Abstract
Background
Salivary gland mucoepidermoid carcinoma (MEC) poses a considerable risk of locoregional and distant metastasis after conventional treatments. There is an evident need for specifying prognostic biomarkers to identify patients who are in need of more intensive and prolonged follow-ups. This study aimed to assess the mucin 1 (MUC1) expression level and its potential regulatory microRNAs in salivary gland MEC and their prognostic potentials.
Materials and Methods
The expression of MUC1 in salivary gland MEC tissues was assessed in 47 samples using immunohistochemistry. Related microRNA (miR-145 and miR-21) were evaluated using quantitative Reverse Transcription PCR. The associations between MUC1 and microRNAs expressions and clinicopathological parameters were investigated.
Results
MUC1 expression levels positively correlated with histologic grade (p < 0.001), clinical stage (p = 0.04), risk of nodal metastasis (p = 0.02), as well as the likelihood of opting for radical treatment (p = 0.01). Increased expression of miR-21 (p < 0.001) and decreased expression of miR-145 (p < 0.001) were observed in MECs compared to normal salivary gland tissue. MiR-145 negatively (p = 0.01) and miR-21 positively (p = 0.01) correlated with MUC1 overexpression. Based on the univariate cox proportional hazard model, histologic grade and MUC1 expression level were significantly associated with disease-free, cancer-specific, and overall survival. However, the multivariable cox proportional hazard model indicated tumor grade as the only prognostic factor associated with disease-free survival.
Conclusion
Our results support the tumor suppressor role of miR-145 and the oncogenic role of miR-21 in salivary gland MEC. Also, MUC1 and miR-145 overexpression, as well as miR-21 suppression, show promising association with histologic tumor grade and clinical stage.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12105-022-01475-0.
Keywords: miR-145, miR-21, mucoepidermoid carcinoma, mucin 1, salivary gland neoplasms
Introduction
Salivary gland tumors (SGTs) are an infrequent and diverse group of tumors that constitutes about 3 to 6% of head and neck neoplasms. According to cancer statistics of GLOBOCAN 2020, the worldwide salivary gland malignancies estimated incidence rates were approximately 0.57 per 100,000 [1]. Among these, mucoepidermoid carcinoma (MEC) is considered to be the most common type of salivary gland malignant tumor.
Despite considerable developments in the diagnosis and treatment of MEC, there is still an increased risk for locoregional and distant failures, which leads to poor overall survival [2]. Furthermore, the exact mechanisms leading to the poor prognosis of anticancer therapeutic approaches have not been elucidated. Thus, there is an absolute necessity to uncover the molecular mechanism underlying the tumor progression, poor prognosis, and unfavorable outcomes [3].
Mucin 1 (MUC1) is a heterodimeric plasma membrane-bound protein that is expressed on the apical plasma membrane of most epithelia. In salivary glands, mucinous cells show positive apical membranous staining for MUC1, while in the intermediate, clear, and epidermoid cells of mucoepidermoid carcinoma, the entire membrane shows positive reactivity for MUC1 [4]. Although specific mucin expression patterns could be preserved during tumoral transformation in an organ-dependent manner, abnormal expression of MUC1 has been identified in up to 95% of adenocarcinomas [5].
Several studies have indicated that increased MUC1 expression is associated with metastasis and poor prognosis in breast, prostate, lungs, and colorectal cancers [6]. In salivary gland carcinomas, few studies have been carried out to investigate the possible link of MUC1 expression to the prognosis, with quite contradictory results [4]. While potential regulatory biomarkers of MUC1 have been observed in ovarian cancer and breast cancer, there is an evident lack of studies focusing on salivary gland malignancies [7, 8].
MicroRNAs are small, 18–25 nucleotide, non-coding RNAs that modulate gene expression at the post-transcriptional level. These classes of RNAs suppress gene expression through translational repression or cleavage of mRNAs targets [3]. There is convincing evidence suggesting that microRNAs have crucial roles in several biological processes as well as oncogenic pathways, including cell cycle, proliferation, migration, cell differentiation, invasion, angiogenesis, and apoptosis [9]. The dysregulation of microRNAs that has been frequently detected in most human cancers is known to play a crucial role in cancer development as either oncogenes or tumor suppressors, determined by their target transcripts. Recent studies showed the profound association between microRNAs and the progression of salivary gland carcinomas [10].
MiR-145 has been described as a vital tumor suppressor microRNA under-expressed in different malignancies, including pancreatic, colorectal, breast, prostate, ovarian cancer, as well as head and neck squamous cell carcinomas [3]. It is shown to play a significant role in inhibiting tumor initiation, growth, invasion, and metastasis in a cell-specific manner [3, 8]. Additionally, lower miR-145 expression is accompanied by poor prognosis in different types of cancers, indicating the need to identify its possible contributions to regulating signaling pathways [8].
MiR-21 is one of the most comprehensively surveyed oncomiRs broadly expressed in various malignancies, such as breast, gastric, colorectal, and lung cancer [9, 11]. The cancerous roles of miR-21 are well studied in the head and neck cancer pathogenesis, especially in squamous cell carcinoma. Although several studies have attempted to uncover its contribution to the initiation and propagation of SGTs, salivary adenoid cystic carcinoma (AdCC) has received the most attention [12]. miR-21 is overexpressed in the salivary gland AdCC, which interferes with apoptosis and enhances cell proliferation, cell invasion, migration, and metastasis [13]. Nevertheless, the potential miR-21 mediated gene regulation process in the development of the most common salivary gland tumor, mucoepidermoid carcinoma, needs to be further investigated.
This study aimed to assess the MUC1 expression level and its potential regulatory microRNAs, miR-21 and miR-145, in salivary gland MEC to determine a possible correlation with tumor prognosis, which might consequently help to enhance the treatments and survival rates.
Materials and methods
Ethics approval
This study was conducted in accordance with the Declaration of Helsinki and the protocol was approved by the Biomedical Research Ethics Committee of Tehran University of Medical Sciences (IR.TUMS.DENTISTRY.REC.1398.011). Written informed consents were obtained from all patients prior to participation.
Case selection
Tumor specimens from 47 patients with major and minor salivary gland malignancies that underwent surgery between 2012 and 2018 were retrieved from the Department of Pathology at the cancer institute of Imam Khomeini hospital complex at Tehran University of Medical Sciences (TUMS). The samples were histologically re-evaluated and graded according to Brandwein classification [14]. Moreover, the clinical staging was assessed based on the 8th edition of the American Joint Committee on Cancer staging system (AJCC) [15]. The patients with multiple synchronous malignancies (except for metastatic MEC) or a history of other types of cancer before MEC were excluded. Surplus salivary gland tissues from the periphery of lesions or at least 1 cm away from the tumor tissue, which did not contain any malignant cells or inflammation, were obtained from six patients as controls. The demographics and clinicopathological features of all patients are presented in Table 1.
Table 1.
Baseline demographics and clinicopathological features of total patients with MEC and MicroRNAs analyzed subpopulation
| Characteristic | MEC patients (n = 47) | MicroRNAs analyzed subpopulation (n = 19) | |
|---|---|---|---|
| Age | 51.47 ± 18.02 | 47.63 ± 21.10 | |
| Gender | |||
| Female | 29 (61.70%) | 13 (68.42%) | |
| Male | 18 (38.30%) | 6 (31.58%) | |
| Anatomic site | |||
| Major salivary glands | |||
| Parotid | 35 (74.47%) | 15 (78.95%) | |
| Submandibular | 5 (10.64%) | 2 (10.53%) | |
| Minor salivary glands | |||
| Skull base | 1 (2.13%) | 0 (0.00%) | |
| Maxilla | 1 (2.13%) | 0 (0.00%) | |
| Pharynx | 3 (6.38%) | 1 (5.26%) | |
| Mandible | 2 (4.26%) | 1 (5.26%) | |
| Histologic grades | |||
| Grade I | 19 (40.42%) | 9 (47.37%) | |
| Grade II | 11 (23.40%) | 3 (15.79%) | |
| Grade III | 17 (36.17%) | 7 (36.84%) | |
| Tumor size | |||
| I | 11 (23.40%) | 6 (31.58%) | |
| II | 15 (31.91%) | 5 (26.32%) | |
| III | 11 (23.40%) | 4 (21.05%) | |
| IVa | 9 (19.15%) | 4 (21.05%) | |
| IVb | 1 (2.13%) | 0 (0.00%) | |
| IVc | 0 (0.00%) | 0 (0.00%) | |
| Lymph node involvement | |||
| N0 | 36 (76.60%) | 17 (89.47%) | |
| N1 | 11 (23.40%) | 2 (10.53%) | |
| Distant metastasis | |||
| M0 | 45 (95.74%) | 18 (94.74%) | |
| M1 | 2 (4.26%) | 1 (5.26%) | |
| Stage | |||
| I | 11 (23.40%) | 6 (31.58%) | |
| II | 11 (23.40%) | 4 (21.05%) | |
| III | 15 (31.91%) | 5 (26.32%) | |
| IVa | 6 (12.76%) | 3 (15.79%) | |
| IVb | 2 (4.26%) | 0 (0.00%) | |
| IVc | 2 (4.26%) | 1 (5.26%) | |
| Treatment modality | |||
| Total parotidectomy | 11 (23.40%) | 4 (21.05%) | |
| Partial parotidectomy | 22 (46.81%) | 11 (57.89%) | |
| Hemi-maxillectomy | 3 (6.38%) | 0 (0.00%) | |
| Total submandibulectomy | 4 (8.51%) | 1 (5.26%) | |
| Segmental mandibulectomy | 4 (8.51%) | 2 (10.53%) | |
| Nasopharynx mass resection | 1 (2.13%) | 0 (0.00%) | |
| Oropharynx mass resection | 2 (4.26%) | 1 (5.26%) | |
| Recurrence | |||
| Recurrent specimens | 10 (21.28%) | 5 (26.32%) | |
| Primary specimens | 37 (78.72%) | 14 (73.68%) | |
| Margins | |||
| Free | 28 (59.57%) | 12 (63.16%) | |
| Involved | 16 (34.04%) | 7 (36.84%) | |
| Unknown* | 3 (6.38%) | 0 (0.00%) | |
Abbreviations: MEC, Mucoepidermoid carcinoma; N0, No regional lymph node metastases; N1, Regional lymph node metastases present; M0, Distant metastasis absent; M1, Distant metastasis present
Note: Data are expressed as mean ± SD or no. (%) unless otherwise noted.
* Margin could not be assessed due to fragmentation of the specimen.
Bioinformatics assessments to identify suitable microRNAs
At this stage, we evaluated MUC1 in the MiRTargetlinker (https://ccb-web.cs.uni-saarland.de/mirtargetlink), MiRTarbase (https://mirtarbase.cuhk.edu.cn), and MienTurnet (http://userver.bio.uniroma1.it/apps/mienturnet) databases and mapped the link between the candidate gene and the microRNAs. The lower the p-value for disease ontology or KEGG analysis, the more significant the term or pathway will be (p < 0.05). (Fig. S1)
Total RNA extraction and cDNA synthesis
The tumor and paired normal tissues of seven shaved cuts (11 μm thickness) from each block were deparaffinized with xylene and ethanol. Total RNA was extracted from Formalin-fixed, paraffin-embedded (FFPE) tissue using TRIzol™ Reagent Ambion by Life Technologies, Thermo Fisher Scientific, Waltham, Massachusetts) according to the manufacturer’s protocol. Extracted RNA was then treated with DNase I (RNase free) (YT9054 Deoxyribonuclease I DNase) to eliminate the contamination of the entire genomic DNA and stored at -80 °C. The concentration and quality of extracted RNA were evaluated using the ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc.) according to absorbance (A) at 260 and 280 nm. The mRNA was reverse transcribed to complementary DNA (cDNA) using miScript II RT Kit (Qiagen®).
Quantitative real-time PCR
Quantitative PCR was carried out in a 7900 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the miScript® SYBR® Green PCR Kit (Qiagen). Universal primer, provided with the Kit, and specific primers for miR-21 and miR-145 were applied. The relative level of microRNA expression was determined using human U6 snRNA as the control. The threshold cycle (Ct) for each microRNA was defined as the number of cycles at which the reaction crossed an arbitrary placed threshold. For statistical analysis, the 2ΔΔCT method was utilized. Primer sequences designed using the BLAST software are presented in Table S1. Each qRT-PCR experiment run was performed in triplicate.
Immunohistochemistry
MEC samples and normal salivary gland tissues were stained using anti-MUC1 antibody (Clone ZM35, Master Diagnostica, Granada, Spain). Detection was performed with the Master Polymer Plus Detection System (Master Diagnostica, Granada, Spain). As a positive control, colon adenocarcinoma was used. For negative control, the primary antibody was substituted by nonimmune serum. The two pathologists who were blinded to clinical outcomes evaluated the IHC results independently by determining the staining intensity (negative = 0, weak = 1, moderate = 2, strong = 3) and counting the percentages of positive tumor cells (0%= 0, 1–10%= 1, 11–50%= 2, 51–80%= 3, 81–100%= 4). Overall immunoreactivity score (IRS) was calculated by multiplying the numeric values of both parameters. In order to determine the optimal cut-off for further statistical analysis, an online tool (https://molpathoheidelberg.shinyapps.io/CutoffFinder_v1/) was used. The patients were divided into two groups; a group with low MUC1 expression levels (IRS 0–4) and another group with high MUC1 expression levels (IRS > 4).
Statistical analysis
Statistical analyses were performed with statistical software IBM SPSS Statistics for Windows, version 23 (IBM Corp., Armonk, N.Y., USA) and MedCalc® Statistical Software version 19.8 (MedCalc Software Ltd, Ostend, Belgium). All statistical tests were two-sided, and P < 0.05 was considered statistically significant. The Shapiro-Wilk W-test was used to check the normality assumption of the continuous variables. Mann-Whitney U-test was used to test the differences between the means of the two groups. The Chi-square test was applied to examine the correlation between the two categorical variables. Disease-free survival was defined as the time from the initial diagnosis to recurrence or death from any cause or to the last follow-up. Cancer-specific survival was considered as the time from the initial diagnosis to death due to mucoepidermoid cancer or to the last follow-up. Overall survival was calculated from diagnosis to the death of the patient for any cause or to the last follow-up. Survival curves were generated using the Kaplan Meier method according to MUC1 expression (low vs. high expression level), grade, and stage and compared using the log-rank test. Univariate Cox’s proportional hazards model was used to assess the association between each selected characteristic and disease-free survival, cancer-specific survival, and overall survival. A stepwise Cox’s proportional hazards model was applied to simultaneously account for variables emerging from univariate analyses with a P-value of < 0.15.
Results
The baseline demographics and clinicopathological features of total patients with MEC and microRNAs analyzed subpopulation are summarized in Table 1. The median follow-up was five years, ranging from 1 to 10 years. There were 14 deaths, of which 13 had shown recurrence. Among 14 deaths, 12 were due to cancer, and two were unrelated. Of the 33 patients that survived, 20 had no evidence of disease, and 13 showed recurrences. Kaplan-Meier curves of overall, cancer-specific, and disease-free survivals of MEC patients are presented in Fig. S2.
MUC1 immunohistochemical findings
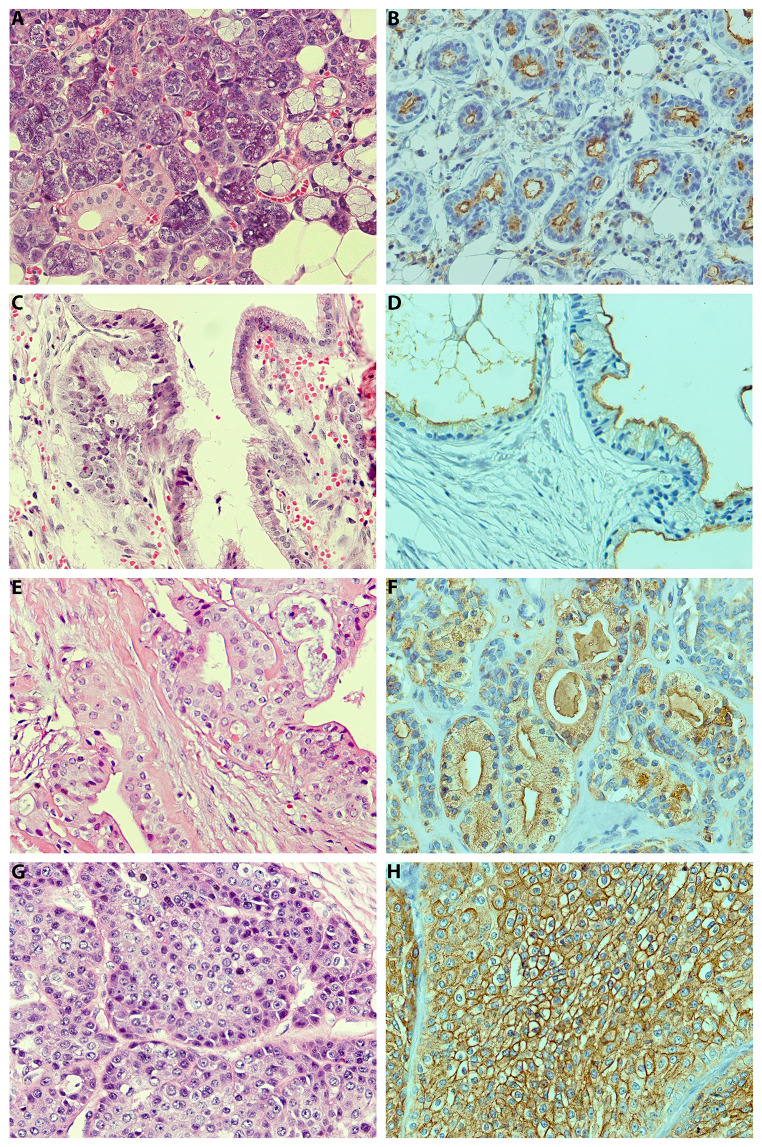
MUC1 immunohistochemical findings of the patients with MEC are presented in Table 2. Representative photomicrographs of normal salivary gland tissues and examples of each tumor grade are depicted in Fig. 1. In grade I, 3 (15.79%) showed apical membranous staining of MUC1, 3 (15.79%) cytoplasmic staining, and 13 (68.42%) both apical membranous and cytoplasmic staining. In grade II, 2 (18.18%) showed entire membranous staining, 1 (9.09%) cytoplasmic staining, and 8 (72.73%) both membranous and cytoplasmic staining. In grade III, the aforementioned patterns were 8 (47.06%), 0 (0.00%) and 9 (52.94%), respectively. Despite no significant differences, more membranous staining was observed in higher grades (Exact p = 0.15).
Table 2.
MUC-1 immunohistochemical findings of patients with MEC
| Characteristic | MEC (n = 47) |
|---|---|
| Extent | |
| Absent | 0 (0.00%) |
| ≤10 | 6 (12.76%) |
| 11–50% | 10 (21.28%) |
| 51–80% | 20 (42.55%) |
| >80% | 11 (23.40%) |
| Intensity | |
| Absent | 0 (0.00%) |
| Week | 15 (31.91%) |
| Moderate | 20 (42.55%) |
| Strong | 12 (25.53%) |
| Immunoreactivity score* | |
| Median (range) | 6 (1 to 12) |
| Cellular compartment | |
| Membrane | 12 (25.53%) |
| Cytoplasm | 4 (8.51%) |
| Membrane + Cytoplasm | 31 (65.96%) |
Abbreviations: MEC, Mucoepidermoid carcinoma
Note: Data are expressed as no. (%) unless otherwise stated.
* The numeric values of Extent and intensity parameters were multiplied, resulting in an immunoreactivity score (IRS) ranging from 0 to 12.
Fig. 1.
Representative hematoxylin-eosin (H&E) (A, C, E, G) and immunohistochemistry for MUC1 (B, D, F, H) in normal salivary gland tissue (A, B), grade I (C, D), grade II (E, F), and grade III (G, H) MECs
Association between MUC1 expression level and the selected patients’ characteristics
There was a significant positive association between MUC1 expression and both histologic grade (p < 0.001) and tumor stage of MEC patients (p = 0.04). IRS was significantly higher in node-positive MEC patients compared to node negative MEC patients (p = 0.02). There was a significant difference between the patients selected for radical treatment versus conservative modalities regarding MUC1 expression (p = 0.01). However, no significant association was found between IRS and tumor size, age, and gender. Furthermore, primary and recurrent specimens were similar in median IRS. (Table 3)
Table 3.
Association between the MEC patients’ characteristics and both immunoreactivity score and relative expression levels of microRNA
| Characteristic | MUC 1 expression level | Relative expression levels of microRNAs | |||||||
|---|---|---|---|---|---|---|---|---|---|
| IRS | P | miR-21 | P | miR-145 | P | ||||
| Age | r = 0.21 | P = 0.15 | r = 0.32 | P = 0.18 | r=-0.08 | P = 0.73 | |||
| Gender | P = 0.58 | P = 0.10 | P = 0.83 | ||||||
| Female | 6 (2 to 8); (1 to 12) | 17.47 (6.28 to 24.22); (2.26 to 70.36) | 0.07 (0.02 to 0.19); (0.01 to 0.45) | ||||||
| Male | 6 (3.75 to 9); (1 to 12) | 39.24 (17.02 to 52.12); (14.19 to 57.15) | 0.06 (0.02 to 0.14); (0.004 to 0.18) | ||||||
| Histologic grades | r = 0.80 | P < 0.0001 | r = 0.63 | P = 0.004 | r=-0.52 | P = 0.02 | |||
| Tumor size | r = 0.28 | P = 0.051 | r = 0.45 | P = 0.052 | r=-0.15 | P = 0.54 | |||
| LN involvement | P = 0.02 | N/D | N/D | ||||||
| N0 | 5 (2 to 6); (1 to 12) | - | - | ||||||
| N1 | 9 (4 to 12); (2 to 12) | - | - | ||||||
| Distant metastasis | N/D | N/D | N/D | ||||||
| M0 | - | - | - | ||||||
| M1 | - | - | - | ||||||
| Tumor stage | r = 0.30 | P = 0.04 | r = 0.46 | P = 0.049 | r=-0.15 | P = 0.54 | |||
| Treatment modality | P = 0.01 | P = 0.10 | P = 0.61 | ||||||
| Conservative | 4 (2 to 6); (1 to 12) | 17.71 (6.61 to 28.11); (2.26 to 70.36) | 0.05 (0.03 to 0.17); (0.01 to 0.45) | ||||||
| Radical | 6 (6 to 9.75); (2 to 12) | 35.18 (19.79 to 53.80); (14.19 to 57.15) | 0.09 (0.01 to 0.16); (0.005 to 0.18) | ||||||
| Recurrence | P = 0.66 | P = 0.96 | P = 0.58 | ||||||
| Recurrent | 6 (2.75 to 9.75); (1 to 12) | 14.19 (7.75 to 60.40); (2.26 to 70.36) | 0.03 (0.01 to 0.32); (0.005 to 0.45) | ||||||
| Primary | 6 (2 to 7); (1 to 12) | 19.38 (8.24 to 37.21); (3.17 to 57.15) | 0.07 (0.03 to 0.14); (0.01 to 0.25) | ||||||
Abbreviations: MEC, Mucoepidermoid carcinoma; IRS, Immunoreactivity score; MUC1, Mucin 1; LN, Lymph node
Note: Data are expressed as median (IQR); range or Spearman’s correlation coefficient.
N/D: No statistical analysis was done due to insufficient data.
Radical treatment modalities: Total parotidectomy, Submandibular gland excision, Hemi-maxillectomy, Segmental mandibulectomy, Hard palate excision.
Conservative treatment modalities: Partial parotidectomy, Nasopharynx mass resection, Oropharynx mass resection, retromolar trigone mass resection.
Relative expression levels of microRNAs in MEC and control groups
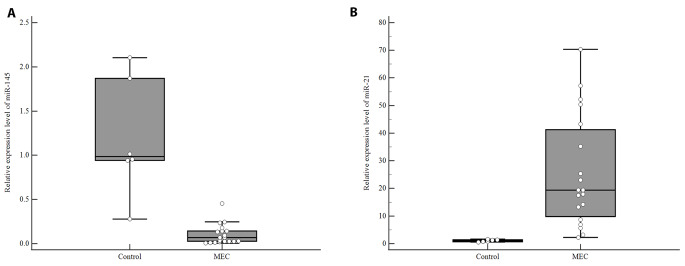
Baseline demographics and clinicopathological features of the patients in microRNAs analyzed subpopulation did not differ from the total MEC patients. Lower expression levels of miR-145 were found in MEC patients compared to the normal salivary gland tissues (p < 0.001, Fig. 2 A, Table S2). The median miR-21 expression level was significantly higher in MEC patients compared to normal salivary gland tissues (p < 0.001; Fig. 2B, Table S2).
Fig. 2.
Relative expression levels of (A) miR-145 and (B) miR-21. Middle point: median; box: interquartile range (25th to 75th percentiles); whisker: range (excluding outliers). Each error bar is constructed using a 95% confidence interval of the mean
Association between miR-145 and miR-21 expression levels and the selected patients’ characteristics
A negative correlation was found between the expression level of miR-145 and the histologic grade of MEC patients (p = 0.004). A positive association was observed between miR-21 expression level and both histologic grade (p = 0.004) and tumor stage of MEC patients (p = 0.04). However, no significant association was found between miR-145 or miR21 expression levels and age, gender, tumor size, being primary or recurrent, and treatment modality. (Table 3)
Association between MUC1 expression level and expression levels of microRNAs
There was a negative association between IRS and miR-145 expression level (r=-0.56 and p = 0.01) and a positive association between IRS and miR-21 expression level (r = 0.56 and p = 0.01).
Association between the selected patients’ characteristics overall, cancer-specific, and disease-free survival
Based on the univariate cox proportional hazard model, histologic grade and MUC1 expression level were significantly associated with the overall, cancer-specific, and disease-free survival. Margins involvement, tumor size, and tumor stage were significantly associated with the overall and cancer-specific survival. There was a significant association between lymph node involvement and cancer-specific survival. Age, anatomic site, primary or recurrent MECs, and treatment modality were not significantly associated with overall, cancer-specific, and disease-free survivals. (Table S3)
Association between MUC1, miR-145, and miR-21 expressions and disease-free, cancer-specific, and overall survival
According to univariate analyses, MUC1 expression level was significantly associated with overall (p = 0.01, HR = 1.22), cancer-specific (p = 0.01, HR = 1.23), and disease-free survival (p = 0.02, HR = 1.13). MiR-145 and miR-21 expressions were not significantly associated with overall, cancer-specific, and disease-free survivals. (Table S3)
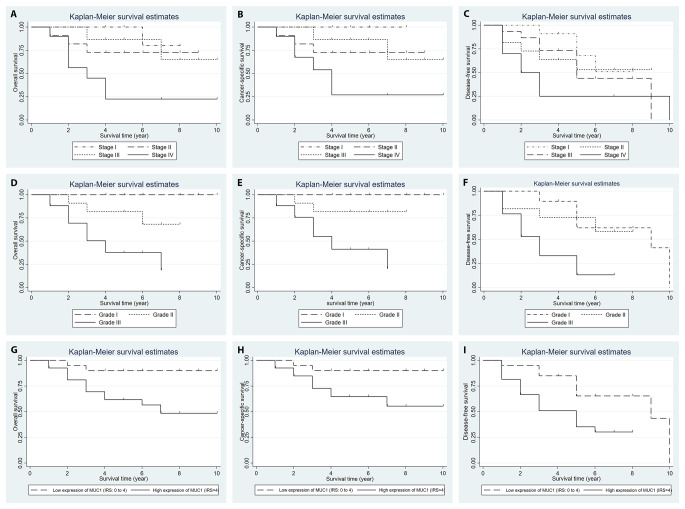
Survival curves generated by the Kaplan Meier method are presented according to grade, stage, and MUC1 expression (Fig. 3).
Fig. 3.
Kaplan-Meier curves of overall survival stratified with (A) tumor stage (P = 0.003), (D) grade (P < 0.001), and (G) MUC1 expression level (Low expression (IRS: 0 to 4) vs. high expression (IRS > 4)) (P = 0.04). Kaplan-Meier curves of cancer-specific survival stratified with (B) tumor stage (P = 0.004), (E) grade (P < 0.001), and (H) MUC1 expression level (Low expression (IRS: 0 to 4) vs. high expression (IRS > 4)) (P = 0.04). Kaplan-Meier curves of disease-free survival stratified with (C) tumor stage (P = 0.16), (F) grade (P < 0.001), and (I) MUC1 expression level (Low expression (IRS: 0 to 4) vs. high expression (IRS > 4)) (P = 0.01). Note: P-values are obtained using log-rank test
Results of multivariable cox proportional hazard model for the association of MUC1, miR-145, expression, and patients’ characteristics with disease-free survival
There was evidence of collinearity among miR-145, MUC1 and grade. To avoid multicollinearity in multivariable cox proportional hazard modeling, three models were fitted and these parameters were alternatively entered into the model. Table S4 represents the results of multivariable cox proportional hazard model for the association of MUC1 and miR-145 expressions and the patients’ characteristics with disease-free survival. Based on multivariable Cox proportional hazard models, the histologic grade was confirmed to be the only significant predictor of disease-free survival (p = 0.002, HR = 4.58).
Discussion
The role of protein-coding genes and their regulatory microRNAs in a variety of cancers has been evaluated [16]. While there are multiple therapeutic benefits in discovering these key regulators, biomarkers capable of identifying poor outcomes in patients need to be further investigated. There are few studies on head and neck cancers that evaluate the association between MUC1 expression and prognosis; however, these studies presented contradictory results [4]. The studies focusing on the molecular regulatory mechanism underlying mucins gene expression and their related data on the prognostic significance of microRNAs in salivary gland malignancies are missing.
MUC1, a glycoprotein involved in the maturation of glandular structures, plays a prominent role in the regulation of various cellular processes, including cell-matrix and cell-cell adhesion, cell proliferation, angiogenesis, invasive growth, cancer metastasis, and apoptotic avoidance [4, 6]. MUC1 overexpression as an oncogene in colon, pancreas, ovarian, breast, and bladder tumors is associated with the epithelial to mesenchymal transition process (EMT) of different cancer cells. Therefore, MUC1 could serve as a critical regulator of metastasis. MUC1 overexpression is accompanied by a significant increase in expression of Vimentin, Snail and Slug transcription factors along with E-cadherin repression, favoring the initiation of EMT, invasion of tumor cells, and augmentation of metastasis [17].
In this study, we found a significant positive association between MUC1 expression and tumor stage and grade. High grade MECs demonstrate the most intense entire membranous MUC1 expression in epidermoid, intermediate, and clear cells, while low-grade tumors showed apical membranous staining of mucinous cells around the lumina with mild cytoplasmic positivity of intermediate cells.
Although recent IHC studies on 40 and 45 salivary MEC found only cytoplasmic expression pattern [4], other investigations evaluating MUC1 expression patterns, in line with our finding, showed both cytoplasmic and membranous staining.
The evaluation of immunohistochemical MUC1 expression in different benign and malignant histologic types of salivary gland neoplasms demonstrated that MUC1 expression could act as a marker of worse prognosis in salivary gland tumors [18]. The evaluation of MUC1 expression in pleomorphic adenoma showed that high expression is correlated with malignant transformation and recurrence [18]. The investigation of 63 MECs by the immunohistochemical staining technique revealed shorter progression-free survival related to elevated MUC1expression [19]. Further studies on 357 cases of MEC using IHC demonstrated that the MUC1 expression was correlated to histologic grade, stage, and nodal involvement [20]. In addition, MUC1 expression in more than 50% [21] and 75% [22] of tumor cells in MECs was strongly associated with a greater histologic grade, risk of metastasis, and worse prognosis. However, a study evaluating mucin genes expression using quantitative PCR techniques showed that elevated MUC1 expression in tumor cells of MECs was related to a less aggressive disease course and a raised survival rate [23]. The present study revealed that overexpression of MUC1 was associated with a greater risk of nodal involvement and lower survival and indicated the need for more radical treatment.
Over the last few years, microRNAs regulation has been identified as one of the most frequent epigenetic modifications. It is known that microRNAs are capable of acting as tumor suppressors or oncogenes by suppressing the translation of related target mRNAs [24]. MiR-145 is a tumor suppressor microRNA known to have an inhibitory effect on various malignancies, considered a principal molecule in the regulation of the EMT process. It can inhibit the propagation of EMT pathways by regulating essential markers of these pathways, including ZEB2, tumor suppressor candidate 3 (TUSC3), and MUC1 [25, 26]. The regulatory effect of MiR-145 on MUC1 has been demonstrated in the pathogenesis of non-neoplastic salivary gland disease, including Sjögren’s Syndrome [27]. Downregulation of miR-145 induced by type I interferon could lead to overexpression of MUC1 and consequently, inflammation and glandular dysfunction [27].
MiR-145 has been demonstrated to be a tumor suppressor in ovarian cancer tissues, cell lines, and serum samples using Northern blot and qRT-PCR analysis, suggesting that its overexpression leads to suppressing cell proliferation, invasion, and tumor growth by negative regulation of MUC1 expression [26].
In addition, using western blot assay in a study on ovarian cancer revealed that the up-regulation of circWHSC1 could lead to MUC1 overexpression. By sponging miR-145, CircWHSC1 could regulate MUC1 oncogene expression that consequently affects ovarian cancer progression [7].
Further in vitro studies on serous epithelial ovarian carcinoma cell line (SKOV3) demonstrated that miR-145 could inhibit MUC1 expression by directly targeting the MUC1 3’-untranslated region that significantly interferes with cell migration and invasion [28]. In the metastatic breast cancer cell line, MUC1 was identified as a direct target of miR-145. Overexpression of MUC1, which induces cell invasion and metastasis, can be suppressed by miR-145 associated with lower β-catenin and oncogenic cadherin 11 expressions [8].
In the head and neck region, the studies that evaluated miR-145 in oral squamous cell carcinoma (OSCC) suggested that miR-145 via suppression of HOXA1 can interfere with the ERK/MAPK signaling pathway resulting in the suppression of tumor progression [29]. Moreover, in OSCC c-Myc and Cdk6 have been identified as the possible target genes of miR-145 at the post-transcriptional level [30]. This is the first study that evaluated miR-145 and one of its known target genes (MUC1) in salivary gland malignancy.
In agreement with previous observations, we recorded significantly lower expression of miR-145 in MEC compared to normal salivary gland tissue, and confirmed a negative correlation between miR-145 expression level and MUC1 expression and histologic grade.
A study on colon cancer CR-HCT-116 cells reported that miR-21 could negatively regulate tumor suppressor miR-145 expression. On the other hand, an elevated level of miR-145 expression can suppress miR-21 through negative feedback. This regulatory mechanism might be due to miR-21 overexpression role in the activation of the RAS Signaling cascade, which results in the repression of miR-145 [31].
Unlike miR-145, the expression of miR-21 in salivary gland malignancies has been evaluated in recent studies, which mainly focus on AdCC. These studies demonstrated that miR-21 overexpressed in AdCC compared to normal salivary gland tissue and also indicated that miR-21 overexpression was associated with metastasis and a worse prognosis [13].
However, few studies concerning the assessment of miR-21 in salivary gland MEC reported that the expression level of miR-21 is significantly higher in MEC compared to normal salivary gland tissue. These findings were in agreement with the results of our study [10]. Additionally, the current study showed that the high expression level of miR-21 was significantly associated with MUC1 expression, grade of differentiation, and clinical stage. High miR-21 and low miR-145 expression in the serum of patients with colorectal cancer were found to be correlated with tumor size, grade, and stage [32].
In conclusion, our findings indicated that MUC1 could act as a useful grade predictor biomarker in salivary gland MEC. MiR-145, a tumor-suppressor microRNA, might be negatively correlated with its potential target MUC1. MiR-21 as an oncomiR may correlate with miR-145 suppression resulting in the positive regulation of MUC1. In future studies, these microRNAs may eventually constitute useful biomarkers as well as therapeutic targets in salivary gland MEC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1(please Unhighlight cells of Muc1, mir145 and mir21 at table S3)
Author contribution
Conceptualization: [Neda Kardouni Khoozestani]; Methodology: [Neda Kardouni Khoozestani, Farid Azmoudeh-Ardalan], Formal analysis and investigation: [Ali Abdolrahmani, Ahmad Reza Shamshiri]; Writing - original draft preparation: [Ali Abdolrahmani]; Writing - review and editing: [Neda Kardouni Khoozestani, Farid Azmoudeh-Ardalan, Ahmad Reza Shamshiri]; Funding acquisition: [Neda Kardouni Khoozestani]; Resources: [Farid Azmoudeh-Ardalan]; Supervision: [Neda Kardouni Khoozestani].
Funding
This study was funded and supported by Tehran University of Medical Sciences, School of Dentistry (grant number #97-02-69-38537).
Availability of data and material
Datasets are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
Declarations
Conflict of interest/Competing interests
The authors declare that they have no conflict of interest.
Ethics approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Biomedical Research Ethics Committee of Tehran University of Medical Sciences (IR.TUMS.DENTISTRY.REC.1398.011).
Consent to participate
Written informed consent was obtained from the patient prior to participation in this study.
Consent for publication
Informed consent was obtained from the patient for publication.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ali Abdolrahmani, Email: a-abdolrahmani@alumnus.tums.ac.ir.
Neda Kardouni Khoozestani, Email: nkardouni@tums.ac.ir.
Farid Azmoudeh-Ardalan, Email: azmoudeh@sina.tums.ac.ir.
Ahmad Reza Shamshiri, Email: arshamshiri@tums.ac.ir.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2021;71(3):209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Son E, Panwar A, Mosher CH, Lydiatt D. Cancers of the Major Salivary Gland. J Oncol Pract. 2018;14(2):99–108. doi: 10.1200/jop.2017.026856. [DOI] [PubMed] [Google Scholar]
- 3.Xu W, Hua Y, Deng F, Wang D, Wu Y, Zhang W, et al. MiR-145 in cancer therapy resistance and sensitivity: A comprehensive review. Cancer Sci. 2020;111(9):3122–31. doi: 10.1111/cas.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson L, van Heerden MB, Ker-Fox JG, Hunter KD, van Heerden WFP. Expression of Mucins in Salivary Gland Mucoepidermoid Carcinoma. Head Neck Pathol. 2021;15(2):491–502. doi: 10.1007/s12105-020-01226-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma J, Yang Y, Huo D, Wang Z, Zhai X, Chen J, et al. LincRNA-RoR/miR-145 promote invasion and metastasis in triple-negative breast cancer via targeting MUC1. Biochem Biophys Res Commun. 2018;500(3):614–20. doi: 10.1016/j.bbrc.2018.04.119. [DOI] [PubMed] [Google Scholar]
- 6.Supruniuk K, Radziejewska I. MUC1 is an oncoprotein with a significant role in apoptosis (Review). Int J Oncol. 2021;59(3). doi:10.3892/ijo.2021.5248. [DOI] [PMC free article] [PubMed]
- 7.Zong ZH, Du YP, Guan X, Chen S, Zhao Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer Res. 2019;38(1):437. doi: 10.1186/s13046-019-1437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sachdeva M, Mo YY. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res. 2010;70(1):378–87. doi: 10.1158/0008-5472.Can-09-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bahreyni A, Alibolandi M, Ramezani M, Sarafan Sadeghi A, Abnous K, Taghdisi SM. A novel MUC1 aptamer-modified PLGA-epirubicin-PβAE-antimir-21 nanocomplex platform for targeted co-delivery of anticancer agents in vitro and in vivo. Colloids Surf B Biointerfaces. 2019;175:231–8. doi: 10.1016/j.colsurfb.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Denaro M, Navari E, Ugolini C, Seccia V, Donati V, Casani AP, et al. A microRNA signature for the differential diagnosis of salivary gland tumors. PLoS ONE. 2019;14(1):e0210968. doi: 10.1371/journal.pone.0210968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, Wang X, Shen H, Deng R, Xue K. Combination of miR-21 with Circulating Tumor Cells Markers Improve Diagnostic Specificity of Metastatic Breast Cancer. Cell Biochem Biophys. 2015;73. doi:10.1007/s12013-015-0573-0. [DOI] [PubMed]
- 12.Cinpolat O, Unal ZN, Ismi O, Gorur A, Unal M. Comparison of microRNA profiles between benign and malignant salivary gland tumors in tissue, blood and saliva samples: a prospective, case-control study. Braz J Otorhinolaryngol. 2017;83(3):276–84. doi: 10.1016/j.bjorl.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan F, Wang C, Li T, Cai W, Sun J. Role of miR-21 in the growth and metastasis of human salivary adenoid cystic carcinoma. Mol Med Rep. 2018;17(3):4237–44. doi: 10.3892/mmr.2018.8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brandwein MS, Ivanov K, Wallace DI, Hille JJ, Wang B, Fahmy A, et al. Mucoepidermoid carcinoma: a clinicopathologic study of 80 patients with special reference to histological grading. Am J Surg Pathol. 2001;25(7):835–45. doi: 10.1097/00000478-200107000-00001. [DOI] [PubMed] [Google Scholar]
- 15.Lydiatt WM, Patel SG, O’Sullivan B, Brandwein MS, Ridge JA, Migliacci JC, et al. Head and neck cancers—major changes in the American Joint Committee on cancer eighth edition cancer staging manual. Cancer J Clin. 2017;67(2):122–37. doi: 10.3322/caac.21389. [DOI] [PubMed] [Google Scholar]
- 16.Dos Santos ES, Normando AGC, Scarini JF, Crescencio LR, de Lima-Souza RA, Mariano FV, et al. Diagnostic and prognostic value of miRNAs on salivary gland tumors: a systematic review and meta-analysis. Oral Maxillofac Surg. 2021;25(4):445–56. doi: 10.1007/s10006-021-00952-0. [DOI] [PubMed] [Google Scholar]
- 17.Roy LD, Sahraei M, Subramani DB, Besmer D, Nath S, Tinder TL, et al. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene. 2011;30(12):1449–59. doi: 10.1038/onc.2010.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahomed F. Recent advances in mucin immunohistochemistry in salivary gland tumors and head and neck squamous cell carcinoma. Oral Oncol. 2011;47(9):797–803. doi: 10.1016/j.oraloncology.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Handra-Luca A, Lamas G, Bertrand JC, Fouret P. MUC1, MUC2, MUC4, and MUC5AC expression in salivary gland mucoepidermoid carcinoma: diagnostic and prognostic implications. Am J Surg Pathol. 2005;29(7):881–9. doi: 10.1097/01.pas.0000159103.95360.e8. [DOI] [PubMed] [Google Scholar]
- 20.Liu S, Ruan M, Li S, Wang L, Yang W. Increased expression of MUC1 predicts poor survival in salivary gland mucoepidermoid carcinoma. J Craniomaxillofac Surg. 2014;42(8):1891–6. doi: 10.1016/j.jcms.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Alos L, Lujan B, Castillo M, Nadal A, Carreras M, Caballero M, et al. Expression of membrane-bound mucins (MUC1 and MUC4) and secreted mucins (MUC2, MUC5AC, MUC5B, MUC6 and MUC7) in mucoepidermoid carcinomas of salivary glands. Am J Surg Pathol. 2005;29(6):806–13. doi: 10.1097/01.pas.0000155856.84553.c9. [DOI] [PubMed] [Google Scholar]
- 22.Siyi L, Shengwen L, Min R, Wenjun Y, Lizheng W, Chenping Z. Increased expression of MUC-1 has close relation with patient survivor in high-grade salivary gland mucoepidermoid carcinoma. J Oral Pathol Med. 2014;43(8):579–84. doi: 10.1111/jop.12170. [DOI] [PubMed] [Google Scholar]
- 23.Shemirani N, Osipov V, Kolker A, Khampang P, Kerschner JE. Expression of mucin (MUC) genes in mucoepidermoid carcinoma. Laryngoscope. 2011;121(1):167–70. doi: 10.1002/lary.21164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov. 2016;6(3):235–46. doi: 10.1158/2159-8290.Cd-15-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang H, Li K, Zheng J, Dou X, Zhao Y, Wang L. microRNA-145 regulates tumor suppressor candidate 3 and mitogen-activated protein kinase pathway to inhibit the progression of colorectal cancer. J Cell Biochem. 2018 doi: 10.1002/jcb.28122. [DOI] [PubMed] [Google Scholar]
- 26.Wu H, Xiao Z, Wang K, Liu W, Hao Q. MiR-145 is downregulated in human ovarian cancer and modulates cell growth and invasion by targeting p70S6K1 and MUC1. Biochem Biophys Res Commun. 2013;441(4):693–700. doi: 10.1016/j.bbrc.2013.10.053. [DOI] [PubMed] [Google Scholar]
- 27.Jara D, Carvajal P, Castro I, Barrera M-J, Aguilera S, González S, et al. Type I Interferon Dependent hsa-miR-145-5p Downregulation Modulates MUC1 and TLR4 Overexpression in Salivary Glands From Sjögren’s Syndrome Patients. Front Immunol. 2021;12. doi:10.3389/fimmu.2021.685837. [DOI] [PMC free article] [PubMed]
- 28.Wang L, Wu X, Wang B, Wang Q, Han L. Mechanisms of miR-145 regulating invasion and metastasis of ovarian carcinoma. Am J Transl Res. 2017;9(7):3443–51. [PMC free article] [PubMed] [Google Scholar]
- 29.Ding J, Sun D, Xie P. Elevated microRNA-145 inhibits the development of oral squamous cell carcinoma through inactivating ERK/MAPK signaling pathway by down-regulating HOXA1. Biosci Rep. 2019;39(6). doi:10.1042/bsr20182214. [DOI] [PMC free article] [PubMed] [Retracted]
- 30.Shao Y, Qu Y, Dang S, Yao B, Ji M. MiR-145 inhibits oral squamous cell carcinoma (OSCC) cell growth by targeting c-Myc and Cdk6. Cancer Cell Int. 2013;13(1):51. doi: 10.1186/1475-2867-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Y, Nangia-Makker P, Farhana L, Levi SGR, Majumdar E. miR-21 and miR-145 cooperation in regulation of colon cancer stem cells. Mol Cancer. 2015;14:98. doi: 10.1186/s12943-015-0372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Q, Yang W, Luo Y, Hu S, Zhu L. Correlation between miR-21 and miR-145 and the incidence and prognosis of colorectal cancer. J buon. 2018;23(1):29–35. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1(please Unhighlight cells of Muc1, mir145 and mir21 at table S3)
Data Availability Statement
Datasets are available from the corresponding author on reasonable request.
Not applicable.