Abstract
Liraglutide, a glucagon-like peptide-1 analog, has beneficial metabolic effects in patients with type 2 diabetes and obesity. Although the high efficacy of liraglutide as an anti-diabetic and anti-obesity drug is well known, liraglutide-induced metabolic alterations in diverse tissues remain largely unexplored. Here, we report the changes in metabolic profiles induced by a 2-week subcutaneous injection of liraglutide in diet-induced obese mice fed a high-fat diet for 8 weeks. Our comprehensive metabolomic analyses of the hypothalamus, plasma, liver, and skeletal muscle showed that liraglutide intervention led to various metabolic alterations in comparison with diet-induced obese or nonobese mice. We found that liraglutide remarkably coordinated not only fatty acid metabolism in the hypothalamus and skeletal muscle but also amino acid and carbohydrate metabolism in plasma and liver. Comparative analyses of metabolite dynamics revealed that liraglutide rewired intertissue metabolic correlations. Our study points to a previously unappreciated metabolic alteration by liraglutide in several tissues, which may underlie its therapeutic effects within and across the tissues.
Keywords: obesity, diabetes, liraglutide, metabolomics, hypothalamus, plasma, liver, skeletal muscle
Abbreviations: DIO, diet-induced obese; GC-MS/MS, gas chromatography–tandem mass spectrometry; GLP-1, glucagon-like peptide-1; GLP-1R, GLP-1 receptor; HFD, high-fat diet; LC-MS/MS, liquid chromatography–tandem mass spectrometry; OPLS-DA, orthogonal partial least squares discriminant analysis; PCA, principal component analysis; VIP, variable influence on projection
Glucagon-like peptide-1 (GLP-1) is a gut-derived incretin hormone that induces insulin secretion, glucagon suppression, and appetite suppression through the signaling cascades of GLP-1 receptor (GLP-1R), which maintains glucose homeostasis and induces weight loss (1, 2, 3, 4). Owing to these beneficial actions, GLP-1 therapies have been applied in clinical development and are widely used for the treatment of type 2 diabetes mellitus and obesity (1, 3, 5). Nevertheless, due to the short biological half-life of native GLP-1 (rapid elimination within 5 min) (5, 6), long-acting GLP-1R agonists have been developed, for example, liraglutide, dulaglutide, albiglutide, and the most recent semaglutide (7). Among the currently available GLP-1R agonists, liraglutide has had the highest market penetration since its first approval by the Food and Drug Administration in 2010 (8, 9). Part of the liraglutide molecule is hydrophobic because palmitate is attached at Lys26, allowing reversible binding to albumin, and this binding extends its biological half-life up to 13 h (10, 11, 12). The clinical efficacy of liraglutide in the treatment of diabetes and obesity has been extensively documented (13, 14, 15, 16, 17, 18, 19). The prominent health benefits of liraglutide include weight loss owing to appetite suppression, reduction of body fat and inflammation, regulation of blood glucose level, induction of brown-fat thermogenesis, and browning of white adipose tissue (16, 20, 21, 22).
In conditions accompanied by diabetes, obesity, and related complications, liraglutide therapy improves the function of multiple tissues (2). Notably, liraglutide suppresses appetite by directly stimulating anorexigenic pathways in the arcuate nucleus of the hypothalamus through GLP-1R, which indirectly suppresses neurotransmission in orexigenic neurons (21). In addition, liraglutide improves impaired hepatic glucose and insulin sensitivity in the liver by enhancing the cyclic AMP signaling pathway, which increases the synthesis of glucose transporter 2 (23). Liraglutide also ameliorates impaired insulin action and increases glucose uptake in the skeletal muscle by increasing the glucose transporter 4 protein level via stimulation of the cyclic AMP signaling pathway (24). However, the metabolic changes induced by liraglutide as the integration of metabolic snapshots are poorly understood.
Metabolomics is a potent approach to surrogate diagnostics, which has advantages in identifying biomarkers and understanding the mechanisms of metabolic disorders such as diabetes and obesity (25, 26, 27). Metabolomics can also be used to assess drug efficacy, that is, therapeutic outcomes (28). Hence, metabolomics analysis of the effects of the known anti-diabetes and anti-obesity therapeutics, such as metformin, vildagliptin, and dipeptidyl peptidase-4 inhibitor, has been performed in patients or in vivo animal models (29, 30, 31, 32). A recent NMR spectroscopy–based urine metabolomics study has detected the effects of liraglutide treatment in diet-induced obese (DIO) mice: liraglutide decreased urine levels of nicotinamide metabolites and taurine and increased those of creatinine and creatine (33). Although urine metabolomics has advantages such as noninvasiveness and the ease of obtaining large volumes of samples, it is difficult to interpret the data on urine metabolites, which are in the final waste products and often do not represent actual changes in various tissues (34). Therefore, the integration of metabolomics data from multiple tissues is required for better understanding the aberrant metabolism at the sites of disease pathogenesis or during clinical challenges (pre- versus post-treatment).
Here, we report a comprehensive metabolomic study to understand the metabolic effects of liraglutide in the hypothalamus, plasma, liver, and skeletal muscle of DIO mice fed high-fat diet (HFD) using liquid chromatography–tandem mass spectrometry (LC-MS/MS) and gas chromatography–tandem mass spectrometry (GC-MS/MS). The major metabolites and metabolic pathways restored by liraglutide in each tissue of DIO mice were also identified in comparison to those in nonobese mice. Our metabolomic results highlight how liraglutide contributes to the therapeutic profile of GLP-1R agonism, offering insights into the clinical efficacy of liraglutide and the understanding of the systems-level effects on metabolic profile associated with metabolic diseases such as obesity and diabetes.
Results
Liraglutide ameliorates HFD-induced obesity
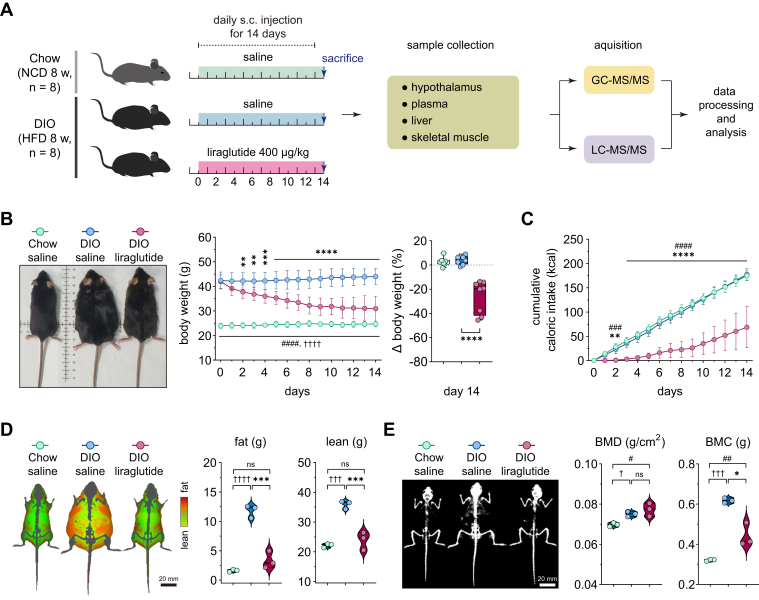
To investigate metabolic changes induced by liraglutide, mice were fed a normal chow diet or HFD for 8 weeks and were given daily subcutaneous injections of saline (hereafter referred to as DIO saline) or 400 μg/kg of liraglutide for 14 days (hereafter referred to as DIO liraglutide). Liraglutide has no effect on body weight, food intake, body fat, and glucose tolerance in nonobese mice, although it changes several pancreatic and adipogenic parameters in those mice (35, 36). We injected nonobese mice with saline as control (hereafter referred to as Chow saline) to investigate the beneficial effects of liraglutide on obese phenotypes and restoration of metabolism in obese animals. After the last injection, the hypothalamus, plasma, liver, and skeletal muscle were extracted and metabolites were analyzed using LC-MS/MS and GC-MS/MS (Fig. 1A).
Figure 1.
Liraglutide ameliorates diet-induced obesity. A, experimental design. A schematic flow diagram of animal experiments and metabolomic analysis. C57BL/6N male mice were fed NCD or HFD for 8 weeks, and liraglutide daily subcutaneous (s.c.) injections were performed for 14 days (n = 8). B, changes in body weight (n = 8). Left panel, representative images of the mice; middle panel, the time course of body weight measured daily; right panel, changes in body weight on day 14 relative to day 0. C, changes in cumulative caloric intake (n = 8). D, differences in fat and lean masses on day 14 (n = 3). Left panel, images of fat and lean masses, scale bar (20 mm). E, differences in bone minerals. Left panel, images of bone minerals (white), scale bar (20 mm). Statistical significance was determined by two-way ANOVA followed by Tukey post hoc test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 in DIO liraglutide versus DIO saline; †p < 0.05, †††p < 0.001, ††††p < 0.0001 in Chow saline versus DIO saline; #p < 0.05, ##p < 0.01, ### p < 0.001, ####p < 0.0001 in DIO liraglutide versus Chow saline. Data are mean ± SD. BMC, bone mineral content; BMD, bone mineral density; DIO, diet-induced obese; HFD, high-fat diet; NCD, normal chow diet.
Liraglutide resulted in a 26.1 ± 4.5% weight loss (Fig. 1B) and significantly reduced caloric intake in DIO mice (Fig. 1C). To assess the effect of liraglutide on body composition, we measured fat and lean masses using dual-energy X-ray absorptiometry after the last dose. Both masses were significantly reduced by liraglutide in DIO mice and to a level similar to that of Chow saline (Fig. 1D). Eight-week HFD significantly increased bone mineral density and contents in comparison with Chow saline. Liraglutide treatment of DIO mice for 14 days reduced bone mineral contents but not density in comparison with DIO saline (Fig. 1E). Collectively, these results indicate that liraglutide ameliorates obese phenotypes in DIO mice.
Liraglutide alters metabolomic profiles in DIO mice
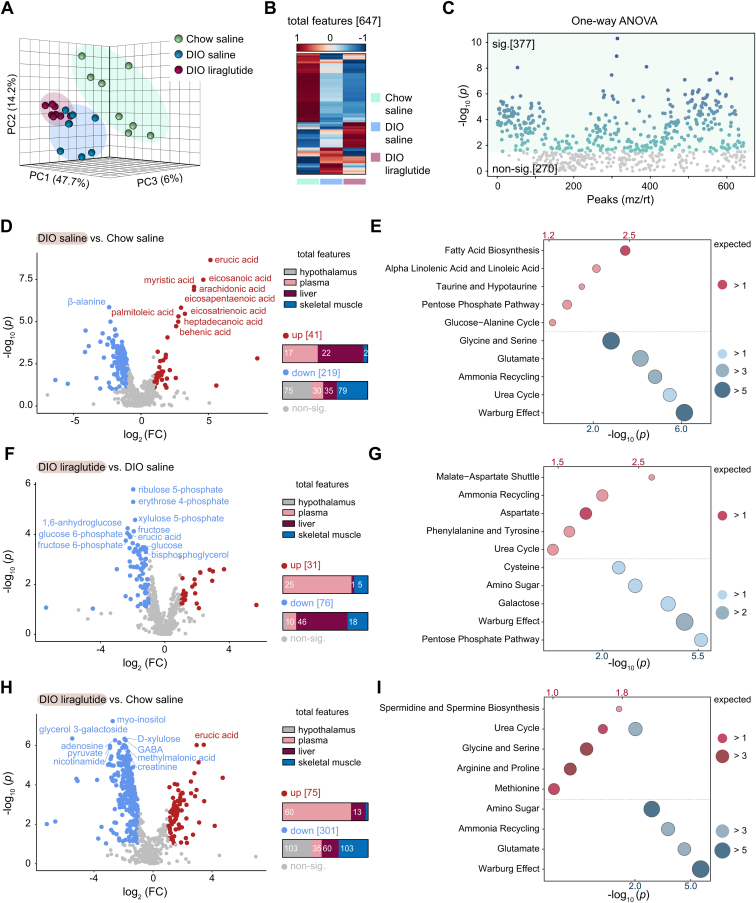
To assess changes in systemic metabolism induced by liraglutide in DIO mice compared to Chow saline mice, we performed targeted and nontargeted metabolomics and detected 647 annotated metabolites in total (125 in the hypothalamus, 178 in plasma, 193 in liver, and 151 in skeletal muscle). Unsupervised principal component analysis (PCA) revealed the effects of HFD and liraglutide on the metabolome. The components containing metabolomic features were clearly distinguishable and did not overlap among the three groups (Chow saline, DIO saline, and DIO liraglutide) (Fig. 2A). In a heatmap, the 647 metabolites displayed a different pattern in each group (Fig. 2B). Among these metabolites, 377 showed a significant difference in response to either HFD or liraglutide (Fig. 2C).
Figure 2.
Comparative metabolomics of the Chow saline, DIO saline, and DIO liraglutide groups. A, principal component analysis (PCA) plot of features. B, heatmap of 647 annotated metabolites. The color bar indicates Z-score (from −1–1). C, one-way ANOVA plot of signature metabolites among the three groups, including 377 metabolites that showed significant differences. D‒I, pairwise comparisons between the indicated groups. D, F, and H, volcano plots. The numbers of upregulated and downregulated metabolites in each tissue are shown as bar graphs. Fold change is abbreviated as FC. E, G, and I, metabolite set enrichment analyses. Red circles, upregulated metabolic pathways; blue circles, downregulated metabolic pathways; the corresponding p values are indicated above and below the plots in the same colors. Circle size indicates the expected enrichment value for a metabolic pathway. sig., significant (darker color indicates higher p values); non-sig., nonsignificant (gray); DIO, diet-induced obese.
To determine changes in the metabolome and metabolic pathways caused by HFD or liraglutide, pairwise comparisons were performed (DIO saline versus Chow saline, DIO liraglutide versus DIO saline, and DIO liraglutide versus Chow saline) (Fig. 2, D‒I). Each group was obviously distinct from the corresponding comparison group in PCA analysis (Fig. S1, A‒C). Supervised orthogonal partial least squares discriminant analysis (OPLS-DA) also showed a clear distinction from the comparison groups (Fig. S1, D‒F). We estimated the variable influence on projection (VIP) scores, which indicate the importance of metabolites to the OPLS-DA model, and listed the scores of the top 10 metabolites (Fig. S1, G‒I). Metabolites with significant differences and their metabolites are shown through volcano plots showing the fold changes and p values (Fig. 2, D, F, and H).
HFD significantly increased 41 metabolites in the plasma, liver, and skeletal muscle (17, 22, and 2 metabolites, respectively), mainly long-chain fatty acids and mono/polyunsaturated fatty acids (Fig. 2D). HFD significantly decreased 219 metabolites, mainly in the hypothalamus and skeletal muscle (75 and 79 metabolites, respectively). To identify the metabolic pathways enriched by HFD, we performed a metabolite set enrichment analysis (top 5) for either increased or decreased metabolites. The increased metabolites impacted the robust enrichment of metabolic pathways involved in fatty acid biosynthesis, alpha linolenic acid/linoleic acid, taurine/hypotaurine, pentose phosphate pathway, and glucose‒alanine cycle metabolism, whereas the decreased metabolites impacted the Warburg effect, urea cycle, ammonia recycling, glutamate, and glycine/serine metabolism (Fig. 2E).
Liraglutide increased 31 metabolites, mostly in plasma (25 metabolites) and more than half of the 76 decreased metabolites were detected in the liver (46 metabolites) (Fig. 2F). Notably, most of the decreased metabolites are the intermediates of the pentose phosphate pathway or Warburg effect in the liver. Consistently, the decreased metabolites were enriched in the pentose phosphate pathway, Warburg effect, galactose, amino sugar metabolism, and cysteine metabolism, while the increased metabolites were enriched in the malate‒aspartate shuttle, ammonia recycling, aspartate, phenylalanine/tyrosine metabolism, and urea cycle (Fig. 2G). Our results suggested that liraglutide decreased the pentose phosphate pathway increased by HFD but elevated the urea cycle and ammonia recycling decreased by HFD (Fig. 2, E and G).
The greatest number of metabolites differed between DIO liraglutide and Chow saline. Metabolites in DIO liraglutide were increased mostly in plasma (60 metabolites) and decreased in the hypothalamus and skeletal muscle (103 metabolites in each) (Fig. 2H). The increased metabolites were enriched in spermidine/spermine biosynthesis and metabolism of amino acids including glycine, serine, arginine, proline, and methionine, whereas the decreased metabolites were highly enriched in the Warburg effect, glutamate, ammonia recycling, and amino sugar metabolism. The urea cycle overlapped with upregulated and downregulated metabolic pathways (Fig. 2I).
Hypothalamus-specific metabolome responses to liraglutide
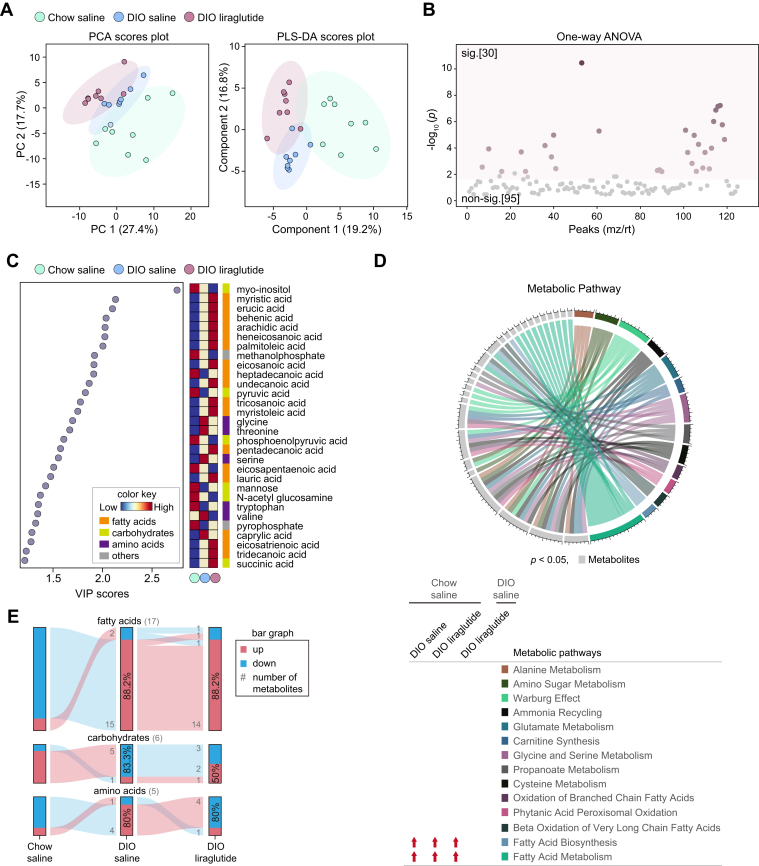
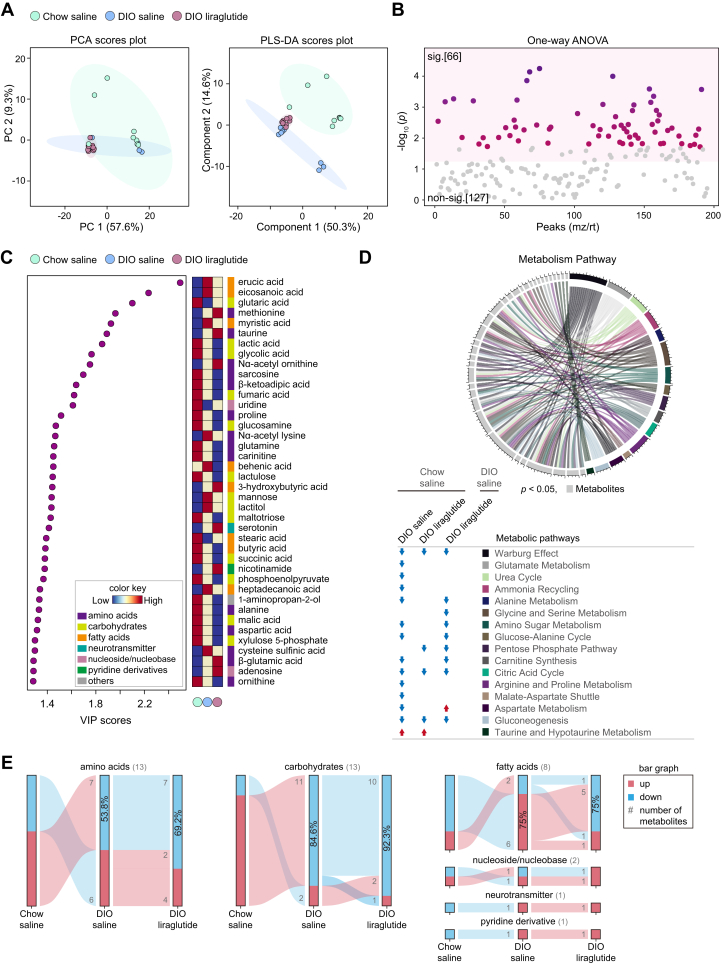
The hypothalamus is a key brain area controlling energy balance and glucose metabolism via numerous metabolic signals such as lipids and amino acids which play a critical role in regulating food intake (37, 38, 39, 40). Although liraglutide reduces appetite by activating GLP-1R in anorexigenic neurons in the hypothalamus (21, 41, 42), the metabolic changes caused by liraglutide in the hypothalamus are poorly understood. Our PCA analysis revealed that the components differed slightly among the three mouse groups, but they did not overlap in partial least squares discriminant analysis (PLS-DA; Fig. 3A). Among 125 annotated metabolites, 30 metabolites showed significantly different responses among the three groups (Fig. 3B). All metabolites were compared in group pairs by OPLS-DA, resulting in clear distinction (Fig. S2, A, C, and E).
Figure 3.
Metabolic alterations induced by liraglutide in the hypothalamus. A, PCA plot of features and partial least squares discriminant analysis (PLS-DA) plot. B, one-way ANOVA plot of signature metabolites among the three groups, including 30 metabolites that showed significant differences. C, variable influence on projection (VIP) score plot (cut-off ≥ 1.2). Color key shows the levels of metabolites and their categories. D, circular plot of metabolic pathways based on metabolite set enrichment analysis (p < 0.05). Metabolites are shown in light gray and metabolic pathways in various colors. The metabolic pathways increased in each group compared to Chow saline or DIO saline are indicated by red arrows. E, flow charts of metabolome changes. Upregulation is shown in red and downregulation in blue. The numbers of upregulated and downregulated metabolites are shown in gray, and the corresponding percentages are shown in bar graphs. sig., significant (darker color indicates higher p values); non-sig., nonsignificant (gray); DIO, diet-induced obese; PCA, principal component analysis.
HFD increased the metabolites of fatty acid metabolism, namely myristic, erucic, behenic, arachidic, heneicosanoic, lignoceric, palmitoleic, eicosanoic, undecanoic, tricosanoic, myristoleic, pentadecanoic, lauric, eicosatrienoic, and tridecanoic acids, which were further increased by liraglutide (Fig. 3C). These results were also consistent between the two pairs of comparison groups (Fig. S2, B, D, and F). Notably, erucic, behenic, arachidic, and lignoceric acids are products of fatty acid biosynthesis in which enzymes synthesize fatty acids from the substrates such as malonyl-CoA, acetyl-CoA, and NADPH. HFD also increased glycine, threonine, and valine, but liraglutide decreased these amino acids (Fig. 3C). Adenosine was decreased in DIO liraglutide compared to both DIO saline and Chow saline (Fig. S2, D, and F).
To identify the metabolic pathways impacted by HFD or liraglutide, we performed metabolite set enrichment analysis for 30 significant metabolites in each group and identified metabolic pathways common to all three groups. Fatty acid biosynthesis and fatty acid metabolism, which includes fatty acid biosynthesis, were specifically increased in DIO saline and DIO liraglutide compared to Chow saline. Liraglutide markedly increased fatty acid metabolism, including biosynthesis, in the hypothalamus (Fig. 3D).
To understand the increase or decrease of metabolite classes by HFD or liraglutide, we displayed the flow charts of metabolites. Among the 28 metabolites that showed significant differences (VIP score ≥ 1.2) in the plasma of the three groups, 88.2% of fatty acids and 80% of amino acids were increased in DIO saline compared to Chow saline; 88.2% of fatty acids were further increased and 80% amino acids were decreased in DIO liraglutide; 83.3% of carbohydrates were decreased in DIO saline compared to Chow saline but 50% of carbohydrates were increased in DIO liraglutide (Fig. 3E). Overall, liraglutide mainly increased fatty acids and decreased amino acids that were increased by HFD in the hypothalamus.
Plasma-specific metabolome responses to liraglutide
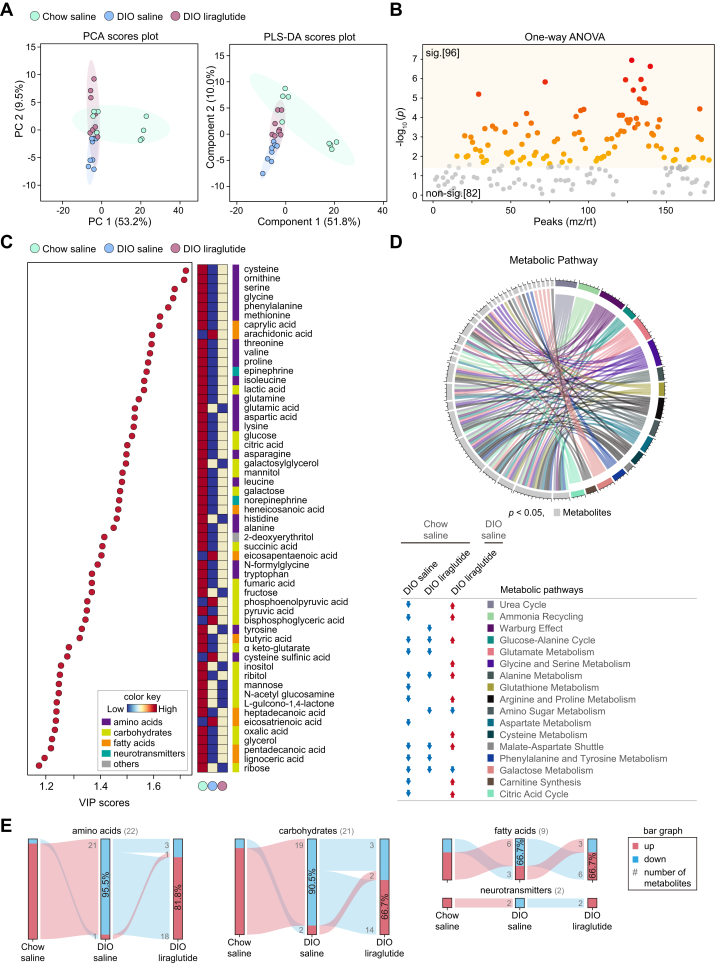
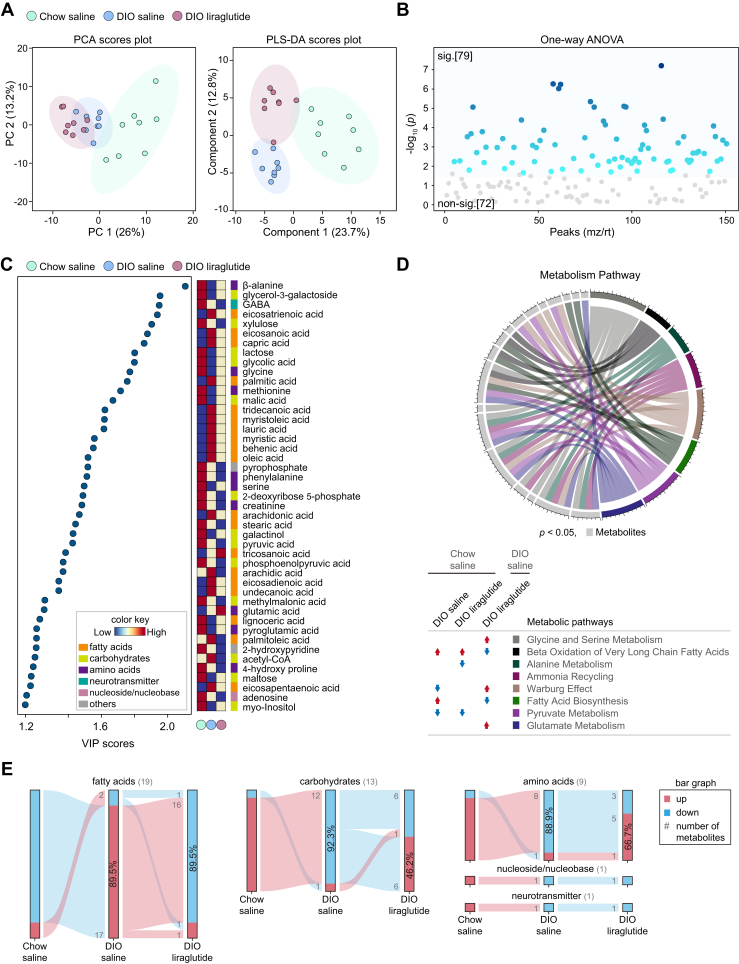
Plasma metabolomics is widely used in clinical and biological studies on metabolic diseases (43). The components were not dramatically distinct among the three groups either in PCA or in PLS-DA (Fig. 4A). Among 178 annotated metabolites, 96 metabolites showed significantly different responses among the three groups (Fig. 4B). OPLS-DA between two groups showed clear differences (Fig. S3, A, C, and E).
Figure 4.
Metabolic alterations in plasma by liraglutide. A, PCA and PLS-DA plots of features. B, one-way ANOVA plot of signature metabolites among the three groups, including 96 metabolites that showed significant differences. C, VIP score plot (cut-off ≥ 1.2). Color key shows the levels of metabolites and their categories. D, circular plot of metabolic pathways based on metabolite set enrichment analysis (p < 0.05). Metabolites are shown in light gray and metabolic pathways in various colors. The metabolic pathways increased and decreased in each group compared to Chow saline or DIO saline are indicated by red and blue arrows, respectively. E, flow chart of metabolome changes. Upregulation is shown in red and downregulation in blue. The numbers of upregulated and downregulated metabolites are shown in gray, and the corresponding percentages are shown in bar graphs. sig., significant (darker color indicates higher p values); non-sig., nonsignificant (gray); DIO, diet-induced obese; PCA, principal component analysis; PLS-DA, partial least squares discriminant analysis; VIP, variable influence on projection.
HFD decreased but liraglutide increased the levels of essential or nonessential amino acids including branched-chain amino acids (Figs. 4C, S3, B, and D). On the contrary, glutamic acid, histidine, and tyrosine, which were decreased by HFD, were further decreased by liraglutide (Fig. 4C). Compared to Chow saline, liraglutide decreased cysteine, glycine, glutamate, serine, methionine, phenylalanine, histidine, glutamine, and tyrosine (Fig. 3F). HFD also decreased the intermediates of glycolysis, citric acid cycle, and epinephrine/norepinephrine compared to Chow saline; interestingly, liraglutide reorganized the levels of these metabolites (Fig. 4C). In addition, both of HFD and liraglutide increased arachidonic acid and taurine compared to those in Chow saline (Fig. S3, B, and F).
Compared to Chow saline, DIO saline tended to decrease almost all metabolic pathways that were common among the three groups, including amino acid metabolism. Intriguingly, liraglutide restored most of the metabolic pathways severely reduced by HFD, except for amino sugar and galactose metabolism, and additionally enhanced cysteine metabolism in the plasma of DIO mice (Fig. 4D).
Among the 54 metabolites that showed significant differences (VIP score ≥ 1.2) in the plasma of the three groups, 95.5% of amino acids, 90.5% of carbohydrates, and 66.7% of fatty acids were decreased in DIO saline compared to Chow saline; 81.8% of amino acids, 66.7% of carbohydrates, and 66.7% of fatty acids were increased in DIO liraglutide compared to DIO saline. The neurotransmitters epinephrine and norepinephrine were reduced in DIO saline compared to Chow saline and increased in DIO liraglutide compared to DIO saline (Fig. 4E). Taken together, these data highlight that liraglutide restores the plasma metabolism of amino acids, carbohydrates, fatty acids, and neurotransmitters that was reduced by HFD.
Liver-specific metabolome responses to liraglutide
Metabolomics analysis in the liver showed that the components of DIO saline and DIO liraglutide mostly overlapped in PCA, but not in PLS-DA (Fig. 5A). Among 193 annotated metabolites, 66 metabolites showed significantly different responses among the three groups (Fig. 5, B). OPLS-DA between two groups showed clear differences (Fig. S4, A, C, and E).
Figure 5.
Metabolic alterations in the liver by liraglutide. A, PCA and PLS-DA plots of features. B, one-way ANOVA plot of signature metabolites among the three groups, including 66 metabolites that showed significant differences. C, VIP score plot (cut-off ≥ 1.2). Color key shows the levels of metabolites and their categories. D, circular plot of metabolic pathways based on metabolite set enrichment analysis (p < 0.05). Metabolites are shown in light gray and in various colors. The metabolic pathways increased and decreased in each group compared to Chow saline or DIO saline are indicated by red and blue arrows, respectively. E, flow chart of metabolome changes. Upregulation is shown in red and downregulation in blue. The numbers of upregulated and downregulated metabolites are shown in gray, and the corresponding percentages are shown in bar graphs. sig., significant (darker color indicates higher p values); non-sig., nonsignificant (gray); DIO, diet-induced obese; PCA, principal component analysis; PLS-DA, partial least squares discriminant analysis; VIP, variable influence on projection.
HFD increased fatty acids such as erucic, eicosanoic, myristic, behenic, and heptadecanoic acids, whereas liraglutide reduced fatty acids (erucic, lauric, myristic, palmitic, arachidic, behenic, heneicosanoic, myristoleic, palmitoleic, oleic, and arachidonic acids) in comparison with DIO liraglutide and DIO saline (Figs. 5C, S4, B, and D). HFD also increased methionine, taurine, Nα-acetyl ornithine, and β-glutamic acid. Notably, these amino acids were further increased by liraglutide. Proline, glutamine, alanine, aspartic acid, and ornithine were decreased by HFD and were further decreased by liraglutide (Fig. 5C). Amino acids such as glutamic acid, aspartic acid, serine, methionine, pyroglutamic acid, alanine, β-alanine, and β-glutamic acid were increased in DIO liraglutide compared to Chow saline. Taurine and nicotinamide were increased in DIO saline and DIO liraglutide compared to Chow saline (Fig. S4F). In carbohydrate metabolism, HFD decreased glycolic acid, glucosamine, lactulose, intermediates of glycolysis such as lactic acid and phosphoenolpyruvate, intermediates of the citric acid cycle such as fumaric, succinic, and malic acids, and xylulose 5-phosphate in the pentose phosphate pathway, whereas liraglutide further decreased all of these metabolites (Fig. 5C). Compared to Chow saline, liraglutide decreased carbohydrates including 1-methylgalactose, glycolic acid, mannitol, succinic acid, and sophorose (Fig. S4F). Serotonin, nicotinamide, and adenosine were increased by HFD and were further increased by liraglutide (Fig. 5C).
Compared to Chow saline, DIO saline tended to decrease most liver metabolic pathways, including amino acid and carbohydrate metabolism, although not taurine/hypotaurine metabolism, which was increased in both DIO saline and DIO liraglutide compared to Chow saline. Compared with DIO saline, liraglutide increased aspartate metabolism, which was decreased by HFD, while other metabolic pathways were further reduced by liraglutide (Fig. 5D).
Among the 38 metabolites that showed significant differences (VIP score ≥ 1.2) in the liver of the three groups, 53.8% of amino acids and 84.6% of carbohydrates were decreased in DIO saline compared to Chow saline; 69.2% of amino acids and 92.3% of carbohydrates were further decreased in DIO liraglutide; 75% of fatty acids were increased in DIO saline compared to Chow saline but 75% of fatty acids were decreased in DIO liraglutide. While adenosine (nucleoside or nucleobase) was increased and uridine was decreased in DIO saline compared to Chow saline, both of them were increased by liraglutide. Serotonin (neurotransmitter) and nicotinamide (pyridine derivative) were increased in DIO saline compared to Chow saline, and they were further increased by liraglutide (Fig. 5E). Overall, liraglutide decreased amino acids, carbohydrates, fatty acids, neurotransmitter, and pyridine increased by HFD in the liver.
Skeletal muscle–specific metabolome responses to liraglutide
Metabolomics analysis in the skeletal muscle showed that the components of DIO saline and DIO liraglutide partially overlapped in PCA but were completely distinct in PLS-DA (Fig. 6A). Among 151 annotated metabolites, 79 metabolites showed significantly different responses among the three groups (Fig. 6B). Paired OPLS-DA between two groups showed clear differences Fig. S5, A, C, and E).
Figure 6.
Metabolic alterations in the skeletal muscle by liraglutide. A, PCA and PLS-DA plots of features. B, one-way ANOVA plot of signature metabolites among the three groups, including 79 metabolites had significant differences C, VIP score plot (cut-off ≥ 1.2). Color key shows the levels of metabolites and their categories. D, circular plot of metabolic pathways based on metabolite set enrichment analysis (p < 0.05). Metabolites are shown in light gray and metabolic pathways in various colors. Color key indicates metabolic pathways. The metabolic pathways increased and decreased in each group compared to Chow saline or DIO saline are indicated by red and blue arrows, respectively. E, flow chart of metabolome changes. Upregulation is shown in red and downregulation in blue. The numbers of upregulated and downregulated metabolites are shown in gray, and the corresponding percentages are shown in bar graphs. sig., significant (darker color indicates higher p values); non-sig., nonsignificant (gray); DIO, diet-induced obese; PCA, principal component analysis; PLS-DA, partial least squares discriminant analysis; VIP, variable influence on projection.
HFD significantly increased fatty acids such as capric, undecanoic, lauric, tridecanoic, myristic, palmitic, behenic, eicosanoic, myristoleic, oleic, palmitoleic, eicosadienoic, eicosatrienoic, arachidonic, and eicosapentaenoic acids (Fig. S5B). Notably, liraglutide markedly reduced all of those fatty acids (Fig. 6C). In amino acid metabolism, HFD decreased glycine, methionine, serine, and phenylalanine, and liraglutide restored these amino acids except phenylalanine. Glutamic acid, which was increased by HFD, was further increased by liraglutide. Conversely, creatinine, which was decreased by HFD, was further decreased by liraglutide (Fig. 6C). Tyrosine, leucine, cysteine, and lysine were increased in DIO liraglutide compared to DIO saline (Fig. S5D). HFD decreased carbohydrates, and liraglutide further decreased intermediates of glycolysis (pyruvate, fructose-1,6-bisphosphate, phosphoenolpyruvate), pentose phosphate pathway (ribose 5-phosphate, sedoheptulose 7-phosphate), and citric acid cycle (succinic acid) (Fig. 6C). Intermediates of glycolysis and pentose phosphate pathway were decreased, but amino acids were increased in DIO liraglutide compared to Chow saline (Fig. S5F).
In the skeletal muscle, acetyl-CoA along with fatty acids that are the metabolites of β-oxidation of very long chain fatty acids (linear fatty acids with more than 18 carbons: eicosatrienoic, eicosanoic, behenic, arachidonic, arachidic, eicosadienoic, and eicosapentaenoic acids) and fatty acid biosynthesis (palmitic, behenic, oleic, arachidic, and palmitoleic acids) were increased in DIO saline compared to Chow saline. On the contrary, these metabolic pathways were reduced in DIO liraglutide compared to DIO saline (Fig. 6D). In addition, liraglutide increased Warburg effect metabolism, which was decreased by HFD and increased amino acids such as glycine, serine and glutamate, which were not changed by HFD (Fig. 6D).
Among the 43 metabolites that showed significant differences (VIP score ≥ 1.2) in the skeletal muscle of the three groups, 89.5% of fatty acids were increased and 92.3% of carbohydrates and 88.9% of amino acids were decreased in DIO saline compared to Chow saline; 89.5% of fatty acids were decreased but 46.2% of carbohydrates and 66.7% of amino acids were increased in DIO liraglutide. Adenosine (nucleoside or nucleobase) and GABA (neurotransmitter) were decreased in DIO saline compared to Chow saline, and they were further decreased by liraglutide (Fig. 6E). Overall, liraglutide dramatically decreased fatty acids increased by HFD in the skeletal muscle and partially restored amino acid and carbohydrate metabolism.
Liraglutide rewires tissue- or group-specific metabolite correlations
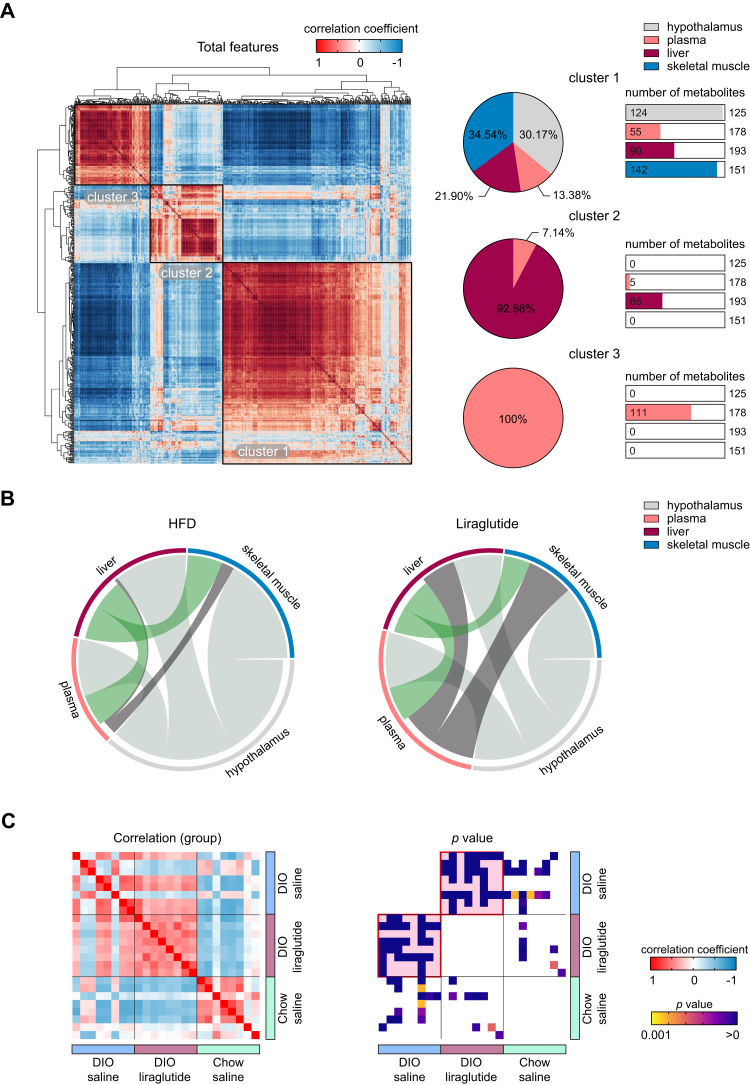
To characterize the metabolic proximity and interrelations among significantly changed metabolites, we compared cross-tissue correlations in the hypothalamus, plasma, liver, and skeletal muscle using Pearson’s correlation coefficients analysis. These 647 metabolites analyzed irrespective of tissues showed three distinct clusters (Fig. 7A). Cluster 1 included 124 (30.17%) metabolites from the hypothalamus, 55 (13.38%) from plasma, 90 (21.90%) from the liver, and 142 (34.54%) from skeletal muscle. In Cluster 1, all tested tissues and plasma metabolites had high correlation with each other. Cluster 2 included 5 (7.14%) metabolites from plasma and 65 (92.86%) from the liver, indicating that plasma and liver metabolites correlated with each other. Cluster 3 consisted of 111 plasma metabolites only.
Figure 7.
Liraglutide rewires correlations of randomized metabolites. A, correlation plot of randomized total metabolites based on Pearson’s correlation coefficients. The percentages of tissue-specific metabolites belonging to clusters 1 to 3 are shown as pie charts. For each cluster, the numbers of metabolites in plasma and each tissue are shown as bar graphs; the total numbers of metabolites detected are shown on the right side of the bar graphs. B, mapping of intertissue metabolite correlations. Ribbons connecting metabolite intertissue classes indicate significant correlations between metabolites. Ribbon thickness indicates the number of significantly correlated metabolites. C, plots of intergroup correlations and p values. Pink background in the p value plot indicates the most significant group between the paired groups.
The intertissue correlation analysis showed that liraglutide mainly enhanced plasma‒liver and plasma‒skeletal muscle correlations compared with those of HFD, but not plasma‒hypothalamus correlations (Fig. 7B). The intergroup correlation analysis showed that DIO liraglutide was less correlated with Chow saline, whereas DIO liraglutide showed greater correlations with DIO saline (Fig. 7C). Liver metabolome had the best intergroup correlation (Fig. S6). These data suggest that liraglutide rewired metabolic correlations and enhanced connectivity within the tested biological matrices.
Beneficial effects of liraglutide on metabolic remodeling in DIO mice
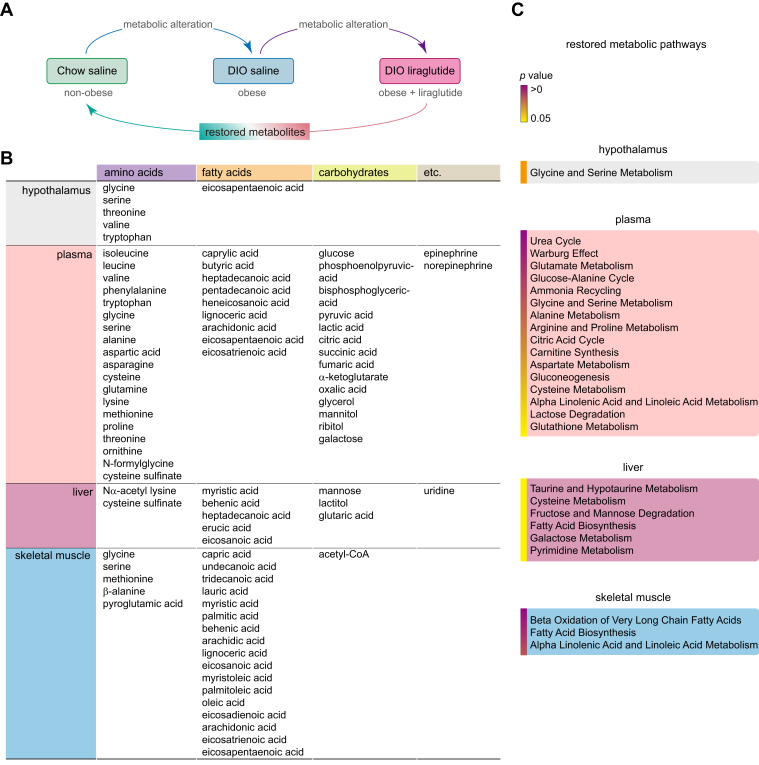
To investigate the beneficial effects of liraglutide on the restoration of metabolism in obese mice, we compared metabolomes in liraglutide-treated DIO mice with those in nonobese mice (Fig. 8A). HFD induced aberrant changes in metabolism compared to that in nonobese mice, whereas liraglutide remodeled metabolomes in diverse tissues and plasma of DIO mice. We summarized the major metabolites and metabolic pathways restored by liraglutide in DIO mice compared to the levels in nonobese mice (Fig. 8, B and C).
Figure 8.
Metabolites and metabolic pathways restored by liraglutide in DIO mice. A, a schematic diagram of comparative metabolomes in Chow saline (nonobese), DIO saline (obese), and DIO liraglutide (obese + liraglutide) groups. B, a table of metabolites restored by liraglutide in DIO mice. Restored metabolites in each tissue and plasma were listed. C, a list of metabolic pathways restored by liraglutide in DIO mice. Restored metabolic pathways in each tissue and plasma were organized. DIO, diet-induced obese.
In the hypothalamus, glycine and serine metabolism (glycine, serine, and threonine) were mainly restored by liraglutide in DIO mice compared to those in nonobese mice (Fig. 8, B and C). In the plasma, amino acids and carbohydrates had the largest number of metabolites restored by liraglutide compared to other tissues. Consequently, individual amino acid metabolism (glutamate, glycine/serine, alanine, arginine/proline, aspartate, and cysteine) and carbohydrate metabolism (Warburg effect, citric acid cycle, gluconeogenesis, and lactose degradation) were mainly restored, along with the restoration of their associated metabolic pathways (urea cycle, glucose-alanine cycle, ammonia recycling, carnitine synthesis, and glutathione metabolism). In addition, α-linolenic acid/linoleic acid metabolism (arachidonic, eicosapentaenoic, and eicosatrienoic acid) were restored by liraglutide (Fig. 8, B and C). In the metabolic pathway analysis of the liver, taurine/hypotaurine metabolism (cysteine sulfinate) and fatty acids biosynthesis (myristic, behenic, heptadecanoic, erucic, and eicosanoic acid) were restored by liraglutide (Fig. 8, B and C). Compared to other tissues or plasma, skeletal muscle had the largest number of fatty acids restored by liraglutide, affecting the related pathways such as β-oxidation of very long chain fatty acids, fatty acid biosynthesis, and α-linolenic acid/linoleic acid metabolism (Fig. 8, B and C).
Overall, liraglutide remodeled metabolic profiling including major metabolites and their metabolic pathways in DIO mice in a tissue-specific manner, which was restored to the level of a nonobese state, suggesting the beneficial effects of liraglutide in altering metabolism in diverse tissues and plasma.
Discussion
Many metabolic signatures have been commonly used in clinical practice, and their drug responses have been studied in multiple target tissues through metabolomics (25). Despite the excellent efficacy of liraglutide in rodents and humans with diabetes or obesity, metabolomic study on the effects of liraglutide in multiple tissues is very rare. A study that used NMR spectroscopy–based metabolomics of the urine of liraglutide-treated DIO mice has shown decreases in NAD+, taurine, and trigonelline and increases in creatine and creatinine (33). Here, we used comprehensive metabolomics, that is, integrated analysis of targeted and untargeted metabolomics using LC-MS/MS or GC-MS/MS, to identify metabolites changed by liraglutide in the hypothalamus, plasma, liver, and skeletal muscle in DIO mice. We compared the metabolic profiles of liraglutide-treated DIO mice with those of nonobese and obese mice to uncover tissue-specific changes in metabolites and metabolomic remodeling among multiple tissues. Liraglutide remarkably coordinated not only fatty acid metabolism in the hypothalamus and skeletal muscle but also amino acid and carbohydrate metabolism in plasma and liver.
Saturated fatty acids such as palmitic and stearic acids lead to dyslipidemia and inflammation in the hypothalamus, and consequently, dysregulate food intake, energy balance, and glucose homeostasis (44, 45). On the contrary, long-chain monounsaturated and polyunsaturated fatty acids such as oleic and palmitoleic acids improve energy homeostasis (46, 47, 48). Our data showed that palmitic, stearic, and oleic acids were not changed by liraglutide, but palmitoleic acid and polyunsaturated fatty acids were significantly elevated by liraglutide in the hypothalamus of obese mice compared to nonobese mice. The elevation of hypothalamic palmitoleic acid by liraglutide may be involved in changes in food intake, energy balance, and glucose homeostasis induced by liraglutide. In the skeletal muscle, liraglutide decreased the levels of fatty acids related to β-oxidation of very long chain fatty acids, which might be due to an increase in β-oxidation. These data are consistent with a previous report that liraglutide promotes β-oxidation in the skeletal muscle of DIO rats (49).
The levels of branched-chain amino acids (leucine, isoleucine, and valine) in the plasma are well known as prominent metabolic features for diabetes and obesity (50). Notably, circulating branched-chain amino acids activate the mTORC1 signaling pathway in pancreatic β cells, which promotes insulin secretion (51, 52). Our results showed that plasma branched-chain amino acids decreased by HFD were restored by liraglutide. Furthermore, our data on the metabolic profiles of numerous essential or nonessential amino acids, which can be used as substrates for the citric acid cycle, urea cycle, and glycolysis or gluconeogenesis, provide a better understanding of the function of amino acids in the liraglutide therapy.
Nicotinamide alleviates liver fibrosis or steatosis in patients with nonalcoholic fatty liver disease by elevating hepatic NAD+ which inhibits lipogenesis and activates fatty acid oxidation (53). Urinary NAD+ metabolism such as N1-methyl-2-pyridone-5-carboxamide and N1-methyl-4-pyridone-3-carboxamide are increased in DIO mice (54) and are decreased by liraglutide (33). We observed an increase in the levels of nicotinamide by HFD in the liver and a further increase by liraglutide. Nicotinamide is catabolized to nicotinic acid, nicotinamide N-oxide, and N1-methyl nicotinamide through the body’s circulation, and these compounds are excreted in the urine (55). Therefore, the levels of nicotinamide metabolites may vary depending on nicotinamide catabolism and tissues affected by liraglutide.
Taurine was dramatically decreased in the urine of liraglutide-treated DIO mice, providing evidence for the amelioration of obesity-induced liver steatosis by liraglutide (33). Increased taurine in the urine indicates liver damage, which increases taurine synthesis in the liver and consequently excretion into the urine (56). On the other hand, other studies found that increased taurine in the liver has beneficial effects, as it lowers lipid levels by increasing the activity of antioxidant enzymes and reducing inflammatory responses (57, 58, 59). Our data showed that taurine levels increased by HFD in the liver were further elevated by liraglutide. Thus, it is noteworthy that liraglutide induced opposite changes in taurine levels in the urine and liver, which is indicative of a beneficial effect of liraglutide in the liver.
Plasma–liver and plasma–skeletal muscle metabolic correlations were enhanced in liraglutide-treated DIO mice, suggesting that circulating liraglutide had the most impact on metabolites in the plasma. Plasma, which mediates the crosstalk between tissues, likely reflected the high correlations of metabolites in the liver and skeletal muscle. Intergroup correlations showed that metabolic profiles of DIO liraglutide resembled those of DIO saline, but not those of Chow saline. This might be due to the fact that liraglutide does not completely revert the obese phenotype to a nonobese state, but makes the phenotype milder (19, 60, 61). Comprehensive studies on intra-tissue and intertissue metabolic correlations, which would consider the spatiotemporal and functional distributions of and communication among intertissue metabolic flows, are needed for elucidation of obesogenic relationships and treatment responses to liraglutide.
Liraglutide mimicked caloric restriction in mice, resulting in dramatic anorexia and weight loss in our study and others (20, 21, 36). Several metabolomic studies on caloric restriction using a very low-calorie diet, a fasting-mimicking diet, or fasting identified metabolomic signatures in the serum and liver (62, 63, 64, 65, 66, 67, 68). Caloric restrictions using these diets or acute fasting markedly increased 3-hydroxybutyric acid, a ketone body, in the serum and liver of both NCD and HFD mice (62, 63, 64, 65, 66). Interestingly, our data showed that liraglutide increased 3-hydroxybutyric acid in the liver but not in the plasma of HFD mice. Caloric restrictions using either a very low-calorie diet or a fasting-mimicking diet decreased the levels of isoleucine, valine, and proline in the serum and glycine and taurine in the liver under both NCD and HFD (62, 65, 66, 67). In contrast, our study showed that liraglutide increased the levels of those amino acids in the plasma and liver of HFD mice, respectively. Taken together, our results suggest that the metabolomic changes by liraglutide reflect integrated outcomes by not only anorexia and weight loss but also multiple beneficial effects of liraglutide. More comprehensive metabolic profiling with a Chow liraglutide group and various types of diet (high carbohydrates, high protein, and etc.) groups will extend our understanding of how information at the metabolite level can be interpreted for the beneficial effects of liraglutide seen in pathological states ranging from diabetes to obesity.
Metabolomics has the potential to enable mapping of biochemical changes induced by drug treatments, which would help to understand how drugs work and what their efficacy profiles are. Our metabolomics study of the effect of liraglutide in DIO mice defined distinct metabolic alterations and signatures within and across multiple tissues. Metabolic alterations caused by liraglutide reflect its efficacy in modifying metabolism, which underlies the responses to this drug. In the future, multiomics integration combined with the data from our study may advance our understanding of the mechanisms of liraglutide action and provide new insights into potential new biomarkers and targets for therapeutic intervention for diabetes and obesity.
Experimental procedures
Animals
C57BL/6N male mice at 8 weeks of age were purchased from Koatech. Adult male mice were housed in a specific pathogen-free animal facility in individually ventilated cages under a 12-h light/dark cycle at 22 ± 3 °C and humidity controlled at 50 ± 10%. Mice had ad libitum access to water and were fed a chow diet (12% kcal from fat; Lab Supply, 5053) or HFD (60% kcal from fat; Envigo, 06414) for 8 weeks. Food intake per cage and individual body weight were measured daily. Total caloric intake (kcal/day) was calculated from the grams of food eaten per day (chow diet, 3.43 kcal/g; HFD, 5.1 kcal/g). All animal studies followed the National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee at the Daegu Gyeongbuk Institute of Science and Technology Laboratory Animal Resource Center.
Subcutaneous injections of liraglutide
Liraglutide (diluted in saline; 400 μg/kg; Cayman 24,727; referred to as DIO liraglutide) or vehicle (saline; referred to as DIO saline) was injected subcutaneously into DIO mice daily for 14 days. Mice fed a normal chow diet were injected only saline (referred to as Chow saline). Administration was performed before the beginning of the dark cycle.
Dual-energy X-ray absorptiometry analysis
Fat and lean masses and bone mineral density and contents were measured using a high-resolution iNSiGHT VET DXA analyzer (Osteosys). Each mouse was measured 4 times for 25 s each under anesthesia using Zoletil 50 (10 mg/kg body weight; Virbac) and Rompun (2.5 mg/kg body weight; Bayer) in 0.9% NaCl. The acquisition results were exported using the built-in software as X-ray attenuation images, bone mineral density images, colorimetric images, and raw data.
Metabolite extraction and sampling
The hypothalamus (14 mg), liver (left lateral lobe, 100 mg), plasma (100 μl), and skeletal muscle (gastrocnemius, 100 mg) were harvested immediately after body composition measurements using DXA analyzer under anesthesia. Collection of all tissues and blood was completed within 3 min per mouse in the order of anesthetized mice. The collected tissues were immediately stored briefly in liquid nitrogen, and blood was collected in EDTA-treated tube and was then centrifuged at 3000g for 15 min. The supernatant was withdrawn and frozen in liquid nitrogen. All samples were stored at −80 °C for 1 day until extraction. Prior to metabolite extraction, tissues were rinsed in DPBS (Corning 23-21-031-CV) to remove residues. Samples were extracted with a mixture of 400 μl of 80% methanol and 200 μl of 60% chloroform containing internal standards (for LC-MS/MS, acetyl-1,2-13C2-CoA, Sigma 658650; for GC-MS/MS, oleic acid-9,10-d2 and 2-isopropylmalic acid, Sigma 616133 and 67407). Tissue samples were then homogenized using a MagNA Lyser (Roche), sonicated (10 cycles, 40 s each, a mid intensity) using a Bioruptor sonicator (Diagenode), and centrifuged at 16,000g for 15 min. The aqueous and organic phase were separated and evaporated to dryness under nitrogen gas (TurboVap, Biotage). For LC-MS/MS, dried samples were reconstituted with 100 μl of initial mobile phase and immediately filtered through a 0.22 μm syringe PTFE filter (Agilent Technologies). For GC-MS/MS, dried samples of the aqueous phase were derivatized with 40 μl of methoxyamine hydrochloride in pyridine (40 mg/ml; Sigma 69479) and 40 μl of MSTFA (Supelco 69479) at 37 °C for 1 h, whereas organic phase samples were reconstituted in hexane with 0.5 M KOH-methanol and incubated at 25 °C for 10 min. Samples were added to 200 μl of 12% w/w BCl3-methanol (12%, w/w; Supelco 91379) for esterification. Reactions were performed at 60 °C for 10 min, the samples were then cooled on ice for 5 min and supplemented with 300 μl of hexane:water, 2:1 (v/v). The supernatants of all samples were transferred into autosampler vials.
GC-MS/MS analysis
GC-MS/MS analysis was carried out in an Agilent 7000B gas chromatography system coupled with an Agilent 7000C triple-quadrupole mass spectrometer. Samples were separated on an Ultra HP-5 ms capillary column (30 m × 0.25 μm, i.d., 0.25 μm film thickness, Agilent J&W Scientific). The inlet temperature was set as follows: inlet, 250 °C; transfer line, 290 °C; the electron impact (70 eV) ion source, 230 °C; the quadrupole, 150 °C. The solvent delay was set at 3.75 min. The gradient was run at a flow rate of 1.2 ml He/min with 4:1 split mode or splitless mode: 100 °C for 2 min, a gradient of 10 °C/min up to 320 °C, and equilibration at 100 °C for 10 min. Multiple reaction monitoring or selected ion mode was applied to targeted analysis, and metabolites were identified with each standard reagent. Scan mode was used in nontargeted analysis, and metabolites were identified using a Fiehn GC-MS library (69). The scan range was from 50 to 900 m/z. Data were normalized to internal standards and targeted metabolites were quantified by the linear calibration curves (R2 ≥ 0.99) using the Mass Hunter software package (Agilent Technologies).
LC-MS/MS analysis
LC-MS/MS analysis was carried out on an Agilent 1290 ultra-performance liquid chromatography system coupled with an Agilent 6490 triple-quadrupole mass spectrometer. Samples (2 μl) were separated on a Phenomenex Synergi HydroRP column (150 mm × 2 mm, 4 μm, 80 Å) maintained at 45 °C with 10 mM ammonium acetate containing 0.1% formic acid in water as buffer A and acetonitrile as buffer B. The flow rate was 0.3 ml/min and the total run time was 9 min. The gradient was as follows: 0 to 2 min at 100% buffer A; 2 to 5 min, linear gradient to 90% buffer B; 5 to 7 min, 90% buffer B; 7 to 7.1 min, return to the initial conditions; 7.1 to 9 min, re-equilibration. The system was run in multiple reaction monitoring mode using optimized collision energy with positive electrospray ionization mode. For accurate peak identification, we compared the precursor/product ion using individual analytical standards (acetyl-CoA, serotonin, GABA, dopamine, epinephrine, and norepinephrine were purchased from Sigma; A2181, H9523, 3835, H8502, PHR1509, and A9512, respectively). Data were normalized to internal standards, and targeted metabolites were quantified by the linear calibration curves (R2 ≥ 0.99) using the Mass Hunter software package (Agilent Technologies).
Metabolomics data processing
Nontargeted GC-MS raw data exported from the Mass Hunter software were converted to analysis base files (“.abf”) using ABF converter (Reifycs), and metabolic features were analyzed using MS-DIAL software v4.80 (RIKEN) (70) with the following parameters: mass scan range, 50 to 900 Da; mass accuracy for centroiding, 0.025 Da; deconvolution EI cut-off, 10 (amplitude); identification, FiehnRI; tolerance, 0.5 Da; score cut-off, 70%; retention time tolerance of alignment, 0.075 min.
Statistical and functional analysis of metabolites
For comprehensive raw data including peak intensities of targeted and nontargeted metabolites, univariate or multivariate statistical analyzes were performed using MetaboAnalyst 5.0, which is based on R packages (71). Analyzes were performed using aggregated or tissue-specific data for metabolites detected in tissues or plasma. The uploaded data file contained 24 samples (8 mice in each of the 3 groups: Chow saline, DIO saline, DIO liraglutide) by 647-peak (mz/rt) data matrix. The raw data were converted to log10 scale by filtering the samples by interquartile range, normalizing to the median relative standard deviation, and then adjusting (auto-scaling) by the SD of each variable. Significance was estimated by one-way ANOVA with multiple comparisons of Fisher’s LSD post hoc test. Significant differences between groups were visualized using PCA or PLS-DA (including OPLS-DA) models. Models were validated using the permutation test (2000 permutations). Hierarchical clustering heatmaps were generated with Euclidean distance and Ward minimum variance linkage algorithm. To identify significant intergroup or intertissue correlations among metabolites, we estimated the correlation coefficients and significance with Pearson r distance. Dimensions of correlation plots were extracted for both features and samples, and the correlation was considered significant at p < 0.001. Volcano plots were calculated to display fold change and t test analysis was used to identify significant features, and the threshold of fold change was defined as 2 or greater. To identify biologically meaningful patterns, we performed an enrichment analysis by converting the names of metabolites corresponding to impact features (VIP score >1.0) into database IDs such as HMDB, PubChem, KEGG, and SMPDB. The results were visualized as a circular plot by separating metabolites and metabolic pathway sets only when p < 0.05.
Quantification and statistical analysis
Quantitative data of metabolic parameters were represented as mean ± SD. Significance was determined by two-way ANOVA with multiple comparisons followed by Tukey post hoc test. Differences between groups were considered statistically significant at p < 0.05. Statistical analysis was performed using GraphPad Prism 9 software.
Data availability
All data are contained within the article and supporting information or are available from the corresponding author, Eun-Kyoung Kim (ekkim@dgist.ac.kr) upon request.
Supporting information
This article contains supporting information (Figs. S1–S6).
Conflict of interest
The authors declare that they have no conflicts of interests with the contents of this article.
Acknowledgments
This work was supported by the National Research Foundation of Korea under Grant Nos. 2018M3C7A1056275 and 2020R1A2B5B02001468.
Author contributions
S. P. and E.-K. K. conceptualization; S. P. and E.-K. K. methodology; S. P. and E.-K. K. formal analysis; S. P. and E.-K. K. data curation; S. P. and S. O. investigation; S. P. and S. O. validation; E.-K. K. supervision; S. P., S. O., and E.-K. K. writing–original draft.
Edited by Qi-Qun Tang
Supporting information
References
- 1.Gribble F.M., Reimann F. Metabolic messengers: glucagon-like peptide 1. Nat. Metab. 2021;3:142–148. doi: 10.1038/s42255-020-00327-x. [DOI] [PubMed] [Google Scholar]
- 2.Drucker D.J. Mechanisms of action and therapeutic application of glucagon-like peptide-1. Cell Metab. 2018;27:740–756. doi: 10.1016/j.cmet.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Madsbad S. Treatment of type 2 diabetes with incretin-based therapies. Lancet. 2009;373:438–439. doi: 10.1016/S0140-6736(08)61247-7. [DOI] [PubMed] [Google Scholar]
- 4.Holst J.J., Deacon C.F., Vilsboll T., Krarup T., Madsbad S. Glucagon-like peptide-1, glucose homeostasis and diabetes. Trends Mol. Med. 2008;14:161–168. doi: 10.1016/j.molmed.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Meier J.J. GLP-1 receptor agonists for individualized treatment of type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2012;8:728–742. doi: 10.1038/nrendo.2012.140. [DOI] [PubMed] [Google Scholar]
- 6.Hui H., Farilla L., Merkel P., Perfetti R. The short half-life of glucagon-like peptide-1 in plasma does not reflect its long-lasting beneficial effects. Eur. J. Endocrinol. 2002;146:863–869. doi: 10.1530/eje.0.1460863. [DOI] [PubMed] [Google Scholar]
- 7.Nauck M.A., Quast D.R., Wefers J., Meier J.J. GLP-1 receptor agonists in the treatment of type 2 diabetes-state of the art. Mol. Metab. 2021;46 doi: 10.1016/j.molmet.2020.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersen A., Lund A., Knop F.K., Vilsboll T. Glucagon-like peptide 1 in health and disease. Nat. Rev. Endocrinol. 2018;14:390–403. doi: 10.1038/s41574-018-0016-2. [DOI] [PubMed] [Google Scholar]
- 9.Parks M., Rosebraugh C. Weighing risks and benefits of liraglutide--the FDA's review of a new antidiabetic therapy. N. Engl. J. Med. 2010;362:774–777. doi: 10.1056/NEJMp1001578. [DOI] [PubMed] [Google Scholar]
- 10.Malm-Erjefalt M., Bjornsdottir I., Vanggaard J., Helleberg H., Larsen U., Oosterhuis B., et al. Metabolism and excretion of the once-daily human glucagon-like peptide 1 analog liraglutide in healthy male subjects and its in vitro degradation by dipeptidyl peptidase IV and neutral endopeptidase. Drug Metab. Dispos. 2010;38:1944–1953. doi: 10.1124/dmd.110.034066. [DOI] [PubMed] [Google Scholar]
- 11.Larsen P.J., Fledelius C., Knudsen L.B., Tang-Christensen M. Systemic administration of the long-acting GLP-1 derivative NN2211 induces lasting and reversible weight loss in both normal and obese rats. Diabetes. 2001;50:2530–2539. doi: 10.2337/diabetes.50.11.2530. [DOI] [PubMed] [Google Scholar]
- 12.Knudsen L.B., Nielsen P.F., Huusfeldt P.O., Johansen N.L., Madsen K., Pedersen F.Z., et al. Potent derivatives of glucagon-like peptide-1 with pharmacokinetic properties suitable for once daily administration. J. Med. Chem. 2000;43:1664–1669. doi: 10.1021/jm9909645. [DOI] [PubMed] [Google Scholar]
- 13.Ostawal A., Mocevic E., Kragh N., Xu W. Clinical effectiveness of liraglutide in type 2 diabetes treatment in the real-world setting: a systematic literature review. Diabetes Ther. 2016;7:411–438. doi: 10.1007/s13300-016-0180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Astrup A., Rossner S., Van Gaal L., Rissanen A., Niskanen L., Al Hakim M., et al. Effects of liraglutide in the treatment of obesity: a randomised, double-blind, placebo-controlled study. Lancet. 2009;374:1606–1616. doi: 10.1016/S0140-6736(09)61375-1. [DOI] [PubMed] [Google Scholar]
- 15.Davies M.J., Bergenstal R., Bode B., Kushner R.F., Lewin A., Skjoth T.V., et al. Efficacy of liraglutide for weight loss among patients with type 2 diabetes: the SCALE diabetes randomized clinical trial. JAMA. 2015;314:687–699. doi: 10.1001/jama.2015.9676. [DOI] [PubMed] [Google Scholar]
- 16.Pi-Sunyer X., Astrup A., Fujioka K., Greenway F., Halpern A., Krempf M., et al. A randomized, controlled trial of 3.0 mg of liraglutide in weight management. New Engl. J. Med. 2015;373:11–22. doi: 10.1056/NEJMoa1411892. [DOI] [PubMed] [Google Scholar]
- 17.Petit J.M., Cercueil J.P., Loffroy R., Denimal D., Bouillet B., Fourmont C., et al. Effect of liraglutide therapy on liver fat content in patients with inadequately controlled type 2 diabetes: the lira-NAFLD Study. J. Clin. Endocrinol. Metab. 2017;102:407–415. doi: 10.1210/jc.2016-2775. [DOI] [PubMed] [Google Scholar]
- 18.Verma S., Poulter N.R., Bhatt D.L., Bain S.C., Buse J.B., Leiter L.A., et al. Effects of liraglutide on cardiovascular outcomes in patients with type 2 diabetes mellitus with or without history of myocardial infarction or stroke. Circulation. 2018;138:2884–2894. doi: 10.1161/CIRCULATIONAHA.118.034516. [DOI] [PubMed] [Google Scholar]
- 19.Lundgren J.R., Janus C., Jensen S.B.K., Juhl C.R., Olsen L.M., Christensen R.M., et al. Healthy weight loss maintenance with exercise, liraglutide, or both combined. N. Engl. J. Med. 2021;384:1719–1730. doi: 10.1056/NEJMoa2028198. [DOI] [PubMed] [Google Scholar]
- 20.Beiroa D., Imbernon M., Gallego R., Senra A., Herranz D., Villarroya F., et al. GLP-1 agonism stimulates brown adipose tissue thermogenesis and browning through hypothalamic AMPK. Diabetes. 2014;63:3346–3358. doi: 10.2337/db14-0302. [DOI] [PubMed] [Google Scholar]
- 21.Secher A., Jelsing J., Baquero A.F., Hecksher-Sorensen J., Cowley M.A., Dalboge L.S., et al. The arcuate nucleus mediates GLP-1 receptor agonist liraglutide-dependent weight loss. J. Clin. Invest. 2014;124:4473–4488. doi: 10.1172/JCI75276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vilsboll T., Zdravkovic M., Le-Thi T., Krarup T., Schmitz O., Courreges J.P., et al. Liraglutide, a long-acting human glucagon-like peptide-1 analog, given as monotherapy significantly improves glycemic control and lowers body weight without risk of hypoglycemia in patients with type 2 diabetes. Diabetes Care. 2007;30:1608–1610. doi: 10.2337/dc06-2593. [DOI] [PubMed] [Google Scholar]
- 23.Rowlands J., Heng J., Newsholme P., Carlessi R. Pleiotropic effects of GLP-1 and analogs on cell signaling, metabolism, and function. Front. Endocrinol. (Lausanne) 2018;9:672. doi: 10.3389/fendo.2018.00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z., Ni C.L., Yao Z., Chen L.M., Niu W.Y. Liraglutide enhances glucose transporter 4 translocation via regulation of AMP-activated protein kinase signaling pathways in mouse skeletal muscle cells. Metabolism. 2014;63:1022–1030. doi: 10.1016/j.metabol.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Park S., Sadanala K.C., Kim E.K. A metabolomic approach to understanding the metabolic link between obesity and diabetes. Mol. Cells. 2015;38:587–596. doi: 10.14348/molcells.2015.0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bain J.R., Stevens R.D., Wenner B.R., Ilkayeva O., Muoio D.M., Newgard C.B. Metabolomics applied to diabetes research: moving from information to knowledge. Diabetes. 2009;58:2429–2443. doi: 10.2337/db09-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cirulli E.T., Guo L., Leon Swisher C., Shah N., Huang L., Napier L.A., et al. Profound perturbation of the metabolome in obesity is associated with health risk. Cell Metab. 2019;29:488–500.e482. doi: 10.1016/j.cmet.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wishart D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016;15:473–484. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- 29.Ryan P.M., Patterson E., Carafa I., Mandal R., Wishart D.S., Dinan T.G., et al. Metformin and dipeptidyl peptidase-4 inhibitor differentially modulate the intestinal microbiota and plasma metabolome of metabolically dysfunctional mice. Can J. Diabetes. 2020;44:146–155.e142. doi: 10.1016/j.jcjd.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Gu X., Al Dubayee M., Alshahrani A., Masood A., Benabdelkamel H., Zahra M., et al. Distinctive metabolomics patterns associated with insulin resistance and type 2 diabetes mellitus. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.609806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aleidi S.M., Dahabiyeh L.A., Gu X., Al Dubayee M., Alshahrani A., Benabdelkamel H., et al. Obesity connected metabolic changes in type 2 diabetic patients treated with metformin. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.616157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomasova P., Buganova M., Pelantova H., Holubova M., Sediva B., Zelezna B., et al. Metabolomics based on MS in mice with diet-induced obesity and type 2 diabetes mellitus: the effect of vildagliptin, metformin, and their combination. Appl. Biochem. Biotechnol. 2019;188:165–184. doi: 10.1007/s12010-018-2899-8. [DOI] [PubMed] [Google Scholar]
- 33.Buganova M., Pelantova H., Holubova M., Sediva B., Maletinska L., Zelezna B., et al. The effects of liraglutide in mice with diet-induced obesity studied by metabolomics. J. Endocrinol. 2017;233:93–104. doi: 10.1530/JOE-16-0478. [DOI] [PubMed] [Google Scholar]
- 34.Bouatra S., Aziat F., Mandal R., Guo A.C., Wilson M.R., Knox C., et al. The human urine metabolome. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mondragon A., Davidsson D., Kyriakoudi S., Bertling A., Gomes-Faria R., Cohen P., et al. Divergent effects of liraglutide, exendin-4, and sitagliptin on beta-cell mass and indicators of pancreatitis in a mouse model of hyperglycaemia. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou J., Poudel A., Chandramani-Shivalingappa P., Xu B., Welchko R., Li L. Liraglutide induces beige fat development and promotes mitochondrial function in diet induced obesity mice partially through AMPK-SIRT-1-PGC1-alpha cell signaling pathway. Endocrine. 2019;64:271–283. doi: 10.1007/s12020-018-1826-7. [DOI] [PubMed] [Google Scholar]
- 37.Blouet C., Jo Y.H., Li X., Schwartz G.J. Mediobasal hypothalamic leucine sensing regulates food intake through activation of a hypothalamus-brainstem circuit. J. Neurosci. 2009;29:8302–8311. doi: 10.1523/JNEUROSCI.1668-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez M., Lage R., Saha A.K., Perez-Tilve D., Vazquez M.J., Varela L., et al. Hypothalamic fatty acid metabolism mediates the orexigenic action of ghrelin. Cell Metab. 2008;7:389–399. doi: 10.1016/j.cmet.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz M.W., Woods S.C., Porte D., Jr., Seeley R.J., Baskin D.G. Central nervous system control of food intake. Nature. 2000;404:661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- 40.Myers M.G., Jr., Affinati A.H., Richardson N., Schwartz M.W. Central nervous system regulation of organismal energy and glucose homeostasis. Nat. Metab. 2021;3:737–750. doi: 10.1038/s42255-021-00408-5. [DOI] [PubMed] [Google Scholar]
- 41.Barreto-Vianna A.R., Aguila M.B., Mandarim-de-Lacerda C.A. Effects of liraglutide in hypothalamic arcuate nucleus of obese mice. Obesity (Silver Spring) 2016;24:626–633. doi: 10.1002/oby.21387. [DOI] [PubMed] [Google Scholar]
- 42.He Z., Gao Y., Lieu L., Afrin S., Cao J., Michael N.J., et al. Direct and indirect effects of liraglutide on hypothalamic POMC and NPY/AgRP neurons - implications for energy balance and glucose control. Mol. Metab. 2019;28:120–134. doi: 10.1016/j.molmet.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dettmer K., Aronov P.A., Hammock B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benoit S.C., Kemp C.J., Elias C.F., Abplanalp W., Herman J.P., Migrenne S., et al. Palmitic acid mediates hypothalamic insulin resistance by altering PKC-theta subcellular localization in rodents. J. Clin. Invest. 2009;119:2577–2589. doi: 10.1172/JCI36714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arruda A.P., Milanski M., Coope A., Torsoni A.S., Ropelle E., Carvalho D.P., et al. Low-grade hypothalamic inflammation leads to defective thermogenesis, insulin resistance, and impaired insulin secretion. Endocrinology. 2011;152:1314–1326. doi: 10.1210/en.2010-0659. [DOI] [PubMed] [Google Scholar]
- 46.Obici S., Feng Z., Morgan K., Stein D., Karkanias G., Rossetti L. Central administration of oleic acid inhibits glucose production and food intake. Diabetes. 2002;51:271–275. doi: 10.2337/diabetes.51.2.271. [DOI] [PubMed] [Google Scholar]
- 47.Lam T.K., Pocai A., Gutierrez-Juarez R., Obici S., Bryan J., Aguilar-Bryan L., et al. Hypothalamic sensing of circulating fatty acids is required for glucose homeostasis. Nat. Med. 2005;11:320–327. doi: 10.1038/nm1201. [DOI] [PubMed] [Google Scholar]
- 48.Nunes E.A., Rafacho A. Implications of palmitoleic acid (Palmitoleate) on glucose homeostasis, insulin resistance and diabetes. Curr. Drug Targets. 2017;18:619–628. doi: 10.2174/1389450117666151209120345. [DOI] [PubMed] [Google Scholar]
- 49.Zhao L., Zhu C., Lu M., Chen C., Nie X., Abudukerimu B., et al. The key role of a glucagon-like peptide-1 receptor agonist in body fat redistribution. J. Endocrinol. 2019;240:271–286. doi: 10.1530/JOE-18-0374. [DOI] [PubMed] [Google Scholar]
- 50.Newgard C.B., An J., Bain J.R., Muehlbauer M.J., Stevens R.D., Lien L.F., et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang J., Dolinger M., Ritaccio G., Mazurkiewicz J., Conti D., Zhu X., et al. Leucine stimulates insulin secretion via down-regulation of surface expression of adrenergic alpha2A receptor through the mTOR (mammalian target of rapamycin) pathway: implication in new-onset diabetes in renal transplantation. J. Biol. Chem. 2012;287:24795–24806. doi: 10.1074/jbc.M112.344259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang S., Zeng X., Ren M., Mao X., Qiao S. Novel metabolic and physiological functions of branched chain amino acids: a review. J. Anim. Sci. Biotechnol. 2017;8:10. doi: 10.1186/s40104-016-0139-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.El-Kady R.R., Ali A.K., El Wakeel L.M., Sabri N.A., Shawki M.A. Nicotinamide supplementation in diabetic nonalcoholic fatty liver disease patients: randomized controlled trial. Ther. Adv. Chronic Dis. 2022;13 doi: 10.1177/20406223221077958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boulange C.L., Claus S.P., Chou C.J., Collino S., Montoliu I., Kochhar S., et al. Early metabolic adaptation in C57BL/6 mice resistant to high fat diet induced weight gain involves an activation of mitochondrial oxidative pathways. J. Proteome Res. 2013;12:1956–1968. doi: 10.1021/pr400051s. [DOI] [PubMed] [Google Scholar]
- 55.McCreanor G.M., Bender D.A. The metabolism of high intakes of tryptophan, nicotinamide and nicotinic acid in the rat. Br. J. Nutr. 1986;56:577–586. doi: 10.1079/bjn19860138. [DOI] [PubMed] [Google Scholar]
- 56.Waterfield C.J., Turton J.A., Scales M.D., Timbrell J.A. Taurine, a possible urinary marker of liver damage: a study of taurine excretion in carbon tetrachloride-treated rats. Arch. Toxicol. 1991;65:548–555. doi: 10.1007/BF01973715. [DOI] [PubMed] [Google Scholar]
- 57.Chen W., Guo J., Zhang Y., Zhang J. The beneficial effects of taurine in preventing metabolic syndrome. Food Funct. 2016;7:1849–1863. doi: 10.1039/c5fo01295c. [DOI] [PubMed] [Google Scholar]
- 58.Song Q., Guo J.X., Zhang Y.Z., Chen W. The beneficial effects of taurine in alleviating fatty liver disease. J. Funct. Foods. 2021;77:104351. [Google Scholar]
- 59.Murakami S., Funahashi K., Tamagawa N., Ning M., Ito T. Taurine ameliorates streptozotocin-induced diabetes by modulating hepatic glucose metabolism and oxidative stress in mice. Metabolites. 2022;12:524. doi: 10.3390/metabo12060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Killion E.A., Wang J., Yie J., Shi S.D., Bates D., Min X., et al. Anti-obesity effects of GIPR antagonists alone and in combination with GLP-1R agonists in preclinical models. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aat3392. [DOI] [PubMed] [Google Scholar]
- 61.Heppner K.M., Marks S., Holland J., Ottaway N., Smiley D., Dimarchi R., et al. Contribution of brown adipose tissue activity to the control of energy balance by GLP-1 receptor signalling in mice. Diabetologia. 2015;58:2124–2132. doi: 10.1007/s00125-015-3651-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim Y.J., Kang D., Yang H.R., Park B.S., Tu T.H., Jeong B., et al. Metabolic profiling of the hypothalamus of mice during short-term food deprivation. Metabolites. 2022;12:407. doi: 10.3390/metabo12050407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kondoh H., Teruya T., Yanagida M. Metabolomics of human fasting: new insights about old questions. Open Biol. 2020;10 doi: 10.1098/rsob.200176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Most J., Redman L.M. Impact of calorie restriction on energy metabolism in humans. Exp. Gerontol. 2020;133 doi: 10.1016/j.exger.2020.110875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xie D., Huang J., Zhang Q., Zhao S., Xue H., Yu Q.Q., et al. Comprehensive evaluation of caloric restriction-induced changes in the metabolome profile of mice. Nutr. Metab. (Lond) 2022;19:41. doi: 10.1186/s12986-022-00674-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Diaz-Ruiz A., Rhinesmith T., Pomatto-Watson L.C.D., Price N.L., Eshaghi F., Ehrlich M.R., et al. Diet composition influences the metabolic benefits of short cycles of very low caloric intake. Nat. Commun. 2021;12:6463. doi: 10.1038/s41467-021-26654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Collet T.H., Sonoyama T., Henning E., Keogh J.M., Ingram B., Kelway S., et al. A metabolomic signature of acute caloric restriction. J. Clin. Endocrinol. Metab. 2017;102:4486–4495. doi: 10.1210/jc.2017-01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guijas C., Montenegro-Burke J.R., Cintron-Colon R., Domingo-Almenara X., Sanchez-Alavez M., Aguirre C.A., et al. Metabolic adaptation to calorie restriction. Sci. Signal. 2020;13 doi: 10.1126/scisignal.abb2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lai Z., Tsugawa H., Wohlgemuth G., Mehta S., Mueller M., Zheng Y., et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Met. 2018;15:53–56. doi: 10.1038/nmeth.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsugawa H., Cajka T., Kind T., Ma Y., Higgins B., Ikeda K., et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Met. 2015;12:523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pang Z., Chong J., Zhou G., de Lima Morais D.A., Chang L., Barrette M., et al. MetaboAnalyst 5.0: nnarrowing the gap between raw spectra and functional insights. Nucl. Acids Res. 2021;49:W388–W396. doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and supporting information or are available from the corresponding author, Eun-Kyoung Kim (ekkim@dgist.ac.kr) upon request.