Abstract
Campylobacter rectus is a periodontal pathogen with a 150-kDa protein on its cell surface. This protein forms a paracrystalline lattice, called the S-layer, surrounding the outer membrane of this gram-negative bacterium. To initiate a genetic analysis of the possible role of the S-layer in the initial interaction of C. rectus with host epithelial cells, C. rectus strains lacking the S-layer protein gene (crsA) were constructed by allelic exchange mutagenesis. Surprisingly, the lack of the S-layer had only a minor effect on the interaction of C. rectus with HEp-2 epithelial cells; CrsA+ cells were 30 to 50% more adherent than were CrsA− bacteria. Since the host cell expression of cytokines appears to play an important role in the pathogenesis of periodontal diseases, the effect of the S-layer on the epithelial cell cytokine response was also examined by quantitative reverse transcriptase PCR and enzyme-linked immunosorbent assay. Although there were no changes in the mRNA levels for the anti-inflammatory cytokines interleukin-1 receptor agonist (IL-1ra), IL-13, and transforming growth factor β, the expression and secretion of the proinflammatory cytokines IL-6, IL-8, and tumor necrosis factor alpha (TNF-α) were significantly induced by both wild-type C. rectus and CrsA− bacteria. Interestingly, the kinetics of cytokine induction differed for the CrsA+ and CrsA− bacteria. At early time points, the HEp-2 cells challenged with CrsA− bacteria produced higher levels of IL-6, IL-8, and TNF-α mRNA and protein than did cells challenged with CrsA+ bacteria. We conclude that C. rectus may help initiate periodontitis by increasing the expression of proinflammatory cytokines and that the S-layer may temper this response to facilitate the survival of C. rectus at the site of infection.
Periodontitis is a chronic inflammatory disease in which destruction of the supporting structures can lead to tooth loss. Although bacteria are needed to initiate disease, it is clear that the host response to the periodontal microbial flora also plays an important role in pathogenesis (12, 31, 46). For example, it has been reported that tumor necrosis alpha (TNF-α) mRNA is found more often in gingival biopsy specimens from patients with chronic adult periodontitis than in specimens from periodontally healthy individuals (35). Similarly, there is a strong correlation between elevated levels of interleukin-1β (IL-1β) in gingival tissues and periodontal disease (18, 34, 41, 42). Recently, Assuma et al. (1) showed that IL-1 and TNF antagonists reduced the inflammatory response in a primate model of periodontitis. All of these results indicated that IL-1 and TNF are important in modulating the tissue damage seen in periodontal disease. Other cytokines, such as IL-6, have also been implicated in gingival tissue destruction, but the data are not as compelling (24).
Campylobacter rectus, a gram-negative, anaerobic rod, appears to be an etiological agent of adult periodontitis (32) and rapidly progressive periodontitis (8). Very little is known about the mechanisms by which this organism induces and/or exacerbates disease. However, it has been reported that C. rectus enhances the production of the proinflammatory cytokines IL-6 and IL-8 in human gingival fibroblasts in vitro (7), although the bacterial molecule inducing this response is unknown. The best candidate for a C. rectus virulence factor that could affect its interaction with host cells is the cell surface layer (S-layer) that covers C. rectus (21, 25). This organism's S-layer is composed of hexagonal paracrystalline arrays of a single 150-kDa polypeptide (5, 6, 29). The C. rectus S-layer protein, encoded by a single-copy gene (crsA), has limited sequence similarity to S-layer proteins from other organisms (47). Thus, the precise role of the C. rectus S-layer in pathogenesis may differ from those of previously described S-layers in other bacteria (37).
Evidence for the virulence potential of the C. rectus S-layer comes from studies with C. rectus cells that lost their S-layer after long-term subculture (5, 19). Low-passage isolates of C. rectus formed larger lesions in a mouse abscess model for soft tissue destruction than did cells which had lost their S-layer after 15 to 17 passages in vitro (19). Additionally, in vitro binding studies indicated that S-layer-containing strains of C. rectus were less adherent to human gingival fibroblasts than were other strains that had lost their S-layer (5). These results suggest that the C. rectus S-layer may help the organism evade host defense mechanisms. However, the strength of this conclusion is diminished by the fact that long-term subculturing may have led to changes in the levels of non-S-layer C. rectus proteins. In addition, some experiments involved comparisons between nonisogenic strains, which are likely to differ in more than their S-layer levels (5).
To test more definitively the role of the C. rectus S-layer in regulating the bacterium-mammalian host cell interaction and in generating cytokine responses, defined isogenic S-layer-positive (CrsA+) and S-layer-negative (CrsA−) strains have been constructed. The isogenic CrsA+ and CrsA− strains were compared for their abilities to bind host cells and to induce cytokines in host cells. Epithelial cells were used in this study because they are the first host cells to come in contact with microbial pathogens. Therefore, they are in a unique position to function as an early signaling system to the immune cells located in the underlying mucosa. Our results indicate that the C. rectus S-layer may play a role in periodontal pathogenesis by enhancing bacterial binding to the epithelium. C. rectus also induces epithelial cells to increase the expression of several proinflammatory cytokines. Interestingly, the presence of the S-layer diminishes this response somewhat.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
C. rectus strain 314, a clinical isolate, was used in these studies (5). The cells were grown in MFF (mycoplasma-formate-fumarate) medium (13) in a Coy anaerobic growth chamber (5% CO2, 10% H2, 85% N2) at 37°C (5, 13). Escherichia coli strain TB-1, used for plasmid propagation, was grown in Luria broth (2) containing spectinomycin or ampicillin, as appropriate, at 50 μg/ml.
Plasmid construction and isolation.
Plasmid pDK619 is a pUC19 derivative that contains a DNA fragment from strain 314 in which the crsA gene is replaced by a spectinomycin resistance (spec) gene (27). Briefly, a 1.5-kb EcoRV fragment containing <10% of the 3′ end of the crs coding region plus 1.2 kb of downstream DNA was cloned into the HindII site of pUC19 to generate an intermediate plasmid called pDK616. A 1.1-kb PCR product containing a spectinomycin resistance gene (27), with EcoRI and BstEII sites engineered onto one end and a BamHI site engineered onto the other, was inserted into pDK616 between the EcoRI and BamHI sites of the multiple-cloning site. Finally, this intermediate plasmid was opened at the unique BstEII site and a 1.4-kb BstEII fragment containing the crsA promoter and region upstream was ligated in to obtain plasmid pDK619 (see Fig. 1A). Small amounts of plasmid were isolated using a miniprep method involving alkali lysis and boiling (2); larger amounts were isolated by an alkali lysis procedure (23). The construction of the plasmid was confirmed by sequencing the fusion junctions using the dideoxy-chain termination method with a T7 Sequenase sequencing kit (Amersham Pharmacia Biotech).
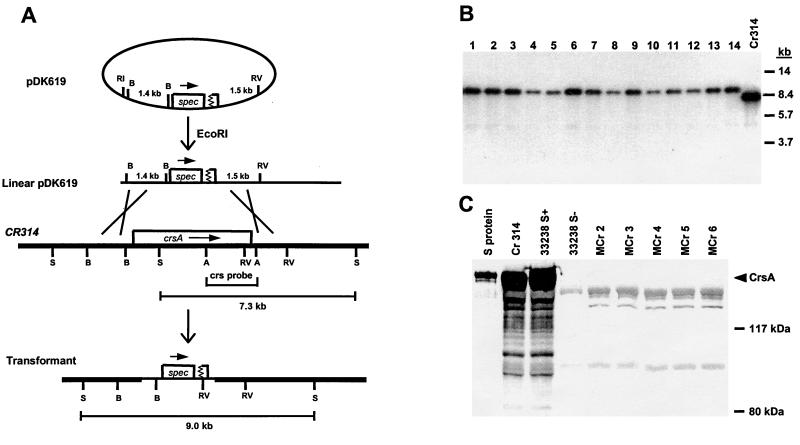
FIG. 1.
Construction of an isogenic CrsA− strain. (A) Schematic representation of the genetic structures expected at the crsA locus in the construction of S-layer-negative strains by recombination of pDK619 with the bacterial chromosome. The 1.4- and 1.5-kb segments in plasmid pDK619 are homologous to the similarly labeled chromosomal segments. Key restriction endonuclease sites are indicated as follows: A, AflII; B, BstEII, RV, EcoRV; S. SacI. The physical maps are not drawn to scale. (B) Chromosomal DNAs isolated from randomly selected transformants (lanes 1 to 14) and 314 (lane Cr314) were digested with SacI and subjected to Southern blot analysis with the crsA probe marked in panel A. The migration positions of the DNA size standards (in kilobases) are indicated on the right. (C) Whole-cell protein samples of randomly selected transformants (Mcr2 to Mcr6) and C. rectus strains 33238/S+, 33238/S− (19), and 314 were subjected to Western blot analysis. A 25-μg sample of each protein sample and 3 μg of purified S-layer protein were electrophoresed on an SDS–7.5% polyacrylamide gel, transferred to a nitrocellulose membrane, and reacted with anti-S-layer serum. The positions of the protein size standards are marked on the right.
DNA transformation by electroporation.
MFF broth (50 ml) inoculated with 15 ml of a fresh overnight culture of strain 314 was grown to an optical density at 660 nm of 0.2 to 0.3. The cells were harvested by centrifugation, washed twice in 10 ml of electroporation buffer (15% glycerol, 0.272 M sucrose, 0.57 mM KH2PO4, 2.43 mM K2HPO4 [pH 7.4]), and resuspended in electroporation buffer (1.5 to 2.0 ml) to an optical density at 660 nm of 6.0. Immediately before electroporation, 600 ng of pDK619 DNA in 3 μl of distilled H2O was added to 60 μl of cells on ice. The mixture of cells and DNA was transferred into an ice-cold electroporation cuvette (2-mm electrode gap) and electroporated at 2.5 kV, 200 Ω, and 25 μF (Gene Pulser; Bio-Rad) to get a time constant of 4.0 to 4.4. Following electroporation, 1 ml of prewarmed MFF was added to the cuvette and the cells were transferred to a sterile tube and grown in an anaerobic chamber for 4 h. Aliquots (0.25 ml) of the cells were then plated onto MFF agar containing spectinomycin (20 μg/ml) and placed in an anaerobic chamber for 3 to 4 days.
Southern blots.
Overnight cultures (10 ml) of C. rectus in MFF broth containing 20 μg of spectinomycin per ml were used for preparing chromosomal DNAs. Genomic DNAs were isolated by a detergent-proteinase K lysis procedure that included treatment with cetyltrimethylammonium bromide to remove polysaccharides and cell debris (2). DNAs were digested with the appropriate restriction endonucleases and electrophoresed for Southern blot analysis as described previously (22). Hybridizations were carried out overnight at 65°C following a 6-h prehybridization. The filter was then washed three times in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 65°C. The crsA gene probe, an isolated AflII fragment which contains the last third of the crsA gene (47), was labeled with [α-32P]ATP using a random-primer DNA-labeling system from Life Technologies (Gaithersburg, Md.)
Western blots.
To prepare whole-cell protein samples for Western blot analysis, C. rectus cell pellets were lysed in sodium dodecyl sulfate (SDS) gel-loading buffer (36) without bromophenol blue and boiled for 5 min. The protein concentration was determined by the bicinchoninic acid protein assay (Pierce Biochemicals, Rockford, Ill.) in accordance with the manufacturer's instructions. Portions (25 μg) of each protein sample and 3 μg of purified S-layer protein (29) were fractionated on an SDS–7.5% polyacrylamide gel, and transferred to a nitrocellulose membrane for 3.5 h at a constant voltage of 50 V. The membrane was washed with 100 mM Tris-buffered saline (TBS) (100 mM Tris-HCl in 0.9% NaCl [pH 7.5]) for 30 min, blocked by a 1-h incubation with BLOTTO (5% nonfat dry milk in 50 mM TBS), and incubated overnight with a 1:500 dilution of anti-S-layer rabbit antiserum (29) (kindly provided by J. Ebersole, Department of Periodontics, University of Texas Health Science Center, San Antonio, Tex.) in BLOTTO. After being washed three times with TBS, the membrane was incubated in a 1:2,000 dilution of horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G (Bio-Rad) for 4 h. The HRP antibody was visualized using an HRP substrate kit.
Adhesion assays.
The HEp-2 cell line, derived from a human larynx epidermal carcinoma (ATCC CCL23), was provided by J. Ebersole. Cells were maintained in complete DMEM (Dulbecco's modification of Eagle's medium [Cellgro] supplemented with 5% fetal calf serum (FCS), 2 mM glutamine, 100 U of penicillin, and 100 μg of streptomycin per ml) in a CO2 incubator at 37°C. Cell cultures were split 1:4 twice a week after detachment of the cells from the flasks with 2.5% trypsin.
The initial qualitative assessment of C. rectus–HEp-2 cell binding was done using acridine orange, a fluorochrome stain (26). Briefly, 106 HEp-2 cells were seeded onto a sterile microscope coverglass (22 mm square) in one well of a six-well tissue culture plate containing 0.9 ml of complete DMEM. After 18 to 20 h of growth, the number of HEp-2 cells in several wells was determined by counting cells in a hemocytometer after their removal from the wells with trypsin. The semiconfluent monolayers were washed with phosphate-buffered saline (PBS; pH 7.4), and then 0.9 ml of DMEM with 2% FCS was added to each well. After 10 to 12 h of growth in MFF, C. rectus cells were harvested by centrifugation and resuspended in antibiotic-free DMEM containing 2% FCS, and 0.1 ml of bacteria was added to the HEp-2 cells at various multiplicities of infection (MOI). After incubation at 37°C in a CO2 incubator for different periods, a coverglass with the HEp-2 monolayer was removed from a well of the tissue culture plate, washed three times with PBS to remove unbound C. rectus, fixed in 100% methanol for 2 min, stained with acridine orange (0.01% acridine orange in sodium acetate buffer) (26), mounted on a slide with a drop of Gel/Mount (Biomeda Corp., Foster City, Calif.), and observed using a fluorescence microscope (D-7082; Carl Zeiss, Oberkochen, Germany) with a FT 510 filter at ×1,000 magnification with an oil immersion objective.
For quantitative adhesion assays, HEp-2 cells (24-h culture) in complete DMEM were seeded in 96-well plates at a density of 5 × 104/well and cultured for 18 to 20 h to get confluent monolayers. The number of HEp-2 cells per well was then determined for four wells by counting cells in a hemocytometer after their removal from the wells with trypsin. C. rectus was labeled by diluting 5 ml of a fresh overnight culture into 200 ml of MFF to which [2,8-3H]adenine (ICN, Costa Mesa, Calif.) had been added to a final concentration of 5 to 10 μCi/ml. After 10 to 12 h of growth, the bacterial cells were harvested, washed three times with 20 ml of MFF to remove unincorporated label, and resuspended in antibiotic-free DMEM containing 2% FCS to get 1.8 × 109 bacteria/ml. Just before starting the adhesion assay, the HEp-2 monolayers were washed with PBS (pH 7.4). A bacterial suspension, at the indicated MOI, was added to each well, and the plates were incubated for different times at 37°C in a 5% CO2 incubator, unless otherwise noted. Subsequently, nonadherent bacteria were removed by washing the monolayer three times with PBS (pH 7.4). Then the monolayer was detached by trypsin treatment and the attached cells were counted in a scintillation counter. The number of bacteria bound per HEp-2 cell was determined as follows. The specific activity of labeled C. rectus cells was determined just before adding the bacteria to the HEp-2 cells. This value was then used to calculate, from the cpm found in a given well, the number of C. rectus cells bound per HEp-2 cell. The significance of differences in binding between samples was determined using the two-tailed Student t test. P < 0.05 was defined as significant.
Quantitative PCR for cytokine expression.
Tissue culture dishes (diameter, 10 cm) were each seeded with 5 × 106 HEp-2 cells in a 10-ml volume of complete DMEM and incubated for 20 h to produce confluent monolayers with 1 × 107 ± 7 × 105 cells per well. C. rectus cells were grown overnight (15 to 16 h) in MFF, harvested, and resuspended in antibiotic-free DMEM containing 2% FCS. Each HEp-2 monolayer was washed with PBS (pH 7.4) and overlaid with 10 ml of a bacterial suspension at an MOI of 1,000 (1,000 bacterial cells per HEp-2 cell). Following incubation for various times in 5% CO2 at 37°C, the bacterial suspension was removed and the monolayers were lysed with buffer containing guanidinium isothiocyanate (RNeasy minikit; Qiagen Inc., Santa Clarita, Calif.). Total RNA was extracted from each HEp-2 cell lysate using the RNeasy mini kit as specified by the manufacturer. The integrity of each RNA sample was checked by gel electrophoresis and ethidium bromide staining.
Reverse transcriptase PCR (RT-PCR) was performed by a previously described method (28) with minor modifications. cDNA was synthesized from 2 μg of each RNA sample at 37°C for 2 h in 20 μl of reverse transcription mix containing 4 μl of 5× reverse transcription buffer (250 mM Tris-HCl[pH 8.3], 375 mM KCl, 15 mM MgCl2), 2 μl of 0.1 M dithiothreitol, 1 μl of RNasin (40 U/μl) (Promega, Madison, Wis.), 2 μl of deoxynucleoside triphosphate mix (2.5 mM each dATP, dCTP, dGTP, and dTTP [New England Biolabs, Inc.]), 2 μl of random hexadeoxynucleotide primers (0.5 μg/μl) (Promega), and 1 μl of Moloney murine leukemia virus RT (200 U/μl) (SuperScript II; Life Sciences, Gaithersburg, Md.). After DNA synthesis, the RT was inactivated by heating the samples at 95°C for 10 min.
To quantitate the levels of various cytokine mRNAs, the cytokine primer pairs shown in Table 1 were used in PCRs with the cDNA samples as templates. Each primer set was designed to direct synthesis across an intron so that any contaminating genomic DNA would give a PCR product distinct from cDNA-derived PCR products. Then 1 μl of each of the cDNAs was added to 24 μl of PCR mix containing 2.5 μl of 10× reaction buffer (0.01% [wt/vol] gelatin, 100 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2), 2 μl of 2.5 mM dNTP mix, 0.125 μl (0.625 U) of Thermus aquaticus DNA polymerase (Taq; Perkin-Elmer, Roche Molecular Systems, Inc., Branchburg, N.J.), 2.5 pmol of 5′ primer labeled with [γ-32P]ATP and T4 polynucleotide kinase, and 2.5 pmol of unlabeled 3′ primer. Each sample was subjected to DNA amplification with a PTC-100 programmable thermal controller (M.J. Research, Inc., Watertown, Mass.) for 3 min at 95°C, 2 min at 55°C, and 2 min at 70°C for the first cycle and 1 min at 95°C, 2 min at 55°C, and 2 min at 70°C for the rest of the cycles. Triplicate determinations were performed for each sample. The linear range of the PCRs for each cytokine and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was determined by running a test set of reactions over a range of cycle values. A cycle number that was in the linear part of the curve was selected for quantitative PCR analyses. The PCR products were resolved on a 5% acrylamide gel, dried, and autoradiographed. The intensities of the labeled PCR products were quantitated using the ImageQuant software program (Molecular Dynamics, Sunnyvale, Calif.). For each sample, the level of a given cytokine PCR product was normalized to the amount of GAPDH PCR product generated from the same sample. The significance of differences between samples was analyzed statistically with a two-tailed Student t test. The identification of the cytokine PCR products was confirmed by sequencing a sample of each after extraction from a gel.
TABLE 1.
Primers used to measure cytokine mRNA by RT-PCR
| Cytokine | Primer sequences | cDNA size (bp) |
|---|---|---|
| IL-1α | 5′-GTTCCTCCATTGATCATCTG-3′ | 204 |
| 5′-GGCTTAAACTCAACCGTCTC-3′ | ||
| IL-1β | 5′-GACACATGGGATAACGAGGC-3′ | 248 |
| 5′-ACGCAGGACAGGTACAGATT-3′ | ||
| IL-6 | 5′-TGTGAAAGCAGCAAAGAGGC-3′ | 236 |
| 5′-TTCTGCAGGAACTGGATCAG-3′ | ||
| IL-8 | 5′-TCTGCAGCTGTGTGAAGGTGCAGTT-3′ | 203 |
| 5′-AACCCTCTGCACCCAGTTTTGCTT-3′ | ||
| TNF-α | 5′-CAAGCCTGTAGCCCATGTTG-3 | 140 |
| 5′-CCTGGGAGTAGATGAGGTAC-3′ | ||
| GAPDH | 5′-AGTCAGCCGCATCTTCTTTTGC-3′ | 291 |
| 5′-CTCCTGGAAGATGGTGATGGGA-3′ |
Detection of cytokines in HEp-2 supernatants by ELISA.
HEp-2 cells (4 × 105 in 1 ml of complete DMEM) were seeded in each well of 24-well plates and incubated for 18 to 20 h to generate monolayers. C. rectus cells were grown overnight in MFF broth, harvested by centrifugation, and resuspended in antibiotic-free DMEM containing 2% FCS. C. rectus cells (1 ml; MOI, 1,000) were then placed on each HEp-2 monolayer, which had been washed with PBS (pH 7.4). The mammalian cell culture supernatants were collected following incubation in 5% CO2 at 37°C for various periods, centrifuged to remove the bacteria, aliquoted, and stored at −70°C for later analysis.
The levels of cytokines in HEp-2 cell supernatants were detected by an enzyme-linked immunosorbent assay (ELISA) as described by Steffen and Ebersole (43) and Ebersole et al. (9). Briefly, Immulon 3 microtiter plates (96 wells) were coated with individual monoclonal antibodies (Genzyme, Cambridge, Mass.) to each cytokine at a concentration of 5 μg/ml in coating buffer (32 mM sodium carbonate, 68 mM sodium bicarbonate, 0.2% sodium azide). Following a 3- to 4-h incubation at 37°C, the plates were blocked by incubation with 1% bovine serum albumin in PBS overnight at 4°C. Undiluted cell culture supernatant samples were then added to the appropriate wells. In addition, serial dilutions of recombinant cytokines (in 1× PBS [pH 7.4], 0.05% Tween 20, 0.1% bovine serum albumin) (Biosource, Camarillo, Calif.) were placed in several wells of each plate as standards. After 1 h of incubation at 37°C, the amount of cytokine bound in each well was assayed using the appropriately diluted anti-cytokine polyclonal rabbit antiserum (Genzyme). This was detected, in turn, by goat anti-rabbit immunoglobulin G–alkaline phosphatase conjugate at a 1:3,000 dilution. The plates were developed using p-nitrophenyl phosphate as the substrate. The reactions were stopped by the addition of 100 μl of 1 N NaOH and read at 405 to 410 nm. A two-tailed Student t test was used to identify statistically significant differences in cytokine levels between samples.
RESULTS
Construction of an isogenic CrsA− strain.
To examine the role of the C. rectus S-layer in bacterium host interactions, a well-defined S-layer-negative (CrsA−) strain was needed. Although S-layer-negative strains are available, they are not ideal for such studies since they have altered levels of other proteins as well (5). Thus, CrsA− mutants that are otherwise identical to the parental CrsA+ strain were constructed by allelic exchange mutagenesis. The S-layer-expressing strain 314 (CrsA+) was transformed with linearized plasmid pDK619 (Fig. 1A) in which >90% of the crsA coding region is replaced by a spectinomycin resistance gene (27). Since the plasmid cannot replicate in C. rectus, spectinomycin-resistant transformants should arise by homologous recombination between DNA on the plasmid and homologous sequences on the C. rectus chromosome through a reciprocal double crossover (Fig. 1A). To prove that the expected recombination had occurred, DNA from 14 randomly selected transformants was digested with SacI and analyzed by Southern blotting with a crsA-specific probe (47). As expected, all the transformants had lost the 7.3-kb wild-type crsA band and gained a 9.0-kb SacI fragment indicative of a gene replacement event (Fig. 1B).
To confirm the DNA analysis, a Western blot analysis with anti-S-layer antibody was done on whole-cell protein from several of these transformants (Fig. 1C). The transformants tested lacked the 150-kDa S-layer protein band found in S-layer-positive strains 314 and 33238. A Coomassie brilliant blue-stained gel showed that the levels of non-S-layer proteins were the same in strain 314 and the transformants (data not shown). Therefore, well-defined S-layer-negative strains of C. rectus which are otherwise isogenic to strain 314 have been constructed. These mutants were designated Mcr2 to Mcr6, and two, Mcr2 and Mcr3, were used for further studies. Interestingly, the CrsA− strains have no significant difference in growth rate or colony morphology in comparison with the parental CrsA+ strain 314, even though the S-layer protein can make up >20% of the total bacterial protein in a wild-type cell.
The S-layer increases the binding of C. rectus to epithelial cells.
The role of the S-layer in the interaction of C. rectus and host cells was investigated using a human epithelial cell line, HEp-2. In preliminary studies, the parameters for optimal binding were evaluated qualitatively using acridine orange staining to visualize the bound bacteria (26). Different MOIs (10:1, 100:1, and 1,000:1) were used in a short-course incubation (5, 15, 30, 45, 60, 90, and 120 min) and in a long-course incubation (2, 4, 6, 8, 10, 12, and 24 h). The bacteria did not affect the viability of the HEp-2 cells during the experiment. The results indicated that the binding of 314 (CrsA+) and Mcr2/3 (CrsA−) to HEp-2 cells was bacterial dose dependent and peaked at 90 to 120 min after the start of the assay (data not shown). Unexpectedly, there appeared to be no large differences in the binding of CrsA+ and CrsA− bacteria to the HEp-2 cells. Quantifying the binding of C. rectus to HEp-2 cells in the acridine orange-stained samples was complicated by the fact that the bacteria tended to bind to the HEp-2 cells in clumps.
To discern possible differences in the binding of the CrsA+ and CrsA− strains to HEP-2 cells, a quantitative adhesion assay was done. Radioactively labeled C. rectus cells were inoculated onto HEp-2 monolayers at MOIs ranging from 30 to 3,000 and incubated for 30, 60, and 90 min. Strains 314 and Mcr2 both showed dose- and time-dependent binding to HEp-2 cells (Fig. 2A). Interestingly, the CrsA+ bacteria consistently bound better to HEp-2 cells than did the mutant Mcr2 bacteria; there can be as much as a twofold difference (MOI = 1,000 in a 90-min incubation). The same results were found when another CrsA− strain (Mcr3) was tested (data not shown), establishing that the binding differences are due to the presence or absence of the S-layer.
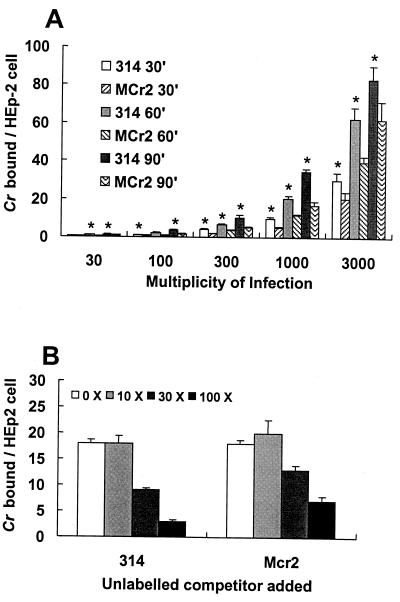
FIG. 2.
Quantitative assessment of CrsA+ and CrsA− strain binding to HEp-2 cells under aerobic conditions. Each bar gives the mean value and standard deviation for four individual samples under each condition. (A) 314 (CrsA+) and Mcr2 (CrsA−) bacteria, labeled to specific activities of 0.036 and 0.045 cpm/cell, respectively, were incubated with HEp-2 cells for different lengths of time and at the MOIs indicated. After the samples were washed, the number of bacteria bound was determined by quantitating the radioactivity associated with the HEp-2 cells. The background binding of labeled bacteria to wells with no HEp-2 cells was low enough to allow the detection of 1 bacterium binding per 250 HEp-2 cells. The asterisks indicate samples in which the binding of 314 cells to HEp-2 monolayers is statistically significantly higher (P < 0.01) than the binding of Mcr2 bacteria to HEp-2 monolayers. (B) Competition of 314 and Mcr2 for the binding of 314 to HEp-2 cells. Labeled 314 bacteria (specific activity, 0.012 cpm/bacterial cell) at an MOI of 1,000 were incubated with HEp-2 cells in the presence of 10×, 30×, and 100× unlabeled 314 or Mcr2 cells. The number of bound radiolabeled cells were determined by scintillation counting.
To test the possibility that the CrsA+ and CrsA− bacteria were binding to different host cell receptors, a competition binding assay was done (Fig. 2B). Labeled strain 314 cells were bound to HEp-2 cells in the presence of increasing numbers of unlabeled 314 or Mcr2 cells. Consistent with the previous binding assays, strain Mcr2 does not compete as well as strain 314. However, both strains did inhibit 314 binding, suggesting that the CrsA+ and CrsA− bacteria were binding to the same receptor on the host cell.
Since C. rectus is an anaerobic bacterium, its binding to HEp-2 cells was also tested under anaerobic conditions. As was found in the aerobic binding assay, Mcr2 was usually 15 to 30% less adherent than 314 to HEp-2 cells (Fig. 3A). This was true at all but the highest MOI and longest incubation time. Thus, the S-layer of C. rectus enhances the interaction between the bacteria and HEp-2 cells. Interestingly, the number of bacteria bound under anaerobic conditions was seven- to ninefold larger than the number bound in the aerobic assay. This suggests either that the differences in metabolism between C. rectus cells under anaerobic versus aerobic conditions can affect bacterial binding to HEp-2 cells or that HEp-2 cells increase their binding capacity under anaerobic conditions. We favor the former possibility, since the binding increase occurs within 30 min of the shift from aerobic to anaerobic conditions, too short a period to allow the synthesis of additional receptors.
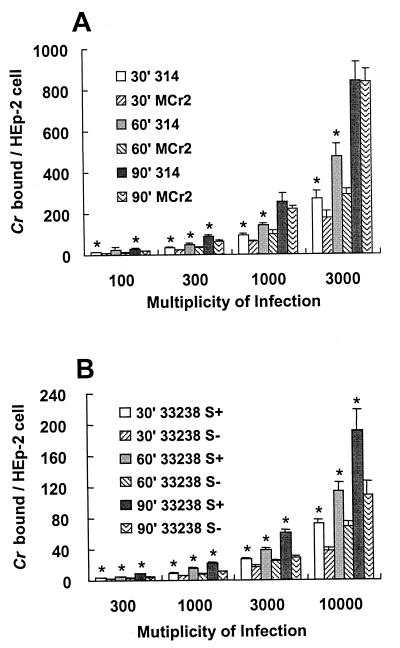
FIG. 3.
Quantitative assessment of CrsA+ and CrsA− strain binding to HEp-2 cells under anaerobic conditions and binding of strain 33238. (A) Binding of strains 314 and Mcr2 to HEp-2 cells under anaerobic conditions. Labeled 314 (CrsA+) and Mcr2 (CrsA−) bacteria were incubated with HEp-2 cells in an anaerobic chamber for different lengths of time and at the MOIs indicated. After the samples were washed, the number of bacteria bound was determined by quantitating the radioactivity associated with the HEp-2 cells. The asterisks indicate samples in which the binding of 314 cells to HEp-2 monolayers is statistically significantly higher (P < 0.05) than the binding of Mcr2 bacteria to HEp-2 monolayers. (B) Binding of strains 33238/S+ and 33238/S− to HEp-2 cells under aerobic conditions. 33238/S+ bacteria are significantly (P < 0.01) more adherent than 33238/S− bacteria to HEp-2 cells at all MOIs and times, as indicated by the asterisks.
Borinski and Holt had previously reported that an S-layer-negative C. rectus strain bound better to fibroblasts than did an S-layer-positive isolate (5). This apparent discrepancy with our results could be because these investigators studied binding to fibroblast cells and not epithelial cells. Alternatively, the difference could be because strain 33238, not 314, was used in the earlier study. To test the latter possibility, an adhesion assay was done on HEp-2 cells with strain 33328 (CrsA+) or its spontaneously derived mutant, 33238/S− (CrsA−) (Fig. 3B). The results showed that the binding pattern of the 33238 strain pair is the same as that of 314 pair, suggesting that the discrepancy between our results and those of Borinski and Holt (5) is due to a difference in the type of mammalian cell used.
C. rectus binding to HEp-2 cells increases certain cytokine levels.
Recent evidence has suggested that the host response to periodontal pathogens is responsible, in part, for the chronic inflammation in periodontal sites that leads to the destruction of the supporting structures of the teeth (31, 48). Certain cytokines such as IL-6, IL-8, and TNF-α appear to be proinflammatory, while others, such as IL-13, IL-1 receptor antagonist (IL-1ra), and transforming growth factor β (TGF-β) are considered to be anti-inflammatory. To determine whether C. rectus and its S-layer might modulate inflammation, the cytokine responses to C. rectus 314 (CrsA+) and Mcr2 (CrsA−) in epithelial cells were investigated. Epithelial cells were chosen since they are the first host cells to come in contact with microbial pathogens. As a preliminary screen for cytokines whose synthesis may be altered by C. rectus or its S-layer, RT-PCR was done on RNA samples from HEp-2 cells that had been incubated with strain 314 or Mcr2 for various lengths of time. Each RNA sample was subjected to RT-PCR for a range of cycles (12 to 40 cycles) using a panel of primers specific for different cytokine mRNAs. The amount of each PCR product was quantitated from the image of an ethidium bromide-stained gel using Molecular Dynamics ImageQuant software. The mRNAs for IL-10, IL-12, IL-13, and IL-1ra were undetectable by RT-PCR in unchallenged or C. rectus-challenged HEp-2 cells (data not shown). On the other hand, signals for the IL-1α, IL-1β, IL-6, IL-8, TNF-α, and TGF-β mRNAs were observed in samples from strain 314- and strain Mcr2-challenged cells, although the TGF-β levels were the same with and without bacterial challenge. Thus, subsequent quantitative experiments focused on IL-1α, IL-1β, IL-6, IL-8, and TNF-α levels, since their levels changed in response to bacterial challenge.
To establish more accurately the effects of C. rectus and its S-layer on the expression of IL-1α, IL-1β, IL-6, IL-8, and TNF-α from HEp-2 cells, the levels of these cytokines were compared among HEp-2 cells challenged with either medium alone, CrsA+ bacteria, or CrsA− bacteria. Two different assays were used; cytokine mRNA levels were measured by quantitative RT-PCR, while the amounts of secreted cytokines were assayed in parallel by ELISAs. In both assays, strain 314 induced increased expression and secretion of IL-6, IL-8, and TNF-α relative to those in HEp-2 cells challenged by medium alone (Fig. 4 and 5). However, the kinetics of the responses, the extent of the inductions, and the effect of challenge with the S-layer-negative strain Mcr2 differed for each cytokine.
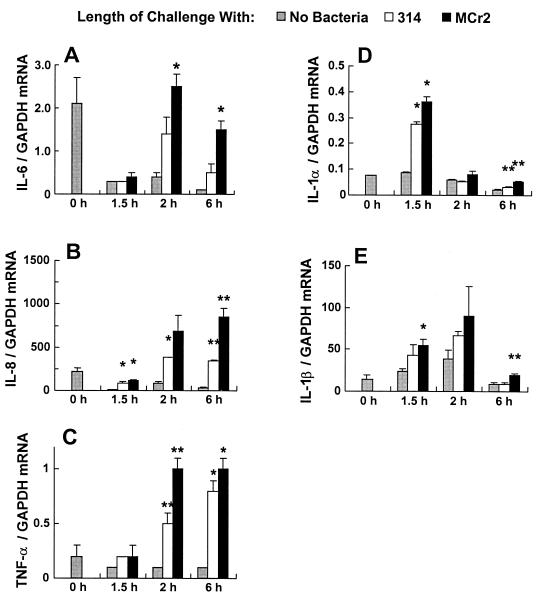
FIG. 4.
Cytokine mRNA expression in HEp-2 cells challenged with C. rectus strains 314 (CrsA+) and Mcr2 (CrsA−). The ordinate represents the amount of specific cytokine RNA normalized to the amount of GAPDH RNA from the same sample. At time zero, the HEp-2 cells were shifted from medium containing 5% FCS to medium containing 2% FCS (No Bacteria), and a sample was taken. The bacteria were then added at an MOI of 1,000, and the HEp-2 cells were harvested for RNA isolation at the indicated times. Each bar gives the mean and standard deviation for three cDNA samples. The asterisks indicate samples in which the cytokine mRNA levels are statistically significantly different from those for medium alone (single asterisk) or from those for the other bacterial strain and medium alone (double asterisk).
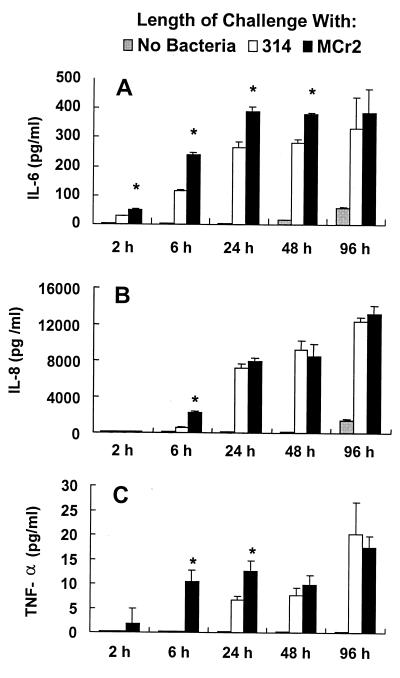
FIG. 5.
ELISAs of cytokine secretion in HEp-2 cells challenged with C. rectus 314 and Mcr2. At time zero, the HEp-2 cells were shifted from medium containing 5% FCS to medium containing 2% FCS (No Bacteria). The bacteria were added to the host cells at an MOI of 1,000, the HEp-2 cell supernatants were collected at the indicated times, and the cytokine levels were determined by ELISA. Each bar gives the mean and standard deviation for three individual samples of each treatment. All of the Mcr2-challenged samples were statistically significantly different from those for the medium alone. The ones marked by an asterisk were also statistically significantly different from the corresponding 314 sample.
A three- to fivefold increase in the IL-6 mRNA level was seen only after 2 h of bacterial challenge (Fig. 4A). The response was about twofold stronger to CrsA− strain Mcr2 than to CrsA+ strain 314. These results were confirmed by the assay for the secretion of IL-6 (Fig. 5A); the release of IL-6 into the supernatants was seen as early as 2 h after C. rectus challenge, and the levels increased up to 24 h after challenge but plateaued at 48 h. Consistent with the mRNA data, higher levels of IL-6 release were seen in the supernatants of Mcr2-challenged cultures than in those of the 314-challenged cultures.
The induction of IL-8 mRNA in HEp-2 cells by C. rectus was rather similar to what was seen for IL-6; there was a four- to eightfold increase after 314 challenge, and Mcr2 bacteria induced an additional 2.5-fold increase (Fig. 4B). The cytokine secretion data for IL-8 were a little different from those for IL-6. Unlike IL-6, IL-8 was not found at detectable levels in the supernatant until 6 h after challenge. Also, the levels induced by CrsA+ and CrsA− bacteria differed at 6 h but were the same at later times (Fig. 5B). These difference in the kinetics of response may or may not be important. Interestingly, IL-8 appears to be the most abundant of all the cytokines tested.
The pattern of the response of TNF-α mRNA to C. rectus was also very similar to those seen for IL-6 and IL-8 (Fig. 4C). TNF-α mRNA was first seen at 1.5 h, and its level was as much as eightfold higher than that in the presence of medium alone. Once again, the Mcr2-challenged HEp-2 cells usually had higher levels of TNF-α mRNA than did the 314-challenged HEp-2 cells, although only the difference at 2 h was statistically significant. Consistent with the mRNA data, the level of released TNF-α increased significantly in HEp-2 cells after the challenge with C. rectus (Fig. 5C). Interestingly, the CrsA− bacteria induced a much stronger TNF-α response than did the wild-type parental bacteria, strain 314, at the early time points (2, 6, and 24 h).
Increased levels of IL-1α and IL-1β mRNAs were detected in bacterially challenged HEp-2 cells (Fig. 4D and E). Unlike the increases in IL-6, IL-8, and TNF-α mRNA levels the increases in IL-1α and IL-1β levels were transient (Fig. 4). The highest levels of IL-1α and IL-1β were found at 1.5 or 2 h after challenge, and they returned to basal level at later times. Unlike what was found with IL-6, IL-8, and TNF-α, the IL-1α and IL-1β mRNA responses did not differ significantly in HEp-2 cells challenged with 314 or Mcr2 bacteria. IL-1β release was not detectable in the ELISA in either bacterially challenged or control cultures at any time points (data not shown). This may reflect the transient nature of the response or may simply indicate that low levels of the cytokine were made. IL-1α release was not assayed, since IL-1α is predominantly a cell-bound cytokine.
Finally, the levels of IL-6, IL-8, and TNF-α mRNA were higher at time zero than at 1.5 h after exposure to medium alone (Fig. 4A to C). This appears to be an artifact of the manner in which the experiment was done. Prior to the experiment, the HEp-2 cells were growing in 5% FCS to maximize growth. However, just before the challenge with C. rectus, the HEp-2 cells were maintained in 2% FCS to maximize C. rectus binding. Thus, at time zero, the levels of IL-6, IL-8, and TNF-α would appear to be induced by a stress response, which decreased once the cells had adapted to the 2% FCS at 1.5 h.
DISCUSSION
As the first host cells to come to contact with microbial pathogens, epithelial cells are in a unique position to function as a barrier to the attachment of bacteria and to serve as an early host signaling system to the immune system cells located in the underlying mucosa. To test whether the C. rectus S-layer can alter these initial responses of epithelial cells, isogenic S-layer-negative strains were constructed and compared to their isogenic S-layer-positive parental cells for the ability to bind to and increase cytokine secretion from a human epithelial cell line, HEp-2. The binding experiments showed that the CrsA+ strain 314 was 30 to 50% more adherent to HEp-2 cells than were CrsA− strains. The relatively small effect of the S-layer on binding to epithelial cells is seen consistently, but it is uncertain whether it is significant during the course of an infection. However, the availability of an isogenic pair of C. rectus strains, one with and one without the S-layer, will facilitate a direct evaluation of the role of the S-layer when tested in an animal model.
S-layers on other bacteria also affect bacterium-host interactions. For example, the S-layer of Aeromonas salmonicida, which is required for the virulence of this fish pathogen (30), increases the adherence of the bacteria by as much as 10-fold to murine (11, 45) and rainbow trout (10, 45) macrophages. Attachment of Clostridium difficile to several host cell types is also dependent upon the S-layer of that organism (37). Similarly, a subcultivated isolate of Lactobacillus acidophilus which had lost its S-layer was less adherent to avian epithelial cells than was the parental S-layer-containing isolate (38). All of these results are similar to what we observed for the C. rectus S-layer, although the C. rectus S-layer, which increased binding to HEp-2 cells by 30 to 50%, did not enhance binding as much as did the S-layers from these other organisms. Interestingly, Graham and McDonald (15) reported that the C. fetus S-layer could cause a small (∼50%) decrease in the binding of C. fetus to HEp-2 cells. Thus, the role of the S-layer in binding to host cells appears complex and is both S-layer and, possibly, host cell type dependent.
Since we have shown that CrsA− C. rectus cells can bind epithelial cells, it is clear that the S-layer is not essential for binding. Nevertheless, the S-layer does enhance the binding of C. rectus to epithelial cells. How might the S-layer increase binding? Reddi and Holt (personal communication) have found that polymyxin B inhibits the binding of C. rectus 33238 to human gingival fibroblast (HGF) cells, suggesting that lipopolysaccharide (LPS) is involved in the binding (40). Perhaps the S-layer helps to organize the outer core or side chain of C. rectus LPS so that it can interact more effectively with host cells and thus increase bacterial binding. This needs to be tested more rigorously.
Breakdown of periodontal attachment is the pathognomonic feature of periodontitis. There is increasing evidence suggesting that periodontal tissue degradation is, in part, the consequence of host cells releasing inflammatory mediators and cytokines in response to periodontal pathogens (3, 16, 17, 20, 44). Therefore, we examined the ability of CrsA+ and CrsA− C. rectus cells to alter the cytokine responses of epithelial cells. Our results showed that C. rectus significantly induced the expression of several cytokine genes and the secretion of the corresponding cytokines. The synthesis and secretion of IL-6, IL-8, and TNF-α were seen with both CrsA+ and CrsA− strains of C. rectus. Increased levels of IL-1 mRNA were also found at some time points after exposure of HEp-2 cells to C. rectus. Interestingly, the kinetics of cytokine induction differed for CrsA+ and CrsA− bacteria. At early time points, the CrsA− bacterium-challenged HEp-2 cells produced higher levels of IL-6, IL-8, and TNF-α mRNA and protein than did the CrsA+ bacterium-challenged host cells. The differences between cytokine secretion induced by CrsA+ and CrsA− bacteria disappeared at later times after bacterial challenge. Since the levels of anti-inflammatory cytokines IL-1ra, IL-13, and TGF-β were not altered by C. rectus challenge of HEp-2 cells, we conclude that a major role of C. rectus appears to be the induction of the proinflammatory cytokines IL-6, IL-8, TNF-α, and, possibly, IL-1. The significantly increased production of the proinflammatory cytokines IL-6, IL-8, and TNF-α by epithelial cells in response to C. rectus challenge is important for recruiting inflammatory cells to the site of infection. The resulting inflammation could potentially have both protective and deleterious effects. Although the C. rectus S-layer ameliorates this response somewhat at early times after challenge, the biological role, if any, of this difference between CrsA+ and CrsA− bacteria is unclear. Perhaps the S-layer may modulate the host cytokine response to a lower level to facilitate the survival of C. rectus at the site of infection. Persistence of the bacteria in periodontal sites could lead to a long-term stimulation of cytokine induction with resulting chronic inflammation involving destruction of the supporting structures of the teeth, as is seen in periodontitis.
C. rectus is a potent inducer of cytokines and mediators in other host cells. Dongari-Bagtzoglou and Ebersole (7) reported that formalin-killed C. rectus induces human gingival fibroblasts to produce IL-6, IL-8, and prostaglandin E2. Reddi et al. (33) found that surface-associated material released from C. rectus stimulates the production of IL-6 and TNF-α in HGF cells and a myelomonocytic cell line. Gillespie et al. (14) also found that purified LPS of Wolinella recta (C. rectus) can induce the production of prostaglandin E and IL-1 in mouse macrophages. Therefore, C. rectus, in general, appears to induce a proinflammatory cytokine response in a variety of host cells. It will be interesting to see if the C. rectus S-layer also modifies the cytokine response in these other host cell types.
Although the physiological importance of the alteration of the cytokine response by the C. rectus S-layer is unproven, components from other bacteria are biologically relevant in suppressing cytokine induction by host cells. Most of these other suppressor molecules are proteins or toxins which inhibit proinflammatory cytokines (49). For example, a plasmid-encoded YopB virulence protein from Yersinia enterocolitica can act as a potent suppressor of both TNF-α expression in macrophages (4) and IL-8 secretion in epithelial cells (39). This modulation of the host cytokine response appears to be important for the survival and proliferation of Y. enterocolitica in host tissue and contributes significantly to the ability of the bacteria to evade antibacterial host defenses. The results of our study suggest that the S-layer protein of C. rectus can be added to the list of cytokine suppressor molecules, since it reduces the cytokine response at certain times after initial bacterium-host cell contact. The mechanism by which the C. rectus S-layer alters the production of proinflammatory cytokines remains to be determined.
Finally, the inflammatory response can be modified by components of the host innate immune defenses, such as neutrophils and the complement pathway. Interestingly, in A. salmonicida, the S-layer is able to protect the bacteria from complement-mediated lysis (37). Similarly, the S-layer of C. fetus can confer serum resistance in the absence of opsonizing antibodies (37). The availability of defined S-layer-negative mutants in C. rectus will allow us to assess whether the C. rectus S-layer has similar functions that might contribute to pathogenesis.
ACKNOWLEDGMENTS
We thank Peter Melby for generously providing the primers for IL-1α, IL-1β, IL-6, and TNF-α and Keith Krolick for sharing the GAPDH primers. Jeff Ebersole kindly supplied anti-S-layer antisera, and Stan Holt furnished the ATCC 33238 strains. We also appreciate helpful discussions with Stan Holt and Jeff Ebersole.
This work was supported by Public Health Service grant DE-10960 from the National Institutes of Health.
REFERENCES
- 1.Assuma R, Oates T, Cochran D, Amar S, Graves D T. IL-1 and TNF antagonist inhibit the inflammatory response and bone loss in experimental periodontitis. J Immunol. 1998;160:403–409. [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1994. [Google Scholar]
- 3.Bartold P M, Haynes D R. Interleukin 6 production by human gingival fibroblasts. J Periodontal Res. 1991;26:339–345. doi: 10.1111/j.1600-0765.1991.tb02072.x. [DOI] [PubMed] [Google Scholar]
- 4.Beuscher H U, Rodel F, Forsberg A, Rolling M. Bacterial evasion of host immune defence: Yersinia enterocolitica encodes a suppressor for tumor necrosis factor alpha expression. Infect Immun. 1995;63:1270–1277. doi: 10.1128/iai.63.4.1270-1277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borinski R, Holt S C. Surface characteristics of Wolinella recta ATCC 33238 and human clinical isolates: correlation of structure with function. Infect Immun. 1990;58:2770–2776. doi: 10.1128/iai.58.9.2770-2776.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dokland T, Olsen I, Farrants G, Johansen B V. Three-dimensional structure of the surface layer of Wolinella recta. Oral Microbiol Immunol. 1990;5:162–165. doi: 10.1111/j.1399-302x.1990.tb00415.x. [DOI] [PubMed] [Google Scholar]
- 7.Dongari-Bagtzoglou A I, Ebersole J L. Production of inflammatory mediators and cytokines by human gingival fibroblasts following bacterial challenge. J Periodontal Res. 1996;31:90–98. doi: 10.1111/j.1600-0765.1996.tb00469.x. [DOI] [PubMed] [Google Scholar]
- 8.Dzink J L, Tanner A C R, Haffajee A D, Socransky S S. Gram negative species associated with active periodontal lesions. J Clin Periodontol. 1985;12:648–659. doi: 10.1111/j.1600-051x.1985.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 9.Ebersole J L, Frey D E, Taubman M A, Smith D J. An ELISA for measuring serum antibodies to Actinobacillus actinomycetemcomitans. J Periodontal Res. 1980;15:621–632. doi: 10.1111/j.1600-0765.1980.tb00321.x. [DOI] [PubMed] [Google Scholar]
- 10.Garduno R A, Kay W W. Interaction of the fish pathogen Aeromonas salmonicida with rainbow trout macrophages. Infect Immun. 1992;60:4612–4620. doi: 10.1128/iai.60.11.4612-4620.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garduno R A, Lee E J Y, Kay W W. S-layer-mediated association of Aeromonas salmonicida with murine macrophages. Infect Immun. 1992;60:4373–4382. doi: 10.1128/iai.60.10.4373-4382.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genco R J. Host responses in periodontal diseases: current concepts. J Periodontol. 1992;63:338–355. doi: 10.1902/jop.1992.63.4s.338. [DOI] [PubMed] [Google Scholar]
- 13.Gillespie J, Holt S C. Growth studies of Wolinella recta, a gram-negative periodontopathogen. Oral Microbiol Immunol. 1987;2:105–111. doi: 10.1111/j.1399-302x.1987.tb00271.x. [DOI] [PubMed] [Google Scholar]
- 14.Gillespie J, Weintraub S T, Wong G G, Holt S C. Chemical and biological characterization of the lipopolysaccharide of the oral pathogen Wolinella recta ATCC 33238. Infect Immun. 1988;56:2028–2035. doi: 10.1128/iai.56.8.2028-2035.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham L L, MacDonald K L. The Campylobacter fetus S layer is not essential for initial interaction with HEp-2 cells. Can J Microbiol. 1998;44:244–250. [PubMed] [Google Scholar]
- 16.Hanazawa S, Hirose K, Ohmori Y, Amano S, Kitano S. Bacteroides gingivalis fimbriae stimulate production of thymocyte activating factor by human gingival fibroblasts. Infect Immun. 1988;56:272–274. doi: 10.1128/iai.56.1.272-274.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heath J K, Atkinson S J, Hembry R M, Reynolds J J, Meikle M C. Bacterial antigens induce collagenase and prostaglandin E2 synthesis in human gingival fibroblasts through a primary effect on circulating mononuclear cells. Infect Immun. 1987;55:2148–2154. doi: 10.1128/iai.55.9.2148-2154.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honig J, Rordorf-Adam C, Siegmund C, Wiedemann W, Erard F. Increased interleukin-1 beta (IL-1β) concentration in gingival tissue from periodontitis patients. J Periodontal Res. 1989;24:362–367. doi: 10.1111/j.1600-0765.1989.tb00883.x. [DOI] [PubMed] [Google Scholar]
- 19.Kesavalu L, Holt S C, Crawley R R, Borinski R, Ebersole J L. Virulence of Wolinella recta in a murine abscess model. Infect Immun. 1991;59:2806–2817. doi: 10.1128/iai.59.8.2806-2817.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kjeldsen M, Holmstrup P, Bendtzen K. Marginal periodontitis and cytokines: a review of the literature. J Periodontol. 1993;64:1013–1022. doi: 10.1902/jop.1993.64.11.1013. [DOI] [PubMed] [Google Scholar]
- 21.Kokeguchi S, Kato K, Kurihara H, Murayama Y. Cell surface protein antigen from Wolinella recta ATCC 33238. J Clin Microbiol. 1989;27:1210–1217. doi: 10.1128/jcm.27.6.1210-1217.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolodrubetz D, Dailey T, Ebersole J, Kraig E. Cloning and expression of the leukotoxin gene from Actinobacillus actinomycetemcomitans. Infect Immun. 1989;57:1465–1469. doi: 10.1128/iai.57.5.1465-1469.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolodrubetz D, Spitznagel J, Jr, Wang B, Phillips L H, Jacobs C, Kraig E. cis element and trans factors are both important in strain-specific regulation of the leukotoxin gene in Actinobacillus actinomycetemcomitans. Infect Immun. 1996;64:3451–3460. doi: 10.1128/iai.64.9.3451-3460.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kono Y, Beagley K W, Fujihashi K, McGhee J R, Taga T, Hirano T, Kishimoto T, Kiyono H. Cytokine regulation of localized inflammation. Induction of actived B cells and IL-6-mediated polyclonal IgG and IgA synthesis in inflamed human gingiva. J Immunol. 1991;146:1812–1821. [PubMed] [Google Scholar]
- 25.Lai C H, Listgarten M A, Tanner A C R, Socransky S S. Ultrastructure of Bacteroides gracilis, Campylobacter concisus, Wolinella recta, and Eikenella corrodens, all from humans with periodontal disease. Int J Syst Bacteriol. 1981;31:465–475. [Google Scholar]
- 26.Lauer B A, Reller L B, Mirrett S. Comparison of acridine orange and Gram stains for detection of microorganisms in cerebrospinal fluid and other clinical specimens. J Clin Microbiol. 1981;14:201–205. doi: 10.1128/jcm.14.2.201-205.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeBlanc D J, Lee L N, Inamine J M. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:1804–1810. doi: 10.1128/aac.35.9.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melby P C, Andrade-Narvaez F J, Darnell B J, Valencia-Pacheco G, Tryon V V, Palomo-Cetina A. Increased expression of proinflammatory cytokines in chronic lesions of human cutaneous leishmaniasis. Infect Immun. 1994;62:837–842. doi: 10.1128/iai.62.3.837-842.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nitta H, Holt S C, Ebersole J L. Purification and characterization of Campylobacter rectus surface layer proteins. Infect Immun. 1997;65:478–483. doi: 10.1128/iai.65.2.478-483.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noonan B, Trust T J. The synthesis, secretion and role in virulence of the paracrystalline surface protein layer of Aeromonas salmonicida and A. hydrophila. FEMS Microbiol Lett. 1997;154:1–7. doi: 10.1111/j.1574-6968.1997.tb12616.x. [DOI] [PubMed] [Google Scholar]
- 31.Page R C. The role of inflammatory mediators in the pathogenesis of periodontal disease. J Periodontal Res. 1991;26:230–242. doi: 10.1111/j.1600-0765.1991.tb01649.x. [DOI] [PubMed] [Google Scholar]
- 32.Rams T E, Feik D, Slots J. Campylobacter rectus in human periodontitis. Oral Microbiol Immunol. 1993;8:230–235. doi: 10.1111/j.1399-302x.1993.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 33.Reddi K, Wilson M, Nair S, Poole S, Henderson B. Comparison of the pro-inflammatory cytokine-stimulating activity of the surface-associated proteins of periodontopathic bacteria. J Periodontal Res. 1996;31:120–130. doi: 10.1111/j.1600-0765.1996.tb00473.x. [DOI] [PubMed] [Google Scholar]
- 34.Roberts F A, Hockett R D, Jr, Bucy R P, Michalek S M. Quantitative assessment of inflammatory cytokine gene expression in chronic adult periodontitis. Oral Microbiol Immunol. 1997;12:336–344. doi: 10.1111/j.1399-302x.1997.tb00735.x. [DOI] [PubMed] [Google Scholar]
- 35.Roberts F A, McCaffery K A, Michalek S M. Profile of cytokine mRNA expression in chronic adult periodontitis. J Dent Res. 1997;76:1833–1839. doi: 10.1177/00220345970760120501. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Sara M, Egelseer E M. Functional aspects of S-layers. In: Sleytr U B, Messner P, Pum D, Sara M, editors. Crystalline bacterial cell surface proteins. San Diego, Calif: Academic Press, Inc.; 1996. pp. 103–131. [Google Scholar]
- 38.Schneitz C, Nuotio L, Lounatma K. Adhesion of Lactobacillus acidophilus to avian intestinal epithelial cells mediated by the crystalline bacterial cell surface (S-layer) J Appl Bacteriol. 1993;74:290–294. doi: 10.1111/j.1365-2672.1993.tb03028.x. [DOI] [PubMed] [Google Scholar]
- 39.Schulte R, Wattiau P, Hartland E L, Robins-Browne R M, Cornelis G R. Differential secretion of interleukin-8 by human epithelial cell lines upon entry of virulent or nonvirulent Yersinia enterocolitica. Infect Immun. 1996;64:2106–2113. doi: 10.1128/iai.64.6.2106-2113.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srimal S, Surolia N, Balasubramanian S, Surolia A. Titration calorimetric studies to elucidate the specificity of the interactions of polymyxin B with lipopolysaccharides and lipid A. Biochem J. 1996;315:679–686. doi: 10.1042/bj3150679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stashenko P, Fujiyoshi P, Obernesser M S, Prostak L, Haffajee A D, Socransky S S. Levels of interleukin 1β in tissue from sites of active periodontal disease. J Clin Periodontol. 1991;18:548–554. doi: 10.1111/j.1600-051x.1991.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 42.Stashenko P, Jandinski J P, Fujiyoshi P, Rynar J, Socransky S S. Tissue levels of bone resorptive cytokines in periodontal disease. J Periodontol. 1991;62:504–509. doi: 10.1902/jop.1991.62.8.504. [DOI] [PubMed] [Google Scholar]
- 43.Steffen M J, Ebersole J L. Sequential ELISA for cytokine levels in limited volumes of biological fluids. BioTechniques. 1996;21:504–509. doi: 10.2144/96213rr04. [DOI] [PubMed] [Google Scholar]
- 44.Tonetti M S, Freiburghaus K, Lang N P, Bickel M. Detection of interleukin-8 and matrix metalloproteinases transcripts in healthy and diseased gingival biopsies by RNA/PCR. J Periodontal Res. 1993;28:511–513. doi: 10.1111/j.1600-0765.1993.tb02114.x. [DOI] [PubMed] [Google Scholar]
- 45.Trust T J, Kay W W, Ishiguro E E. Cell surface hydrophobicity and macrophage association of Aeromonas salmonicida. Curr Microbiol. 1983;9:315–318. [Google Scholar]
- 46.Van Dyke T E, Lester M A, Shapira L. The role of the host response in periodontal disease progression: implications for future treatment strategies. J Periodontol. 1993;64:792–806. doi: 10.1902/jop.1993.64.8s.792. [DOI] [PubMed] [Google Scholar]
- 47.Wang B, Kraig E, Kolodrubetz D. A new member of the S-layer protein family: characterization of the crsA gene from Campylobacter rectus. Infect Immun. 1998;66:1521–1526. doi: 10.1128/iai.66.4.1521-1526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams R C. Periodontal disease. N Engl J Med. 1990;322:373–382. doi: 10.1056/NEJM199002083220606. [DOI] [PubMed] [Google Scholar]
- 49.Wilson M, Seymour R, Henderson B. Bacterial perturbation of cytokine networks. Infect Immun. 1998;66:2401–2409. doi: 10.1128/iai.66.6.2401-2409.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]