Abstract
Targeted protein degradation has recently gained widespread interest as both a novel therapeutic strategy and a useful tool in biomedical research. Targeted protein degraders are often sub-stoichiometric, and do not require strong binding affinity for their targets, enabling access to previously inaccessible targets. Proteolysis-targeting chimeras (PROTACs) are one class of targeted protein degraders that promote degradation by recruiting a target protein to an E3-ligase complex via a heterobifunctional molecule. The modular nature of PROTACs allow for their rational design and systematic optimization. Here we suggest resources and methodologies for the development of PROTAC degraders for researchers that may be new to the field.
Keywords: Targeted Protein Degradation, proteolysis-targeting chimera, PROTAC, compound optimization, medicinal chemistry
INTRODUCTION
Targeted protein degradation (TPD) is an exciting research strategy with expanding applications in chemical probe and therapeutic agent development (Békés, Langley, & Crews, 2022; Chamberlain & Hamann, 2019; Faust & Donovan et al., 2021). Using small molecules called degraders, the ubiquitin-proteasome system (UPS) is “hijacked” and E3-ligases redirected to degrade substrates not endogenously targeted, termed neosubstrates. This is accomplished by molecules promoting the formation of a ternary complex between a protein-of-interest (POI), an E3-ligase component, and the degrader small molecule, which results in ubiquitination of the target substrate and subsequent proteasomal degradation. A major class of small molecules commonly used for TPD are known as proteolysis-targeting chimeras (PROTACs), which consist of a POI-binding ligand and an E3-ligase ligand joined by a linker. To assist groups newly interested in TPD approaches, we provide resources and considerations for the initial design, testing, and validation of PROTAC degraders.
Many publicly accessible databases have compiled the recent work done to develop and characterize small molecule degraders, in addition to ligand information useful for the preliminary design of PROTACs. Though the degrader must be optimized for each specific E3-ligase:target pair, there are numerous published examples of successful PROTAC development which can help inform initial selection of the PROTAC components. If the initial candidate degraders do not exhibit significant target degradation, evaluation of PROTAC-induced target engagement and ternary complex formation can guide optimization of more potent molecules. Following identification of a potent PROTAC, several experiments must be performed to validate its mechanism of action and selectivity. This workflow will aid development of potent, selective PROTAC degraders, along with appropriate controls, suitable for downstream applications as a chemical probe (Fig. 1).
Fig. 1.
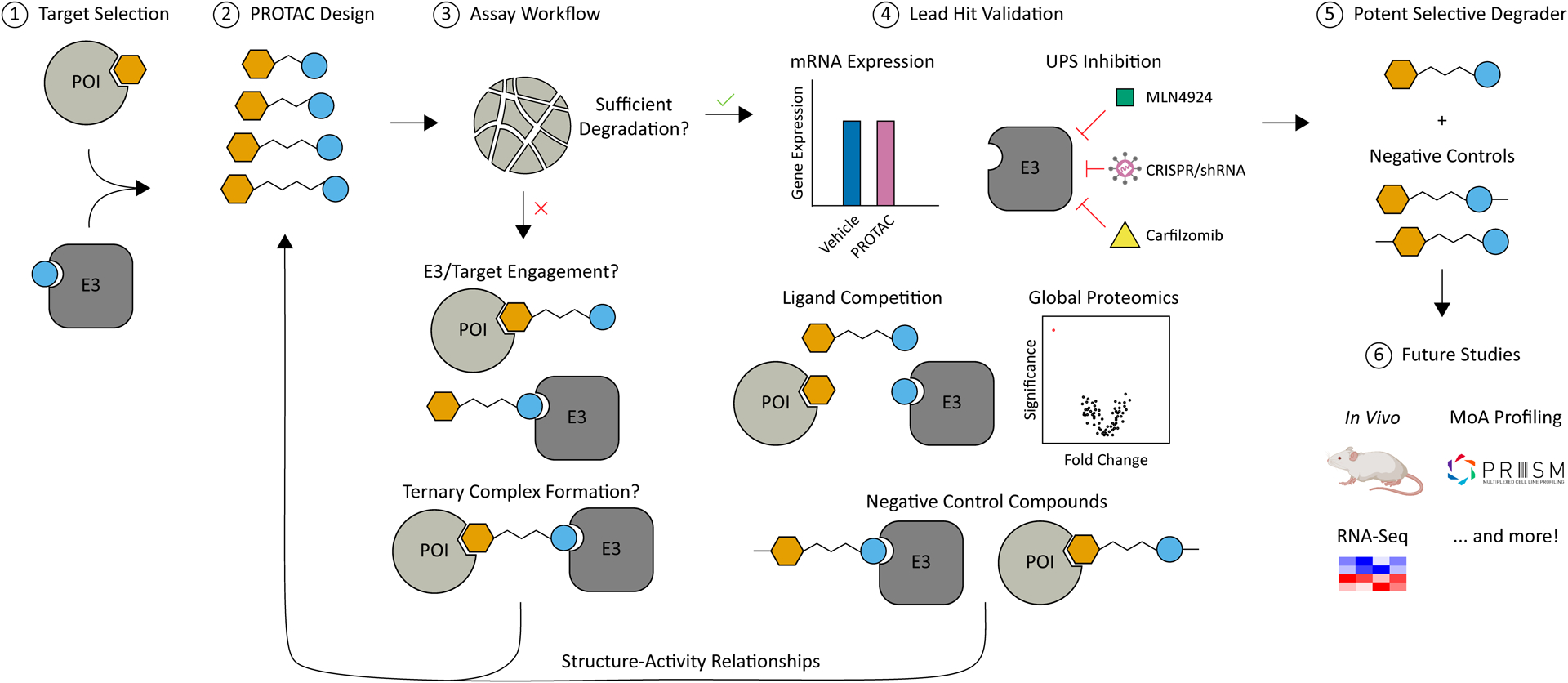
Summary schematic for a typical targeted protein degradation (TPD) project workflow. A target substrate is first selected and preliminary candidate PROTACs are designed based on established E3-ligase binders, commonly used linkers, and potential target ligands. Testing of these compounds for degradation efficiency can result in the identification of a potent degrader. However, if no potent degraders are identified, a workflow can be built around iterative synthesis and testing of target engagement and ternary complex formation to inform the design of more potent PROTAC molecules. Validation of the lead compound is necessary and can further inform design of more selective degraders. Finally, this workflow will afford a potent, selective PROTAC and negative control compounds that can be use in further biological studies.
Getting Started
Although PROTACs hold promise in targeting a subset of the traditionally “undruggable” proteome, there is a minimum requirement of a ligand binder that must be met prior to initial compound development. For informational purposes, this review compiles resources useful for the planning of a TPD project and assumes an existing ligand for the target of interest (Fig. 1). All databases mentioned here are publicly accessible and links to access them are compiled in the Internet Resources section.
Before searching for a suitable ligand, it is advisable to survey the landscape of existing degraders to identify any potential candidates which may fit the needs of your project. Many of these degraders have been compiled, along with their chemical structures, primary targets, and TPD-relevant characteristics such as maximal degradation, in the databases PROTACpedia and PROTAC-DB (Weng et al., 2021; Zaidman et al., 2020).
A handful of databases have been developed to organize and make accessible published ligand-protein data. Each database aggregates its data from different experimental or computational sources, so searching multiple databases is highly recommended. DrugBank and ChEMBL contain the largest sets of drug related information and are suitable first resources (Gaulton et al., 2012; Wishart et al., 2006). BindingDB offers an alternative database to search for ligand-protein interaction data and compiles data from sources not covered by other public databases, such as patents (Liu et al., 2007).
An open-access proteomics resource incorporating quantitative global proteomics experiments of degrader responses for over 200 molecules is offered by the Fischer lab through an online portal. This database features multitargeted degraders to provide early leads for > 220 kinases, all HDACs, and several other protein families such as the C2H2 zinc finger-ome (Donovan et al., 2020; Sievers et al., 2018; Xiong et al., 2021).
As demonstrated by studies utilizing pan- or multitargeted degraders, not all proteins are equally accessible to degradation strategies. To better understand your target’s amenability to TPD, a machine learning model, Model-based Analysis of Protein Degradability (MAPD), was developed to predict the degradability of protein substrates based on protein-intrinsic features (W. Zhang et al., 2021). The results from this study are accessible through an online database of MAPD predictions. Proteins with higher MAPD scores are predicted to be more tractable towards TPD. However, it should be noted that a lower MAPD score does not necessarily indicate greater difficulty in targeting the substrate. Rather, these low-scoring substrates tend to possess less quantity of certain characteristics predicted to be important for TPD i.e. fewer solvent-accessible lysines.
PROTAC Design
PROTACs are composed of three design elements: an E3 ligase recruiting ligand, a target ligand which binds to the protein of interest, and a linker which connects the two (Fig. 2). Despite the large number of known E3 ligases in the human proteome, only a handful have been widely used for small-molecule mediated targeted protein degradation due to the paucity of potent, selective E3-ligase ligands: the Von Hippel–Lindau protein E3 ligase complex (CRL2VHL), the Cereblon E3 ligase complex (CRL4CRBN), the to a lesser extent the Mouse Double Minute 2 homologue (MDM2), and the Cellular Inhibitor of Apoptosis (cIAP) (Ottis & Crews, 2017). Efforts to expand the number of E3-ligase targeting binders are underway. For example, Ward et. al. utilized covalent ligand screening to discover a novel recruiter for RNF4, which was subsequently conjugated to JQ1, a potent inhibitor of the BET family bromodomain proteins, to create an active degrader of BRD4. Ohoka et. al. used 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester, an endogenous metabolite, as a recruiter of the E3 ligase AhR, which when conjugated to JQ1 yielded another active degrader of BRD4 and Li et. al. (2020) have used the DCAF15 binder E7820 to develop BRD4 PROTACs (Fig. 2). As exemplified above, BRD4 is often the first target evaluated for newly developed E3-ligase recruiters, due to its high degradability. However, a recent example developing PROTACs recruiting the E3-ligase KEAP1 has shown that this activity does not always translate to other recruited targets (Du et. al., Cell Chem. Biol., 2022). It has been shown that poor activity of a PROTAC recruiting one E3-ligase can potentially be recovered by switching ligases, therefore surveying multiple ligases in initial designs is advised. Later, following up on hits that recruit an E3-ligase that is abundant in the target tissue can lead to improved efficacy (Lai et al., 2016).
Fig. 2.
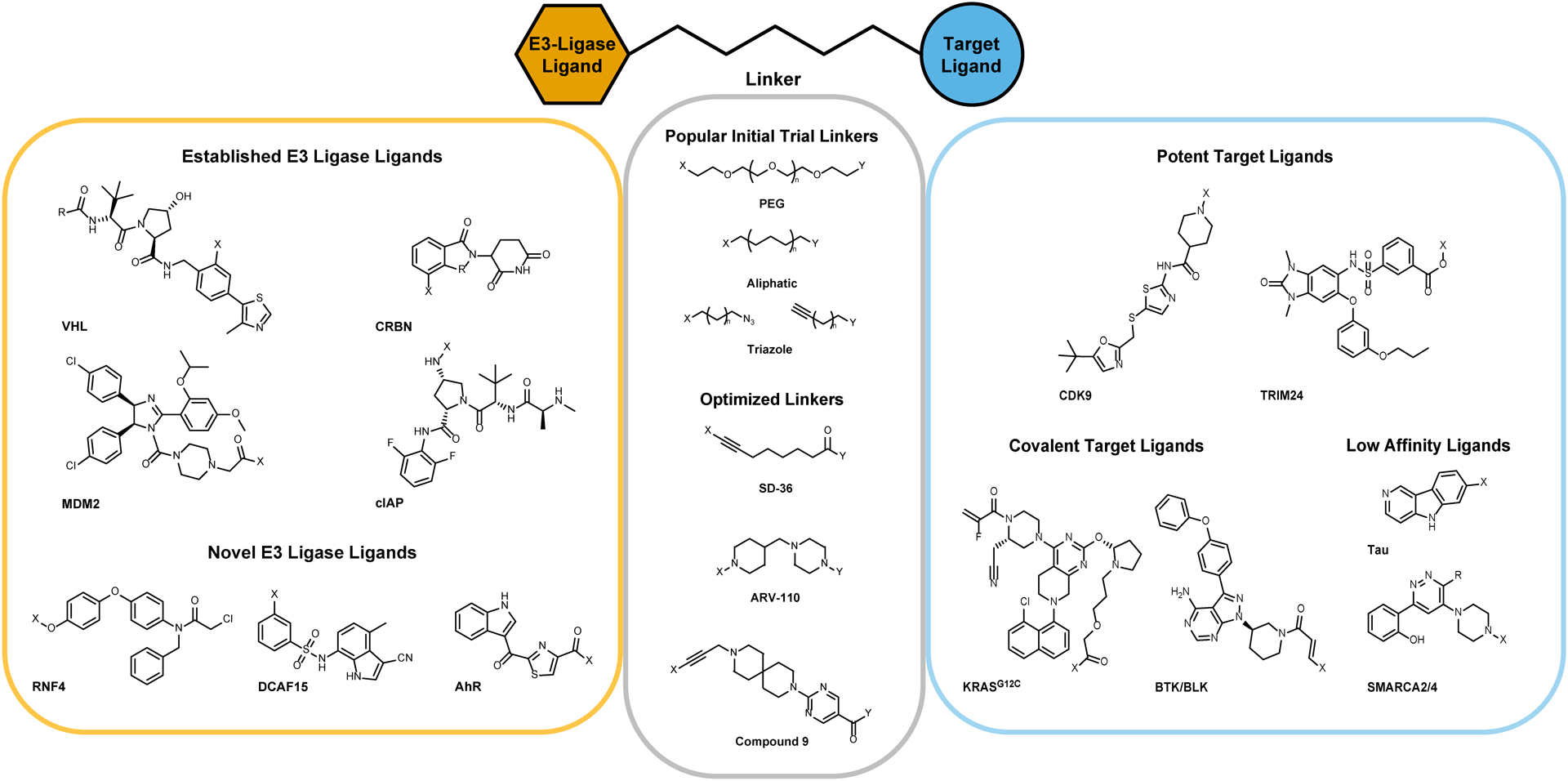
Collection of example chemical scaffolds used in proteolysis-targeting chimera (PROTAC) development. E3-ligase and target binding ligands are designated with their associated E3-ligase or target. Linkers are labeled with their common chemical name or the compound from which they are derived. These structures encompass a wide variety of moieties with different properties that have been used in PROTAC development and are not exhaustive of the ligands that have been incorporated into successful PROTAC molecules.
Selection of a target ligand is specific to one’s desired target, and is not limited to just potent, selective inhibitors. Whereas typical drug design necessitates potent binding to a target’s active site to elicit complete inhibition, PROTACs need only transient target recruitment and do not require access to an active site to trigger degradation (Békés, Langley, & Crews, 2022). QC-01–175 is one such PROTAC developed to effectively degrade a target without a defined catalytic site, in this case aberrant tau, the ligand of which was derived from a positron emission tomography (PET) tracer (Silva et al., 2019). Furthermore, the ligand to recruit the POI does not need to have active pharmacology alone but can be a phenotypically silent binder. For example, IACS-9571 has no anticancer effects despite being a potent binder of the TRIM24 bromodomain, a transcriptional regulator whose overexpression has been associated with oncogenesis in a variety of cancer lineages, including breast, prostate, and lung cancers (Palmer et al., 2015; Gechijian et al., 2018). However, when IACS-7e, derived from IACS-9571, was conjugated to a VHL ligand and used as a PROTAC warhead, it was able to degrade TRIM24 selectively and efficiently in addition to producing an anti-proliferative phenotype (Gechijian et al., 2018).
The linker length and composition can affect both cell permeability and ternary complex formation (E3-ligase-PROTAC-POI), therefore a panel of different linkers should be tried for each E3-ligase/binder pair (Bemis et al., 2021). Popular initial trial linkers include simple hydrocarbon or polyethylene glycol (PEG) chains flanked with a variety of functional groups (Fig. 2). Linker optimization can improve physicochemical properties such as total polar surface area, solubility, and flexibility, such as in the case of Nunes et. al, 2019, in which a long aliphatic hydrocarbon linker was replaced with a spirocyclic pyridine to conformationally restrain the linker, as well as improve solubility (Nunes et al., 2019). Other linker optimization efforts may be aimed more towards inducing a change in the ability to induce cooperativity in a PROTAC. Cooperativity refers to the impact binary complex formation (PROTAC-target or PROTAC-E3-ligase) has on ternary complex formation and can be categorized as positive (binary complex improves ternary complex formation), negative (binary complex hinders ternary complex formation), or non-cooperative (binary complex has no impact on ternary complex formation) (Gadd et al., 2017). This can be simplified as an equation of α = (KDBinary)⁄(KDTernary), whereby values greater than one indicate positive co-operativity, values of 1 indicate non-cooperative complexes, and values less than 1 indicate negative cooperativity (Roy et al., 2019). Testa et al., inspired by the crystal structure of their bromodomain and extra-terminal motif (BET) degrader MZ1 in a ternary complex with VHL and second bromodomain (BD2) of BET, Brd4, developed a macrocyclic PROTAC (macroPROTAC-1) which displayed a newly positive cooperativity with the BD2s of Brd4, Brd3, and Brd2, indicating an improved selectivity over the first bromodomains of the aforementioned proteins. Cooperativity has the additional benefit of dampening the hook effect, a phenomenon where at high concentrations binary interactions overtake ternary complex formation (Churcher, 2018).
Several computational approaches have been developed to leverage protein structural data for rational design of co-operative PROTACs. The Rosetta modeling suite has been used to dock the target and E3 ligase component proteins and subsequently design a linker between the two bound warheads (Bai et al., 2021; Nowak et al., 2018) or iterate between cycles of protein-protein docking and PROTAC conformational sampling (Zaidman et al., 2020). Rosetta is open-source software that can be obtained from the Rosetta Commons website. One limitation of this approach is that the accuracy of the ternary complex docking is reduced when ligand-bound X-ray structures are not available, which necessitates use of modeled input structures.
Despite the growing interest in positive cooperativity, there are also many examples of effective non-cooperative and negatively cooperative PROTACs, such as dBET23, dBET6, and dBET57, all of which have an α of less than 0.4. (Zorba et. al, 2018; Hu & Crews, 2021; Nowak et al., 2018). Regardless, linkers can still confer positive effects to degradation in absence of cooperativity by offering flexibility and thus relief of steric clash such as in the case of a library of Bruton’s tyrosine kinase (BTK) degraders with PEG linkers of varying length were assessed in cellulo and assigned steric scores which revealed that shorter PEG linkers (<11 linker atoms) had greater steric interactions and decreased efficacy than that of longer linkers (Zorba et al., 2018). Another strategy is inducing rigidity to reduce the number of potential non-productive conformations: such as that of IRAK4-PROTAC, the development of which pointed towards the necessity of an inflexible linker which yielded an almost 2-fold increase in potency (Nunes et al., 2019).
Assay Workflow
Following the design of initial candidate(s), measuring target degradation is necessary to identify active degraders. Upon identifying a suitably potent degrader, the next step is to verify the PROTAC mechanism of action (see Lead Compound Validation). Should no active molecules be identified, follow up assays to evaluate cell permeability, target engagement, and ternary complex formation can inform further design (summarized in Table 1). Results from these assays can then be used to inform rational design/optimization to create an improved degrader.
Table 1.
Summary of commonly used techniques to evaluate cellular phenomena important for the optimization of proteolysis-targeting chimera (PROTAC) degraders
| Cellular Phenomenon | Example Assays |
|---|---|
| Degradation |
|
| Cell Permeability |
|
| Target Binding |
|
| Ternary Complex Formation |
|
Degradation
The most commonly used and accessible method of confirming degradation is quantification of cellular protein levels via western blotting, though other strategies such as tagging a target protein with a fluorescent or luminescent reporter (e.g. GFP, HiBiT, NLuc) and tracking loss of signal over time are also popular. Selection of timepoints to assay degradation is crucial. Evaluating degradation at time points longer than 8 h may enable detection of slow or partial degradation by initial unoptimized molecules. However, longer time points run a greater risk of identifying false positives, where the protein levels are reduced due to downstream effects of prolonged target inhibition such as transcriptional differences or changes in the cell cycle. Therefore, many studies perform initial compound testing at both a short (between 4–8 h) and a long (between 12–24 h) time point. Initial compound concentrations should also be carefully considered, as bifunctional molecules may lose activity due to the hook effect at higher concentrations. For this reason, it is recommended that compounds be initially tested across a range of concentrations. Once an optimal time point is elucidated, a dose response curve can be generated, from which a concentration of max degradation (Dmax) and half degradation (D50) can be determined. While there is no standard threshold for potency, ideal candidates for final degraders or further optimization should show degradation at concentrations where low target occupancy is expected, to verify an event driven as opposed to inhibitory pharmacology. As an example, SNS-032, a cyclin-dependent kinase (CDK) inhibitor was shown to have poor selectivity at doses necessary for occupancy driven pharmacology, but when conjugated to thalidomide and used as a PROTAC (Thal-SNS-032), the necessary dose to induce degradation was low enough to target and completely degrade only CDK9 (Olson et al., 2018).
E3-ligase and Target Engagement
PROTAC molecules are high molecular weight and may have low cell membrane permeability, which may present as poor degradation activity. Traditional cell permeability assays used for small molecule drugs, such as Parallel Artificial Membrane Permeability Assays (PAMPA) and Caco-2 cell platforms, can be used to assess PROTACs (Farnaby et al., 2019; Klein et al., 2020). Specialized assays for evaluating PROTAC cellular permeability include those based on competition of the developed degrader library and a well-characterized degrader (H. Huang et al., 2018; Zeng et al., 2020) or fluorescent tracer (Guo et al., 2020) for E3-ligase binding. However, the data measured by these displacement-based methods aggregate both permeability and target binding. If the E3-binder or linker attachment site is modified in the series, it is favorable to perform these assays in both live and permeabilized cells to decouple the two phenomena (Hendrick et al., 2022; Riching et al., 2018).
Cellular competition platforms can be used to measure binary protein-ligand interactions when performed in a permeabilized cell format. However, it is advantageous to perform cell-free biochemical assays to identify suitable target binders and linker attachment sites when sufficient protein is available as these assays provide direct measurements of binding, which may be affected by linker conjugation. For example, fluorescence polarization (FP) is widely used for the rapid screening of large compound libraries and provides a powerful tool for the measurement of binary binding affinities. The FP assay format can be used to quantify the binding affinities for both the monovalent warheads, by attachment of a fluorophore to the ligand (Zoppi et al., 2019), and the developed PROTACs, by competition of a fluorescent probe with the PROTAC (Nowak et al., 2018).
Ternary Complex Formation
Confirmation that target degradation is dependent on ternary complex formation is necessary for optimization efforts that seek to increase ternary complex formation. Assessment of ternary complex formation in cells is made possible by luminescence proximity technologies. Available technologies include NanoBiT, in which two different proteins (SmBit and LgBit) are recombinantly attached to the E3 ligase and target protein, degrader-induced proximity reconstitutes a NanoLuc luciferase generating a quantifiable signal (A. S. Dixon et al., 2016). It is also possible to monitor formation of the ternary complex via co-immunoprecipitations, BRET assays, or affinity pulldowns and subsequent western blot analysis to determine bound species.
Monitoring of complex formation in live cells necessitates the use of an inhibitor of the ubiquitin proteasome system (UPS) to prevent target degradation. Proteasome inhibitors include bortezomib, carfilzomib and MG132, or the NAE inhibitor MLN4924 (X. Zhang et al., 2020).
Lead Compound Validation
Once a potent candidate PROTAC has been identified, its mechanism of action and selectivity must first be validated prior to its use in biological studies (Fig. 1). The first step in validating the mechanism of action for PROTAC molecules is confirming that target depletion is the result of protein degradation rather than reduced transcription. This is accomplished by assessing mRNA levels of the target substrate in compound and vehicle treated cells, with an expected result of no significant change in mRNA expression upon compound treatment (Bondeson et al., 2018; Smith et al., 2019). Competition of the degrader with the parental E3 ligand or target-binding warhead establishes dependence of degradation on the formation of E3-ligase:degrader:target ternary complex (Li et al., 2020; Silva et al., 2019). To further validate E3-dependent target degradation, cellular expression of the E3-ligase component can be ablated by genetic knockout. Additionally, negative control compounds incapable of binding to either the target or the E3-ligase should be synthesized and tested to validate target degradation via E3-ligase recruitment. This is commonly achieved by modifying the E3-ligase or target binding warhead with altered stereochemistry or steric bulk (Hsu et al., 2020; Li et al., 2020; Potjewyd et al., 2020). Finally, inhibition of the neddylation pathway by MLN4924 pre-treatment and the proteasome using Carfilzomib or MG-132 links degradation to Cullin-RING-Ligase (CRL) and proteasome activity.
Global proteomics is crucial to elucidating PROTAC specificity by identifying the downregulated substrates across the cellular proteome. Shorter treatment times (< 6 hours) are used to identify direct targets of PROTAC-induced degradation. Appropriate treatment conditions can be determined empirically by measuring degradation of the target substrate at varying incubation times and compound concentrations. Typically, the shortest treatment condition which exhibits robust degradation of the target substrate and minimal downstream changes is chosen for proteomics selectivity experiments.
Future Considerations
Following development of a potent, selective PROTAC, the degrader is often used as a chemical probe for investigating disease biology or target validation (Fig. 1). Several controls should be included in phenotypic characterization of cells, tissues, or whole organisms to account for potential off-target effects. Additional optimization of pharmacokinetic and pharmacodynamic properties may be needed to enable in vivo experiments. Because of the heterobifunctional nature of PROTACs, it is necessary to deconvolute the cellular effects caused by inhibition of the target substrate from the effects induced by target degradation. This is commonly achieved by modifying the E3-ligase or target binding warhead, yielding a negative control compound incapable of binding to one protein component (Hsu et al., 2020; Li et al., 2020; Potjewyd et al., 2020). Additionally, performing phenotypic screens in both wild-type and E3-ligase knockout cell systems will confirm the dependence of observed phenotypic changes on degradation. Finally, different degradation profiles (potency and selectivity) have been observed for the same molecules in different cell types, and this is incompletely explained by E3-ligase expression levels (Ottis et al., 2019; Yang et al., 2020). Therefore, evaluating degrader activity in the cellular context that phenotypic assays will be performed in is advised if different to the workhorse cell lines of the compound optimization.
The databases and procedures covered here are not exhaustive, and we encourage groups to use this paper as a starting reference with which to explore and assess different methodologies that best fit their projects. More broadly, we hope that this paper will help make the rapidly growing field of TPD more accessible to those interested in leveraging this strategy for the first time.
ACKNOWLEDGEMENTS:
G.L. was supported by an ACS Bridges Fellowship. N.L.T. was supported by an NIH training grant (T32CA009523).
F.M.F. was supported by an Alzheimer’s Association Grant (AARG-22-928354), the Larry L. Hillblom Foundation (2022-A-008-SUP) and the W.M. Keck foundation (8954).
CONFLICT OF INTEREST STATEMENT:
Fleur Ferguson is a scientific co-founder and equity holder in Proximity Therapeutics, a scientific advisory board member (SAB) and equity holder in Triana Biomedicines. Fleur Ferguson is or was recently a consultant or received speaking honoraria from Eli Lilly and Co., RA Capital, Tocris BioTechne and Plexium Inc. The Ferguson lab receives or has received research funding from Ono Pharmaceutical Co. Ltd.
All other authors declare no conflict of interest.
INTERNET RESOURCES:
-
PROTACpedia: https://protacpedia.weizmann.ac.il/ptcb/main
A manually curated database of over 1,000 PROTAC molecules from publications, patents, and other user submission. The database can be searched by inputting a variety of keywords, such as the target name, E3-ligase name, or ligand name. Information provided includes degradation-relevant characteristics, such as DC50, maximal degradation, and binding affinities.
-
PROTAC-DB: http://cadd.zju.edu.cn/protacdb/about
A manually curated database of PROTAC molecules from various sources. The database can be searched using a number of terms, including ligand name, linker name, and target name. In addition to providing degradation-relevant characteristics, this database integrates a computational platform to predict the structure of the ternary E3-ligase:PROTAC:target complex.
-
DrugBank: https://go.drugbank.com/
A database of over 500,000 drugs and their clinical properties. In addition to containing information about a plethora of drugs and drug targets, DrugBank has also developed an API which can help integrate the vast dataset and drug knowledge base into predictive drug discovery platforms.
-
ChEMBL: https://www.ebi.ac.uk/chembl/
A ligand-centric database of numerous compounds which focuses on cellular and in vivo properties. The database can be searched by name of the assay, cell line, compound, and target.
-
BindingDB: https://www.bindingdb.org/rwd/bind/index.jsp
A manually curated database compiling in vitro biophysical properties of many ligands. In addition to gathering data from typical sources, BindingDB provides the advantage of compiling ligand data from sources not covered by other databases, such as patents and less renown journals.
-
Fischer Lab Proteomics Database: https://proteomics.fischerlab.org/
A database actively managed and curated by the Fischer group at Dana-Farber Cancer Institute, which includes quantitative global proteomics data for over 200 degraders and their associated ligands.
-
MAPD: http://mapd.cistrome.org/
A database of MAPD predictions, indicating which protein targets are most inherently easy to degrade, as calculated from a trained machine learning model.
-
Rosetta Commons Website and Rosetta Software Download: https://www.rosettacommons.org/
The RosettaCommons website not only includes access to the source data necessary for compiling the Rosetta modeling software suite, but also provides a useful, active community platform for troubleshooting the various programs contained in the suite.
Contributor Information
Nathan L. Tran, Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, CA.
Georges A. Leconte, Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, CA.
Fleur M. Ferguson, Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, CA; Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, La Jolla, CA.
LITERATURE CITED:
- Bai N, Miller SA, Andrianov G. v, Yates M, Kirubakaran P, & Karanicolas J (2021). Rationalizing PROTAC-Mediated Ternary Complex Formation Using Rosetta. Journal of Chemical Information and Modeling, 61(3), 1368–1382. 10.1021/acs.jcim.0c01451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Békés M, Langley DR, & Crews CM (2022). PROTAC targeted protein degraders: the past is prologue. Nature Reviews Drug Discovery. 10.1038/s41573-021-00371-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis TA, la Clair JJ, & Burkart MD (2021). Unraveling the Role of Linker Design in Proteolysis Targeting Chimeras. Journal of Medicinal Chemistry, 64(12), 8042–8052. 10.1021/acs.jmedchem.1c00482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, & Bourne PE (2000). The Protein Data Bank. Nucleic Acids Research, 28(1), 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond MJ, Chu L, Nalawansha DA, Li K, & Crews CM (2020). Targeted Degradation of Oncogenic KRASG12C by VHL-Recruiting PROTACs. ACS Central Science, 6(8), 1367–1375. 10.1021/acscentsci.0c00411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeson DP, Smith BE, Burslem GM, Buhimschi AD, Hines J, Jaime-Figueroa S, Wang J, Hamman BD, Ishchenko A, & Crews CM (2018). Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell Chemical Biology, 25(1), 78–87.e5. 10.1016/j.chembiol.2017.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K-H, Zengerle M, Testa A, & Ciulli A (2018). Impact of Target Warhead and Linkage Vector on Inducing Protein Degradation: Comparison of Bromodomain and Extra-Terminal (BET) Degraders Derived from Triazolodiazepine (JQ1) and Tetrahydroquinoline (I-BET726) BET Inhibitor Scaffolds. Journal of Medicinal Chemistry, 61(2), 504–513. 10.1021/acs.jmedchem.6b01912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churcher I (2018). Protac-Induced Protein Degradation in Drug Discovery: Breaking the Rules or Just Making New Ones? Journal of Medicinal Chemistry, 61(2), 444–452. 10.1021/acs.jmedchem.7b01272 [DOI] [PubMed] [Google Scholar]
- Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, Butler BL, Binkowski BF, Machleidt T, Kirkland TA, Wood MG, Eggers CT, Encell LP, & Wood K. v. (2016). NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chemical Biology, 11(2), 400–408. 10.1021/acschembio.5b00753 [DOI] [PubMed] [Google Scholar]
- Donovan KA, Ferguson FM, Bushman JW, Eleuteri NA, Bhunia D, Ryu S, Tan L, Shi K, Yue H, Liu X, Dobrovolsky D, Jiang B, Wang J, Hao M, You I, Teng M, Liang Y, Hatcher J, Li Z, … Fischer ES (2020). Mapping the Degradable Kinome Provides a Resource for Expedited Degrader Development. Cell, 183(6), 1714–1731.e10. 10.1016/j.cell.2020.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnaby W, Koegl M, Roy MJ, Whitworth C, Diers E, Trainor N, Zollman D, Steurer S, Karolyi-Oezguer J, Riedmueller C, Gmaschitz T, Wachter J, Dank C, Galant M, Sharps B, Rumpel K, Traxler E, Gerstberger T, Schnitzer R, … Ciulli A (2019). BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nature Chemical Biology, 15(7), 672–680. 10.1038/s41589-019-0294-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, & Overington JP (2012). ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Research, 40(D1), D1100–D1107. 10.1093/nar/gkr777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechijian LN, Buckley DL, Lawlor MA, Reyes JM, Paulk J, Ott CJ, Winter GE, Erb MA, Scott TG, Xu M, Seo H-S, Dhe-Paganon S, Kwiatkowski NP, Perry JA, Qi J, Gray NS, & Bradner JE (2018). Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands. Nature Chemical Biology, 14(4), 405–412. 10.1038/s41589-018-0010-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W-H, Qi X, Yu X, Liu Y, Chung C-I, Bai F, Lin X, Lu D, Wang L, Chen J, Su LH, Nomie KJ, Li F, Wang MC, Shu X, Onuchic JN, Woyach JA, Wang ML, & Wang J (2020). Enhancing intracellular accumulation and target engagement of PROTACs with reversible covalent chemistry. Nature Communications, 11(1), 4268. 10.1038/s41467-020-17997-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick CE, Jorgensen JR, Chaudhry C, Strambeanu II, Brazeau J-F, Schiffer J, Shi Z, Venable JD, & Wolkenberg SE (2022). Direct-to-Biology Accelerates PROTAC Synthesis and the Evaluation of Linker Effects on Permeability and Degradation. ACS Medicinal Chemistry Letters, 13(7), 1182–1190. 10.1021/acsmedchemlett.2c00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JH-R, Rasmusson T, Robinson J, Pachl F, Read J, Kawatkar S, O’ Donovan DH, Bagal S, Code E, Rawlins P, Argyrou A, Tomlinson R, Gao N, Zhu X, Chiarparin E, Jacques K, Shen M, Woods H, Bednarski E, … Bloecher A (2020). EED-Targeted PROTACs Degrade EED, EZH2, and SUZ12 in the PRC2 Complex. Cell Chemical Biology, 27(1), 41–46.e17. https://doi.org/ 10.1016/j.chembiol.2019.11.004 [DOI] [PubMed] [Google Scholar]
- Huang H-T, Dobrovolsky D, Paulk J, Yang G, Weisberg EL, Doctor ZM, Buckley DL, Cho J-H, Ko E, Jang J, Shi K, Choi HG, Griffin JD, Li Y, Treon SP, Fischer ES, Bradner JE, Tan L, & Gray NS (2018). A Chemoproteomic Approach to Query the Degradable Kinome Using a Multi-kinase Degrader. Cell Chemical Biology, 25(1), 88–99.e6. 10.1016/j.chembiol.2017.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, & Ciulli A (2021). E3-ligase Ligands for PROTACs: How They Were Found and How to Discover New Ones. SLAS Discovery, 26(4), 484–502. 10.1177/2472555220965528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein VG, Townsend CE, Testa A, Zengerle M, Maniaci C, Hughes SJ, Chan K-H, Ciulli A, & Lokey RS (2020). Understanding and Improving the Membrane Permeability of VH032-Based PROTACs. ACS Medicinal Chemistry Letters, 11(9), 1732–1738. 10.1021/acsmedchemlett.0c00265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AC, Toure M, Hellerschmied D, Salami J, Jaime-Figueroa S, Ko E, Hines J, & Crews CM (2016). Modular PROTAC Design for the Degradation of Oncogenic BCR-ABL. Angewandte Chemie International Edition, 55(2), 807–810. https://doi.org/ 10.1002/anie.201507634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pinch BJ, Olson CM, Donovan KA, Nowak RP, Mills CE, Scott DA, Doctor ZM, Eleuteri NA, Chung M, Sorger PK, Fischer ES, & Gray NS (2020). Development and Characterization of a Wee1 Kinase Degrader. Cell Chemical Biology, 27(1), 57–65.e9. https://doi.org/ 10.1016/j.chembiol.2019.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Lin Y, Wen X, Jorissen RN, & Gilson MK (2007). BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Research, 35(suppl_1), D198–D201. 10.1093/nar/gkl999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machleidt T, Woodroofe CC, Schwinn MK, Méndez J, Robers MB, Zimmerman K, Otto P, Daniels DL, Kirkland TA, & Wood K. v. (2015). NanoBRET—A Novel BRET Platform for the Analysis of Protein–Protein Interactions. ACS Chemical Biology, 10(8), 1797–1804. 10.1021/acschembio.5b00143 [DOI] [PubMed] [Google Scholar]
- Nowak RP, DeAngelo SL, Buckley D, He Z, Donovan KA, An J, Safaee N, Jedrychowski MP, Ponthier CM, Ishoey M, Zhang T, Mancias JD, Gray NS, Bradner JE, & Fischer ES (2018). Plasticity in binding confers selectivity in ligand-induced protein degradation. Nature Chemical Biology, 14(7), 706–714. 10.1038/s41589-018-0055-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes J, McGonagle GA, Eden J, Kiritharan G, Touzet M, Lewell X, Emery J, Eidam H, Harling JD, & Anderson NA (2019). Targeting IRAK4 for Degradation with PROTACs. ACS Medicinal Chemistry Letters, 10(7), 1081–1085. 10.1021/acsmedchemlett.9b00219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohoka N, Tsuji G, Shoda T, Fujisato T, Kurihara M, Demizu Y, & Naito M (2019). Development of Small Molecule Chimeras That Recruit AhR E3 Ligase to Target Proteins. ACS Chemical Biology, 14(12), 2822–2832. 10.1021/acschembio.9b00704 [DOI] [PubMed] [Google Scholar]
- Ottis P, & Crews CM (2017). Proteolysis-Targeting Chimeras: Induced Protein Degradation as a Therapeutic Strategy. ACS Chemical Biology, 12(4), 892–898. 10.1021/acschembio.6b01068 [DOI] [PubMed] [Google Scholar]
- Ottis P, Palladino C, Thienger P, Britschgi A, Heichinger C, Berrera M, Julien-Laferriere A, Roudnicky F, Kam-Thong T, Bischoff JR, Martoglio B, & Pettazzoni P (2019). Cellular Resistance Mechanisms to Targeted Protein Degradation Converge Toward Impairment of the Engaged Ubiquitin Transfer Pathway. ACS Chemical Biology, 14(10), 2215–2223. 10.1021/acschembio.9b00525 [DOI] [PubMed] [Google Scholar]
- Palmer WS, Poncet-Montange G, Liu G, Petrocchi A, Reyna N, Subramanian G, Theroff J, Yau A, Kost-Alimova M, Bardenhagen JP, Leo E, Shepard HE, Tieu TN, Shi X, Zhan Y, Zhao S, Barton MC, Draetta G, Toniatti C, … Andersen JN (2016). Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor. Journal of Medicinal Chemistry, 59(4), 1440–1454. 10.1021/acs.jmedchem.5b00405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Cho J, & Song EJ (2020). Ubiquitin–proteasome system (UPS) as a target for anticancer treatment. Archives of Pharmacal Research, 43(11), 1144–1161. 10.1007/s12272-020-01281-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potjewyd F, Turner A-MW, Beri J, Rectenwald JM, Norris-Drouin JL, Cholensky SH, Margolis DM, Pearce KH, Herring LE, & James LI (2020). Degradation of Polycomb Repressive Complex 2 with an EED-Targeted Bivalent Chemical Degrader. Cell Chemical Biology, 27(1), 47–56.e15. https://doi.org/ 10.1016/j.chembiol.2019.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Hu Y, Zhou B, Fernandez-Salas E, Yang C-Y, Liu L, McEachern D, Przybranowski S, Wang M, Stuckey J, Meagher J, Bai L, Chen Z, Lin M, Yang J, Ziazadeh DN, Xu F, Hu J, Xiang W, … Wang S (2018). Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression. Journal of Medicinal Chemistry, 61(15), 6685–6704. 10.1021/acs.jmedchem.8b00506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riching KM, Mahan S, Corona CR, McDougall M, Vasta JD, Robers MB, Urh M, & Daniels DL (2018). Quantitative Live-Cell Kinetic Degradation and Mechanistic Profiling of PROTAC Mode of Action. ACS Chemical Biology, 13(9), 2758–2770. 10.1021/acschembio.8b00692 [DOI] [PubMed] [Google Scholar]
- Roy MJ, Winkler S, Hughes SJ, Whitworth C, Galant M, Farnaby W, Rumpel K, & Ciulli A (2019). SPR-Measured Dissociation Kinetics of PROTAC Ternary Complexes Influence Target Degradation Rate. ACS Chemical Biology, 14(3), 361–368. 10.1021/acschembio.9b00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers QL, Petzold G, Bunker RD, Renneville A, Słabicki M, Liddicoat BJ, Abdulrahman W, Mikkelsen T, Ebert BL, & Thomä NH (2018). Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science (New York, N.Y.), 362(6414), eaat0572. 10.1126/science.aat0572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Ferguson FM, Cai Q, Donovan KA, Nandi G, Patnaik D, Zhang T, Huang H-T, Lucente DE, Dickerson BC, Mitchison TJ, Fischer ES, Gray NS, & Haggarty SJ (2019). Targeted degradation of aberrant tau in frontotemporal dementia patient-derived neuronal cell models. ELife, 8, e45457. 10.7554/eLife.45457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BE, Wang SL, Jaime-Figueroa S, Harbin A, Wang J, Hamman BD, & Crews CM (2019). Differential PROTAC substrate specificity dictated by orientation of recruited E3-ligase. Nature Communications, 10(1), 131. 10.1038/s41467-018-08027-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa A, Hughes SJ, Lucas X, Wright JE, and Ciulli A (2020). Structure-Based Design of a Macrocyclic PROTAC. Angewandte Chemie 132, no. 4: 1744–51. 10.1002/ange.201914396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng G, Shen C, Cao D, Gao J, Dong X, He Q, Yang B, Li D, Wu J, & Hou T (2021). PROTAC-DB: an online database of PROTACs. Nucleic Acids Research, 49(D1), D1381–D1387. 10.1093/nar/gkaa807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, & Woolsey J (2006). DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Research, 34(suppl_1), D668–D672. 10.1093/nar/gkj067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Donovan KA, Eleuteri NA, Kirmani N, Yue H, Razov A, Krupnick NM, Nowak RP, & Fischer ES (2021). Chemo-proteomics exploration of HDAC degradability by small molecule degraders. Cell Chemical Biology, 28(10), 1514–1527.e4. 10.1016/j.chembiol.2021.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gao H, Sun X, Sun Y, Qiu Y, Weng Q, & Rao Y (2020). Global PROTAC Toolbox for Degrading BCR–ABL Overcomes Drug-Resistant Mutants and Adverse Effects. Journal of Medicinal Chemistry, 63(15), 8567–8583. 10.1021/acs.jmedchem.0c00967 [DOI] [PubMed] [Google Scholar]
- Zagidullin A, Milyukov V, Rizvanov A, & Bulatov E (2020). Novel approaches for the rational design of PROTAC linkers. Exploration of Targeted Anti-Tumor Therapy, 1(5), 381–390. 10.37349/etat.2020.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidman D, Prilusky J, & London N (2020). PRosettaC: Rosetta Based Modeling of PROTAC Mediated Ternary Complexes. Journal of Chemical Information and Modeling, 60(10), 4894–4903. 10.1021/acs.jcim.0c00589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M, Xiong Y, Safaee N, Nowak RP, Donovan KA, Yuan CJ, Nabet B, Gero TW, Feru F, Li L, Gondi S, Ombelets LJ, Quan C, Jänne PA, Kostic M, Scott DA, Westover KD, Fischer ES, & Gray NS (2020). Exploring Targeted Degradation Strategy for Oncogenic KRASG12C. Cell Chemical Biology, 27(1), 19–31.e6. 10.1016/j.chembiol.2019.12.006 [DOI] [PubMed] [Google Scholar]
- Zhang W, Roy Burman SS, Chen J, Donovan KA, Cao Y, Zhang B, Zeng Z, Zhang Y, Li D, Fischer ES, Tokheim C, & Liu XS (2021). Machine learning modeling of protein-intrinsic features predicts tractability of targeted protein degradation. BioRxiv, 2021.09.27.462040. 10.1101/2021.09.27.462040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Linder S, & Bazzaro M (2020). Drug development targeting the ubiquitin-proteasome system (UPS) for the treatment of human cancers. In Cancers (Vol. 12, Issue 4). MDPI AG. 10.3390/cancers12040902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Bai L, Xu R, Zhao Y, Chen J, McEachern D, Chinnaswamy K, Wen B, Dai L, Kumar P, Yang C-Y, Liu Z, Wang M, Liu L, Meagher JL, Yi H, Sun D, Stuckey JA, & Wang S (2019). Structure-Based Discovery of SD-36 as a Potent, Selective, and Efficacious PROTAC Degrader of STAT3 Protein. Journal of Medicinal Chemistry, 62(24), 11280–11300. 10.1021/acs.jmedchem.9b01530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoppi V, Hughes SJ, Maniaci C, Testa A, Gmaschitz T, Wieshofer C, Koegl M, Riching KM, Daniels DL, Spallarossa A, & Ciulli A (2019). Iterative Design and Optimization of Initially Inactive Proteolysis Targeting Chimeras (PROTACs) Identify VZ185 as a Potent, Fast, and Selective von Hippel–Lindau (VHL) Based Dual Degrader Probe of BRD9 and BRD7. Journal of Medicinal Chemistry, 62(2), 699–726. 10.1021/acs.jmedchem.8b01413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorba A, Nguyen C, Xu Y, Starr J, Borzilleri K, Smith J, Zhu H, Farley KA, Ding W, Schiemer J, Feng X, Chang JS, Uccello DP, Young JA, Garcia-Irrizary CN, Czabaniuk L, Schuff B, Oliver R, Montgomery J, … Calabrese MF (2018). Delineating the role of cooperativity in the design of potent PROTACs for BTK. Proceedings of the National Academy of Sciences, 115(31), E7285–E7292. 10.1073/pnas.1803662115 [DOI] [PMC free article] [PubMed] [Google Scholar]


