Abstract
The oral streptococcal group (mitis phylogenetic group) currently consists of nine recognized species, although the group has been traditionally difficult to classify, with frequent changes in nomenclature over the years. The pneumococcus (Streptococcus pneumoniae), an important human pathogen, is traditionally distinguished from the most closely related oral streptococcal species Streptococcus mitis and Streptococcus oralis on the basis of three differentiating characteristics: optochin susceptibility, bile solubility, and agglutination with antipneumococcal polysaccharide capsule antibodies. However, there are many reports in the literature of pneumococci lacking one or more of these defining characteristics. Sometimes called “atypical” pneumococci, these isolates can be the source of considerable confusion in the clinical laboratory. Little is known to date about the genetic relationships of such organisms with classical S. pneumoniae isolates. Here we describe these relationships based on sequence analysis of housekeeping genes in comparison with previously characterized isolates of S. pneumoniae, S. mitis, and S. oralis. While most pneumococci were found to represent a closely related group these studies identified a subgroup of atypical pneumococcal isolates (bile insoluble and/or “acapsular”) distinct from, though most closely related to, the “typical” pneumococcal isolates. However, a large proportion of isolates, found to be atypical on the basis of capsule reaction alone, did group with typical pneumococci, suggesting that they have either lost capsule production or represent as-yet-unrecognized capsular types. In contrast to typical S. pneumoniae, isolates phenotypically identified as S. mitis and S. oralis, which included isolates previously characterized in taxonomic studies, were genetically diverse. While most of the S. oralis isolates did fall into a well-separated group, S. mitis isolates did not cluster into a well-separated group. During the course of these studies we also identified a number of potentially important pathogenic isolates, which were frequently associated with respiratory disease, that phenotypically and genetically are most closely related to S. mitis but which harbor genes encoding the virulence determinants pneumolysin and autolysin classically associated with S. pneumoniae.
Streptococcus pneumoniae is a common and important human pathogen associated with pneumonia, septicaemia, meningitis, and otitis media. A number of distinct species of naturally transformable viridans or oral streptococci, which are closely related to S. pneumoniae, have now been identified, although the taxonomy and classification of these organisms has long been considered difficult (47). The most closely related species on the basis of 16S rRNA sequence are Streptococcus oralis and Streptococcus mitis, which share over 99% sequence identity with S. pneumoniae, although DNA-DNA similarity values for the entire chromosome are estimated to be less than 60% (28). S. oralis and S. mitis are usually considered to be commensals of the human oral cavity, but in recent years it has become clear that members of these species can be important pathogens. Oral streptococci, including S. mitis and S. oralis, are associated with bacterial endocarditis, especially in patients with prosthetic valves (16). In addition, S. mitis and S. oralis are now recognized as frequent causes of infection in immunocompromised patients, particularly immediately after tissue transplants, and in neutropenic cancer patients (5, 8, 9, 33).
Four phenotypic characteristics are classically used in the diagnostic laboratory for the presumptive identification of S. pneumoniae: colony morphology, optochin sensitivity, bile solubility, and agglutination with antipneumococcal polysaccharide capsule antibodies. Although their colony morphology can be very similar, nonpneumococcal oral streptococci are classically optochin resistant and bile insoluble and do not react with antipolysaccharide antibodies. It is considered important for the laboratory to differentiate between S. pneumoniae and other alpha-hemolytic oral streptococci, notably S. mitis and S. oralis, since misidentification may influence diagnosis and treatment. While these conventional methods allow identification of the majority of pneumococcal isolates, presumptive S. pneumoniae isolates may produce atypical reactions in one or more of the standard tests, and other alpha-hemolytic streptococci may give positive reactions in these tests, leading to difficulties in identification. For example, there are many observations of optochin-resistant pneumococci (1, 30, 37, 38), bile-insoluble S. pneumoniae isolates have been reported (15, 21, 36), and nontypeable, unencapsulated pneumococci have historically been reported to comprise 2% of pneumococci isolated from normally sterile sites (10) and up to 20% of conjunctival isolates (22).
Despite these problems, most diagnostic laboratories still use these conventional identification techniques. A commercially available DNA probe test (AccuProbe Culture Identification Kit for S. pneumoniae) has been reported to show specificity and sensitivity of 100% for pneumococci (13, 14), but it is prohibitively expensive for routine diagnostic use (36). The use of PCR in diagnostic tests, targeting genes considered to be specific targets for the pneumococcus, has been investigated. Two attractive targets for such studies have been genes encoding the putative virulence factors pneumolysin (ply) and the major autolysin (lytA) (25, 40, 41, 43). Such PCR-based diagnostics, while offering some promise, are not, as yet, used frequently in the routine microbiology laboratory setting.
Here we describe the preliminary characterization of a group of “atypical” presumptive isolates of S. pneumoniae which show aberrant reactions to bile and/or do not react with antipneumococcal polysaccharide antibodies. We have used sequencing of housekeeping genes to investigate the genetic background of these organisms in comparison to a selection of “typical” pneumococci and both previously characterized and clinical isolates of S. oralis and S. mitis. In addition, we describe a second group of atypical isolates identified during the course of this study. These organisms appear to be genetically and phenotypically related to S. mitis but harbor genes encoding the virulence factors autolysin and pneumolysin, which are normally associated with pneumococci. Interestingly, many of these isolates were disease associated. The relationships of these organisms to other well-characterized oral streptococci and a preliminary characterization of the genes encoding autolysin and pneumolysin in these organisms are described here.
MATERIALS AND METHODS
Strains.
A complete list of strains used in this study of genetic diversity is provided in Table 1. Isolates were routinely cultured on brain heart infusion (BHI) agar supplemented with 5% (vol/vol) sheep's blood at 37°C and 5% CO2. The origins of typical pneumococcal strains used in the determination of lytA diversity have been described elsewhere (44).
TABLE 1.
Characteristics of strains examined in this study
| Strain no. | Other ID | Identificationa | Originb | Site of isolationc | Latex agglutination/serotyped | Optochin | Bile solubility | Gen- Probee | Presence off:
|
Source or referenceg | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| lytA | lytA101 | ply | ||||||||||
| Group 1 (typical S. pneumoniae) | ||||||||||||
| PC15 | 1011 | S. pneumoniae | UK | Throat | 23F | S | + | + | + | − | + | 35 |
| 11B | 912 | S. pneumoniae | UK | Throat | 15B | S | + | + | + | − | + | 35 |
| PC19 | 1012 | S. pneumoniae | UK | Throat | 35A | S | + | + | + | − | + | 35 |
| CL1 (E226) | 355 | S. pneumoniae | Uruguay | Blood | 14 | S | + | ND | + | − | + | 35 |
| CL26 (Sp8) | 30 | S. pneumoniae | Spain | Blood | 13 | S | + | ND | + | − | + | 35 |
| R6 | S. pneumoniae | US | r | S | + | + | + | − | + | 2 | ||
| 44A | 951 | S. pneumoniae | UK | Throat | 6A | S | + | + | + | − | + | 35 |
| CL10 (KD12) | 312 | S. pneumoniae | Kenya | Throat | 7F | S | + | ND | + | − | + | 35 |
| 86013 | 873 | S. pneumoniae | Kenya | Throat | 8 | S | + | + | + | − | + | 35 |
| 1A | 900 | S. pneumoniae | UK | Throat | 1 | S | + | + | + | − | + | 35 |
| CL13 (Sp9) | 555 | S. pneumoniae | Spain | Blood | 8 | S | + | ND | + | − | + | 35 |
| Group 2a (S. oralis and S. mitis characterized in previous taxonomic studies)h | ||||||||||||
| NCTC 11427 | 542 | S. oralisT | − | R | − | ND | − | − | − | 4, 29 | ||
| NCTC 7864 | 627/571 | S. oralis | − | R | − | − | − | − | − | 4, 29 | ||
| OPA1 | 626 | S. oralis | − | R | − | ND | − | − | − | 4 | ||
| PP53 | 619 | S. oralis | − | R | − | ND | − | − | − | 4 | ||
| HV51 | 621 | S. mitis | − | R | − | ND | − | − | − | 4 | ||
| NCTC 10712 | 567 | S. mitis | − | R | − | − | − | − | − | 4, 29 | ||
| NS51T | 620 | S. mitisT | − | R | − | ND | − | − | − | 4 | ||
| K208 | 622 | S. mitis | − | R | − | ND | − | − | − | 4 | ||
| OS51 | 624 | S. mitis | − | R | − | ND | − | − | − | 4 | ||
| Group 2b (commensal and/ or clinical isolates of S. oralis and S. mitis) | ||||||||||||
| AR37 | 1078 | S. oralis | UK | Blood, endocarditis | − | R | − | ND | − | − | − | D. Beighton |
| COL21 | PN93/447 | Streptococcus sp. | UK | Eye, eye disease | − | R | − | − | − | − | − | This study |
| COL25 | PN93/1003 | S. oralis | UK | Blood, endocarditis | − | R | − | − | − | − | − | This study |
| AR5 | 1072 | S. oralis | UK | Blood, endocarditis | − | R | − | ND | − | − | − | D. Beighton |
| AR13 | 1076 | S. oralis | UK | Endocarditis | − | R | − | ND | − | − | − | D. Beighton |
| AC1372 | 1039 | S. oralis | UK | Blood, endocarditis | − | R | − | ND | − | − | − | D. Beighton |
| C17 | 1073 | S. oralis | UK | Neutropenia | − | R | − | ND | − | − | − | D. Beighton |
| COL19 | PN93/800 | S. oralis | UK | Blood | − | R | − | − | − | − | − | This study |
| COL22 | PN93/1264 | Streptococcus sp. | UK | NK | − | R | − | − | − | − | − | This study |
| M1 | T6(2) 10 | S. mitis | UK | Normal flora | − | R | − | ND | − | − | − | D. Beighton |
| M3 | 1304/16 | S. mitis | UK | Normal flora | − | R | − | ND | − | − | − | D. Beighton |
| M4 | T6(2) 11 | S. mitis | UK | Normal flora | − | R | − | ND | − | − | − | D. Beighton |
| M2 | T6(3) 12 | S. mitis | UK | Normal flora | − | R | − | ND | − | − | − | D. Beighton |
| CL22 | 103 | S. mitis | Spain | Lower respiratory tract | − | R | − | ND | − | − | − | This study |
| AC1374 | 1040 | S. mitis | UK | Blood culture | − | R | − | ND | − | − | − | D. Beighton |
| AC1362 | 1042 | S. oralis | UK | Blood culture | − | R | − | ND | − | − | − | D. Beighton |
| Group 2c (atypical oral streptococci) | ||||||||||||
| COL18 | PN93/454 | Streptococcus sp. | UK | Sputum, chest infection | − | S | − | − | + | − | + | This study |
| COL15 | PN92/1139 | S. mitis | UK | Sputum, chest infection | − | S | − | − | + | − | + | This study |
| COL16 | PN93/952 | Streptococcus sp. | UK | Bronchial lavage, pneumonia, HIV+ | − | S | − | − | + | − | + | This study |
| 806 | S. mitis | UK | NK | − | R | − | − | + | − | + | S. Gillespie | |
| COL28 | PN93/918 | Streptococcus sp. | UK | Sputum, respiratory infection | − | R | − | − | + | − | + | This study |
| COL20 | PN93/776 | Streptococcus sp. | UK | NK | − | S/R | − | − | + | − | + | This study |
| COL24 | PN93/656 | Streptococcus sp. | UK | Sputum | − | S/R | − | − | + | − | + | This study |
| COL17 | PN91/2745 | Streptococcus sp. | UK | NK | − | R | − | − | + | + | + | This study |
| 764 | S. mitis | UK | NK | − | R | − | − | − | − | + | S. Gillespie | |
| Group 3 (putative atypical pneumococci) | ||||||||||||
| 1916 | PN97/3197 | S. pneumoniae | UK | r | S | + | ND | + | − | + | This study | |
| COL1 | PN92/1207 | S. pneumoniae | UK | NK | r | S | + | + | + | − | + | This study |
| COL3 | PN92/944 | S. pneumoniae | UK | Eye, eye disease | r | S | + | + | + | − | + | This study |
| COL6 | PN93/779 | S. pneumoniae | UK | Blood | r | S | + | + | + | − | + | This study |
| COL5 | PN93/832 | S. pneumoniae | UK | Blood | r | S/R | + | + | + | − | + | This study |
| COL7 | PN93/356 | S. pneumoniae | UK | Nasal, respiratory infection | r | S/R | + | + | + | − | + | This study |
| COL8 | PN93/707 | S. pneumoniae | UK | Sputum | − | S/R | + | + | + | − | + | This study |
| COL11 | R93/688 | S. pneumoniae | UK | CSF, meningitis | − | S | + | + | + | − | + | This study |
| COL12 | PN93/950 | S. pneumoniae | UK | Blood | − | S | + | + | + | − | + | This study |
| COL14 | PN93/789 | S. pneumoniae | UK | Blood, pneumonia | − | S | + | + | + | − | + | This study |
| COL9 | PN93/904 | S. pneumoniae | UK | Blood | − | S/R | + | + | + | − | + | This study |
| X158 | Streptococcus sp. | r | S | − | + | + | − | + | This study | |||
| COL27 | PN93/135 | Streptococcus sp. | UK | Sputum, chest infect; HIV+ | − | S/R | − | + | + | − | + | This study |
| COL26 | PN93/403 | Streptococcus sp. | UK | Synovial fluid, septic arthritis | r | R | − | + | + | − | + | This study |
| 101/87 | S. pneumoniaeh | Spain | Blood, pneumonia | − | R | − | ND | − | + | + | 15 | |
| 86027 | 874 | Streptococcus sp. | Kenya | Throat | − | S | −/+ | + | + | − | + | 35 |
Unless stated otherwise, identities are based on the phenotypic tests described in this table in combination with API biotyping and additional biochemical tests when necessary. Where strain profiles were not consistent with any one species or where discordant identities were obtained on separate occasions, strains are not given a species designation. T, type strain.
UK, United Kingdom.
NK, not known; CSF, cerebrospinal fluid; HIV, human immunodeficiency virus.
r, strains which autoagglutinated when latex agglutination-capsular serotyping was performed (rough, nontypeable strains); −, no results against capsular typing sera (smooth, nontypeable strains).
ND, not done.
The presence of lytA, lytA101, and ply was determined by PCR and confirmatory blotting.
D. Beighton, Joint Microbiology Research Unit, Guy's, King's, and St. Thomas' Dental Institute, London, United Kingdom; S. Gillespie, Department of Medical Microbiology, Royal Free Hospital, London, United Kingdom.
Strain phenotypes.
Phenotypes were determined by the standard criteria used in the distinction of oral streptococci: optochin sensitivity, bile solubility, and latex agglutination-capsular serotyping.
(i) Optochin sensitivity.
For the determination of optochin sensitivity, each isolate was cultured in 5 ml of Todd-Hewitt broth. A 1:10 dilution was prepared from the broth culture and used to flood a 5% horse blood agar plate. A 5-μg optochin disk (Mast Diagnostics) was placed on the dried plate which was incubated at 37°C for 18 h. Zone size diameters were then measured (in millimeters) and, as stated by the manufacturer, isolates with zones of >15 mm were considered to be sensitive. Isolates showing zones of 10 to 14 mm were considered to be of reduced sensitivity (illustrated by S/R in Table 1).
(ii) Bile solubility.
Bile solubility was determined by growing organisms in two 5-ml volumes of Todd-Hewitt broth supplemented with 0.5 ml of 20% (wt/vol) glucose. After centrifugation at 3,000 rpm for 20 min the supernatants were discarded, both cell pellets were resuspended in 2.5 ml of phosphate-buffered saline (PBS), and 0.5 ml of 10% (wt/vol) sodium deoxycholate was added to one of the suspensions. These were incubated at 37°C for 15 to 30 min. Complete lysis within 30 min of incubation was indicative of a positive result.
(iii) Latex agglutination and capsular serotyping.
Latex agglutination was performed with organisms grown in 5 ml of Todd-Hewitt broth. The centrifuged deposit was tested with the Slidex Pneumo kit (Biomerieux), which comprises a latex suspension sensitized with antisera to 83 pneumococcal capsular serotypes. The test was performed according to the manufacturer's instructions. Capsular serotyping was performed on cultures grown in 4.5 ml of Todd-Hewitt broth, supplemented with 0.5 ml of 20% (wt/vol) glucose. The cultures were centrifuged at 3,000 rpm for 20 min, and the supernatants were discarded. The pellet (antigen suspension) was vortexed, and slide agglutination was performed against the capsular typing sera (Statens Serum Institut). So-called rough nontypeable strains autoagglutinated with the latex reagent and therefore could not be subject to capsular serotyping, while smooth nontypeable strains did not autoagglutinate but did not react with any of the pooled capsular typing sera.
(iv) DNA probe.
The Accuprobe Streptococcus pneumoniae Culture Identification Test (Gen-Probe, San Diego, Calif.) was used to screen selected isolates. The test is based on the detection of a specific rRNA-encoding sequence found in S. pneumoniae and was performed according to the manufacturer's instructions with four colonies from an overnight culture on 5% blood agar. Any isolate giving a reading of >1,500 photometric light units (PLU) was considered positive, and a reading below this level was considered negative. The test was repeated if the value fell to 1,200 to 1,499 PLU.
(v) Biotyping.
Biotyping was performed using the rapid ID32 Strep (Biomerieux UK, Ltd.). Cultures were grown anaerobically on blood agar at 37°C for 18 h, and suspensions were prepared according to the manufacturer's instructions. The resulting API profile was used to obtain an identification with the Apilab computer key. Additional tests were performed to confirm the identification. These included the catalase reaction, which was performed on an 18-h culture grown on serum glucose agar in 5% CO2, and the test for dextran-levan production (12).
(vi) Assay of hemolytic activity.
Assays of hemolytic activity associated with strains were performed by using a modification of the method described by Benton et al. (6). Cultures were grown to an optical density at 600 nm of 0.4, and assays of both cytoplasmic and extracellular hemolytic activity were performed. Four milliliters of culture was centrifuged for 10 min at 3,000 rpm. The supernatant was retained for analysis of extracellular haemolytic activity, while the pellet was lysed by incubation in 400 μl of lysis buffer (0.01% [wt/vol] sodium dodecyl sulfate, 0.1% [wt/vol] sodium deoxycholate, 0.015 M sodium citrate) at 37°C for 30 min. Hemolytic titers obtained from the supernatant fractions were multiplied by 10 in order to represent numbers of pneumococci comparable with those used to calculate the cytoplasmic titers. In brief, 160 μl of dithiothreitol (DTT) buffer (10 mM DTT–0.1% [wt/vol] bovine serum albumin dissolved in PBS) was added to 96-well V-bottom plates. To the first well of a column 80 μl of culture medium (BHI) or lysed streptococci was added and serially diluted 1:3 across the plate. Control wells consisted of a dilution series of lysate buffer in DTT and BHI alone. Eighty microliters of 2% (vol/vol) horse blood, washed three times and diluted in PBS, was added to all wells. The plates were incubated at 37°C for 30 min and then centrifuged for 10 min at 2,000 rpm. The plates were scored for complete lysis of horse blood, as indicated by the absence of a pellet of cells. The specificity of the assay for pneumolysin was demonstrated by including a D39 derived mutant in which ply had been disrupted by the insertion of an erythromycin resistance cassette and the isogenic parent strain in the assay.
Preparation of chromosomal DNA.
Chromosomal DNA was prepared from all isolates as described previously (45).
Sequence analysis of housekeeping genes.
Fragments of DNA corresponding to the housekeeping genes hexB (DNA mismatch repair protein), recP (transketolase), and xpt (xanthine phosphoribosyltransferase) were amplified by PCR using the primer sets hexBup (5′-CCATTGACGCGGGCTCTA-3′) and hexBdn (5′-CCTGAATACGTCGGAACATCTTT-3′), recPup (5′-ACCGCGACCGCTTTATTCTTTC-3′) and recPdn (5′-ATGCTGACTACGCGGGATTTTTC-3′), and xptup (5′-GAAATTATTAGAAGAA/GCGCATC-3′) and xptdn (5′-TTAGAGATCTGCCTCCA/TTA/GAA-3′), respectively. PCR was performed under standard conditions with 32 cycles of 95°C for 1 min, X°C for 1 min, and 72°C for 1 min, where X°C represents an annealing temperature appropriate for the particular primer set used. Fragments were purified through Qiagen PCR purification columns and sequenced directly by using the same primers and an ABI 373 automated sequencing system. Sequences of 395bp (xptup), 285bp (hexBup), 351bp (hexBdn), 327bp (recPup), and 339bp (recPdn) were obtained from each isolate and used in the analysis presented here.
Phylogenetic analysis.
Preliminary analysis and alignment of sequences was performed in DNAStar. Phylogenetic analysis was performed by using the MEGA suite of programs (31). Dendrograms were constructed by the neighbor-joining method with the Jukes-Cantor correction. The percentage bootstrap confidence levels of internal branches, as defined for MEGA (31), were calculated from 500 random resamplings of the original sequence data with replacement.
Screening of isolates for pneumolysin and autolysin genes.
All isolates were screened for the presence of the pneumolysin (ply), major autolysin (lytA), and atypical autolysin (lytA101) encoding genes by PCR with primer sets plyup-plydn and lytAup-lytAdn as described previously (46) or, in the case of lytA101, the primer lytAup in combination with a novel specific reverse primer lytA101dn (5′-CTACTTCATCGTAATCAAACCGTCAGGTTC-3′). The specificity of PCR products was confirmed by probing with digoxigenin-labeled ply and lytA fragments according to manufacturer's instructions following capillary transfer to a nylon membrane. Probes were obtained after PCR by using the same primer sets, cloning, and confirmatory sequencing of lytA and ply from S. pneumoniae R6 and lytA101 from S. pneumoniae 101/87.
Nucleotide sequence accession numbers.
All sequences obtained in this study have been deposited in EMBL. The housekeeping gene fragments have been assigned the accession numbers AJ240606 to AJ240674 and AJ390850 to AJ391094 and the lytA sequence accession numbers AJ252190 to AJ252196.
RESULTS
Strains included in this study.
A complete list of strains examined in this study and their reactions in classical tests used to distinguish pneumococci from oral streptococci is given in Table 1. One aim of this study was to investigate the genetic relationships between the typical capsular and atypical acapsular and/or bile-insoluble pneumococci sometimes encountered in the diagnostic laboratory. As it became clear that some of these isolates were genetically distinct from pneumococci, isolates of the most closely related streptococci (S. mitis and S. oralis) were included as reference strains. For ease of understanding, the strains in Table 1 have been split into several groups. Group 1 consisted of typical S. pneumoniae isolates fulfilling all the conventional criteria of a pneumococcus being serotypable, optochin sensitive, and bile soluble, and the well-characterized laboratory strain R6. Group 2a contained strains of S. mitis and S. oralis that had been used previously in taxonomic studies (4) included in this study as reference strains. Group 2b consisted of both commensal and pathogenic isolates classified by the suppliers as either S. mitis or S. oralis. All strains in these latter two groups fulfilled conventional criteria for these organisms in being latex agglutination negative, optochin resistant, and bile insoluble. Group 2c consisted of a group of organisms we have called atypical oral streptococci: this designation is based largely upon the presence of pneumococcal virulence factor genes as described later. However, many of these latter isolates were originally obtained because they proved difficult to classify and were considered unusual; many showed some sensitivity to optochin and displayed aberrant biochemical reactions (data not shown). Group 3 consisted of organisms described as atypical pneumococci largely on the basis of being acapsular, either because they autoagglutinated (rough strains) or because they simply failed to react with any pneumococcal antisera. However, some also showed reduced sensitivity to optochin and some were also bile insoluble. The prototype atypical S. pneumoniae strain 101/87, originally described by Diaz et al. (13) on the basis of reactivity with a lytA probe (21), was included within this group as a reference strain.
Genetic relationships between isolates.
In order to determine the genetic relationships between all of the isolates examined in this study, we examined sequence fragments from three housekeeping genes: xpt, recP, and hexB. By using the neighbor-joining method, dendrograms of genetic relationships between isolates were constructed with sequence data from each of the three genes individually to examine tree topology (data not shown). After this, the sequence data of all three genes were combined and treated as a single contiguous fragment in order to obtain an overall picture of genetic relationships between these isolates, as illustrated in Fig. 1.
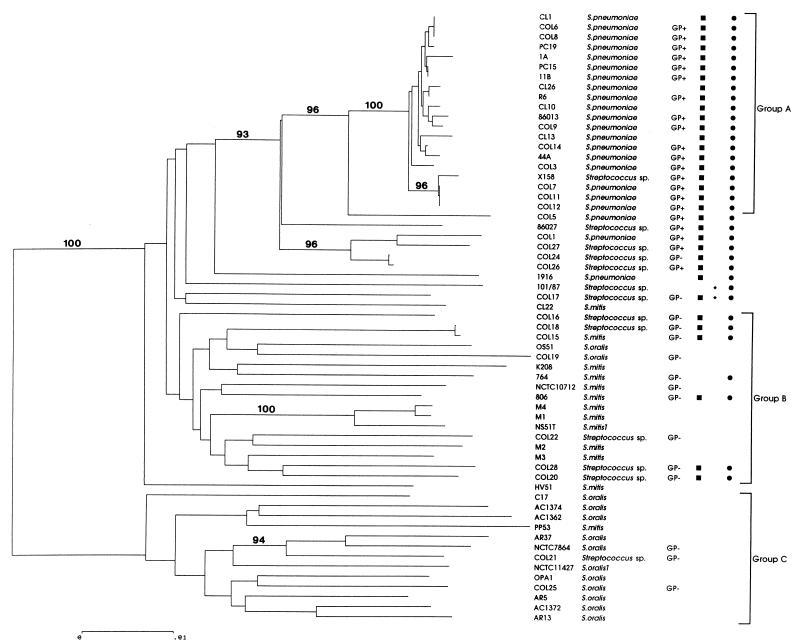
FIG. 1.
Dendrogram of genetic relationships between streptococcal isolates examined in this study constructed from housekeeping gene sequence data by using the neighbor-joining method. Only bootstrap confidence values exceeding 90% are shown. The scale represents the number of nucleotide substitutions per site. Where a consistent identification to species level could not be made on the basis of phenotypic criteria isolates have not been given a species designation. A positive or negative result in the Gen-Probe test where performed is indicated by GP+ or GP−. All isolates were screened for the presence of genes encoding lytA (■), lytA101 (⧫), and ply (●). Groups A, B, and C refer to typical S. pneumoniae, S. mitis, and S. oralis groups, respectively, used to illustrate genetic diversity (see text).
Apart from strains characterized as typical pneumococci, the overall picture provided by the data was one of extensive genetic diversity. Only two major groups were strongly supported by bootstrapping. First, many of the S. pneumoniae isolates fall into a subgroup, labeled A on the dendrogram, containing all isolates characterized as typical pneumococci—the identical group was also supported by bootstrap values of at least 90% when each of the three genes was considered individually. This group is relatively conserved compared with the remaining organisms: the mean nucleotide diversity between members of this group is 1.42% (range, 0 to 3.0%). A second strongly supported group, labeled group C, contained a much more diverse group of organisms predominantly identified as S. oralis and included the S. oralis type strain. With the exception of strain AC1372, which harbored a distant xpt gene, all of these strains fell within the same group in trees constructed by using the three individual housekeeping genes. A feature of this group was the extensive nucleotide diversity relative to that of isolates of typical S. pneumoniae, with a mean diversity of 16.65% (range, 8.7 to 21.2%).
No other major group was strongly supported by bootstrapping with the remainder of the strains forming a diverse group of organisms. Apparently most closely related to, but clearly distinct from, the typical pneumococci are a number of organisms (Col5 to 101/87) classified as atypical pneumococci either on the basis of being acapsular or displaying aberrant reactions to bile and/or optochin. Thus, there appears to be a genuine genetic distinction between typical pneumococci and some of the organisms that have historically been described as atypical pneumococci. The majority of the remaining 20 organisms were identified as S. mitis. This loose association of genetically diverse isolates included commensal S. mitis isolates, isolates associated with invasive disease, isolates previously characterized in taxonomic studies, and the S. mitis type strain. Once again, these organisms were characterized by their extensive genetic diversity. The mean nucleotide diversity within group B (see Fig. 1), which contains most of these organisms, is 16.2% (range, 0.02 to 21.2%).
Genetic relationships of acapsular putative pneumococci.
One initial aim of this study was to examine whether acapsular pneumococci are genetically distinct from typical serotypable pneumococci. Of the 16 acapsular organisms included in group 3, nine were closely related to typical capsular pneumococci and presumably represented organisms that either do not express their capsule or else represent as-yet-unrecognized capsular types. However, the remaining organisms (Col5, Col1, 86027, Col27, Col26, 1916, and 101/87) were clearly genetically distinct from typical capsular organisms.
Relationship of phenotype to genetic background.
Not one of the classical phenotypic criterion was found to satisfactorily distinguish the genetic groups uncovered in this study. As stated above, many of the acapsular pneumococci are clearly typical pneumococci. Many of the atypical pneumococci and the atypical oral streptococci displayed aberrant reactions to optochin and bile. Recently, use of the GenProbe test has been reported to provide accurate and sensitive identification of pneumococci (13, 14), and we therefore used this test on a subset of our isolates. As might be expected, this test, with a nucleic acid probe to a housekeeping gene, provided the most accurate reflection of genetic relationships determined in this study. In addition to all of the typical pneumococci tested, all of the organisms clustering in the atypical pneumococcal group (with the exception of Col24) as far out as the bile-insoluble strain Col26 gave positive GenProbe reactions.
Distribution of the pneumolysin gene.
The distribution of the pnemolysin gene (ply) was studied by using PCR primers plyup and plydn (Table 1, Fig. 1). The specificity of PCR products was confirmed by probing PCR products transferred to a membrane with the S. pneumoniae R6 ply gene at high stringency. All isolates falling into the S. pneumoniae grouping (both typical and atypical) possessed ply and, as expected, isolates within the S. oralis grouping did not contain ply. Unexpectedly, a number of isolates which fell within the broad group containing previously characterized S. mitis isolates were found to possess ply. Interestingly, the majority of these isolates whose history is known are disease-associated isolates. In contrast, none of the S. mitis isolates previously characterized in taxonomic studies (group 2a) or isolates obtained as commensals or from minor infections of the human oral cavity (group 2b) possessed ply (Table 1).
The presence of ply in isolates genetically allied to S. mitis was confirmed by Southern blot by using the R6 ply gene to probe chromosomal DNA (digested with PvuII, which is predicted not to cut within ply on the basis of the published sequence) of some of the group 2c organisms (764, Col15, Col17, Col16, Col18, and Col20) and two of the atypical pneumococci, Col26 and Col27 (data not shown). This probe hybridized with all of the chromosomal DNAs and, in agreement with the extensive genetic diversity among these organisms demonstrated by sequence analysis of housekeeping genes, all isolates displayed distinct restriction profiles. An additional interesting finding was that multiple bands in digests from some of these strains hybridized to the ply probe. Whether these represent the presence of multiple copies of pneumolysin or merely sequence diversity within a single copy of ply requires further investigation.
As final confirmation of the presence of ply in nonpneumococcal oral streptococcal isolates, hemolytic titer assays were performed to compare the hemolytic activity associated with some of the atypical S. mitis strains with both pneumococci and “conventional” S. mitis isolates (Table 2). Both typical pneumococcal controls (strain PC19 and the well-characterized laboratory strain D39) and an atypical pneumococcus (Col27) displayed substantial intracellular and extracellular hemolytic titers. All strains genetically allied to S. mitis, but in which ply had been detected (Col15, Col17, Col20, and 764), also displayed hemolytic activity in both cytoplasmic and extracellular fractions. The specificity of the assay for pneumolysin was confirmed by the inclusion of a ply-deficient D39 mutant strain in these assays. In contrast, S. mitis isolates in which ply was not detected by genetic approaches, as well as several S. oralis isolates, lacked any hemolytic activity under the assay conditions used in this study.
TABLE 2.
Comparison of extracellular and intracellular hemolytic titers of representative isolates examined in this study
| Strain | Strain ID | Hemolytic titera:
|
|
|---|---|---|---|
| Cytoplasmic | Extracellular | ||
| S. pneumoniae | |||
| Typical | PC19 | 729 | 30 |
| D39 | 81 | 30 | |
| D39Δply | – | – | |
| Atypical | Col27 | 27 | 30 |
| S. mitis group | |||
| Typical | NS51T | – | – |
| M3 | – | – | |
| Atypical | Col15 | 243 | 10 |
| Col17 | 729 | 30 | |
| Col20 | 729 | 30 | |
| 764 | 27 | 10 | |
| S. oralis group | NCTC 11427T | – | – |
| Col21 | – | – | |
| Col25 | – | – | |
Hemolytic titer is expressed as the reciprocal of the lowest dilution at which complete lysis of horse blood was seen. Extracellular titers were multiplied by 10 such that they represent a cell number comparable to that used to obtain cytoplasmic titers. –, No hemolytic activity detected.
Distribution of the autolysin gene.
The distribution of the autolysin gene (lytA) was studied by using PCR primers specific for either the typical lytA sequence (primers lytAup and lytAdn) or the atypical lytA101 sequence (primers lytAup and lytA101dn) previously reported from the bile insoluble Spanish S. pneumoniae isolate 101/87 (Table 1). Once again, specificity was confirmed by using either R6 lytA or 101/87 lytA101 to probe membrane-bound PCR products. As expected, all isolates that form part of the pneumococcal group (typical and atypical) were found to possess lytA (Fig. 1). As seen when probing for ply, none of the S. oralis group isolates were found to possess lytA. However, once again, a substantial number of the disease-associated S. mitis isolates were found to possess lytA. Indeed, all but one (764) of the isolates in which ply was detected were also found to possess lytA. No isolates were found to be contain lytA but lack ply. In contrast to lytA, a lytA101-specific product was detected in only the strain S. pneumoniae 101/87, from which this gene was originally isolated (15), and one of the atypical S. mitis strains, Col17.
Sequencing of the lytA gene found in atypical isolates.
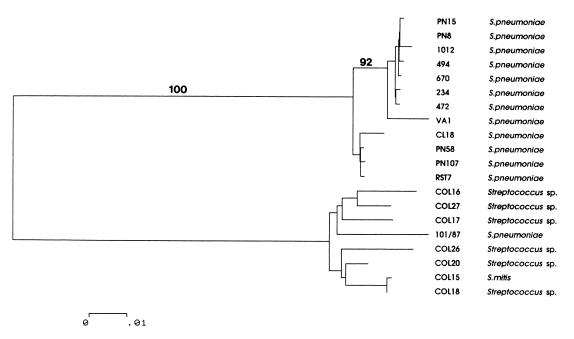
It might be expected that isolates possessing lytA would display the characteristic bile soluble phenotype associated with this gene (42). This was clearly not the case for some isolates of atypical pneumococci (e.g., Col26 and Col27) and the atypical oral streptococci, Thus, in order to examine the relationship of lytA seen in the bile insoluble atypical pneumococci (Col26 and Col27) and some of the atypical oral streptococci (Col15, Col18, Col20, Col16, and Col17) with the known lytA sequence (24), PCR products obtained from these strains were sequenced in full. The relationships between these sequences are illustrated in a dendrogram (Fig. 2) comparing the sequences determined here with the original published sequence of lytA from strain Rst7, nine lytA allelic variants recently reported in typical pneumococci (44), and lytA101 (101/87). All of the sequences were up to 20% divergent from the published Rst7 lytA sequence and other allelic variants seen in typical pneumococci but were much more closely related to lytA101 sequence (5 to 7% divergent). Interestingly, there was between 0.3 and 4.5% divergence within this group, somewhat higher than the levels of divergence than that reported between allelic variants in typical pneumococci (44).
FIG. 2.
Dendrogram of genetic relationships between lytA sequences examined in this study constructed from gene sequence data by using the neighbor-joining method. Only bootstrap values exceeding 90% are shown. The scale represents the number of nucleotide substitutions per site. The upper group of the tree consists of the previously published lytA sequences from strain Rst7, a typical pneumococcal isolate, and a number of allelic variants of lytA recently reported from typical pneumococci (44). The lower group contains the sequence lytA101 from the classical bile-insoluble atypical pneumococcus 101/87 and the lytA sequences determined from atypical pneumococci and atypical oral streptococci examined in this study.
Relationship of genetic background and clinical association.
Many of the S. oralis isolates included in this study were blood isolates obtained from immunocompromised patients. However, these isolates involved in infection appeared to be genetically diverse and widely distributed among the isolates characterized in previous taxonomic studies. Similarly, there was no clear separation of the S. mitis strains containing lytA and ply and associated with disease from the commensal isolates of S. mitis and those characterized previously in taxonomic studies.
DISCUSSION
The primary aim of this study was to examine whether there is a genetic basis to “difficult” or “atypical” isolates sometimes submitted to diagnostic laboratories which have been tentatively identified as pneumococci or oral streptococci but display aberrant phenotypic features. Since the dendrogram of genetic relationships constructed in this study is based on only three housekeeping genes from species that are naturally transformable, we are reluctant to make phylogenetic interpretations based on these data. Recombination is known to occur frequently within pneumococci with the species displaying an epidemic population structure (20, 26, 35). In addition interspecies recombination events between pneumococci and S. oralis and S. mitis have been seen at least in genes under intense selective pressures, such as penicillin-binding-protein encoding genes (17–19). This possibility of recombination makes phylogenetic interpretation potentially hazardous. Having said this, a number of observations are consistent for all three genes examined when considered individually and in the overall analysis, allowing some confidence in them. Two groupings were strongly supported in all trees. These were a subgroup containing the typical pneumococci and the clear separation of the S. oralis group. The atypical organisms (Col5, Col1, Col27, Col24, Col26, 1916, and 86027) were generally placed as the organisms most closely related to pneumococci. In the case of all gene fragments considered individually and the data as a whole the remaining strains (largely identified as S. mitis) formed a relatively diverse group of organisms with few consistent branches.
One obvious feature of the data is the extensive diversity seen within the S. oralis and S. mitis groups when compared to typical pneumococci. This may reflect sampling strategy, although we deliberately selected typical pneumococci thought from previous studies (35) to represent the breadth of genetic diversity within the species. The diversity seen in oral streptococci is consistent with a number of previous reports. For example, among 101 isolates of S. mitis examined, 93 distinct PvuII ribotypes were reported (23). Similarly, restriction fragment length polymorphism analysis of S. mitis populations in and between individuals found limited sharing of genotypes among family members and some 6 to 13 types in individual subjects (27). Repetitive extragenic palindromic PCR has been used to study S. oralis diversity with populations in individuals found to be heterogeneous at a single time point and highly variable when they are monitored longitudinally (3). Recently, PCR-based fingerprinting was also used to examine the relationships between oral streptococci isolated from the blood of neutropenic cancer patients, and all isolates were found to display distinct fingerprint patterns (48).
It is clear from the housekeeping gene sequence data that many isolates that fail to react in the latex agglutination system represent entirely typical pneumococci. It is still not clear why these organisms fail to react with pneumococcal antisera. They may represent as-yet-uncharacterized capsular serotypes, they may possess capsule genes that are nonfunctional or not expressed or, alternatively, they may lack the genes encoding the capsule biosynthetic pathway. We are currently investigating these alternative possibilities using the organisms described in this study. However, a proportion of apparently acapsular isolates did not group with other pneumococci and represent genetically divergent organisms. Thus, there does appear to be a genuine genetic basis to previously described atypical pneumococci which can both fail to react in capsular typing and/or show aberrant biochemical reactions. In light of the extensive genetic diversity within these organisms, it is not surprising that separation on the basis of phenotypic criteria can prove notoriously difficult. The Gen-Probe test was the diagnostic test that appeared to most closely match genetic relationships between isolates with all typical pneumococci and virtually all atypical isolates as far out as Col26 tested giving a positive reaction. Since the basis of this test is hybridization to a housekeeping gene, the correlation with the data presented here is not unexpected. Mundy et al. (36) reported recently that many isolates which show a discordant combination of reactions in capsular typing, optochin sensitivity, and bile solubility tests represent pneumococci on the basis of the Gen-Probe test. In that study the authors suggested the use of Gen-Probe to unequivocally identify isolates showing such discordant phenotypic reactions. Our results indicate that many of the isolates identified as pneumococci by Gen-Probe could actually represent organisms which are genetically rather divergent from typical pneumococci.
Perhaps the most surprising finding of this study was the characterization of isolates phenotypically and genetically allied to S. mitis harboring genes encoding the putative virulence factors pneumolysin and autolysin normally associated with pneumococci. There has been at least one previous report of a lytA probe reacting with optochin-resistant and bile-insoluble streptococci (21), and the use of this gene as a probe to identify atypical pneumococci has been suggested. Our results suggest that using lytA probes to identify difficult organisms as atypical pneumococci could also select organisms which are genetically more closely related to S. mitis. We are not aware of previous reports of the presence of the pneumolysin encoding gene in oral streptococci. Both lytA and ply were absent from previously characterized S. mitis isolates, as well as commensal isolates obtained in this study. Comparative assays of hemolytic activity associated with S. mitis isolates harboring ply and lacking ply (basing the species identity on observed genetic relationships described in this study) suggested that the hemolysin is actively expressed by the ply-containing S. mitis strains. In virtually all cases where a strain history was available, the S. mitis isolates harboring lytA and/or ply were associated with respiratory disease. It is thus tempting to speculate that these genes may enhance the pathogenic potential of these organisms relative to the typical commensal organisms. The detection of ply and lytA in disease-associated putative S. mitis strains provided the impetus for us to include S. oralis strains associated with invasive disease in immunocompromised patients in this study. However, there was no evidence of the presence of lytA or ply in members of this group. It is interesting to consider these findings in the light of virulence studies showing that ply is important in murine intranasal infection (7, 11) and of a recent signature-tagged mutagenesis study (38) which suggested that lytA is important for establishing pneumonia but is not necessary in septicemia. Our data may reflect this in that lytA-containing S. mitis isolates were associated with pneumonia and respiratory tract disease, while isolates of S. oralis from the blood of neutropenic patients did not possess lytA. Perhaps the debilitated state of these patients allows ready access of “commensal” isolates to the bloodstream, bypassing the need for any true invasive step. The detection of “pneumococcal” virulence factor genes in S. mitis also has implications for attempts to use them as diagnostic targets in PCR (25, 40, 41, 43). Clearly, these organisms could cross-react in such tests, and the community needs to be aware that organisms other than “typical” pneumococci may be identified by such tests although, since they may be pathogenic, detection of these organisms is not necessarily an argument against the development of such tests.
We also performed a preliminary characterisation of the ply and lytA genes found in S. mitis isolates. Interestingly, although PCR products were obtained from these strains by using a lytA primer set and, apart from one isolate, not with the lytA101 primer set, their lytA genes were found to be much more closely related to lytA101 than to the typical lytA sequence. The activity of LytA101 is known to be inhibited by sodium deoxycholate, and this property is believed to be responsible for the bile-insensitive phenotype of the host strain 101/87 (15). While the bile insolubility of these strains could reflect their possession of a lytA101-like sequence, other possibilities, such as an autolysin refractory cell wall, cannot be ruled out without further experimentation. Probing Southern blots of the same strains with ply suggested the possibility of substantial diversity of ply within S. mitis isolates despite the fact that evidence to date suggests that ply in S. pneumoniae is a relatively conserved gene (32, 34). An alternative interpretation is that the S. mitis isolates may possess multiple copies of this gene. We are currently investigating both of these possibilities. In light of these results it is interesting to speculate on the evolutionary history of this group of organisms. Clearly, the organisms allied to S. mitis but harboring lytA and ply are genetically diverse, as demonstrated by housekeeping gene sequencing, distinct restriction profiles when probing for ply, and lytA sequencing. This may reflect the fact that these genes are moving freely between organisms with distinct genetic backgrounds. In this respect it is interesting to note all but one strain appeared to harbor both genes and that ply and lytA are closely linked in the pneumococcal genome lying some 7 kb apart. An alternative, though not mutually exclusive, scenario is that these genes represent ancient characteristics that have been lost by some organisms related to the S. mitis group.
We believe it will be important to consider the existence of these atypical organisms in strategies of vaccination against S. pneumoniae. These organisms appear to be of pathogenic potential and may be refractory to immune responses generated by immunogens of typical pneumococci. It is also possible that the atypical organisms we have characterized in this study may act as source of DNA in recombination events, generating new alleles of pneumococcal genes under high selective pressure (17–19). In light of this possibility, it is interesting to note that some of the S. mitis isolates harboring lytA and ply (Col15, Col16, and Col18) have been shown to possess competence-stimulating peptides much more closely related to those of pneumococci than to those reported from other isolates of S. mitis (45). This raises the possibility that these organisms may show an increased likelihood to donate DNA to pneumococci in horizontal gene transfer events simply because their own competence-stimulating peptides may induce a degree of competence in pneumococci. Although this idea requires experimental confirmation, this could mean that these organisms might serve as a pool of variant DNA which could be readily imported into S. pneumoniae genes put under the selective pressure for diversity that inclusion in a potential vaccine might impose. It is thus crucial to examine the nature and extent of genetic diversity in these previously understudied organisms when considering potential vaccine targets in S. pneumoniae.
ACKNOWLEDGMENTS
This work was supported by a project grant from The Wellcome Trust (045171/2/95/Z). A.M.W. is supported by a Wellcome Trust Research Fellowship in Biodiversity.
We are extremely grateful to David Beighton, Rob Whiley, and Steven Gillespie for providing some of the strains included in this study.
REFERENCES
- 1.Austrian R. Random gleanings from a life with the pneumococcus. J Infect Dis. 1975;131:474–484. doi: 10.1093/infdis/131.4.474. [DOI] [PubMed] [Google Scholar]
- 2.Avery O T, MacLeod C M, MacCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from Pneumococcus type III. J Exp Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beighton D, Alum S. Use of repetitive extragenic palindromic PCR (REP-PCR) to study S. oralis. J Dent Res. 1997;76:1026. [Google Scholar]
- 4.Beighton D, Hardie J M, Whiley R A. A scheme for the identification of viridans streptococci. J Med Microbiol. 1991;35:367–372. doi: 10.1099/00222615-35-6-367. [DOI] [PubMed] [Google Scholar]
- 5.Beighton D, Carr A D, Oppenheim B A. Identification of viridans streptococci associated with bacteremia in neutropenic cancer patients. J Med Microbiol. 1994;40:202–204. doi: 10.1099/00222615-40-3-202. [DOI] [PubMed] [Google Scholar]
- 6.Benton K A, Paton J C, Briles D E. Differences in virulence for mice among Streptococcus pneumoniae strains of capsular types 2, 3, 4, 5, and 6 are not attributable to differences in pneumolysin production. Infect Immun. 1997;65:1237–1244. doi: 10.1128/iai.65.4.1237-1244.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry A M, Yother J, Briles D E, Hansman D, Paton J C. Reduced virulence of a defined pneumolysin-negative mutant of Streptococcus pneumoniae. Infect Immun. 1989;57:2037–2042. doi: 10.1128/iai.57.7.2037-2042.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochud P Y, Calandra T, Franciola P. Bacteremia due to viridans streptococci in neutropenic patients: a review. Am J Med. 1994;97:256–264. doi: 10.1016/0002-9343(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 9.Bochud P Y, Eggiman P, Calandra T, van Melle G, Saghafi L, Franciola P. Bacteremia due to viridans streptococci in neutropenic patients with cancer: clinical spectrum and risk factors. Clin Infect Dis. 1994;20:469–470. doi: 10.1093/clinids/18.1.25. [DOI] [PubMed] [Google Scholar]
- 10.Broome C V, Facklam R R. Epidemiology of clinically significant isolates of Streptococcus pneumoniae in the United States. Rev Infect Dis. 1981;3:277–280. doi: 10.1093/clinids/3.2.277. [DOI] [PubMed] [Google Scholar]
- 11.Canvin J R, Marvin A P, Sivakumaran M, Paton J C, Boulnois G J, Andrew P W, Mitchell T J. The role of pneumolysin and autolysin in the pathology of pneumonia and septicemia in mice infected with a type 2 pneumococcus. J Infect Dis. 1995;172:119–123. doi: 10.1093/infdis/172.1.119. [DOI] [PubMed] [Google Scholar]
- 12.Colman G, Ball L C. Identification of streptococci in a medical laboratory. J Appl Bacteriol. 1984;57:1–14. doi: 10.1111/j.1365-2672.1984.tb02351.x. [DOI] [PubMed] [Google Scholar]
- 13.Davis T E, Fuller D D. Direct identification of bacterial isolates in blood cultures by using a DNA probe. J Clin Microbiol. 1991;29:2193–2196. doi: 10.1128/jcm.29.10.2193-2196.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denys G A, Carey R B. Identification of Streptococcus pneumoniae with a DNA probe. J Clin Microbiol. 1992;30:2725–2727. doi: 10.1128/jcm.30.10.2725-2727.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Díaz E, López R, García J L. Role of the major pneumococcal autolysin in the atypical response of a clinical isolate of Streptococcus pneumoniae. J Bacteriol. 1992;174:5508–5515. doi: 10.1128/jb.174.17.5508-5515.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douglas C W I, Heath J, Hampton K K, Preston F E. Identity of viridans streptococci isolated from cases of infective endocarditis. J Med Microbiol. 1993;39:179–182. doi: 10.1099/00222615-39-3-179. [DOI] [PubMed] [Google Scholar]
- 17.Dowson C G, Coffey T J, Kell C, Whiley R A. Evolution of penicillin resistance in Streptococcus pneumoniae; the role of Streptococcus mitis in the formation of a low affinity PBP2B in S. pneumoniae. Mol Microbiol. 1993;9:635–643. doi: 10.1111/j.1365-2958.1993.tb01723.x. [DOI] [PubMed] [Google Scholar]
- 18.Dowson C G, Coffey T J, Spratt B G. Origin and molecular epidemiology of penicillin-binding-protein-mediated resistance to β-lactam antibiotics. Trends Microbiol. 1994;2:361–366. doi: 10.1016/0966-842x(94)90612-2. [DOI] [PubMed] [Google Scholar]
- 19.Dowson C G, Barcus V A, King S, Pickerill P, Whatmore A, Yeo M. Horizontal gene transfer and the evolution of resistance and virulence determinants in Streptococcus. J Appl Microbiol. 1997;83:42S–51S. doi: 10.1046/j.1365-2672.83.s1.5.x. [DOI] [PubMed] [Google Scholar]
- 20.Enright M C, Spratt B G. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 21.Fenoll A, Martinez-Suarez J V, Munoz R, Casal J, Garcia J L. Identification of atypical strains of Streptococcus pneumoniae by a specific DNA probe. Eur J Clin Microbiol Infect Dis. 1990;9:396–401. doi: 10.1007/BF01979468. [DOI] [PubMed] [Google Scholar]
- 22.Finland M, Barnes M W. Changes in the occurrence of capsular serotypes of Streptococcus pneumoniae in Boston City Hospital during selected years between 1935 and 1974. J Clin Microbiol. 1977;5:154–166. doi: 10.1128/jcm.5.2.154-166.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitzsimmons S, Evans M, Pearce C, Sheridan M J, Wientzen R, Bowden G, Cole M F. Clonal diversity of Streptococcus mitis biovar 1 isolates from the oral cavity of human neonates. Clin Diagn Lab Immunol. 1996;3:517–522. doi: 10.1128/cdli.3.5.517-522.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.García P, García J L, García E, López R. Nucleotide sequence and expression of the pneumococcal autolysin gene from its own promoter in Escherichia coli. Gene. 1986;43:265–272. doi: 10.1016/0378-1119(86)90215-5. [DOI] [PubMed] [Google Scholar]
- 25.Gillespie S H, Ullman C, Smith M D, Emery V. Detection of Streptococcus pneumoniae in sputum samples by PCR. J Clin Microbiol. 1994;32:1308–1311. doi: 10.1128/jcm.32.5.1308-1311.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall L M C, Whiley R A, Duke B, George R C, Efstratiou A. Genetic relatedness within and between serotypes of Streptococcus pneumoniae from the United Kingdom: analysis of multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, and antimicrobial resistance patterns. J Clin Microbiol. 1996;34:853–859. doi: 10.1128/jcm.34.4.853-859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hohwy J, Kilian M. Clonal diversity of the Streptococcus mitis biovar 1 population in the human oral cavity and pharynx. Oral Microbiol Immunol. 1995;10:19–25. doi: 10.1111/j.1399-302x.1995.tb00113.x. [DOI] [PubMed] [Google Scholar]
- 28.Kawamura Y, Hou X-G, Sultana F, Miura H, Ezaki T. Determination of 16S rRNA sequences of Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol. 1995;45:406–408. doi: 10.1099/00207713-45-2-406. [DOI] [PubMed] [Google Scholar]
- 29.Kilian M, Mikkelsen L, Henrichsen J. Taxonomic study of viridans streptococci: description of Streptococcus gordonii sp. nov. and emended descriptions of Streptococcus sanguis (White and Niven 1946), Streptococcus oralis (Bridge and Sneath 1982), and Streptococcus mitis (Andrewes and Horder 1906) Int J Syst Bacteriol. 1989;39:471–484. [Google Scholar]
- 30.Kontiainen S, Sivonen A. Optochin resistance in Streptococcus pneumoniae strains isolated from blood and middle ear fluid. Eur J Clin Microbiol. 1987;6:422–424. doi: 10.1007/BF02013101. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S K, Tamura K, Nei M. MEGA: molecular evolutionary genetic analysis software for microcomputers. CABIOS. 1993;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 32.Lock R A, Zhang Q Y, Berry A M, Paton J C. Sequence variation in the Streptococcus pneumoniae pneumolysin gene affecting haemolytic activity and electrophoretic mobility of the toxin. Microb Pathog. 1996;21:71–83. doi: 10.1006/mpat.1996.0044. [DOI] [PubMed] [Google Scholar]
- 33.Lucas V S, Beighton D, Roberts G J, Challacombe S J. Changes in the oral streptococcal flora of children undergoing allogenic bone marrow transplantation. J Infect. 1997;35:135–141. doi: 10.1016/s0163-4453(97)91545-0. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell T J, Mendez F J, Paton J C, Andrew P W, Boulnois G J. Comparison of pneumolysin genes and proteins from Streptococcus pneumoniae types 1 and 2. Nucleic Acids Res. 1990;18:4010. doi: 10.1093/nar/18.13.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mũller-Graf C D M, Whatmore A M, King S J, Trzcinski K, Pickerill A P, Doherty N, Paul J, Griffiths D, Crook D, Dowson C G. Population biology of Streptococcus pneumoniae isolated from oropharyngeal carriage and invasive disease. Microbiology. 1999;145:3283–3293. doi: 10.1099/00221287-145-11-3283. [DOI] [PubMed] [Google Scholar]
- 36.Mundy L S, Janoff E N, Schwebke K E, Shanholtzer C J, Willard K E. Ambiguity in the identification of Streptococcus pneumoniae. Optochin, bile solubility, Quellung and the AccuProbe DNA probe tests. Microbiol Infect Dis. 1998;109:55–61. doi: 10.1093/ajcp/109.1.55. [DOI] [PubMed] [Google Scholar]
- 37.Muñoz R, Fenoll A F, Vicioso D, Casal J. Optochin-resistant variants of Streptococcus pneumoniae. Diagn Microbiol Infect Dis. 1990;13:63–66. doi: 10.1016/0732-8893(90)90056-2. [DOI] [PubMed] [Google Scholar]
- 38.Philips G, Barker R, Brogan O. Optochin-resistant Streptococcus pneumoniae. Lancet. 1988;ii:281. doi: 10.1016/s0140-6736(88)92573-1. [DOI] [PubMed] [Google Scholar]
- 39.Polissi A, Pontiggia A, Feger G, Altieri M, Mottl H, Ferrari L, Simon D. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect Immun. 1998;66:5620–5629. doi: 10.1128/iai.66.12.5620-5629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudolph K M, Parkinson A J, Black C M, Mayer L W. Evaluation of the polymerase chain reaction for the diagnosis of pneumococcal pneumonia. J Clin Microbiol. 1993;31:2661–2666. doi: 10.1128/jcm.31.10.2661-2666.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salo P, Ortqvist A, Leinonen M. Diagnosis of bacteremic pneumococcal pneumonia by amplification of a pneumolysin gene fragment in serum. J Infect Dis. 1995;171:479–482. doi: 10.1093/infdis/171.2.479. [DOI] [PubMed] [Google Scholar]
- 42.Sánchez-Pulles J M, Ronda C, García J L, García P, López R, García E. Searching for autolysin functions. Characterisation of a pneumococcal mutant deleted in the lytA gene. Eur J Biochem. 1986;158:289–293. doi: 10.1111/j.1432-1033.1986.tb09749.x. [DOI] [PubMed] [Google Scholar]
- 43.Ubukata K, Asahi Y, Yamane A, Konno M. Combinational detection of autolysin and penicillin-binding protein 2B genes of Streptococcus pneumoniae by PCR. J Clin Microbiol. 1996;34:592–596. doi: 10.1128/jcm.34.3.592-596.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whatmore A M, Dowson C G. The autolysin encoding gene (lytA) of Streptococcus pneumoniae displays restricted allelic variation despite localized recombination events with genes of pneumococcal bacteriophage encoding cell wall lytic enzymes. Infect Immun. 1999;67:4551–4556. doi: 10.1128/iai.67.9.4551-4556.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whatmore A M, Barcus V A, Dowson C G. Genetic diversity of the streptococcal competence (com) gene locus. J Bacteriol. 1999;181:3144–3154. doi: 10.1128/jb.181.10.3144-3154.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whatmore A M, King S J, Doherty N C, Sturgeon D, Chanter N, Dowson C G. Molecular characterization of equine isolates of Streptococcus pneumoniae: natural disruption of genes encoding the virulence factors pneumolysin and autolysin. Infect Immun. 1999;67:2776–2782. doi: 10.1128/iai.67.6.2776-2782.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whiley R A, Beighton D. Current classification of the oral streptococci. Oral Microbiol Immunol. 1998;13:195–216. doi: 10.1111/j.1399-302x.1998.tb00698.x. [DOI] [PubMed] [Google Scholar]
- 48.Wisplinghoff H, Reinert R R, Cornely O, Seifert H. Molecular relationships and antimicrobial susceptibilities of viridans group streptococci isolated from blood of neutropenic cancer patients. J Clin Microbiol. 1999;37:1876–1880. doi: 10.1128/jcm.37.6.1876-1880.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]