Summary
A variety of anthropogenic organohalide contaminants generated from industry are released into the environment and thus cause serious pollution that endangers human health. In the present study, we investigated the microbial community composition of industrial saponification wastewater using 16S rRNA sequencing, providing genomic insights of potential organohalide dehalogenation bacteria (OHDBs) by metagenomic sequencing. We also explored yet-to-culture OHDBs involved in the microbial community. Microbial diversity analysis reveals that Proteobacteria and Patescibacteria phyla dominate microbiome abundance of the wastewater. In addition, a total of six bacterial groups (Rhizobiales, Rhodobacteraceae, Rhodospirillales, Flavobacteriales, Micrococcales, and Saccharimonadales) were found as biomarkers in the key organohalide removal module. Ninety-four metagenome-assembled genomes were reconstructed from the microbial community, and 105 hydrolytic dehalogenase genes within 42 metagenome-assembled genomes were identified, suggesting that the potential for organohalide hydrolytic dehalogenation is present in the microbial community. Subsequently, we characterized the organohalide dehalogenation of an isolated OHDB, Microbacterium sp. J1-1, which shows the dehalogenation activities of chloropropanol, dichloropropanol, and epichlorohydrin. This study provides a community-integrated multi-omics approach to gain functional OHDBs for industrial organohalide dehalogenation.
Keywords: organohalide, wastewater, metagenome assembled genome, microbial community
Graphical abstract
Public summary
-
•
Many pollutant-degrading microorganisms are hidden in complex environmental microbial communities
-
•
The culture-independent multi-omics approach provides information about the potential key microorganisms and functional genes in industrial saponification wastewater
-
•
The multi-omics approach guides the design of a culture method to obtain the organohalide dehalogenation microorganism
Introduction
Organohalides are a series of organic compounds containing fluorine, chlorine, or bromine.1 Many organohalides are produced in industrial and agricultural processes, including trichloroethene, trichloroethanes, and triclosan.2,3 Organohalides can cause pollution due to their release into the atmosphere or discharge into the environment in industrial wastewater. Organohalides can bioaccumulate through the food chain to damage the human body. Organohalides present a toxic risk because of their endocrine-disrupting effects on animals, making them a serious threat to human health and the environment.3
Several organohalides are produced as byproducts (eg, dichloropropane, dichloroisopropyl ether, and chloropropanol) of propylene oxide production.4 Propylene oxide saponification wastewater also has the characteristics of strong alkalinity, high salt, and high solid suspended matter, which makes the degradation of organohalides in saponification wastewater quite challenging.
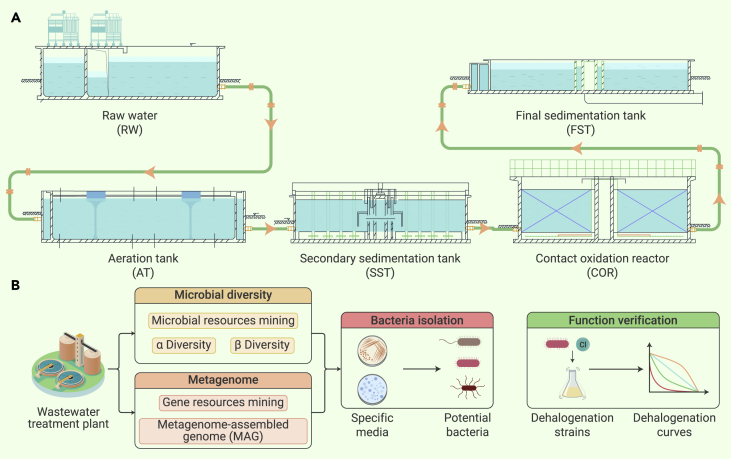
A two-phase biotechnological treatment technology, which consists of activated sludge and contact oxidation, is used to treat organohalides that occur in propylene oxide saponification wastewater (Figure 1A). The aeration tank provides sufficient oxygen for the growth of the activated microbial community in wastewater and the degradation of organic matter. Microorganisms are allowed to flow into the contact oxidation tank to form a biofilm, which continuously reduces the organic matter of the wastewater. An inappropriate microbial community will directly influence the final wastewater treatment efficiency.5
Figure 1.
Overview of the wastewater treatment process and experimental design for the multi-omics analyses
(A) Schematic of the two-phase biotechnological treatment technology used for saponification wastewater. The aeration tank (AT) used activated sludge for the degradation of organic matter. The secondary sedimentation tank (SST) was used to recycle activated sludge from the aeration tank. The contact oxidation reactor (COR) utilized biofilms to degrade organic matter. RW, raw water; FST, final sedimentation tank.
(B) Multi-omics analysis workflow: microbial diversity and metagenomics analyses of the wastewater treatment plant were performed to obtain microbial and genetic profiles that were used as a guide for isolating potential organohalide dehalogenation bacteria and to carry out assays to evaluate their dehalogenation capacity.
Classical cultivation methods (eg, selective nutrient or inhibitors, special physicochemical conditions, and size- or density-based separation) are commonly used to isolate microorganisms from the environment. However, these approaches have failed to identify some populations of microorganisms that are either unculturable or difficult to cultivate. The limitations of classical cultivation methods include lack of knowledge about metabolism (eg, substrate, growth factor, and dormancy), interaction (eg, symbiotic interdependencies), and abundance in the microbial community (eg, rare species).6
Culture-independent amplicon and metagenomic sequencing techniques can improve our understanding of environmental microbial communities. For example, the global core bacterial community of wastewater treatment plants includes 28 operational taxonomic units (OTUs) that were identified using 16S rRNA analysis.7 In addition, metagenomic analysis has been used to identify novel dehalogenases and cofactor synthetic enzymes present in the trichloroethene-degrading microbial community.8 The near-complete or draft genomes of uncultured bacteria were obtained using de novo assembled metagenomic sequences, which may provide the possibility to link taxa with metabolic genes in environmental microbial communities.9 The genome-centric investigation of microorganisms can broaden our understanding of their potential metabolic functions; however, it is also necessary to isolate and culture microorganisms in order to define their microbial features with respect to metabolism and physiology in the real environment for bioremediation.
To circumvent the limitations associated with current cultivation methods, we expanded a multi–omics-guided design of selective media and built a simulated environment suitable for the growth of potential organohalide dehalogenation bacteria (OHDBs); the design was based on the substrate demands and growth factors predicted from the draft genomes of OHDBs in combination with the metabolomic data during a complete wastewater treatment process. In this study, we focused on potential OHDBs for organohalide dehalogenation from the wastewater of a propylene oxide manufacturing plant in Shandong Province, China. 16S rRNA gene sequencing was used to profile the dynamics of the microbial community and to distinguish the microbes that were dominant in key treatment modules. Previous studies of diverse environments like sewage effluent and animal rumen established analyses based on the existence of functional genes for specific substances like antibiotics or lignocellulosic polymers.10,11 These studies provide analysis directions for understanding the degradation capacity of metagenome-assembled genomes (MAGs). In our present study, MAGs were constructed based on metagenomic reads from various phases of the wastewater treatment process, which were used to evaluate the microbial community’s functional capability. An OHDB strain was isolated from the wastewater microbial community via an expanded selective media strategy. Moreover, we performed a series of organohalide dehalogenation capacity characterizations on the OHDB to uncover the potential for organohalide dehalogenation.
Results and discussion
Spatiotemporal variability of wastewater microbial community
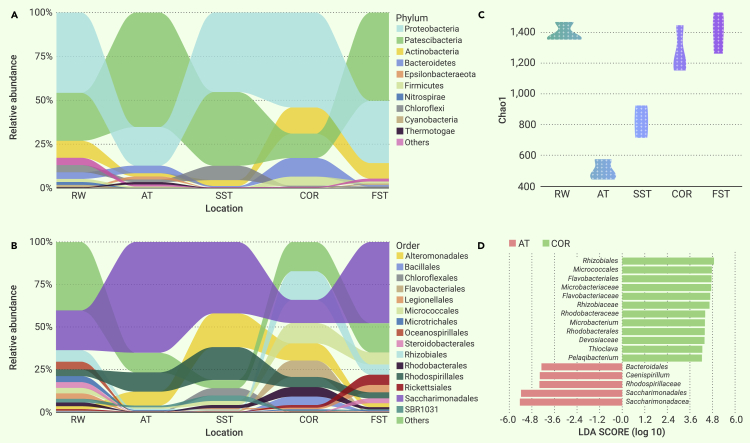
Samples were taken from five distinct phases of the wastewater treatment process. Proteobacteria, Patescibacteria, Actinobacteria, and Bacteroidetes were the dominant phyla in the treatment process (Figure 2A). The bacterial diversity communities of the wastewater treatment process were analyzed using the Chao1 index (Figure 2C). Relatively higher microbial alpha diversity values were found for the raw water (RW) and final sedimentation tank (FST) phases; a sharp decrease in alpha diversity occurred when the wastewater moved from RW to aeration tank (AT). Significant differences in microbial abundance were found in the different phases of wastewater (Adonis, p < 0.001); this was also reflected in the detected variations in community composition (β-diversity; Figure S2).
Figure 2.
Microbial diversity in distinct phases of the wastewater treatment process
(A) Spatiotemporal variability of the top 10 ranking (relative abundance) phyla during the wastewater treatment process (n = 3).
(B) Spatiotemporal variability of the top 15 ranking (relative abundance) orders during the wastewater treatment process (n = 3).
(C) Alpha diversity of the microbial community during the different phases of the wastewater treatment process.
(D) Linear discriminant analysis effect size (LEfSe) analysis of the microbial community during the wastewater treatment process (linear discriminant analysis score>4.2, p < 0.05). These genus with significant differential abundance among distinct phases of the wastewater treatment process.
Changes in environmental factors will influence the structure of microbial communities,12 which were reflected upon the change of temperature (50°C to 35°C–38°C) and pH (11.7 to 6.5–6.9) between RW and AT. Within the top 15 ranking microbial orders (Figure 2B), Saccharimonadales was the most abundant Patescibacteria species during the AT phase. Studies on the specific physiological ecology of Saccharimonadales are scarce, with only a few reports about phosphorus-mining bioindicators and sulfonamide-resistant bacteria available.13,14
Compared with other phases (RW, secondary sedimentation tank [SST], and FST), AT and contact oxidation reactor (COR) showed a more efficient chemical oxygen demand removal ability (Table S1). Thus, identifying the functional taxa of the AT and COR microbial communities should reveal microbial resources for degrading organic chemicals. The linear discriminant analysis effect size (LEfSe) method was used to identify bacterial taxa with significant differential abundance throughout the various phases of the wastewater treatment process (Figure 2D). A total of 17 bacterial taxa were found statistically differentially abundant in the AT and COR phases: five bacteria taxa were found in the AT phase, and 12 bacterial taxa were found in the COR phase. Phylogenetic analysis based on the 16S rRNA gene revealed that these taxa belong to six groups of microorganisms: Flavobacteriales, Rhodospirillales, Rhizobiales, Micrococcales, Rhodobacteraceae, and Saccharimonadales (Figure 2D). We classified these six groups as putative key strains, which were the focus of our follow-up metagenomic sequencing and genome assembly efforts seeking to characterize their dehalogenation potential. The LEfSe results for the taxa of other phases are presented in Figure S1.
OHDBs previously reported under anaerobic conditions (Dehalococcoides, Dehalogenimonas, and Desulfitobacterium) were searched for within the different phases of the wastewater treatment process.15,16,17 In the COR phase, we found that Dehalogenimonas was the most abundant microorganism among these known anaerobic OHDBs; however, its abundance was still less than 0.03%. This indicates that reductive dehalogenation under anaerobic conditions is apparently not the main process through which organohalides are degraded in this wastewater treatment.
Phylogeny of MAGs and identification of functional genes
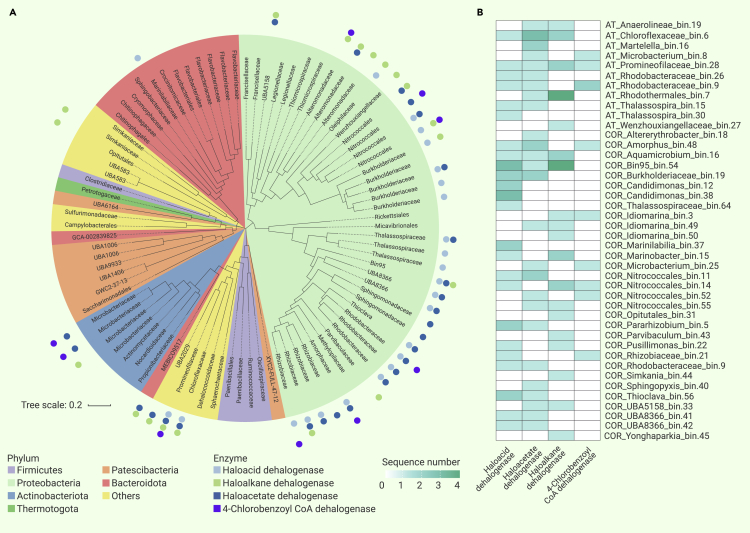
A total of 94 MAGs were obtained and filtered based on metagenomic reads (Figure 3A), which contained MAGs of AT (30) and COR (64). In addition, we found 20 near-perfect MAGs (completeness >95%, contamination <1%) within these MAGs. All MAGs (completeness >70%, contamination <5%) were used for subsequent functional gene and metabolism analyses. The detailed information for completeness and contamination of each MAG quality is shown in Table S3.
Figure 3.
The phylogeny of MAGs and identification of functional genes
(A) Phylogenetic identification of metagenome-assembled genomes (MAGs). These MAGs were selected using CheckM (completeness >70%, contamination <5%) and identified using the Genome Taxonomy Database. Distribution of potential dehalogenases is illustrated by the dots.
(B) Potential dehalogenase sequences from the MAG analysis. The numbers of potential dehalogenase sequences in the various MAGs are indicated with the heatmap values (from 0 to 4).
The Genome Taxonomy Database toolkit was used to analyze MAG taxonomic assignments, and it was found that Proteobacteria, Bacteroidetes, Patescibateria, and Actinobacteria were the dominant phyla (Figure 3A; Tables S4 and S5).18 A phylogenetic tree was constructed using the UBCG pipeline,19 which uses a bacterial core gene set to replace 16S rRNA genes, and the tree was visualized using iTOL,20 with the coloring of the tree clades based on phylum-level taxonomy.
The chloroalkanes and chlorohydrins are the main pollutants in the saponification wastewater treatment process.4 It is important to characterize dehalogenases and to isolate novel OHDBs for improving the organohalide bioremediation capacity. Compared with the substrate promiscuity of oxidative dehalogenase, hydrolytic dehalogenase shows a better substrate specificity, which makes it more favorable for investigation.21,22 Hydrolytic dehalogenases include haloacid dehalogenases; haloalkane dehalogenases; haloacetate dehalogenases; 4-chlorobenzoyl coenzyme A (4-CBCoA) dehalogenases; and halohydrin dehalogenases.21,22 To infer the metabolic potential of MAGs, these hydrolytic dehalogenase genes and specific metabolism were scanned via Prokka and KAAS analysis.23,24
A total of 105 candidate hydrolytic dehalogenase genes were detected in 42 MAGs (Table S6), including 33 identified as haloacid dehalogenases; 32 as haloacetate dehalogenases; 30 as haloalkane dehalogenases; and 10 as 4-CBCoA dehalogenases; halohydrin dehalogenase was not present. The highest number of candidate dehalogenases was from Proteobacteria, indicating that Proteobacteria may play an important role in organohalide removal (Figure 3A). The detailed distribution of dehalogenases observed were mainly encoded by Rhizobiaceae, Rhodobacteraceae, Rhodospirillales, Micrococcales, Sphingomonadales, Chloroflexia, and Burkholderiales and are shown in Figure 3B and Table S6.
Three kinds of predicted dehalogenases were found among the Rhizobiaceae MAGs: three haloacid dehalogenases, one haloacetate dehalogenase, and one 4-CBCoA dehalogenase. Rhizobiaceae reportedly participates in organohalide degradation.25 Examination of three Rhodobacterales MAGs revealed five putative haloacid dehalogenases, four haloacetate dehalogenases, and two 4-CBCoA dehalogenases. The l-haloacid dehalogenase gene (DehRhb) and the chlorothalonil hydrolytic dehalogenase gene (chd) were identified in Rhodobacterales and showed organohalide dehalogenation activity with brominated acid, chlorinated acid, and chlorothalonil.26,27 Rhodospirillales include three MAGs that contained three putative haloacid dehalogenases and one haloacetate dehalogenase. Pentachlorophenol was degraded by Rhodospirillales (Azospirillum) under aerobic conditions.28 There were two putative haloacetate dehalogenases, one haloalkane dehalogenase, and two 4-CBCoA dehalogenases in our Micrococcales MAGs. None of the Flavobacteriales MAGs contained any dehalogenase sequences. Flavobacteriales were found as a significant genus of floc-forming bacteria in flocculent activated sludge.29
Many microorganisms exist in the environmental microbial populations with low abundance, but they may still have a significant influence on substrate degradation; the growth rate of strains and substrate degradation efficiency are not necessarily related.6 Some bacteria with relatively low abundance were found to contain putative dehalogenases. For example, four kinds of dehalogenases were found in MAGs among Hyphomicrobiales, Sphingomonadales, and Chloroflexia (Table S6). Notably, we detected eight predicted haloacid dehalogenases, two haloacetate dehalogenases, and one haloalkane dehalogenase in Burkholderiales MAGs. Chloroflexia are organohalide-respiring bacteria that utilize organohalides via reductive dehalogenases rather than hydrolytic dehalogenases.30,31 Several studies also reported the presence of hydrolytic dehalogenases within Hyphomicrobiales, Sphingomonadales, and Burkholderiales, particularly haloalkane dehalogenases (LinB) and haloacid dehalogenases.32,33,34,35,36 These microorganisms may function as organohalide degraders during the wastewater treatment process.
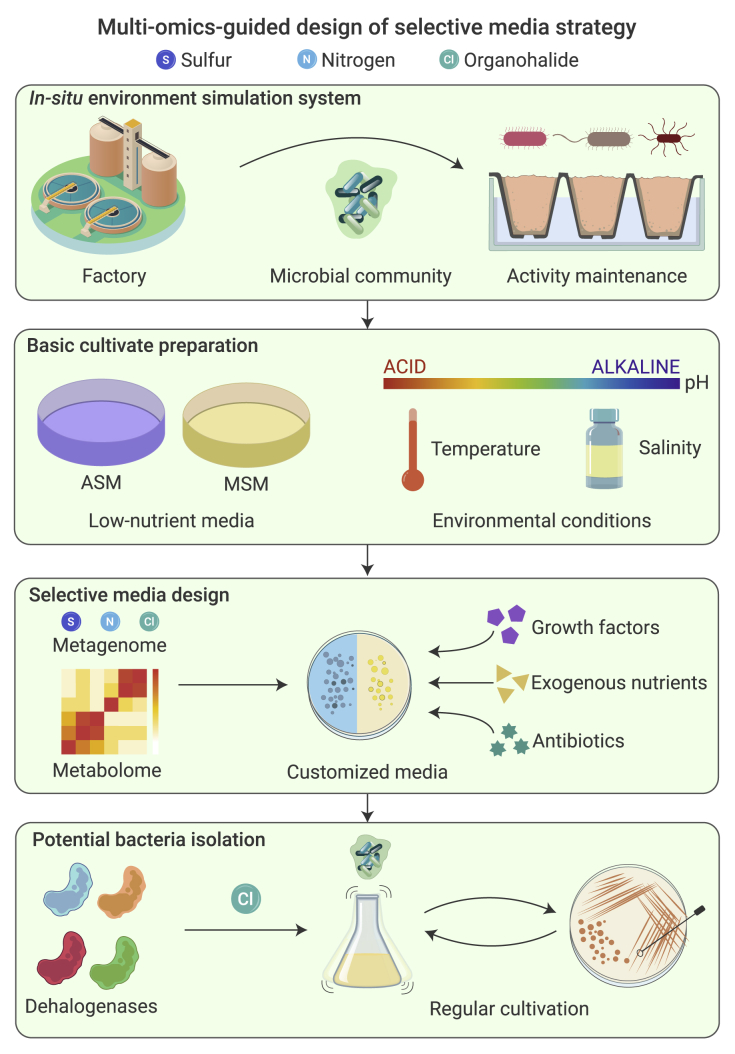
Multi–omics-guided design of selective media strategy
Our predictive analysis of the MAGs revealed potential dehalogenation capacity for Rhizobiaceae, Rhodobacteraceae, Rhodospirillales, Micrococcales, Chloroflexia, Sphingomonadales, and Burkholderiales. Experimental validation of potential OHDBs is required to confirm dehalogenation capacity. There remains a necessity to overcome the obstacles of pure culture isolation, which include (1) the identification of substrates and environmental conditions (temperature, salinity, pH); (2) a requirement for certain growth factors (eg, specific sulfur and nitrogen compounds, metal ions, etc.); and (3) the enrichment of some rare functional taxa in the environment that may have symbiotic interdependencies.6,37,38 The in silico metabolic reconstruction from MAG data can help in identifying potential deficiencies or requirements for successful culture of targeted microorganisms. That is, assessing the genome of a strain can enable inference of which combinations of targeted nutrients should be included in an artificial selective media. For example, the absence of amino acid biosynthesis genes or sulfate metabolic pathways would suggest a requirement for specific nutrients.39 Moreover, metabolomics can be an informative method for investigating metabolites in the environment that can support identification of intermediate metabolites or biomarkers and provide insights into microbial physiology.40 These approaches can reduce trial and error and thereby reduce the time and effort required to determine suitable OHDBs culture conditions.
An illustration of the four stages multi–omics-guided design of selective media strategy we employed is shown in Figure 4. During stage one, an in situ environment simulation system was established to maintain the microbial activity of the in situ microbial community in the original wastewater samples. Activated sludge was added to the activated sludge bioreactor with an aeration device, after which fresh wastewater was periodically added (based on sludge retention time) to remove additional sludge and maintain the stability of the simulated system. These samples were used for subsequent strain isolation. During stage two, the low-nutrient medium (eg, artificial seawater medium, mineral salt medium) was used as the basic medium to support further alterations toward a customized medium. The culture conditions also matched the environmental conditions of the wastewater treatment phases, including 4% salinity, pH 6.5–6.9, and 30°C or 37°C (Table S1).
Figure 4.
The OHDBs from the multi–omics-guided design of selective media strategy
Stage one established an in situ environment simulation system to maintain the activity of the microbial community. Stage two included the preparation of the culture media and physicochemical conditions for further customization. Stage three included the addition of supplementary substances (eg, growth factor, nutrient, and antibiotic) to the media under the guidance of multi-omics data. In stage four, the potential substrates of dehalogenases and the microbial community from the environment simulation system were added to the customized media for subsequent regular cultivation.
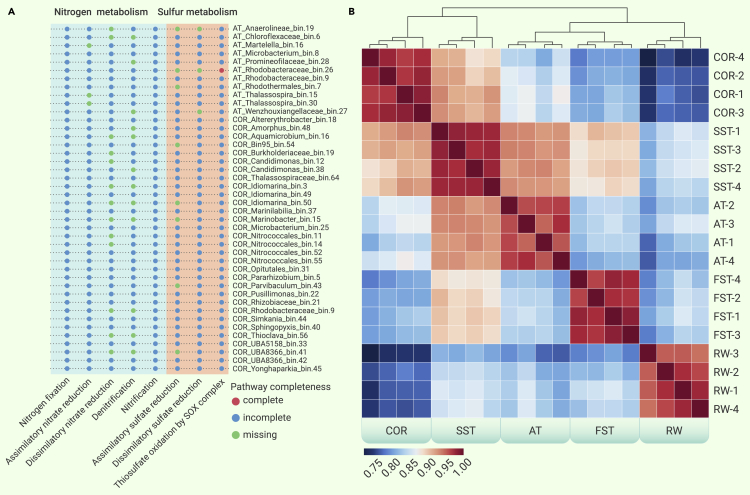
In stage three, the nitrogen and sulfur pathways based on MAGs were used to predict the potential metabolic capabilities of bacteria.9 The metabolic capabilities of potential dehalogenation MAGs for nitrogen and sulfur metabolism are shown in Figure 5A. All 42 of the potential dehalogenation MAGs displayed potential for nitrogen fixation, nitrification, and thiosulfate oxidation. Notably, the Rhodobacteraceae_bin.26 genome contained all the relevant genes of the denitrification pathway. The majority of the potential dehalogenation MAGs (>70%) encoded genes related to nitrate or sulfur reduction.
Figure 5.
The assessment of MAG metabolic capacities
(A) Nitrogen and sulfur metabolism content in MAGs. KEGG pathways are shown in dot form: red dot, the pathway is complete; blue dot, the pathway is incomplete; green dot, the pathway is missing.
(B) Spearman’s correlation analyses of the various phases of the wastewater treatment process. Cluster analysis was performed on each group of samples, which is displayed at the top of the figure (n = 4).
The absence of metabolic pathway genes within identified MAGs may be due to metabolism deficiency of natural microorganisms or draft MAGs. We analyzed metabolic pathways of the potential OHDBs to identify the missing pathways and to determine whether supplemental nutrients should be added to the basic medium (Figure 5A). The SAR11 alpha-proteobacterial clade added exogenous sulfur sources necessary for growth, as evidenced by the deficient assimilation of sulfate reduction in metagenomic datasets. Additional sulfur metabolism supplementation (eg, methionine, cysteine, or thiosulphate) can be added to a customized medium.41
The antibiotic resistance genes detected in the genomes can be used to isolate target organisms, and specific antimicrobial compounds can be used to inhibit the rapid growth of non-target microorganisms.42 The potential MAG antibiotic resistance genes were predicted using CARD (Table S8). The qacJ and qacG (multi-drug resistance efflux pump of quaternary ammonium compounds) genes were found in some MAGs of potential OHDBs (Rhodospirillales, Rhodobacterales, and Burkholderiales).43,44 In contrast, there was an absence of antibiotic resistance genes in the MAGs of Micrococcales, Hyphomicrobiales, and Chloroflexia.
A non-targeted metabolomic was used to detect different metabolites in the wastewater treatment process, with Spearman’s rank correlation coefficient values used to assess the relationships between samples from different phases (Figure 5B). These data indicate that tyrosine metabolism and oxidative phosphorylation are significantly different when wastewater was transferred from RW to AT. The data also indicate that pantothenate and CoA biosynthesis, as well as alanine, aspartate, and glutamate metabolism, were significantly different as the wastewater was moved from AT to COR (Table S7). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the differential metabolite data indicate that succinate is an influencing factor in both the AT and COR phases.
At stage four, we considered known or suspected substrates of hydrolytic dehalogenases.21,45,46,47 The organohalide substrate in our attempts to enrich and isolate OHDBs included dichloropropane, monochloroacetic acid, methyl chloroacetate, and dichloropropanol. The customized medium with specific organohalides was inoculated with the wastewater sample for shaking culture, followed by streak plate on specific media plates. The bacterial clones grown on the plates were picked for the next round of cultivation.
Characterization for organohalide dehalogenation of the isolated OHDB strain
Using this multi-omics strategy to adjust the selective medium to support the growth of OHDBs, Microbacterium sp. J1-1 was isolated from a customized mineral salt medium that contained sodium thiosulfate, disodium succinate, and a substrate mixture including chloroalkane and chlorohydrin without additional antibiotic. The phylogenetic tree of Microbacterium sp. J1-1 based on 16S rRNA is shown in Figure 6A. The morphological characteristics of Microbacterium sp. J1-1 are shown via a scanning electron microscope micrograph (Figure 6B).
Figure 6.
Organohalide dehalogenation assay of Microbacterium sp. J1-1
(A) Phylogenetic tree based on 16S rRNA sequencing.
(B) Scanning electron microscope micrograph of Microbacterium sp. J1-1.
(C) The distribution of dehalogenases in Microbacterium sp. J1-1.
(D) The dehalogenation assay of the resting cell system. Green: the dehalogenation of 1000 μl/l organohalide using Microbacterium sp. J1-1. Orange: the dehalogenation of 1000 μl/l organohalide without bacteria (n = 3, error bar represents the standard error of the mean).
We sequenced the genome of Microbacterium sp. J1-1 and evaluated the presence of candidate hydrolytic dehalogenase genes (Figure 6C). Our assembled genome for Microbacterium sp. J1-1 had each of the hydrolytic dehalogenase types that were annotated in the Micrococcales MAG. Additionally, two haloacid dehalogenases and one halohydrin dehalogenase were found in Microbacterium sp. J1-1. The genome of pure culture OHDB shows more dehalogenases. This may be related to (1) the incompleteness of Micrococcales MAGs (AT_bin.8, COR_bin.25, and COR_bin.45), as we obtained <95% completeness and thus might have missed some functional genes; (2) a portion of the functional genes remain uncharacterized in reference databases or are instead annotated as belonging to a broad protein family (eg, halohydrin dehalogenase belongs to the short-chain dehydrogenase/reductases)48; and (3) the results of the genome binning tools (eg, MetaBat2, MaxBin2, and CONCOCT) are affected by the complexity of the metagenomic dataset and generate a different number and quality of bins for subsequent analyses.49
The dehalogenation ability of Microbacterium sp. J1-1 was determined. Specifically, Microbacterium sp. J1-1 possesses organohalide dehalogenation capacities for chloropropanol, dichloropropanol, and epichlorohydrin (Figure 6D). The putative halohydrin dehalogenase HheA_J1 (Figure S5) from Microbacterium sp. J1-1 shows a high amino acid identity with HheA (98.77%, GenBank: BAA14361.1) and HheA_AD2 (98.36%, GenBank: AAK92100.1), two proteins with reported capacity to degrade chlorohydrin.50,51 We also used organohalides, which are the potential substrates of other dehalogenases in the dehalogenation assay, including monochloroacetic acid, 2-chloropropionic acid, methyl chloroacetate, methyl 2-chloropropionate, and methyl 4-chlorobenzoate. However, none of these were degraded by Microbacterium sp. J1-1. Microbacterium sp. J1-1 is a candidate in the application of further saponification wastewater treatment process.
Conclusion
In the present study, we profiled the dynamics of the microbial community and identified microbes that were dominant in the wastewater treatment process. Proteobacteria, Patescibacteria, Actinobacteria, and Bacteroidetes were the dominant phyla throughout the overall wastewater treatment process. Seven bacterial groups’ MAGs (Rhizobiaceae, Rhodobacteraceae, Rhodospirillales, Micrococcales, Chloroflexia, Sphingomonadales, and Burkholderiales), which have various hydrolytic dehalogenase genes, show potential for dehalogenation in propylene oxide wastewater. Moreover, we subsequently harnessed the multi-omics data and developed a four-stage selective media strategy to facilitate the isolation of OHDBs, which we used to successfully isolate Microbacterium sp. J1-1. After genome assembly for the isolated OHDB, we found that the strain has diverse hydrolytic dehalogenases and possesses the capacity for organohalide dehalogenation. The OHDB is a candidate for further saponification wastewater treatment process. Combining multi-omics prediction information and microbial cultivation will be helpful for obtaining strain resources and genetic resources in more environments in the future.
Materials and methods
Sample collection and DNA extraction
Samples from a WWTP in Shandong Province of China were collected from the following five phases: RW, AT, SST, COR, and FST (Figure 1A). The AT and SST phases comprise the activated sludge module, while the COR and FST phases comprise the contact oxidation module. The wastewater samples have two characteristics of interest: (1) the samples contain various chlorinated organic substances, such as dichloropropane, dichloroisopropyl ether, and chloroacetone, and (2) the water samples have a chloride ion concentration of 23 000–26 000 mg/l, and this level is maintained throughout the wastewater treatment process. More detailed physical and chemical information for each of the different treatment phases can be found in Table S1.
Genomic DNA was extracted from the samples using a DNeasy PowerSoil Kit (QIAGEN), and a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific) was used to measure and evaluate the quantity and quality of the extracted DNA. DNA samples were stored at −20°C.
16S rRNA gene sequencing and metagenome shotgun sequencing
For 16S rRNA gene sequencing, the forward primer 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and the reverse primer 907R (5′-CCGTCAATTCMTTTRAGTTT-3′) were used to target the V4–V5 region. For multiplex sequencing, specific 7-bp barcodes were incorporated into the primers for the various samples. 300-bp paired-end sequencing of the libraries was conducted on the Illumina MiSeq platform using a MiSeq Reagent Kit v.3 (Illumina).
For metagenome shotgun sequencing, the Illumina TruSeq Nano DNA LT Library Preparation Kit was used to construct metagenome shotgun sequencing libraries. The libraries were then sequenced using the PE150 strategy on the Illumina HiSeq X-ten platform. The multi-omics analysis workflow is shown in Figure 1B and was created with BioRender.com.
Sequence analysis
For 16S rRNA gene sequencing, the Quantitative Insights into Microbial Ecology (v.1.8.0) tool was used to denoise and filter the reads data. The OTUs were clustered with a 97% qualified sequence identity threshold.52,53 The taxonomic classifications of OTUs were selected, and the best hit came from the Greengenes Database (release 13.8).54 OTUs with relative abundance below 0.001% among all samples were removed.
For metagenomic shotgun sequencing, raw reads were processed by FastQC (v.0.11.8), Cutadapt (v.1.2.1), and BWA (v.0.7.17).55 Metagenomes were constructed based on qualified reads using IDBA-UD (release 1.1.3). Metagenomic scaffolds (length >300 bp) were used to predict the coding regions based on MetaGeneMark (v.3.26).56,57 With a sequence identity (>90%), coding regions were clustered by CD-HIT (v.4.7) to generate a non-redundant gene catalog.58 DIAMOND (v.0.8.32.94) and KEGG were used to annotate the non-redundant genes catalog for functional predictions.59,60
Microbial diversity analysis
The relative abundance of OTUs at the phylum level were visualized by ggplot2 (v.3.3.3) and ggalluvial (v.0.12.3). The Vegan (v.2.5-7) and Picante (v.1.8.2) R packages were used to calculate the samples’ alpha diversity, including the Chao1 richness estimator, the Shannon diversity index, the Abundance-based coverage estimator index, and the Simpson index. The beta diversity of samples was assessed using Bray-Curtis dissimilarity metrics by non-metric multi-dimensional scaling analysis.61
Identification of discriminative OTUs
LEfSe analysis was conducted to identify discriminative OTUs among the different treatment phases.62 The logarithmic linear discriminant analysis score cutoff was set at 4.2; otherwise, default parameters were used throughout the whole pipeline. The exhaustive LEfSe result is shown in Figure S1.
Generation of MAGs
Metagenomic reads were binned into MAGs via MetaWRAP (v.1.2).63 Initial MAGs were refined and assessed in MetaWRAP. MAGs that met the cutoff of completeness >70% and contamination <5% were used for analysis. The detailed information about completeness and contamination for the MAGs is presented in Table S3.
Phylogenetic analysis
The Genome Taxonomy Database toolkit (v.1.2) was used to make taxonomic assignments of the MAGs based on marker genes.18 Phylogenetic trees were constructed based on the core gene set using UBCG (v.3) and visualized by iTOL.19,20
Metabolism annotation analysis
KO (KEGG Ortholog) of MAGs were assigned by KAAS (v.2.1), and the template gene dataset included the prokaryote representative set and closely related species.24 The presence of specific metabolic pathways was analyzed by KEGG pathway maps (eg, nitrogen metabolism, sulfur metabolism). The antibiotic resistance genes were identified using RGI (v.5.2.1) and CARD (v3.2.3) using the “Perfect and Strict hits only” selection criteria. Functional genes (haloacid dehalogenase, haloalkane dehalogenase, haloacetate dehalogenase, and 4-CBCoA dehalogenase) were obtained from Prodigal (v.2.6.3) and Prokka (v.1.14.5).23,64 Halohydrin dehalogenase was identified from the short-chain dehydrogenase/reductase via MAFFT (v.7.490) based on the special catalytic triads (Ser-Tyr-Arg) and the cofactor binding motif.48 The hypothetical proteins were removed, and the predicted annotations related to dehalogenation were retained for further analysis. The metabolomics data were analyzed via MetaboAnalyst (v.5.0) and KEGG.65
Enrichment and isolation of OHDB
Mineral salt medium consisted of 2.0 g/l KH2PO4, 3.28 g/l Na2HPO4·12H2O, 0.1 g/l MgSO4, 1.0 g/l (NH4)2SO4, and 0.25 g/l FeCl3.66 The optional additional components included disodium succinate (10 mg/l) and sodium thiosulfate (0.1 g/l). Luria-Bertani medium for the resting cell system assay consisted of 10 g/l tryptone, 5 g/l yeast extract, and 10 g/l NaCl. Culture enrichments were incubated at 30°C and 37°C with shaking (200 RPM). Multi–omics-guided design of the selective media strategy is shown in Figure 4 and was created with BioRender.com, adapted from “Microbiology Agar Plates” (2021).
Organohalide dehalogenation determination
The strain was transferred to Luria-Bertani medium supplemented with specific organic halides to induce the dehalogenase genes at 30°C and shaking at 200 RPM. The scanning electron microscope micrograph of Microbacterium sp. J1-1 was obtained via Hitachi S3400N SEM. The bacterial medium was collected to prepare the resting cell system for subsequent dehalogenation experiments. The reaction medium was centrifuged, and the supernatant was transferred into a 25-ml volumetric flask. 2 ml saturated mercury thiocyanate in ethanol and 2 ml 0.25 M ferric ammonium sulfate in nitric acid were added to initiate the chromogenic reaction, and a UV spectrophotometer was used to detect the dehalogenation of organohalides.67
Acknowledgments
This work was supported by a grant (2021YFA0909500) from the National Key R&D Program of China; by the Shanghai Excellent Academic Leaders Program (20XD1421900); by the “Shuguang Program” (17SG09) from the Shanghai Education Development Foundation and Shanghai Municipal Education Commission; and by a grant (32030004) from the National Natural Science Foundation of China.
Author contributions
Y.H. and H.T. designed the study. J.X. and Z.L. collected the samples. W.W. and L.Z. prepared amplicon sequencing and metagenomic sequencing materials. Y.H. performed the amplicon data and metagenomic data analyses. Y.H. and L.W. designed the strain media and conducted the assay. Y.H., L.W., H.H., H.T., and P.X. drafted and reviewed the manuscript.
Declaration of interests
The authors declare no competing interests.
Published Online: November 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xinn.2022.100355.
Web resources
The raw sequence data of the 16S rRNA gene are deposited in NCBI SRA (NCBI: PRJNA785735). The metagenome data are deposited in NCBI SRA (NCBI: PRJNA786644). The bacterial genome of Microbacterium sp. J1-1 (NCBI: PRJNA785372) is also available from NCBI. The metabolomic data are deposited in Metabolights (Database: MTBLS3916).
Lead contact website
Supplemental information
References
- 1.Wang S., Qiu L., Liu X., Xu G., Siegert M., Lu Q., Juneau P., Yu L., Liang D., He Z., Qiu R. Electron transport chains in organohalide-respiring bacteria and bioremediation implications. Biotechnol. Adv. 2018;36:1194–1206. doi: 10.1016/j.biotechadv.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 2.Jugder B.-E., Ertan H., Bohl S., Lee M., Marquis C.P., Manefield M. Organohalide respiring bacteria and reductive dehalogenases: key tools in organohalide bioremediation. Front. Microbiol. 2016;7:249. doi: 10.3389/fmicb.2016.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dann A.B., Hontela A. Triclosan: environmental exposure, toxicity and mechanisms of action. J. Appl. Toxicol. 2011;31:285–311. doi: 10.1002/jat.1660. [DOI] [PubMed] [Google Scholar]
- 4.Nijhuis T.A., Makkee M., Moulijn J.A., Weckhuysen B.M. The production of propene oxide: catalytic processes and recent developments. Ind. Eng. Chem. Res. 2006;45:3447–3459. [Google Scholar]
- 5.LaMartina E.L., Mohaimani A.A., Newton R.J. Urban wastewater bacterial communities assemble into seasonal steady states. Microbiome. 2021;9:116. doi: 10.1186/s40168-021-01038-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis W.H., Tahon G., Geesink P., Sousa D.Z., Ettema T.J.G. Innovations to culturing the uncultured microbial majority. Nat. Rev. Microbiol. 2021;19:225–240. doi: 10.1038/s41579-020-00458-8. [DOI] [PubMed] [Google Scholar]
- 7.Wu L., Ning D., Zhang B., Li Y., Zhang P., Shan X., Zhang Q., Brown M.R., Li Z., Van Nostrand J.D., et al. Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat. Microbiol. 2019;4:1183–1195. doi: 10.1038/s41564-019-0426-5. [DOI] [PubMed] [Google Scholar]
- 8.Brisson V.L., West K.A., Lee P.K.H., Tringe S.G., Brodie E.L., Alvarez-Cohen L. Metagenomic analysis of a stable trichloroethene-degrading microbial community. ISME J. 2012;6:1702–1714. doi: 10.1038/ismej.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan J., Zhou Z., Béjà O., Cai M., Yang Y., Liu Y., Gu J.D., Li M. Genomic and transcriptomic evidence of light-sensing, porphyrin biosynthesis, Calvin-Benson-Bassham cycle, and urea production in Bathyarchaeota. Microbiome. 2020;8:43. doi: 10.1186/s40168-020-00820-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marathe N.P., Berglund F., Razavi M., Pal C., Dröge J., Samant S., Kristiansson E., Larsson D.G.J. Sewage effluent from an Indian hospital harbors novel carbapenemases and integron-borne antibiotic resistance genes. Microbiome. 2019;7:97. doi: 10.1186/s40168-019-0710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gharechahi J., Vahidi M.F., Bahram M., Han J.L., Ding X.Z., Salekdeh G.H. Metagenomic analysis reveals a dynamic microbiome with diversified adaptive functions to utilize high lignocellulosic forages in the cattle rumen. ISME J. 2021;15:1108–1120. doi: 10.1038/s41396-020-00837-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seib M.D., Berg K.J., Zitomer D.H. Influent wastewater microbiota and temperature influence anaerobic membrane bioreactor microbial community. Bioresour. Technol. 2016;216:446–452. doi: 10.1016/j.biortech.2016.05.098. [DOI] [PubMed] [Google Scholar]
- 13.Zhu L., Xu H., Xiao W., Lu J., Lu D., Chen X., Zheng X., Jeppesen E., Zhang W., Wang L. Ecotoxicological effects of sulfonamide on and its removal by the submerged plant Vallisneria natans (Lour.) Hara. Water Res. 2020;170:115354. doi: 10.1016/j.watres.2019.115354. [DOI] [PubMed] [Google Scholar]
- 14.Mason L.M., Eagar A., Patel P., Blackwood C.B., DeForest J.L. Potential microbial bioindicators of phosphorus mining in a temperate deciduous forest. J. Appl. Microbiol. 2020;130:109–122. doi: 10.1111/jam.14761. [DOI] [PubMed] [Google Scholar]
- 15.Krzmarzick M.J., Novak P.J. Removal of chlorinated organic compounds during wastewater treatment: achievements and limits. Appl. Microbiol. Biotechnol. 2014;98:6233–6242. doi: 10.1007/s00253-014-5800-x. [DOI] [PubMed] [Google Scholar]
- 16.Zhao S., Rogers M.J., He J. Abundance of organohalide respiring bacteria and their role in dehalogenating antimicrobials in wastewater treatment plants. Water Res. 2020;181:115893. doi: 10.1016/j.watres.2020.115893. [DOI] [PubMed] [Google Scholar]
- 17.Jing R., Kjellerup B.V. Predicting the potential for organohalide respiration in wastewater: comparison of intestinal and wastewater microbiomes. Sci. Total Environ. 2020;705:135833. doi: 10.1016/j.scitotenv.2019.135833. [DOI] [PubMed] [Google Scholar]
- 18.Chaumeil P.-A., Mussig A.J., Hugenholtz P., Parks D.H. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics. 2019;36:1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Na S.-I., Kim Y.O., Yoon S.-H., Ha S.M., Baek I., Chun J. UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 2018;56:280–285. doi: 10.1007/s12275-018-8014-6. [DOI] [PubMed] [Google Scholar]
- 20.Letunic I., Bork P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oyewusi H.A., Wahab R.A., Huyop F. Dehalogenase-producing halophiles and their potential role in bioremediation. Mar. Pollut. Bull. 2020;160:111603. doi: 10.1016/j.marpolbul.2020.111603. [DOI] [PubMed] [Google Scholar]
- 22.Agarwal V., Miles Z.D., Winter J.M., Eustáquio A.S., El Gamal A.A., Moore B.S. Enzymatic halogenation and dehalogenation reactions: pervasive and mechanistically diverse. Chem. Rev. 2017;117:5619–5674. doi: 10.1021/acs.chemrev.6b00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 24.Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Xie Z., Yan J., He X., Liu W., Sun Y. Enhancement of the thermostability of halohydrin dehalogenase from Agrobacterium radiobacter AD1 by constructing a combinatorial smart library. Int. J. Biol. Macromol. 2019;130:19–23. doi: 10.1016/j.ijbiomac.2019.02.099. [DOI] [PubMed] [Google Scholar]
- 26.Novak H.R., Sayer C., Isupov M.N., Paszkiewicz K., Gotz D., Spragg A.M., et al. Marine Rhodobacteraceae L-haloacid dehalogenase contains a novel His/Glu dyad that could activate the catalytic water. FEBS J. 2013;280:1664–1680. doi: 10.1111/febs.12177. [DOI] [PubMed] [Google Scholar]
- 27.Yue W., Xiong M., Li F., Wang G. The isolation and characterization of the novel chlorothalonil-degrading strain Paracoccus sp. XF-3 and the cloning of the chd gene. J. Biosci. Bioeng. 2015;120:544–548. doi: 10.1016/j.jbiosc.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 28.Kumari P., Kaushik R., Tripathi B.M., et al. Genetic and metabolic profiling of pentachlorophenol utilizing bacteria from agricultural soil irrigated with pulp and paper mill effluent. Indian J. Agric. Sci. 2014;84:124–130. [Google Scholar]
- 29.Lv J., Wang Y., Zhong C., Li Y., Hao W., Zhu J. The microbial attachment potential and quorum sensing measurement of aerobic granular activated sludge and flocculent activated sludge. Bioresour. Technol. 2014;151:291–296. doi: 10.1016/j.biortech.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Yang Y., Sanford R., Yan J., Chen G., Cápiro N.L., Li X., Löffler F.E. Roles of organohalide-respiring Dehalococcoidia in carbon cycling. mSystems. 2020;5 doi: 10.1128/mSystems.00757-19. e00757-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krzmarzick M.J., McNamara P.J., Crary B.B., Novak P.J. Abundance and diversity of organohalide-respiring bacteria in lake sediments across a geographical sulfur gradient. FEMS Microbiol. Ecol. 2013;84:248–258. doi: 10.1111/1574-6941.12059. [DOI] [PubMed] [Google Scholar]
- 32.Skopelitou K., Georgakis N., Efrose R., Flemetakis E., Labrou N.E. Sol–gel immobilization of haloalkane dehalogenase from Bradyrhizobium japonicum for the remediation 1, 2-dibromoethane. J. Mol. Catal. B Enzym. 2013;97:5–11. [Google Scholar]
- 33.Adamu A., Wahab R.A., Aliyu F., Aminu A.H., Hamza M.M., Huyop F. Haloacid dehalogenases of Rhizobium sp. and related enzymes: catalytic properties and mechanistic analysis. Process Biochem. 2020;92:437–446. [Google Scholar]
- 34.Sharma P., Raina V., Kumari R., Malhotra S., Dogra C., Kumari H., Kohler H.P.E., Buser H.R., Holliger C., Lal R. Haloalkane dehalogenase LinB is responsible for β- and δ-hexachlorocyclohexane transformation in Sphingobium indicum B90A. Appl. Environ. Microbiol. 2006;72:5720–5727. doi: 10.1128/AEM.00192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohkouchi Y., Koshikawa H., Terashima Y. Cloning and expression of DL-2-haloacid dehalogenase gene from Burkholderia cepacia. Water Sci. Technol. 2000;42:261–268. [Google Scholar]
- 36.Diez A., Alvarez M.J., Prieto M.I., Bautista J.M., Garrido-Pertierra A. Monochloroacetate dehalogenase activities of bacterial strains isolated from soil. Can. J. Microbiol. 1995;41:730–739. [Google Scholar]
- 37.Zengler K., Zaramela L.S. The social network of microorganisms - how auxotrophies shape complex communities. Nat. Rev. Microbiol. 2018;16:383–390. doi: 10.1038/s41579-018-0004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stams A.J.M., Plugge C.M. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Microbiol. 2009;7:568–577. doi: 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
- 39.Carini P., Steindler L., Beszteri S., Giovannoni S.J. Nutrient requirements for growth of the extreme oligotroph ‘Candidatus Pelagibacter ubique’ HTCC1062 on a defined medium. ISME J. 2013;7:592–602. doi: 10.1038/ismej.2012.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang T., Xu Y., Wang J., Tan X., Zhao X., Zhou P., Kong F., Zhu C., Lu C., Lin H. Evaluation of the differences between biofilm and planktonic Brucella abortus via metabolomics and proteomics. Funct. Integr. Genomics. 2021;21:421–433. doi: 10.1007/s10142-021-00788-7. [DOI] [PubMed] [Google Scholar]
- 41.Tripp H.J., Kitner J.B., Schwalbach M.S., Dacey J.W.H., Wilhelm L.J., Giovannoni S.J. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature. 2008;452:741–744. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- 42.Gutleben J., Chaib De Mares M., van Elsas J.D., Smidt H., Overmann J., Sipkema D. The multi-omics promise in context: from sequence to microbial isolate. Crit. Rev. Microbiol. 2018;44:212–229. doi: 10.1080/1040841X.2017.1332003. [DOI] [PubMed] [Google Scholar]
- 43.Bjorland J., Steinum T., Sunde M., Waage S., Heir E. Novel plasmid-borne gene qacJ mediates resistance to quaternary ammonium compounds in equine Staphylococcus aureus, Staphylococcus simulans, and Staphylococcus intermedius. Antimicrob. Agents Chemother. 2003;47:3046–3052. doi: 10.1128/AAC.47.10.3046-3052.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heir E., Sundheim G., Holck A.L. The qacG gene on plasmid pST94 confers resistance to quaternary ammonium compounds in staphylococci isolated from the food industry. J. Appl. Microbiol. 1999;86:378–388. doi: 10.1046/j.1365-2672.1999.00672.x. [DOI] [PubMed] [Google Scholar]
- 45.Koonin E.V., Tatusov R.L. Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. J. Mol. Biol. 1994;244:125–132. doi: 10.1006/jmbi.1994.1711. [DOI] [PubMed] [Google Scholar]
- 46.Janssen D.B. Evolving haloalkane dehalogenases. Curr. Opin. Chem. Biol. 2004;8:150–159. doi: 10.1016/j.cbpa.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 47.Koudelakova T., Chovancova E., Brezovsky J., Monincova M., Fortova A., Jarkovsky J., Damborsky J. Substrate specificity of haloalkane dehalogenases. Biochem. J. 2011;435:345–354. doi: 10.1042/BJ20101405. [DOI] [PubMed] [Google Scholar]
- 48.Schallmey M., Koopmeiners J., Wells E., Wardenga R., Schallmey A. Expanding the halohydrin dehalogenase enzyme family: identification of novel enzymes by database mining. Appl. Environ. Microbiol. 2014;80:7303–7315. doi: 10.1128/AEM.01985-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yue Y., Huang H., Qi Z., Dou H.M., Liu X.Y., Han T.F., Chen Y., Song X.J., Zhang Y.H., Tu J. Evaluating metagenomics tools for genome binning with real metagenomic datasets and CAMI datasets. BMC Bioinf. 2020;21:334. doi: 10.1186/s12859-020-03667-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Hylckama Vlieg J.E., Tang L., Lutje Spelberg J.H., Smilda T., Poelarends G.J., Bosma T., van Merode A.E., Fraaije M.W., Janssen D.B. Halohydrin dehalogenases are structurally and mechanistically related to short-chain dehydrogenases/reductases. J. Bacteriol. 2001;183:5058–5066. doi: 10.1128/JB.183.17.5058-5066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu F., Nakamura T., Mizunashi W., Watanabe I. Cloning of two halohydrin hydrogen–halide–lyase genes of Corynebacterium sp. strain N-1074 and structural comparison of the genes and gene products. Biosci. Biotechnol. Biochem. 1994;58:1451–1457. doi: 10.1271/bbb.58.1451. [DOI] [PubMed] [Google Scholar]
- 52.Lozupone C.A., Hamady M., Kelley S.T., Knight R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 54.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng Y., Leung H.C.M., Yiu S.M., Chin F.Y.L. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 57.Noguchi H., Park J., Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 60.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clarke K.R. Non-parametric multivariate analyses of changes in community structure. Austral Ecol. 1993;18:117–143. [Google Scholar]
- 62.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uritskiy G.V., DiRuggiero J., Taylor J. MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6:158. doi: 10.1186/s40168-018-0541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hyatt D., Chen G.-L., LoCascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pang Z., Chong J., Zhou G., de Lima Morais D.A., Chang L., Barrette M., Gauthier C., Jacques P.É., Li S., Xia J. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49:W388–W396. doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang W., Li Q., Zhang L., Cui J., Yu H., Wang X., Ouyang X., Tao F., Xu P., Tang H. Genetic mapping of highly versatile and solvent-tolerant Pseudomonas putida B6-2 (ATCC BAA-2545) as a ‘superstar’ for mineralization of PAHs and dioxin-like compounds. Environ. Microbiol. 2021;23:4309–4325. doi: 10.1111/1462-2920.15613. [DOI] [PubMed] [Google Scholar]
- 67.Bergmann J.G., Sanik J. Determination of trace amounts of chlorine in naphtha. Anal. Chem. 1957;29:241–243. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.