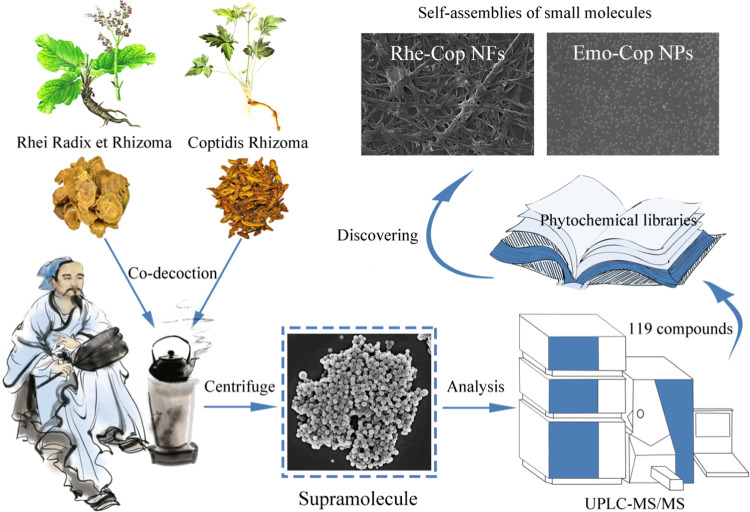
Abstract
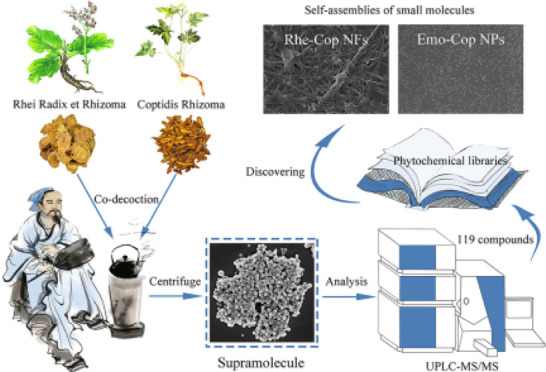
The carrier-free self-assembly of small molecules opens a new window for the development of nanomaterials. This study is dedicated to developing binary small-molecular self-assemblies derived from phytochemicals in traditional Chinese herbal medicine. Among them, Rhei Radix et Rhizoma and Coptidis Rhizoma are a common pair used in clinics for thousands of years. Here, we found that there were numerous spherical supramolecular nanoparticles (NPs) originated from Rhei Radix et Rhizoma and Coptidis Rhizoma decoction. Ultra-performance liquid chromatography/tandem mass spectrometry (UPLC-MS/MS) was used to analyze the composition of the supramolecules, and a total of 119 phytochemicals were identified (23 anthraquinones, 31 alkaloids, 24 organic acids, 8 tannins, and other components). Isothermal titration calorimetry (ITC) showed that the interaction between Rhei Radix et Rhizoma and Coptidis Rhizoma was a spontaneous exothermic reaction, indicating that their phytochemicals had the property of self-assembly and interacted to form supramolecules in the decocting process. Furthermore, scanning electron microscopy (SEM), UV, IR, NMR, and ITC were used to verify that rhein and coptisine could self-assemble into nanofibers (Rhe-Cop NFs), while emodin and coptisine could self-assemble into nanoparticles (Emo-Cop NPs). The formation mechanism analysis of the self-assemblies revealed that they were induced by electrostatic attraction, hydrogen bonding, and π–π stacking, forming nanospheres of about 50 nm and nanofibers. The current study not only provides an idea of discovering carrier-free self-assemblies from traditional herbal medicine decoction but also supplies a reference for the design of binary self-assembly of small molecules in the future.
1. Introduction
Supramolecular refers to the association of two or more kinds of molecules with intermolecular interactions to form a complex and orderly whole with certain integrity, microstructure, and macroscopic characteristics.1 At present, supramolecular self-assembly has shown great prospects in the research and development of nanomaterials.2 It has great potential in the fields of anticancer,3,4 disease diagnosis,5 targeted drug delivery,6,7 and other nanomedicines.8 Self-assembly behavior refers to the process of an orderly arrangement of molecules in a system under the action of noncovalent bonds and finally forming a structure with a certain geometric appearance.9 Numerous self-assembly phenomena from small molecules have been found,10,11 like puerarin hydrogel,12,13 gallic acid gels,14 pomolic acid gels,15 and rhein hydrogel,16,17 and triterpenoids can also self-assemble under suitable solvent conditions,18−20 which exhibit different morphologies and functions, such as oleanolic acid, glycyrrhetinic acid,21 and ursolic acid.22 Our previous studies and other researchers have also found binary assemblers: berberine can self-assemble with baicalin,23 rhein,24 aristolochic acid,25 cinnamic acid,26 curcumin,27 and sennoside A28 to form nanoparticles (NPs). There are also several small-molecule compounds self-assembling with metal ions: luteolin-Fe3+/Ca2+/Mg2+/Zn2+/Cu2+,29,3029,30 tannic acid/catecholamines-Cu2+,31 and baicalin-Al3+.32 Although some progress has been made in this field, the self-assembly of natural small molecules is still at the stage of random, serendipity discovery. And the previous self-assembly system often involves a complex preparation process, high cost, and so on.33,34 The preparation of a self-assembly system is easily affected by external physical and chemical factors (such as solvent,35 pH36,37), resulting in the emergence of a self-assembly system with different structures and functions. Therefore, a goal-oriented design to obtain natural small-molecule self-assembly with a simple strategy is a hot research topic at present (Scheme 1).
Scheme 1. Natural Small-Molecule-Based Carrier-Free Self-Assembly Library Originated from Traditional Chinese Herbal Medicine.
The composition of traditional Chinese medicine (TCM) decoction is complex and diverse, which is a collection of phytochemicals’ libraries.38 The self-assemblies of small molecules in TCM decoction are widespread,39,40 and so it could provide valuable ideas for the accurate discovery of supramolecular components in plants. For example, nanoparticles have been found in Ma-Xing-Shi-Gan-Tang decoction,41 Xue-Fu-Zhu-Yu-Tang decoction,42 Bai-Hu-Tang decoction,43 Ge-Gen-Qin-Lian-Tang decoction,44 San-Huang-Xie-Xin-Tang decoction,45 and others,46,47 which showed fascinating properties and activities. Nanoparticles were not only found in TCM decoction but also in single-flavor decoctions such as Fen-Ge decoction,48,49 Huang-Lian decoction,50 and so on. Most of the nanoparticles in the above decoction had a particle size of 100–600 nm, which exhibited good stability and high bioavailability. Taking Sini decoction51 and Huang-Lian-Jie-Du decoction52 as examples, it was found that the nanoparticles in the decoction were formed by the interaction of many small molecular bioactive components. In addition to the self-assembly through small-molecule interactions in the decoction, some researchers found that polysaccharide53,54 and protein55,56 components in the decoction could also self-assemble and showed good biological activity. Meanwhile, many chemical components in TCM have unique structures and many modification sites, which facilitate self-recognition and self-assembly.10 Therefore, TCM decoction can be used as a carrier-free self-assembly library of small molecules to reduce blindness, contingency, and a green strategy in supramolecular design and construction.
Rhei Radix et Rhizoma and Coptidis Rhizoma are commonly used as herbal pair medicine in TCM. There is obvious turbidity in the decocting process. As one of the rich compound libraries of decoction, Rhein-Ber nanoparticles were found in our previous study.24 Therefore, starting from the nanoaggregate in Rhei Radix et Rhizoma and Coptidis Rhizoma co-decoctions, a total of 119 compounds were identified (23 anthraquinones, 31 alkaloids, 24 organic acids, 8 tannins, and other components). Interestingly, we found that the herbal pair’s main components rhein and coptisine could self-assemble into nanofibers (Rhe-Cop NFs), while emodin and coptisine could self-assemble into nanoparticles (Emo-Cop NPs). Combined with multiple technologies, this study took Emo-Cop NPs and Rhe-Cop NFs as examples to describe the formation mechanism and exploration process of discovering self-assemblies of small molecules without a carrier from Chinese herbal compound decoction.
2. Materials and Methods
2.1. Reagents and Materials
Rhei Radix et Rhizoma and Coptidis Rhizoma were purchased from Beijing Tongrentang (batch number: 20201125, 200301001). Rhein (C15H8O6, Rhe, 98%), emodin (C15H10O5, Emo, 98%), coptisine chloride (C19H14ClNO4, Cop, 98%), and DMSO-d6 were all purchased from Beijing Inokai Technology Co., Ltd. Sodium hydroxide (NaOH), dimethyl sulfoxide (DMSO, C2H6OS), and methanol (CH4O) were all purchased from Beijing Chemical Plant. Ordinary dialysis bag (MWCO: 3000 Da, Shanghai Yuanye Biotechnology Co., Ltd.). Phosphate buffer (Beijing Bairdi Biotechnology Co., Ltd.)
2.2. Sample Preparation
According to the ratio of Rhei Radix et Rhizoma/Coptidis Rhizoma = 1:1, the corresponding weight of herbs was put into a nonwoven bag and 8 times the amount of water was added and boiled together for 40 min. The co-decoction of Rhei Radix et Rhizoma and Coptidis Rhizoma was obtained after filtering out the residue. The co-decoction was centrifuged at 80,000 rpm for 30 min, and the supernatant and supramolecular sites were collected, respectively. All samples were lyophilized.
Coptisine chloride and emodin were precisely weighed at 0.1 mmol and dissolved by adding methanol and DMSO, respectively. After mixing and stirring the two solutions, sodium hydroxide solution was added to adjust the pH to 7.0–7.5. The above solution mixture was added to 60 °C phosphate-buffered saline (PBS) drop by drop. After thorough stirring, water bath ultrasonication was performed for 1 h. The reaction solution was transferred to a dialysis bag for 12 h to remove unreacted raw materials and organic solvents. The dialysate fluid outside should be replaced every 3 h. Emodin-Coptisine nanoparticles (Emo-Cop NPs) were obtained by freeze-drying its dialysate fluid inside. Similarly, Rhein-Coptisine nanofibers (Rhe-Cop NFs) assembly could be prepared.
2.3. Characterization by Dynamic Light Scattering (DLS)
Each sample was dispersed in deionized water and transferred to a colorimetric dish. Using a Malvern particle size analyzer, the mean particle size and polydispersity index (PDI) of the samples were measured in size mode and the ζ-potential of the samples was measured in ζ-mode. The results were repeated three times and averaged.
2.4. Field Emission Scanning Electron Microscopy Observation
The lyophilized powder samples were dispersed in deionized water and 2 μL was transferred to silicon wafers for drying naturally at room temperature. After spraying gold on the surface, a working voltage of 15.0 kV (ZEISS-SUPRA55, ZEISS, Germany) was set to observe the morphology and particle size of each sample.
2.5. Ultra-Performance Liquid Chromatography/Tandem Mass Spectrometry (UPLC-MS/MS)
TC-C18 column (4.6 mm × 250 mm, 5 μm, Agilent) was used. The mobile phase consisted of 0.1% (v/v) aqueous formic acid solution (A) and acetonitrile (B). The gradient elution conditions were 0–30 min, 4–98% B. The flow rate was 0.3 mL/min and the injection volume was 5 μL. The ion source adopted ESI to collect information in positive (ESI+) and negative (ESI–) electrospray ionization mode (EM ACE600, Leica Technology Co., Ltd.). The atomized pressure was 45 psi. The dry gas was nitrogen gas. The capillary and auxiliary gas heater temperatures were both set to 350 °C.
2.6. UV–Visible Absorption Spectrometric Determination
Two lyophilized powders of self-assembly and three monomer components were fully dissolved in methanol separately. The detection wavelength of the UV–visible spectrophotometer (UH5300, HITACHI, Japan) was set at 190–600 nm and methanol was used as a blank solution for full-wavelength scanning.
2.7. Fourier Transform Infrared Spectroscopy Determination
Two lyophilized powders of self-assembly and three monomer components were put into a Fourier transform infrared spectrometer (Tensor 27, Bruker). Later, the infrared spectra of each sample were measured in the range of 400–4000 cm–1 with air as the background.
2.8. Nuclear Magnetic Resonance Hydrogen Spectrometry (1H NMR) Determination
Two lyophilized powders of self-assembly and three monomer components were placed in the nuclear magnetic tube. Then, deuterium dimethyl sulfoxide was added to dissolve thoroughly, and tetramethylsilane (TMS) was used as an internal standard to determine the 1H NMR spectra of each sample in NMR apparatus (Bruker Avance 400 MHz, Bruker, Germany).
2.9. Isothermal Titration Calorimetry (ITC) Determination
All reserve fluid must be filtered with a 0.45 μm microporous membrane before use. The titration threshold was explored through the preliminary experiment, and the titration condition was determined as follows: 10 mg/mL single decoction of Coptidis Rhizoma-titrated 20 mg/mL single decoction of Rhei Radix et Rhizoma; 2.5 mmol/L coptisine chloride-titrated 0.5 mmol/L rhein; 5 mmol/L emodin-titrated 0.5 mmol/L coptisine chloride. The instrument parameters were set as follows: a stirring rate of 250 r/min, titration temperature of 25 °C, and titration interval 180 s (Nano ITC, TA).
3. Results and Discussion
3.1. Morphological Characterization
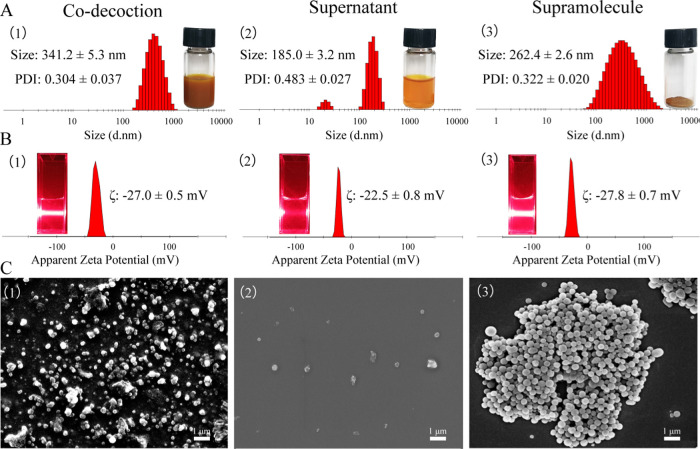
As shown in Figure 1A,B, the co-decoction of Rhei Radix et Rhizoma and Coptidis Rhizoma was turbid. Dynamic light scattering characterization showed that the co-decoction was a homogeneous and stable system with a size of 341.2 nm (PDI = 0.304) and a ζ-potential of −27.0 mV. The Tyndall effect also had a distinct pathway. The co-decoction was centrifuged at 80,000 rpm for 30 min, and then the supernatant and supramolecular site were collected, respectively. After centrifuging, no significant Tyndall effect was observed in the supernatant and its particle size was about 185 nm with uneven dispersion (PDI = 0.483). The collected supramolecular samples were redissolved in water, and it was found that the solution also had a significant Tyndall effect. The particle size of the supramolecular site was mainly 262 nm (PDI = 0.322), and the ζ-potential value was −27.8 mV.
Figure 1.
Morphological characterization of Rhei Radix et Rhizoma and Coptidis Rhizoma co-decoction (1), supernatant (2), and supramolecular portion (3). (A) Particle size and PDI characterization. (B) ζ-Potential and Tyndall effect characterization. (C) Scanning electron microscopy (SEM) characterization.
SEM characterization is shown in Figure 1C; it was observed that the co-decoction mainly contained spherical nanoparticles of 300–500 nm. Rare nanoparticles were observed in the supernatant and their scale distribution was relatively discrete, which was consistent with the DLS characterization. Meanwhile, the supramolecular portion comprised nanospheres of about 300 nm with uniform size and uniform dispersion. SEM results showed that the nanospheres in the co-decoction mainly existed in the supramolecular part.
3.2. Identification of the Constituents in the Supramolecule
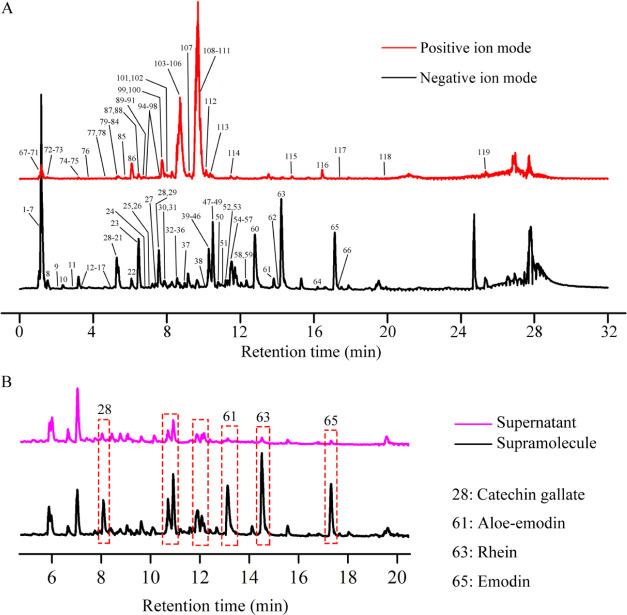
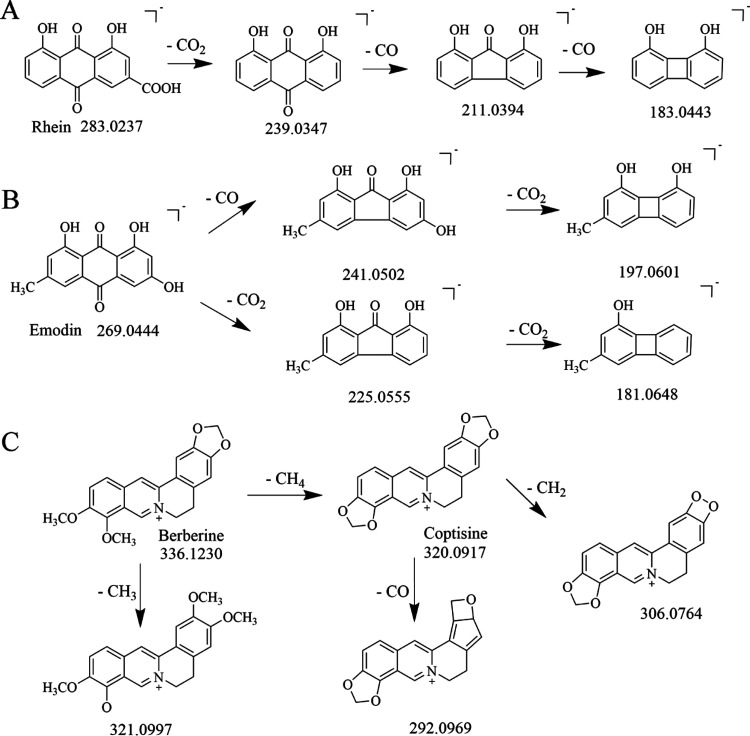
Totally 119 compounds had been tentatively identified in the supramolecular portion (Figure 2A). A total of 23 anthraquinones were identified, including 10 free anthraquinones, 11 bound anthraquinones, and 2 bound anthraones. They were all characteristic constituents of Rhei Radix et Rhizoma and were more likely to produce fragment ions such as [M-H-CO]− and [M-H-CO2]− in negative ion mode. Compounds 63 and 65 were characterized to be rhein and emodin, respectively. Compound 63 (rhein) exhibited a molecular ion at m/z 283.0237 and its fragment ions were [M-H-CO2]− (m/z 239.0347), [M-H-C2O3]− (m/z 211.0394), and [M-H-C3O4]− (m/z 183.0443). Likewise, emodin was produced as a precursor ion at m/z 269.0444 ([M-H]−). The fragment ion at m/z 241.0502 ([M-H-CO]−), m/z 225.0555 ([M-H-CO2]−), m/z 197.0601 ([M-H-C2O3]−), and m/z 181.0648 ([M-H-C2O4]−) originated from the loss of CO moiety and CO2 moiety (Figure 3).
Figure 2.
UPLC-MS/MS characterization. (A) Total ion current chromatograms of the supramolecular site in the positive and negative ion modes. (B) Negative ion current chromatograms of supramolecular and supernatant sites.
Figure 3.
Cracking law of (A) rhein, (B) emodin, and (C) berberine and coptisine.
Alkaloids were the main medicinal ingredients of Coptidis Rhizoma. In this test, 31 alkaloids and their glycoside compounds were detected. For example, Compound 106 (coptisine) produced a precursor ion at m/z 320.0917 ([M]+), yielding the most intense fragment ion at m/z 292.0966 ([M-CO]+). Compound 111 was proposed to be berberine, showing a deprotonated molecule at m/z 336.1230 ([M]+). The major fragment ion at m/z 320.0917 ([M-CH4]+) and m/z 306.0764 ([M-CH4-CH2]+) is attributed to the continuous loss of a CH4 moiety and a CH2 moiety. Other fragments at m/z 321.0997 ([M-CH3]+) and m/z 292.0969 ([M-C2H4O]+) could also be observed in the product ion spectrum. In addition to anthraquinones and alkaloids, there were 24 organic acids, 8 tannins, 8 flavonoids, 5 amino acids, and 7 monosaccharides and polysaccharides in the supramolecular site. Furthermore, there were 13 other components, including phenylbutanone, stilbene glycosides, naphthalene glycosides, and other compounds. The fragment ions and cleavage pathways of some compounds are shown in Table 1, Table S1, and Figure S1.
Table 1. Identification of Compounds at Supramolecular Sites.
| precursor
ion |
||||||||
|---|---|---|---|---|---|---|---|---|
| no. | tR (min) | compound | formula | identity | theoretical (m/z) | experimental (m/z) | Δppm | fragment ion (m/z) |
| 9 | 2.01 | galloylglucose | C13H16O10 | [M-H]− | 331.0659 | 331.0674 | 4.53 | 169.0132 [M-H-C6H10O5]−, 125.0232 [M-H-C7H10O7]− |
| 10 | 2.34 | gallic acid | C7H6O5 | [M-H]− | 169.0131 | 169.0133 | 1.18 | 125.0231 [M-H-CO2]− |
| 27 | 7.37 | rhein glucoside | C21H18O11 | [M-H]− | 445.0765 | 445.0775 | 2.25 | 283.0250 [M-H-C6H10O5]−, 239.0348 [M-H-C7H10O7]−, 211.0396 [M-H-C8H10O8]− |
| 29 | 7.65 | sennoside C/D | C42H40O19 | [M-H]− | 847.2080 | 847.2097 | 2.00 | 685.1560 [M-H-C6H10O5]− |
| 31 | 7.85 | sennoside A/B | C42H38O20 | [M-H]− | 861.1872 | 861.1890 | 2.09 | 699.1338 [M-H-C6H10O5]−, 386.0994 [M-H-C22H19O12]− |
| 35 | 8.70 | aloesin | C19H22O9 | [M-H]− | 393.1180 | 393.1196 | 4.07 | 231.0658 [M-H-C6H10O5]−, 203.0709 [M-H-C7H10O6]− |
| 36 | 8.86 | aloe emodin glucoside | C21H20O11 | [M-H]− | 431.0972 | 431.0984 | 2.78 | 269.0456 [M-H-C6H10O5]− |
| 44 | 10.30 | emodin glucoside | C21H20O11 | [M-H]− | 431.0972 | 431.0984 | 2.78 | 269.0456 [M-H-C6H10O5]− |
| 48 | 10.50 | chrysophanol | C15H10O4 | [M-H]− | 253.0495 | 253.0506 | 4.35 | 225.0555 [M-H-CO]− |
| 54 | 11.42 | physcion | C16H12O5 | [M-H]− | 283.0600 | 283.0612 | 4.24 | 268.0377 [M-H-CH3]−, 240.0425 [M-H-C2H3O]− |
| 55 | 11.47 | physcion glucoside | C22H22O10 | [M-H]− | 445.1129 | 445.1139 | 2.25 | 283.0612 [M-H-C6H10O5]−, 268.0377 [M-H-C7H13O5]−, 240.0425 [M-H-C8H13O6]− |
| 60 | 12.80 | 3-methyl-rhein | C16H10O6 | [M-H]− | 297.0393 | 297.0406 | 4.37 | 253.0505 [M-H-CO2]−, 225.0553 [M-H-C2O3]− |
| 61 | 13.82 | aloe-emodin | C15H10O5 | [M-H]− | 269.0444 | 269.0456 | 4.46 | 241.0498 [M-H-CO]−, 240.0426 [M-H-CHO]−, 223.0397 [M-H-CH2O2]−, 197.0597 [M-H-C2O3]−, 183.0449 [M-H-C3H2O3]− |
| 63 | 14.23 | rhein | C15H8O6 | [M-H]− | 283.0237 | 283.0249 | 4.24 | 239.0347 [M-H-CO2]−, 211.0394 [M-H-C2O3]−, 183.0443 [M-H-C3O4]−, 155.0492 [M-H-C4O5]− |
| 65 | 17.14 | emodin | C15H10O5 | [M-H]− | 269.0444 | 269.0456 | 4.46 | 241.0502 [M-H-CO]−, 225.0552 [M-H-CO2]−, 197.0601 [M-H-C2O3]−, 181.0648 [M-H-C2O4]− |
| 86 | 6.08 | magnoflorine | C20H24NO4+ | [M]+ | 342.1699 | 342.1700 | 0.29 | 297.1122 [M-C2H7N]+, 282.0886 [M-C3H10N]+, 265.0860 [M-C3H11NO]+, 237.0909 [M-C4H11NO2]+ |
| 97 | 7.58 | berberrubine | C19H16NO4+ | [M]+ | 322.1073 | 322.1073 | 0.00 | 307.0839 [M-CH3]+, 294.1124 [M-CO]+ |
| 103 | 8.55 | columbamine | C20H20NO4+ | [M]+ | 338.1386 | 338.1387 | 0.29 | 323.1352 [M-CH3]+, 322.1074 [M-CH4]+, 308.0917 [M-C2H6]+, 306.1130 [M-C2H8]+, 294.1124 [M-C2H4O]+ |
| 104 | 8.64 | epiberberine | C20H18NO4+ | [M]+ | 336.1230 | 336.1229 | –0.30 | 321.0991 [M-CH3]+, 320.0918 [M-CH4]+, 306.0753 [M-C2H6]+, 292.0966 [M-C2H4O]+ |
| 105 | 8.69 | jatrorrhizine | C20H20NO4+ | [M]+ | 338.1386 | 338.1385 | –0.30 | 323.1352 [M-CH3]+, 322.1073 [M-CH4]+, 308.0916 [M-C2H6]+, 306.1127 [M-C2H8]+, 294.1124 [M-C2H4O]+ |
| 106 | 8.75 | coptisine | C19H14NO4+ | [M]+ | 320.0917 | 320.0918 | 0.31 | 292.0966 [M-CO]+ |
| 109 | 9.44 | worenine | C20H16NO4+ | [M]+ | 334.1073 | 334.1073 | 0.00 | 319.0843 [M-CH3]+, 306.1122 [M-CH4]+ |
| 110 | 9.55 | palmatine | C21H22NO4+ | [M]+ | 352.1543 | 352.1542 | –0.28 | 337.1307 [M-CH3]+, 336.1229 [M-CH4]+, 322.1074 [M-C2H6]+, 308.1281 [M-C3H8]+ |
| 111 | 9.67 | berberine | C20H18NO4+ | [M]+ | 336.1230 | 336.1230 | 0.00 | 321.0997 [M-CH3]+, 320.0919 [M-CH4]+, 306.0764 [M-C2H6]+, 292.0969 [M-C2H4O]+, 278.0805 [M-C3H6O]+ |
The contents of anthraquinone and tannin in the supramolecular fraction were much higher than those in the supernatant fraction (Figure 2B). Among them, the content of catechin gallate, 3-methyl-rhein, rhein, and emodin in the supramolecular site accounted for 80 to 90 percent (Table 2), which was much higher than that in the supernatant. It was further speculated that the components of Rhei Radix et Rhizoma and Coptidis Rhizoma interacted with each other during the decocting process, and the products of interaction mainly existed in the supramolecular site but not in the supernatant site.
Table 2. Proportion of Partial Components in Supramolecular and Supernatant Sites.
| peak area (mAU) |
proportion (%) |
||||
|---|---|---|---|---|---|
| no. | compound | supernatant | supramolecule | supernatant | supramolecule |
| 28 | catechin gallate | 150697588 | 710382109 | 17.50 | 82.50 |
| 61 | 3-methyl-rhein | 104630117 | 1060477996 | 8.98 | 91.02 |
| 63 | rhein | 126177173 | 1781611708 | 6.62 | 93.38 |
| 65 | emodin | 84962980 | 1080546999 | 7.29 | 92.71 |
| 103–106 | alkaloids | 24427908772 | 32315248803 | 43.05 | 56.95 |
| 108–111 | 49331489593 | 68356178573 | 41.92 | 58.08 | |
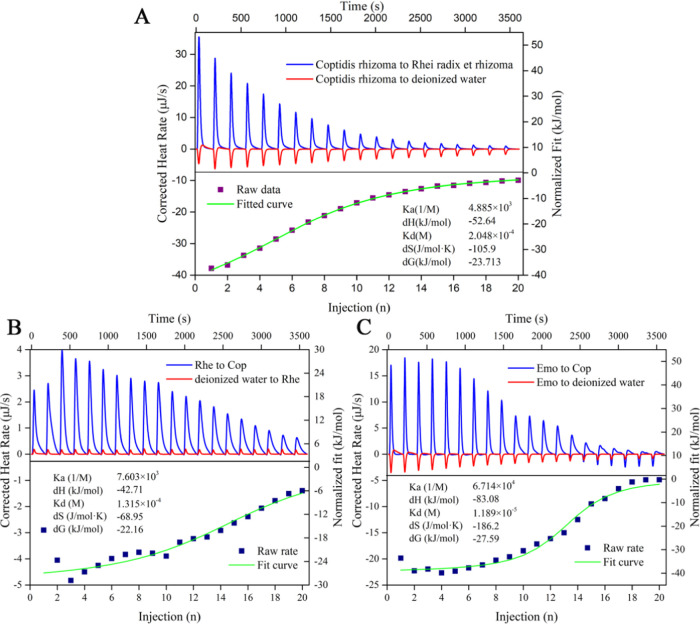
3.3. Isothermal Titration Calorimetry Analysis between the Herbal Pair Medicine
Binding constant (Ka), dissociation constant (Kb), Gibbs free energy (ΔG), enthalpy (ΔH), and entropy (ΔS) could be directly and accurately determined from a single experiment.57,58 ITC has been used by some researchers to analyze the thermodynamic parameters of interaction during co-decocting of medicinal materials.59,60 As shown in Figure 4, the upward peaks of Coptidis Rhizoma-titrated Rhei Radix et Rhizoma indicated an exothermic reaction. In the control group, Coptidis Rhizoma-titrated deionized water presented a downward endothermic characteristic peak (dilution process). The fitted curve showed an approximate S-shape, which was a typical characteristic peak trend of chemical reaction. The results showed that the reaction between the two drugs was thorough, and the effect was strong. Their binding constant Ka was 4.885 × 103.
Figure 4.
ITC characterization: (A) Coptidis Rhizoma-titrated Rhei Radix et Rhizoma. (B) Rhe-titrated Cop. (C) Emo-titrated Cop.
Herein, thermodynamic parameters of titration between the herbal pair medicine were ΔH = −52.64 kJ/mol < 0, ΔS = −105.9 J/mol < 0. According to the second law of thermodynamics, |ΔH| > |−TΔS|, ΔG = −23.713 kJ/mol < 0. It was suggested that the reaction was a spontaneous reaction driven by enthalpy, and the entropy change was unfavorable to the reaction. The results showed that the active components of Rhei Radix et Rhizoma and Coptidis Rhizoma were bonded by noncovalent bonds, such as hydrogen bonds and electrostatic attraction, rather than aggregation driven by physical precipitation.
3.4. Morphological Characterization of Emo-Cop NPs and Rhe-Cop NFs
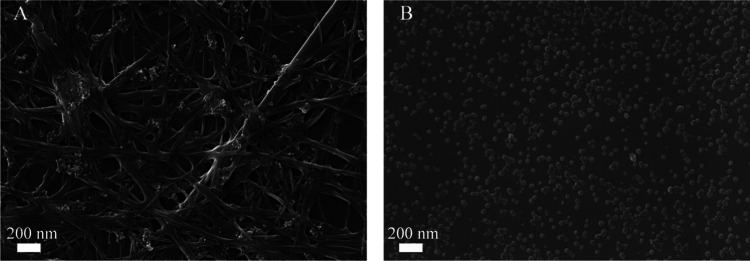
Based on the results shown in Figure 2B, emodin, rhein, and coptidine were selected as the research objects to further reveal the carrier-free self-assembly phenomenon in this drug pair. The prepared Emo-Cop NPs and Rhe-Cop NFs were characterized by SEM (Figure 5). It was found that the morphology of the assembly formed by different components was significantly different. The Emo-Cop NPs were a nanosphere with a particle size between 50 and 100 nm and evenly dispersed in the field of vision. Some scholars have found that protonated rhein tends to aggregate, and so spherical, columnar, and membrane-shaped aggregated micelles could be observed.61 In contrast, the Rhe-Cop NFs presented a fibrous surround state with a fiber width of about 100 nm. By comparing the morphology of the two kinds of self-assembly, the Tyndall Effect and aggregations in the decoction of Rhei Radix et Rhizoma and Coptidis Rhizoma were confirmed.
Figure 5.
Scanning electron microscopy characterization: (A) Rhe-Cop NFs and (B) Emo-Cop NPs.
3.5. Isothermal Titration Calorimetry Analysis
To further reveal the self-assembly mechanism of the herbal pair medicine, we found that the titration process of emodin and coptisine, rhein and coptisine was an exothermic reaction. As shown in Figure 4B,C, Gibbs free energy was −22.61 and −27.59 kJ/mol, respectively. Both of them displayed |ΔH| > |−TΔS|. These results indicated that the reaction of emodin and coptisine, rhein and coptisine was a spontaneous chemical reaction driven by enthalpy, rather than a simple physical process. Current evidence further proved that the assembly process was driven by weak interactions, which was in accordance with the reaction of the herbal pair medicine (Figure 4A).
3.6. UV–Vis Absorption Spectral Analysis
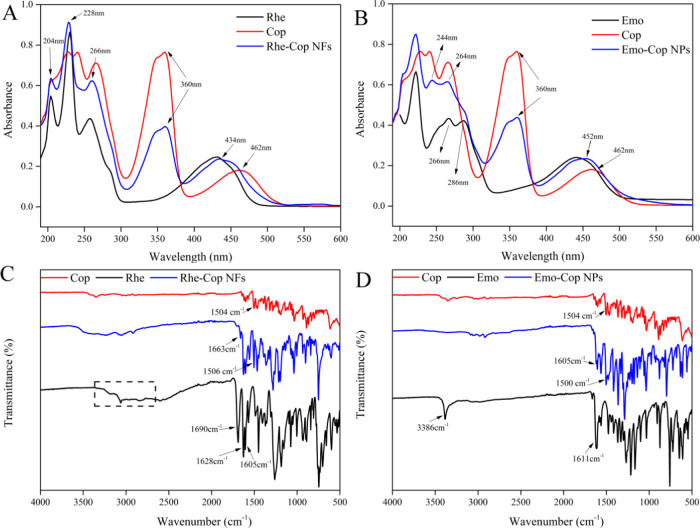
UV–vis absorption spectra reflected the characteristics of some groups in molecules.62 After molecular interaction, the change in the benzene-conjugated system resulted in the shift of the maximum absorption wavelength. Therefore, the law of molecular interaction or the process of self-assembly could be inferred from the changes in UV–vis spectra.63,64
There were five characteristic absorption peaks of coptisine at 226, 242, 266, 360, and 460 nm. The characteristic absorption peaks of rhein were at 204, 230, 258 nm (π → π* transitions), and 430 nm (n → σ* transitions). The characteristic absorption peaks of emodin were at 222 nm (n → σ* transitions), 266, 286 nm (π → π* transitions), and 442 nm (n → σ* transitions). Both Rhe-Cop NFs and Emo-Cop NPs had the characteristic absorption peak of coptisine at 360 nm. Rhe-Cop NFs had the characteristic absorption peaks of rhein at 204, 228, and 268 nm. The characteristic absorption peak at 434 nm of Rhe-Cop NFs showed a blue shift compared with coptisine (from 462 to 434 nm). Compared with emodin, Emo-Cop NPs had characteristic absorption peaks at 244 and 264 nm, while the characteristic absorption peaks of emodin were at 266 and 286 nm (Figure 6A,B).
Figure 6.
(A) UV–visible spectra of Rhe-Cop NFs. (B) UV–visible spectra of Emo-Cop NPs. (C) Fourier infrared spectra of Rhe-Cop NFs. (D) Fourier infrared spectra of Emo-Cop NPs.
Emodin, rhein (anthraquinones), and coptisine (benzyl isoquinoline alkaloid) all had large cyclic conjugated structures. The hydroxyl group and methylene dioxy group of benzene ring as the chromophore made the two kinds of components to appear with strong characteristic absorption in the UV–vis spectrum. These results indicated that the assemblies had characteristic absorption peaks of monomer components and the characteristic peak shift of the assemblies mostly occurred at 260 and 430 nm. It was speculated that the weak intermolecular interaction affected the n−π* and π–π* transitions of intramolecular electrons.65,66
3.7. Fourier Transform Infrared Spectroscopy (FTIR) Characterization Analysis
FTIR was often dominated by the overlap of the doubling frequency and merging frequency of the hydrogen group X–H (X = C, N, O). Because the absorption frequency of different functional groups was different, they were in different positions in the spectrum.67,68
In the infrared spectrum for Rhe-Cop NFs, shown in Figure 6C, the OH stretching vibration band of the COOH of Rhe-Cop NFs appeared at a high wavenumber compared with that in rhein (3000 cm–1). The C=O group vibration band of the COOH group of rhein shifted from 1690 to 1663 cm–1 and the peak intensity decreased significantly. The band at 1504 cm–1 in coptisine shifted to a higher wavenumber of 1506 cm–1 and the peak band intensity increased. The bands 1628 and 1605 cm–1 in the assembly were attributed to the stretching vibration band of the C=O group on rhein’s quinone ring. In the infrared spectrum for Emo-Cop NPs, shown in Figure 6D, the 3386 cm–1 stretching vibration absorption band of 3-OH of emodin disappeared. The C=O group stretching vibration band on the quinone ring of emodin shifted from 1611 to 1605 cm–1 and the peak intensity decreased. The 1504 cm–1 infrared characteristic band of coptisine was an aromatic ring skeleton vibration band, which shifted to a low wavenumber of 1500 cm–1 and the peak intensity increased.
These results indicated that the 3-COOH band on the anthraquinone ring of emodin, the 3-OH band on the anthraquinone ring of emodin, and the aromatic ring skeleton of coptisine significantly changed after assembly, suggesting that there were hydrogen bonding or π–π stacking between rhein and coptisine, emodin and coptisine, respectively.69 Therefore, it was inferred that the interaction between 3-COOH of rhein, 3-OH of emodin, and the quaternary ammonium nitrogen atom of coptisine led to electron delocalization, which reduced the density of the electron cloud and showed that the C=O bond energy of anthraquinone components become weaker and the stretching vibration frequency becomes lower than their single molecules.
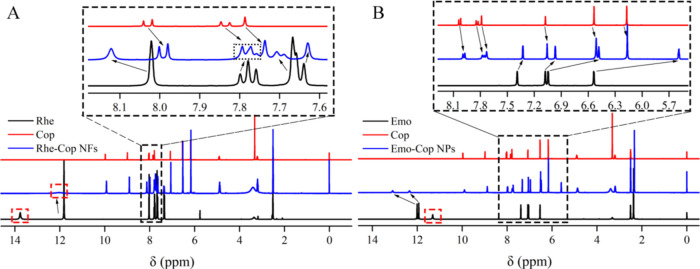
3.8. 1H NMR Characterization Analysis
1H NMR spectra could be used to characterize weak interactions such as hydrogen bonding and π–π stacking.70 Through weak interactions, molecules were stacked and arranged regularly to form assemblies or supramolecules with a certain microscopic morphology. Changes in the spatial position of the molecule affected the density of the electron cloud on the hydrogen atom, leading to fluctuations in the hydrogen signal. Although the fluctuation was not as violent as the reaction of chemical synthesis, it could also accurately determine the site where the weak bond binds to each other.71 Anthracene had three linear benzene rings, which could generate π–π stacking or charge transfer with other electron-rich or electron-poor molecules through π-conjugated groups.72 For example, rhein molecules could self-assemble to form three-dimensional networks composed of nanofibers through noncovalent interactions such as π–π stacking and hydrogen bonding,16 while emodin molecules are stacked into a supramolecular layer through π–π interactions.73
According to the data analysis in Figure 7 and Tables 3–1, the characteristic peaks of 3-COOH of rhein and 3-OH of emodin disappeared, which indicated that rhein and coptisine, emodin and coptisine approached each other under the induction of electrostatic attraction (anthraquinone’s 3-COO– and alkaloids’ 7-N+, anthraquinone’s 3-O– and alkaloids’ s 7-N+). It was also found that the 1,8-OH on both anthraquinone rings fluctuated to the low field direction with large chemical shift values. Therefore, it could be shown that there were obvious hydrogen bonds between rhein molecules and emodin molecules, which formed a layered skeleton structure under the action of intermolecular hydrogen bonds. The π–π stacking between rhein and coptisine was distributed between the anthraquinone layers, resulting in the H-8, H-11, H-12, and H-13 on the benzyl isoquinoline ring moving 0.04–0.11 ppm to the high field. The action of emodin and coptidine was the same as that of rhein and coptisine. Because the skeletal structure of coptidine was similar to that of berberine, the crystal structure of rhein-berberine24 and emodin-berberine73 could be inferred: rhein and coptisine, emodin and coptisine were close to each other under electrostatic action and further formed binary assemblies under the action of π–π stacking.
Figure 7.
Nuclear magnetic hydrogen spectra: (A) Rhe-Cop NFs. (B) Emo-Cop NPs.
Table 3. Chemical Shifts of Emo-Cop NPs, Rhe-Cop NFs, and Their Anthraquinone Monomers.
| Rhe | Rhe-Cop NFs | Δδ (ppm) | Emo | Emo-Cop NPs | Δδ (ppm) |
|---|---|---|---|---|---|
| 13.76 (3-COOH, s) | disappear | 12.01 (1-OH, s) | 13.10 (s) | +1.09 | |
| 11.81 (1,8-OH, s) | 12.02 (brs) | +0.11 | 11.93 (8-OH, s) | 12.35 (s) | +0.42 |
| 8.02 (H-4, s) | 8.12 (s) | +0.10 | 11.31 (3-OH, s) | disappear | |
| 7.78 (H-6, t, J = 8 Hz) | 7.77 (t, J = 8 Hz, J = 4 Hz) | –0.01 | 7.39 (H-5, s) | 7.33 (d, J = 0.8 Hz) | –0.06 |
| 7.67 (H-2, s) | 7.63 (s) | –0.04 | 7.08 (H-7, s) | 6.97 (m) | –0.11 |
| 7.65 (H-5, d, J = 8 Hz) | 7.70 (d, J = 4 Hz) | +0.05 | 7.05 (H-4, d, J = 4 Hz) | 6.48 (m) | –0.57 |
| 7.35 (H-7, d, J = 8 Hz) | 7.35 (d, J = 8 Hz) | 6.54 (H-2, d, J = 4 Hz) | 5.59 (m) | –0.95 | |
| 2.32 (6-CH3, s) | 2.34 (s) | +0.02 |
Table 4. Chemical Shifts of Emo-Cop NFs, Rhe-Cop NFs, and Coptisine.
| Cop | Rhe-Cop NFs | Δδ (ppm) | Emo-Cop NPs | Δδ (ppm) |
|---|---|---|---|---|
| 9.97 (H-8, s) | 9.92 (s) | –0.05 | 9.90 (s) | –0.07 |
| 8.99 (H-13, s) | 8.90 (s) | –0.09 | 8.88 (s) | –0.11 |
| 8.03 (H-11, d, J = 8 Hz) | 7.99 (d, J = 8 Hz) | –0.04 | 7.98 (d, J = 8 Hz) | –0.05 |
| 7.84 (H-12, d, J = 8 Hz) | 7.77 (t, J = 8 Hz, J = 4 Hz) | –0.07 | 7.76 (t, J = 8 Hz, J = 12 Hz) | –0.08 |
| 7.79 (H-1, s) | 7.74 (s) | –0.05 | –0.03 | |
| 7.08 (H-4, s) | 7.05 (s) | –0.03 | 7.05 (s) | –0.03 |
| 6.54 (15-CH2, s) | 6.52 (s) | –0.02 | 6.51 (s) | –0.03 |
| 6.17 (14-CH2, s) | 6.16 (s) | –0.01 | 6.16 (s) | –0.01 |
| 4.90 (6-CH2, t, J = 8 Hz, J = 4 Hz) | 4.88 (t, J = 8 Hz, J = 4 Hz) | –0.02 | 4.87 (s) | –0.03 |
| 3.20 (5-CH2, t, J = 8 Hz, J = 4 Hz) | 3.19 (t, J = 8 Hz, J = 4 Hz) | –0.01 | 3.19 (s) | –0.01 |
4. Conclusions
In this study, spherical supramolecules were found in Rhei Radix et Rhizoma and Coptidis Rhizoma decoction. A total of 119 compounds were identified from the supramolecules by the UPLC-MS/MS method (23 anthraquinones, 31 alkaloids, 24 organic acids, 8 tannins, and other components). Among these, the content of anthraquinones and tannins in the supramolecular was much higher than that in the supernatant, such as rhein and emodin. ITC analysis showed that the interaction between Rhei Radix et Rhizoma and Coptidis Rhizoma was a spontaneous chemical reaction, indicating that active components could form supramolecules through weak bonds. Furthermore, we found that the ingredients of emodin and coptisine could be assembled into NPs of about 50 nm, while rhein and coptidine were assembled into nanofibers. Based on the current evidence, both self-assemblies were mainly induced by electrostatic attraction, hydrogen bonding, and π–π stacking. In this study, the self-assembled entities of small molecules were found based on the herbal pair medicine, which provides a reference for discovery of the natural carrier-free self-assembly from TCM. And it is also beneficial to the design of binary self-assembly of small molecules in the future.
Acknowledgments
This research was funded by the National Natural Science Foundation of China (Nos. 82073974 and 82274072), Beijing Nova program (No. Z201100006820026), the Beijing Municipal Natural Science Foundation (No. 7202116), the Fundamental Research Funds for the Central Universities (2022-XJ-KYQD-008, BUCM-2019-JCRC002, and 2020-JYB-ZDGG-044, China), project of China Association of Chinese Medicine (CACM-2018-QNRC2-B08), and Beijing Key Laboratory for Basic and Development Research on Chinese Medicine (Beijing, 100102).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c04098.
UPLC-MS/MS analysis of supramolecular sites and schematic diagram of the cleavage law of the ingredients (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Webber M. J.; Eric A.; Meijer E.; Robert L. Supramolecular biomaterials. Nat. Mater. 2016, 15, 13–26. 10.1038/nmat4474. [DOI] [PubMed] [Google Scholar]
- Katsuhiko A.; Michihiro N.; Taizo M.; Takeya J.; Shrestha L.; Hill J. Self-assembly as a key player for materials nanoarchitectonics. Sci. Technol. Adv. Mater. 2020, 20, 51–95. 10.1080/14686996.2018.1553108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M. P.; Michael P.; Navidzadeh J. O.; Justin O.; Bobbala S.; Sharan B. Leveraging self-assembled nanobiomaterials for improved cancer immunotherapy. Cancer Cell 2022, 40, 255–276. 10.1016/j.ccell.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H.; Wang W.; Li X.; Han X.; Zhang X.; Wang J.; Liu C.; Huang L.; Gao W. Y. Natural small molecules enabled efficient immunotherapy through supramolecular self-assembly in P53-mutated colorectal cancer. ACS Appl. Mater. Interfaces 2022, 14, 2464–2477. 10.1021/acsami.1c16737. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Cong C.; Zhang J.; Chao J.; Luong H.; Zhao Y.; Wang L. DNA self-assembled Au nanoparticle clusters on silver nanorod arrays for high-sensitive and multiplex detection of cancer-related biomarkers. Nanoscale 2022, 14, 4538–4547. 10.1039/D2NR00133K. [DOI] [PubMed] [Google Scholar]
- Zahra N.; Seyed A.; Sepideh H. A novel self-assembled micelles based on stearic acid modified schizophyllan for efficient delivery of paclitaxel. Colloids Surf., B 2021, 199, 111524 10.1016/j.colsurfb.2020.111524. [DOI] [PubMed] [Google Scholar]
- Mougin J.; Bourgaux C.; Couvreur P. Elongated self-assembled nanocarriers: from molecular organization to therapeutic applications. Adv. Drug Delivery Rev. 2021, 172, 127–147. 10.1016/j.addr.2021.02.018. [DOI] [PubMed] [Google Scholar]
- Shang Y.; Zhi D.; Feng G.; Wang Z.; Mao D.; Guo S.; Liu R.; Liu L.; Zhang S.; Sun S.; Wang K.; Kong D.; Gao J.; Yang Z. Supramolecular nanofibers with superior bioactivity to insulin-like growth factor-I. Nano Lett. 2019, 19, 1560–1569. 10.1021/acs.nanolett.8b04406. [DOI] [PubMed] [Google Scholar]
- Li M.; Wang C.; Di Z.; Li H.; Zhang J.; Xue W.; Zhao M.; Zhang K.; Zhao Y.; Li L. Engineering multifunctional DNA hybrid nanospheres through coordination-driven self-assembly. Angew. Chem., Int. Ed. 2019, 58, 1350–1354. 10.1002/anie.201810735. [DOI] [PubMed] [Google Scholar]
- Zhu J.; Zhang Z.; Wang R.; Zhong K.; Zhang K.; Zhang N.; Liu W.; Feng F.; Qu W. Review of natural phytochemical-based self-assembled nanostructures for applications in medicine. ACS Appl. Nano Mater. 2020, 5, 8537–8556. 10.1021/acsanm.2c01956. [DOI] [Google Scholar]
- Yang X.; Ma C.; Chen Z.; Liu J.; Liu F.; Xie R.; Zhao H.; Deng G.; Chen A.; Gong N.; Yao L.; Zuo P.; Zhi K.; Wang J.; Gao X.; Wang J.; Fan L.; Zhou J. Single small molecule-assembled nanoparticles mediate efficient oral drug delivery. Nano Res. 2019, 12, 2468–2476. 10.1007/s12274-019-2470-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z.; Wei Y.; Wang N.; Zhang J.; Gao Y.; Qian S. Gel formation of puerarin and mechanistic study during its cooling process. Int. J. Pharm. 2018, 548, 625–635. 10.1016/j.ijpharm.2018.07.038. [DOI] [PubMed] [Google Scholar]
- Li W. Supramolecular nanofiber-reinforced puerarin hydrogels as drug carriers with synergistic controlled release and antibacterial properties. J. Mater. Sci. 2020, 55, 6669–6677. 10.1007/s10853-020-04455-3. [DOI] [Google Scholar]
- Huang H.; Gong W.; Wang X.; He W.; Hou Y.; Hu J. Self-assembly of naturally small molecule into supramolecular fibrillar networks for wound healing. Adv. Healthcare Mater. 2022, 11, 2102476 10.1002/adhm.202102476. [DOI] [PubMed] [Google Scholar]
- Hou Y.; Chen M.; Ruan H.; Sun Z.; Wu H.; Xu X.; Yang J.; Ma G.; Zhou X. A new supramolecular natural product gel based on self-assembled pomolic acid from traditional Chinese medicine. Colloid Interface Sci. Commun. 2022, 46, 100583 10.1016/j.colcom.2021.100583. [DOI] [Google Scholar]
- Zheng J.; Fan R.; Wu H.; Yao H.; Yan Y.; Liu J.; Ran L.; Sun Z.; Yi L.; Dang L.; Gan P.; Zheng P.; Yang T.; Zhang Y.; Tang T.; Wang Y. Directed self-assembly of herbal small molecules into sustained release hydrogels for treating neural inflammation. Nat. Commun. 2019, 10, 1604 10.1038/s41467-019-09601-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W.; Zhang X.; Zhang R.; Zhang K.; Li Y.; Xu F. Self-Assembled herbal medicine encapsulated by an oxidation-sensitive supramolecular hydrogel for chronic wound treatment. ACS Appl. Mater. Interfaces 2020, 12, 56898–56907. 10.1021/acsami.0c19492. [DOI] [PubMed] [Google Scholar]
- Bag B. G.; Majumdar R. Self-assembly of renewable nano-sized triterpenoids. Chem. Rec. 2017, 17, 1–34. 10.1002/tcr.201600123. [DOI] [PubMed] [Google Scholar]
- Bag B. G.; Barai A.; Hasan S.; Panja S.; Ghorai S.; Patra S. Terpenoids, nano-entities and molecular self-assembly. Pure Appl. Chem. 2020, 92, 567–577. 10.1515/pac-2019-0812. [DOI] [Google Scholar]
- Zhi K.; Wang J.; Zhao H.; Yang X. Self-assembled small molecule natural product gel for drug delivery: a breakthrough in new application of small molecule natural products. Acta Pharm. Sin. B 2020, 10, 913–927. 10.1016/j.apsb.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Zhao H.; Qiao W.; Cheng J.; Han Y.; Yang X. Nanomedicine-cum-Carrier by co-assembly of natural small products for synergistic enhanced antitumor with tissues protective actions. ACS Appl. Mater. Interfaces 2020, 12, 42537–42550. 10.1021/acsami.0c12641. [DOI] [PubMed] [Google Scholar]
- Fan L.; Zhang B.; Xu A.; Shen Z.; Guo Y.; Zhao R.; Yao H.; Shao J. Carrier-free, pure nanodrug formed by self-assembly of anti-cancer drug for cancer immune therapy. Mol. Pharmaceutics 2018, 15, 2466–2478. 10.1021/acs.molpharmaceut.8b00444. [DOI] [PubMed] [Google Scholar]
- Li T.; Wang P.; Guo W.; Huang X.; Tian X.; Wu G.; Xu B.; Li F.; Yan C.; Liang X.; Lei H. Natural berberine-based Chinese herb medicine assembled nanostructures with modified antibacterial application. ACS Nano 2019, 13, 6770–6781. 10.1021/acsnano.9b01346. [DOI] [PubMed] [Google Scholar]
- Tian X.; Wang P.; Li T.; Huang X.; Guo W.; Yang Y.; Yan M.; Zhang H.; Cai D.; Jia X.; Li F.; Xu B.; Ma T.; Yan C.; Lei H. Self-assembled natural phytochemicals for synergistically antibacterial application from the enlightenment of traditional Chinese medicine combination. Acta Pharm. Sin. B 2020, 10, 1784–1795. 10.1016/j.apsb.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.; Guo W.; Huang G.; Zhen J.; Li Y.; Li T.; Zhao L.; Yuan K.; Tian X.; Huang X.; Feng Y.; Lei H.; Xu A. Berberine-based heterogeneous linear supramolecules neutralized the acute nephrotoxicity of aristolochic acid by the self-assembly strategy. ACS Appl. Mater. Interfaces 2021, 13, 32729–32742. 10.1021/acsami.1c06968. [DOI] [PubMed] [Google Scholar]
- Huang X.; Wang P.; Li T.; Tian X.; Guo W.; Xu B.; Huang G.; Cai D.; Zhou F.; Zhang H.; Lei H. Self-assemblies based on traditional medicine berberine and cinnamic acid for adhesion-induced inhibition multidrug-resistant staphylococcus aureus. ACS Appl. Mater. Interfaces 2020, 12, 227–237. 10.1021/acsami.9b17722. [DOI] [PubMed] [Google Scholar]
- Tian Y.; Tang G.; Gao Y.; Chen X.; Zhou Z.; Li Y.; Li X.; Wang H.; Yu X.; Luo L.; Cao Y. Carrier-free small molecular self-assembly based on berberine and curcumin incorporated in submicron particles for improving antimicrobial activity. ACS Appl. Mater. Interfaces 2022, 14, 10055–10067. 10.1021/acsami.1c22900. [DOI] [PubMed] [Google Scholar]
- Tian X.-h.; Zhang H.; Wang S.; Li T.; Huang X.; Yan M.; Cao X.; Xu B.; Wang P.; Lei H. A new strategy based on acid-alkali complexation for rapidly and accurately fishing phytochemicals in Sennae Folium. Chin. Herb. Med. 2020, 12, 188–194. 10.1016/j.chmed.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Zhao L.; Shen G.; Chang R.; Zhang Y.; Yan X. Coordination self-assembly of natural flavonoids into robust nanoparticles for enhanced in vitro chemo and photothermal cancer therapy. Colloids Surf., A 2020, 598, 124805 10.1016/j.colsurfa.2020.124805. [DOI] [Google Scholar]
- Zheng K.; Xiong Y.; Li Z.; Peng L.; Guo Q.; Li X.; Deng X. ESI-TOF MS analysis and DNA cleavage activity of complexes formed by luteolin and five metal ions in hot water. Inorg. Nano-Met. Chem. 2020, 50, 1181–1188. 10.1080/24701556.2020.1737820. [DOI] [Google Scholar]
- Li X.; Gao P.; Tan J.; Xiong K.; Maitz M.; Pan C.; Wu H.; Chen Y.; Yang Z.; Huang N. Assembly of metal-phenolic/catecholamine networks for synergistically anti-Inflammatory, antimicrobial, and anticoagulant coatings. ACS Appl. Mater. Interfaces 2018, 10, 40844–40853. 10.1021/acsami.8b14409. [DOI] [PubMed] [Google Scholar]
- Jia X.; Yuan Z.; Yang Y.; Huang X.; Han N.; Liu X.; Lin X.; Ma T.; Xu B.; Wang P.; Lei H. Multi-functional self-assembly nanoparticles originating from small molecule natural product for oral insulin delivery through modulating tight junctions. J. Nanobiotechnol. 2022, 20, 116. 10.1186/s12951-022-01260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagalgave S. M.; Sachin D.; Mohammad A.; Duong D.; Bhamidipati K.; Puvvada N.; Rajesh S.; Sidhanath V.; Sheshanath V. Selectivity and bio-compatibility of self-assembled chiral flower-like and helical nanostructures. New J. Chem. 2020, 44, 18092–18101. 10.1039/D0NJ01235A. [DOI] [Google Scholar]
- Lu J.; Hu J.; Jiang Y.; Cui W. The supramolecular organogel formed by self-assembly of ursolic Acid Appended with Aromatic Rings. Materials 2019, 12, 614. 10.3390/ma12040614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. K. Structure- and solvent-triggered influences in the self-assembly of polyoxometalate-steroid conjugates. RSC Adv. 2016, 6, 66431–66437. 10.1039/C6RA13276F. [DOI] [Google Scholar]
- Li D. W.; Tang G. K.; Yao H.; Zhu Y.; Shi G.; Fu Q.; Yang F.; Wang X. Formulation of pH-responsive PEGylated nanoparticles with high drug loading capacity and programmable drug release for enhanced antibacterial activity. Bioact. Mater. 2022, 16, 47–56. 10.1016/j.bioactmat.2022.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontsarik M.; Yaghmur A.; Ren Q.; Maniura K.; Salentinig S. From Structure to Function: pH-Switchable Antimicrobial Nano-Self-Assemblies. ACS Appl. Mater. Interfaces 2019, 11, 2821–2829. 10.1021/acsami.8b18618. [DOI] [PubMed] [Google Scholar]
- Qiao L.; Han M.; Gao S.; Shao X.; Wang X.; Sun L.; Fu X.; Wei Q. Research progress on nanotechnology for delivery of active ingredients from traditional Chinese medicines. J. Mater. Chem. B 2020, 8, 6333–6351. 10.1039/D0TB01260B. [DOI] [PubMed] [Google Scholar]
- Zhi K.; Zhao H.; Yang X.; Zhang J.; Wang J.; Regenstein M. Natural product gelators and general method for obtaining them from organisms. Nanoscale 2018, 10, 3639–3643. 10.1039/C7NR08368H. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Hu K.; Di L.; Wang P.; Liu Z.; Zhang J.; Yue P.; Song W.; Zhang J.; Chen T.; Wang Z.; Zhang Y.; Wang X.; Zhan C.; Cheng Y.; Li X.; Li Q.; Fan J.; Shen Y.; Han J.; Qiao H. Traditional herbal medicine and nanomedicine: converging disciplines to improve therapeutic efficacy and human health. Adv. Drug Delivery Rev. 2021, 178, 113964 10.1016/j.addr.2021.113964. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Gao G.; Chu Q.; Wang H.; Rao P.; Ke L. Chromatographic isolation of nanoparticles from Ma-Xing-Shi-Gan-Tang decoction and their characterization. J. Ethnopharmacol. 2014, 151, 1116–1123. 10.1016/j.jep.2013.12.029. [DOI] [PubMed] [Google Scholar]
- Zhuang Y.; Yan J.; Zhu W.; Chen L.; Liang D.; Xu X. Can the aggregation be a new approach for understanding the mechanism of traditional Chinese medicine. J. Ethnopharmacol. 2008, 117, 378–384. 10.1016/j.jep.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Lü S.; Su H.; Sun S.; Guo Y.; Liu T.; Ping Y.; Li Y. Isolation and characterization of nanometre aggregates from a Bai-Hu-Tang decoction and their antipyretic effect. Sci Rep. 2018, 8, 12209 10.1038/s41598-018-30690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D.; Du Q.; Wang H.; Gao; Zhou J.; Ke L.; Chen T.; Shaw C.; Rao P. Antidiabetic micro-/nanoaggregates from Ge-Gen-Qin-Lian-Tang decoction increase absorption of baicalin and cellular antioxidant activity in vitro. BioMed Res. Int. 2017, 2017, 9217912 10.1155/2017/9217912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.; Chang S.; Yang M.; Chen J.; Chang W. Nanoscale hepatoprotective herbal decoction attenuates hepatic stellate cell activity and chloroform-induced liver damage in mice. Int. J. Nanomed. 2011, 6, 1365–1371. 10.2147/IJN.S19503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.; Zhang Z.; Wang R.; Huang X.; Zhou Y.; Zhang K.; Zhong K.; Gong L.; Li L.; Liu W.; Feng F.; Qu W. Nanoparticles derived from Scutellaria barbata and Hedytois diffusa herb pair and their anti-cancer activity. Pharmacol. Res. 2022, 2, 100048 10.1016/j.prmcm.2022.100048. [DOI] [Google Scholar]
- Zhao G.; Lu H.; Liu M.; Jiang H.; Peng D.; Lu H.; Chen W. Isolation and characterization of natural nanoparticles in naoluo xintong decoction and their brain protection research. Molecules 2022, 27, 1511. 10.3390/molecules27051511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J.; Wua Z.; Yan J.; Pang W.; Liang D.; Xu X. A promising approach for understanding the mechanism of traditional Chinese medicine by the aggregation morphology. J. Ethnopharmacol. 2009, 123, 267–274. 10.1016/j.jep.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Wang G.; Yang C.; Zhang K.; Hu J.; Pang W. Molecular clusters size of Puerariae thomsonii Radix aqueous decoction and relevance to oral absorption. Molecules 2015, 20, 12376–12388. 10.3390/molecules200712376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.; Zhao Q.; Lu J.; Ye D.; Mu S.; Yang X.; Zhang W.; Ma B. Natural nano-drug delivery system in Coptidis Rhizoma extract with modified berberine hydrochloride pharmacokinetics. Int. J. Nanomed. 2021, 16, 6297–6311. 10.2147/IJN.S323685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q.; Chen M.; Yan M.; Wang P.; Lei H.; Xue H.; Ma Q. Comprehensive analysis of Sini decoction and investigation of acid-base selfassembled complexes using cold spray ionization mass spectrometry. Microchem. J. 2021, 173, 107008 10.1016/j.microc.2021.107008. [DOI] [Google Scholar]
- Chen M.; Wang P.; Li T.; Li L.; Li J.; Bai H.; Lei H.; Ma Q. Comprehensive analysis of Huanglian Jiedu decoction: revealing the presence of a self-assembled phytochemical complex in its naturally-occurring precipitate. J. Pharm. Biomed. Anal. 2021, 195, 113820 10.1016/j.jpba.2020.113820. [DOI] [PubMed] [Google Scholar]
- Gao G.; He C.; Wang H.; Guo J.; Ke L.; Zhou J.; Chong P.; Rao P. Polysaccharide nanoparticles from Isatis indigotica Fort. Root decoction: diversity, cytotoxicity, and antiviral activity. Nanomaterials 2022, 12, 30. 10.3390/nano12010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.; Yang Y.; Yuan X.; Xu H.; Chen Q.; Ren R.; Zhang Q.; Hou Z.; Jiao F.; Yin D. Role of particle aggregates in herbal medicine decoction showing they are not useless: considering Coptis chinensis decoction as an example. Food Funct. 2020, 11, 10480–10492. 10.1039/D0FO02179B. [DOI] [PubMed] [Google Scholar]
- Ke L.-j.; Gao G.; Shen Y.; Zhou J.; Rao P. Encapsulation of aconitine in self-assembled licorice protein nanoparticles reduces the toxicity in vivo. Nanoscale Res. Lett. 2015, 10, 449. 10.1186/s11671-015-1155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; Zhang J.; Gao G.; Wang H.; He X.; Chen T.; Ke L.; Rao P.; Wang Q. Boiling Licorice produces self-assembled protein nanoparticles: a novel source of bioactive nanomaterials. J. Agric. Food Chem. 2019, 67, 9354–9361. 10.1021/acs.jafc.9b03208. [DOI] [PubMed] [Google Scholar]
- Prozeller D.; Morsbach S.; Landfester K. Isothermal titration calorimetry as a complementary method for investigating nanoparticle-protein interactions. Nanoscale 2019, 11, 19265. 10.1039/C9NR05790K. [DOI] [PubMed] [Google Scholar]
- Luo Q.; Chen D.; Boom R.; Janssen A. Revisiting the enzymatic kinetics of pepsin using isothermal titration calorimetry. Food Chem. 2018, 268, 94–100. 10.1016/j.foodchem.2018.06.042. [DOI] [PubMed] [Google Scholar]
- Wang H.; Li T.; Xiang H.; Zhang X.; Fang K.; Wu G.; Yan M.; Xue N.; Chen M.; Xie T.; Zhang Y.; Wang P.; Lei H. Origin and formation mechanism investigation of compound precipitation from the traditional Chinese prescription Huang-Lian-Jie-Du-Tang by isothermal titration calorimetry. Molecules 2017, 22, 1456. 10.3390/molecules22091456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.; Chen Z.; Wang Y.; Hao W.; Yuan Q.; Zhou H.; Gao C.; Wang Y.; Wu X.; Wang X. Targeted delivery of Chinese herb pair-based berberine/tannin acid self-assemblies for the treatment of ulcerative colitis. J. Adv. Res. 2021, 11, 263–276. 10.1016/j.jare.2021.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. H.; Zhang X.; Hao Y.; Ren G.; Guo X. Simulation study of the pH sensitive directed self-assembly of rheins for sustained drug release hydrogel. Colloids Surf., B 2020, 195, 111260 10.1016/j.colsurfb.2020.111260. [DOI] [PubMed] [Google Scholar]
- Passos M. L. C.; Saraiva M. Detection in UV–visible spectrophotometry: Detectors, detection systems, and detection strategies. Measurement 2018, 135, 896–904. 10.1016/j.measurement.2018.12.045. [DOI] [Google Scholar]
- Elhabiri M.; Hamacek J.; Bünzli J.; Gary A. Lanthanide homobimetallic triple-stranded helicates: Insight into the self-assembly mechanism. Eur. J. Inorg. Chem. 2004, 2004, 51–62. 10.1002/ejic.200300549. [DOI] [Google Scholar]
- White L. J.; Wark C.; Croucher L.; Draperc E.; Hiscock J. High-throughput characterisation of supramolecular gelation processes using a combination of optical density, fluorescence and UV-Vis absorption measurements. Chem. Commun. 2020, 56, 9557–9560. 10.1039/D0CC04033A. [DOI] [PubMed] [Google Scholar]
- Zeng H.-j.; Liu Z.; Hu G.; Qu L.; Yang R. Investigation on the binding of aloe-emodin with tyrosinase by spectral analysis and molecular docking. Spectrochim. Acta, Part A 2019, 211, 79–85. 10.1016/j.saa.2018.11.045. [DOI] [PubMed] [Google Scholar]
- Corda E.; Hernandez M.; Cortes S.; Sevilla P. Cucurbit[n]urils (n = 6–8) used as host molecules on supramolecular complexes formed with two different drugs: emodin and indomethacin. Colloids Surf., A 2018, 557, 66–75. 10.1016/j.colsurfa.2018.04.068. [DOI] [Google Scholar]
- Olga M.; Patience S. Experimental methods in chemical engineering: Fourier transform infrared spectroscopy—FTIR. Can. J. Chem. Eng. 2019, 98, 25–33. 10.1002/cjce.23664. [DOI] [Google Scholar]
- Martinez-Felipe A.; Brebner F.; Zaton D.; Concellon A.; Ahmadi S.; Piñol 3 M.; Oriol L. Molecular recognition via hydrogen bonding in supramolecular complexes: a fourier transform infrared spectroscopy study. Molecules 2018, 23, 2278. 10.3390/molecules23092278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.; Hu J.; Liang Y.; Cui W. The supramolecular organogel formed by self-assembly of ursolic acid appended with aromatic rings. Materials 2019, 12, 614. 10.3390/ma12040614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittel M.; Biswas P.; Goswami A.; Saha S. Dynamics of hydrogen bonding in three-component nanorotors. Chem. - Eur. J. 2020, 26, 14095–14099. 10.1002/chem.202002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadar M.; Kane D.; Zafrani Y.; Cohen Y. Temperature dependent and pH responsive pillar[5]arenebased complexes and hydrogen bond-based supramolecular pentagonal boxes in water. Chem. - Eur. J. 2020, 26, 11250–11255. 10.1002/chem.202000972. [DOI] [PubMed] [Google Scholar]
- Lu D.; Huang Q.; Wang S.; Wang J.; Huang P.; Du P. The supramolecular chemistry of cycloparaphenylenes and their analogs. Front. Chem. 2019, 7, 668. 10.3389/fchem.2019.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y.; Zhang Y.; Huang Y.; Zhang M.; Lou B. Preparation, crystal structures, and oral bioavailability of two cocrystals of emodin with berberine chloride. Cryst. Growth Des. 2018, 18, 7481–7488. 10.1021/acs.cgd.8b01257. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.