Abstract
The enzymatic pathway of cholesterol biosynthesis has been well characterized. However, there remain several potential interacting proteins that may play ancillary roles in the regulation of cholesterol production. Here, we identified ERG28 (chromosome 14 open reading frame 1 [C14orf1]), a homologue of the yeast protein Erg28p, as a player in mammalian cholesterol synthesis. ERG28 is conserved from yeast to humans but has been largely overlooked in mammals. Using quantitative RT-PCR, luciferase assays, and publicly available chromatin immunoprecipitation sequencing data, we found that transcription of this gene is driven by the transcription factor SREBP-2, akin to most cholesterol synthesis enzymes, as well as identifying sterol-responsive elements and cofactor binding sites in its proximal promoter. Based on a split luciferase system, ERG28 interacted with itself and two enzymes of cholesterol synthesis (NSDHL and SC4MOL). Huh7 ERG28-KO cell lines were generated, revealing reduced total cholesterol levels in sterol-depleted environments. In addition, radiolabeled metabolic flux assays showed a 60–75% reduction in the rate of cholesterol synthesis in the KO versus wild-type cells, which could be rescued by expression of ectopic ERG28. Unexpectedly, KO of ERG28 also impaired the activation of SREBP-2 under sterol-replete conditions, by a yet-to-be defined mechanism. These results indicate that ERG28 is clearly involved in cholesterol synthesis, although the precise role this noncatalytic protein plays in this complex metabolic pathway remains to be fully elucidated. A deeper understanding of ERG28, and other ancillary proteins of cholesterol synthesis, may help inform therapeutic strategies for diseases associated with aberrant cholesterol metabolism.
Supplementary key words: cholesterol, cholesterol biosynthesis, cholesterol metabolism, nuclear receptors/SREBP, lipid biochemistry, molecular biology, transcription, radiolabeled metabolic flux assay
Abbreviations: cDNA, complementary DNA; ChIP-Seq, chromatin immunoprecipitation sequencing; EBP, emopamil-binding protein; ER, endoplasmic reticulum; FCLPDS, lipoprotein-deficient FCS; HEK293T, human embryonic kidney 293T cell line; HMGCR, 3-hydroxy-3-methylglutaryl-CoA reductase; HSD17B7, hydroxysteroid (17-beta) dehydrogenase 7; 25HC, 25-hydroxycholesterol; invSRE, inverse SRE; LDLR, LDL receptor; NF-Y, nuclear factor Y; NSDHL, NAD(P) Dependent Steroid Dehydrogenase-Like; 17-OHP, 17α-hydroxyprogesterone; PBST, PBS with Tween-20; qRT-PCR, quantitative RT-PCR; SC4MOL, sterol-C4-methyl oxidase–like gene; SDS, sodium dodecyl sulfate; SM, squalene monooxygenase; Sp1, specificity protein 1; SQS, squalene synthase; SRE, sterol-response element
Cholesterol is a vital lipid in the human body, and its excess or deficiency can result in a wide array of health problems, such as cardiovascular disease (1) or developmental disorders (2), respectively. The sterol synthesis pathway is highly conserved between yeast and mammals (3), and many enzymes in cholesterol synthesis have homologous enzymes in ergosterol synthesis. Over the decades, insights into the nature and regulation of the yeast ergosterol and mammalian cholesterol synthesis pathways have helped inform each other and advanced our knowledge of both pathways. However, neither the enzymes themselves nor their interactions (4) are absolutely conserved between yeast and mammals. Notably, there is no yeast equivalent of the mammalian DHCR24 enzyme, which performs the final step in cholesterol synthesis, as well as being able to act on many earlier intermediates, and the yeast C24-sterol methyltransferase, Erg6p, is absent in mammals.
ERG28, also known as chromosome 14 open reading frame 1 (C14orf1)/NET51, is a poorly characterized mammalian protein, conserved from yeast, to plants, to humans (supplemental Fig. S1A) (5, 6). Most work has been done in Saccharomyces cerevisiae where Erg28p plays an important role in ergosterol synthesis (5). Although KO strains of Erg28p are not auxotrophic for ergosterol, they grow at a much slower rate and have an ∼70% reduction in ergosterol synthesis compared with wild-type strains (5). In addition to their reduced synthesis of ergosterol, Erg28p KO strains also show a dramatic increase in C4-methylated and C4-keto sterols, suggesting that a deficiency occurs at the C4-demethylation step in ergosterol synthesis (7). Erg28p has been shown (based on multiple yeast two-hybrid system experiments) to have binary interactions with many ergosterol synthesis enzymes and is believed to scaffold these enzymes together into a larger metabolic unit (7, 8, 9). However, proteins that interact in ergosterol synthesis may not necessarily interact in cholesterol synthesis (4).
Despite the 52% sequence similarity between human ERG28 and yeast Erg28p (supplemental Fig. S1B), very little is known about the role ERG28 plays in mammals, or even if it is involved in cholesterol biosynthesis. In humans, ERG28 is located at 14q24.3 (chr14: 75649791–75660876 [GRCh38/hg38]), contains four introns and five exons, and encodes a 140 amino-acid polytopic protein, with a predicted molecular weight of 15.9 kDa. Transcriptomics suggests that ERG28 is expressed in most human tissues (Genotype-Tissue Expression [GTEx] Project, dbGaP accession number: phs000424.vN.pN) and expressed highly in the testis (10) and several cancer cell lines, notably in colon-, leukocyte-, and pancreatic-derived cells (10, 11). The primary subcellular location of mammalian ERG28 is not fully agreed upon. In yeast, Erg28p is localized to the endoplasmic reticulum (ER) (7) and human ERG28 interacts with CLN8, a resident ER protein, in bait-prey systems (12). But in pancreatic cancers, ERG28 demonstrates some plasma membrane localization (11). In 2012, Lee et al. (11) reported the first functional study of human ERG28 where depletion reduced the proliferation of pancreatic cancer cell lines by inhibiting thymidine incorporation. While ERG28 may be assumed to be involved in cholesterol synthesis because of homology with yeast ergosterol synthesis, this has yet to be established. Indeed, there has been very limited work on human ERG28, and any characterization studies conducted in mammalian cells thus far have not examined cholesterol synthesis.
Almost all cholesterol synthesis genes are regulated by the transcription factor SREBP-2 (13). When sterol levels are high in the cell, SREBP-2 is retained in the ER membrane. Conversely, when sterol levels are low, SREBP-2 is cleaved into the nuclear form of nSREBP-2 and SREBP-2 targets, including cholesterol synthesis genes, are transcriptionally upregulated. SREBP-2 is a relatively weak activator of transcription and typically requires the transcriptional cofactors specificity protein 1 (Sp1) or nuclear factor Y (NF-Y) for maximal activation of genes (14, 15).
In this work, we investigated the transcriptional regulation of human ERG28, revealing it is an SREBP-2 target, akin to the bulk of cholesterol synthesis enzymes (13). We explored the protein-protein interactions of ERG28 with several cholesterol synthesis enzymes and found it interacted with members of the C4-demethylation complex. In addition, we generated ERG28 KO human liver cell lines and showed they have reduced production and, under sterol-depleted conditions, when cells depend on de novo cholesterol synthesis, reduced overall levels of cholesterol. Moreover, we found that ERG28 KO cells showed impaired SREBP-2 processing, resulting in a downregulation of many cholesterol synthesis genes. These results reveal for the first time that ERG28 plays an important role in facilitating cholesterol synthesis in mammalian cells.
Materials and Methods
Cell culture
HeLaT cells were maintained in RPMI 1640 medium, and human embryonic kidney 293T (HEK293T) and Huh7 cells were maintained in DMEM high glucose media. All cell lines were maintained with 10% (v/v) FCS and supplemented with penicillin (100 units/ml) and streptomycin (100 μg/ml). For some experiments, cell lines were treated in sterol-depleted media, 10% (v/v) lipoprotein-deficient FCS (FCLPDS), supplemented with penicillin (100 units/ml) and streptomycin (100 μg/ml).
HeLaT cells were gifts from Dr Noel Whitaker (UNSW, Sydney, Australia), Huh7 cells were gifts from the Centre for Vascular Research (UNSW), and HEK293T cells were obtained from the School of Medical Sciences (UNSW).
FCLPDS was prepared inhouse from FCS as described previously (16, 17).
Quantitative real-time PCR
Total RNA was harvested from cells using TRI reagent. Complementary DNA (cDNA) was synthesized from RNA using the SuperScript III First Strand cDNA synthesis kit (Thermo Fisher Scientific).
HeLaT, Huh7, and Be(2)C cDNA sets used in this work were generated previously (18, 19). Briefly, these cell lines were treated for 24 h in sterol-depleted media supplemented with 5 μM compactin (statin), 10 μM 25-hydroxycholesterol (25HC), 10 μM 24(S),25-epoxycholesterol (24,25EC), 10 μM T0-901317 (T-0), or solvent controls. mRNA levels for genes of interest were measured using quantitative RT-PCR (qRT-PCR) and normalized to porphobilinogen deaminase (PBGD) levels. For primer sequences used for qRT-PCR, please refer supplemental Table S1.
Western blotting
Protein lysate for Western blotting was harvested in either 1% (w/v) or 3% (w/v) sodium dodecyl sulfate (SDS) supplemented with 2% (v/v) protease inhibitor cocktail (Sigma-Aldrich). Protein concentration was measured with a BCA assay (Thermo Fisher Scientific), normalized, and separated by 10% or 12% SDS-PAGE. Separated proteins were transferred onto nitrocellulose membranes. Membranes were blocked in 5% (w/v) skim milk/PBS with 0.1% (v/v) Tween-20 (PBST) and probed with the following primary antibodies: anti-α-tubulin (1:50,000 dilution in 5% [w/v] BSA/PBST; Sigma-Aldrich, catalog no.: T5168), anti-GAPDH (1:2,000 dilution in 5% [w/v] BSA/PBST; Cell Signalling Technologies, catalog no.: 2118L), anti-ERG28 (1:1,000 dilution in 5% [w/v] BSA/PBST; Novus Biologicals, catalog no.: nbp2-84865), anti-SREBP2 (1C6) (1:500 dilution in 5% [w/v] skim milk/PBST; Santa Cruz Biotechnology, Inc, catalog no.: sc-13552), and anti-Scap (clone IgG-R139) (1:10,000 dilution in 5% [w/v] skim milk/PBST; a generous gift from Drs Brown and Goldstein [UT Southwestern Medical Center, Dallas, TX]).
Secondary antibodies were diluted 1:10,000 in 5% (w/v) skim milk/PBST (HRP-anti-mouse, Jackson Immunoresearch Laboratories catalog no.: 715-035-150 and HRP-anti-rabbit, Jackson Immunoresearch Laboratories, catalog no.: 711-035-152). HRP-conjugated antibodies were imaged using Immobilon Western HRP Substrate (Merck; catalog no.: WBKLS0500) on an ImageQuant LAS 500.
Access of publicly available SREBP-2 chromatin immunoprecipitation sequencing data
Publicly available chromatin immunoprecipitation sequencing (ChIP-Seq) data from UCSC Genome browser (20) were accessed (August 2020) to investigate SREBP-2 binding in the ERG28 promoter. SREBP-2 ChIP-Seq signal heights were taken from pravastatin-treated HepG2 cells (a human hepatoma cell line) and rabbit IgG anti-SREBP-2 binding in the GM12878 lymphoblast cell line (20, 21).
Plasmid generation
Plasmids generated for this work were cloned using either the Polymerase Incomplete Primer Extension method (22) or restriction enzyme cloning. For primer sequences used for cloning, please refer to supplemental Table S2.
The −1000/+80, −500/+80, −290/+80, −50/+80, and 0/+80 regions of the ERG28 promoter were amplified from HEK293 genomic DNA and cloned into a pGL3-Basic (Promega) backbone to generate Firefly luciferase constructs driven by these promoter regions, creating the −1000/+80 ERG28-luc, −500/+80 ERG28-luc, −290/+80 ERG28-luc, −50/+80 ERG28-luc, and 0/+80 ERG28-luc constructs. Mutations to disrupt predicted SREBP-2, Sp1, and NF-Y binding sites were introduced via site-directed mutagenesis on the −290/+80 ERG28-luc plasmid. Predicted sterol-response elements (SREs) were mutated to xAACAxAAGx, predicted Sp1 sites were mutated to TTTAAA, and predicted NF-Y sites were mutated to AATTCC. To generate the CMV-ERG28-V5 construct, the ERG28 CDS was cloned from HeLaT cDNA into our in-house pcDNA5/FRT construct (23).
NanoBiT® plasmids were cloned following the outlined protocol in the NanoBiT® PPI Starter System (Promega). Genes of interest were amplified with PCR from constructs containing a V5-His epitope tag and subcloned using restriction enzyme cloning into plasmids supplied in the kit (pBiT1.1-C [TK/LgBiT], pBiT2.1-C [TK/SmBiT], pBiT1.1-N [TK/LgBiT], or pBiT2.1-N [TK/SmBiT]).
For all plasmids, sequence was confirmed via Sanger sequencing carried out by the Ramaciotti Centre for Genomics at UNSW Sydney.
Dual luciferase assay
HeLaT cells were transfected with 25 ng phRL-PBGD Renilla luciferase plasmid (14) and 250 ng Firefly luciferase plasmids as listed in figure legends using TransIT-2020 reagent (Mirus Bio) as per the manufacturer’s instructions. For measuring SREBP-2 responsiveness, cells were in addition cotransfected with 25 ng of nSREBP-2 (24), a kind gift from Dr John Shyy, or empty vector plasmid. Cells were transfected for 24 h and then treated as indicated in figure legends. After 24 h of treatment, cells were harvested and assayed for Firefly and Renilla luciferase activity using the Dual-Luciferase Reporter Assay System (Promega) in accordance with the manufacturer’s instructions in the CLARIOstar Plus Microplate reader. Transfection efficiency was accounted for by normalizing Firefly to Renilla luciferase activity in each well, and data were further normalized as stated in figure legends. ERG28-luc plasmids were generated for this publication. DHCR24-luc plasmids were generated previously in the Brown Laboratory (14).
Prediction of transcription factor binding sites in the ERG28 promoter
Putative SREs were predicted in the −290/+10 ERG28 promoter using an updated matrix of SRE sequences previously compiled in house (14, 25) and the Find Individual Motif Occurrences program (26). To calculate scores for each predicted SRE, we took the P value provided by Find Individual Motif Occurrences (26), normalized this to the P value for the LDL receptor (LDLR) SRE (the best match for the consensus sequence), multiplied by 100,000, and log transformed. This value was then multiplied by two, which resulted in numbers closely resembling the scores we have previously reported for predicted SREs (14, 25) using the now-defunct TRED prediction program and gave a score of 10 for the canonical LDLR SRE. Sp1 and NF-Y sites were predicted in the −290/+10 ERG28 promoter using ConSite (27).
NanoBiT® protein-protein interactions of ERG28 and other cholesterol synthesis proteins
HEK293T cells were cotransfected with 350 ng each of two plasmids containing either a LgBiT- or SmBiT-tagged reporter construct using Lipofectamine® LTX reagent (Thermo Fisher Scientific) as per the manufacturer’s instructions in 12-well tissue culture plates. Appropriate LgBiT:HaloTag control pairings were used to ensure that the readouts were specific interactions. After 16 h of transfection, cells were lifted from 12-well plates, and 10% of total cells were plated into polyethyleneimine-coated center wells of white 96-well plates in duplicate. After 20–24 h of adhesion in the wells, media were aspirated and replaced with 50 μl of serum-free Opti-MEM. After a further 30 min–1 h incubation at 37°C to equilibrate temperature, 50 μl of reagent master mix (1 μl of Nano-Glo® Live Cell Substrate, 19 μl of Nano-Glo® LCS Dilution Buffer, and 30 μl of Opti-MEM per well) was added to each well. Plates were mixed by tapping, and luciferase activity was measured over a period of 30 min in a CLARIOstar microplate reader at 37°C. Raw luminosity values were normalized to the signal from the ERG28 LgBiT:HaloTag negative control pair.
Generation of ERG28 KO Huh7 cell lines via CRISPR-Cas9
CHOPCHOP (28, 29, 30) was used to predict guides in the human ERG28 genomic sequence. The two highest ranked guides, (AAGCTCGGACCTTTGGGATCTGG, chr14: 75654942; GGTGCCAGGACGCCAATCGTGGG, chr14: 75651785), were cloned into the PX598 backbone following a modified protocol (31, 32), and sequence was confirmed by Sanger sequencing. Huh7 cells, a human hepatoma cell line, were cotransfected with both guide plasmids for 24 h. Cells were sorted at the UNSW Flow Cytometry Facility on a BD FACSAria™ III Cell Sorter for GFP positivity. Low dilution numbers of the cells were plated and allowed to adhere. After 2–4 weeks, individual colonies were picked, expanded, and screened for disruption of ERG28 genomic DNA and mRNA. Products from these PCRs were sequenced via Sanger sequencing carried out by the Ramaciotti Centre for Genomics at UNSW Sydney to identify the regions of ERG28 deletion in the KOs. Furthermore, qRT-PCR was carried out on cDNA, and Western blotting for endogenous ERG28 protein levels was carried out to confirm reduction in ERG28 levels.
Amplex Red assay to measure cholesterol levels
Cholesterol levels of cellular lysates were measured using the Amplex™ Red Cholesterol Assay Kit (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions, and fluorescence was measured using the CLARIOstar Plus Microplate reader. Briefly, cells were washed twice with PBS, harvested in modified radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% [w/v] SDS, 1.5% [w/v] Nonidet P-40, 0.5% [w/v] sodium deoxycholate, and 2 mM MgCl2 (20)) supplemented with 2% (v/v) protease inhibitor cocktail and subjected to a freeze-thaw cycle to lyse. Lysates were cleared by centrifugation, and cleared lysates were assayed for protein and cholesterol content. Protein concentration was measured with the BCA Protein Assay (Thermo Fisher Scientific), and the total cholesterol content was normalized to protein levels in the sample. In Fig. 4E, the assay was run with and without the addition of cholesterol esterase to measure total and free cholesterol in the samples. From the difference between these values, the percentage of cholesterol stored as cholesteryl ester was calculated.
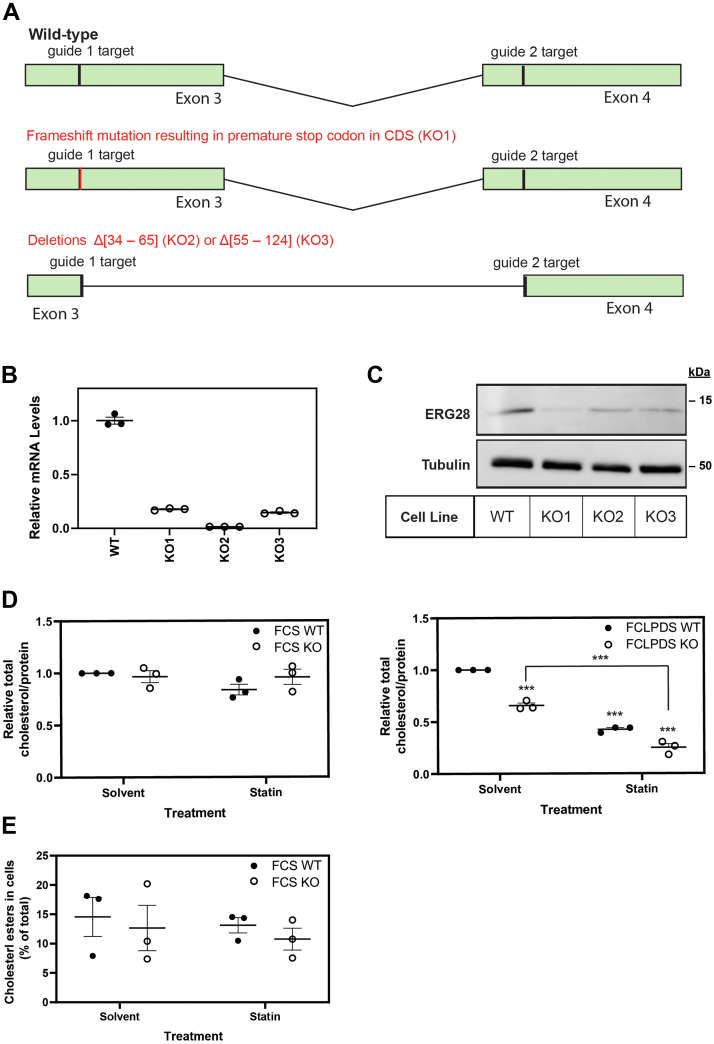
Fig. 4.
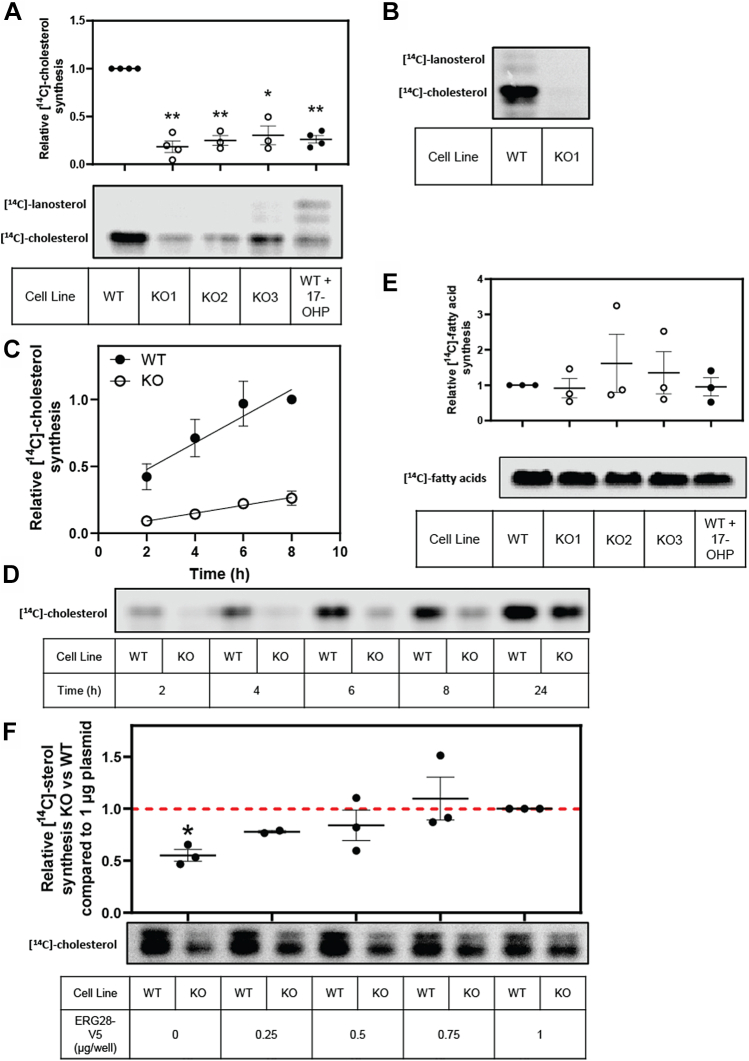
ERG28 KO cell lines have lower total cholesterol levels. A: CRISPR-Cas9 with two guides targeting exons 3 and 4 of human ERG28 was used to generate ERG28 KO cells in Huh7 cells. B: Total RNA was isolated from wild-type and KO cells, and ERG28 mRNA levels were measured using qRT-PCR. An overall reduction in ERG28 mRNA was observed in ERG28 KO cells. C: Protein lysates from ERG28 KO cells were normalized and separated by 12% SDS-PAGE for Western blotting. Membranes were probed for α-tubulin (for housekeeping) and endogenous ERG28 (nbp2-84865). D: Total cholesterol levels were measured in Huh7 wild-type and ERG28 KO cells using the Amplex Red Cholesterol Assay Kit (Invitrogen). Cells were plated in 10% FCLPDS DMEM:HG, and after 24 h, media were replaced and cells were grown for 48 h in either 10% FCS DMEM:HG or 10% FCLPDS DMEM:HG supplemented with 5 μM compactin (statin) or a DMSO solvent control. Cell lysates were harvested, and protein and total cholesterol levels were measured. Cholesterol levels were normalized to the concentration of protein and expressed relative to the wild-type solvent control cholesterol levels in each experiment, which was set to 1. E: Free and total cholesterol levels were measured in Huh7 wild-type and ERG28 KO cells using the Amplex Red Cholesterol Assay Kit (Invitrogen) as per D. Cells were plated in 10% FCLPDS DMEM:HG, and after 24 h, media were replaced and cells were grown for 48 h in either 10% FCS DMEM:HG supplemented with 5 μM compactin (statin) or a DMSO solvent control. Cell lysates were harvested, and protein and free and total cholesterol levels were measured. Cholesterol levels were normalized to the concentration of protein, and the ratio of free to total cholesterol was used to calculate the percentage of cholesterol stored as cholesteryl esters within the cell. Data are presented as the mean ± SEM from n = 3 independent experiments. Statistical significance from the solvent-treated wild-type was determined using a paired two-tailed t-test (∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.005).
Radiolabeling and TLC to measure sterol synthesis
To measure cholesterol synthesis, Huh7 and Huh7 ERG28 KO cells were seeded into 6-well plates and treated as specified in the figure legends. Sterol synthesis was determined by TLC as described previously (33, 34) with minor modifications. Briefly, cells were metabolically labeled for the times listed in the figure legends in 10% FCLPDS DMEM:HG containing 1 or 2 μCi/well [14C]-acetic acid sodium salt (PerkinElmer) or 2 μCi/well [14C]-mevalonate (PerkinElmer). Plates were washed two times with PBS, and cells were lysed in 1 ml of 0.05 M sodium hydroxide. Protein concentration in cell lysates was measured via BCA assay, and lysates were normalized to total cellular protein with 0.05 M sodium hydroxide in a total volume of 1 ml to account for variations in cell numbers. Lysates were saponified in 1 ml 100% (v/v) ethanol, 500 μl 75% (w/v) potassium hydroxide, 1 μl 20 mM butylated hydroxytoluene, and 20 μl 20 mM EDTA, at 70°C for 1 h. Once cooled to room temperature, lipids were extracted from saponified lysates by mixing with 1 ml 100% (v/v) ethanol and 2.5 ml of hexane, centrifuging at 2,000 g for 5 min, and evaporating 2 ml of the upper organic phase to dryness. Dried total nonsaponifiable sterols harvested from the cells were resuspended in hexane and separated via TLC on TLC Silica gel 60 F254 plates using a heptane:ethyl acetate (2:1, v/v) mobile phase. Fatty acids were then harvested from remaining cell lysate by acidification with 1.5 ml 32% hydrochloric acid, mixing with 2.5 ml of hexane, centrifuging at 2,000 g for 5 min, and evaporating 2 ml of the upper organic phase to dryness. Dried fatty acids were resuspended in hexane and separated via TLC on TLC Silica gel 60 F254 plates using a heptane:diethyl ether:acetic acid (90:30:1, v/v) mobile phase. Silica plates were dried, exposed to phosphor screens for 5–14 days, and imaged using the Typhoon FLA 9500 (GE Healthcare).
In this solvent system, the immediate sterol precursor in the Bloch pathways, desmosterol, does not fully resolve from cholesterol (35). As cholesterol is the predominate sterol in cells, this band is labeled as [14C]-cholesterol throughout the text but may contain a mixture of cholesterol and desmosterol.
Transient transfection to overexpress ERG28-V5 in Huh7 cells
To express CMV-ERG28-V5, Huh7 wild-type or Huh7 ERG28 KO cells were seeded in 6-well plates and transfected with the indicated amounts of expression plasmid and 0–1 μg pTK-empty vector DNA (total plasmid amount in the well = 1 μg) and 2 μl Lipofectamine 2000 for 8 h. Cells were then subjected to radiolabeling.
SREBP-2 cleavage assay
To determine the effect of sterol depletion on SREBP-2 cleavage and target gene expression, cells were seeded into 6-well plates. For lipoprotein-depleted conditions, cells were washed thrice with PBS and incubated for 1 h in 10% FCLPDS DMEM:HG containing 1% (w/v) 2-hydroxypropyl-β-cyclodextrin, 10 μM compactin, and 50 μM mevalonate. Next, cells were sequentially washed with PBS, 10% FCLPDS DMEM:HG, and PBS to remove statin and then treated for 5 h with or without 1 μg/ml 25HC in 10% FCLPDS DMEM:HG, with or without 10 μM compactin, and 50 μM mevalonate. For full-serum conditions, cells were refreshed and treated in 10% FCS DMEM:HG PS without additives. During the final 2 h of treatment, medium was supplemented with 25 μg/ml ALLN to preserve the levels of cleaved SREBP-2 (36). To assay SREBP-2 cleavage, total protein was harvested in 1% SDS lysis buffer (10 mM Tris-HCl [pH 7.6], 1% [w/v] SDS, 100 mM NaCl) supplemented with 2% (v/v) protease inhibitor cocktail, passed through a 21-gauge needle until homogenous, and vortexed at room temperature for 15 min. Lysate protein content was quantified using a BCA assay. Samples were analyzed by Western blotting, with SREBP-2 cleavage quantified as a fraction of total SREBP-2 levels and normalized to the statin-treated condition of wild-type cells. To assay SREBP-2 target gene expression, total RNA was harvested and analyzed by qRT-PCR as described above.
Data presentation and statistical analysis
Statistical analysis was conducted as listed in the respective figure legends. Unless specified, statistical significance was determined using a paired two-tailed t-test (∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.005). Densitometric image analysis was carried out in Image Studio Lite (version 5.2.5; LI-COR Biosciences). Graphs were generated in GraphPad Prism 9 (GraphPad Software, Inc). Where applicable, data are presented as mean + SEM or mean + half range.
Results
Human ERG28 is an SREBP-2 gene target
Many metabolic pathways, including cholesterol synthesis, exhibit transcriptional coregulation of their enzymes and associated proteins. The predominant transcription factor driving cholesterol synthesis is SREBP-2 (13). While S. cerevisiae does not have an SREBP homologue as a transcription factor, the majority of ergosterol synthesis-associated genes, including Erg28, are coregulated by the transcription factors Upc2 and Ecm22 (37).
To investigate if ERG28 is coregulated with cholesterol synthesis enzymes, we utilized previously generated cDNA sets (18, 19) from HeLaT, Huh7, and Be(2)C cells that were sterol depleted and treated under a variety of conditions that change expression of SREBP-2 target genes. qRT-PCR indicated an increase in ERG28 mRNA levels when cells were treated with a statin, compactin, and a decrease when cells were treated with the oxysterols 25HC or 24,25EC (Fig. 1A). This pattern is in line with the responses of previously published SREBP-2 targets from these sets (18, 19) and a bona fide SREBP-2 target, one of the terminal cholesterol synthesis enzymes, DHCR24 (14). By contrast, treatment with the liver X receptor agonist T0-901317 did not increase ERG28 (or DHCR24) expression levels. Consistent with other SREBP-2 targets, the response to statins was lowest in Huh7 cells, likely because liver cells have high basal levels of cholesterol synthesis genes, which may be difficult to increase further with statin stimulation (38).
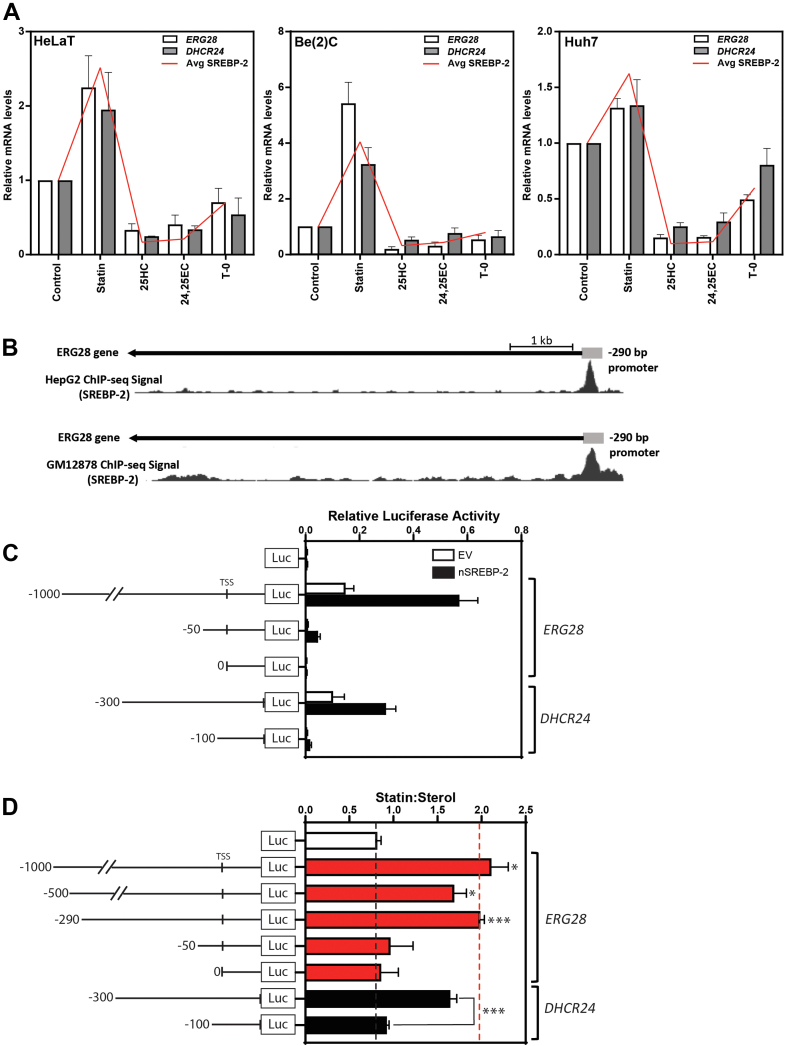
Fig. 1.
ERG28 is transcriptionally responsive to changes in sterol levels and responds like an SREBP-2 target. A: Previously generated cDNA sets (18, 19) from sterol-depleted Huh7, Be(2)C, and HeLaT cells treated for 24 h with 5 μM compactin (statin), 10 μM 25-hydroxycholesterol (25HC), 10 μM 24(S),25-epoxycholesterol (24,25EC), or 10 μM T0-901317 (T-0). mRNA levels for ERG28 and DHCR24 were measured using qRT-PCR and normalized to porphobilinogen deaminase (PBGD). mRNA levels are relative to the solvent control condition, which was set to 1. Data are presented as mean + SEM from n = 3 independent experiments, each performed in triplicate. The average responsiveness for SREBP-2 target genes already published from these cDNA sets (SQLE, LSS, and LDM (19) and HMGCR and TM7SF2 (18)) is represented by the red line (avg SREBP-2). B: ChIP-Seq signals from the UCSC Genome browser (20) of statin-treated HepG2 cells and rabbit IgG anti-SREBP-2 binding in the GM12878 lymphoblast cell line shows SREBP-2 binding in the ERG28 promoter with the −290/+10 bp region outlined. C and D: HeLaT cells were transfected for 24 h with 25 ng Renilla luciferase construct and 250 ng Firefly luciferase constructs as shown. C: Cells were in addition cotransfected with 25 ng nSREBP-2 or empty vector plasmid. Relative luciferase activity represents the ratio of luciferase:renilla activity. D: Cells were treated with 5 μM compactin (statin) or 10 μM 25HC (sterol) in sterol-depleted medium for 24 h and then harvested, and a luciferase assay was performed. The ratio of normalized statin to sterol luciferase activities is presented for each construct. Data are presented as mean + SEM from n = 3–6 independent experiments, each performed in triplicate cultures. ERG28 constructs were compared with luciferase:renilla values for the -luc only construct, and DHCR24 construct activity was compared between the -300/0 and -100/0 DHCR24-luc constructs. Statistical significance was determined using a paired two-tailed t-test (∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.005).
After identifying that ERG28 responds like an SREBP-2 target to statins and sterols, we sought to determine if this was driven by SREBP-2. Using publicly available ChIP-Seq data (20), we observed SREBP-2 binding in the proximal promoter of ERG28, with binding spanning the region between −300/+50 from the ERG28 transcriptional start site (Fig. 1B). In previous work, we found that these ChIP-Seq datasets provide a good indication of the presence and location of SREs in cholesterol synthesis gene promoters (25, 39). To further test SREBP-2 as a regulator of ERG28, constructs containing the proximal ERG28 promoter −1000/+80 driving expression of Firefly luciferase (ERG28-luc) were generated and transiently transfected into human HeLaT cells. When cells were cotransfected with a construct expressing nSREBP-2, the active nuclear form of the transcription factor, luciferase activity increased markedly (Fig. 1C) compared with empty vector, similar to our control DHCR24. This increase in luciferase activity was not observed with −50/+80 or 0/+80 ERG28 constructs, indicating that the SREBP-responsive region is located in the −1,000/−50 region of the promoter.
Next, further truncations of the ERG28-luc construct were generated to identify the minimal sterol-responsive promoter (Fig. 1D). The ratio of luciferase expression when cells were treated with statin (to upregulate SREBP-2 activation) or sterol (to suppress SREBP-2 activation) was used to infer SREBP-2 response (expressed as statin:sterol) (14, 25, 39). Truncations of the ERG28-luc promoter (−1,000/+80, −500/+80, −290/+80, and −50/+80) revealed a loss of responsiveness between the −290 and −50 constructs, suggesting that putative SREs are localized to this region. This responsive region aligns well with the ChIP-Seq peaks of SREBP-2 binding in the ERG28 promoter (Fig. 1B).
Two functional inverse SREs are found in the ERG28 promoter, at the -75 and -13 positions
SREs have a loose 10 bp consensus sequence, reliant on a critical cytosine in the third position (Fig. 2A). We previously generated an in-house weighted positional matrix to predict SREs within promoters (41) and using this identified several putative SREs in the minimally responsive -290/0 region of the ERG28 promoter. The five highest scoring SREs and inverse SREs (invSREs) in the minimally responsive ERG28 promoter were selected for further investigation (Fig. 2B).
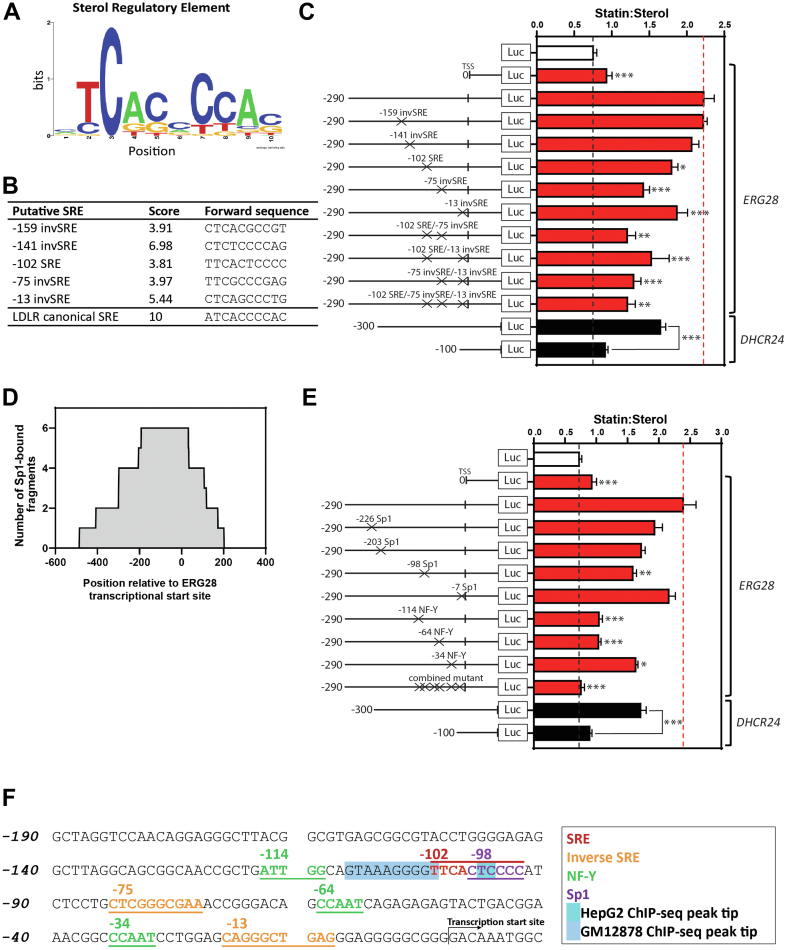
Fig. 2.
Mapping of SREs and other transcription factor binding sites in the ERG28 promoter. A: A weighted positional logo of the SREBP-2 SRE consensus sequence created using WebLogo (40) generated from our in-house matrix (41). B: The top five predicted SREs in the ERG28 promoter from our prediction FIMO (26). The canonical SRE found in the promoter of LDLR is included for reference and given the top available score of 10. C and E: Dual luciferase assay data identifying (C) SREs and (E) Sp1 and NF-Y binding sites in the ERG28 −290/+80 promoter. As for Fig. 1D, HeLaT cells were transfected for 24 h with 25 ng Renilla luciferase construct and 250 ng Firefly luciferase constructs as shown. Cells were treated with 5 μM compactin (statin) or 10 μM 25-HC (sterol) in sterol-depleted medium for 24 h and then harvested, and a luciferase assay was performed. The ratio of normalized statin to sterol luciferase activities is presented for each construct. Data are presented as mean + SEM from n = 4–8 separate experiments, each performed in triplicate cultures. Statistical significance was determined using a paired two-tailed t–test, and comparisons were between the wildtype −290/+80 ERG28-luc constructs and the respective SRE, Sp1, and NF-Y mutants and shown for significant decreases (∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.005). D: Combined ChIP-Seq signal graph showing binding of six Sp1-occupied sequence fragments taken from ENCODE project data (42) within the ERG28 −500 to +200 bp region. The Y-axis shows the number of times that each nucleotide was represented in a sequence fragment. F: A proposed map of transcriptional elements in the −190/+10 ERG28 promoter (compiled from data in Fig. 2C, E). The tips of the SREBP-2 ChIP-Seq peaks obtained from UCSC genome browser are highlighted in shades of blue.
Using site-directed mutagenesis of the −290/+80 ERG28-luc construct, we generated mutant SRE constructs and tested if they were still responsive to manipulation of cellular sterol status (Fig. 2C). We observed a significant decrease in the statin:sterol response in the -102SRE, -75invSRE, and -13invSRE constructs, though the sterol responsiveness was not completely ablated. Combinatorial mutations of two or three putative SREs further reduced the sterol responsiveness of the constructs but did not completely ablate it.
Functional Sp1 and NF-Y sites in the ERG28 promoter
SREBP-2 is a relatively weak activator of transcription, and maximal activation of the promoter is frequently reliant upon the binding of the transcriptional cofactors Sp1 and/or NF-Y in the proximal promoter (43, 44, 45). Using publicly available ChIP-Seq data (42) to investigate Sp1 occupancy at the ERG28 promoter (Fig. 2D), we found that all six available datasets showed Sp1 binding between -192 and -32 bp of the ERG28 transcriptional start site, giving confidence that Sp1 sites reside in this region of the promoter. This strategy has previously been used in our laboratory to identify Sp1 binding regions (46). We have found that the positioning of Sp1 and NF-Y sites in relation to SREs is not consistent (25) and as such could not use distance from the putative SREs to predict functional Sp1 and NF-Y sites.
Using ConSite, we identified several putative Sp1 and NF-Y binding sites in the ERG28 proximal promoter. Sp1 sites were predicted at -226, -203, -98, and -7 relative to the ERG28 transcriptional start site, and NF-Y sites were predicted at -114, -64, and -34. These putative binding sites were mutated in the ERG28-luc −290/+80 construct, and their sterol responsiveness was assayed via luciferase assays (Fig. 2E). This showed that disruption of the predicted NF-Y sites located at -114 and -64 eliminated sterol responsiveness of the constructs (nonsignificant from ERG28-luc −0/+80), and disruption of the Sp1 binding site at -98 or the NF-Y binding site at -34 positions reduced the sterol responsiveness.
The function of the −98 Sp1 or −102 SRE sites are difficult to deconvolute because of the predicted Sp1 site occurring entirely within the -102 SRE. Mutations in this region are likely to disrupt binding of either transcription factor. However, Sp1 occupancy in the promoter of ERG28 (Fig. 2D) indicates this transcription factor is likely to play a role in the transcriptional programming of ERG28.
The −290/+80 ERG28-luc combined mutant construct (Fig. 2E, containing mutations of the -102SRE/-98Sp1, -75inv and -13 inv SREs, and -114 and -64 NF-Y sites) completely ablated the sterol responsiveness of the promoter, suggesting that a combination of these sites is essential for the sterol responsive transcription of ERG28. Compiling the data from these luciferase assays (Fig. 2C, E) and the published SREBP-2 ChIP-Seq peaks (Fig. 1B), we mapped proposed functional elements in the ERG28 proximal promoter (Fig. 2F).
ERG28 interacts with itself, NSDHL, and SC4MOL
After establishing that ERG28 is coregulated with cholesterogenic enzymes by SREBP-2, we sought to determine if the function of the protein is conserved from yeast to humans. Yeast Erg28p interacts strongly with itself and also with a wide number of ergosterol synthesis enzymes, including Erg25p, Erg26p, and Erg27p (the members of the C4-demethylation complex, homologous to sterol-C4-methyl oxidase–like gene [SC4MOL], NAD(P) Dependent Steroid Dehydrogenase-Like [NSDHL], and hydroxysteroid (17-beta) dehydrogenase 7 [HSD17B7], respectively) (7, 47).
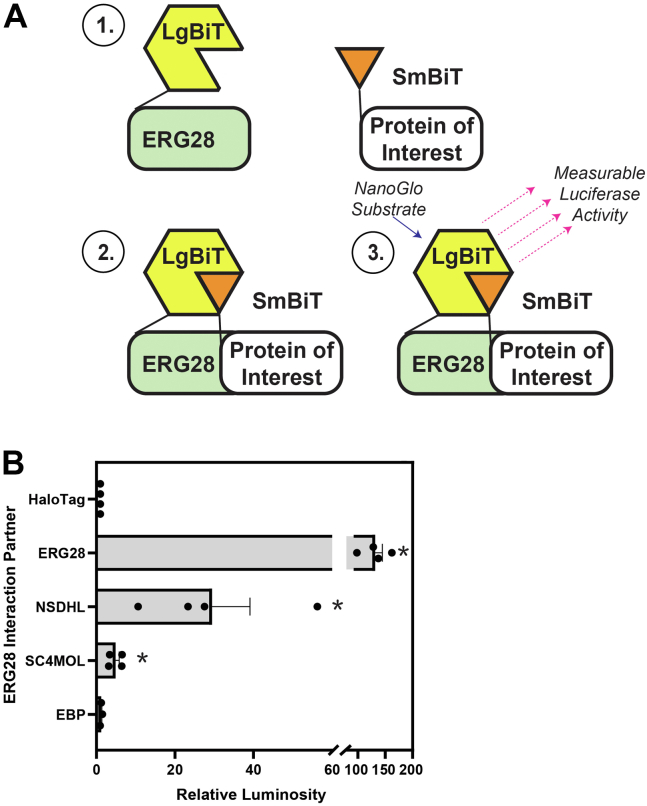
Using the NanoBiT® split luciferase system for detecting protein-protein interactions, plasmids containing the human coding sequence of ERG28, NSDHL, SC4MOL, and emopamil-binding protein (EBP) were generated. These plasmids were cotransfected into HEK293T cells to study the in situ interactions of ERG28 (Fig. 3A). Based on this assay, ERG28 interacts with itself (Fig. 3B), recapitulating the homomeric interaction of Erg28p (8). In addition, ERG28 interacts with both SC4MOL and NSDHL (Fig. 3B), akin to their yeast homologs (Erg25p and Erg26p) (7, 47). While the SC4MOL–ERG28 interaction has been published in BioGrid (48), no interaction between NSDHL and ERG28 has previously been shown. No interactions were detected in situ between EBP and ERG28 (Fig. 3B) despite published BioGrid data (48) indicating these two proteins are interaction partners.
Fig. 3.
ERG28 interacts with itself and NSDHL and SC4MOL but not with EBP. A: An overview of the NanoBiT® luciferase system for protein-protein interactions. 1) Fusion constructs of ERG28 with the LgBiT™ construct and proteins of interest (ERG28, NSDHL, SC4MOL, EBP, or a HaloTag) with the SmBiT™ construct are transfected into HEK293T cells. 2) If the proteins of interest interact with ERG28, the functional NanoLuciferase is formed. 3) When the NanoGlo substrate is supplied to the cells, luciferase activity can be measured and used to infer interaction. B: HEK293T cells were transfected for 24 h with ERG28 LgBiT, and the indicated SmBiT plasmid, split into 96-well plates, and luminosity was measured according to the manufacturer’s protocol. The relative luminosity is normalized to the response of the negative control pair (ERG28 LgBiT and HaloTag™-SmBiT), which was set to 1. Data are presented as mean + SEM of n = 3–4 independent experiments, each conducted in duplicate. Statistical significance from the ERG28 LgBiT:HaloTag SmBiT pairing was determined using a paired one-tailed t-test (∗P < 0.05).
ERG28 KO leads to an impairment in cholesterol synthesis in Huh7 cells
Considering protein-protein interactions of ERG28 partially recapitulate the interactions of Erg28p in yeast (Fig. 3B), we investigated if ERG28 functionally plays a similar role in cholesterol synthesis as Erg28p does in ergosterol synthesis. Therefore, we generated ERG28 KO cell lines in the liver cell line, Huh7, since liver cells are central to cholesterol homeostasis and have high levels of cholesterol synthesis (49). We used CRISPR-Cas9 with two guides targeting exons 3 and 4 of ERG28 and identified three clones (KO1, KO2, and KO3) with disrupted ERG28 by Sanger sequencing (Fig. 4A), measuring mRNA levels (Fig. 4B) and endogenous ERG28 protein levels (Fig. 4C). One clone generated had a 2 base pair deletion after amino acid 54, resulting in a premature stop codon after amino acid 62 (KO1), and two clones had large deletions between the guides, producing a Δ[34–65] (KO2) or Δ[55–124] (KO3) deletion in the coding sequence of ERG28. An overall reduction of 80–98% of ERG28 mRNA (Fig. 4B) and 65–85% of ERG28 protein levels were observed in the three KO clones, with KO1 having the greatest decrease (Fig. 4C). Hence, KO1 was selected for most subsequent work.
As ergosterol synthesis is reduced by Erg28p KO (5), we wanted to know if this function was conserved between yeast and humans and to investigate if ERG28 KO impacted cholesterol levels in human cells. Using Amplex Red assays, we measured the cholesterol levels in wild-type and KO cells treated in either full serum or lipoprotein-deficient serum. There was no statistically significant difference between the total cholesterol levels in Huh7 wild-type and KO cells when grown in full serum media, even after treatment with statin to inhibit cholesterol synthesis (Fig. 4D). When cells were grown in a lipid-deficient environment, where sterol synthesis is upregulated to compensate for a lack of sterol uptake, we observed a ∼35% reduction in the cholesterol pool of the KO compared with wild-type cells. When additionally treated with statin, both the wild-type and ERG28 KO cells showed reductions in their cholesterol pool (Fig. 4D). These results are in line with expectations of ERG28 being involved in sterol synthesis, not uptake. Cholesterol storage is unlikely to be disrupted in ERG28 KO cells as there was no change in the percentage of cholesterol stored as cholesteryl esters in the KO cells compared with wild-type cells grown in full serum (Fig. 4E).
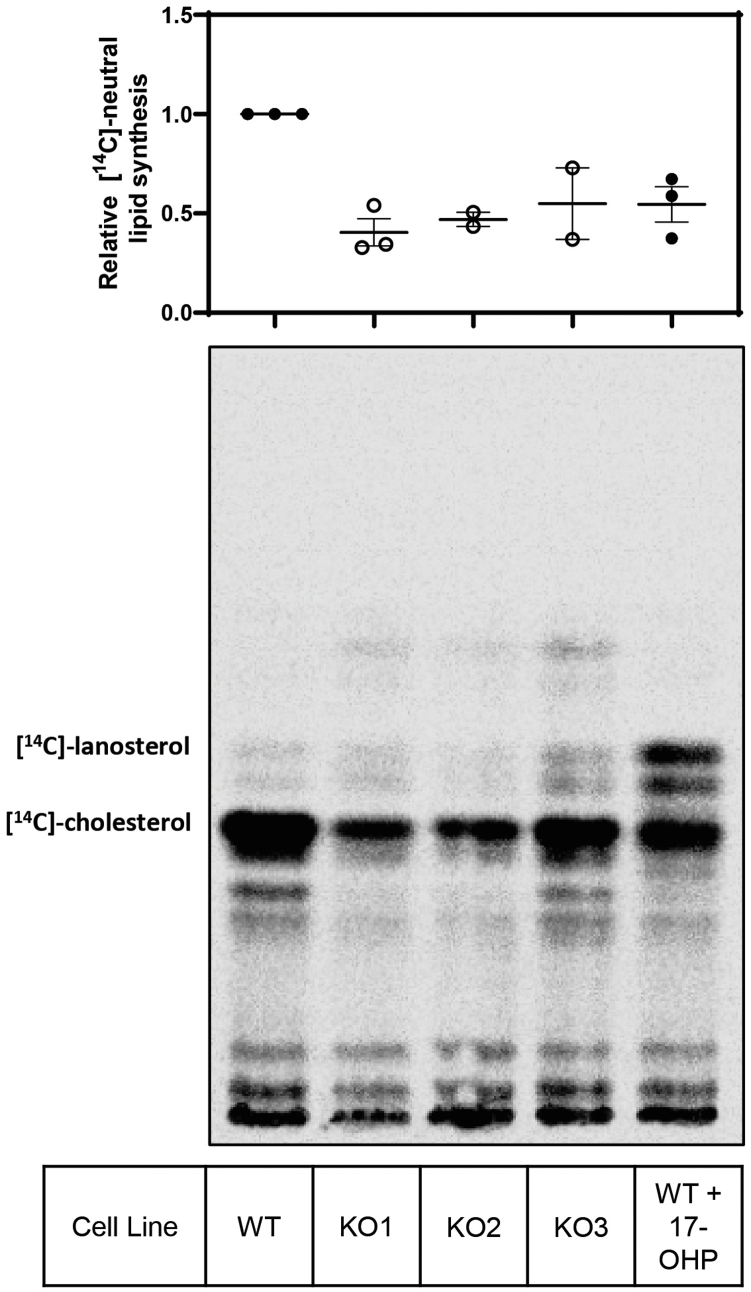
After confirming the ERG28 KO cell lines had less overall cholesterol than the wild-type Huh7 cells, and that this was likely because of a synthetic defect and not reduced uptake, we aimed to directly test if ERG28 is involved in cholesterol synthesis. Using [14C]-acetate to radiolabel wild-type and ERG28 KO cells, the synthesis of cholesterol and other lipids was measured via TLC. The KO cell lines displayed dramatically reduced cholesterol synthesis (Fig. 5A), with clones showing a 60–75% reduction in cholesterol synthesized compared with wild-type when grown in lipoprotein-deficient media. KO cells grown in full serum also showed a reduction in cholesterol synthesis comparative to wild-type (Fig. 5B), suggesting cholesterol synthesis is impaired by ERG28 KO, irrespective of exogenous cholesterol supplementation. KO cells in addition showed a reduced rate of cholesterol production over time (Fig. 5C,D) compared with wild-type Huh7 cells. In addition to reduced cholesterol synthesis, there was also an overall decrease in the synthesis of [14C]-labeled neutral lipids in the KOs (supplemental Fig. S2). No consistent change in the level of fatty acid synthesis was observed in the KO cells, indicating that the rate of uptake of [14C]-acetate was similar between the KO and wild-type cells, and that the inhibitory effect of ERG28 KO was specific for sterol synthesis (Fig. 5E).
Fig. 5.
KO of ERG28 results in reduced cholesterol synthesis. A and B: Wild-type or ERG28 KO cells were pretreated for 16 h in 10% FCLPDS DMEM:HG sterol-deficient media supplemented with 5 μM compactin with or without 10 μM 17-OHP and then radiolabeled with 1 μCi of [14C]-acetate for 4 h in sterol-deficient media with or without 10 μM 17-OHP. Cell lysates were normalized to total cellular protein concentration, and total nonsaponifiable sterols were harvested from the cells (A), dried, and separated on TLC Silica gel 60 F254 plates in a heptane:ethyl acetate (2:1, v/v) mobile phase. Radiolabeled lipids were imaged after ∼ 1 week of exposure using the Typhoon FLA 9500 phosphor imager. Data were normalized to the wild-type and presented as mean ± SEM from n = 3 or 4 independent experiments. Statistical significance was determined using a paired two-tailed t test (∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.005). B: Wild-type or ERG28 KO1 cells were radiolabeled in 10% FCS DMEM:HG with 2 μCi of [14C]-acetate for 4 h and lipids extracted as for A. TLC is representative of n = 2 independent experiments each conducted in triplicate. C–D: Huh7 wild-type cells and ERG28 KO1 cells were pretreated for 16 h in 10% FCLPDS DMEM:HG sterol-deficient media supplemented with 5 μM compactin with or without 10 μM 17-OHP and then radiolabeled with 1 μCi of [14C]-acetate for 2–24 h. Lipids were extracted as per Fig. 5A. Data were normalized to the wild-type 8 h time point and presented as mean ± SEM from n = 3 independent experiments. Statistical significance was determined using a paired two-tailed t-test, all time points were significant P < 0.05. E: After total nonsaponifiable sterols were harvested from the cells (Fig. 5A), the remaining lysate was acidified, and fatty acids were extracted, dried, and separated on TLC Silica gel 60 F254 plates in a heptane:diethyl ether:acetic acid (90:30:1, v/v) mobile phase. F: Huh7 wild-type or ERG28 KO1 cells were transfected for 8 h with increasing amounts of CMV-ERG28-V5 as indicated, pretreated for 16 h in sterol-deficient media supplemented with 5 μM compactin, and then radiolabeled with 1 μCi of [14C]-acetate for 4 h in sterol-deficient media. Lipids were extracted as per Fig. 5A. Data expressed as levels of [14C]-sterol in KO compared with wild-type for each plasmid concentration to account for variations in transfection efficiency, normalized to sterol synthesis for the 1.0 μg condition for each cell line. Data for 0, 0.5, 0.75, and 1.0 μg are from n = 3 independent experiments and presented as mean ± SEM, data for 0.25 μg are from n = 2 independent experiments and presented as mean ± half range.
The phenotype of reduced cholesterol synthesis was able to be reversed in a dose-dependent manner by transiently expressing a CMV-ERG28-V5 construct in the KO cells (Fig. 5F), confirming the reduction in cholesterol synthesis is due to the loss of ERG28.
A major function of Erg28p in yeast is facilitating the effective C4-demethylation of the sterol precursor. As we have shown protein-protein interactions between ERG28 and two enzymes involved in C4-demethlayion, NSDHL and SC4MOL, we sought to investigate if ERG28 played a similar role in mammalian cholesterol synthesis. We treated wild-type Huh7 cells with 17α-hydroxyprogesterone (17-OHP), an inhibitor of HSD17B7 (50), the 3-keto-steroid reductase, which should replicate a synthetic lesion at the C4-demethlyation step. When wild-type cells were treated with 17-OHP there was a reduction in cholesterol synthesis, and accumulation of lanosterol and other neutral lipids (Fig. 5A). However, no such accumulation is observed in ERG28 KO cells (Figs. 5A and S2), indicating that a lesion in cholesterol synthesis occurs at a different location in the pathway than the chemically induced block at HSD17B7.
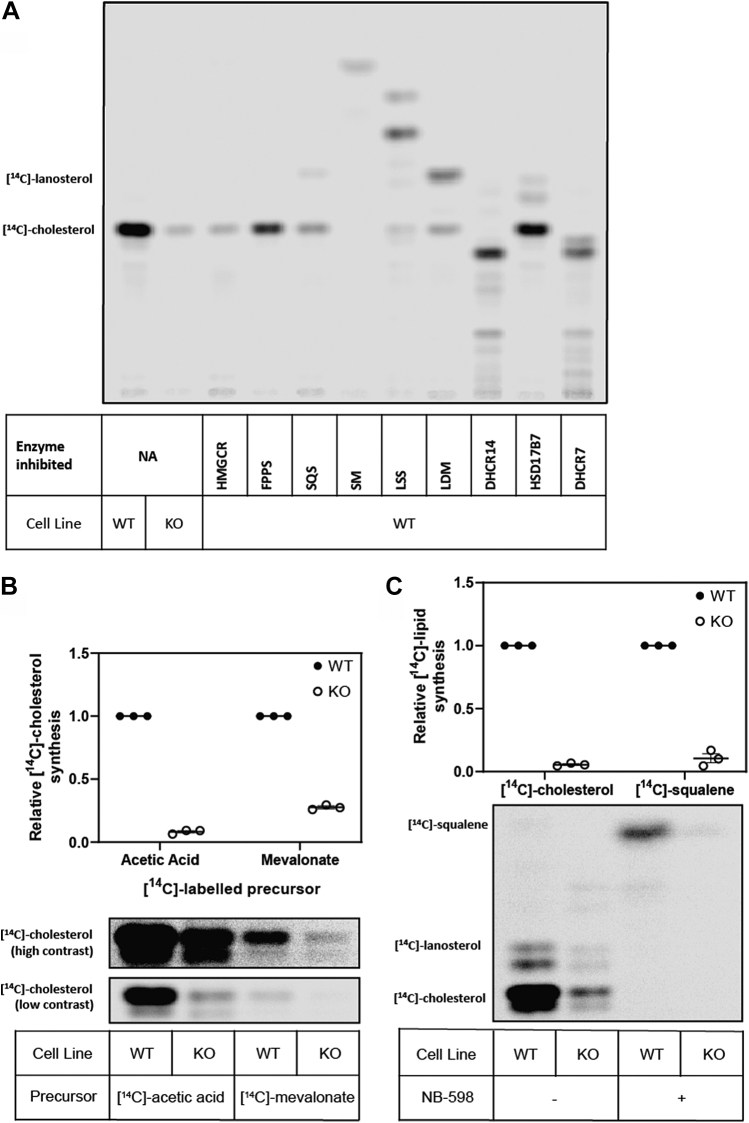
After establishing that the block in cholesterol synthesis driven by ERG28 KO was unlikely to be at the C4-demethylation step, we sought to identify the site of the synthetic lesion. Treatment with a wide variety of cholesterol synthesis inhibitors targeting different enzymes of the pathway failed to produce a pattern of accumulation similar to that observed in the ERG28 KO cells (Fig. 6A). Inhibition of cholesterol synthesis enzymes post-squalene synthase (SQS) produced easily identifiable banding patterns, corresponding to a variety of predicted sterols. However, none of these patterns matched the KO cells indicating that the tested post-SQS enzymes were unlikely to be the site of the synthetic lesion. Inhibition of 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), farnesyl diphosphate synthase (FPPS), and SQS produced a similar reduction of cholesterol synthesis to the KO cells and suggested that the major lesion of cholesterol synthesis in the KO cells was unlikely to be after squalene synthesis.
Fig. 6.
The synthetic lesion in ERG28 KO cells likely occurs between HMGCR and SM. A: Wild-type Huh7 cells were treated with the indicated cholesterol synthesis pathway inhibitors, 5 μM compactin (HMGCR), 100 μM zoledronate (FPPS), 25 μM TAK-475 (SQS), 1 μM NB-598 (SM), 100 nM RO 48-8071 (lanosterol synthase [LSS]), 1 μM GR70585X (lanosterol demethylase [LDM]), 20 μM AY-9944 (14-dehydrocholesterol reductase [DHCR14]), 10 μM 17-OHP (HSD17B7), 1 μM AY-9944 (7-dehydrocholesterol reductase [DHCR7]) and labeled in 10% FCLPDS DMEM:HG with 1 μCi of [14C]-acetate for 24 h. B: Wild-type Huh7 or ERG28 KO1 cells were pretreated for 16 h in 10% FCLPDS DMEM:HG supplemented with 5 μM compactin and then radiolabeled with either 1 μCi of [14C]-acetate or 2 μCi of [14C]-mevalonate for 4 h. C: Wild-type Huh7 or ERG28 KO1 cells were labeled with 1 μCi of [14C]-acetate and treated with or without 1 μM of NB-598 to inhibit SM. A–C: Lipids were extracted, and TLCs were performed as per Fig. 5A. Data are representative of n = 3 independent experiments. Data were normalized to the (B, C) [14C]-cholesterol or (C) [14C]-squalene levels of wild-type cells within each treatment group, which were set to 1.
To determine if the lesion in cholesterol synthesis occurred after the first rate-limiting step, HMGCR, the radiolabeled sterol precursor [14C]-mevalonate (the product of HMGCR), was supplied to cells and sterol synthesis compared with synthesis with [14C]-acetate. While there was an overall reduction in the level of [14C]-cholesterol synthesized when [14C]-mevalonate was used as the labeled precursor (likely because of the low rate of uptake of mevalonate compared with acetate (51)) (Fig. 6B), there was a comparable reduction in [14C]-cholesterol synthesis between the wild-type and KO cells, indicating cholesterol synthesis is inhibited after the first rate-limiting step of cholesterol synthesis, HMGCR.
To confirm the synthetic lesion was before squalene synthesis, squalene monooxygenase (SM) was inhibited in wild-type and KO cells with the SM inhibitor, NB-598 (Fig. 6C). If the primary synthetic lesion occurs at or after SM, we anticipated a comparable accumulation of [14C]-squalene in the wild-type and KO cells. Only the wild-type cells showed a marked accumulation of [14C]-squalene in response to NB-598 treatment (Fig. 6C), with KO cells synthesizing ∼10% compared with the wild-type. This suggests that cholesterol synthesis in ERG28 KO cells is impaired between the two major rate-limiting steps, HMGCR and SM.
KO of ERG28 leads to a general downregulation of cholesterol synthesis genes
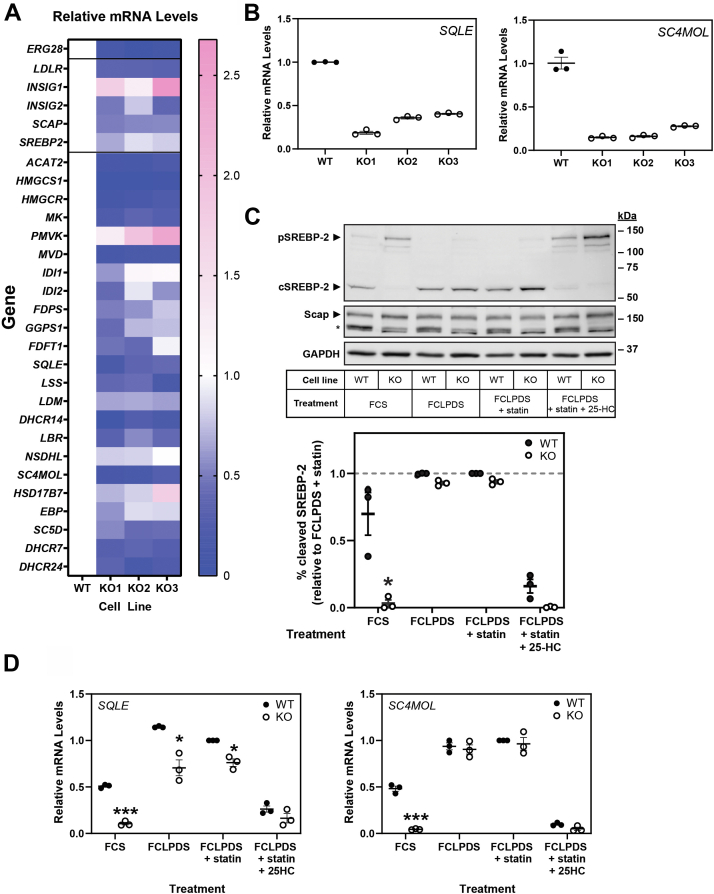
We next speculated that genes in the cholesterol synthesis pathway would be upregulated as a compensatory response to the synthetic lesion because of ERG28 KO and investigated the possibility of transcriptional changes. qRT-PCR performed on RNA extracted from all three ERG28 KO cell lines showed not only were ERG28 mRNA levels reduced in the KOs but so too were the levels of most cholesterol synthesis genes (Fig. 7A), such as SQLE and SC4MOL (Fig. 7B) and many early mevalonate pathway enzymes. In addition to cholesterol synthesis genes, levels of LDLR and SREBP-2, both SREBP-2 targets, were reduced (Fig. 7A). However, INSIG1 and PMVK, also transcriptional targets of SREBP-2 (52), increased (Fig. 7A).
Fig. 7.
SREBP-2 genes are downregulated in ERG28 KO cells. A and B: Previously generated cDNA generated from Huh7 ERG28 KO clones (Fig. 4B) was used to measure the expression of all cholesterol synthesis genes, plus LDLR (as a canonical SREBP-2 target) and INSIG1, INSIG2, SCAP, and SREBP-2 (as components of the SREBP-2 processing machinery). mRNA levels were measured using qRT-PCR and normalized to PBGD. mRNA levels are relative to the wild-type, which was set to 1. A: A heatmap showing overall trends, (B) SQLE and SC4MOL levels showing a reduction in relative gene expression in all three KO clones. C: Huh7 wild-type and ERG28 KO cells were treated in a variety of conditions to investigate SREBP-2 cleavage difference between them. FCS-treated cells (lanes 1 and 2) were treated for 5 h in 10% FCS DMEM:HG PS (parallel to basal conditions in A and B). Lanes 3–8 were treated for 1 h in 10% FCLPDS DMEM:HG containing 1% (w/v) 2-hydroxypropyl-β-cyclodextrin, 10 μM compactin, and 50 μM mevalonate. Next lanes 3–8 were treated for 5 h with or without 1 μg/ml 25-HC in 10% FCLPDS DMEM:HG, with or without 10 μM compactin and 50 μM mevalonate. During the final 2 h of treatment, medium in all lines was supplemented with 25 μg/ml ALLN to preserve the levels of cleaved SREBP-2. Cell lysates were normalized, separated by SDS-PAGE, and protein levels were measured by Western blot using anti-SREBP-2, anti-Scap, and anti-GAPDH. pSREBP-2 refers to the full-length SREBP-2 precursor, cSREBP-2 refers to the C-terminal fragment of cleaved SREBP-2, and ∗ refers to a nonspecific band in the anti-Scap. Western blots are representative of n = 3 independent experiments, SREBP-2 cleavage was quantified as a fraction of total SREBP-2 levels, and normalized to the statin-treated condition of WT cells. D: Cells were treated as per C. Total RNA was harvested, cDNA prepared, and SQLE and SC4MOL mRNA levels were measured using qRT-PCR and normalixed to PBGD. mRNA levels are relative to the wild-type statin-treated condition, which was set to 1. Data are presented as mean ± SEM of n = 3 independent experiments. Comparisons were made between wild-type and KO cells for each treatment condition. Statistical significance was determined using a paired two-tailed t-test (∗P < 0.05 and ∗∗∗P < 0.005).
KO of ERG28 alters SREBP-2 cleavage
The broad downregulation of cholesterol synthesis genes observed in Fig. 7A suggested that SREBP-2, the master transcription factor of cholesterol synthesis, may be affected in ERG28 KO cells.
To investigate this, SREBP-2 cleavage assays were performed (Fig. 7C). In full serum conditions (FCS, lanes 1–2; Fig. 7C), SREBP-2 cleavage decreased in the KO cells (production of cSREBP-2 from its precursor pSREBP-2), consistent with the broad downregulation of SREBP-2 target genes (Fig. 7A,B). However, SREBP-2 cleavage was similar between the KO and wild-type cells in all lipoprotein-deficient conditions (lanes 3–8; Fig. 7C). Levels of Scap were not significantly altered in the KO cells compared with wild-type, suggesting that another element of SREBP-2 cleavage explain the SREBP-2 results observed in KO cells (Fig. 7C). While individual gene expression was reduced in KO cells in full serum, transcript levels were restored nearly to wild-type levels in conditions that favor SREBP-2 activation (Fig. 7D). This confirmed that the pathway was likely to be fully active under the lipoprotein-deficient conditions used to identify the lesion in cholesterol synthesis (Fig. 5A, C, D and Fig. 6A–C).
Discussion
In this work, we have for the first time identified human ERG28 as an important player in cholesterol biosynthesis. Human ERG28 shares some functional homology with its yeast counterpart, Erg28p, in that it is transcriptionally coregulated with the sterol synthesis gene program, interacts with certain sterol synthesis enzymes, and has an important, albeit nonessential, role in sterol synthesis. However, in addition, human ERG28 influences SREBP-2 activation in sterol-replete conditions by a still to be uncovered mechanism.
Transcriptional regulation of ERG28
We confirmed ERG28 is transcriptionally driven by SREBP-2 (Fig. 2A, C) and identified three SREs that aligned with publicly available ChIP-Seq data (Figs. 2B and 3C), three NF-Y, and one Sp1 (Fig. 3E) binding site in the minimal sterol-responsive promoter. Early studies into SREBP-2 coregulated genes (52, 53) suggested that the transcript of ERG28 may be amongst them. However, the sterol-responsive region of the promoter, and the potential role the gene product plays in cholesterol biosynthesis, has not been explored previously. Despite the initial identification of the ERG28 minimal sterol-responsive promoter between -290/-50 (Fig. 1D), mutation of the -13 invSRE also led to a decrease of sterol responsiveness in the ERG28-luc construct (Fig. 2C). It may be that other regulatory elements required for SREBP-2 activation of the -13 invSRE are located within the -290/-50 region (potentially the -64 NF-Y site), and in the −50/+80 construct, the -13 invSRE alone is not strong enough to induce transcription.
The highest scoring predicted SRE in the −290/+80 region of the ERG28 promoter, an invSRE at -141, did not affect sterol responsiveness of the ERG28-luc construct (Fig. 2C). This underscores the loose consensus motif and the need to test multiple predicted SREs and consider additional lines of evidence (Fig. 2A). Our work using previous iterations of this in-house weighted positional matrix also found functional SREs with low scores or not predicted by this method (25, 39). Careful investigation is needed to confidently identify SREs in SREBP-2 gene targets.
While we have identified likely SREBP-2 binding sites, and NF-Y and Sp1 sites in the ERG28 proximal promoter, we cannot exclude the possibility of other regulatory elements in this region contributing to the sterol responsiveness of the ERG28 promoter.
Protein-protein interactions of ERG28
The major role assigned to Erg28p in ergosterol synthesis is as a scaffold for the C4 demethylation complex. This complex comprises Erg25, Erg26p, and Erg27p (three enzymes homologous to NSDHL, SC4MOL, and HSD17B7, respectively), which together facilitate the effective demethylation from the C4 carbon (8, 9). Though, Erg28p also interacts with several other ergosterol synthesis enzymes (7, 9, 47).
The KO of Erg28p primarily leads to an accumulation of C4-methyl sterols, suggesting the C4-demethylation step, that is the sequential removal of the two C4 carbons by Erg25p, Erg26p, and Erg27p, is impacted. Plant ERG28 shares this homologous function of scaffolding the C4-demethylation enzymes together, and a variety of sterol biosynthetic intermediates build up in KO plants (6). We found ERG28 interacts with two members of the C4-demethylation complex, NSDHL and SC4MOL (Fig. 3B), and attempted to recreate this C4-demethylation defect by treating wild-type cells with 17-OHP, an inhibitor of HSD17B7 (the Erg27p homologue). However, the pattern of radiolabeled sterols, notably accumulation of bands corresponding to lanosterol and other sterol intermediates, was very different between the ERG28 KO cells and wild-type cells with inhibition of HSD17B7 (Fig. 5A), arguing against the C4-demethylation step being the primary location of the lesion. Inhibition of SC4MOL or NSDHL should produce a similar sterol profile as HSD17B7 inhibition (Fig. 5A) and a marked accumulation of the substrate of SC4MOL, T-MAS, would be expected.
A study of binary interactions in the human proteome revealed >100 interaction partners of ERG28 including the cholesterol synthesis enzyme EBP (48), which was not detected in our NanoBiT® assays. Moreover, the sterol profiles obtained from our radio-TLCs do not align with the expected results if EBP is the primary lesion in cholesterol synthesis. Preliminary analysis by gas chromatography coupled with mass spectrometry confirms no detectable accumulation of sterol intermediates (data not shown).
Our data (Fig. 3B) and investigation into binary protein-protein interactions in the human proteome show ERG28 interacts with itself (48), indicating ERG28 likely dimerizes (or forms a larger complex containing several ERG28 units). Certain cholesterol synthesis enzymes are known to interact with each other, for example, DHCR7 and DHCR24 (the two terminal enzymes of cholesterol synthesis) functionally and physically interact (54). Also, EBP and DHCR7 interact, together forming the catalytic and regulatory enzyme, cholesterol-5,6-oxide hydrolase (55). In S. cerevisiae, many ergosterol synthesis enzymes interact, including Erg28p interacting with itself (8, 9). However, not every interaction is conserved from yeast to humans: the homologs of HSD17B7 and lanosterol synthase are known to interact in yeast, but this is not conserved in the mammalian system (4). While the current mammalian protein-protein interaction examples are just binary interactions, ERG28 may scaffold numerous cholesterol synthesis enzymes together to facilitate effective conversion of intermediates to cholesterol, which we investigated in our KO cells.
Functional impacts of ERG28 KO
We found ERG28 KO reduces cholesterol levels in the cell (Fig. 4D), through decreased sterol synthesis (Fig. 5A). However, we have yet to fully identify the site(s) of the lesion or the mechanism(s) driving this. While some inferences can be made based on the homology of cholesterol and ergosterol synthesis, the lesion in cholesterol synthesis in ERG28 KOs is not solely at the C4-demethylation step, where the lesion occurs in KO yeast (5) and plants (6). Our collective data suggest that inhibition of cholesterol synthesis occurs in ERG28 KO cells between the two rate-limiting steps of HMGCR (Fig. 6A, B) and SM (Fig. 6C), where the intermediates would not have been extracted in our lipid extraction and visualized on radio-TLC. We speculate that KO of ERG28 may affect multiple steps in cholesterol synthesis.
We identified a general reduction in SREBP-2 target genes in ERG28 KO cells under sterol-replete conditions (Fig. 7A) suggesting impaired SREBP-2 transcriptional programming. Activation of SREBP-2 in full serum was significantly reduced in the ERG28 KO cells (Fig. 7C), potentially explaining the widespread decrease in message levels of SREBP-2 target genes (Fig. 7A, B). By contrast, activation of SREBP-2 was unaffected in sterol-deficient conditions (Fig. 7C), and mRNA levels of SREBP-2 target genes were comparable between the KO and wild-type cells under these conditions (Fig. 7D), indicating the activation mechanism of SREBP-2 is intact. The mechanism(s) driving reduced SREBP-2 activation in ERG28 KO cells remains to be elucidated but does not appear to be due to altered Scap protein levels (Fig. 7C). Radiolabeled tracer experiments (Figs. 5A, C–F and 6) were performed under lipoprotein-deficient conditions where SREBP-2 target gene levels are restored, suggesting that the reduced cholesterol synthesis in ERG28 cells under sterol-depleted conditions is not exclusively because of lower gene expression of synthetic enzymes.
Broader implications of ERG28
Cholesterol synthesis and metabolism is frequently perturbed in a variety of cancers (reviewed in Ref. (56)). Early work on ERG28 identified increased expression of the transcript in colon and leukocyte cancer-derived cells compared with healthy tissue cell lines (10), and the only published function of ERG28 in a mammalian system thus far is related to its upregulation in pancreatic cancers (11). As we have shown ERG28 plays a functional role in cholesterol synthesis, it may be that this increased gene expression in cancers is a driver of, or a result of, perturbations in cholesterol synthesis.
Other published interaction partners of ERG28 include solute carrier family 41 member 1 (57), a magnesium transporter primarily localized to the plasma membrane, and CLN8, an ER-associated membrane protein (12). Even animals auxotrophic for cholesterol synthesis contain homologues of ERG28/Erg28 (58). Despite nematodes being auxotrophic for sterol synthesis and having an incomplete sterol synthesis pathway (59), the homolog of ERG28 in Caenorhabditis elegans, ERG-28, protects the sodium channel, SLO-1, from aspartic protease degradation (60). These additional interaction partners of ERG28, and its conservation in sterol auxotrophic organisms, suggests that beyond its role in cholesterol synthesis, ERG28 may function as a general membrane scaffolding protein in the cell.
Our current work has shown ERG28 plays a significant role in mammalian cholesterol synthesis. ERG28 gene expression is coregulated with cholesterol synthesis enzymes by SREBP-2-mediated transcription. ERG28 protein physically interacts with certain cholesterol synthesis enzymes, and its lack drastically reduces cholesterol synthesis. Further work is required to delineate precisely how ERG28 functions in the mammalian cholesterol synthesis pathway and other roles this protein may play in the cell.
Data availability
The data described in the article are contained within the article.
Supplemental Data
This article contains supplemental data (61, 62, 63, 64).
Conflict of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank members of the Brown Laboratory for critically reviewing this article, Dr Elizabeth Stout and Yvette Aw for technical advice on the generation of CRISPR cell lines, and Jason Sercombe for the assistance with gearing up for radiation work.
Author Contributions
I. M. C.-H. and A. J. B. conceptualization; I. M. C.-H. and A. J. B. methodology; I. M. C.-H. validation; I. M. C.-H. formal analysis; I. M. C.-H., N. M. F., and H. W. C. investigation; I. M. C.-H. writing–original draft; I. M. C.-H., L. J. S. and A. J. B. writing–review & editing; I. M. C.-H., N. M. F., and H. W. C. visualization; L. J. S. and A. J. B. supervision; A. J. B. project administration; L. J. S. and A. J. B. funding acquisition.
Funding and Additional Information
The laboratory of A. J. B. was supported by an Australian Research Council Discovery Project grant (grant no.: DP170101178), New South Wales Health Investigator Development grant, and a University of New South Wales Interlude grant. I. M. C.-H. is supported by a UNSW Scientia PhD Scholarship, and I. M. C.-H., N. M. F., and H. W. C. are supported by the Australian Government Research Training Program Scholarships.
Supplemental data
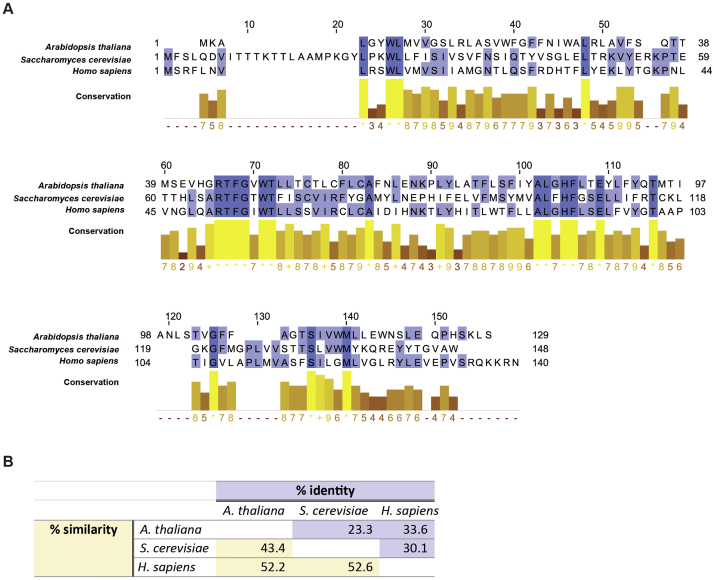
Supplemental Fig. S1ERG28 is conserved between kingdoms. (A and B) Protein sequences for ERG28 from S. cerevisiae, A. thaliana and H. sapiens were retrieved from UniProt (61). (A) Sequences were aligned in MEGA-X (62) using the ClustalW algorithm with default parameters. The resulting alignment was exported into JalView (63) and conservation scores for conserved residues in the alignment were automatically assigned according to (64). (B) Pairwise identity (%) or similarity (%) calculated by Geneious Prime (2020.2.4) from global alignments with free end gaps and a Blosum 62 cost matrix
Supplemental Fig. S2ERG28 KO cells show a reduction in total neutral lipid synthesis compared to wild-type cells. The full plate exposure of Fig. 4B with amplified contrast to visualize faint bands. Wild-type or ERG28 KO cells were pretreated for 16 h in sterol deficient media supplemented with 5 μM compactin with or without 10 μM 17-OHP and then radiolabeled with 1 μCi of [14C]-acetate for 4 h in sterol deficient media with or without 10 μM 17-OHP. Total nonsaponifiable sterols were harvested from the cells, dried and separated on TLC Silica gel 60 F₂₅₄ plates in a heptane: ethyl acetate (2:1) mobile phase. Radiolabeled sterols were imaged after 1 week of exposure using the Typhoon FLA 9500 phosphor imager
References
- 1.Prospective Studies Collaboration Blood cholesterol and vascular mortality by age, sex, and blood pressure: a meta-analysis of individual data from 61 prospective studies with 55 000 vascular deaths. Lancet. 2007;370:1829–1839. doi: 10.1016/S0140-6736(07)61778-4. [DOI] [PubMed] [Google Scholar]
- 2.Platt F.M., Wassif C., Colaco A., Dardis A., Lloyd-Evans E., Bembi B., et al. Disorders of cholesterol metabolism and their unanticipated convergent mechanisms of disease. Annu. Rev. Genomics Hum. Genet. 2014;15:173–194. doi: 10.1146/annurev-genom-091212-153412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sturley S.L. Conservation of eukaryotic sterol homeostasis: new insights from studies in budding yeast. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2000;1529:155–163. doi: 10.1016/s1388-1981(00)00145-1. [DOI] [PubMed] [Google Scholar]
- 4.Taramino S., Teske B., Oliaro-Bosso S., Bard M., Balliano G. Divergent interactions involving the oxidosqualene cyclase and the steroid-3-ketoreductase in the sterol biosynthetic pathway of mammals and yeasts. Biochim. Biophys. Acta. 2010;1801:1232–1237. doi: 10.1016/j.bbalip.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gachotte D., Eckstein J., Barbuch R., Hughes T., Roberts C., Bard M. A novel gene conserved from yeast to humans is involved in sterol biosynthesis. J. Lipid Res. 2001;42:150–154. [PubMed] [Google Scholar]
- 6.Mialoundama A.S., Jadid N., Brunel J., di Pascoli T., Heintz D., Erhardt M., et al. Arabidopsis ERG28 tethers the sterol C4-demethylation complex to prevent accumulation of a biosynthetic intermediate that interferes with polar auxin transport. Plant Cell. 2013;25:4879–4893. doi: 10.1105/tpc.113.115576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mo C., Valachovic M., Randall S.K., Nickels J.T., Bard M. Protein-protein interactions among C-4 demethylation enzymes involved in yeast sterol biosynthesis. PNAS. 2002;99:9739–9744. doi: 10.1073/pnas.112202799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mo C., Bard M. A systematic study of yeast sterol biosynthetic protein-protein interactions using the split-ubiquitin system. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2005;1737:152–160. doi: 10.1016/j.bbalip.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Mo C., Bard M. Erg28p is a key protein in the yeast sterol biosynthetic enzyme complex. J. Lipid Res. 2005;46:1991–1998. doi: 10.1194/jlr.M500153-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Veitia R.A., Ottolenghi C., Bissery M.C., Fellous A. A novel human gene, encoding a potential membrane protein conserved from yeast to man, is strongly expressed in testis and cancer cell lines. Cytogenet. Cell Genet. 1999;85:3–4. doi: 10.1159/000015296. [DOI] [PubMed] [Google Scholar]
- 11.Lee C.N., Heidbrink J.L., McKinnon K., Bushman V., Olsen H., Fitzhugh W., et al. RNA interference characterization of proteins discovered by proteomic analysis of pancreatic cancer reveals function in cell growth and survival. Pancreas. 2012;41:84–94. doi: 10.1097/MPA.0b013e3182236385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Passantino R., Cascio C., Deidda I., Galizzi G., Russo D., Spedale G., et al. Identifying protein partners of CLN8, an ER-resident protein involved in neuronal ceroid lipofuscinosis. Biochim. Biophys. Acta Mol. Cell Res. 2013;1833:529–540. doi: 10.1016/j.bbamcr.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 13.Horton J.D., Goldstein J.L., Brown M.S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zerenturk E.J., Sharpe L.J., Brown A.J. Sterols regulate 3β-hydroxysterol Δ24-reductase (DHCR24) via dual sterol regulatory elements: cooperative induction of key enzymes in lipid synthesis by sterol regulatory element binding proteins. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2012;1821:1350–1360. doi: 10.1016/j.bbalip.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Bennett M.K., Osborne T.F. Nutrient regulation of gene expression by the sterol regulatory element binding proteins: increased recruitment of gene-specific coregulatory factors and selective hyperacetylation of histone H3 in vivo. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6340–6344. doi: 10.1073/pnas.97.12.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein J.L., Basu S.K., Brown M.S. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Met. Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 17.Krycer J.R., Kristiana I., Brown A.J. Cholesterol homeostasis in two commonly used human prostate cancer cell-lines, LNCaP and PC-3. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capell-Hattam I.M., Sharpe L.J., Qian L., Hart-Smith G., Prabhu A.v., Brown A.J. Twin enzymes, divergent control: the cholesterogenic enzymes DHCR14 and LBR are differentially regulated transcriptionally and post-translationally. J. Biol. Chem. 2020;295:2850–2865. doi: 10.1074/jbc.RA119.011323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott N.A., Sharpe L.J., Capell-Hattam I.M., Gullo S.J., Luu W., Brown A.J. The cholesterol synthesis enzyme lanosterol 14α-demethylase is post-translationally regulated by the E3 ubiquitin ligase MARCH6. Biochem. J. 2020;477:541–555. doi: 10.1042/BCJ20190647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navarro Gonzalez J., Zweig A.S., Speir M.L., Schmelter D., Rosenbloom K.R., Raney B.J., et al. The UCSC genome browser database: 2021 update. Nucl. Acids Res. 2021;49:D1046–D1057. doi: 10.1093/nar/gkaa1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevenson J., Krycer J.R., Phan L., Brown A.J. A practical comparison of ligation-independent cloning techniques. PLoS One. 2013;8 doi: 10.1371/journal.pone.0083888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luu W., Sharpe L.J., Stevenson J., Brown A.J. Akt acutely activates the cholesterogenic transcription factor SREBP-2. Biochim. Biophys. Acta. 2012;1823:458–464. doi: 10.1016/j.bbamcr.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 24.Zeng L., Liao H., Liu Y., Lee T.S., Zhu M., Wang X., et al. Sterol-responsive element-binding Protein (SREBP) 2 down-regulates ATP-binding cassette transporter A1 in vascular endothelial cells: a novel role of SREBP in regulating cholesterol metabolism. J. Biol. Chem. 2004;279:48801–48807. doi: 10.1074/jbc.M407817200. [DOI] [PubMed] [Google Scholar]
- 25.Howe V., Sharpe L.J., Prabhu A.V., Brown A.J. New insights into cellular cholesterol acquisition: promoter analysis of human HMGCR and SQLE, two key control enzymes in cholesterol synthesis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862:647–657. doi: 10.1016/j.bbalip.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 26.Grant C.E., Bailey T.L., Noble W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandelin A., Wasserman W.W., Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucl. Acids Res. 2004;32:W249–W252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucl. Acids Res. 2014;42:W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucl. Acids Res. 2016;44:W272–W276. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labun K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucl. Acids Res. 2019;47:W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cong L., Zhang F. In: Chromosomal Mutagenesis. Pruett-Miller S.M., editor. Springer New York; New York, NY: 2015. Genome engineering using CRISPR-Cas9 system BT - chromosomal mutagenesis; pp. 197–217. [Google Scholar]
- 33.Coates H.W., Capell-Hattam I.M., Brown A.J. The mammalian cholesterol synthesis enzyme squalene monooxygenase is proteasomally truncated to a constitutively active form. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong J., Quinn C.M., Brown A.J. Synthesis of the oxysterol, 24(S), 25-epoxycholesterol, parallels cholesterol production and may protect against cellular accumulation of newly-synthesized cholesterol. Lipids Health Dis. 2007;6:10. doi: 10.1186/1476-511X-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zerenturk E.J., Kristiana I., Gill S., Brown A.J. The endogenous regulator 24(S),25-epoxycholesterol inhibits cholesterol synthesis at DHCR24 (Seladin-1) Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2012;1821:1269–1277. doi: 10.1016/j.bbalip.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Kober D.L., Xu S., Li S., Bajaj B., Liang G., Rosenbaum D.M., et al. Identification of a degradation signal at the carboxy terminus of SREBP2: a new role for this domain in cholesterol homeostasis. Proc. Natl. Acad. Sci. U. S. A. 2020;117:28080–28091. doi: 10.1073/pnas.2018578117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vik A., Rine J. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell Biol. 2001;21:6395–6405. doi: 10.1128/MCB.21.19.6395-6405.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blattmann P., Henriques D., Zimmermann M., Frommelt F., Sauer U., Saez-Rodriguez J., et al. Systems pharmacology dissection of cholesterol regulation reveals determinants of large pharmacodynamic variability between cell lines. Cell Syst. 2017;5:604. doi: 10.1016/j.cels.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prabhu A.V., Sharpe L.J., Brown A.J. The sterol-based transcriptional control of human 7-dehydrocholesterol reductase (DHCR7): evidence of a cooperative regulatory program in cholesterol synthesis. Biochim. Biophys. Acta. 2014;1842:1431–1439. doi: 10.1016/j.bbalip.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 40.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharpe L.J., Brown A.J. In: Cholesterol Homeostasis: Methods and Protocols. Gelissen I.C., Brown A.J., editors. Springer New York; New York, NY: 2017. Identifying sterol response elements within promoters of genes; pp. 185–191. [DOI] [PubMed] [Google Scholar]
- 42.Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucl. Acids Res. 2018;46:D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inoue J., Sato R., Maeda M. Multiple DNA elements for sterol regulatory element-binding protein and NF-Y are responsible for sterol-regulated transcription of the genes for human 3-hydroxy-3-methylglutaryl coenzyme A synthase and squalene synthase. J. Biochem. 1998;123:1191–1198. doi: 10.1093/oxfordjournals.jbchem.a022060. [DOI] [PubMed] [Google Scholar]
- 44.Jackson S.M., Ericsson J., Mantovani R., Edwards P.A. Synergistic activation of transcription by nuclear factor Y and sterol regulatory element binding protein. J. Lipid Res. 1998;39:767–776. [PubMed] [Google Scholar]
- 45.Jackson S.M., Ericsson J., Osborne T.F., Edwards P.A. NF-Y has a novel role in sterol-dependent transcription of two cholesterogenic genes. J. Biol. Chem. 1995;270:21445–21448. doi: 10.1074/jbc.270.37.21445. [DOI] [PubMed] [Google Scholar]
- 46.Coates H.W., Chua N.K., Brown A.J. Consulting prostate cancer cohort data uncovers transcriptional control: regulation of the MARCH6 gene. Biochim. Biophys. Acta (Bba) - Mol. Cell Biol. Lipids. 2019;1864:1656–1668. doi: 10.1016/j.bbalip.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Mo C., Valachovic M., Bard M. The ERG28-encoded protein, Erg28p, interacts with both the sterol C-4 demethylation enzyme complex as well as the late biosynthetic protein, the C-24 sterol methyltransferase (Erg6p) Biochim. Biophys. Acta. 2004;1686:30–36. doi: 10.1016/j.bbalip.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 48.Luck K., Kim D.K., Lambourne L., Spirohn K., Begg B.E., Bian W., et al. A reference map of the human binary protein interactome. Nature. 2020;580:402–408. doi: 10.1038/s41586-020-2188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown A.J., Coates H.W., Sharpe L.J. Biochemistry of Lipids, Lipoproteins and Membranes. Elsevier; Amsterdam, Netherlands: 2021. Cholesterol synthesis; pp. 317–355. [Google Scholar]
- 50.Lindenthal B., Holleran A.L., Aldaghlas T.A., Ruan B., Schroepfer G.J., Wilson W.K., et al. Progestins block cholesterol synthesis to produce meiosis-activating sterols. FASEB J. 2001;15:775–784. doi: 10.1096/fj.00-0214com. [DOI] [PubMed] [Google Scholar]
- 51.Chen L., Ma M.Y., Sun M., Jiang L.Y., Zhao X.T., Fang X.X., et al. Endogenous sterol intermediates of the mevalonate pathway regulate HMGCR degradation and SREBP-2 processing. J. Lipid Res. 2019;60:1765–1775. doi: 10.1194/jlr.RA119000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horton J.D., Shah N.A., Warrington J.A., Anderson N.N., Park S.W., Brown M.S., et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cunningham D., Swartzlander D., Liyanarachchi S., Davuluri R.V., Herman G.E. Changes in gene expression associated with loss of function of the NSDHL sterol dehydrogenase in mouse embryonic fibroblasts. J. Lipid Res. 2005;46:1150–1162. doi: 10.1194/jlr.M400462-JLR200. [DOI] [PubMed] [Google Scholar]
- 54.Luu W., Hart-Smith G., Sharpe L.J., Brown A.J. The terminal enzymes of cholesterol synthesis, DHCR24 and DHCR7, interact physically and functionally. J. Lipid Res. 2015;56:888–897. doi: 10.1194/jlr.M056986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kedjouar B., de Médina P., Oulad-Abdelghani M., Payré B., Silvente-Poirot S., Favre G., et al. Molecular characterization of the microsomal tamoxifen binding site. J. Biol. Chem. 2004;279:34048–34061. doi: 10.1074/jbc.M405230200. [DOI] [PubMed] [Google Scholar]
- 56.Huang B., Song B.L., Xu C. Cholesterol metabolism in cancer: mechanisms and therapeutic opportunities. Nat. Metab. 2020;2:132–141. doi: 10.1038/s42255-020-0174-0. [DOI] [PubMed] [Google Scholar]
- 57.Nestler A., Sponder G., Rutschmann K., Mastrototaro L., Weise C., Vormann J., et al. Nature of SLC41A1 complexes: report on the split-ubiquitin yeast two hybrid assay. Magnes Res. 2013;26:56–66. doi: 10.1684/mrh.2013.0339. [DOI] [PubMed] [Google Scholar]
- 58.Veitia R.A., Hurst L.D. Accelerated molecular evolution of insect orthologues of ERG28/C14orf1: a link with ecdysteroid metabolism? J. Genet. 2001;80:17–21. doi: 10.1007/BF02811414. [DOI] [PubMed] [Google Scholar]
- 59.Capell-Hattam I.M., Brown A.J. Sterol evolution: cholesterol synthesis in animals is less a required trait than an acquired taste. Curr. Biol. 2020;30:R886–R888. doi: 10.1016/j.cub.2020.06.007. [DOI] [PubMed] [Google Scholar]
- 60.Oh K.H., Haney J.J., Wang X., Chuang C.F., Richmond J.E., Kim H. ERG-28 controls BK channel trafficking in the ER to regulate synaptic function and alcohol response in C. Elegans. Elife. 2017;6 doi: 10.7554/eLife.24733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.The UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucl. Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livingstone C.D., Barton G.J. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci. 1993;9:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data described in the article are contained within the article.