Abstract
Attention is a core cognitive function that filters and selects behaviourally relevant information in the environment. The cortical mapping of attentional systems identified two segregated networks that mediate stimulus-driven and goal-driven processes, the Ventral and the Dorsal Attention Networks (VAN, DAN). Deep brain electrophysiological recordings, behavioral data from phylogenetic distant species, and observations from human brain pathologies challenge purely corticocentric models. Here, we used advanced methods of functional alignment applied to resting-state functional connectivity analyses to map the subcortical architecture of the Ventral and Dorsal Attention Networks. Our investigations revealed the involvement of the pulvinar, the superior colliculi, the head of caudate nuclei, and a cluster of brainstem nuclei relevant to both networks. These nuclei are densely connected structural network hubs, as revealed by diffusion-weighted imaging tractography. Their projections establish interrelations with the acetylcholine nicotinic receptor as well as dopamine and serotonin transporters, as demonstrated in a spatial correlation analysis with a normative atlas of neurotransmitter systems. This convergence of functional, structural, and neurochemical evidence provides a comprehensive framework to understand the neural basis of attention across different species and brain diseases.
Subject terms: Human behaviour, Attention
Integration of functional, structural, and neurochemical data proposes a framework to comprehensively understand the neural basis of attention across different species and neurological diseases.
Introduction
“Everyone knows what attention is. It is the taking possession by the mind, in clear and vivid form, of one out of what seem several simultaneously possible objects or trains of thought.”1.
Everything we see, feel, or smell is an illusion elaborated by our brain circuits. However, the brain’s capacity is limited. This requires mechanisms for the selection of the most relevant information. The ensemble of cognitive and neural processes involved in capacity limitation and selection underlies ‘attention’ as defined by James1. Behavioral studies have distinguished orienting of attention into a slow, strategic, goal-directed, and voluntary component versus a swift, unexpected, bottom-up, and automatic component2,3. Task-related functional neuroimaging (fMRI) studies segregated these two attentional processes anatomically into a dorsal and ventral attentional network4. The dorsal attention network (DAN) encodes and maintains preparatory signals and modulates top-down sensory (visual, auditory, olfactory and somatosensory) regions.
In contrast, the ventral attention network (VAN) is recruited when attention is re-oriented to novel behaviorally relevant events. Classical core regions of the DAN are the intraparietal sulcus, the superior parietal lobe, and the frontal eye fields. The DAN is considered to have no hemispheric lateralization5–9. In contrast, the temporoparietal junction and the ventrolateral prefrontal cortex constitute the central regions of the VAN. Evidence demonstrates that the VAN is right-lateralized7,8,10. Within their respective networks, DAN and VAN regions have synchronous fMRI signal oscillations at rest8,11–18. Thanks to this synchronization, the two networks have consistently been identified and segregated in resting-state fMRI cortical parcellations14,16–18, although their taxonomy has not always been homogenous in the literature19,20. Hence, the DAN and VAN are organized as independent networks even in the absence of task signals. However, their synchronization can change according to task demands, and they can be acting jointly or separately21,22. Furthermore, DAN and VAN task activations and synchronization levels are modified by focal lesions and correlate with behavioral deficits23–30.
Yet, electrical recording, pathological observations, and phylogenetic comparisons demonstrate that the neuroanatomical framework of attentional mechanisms should extend well beyond a corticocentric model. Electrical recordings in primates showed that subcortical structures have a crucial role in the neural mechanisms of attention. For instance, inactivation of the superior colliculus during motion-change detection markedly disturbs visual attention without affecting the neuronal activity in the visual cortex31. Attentional states also modulate the thalamic pulvinar nuclei32,33 and neuronal discharge patterns in the locus coeruleus34,35. Pathophysiological data from human brain disease supports the critical relevance of deep brain nuclei. Neglect is a clinical syndrome characterized by pathological hemispatial inattention36 and can arise from subcortical lesions in the pulvinar, striatum, or superior colliculus37–40. Patients with attention deficit hyperactivity disorder also present alterations beyond the cortex28, such as in the pulvinar, which is influenced by the severity of the disease and the use of stimulants41.
Additionally, distant phylogenetic species, such as pigeons, have markedly different cortical morphologies but exhibit attention errors and reaction times similar to humans42. With close mammals, such as macaques, relevant functional attention dissimilarities have been described at the cortical level, including the complete absence of a VAN43. Hence, a core phylogenetically relevant subcortical network of areas appears to support the orientation of attention that has been mostly disregarded in the functional neuroimaging literature because of limited field strength or issues arising from average group alignments. Average group alignments of functional neuroimaging maps exclusively based on structural landmarks might typically fail to represent an accurate functional network due to interindividual differences44,45. Specifically, subcortical nuclei are prone to structural misalignment due to their small size, poor contrast in structural MRI, and intersubject cytoarchitectonic variability46–49. In contrast, advanced methods of functional alignment improve structural-functional correspondence across participants50–53. Further, surface interindividual alignment based on morphological features, such as cortical folding, fairly aligns unimodal cortical areas, such as the primary visual and motor cortices, but poorly overlaps higher-order cortical areas50,54. Methods of functional alignment based on fMRI signals during cognitive activation paradigms55,56 and resting-state fMRI connectivity patterns52,57 provided better function matching and have also been used for cross-species functional comparisons58. Functional alignment is different from hyperalignment techniques that project shared neural information beyond the three-dimensional anatomical space, i.e., in high-dimensional spaces59–61. At the subcortical level, our team also demonstrated that functional alignment methods can optimize the group-level mapping of functional networks, improving functional correlations and uncovering a network’s deep brain nuclei components62. However, this method has never been applied to explore the subcortical anatomy of the VAN and the DAN.
Delineating the subcortical components of the DAN and the VAN would allow us to revisit their underlying circuitry through diffusion-weighted imaging tractography that enables in vivo reconstruction of associative, commissural, and projection white-matter tracts63–65. A clearer characterization of the DAN and VAN circuitry will help to better understand brain interactions in healthy and pathological brains66,67.
Subcortical structures also play a critical role within the neurotransmitter systems. Brainstem nuclei are the primary sources of neurotransmitter synthesis and send axonal projections to the cortex and the basal ganglia. The basal ganglia are central targets of the neurotransmitter axonal projections and mediate their physiological effects. Yet the neurochemistry of the DAN and the VAN is limited to primate studies. These studies reported a noradrenergic innervation of regions of the primate attention networks, including the temporoparietal junction and the frontal lobe68–70. Noradrenaline has been proposed as a critical trigger for the reorientation of attention8,70. However, despite its essential neuroscientific and medical importance28, the neurochemical signatures of the VAN and the DAN have never been contrasted in humans. Such an endeavor is now possible thanks to the macroscale mapping of the neurotransmitter receptors and transporters in humans by means of positron emission tomography (PET) and single-photon emission computerized tomography (SPECT) scans71. Accordingly, a normative atlas of nine neurotransmitter systems aligned in the MNI space is now openly available and allows the investigation of the neurochemical signature of brain circuits72–76.
Therefore, we explored the subcortical anatomy of attention networks by aligning the individual resting-state functional maps of the VAN and the DAN in a common functional space. Based on previous electrical recordings, pathological observations, and phylogenetic reports, we hypothesized that basal ganglia and brainstem nuclei, namely the pulvinar, the striatum, the superior colliculi, and the locus coeruleus, are core phylogenetically relevant and functional constituents of the attention networks. Finally, an optimized model of the VAN and the DAN was proposed together with their structural, functional, graph centrality, and neurochemical signature.
Results
VAN anatomical map
The statistical map of the VAN, after functional alignment, is represented in Fig. 1 (left column).
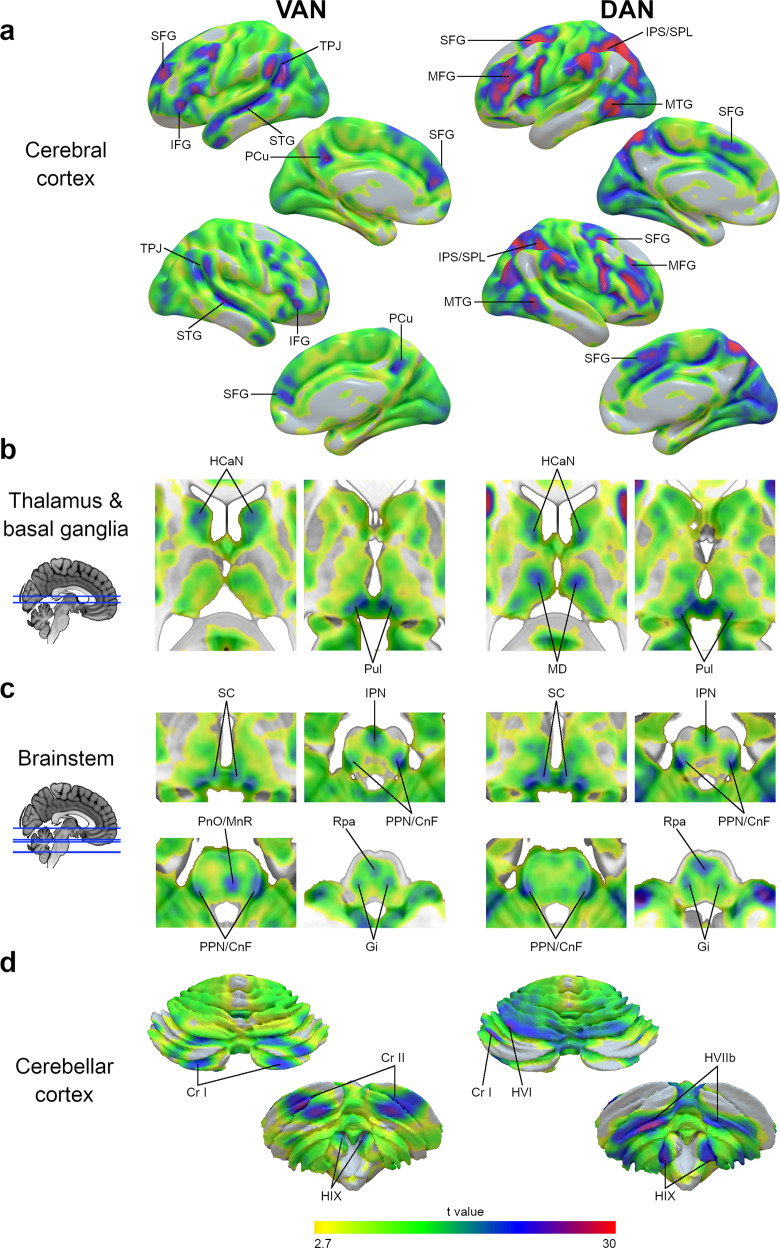
Fig. 1. VAN and DAN maps after functional alignment.
VAN (left) and DAN (right) maps after functional alignment at different anatomical levels, namely the cerebral cortical surface (a), subcortical thalamus and basal ganglia (b), brainstem (c), and cerebellar cortical surface (d). The color gradient represents the t-value distribution (n = 110). CnF cuneiform nucleus, Cr I cerebellar crus I lobule, Cr II cerebellar crus II lobule, DAN dorsal attention network, Gi gigantocellular nucleus, HCaN head of caudate nucleus, HIIb cerebellar lobule IIb, HVI cerebellar lobule VI, HIX cerebellar lobule IX (cerebellar tonsils), IFG inferior frontal gyrus, IPN interpeduncular nucleus, IPS intraparietal sulcus, MnR median raphe nucleus, MD mediodorsal nucleus of the thalamus, MTG middle temporal gyrus, PCu precuneus, PnO nucleus pontis oralis, PPN pedunculopontine nucleus, Pul pulvinar, Rpa raphe pallidus nucleus, SC superior colliculus, SFG superior frontal gyrus, SPL superior parietal lobule, STG superior temporal gyrus, TPJ temporoparietal junction, VAN ventral attention network.
At the cerebral cortical level, the peaks of statistical association were observed in the temporoparietal junction, the inferior frontal gyrus, the anterior part of the superior frontal gyrus, and the superior temporal gyrus (Fig. 1a). Additionally, peaks of statistical association were also present in the crus I, crus II and superior IX cerebellar cortex (Fig. 1d).
A high statistical association was present at the thalamus and basal ganglia level in the head of caudate nuclei and the pulvinar (Fig. 1b). In the brainstem, a high statistical association was observed in voxels overlapping with the superior colliculi, the interpeduncular nucleus, and the pedunculopontine-cuneiform nuclei complex pontis oralis, the gigantocellular nuclei, the raphe pallidus, and median nuclei (Fig. 1c). Table 1 represents the centers of gravity coordinates of the subcortical regions of interest. The VAN statistical and correlation maps are available at https://neurovault.org/collections/XONZLGPJ/.
Table 1.
MNI coordinates of the VAN subcortical regions’ centers of gravity.
| Regions of interest | MNI (X) | MNI (Y) | MNI (Z) |
|---|---|---|---|
| HCaN L | −11 | 8 | 13 |
| Pul L | −4 | −30 | 1 |
| SC L | −9 | −31 | −3 |
| PPN/CnF L | −14 | −29 | −25 |
| Gi L | −10 | −25 | −36 |
| Cr I L | −31 | −73 | −31 |
| Cr II L | −21 | −79 | −42 |
| IPN | 1 | −19 | −21 |
| MnR | −3 | −29 | −28 |
| Rpa | 1 | −28 | −43 |
| HCaN R | 13 | 11 | 12 |
| Pul R | 6 | −29 | 1 |
| SC R | 13 | −30 | −3 |
| PPN/CnF R | 13 | −31 | −25 |
| Gi R | 11 | −24 | −35 |
| Cr I R | 30 | −74 | −30 |
| Cr II R | 24 | −79 | −41 |
CnF cuneiform nucleus, Cr I cerebellar crus I lobule, Cr II cerebellar crus II lobule, Gi gigantocellular nucleus, HCaN head of caudate nucleus, IPN interpeduncular nucleus, L left, MnR median raphe nucleus, PnO nucleus pontis oralis, PPN pedunculopontine nucleus, Pul pulvinar, R right, Rpa raphe pallidus nucleus, SC superior colliculus.
DAN anatomical map
The statistical map of the DAN, after functional alignment, is represented in Fig. 1 (right column).
The peaks of the statistical association at the cerebral cortical level were in the intraparietal sulcus and superior parietal lobule, in the middle and superior frontal gyrus, and in the posterior part of the middle temporal gyrus (Fig. 1a). Peaks of statistical association were also present in the cerebellar cortex’s areas VIIb, inferior IX, left VI, and left I (Fig. 1d).
At the thalamus and basal ganglia level, areas with a high statistical association were located in the head of caudate nuclei and the thalamic pulvinar and mediodorsal nuclei (Fig. 1b). High statistical associations also included voxels overlapping the superior colliculi, the interpeduncular nucleus, the pedunculopontine-cuneiform nuclei complex, the gigantocellular nuclei, and the raphe pallidus nuclei in the brainstem (Fig. 1c). Table 2 represents the centers of gravity of the subcortical regions of interest. The DAN statistical and correlation maps are available at https://neurovault.org/collections/XONZLGPJ/.
Table 2.
MNI coordinates of the DAN subcortical regions’ centers of gravity.
| Regions of interest | MNI (X) | MNI (Y) | MNI (Z) |
|---|---|---|---|
| HCaN L | −10 | 4 | 10 |
| MD L | −9 | −18 | 8 |
| Pul L | −4 | −30 | 1 |
| SC L | −8 | −31 | −3 |
| PPN/CnF L | −14 | −30 | −25 |
| Gi L | −10 | −24 | −35 |
| Cr I L | −39 | −64 | −29 |
| HVI L | −25 | −62 | −24 |
| HVIIb L | −24 | −66 | −49 |
| HIX L | −11 | −51 | −50 |
| IPN | 1 | −19 | −21 |
| Rpa | 1 | −28 | −42 |
| HCaN R | 12 | 7 | 10 |
| MD R | 9 | −16 | 8 |
| Pul R | 6 | −29 | 1 |
| SC R | 11 | −30 | −3 |
| PPN/CnF R | 14 | −30 | −25 |
| Gi R | 11 | −25 | −34 |
| HVIIb R | 25 | −68 | −49 |
| HIX R | 11 | −53 | −51 |
CnF cuneiform nucleus, Cr I cerebellar crus I lobule, Gi gigantocellular nucleus, HCaN head of caudate nucleus, HVI cerebellar lobule VI, HIX cerebellar lobule IX (cerebellar tonsils), IPN interpeduncular nucleus, L left, MD mediodorsal nucleus of the thalamus, PPN pedunculopontine nucleus, Pul pulvinar, R right, Rpa raphe pallidus nucleus, SC superior colliculus.
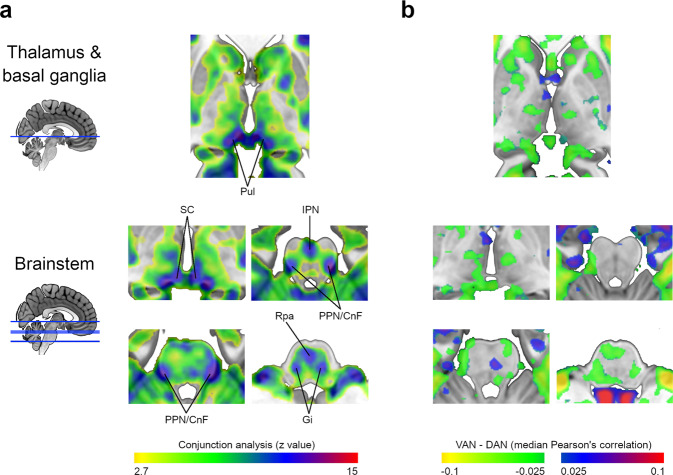
The conjunction analysis showed that most of the subcortical peaks of statistical association were shared by both networks (Fig. 2), explicitly overlapping the pulvinar, the superior colliculi, the interpeduncular nuclei, the pedunculopontine-cuneiform nuclei complex, the gigantocellular nuclei, and the raphe pallidus nuclei.
Fig. 2. VAN and DAN maps similarity.
a Conjunction analysis of the VAN and DAN statistical maps at the thalamus, basal ganglia, and brainstem levels (n = 110). b Difference map resulting from the subtraction of the median DAN Pearson’s correlation map from the VAN. CnF cuneiform nucleus, DAN dorsal attention network, Gi gigantocellular nucleus, IPN interpeduncular nucleus, PPN pedunculopontine nucleus, Pul pulvinar, Rpa raphe pallidus nucleus, SC superior colliculus, VAN ventral attention network. (see maps at https://neurovault.org/collections/XONZLGPJ/).
Structural and functional connectivity of the VAN nodes
The structural connectivity map of the VAN is represented in Fig. 3a.
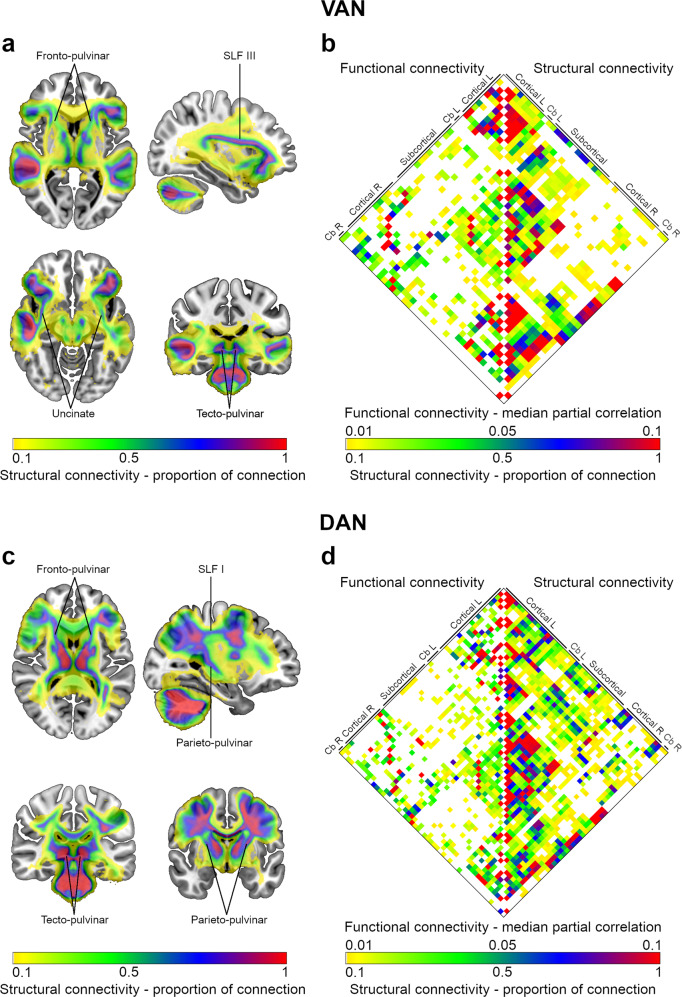
Fig. 3. Structural and functional connectivity of VAN and DAN nodes.
a Structural connectivity map of the VAN. b Matrix with the node-to-node functional and structural connectivity of the VAN, represented on the left and right halves, respectively. c Structural connectivity map of the DAN. d Matrix with the node-to-node functional and structural connectivity of the DAN, represented on the left and right halves, respectively. Nodes of the matrices were labeled in groups according to their anatomical location. A complete list with node labels is available in Supplementary Tables 1, 2. As indicated, color gradients represent the structural connectivity (expressed as the proportion of connection) or the functional connectivity (defined as the median partial correlation). Cb cerebellum, L left, R right, SLF superior longitudinal fasciculus.
The cortical regions of the VAN were connected by the third branch of the Superior Longitudinal Fasciculus (SLF III) and the uncinate fasciculus (Fig. 3a). Fronto-pulvinar and tecto-pulvinar projections established the connections with or between subcortical structures (Fig. 3a). The node-to-node structural and functional connectivity patterns are represented in Fig. 3b.
The maps of the VAN ROIs and the structural connectivity analysis are available at https://neurovault.org/collections/XONZLGPJ/.
Structural and functional connectivity of DAN nodes
The structural connectivity map of the DAN is represented in Fig. 3c.
The cortical regions of the DAN established connections through the first branch of the Superior Longitudinal Fasciculus (SLF I, Fig. 3c). Fronto-pulvinar, parieto-pulvinar, and tecto-pulvinar projections mediated the links with or between subcortical structures (Fig. 3c).
The node-to-node structural and functional connectivity patterns are shown in Fig. 3d.
The maps of the DAN ROIs and the structural connectivity analysis are available at https://neurovault.org/collections/XONZLGPJ/.
Lateralization assessment
Figure 4 illustrates the hemispheric distribution of the structural and functional connectivity measures of the VAN and the DAN.
Fig. 4. VAN and DAN lateralization.
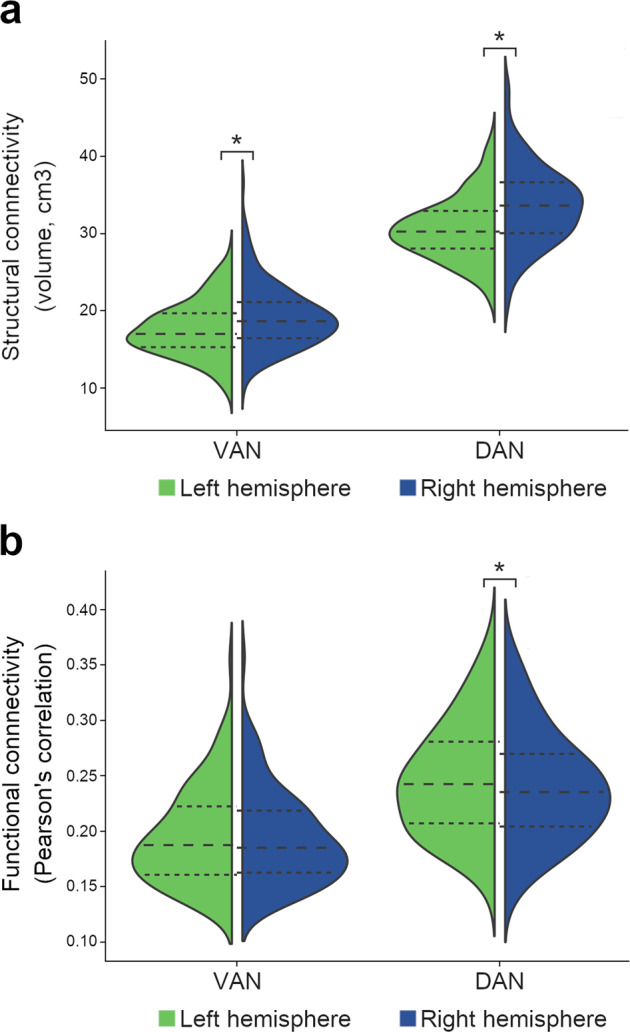
Structural connectivity is expressed in volumes of the structural connection maps (a) and functional connectivity in average Pearson’s correlations (b) across hemispheric nodes. Dashed lines represent the median and the interquartile range; the minimum and maximum correspond to the violin limits. DAN dorsal attention network, VAN ventral attention network. Asterisk (*), significant differences between the right and the left hemispheres (p < 0.05; paired analysis; structural connectivity, n = 177; functional connectivity, n = 110).
The structural connectivity connecting the VAN was significantly larger in the right hemisphere than in the left (right hemisphere 18.7[16.5,21.1]cm3, left hemisphere 17.0[15.3,19.7]cm3; p value <0.001). Pearson’s correlations were not different between the right and left VANs (right hemisphere 0.185[0.162,0.218], left hemisphere 0.188[0.160,0.222]; p value = 0.125).
The DAN’s structural connectivity was also significantly larger in the right hemisphere (right hemisphere 33.6[30.0,36.6]cm3, left hemisphere 30.2[28.0,32.9]cm3; p value <0.001). Pearson’s correlations were significantly higher in the left hemisphere than in the right (right hemisphere 0.240(0.049), left hemisphere 0.246(0.050); p value <0.001).
Graph theory analysis
Figure 5a illustrates the graph theory representation of the VAN and DAN structural connectivity.
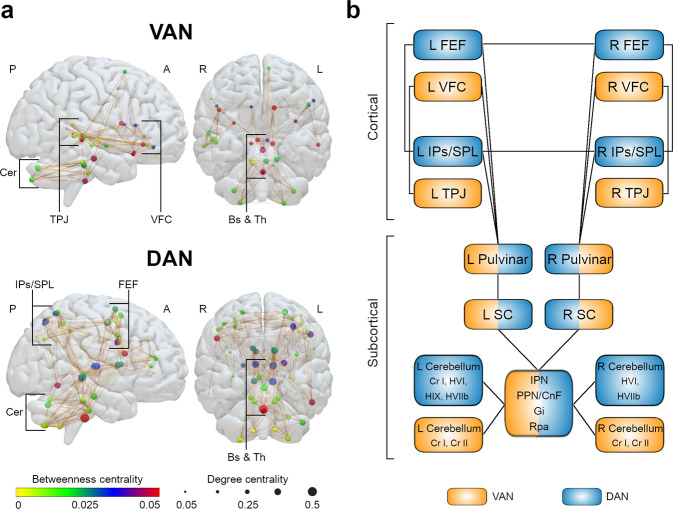
Fig. 5. Graph theory analysis and anatomical model of the VAN and DAN.
a Graph theory analysis of the VAN and DAN structural connectivity. Circles illustrate nodes. Circle colors represent the median betweenness centrality of each node (according to the color gradient), while circle dimensions represent the median degree centrality. Brown lines represent node-to-node structural connections present in at least half of the subjects. b Anatomical model of the VAN and DAN. A anterior, CnF cuneiform nucleus, DAN dorsal attention network, Gi gigantocellular nucleus, IPN interpeduncular nucleus, L left, P posterior, PPN pedunculopontine nucleus, Pul pulvinar, R right, Rpa raphe pallidus nucleus, SC superior colliculus, VAN ventral attention network.
The subcortical structures with the highest median betweenness centrality in the VAN were in the right pulvinar and the left caudate nucleus head (the second and the third highest of all nodes, respectively). The highest median degree of centrality was in the interpeduncular nucleus and the left pedunculopontine-cuneiform nuclei complex (the first and the second highest of all nodes, respectively).
In the DAN, the subcortical structures with the highest median betweenness centrality were the raphe pallidus nucleus and the right mediodorsal nucleus of the thalamus (the first and the seventh highest of all nodes, respectively). The highest median degree of centrality was the raphe pallidus nucleus and the left superior colliculus (the first and the second highest of all nodes, respectively).
Overall, the subcortical structures had high centrality values in both networks. The betweenness centrality and degree centrality values of all nodes in the VAN and the DAN are detailed in Supplementary Tables 3, 4. The anatomical models of the VAN and DAN are illustrated in Fig. 5b.
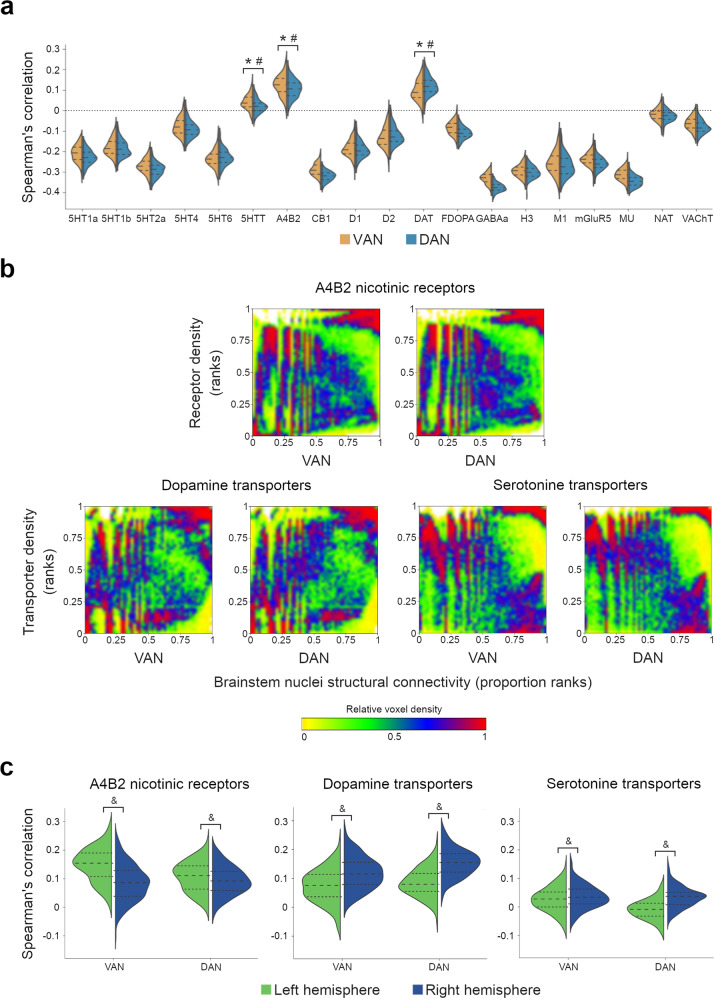
Correlation with the neurotransmitter system
The brainstem nuclei identified in the VAN anatomical map that synthesize neurotransmitters are the pedunculopontine nuclei (cholinergic, glutamatergic, and GABAergic; Benarroch72), the cuneiform nuclei (glutamatergic and GABAergic; Chang et al.73), the gigantocellular nucleus (glutamatergic and GABAergic; Martin et al.74), the raphe nucleus (serotonergic; Van De Kar and Lorens75), and the raphe pallidus nucleus (serotonergic; Heym et al.76). The brainstem nuclei identified in the DAN anatomical map synthesizing neurotransmitters are the pedunculopontine, the cuneiform, the gigantocellular nuclei, and the raphe pallidus nucleus.
The spatial correlations of these brainstem nuclei structural projections with the neurotransmitter systems are represented in Fig. 6. The distributions of acetylcholine α4β2 nicotinic receptors, dopamine transporters, and serotonin transporters were positively correlated with the distribution of VAN and DAN brainstem projections (p < 0.001; Fig. 6a). The scatterplots representing the distributions of the significantly correlated systems are presented in Fig. 6b. Acetylcholine α4β2 nicotinic receptors and serotonin transporters had a higher spatial correlation with the VAN than with the DAN, whereas dopamine transporters had a higher spatial correlation with the DAN (p < 0.001).
Fig. 6. Correlation between the structural projections of the brainstem nuclei and the neurotransmitter systems.
a Distributions of the Spearman’s correlations for the available maps of neurotransmitter receptors and transporters; for the receptors or transporters with two or more maps available, the mean correlation was calculated. Dashed lines represent the median and the interquartile range; the minimum and maximum correspond to the violin limits. b Graphical representation of the statistically significant positive correlations, i.e., the acetylcholine α4β2 nicotinic receptor, dopamine, and serotonin transporter maps. The color map represents the relative voxel density at each graph point. c Spearman’s correlation of the statistically significant positive correlations with the left and right hemispheres. Dashed lines represent the median and the interquartile range; the minimum and maximum correspond to the violin limits. 5HT1a serotonin 1a receptors, 5HT1b serotonin 1b receptors, 5HT2a serotonin 2a receptors, 5HTT serotonin transporters, A4B2 acetylcholine α4β2 nicotinic receptors, CB1 cannabinoid receptors 1, D1 dopamine receptors 1, D2 dopamine receptors 2, DAT dopamine transporters, FDOPA fluorodopa, GABAa GABAa receptors, H3 histamine receptors 3, M1 muscarinic receptors 1, mGluR5 metabotropic glutamate receptors 5, MU mu-opioid receptors, NAT noradrenaline transporters, VAchT vesicular acetylcholine transporters. * Statistically significant positive correlation, corrected for multiple comparisons (p < 0.003); # Statistically significant difference between the VAN and the DAN, corrected for multiple comparisons (p < 0.017); & Statistically significant difference between right and left hemispheres, corrected for multiple comparisons (p < 0.017); n = 177.
The supplemental pairwise correlation analyses between the average VAN and DAN structural projection maps and the neurotransmitter maps revealed similar results: the VAN had a significant positive spatial correlation with acetylcholine α4β2 nicotinic receptors and acetylcholine, dopamine, noradrenaline, and serotonin transporters (Supplementary Table 5); the DAN had a significant positive spatial correlation with acetylcholine α4β2 nicotinic receptors and acetylcholine, dopamine and noradrenaline transporters (Supplementary Table 6).
The correlation of VAN and DAN brainstem projections with the acetylcholine α4β2 nicotinic receptors was significantly higher in the left hemisphere. In contrast, the correlations with the dopamine and serotonin transporters were higher in the right hemisphere (Fig. 6c).
Discussion
This study re-examined the VAN and the DAN neuroanatomy by co-registering individual network maps in a common functional space. We propose a comprehensive model of these networks based on the convergence of functional, structural, and neurochemical findings. First, we confirmed the initial hypothesis that subcortical structures, namely the pulvinar, the superior colliculi, the head of caudate nuclei, and a group of brainstem nuclei, are constituent elements of the attentional networks. Second, we characterized the structural connections underlying functional connectivity. Deep brain nuclei are densely connected and structural network hubs. Third, we showed that the identified brainstem nuclei projections are spatially correlated with the acetylcholine α4β2 nicotinic receptors and serotonin and dopamine transporters.
Pulvinar is a high-order thalamic relay nucleus participating in cortical-thalamocortical circuits that modulate information processing77. Cytoarchitectonically, the pulvinar is divided into four regions: the anterior pulvinar, the inferior pulvinar, the medial pulvinar, and the lateral pulvinar78. The medial pulvinar is particularly important in establishing connections with heteromodal association areas, such as the superior and inferior temporal, the inferior parietal, the dorsolateral prefrontal, and the orbitofrontal cortices79. In our model, the pulvinar regions with the highest statistical level were medial, and we demonstrated that they were structurally connected with VAN cortical areas, through fronto-pulvinar projections, and with DAN cortical areas, by fronto-pulvinar and parieto-pulvinar projections80–82. Pulvinar lesions may induce hemispatial neglect39. Decades ago, Sprague impressively found that hemispherectomy prompted symptoms of hemispatial neglect in cats which were attenuated by removing the contralesional superior colliculus83,84. This effect was later observed in humans85. In our model, the pulvinar connects with the superior colliculi through the tecto-pulvinar fibers86, demonstrating the importance of pulvinar—superior colliculi interactions in attention processes. Therefore, in the context of the so-called Sprague effect, removing the contralesional superior colliculus in cats with hemispatial neglect would damage the spared attentional network and might partially compensate for the imbalance in the attentional processing87,88. Recently, hemispatial neglect was linked to lesions of the human superior colliculus40. The Sprague effect is also mediated by the pedunculopontine nuclei89,90, which is one of the brainstem nuclei included in our model. The pedunculopontine nuclei possess a population of cholinergic neurons in their caudal portion, giving rise to a distinct network that regulates attentional states and enhances the processing of salient stimuli91. The descending projections from these cholinergic neurons innervate the nucleus pontis oralis92 and the gigantocellular nuclei93, while their dorsal ascending projections innervate the colliculi94,95 and several nuclei of the thalamus, including the pulvinar and the mediodorsal nuclei96. The pattern of the pedunculopontine projections closely matches the brainstem and thalamic map evidenced in our analysis. Hence, lesion analyses and axonal tracings studies confirm the validity of our subcortical model of the VAN and the DAN.
The graph theory analysis results are consistent with the subcortical nuclei hub role in the VAN and the DAN organization. Centrality measures indicate how connected a node is with other nodes. These measures are considered surrogates of the node’s relevance for the flow of information and communication within a network97,98. The DAN and the VAN subcortical nuclei had a high degree and betweenness centrality scores, positioning them as networks’ core regions as previously suggested99–101.
The neurotransmitter system correlation analysis reinforced the proposed relationship between the subcortical nuclei of the attention networks. The highest spatial correlation of both networks was with the acetylcholine α4β2 nicotinic receptors. The acetylcholine α4β2 nicotinic receptors have a well-established relationship with sustained attention. Acetylcholine α4β2 nicotinic receptors agonists reduce adult monkey distractibility during matching-to-sample tasks with distractors102 and increase the firing rate of dorsolateral prefrontal neurons during sustained attention tasks, an effect that is reversed by the co-administration of receptor antagonists103. In humans, transdermal nicotine administration improves attentiveness104,105. All these observations in animals and humans support the critical role of the subcortical acetylcholinergic system in attentional processes.
The VAN and DAN brainstem nuclei projections were also spatially correlated with the distribution of dopamine and serotonin transporters. This finding is consistent with the psychopharmacological knowledge about attention. Methylphenidate is the first-line treatment for attention deficit hyperactivity disorder106. Pharmacologically, it is a noradrenaline-dopamine reuptake inhibitor with higher potency for dopamine transporters107,108. Modafinil is a selective inhibitor of dopamine transporters109 and produces attention enhancement effects110,111. Further studies are needed to understand how the interplay between the nicotinic acetylcholine and the dopamine systems occurs in attention networks, but it might be mediated by their interaction at the levels of the striatum112,113 and midbrain114,115. Serotonin reuptake inhibitors also modulate attentional processes116. They increase the perceptual bias towards emotional stimuli117,118 by regulating the activity of visual processing circuits116. Therefore, our improved model of the DAN and VAN functional neuroanatomy appears to reconcile previous neuroimaging and pharmacological findings. As previously suggested8, additional pharmacological studies will be required to understand the preferential association of VAN with acetylcholine α4β2 nicotinic receptors. Similarly, pharmacological studies are required to shed light on the effect of serotonin transporter on the VAN and to reveal the relationship between dopamine transporters and the DAN. Finally, understanding the relationship between the neurochemical signature and hemispheric functional dominance still requires more research in animals and humans8.
Characterizing the human brain’s subcortical anatomy of attention networks fosters the exploration of a common structural-functional attentional framework across species. Attention is far from being a specific cognitive ability of human beings119. Species with either close or distant common ancestors in the phylogenetic tree, such as monkeys, rats, and pigeons, can scan, select and maintain attention to surrounding environmental stimuli42,119–121. A common subcortical attention framework may surpass the challenge of finding the cortical homologs of the human VAN and DAN in other species43. Accordingly, future studies might use the subcortical areas we highlighted to explore comparatively the organization of the VAN and the DAN in non-human species.
In our analysis, VAN and DAN structural connectivity maps were right-lateralized. The right lateralization of the VAN is established in the literature. Evidence demonstrates that the SLF III has a larger volume in the right hemisphere and that its anatomical lateralization correlates with visuomotor processing abilities and the asymmetries of visuospatial task performance7,9,122–125. The SLF I, the main tract connecting DAN cortical regions, does not show a preferential lateralization7,9. However, some DAN areas might be right-lateralized126. The right intraparietal sulcus127 and frontal eye field128 increase their activity for both visual fields, while the left preferentially reacts to contralateral stimulations. The processing of both visual fields in the right hemisphere is corroborated by right hemisphere stroke patients with hemispatial neglect who also present with deficits in goal-driven selective attention for ipsilateral stimuli129. Hence, while the cortical extent of the DAN was not asymmetrical, our structural connectivity analysis, including the cortico-subcortical projection tracts, might have the function-specific dimension of the right lateralization of the DAN.
Regarding functional connectivity, the distribution of VAN was not different between hemispheres, and the DAN was slightly left-lateralized. Task-based fMRI studies indicate right lateralization of the VAN10,11, but the asymmetry might vary according to the nature of the task130. Accordingly, while functional asymmetry is expected for some task-related activations131, resting-state functional connectivity may not capture function-specific asymmetries due to its global nature.
A limitation of our study is the inability to untangle the different roles and dynamic interactions between the proposed subcortical structures. While the cortical regions of the DAN and the VAN are quite neatly segregated132, the subcortical nuclei described in our model probably contributed to both the VAN and the DAN. Future investigations using our model to explore the BOLD signal during task-related fMRI in humans or direct electrical recordings in animals might better dissociate the hierarchical organization and functional role of subcortical regions than resting-state fMRI. In addition, the neurotransmitter systems normative atlas is derived from different samples71. As PET and SPECT tracers are radioactive, it is not possible to map several neurotransmitter systems in the same participants. Although the atlas was replicated in an independent autoradiography dataset and all scans were acquired in healthy volunteers71, the heterogeneity of the data sources may represent a limitation for its interpretation.
In conclusion, this work proposes an improved neuroanatomical model of the VAN and the DAN that includes the pulvinar, the superior colliculi, the head of caudate nuclei, and a group of brainstem nuclei interrelated with the acetylcholine nicotinic and the dopamine and serotonin transporter systems. This comprehensive framework reconciles behavioral, electrophysiological, and psychopharmacological data and provides a shared foundation to explore the neural basis of attention across different species and brain pathologies.
Methods
Resting-state functional imaging (rs-fMRI)
We used 110 7 T resting-state functional MRI datasets from the Human Connectome Project S1200133. Images were preprocessed and registered to the MNI152 space as specified in the Human Connectome Project protocol (http://www.humanconnectome.org/storage/app/media/documentation/s1200/HCP_S1200_Release_Reference_Manual.pdf; Glasser et al. 2013). The Human Connectome Project open access data use terms were followed.
VAN and DAN maps in the structural space
VAN and DAN maps were computed using seed regions of interest defined in the functional cortical parcellation map14. This template includes 23 VAN parcels (11 in the left and 12 in the right hemisphere) and 32 DAN parcels (19 in the left and 13 in the right hemisphere). This parcellation was performed according to resting-state functional connectivity patterns. Each parcel has a homogeneous resting-state functional connectivity signature and is separated from neighboring parcels by abrupt changes in their connectivity profile14.
We calculated functional correlation maps seeded from each VAN cortical parcel using the Funcon-Connectivity tool implemented in the Brain Connectivity and Behavior toolkit (http://toolkit.bcblab.com)134. This tool computes Pearson’s correlation between a seed region’s mean resting-state activity and the brain’s other voxels. Then, the median of the 23 functional connectivity maps (generated from the 23 VAN seeds) was computed to obtain the VAN’s most representative map for each participant. We chose a median because it is less affected by outliers than the mean135. 110 individual VAN maps in the MNI152 were obtained (i.e., one per subject). The same steps were performed to obtain 32 DAN maps.
VAN and DAN maps in the functional space
The 110 individual VAN Pearson’s correlation maps in the MNI152 space were aligned in a functional space to optimize their interindividual alignment of functional areas50–53. We used the Advanced Normalization Tools (ANTs) script “buildtemplateparallel.sh” to perform an iterative (n = 4) diffeomorphic transformation to a common space62,136. Cross-correlation was set as the similarity measure and greedy SyN as the transformation model137,138. The resulting transformation warps were applied to the MNI152 aligned VAN maps, using the ANTs’ script “WarpImageMultiTransform” to represent the 110 individual VAN maps in the functional space. The same steps were performed with the 110 DAN Pearson’s correlations maps. A schematic representation of the functional alignment steps is available in Supplementary Fig. 1.
To calculate group statistical VAN and DAN maps, we performed a permutation inference analysis using FSL’s “randomise” one-sample (5000 permutations) and applied a threshold-free cluster enhancement139. To evaluate the similarity between the VAN and DAN statistical maps, the t-maps were z-transformed, and a conjunction analysis was computed140. A difference map was also calculated by subtracting the median DAN Pearson’s correlation map from the VAN. Illustrations were produced in SurfIce (https://www.nitrc.org/projects/surfice/) and MRIcroGL (https://www.nitrc.org/projects/mricrogl/).
Anatomical validation of the subcortical structures
To identify thalamic nuclei, we visually compared our results with the DISTAL (Deep brain stimulation Intrinsic Template Atlas; Ewert et al. 2018) and the THOMAS (Thalamus Optimized Multi Atlas Segmentation; Su et al. 2019) atlases. The DISTAL atlas is a high-resolution template of subcortical structures in the MNI space used as a reference to localize targets for deep brain stimulation141. The DISTAL atlas segmentation was performed manually, based on histology, structural imaging, and diffusion-weighted imaging141,143. The THOMAS atlas is a template of thalamic nuclei derived from the manual segmentation of 20 White-Matter-Nulled Magnetization Prepared Rapid Gradient Echo (MP-RAGE) 7 T datasets warped to the MNI space142. We used the WIKIBrainStem atlas to identify the brainstem nuclei144. This template is based on mesoscopic T2-weighted and diffusion-weighted images obtained from the ultra-high-field scanning (11.7 T) of an ex vivo human specimen. It provides detailed segmentations of 99 brainstem structures144.
Tractography analysis
We analyzed the structural connectivity of the VAN and DAN, including the new subcortical structures identified in our resting-state functional connectivity analysis. Tractography was computed using 177 diffusion-weighted images from the 7 T dataset of the Human Connectome Project145. The scanning parameters are detailed in Vu et al.145. Preprocessing was performed according to the default Human Connectome Project pipeline (v3.19.0)133. The Human Connectome Project open access data use terms were followed. Tractography processing was prepared as described in Thiebaut de Schotten et al.67 (available at http://opendata.bcblab.com). Briefly, a whole-brain deterministic algorithm was employed using StarTrack (https://mr-startrack.com), applying a damped Richardson-Lucy algorithm optimized for spherical deconvolution146. Then, the individual whole-brain streamline tractograms were registered to the MNI152 space. First, they were converted into density maps, in which the voxel densities corresponded to the number of streamlines crossing each voxel67. Second, individual density maps were aligned to a standard template using the Greedy symmetric diffeomorphic normalization of the Advanced Normalization Tools pipeline136. Third, the resulting template was co-registered to the MNI152 2 mm template using the FSL’s tool “flirt”147. Finally, the resulting transformation warps were applied to the individual whole-brain streamline tractography using Tract Querier148.
Then, we computed the structural connectome of the VAN and DAN models. The cortical nodes were defined according to Gordon et al.14. To determine the subcortical regions of interest, we selected the statistically significant voxels of the subcortical structures identified in the previous sections with a median Pearson’s correlation above r = 0.1. This correlation threshold was applied to avoid including voxels significantly associated with the network but with weak correlations149. The streamlines that crossed at least two ROIs (cortico-cortical, cortico-subcortical, or subcortical-subcortical) were selected using the MRtrix3’s tool “tckedit”150. Afterward, the selected streamlines were converted into streamline density maps using the MRtrix3’s tool ‘tckmap’150. The streamline density maps were binarized, and a group-level overlap map was computed.
ROI-to-ROI structural and functional connectivity analysis
We used MRtrix3’s tool “tck2connectome” to analyze ROI-to-ROI structural connectivity. The cortical and subcortical ROIs were defined as stated in the previous section. Regarding ROI-to-ROI functional connectivity, we computed the partial correlation between the network nodes using the nilearn’s function “ConnectivityMeasure”151. The illustrations of the connectivity matrices were created with Matplotlib 3.4.2152.
Networks lateralization
We assessed the lateralization of the VAN and DAN networks. For functional connectivity, the average Pearson’s correlation across each hemisphere’s VAN and DAN nodes was calculated using the FSL’s function “fslmeants”. The obtained values were compared between the right and left hemispheres. For structural connectivity, we extracted the fiber tracts that crossed two nodes of the same hemisphere. Then, the fiber tracts were converted into volume maps using the MRtrix3’s tool “tckmap”, and the individual volumes were compared between the two hemispheres150. Data were presented as mean (with standard deviations) or median (with interquartile ranges), and paired analyses were performed with paired t-test or Wilcoxon test, according to their distribution.
Graph theory analysis of structural connectivity
To analyze if the newly identified subcortical nuclei would be core regions in the networks, we performed a graph theory analysis of the hub properties of the VAN and DAN nodes. Two measures were used, the degree centrality and the betweenness centrality97. Degree centrality denotes the fraction of nodes connected to the node of interest. Betweenness centrality is the fraction of all-pairs shortest paths that pass through the node of interest97. In graph theory, nodes with high centrality are considered network hubs, i.e., they play a crucial role in the global network function153.
The 177 individual binarized structural connectivity matrices were converted into undirected connectivity graphs, and both measures were calculated using the NetworkX package (https://networkx.org/). ROIs, as defined in the previous sections, constituted the network nodes. The streamlines that crossed at least two ROIs defined network vertices. Considering the conservative parameters of our tractography adjusted over the years to match post-mortem Klingler dissections154–157, there was no threshold for the streamline considered for binarization. Additionally, the streamline count does not accurately reflect the number of axonal projections between regions or the strength of connectivity99,158, and previous work showed that the overall results of the network analysis do not change with modifications in the streamline count binarization threshold159. Then, we calculated the median value of both measures across the 177 network graphs for each node. The illustrations of the network graphs were created with SurfIce (https://www.nitrc.org/projects/surfice/).
Structural correlations with the neurotransmitter system
We studied the relationship between the proposed neuroanatomical models’ subcortical structural projections and the neurotransmitter systems’ spatial distribution. First, we selected the newly identified brainstem nuclei that synthesize neurotransmitters, according to the cytochemical evidence in the literature. Second, we computed the structural projections of these nuclei to the remaining nodes of the VAN and DAN, i.e., we selected the streamlines that crossed the brainstem nuclei of interest and every other node of the network, using the MRtrix3 tool “tckedit”150. Then, we used the MRtrix3 tool “tckmap” to map those streamlines into the MNI space150 and computed the individual Spearman’s correlation between the spatial distribution of the created structural projection map and the neurotransmitter maps provided by Hansen and colleagues using the neuromaps’ tool “compare_images”71,160; https://netneurolab.github.io/neuromaps/). We obtained the correlation values distribution between the 110 individual VAN and DAN maps and each neurotransmitter map. To analyze if the obtained distributions (each composed of 110 correlation values) were significantly higher than zero, a non-parametric statistical test was performed (one-sided Wilcoxon test). The obtained p values were corrected for multiple comparisons using the Bonferroni correction. Finally, we analyzed whether the correlation distributions were different between VAN and DAN, and if they were different between hemispheres (paired t-test or Wilcoxon test, according to data distribution; the Bonferroni correction was also applied). A supplemental pairwise analysis was performed. The average map of the 110 individual VAN and DAN structural projection maps was correlated with the neurotransmitter maps (Spearman’s correlation; neuromaps’ tool “compare_images”; https://netneurolab.github.io/neuromaps/)160. To control for spatial autocorrelations and reduce the risk of false positive results, statistical significance was inferred based on null models generation161–163. Volumetric data were parcellated according to the Automated Anatomical Labeling atlas 3 (AAL3; Rolls et al.164), using the neuromaps’ utility “Parcellater” (https://netneurolab.github.io/neuromaps/)160. AAL3 was chosen because it includes cortical and subcortical parcels. The null parcellations were generated from the average VAN and DAN structural projection maps using the neuromaps’ function “nulls.burt2020” (5000 permutations, generating 5000 null parcellations; https://netneurolab.github.io/neuromaps/)160,162. The graphical representations were created with Matplotlib 3.4.2 and Datashader 0.13.0 (Hunter152; https://datashader.org).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Data
Acknowledgements
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No. 818521; M.T.d.S.), the Marie Skłodowska-Curie program (grant agreement No. 101028551; S.J.F.), a Donders Mohrmann Fellowship (S.J.F., No. 2401512), “Prémio João Lobo Antunes 2018”—SCML (P.N.A.), and “Bolsa de Investigação em Doenças Vasculares Cerebrais 2017”—SPAVC (P.N.A.). Additional funding comes from the University of Bordeaux’s IdEx “Investments for the Future” program RRI “IMPACT,” which received financial support from the French government. M.C. was supported by MIUR—Departments of Excellence Italian Ministry of Research (MART_ECCELLENZA18_01); Fondazione Cassa di Risparmio di Padova e Rovigo (CARIPARO)(Grant Agreement number 55403); Ministry of Health Italy NEUROCONN (RF-2008 -12366899); Celeghin Foundation Padova (CUP C94I20000420007); BIAL foundation grant (No. 361/18); H2020 European School of Network Neuroscience-euSNN, H2020-SC5-2019-2, (Grant Agreement number 869505); H2020 VARCITIES, H2020-SC5-2019-2 (Grant Agreement number 869505); Ministry of Health Italy: EYEMOVINSTROKE (RF-2019-12369300).
Author contributions
P.N.A. implemented part of the methods, performed the analyses, contributed conceptually, and wrote the manuscript. S.J.F. and M.C. contributed conceptually and edited the paper. M.T.d.S. conceived and coordinated the study, reviewed the neuroimaging data, wrote the manuscript, and provided funding.
Peer review
Peer review information
Communications Biology thanks Sandra Hanekamp, Boris Bernhardt and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: George Inglis. Peer reviewer reports are available.
Data availability
The presented brain maps are openly available at https://neurovault.org/collections/XONZLGPJ/, the resting-state functional 7 T MRI datasets in the Human Connectome Project S1200 dataset, and the processed tractographies at http://opendata.bcblab.com. Data were provided by the McDonnell Center for Systems Neuroscience at Washington University. All other data were available from the corresponding author on reasonable request.
Code availability
Analyses were conducted using open software and toolboxes, as specified in the methods. The Funcon-Connectivity code is openly available at https://github.com/chrisfoulon/BCBToolKit; the ANTs scripts “buildtemplateparallel.sh” and “WarpImageMultiTransform” at https://github.com/ANTsX/ANTs; the code where these scripts were applied for the functional alignment performed in this work at https://github.com/Pedro-N-Alves/VAN_DAN_functional_alignment (doi: 10.5281/zenodo.7307027); the “easythresh_conj.sh” code (used for the conjunction analysis) at https://warwick.ac.uk/fac/sci/statistics/staff/academic-research/nichols/; the “tckedit”, “tckmap”, and “tck2connectome” commands’ codes at https://github.com/MRtrix3/mrtrix3; the “ConnectivityMeasure” script at https://github.com/nilearn/nilearn/; the “betweenness_centrality” and “degree_centrality” scripts at https://github.com/networkx/networkx; and the “compare_images”, “Parcellater”, and “nulls.burt202” scripts at https://github.com/netneurolab/neuromaps.
Competing interests
Michel Thiebaut de Schotten is an Editorial Board Member for Communications Biology, but was not involved in the editorial review of, nor the decision to publish this article. The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pedro Nascimento Alves, Email: pedronascimentoalves@gmail.com.
Michel Thiebaut de Schotten, Email: michel.thiebaut@gmail.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-04281-0.
References
- 1.James, W. The Principles of Psychology (Henry Holt and Company, 1890).
- 2.Posner MI. Orienting of attention. Q. J. Exp. Psychol. 1980;32:3–25. doi: 10.1080/00335558008248231. [DOI] [PubMed] [Google Scholar]
- 3.Petersen SE, Posner MI. The attention system of the human brain: 20 years after. Annu. Rev. Neurosci. 2012;35:73–89. doi: 10.1146/annurev-neuro-062111-150525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbetta M, Shulman GL. Control of goal-directed and stimulus-driven attention in the brain. Nat. Rev. Neurosci. 2002;3:201–215. doi: 10.1038/nrn755. [DOI] [PubMed] [Google Scholar]
- 5.Corbetta, M., Kincade, J. M., Ollinger, J. M., Mcavoy, M. P. & Gordon, L. Voluntary orienting is dissociated from target detection in human posterior parietal cortex. Nat. Neurosci. 3, 292–297 (2000). [DOI] [PubMed]
- 6.Buschman TJ, Miller EK. Top-down versus bottom-up control of attention in the prefrontal and posterior parietal cortices. Science. 2007;315:1860–1864. doi: 10.1126/science.1138071. [DOI] [PubMed] [Google Scholar]
- 7.Thiebaut de Schotten M, et al. A lateralized brain network for visuospatial attention. Nat. Neurosci. 2011;14:1245–1247. doi: 10.1038/nn.2905. [DOI] [PubMed] [Google Scholar]
- 8.Corbetta M, Patel G, Shulman GL. The reorienting system of the human brain: from environment to theory of mind. Neuron. 2008;58:306–324. doi: 10.1016/j.neuron.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amemiya K, Naito E, Takemura H. Age dependency and lateralization in the three branches of the human superior longitudinal fasciculus. Cortex. 2021;139:116–133. doi: 10.1016/j.cortex.2021.02.027. [DOI] [PubMed] [Google Scholar]
- 10.Downar J, Crawley AP, Mikulis DJ, Davis KD. A multimodal cortical network for the detection of changes in the sensory environment. Nat. Neurosci. 2000;3:277–283. doi: 10.1038/72991. [DOI] [PubMed] [Google Scholar]
- 11.Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME. Spontaneous neuronal activity distinguishes human dorsal and ventral attention systems. Proc. Natl Acad. Sci. USA. 2006;103:10046–10051. doi: 10.1073/pnas.0604187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vossel S, Weidner R, Driver J, Friston KJ, Fink GR. Deconstructing the architecture of dorsal and ventral attention systems with dynamic causal modeling. J. Neurosci. 2012;32:10637–10648. doi: 10.1523/JNEUROSCI.0414-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szczepanski SM, Pinsk MA, Douglas MM, Kastner S, Saalmann YB. Functional and structural architecture of the human dorsal frontoparietal attention network. Proc. Natl Acad. Sci. USA. 2013;110:15806–15811. doi: 10.1073/pnas.1313903110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon EM, et al. Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb. Cortex. 2016;26:288–303. doi: 10.1093/cercor/bhu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sani I, et al. The human endogenous attentional control network includes a ventro-temporal cortical node. Nat. Commun. 2021;12:1–16. doi: 10.1038/s41467-020-20583-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Power JD, et al. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeo BTT, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaefer A, et al. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb. Cortex. 2018;28:3095–3114. doi: 10.1093/cercor/bhx179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uddin LQ, Yeo BTT, Spreng RN. Towards a universal taxonomy of macro-scale functional human brain networks. Brain Topogr. 2019;32:926–942. doi: 10.1007/s10548-019-00744-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eickhoff SB, Yeo BTT, Genon S. Imaging-based parcellations of the human brain. Nat. Rev. Neurosci. 2018;19:672–686. doi: 10.1038/s41583-018-0071-7. [DOI] [PubMed] [Google Scholar]
- 21.Corbetta, M. & Shulman, G. L. Spatial neglect and attention networks. Ann. Rev. Neurosci.34, 569–599 (2011). [DOI] [PMC free article] [PubMed]
- 22.Luo L, et al. Ten years of nature reviews neuroscience: insights from the highly cited. Nat. Rev. Neurosci. 2010;11:718–726. doi: 10.1038/nrn2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corbetta M, Kincade MJ, Lewis C, Snyder AZ, Sapir A. Neural basis and recovery of spatial attention deficits in spatial neglect. Nat. Neurosci. 2005;8:1603–1610. doi: 10.1038/nn1574. [DOI] [PubMed] [Google Scholar]
- 24.He BJ, et al. Breakdown of functional connectivity in frontoparietal networks underlies behavioral deficits in spatial neglect. Neuron. 2007;53:905–918. doi: 10.1016/j.neuron.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Ptak R, Schnider A. The dorsal attention network mediates orienting toward behaviorally relevant stimuli in spatial neglect. J. Neurosci. 2010;30:12557–12565. doi: 10.1523/JNEUROSCI.2722-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li R, et al. Attention-related networks in Alzheimer’s disease: a resting functional MRI study. Hum. Brain Mapp. 2012;33:1076–1088. doi: 10.1002/hbm.21269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarthy H, et al. Attention network hypoconnectivity with default and affective network hyperconnectivity in adults diagnosed with attention-deficit/hyperactivity disorder in childhood. JAMA Psychiatry. 2013;70:1329–1337. doi: 10.1001/jamapsychiatry.2013.2174. [DOI] [PubMed] [Google Scholar]
- 28.Sanefuji M, et al. Double-dissociation between the mechanism leading to impulsivity and inattention in attention deficit hyperactivity disorder: a resting-state functional connectivity study. Cortex. 2017;86:290–302. doi: 10.1016/j.cortex.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Baldassarre A, et al. Large-scale changes in network interactions as a physiological signature of spatial neglect. Brain. 2014;137:3267–3283. doi: 10.1093/brain/awu297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramsey LE, et al. Normalization of network connectivity in hemispatial neglect recovery. Ann. Neurol. 2016;80:127–141. doi: 10.1002/ana.24690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zénon A, Krauzlis RJ. Attention deficits without cortical neuronal deficits. Nature. 2012;489:434–437. doi: 10.1038/nature11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saalmann YB, Kastner S. Cognitive and perceptual functions of the visual thalamus. Neuron. 2011;71:209–223. doi: 10.1016/j.neuron.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bender DB, Youakim M. Effect of attentive fixation in macaque thalamus and cortex. J. Neurophysiol. 2001;85:219–234. doi: 10.1152/jn.2001.85.1.219. [DOI] [PubMed] [Google Scholar]
- 34.Aston-Jones G, Rajkowski J, Cohen J. Role of locus coeruleus in attention and behavioral flexibility. Biol. Psychiatry. 1999;46:1309–1320. doi: 10.1016/S0006-3223(99)00140-7. [DOI] [PubMed] [Google Scholar]
- 35.Vazey EM, Moorman DE, Aston-Jones G. Phasic locus coeruleus activity regulates cortical encoding of salience information. Proc. Natl Acad. Sci. U. S. A. 2018;115:E9439–E9448. doi: 10.1073/pnas.1803716115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartolomeo, P. Attention Disorders After Right Brain Damage: Living in Halved Worlds (Springer, 2013).
- 37.Healton EB, Navarro C, Bressman S, Brust JCM. Subcortical neglect. Neurology. 1982;32:776–778. doi: 10.1212/WNL.32.7.776. [DOI] [PubMed] [Google Scholar]
- 38.Ferro JM, Martins IP, Távora L. Neglect in children. Ann. Neurol. 1984;15:281–284. doi: 10.1002/ana.410150314. [DOI] [PubMed] [Google Scholar]
- 39.Karnath HO, Himmelbach M, Rorden C. The subcortical anatomy of human spatial neglect: putamen, caudate nucleus and pulvinar. Brain. 2002;125:350–360. doi: 10.1093/brain/awf032. [DOI] [PubMed] [Google Scholar]
- 40.Nyffeler T, Kaufmann BC, Cazzoli D. Visual neglect after an isolated lesion of the superior colliculus. JAMA Neurol. 2021;78:1531–1533. doi: 10.1001/jamaneurol.2021.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov, I. et al. Morphological abnormalities of the thalamus in youths with attention deficit hyperactivity disorder. Am. J. Psychiatry167, 397–408 (2010). [DOI] [PMC free article] [PubMed]
- 42.Blough DS. Visual search in the pigeon: Hunt and peck method. Science. 1977;196:1013–1014. doi: 10.1126/science.860129. [DOI] [PubMed] [Google Scholar]
- 43.Patel GH, et al. Functional evolution of new and expanded attention networks in humans. Proc. Natl Acad. Sci. USA. 2015;112:E5377–E5377. doi: 10.1073/pnas.1420395112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brett M, Johnsrude IS, Owen AM. The problem of functional localization in the human brain. Nat. Rev. Neurosci. 2002;3:243–249. doi: 10.1038/nrn756. [DOI] [PubMed] [Google Scholar]
- 45.Thiebaut de Schotten M, Shallice T. Identical, similar or different? Is a single brain model sufficient? Cortex. 2017;86:172–175. doi: 10.1016/j.cortex.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Carmack PS, et al. Improved agreement between Talairach and MNI coordinate spaces in deep brain regions. Neuroimage. 2004;22:367–371. doi: 10.1016/j.neuroimage.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 47.Amunts K, et al. Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: Intersubject variability and probability maps. Anat. Embryol. 2005;210:343–352. doi: 10.1007/s00429-005-0025-5. [DOI] [PubMed] [Google Scholar]
- 48.Zaborszky L, et al. Stereotaxic probabilistic maps of the magnocellular cell groups in human basal forebrain. Neuroimage. 2008;42:1127–1141. doi: 10.1016/j.neuroimage.2008.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amunts K, et al. BigBrain: an ultrahigh-resolution 3D human brain model. Science. 2013;340:1472–1475. doi: 10.1126/science.1235381. [DOI] [PubMed] [Google Scholar]
- 50.Mueller S, et al. Individual variability in functional connectivity architecture of the human brain. Neuron. 2013;77:586–595. doi: 10.1016/j.neuron.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson EC, et al. MSM: a new flexible framework for multimodal surface matching. Neuroimage. 2014;100:414–426. doi: 10.1016/j.neuroimage.2014.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langs G, Golland P, Ghosh S. Predicting activation across individuals with resting-state functional connectivity based multi-atlas label fusion. Med. Image. Comput. Comput. Assist. Interv. 2015;9350:313–320. doi: 10.1007/978-3-319-24571-3_38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glasser MF, et al. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536:171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fischl B, et al. Cortical folding patterns and predicting cytoarchitecture. Cereb. Cortex. 2008;18:1973–1980. doi: 10.1093/cercor/bhm225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabuncu MR, et al. Function-based intersubject alignment of human cortical anatomy. Cereb. Cortex. 2010;20:130–140. doi: 10.1093/cercor/bhp085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conroy BR, Singer BD, Guntupalli JS, Ramadge PJ, Haxby JV. Inter-subject alignment of human cortical anatomy using functional connectivity. Neuroimage. 2013;81:400–411. doi: 10.1016/j.neuroimage.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nenning KH, et al. Joint embedding: a scalable alignment to compare individuals in a connectivity space. Neuroimage. 2020;222:117232. doi: 10.1016/j.neuroimage.2020.117232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu T, et al. Cross-species functional alignment reveals evolutionary hierarchy within the connectome. Neuroimage. 2020;223:117346. doi: 10.1016/j.neuroimage.2020.117346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haxby JV, Guntupalli JS, Nastase SA, Feilong M. Hyperalignment: modeling shared information encoded in idiosyncratic cortical topographies. Elife. 2020;9:1–26. doi: 10.7554/eLife.56601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haxby JV, et al. A common, high-dimensional model of the representational space in human ventral temporal cortex. Neuron. 2011;72:404–416. doi: 10.1016/j.neuron.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guntupalli JS, et al. A model of representational spaces in human cortex. Cereb. Cortex. 2016;26:2919–2934. doi: 10.1093/cercor/bhw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alves PN, et al. An improved neuroanatomical model of the default-mode network reconciles previous neuroimaging and neuropathological findings. Commun. Biol. 2019;2:370. doi: 10.1038/s42003-019-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Catani, M. & Thiebaut de Schotten, M. Atlas of Human Brain Connections (Oxford Univ. Press, 2012).
- 64.Zhang, F. et al. Quantitative mapping of the brain’s structural connectivity using diffusion MRI tractography: a review. Neuroimage249, 118870 (2022). [DOI] [PMC free article] [PubMed]
- 65.Behrens TEJ, et al. Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nat. Neurosci. 2003;6:750–757. doi: 10.1038/nn1075. [DOI] [PubMed] [Google Scholar]
- 66.Suárez L, Markello R, Betzel R, Misic B. Linking structure and function in macroscale brain. Trends Cogn. Sci. 2020;24:302–315. doi: 10.1016/j.tics.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 67.Thiebaut de Schotten M, Foulon C, Nachev P. Brain disconnections link structural connectivity with function and behaviour. Nat. Commun. 2020;11:5094. doi: 10.1038/s41467-020-18920-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morrison JH, Foote SL. Noradrenergic and serotoninergic innervation of cortical, thalamic, and tectal visual structures in old and new world monkeys. J. Comp. Neurol. 1986;243:117–138. doi: 10.1002/cne.902430110. [DOI] [PubMed] [Google Scholar]
- 69.Foote SL, Morrison JH. Extrathalamic modulation of cortical function. Annu. Rev. Neurosci. 1987;10:67–95. doi: 10.1146/annurev.ne.10.030187.000435. [DOI] [PubMed] [Google Scholar]
- 70.Bouret S, Sara SJ. Network reset: a simplified overarching theory of locus coeruleus noradrenaline function. Trends Neurosci. 2005;28:574–582. doi: 10.1016/j.tins.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 71.Hansen, J. Y. et al. Mapping neurotransmitter systems to the structural and functional organization of the human neocortex. Nat. Neurosci. 25, 1569–1581 (2022). [DOI] [PMC free article] [PubMed]
- 72.Benarroch EE. Pedunculopontine nucleus: functional organization and clinical implications. Neurology. 2013;80:1148–1155. doi: 10.1212/WNL.0b013e3182886a76. [DOI] [PubMed] [Google Scholar]
- 73.Chang SJ, Cajigas I, Opris I, Guest JD, Noga BR. Dissecting brainstem locomotor circuits: converging evidence for cuneiform nucleus stimulation. Front. Syst. Neurosci. 2020;14:1–8. doi: 10.3389/fnsys.2020.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin EM, et al. Molecular and neuroanatomical characterization of single neurons in the mouse medullary gigantocellular reticular nucleus. J. Comp. Neurol. 2011;519:2574–2593. doi: 10.1002/cne.22639. [DOI] [PubMed] [Google Scholar]
- 75.Van De Kar LD, Lorens SA. Differential serotonergic innervation of individual hypothalamic nuclei and other forebrain regions by the dorsal and median midbrain raphe nuclei. Brain Res. 1979;162:45–54. doi: 10.1016/0006-8993(79)90754-6. [DOI] [PubMed] [Google Scholar]
- 76.Heym J, Steinfels GF, Jacobs BL. Activity of serotonin-containing neurons in the nucleus raphe pallidus of freely moving cats. Brain Res. 1982;251:259–276. doi: 10.1016/0006-8993(82)90743-0. [DOI] [PubMed] [Google Scholar]
- 77.Sherman SM. The thalamus is more than just a relay. Curr. Opin. Neurobiol. 2007;17:417–422. doi: 10.1016/j.conb.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stepniewska I, Kaas JH. Architectonic subdivisions of the inferior pulvinar in New World and Old World monkeys. Vis. Neurosci. 1997;14:1043–1060. doi: 10.1017/S0952523800011767. [DOI] [PubMed] [Google Scholar]
- 79.Bridge H, Leopold DA, Bourne JA. Adaptive pulvinar circuitry supports visual cognition. Trends Cogn. Sci. 2016;20:146–157. doi: 10.1016/j.tics.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bos J, Benevento LA. Projections of the medial pulvinar to orbital cortex and frontal eye fields in the rhesus monkey (Macaca mulatta) Exp. Neurol. 1975;49:487–496. doi: 10.1016/0014-4886(75)90103-X. [DOI] [PubMed] [Google Scholar]
- 81.DeVito JL. A horseradish peroxidase-autoradiographic study of parietopulvinar connections in saimiri sciureus. Exp. Brain Res. 1978;32:581–590. doi: 10.1007/BF00239554. [DOI] [PubMed] [Google Scholar]
- 82.Lemaire JJ, et al. White matter anatomy of the human deep brain revisited with high resolution DTI fibre tracking. Neurochirurgie. 2011;57:52–67. doi: 10.1016/j.neuchi.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 83.Sprague JM. Interaction of cortex and superior colliculus in mediation of visually guided behavior in the cat. Science. 1966;153:1544–1547. doi: 10.1126/science.153.3743.1544. [DOI] [PubMed] [Google Scholar]
- 84.Krauzlis, R. J., Lovejoy, L. P. & Zénon, A. Superior colliculus and visual spatial attention. Annu. Rev. Neurosci.36, 165–182 (2013). [DOI] [PMC free article] [PubMed]
- 85.Weddell RA. Subcortical modulation of spatial attention including evidence that the Sprague effect extends to man. Brain Cogn. 2004;55:497–506. doi: 10.1016/j.bandc.2004.02.075. [DOI] [PubMed] [Google Scholar]
- 86.Luppino G, Matelli M, Carey RG, Fitzpatrick D, Diamond IT. New view of the organization of the pulvinar nucleus in Tupaia as revealed by tectopulvinar and pulvinar‐cortical projections. J. Comp. Neurol. 1988;273:67–86. doi: 10.1002/cne.902730107. [DOI] [PubMed] [Google Scholar]
- 87.Vuilleumier P, Hester D, Assal G, Regli F. Unilateral spatial neglect recovery after sequential strokes. Neurology. 1996;46:184–189. doi: 10.1212/WNL.46.1.184. [DOI] [PubMed] [Google Scholar]
- 88.Bartolomeo P, Thiebaut De Schotten M, Doricchi F. Left unilateral neglect as a disconnection syndrome. Cereb. Cortex. 2007;17:2479–2490. doi: 10.1093/cercor/bhl181. [DOI] [PubMed] [Google Scholar]
- 89.Durmer JS, Rosenquist AC. Ibotenic acid lesions in the pedunculopontine region result in recovery of visual orienting in the hemianopic cat. Neuroscience. 2001;106:765–781. doi: 10.1016/S0306-4522(01)00321-9. [DOI] [PubMed] [Google Scholar]
- 90.Valero-Cabré A, Toba MN, Hilgetag CC, Rushmore RJ. Perturbation-driven paradoxical facilitation of visuo-spatial function: revisiting the ‘Sprague effect’. Cortex. 2020;122:10–39. doi: 10.1016/j.cortex.2019.01.031. [DOI] [PubMed] [Google Scholar]
- 91.Mena-Segovia J, Bolam JP. Rethinking the pedunculopontine nucleus: from cellular organization to function. Neuron. 2017;94:7–18. doi: 10.1016/j.neuron.2017.02.027. [DOI] [PubMed] [Google Scholar]
- 92.Mena-Segovia J, Sims HM, Magill PJ, Bolam JP. Cholinergic brainstem neurons modulate cortical gamma activity during slow oscillations. J. Physiol. 2008;586:2947–2960. doi: 10.1113/jphysiol.2008.153874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martinez-Gonzalez C, Van Andel J, Bolam JP, Mena-Segovia J. Divergent motor projections from the pedunculopontine nucleus are differentially regulated in Parkinsonism. Brain Struct. Funct. 2014;219:1451–1462. doi: 10.1007/s00429-013-0579-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jeon C‐J, Spencer RF, Mize RR. Organization and synaptic connections of cholinergic fibers in the cat superior colliculus. J. Comp. Neurol. 1993;333:360–374. doi: 10.1002/cne.903330305. [DOI] [PubMed] [Google Scholar]
- 95.Motts SD, Schofield BR. Sources of cholinergic input to the inferior colliculus. Neuroscience. 2009;160:103–114. doi: 10.1016/j.neuroscience.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Steriade M, Paré D, Parent A, Smith Y. Projections of cholinergic and non-cholinergic neurons of the brainstem core to relay and associational thalamic nuclei in the cat and macaque monkey. Neuroscience. 1988;25:47–67. doi: 10.1016/0306-4522(88)90006-1. [DOI] [PubMed] [Google Scholar]
- 97.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 98.Girvan M, Newman MEJ. Community structure in social and biological networks. Proc. Natl Acad. Sci. USA. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gong G, et al. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb. Cortex. 2009;19:524–536. doi: 10.1093/cercor/bhn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hagmann P, et al. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6:1479–1493. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barabási A-L, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 102.Prendergast MA, et al. Central nicotinic receptor agonists ABT-418, ABT-089, and (–)-nicotine reduce distractibility in adult monkeys. Psychopharmacology. 1998;136:50–58. doi: 10.1007/s002130050538. [DOI] [PubMed] [Google Scholar]
- 103.Sun Y, et al. Nicotinic α4β2 cholinergic receptor influences on dorsolateral prefrontal cortical neuronal firing during a working memory task. J. Neurosci. 2017;37:5366–5377. doi: 10.1523/JNEUROSCI.0364-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Levin ED, et al. Transdermal nicotine effects on attention. Psychopharmacology. 1998;140:135–141. doi: 10.1007/s002130050750. [DOI] [PubMed] [Google Scholar]
- 105.Valentine G, Sofuoglu M. Cognitive effects of nicotine: recent progress. Curr. Neuropharmacol. 2017;16:403–414. doi: 10.2174/1570159X15666171103152136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cortese S, et al. Comparative efficacy and tolerability of medications for attention-deficit hyperactivity disorder in children, adolescents, and adults: a systematic review and network meta-analysis. Lancet Psychiatry. 2018;5:727–738. doi: 10.1016/S2215-0366(18)30269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gatley SJ, Pan D, Chen R, Chaturvedi G, Ding Y-S. Affinities of methylphenidate derivatives for dopamine, norepinephrine and serotonin transporters. Life Sci. 1996;58:231–239. doi: 10.1016/0024-3205(96)00052-5. [DOI] [PubMed] [Google Scholar]
- 108.Faraone SV. The pharmacology of amphetamine and methylphenidate: relevance to the neurobiology of attention-deficit/hyperactivity disorder and other psychiatric comorbidities. Neurosci. Biobehav. Rev. 2018;87:255–270. doi: 10.1016/j.neubiorev.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zolkowska D, et al. Evidence for the involvement of dopamine transporters in behavioral stimulant effects of modafinil. J. Pharmacol. Exp. Ther. 2009;329:738–746. doi: 10.1124/jpet.108.146142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Turner DC, Clark L, Dowson J, Robbins TW, Sahakian BJ. Modafinil improves cognition and response inhibition in adult attention-deficit/hyperactivity disorder. Biol. Psychiatry. 2004;55:1031–1040. doi: 10.1016/j.biopsych.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 111.Repantis D, Schlattmann P, Laisney O, Heuser I. Modafinil and methylphenidate for neuroenhancement in healthy individuals: a systematic review. Pharmacol. Res. 2010;62:187–206. doi: 10.1016/j.phrs.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 112.Zoli M, et al. Identification of the nicotinic receptor subtypes expressed on dopaminergic terminals in the rat striatum. J. Neurosci. 2002;22:8785–8789. doi: 10.1523/JNEUROSCI.22-20-08785.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Exley R, Cragg SJ. Presynaptic nicotinic receptors: a dynamic and diverse cholinergic filter of striatal dopamine neurotransmission. Br. J. Pharm. 2008;153:283–297. doi: 10.1038/sj.bjp.0707510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Forster GL, Blaha CD. Pedunculopontine tegmental stimulation evokes striatal dopamine efflux by activation of acetylcholine and glutamate receptors in the midbrain and pons of the rat. Eur. J. Neurosci. 2003;17:751–762. doi: 10.1046/j.1460-9568.2003.02511.x. [DOI] [PubMed] [Google Scholar]
- 115.Blaha CD, Winn P. Modulation of dopamine efflux in the striatum following cholinergic stimulation of the substantia nigra in intact and pedunculopontine tegmental nucleus-lesioned rats. J. Neurosci. 1993;13:1035–1044. doi: 10.1523/JNEUROSCI.13-03-01035.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Harmer, C. J. & Cowen, P. J. ’It’s the way that you look at it’-a cognitive neuropsychological account of SSRI action in depression. Philos. Trans. R. Soc. B Biol. Sci. 368, 20120407 (2013). [DOI] [PMC free article] [PubMed]
- 117.Browning M, Reid C, Cowen PJ, Goodwin GM, Harmer CJ. A single dose of citalopram increases fear recognition in healthy subjects. J. Psychopharmacol. 2007;21:684–690. doi: 10.1177/0269881106074062. [DOI] [PubMed] [Google Scholar]
- 118.Harmer CJ, Shelley NC, Cowen PJ, Goodwin GM. Increased positive versus negative affective perception and memory in healthy volunteers following selective serotonin and norepinephrine reuptake inhibition. Am. J. Psychiatry. 2004;161:1256–1263. doi: 10.1176/appi.ajp.161.7.1256. [DOI] [PubMed] [Google Scholar]
- 119.Washburn, D. A. & Taglialatela, L. A. in Comparative cognition: Experimental explorations of animal intelligence (eds. Wasserman, E. A. & Zentall, T. R.) Ch. 7 (Oxford Univ. Press, 2006).
- 120.Mackintosh NJ. Selective attention in animal discrimination learning. Psychol. Bull. 1965;64:124–150. doi: 10.1037/h0022347. [DOI] [PubMed] [Google Scholar]
- 121.Wasserman EA, Castro L. Assessing attention in category learning by animals. Curr. Dir. Psychol. Sci. 2021;30:495–502. doi: 10.1177/09637214211045686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Budisavljevic S, et al. Asymmetry and structure of the fronto-parietal networks underlie visuomotor processing in humans. Cereb. Cortex. 2017;27:1532–1544. doi: 10.1093/cercor/bhv348. [DOI] [PubMed] [Google Scholar]
- 123.Cazzoli D, Chechlacz M. A matter of hand: causal links between hand dominance, structural organization of fronto-parietal attention networks, and variability in behavioural responses to transcranial magnetic stimulation. Cortex. 2017;86:230–246. doi: 10.1016/j.cortex.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 124.Chechlacz M, Gillebert CR, Vangkilde SA, Petersen A, Humphreys GW. Structural variability within frontoparietal networks and individual differences in attentional functions: an approach using the theory of visual attention. J. Neurosci. 2015;35:10647–10658. doi: 10.1523/JNEUROSCI.0210-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Howells H, et al. Frontoparietal tracts linked to lateralized hand preference and manual specialization. Cereb. Cortex. 2018;28:1–13. doi: 10.1093/cercor/bhy040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bartolomeo P, Seidel Malkinson T. Hemispheric lateralization of attention processes in the human brain. Curr. Opin. Psychol. 2019;29:90–96. doi: 10.1016/j.copsyc.2018.12.023. [DOI] [PubMed] [Google Scholar]
- 127.Sheremata SL, Silver MA. Hemisphere-dependent attentional modulation of human parietal visual field representations. J. Neurosci. 2015;35:508–517. doi: 10.1523/JNEUROSCI.2378-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Szczepanski SM, Konen CS, Kastner S. Mechanisms of spatial attention control in frontal and parietal cortex. J. Neurosci. 2010;30:148–160. doi: 10.1523/JNEUROSCI.3862-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Snow JC, Mattingley JB. Goal-driven selective attention in patients with right hemisphere lesions: how intact is the ipsilesional field? Brain. 2006;129:168–181. doi: 10.1093/brain/awh690. [DOI] [PubMed] [Google Scholar]
- 130.Doricchi F, MacCi E, Silvetti M, MacAluso E. Neural correlates of the spatial and expectancy components of endogenous and stimulus-driven orienting of attention in the posner task. Cereb. Cortex. 2010;20:1574–1585. doi: 10.1093/cercor/bhp215. [DOI] [PubMed] [Google Scholar]
- 131.Shulman GL, et al. Right hemisphere dominance during spatial selective attention and target detection occurs outside the dorsal frontoparietal network. J. Neurosci. 2010;30:3640–3651. doi: 10.1523/JNEUROSCI.4085-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vossel S, Geng JJ, Fink GR. Dorsal and ventral attention systems: distinct neural circuits but collaborative roles. Neuroscientist. 2014;20:150–159. doi: 10.1177/1073858413494269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Glasser MF, et al. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Foulon C, et al. Advanced lesion symptom mapping analyses and implementation as BCBtoolkit. Gigascience. 2018;7:1–17. doi: 10.1093/gigascience/giy004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kenney, J. Mathematics of Statistics (Chapman & Hall, 1939).
- 136.Avants BB, et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Klein A, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jenkinson, M., Beckmann, C. F., Behrens, T. E. J., Woolrich, M. W. & Smith, S. M. Fsl. Neuroimage62, 782–790 (2012). [DOI] [PubMed]
- 140.Nichols T, Brett M, Andersson J, Wager T, Poline JB. Valid conjunction inference with the minimum statistic. Neuroimage. 2005;25:653–660. doi: 10.1016/j.neuroimage.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 141.Ewert S, et al. Toward defining deep brain stimulation targets in MNI space: a subcortical atlas based on multimodal MRI, histology and structural connectivity. Neuroimage. 2018;170:271–282. doi: 10.1016/j.neuroimage.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 142.Su JH, et al. Thalamus optimized multi atlas segmentation (THOMAS): fast, fully automated segmentation of thalamic nuclei from structural MRI. Neuroimage. 2019;194:272–282. doi: 10.1016/j.neuroimage.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chakravarty MM, Bertrand G, Hodge CP, Sadikot AF, Collins DL. The creation of a brain atlas for image guided neurosurgery using serial histological data. Neuroimage. 2006;30:359–376. doi: 10.1016/j.neuroimage.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 144.Lechanoine, F. et al. WIKIBrainStem: an online atlas to manually segment the human brainstem at the mesoscopic scale from ultrahigh field MRI. Neuroimage236, 118080 (2021). [DOI] [PubMed]
- 145.Vu AT, et al. High resolution whole brain diffusion imaging at 7T for the Human Connectome Project. Neuroimage. 2015;122:318–331. doi: 10.1016/j.neuroimage.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dell’Acqua F, et al. A modified damped Richardson-Lucy algorithm to reduce isotropic background effects in spherical deconvolution. Neuroimage. 2010;49:1446–1458. doi: 10.1016/j.neuroimage.2009.09.033. [DOI] [PubMed] [Google Scholar]
- 147.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1006/nimg.2002.1132. [DOI] [PubMed] [Google Scholar]
- 148.Wassermann D, et al. The white matter query language: a novel approach for describing human white matter anatomy. Brain Struct. Funct. 2016;221:4705–4721. doi: 10.1007/s00429-015-1179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cohen, J. Statistical Power Analysis for the Behavioural Sciences (Lawrence Erlbaum Associates, 1988).
- 150.Tournier, J. D. et al. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage202, 116137 (2019). [DOI] [PubMed]
- 151.Pedregosa F, et al. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 152.Hunter J. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 153.van den Heuvel MP, Sporns O. Network hubs in the human brain. Trends Cogn. Sci. 2013;17:683–696. doi: 10.1016/j.tics.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 154.Thiebaut de Schotten M, et al. Atlasing location, asymmetry and inter-subject variability of white matter tracts in the human brain with MR diffusion tractography. Neuroimage. 2011;54:49–59. doi: 10.1016/j.neuroimage.2010.07.055. [DOI] [PubMed] [Google Scholar]
- 155.Catani M, et al. Short frontal lobe connections of the human brain. Cortex. 2012;48:273–291. doi: 10.1016/j.cortex.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 156.Vergani F, Mahmood S, Morris CM, Mitchell P, Forkel SJ. Intralobar fibres of the occipital lobe: a post mortem dissection study. Cortex. 2014;56:145–156. doi: 10.1016/j.cortex.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 157.Catani, M. The anatomy of the human frontal lobe. Handb. Clin. Neurol. 163, 95–122 (2019). [DOI] [PubMed]
- 158.Jones DK, Knösche TR, Turner R. White matter integrity, fiber count, and other fallacies: the do’s and don’ts of diffusion MRI. Neuroimage. 2013;73:239–254. doi: 10.1016/j.neuroimage.2012.06.081. [DOI] [PubMed] [Google Scholar]
- 159.Shu N, et al. Diffusion tensor tractography reveals disrupted topological efficiency in white matter structural networks in multiple sclerosis. Cereb. Cortex. 2011;21:2565–2577. doi: 10.1093/cercor/bhr039. [DOI] [PubMed] [Google Scholar]
- 160.Markello, R. D. et al. Neuromaps: structural and functional interpretation of brain maps. Nat. Methods19, 1472–1479 (2022). [DOI] [PMC free article] [PubMed]
- 161.Markello RD, Misic B. Comparing spatial null models for brain maps. Neuroimage. 2021;236:118052. doi: 10.1016/j.neuroimage.2021.118052. [DOI] [PubMed] [Google Scholar]
- 162.Burt JB, Helmer M, Shinn M, Anticevic A, Murray JD. Generative modeling of brain maps with spatial autocorrelation. Neuroimage. 2020;220:117038. doi: 10.1016/j.neuroimage.2020.117038. [DOI] [PubMed] [Google Scholar]
- 163.Alexander-Bloch AF, et al. On testing for spatial correspondence between maps of human brain structure and function. Neuroimage. 2018;178:540–551. doi: 10.1016/j.neuroimage.2018.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rolls, E. T., Huang, C. C., Lin, C. P., Feng, J. & Joliot, M. Automated anatomical labelling atlas 3. Neuroimage206, 116189 (2020). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Data
Data Availability Statement
The presented brain maps are openly available at https://neurovault.org/collections/XONZLGPJ/, the resting-state functional 7 T MRI datasets in the Human Connectome Project S1200 dataset, and the processed tractographies at http://opendata.bcblab.com. Data were provided by the McDonnell Center for Systems Neuroscience at Washington University. All other data were available from the corresponding author on reasonable request.
Analyses were conducted using open software and toolboxes, as specified in the methods. The Funcon-Connectivity code is openly available at https://github.com/chrisfoulon/BCBToolKit; the ANTs scripts “buildtemplateparallel.sh” and “WarpImageMultiTransform” at https://github.com/ANTsX/ANTs; the code where these scripts were applied for the functional alignment performed in this work at https://github.com/Pedro-N-Alves/VAN_DAN_functional_alignment (doi: 10.5281/zenodo.7307027); the “easythresh_conj.sh” code (used for the conjunction analysis) at https://warwick.ac.uk/fac/sci/statistics/staff/academic-research/nichols/; the “tckedit”, “tckmap”, and “tck2connectome” commands’ codes at https://github.com/MRtrix3/mrtrix3; the “ConnectivityMeasure” script at https://github.com/nilearn/nilearn/; the “betweenness_centrality” and “degree_centrality” scripts at https://github.com/networkx/networkx; and the “compare_images”, “Parcellater”, and “nulls.burt202” scripts at https://github.com/netneurolab/neuromaps.