Summary
Gene therapy is emerging as a modality in 21st-century medicine. Adeno-associated viral (AAV) gene transfer is a leading technology to achieve efficient and durable expression of a therapeutic transgene. However, the structural complexity of the capsid has constrained efforts to engineer the particle toward improved clinical safety and efficacy. Here, we generate a curated library of barcoded AAVs with mutations across a variety of functionally relevant motifs. We then screen this library in vitro and in vivo in mice and nonhuman primates, enabling a broad, multiparametric assessment of every vector within the library. Among the results, we note a single residue that modulates liver transduction across all interrogated models while preserving transduction in heart and skeletal muscles. Moreover, we find that this mutation can be grafted into AAV9 and leads to profound liver detargeting while retaining muscle transduction—a finding potentially relevant to preventing hepatoxicities seen in clinical studies.
Keywords: adeno-associated virus, gene therapy, capsid, liver toggle, Anc80, hepatotoxicity, AAV9, library, ancestral sequence reconstruction, ASR, hepatotropism
Graphical abstract

Highlights
-
•
Combinatorial synthesis and in vitro and in vivo evaluation of barcoded AAV library
-
•
Multiparametric pharmacokinetic profiling reveals a liver toggle substitution
-
•
Liver toggle decreases liver tropism >500-fold (DNA & RNA) in mice and NHPs
-
•
Liver toggle can be functionally transferred to reprogram other AAV serotypes
Zinn et al. report on experimental approaches to respectively generate and screen libraries of DNA/RNA-barcoded AAV capsids with defined combinatorial diversity. Through these approaches, the authors uncover a substitution capable of detargeting AAV capsids from the liver in mice and nonhuman primates.
Introduction
Adeno-associated viruses (AAVs) are dependoparvoviruses that can be rendered replication defective with utility as vectors in therapeutic in vivo gene transfer. In vivo studies demonstrate broad tissue and cell tropism and the potential of long-term transduction. Dozens of translational studies based on recombinant AAV technology have shown efficacy in preclinical models of hematologic, metabolic, neurosensory, neuromuscular, and neurological disease.1,2,3 Today, clinical studies are ongoing for over 30 of these indications, and 2 AAV gene therapy drugs have been approved by the Food and Drug Administration.2,3,4
Nevertheless, recent clinical studies illustrate potential limitations to the safety and efficacy of currently used AAV technologies. For example, hepatotoxicity following systemic administration of AAV has been reported in several studies5,6,7,8 with adverse events ranging from moderate transaminase elevations to more severe liver toxicity, seemingly in a dose-dependent manner. One high-dose AAV8 X-linked myotubular myopathy study recently reported the death of three subjects due to progressive liver dysfunction following vector administration.8 The etiology of these toxicity events remains largely unresolved, and in the latter case may be related to an underlying liver disease. Still, the dose dependency and the central role of the liver in the toxicity events has led the AAV field to pursue approaches to either reduce dose requirements and/or minimize exposure to the liver.9,10,11
A primary approach to achieve more selective targeting of AAV gene therapies is the use of alternative AAV capsids that can be used to package an identical vector genome encoding the therapeutic cassette. Indeed, key observations with naturally occurring serotypes of AAV demonstrate that capsid structure can influence liver tropism by as much as 100-fold in mouse models.12,13,14 Furthermore, even minimal variations in capsid structure can alter specificity to attachment factors, vector production yields, biodistribution, cellular tropism, and host-response—all properties of direct relevance to clinical translation.15,16,17,18 This kaleidoscopic array of phenotypic variation within a relatively narrow window of structural variation has led to an expansive field that seeks to discover and engineer AAV capsid variants with improved (combinations of) features of clinical or experimental utility.
The T = 1 icosahedral AAV capsid is composed of three structural, C-coterminal viral proteins VP1, 2, and 3, of which VP3 serves as the main structural subunit within the 60-unit capsomer. The core subunit structure is an eight-stranded β barrel conserved across primate AAVs, while nine surface exposed variable regions (denoted VRI-VRIX) have been observed to mediate the functions of particle assembly, infectivity, and transduction specificity among serotypes. Each monomer interfaces with seven neighboring subunits through intricate protein-protein interactions defined by the 2-, 3-, and 5-fold symmetry of the particle structure.19,20
The highly interdependent nature of the AAV capsid has complicated efforts to understand and engineer new phenotypes. Nevertheless, a number of important structure-function studies have identified important regions of various AAV capsids. Identification of the heparin sulfate proteoglycan binding domain of AAV221,22 enabled researchers to study how mutating this domain altered tropism of AAV vectors.16,23 Other structure-function studies have enabled researchers to re-engineer AAV by structurally transferring a galactose binding domain between serotypes,24 engineering capsids that assemble without the aid of a viral cofactor,25 and (dis)engagement of a GPR108, a cognate receptor to most primate AAVs.26
Many studies have also explored mutational or peptide insertion strategies to generate diverse libraries of capsid variants. Functional enrichment is then sought by iterative selective assays in what is referred to as biopanning or directed evolution. These powerful approaches have yielded compelling capsids with unique phenotypes (e.g., AAV7m8, PHP.B, AAV-DJ, and LK-03, among others).27,28,29,30 Unfortunately, the promises of these vectors are not always translated across species,31,32,33,34 a bottleneck the field is trying to address with screening approaches in models thought to be more predictive of the human setting.29,35,36
To address some of the limitations of the current state-of-the-art in AAV development, we performed a gap analysis and developed a set of methods to overcome critical potential shortcomings of AAV library diversification and screening.
First, we sought to build and improve on the power of capsid libraries to identify AAV vectors with desirable biology. Specifically, we aimed to simultaneously maximize library complexity, sequence divergence, and, importantly, functional viability (e.g., capsid assembly competency and infectivity). Previous approaches have yielded highly informative screens by focusing variation within functionally important and mutationally permissive capsid regions based on sequence analysis and structural data.37,38,39 In our approach, we performed maximum-likelihood ancestral sequence reconstruction (ML-ASR) to identify regions of the capsid with evolutionary relevance that served as the basis to curate a fully defined variant library.40 However, in this original study, we were only able to partially characterize this library (776 different clones were evaluated out of a potential of 2,048), and the clones were only screened through a combined vector production/in vitro infectivity approach.
To address the limitations of our previous study, we developed an approach to assemble our library such that each position of variation would be present in combination with every other variant position, using an approach akin to that of CombiGEM,41,42 which we dubbed CombiAAV (Figure S1). Encouraged by the success of other barcoded screens in AAV,43,44,45 and because our positions of variation were distributed throughout the length of the capsid, we also designed the library such that a non-coding “barcode” region would be present as both DNA and RNA. As established by the work of other groups,46,47,48,49 this strategy of coupling our capsid sequences with unique short regions of DNA enabled us to discriminate between and count each barcode with short-read next-generation sequencing (NGS) technologies. Whereas previous approaches have relied on either serial assembly of capsids and barcodes or have confined variation to specific functional domains of the capsid (as in peptide insertion libraries), we designed our approach to paralleled assembly of capsid libraries with combinatorial diversity distributed throughout the capsid coding sequence coupled to individual, unique barcodes.
Finally, we developed an approach to quantitatively measure the abundance of individual capsid variants by using short-read NGS, a technique we call AAVSeq. AAVSeq allows for the functional evaluation of a library across more than one parameter within—or across—assays or models (e.g., parallel screening of multiple tissues in more than one species). This was meant to (1) obtain information on functional trade-offs (e.g. production yield versus liver targeting), (2) interrogate the conservation of a desirable property across models, and eventually in human studies, (3) enable the detection of both loss-of-function and gain-of-function phenotypes (e.g. for targeting specificity), and (4) aggregate larger structure-function datasets with each added screen. We hypothesized that our approach would allow for refined identification of candidate vectors, and also that the datasets produced would inform models, highlight potential mechanism of action, and guide rational design.
Here, the combined use of AAVSeq on a CombiAAV generated library of barcoded AAV capsids within a curated ML-ASR variant library allowed us to identify a conserved structural determinant of liver tropism present in several clinical AAVs. This “liver toggle” motif was further shown to allow functional reprogramming to either enhance or reduce liver targeting while retaining muscle transduction following systemic injection.
Results
Identification of residues potentially involved in liver tropism by ASR
Therapeutic gene transfer of hepatic tissue has been studied extensively to treat inborn errors of metabolism, coagulation disorders, or to provide long-term expression of a therapeutic protein. To this end, AAV has been evaluated in various animal models and clinical studies. From the aggregate of data, we and others have observed that distinct AAV serotypes target the liver with vastly different efficiencies, notwithstanding a relatively high degree of sequence homology.50,51,52 Based on the observation that vectors, such as AAV1 and AAV2, performed substantially less well compared with AAV7, 8, or 9, we hypothesized that a common feature within these classes of AAV capsids was responsible for enhanced liver transduction. These two functionally distinct groups of capsids are monophyletic and likely shared a common ancestor, which we have previously attempted to reconstruct and refer to as Anc80 (Figure 1A). Based on this apparent co-segregation of sequence and function, we further hypothesized that either the enhanced liver tropism of the AAV8 group was acquired from Anc80, or, inversely, that the AAV2 group species had attenuated liver targeting from their ancestral state.
Figure 1.
Approach and substrate
(A) Dendrogram depicting the evolutionary monophyletic origins of AAV vectors with high (orange) or low (blue) hepatotropism in mice. Anc80Lib refers to an AAV variant capsid library derived from ML-ASR. The percent identity to Anc80 (protein) is provided for each extant AAV.
(B) Above, bar graph depicting locations of amino acid differences in the coding sequence of the capsid gene between Anc80 and AAV2. Below, schematic representation of the coding sequence of the resultant barcoded library. The gray rectangle represents the coding sequence of the capsid and the smaller rectangles (either orange or blue) depict the different states at positions of variability within the library (each a single codon).
(C) Circos diagram highlighting structural features of the AAV capsid. The outer circle of the figure is divided into rectangles, each corresponding to an amino acid in the structure of AAV9 (PDB: 3UX1) and are numbered (VP1 numbering). Positions are shaded by the number of predicted quaternary interactions (darker meaning more interactions). Lines drawn between positions on the outer circle indicate predicted interactions. The next set of highlights (purple) correspond to amino acids determined to bind different monoclonal antibodies. The blue highlights identify amino acids involved in glycan binding. The green highlights identify amino acids involved in binding to AAVR. Glyphs identify interactions as being between the 2-fold (green circles), 3-fold (orange triangles), or 5-fold interfaces (blue stars). The inner radius depicts a histogram plotting sequence conservation (Shannon entropy), where green bars indicate high sequence diversity (entropy > 2.0), red bars indicate high conservation (entropy < 1.0), and orange bars indicate moderate diversity (entropy > 1.5). Blue wedges indicate positions of variability within the Anc80 library and are numbered (Anc80VP1/AAV9 VP1). Table S1 provides detailed information on each of the highlighted positions. Note that p1 and p2 (corresponding to positions 168 and 205 by Anc80 VP1 numbering) are not shown due to the lack of structures in the VP1/VP2 unique regions.
Previously, we performed ML-ASR to approximate Anc80 sequences.40 In this work, we developed a library to approximate the sequence of this putative ancestral capsid by identifying 11 amino acids within the capsid, which were determined to be ambiguously ancestral (using a posterior probability of 0.3 as a threshold). Each of these 11 sites has 2 potential states, and these dimorphic variant sites are scattered throughout the capsid sequence (Figure 1B). Previous work had selected and extensively characterized only one member of this library, called Anc80L65, for liver transduction in mice and nonhuman primates (NHPs) and various other target tissues.53,54,55,56 An overlay of the Anc80Lib variable positions in Figure 1D indicated the variation to be distributed in regions of the capsid known to affect capsid assembly, receptor binding, and host-mediated immunity. We thus hypothesized that extensive exploration of the diversity within Anc80Lib would allow us to map discreet or epistatic structural motifs relevant to AAV liver tropism.
Structural exploration of an ASR-informed library
Early alanine scanning and mutational studies highlighted the limited tolerance of the AAV capsid to amino acid substitutions.57,58 More recently, Ogden et al. comprehensively explored the fitness landscape of the AAV2 capsid and reported that between 86% and 97% of mutants were impaired in virion assembly. We hypothesized that the quaternary structural interactions of the icosahedral 60-mer virion reduces its mutational tolerance. In comparison, previous studies on monomeric proteins, such as the bacterial enzyme DNA glycosylase, indicate that 66% of random amino acid chances are tolerated.59 An analysis of the full-particle AAV8 X-ray crystal structure in Figure 1C mapped the intermolecular interactions between VP monomers and identified 36 pairs of amino acids across the 2-fold axis of symmetry, 257 pairs across the 3-fold axis, and 123 pairs across the 5-fold axis. These large numbers of interacting residues support the empirical observations that AAV is highly resistant to mutation, and that even minimal mutations can have large (and often deleterious) effects.15
Design and construction of a barcoded ancestral AAV library
Anc80Lib is a combinatorial sequence space composed of the Anc80 capsid with 11 variant positions that can be occupied by 1 of 2 defined amino acid residues (Table S2). To quantitatively assess each member of the library under various selective conditions and to avoid costly clonal production and characterization, we sought to introduce a barcode identifier that can be sequenced via short-read NGS technologies, such as Illumina.
To assemble all 2,048 defined Anc80Lib capsid variants with a unique barcode, we designed a cloning strategy wherein the library was progressively assembled using rounds of scarless ligation. Figure S1 illustrates the assembly process that, in a stepwise manner, increases library complexity exponentially. To accomplish this, we first designed, synthesized, and pooled synthetic oligonucleotide building blocks in equal ratios, each containing a fragment of the capsid coding sequence on the 3′ end, and a corresponding stretch of nucleotides representing the site of variation (i.e., a fragment of the eventual barcode) flanked by type IIS restriction enzyme recognition sites. Once digested with the type IIS restriction enzyme (either Esp3I or BsaI in alternating rounds of assembly), the resulting fragment is inserted by ligation at the junction of the capsid and barcode coding region of an intermediate plasmid library from a previous round of assembly. At each step, diversity of the library is multiplied by the number of unique sequence-barcode fragments included. In this instance, the assembly process started with a pool of two fragments, then incorporated an additional four fragments in each of the subsequent five rounds of assembly (2 × 45 = 2,048). This process is analogous to the assembly process of CombiGEM41,42 and so we refer to this approach as CombiAAV.
To ascertain the effectiveness of the CombiAAV, we isolated barcodes by polymerase chain reaction (PCR) and subjected them to Illumina NGS to gauge the relative abundance of each mutant within the library. All 2,048 members of Anc80Lib were represented within the sequenced pool (Figure S2). In addition, technical replicates of the isolation and sequencing showed a high degree of reproducibility (Spearman’s rho, ρ = 0.97–99, N = 8), indicating that the process of amplifying and preparing barcodes for sequencing introduced minimal additional noise (Figure S2). Anc80Lib was then subcloned into an ITR-flanked AAV packaging construct expressing Anc80Lib barcode and capsid gene transcripts driven by a CMV promoter (Figure S3).
Vector production screen confirms AAP-interacting capsid residue
Next, an Anc80Lib viral library (referred to as Anc80Lib-BC) was produced by co-transfection of the ITR-Anc80Lib pooled plasmid library under conditions we previously optimized to minimize the occurrence of cross-packaging and/or capsid mosaic formation.60 Since variant position 5 (p5) in Anc80Lib at location 411 (by VP1 numbering) was previously implicated as interacting with the assembly-activating protein (AAP),25 we compared vector production of our library in the presence or absence of AAP from AAV2 (AAP2) in trans (Figure 2A). AAVSeq was performed on the resulting vector preparations (N = 3 with AAP and N = 7 without), and relative enrichment for each library variant was established by normalizing with AAVSeq data from the transfected plasmid pool. Normalized counts were highly correlated within each condition (ρ = 0.93–0.99 without AAP, ρ = 0.90–0.99 with AAP) and all 2048 vector barcodes were detected (Figure S4). In the absence of exogenous AAP2, we observe a 2.87-fold difference in relative enrichment of mutants containing a glutamine at position 5 (p5Q) over those containing a glutamic acid (p5E). In the presence of exogenous AAP, this effect was reduced, with p5Q vectors only being very mildly enriched over p5E vectors (1.17-fold), indicating a role for AAP at this position (Figure 2A). An elastic net regularization approach revealed the role of each site of mutation in our library as any changes of those effects in the presence or absence of exogenous AAP2 (Figure 2B). Because inclusion of exogenous AAP increases production yields (average titer with AAP2: 8.73 × 1012 gc/mL, without AAP2: 5.68 × 1011 gc/mL), AAP2 was included in all further library preparations.
Figure 2.
Library production and in vitro infectivity
(A) A barcoded AAV vector library was produced either with (N = 5) or without (N = 3) addition of an exogenous assembly-activating protein (AAP). Individual dots in MA plots represent distinct barcodes, colors represent the amino acid identity at position 5. To the right of MA plots, eCDF plots with SEM depicted as horizontal error bars.
(B) Results of an independent sites linear elastic net regularization approach show that addition of AAP modifies the impact of certain sites on production of vector.
(C) Huh7 cells were transduced with the barcoded AAV library and DNA/RNA were isolated from those transduced cells (N = 5 per condition). Individual barcodes in MA plots are colored by identity at position 3 within the library. Extant barcoded vectors were also spiked into this transduction mixture. Fold change is plotted as bar graphs (error bars determined by bootstrapping, 1,000 replicates).
(D) Regularization and linear modeling approach reveal potential similarities and differences among our positions of variation with respect to transduction and gene expression in vitro.
Multiplexed in vitro transduction analysis
Then, we evaluated our library of capsids for their ability to transduce the human hepatoma-derived cell line Huh7. To complement the experiment with references, previously characterized AAVs (AAV2, 5, 8, 9, 9-PHP.B, Rh.10, and Rh32.33) were produced in a barcoded manner as in Anc80Lib for compatibility in the NGS readout, and then spiked into Anc80Lib-BC at a ratio of approximately 1:2,000 each (relative to the final library titer). This combined pool of AAVs was then used to transduce Huh7 cells at an average multiplicity of infection (MOI) of 100 vector genomes per cell. Because there were 2,056 individually barcoded vectors within the library, the gross MOI was 2E5 gc/cell. Cells were washed to remove any vector that did not transduce cells, and 3 days after transduction, DNA and RNA were harvested, and barcodes were subsequently sequenced. In this case, individual variant read counts were normalized based on the AAVSeq data on the input viral vector preparation to assess relative enrichment upon in vitro transduction (and not, for instance, viral assembly or packaging). Experiments were performed in five replicates each, and normalized barcode abundances were highly correlated among replicates (ρ = 0.93–0.96 for RNA barcodes, = 0.91–0.92 for DNA barcodes) (Figure S5). Among library variants, we found a large effect at position 3 within our library, wherein the fold-enrichment of p3G capsids was 22.9-fold greater on average than p3A capsids in DNA, and 52.7-fold greater in RNA (Figure 2C). Our included controls were consistent with the literature, where AAV2 and related viruses are known to out-perform many other vectors by large amounts (in RNA) across many cell lines.50 An elastic net regularization approach identified potentially important effects at p6 and p9 within the library, especially in RNA (Figure 2D).
Multiparametric tissue screen in mice and NHPs
We then proceeded to test our approach in vivo by injecting mice (N = 8 per condition) or NHPs (N = 2 per condition) with Anc80Lib-BC spiked with additional separately produced barcoded capsids of commonly used AAVs as reference vectors (doses of 3 × 1011 gc/mouse or 1.6 × 1012 gc/kg in the case of NHPs) (Figure S6). Tissues were collected at an early time point (day 3 for mice, and day 7 for NHPs) or a later time point (day 28) and barcodes were subsequently isolated for AAVSeq analysis. When comparing only the spiked-in reference AAVs, our approach recapitulates many of the findings from other comparative studies of AAV in vivo (Figures S7A and S7B). For instance, at the level of DNA (day 28) in mice, AAV2 and AAV8 appear roughly equivalent in terms of relative enrichment in liver while, at the level of RNA, AAV8 is greatly enriched compared with AAV2. Previous comparative studies of these two vectors have also made similar observations, and have concluded that differential uncoating may be responsible.51 In addition, we observe that AAV9-PHP.B vastly outperforms all other vectors in the mouse hippocampus, consistent with its characterization as a vector especially adept at crossing the blood-brain barrier in C57BL/6 mice.30
Liver toggle effect observed in mice and NHPs
AAVSeq of Anc80Lib in liver samples from both mice and NHPs was highly reproducible from animal to animal (Figures 3A and 3D). Moreover, in both animals, MA plots revealed the emergence of two distinct subpopulations of vectors; one enriched in liver (compared with the injected library), and one reduced in abundance. Further analysis indicated a clear role of Anc80Lib variable position p3 on both liver gene transfer and transduction in both mice and NHPs (Figures 3B and 3E). In mice, AAV capsids within the library with a glycine residue in p3 (p3G) were on average approximately 13.5 times more enriched than those of p3A capsids in DNA, and more than 100-fold greater than p3A in RNA (Figure 3B). In NHPs, a qualitatively similar effect was observed at a modestly reduced difference (∼3.5-fold in DNA and ∼11.9-fold in RNA), although there was substantially more noise in NHPs than there was in mice (Figure 3E), perhaps in part due to the lower number of biological replicates (N = 2 NHPs, N = 8 mice).
Figure 3.
Variation within the Anc80Lib at a single position alters liver tropism of vectors in mice and NHPs
(A) Pairwise comparisons of barcode isolation and counting from livers of three mice at day 3 (N = 8) as DNA (top) or RNA (bottom).
(B) MA plots depicting the fold change of vectors (tissue/vector) colored by amino acid identity at position 3 within our library.
(C) Vectors were sorted by fold-change (top to bottom, positive to negative) and colored by their states at every position (in “fingerprint plots”).
(D) Following administration into NHPs, barcodes are isolated from DNA (top) and RNA (bottom) at day 28 with varying reproducibility (Spearman’s ρ = 0.94 and 0.73 respectively, N = 2 NHPs).
(E) MA plots from NHP data depicting differences among vectors with A/G at position 3.
(F) Fingerprint plots from NHP liver DNA and RNA at day 28.
To further visualize the role of individual variant positions within our library, we sorted the barcodes in each condition by relative enrichment (from most enriched to least from top to bottom) and represented the dimorphic amino acid states as an orange or blue bar (Figures 3C and 3F). This analysis illustrates the dominant effect of p3 on liver targeting within this library across both mice and NHPs. Beyond p3, other effects are noted, such as p1 and p11, which appear to also alter liver barcode enrichment. Combined, these types of analyses may provide signatures of capsid motifs that favor a particular tissue targeting profile.
Systems pharmacokinetics of Anc80Lib in mice versus NHPs
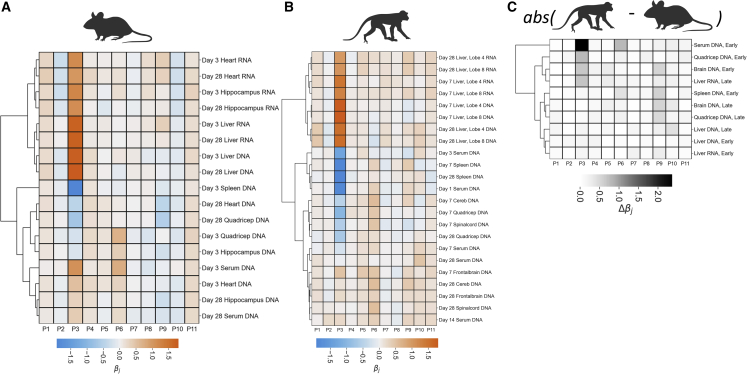
Having observed the role of p3 in the liver of both mice and NHPs, we analyzed the biodistribution of variants in Anc80Lib from liver, spleen, serum, and skeletal muscle (quadriceps). We observed that, although p3G viruses were enriched in livers of both mice and NHPs at this early time point, they were substantially de-enriched in spleens, and (most notably in NHPs) also de-enriched in skeletal muscle, albeit to a lesser degree (Figure 4). In mice p3G capsids persisted in serum longer than p3A capsids; however, in NHPs the opposite was found. Through elastic net regularization and clustering, we find a role for P3A/G across many time points, tissues, and nucleotide types in both mice and NHPs, although not always consistent between the two species (Figures 5A and 5B).
Figure 4.
Pharmacokinetic profiling of barcoded viruses
Barcodes were purified from the DNA of liver, spleen, quadriceps, and serum from either mice (top row, N = 8) or NHPs (bottom row, N = 2) at an early time point (day 3 for mice, day 7 for NHPs, except for serum which is day 3). To illustrate the relationships between these vectors, MA plots were rendered each highlighting the previously identified “liver” toggle and depicted next to a fingerprint plot for each tissue.
Figure 5.
Summary of in vivo tissue tropism studies
(A) Average log2 fold-changes were computed for every RNA/DNA/tissue combination at every time point from the study detailed in Figure S6 (mice N = 8, NHPs N = 2). Data were centered by means subtraction and an independent sites linear model was fit by elastic net regularization to determine the impact of variation at each site within the library on enrichment in each sample. Samples were then clustered by Ward’s method according to the fit coefficients.
(B) The same approach as described in (A) was applied to samples isolated from NHPs.
(C) To compare between species, regularization coefficients were subtracted (cynomolgus macaque – mouse) from one another. The absolute value of the differences were clustered and plotted as in (A and B).
Comparing the role of mutations across time, tissues, and species
Finally, we explored our multiparametric screen through a simple regularization approach. Clustering by regularization coefficients allows us to visualize the roles of individual amino acids within our library across tissues, time points, RNA/DNA, and species. Within the mouse samples that we explored, RNA samples neatly segregated from DNA samples (Figure 5A), and samples from related tissues also clustered together. Similar qualitative patterns emerged in the NHP samples (Figure 5B). To explore the degree to which patterns seen in mouse samples are also found in NHP samples, we plotted the absolute value of the difference in regression coefficients for time point- and tissue-matched samples (Figure 5C). In this approach, it becomes clear that there is a large disparity in the role of positions 3 and 6 within our library in determining the persistence in the serum of these animals, and a possible weaker role of position 9 across several tissues. These differences may be of interest to the field for future study. Importantly, across liver DNA and RNA, we observed minimal differences between mice and NHPs; however, there remain some notable apparent qualitative differences in samples from quadriceps.
Validation of AAVSeq findings
To validate the primary finding on liver tropism and biodistribution, we clonally produced two different capsids from our library, Anc80L1533 (p3G, the mutation associated with the “liver-on” state) and Anc80L1093 (p3A, “liver-off”), each containing an eGFP transgene. Preparations of individual vectors were administered intravenously to five mice each. Relative transduction across tissues is represented in Figure 6. We observed that Anc80L1533 shows an advantage over Anc80L1093 in the mouse liver at the level of both DNA and RNA (2,622-fold in DNA, p < 0.001 Tukey-HSD, 512-fold in RNA, p < 0.001 Tukey-HSD) (Figure 6A). Also consistent with our findings, we observed that Anc80L1533 only shows a small (but significant) advantage over the liver-detargeted Anc80L1093 in mouse quadriceps at the level of gene transfer (2.58-fold, p < 0.001 Tukey-HSD) and expression (2.93-fold, p = 0.048 Tukey-HSD). Within the mouse heart, we observe that Anc80L1093 appears slightly better at gene transfer compared with Anc80L1533 (3.77-fold, p < 0.001 Tukey-HSD); however, expression shows a small but significant disadvantage (−2.99-fold, p < 0.001 Tukey-HSD) (Figure 6A). Taken as a whole, the data confirm our initial findings by AAVSeq of Anc80Lib—that p3A mutants within our library are strongly and significantly detargeted from the livers of mice (in excess of 500-fold) while only modestly decreasing expression in skeletal and cardiac muscle (under 3-fold).
Figure 6.
Clonal confirmation of p3 liver toggle and reprogramming of AAV9 with respect to tissue tropism
(A) Anc80L1533 (p3 = G, Liver On) and Anc80L0193 (p3 = A, Liver Off) were identified from Anc80Lib AAVSeq based on their performance, individually produced with a GFP reporter transgene, and vectors were administered intravenously into five mice per clonal vector. Relative transgene (eGFP) DNA (top) and RNA (bottom) were determined by ddPCR and qRT-PCR, respectively, and represented in box-and-whisker plots for liver (left), quadriceps (middle), and heart (right). After testing for normality and equal variances, significance was determined by an ANOVA followed by a post-hoc test (Tukey’s HSD) (n.s., 0.05 < p < 1, ∗0.01 < p < 0.05, ∗∗0.001 < p < 0.01, ∗∗∗0.0001 < p < 0.001).
(B) Mutations were introduced into AAV9 to confer the observed liver-detargeted phenotype observed in the library and confirmed clonally as in (A). Transgene expression was measured by qRT-PCR, represented, and analyzed statistically as in (A).
Liver toggle reprogramming of clinically relevant AAVs
An alignment of the main AAV serotypes within the Anc80 lineage of hypervariable region 1 of AAV illustrates the conserved state of a glycine within the AAV8 group, and an alanine within the AAV2 group (Table S3). This conservation is in line with the phylogenetic relationship of the liver tropism phenotype (Figure 1A) and suggests the possibility of functionally reprogramming other AAV serotypes with respect to their liver tropism by mutating the liver toggle from glycine to alanine (or vice versa). However, owing to the significant amount of structural variability in VRI of the AAV capsid, the exact homology is somewhat unclear, complicating efforts to determine whether this mutation may also impact non-Anc80 capsids in a similar fashion. Consequently, using our alignment to inform this hypothesis, we aimed to graft this mutation into AAV9, thereby potentially detargeting resultant vectors from the liver.
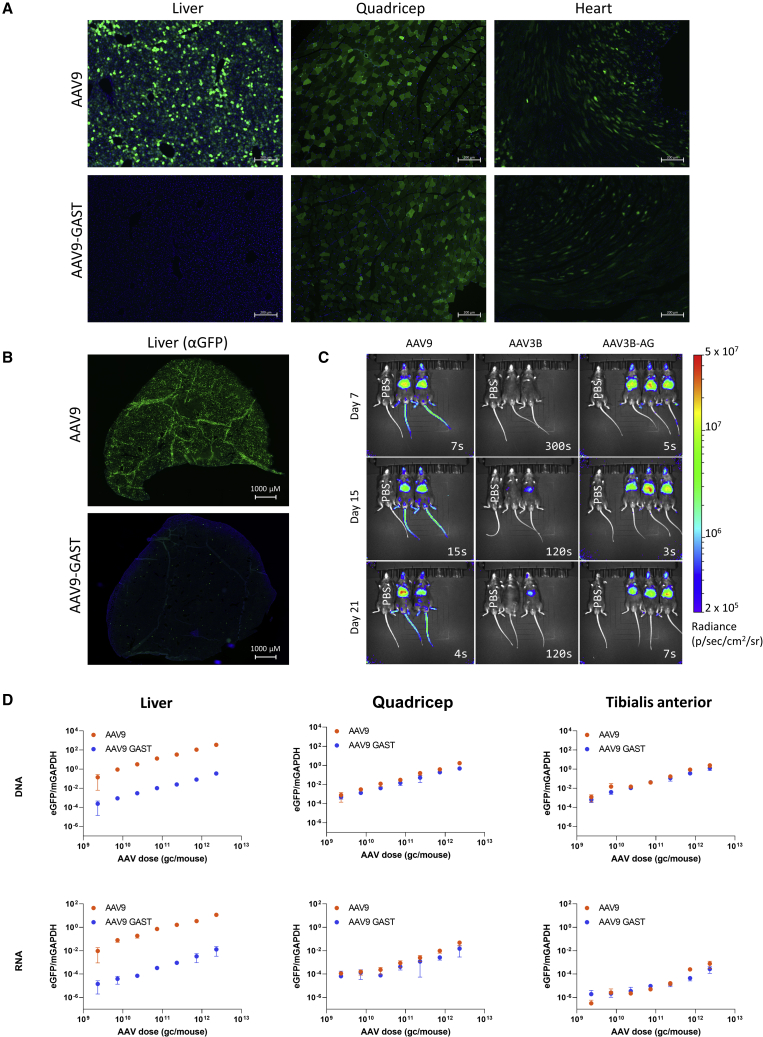
Two AAV9 mutants, AAV9-GA and AAV9-GAST (Table S3), were cloned to mimic the Anc80 liver toggle sequence context. Next, AAV9, AAV9-GA, or AAV9-GAST were evaluated for gene transfer (DNA) and gene expression (RNA) in C57BL/6 mice. Vector preparations were produced, each packaging an eGFP reporter under a CMV promoter. Mice were injected with a dose of 1.87 × 1011 gc/mouse by retro-orbital injection. In line with results from Anc80Lib, AAV9-GA and AAV9-GAST were observed to be significantly detargeted from the liver with respect to both vector genomes (−1,522-fold, AAV9-GA versus AAV9, p < 0.001 Tukey-HSD, −678-fold, AAV9-GAST versus AAV9, p < 0.001 Tukey-HSD) and expression (−363-fold AAV9-GA versus AAV9, p < 0.001 Tukey-HSD, −151-fold AAV9-GAST versus AAV9, p < 0.001 Tukey-HSD), yet we found no significant difference in terms of gene transfer (DNA, p > 0.05) to the quadriceps or heart, and a minor but significant reduction in expression in these tissues (−1.5- and −3.1-fold reductions in quadriceps and heart RNA, respectively, comparing AAV9 and AAV9-GA) (Figure 6B). This effect appeared robust across both single-stranded and self-complementary AAV (Figure S8). The liver-detargeted mutants also appeared to transduce the brain at lower rates than wild-type AAV9 while, remarkably, DNA vector copy numbers were not significantly different (p > 0.05 Tukey-HSD).
We also took sections from AAV9- and AAV9-GAST-injected mice and processed them for GFP histology. The images from these studies further illustrate the distinct reduction of AAV9-GAST expression in liver relative to AAV9, while maintaining transduction of the mouse quadriceps and heart (Figures 7A and 7B). A dose-ranging experiment was performed to determine whether these effects persisted across a large range of doses (2.3 × 109 gc/mouse up to 2.3 × 1012 gc/mouse) using a CB7 promoter driving expression of eGFP. Consistent with previous results and across all doses, AAV9-GAST showed profound de-enrichment in liver across all doses tested (more than 500-fold), while only lagging behind AAV9 in skeletal muscle by at most 5.7-fold (tibialis anterior, RNA, highest dose) (Figure 7D).
Figure 7.
In vivo transduction of reprogrammed AAVs
(A) Mice were injected with either AAV9 or AAV9-GAST containing a self-complementary eGFP transgene (dose of 1.87 × 1011 gc/mouse). Tissues were isolated, preserved, sectioned, and imaged. Scale bars, 200 μm.
(B) Whole mouse livers following injection with a self-complementary eGFP transgene in either AAV9 or AAV9-GAST (dose of 2.65 × 1011 gc/mouse).
(C) IVIS Imaging of mice injected with AAV9, AAV3B, or AAV3B-GA packaging a firefly luciferase transgene. PBS-injected mice are included as negative controls in each image (and labeled accordingly). Optimal exposure time determined by the instrument in the bottom right of each image.
(D) Increasing doses of either AAV9 (teal) or AAV9-GAST (purple) packing an eGFP transgene under a CB7 promoter were injected into C57BL6 Mice (N = 3 per dose). DNA and RNA were isolated from livers, quadriceps, or tibialis anterior (TA) from each mouse and were quantified by ddPCR. Mean values ± SEM are represented.
Having established that these mutations can detarget a liver tropic vector away from the mouse liver, we then hypothesized that we might be able to graft liver-targeting mutations into vectors that have been observed to transduce the mouse liver poorly. To investigate this hypothesis, we created AAV3B-AG by grafting the p3G mutation into AAV3B (Table S3), and in doing so re-created a mutation also investigated by Cabanes-Creus et al.61 (referred to therein as AAV3B-265insG). C57BL/6 mice were injected with either AAV9 (as a positive control), AAV3B, or AAV3B-AG containing a firefly luciferase transgene by retro-oribal injection at a dose of 5 × 1010 genome copies/mouse and assessed for liver transduction by in vivo bioluminescence. In line with our hypothesis and our prior observations with Anc80Lib and AAV9 mutants, along with the findings of Cabanes-Creus et al., modulating the VRI loop of AAV3B in this manner appears to increase luciferase activity in the mouse liver region from an average of 1.02E–5 (p/s/cm2/sr) in AAV3B-injected mice (N = 3) to 2.43E–6 (p/s/cm2/sr) in AAV3B-AG-injected mice (N = 3) (Figures 7C and S7C). AAV9-injected mice averaged 3.36E–6 (p/s/cm2/sr) (N = 3) at day 21.
Discussion
A multitude of preclinical and clinical studies have illustrated the utility of AAV as a therapeutic gene transfer vector. The approval of two AAV-based medicines in the US and other countries further validates the technology and approach.2 However, for many gene therapy indications, current AAV technologies fall short. For example, several clinical programs currently employ systemic administration of AAV to target non-hepatic tissues, such as muscle (e.g., for muscular dystrophies) and spinal cord (e.g., spinal muscular atrophy) (clinicaltrials.gov, NCT03375164, NCT03368742, NCT03362502, and NCT03461289). However, these programs and emerging drugs are complicated by the liver being a primary target for AAV uptake. Safety signals from systemic AAV administration have marred the field for more than a decade, and most recently have been implicated as a contributing factor to the death of three patients in a trial for X-linked myotubular myopathy.8 Liver detargeting of AAV has therefore been an area of active interest to the field.
The safety and efficacy of AAV-based drugs is in part determined by the features conferred by the AAV capsid, which vary between naturally occurring serotypes. Based on these observations, a large emphasis in the field has been enhancing the AAV platform by engineering the protein sequence of the AAV capsid to enhance its pharmacology for specific applications. Engineering of the AAV capsid has been limited to date by our overall sparse understanding of the necessary and sufficient vector-host interactions that lead to productive gene transfer. Screens for small-molecule pharmaceuticals take advantage of common atomic structures to both narrow down candidate molecules and also suggest the mechanism, defining the so-called structure-activity relationship (SAR). Despite the availability of high-resolution atomic structures of the capsid19,20,62,63,64,65,66,67,68 and access to increasingly sophistical analytical approaches, such as machine learning,69 rational design of the AAV particle has been made impractical by the unavailability of rich, quantitative functional datasets across translationally relevant phenotypes that could better define AAV SAR.
The field has thus relied largely on unbiased approaches, such as directed evolution of error-prone PCR or DNA shuffled capsid libraries to develop AAVs with enhanced properties. The complex quaternary structure of AAV requires hundreds of discreet intermolecular protein-protein interfaces for assembly to be retained (Figure 1C). While indeed variation of the capsid structure is tolerated, as is shown by the significant natural diversity, establishing man-made variation has proven more difficult. Recently, an exhaustive single-residue mutagenesis screen showed a majority of mutations to violate assembly requirements imposed by the viral architecture.44 Similarly, screens based on unbiased DNA library designs often result in enrichment for vectors highly similar to existing AAVs.29 In some instances, these variants can dramatically alter the characteristic they were selected for; however, sometimes at steep costs of functional trade-offs, such as manufacturability or species translation.31 These trade-offs are likely in part due to a lack of ability to account for complex epistatic interactions within the capsid that are required for SAR preservation.
Here, we sought to develop an approach that would (1) further the definition of AAV SAR and (2) allow for more a more direct and cost-effective evaluation of the complex in vivo pharmacology of candidate vectors. To address the challenges highlighted by previous efforts, we attacked the problems on multiple levels, in addition to seeking to borrow and optimize elements from previous discovery campaigns. First, a functionally diverse variant library was generated—that is, every member of the library produces a vector capable of being observed in transduction-dependent assays—by leveraging evolutionary ambiguity resultant from an ML-ASR approach. We further hypothesized that this approach would allow the segregation of functional motifs on the capsid. By mutating these variant sites in a full VP library, we aimed to explore unique combinations of phenotypes, epistatic interactions, and functional synergy of motifs. Second, we developed a multiparametric screening methodology with utility across model systems, including in vitro, small animal models, and large animal models (AAVSeq). To enable the study of mutations throughout the 2.4 kilobase capsid gene, we also developed a robust cloning approach (CombiAAV) to generate this barcoded library (Anc80Lib). Finally, an analytical and statistical framework was developed to interrogate individual variants across all screening parameters that allowed for SAR interpretation.
In the proof-of-concept study outlined here, we developed a large, uniquely barcoded AAV variant library called Anc80Lib based on many of the current clinical AAVs. Specifically, Anc80Lib sought to explore a combinatorial sequence space that varied computationally defined capsid residues that segregate AAV2 group capsids from AAV8 group capsids. Anc80Lib was screened in vitro and in vivo in mouse and NHP models, along with commonly used reference AAVs. Based on the divergence in hepatotropism of AAV2 group and AAV8 group vectors, we queried Anc80Lib in an attempt to map structural determinants of the AAV capsid that determine the degree of liver targeting.
Within the library, AAV variants were tagged with a unique identifier (i.e., a barcode) that permits a high-throughput and cost-effective short-read NGS readout. To date, studies of barcoded libraries with diversity distributed through the entire coding sequence of the AAV capsid have been limited in scale by the prior necessity of producing barcoded AAV capsids separately.43,45,70 We overcame this limitation by developing CombiAAV, a pooled barcoding and DNA assembly method that we used to generate complex barcoded capsid libraries at scale. As others have shown, barcoded AAV libraries allow for a quantitative and multiparametric evaluation of many AAV capsids at once, enabling one to gauge functional tradeoffs.43,44,45,70 Here, the multiparametric screening was extended beyond tissues (e.g. liver and muscle) to multiple species (mice and NHPs) for which a readout of gene-transfer (DNA) and transgene expression (mRNA) was obtained. Taken together, our data illustrate an ability to comprehensively assess the systems pharmacology of AAV at the level of both host and the vector.
An illustration of the power of the approach is provided by the identification of a single G-to-A residue change at position 266 to impact liver gene transfer in a manner that is divorced from its targeting and expression in muscle tissue. The liver toggle effect, first observed in systemic screens of Anc80Lib, is functionally conserved when grafted into two clinically relevant vectors: AAV9 to achieve liver detargeting and AAV3B to increase its liver transduction.
AAV3B and the highly homologous LK-03 are part of the AAV2 group and have liver toggle equivalent positions reflecting the predicted liver-off state (alanine). Indeed, in mice, these vectors are poorly hepatotropic and therefore limited for use in some murine disease models. However, AAV3B and LK-03 have been proposed as clinical vectors for liver-directed gene therapy because these vectors efficiently target NHP liver and human hepatocytes grated into immunodeficient mice in vivo, 29,52,71 illustrating that there are additional vector- and/or species-specific mechanisms underlying AAV hepatotropism. Upon mutation of the liver toggle equivalent sequence from an alanine to a glycine in AAV3B (AAV3B-AG), we observed liver transduction to be enhanced by approximately 20-fold. Further studies are needed to determine whether AAV3B-AG improves upon AAV3B’s native hepatotropism in primate models.
Conversely, AAV9, an AAV8 group member with a liver toggle equivalent position reflecting the “liver-on” state (glycine), is often used for therapeutic gene transfer in non-liver tissues following systemic administration.72,73,74 Liver toxicity has been described in both preclinical and clinical studies and is listed as a major side effect of onasemnogene abeparvovec, particularly in high-dose applications, such as those required to efficiently transduce therapeutic targets such as the spinal cord or the musculature.7,8,75 The ability to reduce biodistribution to the liver yet retain transduction of these therapeutic target tissues may increase the therapeutic window for these applications. AAV9-GA and AAV9-GAST substantially reduce vector DNA uptake by over 100-fold in the liver, yet remarkably retain their ability to transduce heart, tibialis anterior, and quadriceps, especially with self-complimentary AAVs, albeit with reduced efficiency. Moreover, when we produced and purified these vectors to high titers we observed that production of AAV9-GAST compared with that of wild-type AAV9 was not impacted (Figure S9).
While this article was initially in submission, a separate group published a body of work also exploring the relationship between VRI sequence variability and hepatotropism in the context of AAV3B.61 In that work, the authors independently derive and test the same AAV3B-AG vector (AAV3B-265insG by their nomenclature) and conclude that insertions into AAV3B modulate liver transduction. The authors also review data that we had previously published in patents as well as presented at conferences, and come to the conclusion that the liver toggle we describe herein may depend on the broader mutational context of VRI, including (crucially) an insertion after position 264 in clade B and C capsids. These two independent studies contextualize each other and together inform an understanding of the role of the VRI loop and its impact on hepatotroprism of AAV vectors. Specifically, when the findings of the two studies are considered together, it appears that the liver toggle we initially identified in our Anc80 library and confirm in AAV9 (a clade F capsid) may alter the structure-function relationship of the VRI loop in a manner that relies upon insertions at position 265 in AAV3B.
Importantly, our data indicate that the liver is not the only tissue that is affected by a modification of the liver toggle but rather has a substantial impact on the overall pharmacological profile of the altered vector (Figures 4, 5, 6, and 7). For example, not unlike the liver, expression of AAV9-GA or AAV9-GAST is reduced in the CNS, yet uptake of vector genomes is much less impacted. Indeed, the Anc80Lib screen in mice and NHPs illustrates a complex, sometimes species-specific, impact on the biodistribution and transduction properties of each of the variant positions, including the p3 liver toggle.
Our data illustrate that the use of multiparametric quantitative screenings of curated libraries like Anc80Lib are powerful in mapping the complex impact on the pharmacology of these modest changes in lineage-conserved residues. Independently, however, they fall short in highlighting the underlying mechanism. We hypothesize that the liver toggle alters the AAV-AAVR interaction in a cell-type-specific manner, as it is positioned within a subset of amino acids on the AAV capsid that interfaces with AAVR, an entry factor for most primate AAVs.76,77 Interestingly, liver-off vectors require AAVR for cell transduction, as both AAV2 and AAV8 group AAVs are dependent on AAVR for transduction based in vitro and in vivo.78,79 In Adachi et al., 2014, the researchers previously identified nine double-alanine mutants of AAV9 between residues 356 and 736, which appeared selectively detargeted from the liver. We recently revisited those data and determined that, of those nine mutants, six of them were either overlapping or within a single amino acid of sites known to interact with AAVR in AAV2 (Figure S10). Three of these implicated sites (W503, P504, and Q590) were previously known to selectively impact liver transduction as identified by Pulicherla et al.9 Taken as a whole, we view these independent findings as suggestive of a possible common mechanism whereby altering the AAV-AAVR interface alters liver tropism. Further studies are therefore needed to interrogate the role of the AAV-AAVR interaction of liver-on versus liver-off AAVs for each of the target tissues within the pharmacological context of an intravenous administration.
In gene therapy, the biology of the target gene is almost always well understood. In contrast, the molecular mechanisms underpinning efficient delivery of a therapeutic gene to a specific target are rarely known. Yet, the gene delivery by a vector is an important component to the safety and efficacy of any gene therapy intervention. This is no different for AAV, one of the leading vector platforms for clinical gene transfer. Here, we describe a quantitative, multiparametric screening approach to AAV libraries, which are diverse within known functional motifs on the capsid, and we apply this approach to interrogate the distinct hepatotropic properties of AAVs in clinical use. This approach now enables us to characterize and select for vectors with attributes fitting therapeutic targets. Furthermore, the rich datasets generated through these screens can further define the SAR of AAV, inspire mechanistic hypotheses, and provide reagents to probe molecular mechanisms. Finally, the approach and resultant dataset informs rational design of AAV. Our methodology is scalable to other vector libraries (through CombiAAV, a pooled barcoded library DNA assembly method), from in vitro to large animal model screens, and in other pharmacological settings (e.g., routes of administration or disease models). Collectively, the datasets, reagents, and methodologies of our work may further the field’s ability to control and direct therapeutic gene delivery.
Limitations of the study
Our work to date represents a systematic evaluation of an evolution-informed library of engineered AAV capsids with respect to several different vector functions both in vitro as well as in vivo. However, there are some important limitations to our study that need to be considered when evaluating our findings. For one, all animal studies reported herein were performed in male animals and as such may not reflect the biological activity in female animals. Moreover, while we report evidence for a liver toggle across a number of library studies in vitro as well as in vivo, we have only partially confirmed these findings and, even then, only in mice and in Huh7 cells. The agreement or disagreement between preclinical models is an open issue in the field and is the basis for ongoing work in our group as well. Finally, while we speculate about a potential role of AAVR in the mechanism of the liver toggle, we do not present any direct evidence herein and as such the molecular mechanism remains an open question.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| ElectroMAX™ Stbl4™ Competent Cells | Invitrogen | Cat# 11635018 |
| AAV-CMV-BC-Anc80Lib | Mass Eye and Ear Gene Therapy Vector Core | N/A |
| AAV-CMV-BC-AAV2 | Mass Eye and Ear Gene Therapy Vector Core | N/A |
| AAV-CMV-BC-AAV5 | Mass Eye and Ear Gene Therapy Vector Core | N/A |
| AAV-CMV-BC-AAV8 | Mass Eye and Ear Gene Therapy Vector Core | N/A |
| AAV-CMV-BC-Rh.10 | Mass Eye and Ear Gene Therapy Vector Core | N/A |
| AAV-CMV-BC-LK-03 | Mass Eye and Ear Gene Therapy Vector Core | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Benzonase | EMD Millipore | Cat# 1016970010 |
| BsaI-HF | New England Biolabs | Cat# R3535 |
| Esp3I | Thermo Scientific | Cat# ER0451 |
| T4 Ligase | New England Biolabs | M0202L |
| Q5® High-Fidelity DNA Polymerase | New England Biolabs | M0491L |
| KAPA HiFi HotStart ReadyMix | Roche | KK2602 |
| Critical commercial assays | ||
| NEBNext® Library Quant Kit for Illumina® | New England Biolabs | Cat# E7630L |
| Nextera XT Index Kit v2 Set A (96 indexes, 384 samples) | Illumina | Cat# FC-131-2001 |
| Deposited data | ||
| Raw Data (Barcode Counts, Imaging, Biodstribution) | Mendeley Data Repository | https://doi.org/10.17632/hd9nnhrttg.1 |
| Experimental models: Cell lines | ||
| HEK293 Cell Line | ATCC | CRL-1573 |
| Huh7 Cell Line | ATCC | PTA-4583 |
| Experimental models: Organisms/Strains | ||
| C57BL/6J mouse strain (Mus musculus) | Jackson Laboratory | Stock#000664 |
| Cynomolgus macaques (Macaca fascicularis) | DaVinci Biomedical Research, Framingham, MA | N/A |
| Oligonucleotides | ||
| DNA Seqencing Fw Primer (F1):TCGTCGGCAGCGTCAGA TGTGTATAAGAGACAGGAGGTCTATATAAGCAGAGCTG GTTTAGTGAACCGT |
This paper | N/A |
| DNA Seqencing Rv Primer (R1): GTCTCGTGGGCTCGGAG ATGTGTATAAGAGACAGGACGAGAACATTTGTTTTGGTA CCTGTCTGCGTAG |
This paper | N/A |
| RNA Sequencing Fw Primer (F2): TCGTCGGCAGCGTCAG ATGTGTATAAGAGACAGGTCAGATCCTGCATGAAGCTT |
This paper | N/A |
| RNA Sequencing Rv Primer (R2): GTCTCGTGGGCTCGGA GATGTGTATAAGAGACAGCTGTCTGCGTAGTTGATCGGC |
This paper | N/A |
| Recombinant DNA | ||
| pSL-CMV-BC-Anc80Lib | This paper | N/A |
| pSL-CMV-BC-AAV2 | This paper | N/A |
| pSL-CMV-BC-AAV5 | This paper | N/A |
| pSL-CMV-BC-AAV8 | This paper | N/A |
| pSL-CMV-BC-AAV9 | This paper | N/A |
| pSL-CMV-BC-AAV9.PHP-B | This paper | N/A |
| pSL-CMV-BC-LK-03 | This paper | N/A |
| pSL-CMV-BC-Rh.10 | This paper | N/A |
| pSL-CMV-BC-Rh32.33 | This paper | N/A |
| pAAP | Vandenberghe Lab | N/A |
| pREP | Vandenberghe Lab | N/A |
| deltaF6 | Vandenberghe Lab | N/A |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com |
| Python 3.7 | Python Software Foundation | https://www.python.org |
| Living Image Version 4.5.5 | Perkin Elmer | https://www.perkinelmer.com/product/li-software-for-lumina-1-seat-add-on-128110 |
| Zen Version 3.3 | Carl Zeiss Microscopy | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html |
| Additional Custom Python Scripts | This paper | Mendeley Repository: https://doi.org/10.17632/hd9nnhrttg.1 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Luk Vandenberghe (luk_vandenberghe@meei.havard.edu).
Materials availability
Plasmid encoding the barcoded Anc80 library generated in this study are available upon request.
Experimental model and subject details
In vitro library experiments
Our in vitro selection experiments were conducted using HuH-7 cells, an immortalized cell line of human hepatic origin. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Thermofisher Cat# 11965084) supplemented with 10% fetal bovine serum (Cytiva, cat no. SH30071.03) in 6-well plates (Corning Product #: 3506) pre-coated with Poly-Ly Lysine (Sigma-Aldrich CAS # 25988-63-0). While the original cells were derived from a biologically male donor, Huh7 cells have a complex karyotype without Y chromosome.80
In vivo library experiments
Mouse
All animal procedures were performed in accordance with protocols approved by the institutional care and use committees (IACUC) at Schepens Eye Research Institute (SERI). Male C57BL/6 mice (6-8 weeks old) were purchased from Jackson Laboratories and allowed to acclimate to the animal care facilities at SERI for a period of one week and were maintained on a regular diet. The barcoded Anc80 library (Anc80BC-Lib) was spiked with singly-produced, barcoded AAVs as in the in vitro infectivity study above (AAV2, 5, 8, 9, PHP-B, LK-03, Rh.10 and Rh.32.33) at a ratio of 1:2053 each (AAV:Total Library) on a vector genome copy basis.
Two days before injection, 50 μL of blood was collected by submandibular bleeds to establish a baseline free of vector for biodistribution studies. On day zero, each mouse was injected retro-orbitally with 3 × 1011 genome copies. On day 3 and 21, eight injected mice (plus one uninjected control) were sacrificed and tissues were collected and flash frozen. Prior to barcode amplification, tissues were mechanically homogenized by beads using either a Geno/Grinder (SPEX SamplePrep) or a Qiagen TissueLyser II (Qiagen Cat No 85300) and nucleotides were isolated using Qiagen DNAEasy Blood and Tissue Kits and RNAEasy Kits (Qiagen Cat No. 74104). The integrity of RNA samples was analyzed and scored by TapeStation prior to proceeding with barcode amplification and extraction.
Nonhuman primate
Experiments with cynomolgus macaques (Macaca fascicularis) were performed at a contract research organization (DaVinci Biomedical Research, Framingham, MA) in accordance with the institutional IACUCs of both DaVinci and SERI. Prior to initiating the study, animals were screened for antibodies against Anc80L65, AAV1, 2, 5, 8, 9 and rh32.33. Based on low antibody titers, four two-year-old male animals (identifiers G62N, G66E, G72E, and H92R) were selected for the study and maintained on a regular diet. None of the animals selected had detectable levels of neutralizing antibodies (titer <⅛) to Anc80L65, or AAV9. The final vector pool was prepared by spiking singly-produced AAV2, 5, 8, Rh.10 and LK-03 into a high-titer prep of Anc80BC-Lib at a ratio of 1:2053 each (AAV:Total Library) on a vector genome copy basis.
Animals were injected intravenously with 4 × 1012 genome copies diluted in 4.75 mL of saline (1.6 × 1012 gc/kg) on day 0. Serum was collected on days 0, 1, 3, and 7. On day 7, two animals (G72E and H92R) were sacrificed and tissues were collected and flash frozen (brain was further dissected prior to freezing). On day 28, the remaining two animals (G62N and G66E) were sacrificed as well. Tissues were prepared as in the mouse experiment prior to barcode amplification and extraction.
In vivo clonal confirmation studies, Anc80
High titer preparations of Anc80L1533 and Anc80L1093 were produced and obtained from the Gene Transfer Vector Core (GTVC) as described above. Five male C57BL/6 mice were each injected with either Anc80L1533 or Anc80L1093 containing an eGFP transgene under a CMV promoter at a dose of 1 × 1011 genome copies per animal by retro orbital administration. After 21 days, animals were sacrificed and tissues were collected and flash frozen. To quantify vector abundances in these tissues, the flash frozen samples were homogenized and DNA/RNA were isolated and quantified as described above. To quantify the number of eGFP DNA molecules/cell, DNA isolated from each cell was quantified by digital-droplet PCR (ddPCR) (QX200 system, BioRad). To quantify the ratio of eGFP RNA molecules/GAPDH, ddPCR was again used.
In vivo clonal confirmation studies, AAV9 and reprogrammed variants
High titer preparations of AAV9, AAV9-GA, and AAV9-GAST were produced and obtained from the Gene Transfer Vector Core (GTVC) as described above. Five male C57BL/6 mice were each injected with either AAV9, AAV9-GA, or AAV9-GAST containing a self-complementary eGFP transgene under a CMV promoter at a dose of 1.87 × 1011 genome copies per animal by retro orbital administration. After 28 days, animals were sacrificed and tissues were collected and flash frozen. A number of samples were also fixed in 4% PFA before being transferred to a sucrose solution for eventual preparation for histology. To quantify vector abundances in these tissues, the flash frozen samples were homogenized and DNA/RNA were isolated and quantified as described above. To quantify the number of eGFP DNA molecules/cell, DNA isolated from each cell was quantified by ddPCR (QX200 system, BioRad). To quantify the ratio of eGFP RNA molecules/GAPDH, RT-qPCR was performed using an Applied Biosystems 7500 Real-Time PCR System using TaqMan PCR master mix reagents (Applied Biosystems) and transgene-specific primer/probes as previously described.81
In vivo luminescence study, AAV3B and reprogrammed variants
High titer preparations of AAV9, AAV3B, and AAV3B-AG were produced and obtained from the GTVC as described above, each containing a firefly luciferase transgene under a CMV promoter. Vectors were administered in a final volume of 100 μL of PBS via retro-orbital venous route into adult male C57BL/6J mice (Jackson Cat. No. 000664) at a dose of 5 × 1010 genome copies/mouse (n = 2–3 per condition). At days 7, 15, and 21, animals were anesthetized and bioluminescent imaging was performed on a IVIS instrument (PerkinElmer Inc, Waltham, MA). Data were analyzed using Living Image Software version 4.7.3.
Method details
Design and assembly of barcoded Anc80Lib
Ancestral capsid sequences were reconstructed, and positions of variation were identified as described extensively in Zinn et al. (2015). To assemble a library of 2048 barcoded capsids based upon these previous reconstructions, first 22 different building-blocks were designed and synthesized commercially by Twist Bioscience (San Francisco, CA), each synthetic building block encompassing a region of the capsid and a corresponding region of the barcode. The building blocks were designed to be assembled in a CombiGEM-like fashion (Figure S1) by repeated rounds of restriction digestion, ligation, bacterial transformation, and purification. In all, five such rounds of assembly were needed to fully assemble our library. Sanger sequencing reactions were performed after each round of assembly to ensure adequate diversity and that barcode sequences remained coupled to capsid sequences.
AAV library vector preparation (for in vitro and in vivo studies)
To produce high-titer preparations of barcoded Anc80-Lib, the Gene Transfer Vector Core (GTVC) at the Grousbeck Gene Therapy Center of Massachusetts Eye and Ear Infirmary performed quadruple plasmid transfections using polyethylenimine (PEI-Max, Polysciences) in HEK293 cells within ten-layer hyperflasks (Corning). The plasmids were pREP (AAV2 Rep genes under the native p5 promoter), pAAP2 (CMV driving expression of AAP2), the library plasmid (CMV-Barcode-Anc80Capsid), and dF6 (containing necessary Adenoviral accessory genes), in a ratio of 10:10:1:20.60 To test the impact of exogenous AAP on library production, AAP was omitted. The ratio of PEI-Max to DNA was maintained at 1.375:1 (w/w). To produce high-titer preparations of eGFP containing vector, vector was produced by traditional triple plasmid transfection using the same HEK293 cells as described previously. Downstream purification was performed as previously described.82 Preparations were tittered by digital-droplet based PCR as described in.83
Barcode amplification and purification
To sequence barcodes from plasmid or virus (DNA), barcodes were first amplified by 15 cycles of PCR using primers designed to amplify the barcode and to incorporate a binding site for the subsequent indexing reaction (key resources table). To sequence barcodes from cellular extracts (DNA or cDNA), barcodes are amplified by 15–35 cycles of PCR, using either primers F1 and R1 (DNA) or F2 and R2 (RNA). These first-round PCR products were loaded onto a 2% Agarose gel and were subsequently visualized, extracted, and purified (Zymoclean Gel DNA Recovery Kit, Cat No. D4007). To render the amplified barcodes compatible with Illumina sequencing technologies, the purified first-round PCR product was then used as template in a subsequent 8-round indexing reaction using Nextera XT Indexing primers (Illumina Cat No FC-131-1001). These second round PCR products were gel purified on a 2% agarose gel and were quantified by QuBit dsDNA HS Assay Kit (ThermoFisher Cat. No. Q32851).
In vitro infectivity study
To a high-titer preparation of barcoded Anc80 library (Anc80BC-Lib), singly-produced barcoded AAVs (AAV2, AAV5, AAV8, AAV9, Rh.10, Rh32.33, LK-03, and AAV9-PHP.B) were spiked at a ratio of 1:2053 each (AAV:Total Library) on a vector genome copy basis. This mixture was added to 6-well dishes confluent with Huh7 cells at a MOI of 200 particles/cell diluted in DMEM without FBS (3 technical replicates) and was allowed to incubate for 1 h at 37°C. To remove excess virus, cells were subsequently washed three times with PBS before DMEM containing 10% FBS was added and cells were returned to 37°C. After 48 h, cells were split into two different tubes. From one tube, DNA was isolated by DNAEasy Blood and Tissue Kits (Qiagen Cat. No. 69504). From the other tube, RNA was isolated by Trizol Extraction, DNA was removed by DNA-Free Removal Kits (ThermoFisher Cat No AM1906) and cDNA was generated by Super-Script IV RT Kits (ThermoFisher Cat No 18090010). Barcodes were amplified, purified as described above before being pooled with PhiX Sequencing Control (Illumina Cat. No. FC-110-3001) and were sequenced on a MiSeq sequencing using a MiSeq v3 150 Cycle Kit (Illumina Cat No. MS-102-3001).
Quantification and statistical analysis
Analysis of NGS data
Data were processed and analyzed using scripts written for this specific purpose (source code and data available in the associated Mendeley data repository). To summarize, the approach begins by summing raw counts obtained from parallel sequencing lanes together, treating each parallel lane as a technical replicate of sequencing. Next, for each sample within an experiment, the reads are normalized for read depth according to Equation 1 where cpm is the counts-per million mapped reads, x is a raw count, s is the sample within the experiment, n is the total number of barcodes within the sample, and b is a barcode within that sample.
| (Equation 1) |
To account for variability across biological replicates within an experiment, samples were log-transformed (base 2) and averaged arithmetically. We add an infinitesimally small quantity to ensure that we never take a log of zero. We refer to the average number of log-transformed normalized counts of a barcode across biological replicates as . To compare this average quantity, we calculate the log-transformed fold-change by subtracting the equivalent quantity derviced from a reference condition as per Equation 2
| (Equation 2) |
Code and data used in this analysis are available in the Mendeley repository.
Regularized regression analysis
To determine the roles of individual sites of variation within our library, we first constructed an independent sites linear model wherein the log-transformed fold-change is related to a series of variables representing all of the positions of variation within our library as seen in Equation 3.
| (Equation 3) |
Individual models were constructed for each sample through an implementation of the Elastic Net employing both an L1 and L2 penalty to account for potential correlational structures among our independent variables as implemented in scikit learn. Hyperparameters dictating mixture of L1 and L2 in each model were derived through an exhaustive search over a grid of hyperparameters also as implemented in scikit-learn. Where specified, data were Z score normalized prior to fitting to account for differences in scale across experimental conditions for purposes of visualization. Code for regularized regression analysis and visualization within the Mendeley repository.
Clonal biodistribution studies
First, abundances of either transgene (eGFP) DNA or transgene RNA were normalized to cell number by calculating the ratio of eGFP to a housekeeper gene (mTfrc in the case of DNA samples, mGAPDH in the case of RNA samples). Next, these ratios were log transformed (base 2) to normalize the distributions. Normality was tested by the Shapiro-Wilk, and homoscedasticity was tested by the Levene test for equal variances. In each of our samples, we confirmed both normality and homoscedasticity, two crucial assumptions for the next step of the statistical analysis. A one-way ANOVA was then conducted for each tissue to identify effects between vectors. To identify individual pairwise effects while correcting for family wise error rates, we used Tukey’s Honestly Significant Difference test (Tukey-HSD).
Structural analysis of AAV9
To generate the circos diagram in Figure 1C, we first loaded a high-resolution structure of AAV9 with monomers separated into discrete chains (PDB accession number 3UX1,66). Next, we iterated through each amino of each chain, searching for amino acids within 4A of any side-chain atoms from separate monomers. This set of potential pairwise interactions were then sorted depending on the axis of symmetry relating the monomers (2-, 3-, or 5-fold) and duplicate interactions were pruned. To plot positions of functional AAV domains identified in non-AAV9 serotypes, we approximated homology by pairwise-alignment of AAV9 with the AAV serotype of interest. The data files used to generate Figure 1C have been uploaded to Mendeley and are publicly available.
Acknowledgments
Funding was provided by Giving/Grousbeck (to L.H.V.) as well as a sponsored research agreement with Lonza Houston (to L.H.V.). A portion of the work also was supported by NIH DP1 OD025432 (to A.J.W.). We thank Ru Xiao and the Gene Transfer Vector Core (https://www.vdb-lab.org/vector-core/) for manufacturing and providing purified, high-titer adeno-associated viral vectors and libraries used in these studies; Yanhe Wen for the initial conceptualization of the fingerprint plots used in this publication; Michael Florea, Dr. Xia Wang, Dr. Weiwei Wang, Dr. Timothy Mitchison, Dr. Debbie Marks, and Dr. Connie Cepko; and our collaborators at Lonza for scientific discussion and helpful suggestions regarding experimental design and data analysis.
Author contributions
Conceptualization, E.Z., H.T.T., P.F.S., C.U., C.T., and L.H.V.; methodology, E.Z., H.T.T., P.F.S., C.U., C.T., B.P., A.J.W., and L.H.V.; validation, E.Z., P.F.S., J. Sanmiguel, E.M., R.G., A.F., and C.D.; formal analysis, E.Z., C.U., N.Z., and J. Sanmiguel; investigation, E.Z., P.F.S., C.U., C.T., B.P., C.R., K.M., and L.H.V.; writing – original draft, E.Z. and L.H.V.; writing – review & editing, E.Z., N.Z., P.F.S., C.U., H.T.T., J. Sanmiguel, A.F., B.P., J. Santos, R.E., E.A.-M., A.J.W., C.T., and L.H.V.; visualization, E.Z., N.Z., E.M., R.G., B.P, and L.H.V.; supervision, A.J.W. and L.H.V.; funding acquisition, A.J.W. and L.H.V.
Declaration of interests
L.H.V. holds equity in Affinia Therapeutics, Akouos, and ciendias bio. He serves on the Board of Directors of Affinia Therapeutics, Addgene, ciendias bio, and Odylia. L.H.V. is an employee of ciendias bio. L.H.V. and E.Z. are inventors of AncAAV technology licensed to Affinia, Akouos, and/or other biopharmaceutical companies from which they may receive royalties. L.H.V. is compensated for his scientific advisory position with Affinia and Akouos. L.H.V. is a SAB member to Akouos, consultant to Affinia and Novartis, and receives research support from Novartis. L.H.V.’s interests were reviewed and are managed by Mass Eye and Ear and Mass General Brigham in accordance with their conflict-of-interest policies. L.H.V., E.Z., C.U., P.F.S., C.T., and H.T.T. are inventors on patent applications relating to AAVSeq, CombiAAV, or the liver toggle technology contained in this publication. A.J.W. and L.H.V. are inventors on patents related to use of AAVs for therapeutic gene delivery in the muscle and other tissues.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 2, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100803.
Supplemental information
Data and code availability
-
•
Raw barcode counts corresponding to every sample included in this publication have been deposited into Mendeley and is publicly available as of the date of publication. Additional raw data corresponding to biodistribution studies and imaging studies have also been deposited into Mendeley and is publicly available as of the date of publication.
-
•
All original code has been deposited in Mendeley and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Li C., Samulski R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020;21:255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 2.Keeler A.M., Flotte T.R. Recombinant adeno-associated virus gene therapy in light of luxturna (and zolgensma and glybera): where are we, and how did we get here? Annu. Rev. Virol. 2019;6:601–621. doi: 10.1146/annurev-virology-092818-015530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudry E., Vandenberghe L.H. Therapeutic AAV gene transfer to the nervous system: a clinical reality. Neuron. 2019;101:839–862. doi: 10.1016/j.neuron.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendell J.R., Al-Zaidy S.A., Rodino-Klapac L.R., Goodspeed K., Gray S.J., Kay C.N., Boye S.L., Boye S.E., George L.A., Salabarria S., et al. Current clinical applications of in vivo gene therapy with AAVs. Mol. Ther. 2021;29:464–488. doi: 10.1016/j.ymthe.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Rasko J., Ozelo M.C., Hoots K., Blatt P., et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 6.Nathwani A.C., Tuddenham E.G.D., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C., et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day J.W., Finkel R.S., Chiriboga C.A., Connolly A.M., Crawford T.O., Darras B.T., Iannaccone S.T., Kuntz N.L., Peña L.D.M., Shieh P.B., et al. Onasemnogene abeparvovec gene therapy for symptomatic infantile-onset spinal muscular atrophy in patients with two copies of SMN2 (STR1VE): an open-label, single-arm, multicentre, phase 3 trial. Lancet Neurol. 2021;20:284–293. doi: 10.1016/S1474-4422(21)00001-6. [DOI] [PubMed] [Google Scholar]
- 8.Philippidis A. After third death, audentes' AT132 remains on clinical hold. Hum. Gene Ther. 2020;31:908–910. doi: 10.1089/hum.2020.29133.bfs. [DOI] [PubMed] [Google Scholar]
- 9.Pulicherla N., Shen S., Yadav S., Debbink K., Govindasamy L., Agbandje-McKenna M., Asokan A. Engineering liver-detargeted AAV9 vectors for cardiac and musculoskeletal gene transfer. Mol. Ther. 2011;19:1070–1078. doi: 10.1038/mt.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D., Li S., Gessler D.J., Xie J., Zhong L., Li J., Tran K., Van Vliet K., Ren L., Su Q., et al. A rationally engineered capsid variant of AAV9 for systemic CNS-directed and peripheral tissue-detargeted gene delivery in neonates. Mol. Ther. Methods Clin. Dev. 2018;9:234–246. doi: 10.1016/j.omtm.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rotundo I.L., Lancioni A., Savarese M., D'Orsi L., Iacomino M., Nigro G., Piluso G., Auricchio A., Nigro V. Use of a lower dosage liver-detargeted AAV vector to prevent hamster muscular dystrophy. Hum. Gene Ther. 2013;24:424–430. doi: 10.1089/hum.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao G., Vandenberghe L.H., Wilson J.M. New recombinant serotypes of AAV vectors. Curr. Gene Ther. 2005;5:285–297. doi: 10.2174/1566523054065057. [DOI] [PubMed] [Google Scholar]
- 13.Grimm D., Pandey K., Nakai H., Storm T.A., Kay M.A. Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J. Virol. 2006;80:426–439. doi: 10.1128/jvi.80.1.426-439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakai H., Fuess S., Storm T.A., Muramatsu S.i., Nara Y., Kay M.A. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J. Virol. 2005;79:214–224. doi: 10.1128/jvi.79.1.214-224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandenberghe L.H., Breous E., Nam H.J., Gao G., Xiao R., Sandhu A., Johnston J., Debyser Z., Agbandje-McKenna M., Wilson J.M. Naturally occurring singleton residues in AAV capsid impact vector performance and illustrate structural constraints. Gene Ther. 2009;16:1416–1428. doi: 10.1038/gt.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Z., Asokan A., Grieger J.C., Govindasamy L., Agbandje-McKenna M., Samulski R.J. Single amino acid changes can influence titer, heparin binding, and tissue tropism in different adeno-associated virus serotypes. J. Virol. 2006;80:11393–11397. doi: 10.1128/jvi.01288-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandenberghe L.H., Wang L., Somanathan S., Zhi Y., Figueredo J., Calcedo R., Sanmiguel J., Desai R.A., Chen C.S., Johnston J., et al. Heparin binding directs activation of T cells against adeno-associated virus serotype 2 capsid. Nat. Med. 2006;12:967–971. doi: 10.1038/nm1445. [DOI] [PubMed] [Google Scholar]
- 18.Zhong L., Li B., Mah C.S., Govindasamy L., Agbandje-McKenna M., Cooper M., Herzog R.W., Zolotukhin I., Warrington K.H., Weigel-Van Aken K.A., et al. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. USA. 2008;105:7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie Q., Bu W., Bhatia S., Hare J., Somasundaram T., Azzi A., Chapman M.S. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nam H.-J., Lane M.D., Padron E., Gurda B., McKenna R., Kohlbrenner E., Aslanidi G., Byrne B., Muzyczka N., Zolotukhin S., Agbandje-McKenna M. Structure of adeno-associated virus serotype 8, a gene therapy vector. J. Virol. 2007;81:12260–12271. doi: 10.1128/jvi.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Opie S.R., Warrington K.H., Jr., Agbandje-McKenna M., Zolotukhin S., Muzyczka N. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 2003;77:6995–7006. doi: 10.1128/jvi.77.12.6995-7006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kern A., Schmidt K., Leder C., Müller O.J., Wobus C.E., Bettinger K., Von der Lieth C.W., King J.A., Kleinschmidt J.A. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 2003;77:11072–11081. doi: 10.1128/jvi.77.20.11072-11081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asokan A., Conway J.C., Phillips J.L., Li C., Hegge J., Sinnott R., Yadav S., DiPrimio N., Nam H.-J., Agbandje-McKenna M., et al. Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nat. Biotechnol. 2010;28:79–82. doi: 10.1038/nbt.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen S., Horowitz E.D., Troupes A.N., Brown S.M., Pulicherla N., Samulski R.J., Agbandje-McKenna M., Asokan A. Engraftment of a galactose receptor footprint onto adeno-associated viral capsids improves transduction efficiency. J. Biol. Chem. 2013;288:28814–28823. doi: 10.1074/jbc.M113.482380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurer A.C., Pacouret S., Cepeda Diaz A.K., Blake J., Andres-Mateos E., Vandenberghe L.H. The assembly-activating protein promotes stability and interactions between AAV's viral proteins to nucleate capsid assembly. Cell Rep. 2018;23:1817–1830. doi: 10.1016/j.celrep.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dudek A.M., Zabaleta N., Zinn E., Pillay S., Zengel J., Porter C., Franceschini J.S., Estelien R., Carette J.E., Zhou G.L., Vandenberghe L.H. GPR108 is a highly conserved AAV entry factor. Mol. Ther. 2020;28:367–381. doi: 10.1016/j.ymthe.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimm D., Lee J.S., Wang L., Desai T., Akache B., Storm T.A., Kay M.A. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008;82:5887–5911. doi: 10.1128/jvi.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalkara D., Byrne L.C., Klimczak R.R., Visel M., Yin L., Merigan W.H., Flannery J.G., Schaffer D.V. In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci. Transl. Med. 2013;5:189ra76. doi: 10.1126/scitranslmed.3005708. [DOI] [PubMed] [Google Scholar]
- 29.Lisowski L., Dane A.P., Chu K., Zhang Y., Cunningham S.C., Wilson E.M., Nygaard S., Grompe M., Alexander I.E., Kay M.A. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deverman B.E., Pravdo P.L., Simpson B.P., Kumar S.R., Chan K.Y., Banerjee A., Wu W.L., Yang B., Huber N., Pasca S.P., Gradinaru V. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hordeaux J., Wang Q., Katz N., Buza E.L., Bell P., Wilson J.M. The neurotropic properties of AAV-PHP.B are limited to C57BL/6J mice. Mol. Ther. 2018;26:664–668. doi: 10.1016/j.ymthe.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hordeaux J., Yuan Y., Clark P.M., Wang Q., Martino R.A., Sims J.J., Bell P., Raymond A., Stanford W.L., Wilson J.M. The GPI-linked protein LY6A drives AAV-PHP.B transport across the blood-brain barrier. Mol. Ther. 2019;27:912–921. doi: 10.1016/j.ymthe.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Q., Chan K.Y., Tobey I.G., Chan Y.A., Poterba T., Boutros C.L., Balazs A.B., Daneman R., Bloom J.M., Seed C., Deverman B.E. Delivering genes across the blood-brain barrier: LY6A, a novel cellular receptor for AAV-PHP.B capsids. PLoS One. 2019;14 doi: 10.1371/journal.pone.0225206. e0225206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramachandran P.S., Lee V., Wei Z., Song J.Y., Casal G., Cronin T., Willett K., Huckfeldt R., Morgan J.I.W., Aleman T.S., et al. Evaluation of dose and safety of AAV7m8 and AAV8BP2 in the non-human primate retina. Hum. Gene Ther. 2017;28:154–167. doi: 10.1089/hum.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulk N.K., Pekrun K., Charville G.W., Maguire-Nguyen K., Wosczyna M.N., Xu J., Zhang Y., Lisowski L., Yoo B., Vilches-Moure J.G., et al. Bioengineered viral platform for intramuscular passive vaccine delivery to human skeletal muscle. Mol. Ther. Methods Clin. Dev. 2018;10:144–155. doi: 10.1016/j.omtm.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paulk N.K., Pekrun K., Zhu E., Nygaard S., Li B., Xu J., Chu K., Leborgne C., Dane A.P., Haft A., et al. Bioengineered AAV capsids with combined high human liver transduction in vivo and unique humoral seroreactivity. Mol. Ther. 2018;26:289–303. doi: 10.1016/j.ymthe.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marsic D., Govindasamy L., Currlin S., Markusic D.M., Tseng Y.S., Herzog R.W., Agbandje-McKenna M., Zolotukhin S. Vector design Tour de Force: integrating combinatorial and rational approaches to derive novel adeno-associated virus variants. Mol. Ther. 2014;22:1900–1909. doi: 10.1038/mt.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Havlik L.P., Simon K.E., Smith J.K., Klinc K.A., Tse L.V., Oh D.K., Fanous M.M., Meganck R.M., Mietzsch M., Kleinschmidt J., et al. Coevolution of adeno-associated virus capsid antigenicity and tropism through a structure-guided approach. J. Virol. 2020;94 doi: 10.1128/jvi.00976-20. e009766-e1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albright B.H., Storey C.M., Murlidharan G., Castellanos Rivera R.M., Berry G.E., Madigan V.J., Asokan A. Mapping the structural determinants required for AAVrh.10 transport across the blood-brain barrier. Mol. Ther. 2018;26:510–523. doi: 10.1016/j.ymthe.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zinn E., Pacouret S., Khaychuk V., Turunen H.T., Carvalho L.S., Andres-Mateos E., Shah S., Shelke R., Maurer A.C., Plovie E., et al. In silico reconstruction of the viral evolutionary lineage yields a potent gene therapy vector. Cell Rep. 2015;12:1056–1068. doi: 10.1016/j.celrep.2015.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong A.S.L., Choi G.C.G., Cui C.H., Pregernig G., Milani P., Adam M., Perli S.D., Kazer S.W., Gaillard A., Hermann M., et al. Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proc. Natl. Acad. Sci. USA. 2016;113:2544–2549. doi: 10.1073/pnas.1517883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong A.S.L., Choi G.C.G., Cheng A.A., Purcell O., Lu T.K. Massively parallel high-order combinatorial genetics in human cells. Nat. Biotechnol. 2015;33:952–961. doi: 10.1038/nbt.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adachi K., Enoki T., Kawano Y., Veraz M., Nakai H. Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nat. Commun. 2014;5:3075. doi: 10.1038/ncomms4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogden P.J., Kelsic E.D., Sinai S., Church G.M. Comprehensive AAV capsid fitness landscape reveals a viral gene and enables machine-guided design. Science. 2019;366:1139–1143. doi: 10.1126/science.aaw2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinmann J., Weis S., Sippel J., Tulalamba W., Remes A., El Andari J., Herrmann A.K., Pham Q.H., Borowski C., Hille S., et al. Identification of a myotropic AAV by massively parallel in vivo evaluation of barcoded capsid variants. Nat. Commun. 2020;11:5432. doi: 10.1038/s41467-020-19230-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown D., Altermatt M., Dobreva T., Chen S., Wang A., Thomson M., Gradinaru V. Deep parallel characterization of AAV tropism and AAV-mediated transcriptional changes via single-cell RNA sequencing. Front. Immunol. 2021;12:730825. doi: 10.3389/fimmu.2021.730825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davidsson M., Wang G., Aldrin-Kirk P., Cardoso T., Nolbrant S., Hartnor M., Mudannayake J., Parmar M., Björklund T. A systematic capsid evolution approach performed in vivo for the design of AAV vectors with tailored properties and tropism. Proc. Natl. Acad. Sci. USA. 2019;116:27053–27062. doi: 10.1073/pnas.1910061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goertsen D., Flytzanis N.C., Goeden N., Chuapoco M.R., Cummins A., Chen Y., Fan Y., Zhang Q., Sharma J., Duan Y., et al. AAV capsid variants with brain-wide transgene expression and decreased liver targeting after intravenous delivery in mouse and marmoset. Nat. Neurosci. 2022;25:106–115. doi: 10.1038/s41593-021-00969-4. [DOI] [PubMed] [Google Scholar]
- 49.Öztürk B.E., Johnson M.E., Kleyman M., Turunç S., He J., Jabalameli S., Xi Z., Visel M., Dufour V.L., Iwabe S., et al. scAAVengr, a transcriptome-based pipeline for quantitative ranking of engineered AAVs with single-cell resolution. Elife. 2021;10 doi: 10.7554/eLife.64175. e64175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ellis B.L., Hirsch M.L., Barker J.C., Connelly J.P., Steininger R.J., Porteus M.H. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virol. J. 2013;10:74. doi: 10.1186/1743-422X-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas C.E., Storm T.A., Huang Z., Kay M.A. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J. Virol. 2004;78:3110–3122. doi: 10.1128/jvi.78.6.3110-3122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L., Bell P., Somanathan S., Wang Q., He Z., Yu H., McMenamin D., Goode T., Calcedo R., Wilson J.M. Comparative study of liver gene transfer with AAV vectors based on natural and engineered AAV capsids. Mol. Ther. 2015;23:1877–1887. doi: 10.1038/mt.2015.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Landegger L.D., Pan B., Askew C., Wassmer S.J., Gluck S.D., Galvin A., Taylor R., Forge A., Stankovic K.M., Holt J.R., Vandenberghe L.H. A synthetic AAV vector enables safe and efficient gene transfer to the mammalian inner ear. Nat. Biotechnol. 2017;35:280–284. doi: 10.1038/nbt.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikeda Y., Sun Z., Ru X., Vandenberghe L.H., Humphreys B.D. Efficient gene transfer to kidney mesenchymal cells using a synthetic adeno-associated viral vector. J. Am. Soc. Nephrol. 2018;29:2287–2297. doi: 10.1681/asn.2018040426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carvalho L.S., Xiao R., Wassmer S.J., Langsdorf A., Zinn E., Pacouret S., Shah S., Comander J.I., Kim L.A., Lim L., Vandenberghe L.H. Synthetic adeno-associated viral vector efficiently targets mouse and nonhuman primate retina in vivo. Hum. Gene Ther. 2018;29:771–784. doi: 10.1089/hum.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hudry E., Andres-Mateos E., Lerner E.P., Volak A., Cohen O., Hyman B.T., Maguire C.A., Vandenberghe L.H. Efficient gene transfer to the central nervous system by single-stranded Anc80L65. Mol. Ther. Methods Clin. Dev. 2018;10:197–209. doi: 10.1016/j.omtm.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu P., Xiao W., Conlon T., Hughes J., Agbandje-McKenna M., Ferkol T., Flotte T., Muzyczka N. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 2000;74:8635–8647. doi: 10.1128/jvi.74.18.8635-8647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lochrie M.A., Tatsuno G.P., Christie B., McDonnell J.W., Zhou S., Surosky R., Pierce G.F., Colosi P. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J. Virol. 2006;80:821–834. doi: 10.1128/jvi.80.2.821-834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo H.H., Choe J., Loeb L.A. Protein tolerance to random amino acid change. Proc. Natl. Acad. Sci. USA. 2004;101:9205–9210. doi: 10.1073/pnas.0403255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmit P.F., Pacouret S., Zinn E., Telford E., Nicolaou F., Broucque F., Andres-Mateos E., Xiao R., Penaud-Budloo M., Bouzelha M., et al. Cross-packaging and capsid mosaic formation in multiplexed AAV libraries. Mol. Ther. Methods Clin. Dev. 2020;17:107–121. doi: 10.1016/j.omtm.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cabanes-Creus M., Navarro R.G., Liao S.H.Y., Baltazar G., Drouyer M., Zhu E., Scott S., Luong C., Wilson L.O.W., Alexander I.E., Lisowski L. Single amino acid insertion allows functional transduction of murine hepatocytes with human liver tropic AAV capsids. Mol. Ther. Methods Clin. Dev. 2021;21:607–620. doi: 10.1016/j.omtm.2021.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lerch T.F., Xie Q., Chapman M.S. The structure of adeno-associated virus serotype 3B (AAV-3B): insights into receptor binding and immune evasion. Virology. 2010;403:26–36. doi: 10.1016/j.virol.2010.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Govindasamy L., Padron E., McKenna R., Muzyczka N., Kaludov N., Chiorini J.A., Agbandje-McKenna M. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J. Virol. 2006;80:11556–11570. doi: 10.1128/jvi.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Govindasamy L., DiMattia M.A., Gurda B.L., Halder S., McKenna R., Chiorini J.A., Muzyczka N., Zolotukhin S., Agbandje-McKenna M. Structural insights into adeno-associated virus serotype 5. J. Virol. 2013;87:11187–11199. doi: 10.1128/jvi.00867-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xie Q., Lerch T.F., Meyer N.L., Chapman M.S. Structure–function analysis of receptor-binding in adeno-associated virus serotype 6 (AAV-6) Virology. 2011;420:10–19. doi: 10.1016/j.virol.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DiMattia M.A., Nam H.-J., Van Vliet K., Mitchell M., Bennett A., Gurda B.L., McKenna R., Olson N.H., Sinkovits R.S., Potter M., et al. Structural insight into the unique properties of adeno-associated virus serotype 9. J. Virol. 2012;86:6947–6958. doi: 10.1128/jvi.07232-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mikals K., Nam H.-J., Van Vliet K., Vandenberghe L.H., Mays L.E., McKenna R., Wilson J.M., Agbandje-McKenna M. The structure of AAVrh32.33, a novel gene delivery vector. J. Struct. Biol. 2014;186:308–317. doi: 10.1016/j.jsb.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Halder S., Van Vliet K., Smith J.K., Duong T.T.P., McKenna R., Wilson J.M., Agbandje-McKenna M. Structure of neurotropic adeno-associated virus AAVrh.8. J. Struct. Biol. 2015;192:21–36. doi: 10.1016/j.jsb.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bryant D.H., Bashir A., Sinai S., Jain N.K., Ogden P.J., Riley P.F., Church G.M., Colwell L.J., Kelsic E.D. Deep diversification of an AAV capsid protein by machine learning. Nat. Biotechnol. 2021;39:691–696. doi: 10.1038/s41587-020-00793-4. [DOI] [PubMed] [Google Scholar]
- 70.Pekrun K., De Alencastro G., Luo Q.J., Liu J., Kim Y., Nygaard S., Galivo F., Zhang F., Song R., Tiffany M.R., et al. Using a barcoded AAV capsid library to select for clinically relevant gene therapy vectors. JCI Insight. 2019;4:131610. doi: 10.1172/jci.insight.131610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li S., Ling C., Zhong L., Li M., Su Q., He R., Tang Q., Greiner D.L., Shultz L.D., Brehm M.A., et al. Efficient and targeted transduction of nonhuman primate liver with systemically delivered optimized AAV3B vectors. Mol. Ther. 2015;23:1867–1876. doi: 10.1038/mt.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bevan A.K., Duque S., Foust K.D., Morales P.R., Braun L., Schmelzer L., Chan C.M., McCrate M., Chicoine L.G., Coley B.D., et al. Systemic gene delivery in large species for targeting spinal cord, brain, and peripheral tissues for pediatric disorders. Mol. Ther. 2011;19:1971–1980. doi: 10.1038/mt.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Foust K.D., Wang X., McGovern V.L., Braun L., Bevan A.K., Haidet A.M., Le T.T., Morales P.R., Rich M.M., Burghes A.H.M., Kaspar B.K. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat. Biotechnol. 2010;28:271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.Kornegay J.N., Li J., Bogan J.R., Bogan D.J., Chen C., Zheng H., Wang B., Qiao C., Howard J.F., Jr., Xiao X. Widespread muscle expression of an AAV9 human mini-dystrophin vector after intravenous injection in neonatal dystrophin-deficient dogs. Mol. Ther. 2010;18:1501–1508. doi: 10.1038/mt.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hinderer C., Katz N., Buza E.L., Dyer C., Goode T., Bell P., Richman L.K., Wilson J.M. Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an adeno-associated virus vector expressing human SMN. Hum. Gene Ther. 2018;29:285–298. doi: 10.1089/hum.2018.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meyer N.L., Hu G., Davulcu O., Xie Q., Noble A.J., Yoshioka C., Gingerich D.S., Trzynka A., David L., Stagg S.M., Chapman M.S. Structure of the gene therapy vector, adeno-associated virus with its cell receptor. Elife. 2019;8 doi: 10.7554/eLife.44707. e44707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang R., Cao L., Cui M., Sun Z., Hu M., Zhang R., Stuart W., Zhao X., Yang Z., Li X., et al. Adeno-associated virus 2 bound to its cellular receptor AAVR. Nat. Microbiol. 2019;4:675–682. doi: 10.1038/s41564-018-0356-7. [DOI] [PubMed] [Google Scholar]
- 78.Dudek A.M., Pillay S., Puschnik A.S., Nagamine C.M., Cheng F., Qiu J., Carette J.E., Vandenberghe L.H. An alternate route for adeno-associated virus (AAV) entry independent of AAV receptor. J. Virol. 2018;92 doi: 10.1128/JVI.02213-17. e02213-e02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pillay S., Meyer N.L., Puschnik A.S., Davulcu O., Diep J., Ishikawa Y., Jae L.T., Wosen J.E., Nagamine C.M., Chapman M.S., Carette J.E. An essential receptor for adeno-associated virus infection. Nature. 2016;530:108–112. doi: 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kasai F., Hirayama N., Ozawa M., Satoh M., Kohara A. HuH-7 reference genome profile: complex karyotype composed of massive loss of heterozygosity. Hum. Cell. 2018;31:261–267. doi: 10.1007/s13577-018-0212-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang L., Calcedo R., Wang H., Bell P., Grant R., Vandenberghe L.H., Sanmiguel J., Morizono H., Batshaw M.L., Wilson J.M. The pleiotropic effects of natural AAV infections on liver-directed gene transfer in macaques. Mol. Ther. 2010;18:126–134. doi: 10.1038/mt.2009.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lock M., Alvira M., Vandenberghe L.H., Samanta A., Toelen J., Debyser Z., Wilson J.M. Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum. Gene Ther. 2010;21:1259–1271. doi: 10.1089/hum.2010.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanmiguel J., Gao G., Vandenberghe L.H. Quantitative and digital droplet-based AAV genome titration. Methods Mol. Biol. 2019;1950:51–83. doi: 10.1007/978-1-4939-9139-6_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw barcode counts corresponding to every sample included in this publication have been deposited into Mendeley and is publicly available as of the date of publication. Additional raw data corresponding to biodistribution studies and imaging studies have also been deposited into Mendeley and is publicly available as of the date of publication.
-
•
All original code has been deposited in Mendeley and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.