Abstract
Vaccines are undoubtedly the most effective means of combating viral diseases like COVID-19. However, there are risks associated with vaccination, such as incomplete viral deactivation or potential adverse effects in humans. However, designing and developing a panel of new drug molecules is always encouraged. In an emergency, drug repurposing research is one of the most potent and rapid options. RdRp (RNA-dependent RNA polymerase) has been discovered to play a pivotal role in viral replication. In this study, FDA-approved drugs bexarotene, diiodohydroxyquinoline, abiraterone, cetilistat, and remdesivir were repurposed against the RdRp by molecular modeling, docking, and dynamic simulation. Furthermore, to validate the potency of these drugs, we compared them to the antiviral remdesivir, which inhibits RdRp. Our finding indicated that the selected drugs have a high potential to be developed as RdRp inhibitors and, with further validation studies, could serve as potential drugs for the treatment of COVID-19.
Keywords: Drug repurposing, Coronavirus, RNA-Dependent RNA polymerase, Docking, Dynamic, Virtual screening
Graphical abstract
1. Introduction
COVID-19 is an infectious disease caused by Coronaviruses (family: Coronaviridae), which has recently been identified as a cause of severe acute respiratory syndromes (SARS Cov-2). Coronaviruses are named as such because their surface is characterized by protrusions resembling crowns [1]. Corona was first reported in China (Wuhan), and COVID-2019 is unlikely to have been transmitted from animal to human in a seafood market [2,3]. The outbreak has been rapidly spread across the countries and territories due to its high human to human contagious nature. Until Oct 13, 2022, 628,556,214 people have been infected, and 6,566,411 have died (https://www.worldometers.info/coronavirus/). Vaccines are without question the most efficient tool for combating viral diseases. However, there are inherent threats with vaccination, including non-complete viral deactivation or potential adverse effects in people; even after a vaccine has been produced, exact quality control is required to ensure the treatment's safety. As a result, developing vaccines is a lengthy process; a vaccine may not be commercially available for years following the emergence of a new disease. The current vaccines are not providing adequate protection against COVID-19 due to logistical difficulties in their distribution, diminishing immunity, and the possibility of transmission from asymptomatic infected patients [4,5]. The advent of vaccines may have broken the transmission chains in this viral pandemic, but there is currently concern about their effectiveness against mutant variants, reported adverse reactions, and possible long-term effects. We must, therefore, find safe, effective, preferably affordable, and most importantly, specific drugs rather than the non-specific drugs currently prescribed in order to save the lives of the infected [6,7].
A computer-assisted drug discovery strategy is the most common method of predicting potential compounds before they are synthesized and tested in vitro [8]. Drug repurposing is the most efficient method of rapidly identifying the unique clinical applications of currently licensed medications to treat COVID-19. Since the toxicity and pharmacokinetics of repurposed pharmaceuticals are already known, this method could minimize the time and expense required to develop a new medicine [9]. The repurposing of existing medications is made possible through molecular docking research against viral protein targets [10]. In terms of mathematics, molecular computation has several forms, some of which can be used in biology and protein research because of their ability to handle a wide range of particle systems. The interactions between these molecules are predicted using molecular docking and molecular dynamics modeling. In silico screening of different potential active chemicals can be accomplished cost- and time-efficient using these strategies [11,12]. Numerous scientific disciplines are using molecular modeling techniques, including drug discovery and design [[13], [14], [15]]. The Coronavirus genome is translated into two classes of proteins within the host cell: structural proteins such as spikes (S), envelopes (E), matrixes (M), and nucleocapsids (N), and nonstructural proteins such as 3-C proteases (3CLpro, NSP5) and RNA Dependent RNA Polymerases (RdRp, NSP12) [16].
Among these proteins, RNA-dependent RNA polymerase (RdRp) has been demonstrated to contribute significantly to the replication of the viral genome (single-stranded RNA) and to the multiplication of the virus in various cells [17,18]. It is essential to inhibit RdRp in order to control the progression of infection in human cells and, as a consequence, the viral load in patients, as RdRp replicates viral genomes by synthesizing new RNA nucleotides. It has been established that RNA-dependent RNA polymerase (RdRp) is an essential enzyme that plays a significant role in the replication and translation of viral genomes; this makes it an excellent therapeutic target for COVID-19. Numerous clinical trials have shown that COVID-19 can be effectively treated with antiviral, antimalarial, and anti-HIV drugs. The FDA has approved Remdesivir (Veklury®) for treating COVID-19 in hospitalized patients aged 12 and older and weighing at least 40 kg. Several viral infections, including SARS-CoV-2, have been successfully treated with an analog of nucleotides (NA) known as Remdesivir. In the presence of NAs, a 5-triphosphate is formed in the cell, which replaces the RdRp substrate and competes with the endogenous nucleotides for incorporation into viral RNA. In a comprehensive two-tier screening approach, Yuan et al. demonstrated that FDA-approved medications cetilistat, abiraterone, diiodohydroxyquinoline, and bexarotene inhibited COVID-19 infection in vitro [19]. As part of this study, we investigated the interaction between four FDA-approved medications, bexarotene (anticancer retinoid), abiraterone (synthetic androstenedione steroid), diiodohydroxyquinoline (antiparasite), and cetilistat (antipancreatic lipase) with SARS-CoV-2 RdRp using docking simulation. In this study, remdesivir (an antiviral drug) was compared to the outcomes obtained with remdesivir. Dynamic simulations also clarify the dynamics of molecule behavior. In light of our findings, we may be able to develop a new method for combating COVID-19.
2. Methods
2.1. Docking of cetilistat, abiraterone, diiodohydroxyquinoline, bexarotene, and remdesivir
2.1.1. Macromolecule (drug target) selection and preparation
The screening of the drugs against RdRp of SARS-CoV-2 was performed by AutoDock Vina [20] with MGL tools 1.5.4 which makes more precise docking calculations and runs faster than AutoDock software [21]. The crystal structure of RdRp (PDB ID: 6NUR) was downloaded from the Protein data bank [22]. The receptor preparation was done by utilizing the AutoDock Tools (v.1.5.4). The addition of Kollman charged atoms to protein crystal structure and the combination of non-polar hydrogen atoms was done by MGL Tools (v.1.5.4). To perform molecular docking analysis the RdRp of SARS-CoV-2 was converted into PDBQT format (.pdbqt). The grid box was set to 100 × 90 × 122 with a grid spacing of 1.00 Å and center parameters of 150.05, 152.94, and 163.03.
2.1.2. Ligands selection and preparation
The 3D structure of bexarotene, diiodohydroxyquinoline, abiraterone, cetilistat, and remdesivir were received from PubChem as SDF files (.sdf) (https://pubchem.ncbi.nlm. nih.gov) and converted to PDB format (.pdb) before perform docking utilizing MGL Tools (v.1.5.4). The BIOVIA Discovery Studio software was employed to visualize the docked results.
2.2. Molecular dynamics (MD) simulations
Because molecular docking results revealed that bexarotene has the best energy in interaction with RdRp of SARS-CoV-2, we investigated the molecular dynamic simulation of this drug with protein. We did the Classical MD simulations [23,24] with the CHARMM27 force field using ran GROMACS 2018.2 software [25,26]. SwissParam [27] was used to generate the ligand topology. The free protein and protein-drug complex were solved using TIP3P water [28] in the cubic box with periodic boundary situations in three directions. The solutes were placed in the box's center, with a minimum distance of 1.0 nm between their surfaces. Markedly, we added Na+ ions to the system to neutralize the charge. The systems were balanced at 300 K and 1 bar, following energy minimization using the steepest descent method. The temperature was kept at 300 K using a modified Berendsen thermostat, and the pressure was kept at 1.0 bar using a Parrinello-Rahman barostat. Bond lengths were calculated using the LINCS algorithm, and long-range electrostatic forces were computed employing the particle-mesh Ewald scheme (PME) (grid spacing 0.16 nm) [29]. For short-range non-bonded interactions, cutoff ratios of 1.0 nm for Coulomb and van der Waals potentials were used. Finally, a 50 ns MD simulation with a time-step of 2 fs was running, with velocities generated randomly using a Maxwell distribution.
3. Results
3.1. Evaluation of the docking performance
Because of its critical role in virus replication, RdRp is considered a key target for developing antiviral drugs [18,30,31]. We recently reported the in-silico screening of bexarotene, diiodohydroxyquinoline, abiraterone, cetilistat, and remdesivir against different SARS-CoV-2 drug targets [32]. However, RdRp, an important drug target, was excluded from this study. The current study used the same strategy to screen the bexarotene, diiodohydroxyquinoline, abiraterone, cetilistat, and remdesivir for their potential to inhibit RdRp of SARS-CoV-2, thereby filling the gap. Molecular docking simulation is widely used in computational drug discovery and design. This technique was used to predict the binding of the selected drugs bexarotene, diiodohydroxyquinoline, abiraterone, cetilistat, and remdesivir with SARS-CoV-2 RdRp-RNA protein. Recently, researchers reported virtual screening inhibitors of SARS-CoV-2 RdRp [[33], [34], [35], [36]]. In our study, the FDA-approved RdRp inhibitor drug remdesivir was used as a comparator drug. Remdesivir is effective against many RNA viruses, including the newly discovered SARS-CoV-2 (COVID-19) and previous coronaviruses [37,38]. We hypothesized that the drugs mentioned above could disrupt SARS-CoV-2 RdRp.
Cetilistat, a pancreatic lipase inhibitor, can be prescribed for obesity treatment. This compound inhibits fat digestion and absorption [39]. Diiodohydroxyquinoline is a quinolone derivative used to treat amoebiasis as a luminal amebicide [40]. When bile is available, diiodohydroxyquinoline is unlikely to be absorbed into the circulation, whereas cetilistat is rapidly hydrolyzed into its metabolites [39,41]. Gastrointestinal complaints have been reported in 15–20% of COVID-19 patients, and some of these individuals have also shown infectious virus particles and identifiable viral RNA [42]. In the intestines of SARS-CoV-2-infected hamsters, high levels of viral nucleocapsid protein, inflammatory processes, and identifiable viral RNA were found [43]. Cetilistat and oral diiodohydroxyquinoline may be effective topical luminal antivirals in suppressing viral shedding in the gastrointestinal system in the same way feces may be a significant source of SARS-CoV-2 infection [44,45]. Nonchemotherapeutic antineoplastic medicines, including bexarotene and abiraterone acetate, have been shown to have minimal immunosuppressive properties [19]. Breast cancer, non-small cell lung cancer (NSCLC), and cutaneous T-cell lymphoma are effectively treated with bexarotene, a third-generation retinoid [[46], [47], [48], [49]]. Abiraterone acetate suppressed the activity of the enzyme CYP17A1, which is responsible for androgen synthesis when coupled with a corticosteroid. Androgen deprivation is the mechanism employed in this therapy to treat resistant prostate cancer [50,51]. As described by Yuan et al. [52], members of the same medication class as bexarotene, such as tamibarotene and AM580, have demonstrated outstanding antiviral effectiveness against several viruses, including influenza, Zika, coronaviruses (SARS-CoV and MERS-CoV), adenoviruses, and enterovirus A71.
According to molecular docking studies, the binding energies of bexarotene, abiraterone, cetilistat, remdesivir, and diiodohydroxyquinoline to the RdRp were - 8.4, - 7.9, −7.6, −7.6 and - 5.5 kcal/mol, respectively, which demonstrate good binding potential towards the RdRp. The binding energy values of the five compounds mentioned with the SARS-CoV-2 RdRp protein are shown in Table 1 .
Table 1.
Interaction energy between RdRp of SARS-CoV-2 and molecules.
| Molecules | ΔG (kcal/mol) |
|---|---|
| Abiraterone | −7.9 |
| Bexarotene | −8.4 |
| Cetilistat | −7.6 |
| Diiodohydroxyquinoline | −5.5 |
| Remdesivir | −7.6 |
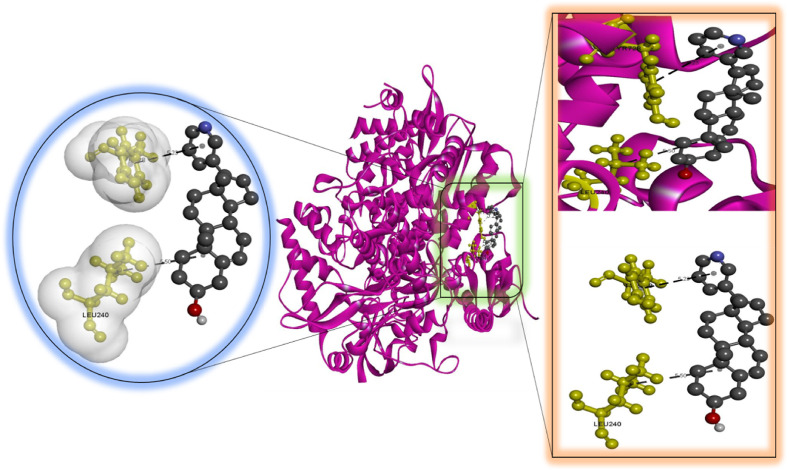
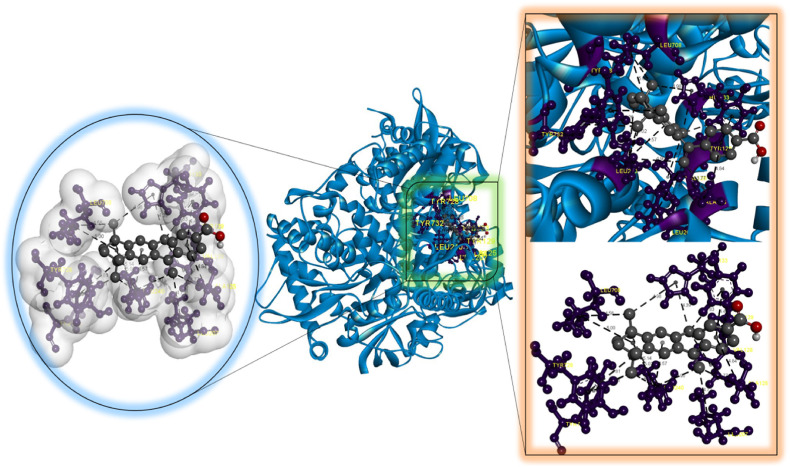
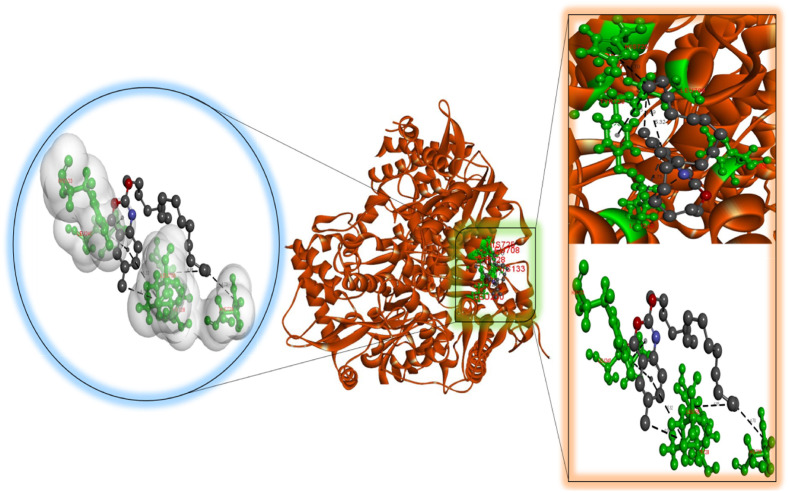
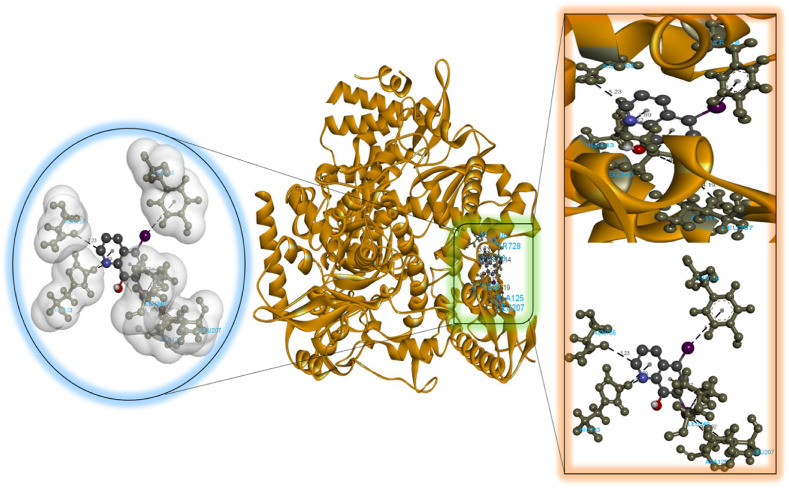
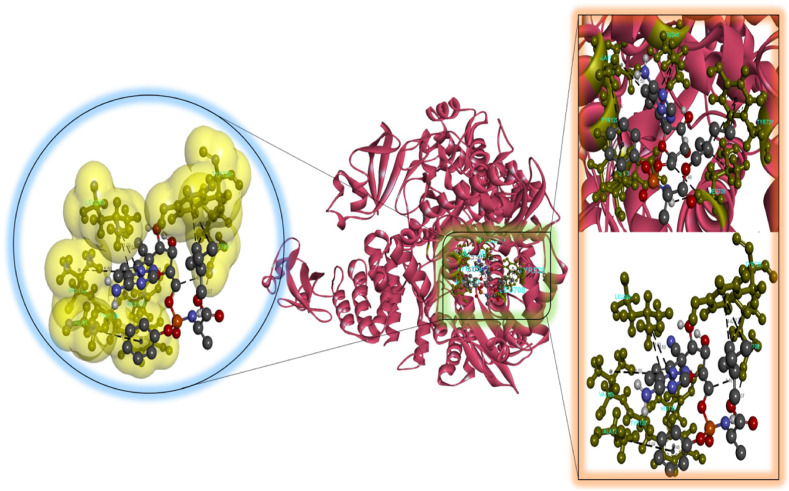
According to our findings, bexarotene was the most effective compound against RdRp, with the lowest binding energy. Also, bexarotene and abiraterone were discovered to have the lowest binding energies than remdesivir. In the meantime, the binding energies of cetilistat are equal to those of remdesivir. Moreover, the binding energy value for diiodohydroxyquinoline (−5.5 kcal/mol) suggests that the ligand-protein complex is stable. Table 2, Table 3, Table 4, Table 5, Table 6 represent the amino acid residues involved, bond lengths, and the type of interactions. Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5 depict the RdRp amino acid residues that interacted with drugs. As evident, The SARS-CoV-2 RdRp protein had hydrophobic interactions with abiraterone and bexarotene. Meanwhile, cetilistat, diiodohydroxyquinoline, and remdesivir interacted with the RdRp of SARS-CoV-2 via hydrogen bonds and hydrophobic interactions. The chosen drugs interacted with various amino acid residues of the RdRp of the SARS-CoV-2 protein (Table 2, Table 3, Table 4, Table 5, Table 6). It is evident from Fig. 2 that the bexarotene interacted with RdRp with LEU240, TYR129, LEU708, VAL128, ALA125, LEU207, TYR129, HIS133, TYR728, and TYR732 amino acids. These interactions can disrupt the biological activity of the SARS-CoV-2 protein RdRp, and, consequently, the viral replication process. Then, we used MD models to confirm the docking poses and analyze the dynamics of interactions between RdRp, RNA, and medicines.
Table 2.
Predicted bonds between interacting atoms of abiraterone and RdRp of SARS-CoV-2.
| S. No. | Amino acid | Amino acid atom | Abiraterone atom | Distance | Nature of interaction |
|---|---|---|---|---|---|
| 1 | TYR728 | Pi-Orbital | Pi-Orbital | 5.22 | Hydrophobic (Pi-Pi) |
| 2 | LEU240 | Alkyl | Alkyl | 5.49 | Hydrophobic (Alkyl) |
Table 3.
Predicted bonds between interacting atoms of bexarotene and RdRp of SARS-CoV-2.
| S. No. | Amino acid | Amino acid atom | Bexarotene atom | Distance | Nature of interaction |
|---|---|---|---|---|---|
| 1 | LEU240 | C–H | Pi-Orbitals | 3.81 | Hydrophobic(Pi-Sigma) |
| 2 | TYR129 | Pi-Orbitals | C10(Alkyl) | 4.47 | Hydrophobic(Pi-Pi) |
| 3 | LEU240 | Alkyl | C10(Alkyl) | 4.26 | Hydrophobic(Alkyl) |
| 4 | LEU240 | Alkyl | C11(Alkyl) | 5.14 | Hydrophobic(Alkyl) |
| 5 | LEU708 | Alkyl | C12(Alkyl) | 5.57 | Hydrophobic(Alkyl) |
| 6 | VAL128 | Alkyl | C18(Alkyl) | 4.94 | Hydrophobic(Alkyl) |
| 7 | ALA125 | Alkyl | C20(Alkyl) | 3.77 | Hydrophobic(Alkyl) |
| 8 | LEU207 | Alkyl | C20(Alkyl) | 4.72 | Hydrophobic(Alkyl) |
| 9 | LEU240 | Alkyl | C20(Alkyl) | 4.90 | Hydrophobic(Alkyl) |
| 10 | LEU240 | Alkyl | Alkyl | 5.02 | Hydrophobic(Alkyl) |
| 11 | LEU708 | Alkyl | Alkyl | 5.00 | Hydrophobic(Alkyl) |
| 12 | ALA125 | Alkyl | Pi-Orbitals | 4.84 | Hydrophobic(Pi-Alkyl) |
| 13 | TYR129 | Pi-Orbitals | C18(Alkyl) | 4.82 | Hydrophobic(Pi-Alkyl) |
| 14 | HIS133 | Pi-Orbitals | C12(Alkyl) | 4.98 | Hydrophobic(Pi-Alkyl) |
| 15 | HIS133 | Pi-Orbitals | C18(Alkyl) | 4.39 | Hydrophobic(Pi-Alkyl) |
| 16 | TYR728 | Pi-Orbitals | C9(Alkyl) | 4.81 | Hydrophobic(Pi-Alkyl) |
| 17 | TYR728 | Pi-Orbitals | C10(Alkyl) | 4.25 | Hydrophobic(Pi-Alkyl) |
| 18 | TYR732 | Pi-Orbitals | C10(Alkyl) | 4.71 | Hydrophobic(Pi-Alkyl) |
Table 4.
Predicted bonds between interacting atoms of cetilistat and RdRp of SARS-CoV-2.
| S. No. | Amino acid | Amino acid atom | Cetilistat atom | Distance | Nature of interaction |
|---|---|---|---|---|---|
| 1 | HIS133 | H-Donor | O3(H-Acceptor) | 2.17 | Hydrogen bond |
| 2 | LEU240 | C–H | Pi-Orbital | 3.61 | Hydrophobic(Pi-Sigma) |
| 3 | LEU240 | C–H | Pi-Orbital | 3.54 | Hydrophobic(Pi-Sigma) |
| 4 | LEU708 | Alkyl | C29(Alkyl) | 4.09 | Hydrophobic(Alkyl) |
| 5 | LEU708 | Alkyl | Pi-Orbital | 5.32 | Hydrophobic(Pi-Alkyl) |
| 6 | HIS725 | Pi-Orbital | C20(Alkyl) | 4.70 | Hydrophobic(Pi-Alkyl) |
| 7 | TYR728 | Pi-Orbital | C20(Alkyl) | 4.85 | Hydrophobic(Pi-Alkyl) |
Table 5.
Predicted bonds between interacting atoms of diiodohydroxyquinoline and RdRp of SARS-CoV-2.
| S. No. | Amino acid | Amino acid atom | Diiodohydroxyquinoline atom | Distance | Nature of interaction |
|---|---|---|---|---|---|
| 1 | HIS133 | H-Donor | N4(H-Acceptor) | 2.22 | Hydrogen bond |
| 2 | ASN705 | H-Acceptor | H-Donor | 3.23 | Hydrogen bond |
| 3 | HIS133 | H-Donor | Pi-Orbital | 2.89 | Hydrogen bond |
| 4 | LEU240 | C–H | Pi-Orbital | 3.79 | Hydrophobic(Pi-Sigma) |
| 5 | ALA125 | Alkyl | I2(Alkyl) | 4.19 | Hydrophobic(Alkyl) |
| 6 | LEU207 | Alkyl | I2(Alkyl) | 4.92 | Hydrophobic(Alkyl) |
| 7 | LEU240 | Alkyl | I2(Alkyl) | 4.73 | Hydrophobic(Alkyl) |
| 8 | TYR728 | Pi-Orbital | I1(Alkyl) | 5.14 | Hydrophobic(Pi-Alkyl) |
Table 6.
Predicted bonds between interacting atoms of remdesivir and RdRp of SARS-CoV-2.
| S. No. | Amino acid | Amino acid atom | Remdesivir atom | Distance | Nature of interaction |
|---|---|---|---|---|---|
| 1 | LEU708 | H-Acceptor | H52(H-Donor) | 3.06 | Hydrogen bond |
| 2 | HIS133 | H-Donor | H-Acceptor | 2.33 | Hydrogen bond |
| 3 | LEU708 | H-Acceptor | C21(H-Donor) | 3.46 | Hydrogen bond |
| 4 | TYR708 | Pi-Orbital | C–H | 3.72 | Hydrophobic(Pi-Sigma) |
| 5 | LEU240 | C–H | Pi-Orbital | 3.70 | Hydrophobic(Pi-Sigma) |
| 6 | LEU240 | C–H | Pi-Orbital | 3.80 | Hydrophobic(Pi-Sigma) |
| 7 | LEU240 | C–H | Pi-Orbital | 3.77 | Hydrophobic(Pi-Sigma) |
| 8 | TYR129 | Pi-Orbital | Pi-Orbital | 4.46 | Hydrophobic(Pi-Pi Stacked) |
| 9 | LEU708 | Alkyl | C37(Alkyl) | 3.91 | Hydrophobic(Alkyl) |
| 10 | ALA125 | Alkyl | Pi-Orbital | 5.04 | Hydrophobic(Pi-Alkyl) |
| 11 | VAL128 | Alkyl | Pi-Orbital | 5.50 | Hydrophobic(Pi-Alkyl) |
| 12 | TYR728 | Pi-Orbital | C37(Alkyl) | 4.42 | Hydrophobic(Pi-Alkyl) |
Fig. 1.
Molecular docking perspective of abiraterone – RdRp of SARS-CoV-2.
Fig. 2.
Molecular docking perspective of bexarotene – RdRp of SARS-CoV-2.
Fig. 3.
Molecular docking perspective of cetilistat – RdRp of SARS-CoV-2.
Fig. 4.
Molecular docking perspective of diiodohydroxyquinoline – RdRp of SARS-CoV-2.
Fig. 5.
Molecular docking perspective of remdesivir – RdRp of SARS-CoV-2.
3.2. Molecular dynamics simulations
We conducted an MD simulation study to evaluate system dynamics and improve the accuracy of the docking computation. For further investigation, bexarotene was chosen as the compound with the highest interaction energy.
3.2.1. Radius of gyration (Rg)
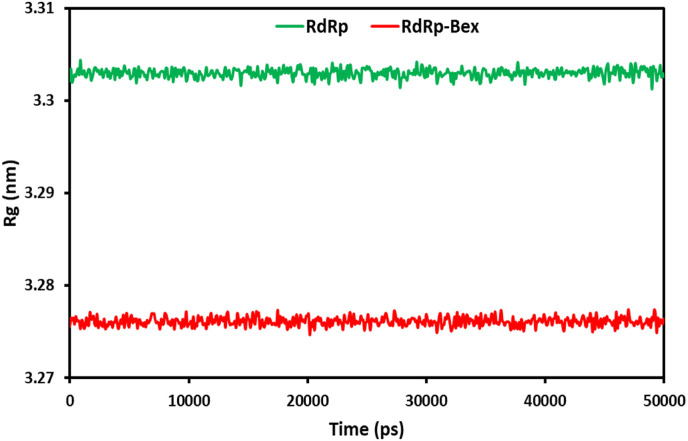
The system compactness can be determined based on the radius of gyration (Rg). The conformational stability of the complex structure during MD modeling is indicated by the graph's stability [53]. Improved structural integrity and folding treatment are indicated by a low Rg value [54]. During a 50ns MD running, the molecule's total extension can be quantified using the protein and ligand-protein complex's gyration radius (Rg) (Fig. 6 ). A constant average Rg of 3.276 nm is maintained by the ligand-protein complex during the whole 50 ns of the MD modeling. Similarly, 3.303 nm was the measured Rg value for the free 6NUR. This demonstrates how more ligand binding increases the protein's stability. The findings from the MD simulations provide unequivocal proof that bexarotene may construct a stable complex with 6NUR, which is an indication of its inhibitory effects on the RdRp receptor.
Fig. 6.
Radius of gyration (Rg) for RdRp of SARS-CoV-2 and bexarotene – (RdRp of SARS-CoV-2) during 50 ns MD simulation.
3.2.2. RMSD
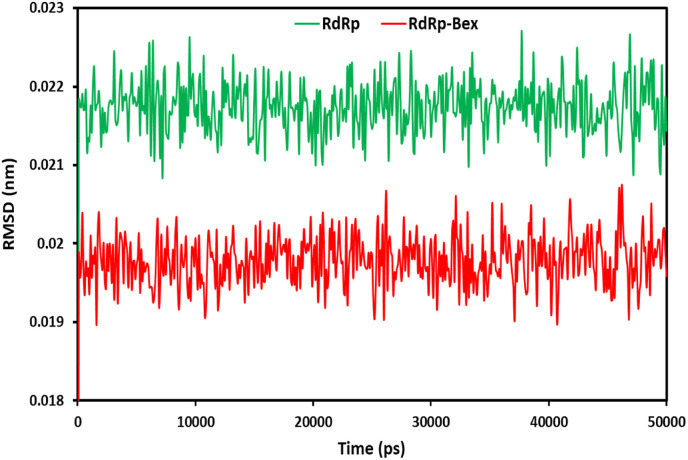
During simulation, the protein's structural alterations and insights from RMSD analysis support the protein's stability and equilibrium. The higher stability of the protein-ligand complex is indicated by the lower RMSD value of the protein backbone [55]. Fig. 7 illustrates the RMSD graph of the backbone atoms in the 6NUR complex containing bexarotene and 6NUR. The RMSD, during the 50ns MD simulation, was determined for the converged 6NUR-Bex structure. According to the findings, after 100 ps, both structures reached a stable state. During the entire 50ns simulation, 6NUR-Bex had a mean of 0.197 A0, while 6NUR had a mean of 0.216 A0. The RMSD value is regarded as acceptable and appropriate if it is less than 1.5 Å. However, it is obviously disregarded if the RMSD number is greater than 3 Å. In addition to studying the conformational changes of the receptors, RMSD measurements are utilized to determine the stability of the receptors with and without ligands [56]. In the MD simulations, the fact that the RMSD value of 6NUR-Bex was significantly lower than that of the protein suggested that bexarotene contributes to the protein's stability.
Fig. 7.
RMSD plots for RdRp of SARS-CoV-2 and bexarotene – (RdRp of SARS-CoV-2) during 50 ns MD simulation.
3.2.3. RMSF
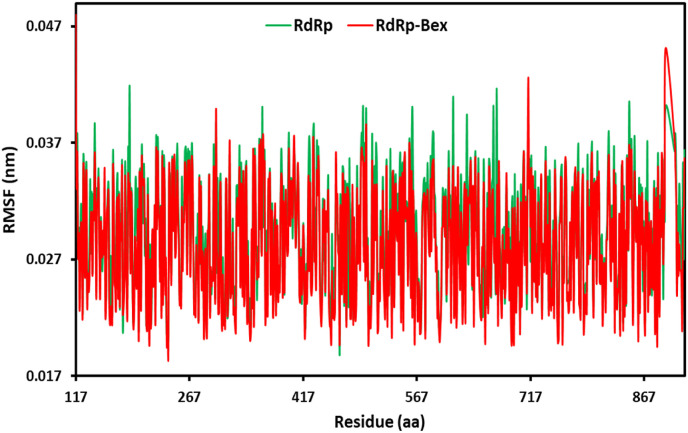
The flexibility of the entire protein concerning its typical structure was assessed using RMSF analysis. High RMSF values showed enhanced flexibility, but low RMSF values showed constricted motions [57]. Energy is released during ligand binding, and the amount of energy released has a direct association with the values of residual fluctuation (RMSF). Fig. 8 displays the RMSF plots generated for each individual residue that makes up the 6NUR complex with bexarotene and 6NUR. The RMSF values for 6NUR-Bex were exceptionally minimal (Average RMSF = 0.28 A0); thus, the ligand-protein complex displayed negligible movement, which demonstrates its stability. Residues inside the loop domain of 6NUR in 6NUR-Bex were found to fluctuate more than alpha-helix and beta-sheet domains during the simulation. This demonstrated that during the 50ns simulation, the protein maintained its stability [58]. The 6NUR residues 895 in the alpha-helix and 714 in the beta-sheet region, except for the loop area, showed considerable fluctuation up to 0.451 A0 and 0.426 A0, respectively. The complex 6NUR-Bex appeared stable overall based on the protein RMSF values [59].
Fig. 8.
RMSF plots for RdRp of SARS-CoV-2 and bexarotene – (RdRp of SARS-CoV-2) during 50 ns MD simulation.
3.3. MM-PBSA binding free energy
The average free binding energy of the 6NUR-Bex was calculated using MmPbSaStat.py (Table 7 ). The information collected from gmmpbsa was used in the calculation of the average free binding energy as well as the standard deviation/error for each of the files. In the drug discovery process, one of the most important factors to consider is the durability of the ligand-receptor contact, which may be evaluated using the binding free energy.
Table 7.
Binding free energy (MM-PBSA) calculations for 6NUR-Bexarotene.
| van der Waal energy | −115.280±0.302 kJ/mol |
| Electrostattic energy | −6.488±0.312 kJ/mol |
| Polar solvation energy | 46.747±0.352 kJ/mol |
| SASA energy | −14.616±0.033 kJ/mol |
| SAV energy | 0.000±0.000 kJ/mol |
| WCA energy | 0.000±0.000 kJ/mol |
| Binding energy | −89.635±0.295 kJ/mol |
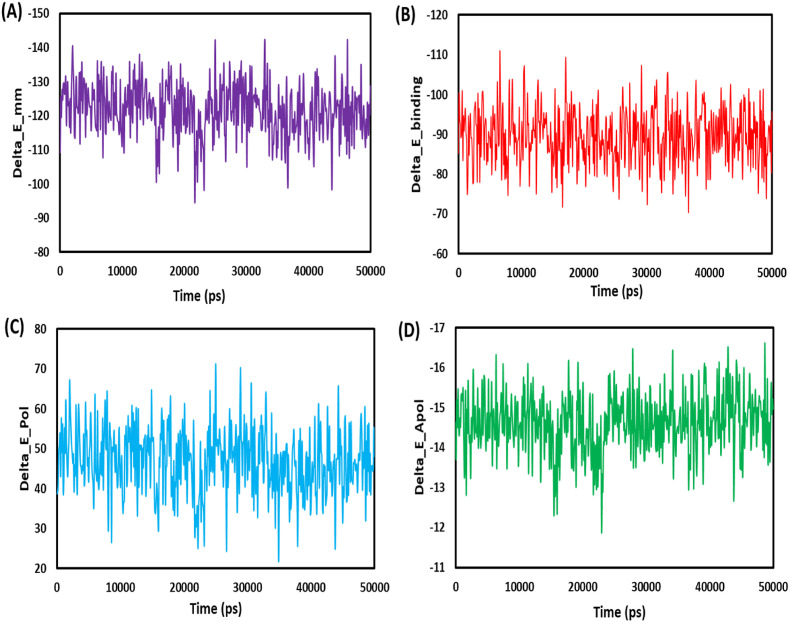
The ligand and protein bind more tightly when the binding energy is lower [60]. To bind 6NUR to bexarotene, the van der Waals, SASA, and electrostatic energies, except for the polar solvation, were utilized. The polar solvation energy was the only energy that was not utilized. The involvement of van der Waals energy to the entire free binding energy was found to be significantly higher than electrostatic contribution energy. The bexarotene linked exclusively with the 6NUR protein, as shown by the binding free energies of 6NUR-Bex, which were found to be −89.635 ± 0.295 kJ/mol. Fig. 9 depict the binding energy versus time plots for 6LU7-Bex. Bexarotene may be a candidate for blocking the RdRp of the SARS-CoV-2 receptor, as shown by the results mentioned above.
Fig. 9.
Graphical representation of the Delta_E_Pol (A) Delta_E_Apol (B) Delta_E_mm (C) Delta_E_binding (D) Bexarotene – RdRp of SARS-CoV-2 during 50 ns MD simulation.
4. Conclusion
Because they have already been evaluated in terms of toxicity and safety in humans for the treatment of various diseases, repurposing FDA-approved medications will be the best option for developing COVID-19 therapies at this time. This research computationally evaluated the inhibitory potential of the FDA-approved drugs bexarotene, diiodohydroxyquinoline, abiraterone, and cetilistat against RdRp of SARS-CoV-2. With further ex vivo and in vivo examinations, our finding suggests that the selected drugs could be the potential drug of choice for the treatment of COVID-19. Gromacs software was used in this study to simulate the interaction of bexarotene with the RdRp of SARS-CoV-2. Furthermore, the RdRp-Bex radius of gyration has values that indicate the systems’ stability. The binding free energy of 6NUR-Bex was determined to be −89.635 ± 0.295 kJ/mol, indicating that the bexarotene interacted with the 6NUR protein uniquely.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors thank the Razi University Research Council for support of this work.
References
- 1.Ashour H.M., Elkhatib W.F., Rahman M.M., Elshabrawy H.A. Insights into the recent 2019 novel coronavirus (SARS-CoV-2) in light of past human coronavirus outbreaks. Pathogens. 2020;9:186. doi: 10.3390/pathogens9030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hui D.S., Azhar E.I., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hernández A.F., Calina D., Poulas K., Docea A.O., Tsatsakis A.M. Safety of COVID-19 vaccines administered in the EU: should we be concerned?, Toxicol. For Rep. 2021;8:871–879. doi: 10.1016/j.toxrep.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anand P., Stahel V.P. The safety of Covid-19 mRNA vaccines: a review. Patient Saf Surg. 2021;15:1–9. doi: 10.1186/s13037-021-00291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asakura H., Ogawa H. Potential of heparin and nafamostat combination therapy for COVID-19. J Thromb Haemostasis. 2020;18:1521–1522. doi: 10.1111/jth.14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Kantar S., Nehmeh B., Saad P., Mitri G., Estephan C., Mroueh M., Akoury E., Taleb R.I. Derivatization and combination therapy of current COVID-19 therapeutic agents: a review of mechanistic pathways, adverse effects, and binding sites. Drug Discov. 2020;25:1822–1838. doi: 10.1016/j.drudis.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gahtori J., Pant S., Srivastava H.K. Modeling antimalarial and antihuman African trypanosomiasis compounds: a ligand-and structure-based approaches. Mol Divers. 2020;24:1107–1124. doi: 10.1007/s11030-019-10015-y. [DOI] [PubMed] [Google Scholar]
- 9.Khan R.J., Jha R.K., Amera G.M., Jain M., Singh E., Pathak A., Singh R.P., Muthukumaran J., Singh A.K. Targeting SARS-CoV-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. J Biomol Struct Dyn. 2021;39:2679–2692. doi: 10.1080/07391102.2020.1753577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kandeel M., Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020;251 doi: 10.1016/j.lfs.2020.117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tazikeh-Lemeski E., Moradi S., Raoufi R., Shahlaei M., Janlou M.A.M., Zolghadri S. Targeting SARS-COV-2 non-structural protein 16: a virtual drug repurposing study. J Biomol Struct Dyn. 2021;39:4633–4646. doi: 10.1080/07391102.2020.1779133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandal S., Mandal S.K. Rational drug design. Eur J Pharmacol. 2009;625:90–100. doi: 10.1016/j.ejphar.2009.06.065. [DOI] [PubMed] [Google Scholar]
- 13.Ansari M., Moradi S., Shahlaei M. A molecular dynamics simulation study on the mechanism of loading of gemcitabine and camptothecin in poly lactic-co-glycolic acid as a nano drug delivery system. J Mol Liq. 2018;269:110–118. [Google Scholar]
- 14.Moradi S., Hosseini E., Abdoli M., Khani S., Shahlaei M. Comparative molecular dynamic simulation study on the use of chitosan for temperature stabilization of interferon αII. Carbohydr Polym. 2019;203:52–59. doi: 10.1016/j.carbpol.2018.09.032. [DOI] [PubMed] [Google Scholar]
- 15.Moradi S., Khani S., Ansari M., Shahlaei M. Atomistic details on the mechanism of organophosphates resistance in insects: insights from homology modeling, docking and molecular dynamic simulation. J Mol Liq. 2019;276:59–66. [Google Scholar]
- 16.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248 doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan S., Chan J.F., Chik K.K., Chan C.C., Tsang J.O., Liang R., Cao J., Tang K., Chen L.-L., Wen K. Discovery of the FDA-approved drugs bexarotene, cetilistat, diiodohydroxyquinoline, and abiraterone as potential COVID-19 treatments with a robust two-tier screening system. Pharmacol Res. 2020;159 doi: 10.1016/j.phrs.2020.104960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh S., Srivastava H.K., Kishor G., Singh H., Agrawal P., Raghava G.P. BioRxiv; 2017. Evaluation of protein-ligand docking methods on peptide-ligand complexes for docking small ligands to peptides. [Google Scholar]
- 22.Ea R. Life Sci; 2020. Remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase RdRp): a molecular docking study) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berendsen H.J., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91:43–56. [Google Scholar]
- 24.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software. 2015;1:19–25. [Google Scholar]
- 25.Bjelkmar P., Larsson P., Cuendet M.A., Hess B., Lindahl E. Implementation of the CHARMM force field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J Chem Theor Comput. 2010;6:459–466. doi: 10.1021/ct900549r. [DOI] [PubMed] [Google Scholar]
- 26.Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I. CHARMM general force field: a force field for drug‐like molecules compatible with the CHARMM all‐atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem. 2011;32:2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 28.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. Chem Phys. 1983;79:926–935. [Google Scholar]
- 29.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅ log (N) method for Ewald sums in large systems. Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 30.Aftab S.O., Ghouri M.Z., Masood M.U., Haider Z., Khan Z., Ahmad A., Munawar N. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J Transl Med. 2020;18:1–15. doi: 10.1186/s12967-020-02439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elfiky A.A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silico perspective. J Biomol Struct Dyn. 2021;39:3204–3212. doi: 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shahabadi N., Zendehcheshm S., Mahdavi M., Khademi F. Inhibitory activity of FDA-approved drugs cetilistat, abiraterone, diiodohydroxyquinoline, bexarotene, remdesivir, and hydroxychloroquine on COVID-19 main protease and human ACE2 receptor: a comparative in silico approach. Inform Med Unlocked. 2021;26 doi: 10.1016/j.imu.2021.100745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebrahimi K.S., Ansari M., Moghaddam M.S.H., Ebrahimi Z., Shahlaei M., Moradi S. In silico investigation on the inhibitory effect of fungal secondary metabolites on RNA dependent RNA polymerase of SARS-CoV-II: a docking and molecular dynamic simulation study. Comput Biol Med. 2021;135 doi: 10.1016/j.compbiomed.2021.104613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh R., Bhardwaj V.K., Purohit R. Potential of turmeric-derived compounds against RNA‐dependent RNA polymerase of SARS‐CoV‐2: an in-silico approach. Comput Biol Med. 2021;139 doi: 10.1016/j.compbiomed.2021.104965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parvez M.S.A., Karim M.A., Hasan M., Jaman J., Karim Z., Tahsin T., Hasan M.N., Hosen M.J. Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach. Int J Biol Macromol. 2020;163:1787–1797. doi: 10.1016/j.ijbiomac.2020.09.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Indu P., Rameshkumar M.R., Arunagirinathan N., Al-Dhabi N.A., Arasu M.V., Ignacimuthu S., Raltegravir, Indinavir Tipranavir. Dolutegravir, and Etravirine against main protease and RNA-dependent RNA polymerase of SARS-CoV-2: a molecular docking and drug repurposing approach. J Infect Public Health. 2020;13:1856–1861. doi: 10.1016/j.jiph.2020.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furuta Y., Gowen B.B., Takahashi K., Shiraki K., Smee D.F., Barnard D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryson A., De La Motte S., Dunk C. Reduction of dietary fat absorption by the novel gastrointestinal lipase inhibitor cetilistat in healthy volunteers. Br J Clin Pharmacol. 2009;67:309–315. doi: 10.1111/j.1365-2125.2008.03311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rickards A. The treatment of amoebiasis with" Diodoquin. Am J Trop Med. 1949;52:33–38. [PubMed] [Google Scholar]
- 41.Padwal R. Cetilistat, a new lipase inhibitor for the treatment of obesity. Curr Opin Invest Drugs. 2008;9:414–421. London, England: 2000. [PubMed] [Google Scholar]
- 42.Cheung K.S., Hung I.F., Chan P.P., Lung K., Tso E., Liu R., Ng Y., Chu M.Y., Chung T.W., Tam A.R. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan J.F.-W., Zhang A.J., Yuan S., Poon V.K.-M., Chan C.C.-S., Lee A.C.-Y., Chan W.-M., Fan Z., Tsoi H.-W., Wen L. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Arch Clin Infect Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hung I., Cheng V., Wu A., Tang B., Chan K., Chu C., Wong M., Hui W., Poon L., Tse D. Viral loads in clinical specimens and SARS manifestations. Emerg Infect Dis. 2004;10:1550. doi: 10.3201/eid1009.040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boehm M.F., Zhang L., Zhi L., McClurg M.R., Berger E., Wagoner M., Mais D.E., Suto C.M., Davies P.J., Heyman R.A. Design and synthesis of potent retinoid X receptor selective ligands that induce apoptosis in leukemia cells. J Med Chem. 1995;38:3146–3155. doi: 10.1021/jm00016a018. [DOI] [PubMed] [Google Scholar]
- 47.Gniadecki R., Assaf C., Bagot M., Dummer R., Duvic M., Knobler R., Ranki A., Schwandt P., Whittaker S. The optimal use of bexarotene in cutaneous T‐cell lymphoma. Br J Dermatol. 2007;157:433–440. doi: 10.1111/j.1365-2133.2007.07975.x. [DOI] [PubMed] [Google Scholar]
- 48.Dragnev K.H., Petty W.J., Shah S.J., Lewis L.D., Black C.C., Memoli V., Nugent W.C., Hermann T., Negro-Vilar A., Rigas J.R. A proof-of-principle clinical trial of bexarotene in patients with non–small cell lung cancer. Clin Cancer Res. 2007;13:1794–1800. doi: 10.1158/1078-0432.CCR-06-1836. [DOI] [PubMed] [Google Scholar]
- 49.Esteva F.J., Glaspy J., Baidas S., Laufman L., Hutchins L., Dickler M., Tripathy D., Cohen R., DeMichele A., Yocum R.C. Multicenter phase II study of oral bexarotene for patients with metastatic breast cancer. Clin Oncol. 2003;21:999–1006. doi: 10.1200/JCO.2003.05.068. [DOI] [PubMed] [Google Scholar]
- 50.Duc I., Bonnet P., Duranti V., Cardinali S., Rivière A., De Giovanni A., Shields-Botella J., Barcelo G., Adje N., Carniato D. In vitro and in vivo models for the evaluation of potent inhibitors of male rat 17α-hydroxylase/C17, 20-lyase. J Steroid Biochem Mol Biol. 2003;84:537–542. doi: 10.1016/s0960-0760(03)00078-5. [DOI] [PubMed] [Google Scholar]
- 51.Arasaratnam M., Crumbaker M., Bhatnagar A., McKay M.J., Molloy M.P., Gurney H. Inter-and intra-patient variability in pharmacokinetics of abiraterone acetate in metastatic prostate cancer. Cancer Chemother Pharmacol. 2019;84:139–146. doi: 10.1007/s00280-019-03862-x. [DOI] [PubMed] [Google Scholar]
- 52.Yuan S., Chu H., Chan J.F.-W., Ye Z.-W., Wen L., Yan B., Lai P.-M., Tee K.-M., Huang J., Chen D. SREBP-dependent lipidomic reprogramming as a broad-spectrum antiviral target. Nat Commun. 2019;10:1–15. doi: 10.1038/s41467-018-08015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kar B., Dehury B., Singh M.K., Pati S., Bhattacharya D. Identification of phytocompounds as newer antiviral drugs against COVID-19 through molecular docking and simulation based study. J Mol Graph Model. 2022;114 doi: 10.1016/j.jmgm.2022.108192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erva R.R., Rajulapati S.B., Potla Durthi C., Bhatia M., Pola M. Molecular dynamic simulations of Escherichia coli L-asparaginase to illuminate its role in deamination of asparagine and glutamine residues. 3 Biotech. 2016;6:1–7. doi: 10.1007/s13205-015-0339-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parihar A., Sonia Z.F., Akter F., Ali M.A., Hakim F.T., Hossain M.S. Phytochemicals-based targeting RdRp and main protease of SARS-CoV-2 using docking and steered molecular dynamic simulation: a promising therapeutic approach for Tackling COVID-19, Comput. Biol Med. 2022;145 doi: 10.1016/j.compbiomed.2022.105468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar D., Kumari K., Vishvakarma V.K., Jayaraj A., Kumar D., Ramappa V.K., Patel R., Kumar V., Dass S.K., Chandra R. Promising inhibitors of main protease of novel corona virus to prevent the spread of COVID-19 using docking and molecular dynamics simulation. J Biomol Struct Dyn. 2021;39:4671–4685. doi: 10.1080/07391102.2020.1779131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Priya R., Sumitha R., Doss C.G.P., Rajasekaran C., Babu S., Seenivasan R., Siva R. Molecular docking and molecular dynamics to identify a novel human immunodeficiency virus inhibitor from alkaloids of Toddalia asiatica. Phcog Mag. 2015;11:S414. doi: 10.4103/0973-1296.168947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prasanth D., Murahari M., Chandramohan V., Panda S.P., Atmakuri L.R., Guntupalli C. In silico identification of potential inhibitors from Cinnamon against main protease and spike glycoprotein of SARS CoV-2. J Biomol Struct Dyn. 2021;39:4618–4632. doi: 10.1080/07391102.2020.1779129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Basit A., Ali T., Rehman S.U. Truncated human angiotensin converting enzyme 2; a potential inhibitor of SARS-CoV-2 spike glycoprotein and potent COVID-19 therapeutic agent. J Biomol Struct Dyn. 2021;39:3605–3614. doi: 10.1080/07391102.2020.1768150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn. 2021;39:3449–3458. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]