Abstract
Ossifying fibroma of the craniofacial bones is a fibro-osseous lesion characterized by varied patterns of bone formation in a fibroblastic stroma. Ossifying fibroma is a putatively benign lesion with no reports of malignant transformation or metastasis. Differentiation from other fibro-osseous lesions can be challenging necessitating synthesis of clinical, radiological and pathological findings. The molecular pathogenesis of ossifying fibroma is poorly understood but recent studies have reported MDM2 gene amplification and chromosomal copy number changes in a subset of ossifying fibromas. MDM2 amplification in ossifying fibroma, if true, presents a diagnostic problem because this genetic event, at least among craniofacial fibro-osseous lesions, was previously considered specific for low-grade osteosarcoma. In the present study, we investigated the utility of MDM2 and CDK4 immunohistochemistry, and fluorescence in situ hybridization for MDM2 gene amplification, in the diagnosis of 44 craniofacial bone ossifying fibromas. Focal MDM2 and CDK4 nuclear immunoreactivity was found in 11 and 1 ossifying fibromas, respectively, but none demonstrated MDM2 amplification by fluorescence in situ hybridization. A single tumor displayed MDM2 amplification without nuclear immunoreactivity to either MDM2 or CDK4. Our data suggest that while focal MDM2 and CDK4 nuclear expression may be detected in a minority of ossifying fibromas, this expression does not correlate with MDM2 amplification. In addition, MDM2 amplification is extremely rare in ossifying fibroma so the detection of this genetic abnormality should continue to raise concern for osteosarcoma.
Keywords: Ossifying fibroma, MDM2 amplification, Immunohistochemistry, Fluorescence in situ hybridization, Osteosarcoma
Introduction
Fibro-osseous lesions of the craniofacial bones are a diverse group of lesions defined by collagenous matrix containing variable amounts of immature osteoid and/or mineralized bone [1, 2]. This group of conditions includes both benign (e.g. ossifying fibroma) and malignant neoplasms (e.g. low-grade osteosarcoma), developmental dysplastic processes (e.g., cemento-osseous dysplasia and fibrous dysplasia) and reactive/inflammatory conditions (e.g., chronic sclerosing osteomyelitis) [2]. Ossifying fibroma (OF) may be further subdivided into three distinct clinicopathological entities: cemento-ossifying fibroma, juvenile trabecular ossifying fibroma, and juvenile psammomatoid ossifying fibroma [2]. As prognosis, management and morbidity differ significantly between the fibro-osseous lesions, accurate diagnosis is essential [3]. There is significant overlap in morphology amongst the fibro-osseous lesions requiring the integration of clinical and radiographic features in addition to pathologic findings to establish the correct classification.
The distinction between the fibro-osseus lesions can be challenging, particularly on a small biopsy. In select cases, immunohistochemistry, genetic or molecular testing may be diagnostically useful especially as the genetics of some fibro-osseous lesions become better understood. For example, fibrous dysplasia harbors an activating mutation in GNAS that appears to be specific for this entity [4]. In OF, rare CTNNB1 and HRPT2 mutations have been described [5, 6]. Perhaps the most important genetic finding in the diagnosis of craniofacial fibro-osseous lesions is amplification of chromosome 12q13-15 subregion that includes the genes MDM2 and CDK4 in low-grade osteosarcomas. MDM2 gene amplification can be identified indirectly by protein overexpression of MDM2 and CDK4 or directly by fluorescent in situ hybridization (FISH), PCR-based or next-generation sequencing-based methods [12, 15]. To date, multiple studies have shown high sensitivity and specificity of MDM2 protein overexpression by immunohistochemistry and/or MDM2 gene amplification to support low-grade osteosarcoma over morphologic mimics [7–12]. However, a recent study reported chromosome 12 long arm rearrangement and amplification of MDM2 and RASAL1 in a subset of juvenile OF using quantitative polymerase chain reaction (qPCR) [13] although MDM2 and CDK4 protein expression were not tested. A separate study used laser dissection and copy number profiling to demonstrate chromosome 12 gains in OF including one OF with gains encompassing MDM2 and CDK4 confirmed by qPCR [14]. These results were not correlated with immunohistochemistry or cytogenetics.
The above results suggest that MDM2 amplification and resulting overexpression can be identified in at least a subset of OF. If confirmed, such a finding raises doubt about the diagnostic utility of MDM2 and CDK4 immunohistochemistry and MDM2 FISH to distinguish between OF and low-grade osteosarcoma [9–12]. In order to further investigate the frequency of MDM2 and CDK4 protein expression and MDM2 amplification in craniofacial OF, we studied a large group of well characterized OF (conventional, juvenile trabecular and juvenile psammomatoid) by immunohistochemistry and FISH.
Materials and Methods
Case Selection and Tissue Microarray Preparation
We initially identified 49 cases of ossifying fibroma from the pathology archives at the University of California San Francisco (UCSF). The clinical and radiographic data were retrieved and reviewed. Of these, 44 cases from 42 patients had available microscopic slides and were further studied. The diagnosis was based on light microscopic, radiographic and clinical features with consensus of all three authors. A tissue microarray (TMA) was prepared using 2 mm cores in triplicate from a formalin fixed paraffin embedded (FFPE) block from 40 cases with available paraffin blocks. For cases with multiple donor blocks, undecalcified blocks were used whenever possible. Furthermore, if decalcification was noted in the gross description, this was recorded. Immunohistochemistry for MDM2, CDK4 and FISH for MDM2 were performed and evaluated on the TMA. Two cases (cases 9 and 17) were stained both with TMA and as a whole slide sections. The remaining four cases (cases 21, 22, 23 and 29) were from the authors’ consultation files with immunohistochemistry on whole sections for MDM2 and CDK4 from the diagnostic workup of these cases but no FFPE blocks were available for FISH.
Immunohistochemistry
Immunostaining was performed on FFPE tissue. Immunohistochemical analysis was carried out following standard protocols. Briefly, 4-µm deparaffinized sections of each tumor were stained for MDM2 (IF2, Life Technologies, New York, USA) and CDK4 (DCS-31, Life Technologies, New York, USA) on a Leica Biosystems’ (Buffalo Grove, IL) Bond III automated immunostainer for 20 min at room temperature. Stained slides were semiquantitatively scored for nuclear staining by three pathologists (DB, AEH, RCJ). Scores were only recorded if all three cores on the TMA contained adequate lesional tissue. Discrepant scores were re-reviewed as a group to arrive at a consensus score. Scoring used previous published methods [10]: a positive result was recorded when at least one cell nucleus was stained per high power field and further stratified based on the percent of positive nuclei: ≤ 10%, 11–25%, 26–50% and > 50%. Only nuclear staining in mononuclear cells was scored as positive. Nuclear staining in osteoclast-type giant cells and cytoplasmic staining were disregarded. Dedifferentiated liposarcoma and normal thymus served as positive and negative controls, respectively [15].
Fluorescence In Situ Hybridization (FISH)
FISH was performed and evaluated following standard protocols. Briefly, 4 μm sections of FFPE tissue were hybridized with fluorescent probes: locus-specific identifier (LSI) MDM2 mapped to chromosome 12q15 (Vysis LSI MDM2 Spectrum Orange Probe) and centromere 12 (Vysis CEP12 Spectrum Green Probe) counterstained with 4, 6-diamidino-2-phenylindole. A minimum of 25 nuclei were visualized per slide for positive cases, 50 nuclei for negative cases. The number of fluorescent signals was evaluated for each nucleus analyzed. If at least one or two bright fluorescent spots per nucleus could not be seen on at least 80% of cells, the result was considered to be non-diagnostic (i.e. hybridization failure). Amplification was defined as a MDM2/CEP12 ratio ≥ 2.0. Dedifferentiated liposarcoma and normal thymus served as positive and negative controls, respectively [15].
Results
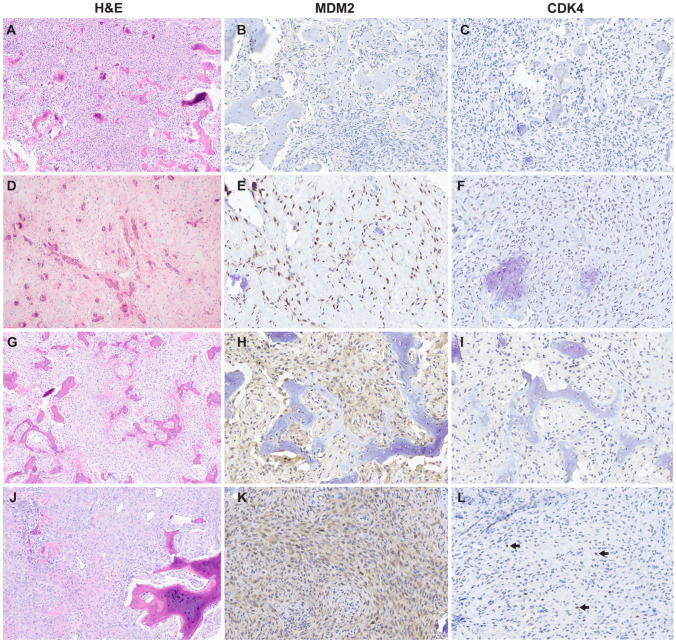
The morphologic, clinical and immunophenotypic characteristics of the OFs are summarized in Table 1. The collection consisted of 44 tumors from 42 patients (11 male (27%) and 31 female (73%) with mean age of 27 years (range 1–68). The majority of the cases were located in the mandible (27, 60%) followed by maxilla (12, 26%). The cases were classified into three subtypes: (conventional) ossifying fibroma (22/44; 49%), juvenile trabecular ossifying fibroma (16/44; 38%), and juvenile psammomatoid ossifying fibroma (6/44; 13%). Figure 1 illustrates representative examples of histology and immunohistochemistry results of each subtype.
Table 1.
Summary of clinical, pathologic and genetic results of ossifying fibromas
| Case# | Age | Gender | Location | Diagnosis | Decalcified | MDM2 IHC | CDK4 IHC | MDM2 FISH |
|---|---|---|---|---|---|---|---|---|
| 1 | 33 | M | Ethmoid | OF | No | – | – | – |
| 2 | 13 | M | Orbit | JPOF | No | – | – | ND |
| 3* | 11 | F | Maxilla | JPOF | Yes | – | – | – |
| 4* | 11 | F | Maxilla | JPOF | No | + | – | – |
| 5 | 26 | F | Mandible | OF | Yes | – | – | – |
| 6 | 9 | F | Mandible | OF | No | + | – | – |
| 7 | 12 | F | Mandible | OF | Yes | – | – | ND |
| 8 | 30 | F | Sphenoid | JPOF | No | ND | – | – |
| 9 | 58 | F | Ethmoid | OF | No | – | – | + |
| 10 | 47 | F | Maxilla | OF | No | + | – | – |
| 11 | 13 | F | Maxilla | JPOF | No | +++ | – | – |
| 12 | 17 | M | Sphenoid | JPOF | No | – | – | ND |
| 13 | 67 | F | Mandible | OF | No | – | – | – |
| 14 | 43 | F | Maxilla | OF | Yes | – | – | – |
| 15** | 12 | M | Mandible | JTOF | No | – | – | – |
| 16** | 12 | M | Mandible | JTOF | No | +++ | – | – |
| 17 | 68 | F | Mandible | OF | No | – | – | – |
| 18 | 45 | F | Mandible | OF | No | – | – | – |
| 19 | 1 | M | Maxilla | JTOF | No | + | – | – |
| 20 | 54 | F | Mandible | OF | Yes | – | – | – |
| 21 | 8 | F | Maxilla | JTOF | No | – | – | NP |
| 22 | 16 | F | Mandible | JTOF | No | – | – | NP |
| 23 | 4 | F | Mandible | JTOF | No | – | – | NP |
| 24 | 12 | F | Mandible | JTOF | No | – | – | – |
| 25 | 33 | F | Mandible | JTOF | No | – | – | – |
| 26 | 31 | F | Mandible | JTOF | Yes | + | – | – |
| 27 | 12 | M | Maxilla | JTOF | No | – | – | – |
| 28 | 12 | M | Maxilla | JTOF | Yes | – | – | – |
| 29 | 46 | F | Mandible | JTOF | Yes | – | – | NP |
| 30 | 31 | F | Mandible | JTOF | Yes | + | – | – |
| 31 | 14 | M | Maxilla | JTOF | Yes | – | – | – |
| 32 | 28 | F | Mandible | JTOF | No | – | – | – |
| 33 | 14 | M | Maxilla | JTOF | No | – | – | – |
| 34 | 29 | M | Mandible | OF | Yes | – | – | – |
| 35 | 41 | F | Mandible | OF | No | + | – | – |
| 36 | 24 | M | Mandible | OF | No | – | – | ND |
| 37 | 12 | F | Mandible | OF | Yes | – | – | ND |
| 38 | 45 | F | Mandible | OF | No | – | – | ND |
| 39 | 26 | F | Mandible | OF | No | – | – | – |
| 40 | 26 | F | Mandible | OF | No | + | + | – |
| 41 | 28 | F | Mandible | OF | No | – | – | – |
| 42 | 25 | F | Mandible | OF | No | – | – | – |
| 43 | 58 | F | Mandible | OF | Yes | + | – | – |
| 44 | 32 | F | Maxilla | OF | No | – | – | – |
MDM2 and CDK immunohistochemistry: –: No staining in any cells; +: ≤10% of positive tumor cells; ++: 11–25% of positive tumor cells; +++: 26–50% of positive tumor cells; ++++: >50% of positive tumor cells
F female; M male; OF ossifying fibroma; JTOF Juvenile trabecular ossifying fibroma; JPOF Juvenile psammomatoid ossifying fibroma; NP not performed; ND non-diagnostic
*Cases 3 and 4 are from the same patient
**Cases 15 and 16 are from the same patient
Fig. 1.
Representative histomorphology and immunohistochemistry results of ossifying fibroma. A–C Ossifying fibroma (case 9) composed of low-grade cellular fibroblastic stroma that invades and replaces woven bone and forms new bone; no nuclear immunoreactivity MDM2 or CDK4. D–F Juvenile psammomatoid ossifying fibroma (case 11) with hyalinized stroma and bone deposits with concentric calcification; nuclear MDM2 positivity in > 26–50% of spindle cells; no nuclear immunoreactivity to CDK4. G–I Juvenile trabecular ossifying fibroma (case 26) with cellular spindled growth and woven bone that presents as anastomosing trabeculae with mineralization; nuclear MDM2 positivity in ≤ 10% of cells (nuclear staining in osteoclast-type giant cells was disregarded- see methods).; no nuclear immunoreactivity to CDK4. J–L Ossifying fibroma (case 41) with bone formation and dense cellular stroma with mineralization and osteoclast-type giant cells; strong cytoplasmic and rare MDM2 nuclear staining in spindle cells; rare faint nuclear CDK4 staining in spindle cells (arrows highlight rare positive nuclei)
MDM2 nuclear immunostaining was observed in 11/43 (25%) OF, of which only 1 case (case 40) also showed CDK4 positivity (1/44, 2%) MDM2 immunostaining, if present, was present ≤ 10% of the cells in the majority of positive cases (9/11, 82%) and ≤ 10% of the cells in the one CDK4 positive case (case 40). Occasional osteoclast-type giant cells demonstrated nuclear MDM2 immunostaining (Fig. 1H) and considered negative for the purposes of scoring.
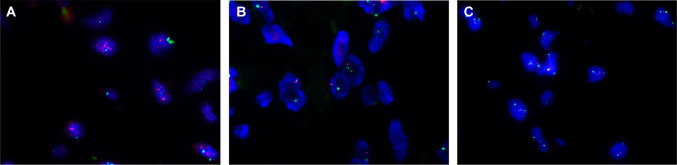
FISH was informative in 34 of the 40 cases with available FFPE tissue. The remaining 6 cases demonstrated insufficient hybridization signals and were considered nondiagnostic by FISH. Representative results are shown in Fig. 2. Only a single case (case 9, Fig. 2A) demonstrated MDM2 amplification (MDM2:CEP12 ratio 5.0:1). This case was negative for MDM2 and CDK4 by IHC on both the TMA and on a whole section (Fig. 1A–C). We identified no correlation between subtype of OF, cellularity, degree of atypia, and MDM2 or CDK4 status. The fraction of MDM2-positive cases was similar in decalcified and non-decalcified samples (3/12 [25%] and 8/28 [29%], respectively).
Fig. 2.
Representative fluorescence in situ hybridization results in ossifying fibroma. A Ossifying fibroma, case 9 was the only case to show MDM2 amplification (MDM2:CEP12 ratio 5.0:1). B, C All remaining ossifying fibromas were negative for MDM2 amplification regardless of MDM2 and CDK4 immunohistochemical status; B JTOF, case 16; C OF, case 1
Discussion
Amplification of 12q13-15 involving MDM2 and CDK4 genes was initially reported as a sensitive and specific finding of parosteal and low-grade central osteosarcomas of long bones [10, 16, 17]. Subsequently, this genetic event was also identified in craniofacial osteosarcomas, detectable by immunohistochemistry or FISH [12]. The above results ostensibly permit distinction of low-grade craniofacial osteosarcomas from other fibro-osseous lesions including OF using immunohistochemistry or genetic methods. However, recently, a chromosome 12 long arm rearrangement covering MDM2 and RASAL1 amplification was reported in a subset of ossifying fibroma using qPCR [13]. Furthermore, using low-output whole genome sequencing, a second study discovered copy number alterations with associated distinct genomic patterns in OF, specifically copy number amplifications of chromosomes 7 and 12 including one case with amplification of the MDM2 gene [14]. These latter two studies raised concern that OF may demonstrate MDM2 amplification and resulting MDM2 and CDK4 expression by immunohistochemistry in OF. If so, these ancillary tests would no longer support osteosarcoma over OF.
To our knowledge, the present series is the largest study of MDM2 and CDK4 expression and MDM2 amplification status in OF. Our results suggest that gene amplification of MDM2 as detected by FISH is extremely rare in OF of the craniofacial bones (3% of OF in this series) while nuclear expression of MDM2 and CDK4 can be seen in a subset of cases (25% and 2%, respectively), but is typically focal (≤ 10% of nuclei staining). These findings are relevant because pathologists may include ancillary immunohistochemistry and/or cytogenetics to distinguish OF from other fibro-osseous lesions, including osteosarcoma. Based on the current study, focal immunopositivity for MDM2 and CDK4 should not be sufficient to warrant a diagnosis of osteosarcoma but may prompt direct testing by FISH. Further, our results suggest that detection of MDM2 amplification by FISH does not exclude OF (positive in 3% of the OF in the present series), but such result should still raise a high index of suspicion for osteosarcoma in which MDM2 amplification is reportedly much more common (67–100%). [8, 16, 17]
The increased frequency of MDM2 amplification in OF detected in recent studies [13, 14] compared to our results and those of prior authors [10–12] is perplexing. Although the discrepancy may be due to different techniques used in detection of MDM2 amplification (i.e. qPCR or FISH), multiple previous studies showed high level of concordance between different molecular assays for MDM2 amplification in adipocytic lesions or for HER2 gene amplification in breast specimens [15, 18]. Thus, the discordant findings should be addressed with optimization of MDM2 testing in bone tumors along with tissue processing protocols. It should also be noted that one previously reported OF with MDM2 amplification recurred multiple times and eventually recurred as osteosarcoma. [14] The above finding raises the possibility that this tumor might have been better classified as osteosarcoma initially since malignant transformation of OF is exceedingly rare in the literature.
Based on the presence of MDM2 amplification, we also considered whether case 9 represented a misdiagnosed low-grade osteosarcoma rather than OF. Low-grade osteosarcoma of the gnathic bones is similar to low-grade central osteosarcoma of long bones in that both frequently demonstrate MDM2 amplification and a fibro-osseous appearance histologically [8, 10–12]. Features that favor low-grade osteosarcoma over OF include radiographically ill-defined margins, elongated parallel trabeculae of woven bone, permeation of the fibrous component into native lamellar bone and mild nuclear atypia (slight nuclear enlargement and hyperchromasia) [2]. In case 9, imaging showed a relatively non-aggressive appearing lesion without cortical destruction or soft tissue extension. Microscopically, this case did not demonstrate any of the above described features of low-grade osteosarcoma. Finally, this patient has not developed a recurrence or progression to higher grade tumor after 15 years of clinical follow-up. In summary, while we cannot exclude that MDM2 amplified case 9 represented a low-grade osteosarcoma, the clinical, radiographic and histomorphologic features argue for OF.
To date, five studies have reported consistently negative IHC results for MDM2 in benign craniofacial fibro-osseous lesions [10–13, 19]. However, in our series, 11 cases (25%) exhibited MDM2 nuclear immunostaining, albeit focal, without MDM2 amplification by FISH. In contrast to other tumors with MDM2 amplification [10, 20, 21], the correlation between IHC and FISH was poor in our study. Acid decalcification may negatively affect antigenicity and DNA-based testing. In the present series, the fraction of cases positive for MDM2 by immunohistochemistry was similar regardless of decalcification status suggesting that decalcification did not have a marked effect on MDM2 antigenicity. However, FISH was inconclusive in some cases, despite the fact that decalcification was not mentioned in the processing of most of these samples. Furthermore, the single case with MDM2 amplification detected by FISH was MDM2 negative by IHC. Therefore we cannot rule out the possibility that these samples were decalcified without having been recorded in the gross description. Because the majority of our cases were informative by FISH and lacked MDM2 amplification, our findings suggests that both negative and positive IHC for MDM2 should be interpreted cautiously. A positive MDM2 and CDK4 IHC result should be supplemented by genetic testing to detect MDM2 gene amplification directly prior to rendering a diagnosis of osteosarcoma. Accordingly, it would be prudent to process fibro-osseous lesions, or at least a portion of the specimen, without acid decalcification to preserve DNA for future genetic testing. If decalcification is unavoidable, the type of decalcifier used should be recorded in the gross description to inform the interpretation of ancillary testing. If possible, EDTA based decalcification is preferred as it appears to have less impact on FISH [22].
A limitation of our study is the risk that the small cores sampled in a TMA may not be representative of whole tumor if there is heterogeneous protein expression. The TMA technique was chosen both to simulate the scant tissue available for diagnosis on small biopsies and to manage financial constraints. However, triplicate cores, obtained from disparate areas of donor blocks showed concordance within a given case providing representation of multiple areas of tumor. Furthermore, all 6 cases stained using whole slides (including case 9, showing MDM2 amplification by FISH) were uniformly negative arguing against heterogeneous or focal expression of MDM2 or CDK4. Finally, while protein expression and immunohistochemical staining may be heterogeneous in a given histologic section, clonal genetic driver events like MDM2 amplification may be more homogeneous in a neoplasm [23].
In summary, we report here that focal (typically ≤ 10% of tumor cells) MDM2 and less frequently CDK4 nuclear expression may be detected in a minority of craniofacial OF. Therefore, positive IHC results for MDM2 and/or CDK4 should not exclude a diagnosis of OF but prompt testing for MDM2 amplification by other methods including FISH. While a positive MDM2 amplification result in a craniofacial fibro-osseous lesion does not completely exclude OF, it should still raise concern for low-grade osteosarcoma because this genetic event is much more common in the latter. Finally, future studies evaluating the complete genomic profiling of OF will be useful to detect specific genetic or molecular events that allow distinction from other fibro-osseous lesions in exceptionally difficult cases.
Author Contributions
All authors confirm they have meaningfully contributed to the research and read and approved the final manuscript.
Funding
The study was funded by the University California at San Francisco Department of Pathology, Clinical Research Endowment.
Data Availability
Possible upon reasonable request, deidentified for maintenance of anonymity and compliance with IRB approval.
Code Availability
Not applicable.
Declarations
Conflict of interest
All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript.
Ethics Approval
This study was approved by the UCSF Human Subjects Institutional Review Board (IRB 11-05361) which did not require informed consent.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Informed Consent
The IRB-approved study was classified as exempt, which does not require informed consent.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.El Mofty SK, Nelson B, Toyosawa S. WHO classification of head and neck tumors. Lyon: IARC Press; 2017. Fibro-osseous and osteochondromatous lesions; pp. 251–5. [Google Scholar]
- 2.Hameed M, Horvai AE, Jordan RCK. Soft tissue special issue: gnathic fibro-osseous lesions and osteosarcoma. Head Neck Pathol. 2020;14(1):70–82. doi: 10.1007/s12105-019-01094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koury ME, Regezi JA, Perrott DH, Kaban LB. “Atypical” fibroosseous lesions: diagnostic challenges and treatment concepts. Int J Oral Maxillofac Surg. 1995;24:162–9. doi: 10.1016/S0901-5027(06)80094-9. [DOI] [PubMed] [Google Scholar]
- 4.Shi RR, Li XF, Zhang R, Chen Y, Li TJ. GNAS mutational analysis in differentiating fibrous dysplasia and ossifying fibroma of the jaw. Mod Pathol. 2013;26:1023–31. doi: 10.1038/modpathol.2013.31. [DOI] [PubMed] [Google Scholar]
- 5.Horvai E, Jordan ACR. Fibro-osseous lesions of the craniofacial bones: β-catenin immunohistochemical analysis and CTNNB1 and APC mutation analysis [published correction appears in Head Neck Pathol. 2014 Sep;8(3):369] Head Neck Pathol. 2014;8(3):291–297. doi: 10.1007/s12105-014-0535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpten JD, Robbins CM, Villablanca A, et al. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat Genet. 2002;32:676–80. doi: 10.1038/ng1048. [DOI] [PubMed] [Google Scholar]
- 7.Junior AT, de Abreu AF, Pinto CA, Carvalho AL, Kowalski LP, Lopes MA. Clinicopathological and immunohistochemical analysis of twenty-five head and neck osteosarcomas. Oral Oncol. 2003;39(5):521–30. doi: 10.1016/S1368-8375(03)00017-4. [DOI] [PubMed] [Google Scholar]
- 8.Lopes MA, Nikitakis NG, Ord RA, Sauk J., Jr Amplification and protein expression of chromosome 12q13-15 genes in osteosarcomas of the jaws. Oral Oncol. 2001;37(7):566–71. doi: 10.1016/S1368-8375(00)00130-5. [DOI] [PubMed] [Google Scholar]
- 9.Guerin M, Thariat J, Ouali M, Bouvier C, Decouvelaere AV, Cassagnau E, et al. A new subtype of high-grade mandibular osteosarcoma with RASAL1/MDM2 amplification. Hum Pathol. 2016;50:70–8. doi: 10.1016/j.humpath.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Dujardin F, Binh MB, Bouvier C, Gomez-Brouchet A, Larousserie F, Muret A, et al. MDM2 and CDK4 immunohistochemistry is a valuable tool in the differential diagnosis of low-grade osteosarcomas and other primary fibro-osseous lesions of the bone. Mod Pathol. 2011;24(5):624–37. doi: 10.1038/modpathol.2010.229. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida A, Ushiku T, Motoi T, Shibata T, Beppu Y, Fukayama M, et al. Immunohistochemical analysis of MDM2 and CDK4 distinguishes low-grade osteosarcoma from benign mimics. Mod Pathol. 2010;23(9):1279–88. doi: 10.1038/modpathol.2010.124. [DOI] [PubMed] [Google Scholar]
- 12.Limbach AL, Lingen MW, McElherne J, et al. The utility of MDM2 and CDK4 immunohistochemistry and MDM2 FISH in craniofacial osteosarcoma. Head Neck Pathol. 2020;14(4):889–98. doi: 10.1007/s12105-020-01139-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tabareau-Delalande F, Collin C, Gomez-Brouchet A, et al. Chromosome 12 long arm rearrangement covering MDM2 and RASAL1 is associated with aggressive craniofacial juvenile ossifying fibroma and extracranial psammomatoid fibro-osseous lesions. Mod Pathol. 2015;28(1):48–56. doi: 10.1038/modpathol.2014.80. [DOI] [PubMed] [Google Scholar]
- 14.Ma M, Liu L, Shi R, et al. Copy number alteration profiling facilitates differential diagnosis between ossifying fibroma and fibrous dysplasia of the jaws. Int J Oral Sci. 2021;13(1):21. doi: 10.1038/s41368-021-00127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sirvent N, Coindre JM, Maire G, et al. Detection of MDM2-CDK4 amplification by fluorescence in situ hybridization in 200 paraffin-embedded tumor samples: utility in diagnosing adipocytic lesions and comparison with immunohistochemistry and real-time PCR. Am J Surg Pathol. 2007;31(10):1476–89. [DOI] [PubMed]
- 16.Tarkkanen M, Böhling T, Gamberi G, et al. Comparative genomic hybridization of low-grade central osteosarcoma. Mod Pathol. 1998;11(5):421–6. [PubMed] [Google Scholar]
- 17.He X, Pang Z, Zhang X, et al. Consistent amplification of FRS2 and MDM2 in low-grade osteosarcoma: a genetic study of 22 cases with clinicopathologic analysis. Am J Surg Pathol. 2018;42(9):1143–55. doi: 10.1097/PAS.0000000000001125. [DOI] [PubMed] [Google Scholar]
- 18.Furrer D, Sanschagrin F, Jacob S, Diorio C. Advantages and disadvantages of technologies for HER2 testing in breast cancer specimens. Am J Clin Pathol. 2015;144(5):686–703. doi: 10.1309/AJCPT41TCBUEVDQC. [DOI] [PubMed] [Google Scholar]
- 19.Kaur H, Kala S, Sood A, et al. Role of MDM2, CDK4, BCL2, parafibromin and galectin 1 in differentiating osteosarcoma from its benign fibro-osseous lesions. Head Neck Pathol. 2022 doi: 10.1007/s12105-022-01434-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binh MB, Sastre-Garau X, Guillou L, et al. MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well-differentiated and dedifferentiated liposarcoma subtypes: a comparative analysis of 559 soft tissue neoplasms with genetic data. Am J Surg Pathol. 2005;29(10):1340–7. doi: 10.1097/01.pas.0000170343.09562.39. [DOI] [PubMed] [Google Scholar]
- 21.Koelsche C, Benhamida JK, Kommoss FKF, et al. Intimal sarcomas and undifferentiated cardiac sarcomas carry mutually exclusive MDM2, MDM4, and CDK6 amplifications and share a common DNA methylation signature. Mod Pathol. 2021;34(12):2122–9. doi: 10.1038/s41379-021-00874-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schrijver W, van der Groep P, Hoefnagel L, et al. Influence of decalcification procedures on immunohistochemistry and molecular pathology in breast cancer. Mod Pathol. 2016;29(12):1460–1470. doi: 10.1038/modpathol.2016.116. [DOI] [PubMed] [Google Scholar]
- 23.Weaver J, Downs-Kelly E, Goldblum JR, et al. Fluorescence in situ hybridization for MDM2 gene amplification as a diagnostic tool in lipomatous neoplasms. Mod Pathol. 2008;21(8):943–9. doi: 10.1038/modpathol.2008.84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Possible upon reasonable request, deidentified for maintenance of anonymity and compliance with IRB approval.
Not applicable.