Abstract
The metabolism of tumor cells is characterized by the regulation of demand, nutrient supply and metabolic enzymes, which are different in cancer tissues from those in corresponding healthy tissues. There is growing evidence that dietary composition influences biological processes that contribute to tumor incidence and progression as much as genetic status. One possibility for specific dietary interventions in cancer patients is to limit methionine intake. The role of methionine metabolism in tumors suggests that interference with the methionine metabolism network by either drug or environmental effects may show substantial therapeutic effects, but the molecular mechanism is not completely clear. In this study, methionine deprivation was found to downregulate cathepsin L (CTSL) and induce proliferation inhibition in glioma cells. We also demonstrated that CTSL is a tumor-related gene, and promotes the proliferation and invasion of glioma. Our results showed that the treatment of methionine metabolism and CTSL related genes in glioma cells may be a novel strategy for glioma therapy in the future.
Keywords: Tumor metabolism, methionine deprivation, cathepsin L, CTSL, glioma
Introduction
Glioma is a common primary intracranial tumor, accounting for approximately 40% of primary brain tumors [1]. Despite comprehensive treatment including surgery, radiotherapy and chemotherapy, the median survival time of patients with glioma is still less than 14.6 months [2,3]. Currently, there is no effective treatment for glioma. Hence, it is urgent to uncover the molecular pathological mechanism of glioma, and to discover potential molecular therapeutic targets. Molecularly targeted tumor therapy is a trend and hot topic in current research [4-6].
Tumor metabolism plays an important role in malignant biological processes. Tumor cells need a high level of nutrients and energy to maintain their high proliferation rate [7,8]. Regulation of the metabolic environment in the tumor cell can significantly change its metabolic activities, resulting in changes in drug sensitivity, proliferation rate and metabolic requirements [9-11]. In recent years, dietary changes have gained increasing attention as a practical way to supplement traditional cancer treatment [12]. Studies have found that the removal of serine and glycine from the diet can regulate the prognosis of cancer [13-16]. The availability of histidine and asparagine mediates response to methotrexate and the progression of breast cancer metastasis, respectively [17,18]. However, there are few reports about which nutrients glioma is sensitive to.
In the clinical diagnosis and treatment of glioma, [11C] methionine positron emission tomography (MET PET) has become an effective diagnostic tool for glioma, due to its lower uptake in normal brain tissue than 18F-FDG (18F-fluorodeoxyglucose) [19-21]. As we know that isotopes have almost identical chemistry, we believe that glioma has a greater need for methionine than normal brain tissue [22]. However, it is still unclear whether methionine can promote the malignant biological behavior of glioma.
Under these circumstances, we carried out a methionine deprivation experiment on tumor-bearing mice, and the results showed that methionine deprivation could prolong the survival time of the mice. Subsequently, proteomic analysis was carried out to study the molecular mechanism by which methionine deprivation inhibits glioma proliferation. Among them, cathepsin L (CTSL), a lysosomal cysteine protease, attracted our attention [23]. CTSL is widely distributed in lysosomes of mammals. Under normal circumstances, CTSL performs biological functions in lysosomes [24,25]. However, changes in expression levels and lysosomal state caused a portion of CTSL being secreted into the extracellular environment [26]. Tumor cells secrete CTSL to the extracellular space, resulting in tumor invasion and metastasis [27-29]. In addition to the secretory type, there is also nuclear CTSL, which may be involved in cell replication and metastasis [30-32]. Upregulation of CTSL is a common phenomenon that occurs in many tumors and is associated with a poor prognosis in patients. Urinary CTSL levels in patients with urothelial carcinoma of the bladder have been shown to predict metastatic spread and tumor recurrence [33]. The serum level of CTSL is significantly elevated in patients with lung, pancreatic and ovarian cancer [34-36]. However, its role in glioma has not been thoroughly studied, so we decided to explore the biological function of CTSL in glioma.
In this study, we first studied the effect of methionine deprivation on glioma growth in vitro and in vivo. Then, the CTSL gene was analyzed in the database of TCGA and CGGA. By upregulating CTSL expression, we confirmed that it can partially reverse the inhibition of methionine deprivation on glioma growth. Survival curves suggested that a high expression level of CTSL predicted poor prognosis of patients with glioma.
Materials and methods
Patients and samples
All glioma tissue specimens (obtained from surgical resection) and nontumor brain tissue specimens (obtained from intracranial decompression surgery in patients with traumatic brain injury) were collected from the Affiliated Hospital of Xuzhou Medical University. None of the patients received chemotherapy, radiation, or immunotherapy. All glioma specimens had a clear diagnosis and were classified according to the WHO criteria. All subjects provided written consent (either the patient or a family member) and were approved by the Ethics Committee of the Affiliated Hospital of Xuzhou Medical University.
Glioma xenograft model and histopathology
BALB/c nude mice (male, 5-6 weeks old) were purchased from Nanjing GemPharmatch Co., Ltd. All experiments were approved by the Ethical Committee of Xuzhou Medical University. To establish glioma-bearing nude mice, 1 × 106 cells (suspended in 5 mL of serum-free media) were administered into the right striatum of mice, as previously described [37]. For H&E staining, the tissue was dehydrated with ethanol and xylene and embedded in paraffin, then a commercial kit (Beyotime Biotechnology, China) was used for staining according to the manufacturer’s instructions. For IHC staining, tissue was also dehydrated, paraffin-embedded and sectioned, followed by staining with the relevant antibodies. A primary antibody against CTSL (1:250, ab203028, Abcam) was used.
Cell culture
The glioma cell lines U251 (RRID: CVCL-0021) and LN229 (RRID: CVCL-0393) were provided by Chinese Academy of Sciences. All cell lines passed the test of DNA profiling (STR). The cells were cultured in DMEM (WISENT) supplemented with 10% FBS (WISENT) and 1% penicillin/streptomycin. All cell lines were cultured in a cell incubator with a 5% CO2 atmosphere under saturated humidity at 37°C.
ITRAQ and data analysis
iTRAQ technology was used to detect the expression of proteins. T tests and fold changes were used to identify differentially expressed proteins, and a P value < 0.05 and a fold change > 1.2 were significant difference proteins. Cluster 3.0 (http://bonsai.hgc.jp/mdehoon/software/Cluster/) was used for bidirectional hierarchical clustering of differentially expressed genes in 6 samples. Hiplot (https://hiplot.com.cn/basic) was used to check the heatmap [38].
Bioinformatics analysis
Clinical data of glioma patients were obtained from two independent portals (TCGA and CGGA). A total of 349 samples from TCGA were used to plot patient survival curves using Betastasis (http://www.betastasis.com/) for clinical correlation analysis. A total of 313 samples were collected from CGGA (http://www. cgga.org.cn/), including 85 GBM samples, to explore the difference in CTSL in glioma with different WHO grades [39].
Cell viability assay
U251 and LN229 cells (5000 cells/well) were seeded into a 96-well plate in DMEM and cultured for 24 h. After incubation with different media (Met (+)/Met (-)) for the indicated times, 10 μl CCK-8 solution was added to each well. Absorbance at 460 nm was detected by a spectrophotometer (BioTek, Winooski, VT, USA) after incubation for 45 min.
EdU assay
U251 and LN229 cells (5000 cells/well) were seeded into a 96-well plate in DMEM and cultured for 24 h. EdU assay was carried out with a commercial kit (RiboBio, Guangzhou, China) according to the manufacturer’s instructions. The images were taken using an EVOSTM FL Auto 2 Imaging System (Thermo Fisher Scientific).
Cell transfection
Lentiviruses sh-NC and sh-CTSL 1-3 and CTSL overexpression plasmid pLenti-CMV-GFP-Puro/CTSL were purchased from Jiangsu Laisen Biotechnology (China). After transfection by the lentiviruses for 48 hours, the cells were cultured with 2.0 μg/ml puromycin. The surviving cells were used in the subsequent experiments. For plasmid transfection, PolyJet (SignaGen) was used according to the manufacturer’s instructions.
Western blot analysis
Total protein was extracted from cells or tissues, and a BCA protein assay kit was used to detect protein concentration. Equal amounts of total protein were loaded to 10% SDS-PAGE gels and then transferred to 0.45 μm pore size PVDF membranes (Millipore). After blocking in 3% BSA solution for 2 hours, the membranes were incubated with primary antibodies at 4°C overnight and secondary antibodies at room temperature for 2 hours. Then, the signals on the membranes were detected using ECL Western blotting Substrate (Thermo Fisher) on Imaging System (Bio Rad, USA). Primary antibodies against the following proteins were used as indicated: CTSL (1:500, ab203028, Abcam), CDK2 (1:2000, ab32147, Abcam), CDK4 (1:1000, P11802, Cell Signaling Technology), Cyclin D1 (1:1000, P24385, Cell Signaling Technology), TGF-β (1:1000, ab215715, Abcam), β-actin (1:10000, No. 66009-1-Ig, Proteintech).
Quantitative real-time PCR (qRT-PCR)
Total RNA extraction was carried out according to our previous paper [40]. SYBR Green PCR Master Mix kit was used to amplify the target gene. qRT-PCR was performed on an Applied Biosystems 7500 (Applied Biosystems, USA). The primer sequences of β-actin and CTSL are shown in Table 1.
Table 1.
The forward and reverse primers of genes
| β-actin | Forward: 5’-CATGTACGTTGCTATCCAGGC-3’ |
| Reverse: 5’-CTCCTTAATGTCACGCACGAT-3’ | |
| CTSL | Forward: 5’-CCTCGAGTTTTATATTGACCCCGATAAAATCA-3’ |
| Reverse: 5’-ATGTCGACCTACTTCCTGCAGCAGGTG-3’ |
Cell cycle analysis
Cell cycle analyses were performed using a commercial kit (Keygentec, Jiangsu, China) according to the manufacturer’s instructions. The cell suspensions were detected by flow cytometry and analyzed using Flow Cytometer (BD Biosciences, San Jose, USA).
Clonogenic cell survival assay
Different cells were seeded into 6-well plates (1000 cells/well) and cultured for 14 days. Finally, the cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet solution.
Wound healing assay
A cell Culture-Insert (No. 80206, ibidi, Germany) was used for this assay. The cells were prepared as a cell suspension at a concentration 5 × 105 cells/ml, and 70 μl was seeded into the well of a Culture-Insert. After incubation for 16 hours, the Culture-Insert was removed and fresh medium was added. Images of three random areas were taken by an EVOSTM FL Auto 2 Imaging System (Thermo Fisher Scientific) at the indicated times (0 hour and 48 hours).
Transwell migration and invasion assays
To assess migration ability, 200 μl cell suspension of serum-free medium (1 × 104 cells/ml) was seeded into the upper chamber. The bottom chamber was infused with 500 μl of DMEM containing 10% FBS. Cells were incubated at 37°C for 36 hours. To assess invasion ability, 10 μg of Matrigel (BD) was used to pre-coat the filters, and was all other steps were the same as those used in the migration assay. Three fields of adherent cells were randomly captured with an EVOSTM FL Auto 2 Imaging System (Thermo Fisher Scientific).
Statistical analysis
The experiments were repeated at least three times independently and the results are shown as the means ± SEM. Statistical comparisons were performed using one-way analysis of variance (ANOVA) for multiple comparisons. P values < 0.05 were considered statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001). SPSS software (version 18.0) and GraphPad Prism 8 were used for statistical analyses.
Result
Methionine deprivation can inhibit tumor growth in vitro and in vivo
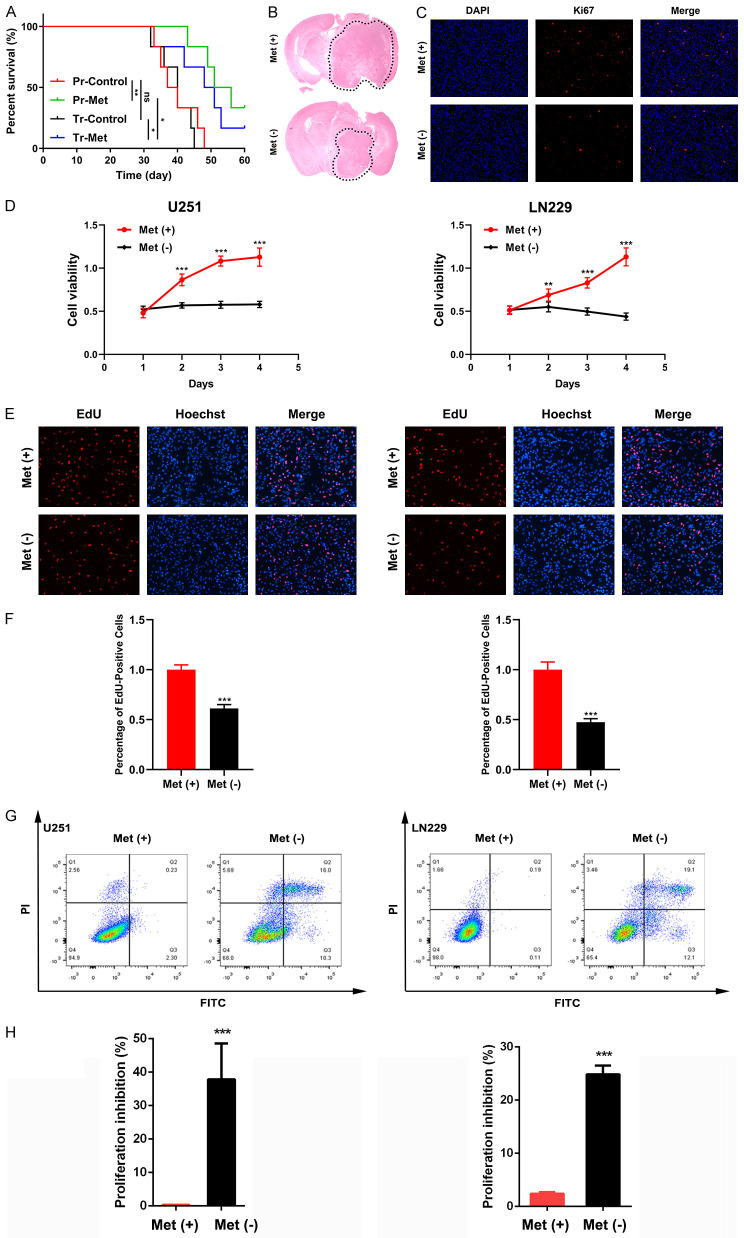
Methionine is an essential amino acid that cannot be synthesized by cells themselves and must be obtained by external ingestion. Therefore, we generated a mouse model of the original xenograft tumors using GL261 glioma cells, which were fed a conventional diet or a methionine deprivation diet. Two different feeding patterns were used, including prevention (Pr) and treatment (Tr). Pr-Met group begin to receive the methionine deprivation diet 14 days before tumor injection, and Tr-Met group start the methionine deprivation at the point of tumor injection. Control groups were fed the conventional diet at all times. The results showed that methionine deprivation could significantly prolong the survival time of tumor bearing mice (Figure 1A). The Pr-Met group showed significant inhibition of glioma growth compared with the Pr-Control group (Figure 1B, 1C). We also cultured glioma cell lines U251 and LN229 in vitro and then assessed the proliferation with and without methionine. The results showed that methionine deprivation significantly reduced the proliferation activity of U251 and LN229 cells in vitro, and the effect was time-dependent (Figure 1D-F). These results suggested that methionine deprivation can inhibit the proliferation of glioma cells in vitro and in vivo. Furthermore, we found that methionine also had a significant effect on apoptosis. We cultured U251 and LN229 cells in vitro and detected the apoptosis rate of cells by flow cytometry. The results showed that methionine deprivation significantly promoted the apoptosis of glioma cells (Figure 1G, 1H). Combined with the above results, we believe that methionine plays an important role in the proliferation of glioma cells and can significantly affect glioma cells.
Figure 1.
Methionine restriction inhibits the proliferation of glioma cells in vitro and in vivo. A. Survival of tumor-bearing mice after different feeding programs. B. Representative H&E-stained brain sections of transplanted tumors. C. Expression of Ki67 protein in glioma after mice were fed with or without methionine. Bar: 200 μm. D. U251 and LN229 cells were cultured with different media and cell viability was assessed by a CCK-8 assay. E and F. EdU assay results of U251 and LN229 cells. Bar: 200 μm. G and H. Flow cytometry results of U251 and LN229 cells. ns = nonsignificance, *P < 0.05, **P < 0.01, ***P < 0.001.
Methionine deprivation downregulate the expression of CTSL in vitro and in vivo
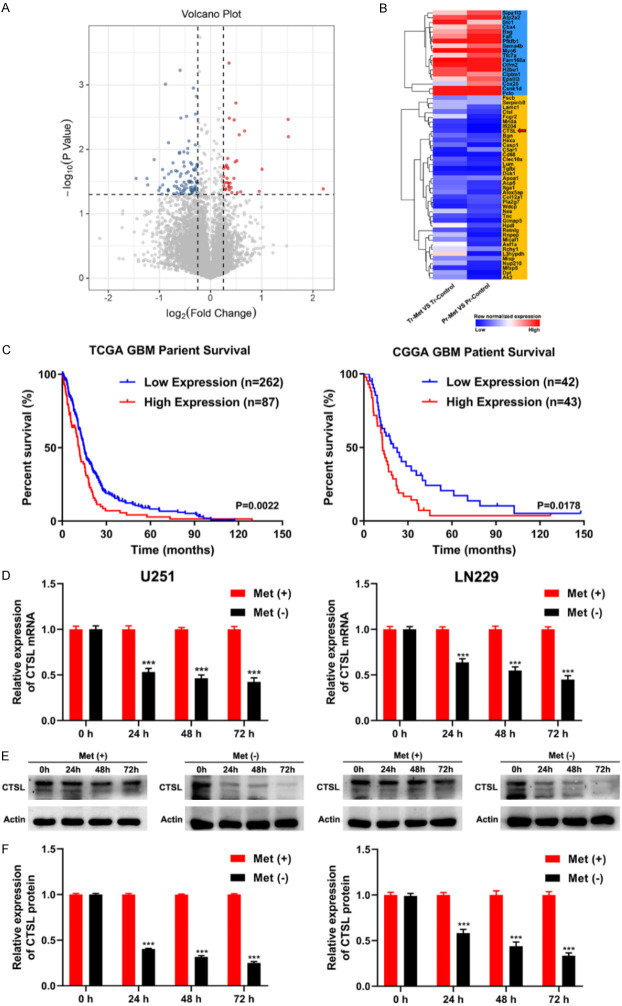
The effect of methionine on glioma prompted us to further study the mechanism involved. Proteomic analysis of previous glioma samples revealed 125 differentially expressed genes (Figures 2A and S1), of which 18 were significantly upregulated and 39 were significantly downregulated (Figure 2B). Afterward, we performed GO analysis and found that most of the GO analysis biological processes were related to enzyme/extracellular matrix/cell adhesion (Figure S2). Next, we used TCGA clinical data to screen out all enzyme/extracellular matrix/cell adhesion genes in glioma patients and map their disease-free survival (DFS) time. Finally, the results showed that CTSL was associated with disease-free survival (Figure 2C). At the same time, we found that after methionine deprivation, the mRNA and protein expression of CTSL gradually decreased over time, demonstrating the expression of downregulation (Figure 2D-F). These data suggest that the expression of CTSL is inhibited after methionine deprivation in vivo and in vitro.
Figure 2.
Methionine restriction downregulate CTSL expression in vitro and in vivo. A. Volcano plot for differentially expressed genes in Pr-groups. B. Heatmap of orthotopically transplanted of glioma tissue. C. Survival analysis was carried out using the date of GBM patients from the database of TCGA and CGGA. D. qRT-PCR showed that methionine restriction significantly decreased the expression of CTSL in U251 and LN229 cells. E and F. Western blot analysis revealed that methionine restriction significantly downregulated CTSL protein expression in U251 and LN229 cells. ***P < 0.001.
Downregulation of CTSL inhibits the growth of glioma in vitro and in vivo
Lentiviruses were used for transfection of glioma cells (Figure S3). Western blot analysis verified the efficiency of downregulation (Figures 3A and S4). The CCK-8 assay also confirmed the decrease in cell viability in U251 and LN229 cells. We further measured the proliferation activity of the cells by EdU assay and the results showed that glioma cells activity was significantly inhibited (Figure 3C, 3D). At the same time, we conducted a colony formation experiment, and the results were consistent with the EdU experiment (Figure S5).
Figure 3.
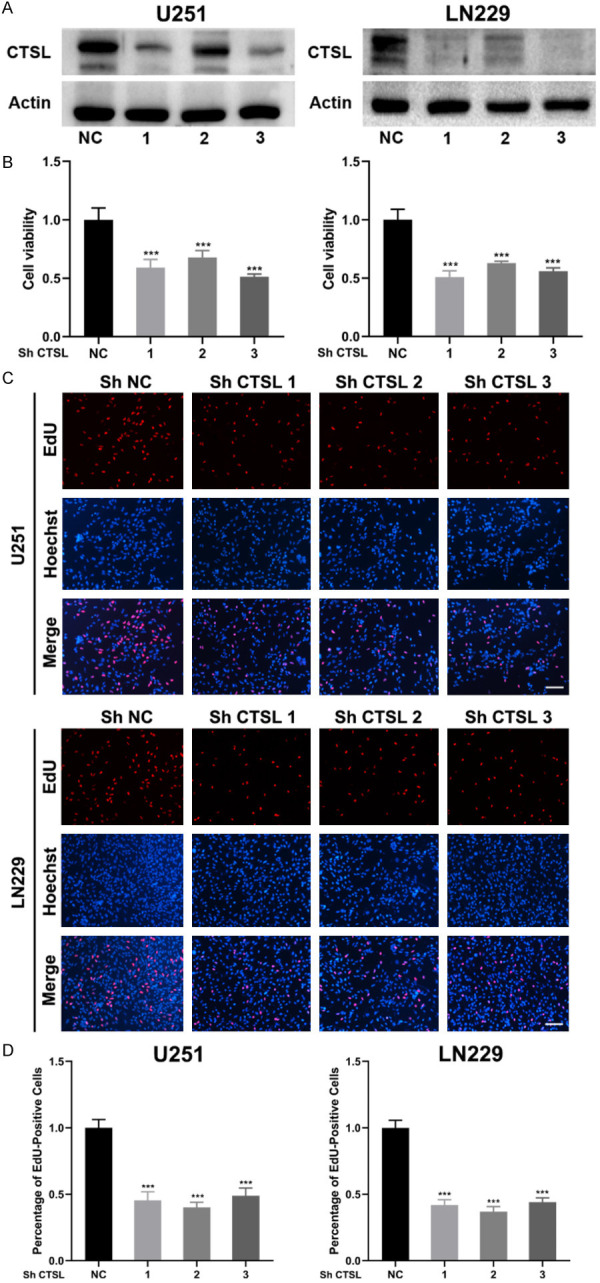
Downregulation of CTSL inhibits glioma proliferation. A. CTSL expression after transfection by lentivirus. B. Cell viability of U251 and LN229 cells after transfection of lentivirus. C and D. EdU assay results of U251 and LN229 cells after transfection by lentivirus. Bar: 200 μm. ***P < 0.001.
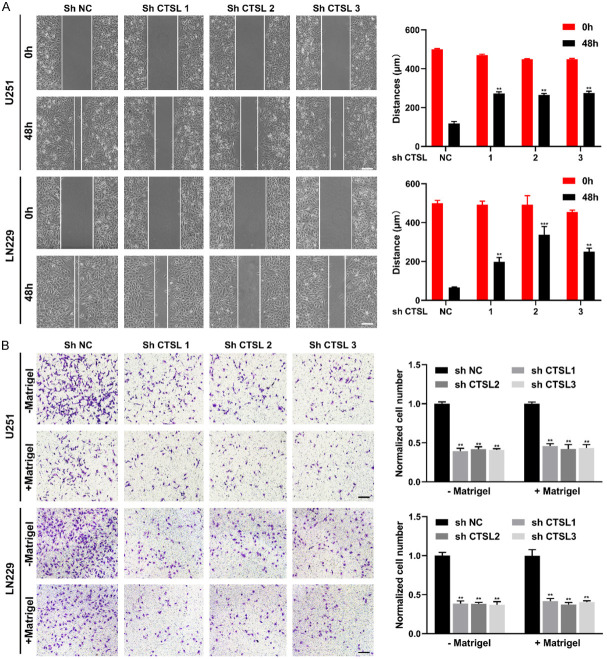
In the wound healing assay, downregulation of CTSL significantly inhibited the migration ability of cells (Figure 4A, 4B), and in the Transwell experiment, downregulation of CTSL also significantly weakened the invasion and migration ability of glioma cells (Figure 4B). These results suggest that downregulation of CTSL can inhibit the proliferation, invasion and migration of glioma cells in vitro.
Figure 4.
CTSL is involved in the regulation of cell migration and invasion. A. Representative images and quantification analysis of the wound healing assay. Bar: 100 μm. B. Representative images and quantification analysis of the Transwell assay. Bar: 100 μm. **P < 0.01, ***P < 0.001.
Furthermore, transfected LN229 glioma cells were injected into the brains of the mice, which were sacrificed 4 weeks later, and brain tissue was used for H&E staining to evaluate glioma proliferation. The results showed that glioma proliferation was significantly inhibited in CTSL downregulated groups (Figure S6). These results suggest that downregulation of CTSL inhibits the invasion and migration of glioma cells.
Methionine restriction inhibits glioma proliferation by downregulating CTSL
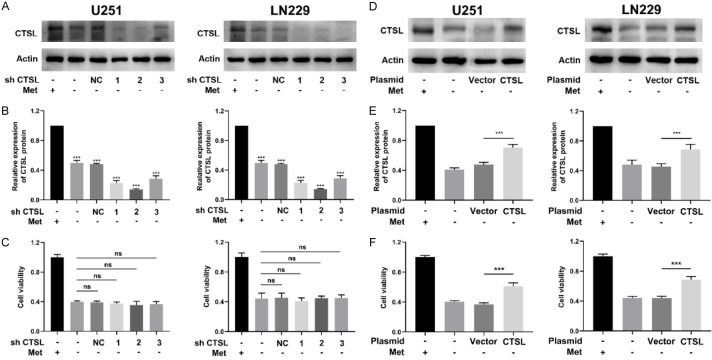
To verify that the inhibition of glioma proliferation was due to the downregulation of CTSL caused by methionine deficiency, glioma cells with downregulation of CTSL were selected and cells were cultured in methionine-free medium. The results showed that CTSL expression was significantly inhibited (Figure 5A, 5B), but cell viability did not differ significantly between the groups cultured with methionine-free medium (Figure 5C). These results suggest that there is no synergistic inhibition of glioma proliferation between the downregulation of CTSL and methionine deprivation. At the same time, we also overexpressed CTSL in glioma cells, and the results showed that overexpression of CTSL could partially decrease the inhibition of methionine deprivation on glioma cells (Figure 5F), and the reexpression of CTSL had a positive effect on cell viability (Figure 5D, 5E). In conclusion, the results show that downregulation of CTSL dose not enhance the inhibitory effect of methionine deprivation on cell proliferation, whereas overexpression of CTSL decrease the inhibition of methionine deprivation, which has the same effect.
Figure 5.
Methionine and CTSL have the same effect on glioma proliferation. A and B. CTSL expression in U251 and LN229 cells after transfection. Cells were cultured as indicated. C. Cell viability of U251 and LN229 cells after transfection. D and E. CTSL expression in U251 and LN229 cells after transfection. Cells were cultured as indicated. F. Cell viability of U251 and LN229 cells after transfection. NS: Nonsignificance, ***P < 0.001.
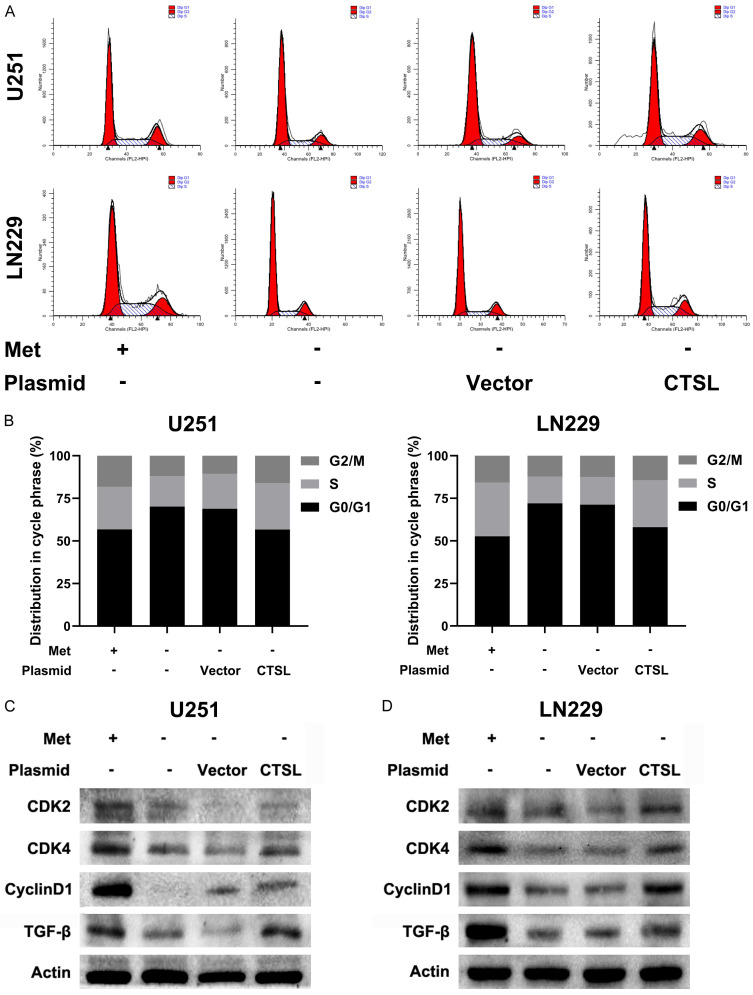
Methionine deprivation/CTSL downregulation induces G1 arrest in glioma cells
Previous studies reported that CTSL can promote tumor cell proliferation by activating CCAAT-displacement protein/cut homeobox (CDP/Cux) transcription factors and accelerating the entry into the S phase of the cell cycle [30]. Therefore, we hypothesized that methionine deprivation affects cell cycle progression. Under these conditions, the distribution of the cell cycle was measured by flow cytometry. These results showed that under methionine deprivation, the percentage of cells in G1 stage increased (Figure 6A, 6B), and CTSL overexpression partially reversed the arrest effect. At the same time, the expression of cell cycle related proteins was consistent with this result (Figure 6C, 6D). These results suggest that methionine deprivation/CTSL downregulation inhibit the G1-to-S transition of glioma cells.
Figure 6.
Methionine deprivation leads to cell cycle arrest of glioma cells. A and B. Flow cytometry was used to analyze cell cycle distribution in U251 and LN229 cells. C and D. Expression of cell-related proteins in U251 and LN229 cells after transfection.
Expression and significance of CTSL in glioma patients
During our study, we found that methionine deprivation could significantly inhibit the expression of CTSL and proliferation, invasion and migration of glioma. However, the role of CTSL in human gliomas remains unclear. We analyzed the CTSL expression data in the CGGA database, and this analysis showed that the expression of CTSL increased with increasing of WHO grade (Figure 7A). In addition, CTSL protein expression was upregulated in human gliomas compared with nontumor tissues (Figure 7B, 7C). These results suggest that CTSL is upregulated in human gliomas.
Figure 7.
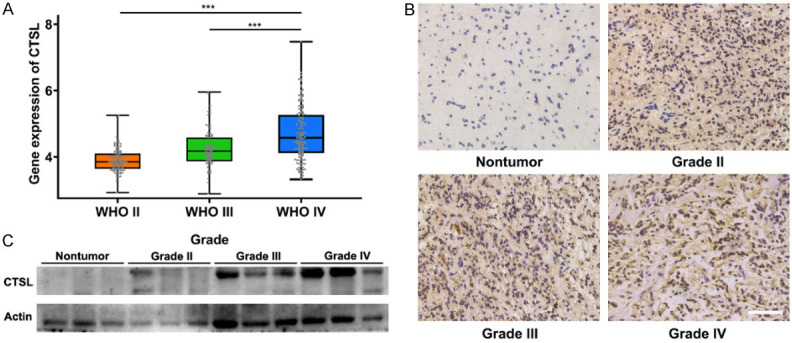
The upregulated expression of CTSL in human gliomas is associated with the prognosis of glioma patients. A. Relative expression of CTSL in CGGA glioma patient tissues. B and C. CTSL expression in different WHO grade human glioma tissues and normal brain tissues. Bar: 100 μm. ***P < 0.001.
Discussion
Alteration of cellular metabolism is an emerging marker of tumor cell proliferation and growth [41-43]. The rapid growth of tumors depends on the external nutrient supply, and the metabolism of tumor cells can be significantly affected by changing the content of substances in the environment. Therefore, finding metabolically sensitive nutrients that target specific tumors should be the key to solving the problem. Inspired by MET PET, we hypothesized that glioma is methionine-dependent. Methionine is an essential amino acid that cannot be synthesized by our body and must be supplied by food. Methionine plays important roles in the human body. Many studies have shown that tumor growth can be significantly inhibited under methionine deprivation condition, but we still do not know how this works. During our research, we found that the growth of glioma was also methionine-dependent, which preliminarily confirmed our hypothesis. Upon screening for differentially expressed genes, we found that the expression of CTSL was significantly downregulated. Through further study and validation of CTSL, we found that it plays an important role in methionine-deprived glioma cells.
One of the characteristics of malignant tumors is their aggressiveness and metastasis. Through metabolic reprogramming, tumor cells express fewer adhesion molecules on their surface and separate from each other [44,45]. At the same time, proteases are secreted to dissolve extracellular matrix components. After passing through the local defect in the basement membrane like amoeba, the tumor cells continue to dissolve interstitial connective tissue and diffuse to more distant areas [46]. CTSL is a lysosomal protease that is involved in terminal degradation of intracellular and endocytic proteins [26,47]. Studies have shown that the expression level of CTSL is strongly correlated with the invasiveness of pancreatic cancer [23,48]. In colorectal cancer, the level and activity of CTSL have been shown to be increased, and the expression of urokinase plasminogen activator -1, its downstream target, has been shown to be increased in colorectal cancer tissues, which is closely related to the incidence of metastasis [49,50]. We have experimentally provided evidence that CTSL expression is upregulated in human glioma tissues, which is consistent with CGGA data. At the same time, we verified that a CTSL knockout inhibited the viability and the invasion and migration abilities of glioma cells in vivo and in vitro. Combined with the above results, we believe that CTSL is essential for the growth of gliomas.
Asanuma et al. have suggested that tumor-secreted cytokines, including VEGF, FGF, PDGF, EGF, NGF, -INFγ and -IL-6, are widely involved in malignant progression and have also been shown to significantly enhance CTSL promoter activity and synthesis [26,51-55]. This suggests that CTSL may be regulated by inflammatory responses. In the tumor microenvironment, the inflammatory response plays an important role in tumorigenesis and tumor development [42]. Inflammatory and immune cells, such as tumor-associated neutrophils (TANs) and tumor-associated macrophages (TAMs), can also induce tumor progression and metastasis [56,57]. An increased number of TANs is associated with a poorer prognosis in patients with hepatocellular carcinoma and renal cell carcinoma [58], and similarly, a high number of TAMs in patients with pancreatic ductal adenocarcinoma is also associated with a poor prognosis [59]. As a way of communicating with tumors, cytokines play a key role in transmitting growth information [60]. Therefore, several cytokines within the tumor tissues were measured, and immune/inflammatory genes were screened by GO analysis and KEGG analysis. Preliminary findings indicated that specific inflammatory factors and potential target genes may be responsible for the upregulation of CTSL. We will continue to explore the function and relationship between inflammation and CTSL.
In conclusion, the downregulation of CTSL inhibited the proliferation, invasion and migration of glioma cells. Mechanistically, methionine deprivation inhibited the expression of CTSL, and overexpression of CTSL partially decreased the inhibition of methionine deprivation in glioma cells. The high dependence of methionine on glioma may be one of the reasons for its malignant biological behavior. This study provides a new idea for the treatment of glioma by dietary modification.
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China [No. 82072796]. This work was also financed by the Social Development Project of Jiangsu Department of Science and Technology [BE2020642] and Postgraduate Research & Practice Innovation Program of Jiangsu Province [KYCX21-2657 and KYCX21-2658].
The consent agreement was signed and approved by the Ethics Committee of The Affiliated Hospital of Xuzhou Medical University. All patients involved in the present study didn’t receive chemotherapy or radiotherapy before the surgery. All animal experiments were approved by the Ethical Committee of Xuzhou Medical University.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Hart MG, Garside R, Rogers G, Stein K, Grant R. Temozolomide for high grade glioma. Cochrane Database Syst Rev. 2013;2013:CD007415. doi: 10.1002/14651858.CD007415.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang P, Shi H, Zhu W, Gui Q, Xu Y, Meng J, Guo X, Gong Z, Chen H. Silver nanoparticles enhance the sensitivity of temozolomide on human glioma cells. Oncotarget. 2017;8:7533–7539. doi: 10.18632/oncotarget.13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu M, Fan Y, Lv S, Xiao B, Ye M, Zhu X. Vincristine and temozolomide combined chemotherapy for the treatment of glioma: a comparison of solid lipid nanoparticles and nanostructured lipid carriers for dual drugs delivery. Drug Deliv. 2016;23:2720–2725. doi: 10.3109/10717544.2015.1058434. [DOI] [PubMed] [Google Scholar]
- 4.Cheng D, Cao N, Chen J, Yu X, Shuai X. Multifunctional nanocarrier mediated co-delivery of doxorubicin and siRNA for synergistic enhancement of glioma apoptosis in rat. Biomaterials. 2012;33:1170–1179. doi: 10.1016/j.biomaterials.2011.10.057. [DOI] [PubMed] [Google Scholar]
- 5.Messaoudi K, Saulnier P, Boesen K, Benoit JP, Lagarce F. Anti-epidermal growth factor receptor siRNA carried by chitosan-transacylated lipid nanocapsules increases sensitivity of glioblastoma cells to temozolomide. Int J Nanomedicine. 2014;9:1479–1490. doi: 10.2147/IJN.S59134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X, Wang Z, Liu N, Cheng Y, Jin W, Zhang P, Wang X, Yang H, Liu H, Zhang Y, Tu Y. Association between SOX9 and CA9 in glioma, and its effects on chemosensitivity to TMZ. Int J Oncol. 2018;53:189–202. doi: 10.3892/ijo.2018.4382. [DOI] [PubMed] [Google Scholar]
- 7.Gao X, Sanderson SM, Dai Z, Reid MA, Cooper DE, Lu M, Richie JP Jr, Ciccarella A, Calcagnotto A, Mikhael PG, Mentch SJ, Liu J, Ables G, Kirsch DG, Hsu DS, Nichenametla SN, Locasale JW. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature. 2019;572:397–401. doi: 10.1038/s41586-019-1437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goncalves MD, Lu C, Tutnauer J, Hartman TE, Hwang SK, Murphy CJ, Pauli C, Morris R, Taylor S, Bosch K, Yang S, Wang Y, Van Riper J, Lekaye HC, Roper J, Kim Y, Chen Q, Gross SS, Rhee KY, Cantley LC, Yun J. High-fructose corn syrup enhances intestinal tumor growth in mice. Science. 2019;363:1345–1349. doi: 10.1126/science.aat8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez PS, O’Prey J, Cardaci S, Barthet VJA, Sakamaki JI, Beaumatin F, Roseweir A, Gay DM, Mackay G, Malviya G, Kania E, Ritchie S, Baudot AD, Zunino B, Mrowinska A, Nixon C, Ennis D, Hoyle A, Millan D, McNeish IA, Sansom OJ, Edwards J, Ryan KM. Mannose impairs tumour growth and enhances chemotherapy. Nature. 2018;563:719–723. doi: 10.1038/s41586-018-0729-3. [DOI] [PubMed] [Google Scholar]
- 10.Lee C, Raffaghello L, Brandhorst S, Safdie FM, Bianchi G, Martin-Montalvo A, Pistoia V, Wei M, Hwang S, Merlino A, Emionite L, de Cabo R, Longo VD. Fasting cycles retard growth of tumors and sensitize a range of cancer cell types to chemotherapy. Sci Transl Med. 2012;4:124ra127. doi: 10.1126/scitranslmed.3003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee C, Safdie FM, Raffaghello L, Wei M, Madia F, Parrella E, Hwang D, Cohen P, Bianchi G, Longo VD. Reduced levels of IGF-I mediate differential protection of normal and cancer cells in response to fasting and improve chemotherapeutic index. Cancer Res. 2010;70:1564–1572. doi: 10.1158/0008-5472.CAN-09-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanarek N, Petrova B, Sabatini DM. Dietary modifications for enhanced cancer therapy. Nature. 2020;579:507–517. doi: 10.1038/s41586-020-2124-0. [DOI] [PubMed] [Google Scholar]
- 13.Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7:1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- 14.Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maddocks ODK, Athineos D, Cheung EC, Lee P, Zhang T, van den Broek NJF, Mackay GM, Labuschagne CF, Gay D, Kruiswijk F, Blagih J, Vincent DF, Campbell KJ, Ceteci F, Sansom OJ, Blyth K, Vousden KH. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature. 2017;544:372–376. doi: 10.1038/nature22056. [DOI] [PubMed] [Google Scholar]
- 16.Pacold ME, Brimacombe KR, Chan SH, Rohde JM, Lewis CA, Swier LJ, Possemato R, Chen WW, Sullivan LB, Fiske BP, Cho S, Freinkman E, Birsoy K, Abu-Remaileh M, Shaul YD, Liu CM, Zhou M, Koh MJ, Chung H, Davidson SM, Luengo A, Wang AQ, Xu X, Yasgar A, Liu L, Rai G, Westover KD, Vander Heiden MG, Shen M, Gray NS, Boxer MB, Sabatini DM. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol. 2016;12:452–458. doi: 10.1038/nchembio.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanarek N, Keys HR, Cantor JR, Lewis CA, Chan SH, Kunchok T, Abu-Remaileh M, Freinkman E, Schweitzer LD, Sabatini DM. Histidine catabolism is a major determinant of methotrexate sensitivity. Nature. 2018;559:632–636. doi: 10.1038/s41586-018-0316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman EM, Nierenberg DW, Santi DV. Selective killing of transformed cells by methotrexate with histidine deprivation or with alpha-amino alcohols. Cancer Res. 1983;43:4703–4708. [PubMed] [Google Scholar]
- 19.Nariai T, Tanaka Y, Wakimoto H, Aoyagi M, Tamaki M, Ishiwata K, Senda M, Ishii K, Hirakawa K, Ohno K. Usefulness Of L-[methyl-11C] methionine-positron emission tomography as a biological monitoring tool in the treatment of glioma. J Neurosurg. 2005;103:498–507. doi: 10.3171/jns.2005.103.3.0498. [DOI] [PubMed] [Google Scholar]
- 20.Singhal T, Narayanan TK, Jacobs MP, Bal C, Mantil JC. 11C-methionine PET for grading and prognostication in gliomas: a comparison study with 18F-FDG PET and contrast enhancement on MRI. J Nucl Med. 2012;53:1709–1715. doi: 10.2967/jnumed.111.102533. [DOI] [PubMed] [Google Scholar]
- 21.Sudo A, Shiga T, Okajima M, Takano K, Terae S, Sawamura Y, Ohnishi A, Nagashima K, Saitoh S. High uptake on 11C-methionine positron emission tomographic scan of basal ganglia germinoma with cerebral hemiatrophy. AJNR Am J Neuroradiol. 2003;24:1909–1911. [PMC free article] [PubMed] [Google Scholar]
- 22.Sadeghi N, Salmon I, Decaestecker C, Levivier M, Metens T, Wikler D, Denolin V, Rorive S, Massager N, Baleriaux D, Goldman S. Stereotactic comparison among cerebral blood volume, methionine uptake, and histopathology in brain glioma. AJNR Am J Neuroradiol. 2007;28:455–461. [PMC free article] [PubMed] [Google Scholar]
- 23.Chauhan SS, Goldstein LJ, Gottesman MM. Expression of cathepsin L in human tumors. Cancer Res. 1991;51:1478–1481. [PubMed] [Google Scholar]
- 24.Poreba M, Rut W, Vizovisek M, Groborz K, Kasperkiewicz P, Finlay D, Vuori K, Turk D, Turk B, Salvesen GS, Drag M. Selective imaging of cathepsin L in breast cancer by fluorescent activity-based probes. Chem Sci. 2018;9:2113–2129. doi: 10.1039/c7sc04303a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueki N, Wang W, Swenson C, McNaughton C, Sampson NS, Hayman MJ. Synthesis and preclinical evaluation of a highly improved anticancer prodrug activated by histone deacetylases and cathepsin L. Theranostics. 2016;6:808–816. doi: 10.7150/thno.13826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudhan DR, Siemann DW. Cathepsin L targeting in cancer treatment. Pharmacol Ther. 2015;155:105–116. doi: 10.1016/j.pharmthera.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denhardt DT, Greenberg AH, Egan SE, Hamilton RT, Wright JA. Cysteine proteinase cathepsin L expression correlates closely with the metastatic potential of H-ras-transformed murine fibroblasts. Oncogene. 1987;2:55–59. [PubMed] [Google Scholar]
- 28.Doherty PJ, Hua L, Liau G, Gal S, Graham DE, Sobel M, Gottesman MM. Malignant transformation and tumor promoter treatment increase levels of a transcript for a secreted glycoprotein. Mol Cell Biol. 1985;5:466–473. doi: 10.1128/mcb.5.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rabin MS, Doherty PJ, Gottesman MM. The tumor promoter phorbol 12-myristate 13-acetate induces a program of altered gene expression similar to that induced by platelet-derived growth factor and transforming oncogenes. Proc Natl Acad Sci U S A. 1986;83:357–360. doi: 10.1073/pnas.83.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goulet B, Sansregret L, Leduy L, Bogyo M, Weber E, Chauhan SS, Nepveu A. Increased expression and activity of nuclear cathepsin L in cancer cells suggests a novel mechanism of cell transformation. Mol Cancer Res. 2007;5:899–907. doi: 10.1158/1541-7786.MCR-07-0160. [DOI] [PubMed] [Google Scholar]
- 31.Goulet B, Truscott M, Nepveu A. A novel proteolytically processed CDP/Cux isoform of 90 kDa is generated by cathepsin L. Biol Chem. 2006;387:1285–1293. doi: 10.1515/BC.2006.159. [DOI] [PubMed] [Google Scholar]
- 32.Moon NS, Zeng WR, Premdas P, Santaguida M, Berube G, Nepveu A. Expression of N-terminally truncated isoforms of CDP/Cux is increased in human uterine leiomyomas. Int J Cancer. 2002;100:429–432. doi: 10.1002/ijc.10510. [DOI] [PubMed] [Google Scholar]
- 33.Svatek RS, Karam J, Karakiewicz PI, Gallina A, Casella R, Roehrborn CG, Shariat SF. Role of urinary cathepsin B and L in the detection of bladder urothelial cell carcinoma. J Urol. 2008;179:478–484. doi: 10.1016/j.juro.2007.09.037. discussion 484. [DOI] [PubMed] [Google Scholar]
- 34.Chen Q, Fei J, Wu L, Jiang Z, Wu Y, Zheng Y, Lu G. Detection of cathepsin B, cathepsin L, cystatin C, urokinase plasminogen activator and urokinase plasminogen activator receptor in the sera of lung cancer patients. Oncol Lett. 2011;2:693–699. doi: 10.3892/ol.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leto G, Tumminello FM, Pizzolanti G, Montalto G, Soresi M, Carroccio A, Ippolito S, Gebbia N. Lysosomal aspartic and cysteine proteinases serum levels in patients with pancreatic cancer or pancreatitis. Pancreas. 1997;14:22–27. doi: 10.1097/00006676-199701000-00004. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Yang HC, Wang Q, Yang ZJ, Chen H, Wang SM, Pan ZM, Tang BJ, Li QQ, Li L. Clinical value of combined detection of serum matrix metalloproteinase-9, heparanase, and cathepsin for determining ovarian cancer invasion and metastasis. Anticancer Res. 2011;31:3423–3428. [PubMed] [Google Scholar]
- 37.Xu H, Han Y, Zhao G, Zhang L, Zhao Z, Wang Z, Zhao L, Hua L, Naveena K, Lu J, Yu R, Liu H. Hypoxia-responsive lipid-polymer nanoparticle-combined imaging-guided surgery and multitherapy strategies for glioma. ACS Appl Mater Interfaces. 2020;12:52319–52328. doi: 10.1021/acsami.0c12971. [DOI] [PubMed] [Google Scholar]
- 38.Wang K, Liu H, Liu J, Wang X, Teng L, Zhang J, Liu Y, Yao Y, Wang J, Qu Y, Chen X, Peng F, Liu H, Wang N, Zhong Y, Hou X, Jiang H, Beylerli O, Liao X, Zhang X, Zhang X, Zhang X, Zhao S. IL1RN mediates the suppressive effect of methionine deprivation on glioma proliferation. Cancer Lett. 2019;454:146–157. doi: 10.1016/j.canlet.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 39.Zhao G, Jia J, Wang L, Zhang Y, Yang H, Lu Y, Yu R, Liu H, Zhu Y. Local delivery of minocycline and vorinostat targets the tumor microenvironment to inhibit the recurrence of glioma. Onco Targets Ther. 2020;13:11397–11409. doi: 10.2147/OTT.S273527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao S, Sha Z, Zhou J, Wu Y, Song Y, Li C, Liu X, Zhang T, Yu R. BYSL contributes to tumor growth by cooperating with the mTORC2 complex in gliomas. Cancer Biol Med. 2021;18:88–104. doi: 10.20892/j.issn.2095-3941.2020.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 43.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 45.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan GJ, Peng ZK, Lu JP, Tang FQ. Cathepsins mediate tumor metastasis. World J Biol Chem. 2013;4:91–101. doi: 10.4331/wjbc.v4.i4.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dennemarker J, Lohmuller T, Muller S, Aguilar SV, Tobin DJ, Peters C, Reinheckel T. Impaired turnover of autophagolysosomes in cathepsin L deficiency. Biol Chem. 2010;391:913–922. doi: 10.1515/BC.2010.097. [DOI] [PubMed] [Google Scholar]
- 48.Cao X, Zhu J, Li X, Ma Y, He Q. Expression of CXCR4 and CXCR7 in papillary thyroid carcinoma and adjacent tissues and their relationship with pathologic indicators of tumor aggressiveness. Endocr J. 2022;69:189–197. doi: 10.1507/endocrj.EJ21-0076. [DOI] [PubMed] [Google Scholar]
- 49.Adenis A, Huet G, Zerimech F, Hecquet B, Balduyck M, Peyrat JP. Cathepsin B, L, and D activities in colorectal carcinomas: relationship with clinico-pathological parameters. Cancer Lett. 1995;96:267–275. doi: 10.1016/0304-3835(95)03930-u. [DOI] [PubMed] [Google Scholar]
- 50.Herszenyi L, Plebani M, Carraro P, De Paoli M, Roveroni G, Cardin R, Tulassay Z, Naccarato R, Farinati F. The role of cysteine and serine proteases in colorectal carcinoma. Cancer. 1999;86:1135–1142. doi: 10.1002/(sici)1097-0142(19991001)86:7<1135::aid-cncr6>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 51.Asanuma K, Shirato I, Ishidoh K, Kominami E, Tomino Y. Selective modulation of the secretion of proteinases and their inhibitors by growth factors in cultured differentiated podocytes. Kidney Int. 2002;62:822–831. doi: 10.1046/j.1523-1755.2002.00539.x. [DOI] [PubMed] [Google Scholar]
- 52.Gallardo E, de Andres I, Illa I. Cathepsins are upregulated by IFN-gamma/STAT1 in human muscle culture: a possible active factor in dermatomyositis. J Neuropathol Exp Neurol. 2001;60:847–855. doi: 10.1093/jnen/60.9.847. [DOI] [PubMed] [Google Scholar]
- 53.Gerber A, Wille A, Welte T, Ansorge S, Buhling F. Interleukin-6 and transforming growth factor-beta 1 control expression of cathepsins B and L in human lung epithelial cells. J Interferon Cytokine Res. 2001;21:11–19. doi: 10.1089/107999001459114. [DOI] [PubMed] [Google Scholar]
- 54.Keerthivasan S, Keerthivasan G, Mittal S, Chauhan SS. Transcriptional upregulation of human cathepsin L by VEGF in glioblastoma cells. Gene. 2007;399:129–136. doi: 10.1016/j.gene.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 55.Lah TT, Hawley M, Rock KL, Goldberg AL. Gamma-interferon causes a selective induction of the lysosomal proteases, cathepsins B and L, in macrophages. FEBS Lett. 1995;363:85–89. doi: 10.1016/0014-5793(95)00287-j. [DOI] [PubMed] [Google Scholar]
- 56.Coffelt SB, Wellenstein MD, de Visser KE. Neutrophils in cancer: neutral no more. Nat Rev Cancer. 2016;16:431–446. doi: 10.1038/nrc.2016.52. [DOI] [PubMed] [Google Scholar]
- 57.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li YW, Qiu SJ, Fan J, Zhou J, Gao Q, Xiao YS, Xu YF. Intratumoral neutrophils: a poor prognostic factor for hepatocellular carcinoma following resection. J Hepatol. 2011;54:497–505. doi: 10.1016/j.jhep.2010.07.044. [DOI] [PubMed] [Google Scholar]
- 59.Diana A, Wang LM, D’Costa Z, Azad A, Silva MA, Soonawalla Z, Allen P, Liu S, McKenna WG, Muschel RJ, Fokas E. Prognostic role and correlation of CA9, CD31, CD68 and CD20 with the desmoplastic stroma in pancreatic ductal adenocarcinoma. Oncotarget. 2016;7:72819–72832. doi: 10.18632/oncotarget.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sielska M, Przanowski P, Wylot B, Gabrusiewicz K, Maleszewska M, Kijewska M, Zawadzka M, Kucharska J, Vinnakota K, Kettenmann H, Kotulska K, Grajkowska W, Kaminska B. Distinct roles of CSF family cytokines in macrophage infiltration and activation in glioma progression and injury response. J Pathol. 2013;230:310–321. doi: 10.1002/path.4192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.