Abstract
Cell wall-anchored polypeptides of the antigen I/II family are produced by many species of oral streptococci. These proteins mediate adhesion of streptococci to salivary glycoproteins and to other oral microorganisms and promote binding of cells to collagen type I and invasion of dentinal tubules. Since infections of the root canal system have a mixed anaerobic bacterial etiology, we investigated the hypothesis that coadhesion of anaerobic bacteria with streptococci may facilitate invasive endodontic disease. Porphyromonas gingivalis ATCC 33277 cells were able to invade dentinal tubules when cocultured with Streptococcus gordonii DL1 (Challis) but not when cocultured with Streptococcus mutans NG8. An isogenic noninvasive mutant of S. gordonii, with production of SspA and SspB (antigen I/II family) polypeptides abrogated, was deficient in binding to collagen and had a 40% reduced ability to support adhesion of P. gingivalis. Heterologous expression of the S. mutans SpaP (antigen I/II) protein in this mutant restored collagen binding and tubule invasion but not adhesion to P. gingivalis or the ability to promote P. gingivalis coinvasion of dentin. An isogenic afimbrial mutant of P. gingivalis had 50% reduced binding to S. gordonii cells but was unaffected in the ability to coinvade dentinal tubules with S. gordonii wild-type cells. Expression of the S. gordonii SspA or SspB polypeptide on the surface of Lactococcus lactis cells endowed these bacteria with the abilities to bind P. gingivalis, penetrate dentinal tubules, and promote P. gingivalis coinvasion of dentin. The results demonstrate that collagen-binding and P. gingivalis-binding properties of antigen I/II polypeptides are discrete functions. Specificity of antigen I/II polypeptide recognition accounts for the ability of P. gingivalis to coinvade dentinal tubules with S. gordonii but not with S. mutans. This provides evidence that the specificity of interbacterial coadhesion may influence directly the etiology of pulpal and periapical diseases.
Infections of the tooth pulp (pulpitis) can develop in several ways: through oral bacterial penetration of dentinal tubules opened by caries, restorative procedures, dental trauma, or tooth wear; from direct bacterial contamination of an exposed pulp subsequent to caries or trauma; or by infection of lateral canals from a deep gingival pocket. Studies of the dynamics of root canal infections show that facultative anaerobes (mainly streptococci) are usually the dominant species in the early stages of infection (13). After longer periods of infection (for example, 6 months or so), facultatively anaerobic bacteria become progressively outnumbered by obligate anaerobes (12, 13). These are the predominant organisms in chronic infections and include species of Fusobacterium, Peptostreptococcus, Prevotella, and Porphyromonas (28, 41, 42). Attempts have been made to correlate the presence of certain bacterial species with clinical symptoms. Although direct relationships are hard to prove, species of Porphyromonas (especially Porphyromonas gingivalis) and Prevotella have been associated with acute symptoms of infection (16, 44).
Studies investigating the bacterial flora of carious coronal dentin and infected root canal dentin consistently show that gram-positive bacteria are predominant (3, 11, 38). In vitro studies have demonstrated that oral streptococci penetrate dentinal tubules over several days and remain viable for prolonged periods (30). The invasion of human dentinal tubules by cells of Streptococcus gordonii and Streptococcus mutans depends upon the production by these bacteria of cell wall-anchored polypeptides of the antigen I/II family. These polypeptides contain approximately 1,500 amino acid (aa) residues and bind salivary components, collagen, and other oral microorganisms (20). Isogenic mutants of S. gordonii that are deficient in production of the antigen I/II polypeptides SspA and SspB are unable to invade dentinal tubules. Likewise, an isogenic mutant of S. mutans deficient in SpaP (antigen I/II) expression is also unable to invade dentin (30). Evidence suggests that the recognition, by antigen I/II family polypeptides, of collagen type I present within the tubules (9) is essential for bacterial invasion and for intratubular growth of streptococci (30).
By contrast to results obtained with gram-positive bacteria, monocultures of gram-negative anaerobic bacteria do not invade dentinal tubules (1, 37). This has led to the notion that dentinal tubules may in some way be selective for streptococci and other facultatively anaerobic bacteria. However, gram-negative bacteria must be able to gain entrance into the root canal system from the oral cavity via the dentinal tubules. This is because these organisms can be found in the root canal systems of teeth that are clinically sound but nonvital owing to trauma (28, 43). Since coadhesion of oral bacteria is recognized as an important mechanism in the development of the dental plaque communities (46), it seemed likely that bacterial cell-cell interactions might play a role also in invasion of dentinal tubules. Accordingly, we hypothesized that a noninvasive organism, such as P. gingivalis, might be able to penetrate tubules in association with a coadhering invasive partner organism, such as S. gordonii.
Coadhesion of P. gingivalis and S. gordonii is reasonably well characterized at the molecular level and involves cell surface proteins on both partners (22, 24). These molecules include the 44-kDa fimbrillin subunit (FimA) and a 35-kDa outer membrane protein of P. gingivalis (22, 25) and the streptococcal antigen I/II polypeptides SspA (1,528 aa residues) and SspB (1,448 aa residues) (5, 20, 24). Expression of SspB on the surface of Enterococcus faecalis cells confers on these cells the ability to bind P. gingivalis (24). Purified recombinant SspA or SspB polypeptide of S. gordonii binds P. gingivalis, whereas recombinant PAc (antigen I/II) from S. mutans does not (5). The P. gingivalis binding domain of SspB is defined by amino acid residues 1167 to 1250 (5) and is identical in SspA (10). By contrast, this region within the S. mutans antigen I/II polypeptide differs in primary sequence and in predicted secondary structure (5).
In this study we have investigated the role of streptococcal antigen I/II family polypeptides in promoting invasion of dentinal tubules by an otherwise nonpenetrating organism. The results demonstrate that adhesion of P. gingivalis to S. gordonii confers on the porphyromonads the ability to coinvade dentinal tubules. Conversely, invasion of dentin by P. gingivalis does not occur in the presence of S. mutans cells or in the presence of S. gordonii sspA sspB cells expressing heterologous SpaP from S. mutans. Thus, specificity of coadhesion may determine the ability of oral bacteria to colonize new environments and influence both the etiology and outcome of oral diseases.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
S. gordonii DL1 (Challis) (35), S. mutans NG8 (kindly provided by A. S. Bleiweis, University of Florida, Gainesville), and isogenic derivatives (27, 30) were cultivated at 37°C in brain heart infusion medium (Difco Laboratories, Detroit, Mich.) containing 0.5% (wt/vol) yeast extract (BHY medium). Kanamycin (250 μg/ml) was included when appropriate to ensure maintenance of replicating plasmid pSMI/II-3 (18) in S. gordonii. Lactococcus lactis MG1363 and derivatives carrying plasmid pTREX1-sspA or pTREX1-sspB (17) were cultured at 30°C in M17 medium (Difco) containing 0.5% (wt/vol) glucose. P. gingivalis ATCC 33277 or YPF1 cells were cultivated in anaerobic BHY medium containing hemin (5 μg/ml) and menadione (1 μg/ml). All cultures were inoculated from stock cell suspensions stored at −80°C in BHY medium containing 15% (vol/vol) glycerol and were grown in closed tubes or bottles without shaking. Bacterial cells were radioactively labeled by growth in BHY medium containing [methyl-3H]thymidine (8 μCi/ml [85 Ci/mmol]; Amersham Corp., Arlington Heights, Ill.) (21) to a specific radioactivity of between 1 × 10−3 and 3 × 10−3 cpm per cell.
Construction of P. gingivalis afimbriate mutant.
P. gingivalis YPF1 fimA was generated by homologous recombination between P. gingivalis ATCC 33277 chromosomal DNA and a suicide plasmid carrying an internal fragment of the fimA gene as follows. The 0.65-kb PstI-HincII internal fragment of the fimA gene (14) was removed and subcloned into pBluescript SK(−) to produce pBlu/FIM. The XbaI-XhoI fragment of this plasmid containing the internal fimA fragment was subcloned into the suicide vector pJRD/ET (47). The resultant plasmid, pJRD/ET/FIM, was then introduced by conjugal transfer into P. gingivalis ATCC 33277 (36). The ensuing gene disruption generated a FimA-deficient mutant (YPF1) as confirmed by Western immunoblot analysis of cell extract proteins with anti-FimA antibodies and by reverse transcription-PCR analysis (data not shown).
Adhesion assays.
Bacterial adhesion to acid-soluble collagen type I immobilized onto the wells of Maxi-Sorb microtiter plates (A/S Nunc, Kamstrup, Denmark) was measured as described previously (30). Binding of radioactively labeled P. gingivalis cells to immobilized cells of streptococci or lactococci was determined by the method of Jenkinson et al. (21). Briefly, streptococcal or lactococcal cells were deposited by centrifugation (800 × g for 5 min) onto the bottom surfaces of microtiter plate wells (107 cells per well). The wells were blocked for 16 h, radioactively labeled P. gingivalis cells (0.05 ml containing 5 × 106 cells) were added to each well, and the numbers bound after incubation with gyratory shaking (100 rpm) at 37°C for 1 h were determined by scintillation counting (21). Fewer than 1% of input P. gingivalis cells bound to blocked (control) wells. All data were analyzed by using Student's t test.
Invasion of dentinal tubules.
Noncarious, unrestored human teeth with single root canals were used for invasion experiments. Two root specimens were prepared from each of >100 teeth collected from numerous subjects, with the specimens being resin-coded so that for any one experiment complementary specimens were avoided. Roots were prepared as previously described (29, 30). For each invasion experiment, six roots were selected at random and incubated at 37°C fully submerged with their pulpal surfaces uppermost in BHY medium containing bacterial cells for 14 days (29). At intervals of 3 days, the culture medium was removed just short of exposing the roots and replaced with fresh medium. The viability of cells and purity of cultures were continually monitored (29). Coinvasion experiments were initiated with cultures containing equal numbers (2 × 108 cells/ml) of each bacterial species. BHY medium containing hemin and menadione (see above) was used for cocultivating P. gingivalis and streptococci, while M17 medium containing glucose, hemin, and menadione was employed for coculturing P. gingivalis and L. lactis at 30°C. Antibiotics were not included in these media. Analysis of bacterial samples, taken during and at the end of experiments, for plasmid-encoded antibiotic resistances (17, 30) and for antigen I/II polypeptide production on Western immunoblots (30) confirmed that plasmids were maintained in the absence of antibiotic selection.
Light microscopy and assessment of invasion.
Infected roots were fixed, demineralized in 10% (wt/vol) formic acid containing 2% (wt/vol) formalin, neutralized, washed, dehydrated, and blocked in wax (29). Consecutive transverse sections (6 μm) were cut from the coronal root dentin cervical area, stained (6), and viewed at a standard magnification (×100). The extent of invasion was expressed as the tubule invasion index (TI), which was calculated as described previously (30). On a linear scale, a TI of <0.2 was classified as nil invasion, while a TI of >2.0 was classified as heavy invasion. Data were analyzed by using Student's t test. Sections that exhibited external root resorption were disregarded, and further sections were cut until the defect was removed. For the immunohistochemical detection of bacterial antigens, sections of wax-blocked specimens were cut, picked up onto gelatin-coated slides, and prepared for incubation with primary antibodies as previously detailed (30). S. gordonii cells were detected with rabbit polyclonal antibodies to a cell surface protein extract of S. gordonii cells (19), and antigen I/II polypeptides were detected with rabbit polyclonal antibodies to SpaP (P1) (from N. A. Jacques, Institute of Dental Research, University of Sydney, Sydney, Australia). P. gingivalis cells were detected with rabbit antibodies raised to whole cells of P. gingivalis (from R. J. Lamont, University of Washington, Seattle). Primary antibodies were diluted in the range of 1:40 to 1:500, and binding of these antibodies was detected following incubation of specimens with peroxidase-conjugated antirabbit antibodies and development with 3,3′-diaminobenzidine (30). Negative controls (no primary antibodies) were included in every experiment.
RESULTS
Role of antigen I/II family polypeptides in adhesion of P. gingivalis to oral streptococci.
S. gordonii strains DL1 and M5 each express two antigen I/II family polypeptides designated SspA and SspB. These are the products of tandemly arranged chromosomal genes that are independently transcribed (10, 20). To determine the role of these polypeptides in promoting adhesion of P. gingivalis to S. gordonii, we utilized isogenic mutants of S. gordonii DL1 defective in production of SspA polypeptide (strain OB220) or in production of both SspA and SspB polypeptides (strain OB219) and compared their binding abilities with that of the wild-type strain DL1.
Approximately 47% of input P. gingivalis ATCC 33277 cells bound to wild-type S. gordonii DL1 cells that had been immobilized onto microtiter plate wells (Table 1). Binding of P. gingivalis ATCC 33277 cells to immobilized S. gordonii OB220 sspA cells was reduced to 73% of the numbers bound to strain DL1, while adhesion to strain OB219 sspA sspB cells was significantly (P < 0.001) reduced further to 60%, compared with adhesion to wild-type streptococci (Table 1). We have not yet succeeded in generating an isogenic sspB mutant. Nevertheless, these results suggest that both SspA and SspB proteins are involved in coadhesion of P. gingivalis cells with S. gordonii cells but also that they are not the sole mediators of this cell-cell interaction. Corresponding data have been obtained showing that both SspA and SspB proteins are involved in binding of S. gordonii cells to collagen type I (30) (Table 1). Adhesion of cells to collagen also appears to involve interactions in addition to those promoted by the antigen I/II polypeptides (30).
TABLE 1.
Specificity of antigen I/II polypeptides in mediating streptococcal cell binding to collagen or P. gingivalis
| Bacterial strain and antigen I/II polypeptide phenotype | Mean no. of cells ± SD (106) adhered to collagen (n = 4)a | Mean no. of cells ± SD (106) bound (n = 4)b
|
|
|---|---|---|---|
| P. gingivalis ATCC 33277 | P. gingivalis YPF1 | ||
| S. gordonii | |||
| DL1 (wild type) (SspA+ SspB+) | 4.68 ± 0.57 | 2.38 ± 0.21 | 1.14 ± 0.10 |
| OB220 sspA | 2.73 ± 0.21 | 1.76 ± 0.19 | 0.93 ± 0.09 |
| OB219 sspA sspB | 1.59 ± 0.17 | 1.43 ± 0.15 | 0.78 ± 0.08 |
| OB573 sspA (SpaP+) | 5.46 ± 0.58 | 1.83 ± 0.20 | 0.95 ± 0.08 |
| OB576 sspA sspB (SpaP+) | 3.72 ± 0.38 | 1.55 ± 0.17 | 0.90 ± 0.07 |
| S. mutans | |||
| NG8 (wild type) (SpaP+) | 1.86 ± 0.15 | 0.95 ± 0.07 | 0.52 ± 0.04 |
| 834 spaP | 0.39 ± 0.02 | 0.69 ± 0.05 | 0.41 ± 0.04 |
Five micrograms of collagen type I digest was immobilized on microtiter plate wells, and the numbers of radioactively labeled streptococcal cells adhered (input, 3 × 107 cells) were measured as described in Materials and Methods.
107 cells of each streptococcal strain were immobilized onto microtiter plate wells, and the numbers of radioactively labeled P. gingivalis cells adhered (input, 5 × 106 cells) were measured as described in Materials and Methods.
Expression of the S. mutans spaP gene from the multicopy plasmid pSMI/II-3 (18) in strain OB220 enhanced the ability of OB220 sspA mutant cells to bind collagen type I (Table 1) to above wild-type binding levels and restored significantly the ability of strain OB219 cells to bind collagen (Table 1) (30). However, expression of SpaP (P1) protein on the surface of these S. gordonii mutants (OB573 and OB576) did not enhance significantly (P > 0.05) their abilities to support adhesion of P. gingivalis ATCC 33277 cells (Table 1). This confirms previous results suggesting that S. mutans Pac protein, which is 99% identical in amino acid sequence to SpaP (P1), is poorly recognized by P. gingivalis cells (5). Levels of binding of P. gingivalis to S. mutans NG8 and 834 spaP were <40% of those to S. gordonii DL1 (Table 1).
P. gingivalis fimbriae mediate coadhesion of cells with S. gordonii.
An afimbriate isogenic mutant of P. gingivalis ATCC 33277 was generated as described in Materials and Methods. Loss of the major surface fimbriae in strain YPF1 resulted in approximately a 50% reduction in numbers of porphyromonad cells bound to S. gordonii DL1 (Table 1). Binding of P. gingivalis YPF1 fimA cells to S. gordonii 219 sspA sspB cells was equivalent to only 32% of the P. gingivalis wild-type numbers bound to S. gordonii DL1. As before, levels of adhesion of P. gingivalis to S. gordonii strains expressing heterologous SpaP (P1) protein were not increased significantly (Table 1). Abrogation of FimA expression also resulted in approximately 50% reduced numbers of P. gingivalis cells bound to S. mutans NG8 cells (Table 1). The slight reduction in numbers of YPF1 cells bound to strain 834 cells, compared with NG8 (Table 1), was not statistically significant (P > 0.05). These data demonstrate that P. gingivalis fimbriae are involved in the binding of porphyromonad cells to S. gordonii and to S. mutans. They also suggest that binding of fimbriae to streptococci occurs independently of interaction of P. gingivalis cells with the SspA and SspB polypeptides.
Invasion of dentinal tubules by P. gingivalis.
Previous studies have established experimental conditions under which cells of oral streptococci reproducibly invade dentinal tubules (29). Randomly selected human tooth roots were incubated with bacterial cultures at 37°C for 14 days and then removed, washed, fixed, sectioned, and visualized by light microscopy. More than 50% of root tubules were invaded by S. gordonii DL1 or S. mutans NG8 cells under these conditions, with penetration of bacterial cells to 200 μm or more (30). The degree of invasion was quantified by assessing microscopically the numbers of tubules infected per unit area and was expressed as the TI (see Materials and Methods). Streptococcal cells were detected either by staining (6) or by incubating sections with antibodies reacting with the S. gordonii cell surface, in which case bound antibodies were detected immunohistochemically (30). The TI values obtained for wild-type S. gordonii cells by the two methods differed somewhat, with the antibody detection method being less sensitive and thus giving lower TI values (Table 2).
TABLE 2.
Coinvasion of dentinal tubules by P. gingivalis and streptococci
| Bacterial strain(s) | Mean TI ± SD (n = 6)a determined by:
|
||
|---|---|---|---|
| Brown-Brenn staining (6)b | Immunohistochemical detectionc with antibodies to:
|
||
| S. gordonii extract or SpaPd | P. gingivalis cells | ||
| S. gordonii DL1 | 2.38 ± 0.21 | 1.69 ± 0.13 | <0.10 |
| P. gingivalis ATCC 33277 | <0.10 | <0.10 | <0.10 |
| P. gingivalis ATCC 33277 and: | |||
| S. gordonii DL1 | 2.21 ± 0.34 | 1.56 ± 0.45 | 1.61 ± 0.24 |
| S. gordonii OB220 | 2.07 ± 0.26 | 1.06 ± 0.32 | 0.91 ± 0.18 |
| S. gordonii OB219 | 0.21 ± 0.12 | 0.15 ± 0.06 | <0.10 |
| S. gordonii OB573 | 2.24 ± 0.19 | 0.67 ± 0.18d | 0.64 ± 0.35 |
| S. gordonii OB576 | 2.26 ± 0.28 | 0.59 ± 0.09d | 0.18 ± 0.03 |
| S. mutans NG8 | 1.67 ± 0.19 | 0.51 ± 0.09d | <0.10 |
| S. mutans 834 | <0.10 | NDe | <0.10 |
| P. gingivalis YPF1 and: | |||
| S. gordonii DL1 | 2.12 ± 0.37 | 1.39 ± 0.23 | 1.31 ± 0.27 |
| S. gordonii OB219 | 0.20 ± 0.08 | <0.10 | <0.10 |
Calculated as described in Materials and Methods. TI of <0.2, nil invasion.
Transverse sections of infected coronal root dentin were stained for bacteria and viewed under ×100 magnification.
Demineralized specimens were fixed, blocked in wax, sectioned, and reacted with primary antiserum, and antibody bound to cells was detected as described in Materials and Methods.
S. gordonii cells expressing SpaP and S. mutans cells were detected with SpaP (P1) antibodies.
ND, not determined.
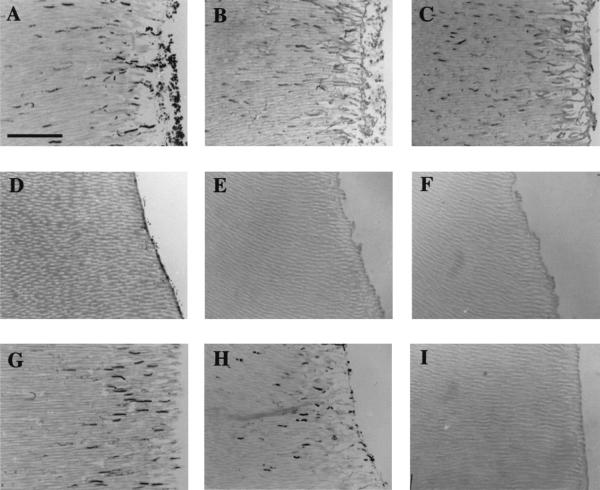
To determine the ability of P. gingivalis cells to invade dentinal tubules, human tooth roots were incubated with P. gingivalis cultures in BHY medium supplemented with hemin (5 μg/ml) and menadione (1 μg/ml). Although this medium supported vigorous growth of P. gingivalis, no penetration of dentinal tubules by P. gingivalis cells was observed following staining of sections (Table 2). We then investigated the ability of P. gingivalis and S. gordonii growing in coculture to penetrate dentinal tubules. After 14 days of incubation, heavy invasion of dentinal tubules by bacteria was evident (TI = 2.21 ± 0.34) as detected by direct staining of microbial cells within sections (Table 2; Fig. 1A). To differentiate clearly between the bacterial types invading the tubules, serial sections were incubated either with S. gordonii cell surface antibodies or with antibodies raised to whole cells of P. gingivalis. Subsequent immunohistochemical detection of bound antibodies demonstrated that infected tubules contained both S. gordonii (Fig. 1B) and P. gingivalis (Fig. 1C) cells. Antibodies to P. gingivalis did not bind to streptococci (Table 2). The TI value for the mixed infection as determined by direct staining was not significantly different from that for S. gordonii alone (Table 2). This indicated that the numbers of tubules infected by bacteria in coculture were similar to those for S. gordonii in monoculture, implying that P. gingivalis and S. gordonii occupied the same tubules. These experiments did not allow accurate estimations of the relative proportions of the two bacterial cell types present within infected dentinal tubules.
FIG. 1.
Transverse sections of human roots showing the invasion of dentinal tubules from the pulpal side by S. gordonii in coculture with P. gingivalis. Bacterial cells were stained (6) (A, D, and G) or detected immunohistochemically with antibodies to S. gordonii surface proteins (B, E, and H) or with antibodies to P. gingivalis cells (C, F, and I). (A, B, and C) Wild-type S. gordonii DL1 cocultured with P. gingivalis ATCC 33277; (D, E, and F) S. gordonii OB219 sspA sspB cocultured with P. gingivalis; (G, H, and I) S. gordonii OB576 sspA sspB expressing SpaP from S. mutans cocultured with P. gingivalis. P. gingivalis cells are detected in panel C only, even though tubule invasion by streptococci is restored in panel G. Bar, 50 μm.
Antigen I/II binding specificity determines P. gingivalis invasive ability.
When P. gingivalis was cocultured with S. gordonii OB220 sspA, invasion levels of both streptococcal and porphyromonad cells were reduced (Table 2). When P. gingivalis cells were cocultured with strain OB219 cells, which were deficient in production of both the SspA and SspB proteins, no invasion of dentinal tubules by either organism was observed (Table 2; Fig. 1D, E, and F). These observations appear to rule out the possibility that streptococcal cell growth simply modifies the environment in some way so as to allow P. gingivalis to penetrate dentin. To determine if P. gingivalis invasion depended upon specificity of antigen I/II binding, cells were cocultured with S. gordonii OB573 or OB576 cells. Expression of S. mutans SpaP (P1) protein by these S. gordonii strains restored their invasive abilities (Table 2; Fig. 1G). Streptococcus-specific antibodies detected the presence within tubules of invading streptococci (Fig. 1H); however, no invasion by P. gingivalis cells was observed (Fig. 1I; Table 2). SpaP antibodies were also utilized in these experiments to demonstrate that SpaP (P1) protein was produced by S. gordonii cells growing within the dentinal tubules (Table 2). These antibodies were less reactive with streptococci than the corresponding cell extract antibodies, and so lower TI values were recorded (Table 2). These results demonstrate that dentinal tubule invasion by P. gingivalis depends upon the specificity of binding to streptococcal antigen I/II polypeptide. In support of this conclusion, P. gingivalis did not invade dentin when cocultured with S. mutans NG8 (Table 2), even though the latter cells showed moderate to high levels of invasion.
Fimbriae are not necessary for P. gingivalis invasion of dentin.
Since coadhesion of P. gingivalis with S. gordonii also involves fimbriae, it was important to assess the role that fimbriae might play in P. gingivalis coinvasion. As might be expected, cells of the afimbriate mutant P. gingivalis YPF1 did not invade dentinal tubules when grown as a monoculture (results not shown). Nevertheless, P. gingivalis YPF1 cells and S. gordonii DL1 cells were able to coinvade dentin and to do so at TI values approaching those obtained for P. gingivalis ATCC 33277 coinvasion (Table 2). As before, no invasion of dentinal tubules by strain YPF1 cells was observed when they were cocultured with S. gordonii OB219 (Table 2). These data support the notion that fimbriae play a significant role in coadhesion of P. gingivalis and S. gordonii but demonstrate that fimbriae are not necessary for coinvasion by P. gingivalis and S. gordonii, which depends upon antigen I/II polypeptide recognition.
Heterologously expressed S. gordonii antigen I/II polypeptides promote coadhesion and coinvasion.
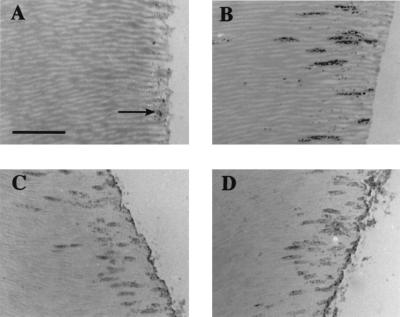
We have previously demonstrated that heterologous expression of S. gordonii SspA or SspB polypeptide on the surface of the food-grade bacterium L. lactis MG1363 confers on these cells the ability to bind salivary glycoproteins and collagen type I (17). Lactococci expressing SspA or SspB were also found to bind twofold-higher numbers of P. gingivalis cells than did L. lactis MG1363. In binding assays, 2.55 × 106 ± 0.21 × 106 P. gingivalis cells bound to 1 × 107 immobilized cells of L. lactis expressing SspA protein, and 2.40 × 106 ± 0.20 × 106 P. gingivalis cells bound to L. lactis cells expressing SspB protein. We then determined if expression of antigen I/II polypeptide in L. lactis might also confer on these cells the ability to invade dentinal tubules. Wild-type lactococci did not invade dentinal tubules, although some cells appeared to attach to the dentin surface (Fig. 2A). Conversely, cells of L. lactis expressing SspA (or SspB) were able to invade dentinal tubules, with a TI of 1.14 ± 0.29 calculated from stained sections (results not shown). Cocultivation of P. gingivalis with L. lactis(pTREX1-sspA) cells expressing SspA demonstrated bacterial invasion of tubules to 100 μm or more (Fig. 2B). Reaction of root sections with antigen I/II polypeptide antibodies demonstrated that lactococcal cells were present within dentinal tubules and that SspA polypeptide was expressed in situ (Fig. 2C). Incubation of root sections with P. gingivalis-specific antibodies showed clearly that P. gingivalis cells coinvaded with lactococci expressing SspA (Fig. 2D). L. lactis(pTREX1-sspB) cells expressing SspB polypeptide also promoted P. gingivalis invasion of dentin (results not shown).
FIG. 2.
Transverse sections of human roots showing the invasion of dentinal tubules by L. lactis strains in monoculture or in coculture with P. gingivalis. (A) Stained wild-type L. lactis MG1363 cells, showing some adhesion of bacteria to the dentin surface (arrow) but no invasion of dentinal tubules; (B) L. lactis cells expressing SspA polypeptide cocultured with P. gingivalis ATCC 33277 and stained (6), showing tubule invasion; (C) as in panel B but reacted with antibodies to SpaP (P1); (D) as in panel B but reacted with antibodies to P. gingivalis cells. Bar, 10 μm.
DISCUSSION
Interbacterial binding is considered to be a significant factor in the development of dental plaque, promoting both selective and temporal acquisition of microorganisms to form complex communities (46). While a considerable amount of information on bacterial cell-cell interactions has been obtained from in vitro studies, it has been difficult to demonstrate the ecological significance of interbacterial adhesion reactions in vivo. Model systems have been developed to approach such considerations, and one of these recently has been used to investigate the effects of oxygen (aerobiosis) on the growth in planktonic or biofilm phase of a defined consortium of oral microorganisms (4). In a two-stage continuous culture, survival of anaerobic bacteria in the second (aerobic) vessel depended upon the ability of Fusobacterium nucleatum to simultaneously coaggregate with both anaerobic (P. gingivalis and Prevotella) and facultatively anaerobic bacteria. In the absence of F. nucleatum, numbers of anaerobic bacteria were significantly reduced, indicating that coaggregation facilitates survival of anaerobes in aerated environments (4). We have now extended the information on the ecological significance of coaggregation (coadhesion) by showing that successful invasion of dentinal tubules by the obligate anaerobe P. gingivalis depends upon adhesion to an invasive partner organism. This process, designated coinvasion, establishes coadhesion as a potential determinant of endodontic disease etiology.
Previous light microscopy studies have demonstrated that several gram-negative anaerobic bacterial species, such as Prevotella melananogenica (1) and Prevotella intermedia (37), were unable to invade dentin. On the other hand, it has been reported that Porphyromonas endodontalis BN11a-f and P. gingivalis ATCC 33277 in monoculture were able to invade bovine dentinal tubules, although very few tubules were infected (40). While the number, distribution, and size of bovine dentinal tubules are similar to those in human dentin, Siqueira et al. (40) removed the cementum from the dentin blocks in their experiments, and this artificially enhances bacterial penetration. Thus, the reported penetration of very few tubules may have occurred by nonspecific incorporation and not through cell division and growth along the tubule length. We have never observed invasion of human dentinal tubules by P. gingivalis cells in monoculture, even over a wide range of different culture conditions of medium, redox potential, and temperature. We have previously provided evidence that the ability of S. gordonii or S. mutans cells to invade dentinal tubules depends upon collagen recognition by antigen I/II polypeptides (30). Although P. gingivalis cells also bind to collagen type I deposited onto hydroxylapatite surfaces (32, 33), it would appear that the ability to bind type I collagen, present within dentinal tubules (9), is not sufficient to promote invasion of tubules by these bacteria in monoculture.
The antigen I/II polypeptides produced by oral streptococci have been shown to have a range of adhesive functions. Sequences within the N-terminal alanine-rich repeat blocks, and sequences C-terminal to the proline-rich repeats in the central region of the polypeptide, bind salivary glycoproteins (20). Sequences within the N-terminal region are also believed to be involved, at least in part, in binding to collagen (39), while an 84-aa sequence downstream of the proline-rich repeats, and present in SspA and SspB proteins from S. gordonii, is implicated in binding to P. gingivalis cells (5, 20). The results in this paper provide further evidence that the regions within the antigen I/II polypeptides that mediate adhesion to collagen or to P. gingivalis are distinct. Binding to collagen appears to be a general property of antigen I/II family polypeptides produced by S. gordonii and S. mutans, while binding to P. gingivalis is a specific property of the SspA and SspB polypeptides (5). There is no evidence to suggest that the SspA and SspB polypeptides differ in their relative affinities of binding to P. gingivalis cells (5). This was confirmed in that similar numbers of P. gingivalis cells bound to lactococcal cells expressing SspA or SspB proteins. On the other hand, the SspA and SspB polypeptides appear to have different binding affinities for salivary agglutinin glycoprotein and for collagen (17).
Previously we have shown that S. gordonii SspA and SspB polypeptides confer on L. lactis cells the ability to bind collagen type I (17). Remarkably, surface expression of these proteins in L. lactis also conferred on lactococci the abilities to penetrate dentinal tubules and to support P. gingivalis coinvasion. These observations support the conclusion that it is the specificity of interaction of P. gingivalis cells with antigen I/II polypeptide that allows coinvasion of dentin by this organism. Bacteria such as S. mutans may colonize tooth surfaces and invade dentinal tubules in an antigen I/II (P1)-dependent mechanism (8), but they do not appear to promote coinvasion. The abilities of other oral streptococcal species, for example Streptococcus oralis, to enhance P. gingivalis invasion of dentin, might therefore depend upon their antigen I/II polypeptide binding specificities. P. gingivalis coaggregates with S. oralis (2, 26), but the role of antigen I/II polypeptide SoaA (20) in this reaction and whether SoaA binds P. gingivalis cells are not known. We are currently investigating the abilities of different streptococcal species to promote coinvasion of dentinal tubules by P. gingivalis and by other periodontal pathogens associated with root canal infections. This will provide information on the extent to which the specificities of other interbacterial adhesion reactions might determine the etiologies of endodontic infections.
Interbacterial coaggregation or coadhesion between P. gingivalis and S. gordonii has been shown recently to enhance colonization of both P. gingivalis and streptococci in a biofilm model of plaque formation (7). S. gordonii outcompetes P. gingivalis for attachment sites in salivary pellicle, and substantial mixed bacterial biofilms develop on saliva-coated glass slides only when deposited S. gordonii cells provide an attachment substrate for P. gingivalis (7). The results in the present paper extend these observations and suggest that specificity of coadhesion may initiate invasive infections of endodontic and periodontal tissues by these organisms. The occurrence of coadhesive interactions in vivo might explain why typically a limited number of species are found in an infected root canal. The presence of gram-negative bacteria in infected dentin (3, 11, 15, 34) and within root canals with intact dentin walls (28, 40, 41, 42, 43) suggests that these bacteria can penetrate dentin in vivo. Associations between gram-negative bacteria and other bacteria that are capable of dentinal tubule invasion may thus allow invasion by the gram-negative organisms.
In summary, invasion of dentinal tubules by streptococci depends upon multiple recognition functions of antigen I/II proteins. When P. gingivalis cells are present in association with streptococci, the ability of P. gingivalis cells to coinvade depends upon the species specificity of antigen I/II recognition, with SspA or SspB proteins on S. gordonii (or on heterologous lactococci) providing attachment sites for P. gingivalis. Binding of P. gingivalis cells to intratubular collagen may assist the invasion process. The production of fimbriae by P. gingivalis, while contributing to coadhesion with S. gordonii in vitro, is not necessary for coinvasion of dentinal tubules with S. gordonii. This is in direct contrast to the requirement for fimbriae in P. gingivalis invasion of gingival epithelial cells (45). Coadhesion of P. gingivalis and S. gordonii therefore functions to extend the invasive potential of P. gingivalis. This property of coinvasion of dentin by oral bacteria has not previously been investigated. Some other gram-negative periodontopathogenic bacteria, such as Actinobacillus actinomycetemcomitans, have a high capacity to invade and replicate within epithelial cells (31), as does P. gingivalis (23, 26). In light of the results presented here, it is possible that coadhesion may also promote coinvasion of epithelial cells, thus enhancing the pathogenic potential of otherwise noninvasive organisms.
ACKNOWLEDGMENTS
We are most grateful to A. S. Bleiweis, N. A. Jacques, R. J. Lamont, and S. F. Lee for providing bacterial strains, plasmids, or antibodies. We thank R. A. Baker and A. Samways for excellent technical assistance and A. R. Holmes and R. J. Lamont for helpful advice and discussions.
The support of the New Zealand Health Research Council, The Wellcome Trust (London, United Kingdom), and NIDCR (grant no. DE12505) is gratefully acknowledged.
REFERENCES
- 1.Akpata E S, Blechman H. Bacterial invasion of pulpal dentin wall in vitro. J Dent Res. 1982;61:435–438. doi: 10.1177/00220345820610021401. [DOI] [PubMed] [Google Scholar]
- 2.Amano A, Fujiwara T, Nagata H, Kuboniwa M, Sharma A, Sojar H T, Genco R J, Hamada S, Shizukuishi S. Porphyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J Dent Res. 1997;76:852–857. doi: 10.1177/00220345970760040601. [DOI] [PubMed] [Google Scholar]
- 3.Ando N, Hoshino E. Predominant obligate anaerobes invading the deep layers of root canal dentine. Int Endod J. 1990;23:20–27. doi: 10.1111/j.1365-2591.1990.tb00798.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradshaw D J, Marsh P D, Watson G K, Allison C. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect Immun. 1998;66:4729–4732. doi: 10.1128/iai.66.10.4729-4732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks W, Demuth D R, Gil S, Lamont R J. Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infect Immun. 1997;65:3753–3758. doi: 10.1128/iai.65.9.3753-3758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown J H, Brenn L. A method for the differential staining of Gram-positive and Gram-negative bacteria in tissue sections. Bull Johns Hopkins Hosp. 1931;48:69–73. [Google Scholar]
- 7.Cook G S, Costerton J W, Lamont R J. Biofilm formation by Porphyromonas gingivalis and Streptococcus gordonii. J Periodontal Res. 1998;33:323–327. doi: 10.1111/j.1600-0765.1998.tb02206.x. [DOI] [PubMed] [Google Scholar]
- 8.Crowley P J, Brady L J, Michalek S M, Bleiweis A S. Virulence of a spaP mutant of Streptococcus mutans in a gnotobiotic rat model. Infect Immun. 1999;67:1201–1206. doi: 10.1128/iai.67.3.1201-1206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai X-F, Ten Cate A R, Limeback H. The extent and distribution of intratubular collagen fibrils in human dentine. Arch Oral Biol. 1991;36:775–778. doi: 10.1016/0003-9969(91)90045-v. [DOI] [PubMed] [Google Scholar]
- 10.Demuth D R, Duan Y, Brooks W, Holmes A R, McNab R, Jenkinson H F. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol Microbiol. 1996;20:403–413. doi: 10.1111/j.1365-2958.1996.tb02627.x. [DOI] [PubMed] [Google Scholar]
- 11.Edwardsson S. Bacteriology of dentin caries. In: Thylstrup A, Leach S A, Qvist V, editors. Dentine and dentine reactions in the oral cavity. Oxford, United Kingdom: IRL Press Ltd.; 1987. pp. 95–102. [Google Scholar]
- 12.Fabricius L, Dahlén G, Holm S E, Möller Å J R. Influence of combinations of oral bacteria on periapical tissues of monkeys. Scand J Dent Res. 1982;90:200–206. doi: 10.1111/j.1600-0722.1982.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 13.Fabricius L, Dahlén G, Öhman A, Möller Å J R. Predominant indigenous oral bacteria isolated from infected root canals after varied times of closure. Scand J Dent Res. 1982;90:134–144. doi: 10.1111/j.1600-0722.1982.tb01536.x. [DOI] [PubMed] [Google Scholar]
- 14.Fujiwara T, Morishima S, Takahashi I, Hamada S. Molecular cloning and sequencing of the fimbrillin gene of Porphyromonas gingivalis strains and characterization of recombinant proteins. Biochem Biophys Res Commun. 1993;197:241–247. doi: 10.1006/bbrc.1993.2467. [DOI] [PubMed] [Google Scholar]
- 15.Haapasalo M. Bacteroides spp. in dental root canal infections. Endod Dent Traumatol. 1989;5:1–10. doi: 10.1111/j.1600-9657.1989.tb00330.x. [DOI] [PubMed] [Google Scholar]
- 16.Hashioka K, Yamasaki M, Nakane A, Horiba N, Nakamura H. The relationship between clinical symptoms and anaerobic bacteria from infected root canals. J Endod. 1992;18:558–561. doi: 10.1016/S0099-2399(06)81214-8. [DOI] [PubMed] [Google Scholar]
- 17.Holmes A R, Gilbert C, Wells J M, Jenkinson H F. Binding properties of Streptococcus gordonii SspA and SspB antigen I/II family polypeptides expressed on the cell surface of Lactococcus lactis MG1363. Infect Immun. 1998;66:4633–4639. doi: 10.1128/iai.66.10.4633-4639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Homonylo-McGavin M K, Lee S F. Role of the C terminus in antigen P1 surface location in Streptococcus mutans and two related cocci. J Bacteriol. 1996;178:801–807. doi: 10.1128/jb.178.3.801-807.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkinson H F. Cell-surface proteins of Streptococcus sanguis associated with cell hydrophobicity and coaggregation properties. J Gen Microbiol. 1986;132:1575–1589. doi: 10.1099/00221287-132-6-1575. [DOI] [PubMed] [Google Scholar]
- 20.Jenkinson H F, Demuth D R. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol Microbiol. 1997;23:183–190. doi: 10.1046/j.1365-2958.1997.2021577.x. [DOI] [PubMed] [Google Scholar]
- 21.Jenkinson H F, Terry S, McNab R, Tannock G W. Inactivation of the gene encoding surface protein SspA in Streptococcus gordonii DL1 affects cell interactions with human salivary agglutinin and oral actinomyces. Infect Immun. 1993;61:3199–3208. doi: 10.1128/iai.61.8.3199-3208.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamont R J, Bevan C A, Gil S, Persson R E, Rosan B. Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol Immunol. 1993;8:272–276. doi: 10.1111/j.1399-302x.1993.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 23.Lamont R J, Chan A, Belton M, Izutso K T, Vasel D, Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1995;63:3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamont R J, Gil S, Demuth D R, Malamud D, Rosan B. Molecules of Streptococcus gordonii that bind to Porphyromonas gingivalis. Microbiology. 1994;140:867–872. doi: 10.1099/00221287-140-4-867. [DOI] [PubMed] [Google Scholar]
- 25.Lamont R J, Hsiao G W, Gil S. Identification of a molecule of Porphyromonas gingivalis that binds Streptococcus gordonii. Microb Pathog. 1994;17:355–360. doi: 10.1006/mpat.1994.1081. [DOI] [PubMed] [Google Scholar]
- 26.Lamont R J, Jenkinson H F. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S F, Progulske-Fox A, Erdos G W, Piacentini D A, Ayakawa G Y, Crowley P J, Bleiweis A S. Construction and characterization of isogenic mutants of Streptococcus mutans deficient in major surface protein antigen P1 antigen I/II. Infect Immun. 1989;57:3306–3313. doi: 10.1128/iai.57.11.3306-3313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Goff A, Bunetel L, Mouton C, Bonnaure-Mallet M. Evaluation of root canal bacteria and their antimicrobial susceptibility in teeth with necrotic pulp. Oral Microbiol Immunol. 1997;12:318–322. doi: 10.1111/j.1399-302x.1997.tb00397.x. [DOI] [PubMed] [Google Scholar]
- 29.Love R M, Chandler N P, Jenkinson H F. Penetration of smeared or nonsmeared dentine by Streptococcus gordonii. Int Endod J. 1996;29:2–12. doi: 10.1111/j.1365-2591.1996.tb01353.x. [DOI] [PubMed] [Google Scholar]
- 30.Love R M, McMillan M D, Jenkinson H F. Invasion of dentinal tubules by oral streptococci is associated with collagen recognition mediated by the antigen I/II family of polypeptides. Infect Immun. 1997;65:5157–5164. doi: 10.1128/iai.65.12.5157-5164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer D H, Mintz K P, Fives-Taylor P M. Models of invasion of enteric and periodontal pathogens into epithelial cells: a comparative analysis. Crit Rev Oral Biol Med. 1997;8:389–409. doi: 10.1177/10454411970080040301. [DOI] [PubMed] [Google Scholar]
- 32.Naito Y, Gibbons R J. Attachment of Bacteroides gingivalis to collagenous substrata. J Dent Res. 1988;67:1075–1080. doi: 10.1177/00220345880670080301. [DOI] [PubMed] [Google Scholar]
- 33.Naito Y, Tohada H, Okuda K, Takazoe I. Adherence and hydrophobicity of invasive and noninvasive strains of Porphyromonas gingivalis. Oral Microbiol Immunol. 1993;8:195–202. doi: 10.1111/j.1399-302x.1993.tb00559.x. [DOI] [PubMed] [Google Scholar]
- 34.Ozaki K, Matsua T, Nakae H, Noiri Y, Yoshiyama M, Ebisu S. A quantitative comparison of selected bacteria in human carious dentine by microscopic counts. Caries Res. 1994;28:137–145. doi: 10.1159/000261635. [DOI] [PubMed] [Google Scholar]
- 35.Pakula R, Walczak W. On the nature of competence of transformable streptococci. J Gen Microbiol. 1963;31:125–133. doi: 10.1099/00221287-31-1-125. [DOI] [PubMed] [Google Scholar]
- 36.Park Y, McBride B C. Characterization of the typ gene product and isolation of a specific protease-deficient mutant of Porphyromonas gingivalis W83. Infect Immun. 1993;61:4139–4146. doi: 10.1128/iai.61.10.4139-4146.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez F, Rochd T, Lodter J-P, Calas P, Michel G. In vitro study of the penetration of three bacterial strains into root dentine. Oral Surg Oral Med Oral Pathol. 1993;76:97–103. doi: 10.1016/0030-4220(93)90302-k. [DOI] [PubMed] [Google Scholar]
- 38.Peters L B, Wesselink P R, Moore W R. The fate and role of bacteria left in root dentinal tubules. Int Endod J. 1995;28:95–99. doi: 10.1111/j.1365-2591.1995.tb00166.x. [DOI] [PubMed] [Google Scholar]
- 39.Sciotti M A, Yamodo I, Klein J P, Ogier J A. The N-terminal half part of the oral streptococcal antigen I/IIf contains two distinct binding domains. FEMS Microbiol Lett. 1997;153:439–445. doi: 10.1111/j.1574-6968.1997.tb12608.x. [DOI] [PubMed] [Google Scholar]
- 40.Siqueira J F, De Uzeda M, Fonseca M E F. A scanning electron microscopic evaluation of in vitro dentinal tubules penetration by selected anaerobic bacteria. J Endod. 1996;22:308–310. doi: 10.1016/S0099-2399(96)80265-2. [DOI] [PubMed] [Google Scholar]
- 41.Sundqvist G. Associations between microbial species in dental root canal infections. Oral Microbiol Immunol. 1992;7:257–262. doi: 10.1111/j.1399-302x.1992.tb00584.x. [DOI] [PubMed] [Google Scholar]
- 42.Sundqvist G. Ecology of the root canal flora. J Endod. 1992;18:427–430. doi: 10.1016/S0099-2399(06)80842-3. [DOI] [PubMed] [Google Scholar]
- 43.Sundqvist G. Taxonomy, ecology, and pathogenicity of the root canal flora. Oral Surg Oral Med Oral Pathol. 1994;78:522–530. doi: 10.1016/0030-4220(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 44.Sundqvist G, Johansson E, Sjögren U. Prevelance of black-pigmented Bacteriodes species in root canal infections. J Endod. 1989;15:13–19. doi: 10.1016/S0099-2399(89)80092-5. [DOI] [PubMed] [Google Scholar]
- 45.Weinberg A, Belton C M, Park Y, Lamont R J. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1997;65:313–316. doi: 10.1128/iai.65.1.313-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whittaker C J, Klier C M, Kolenbrander P E. Mechanisms of adhesion by oral bacteria. Annu Rev Microbiol. 1996;50:513–552. doi: 10.1146/annurev.micro.50.1.513. [DOI] [PubMed] [Google Scholar]
- 47.Xie H, Cai S, Lamont R J. Environmental regulation of fimbrial gene expression in Porphyromonas gingivalis. Infect Immun. 1997;65:2265–2271. doi: 10.1128/iai.65.6.2265-2271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]