Abstract
The COVID-19 pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), remains a global public health crisis. The reduced efficacy of therapeutic monoclonal antibodies against emerging SARS-CoV-2 variants of concern (VOCs), such as omicron BA.5 subvariants, has underlined the need to explore a novel spectrum of antivirals that are effective against existing and evolving SARS-CoV-2 VOCs. To address the need for novel therapeutic options, we applied cell-based high-content screening to a library of natural products (NPs) obtained from plants, fungi, bacteria, and marine sponges, which represent a considerable diversity of chemical scaffolds. The antiviral effect of 373 NPs was evaluated using the mNeonGreen (mNG) reporter SARS-CoV-2 virus in a lung epithelial cell line (Calu-3). The screening identified 26 NPs with half-maximal effective concentrations (EC50) below 50 μM against mNG-SARS-CoV-2; 16 of these had EC50 values below 10 μM and three NPs (holyrine A, alotaketal C, and bafilomycin D) had EC50 values in the nanomolar range. We demonstrated the pan-SARS-CoV-2 activity of these three lead antivirals against SARS-CoV-2 highly transmissible Omicron subvariants (BA.5, BA.2 and BA.1) and highly pathogenic Delta VOCs in human Calu-3 lung cells. Notably, holyrine A, alotaketal C, and bafilomycin D, are potent nanomolar inhibitors of SARS-CoV-2 Omicron subvariants BA.5 and BA.2. The pan-SARS-CoV-2 activity of alotaketal C [protein kinase C (PKC) activator] and bafilomycin D (V-ATPase inhibitor) suggest that these two NPs are acting as host-directed antivirals (HDAs). Future research should explore whether PKC regulation impacts human susceptibility to and the severity of SARS-CoV-2 infection, and it should confirm the important role of human V-ATPase in the VOC lifecycle. Interestingly, we observed a synergistic action of bafilomycin D and N-0385 (a highly potent inhibitor of human TMPRSS2 protease) against Omicron subvariant BA.2 in human Calu-3 lung cells, which suggests that these two highly potent HDAs are targeting two different mechanisms of SARS-CoV-2 entry. Overall, our study provides insight into the potential of NPs with highly diverse chemical structures as valuable inspirational starting points for developing pan-SARS-CoV-2 therapeutics and for unravelling potential host factors and pathways regulating SARS-CoV-2 VOC infection including emerging omicron BA.5 subvariants.
Keywords: SARS-CoV-2 variants of concern, Host-directed antiviral, Human V-ATPase, Human protein kinase C, Human TMPRSS2
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for the ongoing COVID-19 pandemic, a major worldwide public health challenge (McKee and Stuckler, n.d.). As of November 26, 2022, more than 640 million SARS-CoV-2 infections and over 6.6 million deaths have been reported (“COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University,” n.d.; “United States—COVID-19 Overview—Johns Hopkins,” 2022). Control of viral spread has been facilitated by the widespread distribution of several vaccines, along with public health measures including mask wearing and social distancing. However, several variants, termed variants of concern (VOCs), have emerged that have increased transmission capacity; these VOCs cause more severe disease and evade vaccine-mediated and natural immunity (Callaway, 2021a; “SARS-CoV-2 Variants of Concern | CDC,” n.d.). SARS-CoV-2 B.1.617.2 (Delta) is the most pathogenic VOC identified to date with widespread infection and hospitalizations occurring in 2021, even in populations with high vaccination rates (Hacisuleyman et al., 2021; Li et al., 2021; “SARS-CoV-2 Variants of Concern | CDC,” n.d.). A heavily mutated VOC, B.1.1.529 (Omicron) with dramatically reduced neutralization against sera from vaccinated individuals, greater transmissibility, and increased risk of reinfection, emerged at the end of 2021 and continues to evolve and spread worldwide (Callaway, 2021b; Planas et al., 2021; Sievers et al., 2022). This emphasizes the possibility that SARS-CoV-2 is likely to remain a global health threat, and it stresses the need to develop novel effective prophylactic and therapeutic solutions with broad activity against SARS-CoV-2 VOCs (Indari et al., 2021; Meganck and Baric, 2021; Shagufta and Ahmad, 2021).
Natural products (NPs) are a promising but undervalued resource for new antivirals. Compounds derived from diverse sources can encompass structural diversity that falls outside the scope of the chemical spaces found in synthetic chemical compounds, and they have the potential to act via mechanisms distinct from those of conventional therapies. NPs targeting a variety of biological pathways have been highlighted for their antiviral potential against a variety of viruses (Wang and Yang, 2020), and they may also be effective against SARS-CoV-2 infection (Ashhurst et al., 2021; Mani et al., 2020).
To address the need for novel SARS-CoV-2 therapeutics, a cell-based high-content screening (HCS) assay using mNeonGreen (mNG) reporter SARS-CoV-2 virus (Xie et al., 2020) was performed with a diverse library of 373 natural products. Twenty-six of these NPs were found to inhibit SARS-CoV-2 infection in Calu-3 cells, with a <20% reduction in cell viability and with half-maximal effective concentrations (EC50) below 50 μM. We found three NPs isolated from marine organisms—holyrine A (Williams et al., 1999), alotaketal C (Daoust et al., 2013), and bafilomycin D (Carr et al., 2010)—with broad-spectrum activity against SARS-CoV-2 VOCs.
2. Materials and methods
2.1. Cell lines, antibodies and chemicals
Calu-3 cells (ATCC® HTB-55™) and Vero E6 cells (ATCC® CRL-158™) were cultivated according to ATCC recommendations. VeroE6/TMPRSS2 cells (Jochmans et al., 2022) were cultivated according to JCRB recommendations. Huh-7.5.1 cells were kindly provided by Dr. Francis Chisari (Scripps Research Institute)(Moradpour et al., 2004). The SARS-CoV-2 nucleocapsid antibody [HL344] (GTX635679) was kindly provided by GeneTex; mouse anti-dsRNA antibody (J2) was purchased from SCICONS English and Scientific Consulting (10010500); secondary antibodies of goat anti-mouse IgG Alexa Fluor 488 (A-11001) and goat anti-rabbit IgG Alexa Fluor 555 (A-21428) and Hoechst 33342 were obtained from Thermo Fisher Scientific. Prostratin (60857-08-1) and Gö6983 (133053-19-7) were obtained from Sigma. Bryostatin (2383), Gö6976 (2253), PEP005 (4054) and CRT 0066854 (5922) were obtained from Tocris Bioscience. N-0385 was obtained from Drs. Pierre-Luc Boudreault and Richard Leduc (Université de Sherbrooke, QC, Canada) (Shapira et al., 2022).
2.2. SARS-CoV-2 viruses
All SARS-CoV-2 infections were carried out in Biosafety Level 3 (BSL3) facilities (either the University of British Columbia (UBC) Facility for Infectious Disease and Epidemic Research (FINDER) or the Simon Fraser University (SFU) BIO3 laboratory) following the Public Health Agency of Canada and UBC FINDER or SFU BIO3 regulations (UBC BSL3 Permit #B20-0105 and SFU Permit #361–2021). The mNeonGreen SARS-CoV-2 (2019-hCoV/USA-WA1/2020) (Xie et al., 2020) was made by Dr. Shi's laboratory (University of Texas Medical Branch, TX; USA. SARS-CoV-2 VOC (B.1.617.2 Delta and BA.2) was kindly provided by Dr. Mel Krajden (BC Centre for Disease Control, BC, Canada). SARS-CoV-2 Delta was first isolated in VeroE6/TMPRSS2 cells (passage 1) and then passaged in Vero E6 cells (passage 2). Delta virus stocks used in the experiments (passage 3) were propagated in Vero E6 cells (Ogando et al., 2020). SARS-CoV-2 Omicron BA.1 (BC-SFU-OM6) and BA.5 were isolated by Dr. Masahiro Niikura (SFU) from a clinical specimen in VeroE6/TMPRSS2 cells and confirmed as a BA.1 and BA.5 variants by complete genome sequencing (sequence available in GISAID). Omicron subvariants BA.1, BA.2 and BA.5 were amplified in VeroE6/TMPRSS2 cells and used in the experiments at passage 2.
2.3. Natural products
Our natural product screening library contains 373 pure natural products representing a large diversity of chemical scaffolds isolated from a broad spectrum of plants, invertebrates, or microorganisms collected in diverse terrestrial and marine habitats (Table S1). Terrestrial plants and microorganisms were collected in Thailand, Brazil, Canada, and Sri Lanka. Marine invertebrates and microorganisms were collected in ocean waters off the coasts of Canada, Brazil, Italy, Papua New Guinea, Indonesia, Dominica, and Sri Lanka. Most NPs in the screening library were discovered and first reported in the literature by the co-authors. Some of the pure natural products isolated by the coauthors had been previously reported in the literature by other research groups. The structures of all the NPs discovered by the coauthors were elucidated by a detailed analysis of nuclear magnetic resonance (NMR) spectroscopy, mass spectrometry (MS), and/or single-crystal X-ray diffraction data. The chemical structure and purity of our three lead NPs (Holyrine A, alotaketal C, and bafilomycin D) was confirmed by NMR analyses (Fig. S1). Literature references detailing the discovery of the lead 26 active compounds are provided (Table 1 ).
Table 1.
Summary of lead NP candidates.
| Name | Origin | % Inhibition | EC50(μM) | R2 | CC50(μM) | SI | References |
|---|---|---|---|---|---|---|---|
| bafilomycin D | Canada | 92 | 0.04 | 0.29 | 43 | 1064 | (Carr et al., 2010) |
| alotaketal C | Canada | 91 | 0.11 | 0.79 | >100 | 909 | (Daoust et al., 2013) |
| holyrine A | Canada | 93 | 0.28 | 0.76 | >100 | 357 | (Williams et al., 1999) |
| acetyl hyrtiosal | Italy | 97 | 1.97 | 0.78 | >100 | 51 | (Indari et al., 2021, Daoust et al., 2013) |
| dipterocarpol | Thailand | 99 | 3.05 | 0.89 | >100 | 33 | (Chaipukdee et al., 2016) |
| knecorticosanone G | Thailand | 100 | 3.50 | 0.88 | >100 | 29 | (Chu et al., 2020) |
| piperbonin A | Thailand | 100 | 3.61 | 0.82 | >100 | 28 | (Dampalla et al., 2021) |
| asparacemosone B | Thailand | 83 | 4.04 | 0.72 | >100 | 25 | (Tao et al., 2021) |
| chevalone C | Thailand | 100 | 4.14 | 0.85 | 93 | 22 | (Ranganath et al., 2022) |
| chevalone B | Thailand | 95 | 4.47 | 0.94 | >100 | 22 | (Ranganath et al., 2022) |
| giluterrin | Brazil | 88. | 5.44 | 0.86 | 72 | 13 | (Gandhi et al., 2022) |
| territrem B | Thailand | 100 | 6.64 | 0.90 | >100 | 15 | (Li et al., 2022; Huang et al., 2022; Chaipukdee et al., 2016) |
| mollicellin B | Thailand | 100 | 6.79 | 0.93 | >100 | 15 | (Khumkomkhet et al., 2009) |
| terretonin A | Thailand | 82 | 7.31 | 0.94 | >100 | 22 | (Li et al., 2022; Chaipukdee et al., 2016) |
| crotonolide F | Thailand | 97 | 8.21 | 0.81 | >100 | 12 | (Linch et al., 2014; Shyr et al., 2021) |
| phellopterin | Thailand | 96 | 6.35 | 0.83 | 93 | 14 | (Planas et al., 2021; Bao et al., 2018) |
| kaur-16-en-18-oic acid | Thailand | 100 | 15.06 | 0.66 | >100 | 7 | (Kawano et al., 2021; Li et al., 2021) |
| alpinumisoflavone | Thailand | 100 | 16.03 | 0.85 | >100 | 6 | (Stewart 200) |
| 2-butylchrysin | Thailand | 95 | 17.99 | 0.82 | >100 | 6 | Huang et al., 2022 |
| chrysin | Thailand | 99 | 18.80 | 0.93 | >100 | 5 | (Sayed et al., 2020; Schultz et al., 2022) |
| 5-hydroxysophoranone | Thailand | 100 | 20.22 | 0.46 | >100 | 5 | (Moradpour et al., 2004) |
| asparacemosone A | Thailand | 95 | 20.65 | 0.78 | >100 | 5 | (Tao et al., 2021) |
| artahongkongene B | Thailand | 81 | 21.98 | 0.93 | >100 | 5 | (Li et al., 2022) |
| 8-hydroxypinoresinol | Thailand | 82 | 24.35 | 0.93 | >100 | 4 | (Planas et al., 2021) |
| mangostinone | Thailand | 92 | 27.88 | 0.86 | >100 | 4 | (Ashhurst et al., 2021) |
| erysubin F Grey block: hits with EC50 < 10 μM | Thailand | 100 | 45.60 | 0.77 | >100 | 2 | (Tang et al., 2015) |
2.4. SARS-CoV-2 infections and fluorescent staining of intracellular viral biomarkers
Huh-7.5.1 or Calu-3 cells were seeded at concentrations of 104 cells/well in 96-well plates the day before infection. Cells were pretreated for 3 h with fixed (50 μM for screening) or diluted concentrations of compounds (indicated in figure legends) followed by infection with mNeonGreen (mNG) SARS-CoV-2, SARS-CoV-2 Delta, or an Omicronsubvariant (BA.1, BA.2, and BA.5)for 48 h at a multiplicity of infection (MOI) of 1 (Huh-7.5.1 cells) or MOI of 2 (Calu-3 cells). Cells were then fixed with 4% formalin for 30 min to inactivate the virus. The fixative was removed, and the cells were washed with PBS, permeabilized with 0.1% Triton X-100 for 10 min, and blocked with 1% bovine serum albumin (BSA) for 1 h. To quantify nucleocapsid and dsRNA in a subset of experiments, cells were immunostained with rabbit primary antibody HL344 (SARS-CoV-2 nucleocapsid)mouse primary antibody J2 (dsRNA) at working dilutions of 1:1000 overnight at 4 °C. Secondary antibodies were used at a 1:2000dilution and Hoechst was used at 1.5 μg/mLfor 1 h at room temperature in the dark. After washing with PBS, the plates were kept in the dark at 4 °C until imaging on a high-content screening (HCS) platform (CellInsight CX7 HCS, Thermo Fisher Scientific) with a 10X objective orby Leica TCS SP8 laser scanning confocal microscope equipped with HyD detectors, 405 nm laser, and a white light laser, and operated with a Leica Application Suite X (LAS X) software using a 63x/1.40 Oil objective (HC PL APO CS2).
2.5. CellInsight CX7 high-content screening of SARS-CoV-2 infection
Monitoring of the total number of cells (based on nuclei staining) and the number of virus-infected cells (based on mNG expression) was performed using the CellInsight CX7 HCS platform (Thermo Fisher Scientific) as previously described (Olmstead et al., 2012; Shapira et al., 2022). Briefly, nuclei are identified and counted using 350/461 nm wavelength (Hoechst 33342); cell debris and other particles are removed based on a size filter tool. A region of interest (ROI, or “circle”) is then drawn around each host cell and validated against the bright field image to correspond with host cell membranes. The ROI encompasses the “spots” where mNG (485/521 nm wavelength) or another viral marker (dsRNA (485/521 nm wavelength) and nucleocapsid (549/600 nm wavelength) are localized. Finally, the software (HCS Studio Cell Analysis Software, version 4.0) identifies, counts, and measures the pixel area and intensity of the “spots” within the “circle.” The fluorescence measured within each cell (circle) is then added and quantified for each well. The total circle-spot intensity of each well corresponds to intracellular virus levels (Z’>0.6) and is normalized to noninfected cells and infected cells treated with 0.1–1% DMSO. Nine fields were sampled from each well.
2.6. Median effective dose (EC50) curves
Intracellular dose response (EC50values) for selected compounds against mNG SARS-CoV-2 and SARS-CoV-2 VOCs was determined by pretreating Calu-3 cells for 3 h with serially diluted compounds, followed by virus infection for 48 h. Viral infection was detected by mNG fluorescence (mNG SARS-CoV-2) or fluorescent imaging of viral nucleocapsid and dsRNA (SARS-CoV-2 VOCs) as described above (Section 2.5). EC50 experiments were repeated at least three times. Intracellular fluorescent levels were interpolated to negative control (0.1–1% DMSO, no infection) = 0 and positive control (0.1–1% DMSO, with infection) = 100. The GraphPad Prism 9™ (GraphPad Software, Inc.) nonlinear regression fit modeling variable slope was used to generate a dose-response curve [Y = Bottom + (Top-Bottom)/(1 + 10^((LogIC50-X)*HillSlope)], constrained to top = 100, bottom = 0.
2.7. Cytotoxicity assays
Calu-3 cells were seeded with 104 cells/well in 96-well plates. 24 h after seeding, media was aspirated, and serially diluted compounds (described above) were added for an additional 48 h incubation. Cellular viability was assessed with PrestoBlueCell Viability Assay (Thermo Fisher Scientific) according to the manufacturer's instructions. Cells were incubated with 5% PrestoBlue reagent for 2 h before they were read on the SpectraMax Gemini XS spectrofluorometer (Molecular Devices, LLC) set at excitation and emission wavelengths of 555 and 585 nm, respectively. Cellular viability was expressed relative (%) to vehicle-treated cells. Data are from at least three independent experiments.
2.8. Drug combination
Calu-3 cells were seeded at concentrations of 104 cells/well in 96-well plates the day before infection. Cells were pretreated for 3 h with respective combinations of bafilomycin D and N-0385 followed by infection with SARS-CoV-2 Omicron BA.2 as described above for 48 h at a MOI of 2. The two-drug combination was tested using five-fold serial dilutions of bafilomycin D and ten-fold serial dilutions of N-0385 ≤ EC50. The percent inhibition of viral infection for each dose-combination was determined by fluorescent staining of SARS-CoV-2 nucleocapsid as described above. Dose-response percent inhibition matrix of single and combined treatment of bafilomycin D and N-0385 in SARS-CoV-2 infected Calu-3 cells, and 2-D and 3-D interaction landscapes were calculated based on (i) the Loewe additive model, (ii) the zero interaction potency (ZIP) model, (iii) the highest single agent (HSA) model, and (iv) the Bliss model using SynergyFinder V.1 (Tang et al., 2015; Yadav et al., 2015; Zheng et al., 2022). Synergy scores were calculated for each condition and values above 10 were interpreted as a synergistic effect.
3. Results
3.1. Validation of mNG-SARS-CoV-2 as a molecular tool for high-content antiviral drug screening in human cells
The mNG-SARS-CoV-2 reporter virus used in this study was previously described, where it was demonstrated that the mNG transgene is stable and does not affect virus replication in Vero E6 cells (Xie et al., 2020). Although Vero E6 cells are permissive to SARS-CoV-2, these cells are non-human African green monkey kidney-derived cells and consequently their potential applications to provide useful physiological insights into the mechanism of action of new lead molecules in virally infected human cells could be limited by species-specific host-virus interactions (Shapira et al., 2022).
To validate mNG-SARS-CoV-2 as a screening tool in human cells, we first determined the EC50 value of the pro-drug remdesivir (RDV), a direct-acting antiviral (DAA), against mNG-SARS-CoV-2 infection using human Huh-7.5.1 cells. As previously reported, human hepatoma-derived Huh-7.5.1 cells are permissive to SARS-CoV-2 infection (Cagno, 2020; Chu et al., 2020; Fig. S2A) and produce host enzymes that permit a robust intracellular conversion of the pro-drug RDV to the active nucleoside triphosphate, which is misintegrated into viral RNA by the viral RNA-dependent RNA polymerase (Dittmar et al., 2021; Tao et al., 2021). Huh-7.5.1 cells were pretreated with serially diluted RDV for 3 h before mNG-SARS-CoV-2 infection. Using fluorescence-based microscopy, we observed a dose-dependent reduction of our two viral biomarkers’ relative abundance (intracellular mNG and nucleocapsid) in virally infected cells upon treatment with increasing concentration of RDV (Fig. S2B). Relative quantification of fluorescently labeled infected cells was performed using the CellInsight CX7 HCS platform to determine EC50 values (half-maximal effective concentration; drug concentration required to reduce infection by 50%). The EC50 values for RDV using mNG and nucleocapsid staining in Huh-7.5.1 infected cells were 3.6 nM and 7.6 nM, respectively (Fig. S2C), which is consistent with previously published values against the wild-type SARS-CoV-2 [EC50 = 2 nM (Huh-7.5 cells)(Dittmar et al., 2021)].
Human Calu-3 cells are naturally permissive lung epithelial cells that represent a physiologically relevant cell-based system for antiviral drug discovery directed at SARS-CoV-2 VOCs (Shapira et al., 2022). To further validate mNG-SARS-CoV-2 as a screening tool in human cells, we determined the EC50 value of GC376 against mNG-SARS-CoV-2 infection using human Calu-3 lung cells. GC376 is a DAA that inhibits the SARS-CoV-2 main protease (3CLpro) (Fu et al., 2020) and has been demonstrated to have therapeutic efficacy in a mouse model of severe COVID-19 infection(Dampalla et al., 2021). The Calu-3 cells were pretreated with serially diluted GC376 for 3 h before mNG-SARS-CoV-2 infection. Relative quantification of fluorescently labeled infected cells using mNG as a viral biomarker was performed using the CellInsight CX7 HCS platform (Fig. S3A). The EC50 value determined for GC376 against mNG-SARS-CoV-2 is 475 nM (Fig. S3B), which is consistent with previously published values against the wild-type SARS-CoV-2 [EC50 = 230 nM (Vero E6 cells), (Dampalla et al., 2021); EC50 = 700 nM (Vero E6 cells) (Fu et al., 2020)]. These results confirm that mNG-SARS-CoV-2 is a valuable molecular tool for high-content drug screening and for determination of drug efficacy against SARS-CoV-2 using human Calu-3 lung cells.
3.2. Screening of natural products against mNG-SARS-CoV-2
A library of pure NPs representing a large diversity of chemical scaffolds was assembled from extracts of plants, fungi, bacteria, and marine sponges. To identify which NPs exhibit antiviral activity against SARS-CoV-2 infection, we applied a high-throughput screening approach using the mNG-SARS-CoV-2 reporter virus and the CellInsight CX7 High Content Screening (HCS) platform (Fig. 1 A). In this assay, mNG fluorescence and nuclear staining are rapidly quantified and used as a readout for viral infection and compound cytotoxicity, respectively (Shapira et al., 2022). Pretreatment of the cells with NPs for 3 h was followed by infection of Calu-3 cells with the mNG-SARS-CoV-2 reporter virus at a multiplicity of infection (MOI) of 2 for 48 h. The cells were then fixed and the images were processed for quantitative analysis to evaluate the activity of the compounds. A total of 373 NPs at a concentration of 50 μM were screened using these assay conditions. The suitability of the screening assay was measured using Z’ factor calculation (a measure of the dynamic range, defined as the difference between the means of the negative and positive controls (Zhang et al., 1999)); the values of each screening were found to be between 0.5 and 0.7, with an average value of 0.6. Compounds showing 80% or greater inhibition of SARS-CoV-2 infection with less than 20% of cell loss were defined as lead candidates (Fig. 1B). Based on this criterion, 26 compounds were selected for further validation.
Fig. 1.
NP high-content screening with mNG-SARS-CoV-2. A) Graphical representation of the strategy for high-content screening of NPs as antiviral candidates. Illustration was created with BioRender.com. B) Overview of HCS performed in Calu-3 cells. Inhibition (green) was interpolated to the negative control (DMSO) set at 0% inhibition and the positive control (GC376) set at 100% inhibition. Cell loss (blue) was normalized to the negative control. The orange and green lines show the cut-off of inhibition (80%) and cell loss (20%), respectively. Data represent the mean of the results of three independent experiments. The names of the 26 lead compounds are displayed.
3.3. Dose-response analysis of holyrine A, alotaketal C, and bafilomycin D against mNG-SARS-CoV-2 VOCs in human Calu-3 lung cells
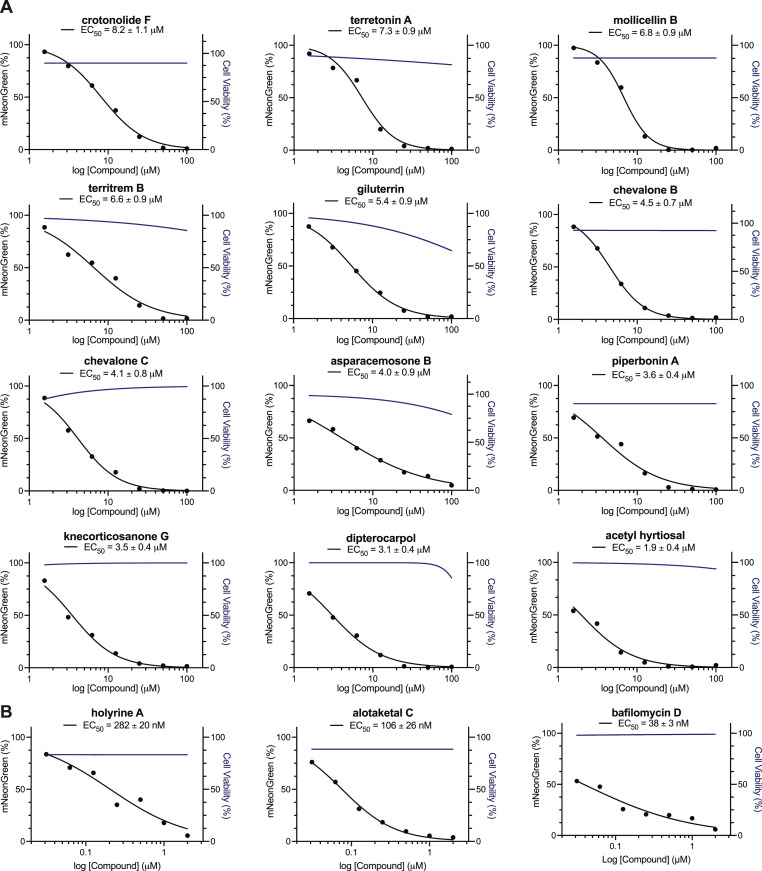
To validate and prioritize the most promising compounds according to their anti-SARS-CoV-2 activity, the 26 lead compounds identified from the primary screening were tested using a broad concentration range (100–0.00064 μM) against mNG-SARS-CoV-2 in Calu-3 cells. The relative quantification of infected cells was used to calculate EC50 values, which ranged from 38 nM to 45.6 μM (Fig. 2A and S4). The half-maximal cytotoxic concentration (CC50) values were also calculated and determined to be > 100 μM for most of the lead compounds (Fig. 2A and S4). The selectivity index (SI) for these 26 compounds, expressed as the ratio of CC50 over EC50 is summarized in Table 1. Importantly, we identified three compounds—holyrine A, alotaketal C, and bafilomycin D (Carr et al., 2010; Daoust et al., 2013; Williams et al., 1999)—with nanomolar activity and high SI values (EC50 = 282 nM, 106 nM, and 38 nM and SI = 357, 909, and 1064, respectively) against mNG-SARS-CoV-2 (Table 1 and Fig. 2 B). A high SI is preferable if a drug is to be viewed as having a favorable safety profile in human Calu-3 cells (Muller and Milton, 2012). Representative images demonstrating the reduction of mNG expression correlating with an increase in NP concentration are shown in Fig. S5.
Fig. 2.
Discovery of 15 antiviral NPs against mNG-SARS-CoV-2. Dose-response curves for infectivity (black) and cell viability (blue) were generated for the indicated serial diluted compounds in Calu-3 cells infected with mNG-SARS-CoV-2 (n = 3). EC50 values were determined using nonlinear regression analysis. The GraphPad Prism 9™ (GraphPad Software, Inc.) nonlinear regression fit modeling variable slope was used to generate a dose-response curve [Y = Bottom + (Top-Bottom)/(1 + 10^((LogIC50-X)*HillSlope)] constrained to top = 100, bottom = 0. The graph shows the average values of two independent experiments. A) 12 NPs with a single-digit micromolar range activity and B) 3 NPs with nanomolar activity against mNG-SARS-CoV-2.
3.4. Pan-SARS-CoV-2 activity of holyrine A, alotaketal C, and bafilomycin D against SARS-CoV-2 VOCs in human Calu-3 lung cells
Variants of concern (VOCs) possess a demonstrated capacity for enhanced transmission, disease severity, and reduced vaccine effectiveness (Callaway, 2021a; 2021b; “SARS-CoV-2 Variants of Concern | CDC,” n.d.). SARS-CoV-2 B.1.617.2 (Delta) is the most severe VOC and may be associated with an increased risk of hospitalization whereas Omicron subvariants (e.g., BA.1, BA.2, BA.5) are linked to greater transmissibility, reduced vaccine efficacy, and increased risk of reinfection (Callaway, 2021a; Hagen, n.d.; Li et al., 2021; Planas et al., 2021). We tested the impact of holyrine A, alotaketal C, and bafilomycin D on Calu-3 cells pretreated with serially diluted compounds for 3 h before SARS-CoV-2 Delta, Omicron BA.1, BA.2 and BA.5 infection. Fluorescent imaging of viral nucleocapsid and dsRNA confirmed viral infection and demonstrated the spheroid-like cell morphology of Delta infection that we previously reported (Shapira et al., 2022), where infected cells appear in small, rounded clusters (Fig. 3 A) in contrast to larger monolayers of uninfected or SARS-CoV-2-Omicron-infected cells (Fig. 3A). The three lead NPs showed a dose-dependent reduction of SARS-CoV-2 Delta and Omicron infection (Fig. 3B-M); relative quantification of virally infected cells was used to calculate their EC50 values. For holyrine A, the EC50 values against SARS-CoV-2 Delta were 600 nM and 1.0 μM for nucleocapsid and dsRNA staining (Fig. 3B), respectively; for SARS-CoV-2 Omicron BA.1, they were 1.7 μM and 1.6 μM, respectively (Fig. 3E), for Omicron BA.2, they were 44 nM and 394 nM for nucleocapsid and dsRNA, respectively (Fig. 3H), and for Omicron BA.5, they were 80 nM and 120 nM for nucleocapsid and dsRNA, respectively (Fig. 3K). For alotaketal C, the EC50 values against SARS-CoV-2 Delta were 4.9 μM and 3.9 μM for nucleocapsid and dsRNA staining, respectively (Fig. 3C); for SARS-CoV-2 Omicron BA.1, they were 700 nM and 400 nM, respectively (Fig. 3F); for Omicron BA.2, they were 700 nM and 700 nM for nucleocapsid and dsRNA, respectively (Fig. 3I) and for Omicron BA.5, they were 300 nM and 700 nM for nucleocapsid and dsRNA, respectively (Fig. 3L). Finally, for bafilomycin D, the EC50 values against SARS-CoV-2 Delta were 2.0 nM and 50 nM for nucleocapsid and dsRNA staining, respectively (Fig. 3D); for SARS-CoV-2 Omicron BA.1, they were 13.4 nM and 67.3 nM, respectively (Fig. 3G); for Omicron BA.2, they were 4.6 nM and 22 nM for nucleocapsid and dsRNA, respectively (Fig. 3J) and for Omicron BA.5, they were 43.9 nM and 38.6 nM for nucleocapsid and dsRNA, respectively (Fig. 3M). Together, these results underline the potential of holyrine A, alotaketal C, and bafilomycin D as broad-spectrum inhibitors against SARS-CoV-2 VOCs.
Fig. 3.
Holyrine A, alotaketal C and bafilomycin D are novel pan-SARS-CoV-2 natural product drug leads. A) Immunofluorescence staining for viral nucleocapsid and dsRNA verified productive SARS-CoV-2 infection by all four VOCs (B.1.617.2 Delta, Omicron BA.1, BA.2 and BA.5) in Calu-3 cells [scale bar = 10 μm]. B-J) Dose-response curves of Calu-3 cells pretreated with the indicated concentrations of holyrine A, alotaketal C and bafilomycin D before infection with SARS-CoV-2 Delta (B.1.617.2), Omicron BA.1, Omicron BA.2 or Omicron BA.5, using nucleocapsid (red circles) and dsRNA (green squares) as infection markers. EC50 values were determined using nonlinear regression analysis. The GraphPad Prism 9™ (GraphPad Software, Inc.) nonlinear regression fit modeling variable slope was used to generate a dose-response curve [Y = Bottom + (Top-Bottom)/(1 + 10^((LogIC50-X)*HillSlope)] constrained to top = 100, bottom = 0. The graph shows the average values of three independent experiments.
3.5. Robust inhibition of SARS-CoV-2 omicron BA.1 infection by protein kinase C (PKC) activators in human Calu-3 lung cells
We previously reported alotaketal C as an activator of protein kinase C (PKC) that is more potent than the well-studied PKC activator prostratin (Wang et al., 2016, 2022). To understand and validate the role of PKC on SARS-CoV-2 infection and the effect of alotaketal C as an activator, we tested the impact of PEP 005 (Hampson et al., 2005; Kedei et al., 2004), bryostatin-1 (Sun and Alkon, 2006), and prostratin (Wang et al., 2016, 2022) natural products known as PKC activators and also the PKC inhibitors Gö6983 (pan-inhibitor) (Wang et al., 2016, 2022), Gö6976 (conventional inhibitor) (Koivunen et al., 2004), and CRT0066854 (atypical inhibitor) (Kjær et al., 2013; Linch et al., 2014) on Calu-3 cells pretreated with serially diluted compounds for 3 h before SARS-CoV-2 BA.1 infection (Fig. 4 A–F). The three PKC activators (prostratin, bryostatin, and PEP 005) showed a dose-dependent reduction of SARS-CoV-2 Omicron BA.1 infection (Fig. 4A–C); relative quantification of virally infected cells was used to calculate their EC50 values. For prostratin, the EC50 values against SARS-CoV-2 Omicron BA.1 were 11.6 μM and 5.0 μM for nucleocapsid and dsRNA staining, respectively (Fig. 4A). For bryostatin, the EC50 values were 222 nM and 523 nM for nucleocapsid and dsRNA staining, respectively (Fig. 4B). For PEP 005, they were 700 nM and 1.5 μM for nucleocapsid and dsRNA, respectively (Fig. 4C). In contrast, the conventional PKC inhibitors enhanced the SARS-CoV-2 Omicron BA.1 infection (Fig. 4D and E) whereas the atypical PKC inhibitor (CRT00668854) showed a dose-dependent reduction of viral infection. The EC50 values against SARS-CoV-2 Omicron BA.1 were 300 nM and 100 nM for nucleocapsid and dsRNA staining, respectively (Fig. 4F).
Fig. 4.
Effect of protein kinase C (PKC) regulation on SARS-CoV-2 Omicron (BA.1) infection. A-F) Dose-response curves of Calu-3 cells pretreated for 3 h with the indicated concentrations of A) prostratin (PKC activator), B) bryostatin (PKC activator), C) PEP 005 (PKC activator), D) Gö6983 (PKC inhibitor), E) Gö6976 (PKC inhibitor) and F) CRT0066854 (PKC inhibitor) before 48 h infection with Omicron BA.1; dsRNA is represented with green squares and nucleocapsid with red circles. EC50 values were determined using nonlinear regression analysis. The GraphPad Prism 9™ (GraphPad Software, Inc.) nonlinear regression fit modeling variable slope was used to generate a dose-response curve [Y = Bottom + (Top-Bottom)/(1 + 10^((LogIC50-X)*HillSlope)] constrained to top = 100, bottom = 0. The graph shows the average values of two independent experiments.
3.6. Synergistic action of bafilomycin D with N-0385 against SARS-CoV-2 omicron BA.2 in human Calu-3 lung cells
In order to investigate further the molecular mechanism of action of bafilomycin D as a pan-SARS-CoV-2 antiviral, we investigated its potential synergistic action when used in combination with N-0385, one of the most potent SARS-CoV-2 pan-variant host-directed antivirals discovered to date (Shapira et al., 2022). N-0385 is a highly potent inhibitor of human TMPRSS2 protease and blocks the TMPRSS2-dependent proteolytical activation of the SARS-CoV-2 spike protein required for viral fusion (Shapira et al., 2022). Calu-3 cells infected with Omicron BA.2 were either treated with bafilomycin D or N-0385 as a single treatment or in combination. The dose-response percent inhibition matrix of single and combined treatment of bafilomycin D and N-0385 in SARS-CoV-2 infected Calu-3 cells is presented in Fig. 5 A. The inhibitory effect of drug combination was higher than the single treatment (Fig. 5A). To analyze the potential synergistic action of bafilomycin D with N-0385 against SARS-CoV-2 omicron BA.2, we used four different reference models (i.e., Loewe, ZIP, HSA, and Bliss). The 2-D (Fig. 5B) and 3-D (Fig. 5C) interaction landscapes between bafilomycin D and N-0385 calculated based on the Loewe additive model (Fig. 5B and C), the ZIP model (Fig. S7; left panel), the HSA model (Fig. S7; middle panel), and the Bliss model (Fig. S7; right panel) show synergy scores of 26.5, 20.9, 26.4, and 23, respectively. All the calculated synergy scores were above 10, which are interpreted here as a synergistic effect of bafilomycin D with N-0385 against SARS-CoV-2 omicron BA.2 in human Calu-3 lung cells. Our results are consistent with the recent finding by Icho et al. (2022), which demonstrated a synergistic effect between the repositioned drug Camostat mesylate (TMPRRS2 inhibitor) and bafilomycin A1, a classic V-ATPase inhibitor against a pre-Omicron SARS-CoV-2 variant (Indari et al., 2021).
Fig. 5.
Synergistic inhibition of SARS-CoV-2 Omicron (BA.2) infection by combined use of bafilomycin D and N-0385. Dose-response curves of inhibition of single and combined treatment of bafilomycin D and N-0385 in SARS-CoV-2 infected Calu-3 cells were used for the analysis using the open-source web application SynergyFinder. A) Dose-response matrix (percentage inhibition) of SARS-CoV-2 infection with combined treatment of bafilomycin D and N-0385. B) Synergy distribution of pairwise combination of bafilomycin D and N-0385 calculated based on Loewe additive model using SynergyFinder. C) Landscape visualization between bafilomycin D and N-0385 calculated based on Loewe additive model using SynergyFinder. Surface is color-coded, red indicates synergistic interactions and green indicates antagonistic interactions.
4. Discussion
The emergence of SARS-CoV-2 has led to an ongoing and evolving global pandemic with serious health, social, and economic consequences. Despite continuing worldwide vaccination campaigns, the recurring appearance of new variants of concern is creating a tremendous need for safe and effective therapeutics and prophylactics against SARS-CoV-2 infection. Importantly, reports on newly emerged SARS-CoV-2 Omicron BA.5 subvariants suggest that current antibody-mediated protection offered by vaccines and monoclonal antibodies is severely reduced compared to protection against SARS-CoV-2 Delta (Callaway, 2021a; Planas et al., 2021). These findings were recently confirmed and resulted in strong recommendations by WHO against the use of two previously approved monoclonal antibodies (sotrovimab and casirivimab-imdevimab) for patients with COVID-19 (“A living WHO guideline on drugs for covid-19,” 2022; Takashita et al., 2022).
So far, the approved direct-acting antivirals (DAAs) (e.g., remdesivir and paxlovid/nirmatrelvir) are proving effective at preventing hospitalization and mortality due to COVID-19 when given early. However, SARS-CoV-2-approved DAAs are all used as monotherapies for which SARS-CoV-2 may develop resistance (Gandhi et al., 2022). For example, several mutations in SARS-CoV-2 viruses sequenced in the population have been identified that may confer resistance to nirmatrelvir, and rebound infection following treatment has been documented (Hu et al., 2022; Jochmans et al., 2022; Ranganath et al., 2022). Moreover, recent findings by Rockett et al. based on genomic surveillance have also revealed co-infection with SARS-CoV-2 Omicron and Delta variants, underlining the need for pan-SARS-CoV-2 antiviral drugs (Rockett et al., 2022). These results underline the need for novel broad-spectrum pan-SARS-CoV-2 therapeutics for developing combination therapy approaches to strengthen treatment capacity against evolving VOCs (Li et al., 2022; Schultz et al., 2022; Shyr et al., 2021).
To address the need for novel therapeutic options for SARS-CoV-2 VOC infection, we applied cell-based high-content screening to a library of NPs with a considerable diversity of chemical scaffolds to identify lead NPs with antiviral activity against fluorescent-tagged mNG-SARS-CoV-2 in human Calu-3 lung cells. From a diverse library of 373 NPs, 73 compounds were identified with >50% inhibition of mNG-SARS-CoV-2 with less than 20% of cell loss, which may be further investigated in future studies (Table S1). We further validated 26 compounds with EC50 values below 50 μM. Sixteen compounds had EC50 values < 10 μM and SI indices >10. Holyrine A, alotaketal C, and bafilomycin D were highly potent with EC50 values in the nanomolar range, and these were subsequently demonstrated to be potent lead antivirals against ancestral SARS-CoV-2, Delta, and Omicron BA.1, BA.2 and BA.5.
Natural products are considered a rich resource for novel antiviral drug development. NPs have the advantage of more favorable toxicological profiles, fewer side effects, and a faster approval process in comparison to chemically engineered drugs (Auth et al., 2021; Wang and Yang, 2020). Along with plant-derived compounds, like Nigella sativa with its inhibitory activity against the hepatitis C virus, several marine products have also been reported to have antiviral activity against different viruses (Barakat et al., 2013; Wang et al., 2014, 2016). This includes NPs that have demonstrated efficacy against coronaviruses, such as SARS-CoV-1 and MERS-CoV (Ashhurst et al., 2021; Mani et al., 2020). NPs have been determined to inhibit viruses through several mechanisms including inhibiting viral entry and/or viral DNA and RNA synthesis, but they can also modulate cellular functions required by different viruses, offering broad-spectrum antiviral activity (Moghadamtousi et al., 2015; Musarra-Pizzo et al., 2021). Previously, some of the NPs in our library (terretonin A, terretonin B, mollicellin B, and phellopterin) were described as presenting anti-inflammatory (Wu et al., 2019), anti-malarial (Chaipukdee et al., 2016; Khumkomkhet et al., 2009), or anti-cancer activities (Bao et al., 2018).
Three antiviral NPs identified in our screening (holyrine A, alotaketal C, and bafilomycin D) showed nanomolar EC50 values of 282 nM, 106 nM, and 38 nM, respectively, against mNG-SARS-CoV-2 (Fig. 2B).
We demonstrated the pan-SARS-CoV-2 activity of these three lead antivirals against SARS-CoV-2 highly transmissible Omicron subvariants (BA.5, BA.2 and BA.1) and highly pathogenic Delta VOCs in human Calu-3 lung cells (Fig. 3). Notably, holyrine A, alotaketal C, and bafilomycin D, are potent nanomolar inhibitors of SARS-CoV-2 Omicron subvariants BA.5 and BA.2 (Fig. 3H-M; Table 2 ). Interestingly, in contrast to bafilomycin D, which presented nanomolar antiviral activity for all SARS-CoV-2 variants tested, we observed discrepancies for the EC50 values determined across the variants for holyrine A and alotaketal C (Table 2). For example, holyrine A is a more potent antiviral against the BA.2 sublineages (Fig. 3H and K; Table 2) than against SARS-CoV-2 BA.1 and Delta (Fig. 3E and B; Table 2). Alotaketal C is a more potent antiviral against highly transmissible Omicron subvariants (BA1, BA2, and BA5) (Fig. 3F, I, 3L; Table 2) than against the highly pathogenic Delta VOC (Fig. 3C; Table 2). Nevertheless, our results confirm the pan-SARS-CoV-2 antiviral activity of holyrine A, alotaketal C, and bafilomycin D against SARS-CoV-2 variants in lung epithelial cells.
Table 2.
SARS-CoV- 2 VOCs inhibition (EC50) for the three lead NPs in Calu-3 cells.
| Name | Delta (EC50) |
BA.1 (EC50) |
BA.2 (EC50) |
BA.5 (EC50) |
||||
|---|---|---|---|---|---|---|---|---|
| Nuca | dsRNA | Nuca | dsRNA | Nuca | dsRNA | Nuca | dsRNA | |
| holyrine A | 600 nM | 1.0 μM | 1.7 μM | 1.6 μM | 44 nM | 394 nM | 80 nM | 120 nM |
| alotaketal C | 4.9 μM | 3.9 μM | 700 nM | 400 nM | 700 nM | 700 nM | 300 nM | 700 nM |
| bafilomycin D | 2.0 nM | 50 nM | 13.4 nM | 67.3 nM | 4.6 nM | 22 nM | 43.9 nM | 38.6 nM |
Nuc = Nucleocapsid.
Holyrine A comes from the indolocarbazole class of alkaloids isolated from a marine actinomycete collected in Canada, and it is closely related to holyrine B, which differs from holyrine A by the addition of an oxygen atom (Williams et al., 1999). Holyrine B was recently described as a potential SARS-CoV-2 3CLpro inhibitor based on virtual screening using docking and molecular simulation (Sayed et al., 2020). However, although holyrine A acted as a pan-SARS-CoV-2 antiviral in our study, neither holyrine A nor B presented activity against 3CLpro in the enzymatic assay (data not shown). These results suggest an alternative mode of action for holyrine A in inhibiting SARS-CoV-2 infection.
Alotaketal C is a sesterterpenoid isolated from the marine sponge Phorbas sp. Collected in Canada (Daoust et al., 2013). We previously reported alotaketal C as an activator of protein kinase C (PKC) that is more potent than the well-studied PKC activator prostratin (Wang et al., 2016, 2022). Our preliminary results of the three natural products acting as PKC activators (prostratin, bryostatin and PEP 005) also demonstrated inhibition of SARS-CoV-2 BA.1 (Fig. 4A–C and S6), consistent with PKC activation inhibiting SARS-CoV-2 infection. We also observed a striking reverse phenomenon using the PKC inhibitors Gö6983 (Gschwendt et al., 1996) and Gö6976 where a dramatic increase in susceptibility of cells to SARS-CoV-2 BA.1 was observed following Gö6983 or Gö6976 treatment (Fig. 4D–E and S6). However, the treatment with an atypical PKC inhibitor showed inhibition of SARS-CoV-2 BA.1 (Fig. 4F and S6). These results strongly suggest that different isoforms of PKC can play a role in the regulation of cellular susceptibility to SARS-CoV-2 infection.
Because many polymorphisms in various PKC genes have been identified and because drugs targeting PKC are under investigation in clinical trials for cancer and other diseases (Callender et al., 2018; Kawano et al., 2021; Ma et al., 2010), future research should explore whether PKC regulation impacts human susceptibility and the severity of SARS-CoV-2 infection. A recent study reported that PKC inhibitors block the replication of ancestral SARS-CoV-2 on A459/ACE2 expressing cells (Huang et al., 2022). The biological effects of PKC are wide-ranging, and even though most PKC isoenzymes are ubiquitous and many cells can co-express different PKC isoforms, some of them are expressed in a tissue-specific manner (Blázquez and Saiz, 2021). We hypothesize that altered susceptibility may be due to these facts as noted in published findings: PKC inhibition by multiple drugs reduced the shedding of ACE2 from the surface of cells (Xiao et al., 2016); ACE2 is the key receptor required for SARS-CoV-2 entry (Hoffmann et al., 2020a); and increased shedding should reduce cell surface ACE2 abundance. Future studies should investigate whether PKC activation by alotaketal C increases ACE2 shedding from human respiratory epithelial cells.
Bafilomycin D is a member of the macrolide antibiotic family, which has been isolated from a Streptomyces sp. obtained from marine sediments collected in Canada (Carr et al., 2010; Kretschmer et al., 1985). bafilomycin A1, B1, and D are inhibitors of vacuolar-type H+ (V)- ATPases (Bowman et al., 1988). Studies have shown that bafilomycin A1 can inhibit both Delta and Omicron variants, independent of TMPRSS2 expression (Indari et al., 2021; Zhao et al., 2022). Interestingly, the use of CRISPR knockout screens has shown that the interference with V-ATPase expression results in the inhibition of SARS-CoV-2 infection (Daniloski et al., 2021; Zhu et al., 2021). Our results confirm the important biological roles of human V-ATPase in the SARS-CoV-2 lifecycle (Indari et al., 2021) and demonstrate that bafilomycin D is a very potent nanomolar pan-SARS-CoV-2 lead antiviral with a high SI value (SI = 1064: Table 1).
In order to investigate further the molecular mechanism of action of bafilomycin D as a pan-SARS-CoV-2 antiviral, we also investigated its potential synergistic action when used in combination with N-0385, a highly potent pan-variant host-directed antiviral (Shapira et al., 2022). We demonstrated a synergistic action of bafilomycin D (a human V-ATPase inhibitor) and N-0385 (a highly potent inhibitor of human TMPRSS2 protease) against SARS-CoV-2 Omicron BA.2. These results suggest that the two highly potent host-directed antivirals, bafilomycin D and N-0385, target two distinct mechanisms of viral entry that depend on distinct host factors, human V-ATPase and human TMPRRS2 (Indari et al., 2021; Kreutzberger et al., 2021). Further studies are needed to confirm the detailed mechanisms of action of bafilomycin D as a pan-SARS-CoV-2 antiviral agent. Importantly, our results demonstrate that the combination of bafilomycin D and N-0385 provides an extremely valuable inspirational starting point for developing urgently needed SARS-CoV-2 multidrug regimen for circulating SARS-CoV-2 Omicron subvariants.
Together, our findings underline the importance of natural product studies not only for identifying potential treatment strategies based on host-directed antivirals but also for unravelling new findings about the biology and pathogenesis of human emerging viruses.
5. Conclusion
In summary, we applied a cell-based fluorescent-screening assay to identify promising anti-SARS-CoV-2 compounds from a diverse NP library. Three lead compounds (holyrine A, alotaketal C, and bafilomycin D) demonstrated nanomolar antiviral potency against mNG-SARS-CoV-2, and we confirmed they had low micromolar to nanomolar pan-SARS-CoV-2 antiviral activity against four VOCs (Delta, Omicron BA.1, BA.2 and BA.5). Further studies may be required to determine the broad-spectrum antiviral activity of the additional 47 NPs not investigated here that demonstrated >50% inhibition at 50 μM using the mNG-SARS-CoV-2, with <20% cell loss. Due to the unique biology of SARS-CoV-2 VOCs in terms of their transmission and pathogenesis, a number of these NPs may present additional promising leads.
Overall, our study provides insight into the potential of NPs with highly diverse chemical structures as valuable inspirational starting points for developing pan-SARS-CoV-2 therapeutics. These therapeutics could be used as part of multidrug regimens to counteract the increasing antiviral drug resistance to the currently limited repertoire of monotherapies against SARS-CoV-2 infections (Li et al., 2022; Schultz et al., 2022; Szemiel et al., 2021). In addition, our antiviral NPs provide new patent-free academia-originated leads for further development as alternatives to patent-protected pharmaceuticals.
Funding acquisition
Novel Coronavirus (COVID-19) Rapid Research Funding program of the Canadian Institutes of Health Research (CIHR) [OV3-170342 (FJ, RJA, PYS, IRN, AC, and NS)]; Novel Coronavirus (COVID-19) Rapid Research Funding program of the Canadian Institutes of Health Research (CIHR) [UBR 322812; VR3-172639 (RL, PLB, and FJ)]; Genome British Columbia/COVID-19 Rapid Response Funding Initiative [COV011 (FJ and RJA)]; NSERC Alliance COVID-19 Grant (AWD-015086 NSERC, 2020; IRN and FJ); Coronavirus Variants Rapid Response Network (CoVaRR-Net) grant (#175622 (FJ, ADO, NS, and IRN); and MITACS Inc. Accelerate Fellowship COVID-19 Award [IT18585 (TS, FJ)]. PYS was supported by NIH grants HHSN272201600013C, U01AI151801, and U19AI171413, and awards from the Sealy & Smith Foundation, the Kleberg Foundation, the John S. Dunn Foundation, the Amon G. Carter Foundation, the Gilson Longenbaugh Foundation, and the Summerfield Robert Foundation. RGSB is supported by FAPESP grant 2019/17721–9.
Supervision
The research was performed under the supervision of FJ.
Author contributions
FJ and RJA conceptualized the initiation of the study. Designed research: FJ, JPV, TS, AO, RJA. Performed research: TS, JPV, IV, CAHT, GG, SE, JDG, AC, DBS. Analyzed data: JPV, AO, FJ. Contributed new reagents or analytical tools: MN, OA, MK, PL, PYS, XX, AF, EM, GN, VFF, JIQB, DIB, JRG, RL, PLB, RGSB, HLT, SL, VS, AR, PM, IP, SC, WJ, SK, KK, CY, BC, DEW, MW, IT, RJA, NS. Confocal labelling, image acquisition and analysis: GG, CAHT, IRN, FJ. JPV and FJ wrote and revised the manuscript with input from all authors. The review of the final manuscript was performed by all authors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors acknowledge the support of the CL3 facility (Facility for Infectious Disease and Epidemic Research (FINDER) of the Life Sciences Institute of the University of British Columbia founded by Dr. François Jean and its biosafety support staff including Dr. Bintou Ahidjo (Research Platform Manager) and T. Dean Airey (FINDER Senior Research Technician). Imaging was performed in the LSI Imaging Core Facility of the Life Sciences Institute at the University of British Columbia, supported by the Life Sciences Institute, the UBC GREx Biological Resilience Initiative. The infrastructure within LSI Imaging Core Facility is funded by the Canadian Foundation for Innovation, BC Knowledge Development Fund, Natural Sciences and Engineering Research Council of Canada Research Tools and Instruments, and UBC Research Facility Support Grants as well as a Strategic Investment Fund (Faculty of Medicine, UBC). We further thank Dr. Alex Ball, Jr., MD, Senior Scientist (GeneTex), for supplying the SARS-CoV-2 (COVID-19) nucleocapsid antibody [HL344] (GTX635679). We also acknowledge a generous donation towards the purchase of the CellInsight CX7 HCS system provided by the Vancouver General Hospital Foundation to Dr. Jean. We also thank Dr. Jill Kelly for proofreading the manuscript, Mike LeBlanc for technical assistance, and Dr. Carl Perez for administrative and logistic support during this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105484.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- A living WHO . 2022. A Living WHO Guideline on Drugs for Covid-19. BMJ o1045. [DOI] [PubMed] [Google Scholar]
- Ashhurst A.S., Tang A.H., Fajtová P., Yoon M.C., Aggarwal A., Bedding M.J., Stoye A., Beretta L., Pwee D., Drelich A., Skinner D., Li L., Meek T.D., McKerrow J.H., Hook V., Tseng C.-T., Larance M., Turville S., Gerwick W.H., O'Donoghue A.J., Payne R.J. Potent anti-SARS-CoV-2 activity by the natural product gallinamide A and analogues via inhibition of cathepsin L. J. Med.Chem.acs. 2021 doi: 10.1021/acs.jmedchem.1c01494. 1c01494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auth J., Fröba M., Große M., Rauch P., Ruetalo N., Schindler M., Morokutti-kurz M., Graf P., Dolischka A., Prieschl-grassauer E., Setz C., Schubert U. Lectin from triticum vulgaris (WGA) inhibits infection with SARS-CoV-2 and its variants of concern alpha and beta. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms221910205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao R., Wang S., Yang X., Chen G., Meng X. Phellopterin-induced caspase-dependent apoptosis through PI3K/AKT pathway inhibition in SMMC-7721 human hepatoma cells. Am. J. Pharm. 2018;37:2498–2501. [Google Scholar]
- Barakat E.M.F., el Wakeel L.M., Hagag R.S. Effects of Nigella sativa on outcome of hepatitis C in Egypt. World J. Gastroenterol. 2013;19:2529–2536. doi: 10.3748/wjg.v19.i16.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez A.B., Saiz J.C. Zika Virus Impact, Diagnosis, Control, and Models. Elsevier; 2021. Protein kinase C as a target in the control of viruses and implication for Zika virus; pp. 409–415. [DOI] [Google Scholar]
- Bowman E.J., Siebers A., Altendorf K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. USA. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagno V. SARS-CoV-2 cellular tropism. Lancet Microbe May. 2020:e2–e3. doi: 10.1016/S2666-5247(20)30008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. Beyond Omicron: what's next for COVID's viral evolution. Nature. 2021;600:204–207. doi: 10.1038/D41586-021-03619-8. [DOI] [PubMed] [Google Scholar]
- Callaway E. Heavily mutated coronavirus variant puts scientists on alert. Nature. 2021 doi: 10.1038/D41586-021-03552-W. [DOI] [PubMed] [Google Scholar]
- Callender J.A., Yang Y., Lordén G., Stephenson N.L., Jones A.C., Brognard J., Newton A.C. Protein kinase Cα gain-of-function variant in Alzheimer's disease displays enhanced catalysis by a mechanism that evades down-regulation. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E5497–E5505. doi: 10.1073/PNAS.1805046115/ASSET/CA0D7F9E-61C8-4A0E-9F3C-072EC6F9278E/ASSETS/GRAPHIC/PNAS.1805046115FIG07.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr G., Williams D.E., Díaz-Marrero A.R., Patrick B.O., Bottriell H., Balgi A.D., Donohue E., Roberge M., Andersen R.J. Bafilomycins produced in culture by Streptomyces spp. isolated from marine habitats are potent inhibitors of autophagy. J. Nat. Prod. 2010;73:422–427. doi: 10.1021/np900632r. [DOI] [PubMed] [Google Scholar]
- Chaipukdee N., Kanokmedhakul K., Kanokmedhakul S., Lekphrom R., Pyne S.G. Two new bioactive iridoids from Rothmannia wittii. Fitoterapia. 2016;113:97–101. doi: 10.1016/J.FITOTE.2016.07.007. [DOI] [PubMed] [Google Scholar]
- Chu H., Chan J.F.-W., Yuen T.T.-T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C.-Y., Tsang J.O.-L., Huang X., Chai Y., Yang D., Hou Y., Chik K.K.-H., Zhang X., Fung A.Y.-F., Tsoi H.-W., Cai J.-P., Chan W.-M., Ip J.D., Chu A.W.-H., Zhou J., Lung D.C., Kok K.-H., To K.K.-W., Tsang O.T.-Y., Chan K.-H., Yuen K.-Y. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/s2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 COVID-19 dashboard by the center for systems science and engineering (CSSE) at Johns Hopkins university [WWW document] https://coronavirus.jhu.edu/map.html n.d. URL. 2.8.21.
- Dampalla C.S., Zheng J., Dinali Perera K., Roy Wong L.-Y., Meyerholz D.K., Nhat Nguyen H., Kashipathy M.M., Battaile K.P., Lovell S., Kim Y., Perlman S., Groutas W.C., Chang K.-O. Proc Natl Acad Sci U S A; 2021. Postinfection Treatment with a Protease Inhibitor Increases Survival of Mice with a Fatal SARS-CoV-2 Infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Jordan T.X., Wessels H.-H., Hoagland D.A., Kasela S., Legut M., Maniatis S., Mimitou E.P., Lu L., Geller E., Danziger O., Rosenberg B.R., Phatnani H., Smibert P., Lappalainen T., tenOever B.R., Sanjana N.E. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell. 2021;184:92–105. doi: 10.1016/j.cell.2020.10.030. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daoust J., Chen M., Wang M., Williams D.E., Chavez M.A.G., Wang Y.A., Merchant C.E., Fontana A., Kieffer T.J., Andersen R.J. Sesterterpenoids isolated from a northeastern pacific phorbas sp. J. Org. Chem. 2013;78:8267–8273. doi: 10.1021/JO4014589/SUPPL_FILE/JO4014589_SI_001.PDF. [DOI] [PubMed] [Google Scholar]
- Dittmar M., Lee J.S., Whig K., Segrist E., Li M., Kamalia B., Castellana L., Ayyanathan K., Cardenas-Diaz F.L., Morrisey E.E., Truitt R., Yang W., Jurado K., Samby K., Ramage H., Schultz D.C., Cherry S. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G.F. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11(1):1–8. doi: 10.1038/s41467-020-18233-x. 2020 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi S., Klein J., Robertson A.J., Peña-Hernández M.A., Lin M.J., Roychoudhury P., Lu P., Fournier J., Ferguson D., Mohamed Bakhash S.A.K., Catherine Muenker M., Srivathsan A., Wunder E.A., Kerantzas N., Wang W., Lindenbach B., Pyle A., Wilen C.B., Ogbuagu O., Greninger A.L., Iwasaki A., Schulz W.L., Ko A.I. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report. Nat. Commun. 2022;13(1):1–8. doi: 10.1038/s41467-022-29104-y. 2022 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwendt M., Dieterich S., Rennecke J., Kittstein W., Mueller H.J., Johannes F.J. Inhibition of protein kinase C μ by various inhibitors. Inhibition from protein kinase c isoenzymes. FEBS Lett. 1996;392:77–80. doi: 10.1016/0014-5793(96)00785-5. [DOI] [PubMed] [Google Scholar]
- Hacisuleyman E., Hale C., Saito Y., Blachere N.E., Bergh M., Conlon E.G., Schaefer-Babajew D.J., DaSilva J., Muecksch F., Gaebler C., Lifton R., Nussenzweig M.C., Hatziioannou T., Bieniasz P.D., Darnell R.B. Vaccine breakthrough infections with SARS-CoV-2 variants. N. Engl.J.Med.NEJMoa. 2021 doi: 10.1056/NEJMoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen A. n.d. How dangerous is the delta variant (B.1.617.2)? [WWW document] https://asm.org/Articles/2021/July/How-Dangerous-is-the-Delta-Variant-B-1-617-2 URL. 8.29.21.
- Hampson P., Chahal H., Khanim F., Hayden R., Mulder A., Assi L.K., Bunce C.M., Lord J.M. PEP005, a selective small-molecule activator of protein kinase C, has potent antileukemic activity mediated via the delta isoform of PKC. Blood. 2005;106:1362–1368. doi: 10.1182/BLOOD-2004-10-4117. [DOI] [PubMed] [Google Scholar]
- Hoffmann Markus, Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Lewandowski E.M., Tan H., Morgan R.T., Zhang X., Jacobs L.M.C., Butler S.G., Mongora M.v., Choy J., Chen Y., Wang J. Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir. bioRxiv. 2022 doi: 10.1101/2022.06.28.497978. 2022.06.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Feng F., Shi Y., Li W., Wang Z., Zhu Y., Yuan S., Hu D., Dai J., Jiang Q., Zhang R., Liu C., Zhang P. Protein kinase C inhibitors reduce SARS-CoV-2 replication in cultured cells. Microbiol. Spectr. 2022 doi: 10.1128/SPECTRUM.01056-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icho S., Rujas E., Muthuraman K., Tam J., Liang H., Landreth S., Liao M., Falzarano D., Julien J.-P., Melnyk R.A. Dual inhibition of vacuolar-ATPase and TMPRSS2 is required for complete blockade of SARS-CoV-2 entry into cells. Antimicrob. Agents Chemother. 2022;66 doi: 10.1128/aac.00439-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indari O., Jakhmola S., Manivannan E., Jha H.C. An update on antiviral therapy against SARS-CoV-2: how far have we come? Front. Pharmacol. 2021:133. doi: 10.3389/FPHAR.2021.632677. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochmans D., Liu C., Donckers K., Stoycheva A., Boland S., Stevens S.K., de Vita C., Vanmechelen B., Maes P., Trüeb B., Ebert N., Thiel V., de Jonghe S., Vangeel L., Bardiot D., Jekle A., Blatt L.M., Beigelman L., Symons J.A., Raboisson P., Chaltin P., Marchand A., Neyts J., Deval J., Vandyck K. bioRxiv; 2022. The Substitutions L50F, E166A and L167F in SARS-CoV-2 3CLpro Are Selected by a Protease Inhibitor in Vitro and Confer Resistance to Nirmatrelvir. 2022.06.07.495116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T., Inokuchi J., Eto M., Murata M., Kang J.H. Activators and inhibitors of protein kinase C (PKC): their applications in clinical trials. Pharmaceutics. 2021;13 doi: 10.3390/PHARMACEUTICS13111748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedei N., Lundberg D.J., Toth A., Welburn P., Garfield S.H., Blumberg P.M. Characterization of the interaction of ingenol 3-angelate with protein kinase C. Cancer Res. 2004;64:3243–3255. doi: 10.1158/0008-5472.CAN-03-3403. [DOI] [PubMed] [Google Scholar]
- Khumkomkhet P., Kanokmedhakul S., Kanokmedhakul K., Hahnvajanawong C., Soytong K. Antimalarial and cytotoxic depsidones from the fungus Chaetomium brasiliense. J. Nat. Prod. 2009;72:1487–1491. doi: 10.1021/np9003189. [DOI] [PubMed] [Google Scholar]
- Kjær S., Linch M., Purkiss A., Kostelecky B., Knowles P.P., Rosse C., Riou P., Soudy C., Kaye S., Patel B., Soriano E., Murray-Rust J., Barton C., Dillon C., Roffey J., Parker P.J., McDonald N.Q. Adenosine-binding motif mimicry and cellular effects of a thieno[2,3- d ]pyrimidine-based chemical inhibitor of atypical protein kinase C isoenzymes. Biochem. J. 2013;451:329–342. doi: 10.1042/BJ20121871. [DOI] [PubMed] [Google Scholar]
- Koivunen J., Aaltonen V., Koskela S., Lehenkari P., Laato M., Peltonen J. Protein kinase C α/β inhibitor Go6976 promotes formation of cell junctions and inhibits invasion of urinary bladder carcinoma cells. Cancer Res. 2004;64:5693–5701. doi: 10.1158/0008-5472.CAN-03-3511. [DOI] [PubMed] [Google Scholar]
- Kretschmer A., Dorgerloh M., Deeg M., Hagenmaier H. The structures of novel insecticidal macrolides: bafilomycins D and E, and oxohygrolidin. Agric. Biol. Chem. 1985;49:2509–2511. doi: 10.1271/bbb1961.49.2509. [DOI] [Google Scholar]
- Kreutzberger A.J.B., Sanyal A., Ojha R., Pyle J.D., Vapalahti O., Balistreri G., Kirchhausen T. Synergistic block of SARS-CoV-2 infection by combined drug inhibition of the host entry factors PIKfyve kinase and TMPRSS2 protease. J. Virol. 2021;95 doi: 10.1128/JVI.00975-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Lou F., Fan H. SARS-CoV-2 Variants of Concern Delta: a great challenge to prevention and control of COVID-19. Signal Transduct. Targeted Ther. 2021;6(1 6):1–3. doi: 10.1038/s41392-021-00767-1. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Wang Y., Lavrijsen M., Lamers M.M., de Vries A.C., Rottier R.J., Bruno M.J., Peppelenbosch M.P., Haagmans B.L., Pan Q. SARS-CoV-2 Omicron variant is highly sensitive to molnupiravir, nirmatrelvir, and the combination. Cell Res. 2022;32(3 32):322–324. doi: 10.1038/s41422-022-00618-w. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linch M., Sanz-Garcia M., Rosse C., Riou P., Peel N., Madsen C.D., Sahai E., Downward J., Khwaja A., Dillon C., Roffey J., Cameron A.J.M., Parker P.J. Regulation of polarized morphogenesis by protein kinase C iota in oncogenic epithelial spheroids. Carcinogenesis. 2014;35:396–406. doi: 10.1093/carcin/bgt313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma R.C.W., Tam C.H.T., Wang Y., Luk A.O., Hu C., Yang X., Lam V., Chan A.W.H., Ho J.S.K., Chow C.C., Tong P.C.Y., Jia W., Ng M.C.Y., So W.Y., Chan J.C.N. Genetic variants of the protein kinase C-beta 1 gene and development of end-stage renal disease in patients with type 2 diabetes. JAMA. 2010;304:881–889. doi: 10.1001/JAMA.2010.1191. [DOI] [PubMed] [Google Scholar]
- Mani J.S., Johnson J.B., Steel J.C., Broszczak D.A., Neilsen P.M., Walsh K.B., Naiker M. 2020. Natural Product-Derived Phytochemicals as Potential Agents against Coronaviruses: A Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee M., Stuckler D. If the world fails to protect the economy, COVID-19 will damage health not just now but also in the future. Nat. Med. 2020 doi: 10.1038/s41591-020-0863-y. [DOI] [PubMed] [Google Scholar]
- Meganck R.M., Baric R.S. Developing therapeutic approaches for twenty-first-century emerging infectious viral diseases. Nat. Med. 2021;27:401–410. doi: 10.1038/s41591-021-01282-0. [DOI] [PubMed] [Google Scholar]
- Moghadamtousi S.Z., Nikzad S., Kadir H.A., Abubakar S., Zandi K. Potential antiviral agents from marine fungi: an overview. Mar. Drugs. 2015;13:4520–4538. doi: 10.3390/MD13074520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpour D., Evans M.J., Gosert R., Yuan Z., Blum H.E., Goff S.P., Lindenbach B.D., Rice C.M. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 2004;78:7400–7409. doi: 10.1128/JVI.78.14.7400-7409.2004/FORMAT/EPUB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P.Y., Milton M.N. The determination and interpretation of the therapeutic index in drug development. Nat. Rev. Drug Discov. 2012;11(10):751–761. doi: 10.1038/nrd3801. 2012 11. [DOI] [PubMed] [Google Scholar]
- Musarra-Pizzo M., Pennisi R., Ben-Amor I., Mandalari G., Sciortino M.T. Antiviral activity exerted by natural products against human viruses. Viruses. 2021;13 doi: 10.3390/V13050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R.W.A.L., van der Meer Y., Caly L., Druce J., de Vries J.J.C., Kikkert M., Barcena M., Sidorov I., Snijder E.J. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020;101:925. doi: 10.1099/JGV.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmstead A.D., Knecht W., Lazarov I., Dixit S.B., Jean F. Human subtilase SKI-1/S1P is a master regulator of the HCV lifecycle and a potential host cell target for developing indirect-acting antiviral agents. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.-H., Porrot F., Staropoli I., Lemoine F., Péré H., Veyer D., Puech J., Rodary J., Baele G., Dellicour S., Raymenants J., Gorissen S., Geenen C., Vanmechelen B., Wawina -Bokalanga T., Martí-Carreras J., Cuypers L., Sève A., Hocqueloux L., Prazuck T., Rey F., Simon-Loriere E., Bruel T., Mouquet H., André E., Schwartz O. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2021:1–7. doi: 10.1038/s41586-021-04389-z. 2021. [DOI] [PubMed] [Google Scholar]
- Ranganath N., O'Horo J.C., Challener D.W., Tulledge-Scheitel S.M., Pike M.L., Michael O'Brien R., Razonable R.R., Shah A. Rebound phenomenon after nirmatrelvir/ritonavir treatment of coronavirus disease-2019 in high-risk persons. Clin. Infect. Dis. 2022 doi: 10.1093/CID/CIAC481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockett R.J., Draper J., Gall M., Sim E.M., Arnott A., Agius J.E., Johnson-Mackinnon J., Fong W., Martinez E., Drew A.P., Lee C., Ngo C., Ramsperger M., Ginn A.N., Wang Q., Fennell M., Ko D., Hueston L., Kairaitis L., Holmes E.C., O'Sullivan M.N., Chen S.C.-A., Kok J., Dwyer D.E., Sintchenko V. Co-infection with SARS-CoV-2 Omicron and Delta variants revealed by genomic surveillance. Nat. Commun. 2022;13(1):1–7. doi: 10.1038/s41467-022-30518-x. 2022 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SARS SARS-CoV-2 variants of concern | CDC [WWW document] https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html n.d. URL. 2.8.21.
- Sayed A.M., Alhadrami H.A., El-Gendy A.O., Shamikh Y.I., Belbahri L., Hassan H.M., Abdelmohsen U.R., Rateb M.E. Microbial natural products as potential inhibitors of SARS-CoV-2 main protease (Mpro) Microorganisms. 2020;8:1–17. doi: 10.3390/microorganisms8070970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D.C., Johnson R.M., Ayyanathan K., Miller J., Whig K., Kamalia B., Dittmar M., Weston S., Hammond H.L., Dillen C., Ardanuy J., Taylor L., Lee J.S., Li M., Lee E., Shoffler C., Petucci C., Constant S., Ferrer M., Thaiss C.A., Frieman M.B., Cherry S. Pyrimidine inhibitors synergize with nucleoside analogues to block SARS-CoV-2. Nature. 2022;604(7904):134–140. doi: 10.1038/s41586-022-04482-x. 2022 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shagufta, Ahmad I. The race to treat COVID-19: potential therapeutic agents for the prevention and treatment of SARS-CoV-2. Eur. J. Med. Chem. 2021;213 doi: 10.1016/j.ejmech.2021.113157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira T., Monreal I.A., Dion S.P., Buchholz D.W., Imbiakha B., Olmstead A.D., Jager M., Désilets A., Gao G., Martins M., Vandal T., Thompson C.A.H., Chin A., Rees W.D., Steiner T., Nabi I.R., Marsault E., Sahler J., Diel D.G., van de Walle G.R., August A., Whittaker G.R., Boudreault P.-L., Leduc R., Aguilar H.C., Jean F. A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature. 2022 doi: 10.1038/s41586-022-04661-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyr Z.A., Cheng Y.S., Lo D.C., Zheng W. Drug combination therapy for emerging viral diseases. Drug Discov. Today. 2021;26:2367. doi: 10.1016/J.DRUDIS.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers B.L., Chakraborty S., Xue Y., Gelbart T., Gonzalez J.C., Cassidy A.G., Golan Y., Prahl M., Gaw S.L., Arunachalam P.S., Blish C.A., Boyd S.D., Davis M.M., Jagannathan P., Nadeau K.C., Pulendran B., Singh U., Scheuermann R.H., Frieman M.B., Vashee S., Wang T.T., Tan G.S. Antibodies elicited by SARS-CoV-2 infection or mRNA vaccines have reduced neutralizing activity against Beta and Omicron pseudoviruses. Sci. Transl. Med. 2022 doi: 10.1126/SCITRANSLMED.ABN7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M.-K., Alkon D.L. Bryostatin-1: pharmacology and therapeutic potential as a CNS drug. CNS Drug Rev. 2006;12:1–8. doi: 10.1111/j.1527-3458.2006.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemiel A.M., Merits A., Orton R.J., MacLean O.A., Pinto R.M., Wickenhagen A., Lieber G., Turnbull M.L., Wang S., Furnon W., Suarez N.M., Mair D., da Silva Filipe A., Willett B.J., Wilson S.J., Patel A.H., Thomson E.C., Palmarini M., Kohl A., Stewart M.E. In vitro selection of Remdesivir resistance suggests evolutionary predictability of SARS-CoV-2. PLoS Pathog. 2021;17 doi: 10.1371/JOURNAL.PPAT.1009929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashita E., Yamayoshi S., Simon V., van Bakel H., Sordillo E.M., Pekosz A., Fukushi S., Suzuki T., Maeda K., Halfmann P., Sakai-Tagawa Y., Ito M., Watanabe S., Imai M., Hasegawa H., Kawaoka Y. Efficacy of antibodies and antiviral drugs against omicron BA.2.12.1, BA.4, and BA.5 subvariants. N. Engl. J. Med. 2022;387:468–470. doi: 10.1056/NEJMc2207519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Wennerberg K., Aittokallio T. What is synergy? The Saariselkä agreement revisited. Front. Pharmacol. 2015;6 doi: 10.3389/fphar.2015.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao S., Zandi K., Bassit L., Ong Y.T., Verma K., Liu P., Downs-Bowen J.A., McBrayer T., LeCher J.C., Kohler J.J., Tedbury P.R., Kim B., Amblard F., Sarafianos S.G., Schinazi R.F. Comparison of anti-SARS-CoV-2 activity and intracellular metabolism of remdesivir and its parent nucleoside. Curr. Res.Pharmacol. Drug Discov. 2021;2 doi: 10.1016/j.crphar.2021.100045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States . 2022. United States - COVID-19 Overview - Johns Hopkins [WWW Document]https://coronavirus.jhu.edu/region/united-states URL. 5.12.22. [Google Scholar]
- Wang M., Sciorillo A., Read S., Divsalar D.N., Gyampoh K., Zu G., Yuan Z., Mounzer K., Williams D.E., Montaner L.J., de Voogd N., Tietjen I., Andersen R.J. Ansellone J, a potent in vitro and ex vivo HIV-1 latency reversal agent isolated from a phorbas sp. marine sponge. J. Nat. Prod. 2022;85:1274–1281. doi: 10.1021/ACS.JNATPROD.1C01225. [DOI] [PubMed] [Google Scholar]
- Wang M., Tietjen I., Chen M., Williams D.E., Daoust J., Brockman M.A., Andersen R.J. Sesterterpenoids isolated from the sponge phorbas sp. activate latent HIV-1 provirus expression. J. Org. Chem. 2016;81:11324–11334. doi: 10.1021/ACS.JOC.6B02312/ASSET/IMAGES/LARGE/JO-2016-02312B_0005.JPEG. [DOI] [PubMed] [Google Scholar]
- Wang S.X., Zhang X.S., Guan H.S., Wang W. Potential anti-HPV and related cancer agents from marine resources: an overview. Mar. Drugs. 2014;12:2019–2035. doi: 10.3390/MD12042019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yang L. Turning the tide: natural products and natural-product-inspired chemicals as potential counters to SARS-CoV-2 infection. Front. Pharmacol. 2020 doi: 10.3389/fphar.2020.01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D.E., Bernan V.S., Ritacco F. v, Maiese W.M., Greenstein M., Andersen R.J. Holyrines A and B, possible intermediates in staurosporine biosynthesis produced in culture by a marine actinomycete obtained from the North Atlantic Ocean. Tetrahedron Lett. 1999;40:7171–7174. [Google Scholar]
- Wu C.-J., Cui X., Xiong B., Yang M.-S., Zhang Y.-X., Liu X.-M. Terretonin D1, a new meroterpenoid from marine-derived Aspergillusï¿¿terreus ML-44. Nat. Prod. Res. 2019;33:2262–2265. doi: 10.1080/14786419.2018.1493583. [DOI] [PubMed] [Google Scholar]
- Xiao F., Zimpelmann J., Burger D., Kennedy C., Hébert R.L., Burns K.D. Protein kinase C-δ mediates shedding of angiotensin-converting enzyme 2 from proximal tubular cells. Front. Pharmacol. 2016;7 doi: 10.3389/FPHAR.2016.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.v., Plante K.S., Weaver S.C., Makino S., LeDuc J.W., Menachery V.D., Shi P.Y. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848. doi: 10.1016/j.chom.2020.04.004. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav B., Wennerberg K., Aittokallio T., Tang J. Searching for drug synergy in complex dose–response landscapes using an interaction potency model. Comput. Struct. Biotechnol. J. 2015;13:504–513. doi: 10.1016/j.csbj.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-H., Chung T.D.Y., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays | enhanced reader [WWW document] J. Biomol. Screen. 1999 doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zhao H., Lu L., Peng Z., Chen L.L., Meng X., Zhang C., Ip J.D., Chan W.M., Chu A.W.H., Chan K.H., Jin D.Y., Chen H., Yuen K.Y., To K.K.W. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg. Microb. Infect. 2022;11:277–283. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Wang W., Aldahdooh J., Malyutina A., Shadbahr T., Tanoli Z., Pessia A., Tang J. SynergyFinder plus: toward better interpretation and annotation of drug combination screening datasets. Dev. Reprod. Biol. 2022 doi: 10.1016/j.gpb.2022.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Yunkai, Feng F., Hu G., Wang Y., Yu Y., Zhu Yuanfei, Xu W., Cai X., Sun Z., Han W., Ye R., Qu D., Ding Q., Huang X., Chen H., Xu W., Xie Y., Cai Q., Yuan Z., Zhang R. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 2021;12:961. doi: 10.1038/s41467-021-21213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.