Abstract
After a myocardial infarction, the boundary between the injured, hypoxic tissue and the adjacent viable, normoxic tissue, known as the border zone, is characterized by an oxygen gradient. Yet, the impact of an oxygen gradient on cardiac tissue function is poorly understood, largely due to limitations of existing experimental models. Here, we engineered a microphysiological system to controllably expose engineered cardiac tissue to an oxygen gradient that mimics the border zone and measured the effects of the gradient on electromechanical function and the transcriptome. The gradient delayed calcium release, reuptake, and propagation; decreased diastolic and peak systolic stress; and increased expression of inflammatory cascades that are hallmarks of myocardial infarction. These changes were distinct from those observed in tissues exposed to uniform normoxia or hypoxia, demonstrating distinct regulation of cardiac tissue phenotypes by an oxygen gradient. Our border-zone-on-a-chip model advances functional and mechanistic insight into oxygen-dependent cardiac tissue pathophysiology.
An engineered oxygen gradient device models heart tissue after a heart attack and integrates quantifiable functional metrics.
INTRODUCTION
A myocardial infarction (MI), commonly known as a heart attack, is often triggered by the occlusion of a coronary artery due to atherosclerotic plaque formation. The occlusion obstructs the flow of blood and oxygen (O2) to a localized site of myocardial tissue, leading to hypoxia and massive cardiomyocyte (CM) cell death (1). Steep O2 gradients develop at the interface between the injured, hypoxic tissue and the surrounding viable, normoxic tissue (2), a region known as the border zone. Over the ensuing days, weeks, and months, the injured myocardium, especially the border zone, undergoes extensive remodeling in its electromechanical properties that can lead to heart failure (3). In vivo and ex vivo models have shown that border zone tissue is a known substrate for arrhythmias (4) and generates less force than remote myocardium (5). These observations suggest that the border zone is especially susceptible to pathophysiological remodeling, likely due to heterogeneities in the cellular microenvironment, including O2 gradients. However, the direct effects of O2 gradients on the structure and function of CMs on molecular, cellular, and tissue levels, including interactions between differentially oxygenated cells, are poorly understood, thereby limiting the development of therapeutic interventions.
In vitro studies are essential for elucidating the effects of O2 and other physical features in the cellular microenvironment on molecular, cellular, and tissue phenotypes in a controlled setting. Conventional approaches for modulating O2 tension in vitro are primarily hypoxia chambers or workstations that globally expose cultured cells to uniform O2 levels. In vitro studies have shown various effects of different durations of uniform hypoxia. For instance, acute uniform hypoxia reduces wavefront propagation (6) and impairs contractility (7). A duration of 5 hours of hypoxia results in internalization of gap junction protein connexin 43 (Cx43) (8). On the scale of 8 to 24 hours of hypoxia, the mRNA of fetal, T-type calcium channel Cav3.2 is up-regulated, which is normally absent in adult myocardium (9). Apoptosis is also induced after 12 to 48 hours (10–12).
The conventional tools for applying uniform hypoxia cannot reproduce the spatial O2 gradients characteristic of the myocardial infarct border zone and thus preclude any insights into the effects of nonuniform O2. Microengineering technologies, such as microelectrode arrays and microfluidic channels, have been used to develop in vitro systems to study myocardial ischemia and injury (6, 13–16). One of these models mimicked a hypoxia gradient, but it was generated using a drug that uncouples mitochondrial oxidative phosphorylation rather than physical O2 tension (13). O2 gradients have also been developed in cardiac organoid models by relying on the transport limitations of passive O2 diffusion into the three-dimensional (3D) tissue. However, this approach offers low spatiotemporal control of O2 and is not practical for rigorously dissecting the effects of O2-dependent cellular cross-talk on cell and tissue physiology due to inherent variability and the limited ability to position and monitor distinct cell types. Organoids also do not recapitulate the native architecture of the myocardium, in which sheets of elongated, anisotropically aligned CMs form an electromechanical syncytium that maximizes electrical signal propagation and force generation in the longitudinal direction. Other diffusion-limited approaches to generate O2 gradients include embedding cells in hydrogels (17) and stacked paper-based culture systems (18), but these models similarly offer little spatiotemporal control and are prone to gradient fluctuations by cellular O2 consumption and cell density. Thus, our understanding of the effects of O2 gradients on cardiac tissue phenotypes has been limited by technical barriers related to controllably and reproducibly exposing aligned cardiac tissues to O2 gradients and subsequently quantifying functional and transcriptional phenotypes in one integrated system.
To overcome these limitations, we engineered a myocardial infarct border-zone-on-a-chip with three key features: (i) aligned cardiac tissues generated via microcontact printing of extracellular matrix protein, (ii) stable microfluidic-controlled spatial O2 gradients generated via continuous gas perfusion, and (iii) integration of assays for on-chip quantifiable functional metrics, including calcium wave propagation velocity and contractility. We then used our device to expose engineered cardiac tissues to uniform normoxia, uniform hypoxia, or an O2 gradient and measured calcium transients, calcium wave propagation velocity, and contractile stress. We also used bulk RNA sequencing (RNA-seq) to examine differentially expressed genes (DEGs) in tissues exposed to uniform and gradient O2. Our functional and transcriptomic analysis revealed that the O2 gradient condition exhibited heterogeneous calcium handling, hypocontractility, and activation of inflammation. These phenotypes were not observed in engineered tissues exposed to global, homogeneous O2 levels, demonstrating the unique impact of the O2 gradient. Thus, our myocardial infarct border-zone-on-a-chip recapitulates select dysfunctional myocardial tissue responses associated with hypoxic cardiac injury and could be implemented to inform and test post-MI therapeutic strategies.
RESULTS
Development of the O2 gradient border-zone-on-a-chip
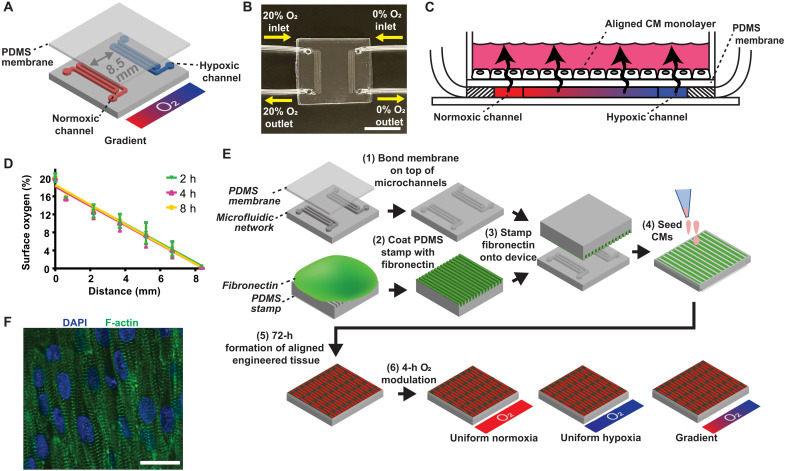
Our goal was to expose engineered cardiac tissue to an O2 gradient similar to the myocardial infarct border zone at the lateral margin of an ischemic area, also known as the intramural border zone (19). To achieve this, we fabricated a polydimethylsiloxane (PDMS) microfluidic device with two serpentine microchannels separated by 8.5 mm, which is in the 2- to 20-mm range of the border zone size measured in humans (20, 21) and other large mammals, such as dogs (22). The channels were interfaced with tubing for gas perfusion and overlaid with a thin gas-permeable PDMS membrane to serve as a surface for cell culture (Fig. 1, A and B). Cells were exposed to an O2 gradient via continuous perfusion of compressed gases through the microchannels that diffused across the membrane on which cells were cultured (Fig. 1C). To generate a 20%-0% linear O2 gradient, one channel was perfused with 5% carbon dioxide (CO2), balanced air and the other was perfused with 5% CO2, balanced nitrogen (N2). The advantage of continuous gas perfusion devices is the maintenance of a stable O2 gradient (Fig. 1D), especially in the presence of cells (23), unlike devices relying on passive O2 diffusion barriers, where the gradient is altered by cell density and O2 consumption (24). To engineer myocardial tissues with native-like alignment, the membrane was microcontact printed with lines of fibronectin and seeded with neonatal rat ventricular myocytes (NRVMs; Fig. 1E). After 3 days, an anisotropic myocardial tissue was formed (Fig. 1F) and subsequently exposed to one of three different O2 conditions for 4 hours: (i) uniform normoxia, (ii) uniform hypoxia, or (iii) 20%-0% O2 gradient. The 4-hour O2 modulation time point is consistent with the duration of ischemia in humans before clinical intervention (25), whereafter the deleterious effects of reperfusion injury contribute to myocardial damage.
Fig. 1. Fabrication and characterization of the O2 gradient myocardial infarct border-zone-on-a-chip.
(A) O2 gradient condition with gas flow through two serpentine microchannel networks separated by 8.5 mm. (B) Fabricated O2 gradient device with gas perfusion microchannels connected to microbore tubing. Scale bar, 10 mm. (C) Cross-sectional schematic showing diffusion of perfused gases across the PDMS membrane to expose the cells to an O2 gradient. (D) Validation of the surface O2 profile. Error bars indicate means ± SD. Solid lines indicate the linear regression. (E) Fabrication and experimental flowchart schematic. (F) Anisotropically aligned engineered cardiac tissue. DAPI (blue) and phalloidin staining of F-actin (green). Scale bar, 20 μm.
An O2 gradient alters calcium transients in anisotropically aligned CMs
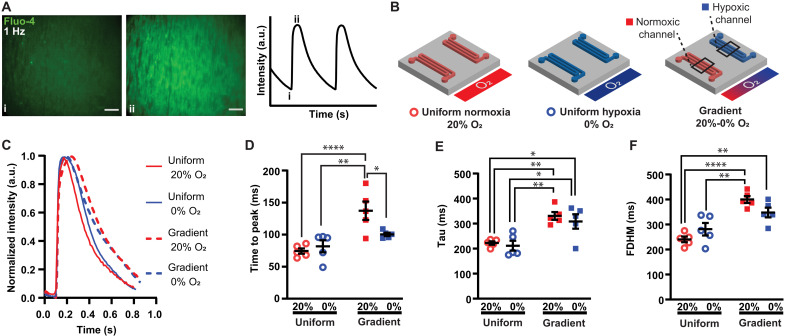
Acute MI and post-MI cardiac dysfunction is associated with calcium handling abnormalities (26). To determine how an O2 gradient alters calcium handling, we measured calcium transients (Fig. 2A) in engineered tissues exposed to 4 hours of uniform normoxia, uniform hypoxia, or an O2 gradient. In the gradient condition, fluorescent traces were analyzed separately in regions above the normoxic channel and hypoxic channel (Fig. 2B).
Fig. 2. Effects of different O2 conditions on CM calcium transients.
(A) Fluo-4 signal in engineered cardiac tissue during 1-Hz electrical pacing, with i and ii corresponding to baseline and peak intensity, respectively. Scale bar, 200 μm. (B) Normoxic (red open circles), hypoxic (blue open circles), and gradient O2 conditions with gas flow through microchannels. Calcium transients from the CMs cultured on the membrane directly above the normoxic channel (red squares) and hypoxic channel (blue squares) were compared in the gradient condition. (C) Representative traces of the calcium transients from the different O2 conditions. Quantification of (D) time to peak, (E) tau time constant of decay, and (F) full duration half maximum (FDHM). Black lines with error bars indicate means ± SEM from n = 5 independent experiments for each condition. *P < 0.05, **P < 0.01, and ****P < 0.0001 (one-way ANOVA, Tukey’s multiple comparison post hoc test).
Calcium transient dynamics were dependent on O2 condition (Fig. 2, C to F). While time to peak (Fig. 2D) was similar in both uniform conditions, it significantly increased in normoxic CMs in the gradient compared to adjacent hypoxic CMs in the gradient as well as uniform hypoxia and uniform normoxia. Tau, the time constant of decay, was higher in normoxic and hypoxic CMs in a gradient as compared to both uniform conditions (Fig. 2E). The full duration half maximum (FDHM) was also elevated in the gradient compared to uniform normoxia (Fig. 2F). Together, these data suggest that an O2 gradient delays calcium release and reuptake in CMs. Time to peak and tau of decay were reported as prolonged in CMs isolated from rats exposed to chronic hypoxia by inspiration of 10% O2 (7). We observed similar trends in the gradient condition but not uniform hypoxia. However, the gradient condition is likely more characteristic of CMs isolated from rats breathing 10% O2 because heterogeneous O2 landscapes arise naturally in the tissue due to O2 transport limitations and diffusion of O2 from blood vessels.
An O2 gradient affects calcium wave propagation velocity
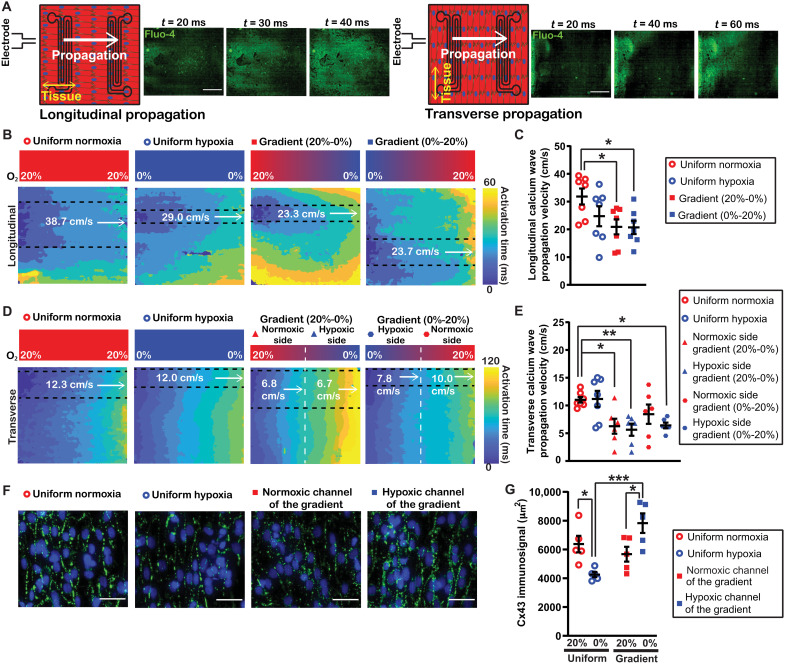
Rapid and uniform propagation of action potentials and subsequent calcium waves in the myocardium is necessary for normal, synchronized excitation-contraction coupling (27). Myocardial conduction velocity is closely linked to calcium propagation velocity due to the important functional relationship between the action potential and the kinetics of the subsequent increase in cytosolic calcium (28). Simultaneous voltage and calcium mapping demonstrates an analogous propagation velocity but with a transient signal peak delay on the scale of tens of milliseconds (29). To assess how an O2 gradient affects calcium wave propagation velocity, we exposed engineered myocardial tissues to the different O2 conditions and generated calcium activation maps in response to point stimulation to quantify propagation velocity (Fig. 3A) (30). Both longitudinal velocity (Fig. 3, B and C) and transverse velocity (Fig. 3, D and E) were measured. Propagation from normoxia to hypoxia and vice versa was examined for tissues exposed to an O2 gradient to determine any effects specific to directionality.
Fig. 3. Regulation of calcium propagation velocity by an O2 gradient.
(A) Tissue orientation relative to electrode position for longitudinal propagation (left) and transverse propagation (right) with example Fluo-4 fluorescent intensity time lapses (inset panels). Scale bar, 2 mm. (B) Activation maps of longitudinal calcium propagation velocity with cropped regions parallel to uniaxial measurement (dashed lines) of the different O2 conditions, including the normoxic control (red open circle), hypoxic control (blue open circle), gradient with normoxic to hypoxic directionality (red square), and gradient with hypoxic to normoxic directionality (blue square). (C) Quantification of longitudinal calcium propagation velocity. (D) Activation maps of transverse calcium propagation velocity from the normoxic control (red open circle), hypoxic control (blue open circle), gradient when the calcium wave propagation was from the normoxic side (left half; red triangle) to the hypoxic side (right half; blue triangle), and gradient when the calcium wave propagation was from the hypoxic side (left half; blue hexagon) to the normoxic side (right half; red hexagon). (E) Quantification of transverse calcium propagation velocity. Black lines with error bars indicate means ± SEM from at least six independent experiments for each condition. *P < 0.05 and **P < 0.01 (one-way ANOVA, comparing the mean of each group with the mean of the uniform normoxia group, Sidak’s multiple comparison post hoc test). (F) Zoomed-in and cropped Cx43 staining (green) and DAPI (blue). Scale bar, 50 μm. (G) Quantification of Cx43 immunosignal. Black lines with error bars indicate means ± SEM from n = 5 for each condition. *P < 0.05 and ***P < 0.001 (one-way ANOVA, Tukey’s multiple comparison post hoc test).
Tissues cultured in uniform hypoxia had slightly, but not significantly, lower calcium propagation velocity in the longitudinal direction compared to uniform normoxia (Fig. 3C). However, longitudinal calcium propagation velocity for tissues in the O2 gradient was significantly lower compared to uniform normoxia (Fig. 3C). In gradient tissues, longitudinal propagation velocity was similar traveling from normoxia to hypoxia and vice versa, indicating that longitudinal propagation velocity was not sensitive to the direction of propagation relative to the O2 gradient.
As expected, transverse calcium propagation velocity was consistently slower than longitudinal. Transverse velocity was similar for tissues exposed to uniform normoxia or uniform hypoxia. In the O2 gradient, transverse velocity was slow enough that it was apparent that propagation velocity was dependent on directionality relative to the gradient. Thus, calcium propagation velocity was calculated independently for the normoxic side and hypoxic side (Fig. 3D). This analysis demonstrated that transverse velocity in the normoxic to hypoxic direction was relatively uniform but significantly slower than the uniform normoxic condition (Fig. 3E).
Transverse velocity in the hypoxic to normoxic direction did not exhibit the same degree of uniformity. Only the hypoxic side was significantly slower than uniform normoxia. Collectively, our data demonstrate that an O2 gradient delays propagation of both longitudinal and transverse calcium waves and is sensitive to directionality in some instances. Therefore, the calcium propagation velocity results in our infarct border zone model were consistent with a previous finding that longitudinal and transverse conduction velocity decreased during acute ischemia (31).
Gap junction protein Cx43 is the primary connexin in ventricular myocardium, mediating cell-to-cell electrical impulse conduction. Uniform hypoxia has previously been shown to decrease the Cx43 signal at gap junctions (32) and increase Cx43 internalization (8). There is a demonstrated relationship between Cx43 abundance and propagation velocity (33–35). To understand whether Cx43 may play a role in the decrease in propagation velocity observed in the O2 gradient, the Cx43 immunosignal was assessed in images of the different O2 conditions (Fig. 3, F and G). The number of CMs, as determined by the nuclei count in each image, was not significantly different between conditions (fig. S1). The Cx43 immunosignal decreased in uniform hypoxia (Fig. 3G). There was also a significant increase in the Cx43 immunosignal in hypoxic CMs in a gradient compared to CMs in uniform hypoxia. In addition, the increased Cx43 immunosignal in the CMs in the region above the hypoxic channel was statistically significant compared to the CMs in the region above the normoxic channel, demonstrating spatial heterogeneity of Cx43 in the engineered tissue exposed to the gradient. Notably, spatial heterogeneity of Cx43 has been associated with conduction block and arrhythmia (36–38).
O2 levels affect contractile stress
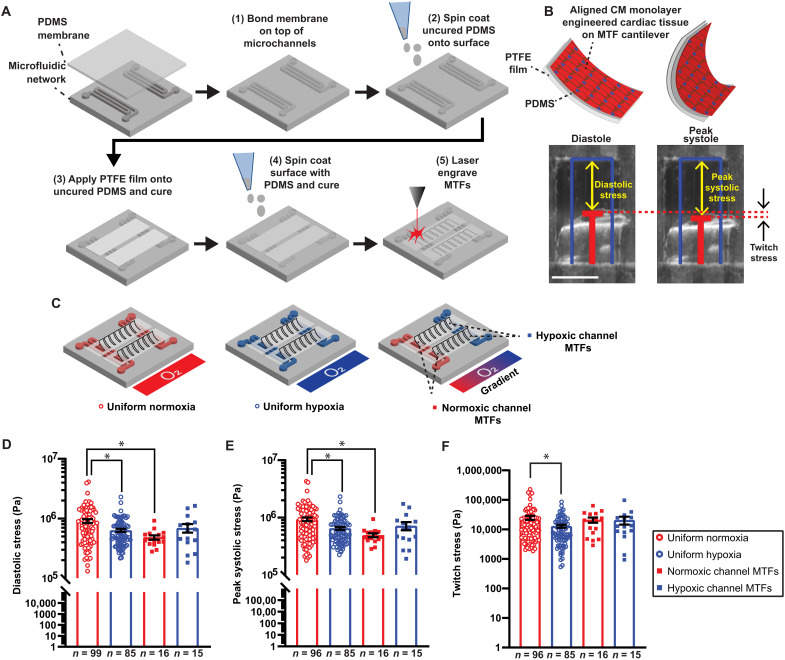
We next investigated whether an O2 gradient also affects the contractile function of engineered cardiac tissue. For this purpose, we integrated the muscular thin film (MTF) assay (39) into our border-zone-on-a-chip microphysiological system to measure contractile stresses generated by the tissues. The O2 control MTF devices were fabricated with an array of polytetrafluoroethylene (PTFE)/PDMS bilayer cantilevers (Fig. 4A). Engineered cardiac tissues were then cultured on top and exposed to different O2 conditions following the protocol previously described (see Fig. 1E). For contractile stress measurements, the precut MTFs were manually peeled away from the O2 control device substrate, allowing them to contract and curl out of plane of the device surface (Fig. 4B). Custom software leveraging a modified Stoney’s equation derived specifically for bilayer MTFs (see Supplementary Text) was used to calculate radii of curvature and contractile stress generation of the MTFs.
Fig. 4. Contractile stresses generated by engineered cardiac tissues exposed to different O2 conditions during 2Hz electrical pacing.
(A) Flowchart of the fabrication of O2 controlled MTF devices. (B) Tissue stress generation reduces the radius of curvature as MTFs contract from diastole to peak systole. Original length of the MTF film (blue) and tracked film edge (red). Scale bar, 1 mm. (C) MTFs in the different O2 conditions: uniform normoxia (red open circle), uniform hypoxia (blue open circle), normoxic channel (red square) of the O2 gradient, and hypoxic channel (blue square) of the O2 gradient. Quantification of (D) diastolic stress, (E) peak systolic stress, and (F) twitch stress. Black lines with error bars indicate means ± SEM. *P < 0.05 (Kruskal-Wallis test, comparing the mean rank of each group with the mean rank of the uniform normoxia group, Dunn’s multiple comparison post hoc test).
Diastolic and peak systolic stresses were determined as well as the difference between them, referred to as twitch stress (Fig. 4B). Contractile stresses from MTFs exposed to three different O2 conditions were compared: (i) uniform normoxia, (ii) uniform hypoxia, and (iii) 20%-0% O2 gradient. In the gradient condition, contractile stresses from MTFs above the normoxic channel and hypoxic channel were determined (Fig. 4C). Plots of the resulting stresses demonstrated that MTFs exposed to uniform hypoxia were significantly weaker than uniform normoxia in diastolic, peak systolic, and twitch stress (Fig. 4, D and F). In addition, diastolic stress and peak systolic stress were suppressed above the normoxic channel in the O2 gradient compared to tissues in uniform normoxia (Fig. 4, D and E). Our findings are consistent with hypocontractile border zone myocardium reported by others (40, 41). Twitch stress in the O2 gradient condition was not significantly different than uniform normoxia.
Our contractile stress measurements echo known systolic dysfunction in pathological conditions, including reduction in systolic pressure after MI (42) and depressed contractility and reduced systolic pressure occurring in heart failure (43). The MTF assay is not as applicable for elucidating diastolic dysfunction due to limitations in decoupling the rate of diastolic relaxation [an important parameter altered in diastolic dysfunction (44)] from the elastic recoil of the PDMS substrate (45). Left ventricular diastolic pressure is known to increase in various pathological cardiac settings, including heart failure (43), acute hypoxia in hypertrophied hearts (46), and MI (47). However, we do not expect to replicate this clinical aspect of MI observed in vivo because the MTF assay does not involve formation of a cardiac chamber. Therefore, it cannot recapitulate certain cardiac physiology parameters, such as pressure-volume relations. However, the measured decrease in the MTF diastolic stress could be explained by a decrease in basal stress caused by a decrease in basal contractile tone due to impairment of the contractile apparatus attributed to O2-dependent cellular and molecular changes in the uniform hypoxia and gradient conditions.
Transcriptomic analysis of engineered cardiac tissues exposed to different O2 landscapes
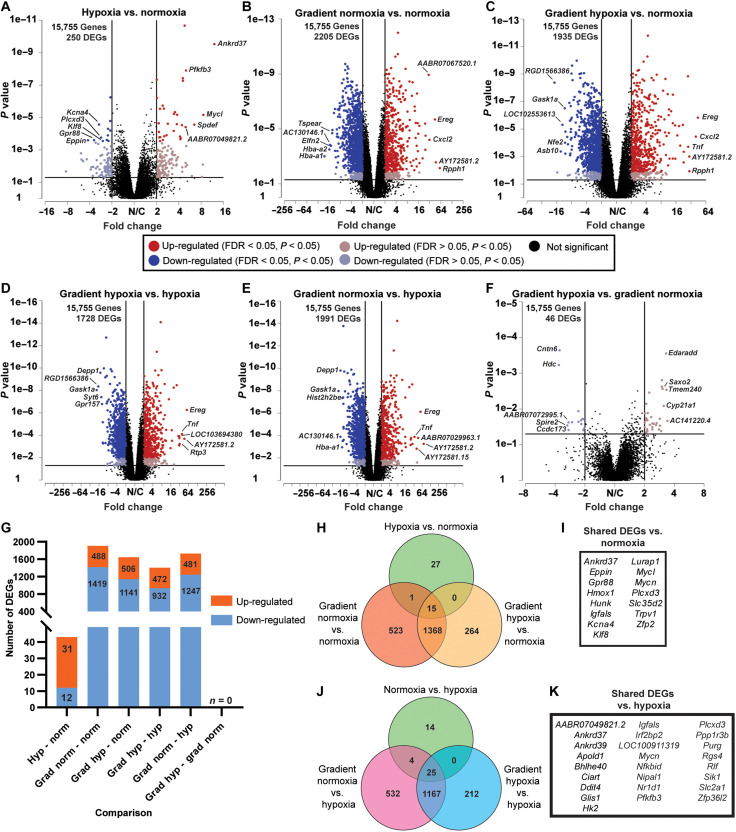
To identify DEGs as a function of O2, the transcriptomes of engineered tissues exposed to normoxia, hypoxia, and the gradient were analyzed using RNA-seq. To distinguish between normoxia and hypoxia in the O2 gradient condition, the device was cut into two equal halves and tissues were isolated separately from both sections, corresponding to the normoxic side and the hypoxic side (here referred to as gradient normoxia and gradient hypoxia, respectively). All six pairwise comparisons of the various O2 conditions were then performed (data S1). Volcano plots of the pairwise comparisons show the up-regulated (red) and down-regulated (blue) DEGs [P < 0.05, absolute value of the fold change (|fold change|) ≥ 2] and top 5 DEGs with the largest |fold change| on the up-regulated and down-regulated side (Fig. 5, A to F). With an included false discovery rate (FDR) < 0.05, a robust count of DEGs was attained (Fig. 5G). The more rigorous cutoff eliminated DEGs in the gradient hypoxia versus gradient normoxia comparison. A Venn diagram shows the overlap of identified DEGs in the comparisons versus uniform normoxia (Fig. 5H) with 15 shared DEGs in those comparisons (Fig. 5I). Comparisons versus uniform hypoxia (Fig. 5J) had 25 shared DEGs (Fig. 5K).
Fig. 5. DEGs in engineered cardiac tissues exposed to different O2 conditions.
(A to F) Volcano plots of pairwise comparisons with down-regulated (blue) and up-regulated (red) genes [P < 0.05, absolute value of the fold change (|fold change|) ≥ 2]. The black horizontal and vertical lines distinguish the fold change and P value cutoff for DEGs. The top 5 down-regulated and up-regulated genes with the highest |fold change| are labeled by gene symbol. (G) Quantification of down-regulated (light blue) and up-regulated (orange) genes with P < 0.05, FDR < 0.05, and |fold change| ≥ 2. (H) Venn diagram indicating the overlap of identified DEGs in the comparisons versus uniform normoxia. (I) List of the 15 shared DEGs in the comparisons with uniform normoxia. (J) Venn diagram indicating the overlap of identified DEGs in the comparisons versus uniform hypoxia. (K) List of the 25 shared DEGs in the comparisons with uniform hypoxia.
In our RNA-seq analysis, many of the top up-regulated or down-regulated genes in the pairwise comparisons correlate well with other hypoxia studies. ANKRD37, which was highly up-regulated in the comparison of hypoxia versus normoxia (Fig. 5A) and a shared DEG in every comparison (Fig. 5, H and J; as listed in Fig. 5, I and K), was also up-regulated in the RKO colon cancer cell line in hypoxia and regulates autophagy (48). DDIT4 was up-regulated in hypoxia (table S1) and was a shared DEG in comparisons versus hypoxia (Fig. 5K). Fittingly, DDIT4 has previously been shown to be under the transcriptional activity of the key hypoxia-inducible transcription factor HIF-1 (49). EREG was one of the top 5 genes up-regulated in the gradient condition (Fig. 5, B to E) and was previously shown to be robustly induced following acute MI in rats (50), underscoring the applicability of our infarct border zone model.
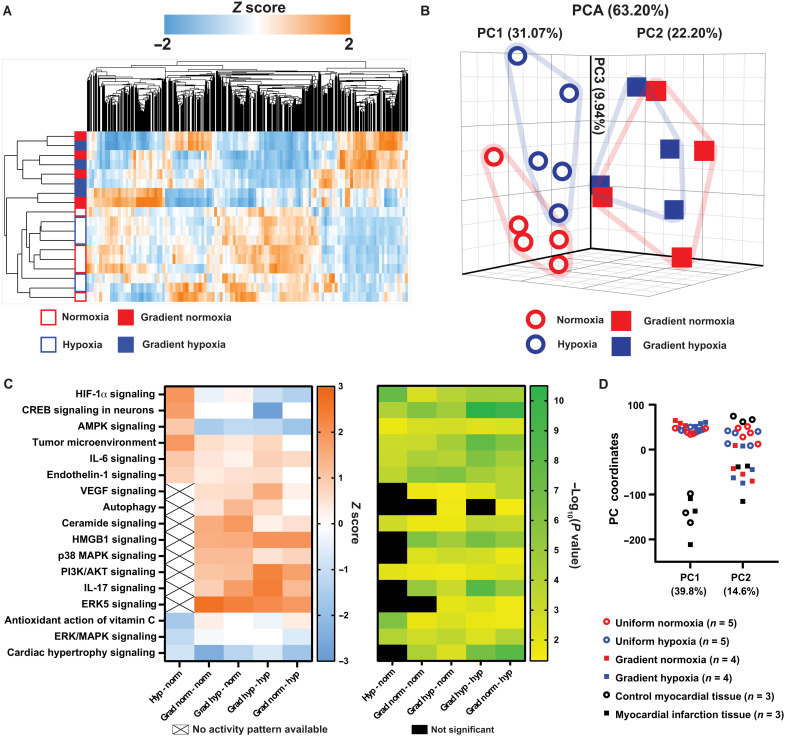
The transcriptional profile of samples from the gradient was clearly distinct from uniform normoxia and hypoxia (Fig. 6A). The hierarchical clustering with a heat map and principal components analysis (PCA) demonstrated clear separation of the gradient samples from uniform normoxia and uniform hypoxia (Fig. 6, A and B). Data S2 lists the component loadings for the first 18 components. However, as apparent in the PCA plot (Fig. 6B), although uniform normoxia and uniform hypoxia cluster separately and away from the gradient, gradient normoxia and gradient hypoxia do not have clear separation from each other (also see fig. S2). The similar transcriptional profile in gradient hypoxia and gradient normoxia samples is likely due to continuity arising from the engineered tissue acting as a syncytium.
Fig. 6. PCA and pathway analysis of the RNA-seq data from the engineered cardiac tissues.
(A) Hierarchical clustering heatmap of the samples exposed to uniform normoxia (open red outline), uniform hypoxia (open blue outline), normoxic side of the gradient (solid red), and hypoxic side of the gradient (solid blue). (B) PCA plot of normoxia (open red circle), hypoxia (open blue circle), normoxic side of the gradient (red squares), and hypoxic side of the gradient (blue squares). The percent of total variance due to the first three PCs is labeled. (C) Heatmap of select canonical pathways and the associated z score (left) and −log10(P value) (right) for the indicated pairwise comparison. (D) PC coordinates of PC1 and PC2 after comparative analysis of the in vitro devices with in vivo rat MI and control myocardium.
To understand the biological functions driving the distinct transcriptional profile of the gradient compared to the uniform controls, Ingenuity Pathway Analysis (IPA) was used to generate a heatmap of the enriched categories of select canonical pathways (tables S2 to S6) and their statistical significance according to the DEGs identified in the various comparisons (Fig. 6C). Activity patterns of several pathways with positive z scores indicate that the normoxic side and hypoxic side of the gradient condition are associated with inflammatory cascades involved in MI, including interleukin-6 (IL-6) signaling (51), IL-17 signaling (52), HMGB1 signaling (53), and p38 mitogen-activated protein kinase (MAPK) signaling (54). Overall, the pathway analysis suggests that the engineered cardiac tissue exposed to an O2 gradient exhibits strong activation of inflammatory responses compared to both uniform normoxia and uniform hypoxia.
To measure the similarity of our model to adult myocardial tissue after acute MI, we compared our transcriptomic data to that of adult myocardium of rats 24 hours after MI caused by surgical ligation and healthy myocardium of sham-operated rats as controls (55). Compared with rat MI transcriptomic data, the DEGs (P < 0.05) of the O2 gradient overlapped with >2000 genes (fig. S3). Data S3 lists 38 up-regulated and 62 down-regulated DEGs that have similar expression profiles in the in vitro gradient and in vivo infarction but a distinct profile in the uniform hypoxia condition. Our list includes the genes HBEGF, AREG, CLEC4E, and IL1B that have previously reported roles in MI or ischemia-reperfusion injury (56–59).
The top PCs (PC1 and PC2) demonstrated clear separation between samples (Fig. 6D). Pathways associated with PC1 and PC2 were determined using IPA analysis of the PC gene loadings as rankings (tables S7 and S8). PC1 stratified in vivo and in vitro samples, likely due to differences in tissue complexity, age (neonatal versus adult), and time point (4-hour O2 modulation versus 24-hour post-MI). For example, pathways associated with non-CM cell types, such as PD-1 signaling, natural killer cell signaling, and leukocyte extravasation (table S7), were represented in PC1. PC2 predominantly grouped the O2 gradient samples with adult myocardial tissue after acute MI while also grouping uniform normoxia and uniform hypoxia with control rat myocardium (Fig. 6D). Pathways represented in PC2 include hypoxia signaling in the cardiovascular system (table S8), supporting our finding that an O2 gradient models in vivo hypoxic cardiac injury more accurately compared to uniform hypoxia.
DISCUSSION
In this study, a microfluidic MI border-zone-on-a-chip was fabricated to expose aligned cardiac tissues to O2 gradients and quantify multimodal physiological responses. This led to the key finding that an O2 gradient regulates calcium handling, contractile stress, and gene expression of engineered cardiac tissues in a manner distinct from tissues exposed to global, uniform O2 levels. Specifically, tissues exposed to the O2 gradient recapitulated hallmarks of arrhythmogenic conditions, such as slower calcium transients, diminished propagation velocity, heterogeneous Cx43 expression, and contractile dysfunction. In addition, only tissues in the gradient expressed transcriptional signatures indicative of inflammation. These findings suggest that an O2 gradient model is vital for understanding how interactions between differentially oxygenated cells contribute to the heterogeneous functional and transcriptional remodeling in the infarct border zone.
The development of arrhythmias due to ischemia or hypoxia-induced cardiac injury has frequently been attributed to fibrosis (60), in which strands or patches of scar tissue can interrupt uniform propagation and induce pinning of spiral waves. However, ventricular arrhythmias in acute ischemia (on the scale of hours) due to coronary artery occlusion are a major cause of sudden cardiac death (61). Our results demonstrating abnormal propagation velocity, which can be arrhythmogenic, support evidence that arrhythmia could develop in peri-infarct myocardial tissue without fibrotic scar formation in early ischemia during acute MI due to nonuniform changes in calcium cycling and propagation in heterogeneous O2 environments.
Although calcium handling and contractility were altered in the gradient, pathway analysis did not reveal any major changes in the expression of genes associated with calcium handling, propagation, contractility, or excitation-contraction coupling. Thus, the observed functional effects in the gradient are likely not regulated at the transcriptional level, but rather the posttranslational or functional level. We observed spatial heterogeneity in the abundance of the gap junction protein Cx43, which plays a crucial role in cell-to-cell electrical impulse conduction. Spatial heterogeneity of Cx43 is associated with arrhythmia and conduction block (36–38). However, Cx43 expression pattern alone does not dictate the extent of electrical coupling, as Cx43 is also prone to phosphorylation events that modulate channel conductance, trafficking, gap junction assembly, and internalization (62). Cx43 has previously been shown to be modified at the posttranslational, but not transcriptional, level in hypoxia (32). Posttranslational modifications, including changes in phosphorylation states, may also be altering the gating properties of Cx43, causing the functional effects observed in the O2 gradient, which could be further investigated with this device.
The increased activity of inflammation-associated signaling in the O2 gradient revealed in the pathway analysis is valuable for understanding the acute inflammatory response in the border zone after MI (63). Although innate immune cells, such as leukocytes and macrophages, have well-defined roles in cardiac inflammation after ischemic injury and MI (64), the contributions of CM-derived mediators to inflammation (65) in the setting of MI are unknown. A better understanding of the inflammatory processes of all cardiac cell types in the border zone could enable therapeutic approaches to target the cardiac immune response beyond the traditionally recognized cells of the innate immune system.
Most of the dysfunctional phenotypes in the O2 gradient were not observed in uniform hypoxia, suggesting that tissue dysfunction in the gradient cannot be accounted for solely by exposure of CMs to low O2 tension. Thus, tissue phenotypes in the gradient must be in part influenced by communication between differentially oxygenated cells. In the gradient condition, a pattern emerged that the normoxic side was typically more dysfunctional than the hypoxic side. Previous studies with conditioned medium have shown that hypoxic CMs secrete factors that can protect other CMs from ischemic stress (66). Thus, in the gradient tissues, hypoxic and normoxic CMs may be continuously exchanging factors that contribute to the observed phenotypes. Establishing the role of ongoing, dynamic communication between differentially oxygenated cells is possible with the device developed in this study but not possible with conventional hypoxia chambers and conditioned medium.
Our study is limited by the use of NRVMs, which lack human relevance. However, NRVMs have higher morphological and physiological relevance by retaining more of the functional characteristics of their original tissue compared to human induced pluripotent stem cell–derived CMs, which tend to have fetal-like phenotypes (67–70). Although adult CMs would be more appropriate than neonatal CMs, adult human CMs are largely unavailable for in vitro studies, and adult rat CMs have limited plasticity and rapidly dedifferentiate in vitro (71–73).
The results of our functional and transcriptional analysis with NRVMs motivate future studies to investigate how intercellular communication across an O2 gradient contributes to myocardial damage and may present therapeutic targets to minimize infarct size, reduce adverse remodeling, and prevent the onset of heart failure. The importance of studying myocardial tissue responses in an O2 gradient is highlighted by our comparison with in vivo tissue, which suggests that hypoxic-normoxic intercellular interactions may be a particularly important source of tissue heterogeneity defining the hypoxic cardiac injury transcriptional profile and phenotype after MI. As there remains a pressing need to improve post-MI clinical outcomes, our in vitro model also has many applications for developing and screening potential drug candidates in the preclinical stage.
MATERIALS AND METHODS
Device fabrication
The microfluidic network layer was designed in AutoCAD (Autodesk, San Rafael, CA) as a 22 mm × 22 mm square containing two serpentine channels (550 μm wide, 100 μm deep, and 175 μm interchannel spacing) separated by an 8.5-mm gap. A 10-cm-diameter silicon master with an array of the 22 mm × 22 mm designs was fabricated using SU-8 2100 photoresist (Kayaku Advanced Materials, Tokyo, Japan) to create the microfeatures via standard photolithography techniques in a class 100 cleanroom. The PDMS microfluidic network layer was fabricated using standard soft lithography procedures. Briefly, Sylgard 184 Silicone Elastomer (Dow, Midland, MI) curing agent was added to the base elastomer at a weight ratio of 1:10. The mixture was mixed and degassed in a planetary centrifugal mixer (Thinky, Laguna Hills, CA) and cast on the silicon master. The PDMS-covered master was placed in a vacuum desiccator to further degas for 1 hour and was then cured overnight in an 80°C benchtop oven. Inlet/outlet ports were punched with a 1.5-mm-diameter biopsy punch (Integra Miltex, Integra Life Sciences, Princeton, NJ).
PDMS membranes were fabricated by spin coating PDMS (10:1 weight ratio of base to curing agent) on a 10-cm-diameter silicon wafer (University Wafer, Boston, MA) at 1000 rpm for 30 s after silanizing the wafer with trichloro(1H,1H,2H,2H-perfluorooctyl) silane (Sigma-Aldrich, St. Louis, MO) overnight in a vacuum desiccator. After curing overnight at 80°C, the 100-μm-thick PDMS membrane was bonded on top of the microchannel layer using a 90-s exposure in a plasma cleaner (Harrick, Ithaca, NY). The PDMS device was then bonded on a 22 mm × 22 mm glass coverslip (Fisher Scientific, Waltham, MA) for MTF and RNA isolation experiments or adhered to the bottom of 35-mm polystyrene dishes using PDMS as an adhesive for calcium imaging experiments.
For MTF experiments, bonded PDMS devices were spin-coated with uncured PDMS at 1000 rpm for 30 s to apply an additional layer of PDMS on top of the existing 100-μm-thick PDMS membrane. The uncured PDMS served as the adhesive to attach two strips of 10-μm PTFE film (Goodfellow, Coraopolis, PA). The PTFE layer was included to overcome inherent PDMS-PDMS attractive forces between the underlying PDMS and the final 15-μm-thick PDMS surface layer. PTFE was chosen for its gas permeability as highlighted by its use in microfluidic bubble traps (74, 75). The devices were placed in a benchtop oven at 37°C for 4 hours to secure the PTFE film in position. Devices were then spin-coated with a final layer of uncured PDMS, covering the PDMS and PTFE film strips, at 4000 rpm for 60 s to deposit a PDMS layer of 15-μm thickness. Devices were cured overnight at 80°C before two rows of seven MTFs (each MTF with dimensions of 2.32 mm length × 1.38 mm width) were laser-engraved into the regions with the underlying PTFE film. Laser engraver settings were chosen to cut through the surface PDMS and underlying PTFE film to achieve bilayer MTF cantilevers with a total thickness of 25 μm (15 μm of PDMS and 10 μm of PTFE film) when manually peeled away from the base substrate.
Oxygen modulation
The two serpentine perfusion channels in the microfluidic network layer were separated by an 8.5-mm gap. During gradient experiments, one channel was continuously perfused with 5% CO2, balanced air and the second channel was continuously perfused with 5% CO2, balanced N2. Similar to a previous study (18), the experimental setup for gas perfusion flowed gas from compressed tanks via two-stage regulators into handheld mini gas regulators (Marsh Bellofram, Newell, WV) and finally into the microchannels of the device. Microbore tubing (Tygon ND-100-80, Cole-Parmer, Vernon Hills, IL) interfaced with the access ports of the device. Manometers (Dwyer Instruments, Michigan City, IN) were connected by three-way valves to monitor the pressure within each of the microchannels. The pressure in the separate microchannels was adjusted individually using the in-line handheld mini gas regulator and maintained at 5-psi pressure to establish and maintain a stable O2 gradient. The headspace surrounding the device submerged in 4 ml of cell culture medium (refer to the cell culture section for composition) or Tyrode’s solution (refer to the calcium transient analysis section for composition) was injected with N2 gas using microbore tubing, which led from a compressed N2 tank into a fabricated PDMS lid enclosure to fit a 35-mm polystyrene dish containing the submerged device.
During uniform hypoxia experiments, devices were housed in a hypoxia chamber (STEMCELL Technologies, Vancouver, Canada) filled with 5% CO2, balanced N2 and maintained in a 37°C cell culture incubator until the device was transitioned to the fluorescent microscope or stereo microscope for calcium imaging experiments or MTF experiments, respectively. The two microchannels were perfused with 5% CO2, balanced N2 at 5-psi pressure during data acquisition. The uniform normoxia experiments were conducted by housing devices in a standard 37°C, 5% CO2 incubator until being transitioned to a microscope. The two microchannels were perfused with 5% CO2, balanced air at 5-psi pressure during data acquisition.
Oxygen gradient validation
The surface O2 profile was validated using fluorescent O2 sensors fabricated by impregnating 100-μm PDMS membranes with platinum(II) octaethylporphyrinketone (PtOEPK) (Frontier Specialty Chemicals, Logan, UT), as detailed elsewhere (76, 77). Briefly, a 35% (w/w) toluene/polystyrene mixture was made before adding PtOEPK (0.5 mg ml−1) to the solution of polystyrene dissolved in toluene. The mixture was spin-coated on a cured 100-μm PDMS membrane on a silicon wafer and left to dry overnight. The dried polystyrene was washed away with isopropanol, leaving behind the PtOEPK-containing PDMS membrane. Sensors were cut from the PDMS and placed on the surface of fabricated devices. The fluorescence of the PtOEPK fluorophore was quenched in the presence of O2. Scans across the sensor while the device was submerged in 4 ml of Dulbecco’s modified Eagle’s medium (DMEM) cell culture medium were used to determine the surface percent O2 profile. Scans of the O2 gradient were taken over an 8-hour period after acquiring calibration measurements of the sensor directly above a microfluidic channel perfused with (i) ambient 20% O2 and (ii) anoxic 0% O2 for 8 hours. The spatial profile of the percent O2 was plotted from the scans of the fluorescent intensity by solving the Stern-Volmer equation. During O2 gradient validation, the hypoxic channel was perfused with 5% CO2, balanced N2 and the normoxic channel was perfused with 5% CO2, balanced air. N2 gas was injected in the headspace surrounding the device submerged in cell culture medium.
Microcontact printing
PDMS stamps for microcontact printing were fabricated using standard soft lithography techniques. A photolithographic chromium mask containing an array of 22 mm × 22 mm squares of 15-μm wide lines separated by 2-μm wide gaps was designed in AutoCAD (Autodesk, San Rafael, CA). The microfeatures were patterned onto a silicon wafer with SU-8 2005 photoresist (Kayaku Advanced Materials, Tokyo, Japan). The silicon master was silanized overnight in a vacuum desiccator with 30 μl of trichloro(1H,1H,2H,2H-perfluorooctyl)silane and was then used as a template for the PDMS stamps. PDMS stamps were sonicated in ethanol for 30 min, dried, and coated with fibronectin (50 μg/ml; BD Biosciences, San Jose, CA) in sterile water for 1 hour. Stamps were dried with compressed air and inverted onto the fabricated devices after an 8-min treatment in a UVO cleaner (Jelight Company, Irvine, CA).
Cell culture
Ventricular tissue was isolated from 2-day-old Sprague-Dawley rats (mixed-sex litters). The care and use of laboratory animals was in accordance with the institutional guidelines of the Institutional Animal Care and Use Committee (IACUC protocol number: 20133). Ventricular tissue was incubated in a trypsin solution (1 mg ml−1) for 12 to 14 hours at 4°C. Four serial solutions of collagenase (1 mg ml−1) were triturated to dissociate the ventricular tissue into a single-cell suspension of NRVMs. The cells were passed through a 40-μm strainer and preplated two times for 45 min each to primarily deplete fibroblasts and reduce otherwise nonmyocyte cell populations. The cell solution was then seeded onto the devices at a concentration of 1.5 × 106 cells per device. Cells were cultured in M199 medium supplemented with 20 mM d-glucose, 10 mM Hepes, 2 mM l-glutamine, 0.1 mM minimum essential medium (MEM) nonessential amino acids, 1.5 μM vitamin B-12, penicillin (50 U ml−1), and 10% heat-inactivated fetal bovine serum (FBS) for 2 days. Then, the medium was changed to supplemented M199 with a reduced concentration of 2% FBS and exchanged every 24 hours. NRVMs were used for experimental studies on days 3 and 4 after seeding.
During all 4-hour O2 modulation experiments, including uniform normoxia, uniform hypoxia, and the O2 gradient, the medium was switched to DMEM without glucose (Gibco, Thermo Fisher Scientific, Waltham, MA) supplemented with d-glucose (0.1 g liter−1), 10 mM Hepes, 0.1 mM MEM nonessential amino acids, 1.5 μM vitamin B-12, and penicillin (50 U ml−1). This low-glucose minus serum condition was used because NRVMs are more resistant to hypoxia than adult rat CMs (78), and low glucose can increase sensitivity to hypoxia (79, 80).
Cell staining
NRVMs on the PDMS membrane of the devices were washed three times with phosphate-buffered saline (PBS) and then fixed with ice-cold methanol for 10 min. After washing three more times with PBS, NRVMs were incubated in 4′,6-diamidino-2-phenylindole (DAPI) and Alexa Fluor 488 phalloidin (Thermo Fisher Scientific, Waltham, MA) with a 1:200 dilution in PBS for 1 hour at room temperature. Cells were washed three times with PBS. Following the final wash, 200 μl of antifade mounting solution (ProLong Gold Antifade Mountant; Thermo Fisher Scientific, Waltham, MA) was added directly to the stained cells. A glass coverslip was laid on top, and the devices were left to cure on a flat surface in the dark for 24 hours before imaging. Devices were placed upside down so that the coverslip was in contact with the slide-holding stage insert and then imaged on a Nikon C2 point-scanning confocal microscope with a 60× oil (n = 1.515) objective.
For Cx43 immunofluorescence staining, NRVMs on the PDMS membrane of the devices were washed three times with PBS and then fixed with 4% paraformaldehyde for 10 min. Following fixation, cells were rinsed twice with PBS and permeabilized with 0.1% Triton X-100 in PBS for 10 min. Cells were kept at room temperature during all incubation steps. Cells were first incubated in 3% bovine serum albumin (BSA) and 0.1% Triton X-100 in PBS for 1 hour. Primary Cx43 antibody (C6219; Sigma-Aldrich, St. Louis, MO) was incubated at a dilution of 1:200 in 3% BSA and 0.1% Triton X-100 in PBS for 1.5 hours. The cells were then washed three times in PBS, with each wash lasting 5 min. Secondary antibody (A-11008 Alexa Fluor 488; Thermo Fisher Scientific, Waltham, MA) and DAPI were used at a dilution of 1:200 in 3% BSA, 5% goat serum, and 0.1% Triton X-100 in PBS for 1.5 hours. The cells on the membrane were rinsed three times in PBS and mounted (ProLong Gold Antifade Mountant; Thermo Fisher Scientific, Waltham, MA) on a glass slide for 24 hours in the dark before imaging. Devices mounted on glass slides were placed in a slide-holding stage insert and imaged with a 40× objective on a Nikon Eclipse Ti inverted fluorescent microscope with additional manual 1.5× magnification and an Andor Zyla Scientific Complementary Metal Oxide Semiconductor (sCMOS) camera.
Calcium transient analysis
Fluo-4 AM dye (Invitrogen) was loaded into NRVMs for calcium transient analysis. NRVMs on devices were incubated with 10 μl of a solution of 50 μg of Fluo-4 AM dissolved in 100 μl of 20% Pluronic F-127 in dimethyl sulfoxide (DMSO; Invitrogen) added to 4 ml of low-glucose Tyrode’s solution [5.0 mM Hepes, 1.0 mM magnesium chloride, 5.4 mM potassium chloride, 135.0 mM sodium chloride, 0.33 mM sodium phosphate, 1.8 mM calcium chloride, and d-glucose (0.1 g liter−1), pH 7.4] for 15 min at 37°C (for a final concentration of approximately 1.14 μM Fluo-4 AM in Tyrode’s solution). Before recordings were obtained, fresh prewarmed low-glucose Tyrode’s solution was added, and cells were allowed to sit for 15 min at 37°C to allow the dye to de-esterify inside the NRVMs. Before data acquisition, the solution was replaced again with 4 ml of fresh, prewarmed low-glucose Tyrode’s solution.
Devices were imaged on a Nikon Eclipse Ti inverted fluorescent microscope within a 37°C incubation chamber. A custom field electrode interfaced with a Myopacer (IonOptix, Westwood, MA) to deliver 10-V electrical pacing at 1 Hz. Image sequences were recorded using a 10× objective lens and an Andor Zyla sCMOS camera at 100 frames per second (FPS) for at least 4 s at 4 binning and a gain of 4. Time to peak, the time constant of decay, and FDHM were quantified using a custom MATLAB code that determined values from fluorescent intensity traces in selected regions of interest in the field of view. Each data point from the uniform controls represented the average of a measured transient parameter from at least four regions of interest in three fields of view for a duration of at least three beats. Each data point from the normoxic and hypoxic channel represented the average of a measured calcium transient parameter from six regions of interest in a field of view for a duration of four beats. Calcium transient data were excluded if the piecewise fit of the trace, as determined in MATLAB, was <0.95. Calcium transient data were also excluded from the first or last activation cycle (beat) if it was less than 80% complete in the captured acquisition.
Calcium wave propagation velocity analysis
Fluo-4 was loaded into NRVMs as outlined in the previous section. A Nikon Eclipse Ti inverted fluorescent microscope with a 37°C incubation chamber was used for image acquisition. A Myopacer (IonOptix, Westwood, MA) was used to deliver 10-V electrical pacing at 1 Hz via a custom platinum point-stimulation electrode. A micromanipulator (PatchMan NP 2; Eppendorf, Framingham, MA) positioned the electrode outside the field of view. Both longitudinal velocity and transverse velocity were measured by adjusting the orientation of the tissue relative to the position of the point electrode. Image sequences were recorded using a 2× objective lens and an Andor Zyla sCMOS camera at 100 FPS for 4 s at 8 binning and a gain of 4. Custom MATLAB software was developed to calculate calcium propagation velocity from image sequences, as previously described (30, 81). Briefly, we generated activation maps, for which each color represents an isochrone and delineates the leading edge of the wavefront and quantified propagation velocity from the linear regression of plots of the x position of each pixel in cropped activation maps versus activation time. A single linear regression of the transverse velocity in an O2 gradient did not result in a close fit. For this reason, a segmental linear regression model was used in the O2 gradient condition for transverse velocity measurement, separating the field into two equal halves deemed the normoxic side and hypoxic side. Each side was subjected to linear regression, and the resulting calcium propagation velocity was plotted.
Each calcium propagation velocity data point measurement represented the average propagation velocity of at least three individual activation cycles and a MATLAB metric of the closeness of fit (robust_s) < 7. Calcium propagation velocity data were excluded from the first or last activation cycle (beat) if it was less than 80% complete in the captured acquisition.
Cx43 image analysis
Quantitative image analysis of Cx43 immunosignal was performed using automated processing with an ImageJ/FIJI macro. Each 16-bit Cx43 image was background-subtracted using a rolling ball radius of 10 pixels. The default Auto Threshold function was used to quantify the Cx43 area in each 416 μm × 351 μm image. Nuclei were counted using the Analyze Particles function on a binary mask of the DAPI image.
MTF analysis of contractile stress
A Nikon SMZ 745T stereo microscope (Nikon Instruments, Melville, NY, USA) with a mounted camera (acA640-120um; Basler AG, Ahrensburg, Germany) was used for live recording of MTF contractions. Devices were placed in a 35-mm-diameter polystyrene petri dish filled with low-glucose Tyrode’s solution [1.8 mM CaCl2, glucose (0.1 g/liter), 5 mM Hepes, 1 mM MgCl, 5.4 mM KCl, 135 mM NaCl, and 0.33 mM NaH2PO4, pH 7.4]. A custom 3D-printed (MakerBot, Brooklyn, NY, USA) field electrode with two platinum wires spaced 1 cm apart was connected to a field stimulator (Myopacer; IonOptix, Westwood, MA, USA). The petri dish was placed in a heating plate (Warner Instruments, Hamden, CT, USA) controlled by the feedback from a thermoprobe, consisting of a cable with a bead thermistor (Warner Instruments, Hamden, CT, USA), in the solution connected to a CL-100 Temperature Controller (Warner Instruments, Hamden, CT, USA) and liquid cooling system (Koolance, Auburn, WA, USA) to maintain the solution at 37°C during the O2 modulation and contractile stress measurement. The precut bilayer laser-engraved MTFs were manually peeled using forceps. Then, deflection of the MTFs was recorded at 2-Hz electrical pacing with an external voltage of 15 V and a collection frame rate of 100 fps.
RNA sequencing
After 4 hours of O2 modulation, engineered cardiac tissues on the devices were lysed using 1 ml of TRIzol reagent (Thermo Fisher Scientific, Waltham, MA). Gradient devices were cut in half to make a separate normoxic side and hypoxic side to be analyzed. The normoxic side and hypoxic side were each lysed using 1 ml of TRIzol reagent. Lysed samples were stored at −80°C until RNA isolation was performed. Total RNA was isolated using the miRNeasy Kit and protocol.
RNA was prepared for sequencing by the Cedars-Sinai Applied Genomics, Computation, and Translational Core. RNA concentration was assessed using a Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA), and RNA quality was assessed using 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA was enriched for mRNA using the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs Inc., Ipswich, MA). Libraries were constructed with the Swift RNA Library Kit (Swift Biosciences, Ann Arbor, MI). Library concentration and size were measured with a Qubit fluorometer and Agilent 4200 TapeStation (Agilent Technologies, Santa Clara, CA), respectively. Libraries were multiplexed and sequenced on NovaSeq 6000 (Illumina, San Diego, CA) using 75–base pair single-end sequencing.
RNA-seq data were submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) data repository (accession number GSE189108; n = 5 uniform normoxia, n = 5 uniform hypoxia, n = 4 normoxic side of O2 gradient, and n = 4 hypoxic side of O2 gradient). Analyses were performed using Partek Flow Genomic Analysis software. Partek Flow workflow tools were used to build the following pipeline: (i) prealignment QA/QC, (ii) bases trimmed from both ends based on a Phred quality score of 20 and a minimum read length of 25 remaining bases, (iii) STAR to align mRNA-seq reads to the rn6 rat genome, (iv) post-alignment QA/QC, (v) quantification to annotation model (Partek E/M), (vi) noise reduction filter where the maximum ≤ 10, (vii) upper-quartile normalization after adding 1.0 to correct for any division by 0 errors, (viii) PCA analysis, and (ix) gene set analysis. Data were assessed via PCA and differential expression analysis with P < 0.05, |fold change| ≥ 2.0, and FDR cutoff = 0.05. Fold-change data from Partek Flow were uploaded to Qiagen IPA software with analysis settings of P < 0.05 and |fold change| ≥ 2.0 to identify canonical pathways.
RNA-seq comparative analysis of in vivo and in vitro MI models
Publicly available RNA-seq datasets of the affected myocardium of rats 24 hours after a surgical ligation MI and control myocardium of rats (n = 3 MI and n = 3 control; GSE201888) were analyzed with Partek Flow Genomic Analysis software. Data were assessed via PCA and differential expression analysis with P < 0.05. PC gene loadings were used as rankings for genes in IPA to interpret the visual separation between samples in the PCA.
The list of up-regulated genes that have similar expression profiles in the in vitro gradient condition and in vivo infarction was compiled with the following requirements: (i) P < 0.05 and the fold change ≥ 2 for gradient normoxia versus normoxia, gradient hypoxia versus normoxia, and MI versus control comparisons and (ii) fold change < 1 for hypoxia versus normoxia. The cutoff for hypoxia versus normoxia ensured that the up-regulated DEGs in the list specifically did not have a similar expression profile in the uniform hypoxia condition and were distinct to the gradient condition (data S3). Similarly, a list of down-regulated genes required the following: (i) P < 0.05 and the fold change ≤ −2 for gradient normoxia versus normoxia, gradient hypoxia versus normoxia, and MI versus control comparisons and (ii) fold change > 1 for hypoxia versus normoxia. Again, the cutoff for hypoxia versus normoxia ensured that the down-regulated DEGs specifically did not have a similar expression profile in the uniform hypoxia condition (data S3).
Statistical analysis
Analyses were performed using Prism 8 by GraphPad (GraphPad Software Inc., San Diego, CA). Calcium transient, calcium propagation velocity, and Cx43 data were determined to be normally distributed by the Shapiro-Wilk test for normality. Statistical significance of time to peak, tau of decay, and FDHM between the different conditions (n = 5 independent experiments for each condition) was assessed by analysis of variance (ANOVA), comparing the mean of each group with the mean of every other group, followed by Tukey’s multiple comparison post hoc test. Data are expressed as means ± SEM. Statistical significance of longitudinal (n = 7 independent experiments) and transverse calcium propagation velocity (n = 7 independent experiments for uniform normoxia and uniform hypoxia and n = 6 independent experiments for the gradient condition) was assessed by ANOVA, comparing the mean of each group with the mean of the uniform normoxia group, followed by Sidak’s multiple comparison post hoc test. Statistical significance of the Cx43 immunosignal (n = 5 for each condition) was assessed by ANOVA, comparing the mean of each group with the mean of every other group, followed by Tukey’s multiple comparison post hoc test. Each data point represents the average area of Cx43 coverage from five different 416 μm × 351 μm size images from a device. Data are expressed as means ± SEM.
For MTF experiments, data were determined to be not normally distributed. Data were analyzed with the nonparametric Kruskal-Wallis test, comparing the mean rank of each group with the mean rank of the uniform normoxia group, followed by Dunn’s multiple comparison post hoc test. Each MTF was treated as an independent sample, consistent with previous literature (39, 82–85). MTF-generated systolic and twitch stress were measured from a sample size of n = 96, n = 85, n = 16, and n = 15 films for uniform normoxia, uniform hypoxia, normoxic channel, and hypoxic channel, respectively. Diastolic stress was measured from a sample size of n = 99, n = 85, n = 16, and n = 15 films for uniform normoxia, uniform hypoxia, normoxic channel, and hypoxic channel, respectively. Film data were collected from eight, seven, eight, and eight independent chips for the different conditions, respectively. For all data, P < 0.05 was considered significant. Data are expressed as means ± SEM.
Acknowledgments
This study used the services of Cedars-Sinai Medical Center Applied Genomics, Computation & Translational Core for RNA sequencing. We acknowledge the USC Nanofabrication Core and W.M. Keck Foundation Photonics Center Cleanroom for photolithography equipment and facilities.
Funding: This work was supported by NIH grant R01 HL153286-01 (to M.L.M. and S.J.P.), American Heart Association Postdoctoral Fellowship 19POST34380814 (to M.L.R.-H.), and USC Viterbi School of Engineering.
Author contributions: Conceptualization: M.L.R.-H. and M.L.M. Methodology: M.L.R.-H. and M.L.M. Fabrication: M.L.R.-H. and N.N.K. Procedures: M.L.R.-H., N.N.K., and S.S.E. Derivation: X.L., J.H., and H.Y. Investigation: M.L.R.-H. Visualization: M.L.R.-H. Supervision: M.L.M. and S.J.P. Writing—original draft: M.L.R.-H. and M.L.M. Writing—review and editing: M.L.R.-H., N.N.K., S.S.E., X.L., J.H., H.Y., S.J.P., and M.L.M.
Competing interests: M.L.R.-H. is an inventor on a patent related to this work filed by the University of Illinois at Chicago (US20130295551A1, filed on 15 March 2013, published on 7 November 2013). M.L.M. is an inventor on a patent related to this work filed by Harvard University (US9857356B2, filed on 10 December 2012, published on 2 January 2018). The authors declare no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data from the myocardial infarct border-zone-on-a-chip devices are available from the NCBI GEO database, under the accession number GSE189108. The in vivo RNA-seq data are available from the NCBI GEO database, under the accession number GSE201888.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S4
Tables S1 to S8
Other Supplementary Material for this : manuscript includes the following:
Data S1 to S3
REFERENCES AND NOTES
- 1.S. D. Prabhu, N. G. Frangogiannis,The biological basis for cardiac repair after myocardial infarction: From inflammation to fibrosis. Circ. Res. 119,91–112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.W. L. Rumsey, M. Pawlowski, N. Lejavardi, D. F. Wilson,Oxygen pressure distribution in the heart in vivo and evaluation of the ischemic “border zone”. Am. J. Physiol. 266,H1676–H1680 (1994). [DOI] [PubMed] [Google Scholar]
- 3.M. A. Pfeffer, E. Braunwald,Ventricular remodeling after myocardial infarction. Experimental observations and clinical implications. Circulation 81,1161–1172 (1990). [DOI] [PubMed] [Google Scholar]
- 4.C. Cabo, J. Yao, P. A. Boyden, S. Chen, W. Hussain, H. S. Duffy, E. J. Ciaccio, N. S. Peters, A. L. Wit,Heterogeneous gap junction remodeling in reentrant circuits in the epicardial border zone of the healing canine infarct. Cardiovasc. Res. 72,241–249 (2006). [DOI] [PubMed] [Google Scholar]
- 5.K. Spaulding, K. Takaba, A. Collins, F. Faraji, G. Wang, E. Aguayo, L. Ge, D. Saloner, A. W. Wallace, A. J. Baker, D. H. Lovett, M. B. Ratcliffe,Short term doxycycline treatment induces sustained improvement in myocardial infarction border zone contractility. PLOS ONE 13,e0192720 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.H. Liu, O. A. Bolonduro, N. Hu, J. Ju, A. A. Rao, B. M. Duffy, Z. Huang, L. D. Black, B. P. Timko,Heart-on-a-chip model with integrated extra- and intracellular bioelectronics for monitoring cardiac electrophysiology under acute hypoxia. Nano Lett. 20,2585–2593 (2020). [DOI] [PubMed] [Google Scholar]
- 7.M. Yadid, J. U. Lind, H. A. M. Ardoña, S. P. Sheehy, L. E. Dickinson, F. Eweje, M. M. C. Bastings, B. Pope, B. B. O’Connor, J. R. Straubhaar, B. Budnik, A. G. Kleber, K. K. Parker,Endothelial extracellular vesicles contain protective proteins and rescue ischemia-reperfusion injury in a human heart-on-chip. Sci. Transl. Med. 12,eaax8005 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.A. Danon, N. Zeevi-Levin, D. Y. Pinkovich, T. Michaeli, A. Berkovich, M. Flugelman, Y. C. Eldar, M. R. Rosen, O. Binah,Hypoxia causes connexin 43 internalization in neonatal rat ventricular myocytes. Gen. Physiol. Biophys. 29,222–233 (2010). [DOI] [PubMed] [Google Scholar]
- 9.P. González-Rodríguez, D. Falcón, M. J. Castro, J. Ureña, J. López-Barneo, A. Castellano,Hypoxic induction of T-type Ca2+ channels in rat cardiac myocytes: Role of HIF-1α and RhoA/ROCK signalling. J. Physiol. 593,4729–4745 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.M. Tanaka, H. Ito, S. Adachi, H. Akimoto, T. Nishikawa, T. Kasajima, F. Marumo, M. Hiroe,Hypoxia induces apoptosis with enhanced expression of Fas antigen messenger RNA in cultured neonatal rat cardiomyocytes. Circ. Res. 75,426–433 (1994). [DOI] [PubMed] [Google Scholar]
- 11.X. Long, M. O. Boluyt, M. L. Hipolito, M. S. Lundberg, J. S. Zheng, L. O’Neill, C. Cirielli, E. G. Lakatta, M. T. Crow,p53 and the hypoxia-induced apoptosis of cultured neonatal rat cardiac myocytes. J. Clin. Invest. 99,2635–2643 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.P. M. Kang, A. Haunstetter, H. Aoki, A. Usheva, S. Izumo,Morphological and molecular characterization of adult cardiomyocyte apoptosis during hypoxia and reoxygenation. Circ. Res. 87,118–125 (2000). [DOI] [PubMed] [Google Scholar]
- 13.L. Ren, W. Liu, Y. Wang, J. C. Wang, Q. Tu, J. Xu, R. Liu, S. F. Shen, J. Wang,Investigation of hypoxia-induced myocardial injury dynamics in a tissue interface mimicking microfluidic device. Anal. Chem. 85,235–244 (2013). [DOI] [PubMed] [Google Scholar]
- 14.X. Ai, W. Lu, K. Zeng, C. Li, Y. Jiang, P. Tu,Microfluidic coculture device for monitoring of inflammation-induced myocardial injury dynamics. Anal. Chem. 90,4485–4494 (2018). [DOI] [PubMed] [Google Scholar]
- 15.M. Häkli, J. Kreutzer, A. J. Mäki, H. Välimäki, H. Lappi, H. Huhtala, P. Kallio, K. Aalto-Setälä, M. Pekkanen-Mattila,Human induced pluripotent stem cell-based platform for modeling cardiac ischemia. Sci. Rep. 11,4153 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.G. Khanal, K. Chung, X. Solis-Wever, B. Johnson, D. Pappas,Ischemia/reperfusion injury of primary porcine cardiomyocytes in a low-shear microfluidic culture and analysis device. Analyst 136,3519–3526 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. Radisic, J. Malda, E. Epping, W. Geng, R. Langer, G. Vunjak-Novakovic,Oxygen gradients correlate with cell density and cell viability in engineered cardiac tissue. Biotechnol. Bioeng. 93,332–343 (2006). [DOI] [PubMed] [Google Scholar]
- 18.B. Mosadegh, B. E. Dabiri, M. R. Lockett, R. Derda, P. Campbell, K. K. Parker, G. M. Whitesides,Three-dimensional paper-based model for cardiac ischemia. Adv. Healthc. Mater. 3,1036–1043 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.C. Mendonca Costa, G. Plank, C. A. Rinaldi, S. A. Niederer, M. J. Bishop,Modeling the electrophysiological properties of the infarct border zone. Front. Physiol. 9,356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.J. T. Lee, R. E. Ideker, K. A. Reimer,Myocardial infarct size and location in relation to the coronary vascular bed at risk in man. Circulation 64,526–534 (1981). [DOI] [PubMed] [Google Scholar]
- 21.S. Nanto, T. Masuyama, Y. J. Lim, M. Hori, K. Kodama, T. Kamada,Demonstration of functional border zone with myocardial contrast echocardiography in human hearts. Simultaneous analysis of myocardial perfusion and wall motion abnormalities. Circulation 88,447–453 (1993). [DOI] [PubMed] [Google Scholar]
- 22.A. J. Buda, R. J. Zotz, K. P. Gallagher,Characterization of the functional border zone around regionally ischemic myocardium using circumferential flow-function maps. J. Am. Coll. Cardiol. 8,150–158 (1986). [DOI] [PubMed] [Google Scholar]
- 23.M. L. Rexius-Hall, J. Rehman, D. T. Eddington,A microfluidic oxygen gradient demonstrates differential activation of the hypoxia-regulated transcription factors HIF-1α and HIF-2α. Integr. Biol. (Camb.) 9,742–750 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Y. Ando, H. P. Ta, D. P. Yen, S. S. Lee, S. Raola, K. Shen,A microdevice platform recapitulating hypoxic tumor microenvironments. Sci. Rep. 7,15233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.E. T. Hasche, C. Fernandes, S. B. Freedman, R. W. Jeremy,Relation between ischemia time, infarct size, and left ventricular function in humans. Circulation 92,710–719 (1995). [DOI] [PubMed] [Google Scholar]
- 26.H. Sutanto, A. Lyon, J. Lumens, U. Schotten, D. Dobrev, J. Heijman,Cardiomyocyte calcium handling in health and disease: Insights from in vitro and in silico studies. Prog. Biophys. Mol. Biol. 157,54–75 (2020). [DOI] [PubMed] [Google Scholar]
- 27.C. J. Fearnley, H. L. Roderick, M. D. Bootman,Calcium signaling in cardiac myocytes. Cold Spring Harb. Perspect. Biol. 3,a004242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.L. D. Gauthier, J. L. Greenstein, R. L. Winslow,Toward an integrative computational model of the Guinea pig cardiac myocyte. Front. Physiol. 3,244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.P. Lee, M. Klos, C. Bollensdorff, L. Hou, P. Ewart, T. J. Kamp, J. Zhang, A. Bizy, G. Guerrero-Serna, P. Kohl, J. Jalife, T. J. Herron,Simultaneous voltage and calcium mapping of genetically purified human induced pluripotent stem cell-derived cardiac myocyte monolayers. Circ. Res. 110,1556–1563 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.A. P. Petersen, D. M. Lyra-Leite, N. R. Ariyasinghe, N. Cho, C. M. Goodwin, J. Y. Kim, M. L. McCain,Microenvironmental modulation of calcium wave propagation velocity in engineered cardiac tissues. Cell. Mol. Bioeng. 11,337–352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.A. G. Kléber, M. J. Janse, F. J. Wilms-Schopmann, A. A. Wilde, R. Coronel,Changes in conduction velocity during acute ischemia in ventricular myocardium of the isolated porcine heart. Circulation 73,189–198 (1986). [DOI] [PubMed] [Google Scholar]
- 32.N. Zeevi-Levin, Y. D. Barac, Y. Reisner, I. Reiter, G. Yaniv, G. Meiry, Z. Abassi, S. Kostin, J. Schaper, M. R. Rosen, N. Resnick, O. Binah,Gap junctional remodeling by hypoxia in cultured neonatal rat ventricular myocytes. Cardiovasc. Res. 66,64–73 (2005). [DOI] [PubMed] [Google Scholar]
- 33.M. L. McCain, T. Desplantez, N. A. Geisse, B. Rothen-Rutishauser, H. Oberer, K. K. Parker, A. G. Kleber,Cell-to-cell coupling in engineered pairs of rat ventricular cardiomyocytes: Relation between Cx43 immunofluorescence and intercellular electrical conductance. Am. J. Physiol. Heart Circ. Physiol. 302,H443–H450 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.P. Beauchamp, K. A. Yamada, A. J. Baertschi, K. Green, E. M. Kanter, J. E. Saffitz, A. G. Kléber,Relative contributions of connexins 40 and 43 to atrial impulse propagation in synthetic strands of neonatal and fetal murine cardiomyocytes. Circ. Res. 99,1216–1224 (2006). [DOI] [PubMed] [Google Scholar]
- 35.B. J. Darrow, V. G. Fast, A. G. Kléber, E. C. Beyer, J. E. Saffitz,Functional and structural assessment of intercellular communication. Increased conduction velocity and enhanced connexin expression in dibutyryl cAMP-treated cultured cardiac myocytes. Circ. Res. 79,174–183 (1996). [DOI] [PubMed] [Google Scholar]
- 36.M. Boulaksil, M. F. Bierhuizen, M. A. Engelen, M. Stein, B. J. Kok, S. C. van Amersfoorth, M. A. Vos, H. V. van Rijen, J. M. de Bakker, T. A. van Veen,Spatial heterogeneity of Cx43 is an arrhythmogenic substrate of polymorphic ventricular tachycardias during compensated cardiac hypertrophy in rats. Front. Cardiovasc. Med. 3,5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.M. Boulaksil, S. K. Winckels, M. A. Engelen, M. Stein, T. A. van Veen, J. A. Jansen, A. C. Linnenbank, M. F. Bierhuizen, W. A. Groenewegen, M. F. van Oosterhout, J. H. Kirkels, N. de Jonge, A. Varró, M. A. Vos, J. M. de Bakker, H. V. van Rijen,Heterogeneous Connexin43 distribution in heart failure is associated with dispersed conduction and enhanced susceptibility to ventricular arrhythmias. Eur. J. Heart Fail. 12,913–921 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Y. Prudat, J. P. Kucera,Nonlinear behaviour of conduction and block in cardiac tissue with heterogeneous expression of connexin 43. J. Mol. Cell. Cardiol. 76,46–54 (2014). [DOI] [PubMed] [Google Scholar]
- 39.A. Grosberg, P. W. Alford, M. L. McCain, K. K. Parker,Ensembles of engineered cardiac tissues for physiological and pharmacological study: Heart on a chip. Lab Chip 11,4165–4173 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.J. F. Wenk, D. Klepach, L. C. Lee, Z. Zhang, L. Ge, E. E. Tseng, A. Martin, S. Kozerke, J. H. Gorman, R. C. Gorman, J. M. Guccione,First evidence of depressed contractility in the border zone of a human myocardial infarction. Ann. Thorac. Surg. 93,1188–1193 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.F. H. Epstein, Z. Yang, W. D. Gilson, S. S. Berr, C. M. Kramer, B. A. French,MR tagging early after myocardial infarction in mice demonstrates contractile dysfunction in adjacent and remote regions. Magn. Reson. Med. 48,399–403 (2002). [DOI] [PubMed] [Google Scholar]
- 42.W. B. Kannel, P. Sorlie, W. P. Castelli, D. McGee,Blood pressure and survival after myocardial infarction: The Framingham study. Am. J. Cardiol. 45,326–330 (1980). [DOI] [PubMed] [Google Scholar]
- 43.W. H. Gaasch,Diagnosis and treatment of heart failure based on left ventricular systolic or diastolic dysfunction. JAMA 271,1276–1280 (1994). [PubMed] [Google Scholar]
- 44.J. J. Thune, S. D. Solomon,Left ventricular diastolic function following myocardial infarction. Curr. Heart Fail. Rep. 3,170–174 (2006). [DOI] [PubMed] [Google Scholar]
- 45.M. L. McCain, S. P. Sheehy, A. Grosberg, J. A. Goss, K. K. Parker,Recapitulating maladaptive, multiscale remodeling of failing myocardium on a chip. Proc. Natl. Acad. Sci. U.S.A. 110,9770–9775 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.L. F. Wexler, B. H. Lorell, S. Momomura, E. O. Weinberg, J. S. Ingwall, C. S. Apstein,Enhanced sensitivity to hypoxia-induced diastolic dysfunction in pressure-overload left ventricular hypertrophy in the rat: Role of high-energy phosphate depletion. Circ. Res. 62,766–775 (1988). [DOI] [PubMed] [Google Scholar]
- 47.G. Diamond, J. S. Forrester,Effect of coronary artery disease and acute myocardial infarction on left ventricular compliance in man. Circulation 45,11–19 (1972). [DOI] [PubMed] [Google Scholar]
- 48.M. Deng, W. Zhang, L. Yuan, J. Tan, Z. Chen,HIF-1a regulates hypoxia-induced autophagy via translocation of ANKRD37 in colon cancer. Exp. Cell Res. 395,112175 (2020). [DOI] [PubMed] [Google Scholar]
- 49.I. Tirado-Hurtado, W. Fajardo, J. A. Pinto,DNA damage inducible transcript 4 gene: The switch of the metabolism as potential target in cancer. Front. Oncol. 8,106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Y. Cai, K. L. Xie, H. L. Wu, K. Wu,Functional suppression of Epiregulin impairs angiogenesis and aggravates left ventricular remodeling by disrupting the extracellular-signal-regulated kinase1/2 signaling pathway in rats after acute myocardial infarction. J. Cell. Physiol. 234,18653–18665 (2019). [DOI] [PubMed] [Google Scholar]
- 51.V. N. Ritschel, I. Seljeflot, H. Arnesen, S. Halvorsen, T. Weiss, J. Eritsland, G. Andersen,IL-6 signalling in patients with acute ST-elevation myocardial infarction. Results Immunol. 4,8–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.M. D. Mora-Ruíz, F. Blanco-Favela, A. K. Chávez Rueda, M. V. Legorreta-Haquet, L. Chávez-Sánchez,Role of interleukin-17 in acute myocardial infarction. Mol. Immunol. 107,71–78 (2019). [DOI] [PubMed] [Google Scholar]
- 53.M. Andrassy, H. C. Volz, N. Riedle, G. Gitsioudis, C. Seidel, D. Laohachewin, A. R. Zankl, Z. Kaya, A. Bierhaus, E. Giannitsis, H. A. Katus, G. Korosoglou,HMGB1 as a predictor of infarct transmurality and functional recovery in patients with myocardial infarction. J. Intern. Med. 270,245–253 (2011). [DOI] [PubMed] [Google Scholar]
- 54.A. J. Muslin,MAPK signalling in cardiovascular health and disease: Molecular mechanisms and therapeutic targets. Clin. Sci. (Lond.) 115,203–218 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.P. M. Docshin, A. A. Karpov, M. V. Mametov, D. Y. Ivkin, A. A. Kostareva, A. B. Malashicheva,Mechanisms of regenerative potential activation in cardiac mesenchymal cells. Biomedicine 10,1283 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.N. Tanaka, K. Masamura, M. Yoshida, M. Kato, Y. Kawai, I. Miyamori,A role of heparin-binding epidermal growth factor-like growth factor in cardiac remodeling after myocardial infarction. Biochem. Biophys. Res. Commun. 297,375–381 (2002). [DOI] [PubMed] [Google Scholar]
- 57.M. Koeppen, J. W. Lee, S. W. Seo, K. S. Brodsky, S. Kreth, I. V. Yang, P. M. Buttrick, T. Eckle, H. K. Eltzschig,Hypoxia-inducible factor 2-alpha-dependent induction of amphiregulin dampens myocardial ischemia-reperfusion injury. Nat. Commun. 9,816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D. Veltman, M. Wu, P. Pokreisz, P. Claus, H. Gillijns, E. Caluwé, M. Vanhaverbeke, W. Gsell, U. Himmelreich, P. R. Sinnaeve, S. P. Janssens,Clec4e-receptor signaling in myocardial repair after ischemia-reperfusion injury. JACC Basic Transl. Sci. 6,631–646 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.N. G. Frangogiannis,Interleukin-1 in cardiac injury, repair, and remodeling: Pathophysiologic and translational concepts. Discoveries (Craiova) 3,e41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.S. de Jong, T. A. van Veen, H. V. van Rijen, J. M. de Bakker,Fibrosis and cardiac arrhythmias. J. Cardiovasc. Pharmacol. 57,630–638 (2011). [DOI] [PubMed] [Google Scholar]
- 61.P. Baumeister, T. A. Quinn,Altered calcium handling and ventricular arrhythmias in acute ischemia. Clin. Med. Insights Cardiol. 10,61–69 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.J. L. Solan, P. D. Lampe,Connexin43 phosphorylation: Structural changes and biological effects. Biochem. J. 419,261–272 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.R. J. Vagnozzi, M. Maillet, M. A. Sargent, H. Khalil, A. K. Z. Johansen, J. A. Schwanekamp, A. J. York, V. Huang, M. Nahrendorf, S. Sadayappan, J. D. Molkentin,An acute immune response underlies the benefit of cardiac stem cell therapy. Nature 577,405–409 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.F. K. Swirski, M. Nahrendorf,Leukocyte behavior in atherosclerosis, myocardial infarction, and heart failure. Science 339,161–166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.T. Aoyagi, T. Matsui,The cardiomyocyte as a source of cytokines in cardiac injury. J. Cell Sci. Ther. 2012,003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.B. Chanyshev, A. Shainberg, A. Isak, Y. Chepurko, E. Porat, E. Hochhauser,Conditioned medium from hypoxic cells protects cardiomyocytes against ischemia. Mol. Cell. Biochem. 363,167–178 (2012). [DOI] [PubMed] [Google Scholar]
- 67.X. Yang, L. Pabon, C. E. Murry,Engineering adolescence: Maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 114,511–523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.F. B. Bedada, M. Wheelwright, J. M. Metzger,Maturation status of sarcomere structure and function in human iPSC-derived cardiac myocytes. Biochim. Biophys. Acta 1863,1829–1838 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.A. M. da Rocha, K. Campbell, S. Mironov, J. Jiang, L. Mundada, G. Guerrero-Serna, J. Jalife, T. J. Herron,hiPSC-CM monolayer maturation state determines drug responsiveness in high throughput pro-arrhythmia screen. Sci. Rep. 7,13834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.B. M. Ulmer, T. Eschenhagen,Human pluripotent stem cell-derived cardiomyocytes for studying energy metabolism. Biochim. Biophys. Acta Mol. Cell Res. 1867,118471 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.S. Hein, S. Kostin, J. Schaper,Adult rat cardiac myocytes in culture: ‘Second-floor’ cells and coculture experiments. Exp. Clin. Cardiol. 11,175–182 (2006). [PMC free article] [PubMed] [Google Scholar]
- 72.F. Nippert, R. Schreckenberg, K. D. Schlüter,Isolation and cultivation of adult rat cardiomyocytes. J. Vis. Exp. 2017,56634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.M. R. Glick, A. H. Burns, W. J. Reddy,Dispersion and isolation of beating cells from adult rat heart. Anal. Biochem. 61,32–42 (1974). [DOI] [PubMed] [Google Scholar]
- 74.C. Liu, J. A. Thompson, H. H. Bau,A membrane-based, high-efficiency, microfluidic debubbler. Lab Chip 11,1688–1693 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.J. K. Yip, M. Harrison, J. Villafuerte, G. E. Fernandez, A. P. Petersen, C. L. Lien, M. L. McCain,Extended culture and imaging of normal and regenerating adult zebrafish hearts in a fluidic device. Lab Chip 20,274–284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.M. L. Rexius-Hall, G. Mauleon, A. B. Malik, J. Rehman, D. T. Eddington,Microfluidic platform generates oxygen landscapes for localized hypoxic activation. Lab Chip 14,4688–4695 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.E. Sinkala, D. T. Eddington,Oxygen sensitive microwells. Lab Chip 10,3291–3295 (2010). [DOI] [PubMed] [Google Scholar]
- 78.D. Sedmera, P. Kucera, E. Raddatz,Developmental changes in cardiac recovery from anoxia-reoxygenation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 283,R379–R388 (2002). [DOI] [PubMed] [Google Scholar]
- 79.K. R. Carraway, E. M. Johnson, T. C. Kauffmann, N. J. Fry, K. D. Mansfield,Hypoxia and hypoglycemia synergistically regulate mRNA stability. RNA Biol. 14,938–951 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.R. Malhotra, F. C. Brosius,Glucose uptake and glycolysis reduce hypoxia-induced apoptosis in cultured neonatal rat cardiac myocytes. J. Biol. Chem. 274,12567–12575 (1999). [DOI] [PubMed] [Google Scholar]
- 81.A. P. Petersen, N. Cho, D. M. Lyra-Leite, J. W. Santoso, D. Gupta, N. R. Ariyasinghe, M. L. McCain,Regulation of calcium dynamics and propagation velocity by tissue microstructure in engineered strands of cardiac tissue. Integr. Biol. (Camb.) 12,34–46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.G. Wang, M. L. McCain, L. Yang, A. He, F. S. Pasqualini, A. Agarwal, H. Yuan, D. Jiang, D. Zhang, L. Zangi, J. Geva, A. E. Roberts, Q. Ma, J. Ding, J. Chen, D. Z. Wang, K. Li, J. Wang, R. J. Wanders, W. Kulik, F. M. Vaz, M. A. Laflamme, C. E. Murry, K. R. Chien, R. I. Kelley, G. M. Church, K. K. Parker, W. T. Pu,Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 20,616–623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.M. L. McCain, A. Agarwal, H. W. Nesmith, A. P. Nesmith, K. K. Parker,Micromolded gelatin hydrogels for extended culture of engineered cardiac tissues. Biomaterials 35,5462–5471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.A. P. Nesmith, M. A. Wagner, F. S. Pasqualini, B. B. O’Connor, M. J. Pincus, P. R. August, K. K. Parker,A human in vitro model of Duchenne muscular dystrophy muscle formation and contractility. J. Cell Biol. 215,47–56 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.M. L. Rexius-Hall, N. N. Khalil, A. M. Andres, M. L. McCain,Mitochondrial division inhibitor 1 (mdivi-1) increases oxidative capacity and contractile stress generated by engineered skeletal muscle. FASEB J. 34,11562–11576 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S4
Tables S1 to S8
Data S1 to S3