Abstract
Background:
Hypertrophic cardiomyopathy (HCM), a disease with myocardial fibrosis manifestation, is a common cause of sudden cardiac death (SCD) due to ventricular arrhythmias (VA). Current clinical risk stratification criteria are inadequate in identifying patients who are at risk for VA and in need of an implantable cardioverter defibrillator (ICD) for primary prevention.
Objective:
We aimed to develop a risk prediction approach based on imaging biomarkers from the combination of late gadolinium contrast-enhanced (LGE) MRI and T1 mapping. We then aimed to compare the prediction to a virtual heart computational risk assessment approach based on LGE-T1 virtual heart models.
Methods:
The methodology involved combining short-axis LGE-MRI with post-contrast T1 maps to define personalized thresholds for diffuse and dense fibrosis. The combined LGE-T1 maps were used to evaluate imaging biomarkers for VA risk prediction. The risk prediction capability of the biomarkers was compared with that of the LGE-T1 virtual heart arrhythmia inducibility simulation. VA risk prediction performance from both approaches was compared to clinical outcome (presence of clinical VA).
Results:
Image-based biomarkers, including hypertrophy, signal intensity heterogeneity, and fibrotic border complexity, could not discriminate high vs low VA risk. LGE-T1 virtual heart technology outperformed all the image-based biomarker metrics and was statistically significant in predicting VA risk in HCM.
Conclusions:
We combined two MR imaging techniques to analyze imaging biomarkers in HCM. Raw and processed image-based biomarkers cannot discriminate patients with VA from those without VA. Hybrid LGE-T1 virtual heart models could correctly predict VA risk for this cohort and may improve SCD risk stratification to better identify HCM patients for primary preventative ICD implantation.
Keywords: Digital twin, Hypertrophic cardiomyopathy, Diffuse fibrosis, Arrhythmogenesis
Introduction
Hypertrophic cardiomyopathy (HCM) is the most common genetic cardiac disease with an incidence of 1 in 500, resulting in substantial morbidity, especially in young patients, and decreased quality of life [1]. The disease presents with fibrosis proliferation of the ventricular myocardium which can create substrates for ventricular arrhythmias (VA) leading to sudden cardiac death (SCD) in patients who are often asymptomatic [2]. Prophylactic implantable cardioverter defibrillator (ICD) deployment is used to prevent SCD due to VA in patients with HCM [3]. These procedures carry the risk of potential complications as well as morbidity. However, current risk stratification criteria, such as those adopted by the American Heart Association and European Society of Cardiology fail to accurately identify all patients at risk for SCD, leading to suboptimal rates of appropriate ICD implantation [4–6]. Thus, many HCM patients receive ICDs without gaining any health benefits, while others are not effectively safeguarded. Additionally, these risk criteria do not consider the extent of myocardial fibrosis caused by the disease.
Our group has previously developed virtual-heart technology to predict SCD risk due to VA for ischemic and non-ischemic cardiomyopathies [7–10]. In the ischemic population, the technology is based on assessing the arrhythmogenic propensity of the disease-induced focal fibrotic substrate derived from contrast-enhanced late gadolinium enhancement (LGE) cardiovascular magnetic resonance (MRI) [11]. In patients with cardiac sarcoidosis, positron emission tomography (PET) imaging was combined with the LGE-MRI data to represent the disease-specific active myocardial inflammation that contributes to arrhythmogenesis [7]. In the HCM population, we combined post-contrast T1 mapping and LGE-MRI data to represent the fibrotic remodeling that occurs in HCM [9].
Patient-specific myocardial diffuse fibrosis forms in patients with HCM as a result of hypertrophic remodeling. Fibrosis and image-based features can be quantified in various ways, including the amount, distribution, randomness, and complexity. The amount and distribution of fibrosis has been associated with arrhythmogenic risk [12]. Entropy has been used to measure ventricular myocardial tissue heterogeneity as a predictor of major adverse cardiac events [7,13]. Additionally, fractal dimension analysis, a measure of geometric complexity, has been used to characterize cardiac fibrosis and stratify arrhythmia risk [7]. However, an analysis of combined LGE-MRI imaging with post-contrast T1 mapping (LGE-T1), has never been assessed before, and it remains unclear whether these imaging biomarkers could be indicative of risk of VA.
The aim of this article is to evaluate two novel approaches for arrhythmia risk prediction in HCM patients: 1) using imaging biomarkers from the combination of LGE-MRI with post-contrast T1 mapping (LGE-T1) and 2) using personalized mechanistic heart models reconstructed from the same combined images. In this retrospective study, we also assess the predictive capability of these two approaches compared with that of the current clinical metrics.
Methods
Study population and imaging data
In this retrospective study, we included 26 patients diagnosed with HCM based on the presence of left-ventricular (LV) wall thickness ≥ 15 mm in the absence of other ventricular diseases [12]. All 26 HCM patients in our study were deemed at high risk for SCD by AHA criteria and received ICDs for primary prevention, but only 13 patients, i.e., 50% of the cohort, actually experienced VA determined by appropriate ICD firing. Short-axis LGE-MRI and post-contrast T1 maps were acquired under institutional review board approval for all 26 patients, previously described in full detail [9].
Combining LGE-MRI with post-contrast T1-mapping
For each patient, we incorporated the information of the single post-contrast T1 map to the short-axis LGE-MRI to find personalized thresholds of diffuse and dense fibrosis as shown in Fig. 1, previously described in full detail [9]. Briefly, regions of non-fibrotic myocardium (relaxation time (RT): >450 ms) and diffuse (RT: 350–450 ms) and dense fibrosis (RT: <350 ms) of each mid-myocardial T1 map were delineated on the LV segmentation based on histopathological evidence [2]. Next, the distribution of signal intensity of the corresponding short-axis LGE-MRI myocardium was binarized into regions of high and low intensity, as we have done previously [7], with the mean of the lower-intensity region chosen as the reference of the non-fibrotic myocardium. We then used the amount of diffuse and dense fibrosis from the T1 map to calculate new, personalized thresholds on the corresponding LGE-MRI signal intensity, represented by standard deviations above the reference mean. This relationship was then applied to the entire myocardium of the LGE-MRI short-axis sequence to create a personalized, volumetric representation of non-fibrotic myocardium, diffuse fibrosis, and dense fibrosis. The personalized thresholds for diffuse and dense fibrosis were unique to each patient.
Fig. 1.
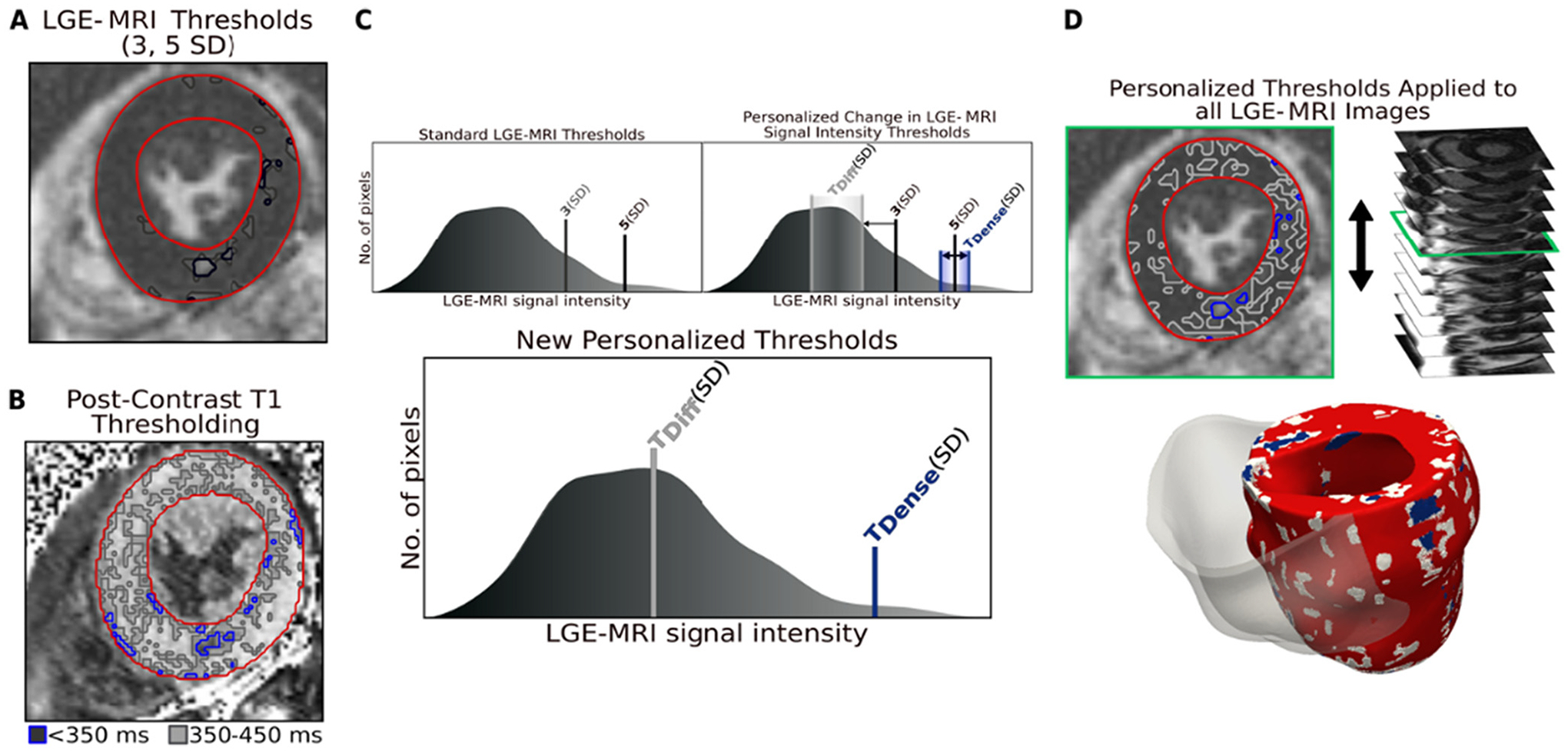
Combining LGE-MRI with Post-Contrast T1 Mapping. A. Segmentation of short-axis LGE-MRI myocardium. B. Segmentation of T1 map myocardium and thresholding of diffuse fibrosis (gray) and dense fibrosis (blue). C. Top left: Standard thresholds for gray zone and focal scar in the ischemic population, applied to the LGE-MRI segmentation from the mean of the low signal myocardium in (A). Top right: Representation of the change in thresholds based on distribution in (B). Bottom: The new personalized thresholds for fibrosis for this patient. D. Top: Application of personalized thresholds to each image in the LGE-MRI short-axis. Bottom: Personalized geometrical reconstruction of the LGE-T1 virtual heart. SD: standard deviation; LGE-MRI: late gadolinium enhanced magnetic resonance imaging.
Image-based biomarkers of the combined LGE-T1 images were then evaluated for each patient.
Risk prediction approach 1: image biomarker selection
We hypothesized that features extracted from the raw (LGE-MRI, T1) and processed (LGE-T1) images might provide prognostic information regarding risk of VA. The entropy and fractal dimension were chosen because previous studies relied on these biomarkers. The amount of fibrotic remodeling and distribution were included for comparison as previously described [9].
The entropy of an MR image quantifies the heterogeneity of the signal intensity and has previously been used to predict SCD risk in ischemic cardiomyopathy. Using the raw MR images and segmentation masks, we calculated the entropy of the volumetric LGE-MRI short-axis volume, intra-patient short-axis slices, and post-contrast T1 maps for each patient’s LV myocardium. Entropy was computed according to the following formula [14] using the normalized histogram of image intensity
| (1) |
where k is the number of gray levels in the image, and pk is the probability associated with each gray level.
Fractal dimension analysis quantifies the complexity of a geometry and has been previously used to characterize cardiac fibrosis in patients with cardiac sarcoidosis, another non-ischemic cardiac disease [7]. Using an interpolated mask of the segmentation with fibrotic boundaries, we quantified the complexity, or fractal dimensions of the diffuse and dense fibrotic borders, individually and together. We also investigated the complexity of the fibrotic boundaries at the base, mid-myocardium, and apex. We used the box counting method for calculating the fractal dimension (FD) as follows
| (2) |
where N is the number of boxes that cover the pattern and r is the magnification of the box size.
Risk prediction approach 2: personalized LGE-T1 virtual heart models
Geometrical reconstructions of the LV myocardium with fibrotic remodeling were constructed using the combined LGE-T1 images for assessing VA risk via simulations of rapid pacing as previous described in full detail [9]. Briefly, high resolution finite-element tetrahedral meshes were constructed directly from the ventricular segmentations and fiber orientations were applied to each mesh using a previously validated approach [15,16]. Regions of non-fibrotic myocardium, diffuse fibrosis, and dense fibrosis were defined using the combined LGE-T1 data. Regions of non-fibrotic myocardium were represented by the human ventricular myocyte model by ten Tusscher et al [17] at the cellular level as done in our previous studies [7,8,11,18]. For regions of diffuse fibrosis, we modified the ionic conductances of the ten Tusscher model based on HCM-specific data reported by Coppini et al. [19] At the tissue level, conductivity values along the longitudinal and transverse fiber directions in fibrotic and non-fibrotic myocardium were the same as previously implemented for non-ischemic patient heart models [8]. Dense fibrosis was considered electrically inexcitable.
Full details regarding simulated electrical activity in the models can be found in previous publications [7,8,18,20]. Each virtual heart was paced sequentially from seven uniformly distributed endocardial LV locations using a validated rapid pacing protocol described in detail in previous studies [10,11,18]. Similar to our work on VA risk stratifications for patients with ischemic cardiomyopathy [11], simulation results were analyzed to determine whether reentrant VA was induced in the LV HCM models following rapid pacing from any of the sites. If VA was induced from at least one pacing site in a given personalized HCM virtual heart, the patient was then considered at risk of VA. Simulation results were blind to clinical outcome.
The evaluation of the image-based biomarkers and HCM virtual heart technology were then compared to the clinical outcome for all patients.
Results
Overview of the two VA risk predictors
We introduce two approaches for VA risk prediction in HCM using a combination of LGE-MRI and post-contrast T1 maps. In the first approach, we extract and evaluate image-based biomarkers from the individual and personalized combination of LGE-MRI and post-contrast T1 maps for each patient. In the second approach, each patient’s propensity to arrhythmia is non-invasively assessed using personalized LGE-T1 mechanistic heart models reconstructed using the same combined images. For each risk predictor, we compare the clinical outcome (i.e., presence of clinically diagnosed VA) to the assessment of the extracted features. Full details regarding the patients’ clinical characteristics can be found in our previous publication [9].
Approach 1: evaluation of the LGE-T1 image biomarkers risk prediction performance
Full analysis of the image-based biomarker approach can be found in Table 1.
Table 1.
Overview of image-based biomarkers and LGE-T1 virtual heart technology.
| Without VA | With VA | P-value | |
|---|---|---|---|
| Image-Based Biomarkers | |||
| Dense Fibrosis (%) | 3.36 ± 1.23 | 2.99 ± 1.49 | 0.53 |
| Diffuse Fibrosis (%) | 39.49 ± 8.26 | 41.38 ± 10.59 | 0.59 |
| Volumetric LGE-MRI Entropy | 0.43 ± 0.07 | 0.39 ± 0.09 | 0.21 |
| Entropy – Base | 0.54 ± 0.09 | 0.49 ± 0.12 | 0.16 |
| Entropy – Mid-Myocardium | 0.43 ± 0.07 | 0.41 ± 0.10 | 0.51 |
| Entropy – Apex | 0.28 ± 0.09 | 0.22 ± 0.08 | 0.08 |
| Entropy – Post-Contrast T1 Map | 0.14 ± 0.03 | 0.14 ± 0.06 | 0.93 |
| Fractal Dimension – Fibrotic Volume | 2.23 ± 0.08 | 2.28 ± 0.08 | 0.08 |
| FD – Diffuse Fibrosis | 2.20 ± 0.08 | 2.26 ± 0.09 | 0.09 |
| FD – Dense Fibrosis | 1.77 ± 0.15 | 1.80 ± 0.14 | 0.60 |
| FD – Base (diffuse & dense) | 2.12 ± 0.09 | 2.18 ± 0.10 | 0.10 |
| FD – Mid-Myocardium (diffuse & dense) | 2.10 ± 0.10 | 2.14 ± 0.10 | 0.28 |
| FD – Apex (diffuse & dense) | 2.00 ± 0.11 | 2.01 ± 0.09 | 0.90 |
| LGE-T1 Virtual Heart Model | |||
| Simulated VAs | 0.46 ± 0.77 | 2.10 ± 1.29 | < 0.01 |
| Unique Simulated VAs | 0.38 ± 0.65 | 1.54 ± 0.97 | < 0.01 |
LGE-MRI: late gadolinium enhanced magnetic resonance imaging; VA: ventricular arrhythmia; FD: fractal dimension.
Entropy
To evaluate the heterogeneity of the myocardial signal intensity, we used the entropy function described by Eq. (1). Fig. 2 presents a comparison of the entropy between patients with and without clinical VA. Fig. 2A, left, shows the comparison of entropy calculated from the entire short-axis LGE-MRI myocardial volume. There was not a statistically significant difference between the patients with and without clinical VA, although the heterogeneity of the signal intensity of the patients without VA was 9.03% higher on average. This suggests that the distribution of fibrotic substrate and non-fibrotic myocardium may play a role in arrhythmogenesis. Fig. 2A, right, investigates the entropy of the LGE-MRI at each location in along the short-axis slices, with the greatest difference occurring near the basal section of the heart. While not statistically significant, the entropy of the patients without VA maintains consistently greater values of heterogeneity. Fig. 2B shows the entropy comparison of the post-contrast T1 maps between the patients with and without VA. The relationship is statistically insignificant, which is expected since each T1 map only represents one cross-section of the short-axis myocardium. In addition, the post-contrast T1 maps were acquired from the mid-myocardial region for each patient which also corresponds to the least difference of the mid-myocardial LGE-MRI in Fig. 2A, right.
Fig. 2.
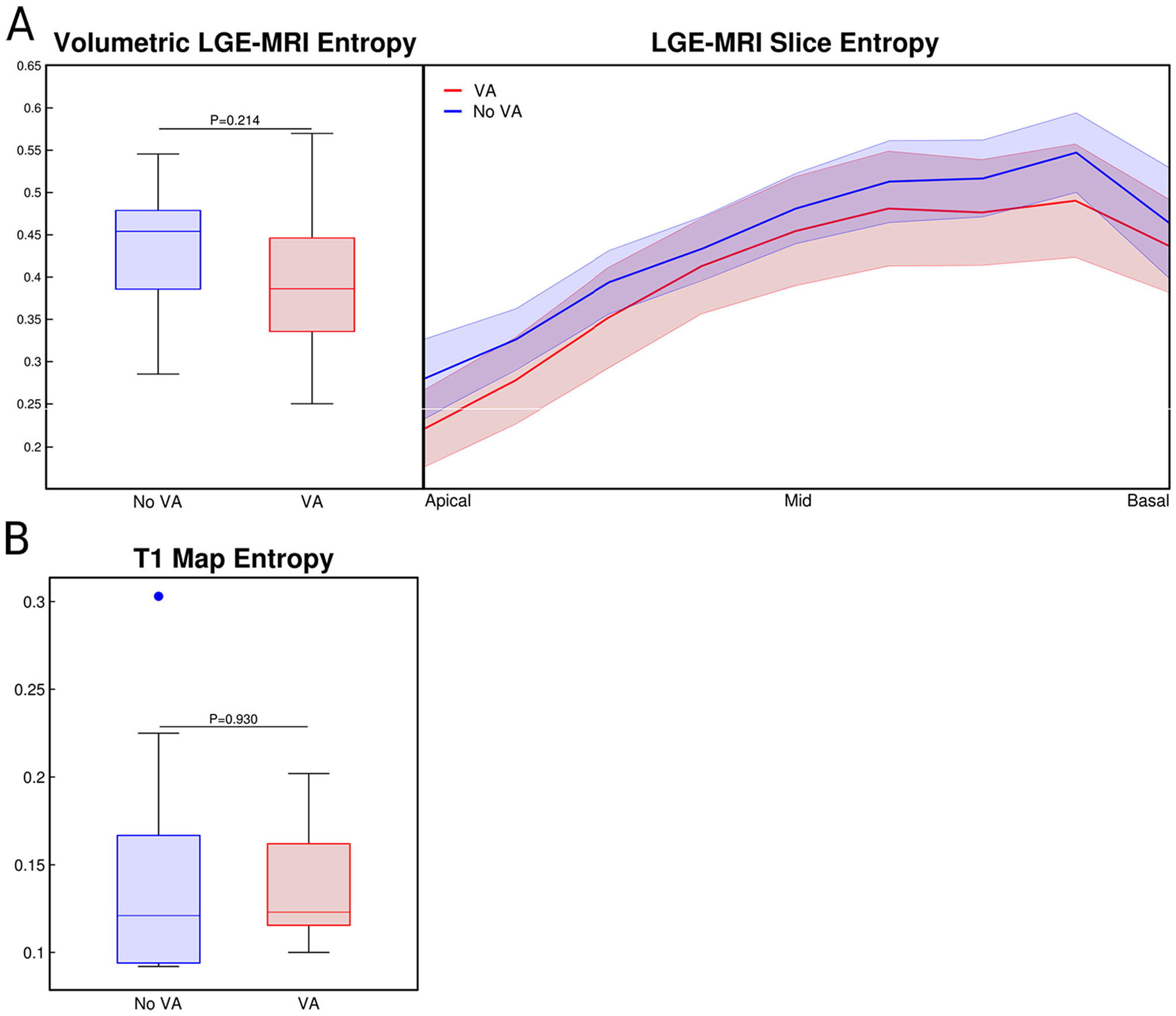
Entropy of MR Signal Intensity. A. Left: Comparison of the volumetric entropy of the LGE-MRI signal intensity between patients with and without VA. Right: Analysis of the short-axis, slice-by-slice entropy from the volumetric LGE-MRI. B. Comparison of the single post-contrast T1 map entropy between patients with and without VA. LGE-MRI: late gadolinium enhanced magnetic resonance imaging; VA: ventricular arrhythmia.
Fractal dimensions
The complexity of the fibrotic substrate, diffuse and dense fibrosis, was evaluated using the fractal dimension described by Eq. (2). Fig. 3 shows the comparison of border complexity for the combined fibrotic substrate, left, and the individual analyses of diffuse, center, and dense fibrosis, right. The fractal dimension was greater in the patients with VA for all three scenarios, but without statistical significance. Interestingly, the complexity of the combined fibrotic border was more significant (p = 0.080) than the complexity of the diffuse fibrosis (p = 0.095) and dense fibrosis (p = 0.593), individually. The amount of diffuse fibrosis is significantly greater than the amount of dense fibrosis, resulting in a similar probability of complexity as the combined fibrotic substrate.
Fig. 3.
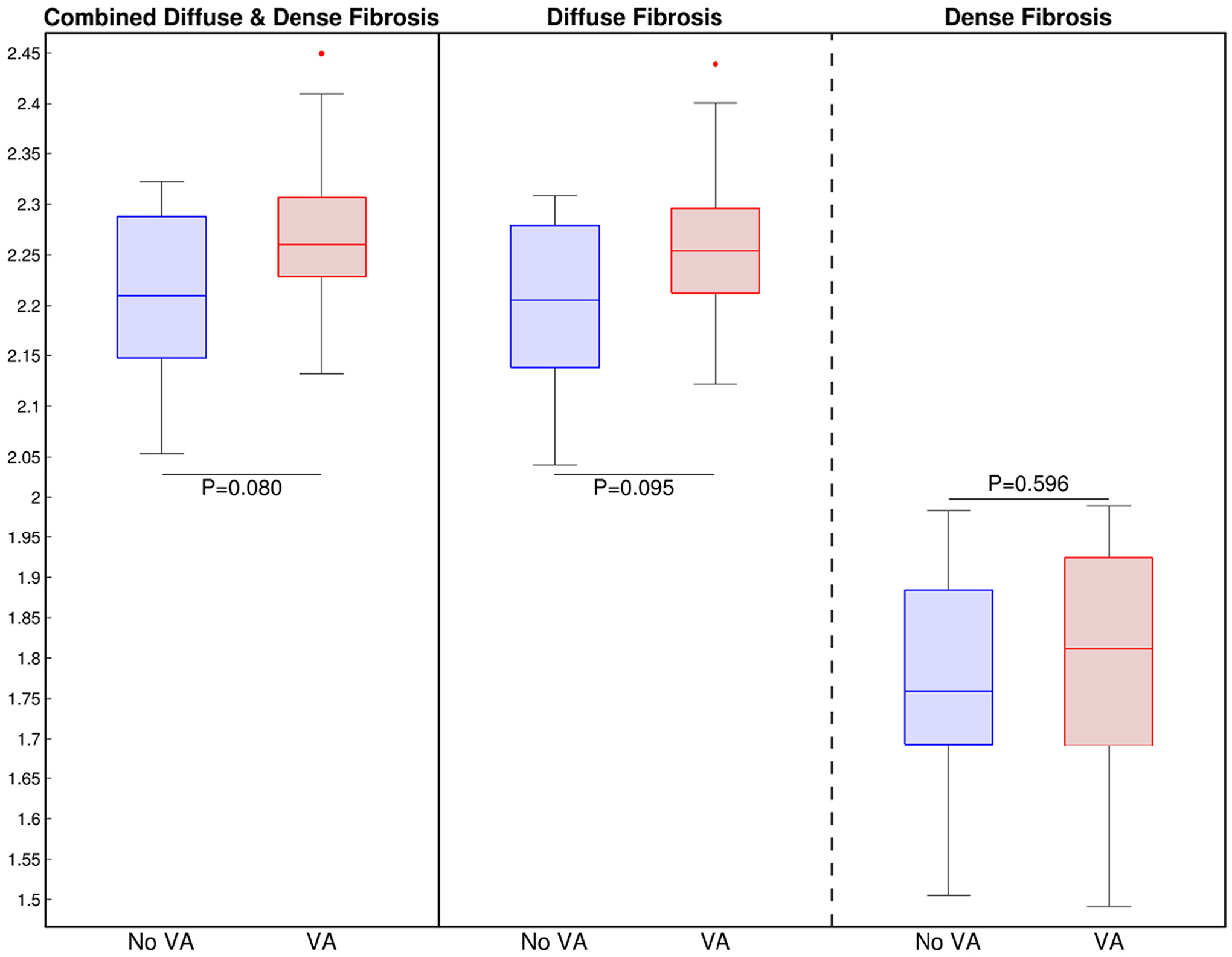
Fractals of the Fibrotic Substrate. Left: Comparison of the fractal dimension of the combined fibrosis (diffuse and dense fibrosis) borders between patients with and without VA. Middle: Comparison of the fractal dimension of only the diffuse fibrosis borders between patients with and without VA. Right: Comparison of the fractal dimension of only the dense fibrosis borders between patients with and without VA. VA: ventricular arrhythmia.
No statistical differences were found in the complexity of the combined fibrotic borders at the base, mid-myocardium, and apex (p = 0.10, 0.28, 0.90) as shown in Table 1. The probability was greatest for the basal region, which may be explained by the greater extent of hypertrophy and fibrosis across all patients.
The amounts of diffuse and dense fibrosis, and level of hypertrophy were all found to be statistically insignificant, as previously reported [9].
Approach 2: evaluation of the LGE-T1 virtual heart technology
Table 1 also shows the average number of VAs induced via rapid pacing per patient from all seven pacing locations. Patients without VA had an average of 0.46 ± 0.77 VAs induced whereas patients with VA had an average of 2.10 ± 1.29 (p < 0.01). We then considered the uniqueness of the VAs, or repetitive reentrant morphologies from different pacing locations within each patient heart model, and the patients without VA had an average of 0.38 ± 0.65 VAs induced whereas patients with VA had an average of 1.45 ± 0.97 (p < 0.01).
Finally, we compared the performance of two approaches for VA risk prediction in HCM: image-based biomarkers and LGE-T1 virtual heart technology, both developed from the personalized incorporation of LGE-MRI and T1 mapping. While statistically insignificant, the image-based biomarkers offer insights into the role of fibrotic remodeling towards arrhythmogenesis. The approach of the LGE-T1 virtual heart technology outperformed all the image-based biomarkers, correctly predicting VA risk in 21 out of the 26 patients.
Discussion
In this study, we present two approaches for assessing VA risk prediction in patients with HCM, in the hope that they may be used to guide prophylactic ICD implantation. LGE-MRI and T1 mapping provide different information about ventricular structure and remodeling, thus we hypothesized that combining them might be predictive of VA risk, as both diffuse and focal fibrosis can be a substrate for arrhythmogenesis. The LGE-T1 combination provides a personalized adjustment of the threshold used to calculate the patient-specific diffuse fibrosis hallmark to HCM. For the first time, we present a methodology for analyzing combined LGE-T1 imaging biomarkers and compare the resulting efficacy to that of the LGE-T1 virtual heart technology.
The image-based biomarkers were not significantly associated with VA risk for this cohort. However, certain insights can be gained from the analysis of the LGE-MRI signal intensity heterogeneity and fibrotic border zone complexity. Fractals of the fibrotic substrate showed more significance when considering the combined regions of diffuse and dense fibrosis versus discriminating either of them. The entropy for the patients without VA was consistently greater than the entropy of the patients with VA, for both the whole LGE-MRI volume and along the short axis of the LV. This suggests that VA inducibility may require coexistence of both non-injured and injured myocardium to produce an arrhythmogenic substrate. Previous studies have shown that entropy is independently associated with VAs, in patients with nonischemic and post-infarct related arrhythmias [13,14].
Our LGE-T1 virtual heart technology uses multiscale computational models of patients’ hearts reconstructed from LGE-MRI and T1 mapping. The HCM virtual-heart technology’s ability to comprehensively evaluate substrate arrhythmogenicity with electrophysiological characteristics underlies its superior performance. The virtual heart models represent the electrophysiology based on the combination of the imaging, with both the structural and functional properties of the myocardium. Furthermore, the virtual heart models are capable of uncovering emergent phenomena, such as the formation of reentrant VAs at specific locations, which may differ depending on the pacing location. The combination of electrophysiology and personalized structural remodeling, as well as the comprehensive evaluation of the substrate by pacing from different locations renders the approach of higher predictive capability. Our LGE-T1 virtual heart technology provided a much-improved risk assessment in this cohort compared to both the image biomarkers assessed herein and to the current clinical risk stratification methods [9].
Our study was limited by a small sample size, including the fact that a few LGE-MRI scans of HCM patients had imaging artifact, which prevented us from including more patients with LGE-MRI and T1 data.
Conclusions
For the first time, we combined two MR imaging techniques to analyze imaging biomarkers in HCM. Raw and processed image-based biomarkers cannot discriminate patients with VA from those without VA. The two imaging modalities were also the basis for constructing hybrid LGE-T1 technology. The LGE-T1-based virtual heart models could correctly predict VA risk for this cohort and may improve SCD risk stratification to better identify HCM patients for primary preventative ICD implantation.
Funding
NIH Grant R01HL142496 to NAT, NSF DGE-1746891 to RPO
Footnotes
CRediT authorship contribution statement
Ryan P. O’Hara: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Visualization, Project administration. Adityo Prakosa: Conceptualization, Methodology, Software, Investigation, Project administration. Edem Binka: Methodology, Data curation. Audrey Lacy: Investigation. Natalia A. Trayanova: Conceptualization, Resources, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors have no conflicts to disclose.
References
- [1].Maron BJ. Hypertrophic cardiomyopathy: an important global disease. Am J Med 2004;116:63–5. 10.1016/j.amjmed.2003.10.012. [DOI] [PubMed] [Google Scholar]
- [2].Galati G, Leone O, Pasquale F, Olivotto I, Biagini E, et al. Histological and histometric characterization of myocardial fibrosis in end-stage hypertrophic cardiomyopathy: a clinical-pathological study of 30 explanted hearts. Circ Heart Fail 2016;9:e003090. 10.1161/CIRCHEARTFAILURE.116.003090. [DOI] [PubMed] [Google Scholar]
- [3].Lambiase PD, Gold MR, Hood M, Boersma L, Theuns DAMJ, et al. Evaluation of subcutaneous ICD early performance in hypertrophic cardiomyopathy from the pooled EFFORTLESS and IDE cohorts. Heart Rhythm 2016;13:1066–74. 10.1016/j.hrthm.2016.01.001. [DOI] [PubMed] [Google Scholar]
- [4].Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, et al. American College of Cardiology Foundation/American Heart Association task force on practice guidelines. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: a report of the American College of Cardiology Foundation/American Heart Association task force on practice guidelines. Developed in collaboration with the American Association for Thoracic Surgery, American Society of Echocardiography, American Society of Nuclear Cardiology, Heart Failure Society of America, Heart Rhythm Society, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. J Am Coll Cardiol 2011;58:e212–60. 10.1016/j.jacc.2011.06.011. [DOI] [PubMed] [Google Scholar]
- [5].O’Mahony C, Jichi F, Pavlou M, Monserrat L, Anastasakis A, et al. A novel clinical risk prediction model for sudden cardiac death in hypertrophic cardiomyopathy (HCM risk-SCD). Eur Heart J 2014;35:2010–20. 10.1093/eurheartj/eht439. [DOI] [PubMed] [Google Scholar]
- [6].Schinkel AFL, Vriesendorp PA, Sijbrands EJG, Jordaens LJLM, ten Cate FJ, et al. Outcome and complications after implantable cardioverter defibrillator therapy in hypertrophic cardiomyopathy. Circ Heart Fail 2012;5:552–9. 10.1161/CIRCHEARTFAILURE.112.969626. [DOI] [PubMed] [Google Scholar]
- [7].Shade JK, Prakosa A, Popescu DM, Yu R, Okada DR, et al. Predicting risk of sudden cardiac death in patients with cardiac sarcoidosis using multimodality imaging and personalized heart modeling in a multivariable classifier. Sci Adv 2021;7:eabi8020. 10.1126/sciadv.abi8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shade JK, Cartoski MJ, Nikolov P, Prakosa A, Doshi A, et al. Ventricular arrhythmia risk prediction in repaired tetralogy of fallot using personalized computational cardiac models. Heart Rhythm 2020;17:408–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].O’Hara RP, Binka E, Prakosa A, Zimmerman SL, Cartoski MJ, et al. Personalized computational heart models with T1-mapped fibrotic remodeling predict sudden death risk in patients with hypertrophic cardiomyopathy. Elife 2022;11. 10.7554/eLife.73325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cartoski MJ, Nikolov PP, Prakosa A, Boyle PM, Spevak PJ, et al. Computational identification of ventricular arrhythmia risk in pediatric myocarditis. Pediatr Cardiol 2019;40:857–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Arevalo HJ, Vadakkumpadan F, Guallar E, Jebb A, Malamas P, et al. Arrhythmia risk stratification of patients after myocardial infarction using personalized heart models. Nat Commun 2016;7:11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chu LC, Corona-Villalobos CP, Halushka MK, Zhang Y, Pozzessere C, et al. Structural and functional correlates of myocardial T1 mapping in 321 patients with hypertrophic cardiomyopathy. J Comput Assist Tomogr 2017;41:653–60. 10.1097/RCT.0000000000000564. [DOI] [PubMed] [Google Scholar]
- [13].Antiochos P, Ge Y, van der Geest RJ, Madamanchi C, Qamar I, et al. Entropy as a measure of myocardial tissue heterogeneity in patients with ventricular arrhythmias. JACC Cardiovasc Imaging 2022;15:783–92. 10.1016/j.jcmg.2021.12.003. [DOI] [PubMed] [Google Scholar]
- [14].Androulakis AFA, Zeppenfeld K, Paiman EHM, Piers SRD, Wijnmaalen AP, et al. Entropy as a novel measure of myocardial tissue heterogeneity for prediction of ventricular arrhythmias and mortality in post-infarct patients. JACC Clin Electrophysiol 2019;5:480–9. 10.1016/j.jacep.2018.12.005. [DOI] [PubMed] [Google Scholar]
- [15].Bayer JD, Blake RC, Plank G, Trayanova NA. A novel rule-based algorithm for assigning myocardial fiber orientation to computational heart models. Ann Biomed Eng 2012;40:2243–54. 10.1007/s10439-012-0593-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Prassl AJ, Kickinger F, Ahammer H, Grau V, Schneider JE, et al. Automatically generated, anatomically accurate meshes for cardiac electrophysiology problems. IEEE Trans Biomed Eng 2009;56:1318–30. 10.1109/TBME.2009.2014243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].ten Tusscher KHWJ, Panfilov AV. Alternans and spiral breakup in a human ventricular tissue model. Am J Physiol Heart Circ Physiol 2006;291:H1088–100. 10.1152/ajpheart.00109.2006. [DOI] [PubMed] [Google Scholar]
- [18].Prakosa A, Arevalo HJ, Deng D, Boyle PM, Nikolov PP, et al. Personalized virtual-heart technology for guiding the ablation of infarct-related ventricular tachycardia. Nat Biomed Eng 2018;2:732–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Coppini R, Ferrantini C, Yao L, Fan P, Del Lungo M, et al. Late sodium current inhibition reverses electromechanical dysfunction in human hypertrophic cardiomyopathy. Circulation 2013;127:575–84. 10.1161/CIRCULATIONAHA.112.134932. [DOI] [PubMed] [Google Scholar]
- [20].Vigmond E, Weber dos Santos R, Prassl A, Deo M, Plank G. Solvers for the cardiac bidomain equations. Prog Biophys Mol Biol 2008;96:3–18. 10.1016/j.pbiomolbio.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]


