Abstract
Introduction:
Prior studies have shown inconsistent findings of association between depression and epigenetic aging. DNA methylation (DNAm) age acceleration can measure biological aging. We adopted a robust co-twin control study design to examine whether depression is associated with DNAm age acceleration after accounting for the potential confounding influences of genetics and family environment.
Methods:
We analyzed data on a sub-cohort of the Vietnam Era Twin Registry. A total of 291 twins participated at baseline and 177 at follow-up visit after a mean of 11.7 years, with 111 participants having DNA samples for both time points. Depression was measured using the Beck Depression Inventory II (BDI-II). Six measures of DNAm age acceleration were computed at each time point, including Horvath’s DNAm age acceleration (HorvathAA), intrinsic epigenetic age acceleration (IEAA), Hannum’s DNAm age acceleration (HannumAA), extrinsic epigenetic age acceleration (EEAA), GrimAge acceleration (GrimAA), and PhenoAge acceleration (PhenoAA). Mixed-effects modeling was used to assess the within-pair association between depression and DNAm age acceleration.
Results:
At baseline, a ten-unit higher BDI-II total score was associated with HannumAA (0.73 years, 95% CI 0.13–1.33, p=0.019) and EEAA (0.94 years, 95% CI 0.22–1.66, p=0.012). At follow-up, ten-unit higher BDI-II score was associated with PhenoAA (1.32 years, 95% CI 0.18–2.47, p=0.027).
Conclusion:
We identified that depression is associated with higher levels of DNAm age acceleration. Further investigation is warranted to better understand the underlying mechanisms for the potential causal relationship between depression and accelerated aging.
Keywords: Depression, Epigenetic Age Acceleration, Epigenetic Aging, DNA methylation
Introduction
Depression is a common and long-term disorder of high prevalence across many countries, with a lifetime prevalence of 19.2% in the US, and a median age of onset of 22.7 years(Kessler & Bromet, 2013). Worldwide, depression is currently affecting 350 million people(Smith, 2014), with common clinical symptoms such as fatigue, depressed mood, pessimism, low self-esteem and suicide ideation(Wong & Licinio, 2001). The underlying etiology is complex and may involve interactions between genetic susceptibility and environmental factors such as stressful events(Wong & Licinio, 2001).
DNA methylation (DNAm) is an epigenetic mechanism that cells use to regulate gene expression levels, and the epigenetic changes play a critical role in the aging process(Jones, Goodman, & Kobor, 2015; Xiao, Wang, & Kong, 2019). A number of epigenome-wide association studies have identified epigenetic loci that are indicators of biological aging(Bocklandt et al., 2011; Teschendorff et al., 2010). In recent years, measures of DNAm aging and age acceleration comparing biological to chronological aging have been widely used to measure the cumulative effect of age-related health decline(Horvath, 2013). Such “epigenetic clocks” are helpful for unpacking many chronic disease mechanisms such as cancer(Dugue et al., 2018) and cardiovascular disease(Roetker, Pankow, Bressler, Morrison, & Boerwinkle, 2018), but have also been linked to neuropsychopathology like cognitive decline and Alzheimer’s disease(McCartney et al., 2018; Vaccarino et al., 2021), and posttraumatic stress disorder (PTSD)(Wang et al., 2021).
Consistent findings have suggested that depressed patients have a shorter lifespan(Walker, McGee, & Druss, 2015; Zivin et al., 2012). Premature biological aging could be a possible mechanism of reduced life expectancy among depressed patients. Consistent with this hypothesis, some studies have suggested that depression may play a role in DNAm age acceleration, but the evidence remains mixed. Han et al. derived a DNAm age acceleration based on chronological age, which is a similar approach to the Horvath’s method, and reported that the individuals with depression had higher epigenetic aging in blood and brain tissue compared with controls without depression, with a dose response with severity of depression measured by the 30-item Inventory of Depressive Symptomatology(Han et al., 2018). In contrast, two other studies of epigenetic aging derived from the Horvath-based algorithm showed inconsistent findings. Li et al. provided evidence against an association between epigenetic aging acceleration and depression in a study of brain tissue(Li, He, Ma, & Chen, 2018). The other study reported an association of epigenetic age acceleration with reduced “positive affect” but no association with total depressive symptom scores measured by the 20-item Center of Epidemiological Studies-Depression(Beydoun et al., 2019). Findings from another recent study suggested greater age acceleration, measured with DNAm GrimAge (Lu et al., 2019), among untreated depressed patients compared to matched healthy controls(Protsenko et al., 2021).
Due to these inconsistent findings, we investigated the association between depression and DNAm age acceleration using a robust co-twin control study design. This design allowed us to examine the role of depression on age acceleration while controlling for shared genetic material and other potential confounding familial factors. Further, we explored the potential temporal relationships underlying such association under a longitudinal cross-lagged co-twin control framework.
Materials and Methods
Study Participants
The Vietnam Era Twin Registry is one of the largest national cohorts of adult twins in the United States. Monozygotic (MZ) and dizygotic (DZ) male-male twin pairs who served in the military during the Vietnam conflict in 1964–1975 were included in the Registry. The study design and enrollment details were previously described in detail (Forsberg et al., 2020). Participants included in the present study were from the Emory Twin Study, which is a subset of the Vietnam Era Twin Registry. Participants had a baseline in-person visit in 2002–2010(Vaccarino et al., 2013) and another follow-up visit after a mean of 11.7 years, with health questionnaires completed and blood samples collected at both time points. Samples of whole blood were returned to the Vietnam Era Twin Registry overnight, and DNA was extracted and banked for future research purposes. For the present study, we analyzed baseline data on 291 participants with both DNAm and depression data, including 130 twin pairs (86 MZ, 44 DZ) and 31 unpaired twins. Among the 130 pairs, thirty pairs were combat veterans, thirty-one pairs were non-combat veterans, and 69 pairs were discordant in combat exposure. We also analyzed follow-up data from 177 participants with both DNAm and depression data, including 77 twin pairs (50 MZ, 27 DZ) and 23 unpaired twins. Among the 77 pairs, eighteen pairs were combat veterans, twenty-five pairs were non-combat veterans, and 34 pairs were discordant in combat exposure. A total of 111 participants had both baseline and follow-up visit, including 33 MZ, 12 DZ twin pairs and 21 single twins.
Measures of Depression and Risk Factor Variables
We used the Beck Depression Inventory II (BDI-II) questionnaire for the assessment of depression at in-person visits for both baseline and follow-up. The BDI-II has 21 items for self-evaluation of symptoms of depression, with a total score range from 0–63; a higher score indicates higher severity of depression(Aaron T. Beck, Steer, & Carbin, 1988; A. T. Beck, Ward, Mendelson, Mock, & Erbaugh, 1961; Steer, Ball, Ranieri, & Beck, 1999). In addition, the Structured Clinical Interview for psychiatric disorders (SCID) according to the Diagnostic and Statistical Manual for Psychiatric Disorders IV (DSM-IV) was performed at both time points to obtain a clinical diagnosis of current PTSD at both in-person visits, which was included in the analysis as it is a common comorbidity of depression.
Information on medical history and traditional cardiovascular risk factors was obtained by interview, blood pressure and anthropometric measurements and analysis of blood samples. We calculated the body mass index (BMI), and obtained information on current smoking, number of alcoholic drinks per week, and history of coronary heart disease. The Baecke questionnaire was used to evaluate habitual physical activity(Pols et al., 1995).
DNA Methylation Data Collection and Processing
The Illumina Infinium MethylationEPIC (850K) BeadChip(Moran, Arribas, & Esteller, 2016) was used for DNA methylation data collection. Over 850,000 genome-wide methylation sites in peripheral blood lymphocytes were measured. Building upon the HumanMethylation 450K BeadChip, the 850K chip measures more than 90% of the original DNAm sites and additional 350,000 sites in enhancer regions from FANTOM5 and ENCODE(Lizio et al., 2015; Siggens & Ekwall, 2014). The software BeadStudio was used for calculating the beta values ranged from 0 to 1, which are proportions of methylation intensity. For quality control, four samples with >10% missing rate or detection p-value >0.01 were excluded. The R package “minfi” was used for batch effect correction and data normalization(Lehne et al., 2015).
DNA Methylation Age Acceleration Calculation
A total of 4 types of DNAm age were estimated for the participants based on Horvath’s(Horvath, 2013), Hannum’s(Hannum et al., 2013), DNAm GrimAge(Lu et al., 2019), and DNAm PhenoAge(Levine et al., 2018) analytical strategies. Horvath’s and Hannum’s DNAm estimates use elastic net regression models to calculate methylome aging, which is a biomarker highly associated with chronological age and mortality. The optimized algorithms of Horvath’s and Hannum’s DNAm age incorporate a total of 353 and 71 epigenetic markers, with 334 and 65 markers available in our study, respectively(Chen et al., 2016; Hannum et al., 2013; Horvath, 2013; Marioni et al., 2015; Perna et al., 2016). Horvath’s algorithm was trained from data representative of a wide spectrum of tissue types, thus can result in a highly accurate estimate without adjustment for most tissues(Horvath, 2013), while Hannum’s estimate is highly accurate in blood-based data(Hannum et al., 2013). The DNAm GrimAge is a composite measure of DNAm age estimated using 1,030 epigenetic markers that incorporate the effects of smoking and 7 plasma proteins, and was shown to be highly associated with coronary heart disease, cancer, and mortality(Lu et al., 2019). The DNAm PhenoAge algorithm including 513 epigenetic markers, which were all available in our study, was derived from various aging phenotypes, and has excellent prediction of aging outcomes, such as mortality, cancer, and physical functioning(Levine et al., 2018).
Based on the 4 types of DNAm age estimates, a total of 6 measures of DNAm age acceleration were derived from the residuals of regression between chronological age and DNAm age: Horvath’s DNAm age acceleration (HorvathAA), intrinsic epigenetic age acceleration (IEAA), Hannum’s DNAm age acceleration (HannumAA), extrinsic epigenetic age acceleration (EEAA), GrimAge acceleration (GrimAA), and PhenoAge acceleration (PhenoAA). We analyzed all 6 DNAm age acceleration measures to explore how they affect results. Among the 6 measures, HorvathAA, HannumAA, GrimAA and PhenoAA were calculated without adjusting for cell type proportions. IEAA was derived from the residuals of HorvathAA adjusting for cell types, and similarly, EEAA was derived from HannumAA adjusting for cell types(Chen et al., 2016). All DNAm age and age acceleration were derived using the online calculator (https://dnamage.genetics.ucla.edu) created by Horvath’s research group(Horvath, 2013).
Statistical Analysis
For the 291 participants at baseline and 177 participants at follow-up visit separately, demographic factors and measure of depression were reported as means and standard deviations for continuous variables, and counts and percentages for categorical variables. These descriptive data for the 111 participants who had both visits were also reported.
The DNAm age acceleration at the baseline and the follow-up visit was assessed using linear mixed effect models with random intercepts of each twin pair separately for the MZ and DZ zygosity groups to incorporate within pair correlation and different levels of clustering by zygosity. The DNAm age acceleration measures were treated as dependent variables and the visit (follow-up vs baseline) was treated as independent variable. The least squared means of DNAm age acceleration for each time point were calculated.
For our main analysis using within-twin effect models, we focused on the within-pair difference to control for shared genetic material and other confounding factors such as age and shared environmental exposures(McGue, Osler, & Christensen, 2010). For each twin pair, we first calculated the mean of BDI-II total score and the 6 DNAm age acceleration measures. Then, we calculated the within-pair difference in these metrics. Separately for baseline and follow-up visits, the association between within-pair difference of BDI-II total score and DNAm age acceleration was investigated using linear mixed effect models, which incorporated random intercepts of each twin pair separately for the MZ and DZ zygosity groups. We treated each one of the 6 within-pair difference of DNAm age acceleration measures as separate dependent variables, and the within-pair difference of BDI-II total score as independent variable. A total of 4 hierarchical models were adopted: Model 1 only considered zygosity; Model 2 additionally adjusted for current smoking and coronary heart disease history; Model 3 additionally adjusted for BMI, number of alcoholic drinks per week, and Baecke score of physical activity; Model 4 additionally adjusted for PTSD, which is a frequent comorbidity of depression. Sensitivity analysis with additional adjustment for blood cell composition including CD8+ T cells, CD4+ T cells, natural killer cells, B cells, monocytes, and granulocytes was also performed.
Separately for baseline and follow-up visits, we also examined the cross-sectional association between BDI-II total score and DNAm age acceleration using similar aforementioned models, treating the twins as individuals and incorporating random intercepts of each twin pair separately for the MZ and DZ zygosity groups.
Using the longitudinal data of both baseline and follow-up visits, we further explored the association between BDI-II total score and DNAm age acceleration using a cross-lagged co-twin control study design(Huang et al., 2019) with the goal of understanding the temporal relationship. Under this approach, we examined the association of the within-pair difference of BDI-II total score at baseline with the within-pair difference of DNAm age acceleration at follow-up using aforementioned linear mixed effect models. Additional analyses were conducted treating the twins as individuals. All analyses were performed in SAS 9.4 (SAS Institute, Cary NC). Due to the exploratory nature of the study, we did not perform correction for multiple testing. A raw p value < 0.05 was considered statistically significant.
Results
Among the 291 twins who underwent the baseline visit and the 177 participants underwent the follow-up visit, around 65% were monozygotic; the mean baseline and follow-up age was 56.0 and 67.6 years, respectively (Table 1). Among twins with data at both visits, the Baecke score of physical activity (baseline 7.4; follow-up 8.0) and the proportion with PTSD (baseline 15.3%; follow-up 29.7%) increased at the second visit compared to the first. The proportion of current smokers (baseline 29.7%; follow-up 21.6%), and the number of alcoholic drinks consumed per week (baseline 5.6; follow-up 1.6) was lower at the second visit compared to the first, whereas BMI, coronary heart disease history and BDI-II total score remained the same. Among the 6 DNAm age acceleration measures, none of them showed a statistically significant DNAm age deceleration or acceleration at either time points, with 95% CIs covering the null (Table 2).
Table 1.
Demographic characteristics at the baseline and at the follow-up visit.
| Overall Sample | Sample with Both Baseline and Follow-up Assessments | |||
|---|---|---|---|---|
| Variable | Baseline (N=291) | Follow-up (N=177) | Baseline (N=111) | Follow-up (N=111) |
| Demographics | ||||
| Monozygotic, n (%) | 190 (65.3) | 114 (64.4) | 78 (70.3) | 78 (70.3) |
| In combat, n (%) | 141 (48.5) | 80 (45.2) | 49 (44.1) | 49 (44.1) |
| Age in years, mean (SD) | 56.0 (3.2) | 67.6 (2.5) | 55.9 (3.2) | 67.6 (2.5) |
| BMI in kg/m2, mean (SD) | 29.9 (5.1) | 29.5 (4.3) | 29.9 (4.5) | 29.9 (4.5) |
| Current smoking, n (%) | 87 (29.9) | 34 (19.3) | 33 (29.7) | 24 (21.6) |
| Alcoholic drinks/week, mean (SD) | 5.4 (10.9) | 1.8 (3.5) | 5.6 (12.0) | 1.6 (3.2) |
| Baecke score of physical activity, mean (SD) | 7.2 (1.8) | 8.0 (1.5) | 7.4 (1.8) | 8.0 (1.5) |
| History of coronary heart disease, n (%) | 42 (14.4) | 16 (9.0) | 9 (8.1) | 9 (8.1) |
| Current PTSD, n (%) | 55 (18.9) | 44 (24.9) | 17 (15.3) | 33 (29.7) |
| Measure of depression | ||||
| BDI-II total score, mean (SD) | 7.0 (8.3) | 5.8 (6.4) | 6.2 (8.5) | 6.5 (6.8) |
Abbreviation: SD, standard deviation.
Table 2.
Estimated DNAm age acceleration (in years) at the baseline and at the follow-up visit.
| DNAm Age Acceleration (years) |
Baseline (N=111) Mean (95% CI) |
Follow-up (N=111) Mean (95% CI) |
|---|---|---|
| HorvathAA | 0.0066 (−0.92, 0.93) |
0.29 (−0.64, 1.21) |
| IEAA | 0.14 (−0.69, 0.96) |
0.18 (−0.64, 1.00) |
| HannumAA | −0.42 (−1.30, 0.46) |
0.44 (−0.44, 1.32) |
| EEAA | −0.79 (−1.93, 0.35) |
0.59 (−0.55, 1.73) |
| GrimAA | −0.98 (−2.09, 0.13) |
0.045 (−1.07, 1.16) |
| PhenoAA | −0.61 (−1.63, 0.40) |
−0.19 (−1.20, 0.82) |
Abbreviation: CI, confidence interval.
The association of BDI-II total score with DNAm age acceleration (in years) at the baseline and at the follow-up visit are depicted as scatterplots shown in Supplemental Figure 1. The most prominent positive association can be observed between BDI-II total score and GrimAA.
Using the within-twin pair effect models, ten-unit higher in BDI-II total score at baseline was associated with 0.73 (95% CI 0.13–1.33, p=0.019) years of age acceleration measured with HannumAA and 0.94 (95% CI 0.22–1.66, p=0.012) years of age acceleration measured with EEAA with fully adjustment of covariates including PTSD in Model 4 (Table 3, Figure 1). At follow-up, ten-unit higher in BDI-II total score was associated with PhenoAA (1.32 years of age acceleration, 95% CI 0.18–2.47, p=0.027). A borderline association with GrimAA (0.99 years of age acceleration, 95% CI −0.011–1.99, p=0.052) was also observed. Sensitivity analysis with additional adjustment for blood cell composition showed similar findings (Supplemental Table 1).
Table 3.
Within-pair association of BDI-II total score (per 10 units higher score) with DNAm age acceleration (in years) at the baseline and at the follow-up visit, using within-twin effect models.
| Visit | DNAm Age Acceleration (years) |
Model 1 Baseline N=260 Follow-up N=154 |
Model 2 Baseline N=260 Follow-up N=152 |
Model 3 Baseline N=246 Follow-up N=144 |
Model 4 Baseline N=246 Follow-up N=144 |
||||
|---|---|---|---|---|---|---|---|---|---|
| Estimate (95% CI) |
P | Estimate (95% CI) |
P | Estimate (95% CI) |
P | Estimate (95% CI) |
P | ||
| Baseline | HorvathAA | 0.15 (−0.47, 0.77) |
0.632 | 0.11 (−0.52, 0.73) |
0.739 | 0.076 (−0.56, 0.71) |
0.817 | −0.049 (−0.70, 0.61) |
0.885 |
| IEAA | 0.070 (−0.53, 0.67) |
0.818 | 0.020 (−0.58, 0.62) |
0.948 | −0.052 (−0.67, 0.57) |
0.870 | −0.16 (−0.80, 0.47) |
0.613 | |
| HannumAA |
0.91
(0.36, 1.47) |
0.002 |
0.88
(0.32, 1.44) |
0.003 |
0.81
(0.23, 1.40) |
0.008 |
0.73
(0.13, 1.33) |
0.019 | |
| EEAA |
1.11
(0.44, 1.78) |
0.002 |
1.07
(0.40, 1.75) |
0.002 |
1.05
(0.35, 1.75) |
0.004 |
0.94
(0.22, 1.66) |
0.012 | |
| GrimAA | 0.56 (−0.12, 1.24) |
0.110 | 0.40 (−0.24, 1.05) |
0.225 | 0.40 (−0.26, 1.06) |
0.237 | 0.45 (−0.23, 1.14) |
0.198 | |
| PhenoAA |
0.75
(0.029, 1.47) |
0.044 | 0.63 (−0.10, 1.35) |
0.094 | 0.61 (−0.14, 1.37) |
0.115 | 0.64 (−0.14, 1.42) |
0.112 | |
| Follow-up | HorvathAA | 0.11 (−0.81, 1.03) |
0.818 | 0.11 (−0.82, 1.03) |
0.823 | 0.016 (−0.94, 0.97) |
0.975 | 0.013 (−0.95, 0.97) |
0.979 |
| IEAA | 0.34 (−0.57, 1.25) |
0.468 | 0.34 (−0.57, 1.25) |
0.465 | 0.29 (−0.65, 1.23) |
0.547 | 0.28 (−0.67, 1.23) |
0.565 | |
| HannumAA | 0.26 (−0.44, 0.96) |
0.467 | 0.27 (−0.43, 0.98) |
0.447 | 0.27 (−0.47, 1.00) |
0.481 | 0.28 (−0.46, 1.03) |
0.455 | |
| EEAA | 0.20 (−0.69, 1.08) |
0.664 | 0.22 (−0.67, 1.10) |
0.629 | 0.15 (−0.78, 1.07) |
0.757 | 0.16 (−0.76, 1.09) |
0.729 | |
| GrimAA |
1.20
(0.18, 2.22) |
0.023 |
1.01
(0.048, 1.97) |
0.043 |
1.00
(0.024, 1.98) |
0.049 | 0.99 (−0.011, 1.99) |
0.052 | |
| PhenoAA |
1.48
(0.37, 2.58) |
0.011 |
1.44
(0.34, 2.54) |
0.012 |
1.33
(0.19, 2.47) |
0.025 |
1.32
(0.18, 2.47) |
0.027 | |
Abbreviation: CI, confidence interval.
Model 1: adjusted for zygosity;
Model 2: Model 1 + current smoking, coronary heart disease history;
Model 3: Model 2 + BMI, number of alcoholic drinks per week, Baecke score of physical activity;
Model 4: Model 3 + PTSD.
Figure 1.
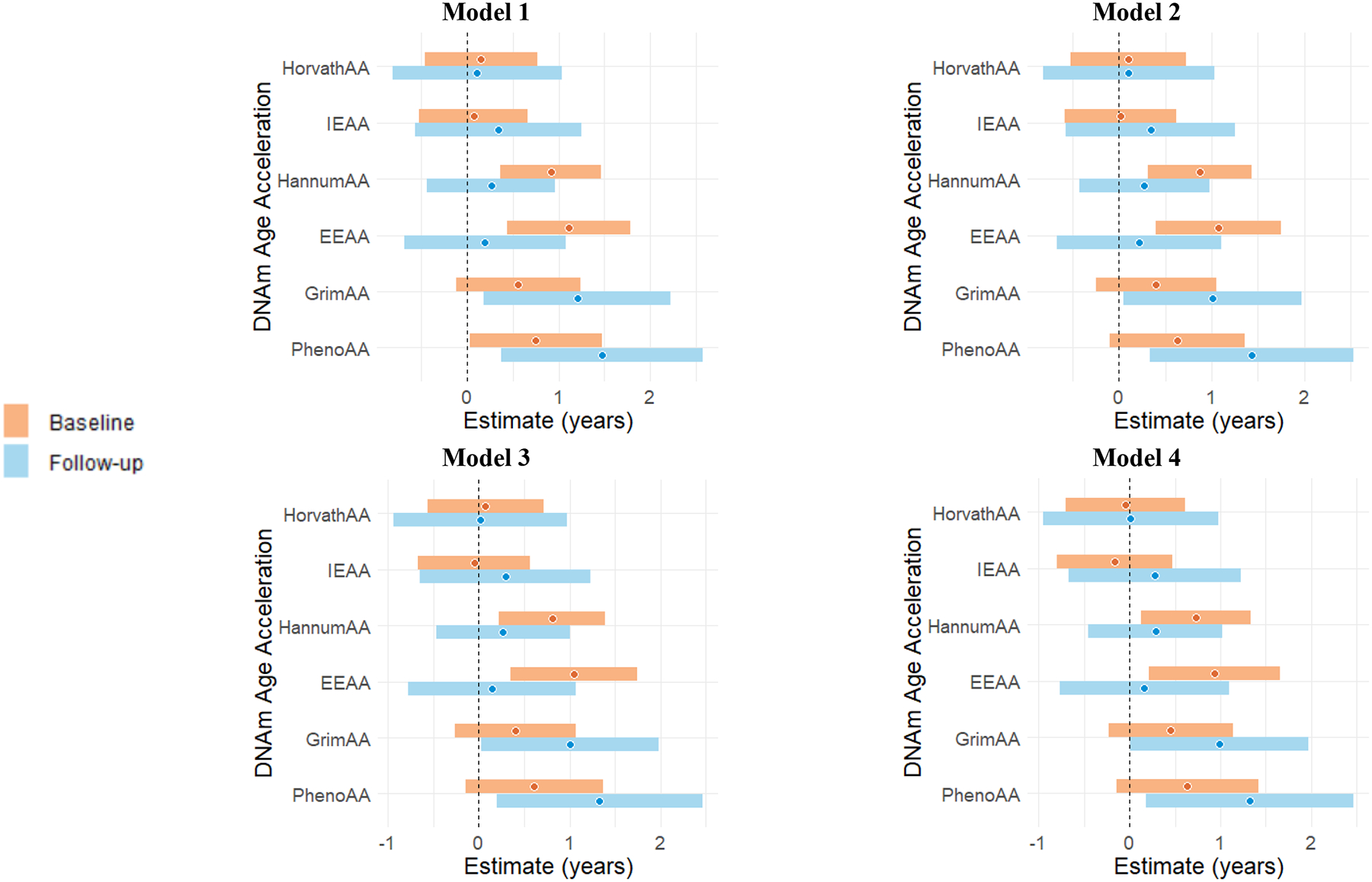
Within-pair association of BDI-II total score (per 10 units higher score) with DNAm age acceleration (in years) at the baseline and at the follow-up visit, using within-twin effect models.
Model 1: adjusted for zygosity;
Model 2: Model 1 + current smoking, coronary heart disease history;
Model 3: Model 2 + BMI, number of alcoholic drinks per week, Baecke score of physical activity;
Model 4: Model 3 + PTSD.
For the analysis treating twins as individuals, at baseline, a higher BDI-II total score was associated with age acceleration measured with EEAA in Model 2 and GrimAA in Model 1 (Supplemental Table 2). At the follow-up visit, a higher BDI-II total score was associated with GrimAA in Model 1. However, these associations attenuated after further adjustment.
For the cross-lagged within-twin pair effect models, we observed null associations between BDI-II total score at baseline and the age acceleration measures at follow-up visit (Supplemental Table 3A). When treating twins as individuals, a higher BDI-II total score at baseline was associated with age acceleration measured with GrimAA at follow-up in Model 1, but the association attenuated after adjustment (Supplemental Table 3B).
Discussion
Using a robust co-twin control study design, we reported associations between depression and specific measures of DNAm age acceleration. We analyzed data from blood samples, and the measures of DNAm age acceleration used were derived from blood-based data except for Horvath’s estimate of multi-tissue samples. Each epigenetic clock represents a unique fraction of biological aging profile. Horvath’s and Hannum’s clocks were based on the selected epigenetic markers associated with chronological age, thus have high correlations with chronological age(McCartney et al., 2018). Given the high prevalence of smoking of ~20% to 30% in our study cohort, GrimAA incorporating the epigenetic markers of smoking is a suitable measure that could potentially capture the intrinsic differences in epigenetic change due to smoking between participants. Additionally, PhenoAA takes the markers of phenotypic aging measures into account, and incorporates variation in physiological status and morbidity profile among the participants in our study. A positive association was identified between BDI-II total score and 3 DNAm age acceleration measures, including HannumAA, EEAA and PhenoAA, either at baseline or at follow-up, after adjusting for potential confounding factors. This within pair association suggests that it is not due to confounding by genetic or familial factors. The smaller sample size at the follow-up visit might explain why somewhat fewer associations were statistically significant at follow-up compared with baseline. However, the effects were aligned in the same direction.
Our study contributes to growing evidence supporting the association between depression and epigenetic aging. Our results are consistent with the findings of higher DNAm aging in patients with depression in a previous case-control study(Han et al., 2018). The direction of our findings is also in line with the study of depressive symptoms by Beydoun et al., particularly regarding the “positive affect” domain of the depression measure(Beydoun et al., 2019). In addition, the suggestive association with GrimAA we identified was consistent with a prior study among depressed cases compared to controls(Protsenko et al., 2021). GrimAA was shown to be associated with alcohol consumption in a recent study(Kresovich et al., 2021), which may indicate that GrimAA can be a marker of lifestyle factors such as alcohol consumption and smoking that predispose to premature mortality.
The study of temporal mechanism using the longitudinal cross-lagged approach showed weak and insignificant longitudinal associations between BDI-II and DNAm. This would argue against a temporal relationship linking depressive symptoms to accelerated biological aging. However, this analysis was limited by the small sample size and thus is not conclusive. Future studies, with a larger sample size, are warranted to understand the mechanisms through which depression may speed up aging on a molecular basis, and eventually result in various chronic diseases.
Previous studies have reported the association between epigenetic aging and several lifestyle factors including BMI, alcohol consumption, smoking and physical activity(Gao, Zhang, Breitling, & Brenner, 2016; Quach et al., 2017; Wu et al., 2019), which are also known factors associated with depression(Fluharty, Taylor, Grabski, & Munafo, 2017; Jantaratnotai, Mosikanon, Lee, & McIntyre, 2017; Sarris et al., 2020). In our study, we considered the potential confounding effects of these factors, as well as a history of coronary heart disease, which is associated with epigenetic aging(Roetker et al., 2018). The cross-sectional association between depression and DNAm age acceleration measures persisted after adjustment, leading to the hypothesis that mechanisms other than these factors may be involved. In addition, most of the associations found were also independent of PTSD status, which has been previously reported as being associated with DNAm age acceleration(Wang et al., 2021) and is frequently comorbid with depression. There has been increasing evidence suggesting strong associations between depression and multimorbidity of chronic conditions(Read, Sharpe, Modini, & Dear, 2017), and such associations are likely to be bidirectional(Katon, 2011). A biological aging process could be the underlying mechanism, and could involve changes in activities of gene transcription, shedding light on potential intervention targets if a causal pathway can be established.
Our study has several strengths, including the co-twin control study design, replication of the overall findings at two time points and consideration of various risk factors as potential confounders. The within-twin pair effect modeling is a strategy that can help explore the causal association between depression and DNAm age acceleration through a counterfactual framework that can minimize bias. This counterfactual framework can help establish causal inference in observational studies, given that the genetic profile and early environmental exposures are shared within twin pairs, and the observed phenotypic difference between twins are likely caused by non-shared environmental factors(McGue et al., 2010). The study was subject to several potential limitations. Only a subgroup of participants had data on DNAm age acceleration at both visits, which limited the statistical power especially for the longitudinal analysis. Future studies using a larger sample size with longitudinal data are needed to better elucidate the possible causal association between depression and DNAm age acceleration. The information of depression onset, remission and relapse was not collected, and future studies should investigate the association between trajectories of the change in depression status and epigenetic aging. The study is also subject to residual confounding. Specifically, the measurement of smoking behavior may not accurately measure the exposure, since a more detailed measurement of smoking status such as pack-years was not used. Further, the potential confounding factors considered may not capture the overall health status of the participants. In addition, this present study was conducted in a male-only cohort, even though women have a higher rate of depression compared to men, which is possibly due to biological factors such as stress responsiveness and impact of sex hormones on the neurotransmitter system(Parker & Brotchie, 2010; Weissman et al., 1996). Further investigations should be conducted among female participants to assess the generalizability of our findings.
Conclusion
We found an association between depression, assessed as a continuous measure of depressive symptom severity, and some (but not all) biomarkers of DNAm age acceleration, independent of lifestyle factors, medical history of heart disease and PTSD. Because the association was present within twin pairs, the mechanism is not likely due to shared genetic or familial exposures. Further studies are needed to assess the causal pathways involved, and whether treating depression can slow down or reverse biological aging.
Supplementary Material
Supplemental Figure 1. Scatterplot for the association of BDI-II total score with DNAm age acceleration (in years) at the baseline and at the follow-up visit. The blue line indicates the linear association, and the gray area indicates the 95% confidence interval.
Acknowledgements:
The study received financial support from the National Institutes of Health (grants R01 HL68630, R01 AG026255, R01 HL125246, 2K24 HL077506, and R01 HL136205).
The United States Department of Veterans Affairs has provided financial support for the development and maintenance of the Vietnam Era Twin (VET) Registry. Numerous organizations have provided invaluable assistance in the conduct of this study, including: Department of Defense; National Personnel Records Center, National Archives and Records Administration; the Internal Revenue Service; National Institutes of Health; National Opinion Research Center; the National Research Council, National Academy of Sciences; the Institute for Survey Research, Temple University. Most importantly, the authors gratefully acknowledge the continued cooperation and participation of the members of the VET Registry and their families. Without their contribution, this research would not have been possible.
Footnotes
Disclaimer Statement:
All statements and opinions are solely of the authors and do not necessarily reflect the position or policy of the VET Registry, VA, or United States Government.
Conflict of Interest:
The authors claim no conflict of interest.
References:
- Beck AT, Steer RA, & Carbin MG (1988). Psychometric properties of the Beck Depression Inventory: Twenty-five years of evaluation. Clinical Psychology Review, 8(1), 77–100. doi: 10.1016/0272-7358(88)90050-5 [DOI] [Google Scholar]
- Beck AT, Ward CH, Mendelson M, Mock J, & Erbaugh J (1961). An inventory for measuring depression. Arch Gen Psychiatry, 4, 561–571. doi: 10.1001/archpsyc.1961.01710120031004 [DOI] [PubMed] [Google Scholar]
- Beydoun MA, Hossain S, Chitrala KN, Tajuddin SM, Beydoun HA, Evans MK, & Zonderman AB (2019). Association between epigenetic age acceleration and depressive symptoms in a prospective cohort study of urban-dwelling adults. J Affect Disord, 257, 64–73. doi: 10.1016/j.jad.2019.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocklandt S, Lin W, Sehl ME, Sanchez FJ, Sinsheimer JS, Horvath S, & Vilain E (2011). Epigenetic predictor of age. PLoS One, 6(6), e14821. doi: 10.1371/journal.pone.0014821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai PC, … Horvath S (2016). DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY), 8(9), 1844–1865. doi: 10.18632/aging.101020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugue PA, Bassett JK, Joo JE, Jung CH, Ming Wong E, Moreno-Betancur M, … Milne RL (2018). DNA methylation-based biological aging and cancer risk and survival: Pooled analysis of seven prospective studies. Int J Cancer, 142(8), 1611–1619. doi: 10.1002/ijc.31189 [DOI] [PubMed] [Google Scholar]
- Fluharty M, Taylor AE, Grabski M, & Munafo MR (2017). The Association of Cigarette Smoking With Depression and Anxiety: A Systematic Review. Nicotine Tob Res, 19(1), 3–13. doi: 10.1093/ntr/ntw140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg C, Liu C, Mori A, Tsai M, Sporleder J, Moore K, … Smith N (2020). Cohort Profile: The Vietnam Era Twin Registry (VET Registry). Int J Epidemiol, 49(1), 22–22d. doi: 10.1093/ije/dyz217 [DOI] [PubMed] [Google Scholar]
- Gao X, Zhang Y, Breitling LP, & Brenner H (2016). Relationship of tobacco smoking and smoking-related DNA methylation with epigenetic age acceleration. Oncotarget, 7(30), 46878–46889. doi: 10.18632/oncotarget.9795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han LKM, Aghajani M, Clark SL, Chan RF, Hattab MW, Shabalin AA, … Penninx B (2018). Epigenetic Aging in Major Depressive Disorder. Am J Psychiatry, 175(8), 774–782. doi: 10.1176/appi.ajp.2018.17060595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, … Zhang K (2013). Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell, 49(2), 359–367. doi: 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S (2013). DNA methylation age of human tissues and cell types. Genome Biol, 14(10), R115. doi: 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Su S, Goldberg J, Miller AH, Levantsevych OM, Shallenberger L, … Vaccarino V (2019). Longitudinal association of inflammation with depressive symptoms: A 7-year cross-lagged twin difference study. Brain Behav Immun, 75, 200–207. doi: 10.1016/j.bbi.2018.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantaratnotai N, Mosikanon K, Lee Y, & McIntyre RS (2017). The interface of depression and obesity. Obes Res Clin Pract, 11(1), 1–10. doi: 10.1016/j.orcp.2016.07.003 [DOI] [PubMed] [Google Scholar]
- Jones MJ, Goodman SJ, & Kobor MS (2015). DNA methylation and healthy human aging. Aging Cell, 14(6), 924–932. doi: 10.1111/acel.12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katon WJ (2011). Epidemiology and treatment of depression in patients with chronic medical illness. Dialogues Clin Neurosci, 13(1), 7–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, & Bromet EJ (2013). The epidemiology of depression across cultures. Annu Rev Public Health, 34, 119–138. doi: 10.1146/annurev-publhealth-031912-114409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresovich JK, Martinez Lopez AM, Garval EL, Xu Z, White AJ, Sander DP, & Taylor JA (2021). Alcohol consumption and methylation-based measures of biological age. J Gerontol A Biol Sci Med Sci. doi: 10.1093/gerona/glab149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehne B, Drong AW, Loh M, Zhang W, Scott WR, Tan ST, … Chambers JC (2015). A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol, 16, 37. doi: 10.1186/s13059-015-0600-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, … Horvath S (2018). An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY), 10(4), 573–591. doi: 10.18632/aging.101414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, He Y, Ma X, & Chen X (2018). Epigenetic age analysis of brain in major depressive disorder. Psychiatry Res, 269, 621–624. doi: 10.1016/j.psychres.2018.09.001 [DOI] [PubMed] [Google Scholar]
- Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S, … consortium F (2015). Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol, 16, 22. doi: 10.1186/s13059-014-0560-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, … Horvath S (2019). DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY), 11(2), 303–327. doi: 10.18632/aging.101684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, … Deary IJ (2015). DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol, 16, 25. doi: 10.1186/s13059-015-0584-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney DL, Stevenson AJ, Walker RM, Gibson J, Morris SW, Campbell A, … Marioni RE (2018). Investigating the relationship between DNA methylation age acceleration and risk factors for Alzheimer’s disease. Alzheimers Dement (Amst), 10, 429–437. doi: 10.1016/j.dadm.2018.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Osler M, & Christensen K (2010). Causal Inference and Observational Research: The Utility of Twins. Perspect Psychol Sci, 5(5), 546–556. doi: 10.1177/1745691610383511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran S, Arribas C, & Esteller M (2016). Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics, 8(3), 389–399. doi: 10.2217/epi.15.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker G, & Brotchie H (2010). Gender differences in depression. Int Rev Psychiatry, 22(5), 429–436. doi: 10.3109/09540261.2010.492391 [DOI] [PubMed] [Google Scholar]
- Perna L, Zhang Y, Mons U, Holleczek B, Saum KU, & Brenner H (2016). Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenetics, 8, 64. doi: 10.1186/s13148-016-0228-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols MA, Peeters PH, Bueno-De-Mesquita HB, Ocke MC, Wentink CA, Kemper HC, & Collette HJ (1995). Validity and repeatability of a modified Baecke questionnaire on physical activity. Int J Epidemiol, 24(2), 381–388. doi: 10.1093/ije/24.2.381 [DOI] [PubMed] [Google Scholar]
- Protsenko E, Yang R, Nier B, Reus V, Hammamieh R, Rampersaud R, … Wolkowitz OM (2021). “GrimAge,” an epigenetic predictor of mortality, is accelerated in major depressive disorder. Transl Psychiatry, 11(1), 193. doi: 10.1038/s41398-021-01302-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, … Horvath S (2017). Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY), 9(2), 419–446. doi: 10.18632/aging.101168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read JR, Sharpe L, Modini M, & Dear BF (2017). Multimorbidity and depression: A systematic review and meta-analysis. J Affect Disord, 221, 36–46. doi: 10.1016/j.jad.2017.06.009 [DOI] [PubMed] [Google Scholar]
- Roetker NS, Pankow JS, Bressler J, Morrison AC, & Boerwinkle E (2018). Prospective Study of Epigenetic Age Acceleration and Incidence of Cardiovascular Disease Outcomes in the ARIC Study (Atherosclerosis Risk in Communities). Circ Genom Precis Med, 11(3), e001937. doi: 10.1161/CIRCGEN.117.001937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris J, Thomson R, Hargraves F, Eaton M, de Manincor M, Veronese N, … Firth J (2020). Multiple lifestyle factors and depressed mood: a cross-sectional and longitudinal analysis of the UK Biobank (N = 84,860). BMC Med, 18(1), 354. doi: 10.1186/s12916-020-01813-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggens L, & Ekwall K (2014). Epigenetics, chromatin and genome organization: recent advances from the ENCODE project. J Intern Med, 276(3), 201–214. doi: 10.1111/joim.12231 [DOI] [PubMed] [Google Scholar]
- Smith K (2014). Mental health: a world of depression. Nature, 515(7526), 181. doi: 10.1038/515180a [DOI] [PubMed] [Google Scholar]
- Steer RA, Ball R, Ranieri WF, & Beck AT (1999). Dimensions of the Beck Depression Inventory-II in clinically depressed outpatients. J Clin Psychol, 55(1), 117–128. doi: [DOI] [PubMed] [Google Scholar]
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, … Widschwendter M (2010). Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res, 20(4), 440–446. doi: 10.1101/gr.103606.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccarino V, Goldberg J, Rooks C, Shah AJ, Veledar E, Faber TL, … Bremner JD (2013). Post-traumatic stress disorder and incidence of coronary heart disease: a twin study. J Am Coll Cardiol, 62(11), 970–978. doi: 10.1016/j.jacc.2013.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccarino V, Huang M, Wang Z, Hui Q, Shah AJ, Goldberg J, … Sun YV (2021). Epigenetic Age Acceleration and Cognitive Decline: A Twin Study. J Gerontol A Biol Sci Med Sci. doi: 10.1093/gerona/glab047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker ER, McGee RE, & Druss BG (2015). Mortality in mental disorders and global disease burden implications: a systematic review and meta-analysis. JAMA Psychiatry, 72(4), 334–341. doi: 10.1001/jamapsychiatry.2014.2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hui Q, Goldberg J, Smith N, Kaseer B, Murrah N, … Sun YV (2021). Association between Posttraumatic Stress Disorder and Epigenetic Age Acceleration in a Sample of Twins. Psychosom Med. doi: 10.1097/PSY.0000000000001028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman MM, Bland RC, Canino GJ, Faravelli C, Greenwald S, Hwu HG, … Yeh EK (1996). Cross-national epidemiology of major depression and bipolar disorder. JAMA, 276(4), 293–299. [PubMed] [Google Scholar]
- Wong ML, & Licinio J (2001). Research and treatment approaches to depression. Nat Rev Neurosci, 2(5), 343–351. doi: 10.1038/35072566 [DOI] [PubMed] [Google Scholar]
- Wu X, Huang Q, Javed R, Zhong J, Gao H, & Liang H (2019). Effect of tobacco smoking on the epigenetic age of human respiratory organs. Clin Epigenetics, 11(1), 183. doi: 10.1186/s13148-019-0777-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao FH, Wang HT, & Kong QP (2019). Dynamic DNA Methylation During Aging: A “Prophet” of Age-Related Outcomes. Front Genet, 10, 107. doi: 10.3389/fgene.2019.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivin K, Ilgen MA, Pfeiffer PN, Welsh DE, McCarthy J, Valenstein M, … Kales HC (2012). Early mortality and years of potential life lost among Veterans Affairs patients with depression. Psychiatr Serv, 63(8), 823–826. doi: 10.1176/appi.ps.201100317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Scatterplot for the association of BDI-II total score with DNAm age acceleration (in years) at the baseline and at the follow-up visit. The blue line indicates the linear association, and the gray area indicates the 95% confidence interval.


