Abstract
Acute myeloid leukemia (AML) is a type of leukemia with an aggressive phenotype, that commonly occurs in adults and with disappointing treatment outcomes. Genetic alterations were implicated in the etiology of cancers and form the basis for defining patient prognoses and guiding targeted therapies. In the present study, we leveraged bulk and single-cell RNA sequencing datasets from AML patients to determine the clinical significance of Fms-related receptor tyrosine kinase 3 (FLT3) alterations on the T-cell phenotype and immune response of AML patients. Subsequently, we evaluated the therapeutic potential of Lwk-n019, a novel small-molecule derivative of thiochromeno[2,3-c]quinolin-12-one. Our results suggested that FLT3 plays an important role in the progression, aggressive phenotype, and worse immune response of patients. An FLT3 mutation was associated with dysfunctional T-cell phenotypes, and high risk and shorter survival of AML patients. Our findings further suggested that the aggressiveness of AML and the prognostic role of FLT3 are associated with the co-occurrence of NPM1 and DNMT3A mutations. Lwk-n019 demonstrated dose-dependent anticancer activities against various leukemia cancer cell lines. Lwk-n019 demonstrated highly selective kinase inhibitory activities against the wild-type FLT3 (D835V) and mutant FLT3 (internal tandem duplication (ITD), D835V) with >95% and 99% inhibitory levels, respectively. Moreover, the compound demonstrated the best binding constant (Kd value) of 0.77 µM against FLT3 (ITD, 835V). In addition, Lwk-n019 significantly inhibited the activities of both the topoisomerase I (TOPI) and TOPII enzymes, with higher TOPI inhibitory activity than camptothecin, a clinical inhibitor. While the jejunum, duodenum, cecum, and colon were prime sites of absorption, Lwk-n019 achieved maximum concentration (Cmax), Vd, blood/plasma ratio, time to maximum concentration (Tmax), area under the receiver operating concentration curve (AUC)(0-24), and AUC(0-∞) values of 0.665 µg/mL, 5.21 Vc, L/kg, 1.5 h, 6634.7, and 6909.2, respectively. In conclusion, Lwk-n019 demonstrated anticancer activities via multi-target inhibition of TOPs and kinases with high inhibition preference for mutant ITD-FLT3. The present pioneer study provides a basis for advanced optimization of drug potency, selectivity, specificity, and other properties desired of anticancer drug leads. Studies are ongoing to determine the full therapeutic properties of Lwk-n019 and the detailed mechanisms of FLT3 in TOP inhibition.
Keywords: Acute myeloid leukemia, topoisomerase, immune infiltration, FLT3, Lwk-n019
Introduction
Leukemia is the most prevalent cancer in children and accounts for about 28% of childhood cancers, followed by cancers of the brain and other nervous system cancers which account for 27% of cases [1]. However, more than 25% of these malignancies are borderline/benign. In adolescents, the types and distribution of these cancers differ from those found in children [2]; for instance, in cancers of the brain/nervous system, more than 50% of borderline/benign malignancies exhibit the highest prevalence (21%), followed by lymphomas (19%) [1]. In addition, non-Hodgkin lymphomas exhibit almost twice the prevalence in adolescents, compared to children [1,3].
Acute myeloid leukemia (AML) is a type of leukemia that develops quickly and has an aggressive phenotype; it is characteristically fatal within weeks or months if left untreated. It is one of the most prevalent leukemias among adults but is rarely diagnosed in people under 40 years of age. In 2021, AML affected about 20,240 people, and resulted in 11,400 deaths (1.9% of all cancer case) globally [4]. Females are less affected than males, and the 5-year survival rate is disappointing (35%) in patients below 60 years of age and more disappointing (10%) in older patients (>60 years) [5]. In fact older people with a poor health status exhibited disappointing survival of about 5-10 months [5]. The underlying mechanism involves displacement of normal bone marrow by leukemic cells, which results in decreased platelet, white blood cell (WBC), and red blood cell (RBC) counts.
Adequate use of antileukemic drugs coupled with rigorous use of prognostic factors for risk-based therapy in clinical trials has ensured stable treatment outcomes in younger patients [6,7]. However, treatment outcomes in older patients have been far less satisfactory with <40% cure rates [7,8], despite the use of various therapeutic strategies including hematopoietic stem-cell (HSC) transplantation [8-12]. The poor prognoses of adult patients were associated with increased rates of high-risk leukemia with increased therapeutic resistance, poorer tolerance of side effects, less-effective therapeutic regimens, and poorer tolerance and compliance to treatment compared to younger patients [8].
Genetic alterations were implicated in the development and progression of cancers and form the basis for defining patient prognoses and guiding targeted therapy [13]. The identification of high expression levels of Fms-related receptor tyrosine kinase 3 (FLT3) in AML spurred the development of FLT3 inhibitors [14]. However, there is a need to identify and develop new targeted therapies to address worse outcomes of patients.
FLT3 is a cytokine receptor that plays a vital role in normal hematopoiesis [15]. FLT3 mutations have high prevalence rates in AML, and the World Health Organization recommends FLT3 mutation screening of AML patients, particularly in cytogenetically normal (CN)-AML [15,16]. FLT3 mutations often involve internal tandem duplications (ITDs) of AA within the juxtamembrane region of the receptor resulting in the constitutive activation of tyrosine kinase [17]. The co-occurrence of FLT3 mutations with other gene mutations, such as nucleophosmin (NPM1), has been widely observed and is associated with unfavorable prognoses of CN-AML patients [18]. Among FLT3 mutations, the worse prognosis of patients is attributed to the presence of FLT3-ITD mutations, while the prognostic impacts of FLT3-tyrosine kinase domain (TKD) mutations remain elusive [15,19].
Topoisomerases (TOPs) are essential regulators of DNA topology that ensure adequate DNA replication, recombination, transcription, repair, and other vital cellular processes [20]. There are two classes of TOPs: type I (TOPI) enzymes function to prevent excess negative supercoiling [21,22], while type II (TOPII) enzymes cut and rejoin DNA double strands during catalysis [22]. Type I TOPs are divided into type IA (Top3a and Top3b) and type IB (Top1 and Top1mt), while type II TOPs consist solely of Top2a and Top2b [23]. Due to the essential nature of DNA TOPs in replication, cell division, and cell-cycle progression, several inhibitors of TOPs have been developed into important anticancer drugs. However, their efficacy comes with significant side effects including febrile neutropenia, sepsis, infection, abdominal pain, alopecia, anemia, asthenia, constipation, dyspnea, fatigue, fever, headache, leukopenia, nausea, neutropenia, stomatitis, thrombocytopenia, vomiting, granulocytopenia, diarrhea, and anorexia [24]. In this study, Lwk-n019, a novel thiochromeno[2,3-c]quinolin-12-one derivative was explored as having anti-leukemia activities via multi-target inhibition of TOPs and kinases with a high inhibitory preference for mutant FLT3.
Materials and methods
Subcellular localization and RNA expression of FLT3 in various cell lines
We evaluated the expression profile of FLT3 using Human Protein Atlas (HPA) RNA-sequencing (RNA-Seq) data from a total of 69 cell lines, 52 human tissues, and 18 blood cell types as well as total peripheral blood mononuclear cells (PBMCs). RNA expression data as normalized transcript per million (nTPM) expression values were visualized using a bar graph. We characterized the intracellular localization of FLT3 via indirect immunofluorescence staining of REH cells with the HPA047539 antibody from the HPA database. Accordingly, cells were stained with reference markers: an anti-tubulin antibody for microtubules, DAPI for nuclei, and an anti-calreticulin antibody for endoplasmic reticula (ERs). Stained marker cells were merged with the FLT3 antibody, and co-localized subcellular sections were mapped and captured.
FLT3 genetic alterations and their effects on AML prognoses
We explored the CBioPortal server for pan-cancer AML datasets consisting of 200 patients with AML. Using patients and samples from these datasets, we evaluated the frequency of genetic alterations, mutation profiles, the co-occurrence of genetic alterations, and the prognostic role of FLT3. Patients were divided into two groups based on the presence or absence of FLT3 alterations, and overall survival was visualized using a Kaplan-Meir plot.
FLT3’s association with the immune response against AML
We used the TIDE tool to decipher correlations and roles of FLT3 expression and mutations on levels of cytotoxic lymphocytes (CTLs) and dysfunctional T-cell phenotypes, and the risk of death due to AML. Dysfunctional T-cell phenotypes were defined as the presence of high amounts of inactive CTLs that induced a poor immune response in patients. Dysfunctional T cells were represented by Z scores, where positive and negative Z score respectively indicated dysfunctional and active T-cell states.
Leveraging single-cell (sc) transcriptomic datasets for FLT3 profiling
The scRNA-Seq dataset analytical tool, CancerSEA, was used to decode distinct functional states of leukemic tumor cells at a single-cell resolution [25]. We also analyzed a single-cell sequencing dataset (GSE76312) from patients with chronic myeloid leukemia (CML) for FLT3 expression across each single cell. We also evaluated the functional relevance of the gene by correlating single-cell FLT3 expression levels with the stemness level and metastasis of patients. Furthermore, we used the Tumor Immune Single-Cell Hub (TISCH) [26], to characterize FLT3 expression in relation to the tumor immune microenvironment of AML patients (AML_GSE116256) at a single-cell resolution.
Reagents, drugs, and chemicals
A 10 mM stock solution of Lwk-n019 was prepared in dimethyl sulfoxide (DMSO) and kept at -20°C. Roswell Park Memorial Institute (RPMI) medium was procured from ThermoFisher Scientific (Waltham, MA, USA). TOPI and TOPII, supercoiled pRYG DNA, supercoiled pHOT1 DNA, camptothecin, and VP-16 were obtained from TopoGEN. Phosphate-buffered saline (PBS), sulforhodamine B (SRB), DMSO, acetic acid, fetal bovine serum (FBS), TRIS base, and organic solvents used for the synthesis of Lwk-n019 were procured from Sigma Aldrich (St. Louis, MO, USA).
Cell lines and culture
Human leukemia cell lines, including CCRF-CEM, HL-60(TB), K-562, MOLT-4, RPMI-8226, and SR, were acquired from the US National Cancer Institute, while TFI and kasumi1 were obtained from American Type Culture Collection (ATCC. Manassas, VA, USA). Cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with penicillin (25 U/mL), streptomycin (25 U/mL), and 10% FBS and incubated in 95% humidity at 37°C in a 5% CO2 condition. Cells were re-suspended in fresh medium after 44-72 h and subcultured at 75%-85% confluence.
In vitro antiproliferative assay
In vitro antiproliferative activities of Lwk-n019 against the various leukemia cell lines were evaluated using the SRB reagent protocol [27]. About 5,000-40,000 viable cells were seeded in each well of 96-well plates for 24 h. After 24 h of incubation, the medium was replaced, and cells were treated with Lwk-n019 at different concentrations of 0.1-10 μM for 48 h. After incubation, cells were washed with 1% PBS and incubated with 10% trichloroacetic acid (TCA) at room temperature for 1 h [28]. Plates were washed with double-distilled (dd) H2O, and further incubated with 0.4% SRB for 60 min. Plates were washed with 1% acetic acid to remove any unbound SRB dye. The plates were air-dried, and cells were re-dissolved in a 20 mM Tris-based solution for 15 min under constant agitation. Cell viabilities with different Lwk-n019 treatments were monitored at 515 nm. The 50% maximal inhibitory concentrations (IC50) was calculated as described previously [29,30].
Kinase inhibition assays (KINOMEscan™)
KINOMEscan™ Profiling was conducted by LeadHunter™ Discovery Services (Discove Rx) against a panel of 468 kinases. The detailed protocol is provided in the Supplementary File. Lwkn-019 was screened at a concentration of 10 µM, and primary screen binding interactions are reported as the % Ctrl, where lower numbers indicate stronger hits in the matrix: % Ctrl = Lwk-n019 signal-positive control signal; Negative control signal - positive control signal.
Where the negative control was DMSO (100% Ctrl), and the positive control was the control compound (0% Ctrl).
TOPI and TOPII DNA assays
TOP1 and TOPII DNA activities were analyzed using TOP assay kits (TopoGEN, USA) as described in previous studies [31-33]. In the TOPI assay, 4 units of recombinant human DNA TOPI was incubated with 0.5 μg of plasmid pHOT DNA in relaxation buffer (10 mM Tris-HCl (pH 7.9), 0.15 M NaCl, 0.1% bovine serum albumin (BSA), 1 mM EDTA, 5% glycerol, and 0.1 mM spermidine). For TOPII activity, 4 units of human TOPII was incubated with 0.5 μg of supercoiled pRYG DNA in cleavage buffer (30 mM Tris-HCl (pH 7.8), 50 mM KCl, 10 mM MgCl2, 3 mM ATP, and 15 mM mercaptoethanol). Lwk-n019 at varying concentrations (1-50 µM) was incubated with the reaction mixture at 37°C for 1 h followed by the addition of proteinase K (50 μg/mL) and 1% sodium dodecylsulfate (SDS), and subjected to 0.8% agarose gel electrophoresis. Gels were stained with ethidium bromide and photographed under ultraviolet (UV) light. Camptothecin at 50 μM was used as a reference inhibitor for the TOP1 enzyme, while etoposide (VP-16) (50 μM) was used as the positive control for the TOPII enzyme.
In vivo pharmacokinetic (PK) and regional gastrointestinal tract (GIT) absorption analysis of Lwk-n019 in rats
To assess the PK properties of Lwk-n019, male rats were administered 5 mg/kg body weight (BW) of Lwk-n019, and blood samples were collected in Na2EDTA vaporized tubes at 1-h intervals for 24 h. Blood was centrifuged, and plasma was separated as described in a previous studies [34,35]. Plasma samples were quantified for Lwk-n019 using a previously validated ultra-precision liquid chromatographic (UPLC) method [36]. The drug concentration-time profile and various PK parameters, including the area under the receiver operating characteristic curve from 0 to 24 h (AUC0→24), AUC from 0 to infinity (AUC0→∞), maximum concentration in serum (Cmax), and duration (time) needed to achieve Cmax (Tmax), were estimated using GastroPlus™ software (vers. 9.0, Simulation Plus, Lancaster, PA, USA). In addition, regional drug absorption characteristics from nine GIT compartments, including the cecum, colon, stomach, jejunum-1, jejunum-2, duodenum, ileum-1, ileum-2, and ileum-3, were determined.
Molecular docking analysis
The three-dimensional (3D) crystal structures of the protein targets of TOP1 (PDB: 1AB4), TOPII (PDB: 1EJ9), and FLT3 were downloaded from the PDB server and processed to generate PDBQT files of the proteins via AutoDock Vina software (vers. 0.8) [37]. The MOL2 file of the ligand (Lwk-0n19) in it 3D shape was obtained through the Avogadro molecular builder tool [38], and processed to PEBQT format via stepwise preparation using the PyMOL and AutoDock tools. Further preparation of the protein with respect to H2, H2O, and charge placement were conducted via the AutoDock Vina tool as described in previous studies [39,40]. Protein-ligand docking was conducted using default settings of AutoDock Vina as described previously [30]. Docked complexes were visualized and analyzed for various interactions using the PyMOL and Discovery studio tools [41], and an online protein-ligand interaction profiler (PLIP) program.
Statistical data analysis
Analyses were carried out with various replicates, and data were uploaded and analyzed via GraphPad Prism vers. 8.0. Data are presented as the mean ± standard deviation (SD). Student’s t-test was used for statistical comparisons between different groups/treatment doses. Statistical significance was considered at and represented as * P<0.05, ** P<0.001 and *** P<0.001.
Results
Expression and prognosis of FLT3 in AML patients
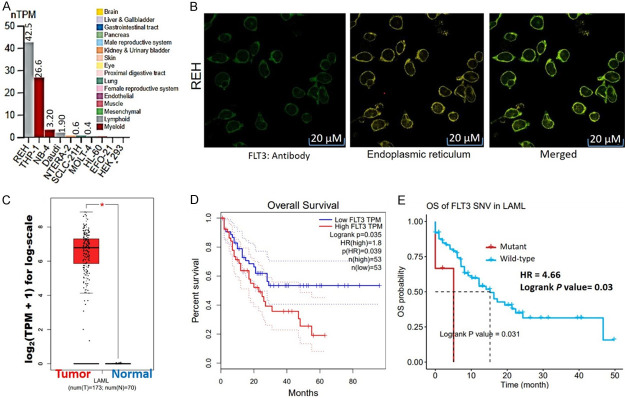
We evaluated the expression profile of FLT3 using HPA RNA-Seq data. Our results revealed that out of 69 cell lines, FLT was mainly expressed in myeloid and lymphoid cells with the highest transcript levels in the REH (42.5%) and THP-1 cell lines (26.6%), while the NB-4, Daudi, NTERA-2, SCLC-21H, MOLT-4, HL-60, EFO-21, and HEK293 cell lines showed very low expression levels (Figure 1A). Furthermore, we queried the subcellular localization of the gene using the cell line (REH) with the greatest expression and found that the gene was mainly localized to the ER (Figure 1B). The differential expression levels of FLT3 in AML patients revealed that FLT3 was highly overexpressed in AML tissues, compared to matched TCGA normal tissues (Figure 1C). In addition, AML patients with higher expression levels and mutant variants of FLT3 exhibited shorter survival (P<0.05; and hazard ratios (HR = 1.8 and 4.66)) compared to cohorts with low expression (Figure 1D) and the wild-type (WT) variant of FLT3 (Figure 1E).
Figure 1.
A. Bar graph of mRNA expression of FLT3 across various cell lines as normalized transcript per million (nTPM) expression values. B. Subcellular localization of FLT3 based on indirect immunofluorescence staining of REH cells with the HPA047539 antibody from the Human Protein Atlas (HPA) database. C. Bar graph of differential expression profiles of FLT3 between tumor samples from acute myeloid leukemia (AML) patients and normal tissues. D. Kaplan-Meir plot of overall survival between AML patients with higher vs. lower expression levels of FLT3. E. Kaplan-Meir plot of overall survival between AML patients with mutant vs. wide-type variants of FLT3.
FLT3 genetic alterations occur most frequently in AML patients and are associated with worse prognoses
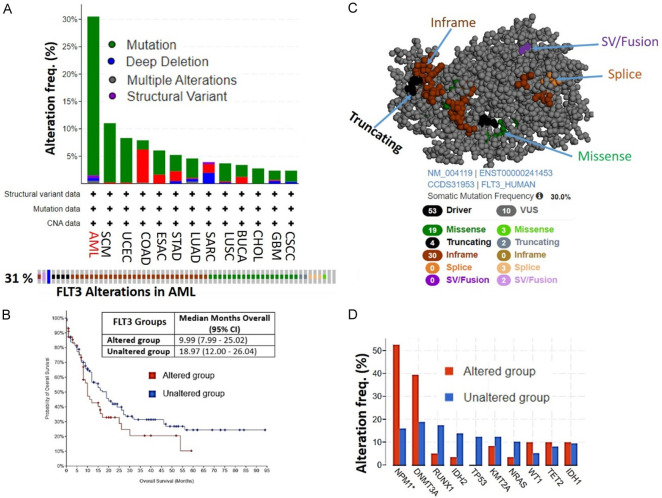
Our analysis of FLT3 genetic alterations revealed that FLT3 was mostly altered in AML compared to other TCGA cancer types. Out of a total of 200 AML patients/samples, FLT3 was altered in 61 (31%) of queried patients/samples. The frequency of FLT3 alterations was greater than three-times higher in AML compared to other cancer types. Genetic mutations (29%) were the most frequent type of genetic alteration of FLT3, while structural variants (0.5%), deep deletions (0.5%), and multiple alterations (0.5%) occurred at lower frequencies (Figure 2A). Consequently, AML patients with an FLT3 genetic alteration exhibited shorter survival periods (9.99 months; 95% CI) compared to AML patients with no FLT3 alterations (18.97 months; 95% CI) (Figure 2B). Among the mutations, in-frame and missense mutations were the most frequent somatic mutations of FLT3 in AML patients (Figure 2C). These mutations of FLT3 were associated with mutation co-occurrences with other genes including NPM1 (>50%) and DNMT3A (40%) (Figure 2D).
Figure 2.
FLT3 genetic alterations occur most frequently in acute myeloid leukemia (AML) patients and are associated with worse prognoses. A. Bar plot of FLT3 genetic alterations across different cancer types (upper panel). The lower panel shows individual types of FLT3 genetic alterations in 31% of AML cases in TCGA database. B. Kaplan-Meir plot of survival differences between AML patients with FLT3 alterations vs. patients with wild-type FLT3. C. Three-dimensional view of FLT3 showing specific mutations in AML patients. D. Frequencies of the co-occurrence of FLT3 genetic alteration with other genes in AML patients in TCGA.
FLT3 mutations are associated with dysfunctional T-cell phenotypes, high risk, and shorter survival of AML patients
Having observed that FLT3 is frequently mutated in leukemia patients, we evaluated the significance of these mutations in the immune response of patients via analyzing levels of CTLs and their functional phenotypes in AML cohorts (Figure 3A-D). Interestingly, we found that FLT3 mutations were inversely associated with levels of CTLs (r = -0.376, P = 0.003) and mediated T-cell dysfunctional phenotypes (Z score = 0.798) where high levels of CTLs in FLT3 mutant-bearing patients were inactive and induced a low immune response, while high CTL levels in FLT3 WT-bearing patients induced a good immune response (Figure 3C). Furthermore, these FLT3 mutant-bearing patients exhibited a significantly higher death risk (Z score = 2.22, P = 0.0026) than FLT3 WT-bearing patients. Therefore, we summarized that FLT3 mutations are associated with worse immune responses, dysfunctional T-cell phenotypes, high risk, and shorter survival of AML patients.
Figure 3.
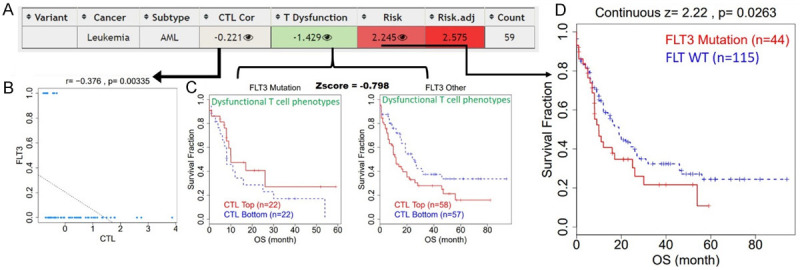
FLT3 mutations are associated with dysfunctional T-cell phenotypes, high risk, and shorter survival of acute myeloid leukemia (AML) patients. A. Summary plot of immune responses of FLT3 mutant-bearing AML patients. B. Scatterplot of the correlation between FLT3 mutations and levels of active cytotoxic T cells. C. Plot of dysfunctional T-cell phenotypes showing that mutant FLT3 expression induced a state of dysfunctional T cells. D. Kaplan-Meir plot of differences in immune responses between FLT3 mutant-bearing vs. FLT3-wild-type (WT)-bearing AML patients.
Single-cell transcriptomic datasets revealed the functional immune-pathological role of FLT3 in leukemia
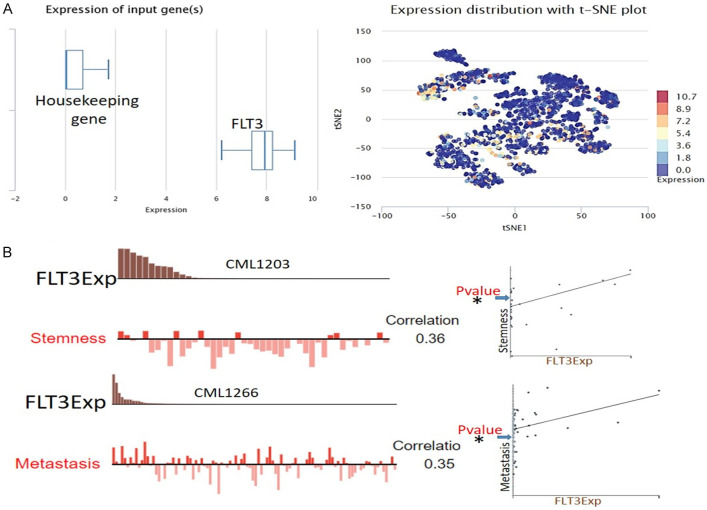
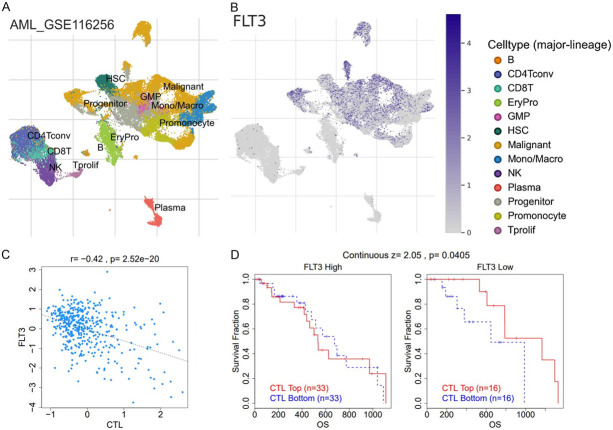
We analyzed a single-cell sequencing dataset (GSE76312) from patients with chronic myeloid leukemia (CML) and found that FLT3 was highly overexpressed and widely distributed in each single cell in the tumor microenvironment (TME) of patients (Figure 4A). We also evaluated the functional relevance in different cell groups and found that single-cell expression levels of FLT3 showed positive correlations with stemness (r = 0.36, P<0.05) and metastasis (r = 0.35, P<0.05) of patients with chronic leukemia (Figure 4B). We also queried FLT3 expression within the tumor immune microenvironment using a single-cell sequencing dataset (AML_GSE116256) consisting of 21 patients with primary AML. Using the single-cell sequencing dataset of 21 patients, 38,348 cells in total comprising B cells, cluster of differentiation (CD)4 T cells, CD8 T cells, natural killer (NK) cells, monocytes/macrophages, progenitors, granulocyte-monocyte progenitors (GMPs), hematopoietic stem cells (HSCs), plasma, and malignant cells were obtained (Figure 5A). Interestingly, we found that FLT3 was highly expressed in monocytes/macrophages, promonocytes, GMPs, progenitor cells, and malignant cells (Figure 5B). In contrast, we found that FLT3 was not expressed by CD4 T cells, CD8 T cells, NK cells, or proliferative T cells. This suggests that FLT3 expression within the TME is associated with the exclusion of active cytotoxic cells. Coherently, we analyzed the relationship between bulk FLT3 expression and levels of active cytotoxic lymphocytes and their prognostic role in AML patients. Our results revealed that FLT3 expression levels were negatively associated (r = 0.42, P = 2.52 × 10-20) with the level of active CTLs (Figure 5C), and induced a state of dysfunctional T-cell phenotypes (Z score = 2.05, P = 0.00405) and worse prognoses of AML patients (Figure 5D).
Figure 4.
Single-cell sequencing profile of the expression and functional role of FLT3 in leukemia. A. The box diagram indicates the expression distribution of FLT3 in cells of the datasets. B. T-SNE describes the distribution of cells, every point represents a single cell, and the color of the point represents the expression level of the gene in the cell.
Figure 5.
Single-cell sequencing profile of FLT3 expression in the tumor immune microenvironment of leukemia patients. A. T-Distributed stochastic neighbor embedding plot of single-cell RNA-sequencing data of immune and malignant cells in leukemia patients. Identified clusters are represented by different colors. B. Expression distributions of FLT3 in different cell types. C. Scatterplot of the correlations between the expression levels of FLT3 and levels of active cytotoxic T cells. D. Plot of dysfunctional T-cell phenotypes showing that high FLT3 expression.
Overview of Lwkn-019, a novel anti-leukemia small molecule
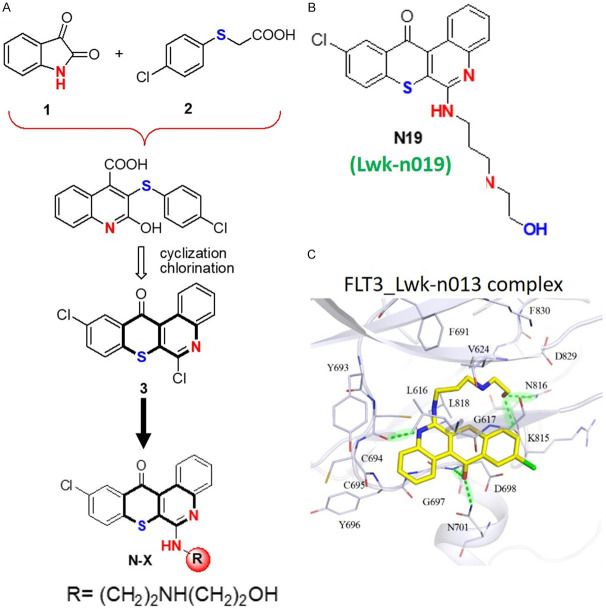
We report the biological activity of a novel anti-leukemia small molecule, Lwk-n019 (10-chloro-6-(4-(3-(piperidin-4-yl)propyl)piperidin-1-yl)-12H-thiochromeno[2,3-c]quinolin-12-one), via multitarget inhibition of TOPs and kinases with a high inhibitory preference for mutant FLT3. The synthesis of Lwk-n019 followed a three-process protocol strategy (Figure 6A) described in our previous studies and patent [42]. The strategy entailed a three-process protocol. Cyclization and chlorination of isatin 1 with p-chlorophenylthioacetic acid 2 by Pfitzinger reaction gave key intermediate 3. Coupling and amination of 3 with appropriate primary amines by fragment-based drug discovery (FBDD) furnished Lwk-n019 (Figure 6B). Subsequently, we conducted a molecular docking simulation to gain insights into the potential binding affinity of Lwk-n019 with FLT3 and found strong interactions within the Lwk-n019-FLT3 complex (Figure 6C). The ring and chain groups of Lwk-n019 respectively formed three and two hydrogen bonds with FLT3 residues. The ring group also produced stable van der Waals interactions with the binding site of the target.
Figure 6.
Overview of Lwkn-019 (N19), a novel anti-leukemia small molecule. A. Overview of the synthesis of Lwkn019 (10-chloro-6-(4-(3-(piperidin-4-yl)propyl)piperidin-1-yl)-12H-thiochromeno[2,3-c]quinolin-12-one) [42]. B. Chemical structure of Lwk-n019. C. Binding conformation of N19 with FLT3. The ring and chain groups of Lwkn-09 respectively formed three and two hydrogen bonds with FLT3 residues. Hydrogen bonds between residues and Lwkn-09 are represented as light-green dashes. The ring group also produced stable van der Waals interactions with the binding target.
Lwk-n019 demonstrated in vitro antiproliferative properties against leukemia
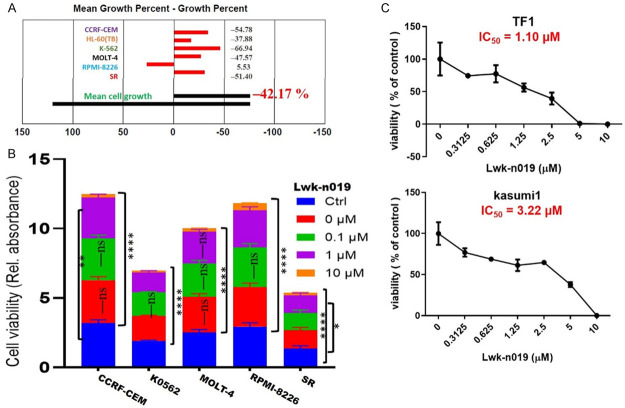
We evaluated the antiproliferative activities of Lwk-n019 against six human cell line subsets of leukemia (Figure 7). Analysis of the anticancer profile of Lwk-n019 with a single-dose screening revealed that the compound exhibited cytotoxic profiles against four NCI cell lines including CCRF-CEM, K0562, MOLT-4, and SR, while demonstrating antiproliferative effects against RPMI-8226 (Figure 7A). The compound demonstrated dose-dependent anticancer activities against the NCI-60 cell line subset of leukemia including CCRF-CEM (IC50 = 1.83 µM), K0562 (IC50 = 1.73 µM), MOLT-4 (IC50 = 1.88 µM), RPMI-8226 (IC50 = 1.84 µM), and SR (IC50 = 1.98 µM) (Figure 7B). Furthermore, our cell viability assays revealed that the drug also exhibited dose-related antiproliferative activities against other leukemia cell lines including the TF1, and kasumi 1 cell lines with respective IC50 values of 1.10 and 3.22 µM (Figure 7C). Altogether, our study points out the potential role of Lwk-n019 in treating leukemia.
Figure 7.
Anticancer activities of Lwk-n019 against leukemia cancer cell lines. High-throughput (A) single-dose activity profiling and (B) dose-dependent anticancer activities of Lwk-n019 against the NCI-60 cell line subsets of human leukemia. (C) Antiproliferative effect of Lwk-n019 against non-NCI-60 leukemia cell lines. * P<0.05, ** P<0.001, **** P<0.0001.
Kinases and TOPs are mechanistic target fingerprints for Lwk-n019
Based on the in vitro anticancer activities of Lwk-n019 against a panel of well-characterized NCI cell lines, the anticancer mechanistic fingerprint of the compound was identified revealing correlations with several anti-leukemia agents, kinase (FLT3, RET, phosphatidylinositol 3-kinase (PI3K), and tyrosine kinase (TK)) inhibitors, and TOP inhibitors (Table 1). These findings hinted at the biological processes and potential mechanistic targets of Lwk-n019 for experimental validation.
Table 1.
Anticancer fingerprints of Lwk-n019 based on NCI-investigated drugs
| Correlation | CCLC | Target NSC | Target descriptor | |
|---|---|---|---|---|
| 0.42 | 57 | 818434 | LOXO-292 | RET inhibitor |
| 0.42 | 57 | 759224 | IDELALISIB | PI3K inhibitor |
| 0.4 | 57 | 811429 | CT-BLU667 | RET inhibitor |
| 0.39 | 57 | 789102 | IVOSIDENIB | Anti-leukemia |
| 0.39 | 57 | 775772 | GLASDEGIB | Anti-leukemia |
| 0.38 | 58 | 772469 | INK-1197 | PI3K |
| 0.36 | 56 | 760766 | VANDETANIB | TKI |
| 0.35 | 58 | 789300 | Pexidartinib (PLX3397) | FLT3 inhibitor |
| 0.34 | 58 | 141540 | VP-16 (etoposide) | Top II |
| 0.12 | 58 | 125066 | bleomycin | Anti-leukemia |
| 0.12 | 57 | 122819 | VM-26 (teniposide) | TOP II |
| 0.12 | 57 | 609699 | topotecan | TOP1 |
CCLC, Common Cell Line Count; Correlation, Pearson’s Correlation Coefficient.
Lwk-n019 demonstrated potent inhibitory effects against TOPs
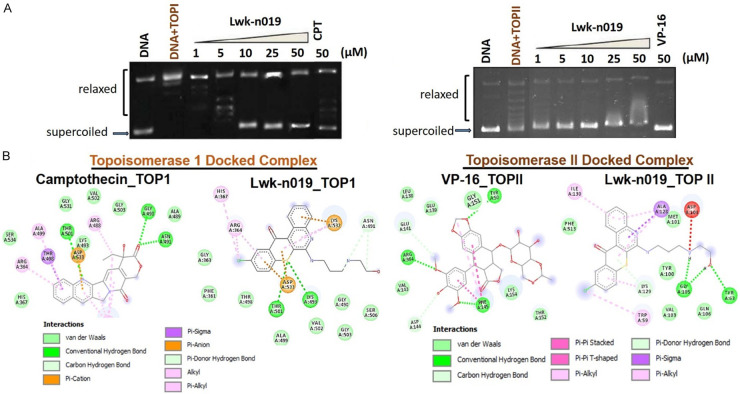
We assessed the activities of Lwk-019 against TOPI and TOPII and found that treatment with Lwk-n019 significantly inhibited the activities of both TOPI and TOPII (Figure 8A) enzymes. Lwk-n019 demonstrated higher TOPI inhibitory activity than camptothecin, a clinical TOP1 inhibitor (Figure 8A), at the same concentration of 50 µM. A molecular docking study showed that Lwk-0n19 docked well to the binding domains of TOPI and TOPII with respective binding efficacies of -6.7 and -7.6 kcal/mol, while standard drugs (camptothecin and VP-16) showed respective binding affinities of -8.8 and -7.4 kcal/mol against TOPI and TOPII enzymes (Figure 8B). Several non-covalent interactions including H-bonds, alkyl interactions, pi-interactions, and van der Waal forces were observed between the Lwk-n019-TOPI/II complexes (Figure 8B). Summing up the above data, it was confirmed that Lwk-n019 is a potent and non-selective inhibitor of TOPI and TOPII enzymes.
Figure 8.
Topoisomerase (TOP) inhibitory activities of Lwk-n019. Effect of Lwk-n019 on (A) DNA TOPI and TOPII activities. (B) Molecular docking simulation of Lwk-n019 interactions with TOP1 and TOPII.
Biochemical kinase profile of Lwk-n019 against a large panel of kinases
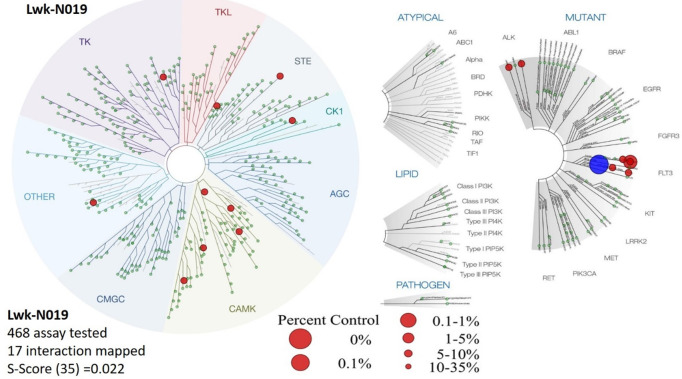
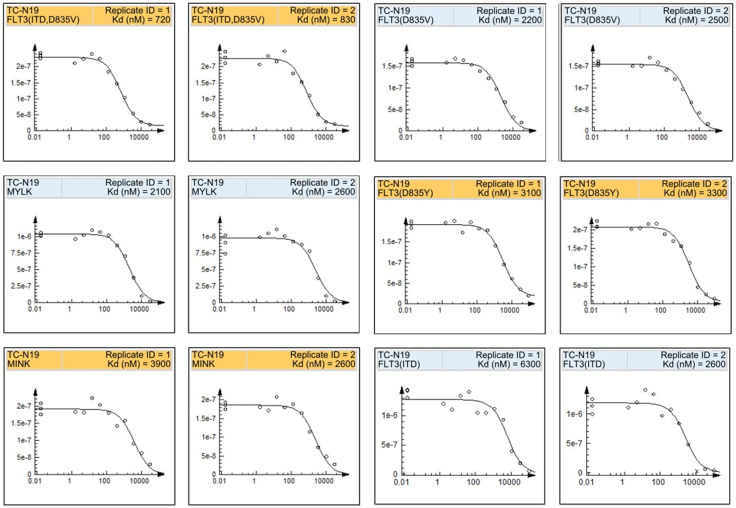
We evaluated the in vitro biochemical activities of Lwk-n019 against a large panel of 468 human kinases (Figure 9; Table 2). Among the large panel, only 16 wild-type (WT) and mutated kinases exhibited ≤35 activities after Lwk-n019 treatment (Figure 9; Table 2). The compound also exhibited a selectivity score (S30) of 0.022. Interestingly, the drug exhibited highly selective kinase-inhibitory activities against WT FLT3 (D835V) and mutant FLT3 (ITD, D835V). These two kinases were respectively inhibited by >95% (3.2% remaining kinase) and 99% (0.85% remaining kinase) by 10 µmol of Lwk-n019. More-over, the compound demonstrated the best binding constant (Kd) of 0.77 µM (770 nM) against FLT3 (ITD, 835V). The plots for the dissociation constants for Lwk-n019-kinase interactions for the top six hits are displayed in Figure 10.
Figure 9.
Kinome-wide inhibition profile of Lwk-n019 at 10 µmol. Kinases showing ≤35 activities after Lwk-n019 treatment were mapped on a kinome tree. The sizes of the dots indicate the remaining activity of each kinase after inhibition. TREEspot™ was used to visualize interaction maps, and red circles indicate bound kinases. Larger circle sizes indicate higher binding affinities. S-score, selectivity score.
Table 2.
In vitro inhibitory profiling of Lwk-n019 against human kinases
| Target | Lwk-n019 | Target | Lwk-n019 |
|---|---|---|---|
|
| |||
| Gene Symbol | % Ctrl | Gene symbol | Kd (µM) |
| ALK | 29 | ALK | 7.6 |
| ALK (C1156Y) | 31 | ALK (C1156Y) | 4.7 |
| ALK (L1196M) | 70 | ALK (L1196M) | 9.3 |
| CSNK1A1L | 26 | CSNK1A1L | 6.2 |
| CSNK2A1 | 25 | CSNK2A1 | 9.9 |
| FLT3 | 74 | FLT3 | 12 |
| FLT3 (D835H) | 32 | FLT3 (D835H) | 8.4 |
| FLT3 (N841I) | 57 | FLT3 (N841l) | 14 |
| FLT3 (D835V) | 3.2 | FLT3 (D835V) | 2.3 |
| FLT3 (D835Y) | 18 | FLT3 (D835Y) | 3.2 |
| FLT3 (ITD) | 12 | FLT3 (ITD) | 4.5 |
| FLT3 (ITD, D835V) | 0.85 | FLT3 (ITD, D835V) | 0.77 |
| FLT3 (ITD, F691L) | 25 | FLT3 (ITD, F691L) | 6.3 |
| MINK | 15 | MINK | 3.3 |
| MKNK2 | 31 | MKNK2 | 4.6 |
| MYLK | 17 | MYLK | 2.4 |
| STK33 | 31 | STK33 | 7.2 |
| PAK3 | 35 | ||
| PIM3 | 34 | ||
| ARK5 | 33 | ||
Lwk-N019 was screened at 10 µM. % Ctrl, results of primary screen binding interactions where lower numbers indicate stronger hits in the matrix. Kd, dissociation constant for Lwk-n019-kinase interactions.
Figure 10.
Representative plot of the dissociation constants for Lwk-n019-kinase interactions for the top six hits. The kinase activity based on the qPCR signal (y-axis) is plotted against the log10 concentration of Lwk-n019 (x-axis).
In vivo PK properties of Lwk-n019
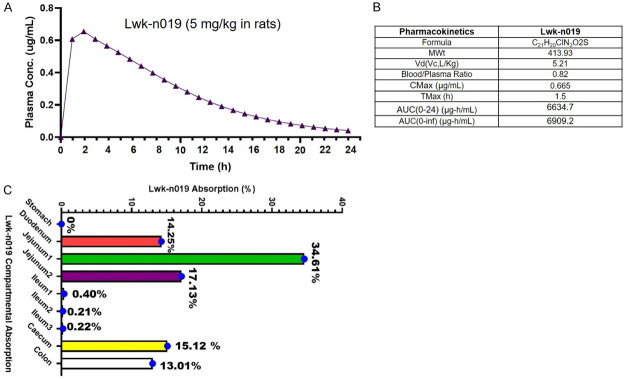
We evaluated the PK properties of Lwk-n019 using an in vivo animal model. Interestingly, plasma Lwk-n019 concentrations following i.v. administration peaked at 0.608 and 0.655 µg/mL after 1 and 2 h, respectively. After that, a progressive decrease in the plasma concentration with increased time was observed. The least drug concentrations of 0.00409 µg/mL was obtained at 24 h (Figure 11A). Lwk-n019 achieved Cmax, Vd, blood/plasma ratio, Tmax, AUC0-24, and AUC0-∞ values of 0.665 µg/mL, 5.21 Vc, L/kg, 1.5 h, 6634.7, and 6909.2, respectively (Figure 11B). Values of the percent absorption from the various regional segments are illustrated in Figure 11C. The duodenum (14.25%), jejunum 1 (34.61%), jejunum 2 (17.13%), cecum (15.12%), and colon (13.01%) were prime sites of Lwk-n019 absorption. The least absorption occurred in the ileum 1 (0.40%), ileum 2 (0.21 %), and ileum 3 (0.22%), and no absorption was detected in the stomach (Figure 11C).
Figure 11.
Pharmacokinetic parameters of orally administered Lwk-n019. A. Time vs. plasma concentrations of Lwk-n019 after injection into a rat. B. Pharmacokinetic parameters of Lwk-n019. C. Compartmental absorption of Lwk-n019. Cmax, maximum concentration in plasma; Tmax, time to reach Cmax; AUC, area under the receiver operating characteristic plasma concentration-time curve.
Discussion
Our analysis of bulk and single-cell RNA sequencing data of the AML TME revealed that FLT3 was expressed at very high levels in AML patients and was associated with a high risk of death, shorter survival durations, and worse prognoses of patients, i.e., AML patients with higher expression levels and mutant variants of FLT3 exhibited shorter survival compared to counterparts with low expression and the WT variant of FLT3. In line with our observations, a previous study indicated that FLT3 is involved in the normal development of stem cells and the immune system [17], and hence plays important roles in the progression, aggressive phenotypes, and worse immune responses of patients. FLT3 exhibited its oncogenic role via synergizing with other oncogenes and growth factors to stimulate proliferation of stem, dendritic, progenitor, and NK cells, and conferred an aggressive tumor phenotype [17]. Altogether, the present study revealed that FLT3 is an oncogenic driver and represents an important biomarker for the diagnosis and prognosis of AML. Our analysis of FLT3 genetic alterations revealed that approx. 31% of AML patients harbored genetic alterations of FLT3, out of which genetic mutations (29%) constituted the greatest proportion. These findings agreed well with a study by Gilliland and Griffin [17] which reported that 30% of AML patients had mutations of FLT3. Activation of mutations in FLT3 was also reported in 25% of acute lymphoid leukemia patients [43]. Several clinical and retrospective studies indicated that FLT3 mutations are independent variable that confer a poor prognosis in AML [44-47]. However, in studies involving only elderly patients with AML, it was found that FLT3 mutations did not confer worse prognoses compared to patients with WT FLT3 [48,49]. It is therefore reasonable to assume that because the overall prognosis in elderly AML patients is generally worse [17] irrespective of mutations, FLT3-ITD is not as important a prognostic marker in that patient group as it is in younger patients. Our analysis further revealed that more than 50% and about 40% of AML patients with FLT3 mutations also respectively exhibited NPM1 and DNMT3A mutations. The co-occurrence of FLT3 mutations with other gene mutations such as NPM1 has been widely observed and is associated with unfortunate prognoses of CN-AML patients [18]. The prognostic role of NPM1 strictly depends on the presence of FLT3-ITD mutations, as patients with NPM1 mutations in the absence of FLT3-ITD demonstrated better risk profiles and prognoses [18]. Our findings therefore suggested that the aggressiveness of AML and the prognostic role of FLT3 are associated with the co-occurrence of NPM1 and DNMT3A mutations. Collectively, our integrated bioinformatics analysis suggested that FLT3 in conjunction with NPM1 and DNMT3A is an important onco-immunogenic signature of aggressive AML. Hopefully, this potential biomarker signature can be explored for better diagnosis, prognosis, and monitoring of AML [50], and serve as an attractive therapeutic target for kinase inhibitors for treating AML.
Over the years, our research group has made significant progress in drug discovery via in-house synthesized small-molecule multi-target drugs with translational relevance for treating inflammation, cancers, and immune-related disorders [28,30,51-69]. Herein, we reported the PKs and therapeutic efficacy of Lwk-n019 for treating leukemia via inhibition of FLT3 and TOPs. We found that Lwk-n019 demonstrated dose-dependent in vitro antiproliferative properties against several leukemia cell lines, hence revealing the potential role of Lwk-n019 in treating leukemia.
After establishing Lwk-n019’s inhibitory effects on leukemia cell lines, we further explored potential kinases targeted by Lwk-n019 via a kinome analysis. We found that Lwk-n019 had a high binding affinity and inhibitory effect on FLT3. The profiling demonstrated that Lwk-n019 is a potent inhibitor of mutant FLT3 and might be a candidate for testing as an antileukemic agent in AML patients with mutant FLT3 receptors. Interestingly, Lwk-n019 displayed a higher level of selectivity (S(10) score of 0.0043) toward FLT3 and FLT3 mutants than crenolanib (S(10) score of 0.12). Moreover, the compound demonstrated the best binding constant (Kd) of 0.77 µM (770 nM) against FLT3 (ITD, 835V). Several FLT3 inhibitors, including SMKIs, AC220, lestaurtinib, midostaurin, tandutinib, AT-9283, LS-104, dasatinib, sorafenib, and sunitinib, were developed for AML therapy [70]; however, satisfaction with these drugs is limited by high toxicity and severe side effects [71]. It is therefore noteworthy that the present study identified a new FLT3 inhibitor with high selectivity for mutant variants, which would be a very useful therapeutic agent for improving the prognosis of AML patients, especially patients harboring FLT3 mutations.
TOPs are essential regulators of DNA replication, transcription, and repair, and other vital cellular processes [20]. Several inhibitors of TOPs were developed into important anticancer drugs. However, their efficacy comes with significant side effects [24]. To our delight, we found that Lwk-n019, demonstrated an inhibitory role against DNA TOPI and TOPII activities at a similar potency compared to their respective clinical inhibitors. Lwk-n019 therefore represents a new class of TOP inhibitors for treating cancer.
Our data so far suggested that Lwk-n019 not only serves as a selective potent inhibitor of FLT3 mutants but also inhibits the TOP1 and TOPII enzymes. These multi-target activities of Lwk-n019 are important properties desired of a durable anticancer agent to minimize the tendency to lose efficacy and optimal therapeutic outcomes. Studies are ongoing to determine the full therapeutic properties and detailed mechanisms of FLT3 and TOP inhibition.
Drug PK properties are an important driving factor of successful drug discovery, and several drugs failed in the clinic due to poor PK properties [56,58,72,73]. Drug PKs also aid in selecting drug dosages and treatment regimens [74,75]. Consequently, we provide in vivo evidence of the good PKs of Lwk-n019 in rats. While the duodenum, jejunum, cecum, and colon were prime sites of Lwk-n019 absorption, the achieved Cmax, Vd, blood/plasma ratio, Tmax, AUC, and consistent time-dependent clearance of Lwkn-019 strongly suggested that i.v. administration of Lwk-n019 induced no apparent drug accumulation, and that it was rapidly cleared from the circulation [76]. These studies demonstrated a useful point of reference for subsequent drug optimization of higher potency, selectivity, and other characteristics of a good drug lead.
Conclusions
On a concluding note, our analysis of bulk and single-cell RNA sequencing datasets from AML patients revealed that FLT3 is an oncogenic modulator of the immune system and plays important roles in the progression, aggressive phenotype, and worse immune responses of patients. Our findings further suggested that the aggressiveness of AML and the prognostic role of FLT3 were associated with the co-occurrence of NPM1 and DNMT3A mutations. Lwk-n019, a novel small molecule, demonstrated anticancer activities via multi-target inhibition of topoisomerases and kinases with a high inhibition preference for mutant ITD-FLT3. The present study pioneered the basis for advance optimization of drug potency, selectivity, specificity, and other properties desired of an anticancer drug lead. Studies are ongoing to determine the full therapeutic properties and detailed mechanisms of FLT3 and TOP inhibition.
Acknowledgements
HS.Huang was funded by NSTC (National Science and Technology Council): NSTC111-2314-B-038-017 and MOST111-2314-B-038-122. Alexander TH Wu is funded by the following grants: 111-2314-B-038 -142 and 111-2314-B-038-098, from National Science and Technology Council (NSTC), Taiwan.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Dong Y, Shi O, Zeng Q, Lu X, Wang W, Li Y, Wang Q. Leukemia incidence trends at the global, regional, and national level between 1990 and 2017. Exp Hematol Oncol. 2020;9:14. doi: 10.1186/s40164-020-00170-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thacker N, Abla O. Epidemiology of non-hodgkin lymphomas in childhood and adolescence. In: Abla O, Attarbaschi A, editors. Non-hodgkin’s lymphoma in childhood and adolescence. Cham: Springer International Publishing; 2019. pp. 15–22. [Google Scholar]
- 4.“Acute Myeloid Leukemia-Cancer Stat Facts”. NCI. Retrieved 04 April 2022 [Google Scholar]
- 5.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 6.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 7.Jabbour E, O’Brien S, Konopleva M, Kantarjian H. New insights into the pathophysiology and therapy of adult acute lymphoblastic leukemia. Cancer. 2015;121:2517–2528. doi: 10.1002/cncr.29383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 9.Larson R. The US trials in adult acute lymphoblastic leukemia. Ann Hematol. 2004;83:S127–S128. doi: 10.1007/s00277-004-0850-2. [DOI] [PubMed] [Google Scholar]
- 10.Annino L, Vegna ML, Camera A, Specchia G, Visani G, Fioritoni G, Ferrara F, Peta A, Ciolli S, Deplano W, Fabbiano F, Sica S, Di Raimondo F, Cascavilla N, Tabilio A, Leoni P, Invernizzi R, Baccarani M, Rotoli B, Amadori S, Mandelli F GIMEMA Group. Treatment of adult acute lymphoblastic leukemia (ALL): long-term follow-up of the GIMEMA ALL 0288 randomized study. Blood. 2002;99:863–871. doi: 10.1182/blood.v99.3.863. [DOI] [PubMed] [Google Scholar]
- 11.Hunault M, Harousseau JL, Delain M, Truchan-Graczyk M, Cahn JY, Witz F, Lamy T, Pignon B, Jouet JP, Garidi R, Caillot D, Berthou C, Guyotat D, Sadoun A, Sotto JJ, Lioure B, Casassus P, Solal-Celigny P, Stalnikiewicz L, Audhuy B, Blanchet O, Baranger L, Béné MC, Ifrah N GOELAMS (Groupe Ouest-Est des Leucémies Airguës et Maladies du Sang) Group. Better outcome of adult acute lymphoblastic leukemia after early genoidentical allogeneic bone marrow transplantation (BMT) than after late high-dose therapy and autologous BMT: a GOELAMS trial. Blood. 2004;104:3028–3037. doi: 10.1182/blood-2003-10-3560. [DOI] [PubMed] [Google Scholar]
- 12.Kantarjian H, Thomas D, O’Brien S, Cortes J, Giles F, Jeha S, Bueso-Ramos CE, Pierce S, Shan J, Koller C, Beran M, Keating M, Freireich EJ. Long-term follow-up results of hyperfractionated cyclophosphamide, vincristine, doxorubicin, and dexamethasone (Hyper-CVAD), a dose-intensive regimen, in adult acute lymphocytic leukemia. Cancer. 2004;101:2788–2801. doi: 10.1002/cncr.20668. [DOI] [PubMed] [Google Scholar]
- 13.Dohner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson RA, Lo-Coco F, Naoe T, Niederwieser D, Ossenkoppele GJ, Sanz MA, Sierra J, Tallman MS, Löwenberg B, Bloomfield CD European LeukemiaNet. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 14.Brown P, Levis M, Shurtleff S, Campana D, Downing J, Small D. FLT3 inhibition selectively kills childhood acute lymphoblastic leukemia cells with high levels of FLT3 expression. Blood. 2005;105:812–820. doi: 10.1182/blood-2004-06-2498. [DOI] [PubMed] [Google Scholar]
- 15.Safran M, Rosen N, Twik M, BarShir R, Stein TI, Dahary D, Fishilevich S, Lancet D. Practical Guide to Life Science Databases. Springer; 2021. The genecards suite; pp. 27–56. [Google Scholar]
- 16.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 17.Kayser S, Schlenk RF, Londono MC, Breitenbuecher F, Wittke K, Du J, Groner S, Späth D, Krauter J, Ganser A, Döhner H, Fischer T, Döhner K German-Austrian AML Study Group (AMLSG) Insertion of FLT3 internal tandem duplication in the tyrosine kinase domain-1 is associated with resistance to chemotherapy and inferior outcome. Blood. 2009;114:2386–2392. doi: 10.1182/blood-2009-03-209999. [DOI] [PubMed] [Google Scholar]
- 18.O’Donnell MR, Abboud CN, Altman J, Appelbaum FR, Arber DA, Attar E, Borate U, Coutre SE, Damon LE, Goorha S, Lancet J, Maness LJ, Marcucci G, Millenson MM, Moore JO, Ravandi F, Shami PJ, Smith BD, Stone RM, Strickland SA, Tallman MS, Wang ES, Naganuma M, Gregory KM. Acute myeloid leukemia. J Natl Compr Canc Netw. 2012;10:984–1021. doi: 10.6004/jnccn.2012.0103. [DOI] [PubMed] [Google Scholar]
- 19.Levis M, Small D. FLT3: ITDoes matter in leukemia. Leukemia. 2003;17:1738–1752. doi: 10.1038/sj.leu.2403099. [DOI] [PubMed] [Google Scholar]
- 20.Chen SH, Chan NL, Hsieh TS. New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem. 2013;82:139–170. doi: 10.1146/annurev-biochem-061809-100002. [DOI] [PubMed] [Google Scholar]
- 21.Drlica K. Control of bacterial DNA supercoiling. Mol Microbiol. 1992;6:425–433. doi: 10.1111/j.1365-2958.1992.tb01486.x. [DOI] [PubMed] [Google Scholar]
- 22.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 23.Moukharskaya J, Verschraegen C. Topoisomerase 1 inhibitors and cancer therapy. Hematol Oncol Clin North Am. 2012;26:507–525. vii. doi: 10.1016/j.hoc.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 24.You F, Gao C. Topoisomerase inhibitors and targeted delivery in cancer therapy. Curr Top Med Chem. 2019;19:713–729. doi: 10.2174/1568026619666190401112948. [DOI] [PubMed] [Google Scholar]
- 25.Yuan H, Yan M, Zhang G, Liu W, Deng C, Liao G, Xu L, Luo T, Yan H, Long Z, Shi A, Zhao T, Xiao Y, Li X. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019;47:D900–D908. doi: 10.1093/nar/gky939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun D, Wang J, Han Y, Dong X, Ge J, Zheng R, Shi X, Wang B, Li Z, Ren P, Sun L, Yan Y, Zhang P, Zhang F, Li T, Wang C. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021;49:D1420–D1430. doi: 10.1093/nar/gkaa1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orellana EA, Kasinski AL. Sulforhodamine B (SRB) assay in cell culture to investigate cell proliferation. Bio Protoc. 2016;6:e1984. doi: 10.21769/BioProtoc.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawal B, Kuo YC, Wu ATH, Huang HS. BC-N102 suppress breast cancer tumorigenesis by interfering with cell cycle regulatory proteins and hormonal signaling, and induction of time-course arrest of cell cycle at G1/G0 phase. Int J Biol Sci. 2021;17:3224–3238. doi: 10.7150/ijbs.62808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyd MR, Paull KD. Some practical considerations and applications of the National Cancer Institute in vitro anticancer drug discovery screen. Drug Dev Res. 1995;34:91–109. [Google Scholar]
- 30.Lawal B, Lee CY, Mokgautsi N, Sumitra MR, Khedkar H, Wu ATH, Huang HS. MTOR/EGFR/iNOS/MAP2K1/FGFR/TGFB1 are druggable candidates for N-(2,4-difluorophenyl)-2’,4’-difluoro-4-hydroxybiphenyl-3-carboxamide (NSC765598), with consequent anticancer implications. Front Oncol. 2021;11:656738. doi: 10.3389/fonc.2021.656738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen TC, Yu DS, Chen SJ, Chen CL, Lee CC, Hsieh YY, Chang LC, Guh JH, Lin JJ, Huang HS. Design, synthesis and biological evaluation of tetracyclic azafluorenone derivatives with topoisomerase I inhibitory properties as potential anticancer agents. Arab J Chem. 2019;12:4348–4364. [Google Scholar]
- 32.Bielawski K, Winnicka K, Bielawska A. Inhibition of DNA topoisomerases I and II, and growth inhibition of breast cancer MCF-7 cells by ouabain, digoxin and proscillaridin A. Biol Pharm Bull. 2006;29:1493–1497. doi: 10.1248/bpb.29.1493. [DOI] [PubMed] [Google Scholar]
- 33.Das P, Jain CK, Roychoudhury S, Majumder HK, Das S. Design, synthesis and in vitro anticancer activity of a Cu(II) complex of carminic acid: a novel small molecule inhibitor of human DNA topoisomerase I and topoisomerase II. ChemistrySelect. 2016;1:6623–6631. [Google Scholar]
- 34.Sangodele JO, Inuwa Z, Lawal B, Adebayo-Gege G, Okoli BJ, Mtunzi F. Proxeed plus salvage rat testis from ischemia-reperfused injury by enhancing antioxidant’s activities and inhibition of iNOS expression. Biomed Pharmacother. 2021;133:111086. doi: 10.1016/j.biopha.2020.111086. [DOI] [PubMed] [Google Scholar]
- 35.Shittu OK, Lawal B, Alozieuwa BU, Haruna GM, Abubakar AN, Berinyuy EB. Alteration in biochemical indices following chronic administration of methanolic extract of Nigeria bee propolis in Wistar rats. Asian Pac J Trop Dis. 2015;5:654–657. [Google Scholar]
- 36.Hussain A, Alshehri S, Ramzan M, Afzal O, Altamimi ASA, Alossaimi MA. Biocompatible solvent selection based on thermodynamic and computational solubility models, in-silico GastroPlus prediction, and cellular studies of ketoconazole for subcutaneous delivery. J Drug Deliv Sci Technol. 2021;65:102699. [Google Scholar]
- 37.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminform. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawal B, Liu YL, Mokgautsi N, Khedkar H, Sumitra MR, Wu ATH, Huang HS. Pharmacoinformatics and preclinical studies of nsc765690 and nsc765599, potential stat3/cdk2/4/6 inhibitors with antitumor activities against nci60 human tumor cell lines. Biomedicines. 2021;9:92. doi: 10.3390/biomedicines9010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shih ML, Lawal B, Cheng SY, Olugbodi JO, Babalghith AO, Ho CL, Cavalu S, Batiha GE, Albogami S, Alotaibi SS, Lee JC, Wu ATH. Large-scale transcriptomic analysis of coding and non-coding pathological biomarkers, associated with the tumor immune microenvironment of thyroid cancer and potential target therapy exploration. Front Cell Dev Biol. 2022;10:923503. doi: 10.3389/fcell.2022.923503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Visualizer DS. BIOVIA, Dassault Systèmes, BIOVIA Workbook, Release 2020; BIOVIA Pipeline Pilot, Release 2020, San Diego: Dassault Systèmes. 2020 [Google Scholar]
- 42.Chen TC, Wu CL, Lee CC, Chen CL, Yu DS, Huang HS. Structure-based hybridization, synthesis and biological evaluation of novel tetracyclic heterocyclic azathioxanthone analogues as potential antitumor agents. Eur J Med Chem. 2015;103:615–627. doi: 10.1016/j.ejmech.2014.09.050. [DOI] [PubMed] [Google Scholar]
- 43.Armstrong SA, Mabon ME, Silverman LB, Li A, Gribben JG, Fox EA, Sallan SE, Korsmeyer SJ. FLT3 mutations in childhood acute lymphoblastic leukemia. Blood. 2004;103:3544–3546. doi: 10.1182/blood-2003-07-2441. [DOI] [PubMed] [Google Scholar]
- 44.Kiyoi H, Naoe T, Nakano Y, Yokota S, Minami S, Miyawaki S, Asou N, Kuriyama K, Jinnai I, Shimazaki C. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood. 1999;93:3074–3080. [PubMed] [Google Scholar]
- 45.Meshinchi S, Woods WG, Stirewalt DL, Sweetser DA, Buckley JD, Tjoa TK, Bernstein ID, Radich JP. Prevalence and prognostic significance of Flt3 internal tandem duplication in pediatric acute myeloid leukemia. Blood. 2001;97:89–94. doi: 10.1182/blood.v97.1.89. [DOI] [PubMed] [Google Scholar]
- 46.Iwai T, Yokota S, Nakao M, Okamoto T, Taniwaki M, Onodera N, Watanabe A, Kikuta A, Tanaka A, Asami K, Sekine I, Mugishima H, Nishimura Y, Koizumi S, Horikoshi Y, Mimaya J, Ohta S, Nishikawa K, Iwai A, Shimokawa T, Nakayama M, Kawakami K, Gushiken T, Hyakuna N, Fujimoto T, et al. Internal tandem duplication of the FLT3 gene and clinical evaluation in childhood acute myeloid leukemia. The children’s cancer and leukemia study group, Japan. Leukemia. 1999;13:38–43. doi: 10.1038/sj.leu.2401241. [DOI] [PubMed] [Google Scholar]
- 47.Kottaridis PD, Gale RE, Frew ME, Harrison G, Langabeer SE, Belton AA, Walker H, Wheatley K, Bowen DT, Burnett AK, Goldstone AH, Linch DC. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98:1752–1759. doi: 10.1182/blood.v98.6.1752. [DOI] [PubMed] [Google Scholar]
- 48.Stirewalt DL, Kopecky KJ, Meshinchi S, Appelbaum FR, Slovak ML, Willman CL, Radich JP. FLT3, RAS, and TP53 mutations in elderly patients with acute myeloid leukemia. Blood. 2001;97:3589–3595. doi: 10.1182/blood.v97.11.3589. [DOI] [PubMed] [Google Scholar]
- 49.Rombouts WJ, Blokland I, Löwenberg B, Ploemacher RE. Biological characteristics and prognosis of adult acute myeloid leukemia with internal tandem duplications in the Flt3 gene. Leukemia. 2000;14:675–683. doi: 10.1038/sj.leu.2401731. [DOI] [PubMed] [Google Scholar]
- 50.Li W, Zhong C, Jiao J, Li P, Cui B, Ji C, Ma D. Characterization of hsa_circ_0004277 as a new biomarker for acute myeloid leukemia via circular RNA profile and bioinformatics analysis. Int J Mol Sci. 2017;18:597. doi: 10.3390/ijms18030597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawal B, Kuo YC, Tang SL, Liu FC, Wu ATH, Lin HY, Huang HS. Transcriptomic-based identification of the immuno-oncogenic signature of cholangiocarcinoma for HLC-018 multi-target therapy exploration. Cells. 2021;10:2873. doi: 10.3390/cells10112873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang HS, Chiu HF, Lee AL, Guo CL, Yuan CL. Synthesis and structure-activity correlations of the cytotoxic bifunctional 1,4-diamidoanthraquinone derivatives. Bioorg Med Chem. 2004;12:6163–6170. doi: 10.1016/j.bmc.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 53.Huang HS, Chiu HF, Yeh PF, Yuan CL. Structure-based design and synthesis of regioisomeric disubstituted aminoanthraquinone derivatives as potential anticancer agents. Helv Chim Acta. 2004;87:999–1006. [Google Scholar]
- 54.Lawal B, Wu ATH, Huang HS. Leveraging bulk and single-cell RNA sequencing data of NSCLC tumor microenvironment and therapeutic potential of NLOC-15A, a novel multi-target small molecule. Front Immunol. 2022;13:872470. doi: 10.3389/fimmu.2022.872470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khedkar HN, Wang YC, Yadav VK, Srivastava P, Lawal B, Mokgautsi N, Sumitra MR, Wu ATH, Huang HS. In-silico evaluation of genetic alterations in ovarian carcinoma and therapeutic efficacy of NSC777201, as a novel multi-target agent for TTK, NEK2, and CDK1. Int J Mol Sci. 2021;22:5895. doi: 10.3390/ijms22115895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawal B, Kuo YC, Sumitra MR, Wu ATH, Huang HS. In vivo pharmacokinetic and anticancer studies of HH-N25, a selective inhibitor of topoisomerase I, and hormonal signaling for treating breast cancer. J Inflamm Res. 2021;14:4901–4913. doi: 10.2147/JIR.S329401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lawal B, Liu YL, Mokgautsi N, Khedkar H, Sumitra MR, Wu ATH, Huang HS. Pharmacoinformatics and preclinical studies of NSC765690 and NSC765599, potential STAT3/CDK2/4/6 inhibitors with antitumor activities against NCI60 human tumor cell lines. Biomedicines. 2021;9:92. doi: 10.3390/biomedicines9010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lawal B, Wang YC, Wu ATH, Huang HS. Pro-oncogenic c-Met/EGFR, biomarker signatures of the tumor microenvironment are clinical and therapy response prognosticators in colorectal cancer, and therapeutic targets of 3-phenyl-2H-benzo[e][1,3]-oxazine-2,4(3H)-dione derivatives. Front Pharmacol. 2021;12:691234. doi: 10.3389/fphar.2021.691234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee CC, Huang KF, Chang DM, Hsu JJ, Huang FC, Shih KN, Chen CL, Chen TC, Chen RH, Lin JJ, Huang HS. Design, synthesis and evaluation of telomerase inhibitory, hTERT repressing, and anti-proliferation activities of symmetrical 1,8-disubstituted amidoanthraquinones. Eur J Med Chem. 2012;50:102–112. doi: 10.1016/j.ejmech.2012.01.044. [DOI] [PubMed] [Google Scholar]
- 60.Lee CC, Liu FL, Chen CL, Chen TC, Chang DM, Huang HS. Discovery of 5-(2’,4’-difluorophenyl)-salicylanilides as new inhibitors of receptor activator of NF-κB ligand (RANKL)-induced osteoclastogenesis. Eur J Med Chem. 2015;98:115–126. doi: 10.1016/j.ejmech.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 61.Lee CC, Liu FL, Chen CL, Chen TC, Liu FC, Ahmed Ali AA, Chang DM, Huang HS. Novel inhibitors of RANKL-induced osteoclastogenesis: design, synthesis, and biological evaluation of 6-(2,4-difluorophenyl)-3-phenyl-2H-benzo[e][1,3]oxazine-2,4(3H)-diones. Bioorg Med Chem. 2015;23:4522–4532. doi: 10.1016/j.bmc.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 62.Lee JC, Wu ATH, Chen JH, Huang WY, Lawal B, Mokgautsi N, Huang HS, Ho CL. HNC0014, a multi-targeted small-molecule, inhibits head and neck squamous cell carcinoma by suppressing c-Met/STAT3/CD44/PD-L1 oncoimmune signature and eliciting antitumor immune responses. Cancers (Basel) 2020;12:3759. doi: 10.3390/cancers12123759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu FC, Lu JW, Chien CY, Huang HS, Lee CC, Lien SB, Lin LC, Chen LW, Ho YJ, Shen MC, Ho LJ, Lai JH. Arthroprotective effects of Cf-02 sharing structural similarity with quercetin. Int J Mol Sci. 2018;19:1453. doi: 10.3390/ijms19051453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Madamsetty VS, Pal K, Dutta SK, Wang E, Thompson JR, Banerjee RK, Caulfield TR, Mody K, Yen Y, Mukhopadhyay D, Huang HS. Design and evaluation of PEGylated liposomal formulation of a novel multikinase inhibitor for enhanced chemosensitivity and inhibition of metastatic pancreatic ductal adenocarcinoma. Bioconjug Chem. 2019;30:2703–2713. doi: 10.1021/acs.bioconjchem.9b00632. [DOI] [PubMed] [Google Scholar]
- 65.Mokgautsi N, Wang YC, Lawal B, Khedkar H, Sumitra MR, Wu ATH, Huang HS. Network pharmacological analysis through a bioinformatics approach of novel NSC765600 and NSC765691 compounds as potential inhibitors of CCND1/CDK4/PLK1/CD44 in cancer types. Cancers. 2021;13:2523. doi: 10.3390/cancers13112523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mokgautsi N, Wen YT, Lawal B, Khedkar H, Sumitra MR, Wu ATH, Huang HS. An integrated bioinformatics study of a novel niclosamide derivative, nsc765689, a potential gsk3β/β-catenin/stat3/cd44 suppressor with anti-glioblastoma properties. Int J Mol Sci. 2021;22:2464. doi: 10.3390/ijms22052464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen CJ, Lin PL, Lin HC, Cheng YW, Huang HS, Lee H. RV-59 suppresses cytoplasmic Nrf2-mediated 5-fluorouracil resistance and tumor growth in colorectal cancer. Am J Cancer Res. 2019;9:2789–2796. [PMC free article] [PubMed] [Google Scholar]
- 68.Yadav VK, Huang YJ, George TA, Wei PL, Sumitra MR, Ho CL, Chang TH, Wu ATH, Huang HS. Preclinical evaluation of the novel small-molecule MSI-N1014 for treating drug-resistant colon cancer via the LGR5/β-catenin/miR-142-3p network and reducing cancer-associated fibroblast transformation. Cancers (Basel) 2020;12:1590. doi: 10.3390/cancers12061590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu ATH, Huang HS, Wen YT, Lawal B, Mokgautsi N, Huynh TT, Hsiao M, Wei L. A preclinical investigation of GBM-N019 as a potential inhibitor of glioblastoma via exosomal mTOR/CDK6/STAT3 signaling. Cells. 2021;10:2391. doi: 10.3390/cells10092391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 71.Simon T, Tomuleasa C, Bojan A, Berindan-Neagoe I, Boca S, Astilean S. Design of FLT3 inhibitor-gold nanoparticle conjugates as potential therapeutic agents for the treatment of acute myeloid leukemia. Nanoscale Res Lett. 2015;10:466. doi: 10.1186/s11671-015-1154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Leon J, Ruan CJ, Schoretsanitis G, De las Cuevas C. A rational use of clozapine based on adverse drug reactions, pharmacokinetics, and clinical pharmacopsychology. Psychother Psychosom. 2020;89:200–214. doi: 10.1159/000507638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nichols AI, Focht K, Jiang Q, Preskorn SH, Kane CP. Pharmacokinetics of venlafaxine extended release 75 mg and desvenlafaxine 50 mg in healthy CYP2D6 extensive and poor metabolizers. Clin Drug Investig. 2011;31:155–167. doi: 10.2165/11586630-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 74.Gallo JM, Vicini P, Orlansky A, Li S, Zhou F, Ma J, Pulfer S, Bookman MA, Guo P. Pharmacokinetic model-predicted anticancer drug concentrations in human tumors. Clin Cancer Res. 2004;10:8048–8058. doi: 10.1158/1078-0432.CCR-04-0822. [DOI] [PubMed] [Google Scholar]
- 75.White RE, Manitpisitkul P. Pharmacokinetic theory of cassette dosing in drug discovery screening. Drug Metab Dispos. 2001;29:957–966. [PubMed] [Google Scholar]
- 76.Clausen DM, Guo J, Parise RA, Beumer JH, Egorin MJ, Lazo JS, Prochownik EV, Eiseman JL. In vitro cytotoxicity and in vivo efficacy, pharmacokinetics, and metabolism of 10074-G5, a novel small-molecule inhibitor of c-Myc/Max dimerization. J Pharmacol Exp Ther. 2010;335:715–727. doi: 10.1124/jpet.110.170555. [DOI] [PMC free article] [PubMed] [Google Scholar]