (Wiegert et al. 2013).
Secondly, E. deses and S. maxima—both representing Euglenaceae, which were proposed to have ancestrally lost the inverted rDNA repeats—do, in fact, possess IRs. This finding is particularly puzzling because of the position of these two species on the phylogenetic tree, indicating that, for the observed IR loss and retention pattern to appear, there must have been six independent IR losses within the Euglenaceae alone: in the ancestor of E. gracilis/hiemalis/longa/agilis/undulata/clara/viridis, in Euglena mutabilis, in the genus Trachelomonas, in the genus Colacium, in the genus Euglenaformis, and in the ancestor of the genera Monomorphina, Cryptoglena, and Euglenaria. Bearing this in mind, the ancestral IR loss in Euglenaceae (as proposed by Karnkowska et al. 2018; see fig. 2), followed by two independent regains in E. deses and S. maxima, would be a substantially more parsimonious explanation for the pattern observed in the extant species.
Last, but not least, we found Flexiglena variabilis—a representative of the newest described euglenophyte genus, Flexiglena (Łukomska-Kowalczyk et al. 2021)—to possess a single rDNA copy, even though its phylogenetic position among predominantly IR-bearing taxa (Lepocinclis and Discoplastis spp.; see fig. 2) suggested that it is rather likely to carry IRs as well. In contrast with the other freshwater euglenophyte family, in Phacaceae the observed IR loss and retention pattern has two comparably likely explanations: either two independent IR losses in Phacus and Flexiglena and their retention in Lepocinclis and Discoplastis, or two independent gains in Lepocinclis and Discoplastis and retention of the ancestrally IR-less state in Phacus and Flexiglena. However, assuming that Euglenaceae were ancestrally IR-less as outlined above, a single loss in the ancestor of freshwater euglenophytes (Euglenales—Euglenaceae + Phacaceae; see the systematics in Kostygov et al. 2021) and subsequent regains (twice in Euglenaceae and twice in Phacaceae) would be a possible hypothetical scenario of the evolutionary path of the rDNA operon copies in the investigated group.
The Original ptDNA Organization in the Euglenophyte Ancestor and the Revised History of Transitions
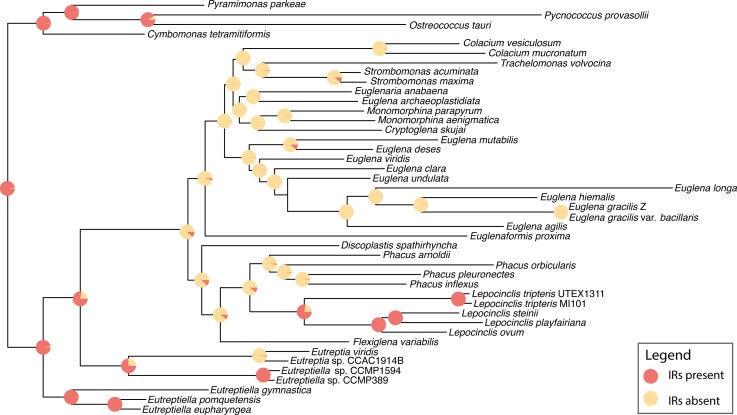
To unravel the convoluted history of ribosomal operon organization in euglenid plastids, we performed computational reconstruction of the ancestral states on the investigated group's phylogeny (fig. 3). Among the four tested models for transition rates between IR presence and absence, the model with unequal transition rate has been selected as best-fitting for the dataset with Akaike information criterion (AIC) value of 45.04, closely followed by a model with equal transition rate (AIC = 46.91), while unidirectional transition models were significantly worse (both with AIC = 2 × 105). The reconstruction produced no ambiguous ancestral states on any node on the tree, with the IR presence in the ancestor of all euglenid plastids reconstructed at >99% probability.
Fig. 3.
Ancestral state reconstruction of plastid genome organization in Euglenophyta, mapped onto the group's phylogeny (see fig. 2). State transition rate was preset as unequal (ARD model), and the transition rates were calculated based on empirical data. Pie charts at the nodes represent the calculated probabilities of the respective states.
Moreover, the reconstructed states at the other nodes point toward the hypothesis outlined before: that the IRs were lost in euglenophytes only twice—in the ancestor of all freshwater euglenids (Euglenales), and in the ancestor of the genus Eutreptia (see fig. 3)—and were subsequently regained independently by E. deses, S. maxima, Discoplastis spathirhyncha (or the genus Discoplastis—however, the lack of ptDNA sequences of its other species makes it impossible to determine), and the genus Lepocinclis. This stands in opposition to the hypothesis that IR gains, in contrast to losses, are very rare, and the only piece of irrefutable evidence for such an occurrence ever taking place comes from the green algal genus Chamaetrichon, whose peculiar plastid genome possesses three IRs (Turmel et al. 2015, 2017). Still, it is worth noting that IR regains in plastid genomes have been proposed in recent studies of land plants (particularly the genera Medicago and Melilotus), some of which even suggest specific molecular mechanisms, such as double-strand break repair systems, to be responsible for this occurrence (Choi et al. 2019; Wu et al. 2021).
Nonetheless, it is necessary to underscore that our findings do not suggest the IR losses and gains to be equally likely events. Quite the contrary, we provide statistically supported estimation of the likelihood of IR gain compared to IR loss, which, according to the empirically-determined state transition rate in the ancestral state reconstruction using ARD model (yielding the lowest AIC value; see the first paragraph of this chapter), is approximately 0.766:1. Bearing in mind that an IR gain is most certainly a more mechanically complex occurrence than IR loss, this ratio might seem unreasonably high; however, this value was obtained in an analysis of only 43 data points with a total of six state transitions. Therefore, with more plastid-bearing lineages taken into account, the result might be substantially different, especially considering that the molecular mechanism underlying the transitions in rDNA arrangement has never been observed in action in any lineage; until such a mechanism is documented, any state transition likelihood can only be treated as a theoretical approximation.
On the other hand, the IRs of euglenids are substantially different from their counterparts in primary plastids of plants or green algae. While the IRs in plastid genomes usually flank two single-copy regions containing most of the protein-coding genes and tRNA genes, those in euglenophytes are almost always adjacent to each other—the small single-copy region is very short and has no coding content, and the entire protein-coding gene repertoire is located in the large single-copy region (Karnkowska et al. 2018; Maciszewski et al. 2022). Only one exception has been documented so far—Eutreptiella gymnastica, where one of the repeats is split, and a six-gene insertion separates the rrn16 and rrn23 genes, thus constituting a unique kind of a small single-copy region (Hrdá et al. 2012). Nonetheless, despite the presence of IRs in some lineages, none of the euglenid plastid genomes actually carries a true quadripartite structure.
Additionally, all euglenid IRs have identical gene content of the ribosomal operon and two tRNA genes, in contrast with the substantially more complex IRs known from green algae or raphidophytes, which encompass a wide array of protein-coding genes (Turmel et al. 2015, 2017; Lemieux et al. 2016; Kim et al. 2022). Although it is quite difficult to state whether the euglenophyte plastids’ IRs are secondarily simplified, or that those in extant plants and algae gained their complexity late in their evolution and the simplicity is actually plesiomorphic, the adjacent position and small size of euglenid IRs makes it more plausible that they are products of numerous independent duplications. Moreover, this complete lack of content diversity among euglenid IRs does not corroborate the hypothesis that incorporation of foreign genetic elements (e.g., viral genes or group II introns) into the IRs might be a factor involved in their destabilization and loss (Lemieux et al. 2016).
Finally, studies of the plastid genomes of the lycophyte genus Selaginella have unveiled the possibility of IR reinversion, forming direct repeats (DR), where the rDNA operon copies are present in the same orientation, but do not lie consecutively in the genome, as in case of euglenid TRs (Mower et al. 2019; Zhang et al. 2019). It would be tempting to hypothesize that IR regains in euglenophyte plastomes might in fact be TR reinversions, or vice versa—TR acquisition in euglenids may be an analogous occurrence to DR acquisition in Selaginella, especially considering that all repeat variants in euglenid ptDNA are adjacent to one another. However, in contrast with the lycophytes, there are no IR-bearing taxa sister to TR-bearing ones among euglenophytes—that is the IR- and TR-bearing species are phylogenetically separated by taxa bearing single rDNA copies (see fig. 1)—which, unfortunately, makes the elegant hypothesis of IR/TR transition fluidity poorly supported in this particular group.
IR Presence Does Not Impact the Rate of Evolution of Either Protein-coding Genes or rRNA Genes
As the organization of euglenophyte ptDNA seems to have undergone numerous rearrangements over time, there does not seem to be strong evolutionary pressure for retention of any particular organization type. Previous studies have shown that the IRs are conserved in plastid genomes due to their stabilizing activity as homologous recombination sites, as is evident from the substantially increased rate of sequence evolution and genomic rearrangements observed in IR-less taxa (Palmer and Thompson 1982; Zhu et al. 2016; Claude et al. 2022). However, these analyses took only the data from primary plastids into account, leaving the diverse secondary plastid-bearing taxa understudied in this regard. Thus, given that there is a growing body of evidence for dissimilarity of evolutionary dynamics and selection intensity between primary and complex plastids (Uthanumallian et al. 2021), the missing piece of the puzzle may be tremendously important. To fill this information void, we have undertaken an analysis of the sequence evolution rate in ribosomal subunit genes, which form the bulk of the length of the plastid IRs, as well as in protein-coding genes encoded outside of the IRs in the IR-bearing and IR-less plastomes of Euglenophyta.
We calculated dN/dS ratios based on the sequences of 58 plastid protein-coding genes in euglenid plastids, and obtained mean and standard deviation values for IR-bearing and IR-less taxa: dN/dS = 0.158 ± SD = 0.0378, and dN/dS = 0.197 ± SD = 0.0653, respectively. These values were compared using Mann–Whitney U test, which yielded the z-score of −0.159 and the P-value of 0.436, which, quite interestingly, can be clearly interpreted as no difference between the investigated groups. Furthermore, our analysis of the rate of rDNA evolution—the small and large ribosomal subunit genes, which form the IRs—produced congruent results in both studied variants of the rDNA-based phylogeny (table 2), indicating no differences in the rate of evolution between the ribosomal subunit genes enclosed within IRs and ones present in single copies. These results together, being contradictory with analogical studies of green algae (Zhu et al. 2016) and land plants (Ping et al. 2021; Claude et al. 2022), constitute yet another key difference in the evolutionary paths of primary and secondary plastids and their genomes, pointing towards the loss of the stabilizing function of the IRs in this particular secondary plastid-bearing lineage.
Table 2.
Comparison of the Rates of Evolution of rRNA Genes in IR-bearing and IR-less Euglenid Plastid Genomes Estimated via Phylogenetic Tree Branch Length Analysis. P-values represented in bold.
| Constrained phylogeny | De novo-reconstructed phylogeny | |
| Mean branch length ± SD | ||
| IR-bearing | 0.0492 ± 0.0644 | 0.0473 ± 0.0604 |
| IR-less | 0.0543 ± 0.0577 | 0.0539 ± 0.0570 |
| Mann–Whitney U test's z-score; P-value | ||
| 0.309; 0.757 | 0.127; 0.897 | |
If the presence of the IRs does not impact the rate of evolution of both protein-coding and rRNA genes in the analyzed plastid genomes, it is only logical to assume that there is no discernible consequence to IR loss or retention in euglenophytes at all. We are therefore inclined to hypothesize that, contrary to primary plastid-bearing taxa, euglenid plastid genome organization follows the path of neutral evolution, with spontaneous rearrangements, such as rDNA copy gains or losses, being only retained as a result of the absence of the selective pressure to keep a specific genome architecture or, as suggested in recent works, as an artifact of ptDNA replication (Choi et al. 2019). Naturally, a conclusion that secondary plastids’ genome structure only evolves neutrally would be unsupported due to the lack of published results of analogical rate of evolution versus ribosomal operon organization analyses for taxa other than euglenids—to determine that, further studies are necessary.
Still, the euglenid plastid genome IRs remain a rather puzzling evolutionary peculiarity when a broader context is considered. Previous studies have demonstrated that IRs are diverse genetic elements, ranging from several to thousands of nucleotides in length, spread across the entire tree of life, with multifaceted influence on the genome structure and evolution due to their capabilities for forming hairpins, and constituting the sites for flip-flop recombination and template switching during DNA replication. As a result, certain forms of IRs can have either generally stabilizing influence on the genome as constituents of DNA repair systems, as demonstrated in plant plastids, or, on the contrary, destabilizing impact as hotspots for mutations, as shown in bacteria and eukaryotic nuclei (Maréchal and Brisson 2010; Turmel et al. 2017; Lavi et al. 2018). Therefore, it comes as a great surprise that in a certain genetic setting, IRs can have no noticeable impact on the genome's evolution whatsoever. Moreover, taking into account that bacterial genomes can carry thousands of IR pairs, albeit very short, the curiosity lies not just in their retention in ptDNA, but in the fact that only a single pair is retained (Lavi et al. 2018).
It is also worth noting that IR losses in certain complex plastid-bearing algae, such as cryptophytes, have been proposed to be concomitant with loss of photosynthesis. This hypothesis is particularly interesting due to the presence of numerous parallels between euglenid and cryptophyte plastids, despite their different origins and vast evolutionary distance between the host lineages—for example, independent group II intron expansion and acquisition, followed by partial degeneration, of intron-encoded maturase genes (Maciszewski et al. 2022; Suzuki et al. 2022). A connection between IR decay and a shift to heterotrophy is not likely in the case of euglenids, since most of them lost IRs, while losses of photosynthesis in this lineage are comparably scarce. However, a possible link between the accumulation of a novel kind of dispersed repeats in the form of group II introns, and IR degeneration due to induced plastome instability—and, additionally, the acquisition of new homologous recombination sites which turned IRs redundant—is certainly a plausible explanation for the cooccurrence of these two rather uncommon traits both in euglenid and cryptophyte plastids (Lee et al. 2021; Suzuki et al. 2022). Unfortunately, a solid proof for a link between group II intron expansion and IR redundancy is currently out of our reach, as it would require reference data from intron-less euglenid ptDNA, which have never been identified, while using data from pyramimonadalean green algae (the closest extant relatives of the euglenid plastid) could lead to erroneous conclusions due to the documented shift in the rate of plastid genome evolution following an endosymbiotic event (Uthanumallian et al. 2021).
Conclusions
In the presented work, we obtained full plastid genome sequences of seven species of freshwater photosynthetic euglenids, selected according to their phylogenetic positions within or adjacent to taxa known to have undergone ptDNA structure rearrangements in order to investigate the evolutionary dynamics of the genome organization within this model secondary plastid-bearing group. Only Phacus arnoldii simply shared the plastome structure of their closest relatives; however, we found many of the studied species to have divergent ribosomal operon organization—E. agilis and E. undulata, members of a TR-bearing clade, have a single rDNA copy, while C. mucronatum, located within a TR-less clade, has secondarily acquired tandem repeats. Similarly, E. deses and S. maxima, both located within IR-less clades, possess IRs, while F. variabilis, branching within a predominantly IR-bearing clade, does not have them.
Our findings have demonstrated that the variability of ptDNA organization in euglenophytes, despite being studied before, is even more immense than previously thought, and that the scenario of independent rDNA IR regains in this lineage should not be ruled out. The proposed scenario of up to four independent secondary acquisitions of IRs not only challenges the current status quo on the unlikelihood of formation of IRs de novo, but also impacts the more general theory of progressing reductive evolution of organellar genomes by constituting a prominent example of an increase, and not decrease, in an organellar genome's complexity.
We therefore suggest that the reason behind the tremendous diversity in the architecture of the repeated ribosomal operon sequences lies in their partial loss of function: the secondary plastid of euglenophytes did not inherit the IR recombination-based repair mechanism, acting in the primary plastids of the Archaeplastida, and therefore the retention of IRs themselves offers little, if any, selective advantage. As a result, the photosynthetic organelle and its host do not bear any serious evolutionary consequences of gains and losses of additional copies of short RNA-encoding genes, leading to formation of a multitude of divergent forms. Moreover, if other recombination sites, such as group II introns, have been introduced into the genome, the loss of redundant IRs might even be beneficial.
Our understanding of the processes shaping the organellar genome structure and contents is, however, still limited. The next step worth taking to deepen it would be the broadening of the scope of the research subject by analyzing the genome structure and rates of evolution in other, especially more diverse complex plastid-bearing taxa, such as haptophytes or dinoflagellates. Subsequently, once more data is available, the plastid genome dynamics could be cross-referenced with the unique biological, physiological and genetic traits of different organisms with independently acquired plastids, which would help unravel the real significance of the evolutionary transformations investigated in this study. Furthermore, the intertwined influences of intron expansion, IR degeneration and photosynthesis losses would undoubtedly merit further investigation, and the mechanism which would compensate for the loss of the postulated key ptDNA repair system in other, intron-less plastid genomes still awaits discovery.
Materials and Methods
Research Subjects, Cultivation and Isolation of the Genetic Material
For the purpose of this work, we cultivated seven strains of photosynthetic freshwater euglenids: E. agilis ACOI 2790, E. deses CCAP 1224/20, E. undulata MI03, S. maxima ACOI 2992, C. mucronatum SAG 1211-1, P. arnoldii ASW08064, and F. variabilis Boża Wola strain (environmental isolate). Optimal growth of the microorganisms was observed on liquid S2T2 medium prepared according to the recipe on the ACOI culture collection website (http://acoi.ci.uc.pt), supplemented with 4 μg/ml of vitamin B12 and a single autoclaved pea seed (Pisum sativum), maintained in a room temperature light cabinet with 16/8 h light/dark cycle in 10-ml glass tubes. Satisfactory culture density was determined by microscopic observations, after which the cultures were centrifuged for 3 min at 3,000 rpm.
Total DNA isolation from cell pellets was performed using DNeasy Blood & Tissue Kit (QIAGEN, USA) according to the manufacturer's protocol, including an additional step of RNA digestion using RNase A. Quality control of the obtained isolates was carried out via spectrophotometric analysis using an Implen NP80 NanoPhotometer (Implen GmbH, Germany).
High-throughput DNA Sequencing and Plastid Genome Assembly
Total DNA samples of the seven euglenid strains were handed to an external company (Genomed S.A., Warsaw, Poland) for high-throughput sequencing using Illumina MiSeq platform. The sequencing yielded paired-end reads of different lengths, depending on the library preparation method used: 250 bp for E. agilis (approximately 7.2 million reads), E. deses (4.4 million reads), P. arnoldii (8.2 million reads), and S. maxima (4.0 million reads), and 300 bp for C. mucronatum (5.6 million reads), E. undulata (4.2 million reads), and F. variabilis (3.5 million reads). Quality control of the sequencing libraries was carried out using FastQC v0.11.6 tool (Andrews, 2010), and data trimming (i.e., removal of the Illumina Universal Adapter sequences) was performed using Trimmomatic v0.39 tool (Bolger et al. 2014).
Initial genome assembly of all datasets was performed using SPAdes v3.15.2 (Prjibelski et al. 2020), followed by identification of plastid genome fragments among the assembled contigs via the BLASTN algorithm (Altschul et al. 1990) using publicly available euglenid ptDNA sequences as queries. Largest contigs identified as plastid genome fragments were extracted and used as seed sequences for plastid genome assembly using NOVOPlasty v4.3.1 (Dierckxsens et al. 2017). Although all plastid genomes have been successfully circularized, additional quality control was employed to verify their completeness: all plastid genome hits were extracted and visualized in Bandage v0.8.1 software (Wick et al. 2015) to confirm the circularization of the assembly; additionally, raw reads were mapped onto the NOVOPlasty assemblies using Bowtie v2.2.6 (Langmead and Salzberg 2012) and Samtools v1.6 (Li et al. 2009) and the coverage per nucleotide position was calculated using Bedtools v2.25.0 (Quinlan and Hall 2010) to detect putative low-coverage regions which would indicate misassembly.
Plastid Genome Annotation and Visualization
Annotation of the obtained ptDNA sequences was carried out in Geneious Prime v2022.1.1 software (https://www.geneious.com) using Live Annotate & Predict toolkit (Find ORFs and Annotate From… features), utilizing a manually constructed database of all published euglenid plastid genomes as reference data for gene annotations. Identities and exon boundaries of all protein-coding genes were confirmed by cross-referencing with the NCBI non-redundant protein database (NCBI-nr) via the BLASTX algorithm (Altschul et al. 1990) and the PFAM 35.0 protein families’ database (pfam.xfam.org) using the browser-accessible internal HMM search feature (Mistry et al. 2021). Plastid genome maps were generated using the OGDraw v1.3.1 online tool (Greiner et al. 2019).
Plastid Genome-based Phylogenomic Analysis
58 protein-coding genes of non-ambiguous origins and function (i.e., excluding intron maturase genes, such as roaA, ycf13, or mat2/4/5/6/7) were extracted from the annotated ptDNA sequences, translated to amino acid sequences, and combined with their homologs from 32 published plastid genome sequences of Euglenophyta and four published plastid genome sequences of Pyramimonadales (Chlorophyta). Protein sequences were aligned using the L-INS-i method in MAFFT v7.310 (Katoh and Standley 2013), and the single-gene alignments were concatenated using catsequences script (https://github.com/ChrisCreevey/catsequences) to produce a data matrix with total length of 18,143 amino acids.
The concatenated alignment was used as the input for phylogenetic analyses via a maximum likelihood method implemented in IQ-TREE v2.0.6 software (Minh et al. 2020), and via the Bayesian inference method implemented in MrBayes v3.2.6 (Ronquist et al. 2012). Maximum likelihood phylogeny reconstruction used a partitioned matrix with automatic substitution model selection for each partition (-m TEST parameter), and 1,000 non-parametric bootstrap replicates. Bayesian reconstruction used a non-partitioned dataset with preset sequence evolution model (cpREV), with 1,000,000 generations (including 250,000 generations burn-in), after which convergence of the Markov chains was achieved. Both methods yielded fully congruent tree topology.
Ancestral State Reconstruction
Reconstruction of ancestral states of plastid genome organization (IR-bearing vs. IR-less) was carried out using corHMM v2.7 package (Beaulieu et al. 2013) in R v4.1.3, implemented in R Studio 2022.02.0 Build 443 (RStudio Team, 2020). IR presence was encoded as a binary trait, mapped (and plotted via plotRECON command) onto the tree topology obtained via Bayesian reconstruction. Four manually constructed substitution matrices were tested: with equal rate of state transition, with unequal rate of state transition, with only 0→1 transition possible, and with only 1→0 transition possible.
Rate of Evolution Estimation
For protein-coding genes, codon alignments for all single gene clusters were prepared using PAL2NAL v14 software (Suyama et al. 2006). Rates of synonymous and non-synonymous substitutions (dN/dS) for all gene alignments were calculated using CodeML tool implemented in PamlX v1.3.1 toolkit (Xu and Yang 2013). Mean dN/dS values were calculated for two groups of euglenophytes: IR-bearing (13 taxa) and IR-less (26 taxa) for all 58 genes, and compared using two-sided Mann–Whitney U test implemented in an online Social Science Statistics calculator (https://www.socscistatistics.com/tests/mannwhitney/).
For rRNA genes, nucleotide sequence alignments were prepared using L-INS-i method in MAFFT v7.310 (Katoh and Standley 2013), and the two alignments (rrn16, rrn23) were concatenated using catsequences script (https://github.com/ChrisCreevey/catsequences) to produce a data matrix with total length of 5,954 nucleotides. The concatenated alignment was used as the input for phylogenetic analysis via maximum likelihood method implemented in IQ-TREE v2.0.6 software (Minh et al. 2020) with automatic substitution model selection (-m TEST parameter), and 1,000 non-parametric bootstrap replicates, in two variants: with no constraints, and with constrained topology based on the results of the plastid protein-coding genes-based phylogeny. For both phylogenies, mean branch length values were calculated for branches divided into two groups: reconstructed as IR-bearing (19 branches in total) and reconstructed as IR-less (52 branches in total). Branches at which state transitions occurred were not taken into account. Mean values were subsequently compared using two-sided Mann–Whitney U test implemented in an online Social Science Statistics calculator (https://www.socscistatistics.com/tests/mannwhitney/).
Supplementary Material
Acknowledgments
This work was supported by National Science Centre, Poland (Preludium grant 2018/31/N/NZ8/01840 to KM), Ministry of Science and Higher Education, Poland (Excellence Initiative—Research University/IDUB grant to AF), and the European Molecular Biology Organization (EMBO Installation Grant 4150 to AK and Ministry of Education and Science, Poland). We would like to sincerely thank our colleagues from the Institute of Evolutionary Biology (University of Warsaw, Poland): Jakub Baczyński, for his invaluable support in ancestral state reconstruction analyses, and Bożena Zakryś, for supporting us with her great experience and expertise in maintenance of algal cultures.
Contributor Information
Kacper Maciszewski, Institute of Evolutionary Biology, Faculty of Biology, Biological and Chemical Research Centre, University of Warsaw, Żwirki i Wigury 101, 02-089 Warsaw, Poland.
Alicja Fells, Institute of Evolutionary Biology, Faculty of Biology, Biological and Chemical Research Centre, University of Warsaw, Żwirki i Wigury 101, 02-089 Warsaw, Poland.
Anna Karnkowska, Institute of Evolutionary Biology, Faculty of Biology, Biological and Chemical Research Centre, University of Warsaw, Żwirki i Wigury 101, 02-089 Warsaw, Poland.
Supplementary material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
K.M. and A.K. conceptualized the study, obtained the funding, interpreted the results, and prepared the final version of the manuscript; A.F. assembled and annotated the plastid genome of Flexiglena variabilis; K.M. formulated the hypotheses, assembled, and annotated the other six plastid genomes, performed the bioinformatic analyses, and prepared the draft version of the manuscript with figures and tables; A.K. supervised the work.
Data Availability Statement
Euglenid plastid genome sequences obtained in this study have been deposited in the NCBI GenBank database under accession numbers: OP179277-OP179283. Additional supplementary data, Supplementary Material online, including the plastid genome-derived concatenated protein alignments and morphological data matrix used in this study, have been deposited in FigShare repository: https://doi.org/10.6084/m9.figshare.20486148.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: A quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [WWW Document].
- Archibald JM. 2015. Genomic perspectives on the birth and spread of plastids. Proc Natl Acad Sci USA. 112:10147–10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, O’Meara BC, Donoghue MJ. 2013. Identifying hidden rate changes in the evolution of a binary morphological character: the evolution of plant habit in campanulid angiosperms. Syst Biol. 62:725–737. [DOI] [PubMed] [Google Scholar]
- Bennett MS, Wiegert K, Triemer RE. 2012. Comparative chloroplast genomics between Euglena viridis and Euglena gracilis (Euglenophyta). Phycologia. 51:711–718. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Imanian B, Hehenberger E, Hirakawa Y, Maruyama S, Keeling PJ. 2014. Endosymbiotic gene transfer in tertiary plastid-containing dinoflagellates. Eukaryot. Cell. 13:246–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi IS, Jansen R, Ruhlman T. 2019. Lost and found: return of the inverted repeat in the legume clade defined by its absence. Genome Biol Evol. 11:1321–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claude SJ, Park S, Park S. 2022. Gene loss, genome rearrangement, and accelerated substitution rates in plastid genome of Hypericum ascyron (Hypericaceae). BMC Plant Biol. 22:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Archibald JM. 2017. Endosymbiosis: did plastids evolve from a freshwater cyanobacterium? Curr Biol. 27:R103–R105. [DOI] [PubMed] [Google Scholar]
- de Vries J, Archibald JM. 2018. Plastid genomes. Curr Biol. 28:R336–R337. [DOI] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gockel G, Hachtel W. 2000. Complete gene map of the plastid genome of the nonphotosynthetic euglenoid flagellate Astasia longa. Protist. 151:347–351. [DOI] [PubMed] [Google Scholar]
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47:W59–W64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallick RB, Hong L, Drager RG, Favreau MR, Monfort A, Orsat B, Spielmann A, Stutz E. 1993. Complete sequence of Eugena gracilis chloroplast DNA. Nucleic Acids Res. 21:3537–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han KY, Maciszewski K, Graf L, Yang JH, Andersen RA, Karnkowska A, Yoon HS. 2019. Dictyochophyceae plastid genomes reveal unusual variability in their organization. J Phycol. 55:1166–1180. [DOI] [PubMed] [Google Scholar]
- Hewadikaramge ME, Linton E. 2018. Intrageneric chloroplast genome comparison in the genus Euglena (Phylum: Euglenophyta) with annotated chloroplast genomes of Euglena hiemalis and Euglena clara. J Appl Phycol. 30:3167–3177. [Google Scholar]
- Hovde BT, Starkenburg SR, Hunsperger HM, Mercer LD, Deodato CR, Jha RK, Chertkov O, Jr MR, Cattolico RA. 2014. The mitochondrial and chloroplast genomes of the haptophyte Chrysochromulina tobin contain unique repeat structures and gene profiles. BMC Genom. 15:604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe CJ, Barbrook AC, Nisbet RER, Lockhart PJ, Larkum AWD. 2008. The origin of plastids. Philos Trans R Soc B: Biological Sciences. 363:2675–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdá Š, Fousek J, Szabová J, Hampl V, Vlček Č. 2012. The plastid genome of Eutreptiella provides a window into the process of secondary endosymbiosis of plastid in euglenids. PLoS ONE. 7:e33746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C, Knoll AH, Chan CX, Verbruggen H. 2018. Plastid phylogenomics with broad taxon sampling further elucidates the distinct evolutionary origins and timing of secondary green plastids. Sci Rep. 8:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DM, Wicke S, Gan L, Yang JB, Jin JJ, Yi TS. 2020. The loss of the inverted repeat in the putranjivoid clade of Malpighiales. Front Plant Sci. 11:942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikawa R, Tanifuji G, Kawachi M, Miyashita H, Hashimoto T, Inagaki Y. 2015. Plastid genome-based phylogeny pinpointed the origin of the green-colored plastid in the dinoflagellate Lepidodinium chlorophorum. Genome Biol Evol. 7:1133–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnkowska A, Bennett MS, Triemer RE. 2018. Dynamic evolution of inverted repeats in Euglenophyta plastid genomes. Sci Rep. 8:16071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnkowska A, Bennett MS, Watza D, Kim JI, Zakryś B, Triemer RE. 2014. Phylogenetic relationships and morphological character evolution of photosynthetic euglenids (Excavata) inferred from taxon-rich analyses of five genes. J Eukaryot Microbiol. 62:362–373. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. 2010. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc B: Biological Sciences. 365:729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Jo BY, Park MG, Yoo YD, Shin W, Archibald JM. 2022. Evolutionary dynamics and lateral gene transfer in raphidophyceae plastid genomes. Front Plant Sci. 13:896138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Linton EW, Shin W. 2015. Taxon-rich multigene phylogeny of the photosynthetic euglenoids (Euglenophyceae). Front Ecol Evol. 3:98. [Google Scholar]
- Kostygov AY, Karnkowska A, Votýpka J, Tashyreva D, Maciszewski K, Yurchenko V, Lukeš J. 2021. Euglenozoa: taxonomy, diversity and ecology, symbioses and viruses. Open Biol. 11:200407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavi B, Karin EL, Pupko T, Hazkani-Covo E. 2018. The prevalence and evolutionary conservation of inverted repeats in Proteobacteria. Genome Biol Evol. 10:918–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Choi IS, Cardoso D, de Lima HC, de Queiroz LP, Wojciechowski MF, Jansen RK, Ruhlman TA. 2021. The chicken or the egg? Plastome evolution and an independent loss of the inverted repeat in papilionoid legumes. Plant J. 107:861–875. [DOI] [PubMed] [Google Scholar]
- Lemieux C, Otis C, Turmel M. 2016. Comparative chloroplast genome analyses of streptophyte green algae uncover major structural alterations in the Klebsormidiophyceae, Coleochaetophyceae and Zygnematophyceae. Front Plant Sci. 7:697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton EW, Karnkowska-Ishikawa A, Kim JI, Shin W, Bennett MS, Kwiatowski J, Zakryś B, Triemer RE. 2010. Reconstructing euglenoid evolutionary relationships using three genes: nuclear SSU and LSU, and chloroplast SSU rDNA sequences and the description of Euglenaria gen. nov. (Euglenophyta). Protist. 161:603–619. [DOI] [PubMed] [Google Scholar]
- Łukomska-Kowalczyk M, Chaber K, Fells A, Milanowski R, Zakryś B. 2021. Description of Flexiglena gen. nov. and new members of Discoplastis and Euglenaformis (Euglenida). J Phycol. 57:766–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciszewski K, Dabbagh N, Preisfeld A, Karnkowska A. 2022. Maturyoshka: a maturase inside a maturase, and other peculiarities of the novel chloroplast genomes of marine euglenophytes. Mol Phylogenet Evol. 170:107441. [DOI] [PubMed] [Google Scholar]
- Maier RM, Schmitz-Linneweber C. 2004. Plastid genomes. In: Daniell H and Chase C, editors. Molecular biology and biotechnology of plant organelles. Dordrecht: (The Netherlands: ): Springer. p. 115–150. [Google Scholar]
- Maréchal A, Brisson N. 2010. Recombination and the maintenance of plant organelle genome stability. New Phytol. 186:299–317. [DOI] [PubMed] [Google Scholar]
- Marin B, Palm A, Klingberg M, Melkonian M. 2003. Phylogeny and taxonomic revision of plastid-containing euglenophytes based on SSU rDNA sequence comparisons and synapomorphic signatures in the SSU rRNA secondary structure. Protist. 154:99–145. [DOI] [PubMed] [Google Scholar]
- Mathur V, Kolísko M, Hehenberger E, Irwin NAT, Leander BS, Kristmundsson Á, Freeman MA, Keeling PJ. 2019. Multiple independent origins of apicomplexan-like parasites. Curr Biol. 29:2936–2941.e5. [DOI] [PubMed] [Google Scholar]
- Méndez-Leyva AB, Guo J, Mudd EA, Wong J, Schwartz JM, Day A. 2019. The chloroplast genome of the marine microalga Tisochrysis lutea. Mitochondrial DNA Part B. 4:253–255. [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R, Teeling E. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37:1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, et al. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res. 49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Ma PF, Grewe F, Taylor A, Michael TP, VanBuren R, Qiu YL. 2019. Lycophyte plastid genomics: extreme variation in GC, gene and intron content and multiple inversions between a direct and inverted orientation of the rRNA repeat. New Phytol. 222:1061–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák Vanclová AMG, Zoltner M, Kelly S, Soukal P, Záhonová K, Füssy Z, Ebenezer TE, Lacová Dobáková E, Eliáš M, Lukeš J, et al. 2020. Metabolic quirks and the colourful history of the Euglena gracilis secondary plastid. New Phytol. 225:1578–1592. [DOI] [PubMed] [Google Scholar]
- Oborník M, Lukeš J. 2015. The organellar genomes of Chromera and Vitrella, the phototrophic relatives of apicomplexan parasites. Annu Rev Microbiol. 69:129–144. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Thompson WF. 1982. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell. 29:537–550. [DOI] [PubMed] [Google Scholar]
- Ping J, Hao J, Li J, Yang Y, Su Y, Wang T. 2021. Loss of the IR region in conifer plastomes: changes in the selection pressure and substitution rate of protein-coding genes. Ecol Evol. 12:e8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce-Toledo RI, López-García P, Moreira D. 2019. Horizontal and endosymbiotic gene transfer in early plastid evolution. New Phytol. 224:618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using SPAdes de novo assembler. Curr Protoc Bioinformatics. 70:e102. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team . 2020. RStudio: integrated development for R. Boston: (MA: ): RStudio, PBC. [Google Scholar]
- Smith DR, Keeling PJ. 2015. Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci USA. 112:10177–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Matsuzaki R, Yamaguchi H, Kawachi M. 2022. What happened before losses of photosynthesis in cryptophyte algae? Mol Biol Evol. 39(2):msac001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. 2015. Dynamic evolution of the chloroplast genome in the green algal classes Pedinophyceae and Trebouxiophyceae. Genome Biol Evol. 7:2062–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. 2017. Divergent copies of the large inverted repeat in the chloroplast genomes of ulvophycean green algae. Sci Rep. 7:994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uthanumallian K, Iha C, Repetti SI, Chan CX, Bhattacharya D, Duchene S, Verbruggen H. 2021. Tightly constrained genome reduction and relaxation of purifying selection during secondary plastid endosymbiosis. Mol Biol Evol. 39:msab295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31:3350–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegert KE, Bennett MS, Triemer RE. 2012. Evolution of the chloroplast genome in photosynthetic euglenoids: a comparison of Eutreptia viridis and Euglena gracilis (Euglenophyta). Protist. 163:832–843. [DOI] [PubMed] [Google Scholar]
- Wiegert KE, Bennett MS, Triemer RE. 2013. Tracing patterns of chloroplast evolution in euglenoids: contributions from Colacium vesiculosum and Strombomonas acuminata (Euglenophyta). J Eukaryot Microbiol. 60:214–221. [DOI] [PubMed] [Google Scholar]
- Wu S, Chen J, Li Y, Liu A, Li A, Yin M, Shrestha N, Liu J, Ren G. 2021. Extensive genomic rearrangements mediated by repetitive sequences in plastomes of Medicago and its relatives. BMC Plant Biol. 21:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Yang Z. 2013. Pamlx: a graphical user interface for PAML. Mol Biol Evol. 30:2723–2724. [DOI] [PubMed] [Google Scholar]
- Zhang HR, Zhang XC, Xiang QP. 2019. Directed repeats co-occur with few short- dispersed repeats in plastid genome of a spikemoss, Selaginella vardei (Selaginellaceae, Lycopodiopsida). BMC Genom. 20:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Guo W, Gupta S, Fan W, Mower JP. 2016. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 209:1747–1756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Euglenid plastid genome sequences obtained in this study have been deposited in the NCBI GenBank database under accession numbers: OP179277-OP179283. Additional supplementary data, Supplementary Material online, including the plastid genome-derived concatenated protein alignments and morphological data matrix used in this study, have been deposited in FigShare repository: https://doi.org/10.6084/m9.figshare.20486148.