Abstract
Objective: This study aimed to evaluate the resistance of Helicobacter pylori (H. pylori) to common antibiotics in Shanghai. Methods: A total of 1171 eligible subjects participated in the study. Antibiotic susceptibility to six common antibiotics was examined with the disk diffusion method. Mutations in resistant-related genes were identified via Sanger sequencing analysis. Results: Overall, the resistance rates of strains to amoxicillin, clarithromycin, levofloxacin, metronidazole, tetracycline, and furazolidone were 0.1%, 27.8%, 31.1%, 79.9%, 0.1%, and 0.5%, respectively. Compared with untreated patients, resistance rates of clarithromycin (P < 0.01), levofloxacin (P < 0.01), and metronidazole were significantly higher in re-treated patients (P < 0.05). The total multiple resistance rate was 40.5%. Age (levofloxacin), gender (clarithromycin, levofloxacin, and metronidazole) and endoscopic findings (clarithromycin and levofloxacin) were independent factors influencing antibiotic resistance. High correlation was observed between the drug susceptibility test and molecular test for the resistance to clarithromycin and levofloxacin. Conclusions: The resistance rates of H. pylori to amoxicillin, tetracycline, and furazolidone were low, whereas the resistance rates of H. pylori to clarithromycin, levofloxacin, and metronidazole were high, especially in re-treated patients. Our results indicate that the clinical resistance patterns of clarithromycin and levofloxacin could be guided by relevant gene mutations.
Keywords: Antibiotic, gene mutations, Helicobacter pylori, resistance, Shanghai
Introduction
Helicobacter pylori (H. pylori) is a Gram-negative spiral bacterium that colonizes the human stomach and duodenal mucosa and it affects 50% of the global population [1,2]. The infection rate ranges from 25% to 50% in developed countries, whereas over 90% of the population is infected in developing countries [3]. H. pylori infection has become a growing concern, closely related to the development and occurrence of gastritis, ulcers, and tumors [4]. To prevent and treat these diseases, successfully eradicating H. pylori is essential [5-7]. In 1994, H. pylori was identified as a category I carcinogen for gastric cancer and recommended for eradication by the World Health Organization (WHO) [8].
Triple therapy, comprising a proton pump inhibitor (PPI) and two antibiotics (amoxicillin plus clarithromycin or metronidazole) has been widely applied as an eradication therapy [9]. Nevertheless, the effectiveness of this therapy has been severely compromised by increasing antibiotic resistance, with several studies showing eradication rates falling to less than 80% [10-13]. It is thus critical to determine the mechanisms of H. pylori resistance to common antibiotics, primarily those induced by numerous chromosomal point mutations to establish rational combinations of antibiotics for therapy [14,15].
To determine the antibiotic resistance of H. pylori, traditional antibiotic susceptibility tests or molecular tests with PCR and sequencing can be used. Antibiotic susceptibility testing is effective, but difficult to isolate and culture. Compared with antibiotic susceptibility tests, molecular tests are inexpensive and efficient in that a large number of DNA sequences can be generated within a short time to clarify antibiotic resistance. The resistance of different H. pylori strains to common antibiotics could be attributed to the mutations of related genes, such as penicillin-binding protein 1 (PBP1) (amoxicillin), 23S rRNA (clarithromycin), gyrA (levofloxacin), rdxA (metronidazole), 16S rRNA (tetracycline), porD, and oorD (furazolidone) [11,15-19]. Antibiotic resistance varies widely between geographical areas [14,20,21]. Therefore, antibiotics involved in eradication regimens could be selected according to local epidemiologic resistance characteristics.
Our study aimed to evaluate the antibiotic resistance of H. pylori, and we are the first to comprehensively analyze the relationship between the resistance of H. pylori to six common antibiotics and specific mutations of associated genes in Shanghai.
Patients and methods
Patients and tissue samples
From August 2019 to May 2020, 1171 patients undergoing upper endoscopy at the Endoscopy Center of Songjiang Hospital, Shanghai Jiaotong University School of Medicine, were enrolled. One biopsy from the gastric corpus and two pieces of tissue from the gastric antrum were collected from each patient for subsequent testing.
Inclusion and exclusion criteria
Inclusion criteria were as follows: (1) patients aged 18-80 years and, (2) patients positive for the 13C-urea breath test. Exclusion criteria were as follows: (1) patients who had taken PPI, H2 receptor blockers, bismuth or Chinese herbal medicine no more than 2 weeks before enrolment, (2) patients who had received antibiotics no more than 4 weeks before enrolment, (3) patients who previously underwent gastrointestinal surgery, (4) patients with gastrointestinal malignancy, (5) patients with serious heart, lung, liver or renal dysfunction or significant mental disorders, (6) pregnant or lactating women, and (7) patients who participated in other clinical trials no more than last three months before enrollment. This study was approved of by the Ethics Committee at Songjiang Hospital, Shanghai Jiaotong University School of Medicine (201901). All subjects signed an informed consent.
Culture and identification of H. pylori
The biopsies were ground, and then inoculated directly onto a Columbia Agar plate containing 5% defibrinated sheep blood, and biopsies of the gastric antrum and body were marked. Plates were incubated microaerophilically (5% O2, 10% CO2, and 85% N2) for 72 h at 37°C. If the incubation failed, we extended the incubation time to 7-10 days to judge whether the strain was successfully cultivated. Suspected colonies were confirmed as H. pylori if bacteria were positive for catalase, urease, and oxidase reactions, and appeared as Gram-negative bacilli under the microscope.
Antibiotic susceptibility testing
The inhibition zones of common antibiotics were determined by the disc diffusion method. The amount of amoxicillin, clarithromycin, levofloxacin, metronidazole, tetracycline, and furazolidone in each dish was 10 μg, 15 μg, 5 μg, 5 μg, 30 μg, and 100 μg, respectively. The inhibition zone diameter breakpoints were formulated with reference to the antimicrobial susceptibility test criteria established by the China Pharmaceutical and Biological Products Appraisal Institute. A strain was considered resistant to amoxicillin, clarithromycin, levofloxacin, metronidazole, tetracycline, and furazolidone with inhibition zones of < 14 mm, 13 mm, 13 mm, 16 mm, 14 mm, and 14 mm, respectively [22] (Supplementary Table 1).
DNA extraction
DNA was extracted by Magen HiPure bacterial DNA kits (Magen Biotech Corporation, Guangdong, China), and stored at -20°C until use.
Mutational analysis of resistance genes
We detected specific site mutations of the following genes: penicillin-binding protein 1 (PBP1), 23S rRNA, porD, oorD, 16S rRNA, gyrA, and rdxA. To do so, a PCR-based sequencing approach was conducted on susceptible and resistant strains. The primer sequences of target genes are presented in Table 1 [22]. Both strands of amplified products were sequenced via the Sanger method using a generation sequencer (ThermoFisher, Shanghai, China). Chromas and Sequencing Analysis 5.2 software was used for comparative sequence analyses of resistant and susceptible strains.
Table 1.
Genes and primer sequences for detection of mutations associated with antibiotic resistance
| Antibiotic | Gene | Mutation site | Primer sequences (5’-3’) | Product size |
|---|---|---|---|---|
| CAM | 23S rRNA | A2142; A2143 | F: AATGGCGTAACGAGATGGGAG | 501 bp |
| R: TCCATAAGAGCCAAAGCCCTTAC | ||||
| AMX | PBP1 | 320Ala; 366Phe; 369Ala; 374Val; 414Ser; 423Leu; 556Thr; 562Asn; 593Thr | F: GAAAAAATCGCTAAAGAAGAGCCAA | 851 bp; 549 bp |
| R: TACGCATGAAATACGAATACACAGG | ||||
| FUR | porD; oorD | G353A; A356G; C357T; A41G; A122G; A335G | F: GGCGTGGATTATTCTCATTGTAAAGG | 427 bp; 789 bp |
| R: GGATAGGCTGCGATGACATCAATT | ||||
| F: CATGCTTTCAGCGCGACTTATA | ||||
| R: CCCACTTCAATCGCCGCTTTA | ||||
| TCN | 16S rRNA | AGA926-928 | F: GTGCAAGCGTTACTCGGAATCA | 366 bp |
| R: AACCCAACATCTCACGACACGA | ||||
| LEV | gyrA | N87I; N87K; D91N; D91Y; D91G; N87Y; N87T; N87H | F: ATTCCATGAGCGTGATCATAGG | 602 bp |
| R: GTTATCGCCATCAATAGAGCCAAA | ||||
| MNZ | rdxA | A61; T62; A91; C92; G392; A610; C92; A614 | F: TGAGCATGGGGCAGATTTTAAGC | 959 bp |
| R: TTGAAAACACCCCTAAAAGAGCG |
AMX, amoxicillin; CAM, clarithromycin; LEV, levofloxacin; MNZ, metronidazole; TCN, tetracycline; FUR, furazolidone.
Statistical analysis
Categorical data are presented as percentages. The chi-squared or Fisher’s exact was used to compare antibiotic resistance rates between the untreated and retreated patients, and the gene mutation rates between antibiotic-susceptible and -resistant strains. The factors that could influence the antibiotic resistance were analyzed by univariate analyses. Thereafter, the relationships between independent variables and dependent variables (antibiotic susceptibility or resistance) were analyzed by binary logistic regression, and the odds ratios (ORs) and 95% confidence intervals (CIs) were calculated. A two-sided p-value < 0.05 indicated statistical significance. SPSS 19.0 software was used for all statistical analyses.
Results
Patient characteristics
Overall, 1116 (95.3%, 1116/1171) H. pylori isolates were cultured successfully from endoscopic biopsy tissues from 1171 patients. Among these, 1060 (90.5%, 1060/1171) patients in the untreated group and 106 (9.1%, 106/1171) in the re-treated group that had failed in previous H. pylori eradication treatment were included. The remaining 5 (0.4%, 5/1171) patients were from the recurrence group of patients who retested positive for H. pylori after confirmation of the success of H. pylori eradication treatment (Table 2).
Table 2.
Characteristics of patients
| Characteristics | Untreated group | Re-treated group | Recurrence group | Total |
|---|---|---|---|---|
| n = 1060 | n = 106 | n = 5 | n = 1171 | |
| Isolation strains, N (%) | 1014 (95.7) | 98 (92.5) | 4 (80.0) | 1116 (95.3) |
| Age (X±S) | 45.63±13.61 | 46.79±12.51 | 45.00±10.61 | 45.73±13.50 |
| Male, N (%) | 489 (46.1) | 43 (40.6) | 3 (60.0) | 535 (45.7) |
| Endoscopic diagnosis, N (%) | ||||
| Chronic gastritis | 822 (77.6) | 93 (87.7) | 5 (100) | 920 (78.6) |
| Gastric ulcer | 58 (5.5) | 4 (3.8) | 0 | 62 (5.3) |
| Duodenal ulcer | 180 (17.0) | 9 (8.5) | 0 | 189 (16.1) |
Prevalence of antibiotic resistance
Figure 1 shows that the overall resistance rates were 79.9% for metronidazole (892/1116), 31.1% for levofloxacin (347/1116), 27.8% for clarithromycin (310/1116), 0.5% for furazolidone (6/1116), 0.1% for amoxicillin (1/1116), and 0.1% for tetracycline (1/1116). Compared with untreated patients, the re-treated patients showed significantly higher resistance rates for clarithromycin (67.4%, 66/98 vs 23.8%, 241/1014, P < 0.01), levofloxacin (45.9%, 45/98 vs 29.7%, 301/1014, P < 0.01), and metronidazole (91.8%, 90/98 vs 78.7%, 798/1014, P < 0.05).
Figure 1.
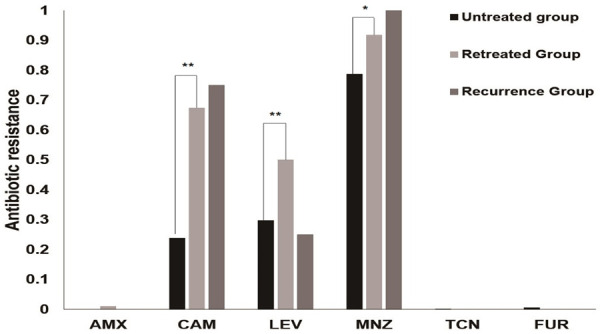
Antibiotic resistance rates of H. pylori strains. AMX, amoxicillin; CAM, clarithromycin; LEV, levofloxacin; MNZ, metronidazole; TCN, tetracycline; FUR, furazolidone. *, P < 0.05; **, P < 0.01.
Multiple antibiotic resistance
Among the 1116 strains tested, only 153 (13.7%, 153/1116) strains were sensitive to all antibiotics, while 511 (45.8%, 511/1116), 314 (28.1%, 314/1116), 137 (12.3%, 137/1116), and 1 (0.1%, 1/1116) were resistant to one, two, three, and four antibiotics, respectively. Metronidazole/levofloxacin and metronidazole/clarithromycin were the predominant dual-resistance patterns in the untreated and re-treated groups, respectively. Triple resistance to levofloxacin, clarithromycin, and metronidazole, was the predominant pattern in both groups. In the re-treated group, a quadruple resistance pattern to amoxicillin, levofloxacin, clarithromycin, and metronidazole was observed. Compared with untreated patients, the multiple antibiotic resistance rate was significantly higher in the untreated group (71.4%, 70/98 vs 37.4%, 379/1014, P < 0.01) (Table 3).
Table 3.
Multiple patterns of resistance of H. pylori strains to all tested antibiotics
| Resistance Profiles, N (%) | Untreated group | Re-treated group | Recurrence group | All | P (untreated vs re-treated) |
|---|---|---|---|---|---|
| n = 1014 | n = 98 | n = 4 | n = 1116 | ||
| All susceptible | 151 (14.9) | 2 (2.0) | 0 | 153 (13.7) | 0.001 |
| Mono resistance | 484 (47.7) | 26 (26.5) | 1 (25.0) | 511 (44.8) | 0.01 |
| MNZ | 429 (42.3) | 20 (20.4) | 1 (25.0) | 450 (40.3) | 0.01 |
| CAM | 26 (2.6) | 6 (6.1) | 0 | 32 (2.9) | 0.090 |
| LVX | 28 (2.8) | 0 | 0 | 28 (2.5) | 0.184 |
| Dual resistance | 277 (27.3) | 35 (35.7) | 2 (50.0) | 314 (28.1) | 0.077 |
| CAM+MNZ | 102 (10.1) | 25 (25.5) | 2 (50.0) | 129 (11.6) | 0.01 |
| LVX+MNZ | 160 (15.8) | 9 (9.2) | 0 | 169 (15.1) | 0.082 |
| CAM+LVX | 12 (1.2) | 1 (1.0) | 0 | 13 (1.2) | 1.00 |
| FUR+LVX | 3 (0.3) | 0 | 0 | 3 (0.3) | 1.00 |
| Triple resistance | 102 (10.1) | 34 (34.7) | 1 (25.0) | 137 (12.3) | 0.01 |
| CAM+LVX+MNZ | 99 (9.8) | 34 (34.7) | 1 (25.0) | 134 (12.0) | 0.01 |
| LVX+FUR+MNZ | 2 (0.2) | 0 | 0 | 2 (0.2) | 1.00 |
| CAM+TCN+MNZ | 1 (0.1) | 0 | 0 | 1 (0.1) | 1.00 |
| Quadruple resistance | 0 | 1 (1.0) | 0 | 1 (0.1) | 0.004 |
| CAM+LVX+AMX+MNZ | 0 | 1 (1.0) | 0 | 1 (0.1) | 0.004 |
AMX, amoxicillin; CAM, clarithromycin; LEV, levofloxacin; MNZ, metronidazole; TCN, tetracycline; FUR, furazolidone.
Factors influencing H. pylori antibiotic resistance
Univariate analysis showed that the resistance rates to clarithromycin, levofloxacin, and metronidazole were significantly different between men and women as well as in different age groups. Moreover, the resistance rates to levofloxacin and clarithromycin were significantly different between patients with peptic ulcer and chronic gastritis. Multivariate analysis revealed the following independent factors: age for levofloxacin (OR 1.030, 95% CI: 1.020-1.041, P < 0.01), gender for clarithromycin (OR 1.420, 95% CI: 1.080-1.868, P = 0.012), levofloxacin (OR 1.694, 95% CI: 1.293-2.219, P < 0.01), and metronidazole (OR 1.377, 95% CI: 1.021-1.857, P = 0.036), and endoscopic findings for clarithromycin (OR 0.415, 95% CI: 0.282-0.611, P < 0.01) and levofloxacin (OR 0.707, 95% CI: 0.502-0.996, P = 0.035) (Table 4).
Table 4.
Potential factors influencing antibiotic resistance of H. pylori
| Resistance rate, N (%) | AMX | CAMa | LVXb | MNZc | TCN | FUR |
|---|---|---|---|---|---|---|
| Age | ||||||
| ≤ 20 | 0 | 5 (25.0) | 2 (10.0) | 12 (60.0) | 0 | 0 |
| 21-30 | 0 | 26 (19.4) | 18 (13.4) | 105 (78.4) | 0 | 2 (1.5) |
| 31-40 | 0 | 89 (31.5) | 68 (24.0) | 216 (76.3) | 0 | 2 (0.7) |
| 41-50 | 1 | 67 (26.4) | 99 (39.0) | 216 (85.0) | 0 | 1 (0.4) |
| 51-60 | 0 | 68 (27.8) | 85 (34.7) | 198 (80.8) | 0 | 0 |
| 61-70 | 0 | 46 (32.2) | 6 (42.0) | 118 (82.5) | 1 (0.7) | 1 (0.7) |
| 71-80 | 0 | 9 (24.3) | 15 (40.5) | 27 (73.0) | 0 | 0 |
| P value | 0.758 | 0.023 | < 0.001 | 0.037 | 0.339 | 0.645 |
| Gender | ||||||
| male | 0 | 117 (22.9) | 123 (24.0) | 393 (76.7) | 1 (0.2) | 1 (0.2) |
| female | 1 (0.2) | 193 (32.0) | 224 (37.1) | 499 (82.7) | 0 | 5 (0.8) |
| P value | 1.00 | 0.001 | < 0.001 | 0.015 | 0.459 | 0.303 |
| Endoscopic diagnosis | ||||||
| Chronic gastritis | 1 (0.1) | 274 (31.3) | 291 (33.3) | 708 (80.9) | 1 (0.1) | 6 (0.7) |
| Peptic ulcer | 0 | 36 (14.9) | 56 (23.2) | 184 (76.4) | 0 | 0 |
| P value | 1.00 | < 0.001 | 0.03 | 0.117 | 1.00 | 0.429 |
AMX, amoxicillin; CAM, clarithromycin; LEV, levofloxacin; MNZ, metronidazole; TCN, tetracycline; FUR, furazolidone.
significant differences in clarithromycin resistance rate in relation to gender (OR 1.420, 95% CI: 1.080-1.868, P = 0.012) and endoscopic diagnosis (OR 0.415, 95% CI: 0.282-0.611, P < 0.01).
significant differences in levofloxacin resistance rate in relation to age (OR 1.030, 95% CI: 1.020-1.041, P < 0.01), gender (OR 1.694, 95% CI: 1.293-2.219, P < 0.01) and endoscopic diagnosis (OR 0.707, 95% CI: 0.502-0.996, P = 0.035).
significant differences in metronidazole resistance rate in relation to gender (OR 1.377, 95% CI: 1.021-1.857, P = 0.036).
Detection of H. pylori antibiotic resistance-related gene mutations
Sanger sequencing on gastric biopsies from 1171 patients revealed that 1163 patients had H. pylori genes (99.3%, 1163/1171), of which 1116 were successfully cultured (95.3%, 1116/1171). And the chromatograph images of Sanger’s sequencing were shown in Supplementary Figure 1. The amoxicillin-resistant strain (0.1%, 1/1116) was not found to have amino acid substitutions in PBP1, while 46 (4.1%, 46/1115) among 1115 susceptible strains revealed PBP1 amino acid substitutions, in particular, 562Asn/Tyr (43.5%, 20/46). No 16S rRNA mutations were detected in the tetracycline-resistant strain (0.1%, 1/1116), while 330 (29.6%, 330/1115) tetracycline-sensitive strains displayed 16S rRNA mutations, especially A928C (47.3%, 156/330). Based on 23S rRNA sequences among the 310 strains resistant to clarithromycin, 279 (90.0%, 279/310) exhibited a point mutation. Additionally, A2143G (99.3%, 277/279) was the most commonly identified variant (Figure 2). Additionally, 110 (13.6%, 110/806) clarithromycin-sensitive strains exhibited a point mutation. The kappa value of consistency between the drug sensitivity and molecular tests was 0.708. As shown in Figure 3, 317 (91.4%, 317/347) levofloxacin-resistant strains contained amino acid variants of the gyrA subunit-in particular, N87K (49.8%, 158/317)-while 79 (10.3%, 79/769) levofloxacin-sensitive strains had amino acid substitutions, leading to a kappa value of 0.781. Overall, 890 (99.8%, 890/892) strains resistant to metronidazole harbored rdxA gene mutations, with the most common variation type identified as a combination of A91G, C92A, G392A and A610G (78.7%, 700/890; Figure 4). In addition, all metronidazole-susceptible strains (100%, 224/224) harbored an rdxA gene mutation. DNA sequence analysis of porD and oorD of six strains resistant to furazolidone revealed that three (50.0%, 3/6) exhibited mutations, in particular, a combination of mutations at sites G353A, A356G, C357T, A41G, and A335G (66.7%, 2/3) (Figure 5), leading to the substitutions of Thr by Val, Ala by Thr, Thr by Val, Asp by Lys, and Asp by Gly, respectively. Notably, 431 (38.8%, 431/1110) furazolidone-sensitive strains additionally contained point mutations. Compared with susceptible strains, strains resistant to clarithromycin and levofloxacin showed higher mutation rates of 23S rRNA and gyrA genes, respectively (all P < 0.01).
Figure 2.

Gene mutations related to clarithromycin resistance.
Figure 3.

Amino acid variations in the QRDR region of gyrA related to levofloxacin resistance.
Figure 4.

Mutation types of rdxA related to metronidazole resistance.
Figure 5.
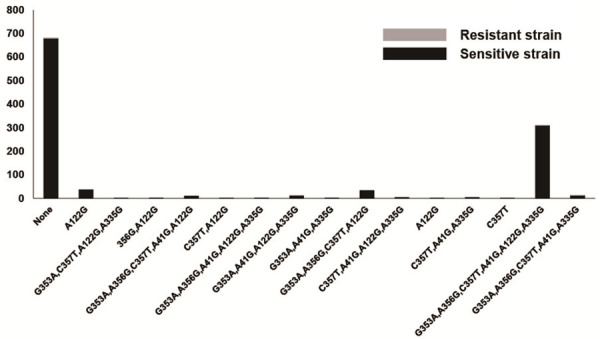
Mutation types of porD and oorD genes related to furazolidone resistance.
Discussion
The Kyoto Consensus proposes that gastritis-induced H. pylori is a contagious disease and requires eradication [23]. The key to implementing effective empirical first-line eradication therapy is to understand the local antibiotic resistance.
Clarithromycin is a major antibiotic for managing H. pylori infection, and increased resistance significantly affects the pathogen eradication rate [24,25]. The resistance rate to clarithromycin in our study (27.8%, 310/1116) was higher than previous reports in China [26,27]. The Maastricht V/Florence Consensus Report suggests that in cases where more than 15% of stains are resistant to clarithromycin, triple therapy including clarithromycin should be avoided in the absence of prior susceptibility testing [28]. Thus, regimens containing clarithromycin should be adopted with caution. Resistance to clarithromycin is mainly induced by point mutations at positions 2142 and 2143, of the V domain of 23S rRNA [10,16]. Wueppenhorst et al. [29] determined A2142G, A2142C, and A2143G as the three most common mutations responsible for more than 90% of cases resistant to clarithromycin. The mononucleotide substitution from A to G was observed in 279 (90%, 279/310) clarithromycin-resistant strains, the majority with A2143G (99.3%, 277/279) and only two (0.7%, 2/279) with A2142G. Accordingly, we speculate that the A2143G mutation is a key contributory factor to clarithromycin resistance. Other resistance mechanisms, for example, the presence of an efflux pump, may be prevalent in remaining resistant strains without 23S rRNA gene mutations [30]. Compared with strains sensitive to clarithromycin, the resistant strains showed a significantly higher mutation rate (P < 0.01). The results of the molecular test were consistent with those of the antibiotic susceptibility test, with a kappa coefficient of 0.708, which can provide a reference for the clinical application of clarithromycin.
Levofloxacin, widely used for urinary infection treatment, was recommended as a remedy after regimens based on clarithromycin. The resistance rate of levofloxacin reached 31.1% (347/1116) in our study, surpassing that of clarithromycin, and greatly reducing the efficacy of levofloxacin-based eradication regimens. Considering the high resistance rates to clarithromycin and levofloxacin, tailored regimens based on antibiotic susceptibility tests could be an alternative option H. pylori resistance to levofloxacin is mainly induced by amino acid substitution in the quinolone resistance-determining regions of gyrA [17]. In this study, we identified 14 amino acid substitutions related to gyrA mutations in 317 (91.4%, 317/347) resistant strains, predominantly at positions N87K (49.8%, 158/317), D91N (16.1%, 51/317) and 91 (9.2%, 29/317). The mutation rate of gyrA in levofloxacin-resistant strains was significantly higher than that in sensitive strains (P < 0.01). The results of the molecular test were consistent with those of the antibiotic susceptibility test, with a kappa coefficient of 0.781, supporting the detection of a specific locus for GyrA to determine the resistance of levofloxacin.
The prevalence of H. pylori resistant to metronidazole is very high in some countries (e.g., 82.4% for Iran, 83.8% for India, and 82.7% for China), due to its widespread use in the treatment of various infectious diseases, including parasitic diseases, female genital diseases and dental infections [14,16,20,21,27,28,31]. Consistent with previous reports in China, we found a high resistance rate of metronidazole (78.2%, 892/1116) [20,25,26]. Although metronidazole resistance can be partly overcome by increasing doses or extending the duration of treatments, metronidazole should be left out as a first-line empirical treatment regimen without antibiotic susceptibility tests. Previous studies have reported that the H. pylori resistance to metronidazole is mainly induced by rdxA gene mutations [11,32,33]. RdxA gene variations were identified in 890 (99.8%, 890/892) metronidazole-resistant strains in this study, the most common being a combination of A91G, C92A, G392A and A610G (78.7%, 700/890). Additionally, metronidazole-sensitive and -resistant strains showed highly similar mutation points. Thus, molecular detection may not be suitable for metronidazole resistance detection.
Data from the present study showed negligible resistance to amoxicillin, tetracycline, and furazolidone (< 1%), indicating that these antibiotics could be effectively adopted for empirical first-line eradication therapies. However, clinical use of tetracycline and furazolidone often leads to a number of adverse effects, such as bone marrow suppression, exfoliative dermatitis, and staining of teeth. Additionally, these two antibiotics are not readily available in China. H. pylori resistance to amoxicillin is most related to PBP gene mutations, including PBP1, PBP2, PBP3, and PBP4 [15,34,35]. In our study, the amoxicillin-resistant strain contained no point mutations at PBP1 while 4.1% (46/1115) sensitive strains showed amino acid substitution at PBP1. The major mechanism of tetracycline resistance is mutations at bases 926-928 on the 16S rRNA gene [18]. A previous study reported the requirement for three adjacent mutations to achieve a high level of resistance [36]. No 16S rRNA gene mutations were observed in the tetracycline-resistant strain, suggesting that other mechanisms (for example, changes in membrane permeability) may be responsible for resistance [37]. In contrast, 330 (29.6%, 330/1115) sensitive strains displayed gene mutations, and all displayed one or two base mutations (A926T and A928C: 27.3% (90/330), A928C: 47.3% (156/330), A926C: 14.9% (49/330), and A926G: 9.4% (31/330)), which could have a limited impact on resistance. The mechanisms underlying resistance to furazolidone are similar to those for metronidazole, and Kwon et al. [19] demonstrated the involvement of porD and oorD gene mutations. Three (50%, 3/6) furazolidone-resistant strains exhibited mutations, the most common being a combination of G353A, A356G, C357T, A41G, and A335G (66.7%, 2/3). Notably, however, 431 (38.8%, 431/1110) furazolidone-sensitive strains exhibited similar point mutations. The mechanism of H. pylori resistance to furazolidone appears to be complex.
A higher rate of multiple resistance (40.5%, 452/1116) was found in our study than that in previous reports [26,27]. It is important to exercise caution to select effective antibiotic combinations as first-line regimens based on antibiotic susceptibility tests, especially for re-treated patients. Combinations of metronidazole/levofloxacin and metronidazole/clarithromycin should be avoided as much as possible. In addition, we found that the previous use of clarithromycin, levofloxacin, and metronidazole predisposes patients to resistance, because resistance rates to these antibiotics of re-treated patients were significantly higher than those of untreated patients (P < 0.05).
Previous studies have reported that several clinical factors were associated with antibiotic resistance of H. pylori, such as gender, patient age, and endoscopic diagnosis [38,39]. Multivariate analyses revealed gender and endoscopic diagnosis as independent risk factors influencing resistance to clarithromycin. The resistance rate to clarithromycin in females was significantly higher than that in males (P < 0.05). Moreover, patients with chronic gastritis showed higher resistance to clarithromycin than those with peptic ulcer (P < 0.01), leading to the conclusion that use of clarithromycin in females and patients with chronic gastritis should be considered carefully. Independent risk factors for levofloxacin resistance include gender, age and endoscopic diagnosis. Patients under 30 years of age showed significantly lower resistance rates of levofloxacin than those over 30 years (P < 0.01). The resistance rate of levofloxacin was highest in patients aged 61 to 70 years (42.0%, 60/143), and significantly higher in females than males (P < 0.01), which may be related to more frequent use in women and elderly patients with a high incidence of urinary infections. Moreover, patients with chronic gastritis have higher resistance rates of levofloxacin than those with peptic ulcer. Treatment with levofloxacin should therefore be considered with caution, especially for the elderly, female patients and patients with chronic gastritis. Gender was determined as an independent risk factor influencing resistance to metronidazole. In addition, the resistance rate was significantly higher in females than males (P < 0.01), which may be explained by the frequent use of metronidazole to treat gynecological diseases.
Conclusion
Resistance of H. pylori to common antibiotics is an ongoing concern in Shanghai. Clarithromycin-, levofloxacin-, and metronidazole-based regimens are not suitable as first-line antibiotics due to high resistance rates, while amoxicillin, tetracycline and furazolidone present good options as components of eradication therapy. Data obtained from molecular detection analyses of clarithromycin and levofloxacin resistance could provide effective guidance for clinical medication. In further studies, we will evaluate the impact of specific point mutations of associated genes on antibiotic resistance by analyzing the efficacy of tailored regimens.
Acknowledgements
We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript, and the Advanced Appropriate Technology Promotion Project of the Shanghai Municipal Health Commission for supporting the study (2019SY067).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Backert S, Neddermann M, Maubach G, Naumann M. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2016;21(Suppl 1):19–25. doi: 10.1111/hel.12335. [DOI] [PubMed] [Google Scholar]
- 2.Lopes D, Nunes C, Martins MC, Sarmento B, Reis S. Eradication of Helicobacter pylori: past, present and future. J Control Release. 2014;189:169–186. doi: 10.1016/j.jconrel.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 3.Sjomina O, Pavlova J, Niv Y, Leja M. Epidemiology of Helicobacter pylori infection. Helicobacter. 2018;23(Suppl 1):e12514. doi: 10.1111/hel.12514. [DOI] [PubMed] [Google Scholar]
- 4.Graham DY. Illusions regarding Helicobacter pylori clinical trials and treatment guidelines. Gut. 2017;66:2043–2046. doi: 10.1136/gutjnl-2017-314744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, Malfertheiner P, Graham DY, Wong VWS, Wu JCY, Chan FKL, Sung JJY, Kaplan GG, Ng SC. Global prevalence of helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology. 2017;153:420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 6.Plummer M, de Martel C, Vignat J, Ferlay J, Bray F, Franceschi S. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health. 2016;4:e609–616. doi: 10.1016/S2214-109X(16)30143-7. [DOI] [PubMed] [Google Scholar]
- 7.Nagy P, Johansson S, Molloy-Bland M. Systematic review of time trends in the prevalence of Helicobacter pylori infection in China and the USA. Gut Pathog. 2016;8:8. doi: 10.1186/s13099-016-0091-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.IARC working group on the evaluation of carcinogenic risks to humans: some industrial chemicals. Lyon, 15-22 February 1994. IARC Monogr Eval Carcinog Risks Hum. 1994;60:1–560. [Google Scholar]
- 9.Mahachai V, Vilaichone RK, Pittayanon R, Rojborwonwitaya J, Leelakusolvong S, Maneerattanaporn M, Chotivitayatarakorn P, Treeprasertsuk S, Kositchaiwat C, Pisespongsa P, Mairiang P, Rani A, Leow A, Mya SM, Lee YC, Vannarath S, Rasachak B, Chakravuth O, Aung MM, Ang TL, Sollano JD, Trong Quach D, Sansak I, Wiwattanachang O, Harnsomburana P, Syam AF, Yamaoka Y, Fock KM, Goh KL, Sugano K, Graham D. Helicobacter pylori management in ASEAN: the Bangkok consensus report. J Gastroenterol Hepatol. 2018;33:37–56. doi: 10.1111/jgh.13911. [DOI] [PubMed] [Google Scholar]
- 10.Zerbetto De Palma G, Mendiondo N, Wonaga A, Viola L, Ibarra D, Campitelli E, Salim N, Corti R, Goldman C, Catalano M. Occurrence of mutations in the antimicrobial target genes related to levofloxacin, clarithromycin, and amoxicillin resistance in Helicobacter pylori isolates from Buenos Aires City. Microb Drug Resist. 2017;23:351–358. doi: 10.1089/mdr.2015.0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miftahussurur M, Syam AF, Nusi IA, Makmun D, Waskito LA, Zein LH, Akil F, Uwan WB, Simanjuntak D, Wibawa ID, Waleleng JB, Saudale AM, Yusuf F, Mustika S, Adi P, Maimunah U, Maulahela H, Rezkitha YA, Subsomwong P, Nasronudin , Rahardjo D, Suzuki R, Akada J, Yamaoka Y. Surveillance of Helicobacter pylori antibiotic susceptibility in indonesia: different resistance types among regions and with novel genetic mutations. PLoS One. 2016;11:e0166199. doi: 10.1371/journal.pone.0166199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gisbert JP, Pajares JM. Review article: Helicobacter pylori “rescue” regimen when proton pump inhibitor-based triple therapies fail. Aliment Pharmacol Ther. 2002;16:1047–1057. doi: 10.1046/j.1365-2036.2002.01276.x. [DOI] [PubMed] [Google Scholar]
- 13.Xie T, Cui X, Zheng H, Chen D, He L, Jiang B. Meta-analysis: eradication of Helicobacter pylori infection is associated with the development of endoscopic gastroesophageal reflux disease. Eur J Gastroenterol Hepatol. 2013;25:1195–1205. doi: 10.1097/MEG.0b013e328363e2c7. [DOI] [PubMed] [Google Scholar]
- 14.Farzi N, Yadegar A, Sadeghi A, Asadzadeh Aghdaei H, Marian Smith S, Raymond J, Suzuki H, Zali MR. High prevalence of antibiotic resistance in Iranian Helicobacter pylori isolates: importance of functional and mutational analysis of resistance genes and virulence genotyping. J Clin Med. 2019;8:2004. doi: 10.3390/jcm8112004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao C, Du SY, Fang L, Fan YH, Song AP, Chen H. Eradication treatment of Helicobacter pylori infection based on molecular pathologic antibiotic resistance. Infect Drug Resist. 2020;13:69–79. doi: 10.2147/IDR.S232169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ducournau A, Benejat L, Sifre E, Bessede E, Lehours P, Megraud F. Helicobacter pylori resistance to antibiotics in 2014 in France detected by phenotypic and genotypic methods. Clin Microbiol Infect. 2016;22:715–718. doi: 10.1016/j.cmi.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Moore RA, Beckthold B, Wong S, Kureishi A, Bryan LE. Nucleotide sequence of the gyrA gene and characterization of ciprofloxacin-resistant mutants of Helicobacter pylori. Antimicrob Agents Chemother. 1995;39:107–111. doi: 10.1128/aac.39.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerrits MM, de Zoete MR, Arents NL, Kuipers EJ, Kusters JG. 16S rRNA mutation-mediated tetracycline resistance in Helicobacter pylori. Antimicrob Agents Chemother. 2002;46:2996–3000. doi: 10.1128/AAC.46.9.2996-3000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon DH, Lee M, Kim JJ, Kim JG, El-Zaatari FA, Osato MS, Graham DY. Furazolidone- and nitrofurantoin-resistant Helicobacter pylori: prevalence and role of genes involved in metronidazole resistance. Antimicrob Agents Chemother. 2001;45:306–308. doi: 10.1128/AAC.45.1.306-308.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Q, Long X, Ji Y, Liang X, Li D, Gao H, Xu B, Liu M, Chen Y, Sun Y, Zhao Y, Xu G, Song Y, Yu L, Zhang W, Liu W, Graham DY, Lu H. Randomised controlled trial: susceptibility-guided therapy versus empiric bismuth quadruple therapy for fi rst-line Helicobacter pylori treatment. Aliment Pharmacol Ther. 2019;49:1385–1394. doi: 10.1111/apt.15273. [DOI] [PubMed] [Google Scholar]
- 21.Ortiz V, Estevez-Ordonez D, Montalvan-Sanchez E, Urrutia-Argueta S, Israel D, Krishna US, Romero-Gallo J, Wilson KT, Peek RM, Dominguez R, Morgan DR. Helicobacter pylori antimicrobial resistance and antibiotic consumption in the low-resource Central America setting. Helicobacter. 2019;24:e12595. doi: 10.1111/hel.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong Z, Zhang Z, Wang J, Hu Y, Mi Y, He B, Zhang Y, Zhang X, Xia X, Huang H, Lai Y, Lin M, Su C, Zhang Z, Wu Z, Lu L, Zhang B, Huang S, Zhong C, Zeng X, Peng Y, Chen G, Zhang H, Zhou G, Liu S, Yang C, Yan L, Chen A, Zhang G, Xu P, Wang S, Zheng P, Xu S, Gao H. A retrospective study of the antibiotic-resistant phenotypes and genotypes of Helicobacter pylori strains in China. Am J Cancer Res. 2021;11:5027–5037. [PMC free article] [PubMed] [Google Scholar]
- 23.Sugano K, Tack J, Kuipers EJ, Graham DY, El-Omar EM, Miura S, Haruma K, Asaka M, Uemura N, Malfertheiner P faculty members of Kyoto Global Consensus Conference. Kyoto global consensus report on Helicobacter pylori gastritis. Gut. 2015;64:1353–1367. doi: 10.1136/gutjnl-2015-309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Djennane-Hadibi F, Bachtarzi M, Layaida K, Ali Arous N, Nakmouche M, Saadi B, Tazir M, Ramdani-Bouguessa N, Burucoa C. High-level primary clarithromycin resistance of Helicobacter pylori in Algiers, Algeria: a prospective multicenter molecular study. Microb Drug Resist. 2016;22:223–226. doi: 10.1089/mdr.2015.0209. [DOI] [PubMed] [Google Scholar]
- 25.Molina-Infante J, Gisbert JP. Optimizing clarithromycin-containing therapy for Helicobacter pylori in the era of antibiotic resistance. World J Gastroenterol. 2014;20:10338–10347. doi: 10.3748/wjg.v20.i30.10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han R, Lu H, Jiang MW, Tan KW, Peng Z, Hu JL, Fang DC, Lan CH, Wu XL. Multicenter study of antibiotic resistance profi le of H. pylori and distribution of CYP2C19 gene polymorphism in rural population of Chongqing, China. Gastroenterol Res Pract. 2016;2016:8547686. doi: 10.1155/2016/8547686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji Z, Han F, Meng F, Tu M, Yang N, Zhang J. The association of age and antibiotic resistance of Helicobacter pylori: a study in Jiaxing City, Zhejiang Province, China. Medicine (Baltimore) 2016;95:e2831. doi: 10.1097/MD.0000000000002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malfertheiner P, Megraud F, O’Morain CA, Gisbert JP, Kuipers EJ, Axon AT, Bazzoli F, Gasbarrini A, Atherton J, Graham DY, Hunt R, Moayyedi P, Rokkas T, Rugge M, Selgrad M, Suerbaum S, Sugano K, El-Omar EM European Helicobacter and Microbiota Study Group and Consensus panel. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut. 2017;66:6–30. doi: 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- 29.Wueppenhorst N, Stueger HP, Kist M, Glocker E. Identifi cation and molecular characterization of triple- and quadruple-resistant Helicobacter pylori clinical isolates in Germany. J Antimicrob Chemother. 2009;63:648–653. doi: 10.1093/jac/dkp003. [DOI] [PubMed] [Google Scholar]
- 30.Yonezawa H, Osaki T, Hanawa T, Kurata S, Ochiai K, Kamiya S. Impact of Helicobacter pylori biofi lm formation on clarithromycin susceptibility and generation of resistance mutations. PLoS One. 2013;8:e73301. doi: 10.1371/journal.pone.0073301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandya HB, Agravat HH, Patel JS, Sodagar NR. Emerging antimicrobial resistance pattern of Helicobacter pylori in central Gujarat. Indian J Med Microbiol. 2014;32:408–413. doi: 10.4103/0255-0857.142256. [DOI] [PubMed] [Google Scholar]
- 32.Gerrits MM, van der Wouden EJ, Bax DA, van Zwet AA, van Vliet AH, de Jong A, Kusters JG, Thijs JC, Kuipers EJ. Role of the rdxA and frxA genes in oxygen-dependent metronidazole resistance of Helicobacter pylori. J Med Microbiol. 2004;53:1123–1128. doi: 10.1099/jmm.0.45701-0. [DOI] [PubMed] [Google Scholar]
- 33.Binh TT, Suzuki R, Trang TT, Kwon DH, Yamaoka Y. Search for novel candidate mutations for metronidazole resistance in Helicobacter pylori using next-generation sequencing. Antimicrob Agents Chemother. 2015;59:2343–2348. doi: 10.1128/AAC.04852-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLoney CR, Schiller NL. Competition of various beta-lactam antibiotics for the major penicillin-binding proteins of Helicobacter pylori: antibacterial activity and effects on bacterial morphology. Antimicrob Agents Chemother. 1999;43:2702–2709. doi: 10.1128/aac.43.11.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krishnamurthy P, Parlow MH, Schneider J, Burroughs S, Wickland C, Vakil NB, Dunn BE, Phadnis SH. Identifi cation of a novel penicillin-binding protein from Helicobacter pylori. J Bacteriol. 1999;181:5107–5110. doi: 10.1128/jb.181.16.5107-5110.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trieber CA, Taylor DE. Mutations in the 16S rRNA genes of Helicobacter pylori mediate resistance to tetracycline. J Bacteriol. 2002;184:2131–2140. doi: 10.1128/JB.184.8.2131-2140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan R, Nahar S, Mukhopadhyay AK, Berg DE, Ahmad MM, Okamoto K, Nair GB, Rahman M. Isolation of tetracycline-resistant clinical Helicobacter pylori without mutations in 16S rRNA gene in Bangladesh. Microbiol Immunol. 2008;52:508–511. doi: 10.1111/j.1348-0421.2008.00062.x. [DOI] [PubMed] [Google Scholar]
- 38.Bai P, Zhou LY, Xiao XM, Luo Y, Ding Y. Susceptibility of Helicobacter pylori to antibiotics in Chinese patients. J Dig Dis. 2015;16:464–470. doi: 10.1111/1751-2980.12271. [DOI] [PubMed] [Google Scholar]
- 39.Song Z, Zhang J, He L, Chen M, Hou X, Li Z, Zhou L. Prospective multi-region study on primary antibiotic resistance of Helicobacter pylori strains isolated from Chinese patients. Dig Liver Dis. 2014;46:1077–1081. doi: 10.1016/j.dld.2014.08.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


