Abstract
Sf6 is a bacterial virus that infects the human pathogen Shigella flexneri. Here, we describe the cryo–electron microscopy structure of the Sf6 tail machine before DNA ejection, which we determined at a 2.7-angstrom resolution. We built de novo structures of all tail components and resolved four symmetry-mismatched interfaces. Unexpectedly, we found that the tail exists in two conformations, rotated by ~6° with respect to the capsid. The two tail conformers are identical in structure but differ solely in how the portal and head-to-tail adaptor carboxyl termini bond with the capsid at the fivefold vertex, similar to a diamond held over a five-pronged ring in two nonidentical states. Thus, in the mature Sf6 tail, the portal structure does not morph locally to accommodate the symmetry mismatch but exists in two energetic minima rotated by a discrete angle. We propose that the design principles of the Sf6 tail are conserved across P22-like Podoviridae.
Cryo-EM reveals the building blocks, assembly, and symmetry mismatches of the Shigella virus Sf6 tail apparatus.
INTRODUCTION
Infections caused by the gram-negative pathogen Shigella (or shigellosis) are a leading cause of morbidity and mortality worldwide. Four species of Shigella responsible for shigellosis are known (e.g., Shigella dysenteriae, Shigella boydii, Shigella flexneri, and Shigella sonnei) and represent a substantial burden to public health (1), especially in children under the age of 5 in developing countries where the mortality rate can be as high as 2.9 deaths per 100,000 infections (2). The emergence of antibiotic-resistant Shigella strains together with the lack of efficient vaccines have made Shigella a priority organism for the World Health Organization (3). As an alternative to combat shigellosis, phage therapy has also regained attention in recent years (4, 5).
Bacteriophage Sf6 infects S. flexneri and is a member of the P22-like family of short-tailed bacteriophages (Podoviridae), initially identified for its ability to convert S. flexneri group antigens (6, 7). Sf6 expresses an O-antigen acetyltransferase (8) that catalyzes 2-O-acetylation of the host O-antigen backbone, which results in serotype conversion (9). Morphologically, Sf6 has an icosahedral T = 7laevo capsid with a maximal diameter of 690 Å and a short tail, similar to the Salmonella phage P22 (9). An icosahedral reconstruction of Sf6 (10) revealed that the capsid is built by 415 copies of the 425-residue-coat protein (gp5) that adopts a classical HK97 fold (11). Sf6 coat proteins assemble via noncovalent interactions, generating a flexible and sturdy nanocontainer, only 12 Å at the thinnest part (12). Sf6 capsids copurify with lipid vesicles containing two host outer membrane proteins, OmpA and OmpC, that in vivo and in vitro studies have demonstrated to function as host receptors (13, 14).
A unique fivefold vertex of the Sf6 capsid is occupied by a short, noncontractile tail apparatus that breaks the icosahedral symmetry (15–17). The tail assembles onto a dodecameric portal protein that replaces a capsid penton at the unique vertex. The portal protein functions as a bidirectional gateway through which a virus can both package and eject DNA (18). During assembly, the portal protein recruits a large terminase subunit (TerL) (19) that assembles into a genome-packaging motor (20), which is aided by a DNA recognition small terminase subunit (TerS) (21). The Sf6 ∼39–kilo–base pair genome is packaged into an immature precursor capsid (or procapsid) using a headful DNA packaging mechanism similar to phage P22 (22, 23). Genome packaging is terminated by endonucleolytic cleavage of the newly encapsidated DNA by the headful packaging nuclease (24), which makes up the C-terminal domain of TerL. Concurrently, the portal protein recruits three tail factors that displace TerL and stabilize the newly packaged particles, preventing leakage of the genome (25). The first factor, known as the head-to-tail adaptor (gp7) (26), bridges the portal to the conserved tail hub (gp8) (7). gp7 and gp8 extend the portal channel, and the latter recruits six copies of the trimeric tailspike (27), an endorhamnosidase that cleaves the lipopolysaccharide (LPS) O-antigen receptor during phage adsorption to the cell surface (28). The tail hub is closed by a trimeric tail needle (gp9) (29) that is similar to P22 gp26 (30, 31) (table S1).
Genome ejection, the mirror process of genome packaging, is triggered by the interaction of an Sf6 virion with primary (LPS) and secondary (outer membrane proteins OmpA and OmpC) receptors on the Shigella surface (13). This initial recognition event is followed by the ejection of internal virion proteins (or ejection proteins) that extend the tail (32, 33). Only one Sf6 ejection protein, gp12, similar to T7 gp15 (33, 34), has been characterized (35). On the basis of genomic location and sequence conservation, Sf6 ejection proteins are similar to P22 (32) and may exist in two conformations: loosely bound to the portal barrel inside the virion (36) and assembled into a transenvelope channel in the host cell envelope after ejection (37). In vitro, genome ejection can be mediated by LPS and OmpA and OmpC, albeit this ejection is slower than observed during natural infection (13). Furthermore, the Sf6 tail needle contains a trimeric knob at its distal tip, similar to the adenovirus knob (38), possibly implicated in controlling the rate of genome ejection in Shigella (39).
This paper describes the high-resolution structure of the phage Sf6 tail machine before genome delivery that we characterized using cryo–electron microscopy (cryo-EM). We report that the Sf6 tail apparatus exists in two structurally distinct conformations. The structures presented here shed light on the conformational dynamics of a Podoviridae tail, and the role of symmetry mismatches in virus assembly.
RESULTS
In situ structure of Sf6 noncontractile tail machine
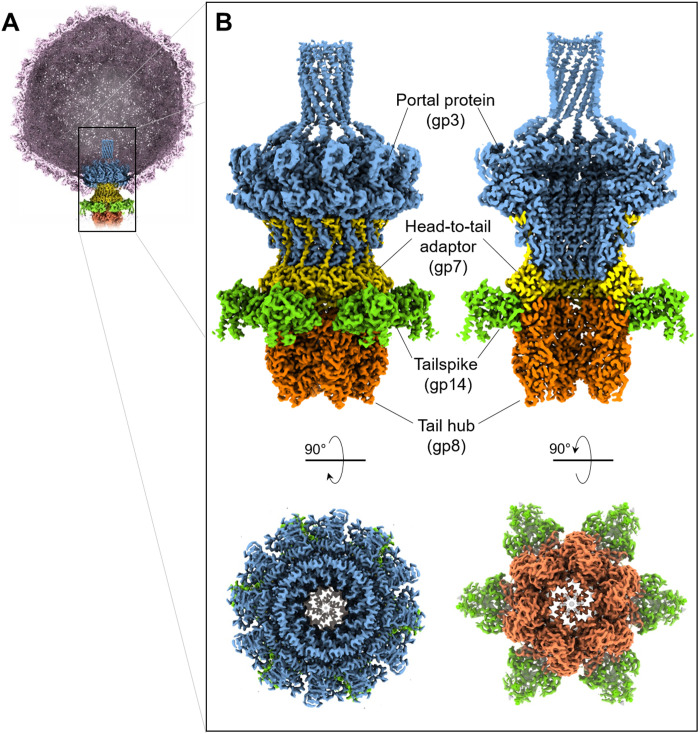
We used localized reconstruction to visualize the Sf6 tail (Fig. 1, A and B, and fig. S1). Aiming at a high-resolution reconstruction of this nonicosahedral capsid component, we collected a large dataset (7977 movies) of Sf6 mature virions using a 300-kV Titan Krios electron microscope equipped with a Gatan K3 direct detector. After identifying a three-dimensional (3D) class that contained the phage tail, the density for the unique vertex was subjected to 12- and 6-fold rotational averaging, yielding excellent maps at 2.7- and 2.8-Å resolution, which are referred to as C12 and C6 maps, respectively (fig. S2). The C12 map (fig. S3A) was used to build de novo models of the dodecameric portal protein (residues 1 to 671) bound to 12 copies of the head-to-tail adaptor (residues 1 to 150). Similarly, the C6 map (fig. S3B) was used to build six copies of the tail hub (residues 2 to 472) and six trimeric tailspike N termini (residues 4 to 123). In total, we built 14,310 amino acids in symmetrized localized reconstructions (Fig. 2A), which were real space–refined to a final correlation coefficient (CC) of ~0.9, indicating an excellent model-to-map fit (Table 1). Unexpectedly, the tailspike body (residues 124 to 623) and tail needle (residues 1 to 282) were poorly visible in symmetrized maps because of their flexibility when bound to the tail machine. The needle could be identified and modeled in a lower-resolution asymmetric (C1) map, whereas for the tailspike, a focused reconstruction from symmetry expansion of the C6 map revealed two equally populated conformations of the C-terminal body (fig. S2), related by a hinge movement. We modeled 1862 residues of the full-length Sf6 tailspike, which were refined to a final CC of ~0.84- at 3.6-Å resolution (Table 1).
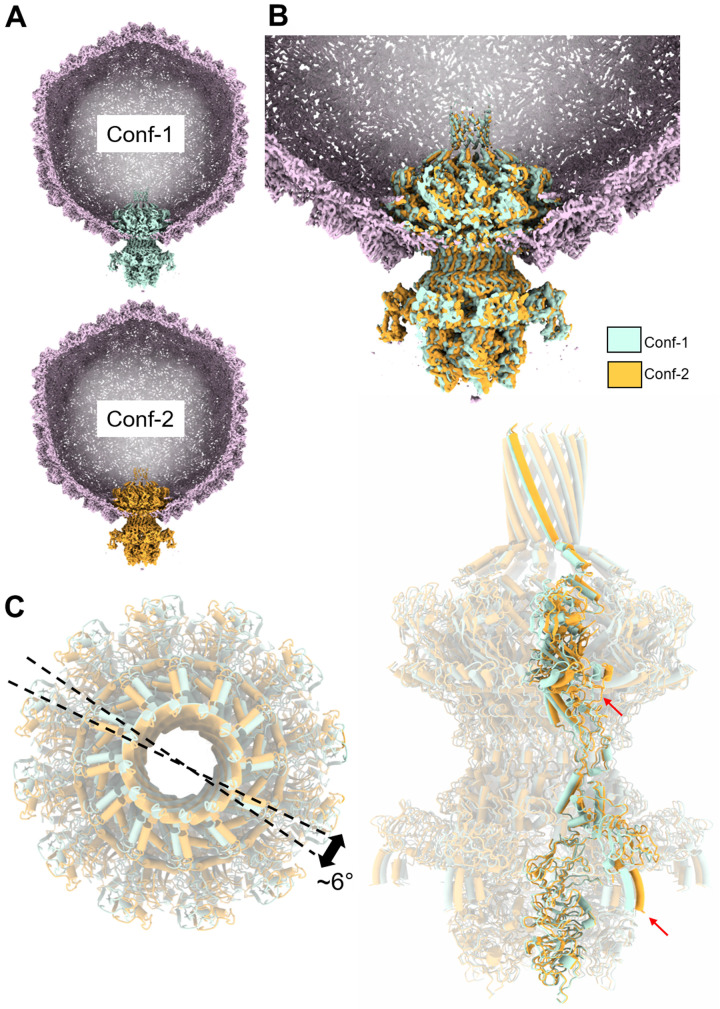
Fig. 1. Localized reconstruction of phage Sf6 tail machine.
(A) A section through the Sf6 virion was visualized at ~1.6σ with the coat protein colored in light magenta. The C6-symmetrized localized reconstruction was overlaid to the unique vertex and is magnified in (B). Tail factors visible at high contour are the portal protein (blue), head-to-tail adaptor (yellow), tailspike N termini (green), and tail hub (orange).
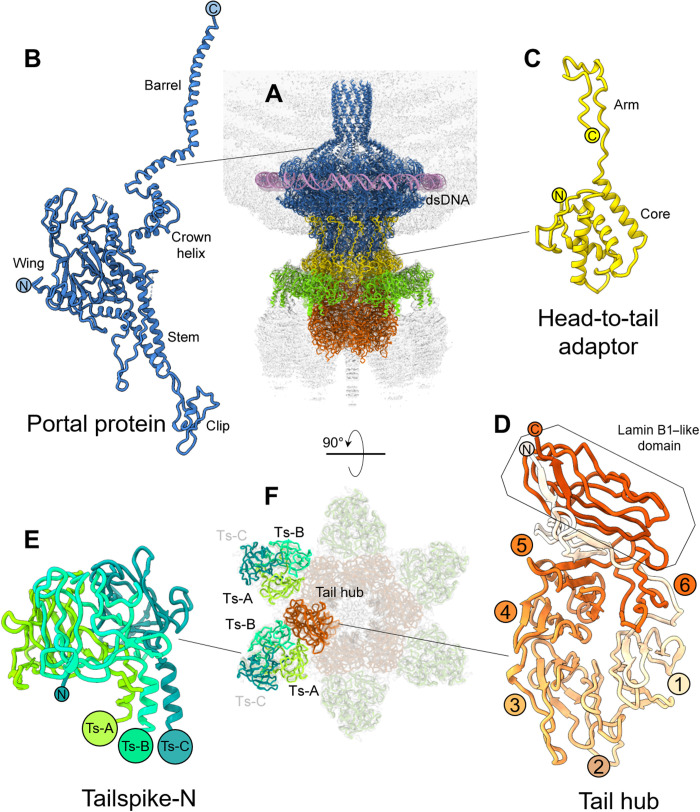
Fig. 2. Atomic structures of Sf6 tail factors.
(A) A ribbon diagram of all tail factors builtde novo in the symmetrized localized reconstruction overlaid to the C6 map of the whole tail displayed at low contour (light gray). Individual tail factors were color-coded, as in Fig. 1. Protomer ribbon diagram for (B) portal protein, (C) head-to-tail adaptor, (D) tail hub, and (E) tailspike N termini (residues 1 to 123) with the three promoters shaded in different tones of green. (F) A bottom view of the symmetrized localized reconstruction in (A) reveals the relative arrangement of two tailspike trimers (each consisting of three subunits Ts-A, Ts-B, and Ts-C) with respect to a tail hub protomer.
Table 1. Map and model refinement statistics.
| Data collection | Shigella phage Sf6 virion | |||||
|---|---|---|---|---|---|---|
| Magnification | 81,000× | |||||
| Voltage (kV) | 300 | |||||
| Electron exposure (e−/Å2) | 50 | |||||
| Defocus range (um) | −0.5 to −1.5 | |||||
| Pixel size (Å) | 0.561 (1.122) | |||||
| Initial particle images (no.) | 67,439 | |||||
| Final particle image (no.) | 39,771 (C12/C6) | 20,320 (C1 Conf-1) | 19,148 (C1 Conf-2) | 67,439 (C1 tailspike) | ||
| Processing and refinement | ||||||
| PDB entry code | 7SFS | 7SG7 | 7SPU | 7SP4 | 7UKJ | |
| Symmetry | C12 | C6 | C1 (Conf-1) | C1 (Conf-2) | C1 | |
| Model | Portal:head-to-tail | Tail hub: tailspike-N | Portal:coat:head-to-tail | Tailspike (full length) | ||
| Map resolution (Å) | 2.7 | 2.8 | 3.8 | 3.7 | 3.6 | |
| Fourier Shell Correlation threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | |
| Initial model used (PDB code) | De novo | De novo | De novo | De novo | De novo + 2VBE | |
| Correlation Coefficient (map-model) | 0.85 | 0.88 | 0.86 | 0.87 | 0.84 | |
| Model composition | Number of chains | 24 | 24 | 54 | 54 | 3 |
| Nonhydrogen atoms | 73,764 | 38,802 | 169,894 | 169,214 | 14,196 | |
| Residues | 9,336 | 4,974 | 21,977 | 21,891 | 1,862 | |
| RMS deviations | Bond lengths (Å) | 0.003 (0) | 0.003 (0) | 0.003 (0) | 0.007 (0) | 0.003 (0) |
| Bond angles (°) | 0.5 (2) | 0.6 (14) | 0.6 (44) | 0.7 (67) | 0.6 (1) | |
| Validation | MolProbity score | 1.59 | 1.76 | 1.63 | 2.03 | 2.38 |
| Clashscore | 4.85 | 4.46 | 5.83 | 8.92 | 11.99 | |
| Rotamer outliers (%) | 0.00 | 0.00 | 0.03 | 0.01 | 1.97 | |
| Ramachandran plot | Favored (%) | 95.08 | 90.32 | 95.54 | 89.97 | 89.44 |
| Allowed (%) | 4.92 | 9.68 | 4.46 | 9.97 | 10.56 | |
| Outliers (%) | 0.00 | 0.00 | 0.00 | 0.06 | 0.00 | |
Sf6 portal protein and head-to-tail adaptor at a 2.7-Å resolution
The Sf6 portal protomer adopts the classic portal fold (18), with five distinct subregions known as the wing, clip, stem, crown, and a C-terminal helical barrel (Fig. 2B and fig. S4). The wing, clip, and stem are structurally continuous and form the portal body, whereas the crown and barrel do not make intramolecular contact with the rest of the protomer tertiary structure and are highly dynamic, possibly moving like a lever (40). A Dali (41) search identified the related phage P22 portal protein (42–44) as the most similar to the Sf6 portal protomer described here (table S1). The root mean square deviation (RMSD) between these two proteins is 3.7 Å despite less than 22% sequence identity. In the quaternary structure of the Sf6 portal, each portal protomer spirals ~145° around the channel axis, over a total length of ~185 Å (fig. S5A). The body of each protomer is nearly parallel to the 12-fold axis running along the central channel, while the stem and the barrel helices are tilted by ~20°. The C-terminal helical barrel (45) was exceptionally well resolved in the reconstruction, allowing unambiguous assignment of all side chains. We found that the interface between two neighboring barrel helices (residues 600 to 671) is stabilized by four hydrogen bonds and 164 van der Waals (hydrophobic) contacts (fig. S5B). This modest binding interface suggests that isolated barrel helices do not form a stable quaternary structure in solution (40) or procapsid (44) but assemble into a tunnel when stabilized by surrounding DNA and possibly ejection proteins (36). As reported for other phages, a ring of electron density around the portal perimeter (44, 46) corresponding to double-stranded DNA (dsDNA) (Fig. 2A) revealed that the portal N termini, which reside in the wing domain, make direct contact with the DNA backbone (fig. S5C). This region of the portal protein was hypothesized to be part of the sensing mechanism that signals headful packaging (44, 46).
Attached to the portal protein, we identified 12 copies of the head-to-tail adaptor gp7. The C12 map had a strong density for the first 150 residues (Fig. 2C and figs. S3A and S4) but no discernable signal for the C-terminal residues 151 to 160. The structure of the Sf6 head-to-tail adaptor is similar but not identical (RMSD of ~4.9 Å) to the crystallographic structure, which crystallized as a monomer (26). The most significant difference between the in situ cryo-EM structure and the x-ray model lies in residues 16 to 36 that are rotated by ~65° in the x-ray model. Sf6 head-to-tail adaptor consists of a helical globular domain and a C-terminal arm inserted at the interface between two portal protein protomers (Fig. 2C and fig. S6). Each head-to-tail protomer makes a handful of contacts with the portal that account for a buried binding interface of ~850 A2 (table S2). In P22, the homologous head-to-tail factor gp4 is monomeric in solution but oligomerizes only upon binding to the portal protein (42, 47), which is also a likely scenario in Sf6, given the 33% sequence identity between head-to-tail adaptors (table S1).
De novo structure of the tail hub and tailspike N termini at a 2.8-Å resolution
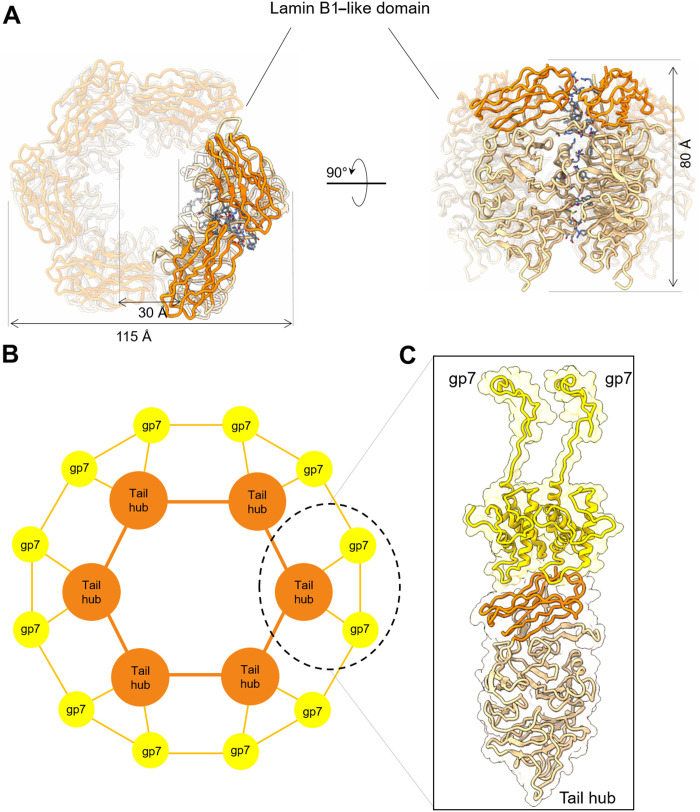
The lower part of the Sf6-localized reconstruction was best visible in the C6 map, consistent with the quaternary structure of the tail hub (gp8) and stoichiometry of tailspikes (gp14) (Fig. 2A) (9). We built a complete atomic model of the tail hub, for which a structure has not been solved previously for any of the P22-like phages (7). The tail hub protomer folds into an interrupted β-propeller, built by six four-stranded blades and an apical insertion domain (Fig. 2D and figs. S3B and S4). The latter domain is formed by N- and C-terminal extensions emanating from the first and sixth blades that come together to generate a lamin B1–like fold, as suggested by Dali (41). Similarly, Dali identified the human E3 ubiquitin-protein ligase WD40 repeat of RFWD3 [Protein Data Bank identifier (PDB ID): 6CVZ] as the known structure with the highest similarity to the Sf6 tail hub gp8 (RMSD = 3.7 Å and Dali Z score = 18.3) (41) despite the limited sequence identity of ~11%. The β-propeller is a ubiquitous protein fold, highly functionally diversified, and found in all kingdoms of life (48) in proteins that do not share a common ancestor. It is unlikely that the tail hub β-propeller evolved to bind DNA but serves only a structural role, which is well conserved in P22-like phages (7). The in situ structure showed six tail hub protomers assembled into a hexameric quaternary structure of 115 Å by 115 Å by 80 Å with the individual β-propellors laying vertically, facing each other (Fig. 3A). Each tail hub:tail hub dimeric interface is stabilized by six hydrogen bonds, one salt bridge, and 16 van der Waals contacts (table S2), overalla modest binding interface for such a large protein, suggesting that the tail hub may not be oligomeric in solution in the absence of a tail. Accordingly, the P22 tail hub ortholog, gp10, which is ~90% identical in the Salmonella/Escherichia/Shigella P22-like phages (7), is monomeric in solution but oligomerizes only during assembly (49, 50).
Fig. 3. Sf6 tail hub quaternary structure and assembly to the head-to-tail adaptor.
(A) Ribbon diagram of the isolated tail hub hexamer with subunits colored in sandy brown and the lamin B1–like domain in orange-red. Residues stabilizing the hexameric assembly in 3.5-Å bonding distance are colored in slate blue. (B) Schematic diagram of the symmetry mismatch between six copies of the tail hub and 12 copies of the head-to-tail adaptor (labeled as gp7). The 1:2 binding interface containing one tail hub protomer bound to two gp7 is circled and shown as a ribbon diagram in (C).
The tail hub interior lumen in the Sf6 tail has a diameter of 30 Å, and the second blade from each propeller forms the bottom of the tail hub channel (Fig. 2D) that contacts the tail needle. However, the density for the tail needle was partially smeared in the C6 map and could be identified only in an asymmetric reconstruction. The binding interface between the hexameric tail hub and the dodecameric head-to-tail adaptor also involves a symmetry mismatch, which is resolved by a 1:2 binding stoichiometry. Each tail hub protomer contacts two head-to-tail adaptors (Fig. 3B), burying a surface of ~1010 Å2 (table S2). The tail hub lamin B1–like domain binds at the interface between two head-to-tail adaptors (Fig. 3C), explaining how a tail hub monomer can only assemble onto a preoligomerized dodecameric ring of head-to-tail factors. This structural observation explains why, in phage P22, the ortholog factors gp10 (tail hub) and gp4 (head-to-tail adaptor) are monomeric and do not significantly interact with each other in solution, but gp10 assembles onto gp4 only when added sequentially to an immobilized portal protein:gp4 complex (49, 50). Thus, the atomic structure of the Sf6 tail explains the sequential and cooperative process by which tail factors self-organize themselves into a molecular machine, previously characterized for P22 (49, 50).
A quasi-twofold symmetric binding interface between tailspike and tail hub
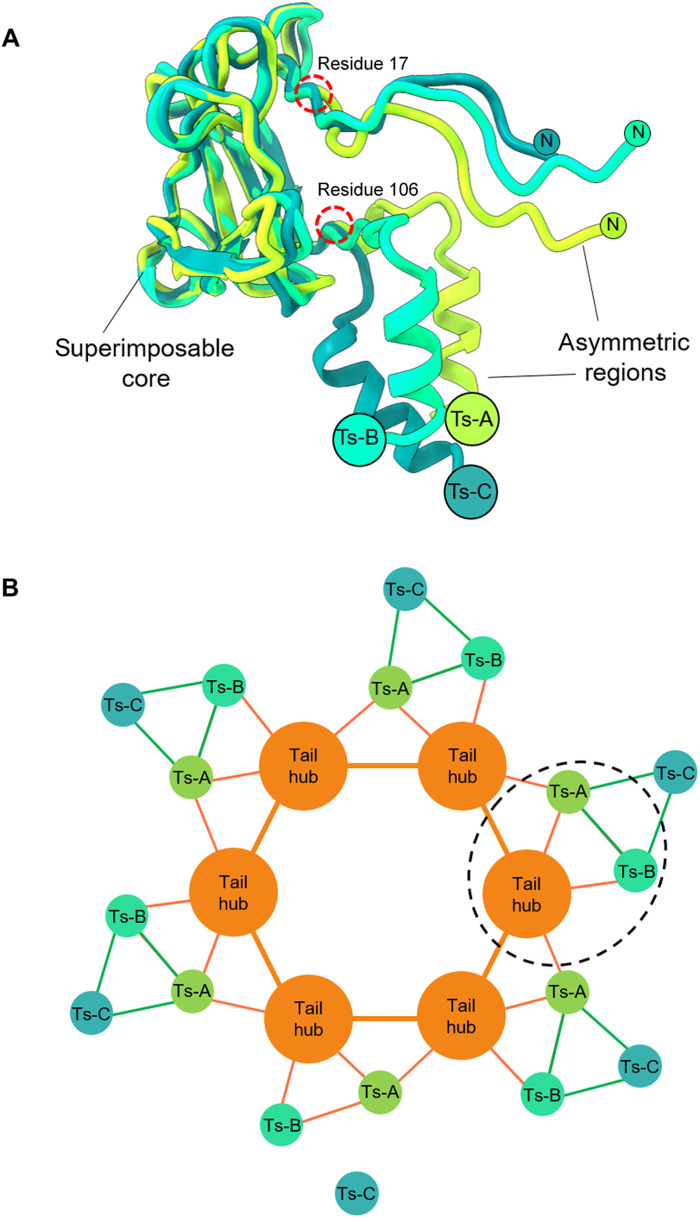
The C6-localized reconstruction also had excellent density for the tailspike N termini that attach laterally to the tail hub (Fig. 2E and fig. S4). We built de novo structures of the tailspike residues 1 to 123, while the tailspike body (residues 124 to 623) was poorly visible in the C6 map because of its flexibility. In Sf6, the tailspikes do not make lateral contacts with the tail hub and thus deviate from perfect sixfold rotational symmetry (9). However, the N-terminal domain of the tailspike, also referred to as the head-binding domain, was perfectly sixfold symmetric. The de novo model of the tailspike N termini revealed an asymmetric homotrimer shaped similar to a clubhead (Fig. 2E), markedly different from the mushroom-like arrangement shown by a previous crystal structure of bacteriophage P22’s tailspike protein, which is 62% identical (table S1) (51). The overall RMSD between protomers varies between 4.7 and 6.0 Å, underscoring the intrinsic asymmetry of the Sf6 tailspike N domains. Tailspike head binding domain protomers deviate in the N-terminal residues 4 to 16 and C-terminal residues 107 to 123, which link the head-binding domain to the homotrimeric tailspike core but are nearly identical between residues 17 and 106 (Fig. 4A).
Fig. 4. Sf6 tailspike N termini asymmetry and assembly to the head hub.
(A) Secondary structure superimposition of the three tailspike-N protomers reveals a superimposable core between residues 17 to 106. Both N and C termini deviate significantly in the three protomers (Ts-A, Ts-B, and Ts-C). (B) Schematic diagram of the symmetry mismatch between the tail hub and a trimer of tailspike N-terminal protomers. The pseudo–2:1 binding interface containing one tail hub protomer bound to Ts-A and Ts-B is circled. Ts-A binds two head-to-tail equivalents intermolecularly, whereas Ts-B binds only one head-to-tail adaptor intramolecularly.
The asymmetry in the tailspike N termini explains the symmetry mismatch between the trimeric tailspike and the hexameric tail hub core. Each tailspike N trimer contains a dummy protomer (Ts-C in Fig. 4B) that points outward and does not contact the tail hub. Instead, the tailspike protomers A and B make direct yet distinct contacts with the tail hub. The tailspike protomer B (Ts-B) contacts just one juxtaposed tail hub protomer, making three hydrogen bonds, two salt bridges, and 25 van der Waals contacts. In contrast, protomer A (Ts-A) makes lateral contacts with two tail hub protomers for a total of eight hydrogen bonds, two salt bridges, and 63 van der Waals interactions (Fig. 4B and table S2). Thus, the built-in asymmetry of tailspike protomers generates a quasi-twofold binding interface that recruits two neighboring tail hub protomers, resolving the symmetry mismatch between a trimer and a hexamer. Given the tail hub conservation (7), we posit that this binding topology is conserved in P22-like phages and perhaps more broadly in Podoviridae.
Sf6 tail exists in two distinct conformations
To identify features that deviate from sixfold symmetry, we also determined an asymmetric reconstruction of the tail apparatus, followed by a new round of 3D classification (fig. S1). Unexpectedly, we identified two distinct conformations of the tail apparatus, Conf-1, and Conf-2, roughly equally represented in our dataset (e.g., 20,000 and 19,000 particles, respectively). The two maps have a cross-CC of 0.87, displaying significant differences in the tail-to-head adaptors (fig. S7). The two conformations were refined to 3.8- and 3.7-Å resolution, respectively (figs. S2 and S3, C and D), revealing near-atomic density features for all components in the tail (Fig. 5, A and B). To rationalize how the dodecameric portal protein assembles at the fivefold vertex, we docked all atomic models of tail factors built in the C12 and C6 maps (Fig. 2A) and the previously determined coat protein (10) in the two conformations. The composite models, ~21,977 and 21,891 residues for Conf-1 and Conf-2, respectively, were subjected to rigid and real-space refinement, yielding final CC = 0.86 and 0.87, respectively (Table 1). The tails were then superimposed, fixing the position of the coat protein layer that serves as a stator, revealing a ~6° rotation between the two conformations (Fig. 5C and fig. S8). The most significant deviations were observed in the portal wing and tailspike N-terminal end (red arrows in Fig. 5C), possibly explaining why the tailspike body was smeared in the C6 map.
Fig. 5. Two conformations of the tail apparatus.
(A) Slice-through view of Conf-1 and Conf-2 with the tail colored in cyan and orange, respectively. The two C1 maps are displayed at high contour (~2.5σ), and no density is visible for the tailspike body, tail needle, and part of the portal barrel. (B) Overlay of the two tail conformations placed at the unique vertex, color-coded as in (A). (C) Overlay of the two isolated tail conformers shown in top (left) and side (right) views. Only one protomer of the portal, head-to-tail adaptor, tail hub, and tailspike N termini is highlighted. The rotation angle between conformers is approximately 6°.
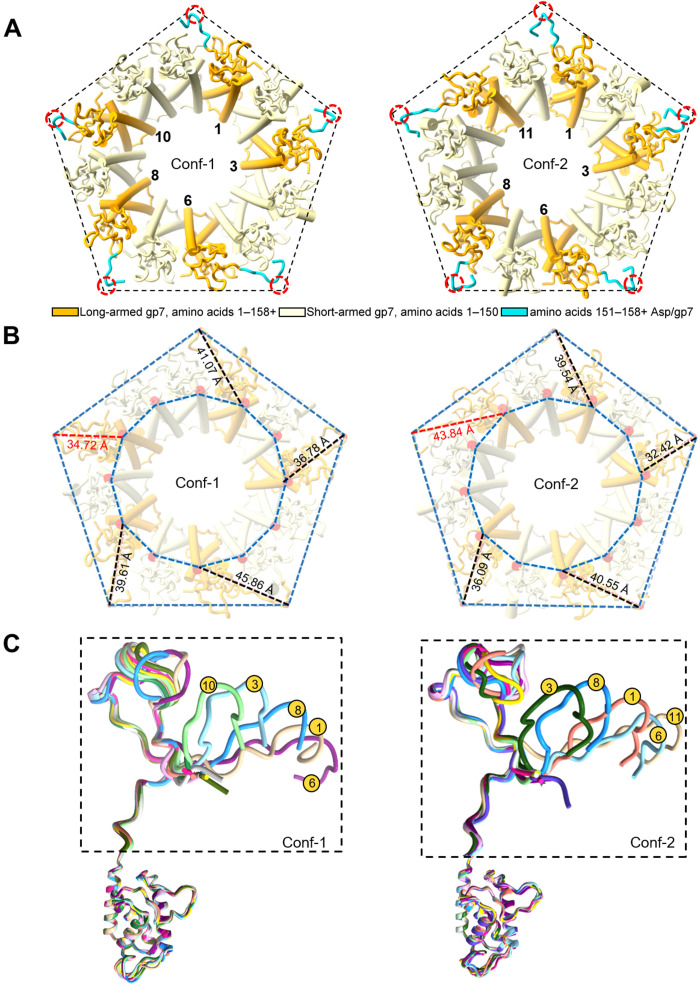
We inspected all bonds formed by the portal and head-to-tail adaptor with coat subunits to rationalize why the tail exists in two equivalent states. The asymmetric map revealed a key feature lost in the C6 map. Five protomers of the head-to-tail adaptor gp7 had extended C-terminal arms (“long-armed protomers”) with residues 151 to 160, making contact with the coat protein (colored in cyan in Fig. 6). The remainder seven head-to-tail adaptors had short arms with no density after residue 150 (Fig. 6A), as seen in the C6 map (“short-armed protomers”), following the perfect sixfold rotational symmetry (Fig. 2C). The five long-armed subunits of the head-to-tail adaptor directly contact the coat protein via C-terminal prongs (fig. S9), effectively obeying the fivefold capsid symmetry. However, the two tail conformations differ in how long- and short-armed protomers are arranged (Fig. 6A). In Conf-1, there was only one short-armed subunit between protomers 8 and 10, whereas, in Conf-2, two short-armed protomers lay between the long-armed protomers 8 and 11. Accordingly, the distances between the long-armed protomers 10 and 11 from a fixed point in the coat varied by ~10 Å in the two conformers (Fig. 6B and fig. S8), generating a ~6° rotation angle between the two superimposed tail conformations (Fig. 5C). Comparing all head-to-tail adaptors in the two conformations revealed that the five long-armed protomers of the head-to-tail adapter have structurally divergent C-terminal prongs (Fig. 6C) that make up most of the contacts with the coat subunits (fig. S9). The overall RMSD between head-to-tail factors in the two states is 2.3 Å, consistent with different structures. The portal protein is identical in the two tail conformers (Fig. 5A) but differs solely in how it inserts at the fivefold vertex due to the ~6° rigid body rotation of Conf-2 with respect to Conf-1. The entire tail apparatus is rigidly held into the fivefold vertex by the head-to-tail C termini, similar to a diamond that can be held in two nonidentical states by the prongs of a ring.
Fig. 6. Five long-armed head-to-tail protomers make direct contacts with the fivefold vertex.
(A) Schematic diagram of the head-to-tail adaptors protomers viewed from the bottom at the capsid fivefold vertex (shown as a pentagon). Five long-armed head-to-tail protomers carrying a C-terminal extension are shown in orange with residues 151 to 158 colored in cyan, while the other seven protomers (residues 1 to 150) are in light yellow. Conf-1 and Conf-2 differ by the pattern of bonding between long-armed head-to-tail adaptors and the coat protein. The dashed red circles point to the contact of gp7 Asp156 with the coat protein. (B) Angular view of the symmetry mismatch shown in (A), illustrating the different distances of head-to-tail protomers from the pentamer vertices in the two tail conformations. The distances between fixed points in the coat and head-to-tail protomers are compared in the two conformers. (C) Overlay of all head-to-tail protomers in Conf-1 and Conf-2 reveals significant variations in the length and conformation of the long-armed subunits C termini.
The mismatched interface between the dodecameric portal and fivefold coat protein
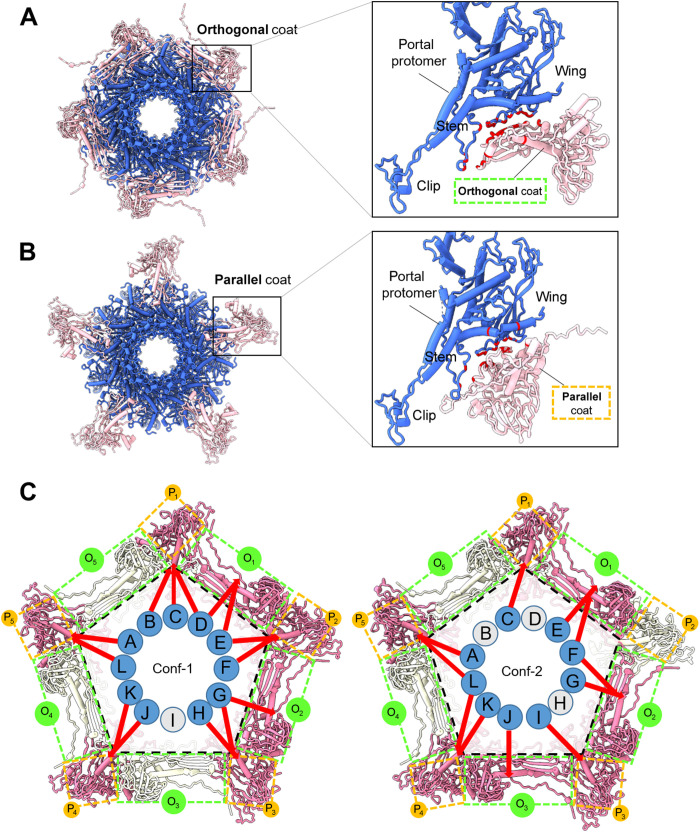
We next analyzed the mismatched interface between the portal and the coat protein in Conf-1 and Conf-2, trying to decipher how this symmetry mismatch is resolved in the mature virion tail. We found that the portal makes many direct contacts with the coat protein lattice within a 5-Å range, but these contacts are different in the two conformations. Specifically, 10 copies of coat proteins are in bonding distance with the portal ring (Fig. 7, A and B). Five coat proteins are roughly orthogonal to the portal wing (Fig. 7A), while the other five run parallel to the outer portal rim (Fig. 7B). In both cases, the portal wing loops make contact with the coat (at portal, residues 11 to 16, 29 to 44, 95 to 106, 203 to 209, and 358 to 396), although the bonding pattern is different in the two conformations. A superimposition of the 12 portal protomers in each conformer revealed no significant conformational changes at the tertiary structure level (RMSD of ~0.1 Å), suggesting that the portal remains homosymmetric in both tail conformers. Similarly, the quaternary structures of portal proteins from Conf-1 and Conf-2 were identical when superimposed (fig. S7). Thus, the portal protein does not change its structure or morph locally to resolve the mismatch (52). Instead, in the two tail conformers, the portal is cemented differently at the fivefold and makes different interactions with the coat lattice. In Conf-1, 11 of 12 portal subunits contact seven coat proteins (five parallel and two orthogonal) of the 10 coat subunits, making 280 bonds that include 11 hydrogen bonds, three salt bridges, and 266 van der Waals contacts (Fig. 7C). In Conf-2, nine portal protomers contact seven coat proteins (four parallel and three orthogonal) , making 353 bonds that include five hydrogen bonds, five salt bridges, and 345 van der Waals contacts (Fig. 7C). Thus, the symmetry mismatch between the dodecameric portal protein and the coat proteins surrounding the portal vertex (located in a fivefold vertex of the icosahedral lattice) is resolved by the quasi-fivefold arrangement of the head-to-tail adaptor C-terminal prongs that create two energetic minima rotated by a discrete angle. The portal is similar to a diamond held over a five-pronged ring.
Fig. 7. The portal protein:coat-mismatched binding interface.
(A and B) Top view of the portal dodecamer (blue) and surrounding coat protein subunits (pink) in bonding distance with the portal dodecamer at the fivefold vertex. Coat subunits can be “orthogonal” (A) or “parallel” (B) to the portal protomer, as shown in the magnified panels on the right-hand side, where bonding residues in the portal and coat are colored in red. (C) A schematic representation of the portal dodecamer at the fivefold vertex. Portal subunits (identified as A to L) making contact with the coat are colored blue, while noncontacting are in gray. Similarly, coat subunits are colored in pink (bonding with portal protomers) or gray (nonbonding). Orthogonal (O1–5) and parallel (P1–5) coat subunits are indicated by dashed lines colored green and orange, respectively. Red arrows indicate the exact portal subunits, making direct contact with coat subunits.
Tail needle and tailspike deviate from perfect sixfold rotational symmetry
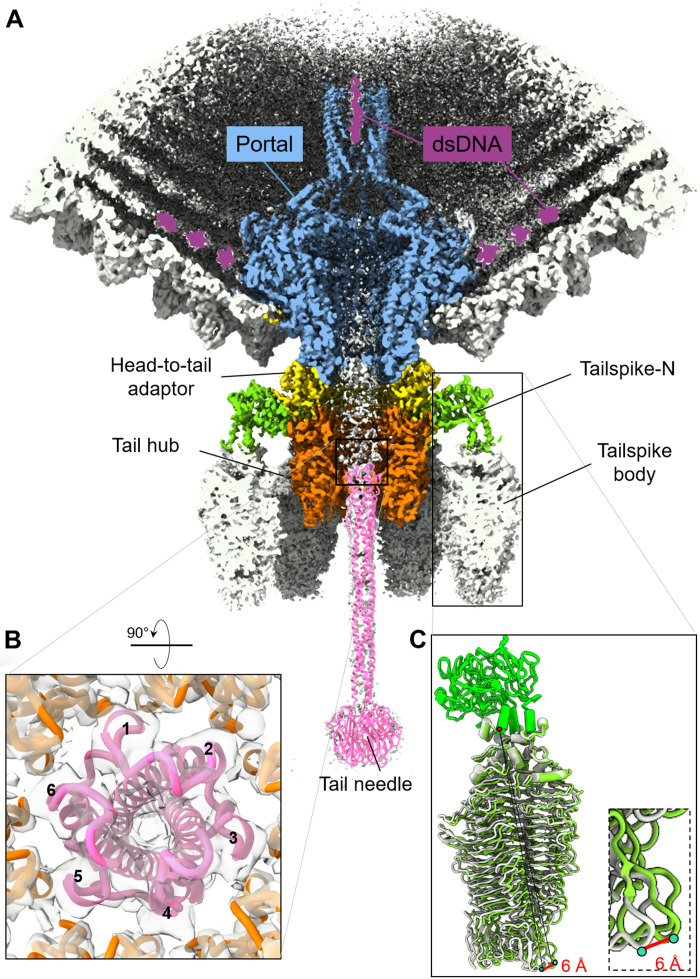
The tail needle (gp9) and tailspike body (residues 124 to 623) had weak density in the C6 map, suggesting deviation from perfect rotational symmetry. Both proteins were visible in the C1 maps, but their density was weaker than for other tail factors. We generated a composite model for the tail needle based on a crystal structure of the Sf6 C-terminal knob (38) and knowledge of the P22 tail needle helical core (29, 31) (table S1). We docked the full-length Sf6 tail needle model into the C1 density and subjected it to real-space refinement (Fig. 8A and fig. S10). The distal knob provided an excellent reference point to model the tail needle in the density and align the N-terminal tip proximal to the tail hub channel. We found that the mismatched interface between the hexameric tail hub channel and trimeric tail needle is resolved by a quasi-sixfold symmetric conformation of the tail needle N termini, whereby a helical hairpin generates a quasi–six-helix bundle symmetric to the tail hub subunits (Fig. 8B).
Fig. 8. A global view of the Sf6 unique vertex.
(A) Magnified view of Sf6 asymmetric reconstruction Conf-2 at a 3.7-Å resolution color-coded as in Fig. 1. At a lower contour level (~1.5σ), the density shows features for dsDNA running around the portal wings (colored in purple), the tailspike body, and the tail needle (gray). An atomic model for the tail needle (colored in magenta) was rigid body–refined against the C1 density and is overlaid to the density. (B) A rotated and magnified view of the tail needle N terminus plugging the tail hub hexameric channel. The tail needle generates a quasi-sixfold binding surface for the hexameric tail hub: The tail needle vertices are labeled 1 to 6. (C) superimposition of the tailspike conformations colored in light green and gray. The zoom-in panel shows the region of maximum displacement.
More complex was the interpretation of the tailspike body, smeared in the C6 map (Fig. 1B). A focused reconstruction of the tailspike core revealed two 3D classes of the spike body, which we refined to a 3.6-Å resolution (Table 1 and fig. S11, A and B). The two classes have identical N termini (residues 1 to 123), but the triple–β helix body (residues 124 to 678) exists in two superimposable conformations (RMSD of 1.1 Å), showing a maximum displacement of ~6 Å at the distal tip (Fig. 8C) (27). The spike body consists of a right-handed β-helix (fig. S11C), which harbors a catalytic site at the interface between subunits with putative catalytic residues Asp399 and Glu366. The C terminus of each tailspike protomer contains a β-sandwich domain that forms a left-handed, coiled β coil (fig. S11C). A maximum displacement of ~6 Å between tailspike conformations occurs at the distal tip of the β-sandwich domain (Fig. 8C). Modeling all six tailspikes in the Sf6 tail, each with its discrete motion of the endorhamnosidase body, suggests that the tailspikes are not static anchors but can fluctuate (fig. S11D) similarly to a scanning device to facilitate host attachment and cleavage of the bacterium LPS, as proposed for Staphylococcus aureus phage P68 tail fibers (53, 54).
Lastly, a distinct density for dsDNA is visible inside the top portion of the portal barrel and the surrounding perimeter (colored in purple in Fig. 8A). Additional density, disconnected from the DNA density in the barrel, is visible at a low contour level at the bottom of the tail channel (fig. S12). Notably, this density has the same shape and size as the Sf6 ejection protein gp12 (35), suggesting that one or more of the ejection proteins may reside inside the tail channel (37).
DISCUSSION
Here, we have described the atomic structure of the Sf6 tail machine, which was solved by combining localized and asymmetric single-particle reconstructions. The total length of the Sf6 tail machine is 300 Å, from the top of the barrel to the tail needle anchoring point in the tail hub. The internal diameter ranges from ~19 Å in the middle of the tunnel loops to ~45 Å in the portal vestibule, right under the barrel (Fig. 8A). A Coulombic electrostatic potential shows that the DNA channel is mildly negatively charged and accommodates ~100 bases of dsDNA (fig. S13). Overall, our work details the whole architecture of a Podoviridae noncontractile tail machine, resolving four symmetry mismatches and revealing the existence of two conformations of the tail apparatus.
The existence of a symmetry mismatch between the dodecameric portal protein and the fivefold vertex has intrigued scientists for over half a century (55). With the advent of high-resolution cryo-EM and localized reconstruction techniques (56), it has been possible to visualize the structure of the portal protein in situ and its binding association with the coat protein (56, 57). Fang et al. (52) reported the in situ structure of a symmetry-mismatched viral portal vertex from phage T4. These authors determined that the portal subunits underwent local conformational changes, reaching out to nearby capsid subunits and forming similar interactions in different capsid environments. The reported morphing of portal protein subunits was limited to regions of the portal wing facing the capsid that underwent subtle motion, while the rest of the portal structure remained in strict 12-fold symmetry. On the basis of these findings, Fang et al. (52) speculated that the structural morphing of portal subunits compensates for structural and symmetry conflicts, creating a dynamic vertex that drives distinct processes. However, this work was based on a medium-resolution (~4.5 Å) reconstructionof a T4 mutant consisting of a mature head emptied of DNA and lacking the tail (52). Similarly, a cryo-EM procapsid structure from the thermostable phage P23-45 revealed local deviations in the portal protein that allowed it to fit into the coat lattice. Regions within the portal wing and clip domains were found to undergo local conformational changes to adjust and make specific contacts with the major capsid protein (58, 59), although the overall portal topology remained 12-fold symmetric. Thus, the notion of portal morphing to resolve the symmetry mismatch with the coat is likely true for a precursor capsid (e.g., procapsid) or a tailless mature virion mutant, as in T4, lacking the tail-to-head adaptor. In the absence of this factor, the portal has greater freedom to move inside the capsid and possibly morph locally to resolve the symmetry mismatch with the coat.
Our work on the phage Sf6 tail reveals a different strategy to assemble a sixfold symmetric tail apparatus at a fivefold vertex of a mature, tailed virion. We found that, as part of the tail apparatus, the portal exists in two discrete conformations, identical to each other but rotated by ~6°. The two tail conformations are cemented at the fivefold unique vertex by the C-terminal prongs of five unique head-to-tail adaptors that generate two combinations of contacts with the coat lattice due to a nondivisible relationship between 12 (number of head-to-tail adaptors) and 5 (number of vertices at the fivefold). The head-to-tail prongs anchor two structurally identical tail complexes at the unique vertex, rotated by ~6° around the capsid that functions as a stator. Portal protomers make different bonds in the two conformers with distinct coat subunits, mainly via the wing region. Thus, the portal does not morph but is held similar to a diamond onto a five-pronged ring in two distinct positions, reflecting energetically equivalent states. A dynamic equilibrium is unlikely between these two states that are thermodynamic end points in the tail assembly. Our work on the Sf6 tail machine extends Caspar and Klug’s principle of quasi-equivalence (60), well established for coat proteins in the capsid lattice, to the head-to-tail adaptor gp7, which is a highly structurally conserved tail factor in the P22-like bacteriophages (7, 26). The principle of quasi-equivalence explains how viral coat proteins can adopt locally similar but nonidentical positions across the capsid surface, effectively allowing a single protein to close into an icosahedral cage. Here, we show that the quasi-equivalence of Sf6 head-to-tail adaptors at the level of the C-terminal prongs determines two ways to cement the tail apparatus at the fivefold vertex, solving the mismatched symmetry interface between the portal and coat protein. The head-to-tail adaptor binds the portal protein to initiate displacement of TerL, marking the end of genome packaging and stabilizing the genome inside the capsid (25).
We also resolved a second symmetry mismatch between the sixfold symmetrictail hub, which we built de novo at a 2.8-Å resolution, and the trimeric tailspike head domain. We found that the tailspike N termini are profoundly asymmetric and one of the three subunits does not make contact with the tail hub. The tailspike N termini, also known as the head-binding domain (51), can be considered a pseudo-twofold symmetric trimer that uses only two protomers to contact a tail hub monomer. Lastly, our structure sheds light on a simpler symmetry mismatch between the dodecameric head-to-tail adapter and the hexameric tail hub, whereby the lamin B1–like domain formed from the N and C termini of the tail hub inserts at the dimerization interface between two head-to-tail adaptors, making symmetric 2:1 contacts. Similarly, we deciphered how the tail needle can plug the tail hub tunnel. The N-terminal α-helical hairpin of the Sf6 tail needle is highly conserved in all P22-like phages (29, 50, 61), where it generates a quasi-sixfold plug that makes symmetric contacts with the hexameric tail hub channel.
The results presented here, together with a wealth of data available in the literature on P22-like phages (9), allow us to propose a model for the Shigella phage Sf6 assembly. The Sf6 procapsid self-assembles in infected cells from the coat, scaffolding, and portal protein (62). Each of these three proteins exists in procapsids, an immature conformation poised for maturation upon DNA packaging. The portal dodecamer adopts a metastable, asymmetric structure with individual portal protomers arranged in a quasi-fivefold conformation (44), which, we suggest, morph locally to bond nearby coat subunits (52). The procapsid portal recruits TerL, which oligomerizes to form a pentameric motor and newly replicated concatemeric Sf6 DNA in a reaction mediated by TerS. TerL packages the viral genome, possibly using a helical-to-planar ratchet mechanism (20), which results in the expansion and maturation of the Sf6 capsid (10). As the head fills up and the packaging motor slows down, the headful nuclease located at the C terminus of TerL cleaves the viral genome (24), concomitant with the head-to-tail gp7 oligomerization upon binding to the portal (47) that competes off TerL.
Our data, presented here, indicate two ways to cement the portal dodecamer at the fivefold vertex dictated by the nature of the head-to-tail, which makes simultaneous contacts with the portal and coat protein. The portal can exist in two bonding interactions with the coat lattice at the fivefold vertex, retaining an identical structure in the two states but making different bonds. We propose that the two conformations are generated by the fact that the head-to-tail factor does not assemble onto the portal as a preformed dodecamer but oligomerizes sequentially upon binding (47), possibly from different positions of the portal protein that concomitantly undergoes maturation from an asymmetric procapsid conformation to a symmetric dodecamer (44). The head-to-tail dodecameric surface is then attached by six copies of the tail hub, which also assembles sequentially from monomeric protomers into a hexameric ring (49), extending the DNA channel. Because the binding of both tail factors (e.g., head-to-tail onto portal and tail hub onto head-to-tail) involves the oligomerization of monomers into an oligomer, the assembly reaction is essentially unidirectional, as monomer dissociation would be energetically less favored than binding and oligomerization. The assembly reaction is not complete until the tail needle seals the tail channel using an N-terminal quasi-sixfold symmetric helical hairpin (63) that plugs the sixfold symmetric channel, stabilizing the newly packaged DNA (25). The trimeric tailspike is the last factor to complete the Sf6 virion. The symmetry mismatch of six trimeric tailspike assembling onto a tail hub hexamer is resolved by the asymmetric conformation of the tailspike N domain, where only two tailspike protomers contact one tail hub, leaving a third subunit solvent exposed. A hinge between the Sf6 tailspike N terminus and core makes this spike very flexible, unlike P22 (9). Because there is a density that we ascribe to protein in the bottom of the tail channel, we propose that once the tail channel is fully assembled and sealed, some of the ejection proteins encapsidated in the procapsid may move into the tail channel, ready to be ejected into the host cell envelope upon attachment (32). The end of the Sf6 genome is visible in the upper part of the portal, possibly bound or threaded through other ejection proteins clustered in the proximity of the portal barrel (36).
In summary, we have deciphered the architecture and design principles of a phage tail machine using the power of localized reconstruction. The most unexpected finding of this work is the unexpected combination of pseudo-symmetric interfaces (e.g., tail hub:tailspike-N domain, tail hub:tail needle, etc.) and asymmetric features (e.g., head-to-tail C-terminal prongs and tailspike-N) that plastically accompany the symmetry fallout from a dodecameric portal protein to the trimeric tail needle. Our work explains the hierarchical sequential and highly cooperative assembly of the head-to-tail adaptor, tail hub, and tail needle previously observed biochemically for P22 (47, 49, 50, 64). We propose that these structural principles of virus assembly are conserved in other short-tailed bacteriophages of the Podoviridae superfamily.
MATERIALS AND METHODS
Sf6 phage generation for cryo-EM
S. flexneri serotype Y strain PE577 was grown at 30°C to midlog (2 × 108 cells/ml) in superbroth and infected with Sf6 phage at a multiplicity of infection of 0.07. Growth was continued until cell lysis or for up to 6 hours before addition of 0.6% chloroform. Cell debris was pelleted at 5422g for 10 min in a Sorvall Lynx 6000 centrifuge (ThermoFisher Scientific, Waltham MA) using an F9-6 x 1000 LEX rotor (Thermo Fisher Scientific). The supernatant was treated with 7% polyethylene glycol (molecular weight, 6000 to 8000) and 0.5 M NaCl. The solution was stirred for ~16 hours at 4°C before pelleting at 13,881g using the same centrifuge and rotor as above. The pellets were resuspended in 20 mM tris and 100 mM MgCl2 (pH 7.6) and centrifuged at 8228g for 10 min in an F20-12 x 50 LEX rotor (Thermo Fisher Scientific) to remove debris. The supernatant was centrifuged at 146,300g for 45 min in a Sorvall WX+ ultracentrifuge Series (Thermo Fisher Scientific) using a Thermo Fisher Scientific T-865 rotor to pellet phage. Pellets were resuspended in 20 mM tris and 100 mM MgCl2 on a reciprocal shaker (Eberbach, Ann Arbor MI), operating at 180 revolutions per minute for ~16 hours at 4°C. The phage and procapsid mixture was separated using cesium chloride gradients. The mixture was applied to the top of the gradient formed from a 25% sucrose solution (1.4 grams per cubic centimeter) and cesium chloride solution (1.6 grams per cubic centimeter; top to bottom). Sucrose and cesium chloride solutions were made using 20 mM sodium phosphate (pH 7.6). Gradients were centrifuged at 90,980g for 45 min in a Sorvall MX 120 micro-ultracentrifuge (Thermo Fisher Scientific) using a S50ST rotor. The phage band sediments at the interface between the 1.4 and 1.6 grams per cubic centimeter steps were extracted and dialyzed against 10 mM tris and 25 mM MgCl2 (pH 7.6).
Vitrification and data collection
Sf6 mature virions (2.5 μl) at 1 × 1014 phages per ml were applied to a 200-mesh copper Quantifoil R 2/1 holey carbon grid (EMS) previously glow discharged negatively for 60 s at 15 mA using an easiGlow (PELCO). The grid was blotted for 7 s at a blot force of 1 and frozen immediately in liquid ethane using a Vitrobot Mark IV (FEI). Micrographs were screened on a 200-kV Glacios equipped with a Falcon4 detector at Thomas Jefferson University. EPU software was used for data collection using accurate positioning mode. For high-resolution data collection, micrographs were collected on a Titan Krios microscope operated at 300 kV and equipped with a K3 direct electron detector camera (Gatan) at the National Cryo-EM Facility at the Frederick National Laboratory, MD. Micrographs were collected in super-resolution mode plus energy filter with an image pixel size of 0.561 Å, a nominal 81,000× magnification, a total dose of 50 e-/Å2, 40 frames, and a defocus range of −0.5 to −1.5 μm. Additional collection parameters are in Table 1.
Cryo-EM single-particle analysis
Two sets of micrographs were combined after motion correction with MotionCor2 (65), yielding 7977 micrographs. RELION’s implementation of motion correction was applied to the micrographs with options of dose-weighted averaged micrographs and sum of non–dose-weighted power spectra every 4 e−/Å2. CTF (contrast transfer function) estimation was carried out using CTFFIND4 (66). After an initial round of reference picking and 2D classification, 67,000 virion particles were subjected to a reference-free low-resolution reconstruction without imposing symmetry. The particles were then 3D classified into four classes with I3 symmetry imposed. Of the four classes, one class of 53,000 particles had the most resolved reconstruction and was subjected to 3D autorefined to align the particles finely. The particles were then expanded according to I3 symmetry using RELION’s relion_particle_symmetry_expand function to obtain 60 times the 53,000 particles. A cylindrical mask (r = 200 Å) was generated using Scipion 3.0 (67) and then resampled onto a reference map covering the fivefold vertex in UCSF Chimera (68). The cylindrical mask was then used for nonsampling 3D classification without imposing symmetry to search for the tail. Locally aligned particles were then combined from three classes, and duplicates were removed, yielding 39,000 particles. The initial localized reference map was reconstructed directly from one of the classes using RELION’s relion_reconstruct function. Selected 3D classes were further autorefined using C5 symmetry, followed by fivefold symmetry particle expansion. The expanded particles were subjected to a third 3D classification, and the map was symmetrized by imposing C12 and C6, which gave the best density for the portal protein/head-to-tail adaptor and tail hub/tailspike, respectively. After the fourth 3D classification using C6 symmetry expanded particles, local maps with two tail apparatus conformations were obtained. Full virion maps with two conformations were also traced after the former pool of particles were binned to two-thirds. All steps of single-particle analysis (SPA), including 2D/3D classification, 3D refinement, CTF refinement, particle polishing, postprocessing, and local resolution calculation, were carried out using RELION 3.1.2 (69, 70). The final densities were sharpened using phenix.auto_sharpen (71). RELION_postprocess (69, 70) was used for local-resolution estimation, and drawings of electron density maps and local resolution maps were generated using UCSF ChimeraX (72). A detailed workflow of SPA is shown in fig. S1.
De novo model building, oligomer generation, and refinement
The 2.7-Å C12-averaged localized reconstruction was used to build a model of dodecameric portal protein (residues 1 to 671) bound to 12 copies of the head-to-tail adaptor (residues 1 to 150). Sf6 portal barrel was modeled between 600 and 671, while C-terminal residues 672 to 708 had no discernable density. Similarly, the head-to-tail adaptor C-terminal residues 151 to 160 were not visible in the C12 map. Using the 2.8-Å C6-averaged localized reconstruction, we built a complete model of the hexameric tail hub (residues 2 to 472) bound to six copies of the trimeric tailspike N termini (residues 4 to 123). The tailspike body (residues 124 to 623) was modeled in a focused asymmetric reconstruction generated from the C6 symmetry expanded map, using a cylindrical mask covering only one tailspike (fig. S11A). All de novo atomic models (except the tail needle) shown in this paper were built manually using Coot (73) or Chimera (68) and refined using several rounds of rigid-body, real-space, and B factor refinement using phenix.real_space_refinement (74). The final models of the portal:head-to-tail adaptor (24 chains) and tail hub:tailspike-N (24 chains) yielded a final CC of 0.85 and 0.88, respectively, and excellent stereochemistry (Table 1). After obtaining accurate atomic models from the C12/C6-symmetrized local reconstruction, the portal head-to-tail adaptor, tail hub, and tailspike models were placed in the C1 maps, revealing additional densities for the C-terminal residues 151 to 160 of five head-to-tail adaptors. The models were extended by hand in Coot (73), and all tail components corresponding to ~21,977 and 21,891 residues for Conf-1 and Conf-2, respectively, were subjected to rigid body and real-space refinement, yielding a final CC = 0.86 and 0.87, respectively (Table 1). The full-length tailspike was modeled by combining N-terminal residues 4 to 123 (build de novo in the C6 map) and residues 124 to 623 in the tailspike body, which are similar to the crystal structure (PDB ID: 2VBE) (27). The model was placed in the 3.6-Å asymmetric focused map and real space–refined, yielding a final CC = 0.84 (Table 1). Last, the tail needle knob (PDB ID: 3RWN) (38) and a model of the full-length Sf6 tail needle (50) were placed in the C1 density refined using the fit-into-map command in Chimera (68). All final models were validated using MolProbity (75) (Table 1).
Structure analysis
All ribbon and surface representations were generated using ChimeraX (72) and PyMOL (76). Structural neighbors and flexible regions were identified using the Dali server (41). Binding interfaces were analyzed using PISA (77) and PDBsum (78) to determine bonding interactions, interatomic distances, and type of bonds. Sequence and secondary structure alignment were done using PDBsum (78). DynDom (79) was used to identify domain movements. RMSD between superimposed PDBs was calculated using SuperPose version 1.0 (superpose.wishartlab.com) (80). dsDNA was generated using Coot (73). The Coulombic electrostatic potential was calculated and displayed with surface coloring using ChimeraX (72).
Acknowledgments
We thank the National Cryo-EM Facility staff at National Cancer Institute (NCI) Frederick National Laboratory for assistance in data collection.
Funding: This work was supported by the National Institutes of Health grants R01 GM100888, R35 GM140733, and S10 OD030457 to G.C. and R01 GM076661 to C.M.T. Research in this publication includes work carried out at the Sidney Kimmel Cancer Center X-ray Crystallography and Molecular Interaction Facility at Thomas Jefferson University, which is supported in part by NCI Cancer Center Support Grant P30 CA56036. This research was partly supported by the NCI’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research under contract HSSN261200800001E.
Author contributions: F.L., C.-F.D.H., and R.Y. performed all steps of cryo-EM and structural analysis, deposition of atomic coordinates and maps, and figure preparation. R.W. purified the Sf6 virion. G.C. and C.M.T. supervised the project and wrote the paper. All authors contributed to the writing and editing of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Atomic coordinates for Sf6 portal:head-to-tail complex (C12), tail hub:tailspike-N (C6), portal:head-to-tail:coat from Conf-1 (C1), portal:head-to-tail:coat from Conf-2 (C1), and full-length tailspike (C1) have been deposited in the PDB with accession codes 7SFS, 7SG7, 7SPU, 7SP4, and 7UKJ; the cryo-EM density maps have been deposited in the Electron Microscopy Data Bank with accession codes EMD-25101, EMD-25106, EMD-23372, EMD-25365, and EMD-26582.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Tables S1 and S2
REFERENCES AND NOTES
- 1.S. S. Tang, S. K. Biswas, W. S. Tan, A. K. Saha, B. F. Leo,Efficacy and potential of phage therapy against multidrug resistant Shigella spp. PeerJ 7,e6225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.I. A. Khalil, C. Troeger, B. F. Blacker, P. C. Rao, A. Brown, D. E. Atherly, T. G. Brewer, C. M. Engmann, E. R. Houpt, G. Kang, K. L. Kotloff, M. M. Levine, S. P. Luby, C. A. MacLennan, W. K. Pan, P. B. Pavlinac, J. A. Platts-Mills, F. Qadri, M. S. Riddle, E. T. Ryan, D. A. Shoultz, A. D. Steele, J. L. Walson, J. W. Sanders, A. H. Mokdad, C. J. L. Murray, S. I. Hay, R. C. Reiner Jr.,Morbidity and mortality due to Shigella and enterotoxigenic Escherichia coli diarrhea: The Global Burden of Disease Study 1990–2016. Lancet Infect. Dis. 18,1229–1240 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.E. Tacconelli, E. Carrara, A. Savoldi, S. Harbarth, M. Mendelson, D. L. Monnet, C. Pulcini, G. Kahlmeter, J. Kluytmans, Y. Carmeli, M. Ouellette, K. Outterson, J. Patel, M. Cavaleri, E. M. Cox, C. R. Houchens, M. L. Grayson, P. Hansen, N. Singh, U. Theuretzbacher, N. Magrini; WHO Pathogens Priority List Working Group ,Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18,318–327 (2018). [DOI] [PubMed] [Google Scholar]
- 4.L. D. Goodridge,Bacteriophages for managing Shigella in various clinical and non-clinical settings. Bacteriophage 3,e25098 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.A. Llanos-Chea, R. J. Citorik, K. P. Nickerson, L. Ingano, G. Serena, S. Senger, T. K. Lu, A. Fasano, C. S. Faherty,Bacteriophage therapy testing against Shigella flexneri in a novel human intestinal organoid-derived infection model. J. Pediatr. Gastroenterol. Nutr. 68,509–516 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.P. Gemski Jr., D. E. Koeltzow, S. B. Formal,Phage conversion of Shigella flexneri group antigens. Infect. Immun. 11,685–691 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.S. R. Casjens, P. A. Thuman-Commike,Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology 411,393–415 (2011). [DOI] [PubMed] [Google Scholar]
- 8.N. K. Verma, J. M. Brandt, D. J. Verma, A. A. Lindberg,Molecular characterization of the O-acetyl transferase gene of converting bacteriophage SF6 that adds group antigen 6 to Shigella flexneri. Mol. Microbiol. 5,71–75 (1991). [DOI] [PubMed] [Google Scholar]
- 9.K. N. Parent, E. B. Gilcrease, S. R. Casjens, T. S. Baker,Structural evolution of the P22-like phages: Comparison of Sf6 and P22 procapsid and virion architectures. Virology 427,177–188 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.H. Zhao, K. Li, A. Y. Lynn, K. E. Aron, G. Yu, W. Jiang, L. Tang,Structure of a headful DNA-packaging bacterial virus at 2.9 A resolution by electron cryo-microscopy. Proc. Natl. Acad. Sci. U.S.A. 114,3601–3606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.W. R. Wikoff, L. Liljas, R. L. Duda, H. Tsuruta, R. W. Hendrix, J. E. Johnson,Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289,2129–2133 (2000). [DOI] [PubMed] [Google Scholar]
- 12.W. Jiang, L. Tang,Atomic cryo-EM structures of viruses. Curr. Opin. Struct. Biol. 46,122–129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.K. N. Parent, M. L. Erb, G. Cardone, K. Nguyen, E. B. Gilcrease, N. B. Porcek, J. Pogliano, T. S. Baker, S. R. Casjens,OmpA and OmpC are critical host factors for bacteriophage Sf6 entry in Shigella. Mol. Microbiol. 92,47–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.H. Zhao, R. D. Sequeira, N. A. Galeva, L. Tang,The host outer membrane proteins OmpA and OmpC are associated with the Shigella phage Sf6 virion. Virology 409,319–327 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.S. R. Casjens, I. J. Molineux,Short noncontractile tail machines: Adsorption and DNA delivery by podoviruses. Adv. Exp. Med. Biol. 726,143–179 (2012). [DOI] [PubMed] [Google Scholar]
- 16.K. N. Parent, J. R. Schrad, G. Cingolani,Breaking symmetry in viral icosahedral capsids as seen through the lenses of x-ray crystallography and cryo-electron microscopy. Viruses 10,67 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A. Bhardwaj, A. S. Olia, G. Cingolani,Architecture of viral genome-delivery molecular machines. Curr. Opin. Struct. Biol. 25,1–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.C. L. Dedeo, G. Cingolani, C. M. Teschke,Portal protein: The orchestrator of capsid assembly for the dsDNA tailed bacteriophages and herpesviruses. Annu. Rev. Virol. 6,141–160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.H. Zhao, T. E. Christensen, Y. N. Kamau, L. Tang,Structures of the phage Sf6 large terminase provide new insights into DNA translocation and cleavage. Proc. Natl. Acad. Sci. U.S.A. 110,8075–8080 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.M. Woodson, J. Pajak, B. P. Mahler, W. Zhao, W. Zhang, G. Arya, M. A. White, P. J. Jardine, M. C. Morais,A viral genome packaging motor transitions between cyclic and helical symmetry to translocate dsDNA. Sci. Adv. 7,eabc1955 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.H. Zhao, C. J. Finch, R. D. Sequeira, B. A. Johnson, J. E. Johnson, S. R. Casjens, L. Tang,Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proc. Natl. Acad. Sci. U.S.A. 107,1971–1976 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.S. Casjens, D. A. Winn-Stapley, E. B. Gilcrease, R. Morona, C. Kuhlewein, J. E. Chua, P. A. Manning, W. Inwood, A. J. Clark,The chromosome of Shigella flexneri bacteriophage Sf6: Complete nucleotide sequence, genetic mosaicism, and DNA packaging. J. Mol. Biol. 339,379–394 (2004). [DOI] [PubMed] [Google Scholar]
- 23.J. C. Leavitt, E. B. Gilcrease, K. Wilson, S. R. Casjens,Function and horizontal transfer of the small terminase subunit of the tailed bacteriophage Sf6 DNA packaging nanomotor. Virology 440,117–133 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.A. Roy, G. Cingolani,Structure of p22 headful packaging nuclease. J. Biol. Chem. 287,28196–28205 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.H. Strauss, J. King,Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J. Mol. Biol. 172,523–543 (1984). [DOI] [PubMed] [Google Scholar]
- 26.L. Liang, H. Zhao, B. An, L. Tang,High-resolution structure of podovirus tail adaptor suggests repositioning of an octad motif that mediates the sequential tail assembly. Proc. Natl. Acad. Sci. U.S.A. 115,313–318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.J. J. Muller, S. Barbirz, K. Heinle, A. Freiberg, R. Seckler, U. Heinemann,An intersubunit active site between supercoiled parallel β helices in the trimeric tailspike endorhamnosidase of Shigella flexneri Phage Sf6. Structure 16,766–775 (2008). [DOI] [PubMed] [Google Scholar]
- 28.J. E. H. Chua, P. A. Manning, R. Morona,The Shigella flexneri bacteriophage Sf6 tailspike protein (TSP)/endorhamnosidase is related to the bacteriophage P22 TSP and has a motif common to exo- and endoglycanases, and C-5 epimerases. Microbiology (Reading) 145 (Pt 7),1649–1659 (1999). [DOI] [PubMed] [Google Scholar]
- 29.A. Bhardwaj, N. Walker-Kopp, S. R. Casjens, G. Cingolani,An evolutionarily conserved family of virion tail needles related to bacteriophage P22 gp26: Correlation between structural stability and length of the alpha-helical trimeric coiled coil. J. Mol. Biol. 391,227–245 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D. Andrews, J. S. Butler, J. Al-Bassam, L. Joss, D. A. Winn-Stapley, S. Casjens, G. Cingolani,Bacteriophage P22 tail accessory factor GP26 is a long triple-stranded coiled-coil. J. Biol. Chem. 280,5929–5933 (2005). [DOI] [PubMed] [Google Scholar]
- 31.A. S. Olia, S. Casjens, G. Cingolani,Structure of phage P22 cell envelope-penetrating needle. Nat. Struct. Mol. Biol. 14,1221–1226 (2007). [DOI] [PubMed] [Google Scholar]
- 32.N. A. Swanson, C. D. Hou, G. Cingolani,Viral ejection proteins: Mosaically conserved, conformational gymnasts. Microorganisms 10,504 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.N. A. Swanson, R. K. Lokareddy, F. Li, C.-F. D. Hou, S. Leptihn, M. Pavlenok, M. Niederweis, R. A. Pumroy, V. Y. Moiseenkova-Bell, G. Cingolani,Cryo-EM structure of the periplasmic tunnel of T7 DNA-ejectosome at 2.7 A resolution. Mol. Cell 81,3145–3159.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M. Perez-Ruiz, M. Pulido-Cid, J. R. Luque-Ortega, J. M. Valpuesta, A. Cuervo, J. L. Carrascosa,Assisted assembly of bacteriophage T7 core components for genome translocation across the bacterial envelope. Proc. Natl. Acad. Sci. U.S.A. 118,e2026719118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.H. Zhao, J. A. Speir, T. Matsui, Z. Lin, L. Liang, A. Y. Lynn, B. Varnado, T. M. Weiss, L. Tang,Structure of a bacterial virus DNA-injection protein complex reveals a decameric assembly with a constricted molecular channel. PLOS ONE 11,e0149337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.W. Wu, J. C. Leavitt, N. Cheng, E. B. Gilcrease, T. Motwani, C. M. Teschke, S. R. Casjens, A. C. Steven,Localization of the houdinisome (ejection proteins) inside the bacteriophage P22 virion by bubblegram imaging. MBio 7,e01152-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.C. Wang, J. Tu, J. Liu, I. J. Molineux,Structural dynamics of bacteriophage P22 infection initiation revealed by cryo-electron tomography. Nat. Microbiol. 4,1049–1056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.A. Bhardwaj, I. J. Molineux, S. R. Casjens, G. Cingolani,Atomic structure of bacteriophage Sf6 tail needle knob. J. Biol. Chem. 286,30867–30877 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J. C. Leavitt, L. Gogokhia, E. B. Gilcrease, A. Bhardwaj, G. Cingolani, S. R. Casjens,The tip of the tail needle affects the rate of DNA delivery by bacteriophage p22. PLOS ONE 8,e70936 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.C. F. David Hou, N. A. Swanson, F. Li, R. Yang, R. K. Lokareddy, G. Cingolani,Cryo-EM structure of a kinetically trapped dodecameric portal protein from the pseudomonas-phage PaP3. J. Mol. Biol. 434,167537 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.L. Holm, P. Rosenstrom,Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38,W545–W549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.A. S. Olia, P. E. Prevelige Jr., J. E. Johnson, G. Cingolani,Three-dimensional structure of a viral genome-delivery portal vertex. Nat. Struct. Mol. Biol. 18,597–603 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.H. Zheng, A. S. Olia, M. Gonen, S. Andrews, G. Cingolani, T. Gonen,A conformational switch in bacteriophage p22 portal protein primes genome injection. Mol. Cell 29,376–383 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R. K. Lokareddy, R. S. Sankhala, A. Roy, P. V. Afonine, T. Motwani, C. M. Teschke, K. N. Parent, G. Cingolani,Portal protein functions akin to a DNA-sensor that couples genome-packaging to icosahedral capsid maturation. Nat. Commun. 8,14310 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.J. Tang, G. C. Lander, A. Olia, R. Li, S. Casjens, P. Prevelige Jr., G. Cingolani, T. S. Baker, J. E. Johnson,Peering down the barrel of a bacteriophage portal: The genome packaging and release valve in p22. Structure 19,496–502 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.G. C. Lander, L. Tang, S. R. Casjens, E. B. Gilcrease, P. Prevelige, A. Poliakov, C. S. Potter, B. Carragher, J. E. Johnson, The Structure of an infectious p22 virion shows the signal for headful DNA packaging. Science 312, 1791–1795 (2006). [DOI] [PubMed] [Google Scholar]
- 47.A. S. Olia, J. Al-Bassam, D. A. Winn-Stapley, L. Joss, S. R. Casjens, G. Cingolani,Binding-induced stabilization and assembly of the phage P22 tail accessory factor gp4. J. Mol. Biol. 363,558–576 (2006). [DOI] [PubMed] [Google Scholar]
- 48.K. O. Kopec, A. N. Lupas,β-Propeller blades as ancestral peptides in protein evolution. PLOS ONE 8,e77074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.A. S. Olia, A. Bhardwaj, L. Joss, S. Casjens, G. Cingolani,Role of gene 10 protein in the hierarchical assembly of the bacteriophage P22 portal vertex structure. Biochemistry 46,8776–8784 (2007). [DOI] [PubMed] [Google Scholar]
- 50.A. Bhardwaj, A. S. Olia, N. Walker-Kopp, G. Cingolani,Domain organization and polarity of tail needle GP26 in the portal vertex structure of bacteriophage P22. J. Mol. Biol. 371,374–387 (2007). [DOI] [PubMed] [Google Scholar]
- 51.A. Seul, J. J. Muller, D. Andres, E. Stettner, U. Heinemann, R. Seckler,Bacteriophage P22 tailspike: Structure of the complete protein and function of the interdomain linker. Acta Crystallogr. D Biol. Crystallogr. 70,1336–1345 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Q. Fang, W. C. Tang, P. Tao, M. Mahalingam, A. Fokine, M. G. Rossmann, V. B. Rao,Structural morphing in a symmetry-mismatched viral vertex. Nat. Commun. 11,1713 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.C. Koc, G. Xia, P. Kuhner, S. Spinelli, A. Roussel, C. Cambillau, T. Stehle,Structure of the host-recognition device of Staphylococcus aureus phage ϕ11. Sci. Rep. 6,27581 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D. Hrebik, D. Stverakova, K. Skubnik, T. Fuzik, R. Pantucek, P. Plevka,Structure and genome ejection mechanism of Staphylococcus aureus phage P68. Sci. Adv. 5,eaaw7414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.R. W. Hendrix,Symmetry mismatch and DNA packaging in large bacteriophages. Proc. Natl. Acad. Sci. U.S.A. 75,4779–4783 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. L. Ilca, A. Kotecha, X. Sun, M. M. Poranen, D. I. Stuart, J. T. Huiskonen,Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun. 6,8843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.V. Abrishami, S. L. Ilca, J. Gomez-Blanco, I. Rissanen, J. M. de la Rosa-Trevin, V. S. Reddy, J. M. Carazo, J. T. Huiskonen,Localized reconstruction in Scipion expedites the analysis of symmetry mismatches in cryo-EM data. Prog. Biophys. Mol. Biol. 160,43–52 (2021). [DOI] [PubMed] [Google Scholar]
- 58.O. W. Bayfield, A. C. Steven, A. A. Antson,Cryo-EM structure in situ reveals a molecular switch that safeguards virus against genome loss. eLife 9,e55517 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O. W. Bayfield, E. Klimuk, D. C. Winkler, E. L. Hesketh, M. Chechik, N. Cheng, E. C. Dykeman, L. Minakhin, N. A. Ranson, K. Severinov, A. C. Steven, A. A. Antson,Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids. Proc. Natl. Acad. Sci. U.S.A. 116,3556–3561 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.D. L. Caspar, A. Klug,Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 27,1–24 (1962). [DOI] [PubMed] [Google Scholar]
- 61.A. Bhardwaj, R. S. Sankhala, A. S. Olia, D. Brooke, S. R. Casjens, D. J. Taylor, P. E. Prevelige Jr., G. Cingolani,Structural plasticity of the protein plug that traps newly packaged genomes in podoviridae virions. J. Biol. Chem. 291,215–226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.T. Motwani, R. K. Lokareddy, C. A. Dunbar, J. R. Cortines, M. F. Jarrold, G. Cingolani, C. M. Teschke,A viral scaffolding protein triggers portal ring oligomerization and incorporation during procapsid assembly. Sci. Adv. 3,e1700423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.A. S. Olia, S. Casjens, G. Cingolani,Structural plasticity of the phage P22 tail needle gp26 probed with xenon gas. Protein Sci. 18,537–548 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.K. Lorenzen, A. S. Olia, C. Uetrecht, G. Cingolani, A. J. Heck,Determination of stoichiometry and conformational changes in the first step of the P22 tail assembly. J. Mol. Biol. 379,385–396 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.S. Q. Zheng, E. Palovcak, J. P. Armache, K. A. Verba, Y. Cheng, D. A. Agard,MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14,331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.A. Rohou, N. Grigorieff,CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192,216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.P. Conesa Mingo, J. Gutierrez, A. Quintana, J. M. de la Rosa Trevin, A. Zaldivar-Peraza, J. Cuenca Alba, M. Kazemi, J. Vargas, L. Del Cano, J. Segura, C. O. S. Sorzano, J. M. Carazo,Scipion web tools: Easy to use cryo-EM image processing over the web. Protein Sci. 27,269–275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, T. E. Ferrin,UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25,1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 69.S. H. Scheres,RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180,519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.J. Zivanov, T. Nakane, B. O. Forsberg, D. Kimanius, W. J. Hagen, E. Lindahl, S. H. Scheres,New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7,e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.T. C. Terwilliger, O. V. Sobolev, P. V. Afonine, P. D. Adams,Automated map sharpening by maximization of detail and connectivity. Acta Crystallogr. D Struct. Biol. 74,545–559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.T. D. Goddard, C. C. Huang, E. C. Meng, E. F. Pettersen, G. S. Couch, J. H. Morris, T. E. Ferrin,UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27,14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.P. Emsley, K. Cowtan,Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60,2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 74.P. V. Afonine, B. K. Poon, R. J. Read, O. V. Sobolev, T. C. Terwilliger, A. Urzhumtsev, P. D. Adams,Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74,531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.I. W. Davis, A. Leaver-Fay, V. B. Chen, J. N. Block, G. J. Kapral, X. Wang, L. W. Murray, W. B. Arendall III, J. Snoeyink, J. S. Richardson, D. C. Richardson,MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35,W375–W383 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.W. L. DeLano, The PyMOL molecular graphics system, version 1.8 Schrödinger, LLC. (2002).
- 77.E. Krissinel, K. Henrick,Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372,774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 78.R. A. Laskowski,PDBsum new things. Nucleic Acids Res. 37,D355–D359 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.S. Hayward, H. J. Berendsen,Systematic analysis of domain motions in proteins from conformational change: New results on citrate synthase and T4 lysozyme. Proteins 30,144–154 (1998). [PubMed] [Google Scholar]
- 80.R. Maiti, G. H. Van Domselaar, H. Zhang, D. S. Wishart,SuperPose: A simple server for sophisticated structural superposition. Nucleic Acids Res. 32,W590–W594 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Tables S1 and S2